Abstract
Linezolid and tedizolid are oxazolidinones with established clinical utility for the treatment of Gram-positive pathogens. Over time it has become apparent that even modest structural changes to the core phenyl oxazolidinone leads to drastic changes in biological activity. Consequently, the structure–activity relationship around the core oxazolidinone is constantly evolving, often reflected with new structural motifs present in nascent oxazolidinones. Herein we describe the use of cryo-electron microscopy to examine the differences in binding of several functionally different oxazolidinones in the hopes of enhanced understanding of their SAR. Tedizolid, radezolid, T145, and contezolid have been examined within the peptidyl transferase center (PTC) of the 50S ribosomal subunit from methicillin resistant Staphylococcus aureus. The ribosome–antibiotic complexes were resolved to a resolution of around 3 Å enabling unambiguous assignment of how each antibiotic interacts with the PTC.
Keywords: antibiotics, linezolid, contezolid, oxazolidinones, cryo-EM, ribosome
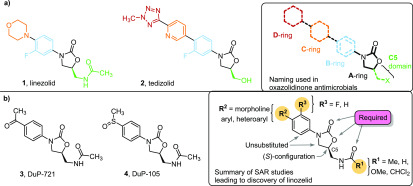
Linezolid (1) was the first oxazolidinone antimicrobial agent developed, and is the first purely synthetic antibiotic known.1−3 Its potency against all major Gram-positive pathogens, including newer multidrug resistant strains such as methicillin resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE), have ensured it has gained a significant place as a last line antibiotic.4 Oxazolidinones act as protein synthesis inhibitors, binding in the 23S rRNA region adjacent to the PTC within the 50S subunit.5 They bind directly in the A-site position of where the 5′ amino acylated tRNA docks, thereby sterically hindering any incoming tRNA substrates into the enzymatic core of the ribosome.6 This novel mode of action results in a remarkable absence of cross-resistance between linezolid (1) and other antimicrobials.7 This lack of cross-resistance is paramount to the success of linezolid (1) given the emergence of multidrug resistant pathogens. Despite such promising features, safety limitations surrounding linezolid (1) have limited the expansion of the oxazolidinone family of antimicrobials.7−9 The reasoning behind this is attributed to the homology between the 23S ribosomal target and the closely related mitochondrial protein synthesis processes in mammals,5,7,10,11 thereby resulting in adverse effects such as myelosuppression and monoamine oxidase (MAO) inhibition.7,10,12 Despite these limitations over the last decades a number of oxazolidinone antimicrobials have been developed based on linezolid (1) with modifications in the C5-domain, B-ring, C-ring, and addition of a D-ring (Figure 1a).
Figure 1.
(a) Structure of the two clinically approved oxazolidinone antibiotics, linezolid (1) and tedizolid (2). On the right is the naming convention of the oxazolidinone chemical extensions. (b) The two precursor DuPont compounds DuP-721 (3) and DuP-105 (4), on the right is the general SAR approach for the first generation of oxazolidinone antibiotics.
Since the discovery of linezolid (1) over two decades ago, the only other oxazolidinone to be clinically approved is tedizolid (2).13 Considering the manifold efforts into the discovery of oxazolidinone antimicrobials it is somewhat surprising that only two successful analogues have been uncovered. Of the thousands of oxazolidinones synthesized many boast greater antibacterial properties than linezolid (1) and even tedizolid (2).14 However, increased toxicity due to mitochondrial binding is often associated with greater antimicrobial potency, and is rarely addressed. In more recent times the focus around the synthesis of oxazolidinones has shifted from potency, prioritizing an improved safety profile.15
Structure–activity relationships (SAR) are an essential tool in the discovery of new antimicrobials. Over the extended period in which oxazolidinone antimicrobials have been developed the SAR has evolved, presenting new opportunities for the introduction of functional motifs with, increasingly, a focus on reducing toxicity. Long before linezolid (1) was received clinically, the SAR of oxazolidinone based antimicrobials had been explored by DuPont.16,17 The pioneering work culminated with the first clinical candidate oxazolidinones DuP-721 (3) and DuP-105 (4).16,17 However, DuPont discontinued research into these progenitor oxazolidinones due to concerns with animal toxicity. Scientists at Upjohn Co. later continued the investigation culminating in the initial discovery of linezolid (1) which received regulatory approval in the early 2000s (Figure 1b).13
DuPont’s extensive SAR research, allowed the eventual identification of linezolid (1) as a therapeutic agent. The SAR gave a generalized guide for the synthesis for novel oxazolidinones that remains important to this day. Specifically, it was demonstrated that the A-ring oxazolidinone core should have (S) chirality, while acetamide or substituted acetamide (R1) were the optimal substituents for the C5-domain with larger structures reducing biological activity.17,18 The B-ring system often retains substituents meta and para to the amine, while a C-ring (R2) can be tolerated.
Some 14 years after the discovery of linezolid (1) the next successful clinical candidate, tedizolid, was developed. First synthesized in 2011, tedizolid (2) is a second generation oxazolidinone and one of a few pyridyl phenyl derivatives found to have improved biologically activity.19 However, these analogues often suffer from low bioavailability and insolubility requiring alteration of the C5-domain to incorporate a phosphate prodrug approach.20 The pyridyl phenyl motifs extended typical C-ring modification by the addition of a D-ring. Yoon et al. demonstrated that these D-ring systems impacted the C5-domain thereby allowing functionality other than the archetypal acetamide reported by DuPont to be introduced (see Figure 1).20
Before the discovery of tedizolid (2) problems with toxicity had become apparent with oxazolidinone antimicrobials. In 2005 Barbachyn reported the first example of B-ring modifications leading to an improved safety profile.1,7 However, altered B-ring oxazolidinones had previously made little progress as potential therapeutics. It wasn’t until 2014 when Gordeev et al. demonstrated the potential of fluorinated B-ring systems which lead to the discovery of contezolid.7 This novel oxazolidinone maintained anti MRSA potency while having an improved safety profile, halving myelosuppression and reducing MAO inhibition by a third.1,7 Accelerated development of contezolid allowed entry to phase III trials in late 2019 with promising results and the potential to replace linezolid (1). Studies by Gordeev et al. showed that trisubstituted B-ring systems often suffer from a reduction in antimicrobial activity.7 However, by simultaneously modifying the C-ring and C-5 domain, improved minimum inhibitory concentration (MIC) values could be retained along with the desired safety profile.
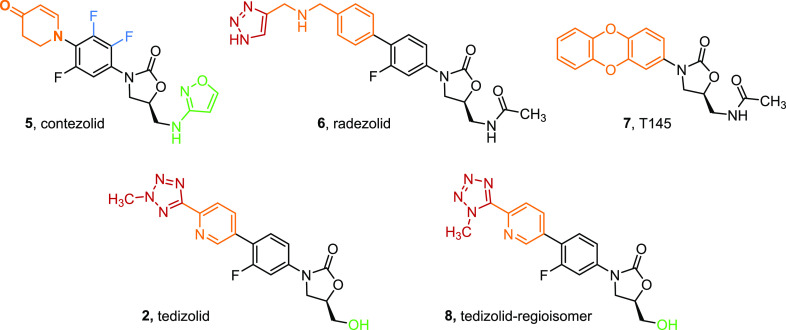
The development of oxazolidinones antimicrobials presents a challenge with a delicate balance between antimicrobial activity and toxicity required. The addition of a D-ring enables changes to the C-5 domain invoking a different mode of binding within the ribosomal subunit. This is also apparent for trisubstituted B-ring systems such as contezolid (5), where the C5-domain acetamide was changed to an isoxazaole to retain its MIC. Furthermore, D-ring systems or indeed extended C-ring systems such as those in tedizolid (2) and radezolid (6) exhibit binding to the periphery of the PTC previously unseen. Thus, while the oxazolidinone antimicrobials retain a common mode of action their interactions with the PTC appear to be suitably perturbed by various structural changes. To gain both a better understanding of how oxazolidinone structure impacts PTC binding, and to further develop the use of cryo-EM as a tool in structure-based drug design, we have examined the oxazolidinones, contezolid (5), radezolid (6), T145 (7), tedizolid (2), and a regioisomer of tedizolid (8), using cryo-EM (Figure 2). From this study it has been possible to clearly map the main sites of interaction, and highlight chemical differences in these drugs and how they lead to different interactions in the binding site. Moreover, our results show that by comparing the binding mode of four different oxazolidinones we can more accurately determined the “core” residues that compose the binding pocket for this important class of drugs.
Figure 2.
Oxazolidinone compounds synthesized and used in the cryo-EM study.
Results and Discussion
Synthesis of Oxazolidione Compounds
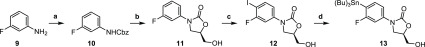
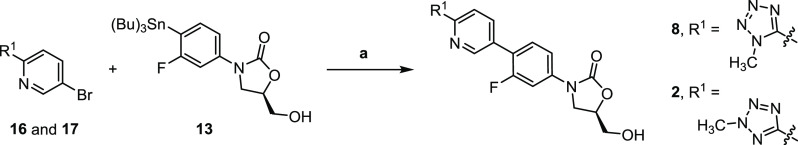
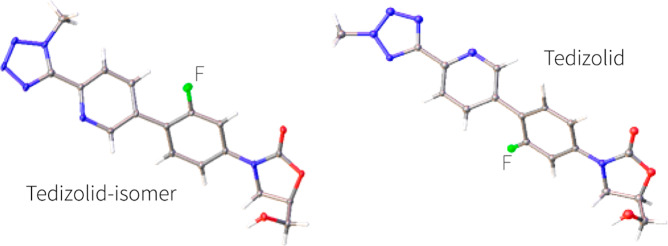
To allow a comprehensive cryo-EM study into oxazolidinone antimicrobials most of these needed to be synthesized. Tedizolid (2) was prepared following a synthetic procedure described by Yoon et al.20 with minor modifications. Specifically, coupling of stannane 13 (Scheme 1) with a mixture of tetrazoles 16 and 17, themselves prepared in two steps from pyridine 14 (Schemes 2 and 3), gave tedizolid (2) and the regioisomeric compound 8. While originally intended to deliver tedizolid alone, formation of 8 was considered fortuitous as it would allow the impact the site of methylation has on ribosome binding to be examined. To unambiguously determine the structures of tedizolid (2) and its isomer 8, two X-ray crystal structures were determined to ensure the integrity of the final compounds used for the cryoEM studies (Figure 3).
Scheme 1. Synthesis of Oxazolidinone 13 by a Procedure Modified from Yoon et al.
Reagents and conditions: (a) K2CO3, Cbz-Cl, THF/H2O, 0 °C to rt; (b) n-BuLi, (R)-glycidyl butyrate, THF, −78 °C to rt; (c) NIS, TFA, rt; (d) (Bu)3Sn–Sn(Bu)3, Pd(PPh3)2Cl2, dioxane, 100 °C.20
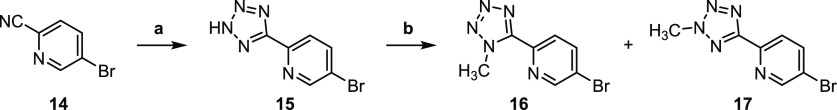
Scheme 2. Synthesis of Tetrazoles 16 and 17 by a Procedure Modified from Yoon et al.
Reagents and conditions: (a) NaN3, ZnCl2, pyridine, reflux; (b) CH3I, NaOH, DMF, 0−40 °C.20
Scheme 3. Synthesis of Tedizolid (2) and Regiosiomer 8 by a Procedure Modified from Yoon et al.
Reagents and conditions: (a) Pd(PPh3)2Cl2, LiCl, NMP, 120 °C.20
Figure 3.
X-ray crystal structures from this study of tedizolid (2) (right) and its regioisomer 8 (left), both compounds are very close in structure except for the relative position of the fluorinated benzene in the B-ring which is rotated 180° between each structure.
Radezolid (6) was synthesized following procedures defined by Shili et al.21 albeit with the key Suzuki coupling requiring an increased catalyst loading. While the synthesis of contezolid (4) was achieved following a sequence described by Gordeev et al.,7 oxazolidinone formation was achieved using LiHMDS instead of LiOtBu. Finally, oxazolidinone T145 (7) was prepared using the procedure proposed by Ippoliti et al.;22 however, amide formation exploited a Gabriel synthesis rather than working from an azide (Scheme 4).
Scheme 4. Proposed Synthesis of T145 (7) by Ippoliti et al. Showing Modifications in the Sequence.
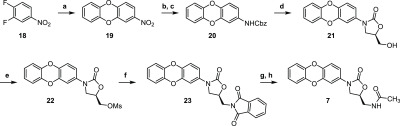
Reagents, and conditions: (a) Catechol, K2CO3, DMF, 100 °C; (b) Pd/C, H2, THF, rt; (c) Cbz-Cl, NaHCO3, THF/H2O, rt; (d) LiHMDS, (R)-glycidyl butyrate, THF, −78 °C to rt; (e) MsCl, Et3N, CH2Cl2, 0 °C to rt; (f) K-Phth, DMF, 90 °C; (g) N2H4·H2O, EtOH, reflux; (h) AcO2, Et3N, CH2Cl2, 0 °C to rt.22
Cryo-EM Determination of Ribosome/Antibiotic Complexes
To better understand the precise binding mode of these members of the oxazolidinone family of antibiotics with their cognate target, the bacterial ribosome, we employed single particle cryo-EM. The structures were achieved by combining dimethyl sulfoxide (DMSO) solubilized antibiotic (final concentration ∼10 μM) to freshly thawed 70S ribosomes isolated from MRSA. This solution was allowed to incubate at 37 °C for 15 min, on ice for 1 h, then immediately applied to glow discharged TEM grids and plunge frozen with liquid ethane. All specimens were imaged at an acceleration voltage of 200 kV. While specimens contained the 70S ribosomes, only the 50S ribosomal subunit was subjected to final refinement in RELION or cryoSPARC (see methods section), in order to achieve the highest possible resolution.
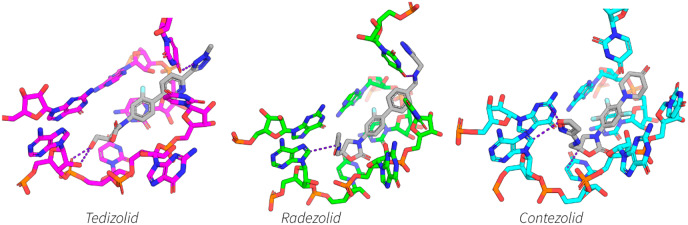
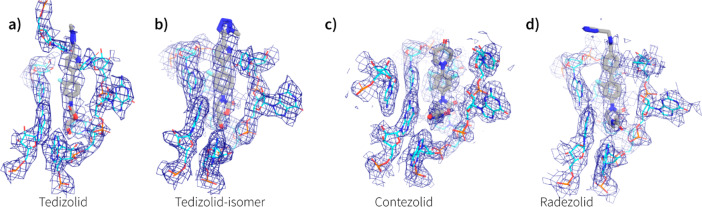
The density observed around the oxazolidinone binding site allowed for unambiguous placement of every antibiotic (see Figure 4) except for T145, which only showed density for the lower part of the B-ring and C5-domain (see Supporting Information, Figure S2). For this reason, T145 will be left out of further discussions. The binding site of the oxazolidinones is remarkably similar to that previously described4,6,23 and is at the A-site of the PTC, completely encapsulated by rRNA (see Figures 4 and 5).
Figure 4.
Calculated densities around the antibiotic binding site: (a) tedizolid; (b) tedizolid-regioisomer; (c) contezolid; and (d) radezolid. All density is drawn at 4 σ and the binding site is drawn from the same relative ribosome orientation.
Figure 5.
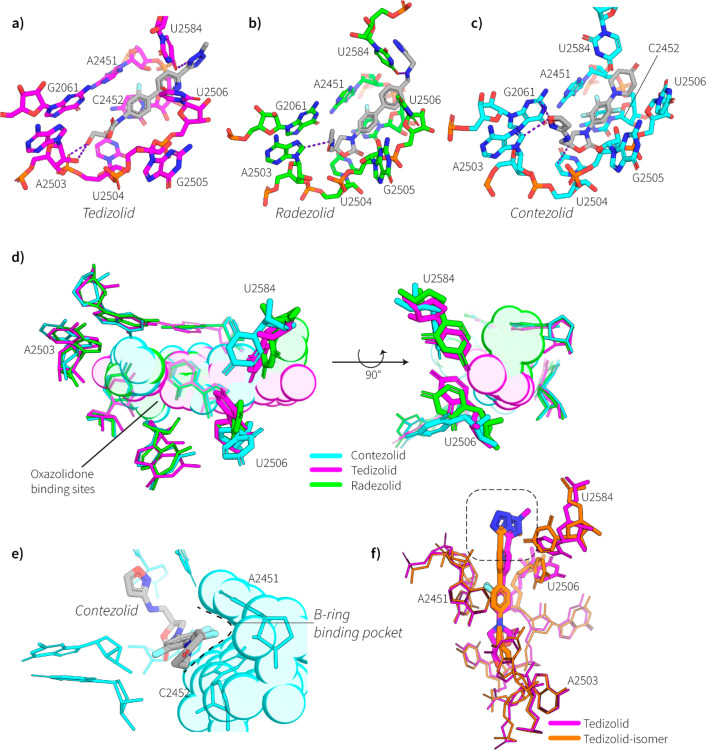
Cartoon representation of the binding site of the different oxazolidinone derivatives in the PTC of the 50S subunit of the MRSA ribosome. (a) Binding site of tedizolid (2), showing hydrogen bonds as dashed blue lines between the tedizolid hydroxyl and the backbone of A2503 and an interaction between the tedizolid tetrazole and U2584. (b) Binding site of radezolid (6) showing a van-der Waals interaction between the radezolid acetyl group and the purine nucleobase A2503, and a hydrogen bond with the secondary amine and U2584, shown as dashed blue lines. (c) Binding site of contezolid (5) highlighting π–π interactions between the isoxazole in the C5-domain of contezolid with G2061 and a van der Waals interaction with A2503, shown as a dashed blue line is also a tighter interaction of the oxazolidinone with the nucleobase of U2504. (d) Overlay of binding site and relative orientations of the rRNA nucleotides of contezolid (cyan), radezolid (green), and tedizolid (magenta), the oxazolidinones are shown as a translucent cloud. Highlighted in bolder sticks are the nucleotides that exhibit the greatest conformational change depending on the oxazolidinone bound. (e) View of the B-ring binding pocket defined by A2451 and C2452 which nestles the fluorinated benzene of contezolid (5). (f) Overlay of both the tedizolid (2) (magenta) and tedizolid-regioisomer 8 (orange) ribosome binding site. Highlighted in bolder sticks is the two nucleobases that change the most in conformation between the two structures (U2584 and U2506), which are presumably disturbed by the position of the methyl group (highlighted in dashed box) which slightly offsets the binding position of the isomer relative to clinically used, tedizolid (2).
Tedizolid (2) with the extended C-ring consisting of a pyridine and methylated tetrazole still retains a classical oxazolidinone binding pose, but with subtle differences. The tetrazole enables a close lone-pair interaction (3 Å away) between the tetrazole and U2584 (E. coli rRNA numbering used throughout) (see Figure 5a), and as the C5-domain substituent is only a hydroxyl group it forms two possible hydrogen-bond contacts with the phosphate oxygen and the ribose 2′-OH of A2503. The comparison of tedizolid (2) with the methyl tetrazole regioisomer 8 is also of interest. The regioisomer of tedizolid places the methyl group in an unfavorable position which abolishes the lone-pair interaction with U2584, causing rearrangement of both U2584 and U2506, which moves in closer to the binding site of the tedizolid-regioisomer 8 (see Figure 5f).
Radezolid (6) also has a C-ring extension and binds in a very similar pose to tedizolid (2), and utilizes the secondary amine in the C-ring to form a hydrogen bond with the uracil nucleobase U2584, which in turn brings the nucleotide closer to the aromatic triazole moiety opening up the potential for π–π interactions between U2584 and the triazole (see Figure 5b,d). The acetyl substituent in the C5-domain adopts a similar pose as compounds previously studied in our group,4 which directs toward A2503 for a van-der Waals interaction with the adenosine nucleobase.
Contezolid (5), while not having such a long C-ring modification, replaces the morpholine (as found in linezolid (1)) with a piperidinone which does not appear to make any extra contacts with either U2584 or U2506, both of which seem to change conformations significantly when contezolid (5) is bound (see Figure 5c,d), both nucleotides orientating further from the oxazolidinone binding pocket. However, the C-ring also has a trifluorinated benzyl group which nestles in further to the hydrophobic pocket defined by the nucleotides A2451 and C2452 (see Figure 5e), also the isoxazole in the C5-domain makes new contacts with both G2061 (a π–π interaction) and A2503 (a van der Waals interaction).
Oxazolinones represent a crucial chemical functionality in the development of new antimicrobial agents. Their use however has been marred by toxicity and relatively high minimatory inhibition concentrations (MIC). Efforts to improve on linezolid (1) have led to compounds like tedizolid (2) and radezolid (6) which sought to improve efficacy by expanding the C-ring portion of the chemical in an effort to maximize contacts within the ribosomal binding pocket. This study shows that these efforts have yielded results with tedizolid having over a 10 time better MIC than linezolid (1)24 which is most likely due to the increased contacts that the C-ring has with both U2506 and U2584 as well as the extra hydrogen bonds between the 5′-OH and A2503. The enhanced efficacy of radezolid (6)25 compared to linezolid (1) is also most likely due to the extra contacts the C-ring triazole extension makes with U2584 as well as the secondary amine.
It seems that the regioisomer of tedizolid while binding a very similar pocket to its clinical counterpart, perhaps makes some deleterious interactions due to the positioning of the methyl group on the tetrazole, most likely the reason why this compound was abandoned. Tedizolid (2) also shows that extensions in the C5-domain may be unnecessary for efficacy, but when compared to contezolid (5), perhaps plays a part in toxicity and selective binding to bacterial vs mitochondrial ribosomes.
Design choices during the development of contezolid (5), one of the newest members of the oxazolidione family, seem to have avoided inclusion of C-ring expansions. This is most likely due to the poor solubility associated with greater aromatic functionality which seems critical to binding, and adjustments have been made to the B-ring, by adding extra fluorines to the core benzyl group. Our cryo-EM results suggest that this allows this portion of the drug to nestle in a hydrophobic pocket defined by A2451 and C2452. While this interaction is not novel compared to other members of the family, combination with the isoxazole in the C5-domains clearly allows this compound to bind effectively (4 fold enhanced MIC than linezolid (1)) and endows the drug with a higher efficacy compared to linezolid (1) with seemingly less associated toxicity. When contezolid (5) binds to the ribosome it seems to reflect that making interactions with U2584 and U2506 is not crucial for oxazolidinone function and that the core binding site consists of the residues G2061, A2503, U2504, G2505, A2451, and C2452. Any new oxazolidinone drugs should potentially focus efforts in improving interactions with these core residues.
Materials and Methods
Purification of Ribosomes from MRSA
Ribosomes were isolated as previously described.4
X-ray Crystallography
Single crystals of tedizolid and tedizolid-regioisomer were grown by the vapor diffusion method, diffusing n-pentane into a dichloromethane/methanol (1:1) solution of the antibiotic. A suitable crystal was selected and collected on a XtaLAB Synergy, Dualflex, HyPix diffractometer. The crystal was kept at 123.00(10) K during data collection. Using Olex2,26 the structure was solved with the ShelXT structure solution program using Intrinsic Phasing and refined with the ShelXL27,28 refinement package using Least Squares minimization. See Table S2 for crystallographic parameters.
Electron Microscopy
Cryo-EM samples were prepared by plunge vitrification in liquid ethane on a Vitrobot Mark IV (Thermo Fisher Scientific) with a blotting chamber set to 4 °C and 100% humidity. A 3 μL aliquot of the sample solution was applied on Quantifoil R1.2/1.3 Cu 200 mesh (Quantifoil) glow-discharged grids and blotted for 1.5 s before the grid was plunged. Data were collected on a Glacios or Artica (see Supporting Information, Table S1) (Thermo Fisher Scientific) 200 kV electron microscope equipped with a Falcon 3 direct electron detector. The microscope was set with a 50 μm condenser aperture, no objective aperture, spot at 6, and beam diameter of 2.4 μm. The equivalent pixel size on the detector was either in the range between 0.874–1.14 Å/pixel depending on the specimen (see Supplemental Table S1). The total electron dose was either 47 or 50 e.Å–2 with an expose time of either 47 or 50 s. Multiframe movies were automatically acquired with the EPU software package (Thermo Fisher Scientific) in gain-normalized uncompressed MRC format with a 9-position beam-image shift data acquisition scheme and target defocus range of 0.4 to 1.6 μm.
Data Processing
Movies were motion-corrected, dose-weighted, and integrated using UCSF MotionCor2.29,30 This was followed by CTF estimation using the GCTF31 software package. Particles were picked from the micrographs using the automated procedure in the crYOLO software package.32,33 Particle extraction and reference-free 2D classification was carried out in RELION (version 3.0.7).34 Initial 3D references were used from a previous study4 and used for 3D classification in RELION. A homogeneous subset of particles was then subjected to cycles of Bayesian particle polishing and CTF refinement as implemented in RELION. This homogeneous subset of polished particles was used for a 3D refinement in RELION and was further classified into 3D classes with a fine grain angular sampling only allowing for local Euler angle searches. Particles belonging to the 3D class were further refined in RELION (version 3.1) where their higher order CTF parameters were rerefined taking into account particles belonging to each image shift group. Further 3D refinements where the 30S subunit was masked and a final 3D refinement was carried out in RELION (version 3.1), yielding consensus maps of the complex at a global resolution (FSC = 0.143) of between 2.9 and 3.2 Å (see Table S1). For the tedizolid regioisomer 8, the final refinement was carried out in cryoSPARC35 using their nonuniform refinement strategy, with a mask that covered the 50S subunit.
Atomic Model Refinement
The 50S models were all based on PDB: 6DDD(4) which was used as the initial PDB template. All models were flexibly fitted into the EM density maps using the molecular dynamics flexible fitting (MDFF) simulation with nanoscale molecular dynamics (NAMD).36 These models were then subjected to real-space refinement, as implemented in the PHENIX software package.37 Refinement restraints for each antibiotic were generated in eLBOW routine in PHENIX and placed and manually refined in coot.38 After iterative refinement and manual adjustments, comprehensive validation implemented in PHENIX was performed to assess the model quality as presented in Table S1.
Structure statistics are detailed in Table S1.
Figures
All figures were generated in PyMOL.39
Data Availability
The cryo-EM density maps were deposited in the Electron Microscopy Data Bank under accession codes EMD-21872, EMD-21873, EMD-21887, and EMD-21888 for contezolid-50S, radezolid-50S, tedizolid-50S, and tedizolid-regioisomer-50S, respectively. The PDB coordinates were deposited in the PDB database under the accession codes 6WQN, 6WQQ, 6WRS, and 6WRU for contezolid-50S, radezolid-50S, tedizolid-50S, and tedizolid-regioisomer-50S, respectively.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00041.
Full synthetic methods for the antibiotic synthesis, tables containing the cryo-EM and X-ray structure statistics, and figures for the Fourier shell correlation (FSC) curves of each structure (PDF)
This work was supported by a grants from US-DOD (PRMRP: W81XWH1910126) for M.J.B. and by the National Health & Medical Research Council of Australia (Program Grant 1092262 to T.L.) and the Australia Research Council (DP 170103567 to D.W.L.). The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication. We acknowledge the support of the Ramaciotti Center for Electron Microscopy for the data collection on the Artica and for Thermo Scientific for the Glacios housed in Prof. Patrick Sexton’s laboratory. This work was supported by the MASSIVE HPC facility (www.massive.org.au).
The authors declare no competing financial interest.
Supplementary Material
References
- Fisher J. F., Mobashery S., and Miller M. J (2018) Antibacterials, Vol. II, Topics in Medicinal Chemistry , (26), , Springer. [Google Scholar]
- Brickner S. J.; Barbachyn M. R.; Hutchinson D. K.; Manninen P. R. (2008) Linezolid (ZYVOX), the first member of a completely new class of antibacterial agents for treatment of serious gram-positive infections. J. Med. Chem. 51 (7), 1981–90. 10.1021/jm800038g. [DOI] [PubMed] [Google Scholar]
- Ford C. W.; Zurenko G. E.; Barbachyn M. R. (2001) The discovery of linezolid, the first oxazolidinone antibacterial agent. Curr. Drug Targets: Infect. Disord. 1 (2), 181–99. 10.2174/1568005014606099. [DOI] [PubMed] [Google Scholar]
- Belousoff M. J.; Venugopal H.; Wright A.; Seoner S.; Stuart I.; Stubenrauch C.; Bamert R. S.; Lupton D. W.; Lithgow T. (2019) cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria. ChemMedChem 14, 527. 10.1002/cmdc.201900042. [DOI] [PubMed] [Google Scholar]
- Belousoff M. J.; Eyal Z.; Radjainia M.; Ahmed T.; Bamert R. S.; Matzov D.; Bashan A.; Zimmerman E.; Mishra S.; Cameron D.; Elmlund H.; Peleg A. Y.; Bhushan S.; Lithgow T.; Yonath A. (2017) Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome. mBio 8 (3), 821–32. 10.1128/mBio.00395-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Z.; Matzov D.; Krupkin M.; Wekselman I.; Paukner S.; Zimmerman E.; Rozenberg H.; Bashan A.; Yonath A. (2015) Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 112 (43), E5805–14. 10.1073/pnas.1517952112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordeev M. F.; Yuan Z. Y. (2014) New potent antibacterial oxazolidinone (MRX-I) with an improved class safety profile. J. Med. Chem. 57 (11), 4487–97. 10.1021/jm401931e. [DOI] [PubMed] [Google Scholar]
- Nagiec E. E.; Wu L.; Swaney S. M.; Chosay J. G.; Ross D. E.; Brieland J. K.; Leach K. L. (2005) Oxazolidinones inhibit cellular proliferation via inhibition of mitochondrial protein synthesis. Antimicrob. Agents Chemother. 49 (9), 3896–902. 10.1128/AAC.49.9.3896-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee E. E.; Ferguson M.; Bentley A. T.; Marks T. A. (2006) Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob. Agents Chemother. 50 (6), 2042–9. 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger C.; Roberts J. A.; Muller L. (2018) Clinical Pharmacokinetics and Pharmacodynamics of Oxazolidinones. Clin. Pharmacokinet. 57 (5), 559–575. 10.1007/s40262-017-0601-x. [DOI] [PubMed] [Google Scholar]
- De Vriese A. S.; Coster R. V.; Smet J.; Seneca S.; Lovering A.; Van Haute L. L.; Vanopdenbosch L. J.; Martin J. J.; Groote C. C.; Vandecasteele S.; Boelaert J. R. (2006) Linezolid-induced inhibition of mitochondrial protein synthesis. Clin. Infect. Dis. 42 (8), 1111–7. 10.1086/501356. [DOI] [PubMed] [Google Scholar]
- Lee E. Y.; Caffrey A. R. (2018) Thrombocytopenia with Tedizolid and Linezolid. Antimicrob. Agents Chemother. 62 (1), 1453-17 10.1128/AAC.01453-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner S. J.; Hutchinson D. K.; Barbachyn M. R.; Manninen P. R.; Ulanowicz D. A.; Garmon S. A.; Grega K. C.; Hendges S. K.; Toops D. S.; Ford C. W.; Zurenko G. E. (1996) Synthesis and Antibacterial Activity of U-100592 and U-100766, Two Oxazolidinone Antibacterial Agents for the Potential Treatment of Multidrug-Resistant Gram-Positive Bacterial Infections. J. Med. Chem. 39, 673–679. 10.1021/jm9509556. [DOI] [PubMed] [Google Scholar]
- Komine T.; Kojima A.; Asahina Y.; Saito T.; Takano H.; Shibue T.; Fukuda Y. (2008) Synthesis and Structure - Activity Relationship Studies of Highly Potent Novel Oxazolidinone Antibacterials. J. Med. Chem. 51, 6558–6562. 10.1021/jm800800c. [DOI] [PubMed] [Google Scholar]
- Das B.; Rudra S.; Yadav A.; Ray A.; Rao A. V.; Srinivas A. S.; Soni A.; Saini S.; Shukla S.; Pandya M.; Bhateja P.; Malhotra S.; Mathur T.; Arora S. K.; Rattan A.; Mehta A. (2005) Synthesis and SAR of novel oxazolidinones: discovery of ranbezolid. Bioorg. Med. Chem. Lett. 15 (19), 4261–7. 10.1016/j.bmcl.2005.06.063. [DOI] [PubMed] [Google Scholar]
- Renslo A. R.; Luehr G. W.; Gordeev M. F. (2006) Recent developments in the identification of novel oxazolidinone antibacterial agents. Bioorg. Med. Chem. 14 (12), 4227–40. 10.1016/j.bmc.2006.01.068. [DOI] [PubMed] [Google Scholar]
- Brickner S. J. (1996) Oxazolidinone antibacterial agents. Curr. Pharm. Des. 2, 175–194. [Google Scholar]
- Chellat M. F.; Raguz L.; Riedl R. (2016) Targeting Antibiotic Resistance. Angew. Chem., Int. Ed. 55 (23), 6600–26. 10.1002/anie.201506818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G. G.; Love R.; Adam H.; Golden A.; Zelenitsky S.; Schweizer F.; Gorityala B.; Lagace-Wiens P. R.; Rubinstein E.; Walkty A.; Gin A. S.; Gilmour M.; Hoban D. J.; Lynch J. P. 3rd; Karlowsky J. A. (2015) Tedizolid: a novel oxazolidinone with potent activity against multidrug-resistant gram-positive pathogens. Drugs 75 (3), 253–70. 10.1007/s40265-015-0352-7. [DOI] [PubMed] [Google Scholar]
- Im W. B.; Choi S. H.; Park J. Y.; Choi S. H.; Finn J.; Yoon S. H. (2011) Discovery of torezolid as a novel 5-hydroxymethyl-oxazolidinone antibacterial agent. Eur. J. Med. Chem. 46 (4), 1027–39. 10.1016/j.ejmech.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Shili C., R. L, Yusheng W. U., Jiacheng Z., and Roger H.. Process for the synthesis of triazoles using heterocyclization, reductive amination and cross-coupling reactions. 2006.
- Ebner D. C.; Culhane J. C.; Winkelman T. N.; Haustein M. D.; Ditty J. L.; Ippoliti J. T. (2008) Synthesis of novel oxazolidinone antimicrobial agents. Bioorg. Med. Chem. 16 (5), 2651–6. 10.1016/j.bmc.2007.11.040. [DOI] [PubMed] [Google Scholar]
- Belousoff M. J.; Eyal Z.; Radjainia M.; Ahmed T.; Bamert R. S.; Matzov D.; Bashan A.; Zimmerman E.; Mishra S.; Cameron D.; Elmlund H.; Peleg A. Y.; Bhushan S.; Lithgow T.; Yonath A. (2017) Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome. mBio 8 (3), 395-17 10.1128/mBio.00395-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan S.; McKee E. E.; Das D.; Tulkens P. M.; Hosako H.; Fiedler-Kelly J.; Passarell J.; Radovsky A.; Prokocimer P. (2015) Nonclinical and pharmacokinetic assessments to evaluate the potential of tedizolid and linezolid to affect mitochondrial function. Antimicrob. Agents Chemother. 59 (1), 178–85. 10.1128/AAC.03684-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.; Chen Z.; Lin Z.; Sun X.; Bai B.; Xu G.; Chen J.; Yu Z.; Qu D. (2020) Radezolid Is More Effective Than Linezolid Against Planktonic Cells and Inhibits Enterococcus faecalis Biofilm Formation. Front. Microbiol. 11, 196. 10.3389/fmicb.2020.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolomanov O. V.; Bourhis L. J.; Gildea R. J.; Howard J. A. K.; Puschmann H. (2009) OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 42, 339–341. 10.1107/S0021889808042726. [DOI] [Google Scholar]
- Sheldrick G. M. (2015) Crystal structure refinement with SHELXL. Acta Crystallogr., Sect. C: Struct. Chem. 71, 3–8. 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. M. (2015) SHELXT - Integrated space-group and crystal-structure determination. Acta Crystallogr., Sect. A: Found. Adv. 71, 3–8. 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Mooney P.; Zheng S.; Booth C. R.; Braunfeld M. B.; Gubbens S.; Agard D. A.; Cheng Y. (2013) Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10 (6), 584–90. 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S. Q.; Palovcak E.; Armache J. P.; Verba K. A.; Cheng Y.; Agard D. A. (2017) MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14 (4), 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. (2016) Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193 (1), 1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.; Raunser S. (2020) The evolution of SPHIRE-crYOLO particle picking and its application in automated cryo-EM processing workflows. Commun. Biol. 3 (1), 61. 10.1038/s42003-020-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T.; Merino F.; Stabrin M.; Moriya T.; Antoni C.; Apelbaum A.; Hagel P.; Sitsel O.; Raisch T.; Prumbaum D.; Quentin D.; Roderer D.; Tacke S.; Siebolds B.; Schubert E.; Shaikh T. R.; Lill P.; Gatsogiannis C.; Raunser S. (2019) SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218. 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J.; Nakane T.; Forsberg B. O.; Kimanius D.; Hagen W. J.; Lindahl E.; Scheres S. H. (2018) New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, 42166 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A.; Rubinstein J. L.; Fleet D. J.; Brubaker M. A. (2017) cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14 (3), 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26 (16), 1781–802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D.; Afonine P. V.; Baker M. L.; Bunkoczi G.; Chen V. B.; Croll T. I.; Hintze B.; Hung L. W.; Jain S.; McCoy A. J.; Moriarty N. W.; Oeffner R. D.; Poon B. K.; Prisant M. G.; Read R. J.; Richardson J. S.; Richardson D. C.; Sammito M. D.; Sobolev O. V.; Stockwell D. H.; Terwilliger T. C.; Urzhumtsev A. G.; Videau L. L.; Williams C. J.; Adams P. D. (2019) Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct Biol. 75 (Pt 10), 861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66 (4), 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps were deposited in the Electron Microscopy Data Bank under accession codes EMD-21872, EMD-21873, EMD-21887, and EMD-21888 for contezolid-50S, radezolid-50S, tedizolid-50S, and tedizolid-regioisomer-50S, respectively. The PDB coordinates were deposited in the PDB database under the accession codes 6WQN, 6WQQ, 6WRS, and 6WRU for contezolid-50S, radezolid-50S, tedizolid-50S, and tedizolid-regioisomer-50S, respectively.