Abstract
The nucleocapsid protein is significant in the formation of viral RNA of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), accounting for the largest proportion of viral structural proteins. Here, we report for the first time that the 11S proteasomal activator PA28γ regulates the intracellular abundance of the SARS-CoV-2 N protein (nCoV N). Furthermore, we have identified proteasome activator PA28γ as a nCoV N binding protein by co-immunoprecipitation assay. As a result of their interaction, nCoV N could be degraded by PA28γ-20S in vitro degradation assay. This was also demonstrated by blocking de novo protein synthesis with cycloheximide. The stability of nCoV N in PA28γ-knockout cells was greater than in PA28γ-wildtype cells. Notably, immunofluorescence staining revealed that knockout of the PA28γ gene in cells led to the transport of nCoV N from the nucleus to the cytoplasm. Overexpression of PA28γ enhanced proteolysis of nCoV N compared to that in PA28γ-N151Y cells containing a dominant-negative PA28γ mutation, which reduced this process. These results suggest that PA28γ binding is important in regulating 20S proteasome activity, which in turn regulates levels of the critical nCoV N nucleocapsid protein of SARS-CoV-2, furthering our understanding of the pathogenesis of COVID-19.
Keywords: PA28γ, SARS-CoV-2, nCoV N, Protein degradation, COVID-19
Highlights
-
•
PA28γ can bind nucleocapsid protein of SARS-CoV-2 in cells and in vitro.
-
•
SARS-CoV-2 nucleocapsid protein can be degraded by PA28γ-20S proteasome system.
-
•
PA28γ can regulate SARS-CoV-2 nucleocapsid protein.
1. Introduction
As of May 31, 2020, nearly 6 million cases of coronavirus disease 2019 (COVID-19) and over 350,000 deaths from the disease, have been reported worldwide [1]. A novel coronavirus is the cause of COVID-19. Taxonomically, SARS-CoV-2 forms a clade within the subgenus sarbecovirus, orthocoronavirinae subfamily [2]. SARS-CoV-2 has a positive single-stranded RNA genome, approximately 29.8 kb, including a various number (from 6 to 11) of open reading frames (ORFs) [3]. The first ORF, representing over 60% of the entire genome, encodes 16 non-structural proteins, while the remaining ORFs encode auxiliary proteins and four structural proteins [4]. The four structural proteins are the small envelope protein (E), matrix protein (M), spike surface glycoprotein (S), and nucleocapsid protein (N) [5].
The SARS-CoV-2 nucleocapsid protein (hereafter, referred to as nCoV N) accounts for the largest proportion of viral structure proteins and is the most abundant protein in virus-infected cells. Its primary function is to package the viral RNA genome into a ribonucleoprotein complex, the capsid [6]. The nucleocapsid protein encoded by SARS-CoV-2 can act as a viral inhibitory factor of RNA interference in cells [7]. Furthermore, it has been shown that the N protein of SARS-CoV can modulate the host cellular machinery and it may serve in a regulatory role during the viral life cycle [8]. Therefore, the nucleocapsid protein is a crucial multifunctional protein, involved in the process of virus infection, replication, and packaging [9].
In general, viral nuclear proteins can enter the host nucleus and interact with a variety of host proteins to interfere with the life cycle of the host cell. It has been shown that the coronavirus N protein is not only localized in the cytosol but also, to a certain extent, translocated into the nucleus where it may interact with various cellular proteins that modulate cellular functions [10]. This process may depend on interaction of the N protein with the proteasome activator PA28γ, which is localized in the nucleus. PA28γ could be critical for degrading the SARS-CoV-19 nCoV N protein in the nucleus as part of the 20S proteasome, which acts to degrade proteins in a ubiquitin-independent manner, such as seen in the hepatitis C virus (HCV) core protein [11].
The proteasome has an important role in the degradation of unneeded or damaged proteins by proteolysis. Two distinct proteasomes differentially target proteins for degradation. The 26S proteasome, formed by association of the 20S catalytic core (composed of α and β subunits) with the 19S regulator, is responsible for degradation of the majority of proteins through a ubiquitin (Ub)- and ATP-dependent pathway [12]. Additionally, the 20S proteasome, which is required for the Ub- and ATP-independent degradation of specific target substrates, is generated by a combination of one 20S catalytic core and one proteasomal activator 28 (PA28) member [13]. Of the three PA28 family members, PA28γ (also called REGγ, 11Sγ, PSME3, or Ki antigen) is implicated in tumorigenesis because it regulates cell proliferation and apoptosis, and it predominantly exerts its function through nuclear proteolysis [14]. PA28γ has been known to target numerous intact proteins directly through proteasomal degradation. This establishes the function of PA28γ in a variety of biological processes with physiological and pathological relevance. In addition, PA28γ can also regulate some viruses such as the HCV core protein, hepatitis B virus X protein, and human immunodeficiency virus type 1 Tat [15]. The nuclear retention and stability of PA28γ are regulated via a PA28γ-dependent pathway through which HCV pathogenesis may be exerted [16]. Moreover, the HCV core protein can decrease 20S proteasome activity in the presence of PA28γ [17].
It has been proposed that the ubiquitin-proteasome system plays a critical role during various stages of the coronavirus infection cycle [18]. In turn, the proteasomal inhibitor MG132 strongly inhibits SARS-CoV replication by interfering with early steps of the viral life cycle [19]. However, the potential role of the Ub- and ATP-independent degradation pathway in the field of coronavirus research is unknown. A previous study found that the SARS-CoV N protein can interact with the host cell proteasome subunit p42, a subunit of the 26S proteasome [20]. The two activators, PA28 and 26S, can bind to the Ub- and ATP-independent 20S proteasome simultaneously [21]. Thus, there may be a connection between the SARS-CoV N protein and PA28. Owing to over 90% amino acid sequence similarity between SARS-CoV N protein and the SARS-CoV-2 N protein [22], the latter nCoV N protein may be presumed to play the same role in this process. The precise role of PA28γ in the degradation of coronavirus proteins is still unclear. In the present study, we found that the N protein of SARS-CoV-2 could be degraded by PA28γ in vitro. This may indicate that PA28γ is a regulator for SARS-CoV-2 N protein degradation. We also investigated the interaction between SARS-CoV-2 N protein and the PA28γ-20S proteasome system through a co-immunoprecipitation assay. This new finding provides a clue for understanding the previously unresolved physiological roles of the proteasome-dependent degradation of the SARS-CoV-2 N protein during pathogenesis of COVID-19.
2. Materials and methods
2.1. Plasmids and reagents
Plasmid Flag-PA28γ, FRT-PA28γ, and FRT-PA28γN151Y were previously generated [23]. HA-2019-nCoV-N was generated by polymerase chain reaction using the primers Forward: 5′-CCGCTCGAGATGAGCGATAACGGTCCGC-3′ and Reverse: 5′-CGCGGATCCTTACGCTTGGGTGCTATCCGC-3′ and was inserted into the pSG5 vector. The template gene, pET-32a-N-protein, was kindly provided by Dr. Tao from Shanghai Center for Systems Biomedicine, Key Laboratory of Systems Biomedicine (Ministry of Education), Shanghai Jiao Tong University. The clone was constructed following standard molecular cloning technology and confirmed by DNA sequence analysis.
The sources of the following antibodies and reagents were as follows: rabbit anti-HA (HUABIO, Hangzhou, China), anti- mouse Flag (MBL, Beijing, China), mouse anti-β-actin (MBL), and PA28γ (Abcam, Cambridge, UK); Flag-M2 beads, HA-beads, and cycloheximide (Sigma-Aldrich, St. Louis, MO).
2.2. Cell culture and transfection
The 293T cells were purchased from ATCC; the 293T (PA28γ-knockout) cell was constructed by TALENT. Cells were cultured in Dulbecco’s modified Eagle’s medium. Fetal bovine serum 10% and antibiotics were added to the media.
The 293T cells (PA28γ WT and PA28γ knockout) were transiently transfected with the plasmid HA-pSG5-nCoV-N, and Lipofectamine 2000 transfection reagent (Invitrogen) was used depending on the manufacturer’s instructions.
2.3. Protein degradation in vitro assay
The PA28γ and 20S proteasome proteins were purified well [24]. The SARS-CoV-2 N protein was created by in vitro translation using the TNT kit (Promega).
2.4. Immunofluorescence (IF) staining
The day before the experiment, the cells were inserted into 60-mm dishes covered with sterile slides and maintained at 37 °C in 5% CO2 until they were attach to the plate about 24 h later. The cells were then fixed using 4% paraformaldehyde for 10 min at room temperature. After washing with PBS. Again, cells were blocked using 4% serum for 1 h. Next, anti-HA was added and the cells were incubated at 4 °C overnight. Subsequently, cells were incubated with fluorescent secondary antibody for 1 h at room temperature. Cell images were visualized using a fluorescence microscope.
2.5. Co-immunoprecipitation (Co-IP)
Cells were collected and lysed on ice using the following lysis buffer. The 10% cell lysate was collected to be the input of Co-IP assay.
3. Results
3.1. PA28γ interacts with nCoV N
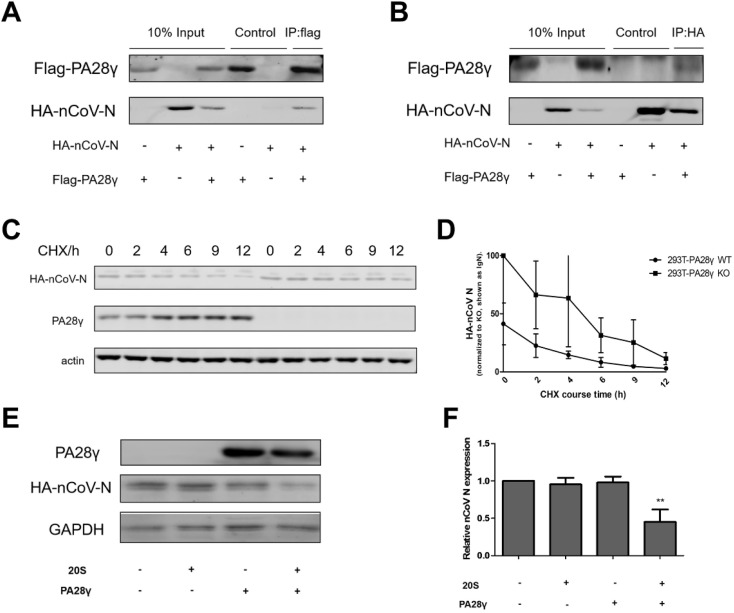
The previous studies have demonstrated that PA28γ can bind to the core protein of HCV and promote its degradation [11]. Thus, to investigate whether exogenous PA28γ can interact with nCoV N, we carried out a co-IP assay. The data support an interaction between PA28γ and nCoV N. Both plasmids Flag-PA28γ and HA-nCoV-N were transfected into 293T cells. PA28γ was immunoprecipitated with an anti-Flag antibody (Fig. 1 A). nCoV N could easily be observed in the immunoprecipitate by Western blot analysis. Similarly, PA28γ was readily detected using an anti-HA antibody (Fig. 1B). Our results demonstrate that exogenous PA28γ can physically interact with nCoV N.
Fig. 1.
PA28γ can regulate nCoV N by binding and degradation. (A) The 293T cell lines were co-transfected with HA-nCoV-N and Flag-PA28γ plasmids and then subjected to immunoprecipitation with conjugated anti-Flag beads. (B) The 293T cell lines were co-transfected with the indicated plasmids and then subjected to immunoprecipitation with conjugated anti-HA beads. All samples were analyzed by western blotting using the indicated antibodies. (C) Degradation dynamics of nCoV N following a time-course treatment with cycloheximide (CHX, protein synthesis inhibitor, 100 μg/ml) in 293T PA28γ wild-type and 293T PA28γ knockout cell lines. HA-nCoV-N means the expression of nCoV N after the transfection of the plasmid HA-pSG5-nCoV N, PA28γ means the endogenous PA28γ expression in 293T cell lines (it does not express in KO cell line), actin is used as a reference. (D) According to Western blot bands intensity, the expression of nCoV N in 293T WT and KO could be calculated. Normalized with actin, the quantification of nCoV N degradation was demonstrated. WT, wild-type PA28γ; KO, knockout PA28γ. (E) In vitro proteolytic analysis of PA28γ-mediated degradation of nCoV N. Purified PA28γ, 20S proteasome, and in vitro-translated nCoV N were incubated as indicated and described in materials and methods. PA28γ means the PA28γ protein expression, HA-nCoV-N means the nCoV N translated protein expression and GAPDH means the reference. (F) Quantification of nCoV N relative expression was normalized with the vitro-translated nCoV N. Results presented as the means ± SEM. ∗p < 0.05; ∗∗p < 0.01 versus control, n = 3.
3.2. nCoV N regulation in PA28γ-WT and PA28γ-KO cell lines
Next we determined whether endogenous PA28γ plays a role in the degradation of nCoV N. The 293T cells (PA28γ-WT and PA28γ-KO) were transfected with the plasmid HA-pSG5-nCoV N and then treated with cycloheximide (CHX) for the indicated times. We found that after 12 h, the expression of nCoV N was higher in PA28γ-KO cells than in WT cells (Fig. 1C), indicating that the protein was more stable in the absence of PA28γ in the cells. Moreover, the stability of nCoV N protein in PA28γ-WT was lower than that in the PA28γ-KO cell line over time (Fig. 1D). Taken together, the data show that endogenous PA28γ may have an effect on the degradation of nCoV N protein through interaction.
3.3. PA28γ promotes the degradation of nCoV N
PA28γ has been known to direct proteasomal degradation of a number of complete proteins. And the 20S proteasome is responsible for Ub- and ATP-independent degradation of specific target substrates, in combination with PA28γ. Therefore, to further validate the role of PA28γ in the degradation of nCoV N, we performed an in vitro proteolytic analysis of nCoV N. We found that nCoV N exhibits remarkable down-regulation in the presence of PA28γ and the 20S proteasome, while PA28γ or 20S proteasome alone serves no function in the degradation of nCoV N (Fig. 1E and F). These data indicate that PA28γ can promote the degradation of nCoV N in a 20S proteasome- dependent manner.
3.4. PA28γ-mediated degradation of nCoV N
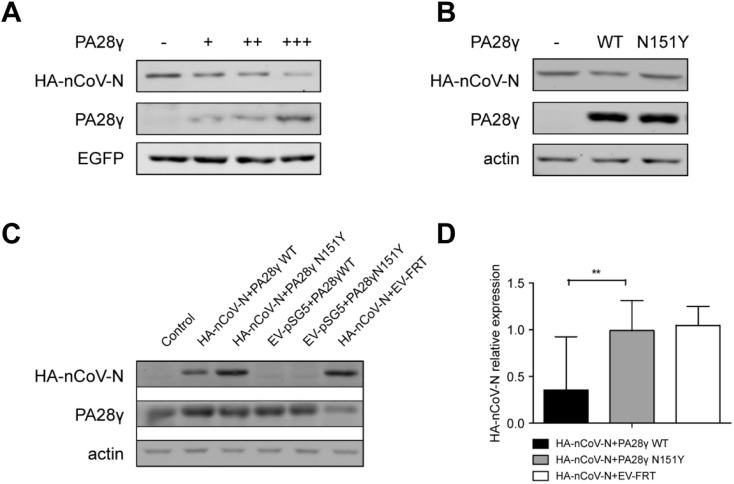
To verify the effect of PA28γ on the degradation of nCoV N, we co-transfected PA28γ and HA-nCoV N into 293T-PA28γ KO cell lines. EV-pSG5+PA28γWT, EV-pSG5+PA28γ, N151Y, and HA-nCoV + EV-FRT were transfected into cells as negative controls for comparison. As expected, the level of nCoV N was significantly reduced when exogenous PA28γ was normally expressed in the cell. In addition, when the inactive mutant N151Y PA28γ was transfected into the cells with HA-nCoV, we found that the level of nCoV N remained relatively equal to that of the control (Fig. 2 B, C and D). This confirmed that the degradation of nCoV N could be mediated by PA28γ. We further investigated the effectiveness of the expression of PA28γ on the degradation of nCoV N by adding an equal amount of EGFP expression plasmid to detect the expression efficiency. We found that the increased expression level of PA28γ resulted in a marked reduction in nCoV N protein in a dose dependent manner (Fig. 2A), confirming that it was the PA28γ that promotes the degradation of nCoV N and its effectiveness is related to dosage.
Fig. 2.
PA28γ-mediated degradation of nCoV N in 293T cells. (A) The 293T cells were co-transfected HA-nCoV-N with increasing amounts of PA28γ. The nCoV N protein levels were determined by western blotting. A constant amount of an enhanced green fluorescent protein (EGFP) expression plasmid was included to monitor transfection efficiency. (B) The 293T PA28γ KO cells were transfected the same amount of only HA-nCoV-N, HA-nCoV-N with FRT-PA28γ wt and HA-nCoV-N with FRT-PA28γN151Y. (C) Cells were transient transfected with the same amount of HA-nCoV-N, FRT-PA28γ wild-type, FRT-PA28γ N151Y, HA-pSG5 vector, and FRT vector using Lipo2000 transfection reagent for 48 h. Protein expression was detected using anti-PA28γ, anti-HA, and anti-actin antibodies. Non-transfected 293T cells were used as a control. (D) Quantification of nCoV N relative expression. Data are presented as means ± SEM. ∗p < 0.05; ∗∗p < 0.01 versus control, n = 3.
3.5. Location and expression of nCoV N in 293T cells
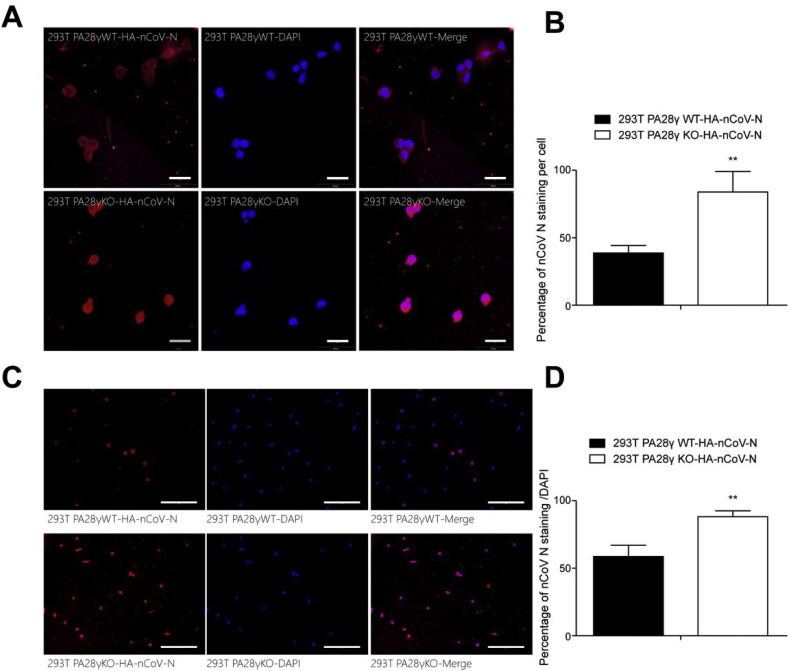
To more intuitively reflect the promoting effect of PA28γ on nCoV N degradation, immunofluorescence staining assay was used. The same amount of HA-nCoV-N was transfected into 293T cells lines (PA28γ-WT and PA28γ-KO). The expression level of nCoV N protein (red) in the PA28γ-KO cell lines was observed to be prominently higher than that in the PA28γ-WT cell line (Fig. 3 C and D), confirming that PA28γ deficiency promotes nCoV N in the cell line.
Fig. 3.
Location and expression of nCoV N in 293T PA28γWT and PA28γKO cells. (A) Cells were transiently transfected with HA-nCoV-N, fixed, and immunostained with anti-HA (red color). Cell nuclei were stained with DAPI (blue color). Scale bar: 50 μm. (B) Quantification analysis of nCoV N expression as a percentage per cell of three independent experiments (n = 100; values represent the mean ± SEM; ∗p < 0.05; ∗∗p < 0.01 versus control). (C) Cells were transiently transfected with the same amount of HA-nCoV-N plasmid, fixed, immune-stained with anti-HA (red color), and stained with DAPI (blue color). Scale bar: 100 μm. (D) Quantification analysis of nCoV N expression as a percentage in the same cell area. Each data set represents three independent experiments (n = 300 . values represent the mean ± SEM; ∗p < 0.05; ∗∗p < 0.01 versus control). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We then investigated the cellular localization and expression of nCoV N in 293T cells under the regulation of PA28γ by immunofluorescence staining. The same amount of nCoV N was transfected into 293T cell lines (PA28γ-WT and PA28γ-KO). In the PA28γ-WT cell lines, the nCoV N protein (red) was prominently expressed in the cytoplasm, while in the PA28γ-KO cell lines it existed both in the nucleus and the cytoplasm (Fig. 3A). In addition, the total expression of nCoV N was quantified and, as expected, PA28γ-WT cell lines exhibited a decrease in nCoV N (Fig. 3B). PA28γ has been known to express in the nucleus [11]. Therefore, our results indicate that the nCoV N in the nucleus was degraded under the regulation of PA28γ, while that in the cytoplasm was not.
3.6. Cartoon depicting the nCoV N protein degradation process through the PA28γ-20S system
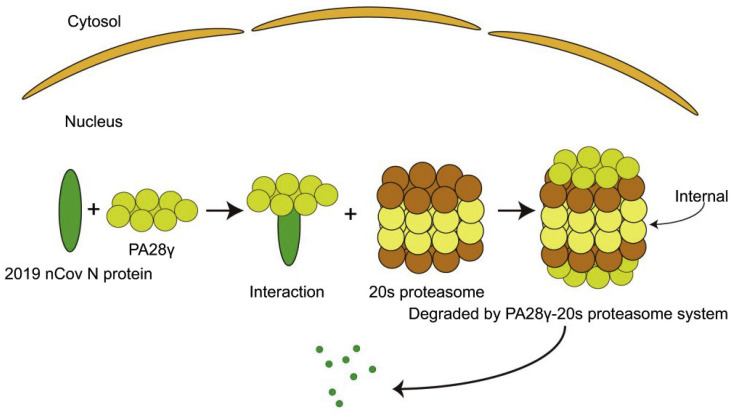
A schematic diagram of the degradation process shows that PA28γ first binds with nCoV N and transports it to the 20S proteasome subunit where internal degradation occurs (Fig. 4 ). Thus, the time of existence of nCoV N in the nucleus is much shorter compared to that in the cytoplasm.
Fig. 4.
Schematic diagram of the progress of PA28γ-20S degradation of nCoV N. nCoV N enters the nucleus, binds to PA28γ, and is then degraded internally under the action of the 20 S proteasome. nCoV N is internally after 20S proteasome binding to PA28γ.
4. Discussion
An increasing number of studies have shown that the proteasome is associated with viral infection, and there is evidence that HCV core proteins can be degraded through the PA28γ-20S system. That is, the nuclear retention and stability of HCV core proteins are regulated by PA28γ-dependent pathways such that the pathogenicity of HCV may be achieved through this pathway [16]. In addition, the human immunodeficiency virus-1 (HIV-1) Tat protein can inhibit the peptidase activity of the 20S proteasome by competing with the 11S/PA28 regulator (REG) for binding at the REG/Tat-proteasome-binding (RTP) site and interfering with antigen processing [25]. It has been shown that the hepatitis B virus X protein-derived polypeptide, harboring the α4 proteasome subunit binding motif, impairs the activation of 20S proteasomes by PA28 [26]. The coxsackievirus infection can be enhanced by proteasome activator PA28γ promoting p53 degradation [27], similar to the mechanism of degradation observed in the HBx virus [28]. Furthermore, protein p30 interactions with PA28γ may also affect ATM functions and increase cell survival [29]. On the other hand, PA28γ acts as a co-repressor of HTLV-1 p30 to suppress virus replication and is required for the maintenance of control viral latency [33]. Therefore, previous studies have shown that PA28γ is closely related to the HTLV-1 virus and plays an indelible role in the formation and spread of the virus. Additionally, the ability of PA28γ to promote viral protein degradation suggests its involvement in viral pathogenesis.
Studies also have demonstrated that the novel coronavirus nucleocapsid protein (N) shares nearly 90% amino acid sequence homology with SARS coronavirus. SARS coronavirus N protein antibodies can cross-react with novel coronavirus, but cannot provide cross-immunity. Similar to SARS-CoV, nCoV N proteins can inhibit RNA interference (RNAi) to overcome the host defense [32]. Early research revealed the involvement of the ubiquitin-proteasome system (UPS) in multiple steps of the coronavirus infection cycle and identified UPS as a potential drug target to modulate the significance of coronavirus infection [18].
In the present study, we found that a novel ubiquitination-independent pathway could regulate the protein levels of nCoV N, and PA28γ could control the abundance of nCoV N by regulating its stability. In addition, we showed that PA28γ interacts with nCoV N and promotes its intracellular degradation. SARS coronavirus nucleocapsid protein is a very important viral structural protein and an important indicator in early diagnosis due to its abundance and high conservation in cells. Studying nucleocapsid protein structural function, how it participates in transcription and translation, the molecular mechanism in virus-infected cells, and the regulation of gene expression will help to thoroughly understand SARS coronaviruses and find effective methods for prevention and treatment of related diseases [6]. When SARS-CoV-2 invades the human body, it makes contact with human immune cells, thus causing the immune cells to produce massive IFN-γ, and IFN-γ can induce PA28γ [32]. This in turn stimulates proteasome activity resulting in degradation of the coronavirus N protein. As a result, virus production is blocked and proliferation and diffusion are greatly amelioraten.
In conclusion, our study substantiates that PA28γ could mediate the degradation of nCoV N. These results suggest that the PA28γ interaction has an important role in regulating 20S proteasome activity and furthers our understanding of the pathogenesis of 2019-nCoV. However, this interaction correlates to the of truncated nCoV N, further work is necessary to locate the accurate region of nCoV N interact with PA28γ. In addition, some mechanisms may be found. Understanding the precise function of PA28γ may give us new insight into virus-cell interactions and lead to a greater understanding of the pathogenicity of 2019-nCoV infection.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Shanghai Committee of Science and Technology. (no. 16DZ2348900).
References
- 1.WHO. Coronavirus Disease 2019 (COVID-19): Situation Report—132. May 31, 2020. [Google Scholar]
- 2.Zhu N., Zhang D.Y., Wang W.L., Li X.W., Yang B., Song J.D., Zhao X., Huang B.Y., Shi W.F., Lu R.J., Niu P.H., Zhan F.X., Ma X.J., Wang D.Y., Xu W.B., Wu G.Z., Gao G.G.F., Tan W.J., Coronavirus C.N. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song Z.Q., Xu Y.F., Bao L.L., Zhang L., Yu P., Qu Y.J., Zhu H., Zhao W.J., Han Y.L., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses-Basel. 2019;vol. 11 doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu A.P., Peng Y.S., Huang B.Y., Ding X., Wang X.Y., Niu P.H., Meng J., Zhu Z.Z., Zhang Z., Wang J.Y., Sheng J., Quan L.J., Xia Z.X., Tan W.J., Cheng G.H., Jiang T.J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein - forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu J., Xu J., Zhang L., Shu T., Wu D., Huang M., Ren Y., Li X., Geng Q., Xu Y., Qiu Y., Zhou X. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci. China Life Sci. 2020 doi: 10.1007/s11427-020-1692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surjit M., Liu B.P., Chow V.T.K., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu J., Chen R., Hu J., Qu H., Zhao Y., Cao S., Wen X., Wen Y., Wu R., Zhao Q., Ma X., Huang X. Identification of a novel linear B-cell epitope on the nucleocapsid protein of porcine deltacoronavirus. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21020648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timani K.A., Liao Q.J., Ye L.B., Zeng Y.C., Liu J., Zheng Y., Ye L., Yang X.J., Kong L.B., Gao J.R., Zhu Y. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114:23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki R., Moriishi K., Fukuda K., Shirakura M., Ishii K., Shoji I., Wakita T., Miyamura T., Matsuura Y., Suzuki T. Proteasomal turnover of hepatitis C virus core protein is regulated by two distinct mechanisms: a ubiquitin-dependent mechanism and a ubiquitin-independent but PA28 gamma-dependent mechanism. J. Virol. 2009;83:2389–2392. doi: 10.1128/JVI.01690-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coux O., Tanaka K., Goldberg A.L. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 13.Ma C.P., Slaughter C.A., DeMartino G.N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain) J. Biol. Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 14.Mao I., Liu J., Li X., Luo H. REGgamma, a proteasome activator and beyond? Cell. Mol. Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivakumaran H., van der Horst A., Fulcher A.J., Apolloni A., Lin M.H., Jans D.A., Harrich D. Arginine methylation increases the stability of human immunodeficiency virus type 1 Tat. J. Virol. 2009;83:11694–11703. doi: 10.1128/JVI.00499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriishi K., Okabayashi T., Nakai K., Moriya K., Koike K., Murata S., Chiba T., Tanaka K., Suzuki R., Suzuki T., Miyamura T., Matsuura Y. Proteasome activator PA28gamma-dependent nuclear retention and degradation of hepatitis C virus core protein. J. Virol. 2003;77:10237–10249. doi: 10.1128/JVI.77.19.10237-10249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng Y., Shimamoto S., Maruno T., Kobayashi Y., Matsuura Y., Kawahara K., Yoshida T., Ohkubo T. N-terminal HCV core protein fragment decreases 20S proteasome activity in the presence of PA28gamma. Biochem. Biophys. Res. Commun. 2019;509:590–595. doi: 10.1016/j.bbrc.2018.12.167. [DOI] [PubMed] [Google Scholar]
- 18.Raaben M., Posthuma C.C., Verheije M.H., te Lintelo E.G., Kikkert M., Drijfhout J.W., Snijder E.J., Rottier P.J.M., de Haan C.A.M. The ubiquitin-proteasome system plays an important role during various stages of the coronavirus infection cycle. J. Virol. 2010;84:7869–7879. doi: 10.1128/JVI.00485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider M., Ackermann K., Stuart M., Wex C., Protzer U., Schatzl H.M., Gilch S. Severe acute respiratory syndrome coronavirus replication is severely impaired by MG132 due to proteasome-independent inhibition of M-calpain. J. Virol. 2012;86:10112–10122. doi: 10.1128/JVI.01001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q., Li C.A., Zhang Q.F., Wang T., Li J.D., Guan W.X., Yu J.S., Liang M.F., Li D.X. Interactions of SARS Coronavirus Nucleocapsid Protein with the host cell proteasome subunit p42. Virol. J. 2010;7 doi: 10.1186/1743-422X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendil K.B., Khan S., Tanaka K. Simultaneous binding of PA28 and PA700 activators to 20 S proteasomes. Biochem. J. 1998;332(Pt 3):749–754. doi: 10.1042/bj3320749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannan S., Ali P.S.S., Sheeza A., Hemalatha K. COVID-19 (Novel Coronavirus 2019) - recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020;24:2006–2011. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- 23.Zuo Q., Cheng S., Huang W., Bhatti M.Z., Xue Y., Zhang Y., Zhang B., Li L., Wu L., Fu J., Chen J., Li X. REGgamma contributes to regulation of hemoglobin and hemoglobin delta subunit. Oxid. Med. Cell. Longev. 2017;2017:7295319. doi: 10.1155/2017/7295319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao X., Li J., Pratt G., Wilk S., Rechsteiner M. Purification procedures determine the proteasome activation properties of REG gamma (PA28 gamma) Arch. Biochem. Biophys. 2004;425:158–164. doi: 10.1016/j.abb.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Huang X.H., Seifert U., Salzmann U., Henklein P., Preissner R., Henke W., Sijts A.J., Kloetzel P.M., Dubiel W. The RTP site shared by the HIV-1 Tat protein and the 11 S regulator subunit alpha is crucial for their effects on proteasome function including antigen processing. J. Mol. Biol. 2002;323:771–782. doi: 10.1016/s0022-2836(02)00998-1. [DOI] [PubMed] [Google Scholar]
- 26.Stohwasser R., Holzhutter H.G., Lehmann U., Henklein P., Kloetzel P.M. Hepatitis B virus HBx peptide 116-138 and proteasome activator PA28 compete for binding to the proteasome alpha4/MC6 subunit. Biol. Chem. 2003;384:39–49. doi: 10.1515/BC.2003.005. [DOI] [PubMed] [Google Scholar]
- 27.Gao G., Wong J., Zhang J., Mao I., Shravah J., Wu Y., Xiao A., Li X., Luo H. Proteasome activator REGgamma enhances coxsackieviral infection by facilitating p53 degradation. J. Virol. 2010;84:11056–11066. doi: 10.1128/JVI.00008-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeom S., Jeong H., Kim S.S., Jang K.L. Hepatitis B virus X protein activates proteasomal activator 28 gamma expression via upregulation of p53 levels to stimulate virus replication. J. Gen. Virol. 2018;99:655–666. doi: 10.1099/jgv.0.001054. [DOI] [PubMed] [Google Scholar]
- 29.Anupam R., Datta A., Kesic M., Green-Church K., Shkriabai N., Kvaratskhelia M., Lairmore M.D. Human T-lymphotropic virus type 1 p30 interacts with REGgamma and modulates ATM (ataxia telangiectasia mutated) to promote cell survival. J. Biol. Chem. 2011;286:7661–7668. doi: 10.1074/jbc.M110.176354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz K., van Den Broek M., Kostka S., Kraft R., Soza A., Schmidtke G., Kloetzel P.M., Groettrup M. Overexpression of the proteasome subunits LMP2, LMP7, and MECL-1, but not PA28 alpha/beta, enhances the presentation of an immunodominant lymphocytic choriomeningitis virus T cell epitope. J. Immunol. 2000;165:768–778. doi: 10.4049/jimmunol.165.2.768. [DOI] [PubMed] [Google Scholar]
- 33.Ko N.L., Bellon Taylor J.M., Bai M., Shevtsov X.T., Dundr S.P., C. M. Nicot. Blood. 2013;121 doi: 10.1182/blood-2012-03-420414. [DOI] [PMC free article] [PubMed] [Google Scholar]