Upstream activation factor (UAF) is a multifunctional transcription factor in Saccharomyces cerevisiae that plays dual roles in activating RNA polymerase I (Pol I) transcription and repression of Pol II. For Pol I, UAF binds to a specific upstream element in the ribosomal DNA (rDNA) promoter and interacts with two other Pol I initiation factors, the TATA-binding protein (TBP) and core factor (CF). We used an integrated combination of chemical cross-linking mass spectrometry (CXMS), molecular genetics, protein biochemistry, and structural modeling to understand the topological framework responsible for UAF complex formation.
KEYWORDS: cross-linking, integrated modeling, mass spectrometry, RNA polymerase I, transcription, UAF
ABSTRACT
Upstream activation factor (UAF) is a multifunctional transcription factor in Saccharomyces cerevisiae that plays dual roles in activating RNA polymerase I (Pol I) transcription and repression of Pol II. For Pol I, UAF binds to a specific upstream element in the ribosomal DNA (rDNA) promoter and interacts with two other Pol I initiation factors, the TATA-binding protein (TBP) and core factor (CF). We used an integrated combination of chemical cross-linking mass spectrometry (CXMS), molecular genetics, protein biochemistry, and structural modeling to understand the topological framework responsible for UAF complex formation. Here, we report the molecular topology of the UAF complex, describe new structural and functional domains that play roles in UAF complex integrity, assembly, and biological function, and provide roles for previously identified UAF domains that include the Rrn5 SANT and histone fold domains. We highlight the role of new domains in Uaf30 that include an N-terminal winged helix domain and a disordered tethering domain as well as a BORCS6-like domain found in Rrn9. Together, our results reveal a unique network of topological features that coalesce around a histone tetramer-like core to form the dual-function UAF complex.
INTRODUCTION
Saccharomyces cerevisiae RNA polymerase I (Pol I) is responsible for the transcription of the 35S pre-rRNA (rRNA), one of the first and highly regulated steps of the ribosome biogenesis pathway that maintains proper ribosome levels in the cell. Four initiation factors coordinate the recruitment of Pol I to the rDNA promoter, including the upstream activating factor (UAF), TATA-binding protein (TBP), core factor (CF), and Rrn3 (1–11). The least is known about UAF, which contains six different proteins that include Rrn9, Rrn5, Uaf30, Rrn10, and histones H3 and H4 (12–14). UAF binds to a large cis-regulatory element, called the upstream control element (UCE), from positions −155 to −60 relative to the +1 transcription start site and interacts with Pol I factors TBP and CF (9, 15–17).
UAF is not necessary for in vitro Pol I transcription but is required for in vivo Pol I activity, as deletion of rrn5, rrn9, or rrn10 reduces yeast growth and rRNA transcription (1, 2, 12, 18–20). A particularly interesting function of UAF is its ability to repress upstream-initiated Pol II transcription at the ribosomal DNA (rDNA) promoter, where it acts as a Pol II barrier (19, 20). In yeast cells, Pol II gains the ability to transcribe rDNA when UAF activity is disrupted, resulting in a phenomenon called the polymerase switching phenotype (19, 20). UAF is not restricted to the site of Pol I transcription and early ribosome biogenesis in the nucleolus but can directly bind to the SIR2 locus to repress nuclear Pol II-dependent SIR2 transcription in response to rDNA copy loss (21, 22). These studies suggest that UAF plays an important role in helping yeast cells maintain rDNA copy number through its dual Pol I-activating and Pol II-repressive functions (21, 22).
UAF reconstitution studies of transcriptionally active complex recently revealed new insights into UAF assembly. It was shown that all the UAF-specific subunits interact with histone H3 (23). Interestingly, no single UAF subunit, including the histone subunits, was required for complex integrity, indicating an interconnected network of protein-protein interaction forms the complex (23). Stoichiometry studies also revealed that UAF contains two copies of histone H3 while the remaining subunits are found in single copies, suggesting that UAF contains a novel tetramer-like subcomplex with the Rrn5 histone fold domain (23).
Here, we analyzed the molecular topology of the UAF complex by combined chemical cross-linking and mass spectrometry (CXMS). We integrated our CXMS analysis with molecular genetics and biochemical assays to precisely map the biologically relevant domains and regions of the UAF subunits. We highlight new domains for the UAF subunits and describe their roles in complex integrity, assembly, and biological function. Together, our results reveal a unique network of topological and functional domains that coalesce to form the dual-function UAF complex.
RESULTS
Predicted structural domains within UAF subunits.
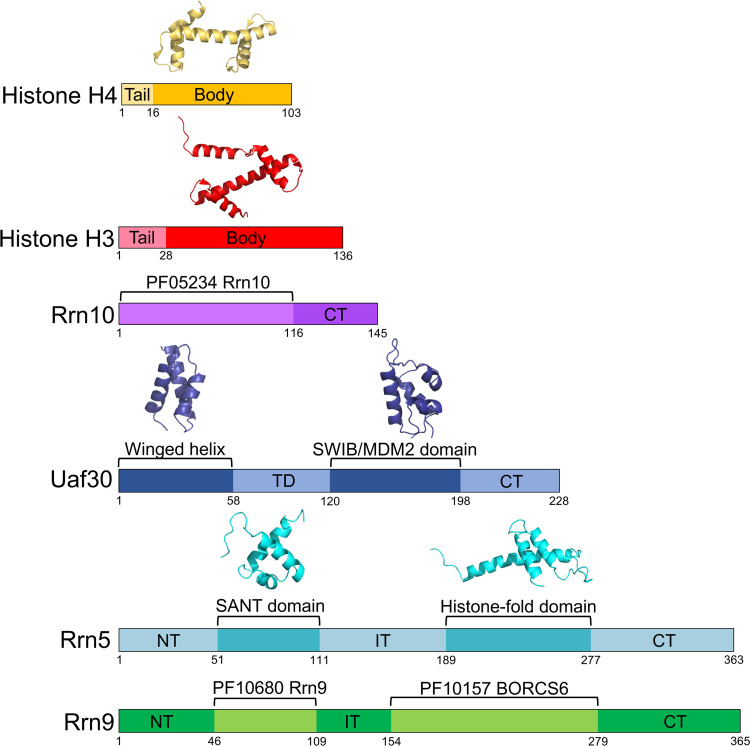
Aside from the known structures of histones H3 and H4, there are no structures available for the remaining UAF subunits. We analyzed the UAF protein sequences by HHpred to find additional structural and/or functional sequence signatures. HHpred is a powerful remote homology detection server that often yields clues on new domains and/or functions that remain undetected by less sensitive homology detection servers (24–26). Uaf30 contains two predicted domains that include an N-terminal helix-turn-helix winged helix domain and a more central SWIB/MDM2 domain previously identified in earlier reports (Fig. 1) (27). Rrn5 is also predicted to contain two identifiable domains that include a SANT domain in the N-terminal half of the protein as well as a histone fold (HF) domain in the C-terminal half (Fig. 1) (23, 27). Rrn9 and Rrn10 lack homology to proteins with a known structure in the Protein Data Bank (PDB) database but contain regions homologous to sequences in the Protein Family (Pfam) database of protein sequence signatures (Fig. 1). For instance, the N-terminal two-thirds of Rrn10 represents a conserved Rrn10 Pfam signature found in the other Rrn10 homologs, whereas Rrn9 contains two conserved Pfam homology regions. These include an Rrn9 protein family signature in its N-terminal half and a Pfam signature belonging to the BORCS6-like protein family (Fig. 1). As shown below, several of these predicted domains play important roles in UAF complex assembly and biological function.
FIG 1.
Predicted domain organization of UAF subunits. Domain maps of each UAF subunit are shown and colored accordingly. Predicted functional and/or structural domains are labeled. Structural models of available domains are depicted above the domain maps. NT, N terminal; CT, C terminal; TD, tethering domain; ID, internal domain. Domain boundaries are denoted with residue numbers below the maps.
Chemical cross-linking and mass spectrometry of UAF.
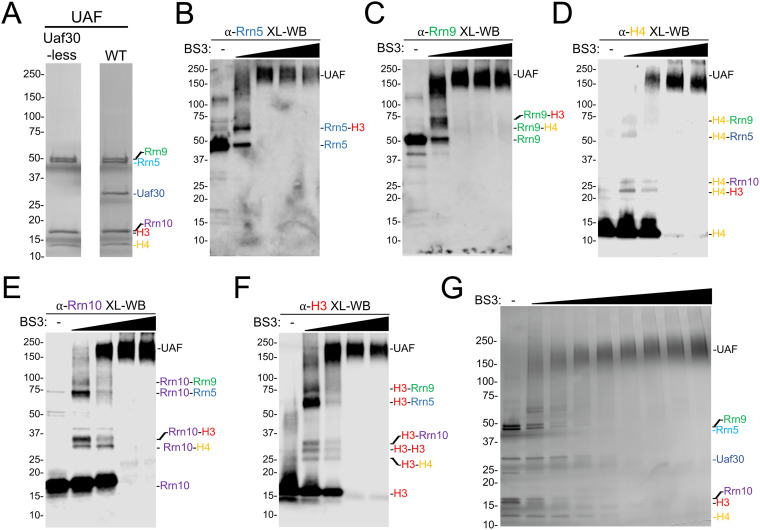
To understand how the core UAF subunits connect to each other, we first cross-linked a UAF subcomplex lacking Uaf30 (Fig. 2A) and analyzed the cross-linked complexes by Western blotting to examine cross-linking intermediates. Uaf30 is not required for UAF complex integrity, it more readily dissociates from the complex based on our previous native MS experiments (19, 23), and we lack a native antibody for the Uaf30 subunit, unlike the other UAF subunits. For each antibody tested, we detected three types of protein species that include the un-cross-linked protein, fully cross-linked complex, and various cross-linking intermediates (Fig. 2B to F). For the two larger proteins in the complex, Rrn5 and Rrn9, we resolved fewer intermediates, likely due to their size and limited gel resolution. For the Rrn5 and Rrn9 antibodies, we resolved intermediates of Rrn5 cross-linked to H3 and Rrn9 cross-linked to H3 and H4 (Fig. 2B and C).
FIG 2.
BS3 cross-linking of UAF and Uaf30-less subcomplex. (A) Coomassie blue-stained SDS-PAGE gel of purified holo-UAF (right) and Uaf30-less UAF (left) used in our cross-linking experiments. (B) Chemical cross-linking of recombinant holo-UAF. Products of incubation of the increasing concentrations from 0.2 to 10 mM BS3 are shown, visualized by SDS-PAGE and silver staining. (C to G) Western blot analysis of BS3 cross-linked Uaf30-less UAF. Recombinant UAF lacking Uaf30 was either left untreated or treated with increasing concentrations of BS3 cross-linker (0.2 to 4 mM) and then analyzed by Western blotting with various UAF subunit antibodies. Antibodies included histone H3 (C), histone H4 (D), Rrn10 (E), Rrn5 (F), and Rrn9 (G). Predicted UAF cross-linked intermediates, based on their calculated molecular weights, are labeled on the right of each Western blot image.
UAF antibodies against the smaller proteins in the complex were particularly informative. For instance, when using the H4 antibody, we detected cross-linking intermediates for H4 cross-linked to H3, Rrn5, Rrn9, and Rrn10 (Fig. 2D). Likewise, for the Rrn10 antibody, we detected cross-linking intermediates for Rrn10 cross-linked to H3, H4, Rrn5, and Rrn9 (Fig. 2E). Finally, for the H3 antibody, we detected cross-linking intermediates with each UAF subunit along with an additional intermediate of H3 cross-linked to itself (Fig. 2F), suggesting that there are two copies of the histone H3 subunit in the UAF complex, which is consistent with previous conclusions from native MS findings (23).
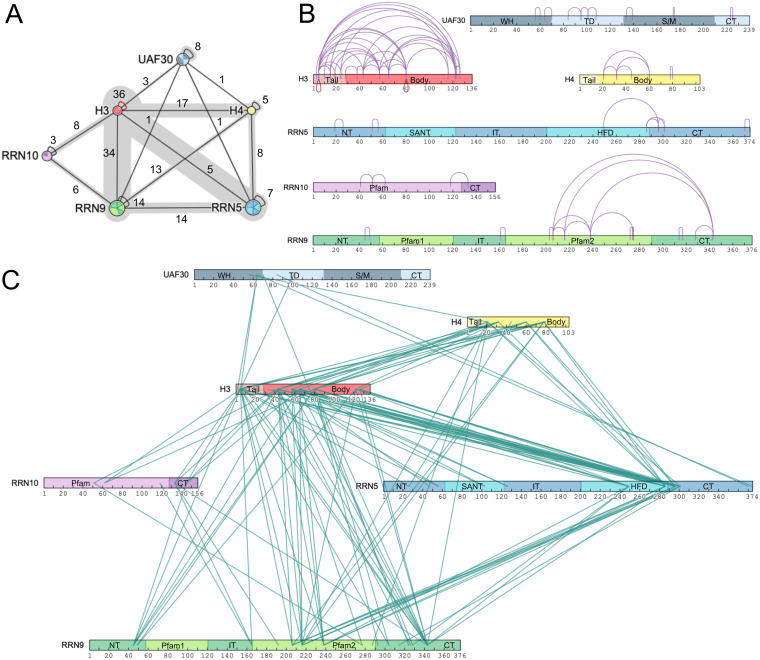
We next used combined chemical cross-linking and mass spectrometry (CXMS) to map potential protein-protein interactions with UAF and their potential topological orientation. We reconstituted UAF by coexpressing all six subunits in two compatible vectors in bacteria for use in our cross-linking analysis (Fig. 2A). We cross-linked the UAF complex with the homo-bifunctional and primary amine-reactive cross-linker BS3. Each UAF subunit contains 10 or more primary amine-reactive lysines well distributed throughout the proteins (see Fig. S1A and B in the supplemental material). UAF was incubated with increasing amounts of BS3 cross-linker and resulted in a cross-linked complex that migrates near the predicted mass sum of the entire complex (∼175 kDa) (23) (Fig. 2G), indicating our experimental conditions primarily cross-link monomeric UAF. BS3 cross-linked UAF was digested with trypsin, and the resulting peptides were analyzed by MS and by the Plink2 cross-link search database to identify cross-linked peptides and their site of cross-linking (28). Identified cross-links were used to generate a linkage map within and between the UAF subunits. CXMS analysis detected a total of 202 cross-links subdivided into 73 intramolecular and 132 intermolecular cross-links within and between the UAF subunits, respectively (Fig. 3 and Tables S2 and S3). We detected intramolecular cross-linking within each UAF subunit (Fig. 3A and B), and, strikingly, we observed two histone H3 cross-links with itself to the same peptide (Fig. 3B). This could be two complexes cross-linking together, but this more likely indicates that there are two copies of histone H3 in the UAF complex (23).
FIG 3.
Linkage map of cross-linked UAF lysine residues. (A) Protein-protein linkage map of UAF. The numbers of inter- and intramolecular cross-links between and within the proteins are listed. (B) Intramolecular cross-linking within the UAF subunits. (C) Intermolecular cross-linking between UAF subunits. Intra- and intermolecular cross-links are depicted as purple loops and teal lines, respectively. Red loops denote intramolecular cross-links where the same lysine in the same or similar peptides cross-link to each other. Domains are labeled and colored as described for Fig. 1.
The most extensive intermolecular cross-linking was observed with histone H3 and Rrn9, where they cross-linked to each of the five other UAF subunits, while histone H4, Rrn5, and Uaf30 cross-linked to only four other UAF subunits (Fig. 3A). We observed the fewest intermolecular cross-links with Rrn10, which only cross-linked to H3 and Rrn9 (Fig. 3A). Histone H3 exhibited the most intermolecular cross-linking, especially with histone H4 and Rrn9, and, likewise, we also detected extensive cross-linking between Rrn9 with histone H4 and Rrn5 (Fig. 3A and C). Together, these findings suggest that Rrn5, Rrn9, and histones H3 and H4 form the core of the UAF complex. We also mapped these cross-links onto our UAF domain predictions (Fig. 3B and C). Each UAF domain cross-links to one or more domains in the other subunits, except for four domains that include the histone H4 tail domain, the Uaf30 SWIB/MDM2 and CT domains, and the Rrn9 Pfam1 domain (Fig. 3B and C).
UAF domains important for yeast cell growth.
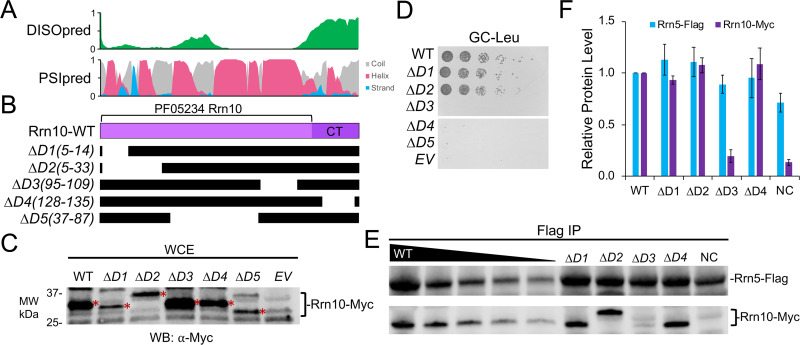
At the most basic level, cross-linking provides proximity information on the cross-linked sites, where the presence of cross-links with a subunit or between subunits of a complex does not necessarily indicate a direct protein-protein interaction and the lack of cross-links does not indicate the absence of a direct interaction. To systematically examine the domains and regions important for UAF function and validate our CXMS analysis, we generated a series of internal deletions within all four UAF-specific subunits (Rrn5, Rrn9, Rrn10, and Uaf30). We used a combination of secondary structure prediction programs that include PSIpred and DISOpred to predict regions containing secondary structure and disorder (29, 30), respectively. Our protein sequence analysis helped guide the removal of specific structural elements and reduce the chance of creating unstable protein variants. For instance, we removed entire secondary structure elements that include alpha-helical or beta sheet segments so that coils were fused with coil segments. We also inserted a glycine-serine-glycine linker for the internal deletion variants in place of the removed domain to further enhance protein stability (31). All of our UAF subunit deletion variants were N-terminally Myc tagged, expressed in yeast cells, and examined for protein expression by Western blotting compared to their respective wild-type (WT) forms. This was followed by determining the growth phenotypes of these UAF deletion variants in their respective deletion strains.
Five different Rrn10 deletion variants were generated and tested for expression in yeast compared to that of the wild type (Fig. 4A and B). All five Rrn10 deletion variants were expressed in yeast at or below wild-type levels (Fig. 4C). Rrn10 deletions D1(5-14) and D2(5-33), which removed the N terminus, were viable and grew similarly to wild-type Rrn10, whereas deletions removing the central half and the disordered C-terminal domain in Rrn10 deletion variants D3 to D5 resulted in severe slow-growth phenotypes (Fig. 4A and D). Most notable is that both the central region and C terminus of Rrn10 cross-link to both Rrn9 and histone H3 (Fig. 3C), suggesting that these regions are important for protein-protein interaction and association with UAF.
FIG 4.
Functional analysis of UAF-specific subunit Rrn10. (A) Immediately above the Rrn10 protein diagram is the DISOpred disorder, shown in green, followed by the PSIpred secondary structure predictions (helix, red; beta strand, blue; coil, gray). The scale bar on the y axis from 0 to 1 indicates the confidence of the predictions from low to high, respectively. (B) Below the Rrn10 protein diagram are the schematics of the various deletion constructs. Black bars denote regions included in the deletion construct. (C) Protein expression levels of N-terminally Myc-tagged Rrn10 deletion variants. The wild type and the indicated deletion variant were expressed in yeast, and whole-cell extracts were analyzed by Western blotting (WB) using the Myc epitope tag antibody. The Rrn10 protein bands are labeled with asterisks on their right side. For unknown reasons, Rrn10 deletion mutant D2 has a reproducibly anomalous mobility when expressed in bacteria and in yeast. MW, molecular weight. (D) An Δrrn10 yeast strain containing a plasmid that expresses the rDNA locus under the control of the GAL7 promoter was transformed with the indicated RRN10 expression constructs and spotted onto glucose complete (GC) plates lacking leucine. (E) The wild type and the indicated Rrn10 deletions were transformed into an Rrn5-Flag-tagged yeast strain. Western blot analysis of the Rrn10 wild type and deletion variants immunoprecipitated by Flag-tagged Rrn5, shown using the indicated antibodies. (F) Relative protein levels of the Flag-Rrn5 pulldown are normalized against the WT, which was set at 1. Pulldown assays were performed in biological duplicate, and representative assay results are shown. Error bars denote standard deviations. NC, negative control; EV, empty vector.
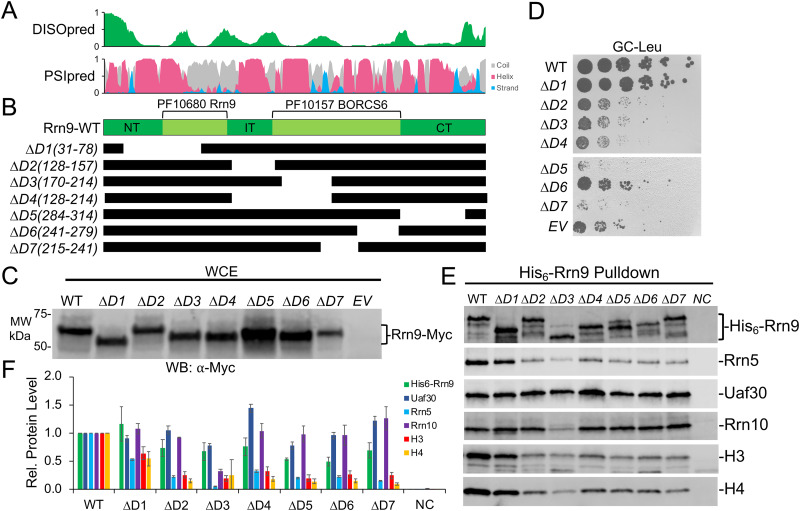
We designed seven Rrn9 deletion variants (Fig. 5A and B), and all of them were expressed at levels at or near wild-type Rrn9 levels in yeast (Fig. 5C). We observed a range of growth phenotypes for the Rrn9 deletion variants. For instance, the Rrn9 deletion mutants D2 to D7 all exhibited slow-growth phenotypes, with the exception of D1(31-78), which grew similarly to the wild type (Fig. 5D). Rrn9 deletion mutant D6(241-279) had a moderate slow-growth phenotype, while D2 to D4 were similar to cells lacking Rrn9 expression (Fig. 5D). We also observed dominant-negative growth phenotypes with Rrn9 deletion mutants D5(284-341) and D7(214-241), where they grew markedly slower than cells lacking Rrn9 (Fig. 5D). Both Rrn9 deletion mutants D5 and D7 removed a central portion of the Rrn9 Pfam BORCS6-like domain and the C-terminal (CT) domain (Fig. 5B), which exhibited extensive intramolecular cross-linking within Rrn9 and intermolecular cross-linking with all of the other UAF subunits (Fig. 3). However, Rrn9 deletion variant D1(31-78) removed the nonessential N-terminal region that also contains an intermolecular cross-linking cluster that is likely not functionally relevant for Rrn9 biological function. Together, these Rrn9 growth assay results indicate that all of the Rrn9 domains, with the exception of the N-terminal (NT) domain, are important for UAF function.
FIG 5.
Functional analysis of UAF-specific subunit Rrn9. (A) Immediately above the Rrn9 protein diagram is the DISOpred disorder, shown in green, followed by the PSIpred secondary structure predictions (helix, red; beta strand, blue; coil, gray). The scale bar on the y axis from 0 to 1 indicates the confidence of the predictions from low to high, respectively. (B) Below the Rrn9 protein diagram are the schematics of the various deletion constructs. Black bars denote regions included in the deletion construct. (C) Protein expression levels of N-terminally Myc-tagged Rrn9 deletion variants. The wild type and the indicated deletion variant were expressed in yeast, and whole-cell extracts were analyzed by Western blotting using the Myc epitope tag antibody. (D) A Δrrn9 yeast strain containing a plasmid that expresses the rDNA locus under the control of the GAL7 promoter was transformed with the indicated Rrn9 expression constructs and spotted onto glucose complete (GC) plates lacking leucine. (E) The wild type and the indicated Rrn9 deletions were coexpressed in bacteria with all remaining UAF subunits. Western blot analysis of isolated UAF complexes precipitated by the His6-tagged Rrn9 wild type or deletion variants is shown using the indicated antibodies. (F) Relative (Rel.) protein levels of the His6-Rrn9 pulldown are normalized against the WT, which was set at 1. Pulldown assays were performed in biological duplicate, and representative assay results are shown. Error bars denote standard deviations. NC, negative control; EV, empty vector.
For Rrn5, we were unable to utilize the Rrn5 deletion strain for our growth assays, because it would rapidly undergo a polymerase switch phenotype where Pol II rather than Pol I transcribes the rDNA locus and suppresses the UAF deletion phenotype (19, 20). The polymerase switch phenotype was not an issue for the Rrn9 and Rrn10 strains. To circumvent this issue, we engineered a double deletion strain of Rrn5 and Rpd3, the histone deacetylase that was previously shown to repress the polymerase switch phenotype, which is synthetically lethal when combined with Rrn5 (32, 33).
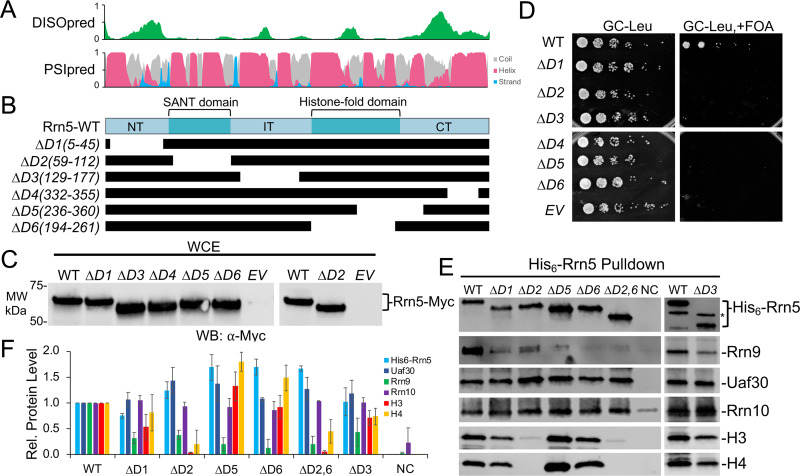
We designed six Rrn5 deletion mutants spanning the entire protein length (Fig. 6A and B), and we found that all were expressed at or near wild-type levels in yeast (Fig. 6C). We also found that all six Rrn5 deletion mutants exhibited lethal growth phenotypes (Fig. 6D), indicating that all Rrn5 domains, including the SANT and histone fold domains, are essential for yeast growth in the context of the double deletion strain. This is consistent with our cross-linking that shows all five Rrn5 domains cross-link with all other UAF subunits except Rrn10 (Fig. 3), suggesting that each Rrn5 domain plays an important role in UAF assembly and/or function.
FIG 6.
Functional analysis of UAF-specific subunit Rrn5. (A) Immediately above the Rrn5 protein diagram is the DISOpred disorder, shown in green, followed by the PSIpred secondary structure predictions (helix, red; beta strand, blue; coil, gray). The scale bar on the y axis from 0 to 1 indicates the confidence of the predictions from low to high, respectively. (B) Below the Rrn5 protein diagram are the schematics of the various deletion constructs. Black bars denote regions included in the deletion construct. (C) Protein expression levels of N-terminally Myc-tagged Rrn5 deletion variants. The wild type and the indicated deletion variants were expressed in yeast, and whole-cell extracts were analyzed by Western blotting using the Myc epitope tag antibody. (D) Growth of yeast strain with an Rrn5 plasmid shuffle strain (Δrrn5/Δrpd3). Equivalent amounts of yeast cells were serially diluted and spotted onto glucose complete (GC) medium lacking leucine, with or without 5-FOA. (E) The wild type and the indicated Rrn5 deletions were coexpressed in bacteria with all remaining UAF subunits. Western blot analysis of isolated UAF complexes precipitated by the His6-tagged Rrn5 wild type or deletion variants is shown using the indicated antibodies. (F) Relative (Rel.) protein levels of the His6-Rrn5 pulldown are normalized against the WT, which was set at 1. Pulldown assays were performed in biological duplicate, and representative assay results are shown. Error bars denote standard deviations. NC, negative control; EV, empty vector.
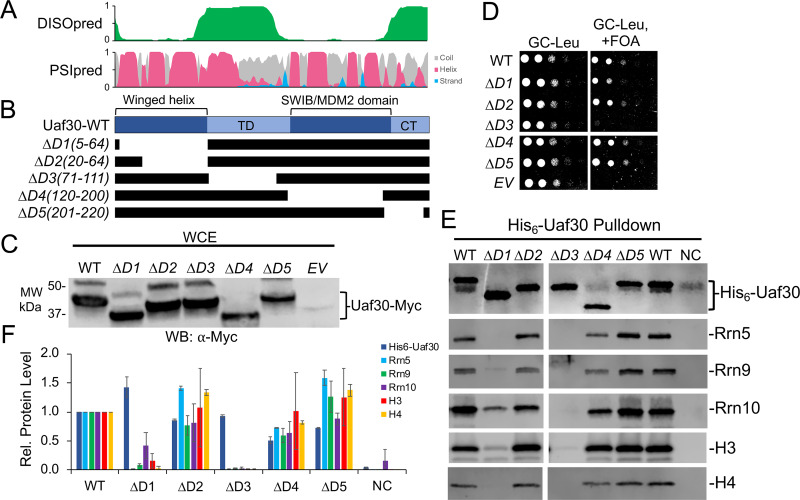
Finally, for Uaf30 we designed five different deletion variants (Fig. 7A and B). All five Uaf30 deletion variants were expressed at levels similar to those of the wild type (Fig. 7C). We were unable to test the effect of the Uaf30 domain deletions on yeast growth, as Uaf30 is nonessential and has no discernible growth phenotype when deleted (data not shown). However, Uaf30 is synthetically lethal with Rad50, a subunit of the MRX complex that is important for DNA damage repair, telomere maintenance, and nonhomologous end joining (34).
FIG 7.
Functional analysis of UAF-specific subunit Uaf30. (A) Immediately above the Uaf30 protein diagram is the DISOpred disorder, shown in green, followed by the PSIpred secondary structure predictions (helix, red; beta strand, blue; coil, gray). The scale bar on the y axis from 0 to 1 indicates the confidence of the predictions from low to high, respectively. (B) Below the Uaf30 protein diagram are the schematics of the various deletion constructs. Black bars denote regions included in the deletion construct. (C) Protein expression levels of N-terminally Myc-tagged Uaf30 deletion variants. The wild type and the indicated deletion variants were expressed in yeast, and whole-cell extracts were analyzed by Western blotting using the Myc epitope tag antibody. (D) Growth of yeast strain with a Uaf40 plasmid shuffle strain (Δuaf30/Δrad50). Equivalent amounts of yeast cells were serially diluted and spotted onto glucose complete (GC) medium lacking leucine, with or without 5-FOA. (E) The wild type and the indicated Uaf30 deletions were coexpressed in bacteria with all remaining UAF subunits. Western blot analysis of isolated UAF complexes precipitated by the His6-tagged Uaf30 wild type or deletion variants is shown using the indicated antibodies. (F) Relative (Rel.) protein levels of the His6-Uaf30 pulldown are normalized against the WT, which was set at 1. Pulldown assays were performed in biological duplicate, and representative assay results are shown. Error bars denote standard deviations. NC, negative control; EV, empty vector.
Therefore, we created a Uaf30 plasmid shuffle strain in a yeast background containing a Rad50 deletion. Using this strain, we observed slightly reduced growth phenotypes for Uaf30 deletion mutant D1(5-64) and a near-lethal growth phenotype of Uaf30 deletion mutant D3(71-11), while the remaining mutants grew similarly to wild-type Uaf30-expressing cells (Fig. 7D). Remarkably, our cross-linking shows that Uaf30 deletion mutants D1 and D3 cross-link to four UAF subunits, including Rrn5, Rrn9, and histones H3 and H4, demonstrating that these cross-linked regions are important for UAF function.
Domains important for UAF complex integrity.
Given the low expression levels of UAF in the cell, it is often difficult to perform immunoprecipitations from yeast cells in a manner that allows definitive detection of all the UAF subunits without extensive scale-up. To test the importance of topological domains in UAF complex integrity, we used our recombinant system to reconstitute UAF in bacteria. We expressed all the UAF subunits from three compatible coexpression vectors where one of the UAF subunits was His6 tagged in either its wild-type or deletion variant form. After nickel-agarose affinity purification, UAF complexes were analyzed by SDS-PAGE and Western blotting using available native and epitope tag antibodies for the recombinant UAF subunits.
We first examined the domains of Uaf30 necessary for association with the UAF complex. Previous studies have shown that Uaf30 is not required for UAF complex integrity (19). Likewise, Uaf30 is also easily dissociable from the complex, unlike other UAF subunits, as revealed by our prior native-MS analysis (23), indicating that Uaf30 lies on the periphery of the complex. Two Uaf30 domains were necessary for its association in the UAF complex. The removal of the N-terminal winged helix domain and the region between the winged helix and SWIB/MDM2 domain reduces the association of Uaf30 with the other UAF subunits (Fig. 7E and F). Both of these domains also show extensive cross-linking with other UAF subunits, which agrees well with their role in UAF association. Interestingly, DISOpred predicts the latter region is disordered, suggesting that it acts more like an extended tethering domain, since it also cross-links to multiple UAF subunits (Fig. 7A). The remaining Uaf30 domains, which include the SWIB/MDM2 and C-terminal domains, can be removed with little consequence to UAF complex integrity and cell growth (Fig. 7D to F). Note that we did not detect any cross-links with other UAF subunits in the SWIB/MDM2 and C-terminal domains, further suggesting that these Uaf30 domains reside on the periphery further away from the core of the UAF complex.
We next examined the Rrn9 deletion mutants for their effect on UAF complex integrity. All seven Rrn9 deletions were partially defective, in association with the other UAF subunits, except for Rrn9 deletion mutant D1(31-78), which copurified at wild-type levels of the other UAF subunits (Fig. 5E and F), as expected given its wild-type growth phenotype (Fig. 5D). For the remaining Rrn9 deletion mutants, we noticed modest reductions in Rrn5 and histone H3 and H4 association (Fig. 5E and F). This is consistent with our yeast growth results, showing that most of the Rrn9 protein besides the N terminus is necessary for growth, along with our cross-linking data, indicating the other UAF subunits cross-link the central and C-terminal regions of Rrn9. Uaf30 retained nearly wild-type association with all the Rrn9 deletion mutants, while Rrn10 also retained nearly wild-type association, except for the D3(71-111) mutant, which removed a portion of an internal domain between the Rrn9 Pfam domains as well as the N terminus of the Pfam BORC6-like domain, both of which cross-link to Rrn10 (Fig. 5E and F).
For Rrn5, we tested the importance of several domains for UAF complex integrity. All the Rrn5 mutants showed reduced association with Rrn9 (Fig. 6E and F), which was expected for the deletions removing the Rrn5 histone fold and C-terminal domain, which cross-link extensively to Rrn9. Uaf30 and Rrn10 remain associated at nearly wild-type levels for all the Rrn5 deletions tested (Fig. 6E and F). Interestingly, the removal of the Rrn5 SANT domain reduces histone H3 and H4 association (Fig. 6E and F). The SANT domain cross-links to histone H3 tail and body domains, suggesting they are in close proximity, and this agrees well with the role of SANT domain-containing protein in interacting with histone proteins (35, 36). Unexpectedly, we did not observe reduced association of the histone subunits with deletion D6(194-261), which removes the Rrn5 histone fold domain (Fig. 6E and F). We also combined the Rrn5 deletions D2(59-112) and D6(194-261) to simultaneously remove the SANT and histone fold domains, which results in a biochemical phenotype similar to that of Rrn5-D2 deletion alone (Fig. 6E and F). Although we observe extensive cross-linking between the Rrn5 histone fold domain and histones H3 and H4, it is not required for their association with the UAF complex. However, we observed reduced Rrn9 association, with Rrn5 lacking the histone fold domain, suggesting it is necessary for overall UAF complex integrity.
We were unable to use our recombinant expression system to examine the effect of the Rrn10 deletion variants on UAF complex integrity due to the incompatibility of the coexpression vectors (data not shown). Instead, we transformed our C-terminally Myc-tagged Rrn10 variants into a yeast strain where Rrn5 is Flag tagged in the chromosome. We examined several Rrn10 mutants, with the exception of Rrn10 deletion mutant D5, which expresses poorly (Fig. 4C). When we immunoprecipitated Rrn5 from these strains, we found that Rrn10 deletion mutant D3(95-109) no longer immunoprecipitated with Rrn5, indicating that this region is important for the association of Rrn10 with the UAF complex, where the other Rrn10 mutants tested precipitated similarly with Rrn5 (Fig. 4E and F). Rrn10 deletion mutant D3 removes a region important for cell growth yet lacks cross-linking with other UAF subunits, which may be explained by the fact that there is only a single residue in this region. However, the Rrn10-D3 region is immediately flanked by two cross-linking patches on either side. Rrn10 mutant D4(128-135) removes a portion of the C terminus important for cell growth and cross-links with other UAF subunits, yet it retained association with Rrn5 (Fig. 4E and F). This suggests that this region is in close proximity to these other UAF subunits; however, the C terminus may not be important for UAF complex integrity but serves another UAF function. We attempted Western analysis for the other UAF subunits that include Rrn9 and histones H3 and H4, but these native antibodies lacked sufficient sensitivity, unlike the multicopy epitope tags (data not shown).
Modeling of UAF topology.
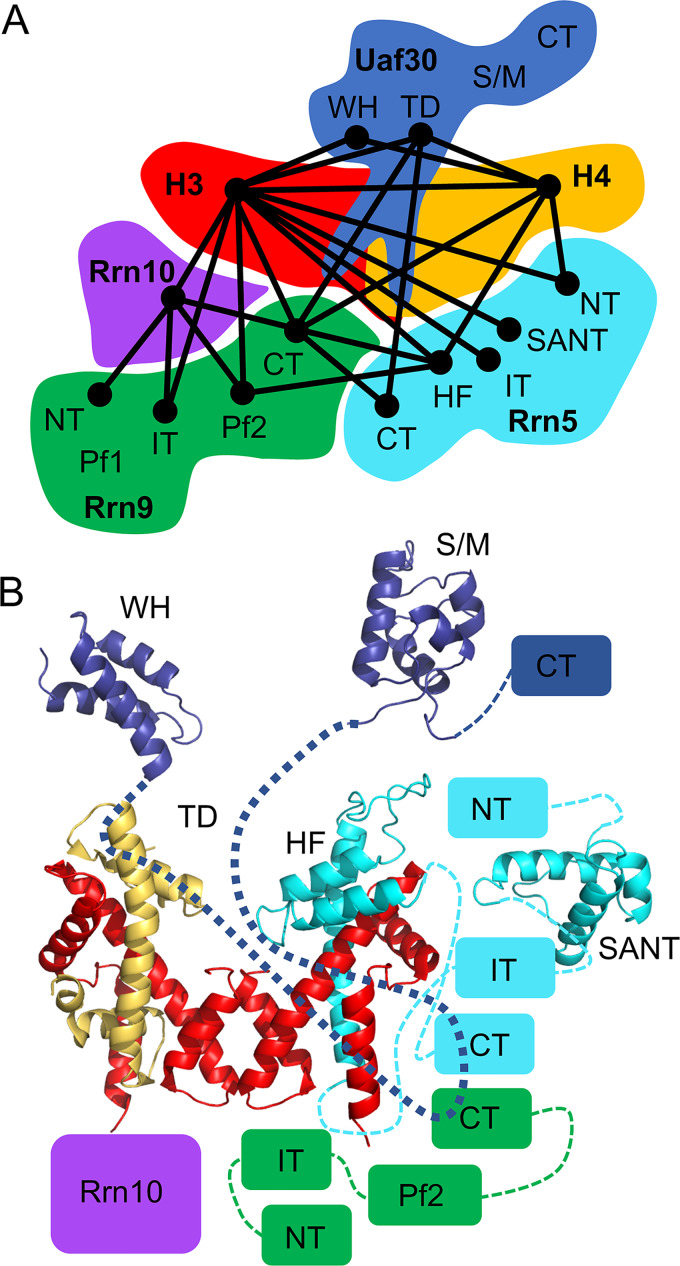
To understand the topology of the UAF complex, we used a combination of bioinformatics, protein modeling, and CXMS data to derive a topological model of potential domain-domain contacts. Interdomain cross-links were to a single connection, and their position and proximity were based on their potential interaction and/or known interaction (Fig. 8A). The topological domain-domain contact model shows an interconnected network of domain interactions centered around the histone subunits.
FIG 8.
Molecular topological model of UAF. (A) Cartoon representation of the domain contacts within UAF. Lines between UAF domains indicate a potential domain contact identified by cross-linking. (B) Topological linkage map of UAF depicting all the UAF structural and functional domains. Domains are positioned using the cross-linking, genetic, and biochemical results as a guide. Domains that lack known structure are depicted as boxes. Dashed lines connect each domain within a protein. TD, tethering domain; HF, histone fold domain; S/M, SWIB/MDM2 domain; WH, winged helix domain; NT, N-terminal domain; IT, internal domain; CT, C-terminal domain.
We built models for the predicted structured domains of the UAF subunits using HHpred alignments with known PDB structures, followed by comparative modeling with Modeller (Fig. 1). Atomic structures of histone H3 and H4 are known, and together with the new homology models, we can build models for covering ∼40% of the UAF protein sequences. Together with our previous results and those presented here, UAF contains two copies of histone H3 and a single copy of histone H4. We predict that the UAF histones assemble into a tetramer arrangement that lacks one of the copies of histone H4. To complete the hybrid tetramer-like structure, we replace the missing histone H4 copy with a model of the Rrn5 histone fold domain (Fig. 1 and 8B).
We next mapped the cross-links onto the structural models and measured their C-α–C-α distances. The BS3 cross-linker has an 11.4-Å linker arm and has a theoretical maximum allowable distance between C-α atoms of cross-linked lysine residues of 34 Å. A total of 22 intramolecular cross-links are within structured domains with either known or predicted structures. When mapped onto the domain models, 18 of 22 cross-links have C-α–C-α distances of 34 Å or less (37) (Fig. S2A and B). Given that there are two copies of histone H3 in the complex, CXMS cannot distinguish between an interlink between two copies or an intralink within one copy. Therefore, measurable C-α–C-α distances for cross-links between H3-H3, H3-R5, and H3-H4 body domains were measured from both copies in their respective dimer or tetramer positions. Overall, we found that ∼76% of the H3 interlinks measured were within the cross-linker distance constraints if H3 was in the dimer position, whereas only ∼13% were within the distance constraints when measured in the tetramer position (Fig. S2A, C, and D). It is also likely that cross-linking occurs within histone dimer pairs more frequently than between tetramer pairs because of the interwoven interaction between dimerized subunits versus their tetramer interaction interface.
One of the H3 intralinks within the same peptide was measurable, and its distance of >55 Å was well above the cross-linker distance maximum. This suggests that the UAF tetramer is oriented differently from that of the canonical nucleosomal tetramer. We were also able to measure the C-α–C-α distance between histone H4 and the Rrn5 histone fold domain, and all of them exceeded the cross-linker distance constraints (Fig. S2B), but we note that the Rrn5 lysine cross-linked within this domain is highly mobile, as it is located in an extended and flexible loop. Taken together, these results reveal an increasingly intricate and interconnected network of protein-protein interactions where the UAF-specific subunits surround the histone tetramer-like core. Furthermore, the UAF tetramer-like core may be in a unique UAF-specific arrangement that is different from that of the canonical H3-H4 tetramer.
DISCUSSION
Here, we resolved the molecular and functional topology of the Pol I transcription activator UAF. We found that UAF assembles through an interconnected network of protein-protein interaction around a unique hybrid tetramer-like core. We found several new and biologically important structural domains, identified by deep protein sequence analysis, that participate in the formation of the UAF complex. Our results provide a topological framework to further explore the functional features of the UAF complex. We highlight some of these structural and functional features below and discuss their potential contribution to the UAF complex.
We found two new functional domains within the N-terminal half of Uaf30 that include the winged helix domain and the tethering domain. The winged helix domain belongs to a subtype of the helix-turn-helix protein family that functions in both protein-protein and protein-DNA interactions and is a common motif found in transcription factors (38, 39). Our cross-linking and functional assays support a role for the Uaf30 winged helix domain as a protein-protein interaction interface, since a portion is necessary for Uaf30 association into the UAF complex. However, the Uaf30 winged helix domain may also contribute to the interaction with the rDNA promoter. In the absence of Uaf30, the UAF complex results in a smaller DNase I footprint at the rDNA promoter, in vitro DNA binding activity of UAF is reduced, and it no longer stably associates with chromatin in vivo (40), suggesting a role for Uaf30 in protein-rDNA interaction. We also identified a unique disordered region we called the tethering domain between the winged helix and SWIB/MDM2 domain of Uaf30. Several UAF subunits cross-link to this region, and its absence results in dissociation of Uaf30 from the complex, suggesting that it helps anchor Uaf30 to the complex.
Strikingly, we did not detect cross-linking within the SWIB/MDM2 domain, and it was not required for UAF association. The SWIB/MDM2 domain belongs to a conserved protein fold family, where it is found in a subunit of the SWI/SNF chromatin remodeling complex and the negative regulator of the p53 tumor suppressor MDM2, which participate in protein-protein and chromatin-related interactions (41). For UAF, it appears the Uaf30 SWIB/MDM2 and C-terminal domain are not required for Uaf30 association with UAF and may reside at the periphery of the complex, where they are accessible to interact with other transacting factors among the Pol I initiation and/or regulatory factors or possibly rDNA. We also note that the C-terminal domain of Uaf30 is phosphorylated (42–44). Although its functional and/or regulatory significance remains unclear, a peripheral location in the complex is ideal for phosphorylation.
Our topological model of UAF also suggests an important role of the Rrn5 SANT domain. SANT domains are multifunctional and participate in both protein-protein and protein-DNA interactions (35, 36, 45) that consist of three tandem alpha helices arranged in helix-turn-helix assembly (36, 45). When the essential Rrn5 SANT domain is deleted, Rrn5 no longer associates with UAF, indicating its importance in UAF complex integrity. Besides interacting with DNA, SANT domains also interact with unmodified histone tails (35, 36). The SANT domain of UAF has the opportunity to interact with the tail domains of histone H4 and two copies of histone H3. Additional focus on the role of the Rrn5 SANT domain in the UAF complex will be necessary to investigate the possibilities of this important topological and functional feature.
The remaining UAF subunits Rrn9 and Rrn10 remain a mystery in terms of their protein structure, but we identified several Pfam domains. The most informative identified Pfam domain is in Rrn9; it matches a domain similar to the BORC6 subunit of the octameric BORC complex, which plays a role in lysosome movement and localization to the cytosolic face of the cell periphery (46, 47). This domain contains coil-coiled segments (47), indicating a role in protein-protein interactions for the assembly and integrity of both BORC and UAF. In addition, Rrn9 BORCS6 also may be important for TBP and Rrn10 interaction (10). Rrn9 point mutations (L185S and F186S) in this region reduce the genetic interaction between Rrn9 with both TBP and Rrn10 in two-hybrid assays (10). Furthermore, removal of the N-terminal portion of the Rrn9 BORCS6-like domain, which is required for Rrn10 association in our reconstituted assay, overlaps the Rrn9 point mutations, which reduce both TBP and Rrn10 interactions (10). This further demonstrates that the Rrn9 BORCS6 domain is biologically relevant for UAF function.
Recent genetic screening for SIR2 repressors identified all four UAF-specific subunits, including Rrn5, Rrn9, Rrn10, and Uaf30, in the repression of SIR2 at the promoter (22). Each of these suppressor mutations were mapped in many of the domains we identified in the analysis presented here. More specifically, suppressor mutations were mapped in all of the Rrn5 domains, where each domain is necessary for biological function (22). For Rrn9, suppressor mutations were found in all of the Rrn9 domains, with the exception of the nonessential N-terminal domain (22). Rrn10 contains suppressor mutations in the Pfam and C-terminal domains (22), which are both required for yeast cell growth and UAF function. Finally, Uaf30 contained suppressor mutations in the tethering domain and SWIB/MDM2 domain (22), again consistent with our analysis, and reveals a biological relevance to the SWIB/MDM2 domain for UAF function. Together, our results are in nearly perfect agreement with domains that were identified as important for UAF in rDNA copy number maintenance and SIR2 repression.
Ultimately, high-resolution structures of the UAF complex alone and in complex with DNA and other protein factors is the next logical step in understanding the role of these biologically relevant topological features. High-quality yeast structures of minimal preinitiation complexes (PICs) containing Pol I, Rrn3, and CF are already available and have informed us of the conserved and unique properties of the Pol I transcription system (48–53). Future assemblies that include Pol I factors such as TBP, CF, and/or the entire Pol I PIC, as well as assemblies at the SIR2 locus, will help us further understand the functionality of UAF and its unique domains in Pol I activation and Pol II repression. The work presented here will help complement and extend future UAF structural studies.
MATERIALS AND METHODS
UAF sequence analysis and structural modeling.
DISOpred was used to predict UAF subunit disordered regions (30), and PSIpred was used to predict UAF subunit secondary structure (29). Confidence values for the predictions were plotted as area graphs for each residue position. Structure similarity searches were performed by HHpred (http://toolkit.tuebingen.mpg.de/hhpred/) with HMM PDB and Pfam databases under default settings and thresholds (25, 54, 55). UAF subunits Rrn5, Rrn9, Rrn10, and Uaf30 were searched by HHpred to identify and align homologous proteins of known structure and/or function. PIR alignments of the query and target sequences were generated by HHPred with the “create model” option on the HHPred results page and manually converted to FASTA format. Templates were selected based on the highest probability score and highest protein identity. These alignments were used to generate structural homology models by Modeller 9v10 with the UCSF chimera interface (56, 57). Five models of each domain were generated, and the best-scoring model was chosen. The following sequences and templates were used for structural modeling: Uaf30 winged helix domain residues 1 to 58 aligned with human DEK residues 11 to 69 (PDB entry 1Q1V_A) (58), Uaf30 SWIB/MDM2 residues 114 to 204 aligned with Arabidopsis thaliana SWI/SNF complex subunit RAFL11 residues 2 to 93 (PDB entry 1V31_A), Rrn5 SANT domain residues 52 to 111 aligned with human Myb SANT domain residues 1 to 63 (PDB entry 1WGX_A), and Rrn5 histone fold domain residues 190 to 277 aligned with human NF-Y residues 25 to 93 (PDB entry 4CSR_A).
UAF expression and purification.
Expression and purification strategies were adapted and modified as previously described (23). Briefly, we used two compatible bacterial coexpression vectors to reconstitute UAF, the pET-Duet vector containing His6 epitope-tagged RRN9, UAF30, and RRN10, and the pCDF-Duet vector containing histones H3 and H4 and His6-tagged RRN5. UAF was expressed by autoinduction in LOBSTR BL21(DE3) RIL cells (Kerafast) in TB medium (0.024% yeast extract, 0.012% tryptone, and 0.4% glycerol) supplemented with autobase (0.17 M KH2PO4 and 0.72 M K2HPO4), 5052 (0.1% alpha-lactose monohydrate, 0.25% glycerol, and 0.025% glucose), and MgSO4 (2 mM). After inoculation, cells were grown at 37°C until an optical density at 600 nm (OD600) of ∼0.5, cooled on ice, and grown at 20°C for 18 h. Cells were harvested by centrifugation and washed once with UAF extraction buffer [200 mM Tris-HCl (pH 8.0), 400 mM (NH4)2SO4, 20 mM imidazole, 0.1% Tween 20, and 10% glycerol] supplemented with protease inhibitors and 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP). Pellets were resuspended in extraction buffer supplemented with 1 mg/ml lysozyme on ice for 30 min and then lysed by sonication. Lysates were cleared and incubated with nickel-Sepharose beads (G-Biosciences) overnight at 4°C. UAF-bound beads were washed three times with wash buffer (20 mM Tris-HCl [pH 8.0], 20% glycerol, 0.1% Tween 20, 20 mM imidazole, and 450 mM KCl). UAF was eluted in batch with wash buffer supplemented with 250 mM imidazole. Elutions were pooled and incubated overnight with SP Sepharose beads (GE Healthcare) at 4°C. UAF-bound beads were washed three times with wash buffer. UAF was eluted in batch with SP elution buffer (20 mM Tris-HCl [pH 8.0], 20% glycerol, 0.1% Tween 20, and 750 mM KCl). UAF was concentrated on a 100-kDa-cutoff Centricon filter (Millipore) and then further purified by size exclusion chromatography on a Superose 6 10/300 column (GE Healthcare) equilibrated in SEC buffer (20 mM Tris-HCl [pH 8.0], 20% glycerol, 0.1% Tween 20, 450 mM KCl, 20 mM l-arginine) at a flow rate of 0.5 ml/min. Peak fractions were concentrated again on a 100-kDa-cutoff Centricon filter (Millipore) and stored at –80°C.
BS3 cross-linking.
Fifty micrograms of UAF was cross-linked with BS3 [bis(sulfosuccinimidyl) suberate; Thermo Scientific] at final concentrations of 2 mM and 4 mM BS3 for 2 h at room temperature, stored overnight at 4°C in 200-μl reaction mixtures, and further processed as previously described (4). Briefly, an equal volume of trifluoroethanol was added to the BS3 cross-linked protein complex, and proteins were denatured at 60°C for 30 min and then reduced by addition of 5 mM TCEP for 30 min at 37°C. The sample next was alkylated with iodoacetamide at a 10 mM final concentration for 30 min in the dark at room temperature, followed by 10-fold dilution with 20 mM triethanolamine and digestion with 1 μg of trypsin overnight at 37°C. The peptides were further purified on C18 spin columns (Nest Group), dried, and stored at 20°C.
Mass spectrometry sample preparation.
Dried peptides were resuspended in 5% acetonitrile–0.1% trifluoroacetic acid solution and analyzed on a Thermo Scientific Orbitrap Elite or Lumos with higher-energy collisional dissociation (HCD) fragmentation as previously described (4). Briefly, serial MS events included one FTMS1 event at 30,000 resolution followed by ten FTMS2 events at 15,000 resolution. Additional settings were as follows: MS mass range greater than 1,800; m/z value as masses enabled; charge state rejection of +1, +2, and unassigned charges; monoisotopic precursor selection enabled; dynamic exclusion enabled; repeat count of 1; exclusion list size of 500; exclusion duration of 30 s; HCD normalized collision energy of 35%; isolation width of 3 Da; minimum signal count of 5,000; and FTMS MSn AGC target of 50,000. The RAW files were converted to mgf files and analyzed by the pLink2 database-searching algorithms (28). The following pLink2 parameters settings were used: (i) up to three miscleavages, (ii) static modification on cysteines (+57.0215 Da), (iii) differential oxidation modification on methionines (+15.9949 Da), (iv) differential modification on the peptide N-terminal glutamate residues (−18.0106 Da) or N-terminal glutamine residues (−17.0265 Da), and (v) 2% false discovery rate (FDR). All possible tryptic peptide pairs within 20 ppm of the precursor mass are used for cross-linked peptide searches. The cross-linked peptides were considered confidently identified if at least four consecutive b or y ions for each peptide were observed. The xiNET cross-linking viewer was used to visualize UAF cross-linking linkage maps (http://www.crosslinkviewer.org) (59).
Yeast growth assays.
The genetic backgrounds for the Rrn9 and Rrn10 strains were the following: Rrn9, MATa ade2-1 ade3::hisG ura3-1 his3-11 trp1-1 leu2-112 can1 rrn9Δ::HIS3 pNOY103; Rrn10, MATa ade2-1 ade3::hisG ura3-1 his3-11 trp1-1 leu2-112 can1 rrn10Δ::HIS3 (60, 61). These strains are deleted for RRN9 or RRN10 and pNOY103, which expresses the 35S rRNA transcript under the control of the GAL7 promoter (60, 61). These yeast strains were transformed with either wild-type or mutant UAF subunit deletion variants. Cells were grown in galactose complete medium lacking leucine and then washed extensively with water to remove residual galactose. Cells next were serially diluted and spotted on glucose-containing medium lacking leucine. Plates were incubated for 2 to 3 days at 30°C and then assessed for growth relative to that of the wild type. For Rrn5 and Uaf40, yeast cell viability assays were performed on glucose complete plates containing 1 g/liter 5-fluoroorotic acid (5-FOA). The genetic backgrounds for the Rrn5 and Uaf30 deletion strains are the following: Rrn5, MATa ade2-1 ade3::hisG ura3-1 his3-11 trp1-1 leu2-112 can1 rpd3::NatX rrn5Δ::HphB pRS316-RRN5; Uaf30, MATa ade2-1 ade3::hisG ura3-1 his3-11 trp1-1 leu2-112 can1 rad50Δ::HIS3 uaf30Δ::KanMX pRS316-UAF30. Yeast Myc-tagged pRS425 PGK expression vectors used for growth assays are listed in Table S1 in the supplemental material.
Yeast protein expression levels.
Whole-cell extracts (WCE) were prepared, with minor modifications, as described in references 4 and 62. In brief, 5 ml of yeast cells was grown in glucose complete minimal medium lacking leucine to an OD of 0.8 to 1.0. Cells were washed with water, pelleted, and resuspended in 0.1 M NaOH for 5 min at room temperature. Cells were pelleted and resuspended in 1× SDS sample buffer (Bio-Rad) and heated at 95°C for 5 min. Cell debris was pelleted, and soluble extract was collected and analyzed by SDS-PAGE and Western blotting. Proteins were resolved on Bio-Rad 4 to 20% Tris-glycine polyacrylamide gradient gels in 1× Tris-glycine SDS buffer, transferred to low-fluorescence polyvinylidene difluoride (Millipore), and probed with anti-MYC antibody (BioLegend). The UAF subunit variants were expressed in their appropriate genetic backgrounds, listed above, with the exception of the Uaf30 variants, which were expressed in the wild-type background: MATa ade2-1 ade3::hisG ura3-1 his3-11 trp1-1 leu2-112 can1.
UAF complex integrity assays.
UAF bacterial expression plasmids used for these complex integrity assays are listed in Table S1. Three compatible UAF bacterial expression vectors in pET-DUET, pCDF-DUET, and pACYC-DUET were cotransformed into BL21(DE3) cells and expressed by autoinduction in TB medium. After inoculation, cells were grown at 37°C until an OD600 of ∼0.5 and grown at 20°C for 16 to 18 h. Cells were harvested by centrifugation, washed once with UAF extraction buffer, and frozen at –80°C. To perform the pulldown experiment, the cell pellets were thawed and lysed with sonication, and soluble extract was prepared as described above for UAF. Cleared lysates were incubated with nickel-Sepharose beads (G-Biosciences) overnight at 4°C. Protein-bound beads were washed four times with wash buffer (20 mM Tris-HCl [pH 8.0], 20% glycerol, 0.1% Tween 20, and 450 mM KCl) and eluted three times with two bead volumes of elution buffer (wash buffer supplemented with 250 mM imidazole). Eluted proteins were separated on 4 to 20% Tris-glycine SDS-PAGE gel (Bio-Rad) and analyzed by Western blotting using the following antibodies: anti-histone H3 (ab46765; Abcam), anti-histone H4 (ab10158; Abcam), anti-Flag (F3165; Sigma) for Uaf30, and rabbit polyclonal antibodies against Rrn10, Rrn5, or Rrn9 (Knutson laboratory via Reeder laboratory).
Flag immunoprecipitations were performed from 1-liter mid-log-phase cultures grown in glucose complete (GC) medium lacking leucine. A Flag-tagged Rrn5 yeast strain was used for these assays with the following genetic background: MATα ade2-1 ade3::hisG ura3-1 his3-11 trp1-1 leu2-112 can1 RRN5-3×Flag::NatX. Cells were collected by centrifugation and lysed by bead beating (Omni Bead Ruptor Elite) in lysis buffer (100 mM Tris-HCl [pH 7.9], 250 mM ammonium sulfate, 1 mM EDTA, and 10% glycerol) supplemented with 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitors. Lysates were cleared by centrifugation, and approximately 15 mg of lysate was diluted with two volumes of IP dilution buffer (25 mM HEPES [pH 7.5], 50 mM NaCl, and 1 mM EDTA) and incubated overnight at 4°C with 100 μl of anti-Flag affinity agarose (Bimake). Beads were washed four times in 1 ml Tris-buffered saline (TBS) (50 mM Tris [pH 7.5], and 150 mM) containing 0.1% Tween and two times with TBS. Proteins were eluted three times in TBS containing 150 ng/ml 3×Flag peptide (Sigma) at room temperature. Eluted proteins were separated on 4% to 20% Tris-glycine SDS-PAGE gel (Bio-Rad) and analyzed by Western blotting using the anti-Flag and anti-Myc antibodies described above.
Supplementary Material
ACKNOWLEDGMENTS
We thank Knutson laboratory members for their critical review of the manuscript. We also thank Phil Gafkin at the Fred Hutchison Cancer Research Center proteomic facility and Ebbing De Jong at the SUNY-Upstate proteomics facility for their help with mass spectrometry.
The work was supported by grants to B.A.K. from the U.S. National Institutes of Health (NCI 5K22CA184235) and a Sinsheimer Scholar award from the Alexandrine and Alexander L. Sinsheimer Fund, Central New York Community Foundation, Joseph C. George Fund, Virginia Simons & C. Adele Brown Fund, and Carol Baldwin Research Foundation.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Aprikian P, Moorefield B, Reeder RH. 2000. TATA binding protein can stimulate core-directed transcription by yeast RNA polymerase I. Mol Cell Biol 20:5269–5275. doi: 10.1128/MCB.20.14.5269-5275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedwell GJ, Appling FD, Anderson SJ, Schneider DA. 2012. Efficient transcription by RNA polymerase I using recombinant core factor. Gene 492:94–99. doi: 10.1016/j.gene.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cormack BP, Struhl K. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 4.Knutson BA, Luo J, Ranish J, Hahn S. 2014. Architecture of the Saccharomyces cerevisiae RNA polymerase I core factor complex. Nat Struct Mol Biol 21:810–816. doi: 10.1038/nsmb.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lalo D, Steffan JS, Dodd JA, Nomura M. 1996. RRN11 encodes the third subunit of the complex containing Rrn6p and Rrn7p that is essential for the initiation of rDNA transcription by yeast RNA polymerase I. J Biol Chem 271:21062–21067. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- 6.Lin CW, Moorefield B, Payne J, Aprikian P, Mitomo K, Reeder RH. 1996. A novel 66-kilodalton protein complexes with Rrn6, Rrn7, and TATA-binding protein to promote polymerase I transcription initiation in Saccharomyces cerevisiae. Mol Cell Biol 16:6436–6443. doi: 10.1128/MCB.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milkereit P, Tschochner H. 1998. A specialized form of RNA polymerase I, essential for initiation and growth-dependent regulation of rRNA synthesis, is disrupted during transcription. EMBO J 17:3692–3703. doi: 10.1093/emboj/17.13.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddiqi I, Keener J, Vu L, Nomura M. 2001. Role of TATA binding protein (TBP) in yeast ribosomal DNA transcription by RNA polymerase I: defects in the dual functions of transcription factor UAF cannot be suppressed by TBP. Mol Cell Biol 21:2292–2297. doi: 10.1128/MCB.21.7.2292-2297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steffan JS, Keys DA, Dodd JA, Nomura M. 1996. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev 10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 10.Steffan JS, Keys DA, Vu L, Nomura M. 1998. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol Cell Biol 18:3752–3761. doi: 10.1128/MCB.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto RT, Nogi Y, Dodd JA, Nomura M. 1996. RRN3 gene of Saccharomyces cerevisiae encodes an essential RNA polymerase I transcription factor which interacts with the polymerase independently of DNA template. EMBO J 15:3964–3973. doi: 10.1002/j.1460-2075.1996.tb00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keys DA, Lee BS, Dodd JA, Nguyen TT, Vu L, Fantino E, Burson LM, Nogi Y, Nomura M. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev 10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- 13.Keener J, Dodd JA, Lalo D, Nomura M. 1997. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc Natl Acad Sci U S A 94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiqi IN, Dodd JA, Vu L, Eliason K, Oakes ML, Keener J, Moore R, Young MK, Nomura M. 2001. Transcription of chromosomal rRNA genes by both RNA polymerase I and II in yeast uaf30 mutants lacking the 30 kDa subunit of transcription factor UAF. EMBO J 20:4512–4521. doi: 10.1093/emboj/20.16.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe SY, Schultz MC, Reeder RH. 1992. In vitro definition of the yeast RNA polymerase I promoter. Nucleic Acids Res 20:279–285. doi: 10.1093/nar/20.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musters W, Knol J, Mass P, Dekker AF, van Heerikhuizen H, Planta RJ. 1989. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res 17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulkens T, Riggs DL, Heck JD, Planta RJ, Nomura M. 1991. The yeast RNA polymerase I promoter: ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res 19:5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keener J, Josaitis CA, Dodd JA, Nomura M. 1998. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J Biol Chem 273:33795–33802. doi: 10.1074/jbc.273.50.33795. [DOI] [PubMed] [Google Scholar]
- 19.Oakes M, Siddiqi I, Vu L, Aris J, Nomura M. 1999. Transcription factor UAF, expansion and contraction of ribosomal DNA (rDNA) repeats, and RNA polymerase switch in transcription of yeast rDNA. Mol Cell Biol 19:8559–8569. doi: 10.1128/MCB.19.12.8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vu L, Siddiqi I, Lee BS, Josaitis CA, Nomura M. 1999. RNA polymerase switch in transcription of yeast rDNA: role of transcription factor UAF (upstream activation factor) in silencing rDNA transcription by RNA polymerase II. Proc Natl Acad Sci U S A 96:4390–4395. doi: 10.1073/pnas.96.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iida T, Kobayashi T. 2019. How do cells count multi-copy genes? “Musical chair” model for preserving the number of rDNA copies. Curr Genet 65:883–885. doi: 10.1007/s00294-019-00956-0. [DOI] [PubMed] [Google Scholar]
- 22.Iida T, Kobayashi T. 2019. RNA polymerase I activators count and adjust ribosomal RNA gene copy number. Mol Cell 73:645–654. doi: 10.1016/j.molcel.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 23.Smith ML, Cui W, Jackobel AJ, Walker-Kopp N, Knutson BA. 2018. Reconstitution of RNA polymerase I upstream activating factor and the roles of histones H3 and H4 in complex assembly. J Mol Biol 430:641–654. doi: 10.1016/j.jmb.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alva V, Nam SZ, Soding J, Lupas AN. 2016. The MPI bioinformatics toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, Guo A, Boukhgalter B, Van Den Heuvel K, Tripp M, Pape L. 2002. Characterization of the fission yeast ribosomal DNA binding factor: components share homology with upstream activating factor and with SWI/SNF subunits. Nucleic Acids Res 30:5347–5359. doi: 10.1093/nar/gkf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZL, Meng JM, Cao Y, Yin JL, Fang RQ, Fan SB, Liu C, Zeng WF, Ding YH, Tan D, Wu L, Zhou WJ, Chi H, Sun RX, Dong MQ, He SM. 2019. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat Commun 10:3404. doi: 10.1038/s41467-019-11337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 30.Jones DT, Cozzetto D. 2015. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Zaro JL, Shen WC. 2013. Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65:1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandmeier JJ, French S, Osheim Y, Cheung WL, Gallo CM, Beyer AL, Smith JS. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J 21:4959–4968. doi: 10.1093/emboj/cdf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oakes ML, Siddiqi I, French SL, Vu L, Sato M, Aris JP, Beyer AL, Nomura M. 2006. Role of histone deacetylase Rpd3 in regulating rRNA gene transcription and nucleolar structure in yeast. Mol Cell Biol 26:3889–3901. doi: 10.1128/MCB.26.10.3889-3901.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gobbini E, Cassani C, Villa M, Bonetti D, Longhese MP. 2016. Functions and regulation of the MRX complex at DNA double-strand breaks. Microb Cell 3:329–337. doi: 10.15698/mic2016.08.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell 10:935–942. doi: 10.1016/S1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 36.Boyer LA, Latek RR, Peterson CL. 2004. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol 5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 37.Merkley ED, Rysavy S, Kahraman A, Hafen RP, Daggett V, Adkins JN. 2014. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci 23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harami GM, Gyimesi M, Kovacs M. 2013. From keys to bulldozers: expanding roles for winged helix domains in nucleic-acid-binding proteins. Trends Biochem Sci 38:364–371. doi: 10.1016/j.tibs.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Teichmann M, Dumay-Odelot H, Fribourg S. 2012. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription 3:2–7. doi: 10.4161/trns.3.1.18917. [DOI] [PubMed] [Google Scholar]
- 40.Hontz RD, French SL, Oakes ML, Tongaonkar P, Nomura M, Beyer AL, Smith JS. 2008. Transcription of multiple yeast ribosomal DNA genes requires targeting of UAF to the promoter by Uaf30. Mol Cell Biol 28:6709–6719. doi: 10.1128/MCB.00703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett-Lovsey R, Hart SE, Shirai H, Mizuguchi K. 2002. The SWIB and the MDM2 domains are homologous and share a common fold. Bioinformatics 18:626–630. doi: 10.1093/bioinformatics/18.4.626. [DOI] [PubMed] [Google Scholar]
- 42.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J. 2013. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 10:676–682. doi: 10.1038/nmeth.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soulard A, Cremonesi A, Moes S, Schutz F, Jeno P, Hall MN. 2010. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell 21:3475–3486. doi: 10.1091/mbc.e10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. 2008. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aasland R, Stewart AF, Gibson T. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci 21:87–88. doi: 10.1016/0968-0004(96)30009-1. [DOI] [PubMed] [Google Scholar]
- 46.Langemeyer L, Ungermann C. 2015. BORC and BLOC-1: shared subunits in trafficking complexes. Dev Cell 33:121–122. doi: 10.1016/j.devcel.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 47.Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell 33:176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engel C, Neyer S, Cramer P. 2018. Distinct mechanisms of transcription initiation by RNA polymerases I and II. Annu Rev Biophys 47:425–446. doi: 10.1146/annurev-biophys-070317-033058. [DOI] [PubMed] [Google Scholar]
- 49.Engel C, Gubbey T, Neyer S, Sainsbury S, Oberthuer C, Baejen C, Bernecky C, Cramer P. 2017. Structural basis of RNA polymerase I transcription initiation. Cell 169:120–131. doi: 10.1016/j.cell.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Han Y, Yan C, Nguyen THD, Jackobel AJ, Ivanov I, Knutson BA, He Y. 2017. Structural mechanism of ATP-independent transcription initiation by RNA polymerase I. Elife 6:e27414. doi: 10.7554/eLife.27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khatter H, Vorlander MK, Muller CW. 2017. RNA polymerase I and III: similar yet unique. Curr Opin Struct Biol 47:88–94. doi: 10.1016/j.sbi.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 52.Sadian Y, Baudin F, Tafur L, Murciano B, Wetzel R, Weis F, Muller CW. 2019. Molecular insight into RNA polymerase I promoter recognition and promoter melting. Nat Commun 10:5543. doi: 10.1038/s41467-019-13510-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadian Y, Tafur L, Kosinski J, Jakobi AJ, Wetzel R, Buczak K, Hagen WJ, Beck M, Sachse C, Muller CW. 2017. Structural insights into transcription initiation by yeast RNA polymerase I. EMBO J 36:2698–2709. doi: 10.15252/embj.201796958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Webb B, Sali A. 2014. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 58.Devany M, Kotharu NP, Matsuo H. 2004. Solution NMR structure of the C-terminal domain of the human protein DEK. Protein Sci 13:2252–2259. doi: 10.1110/ps.04797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Combe CW, Fischer L, Rappsilber J. 2015. xiNET: cross-link network maps with residue resolution. Mol Cell Proteomics 14:1137–1147. doi: 10.1074/mcp.O114.042259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nogi Y, Vu L, Nomura M. 1991. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 88:7026–7030. doi: 10.1073/pnas.88.16.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nogi Y, Yano R, Nomura M. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci U S A 88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang T, Lei J, Yang H, Xu K, Wang R, Zhang Z. 2011. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28:795–798. doi: 10.1002/yea.1905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.