Abstract
Background
A prognostic model combining biomarkers of metaphase-anaphase transition of the cell cycle was developed for invasive breast cancer. The prognostic value and clinical applicability of the model was evaluated in comparison with the routine prognosticators of invasive breast carcinoma.
Methods
The study comprised 1135 breast cancer patients with complete clinical data and up to 22-year follow-up. Regulators of metaphase-anaphase transition were detected immunohistochemically and the biomarkers with the strongest prognostic impacts were combined into a prognostic model. The prognostic value of the model was tested and evaluated in separate patient materials originating from two Finnish breast cancer centers.
Results
The designed model comprising immunoexpressions of Securin, Separase and Cdk1 identified 8.4-fold increased risk of breast cancer mortality (p < 0.0001). A survival difference exceeding 15 years was observed between the majority (> 75%) of patients resulting with favorable as opposed to unfavorable outcome of the model. Along with nodal status, the model showed independent prognostic impact for all breast carcinomas and for subgroups of luminal, N+ and N- disease.
Conclusions
The impact of the proposed prognostic model in predicting breast cancer survival was comparable to nodal status. However, the model provided additional information in N- breast carcinoma in identifying patients with aggressive course of disease, potentially in need of adjuvant treatments. Concerning N+, in turn, the model could provide evidence for withholding chemotherapy from patients with favorable outcome.
Keywords: Breast cancer, Prognosis, Proliferation, Cell cycle, Securin, Separase
Background
Cell proliferation, hormone-regulation and HER2 amplification are considered the main biological processes driving breast cancer progression. Although proliferation has been shown a valid prognosticator in all subtypes, particularly triple-negative breast carcinoma (TNBC) has been characterized by high expression of proliferation-related genes [1, 2]. The prognostic value of proliferation is acknowledged in the clinical pathology practice as part of the traditional histological grading as well as in intrinsic classification and in modern personalized signatures retrieved from microarray-based expression-profiling [3–5]. However, the impact of deregulated proliferation is still not accurately reflected in the routine clinical parameters and pathological markers applied to treatment decisions of breast cancer patients.
Genetic integrity of the dividing cell is ensured by complex and intricately monitored cellular events at the metaphase-anaphase transition of the cell cycle [6]. Dysfunction of these regulators can lead into missed sister chromatid separation, chromosomal instability and aneuploidy. Premature sister chromatid separation is prevented by the highly controlled sequential activation and inactivation of a cascade of regulatory proteins, particularly Cdc20 (cell division cycle 20), Cohesin, Separase (Extra Spindle Pole Bodies Like protein 1, ESPL1), Securin (Pituitary tumor-transforming gene 1, Pttg1), Pttg1IP (Pituitary tumor-transforming gene 1 interacting protein, also Pituitary tumor-transforming gene 1 binding factor, PBF), Cdk1 (Cyclin-Dependent Kinase protein 1) and CyclinB1 (G2/mitotic-specific cyclin-B1). In more detail, correct segregation of the chromosomes is triggered at the Spindle Assembly Checkpoint (SAC) controlled by Cdc20 activating Anaphase-Promoting Complex / Cyclosome (APC/C) to create the APC/CCdc20 complex. Throughout metaphase, contact between the chromatids is maintained by rings of Cohesin. At the initiation of anaphase, Cohesin is removed triggered by Separase and activated by degradation of Securin and/or the Cdk1/CyclinB1 complex, leading to separation of the sister chromatids [7, 8].
In our previous research, PTTG1, the gene of human Securin, was detected with the most significant expression difference between human breast cancer and normal breast glandular tissue on basis of a cDNA microarray analysis involving 4000 cancer related genes [9]. In addition to Securin, also several other regulators of metaphase-anaphase transition have been shown with independent prognostic impacts in breast cancer [10–18]. In the present study, we introduce on basis of a total of 1135 patients with up to 22-year follow-up, a clinically applicable combination of biomarkers of metaphase-anaphase transition leading to optimal detection of aggressive course of disease and cancer mortality in invasive breast cancer.
Methods
Patient materials
The study comprises patients diagnosed and treated with unilateral invasive breast carcinoma in two different institutions (Table 1). The first cohort (I) (n = 781) originated from the Central Hospital of Central Finland, Jyväskylä, Finland, from years 1987–1997 resulting in maximum follow-up time of 22.7 years. The second cohort (II) (n = 354) was collected from Turku University Hospital and Auria biobank, Turku, Finland. This cohort was classified into intrinsic subgroups comprising 208 patients with luminal and 148 patients with triple-negative breast carcinomas diagnosed and treated during 2005–2015 resulting in maximum follow-up times of 14 and 17.8 years, respectively.
Table 1.
Summary of patient cohorts with clinico-patohologic characteristics
| Cohort I | Cohort II | ||
|---|---|---|---|
| All subtypes (n = 781) |
Luminal (n = 208) |
TNBC (n = 146) |
|
| Mean age at diagnosis (range) (years) | 61 (28–95) | 62 (42–76) | 60 (39–78) |
| Axillary lymph node positive (%) | 44.8 | 22.8 | 32.1 |
| Mean tumor size (range) (cm) | 2.3 (0.2–16.0) | 1.9 (0.4–7.0) | 2.5 (0.2–18.0) |
| Histological type (%) | |||
| Infiltrating ductal NOS | 75. 4 | 72.3 | 100 |
| Special type | 24.6 | 27.2 | 0 |
| Intrinsic subtype (%) | |||
| Luminal | 67.6 | 100 | – |
| Her2-amplified | 18.6 | – | – |
| Triple-negative | 13.8 | – | 100 |
| Histological grade (%) | |||
| Low (1-2) | 79.6 | 81.6 | 0 |
| High (3) | 20.4 | 18.4 | 100 |
| Median follow-up time (max) (years) | 12.4 (22.7) | 11.8 (14.0) | 6.9 (17.8) |
| Dead of breast cancer (%) | 30.7 | 10.7 | 22.6 |
For all patients, the biomarkers of the metaphase-anaphase transition were immunohistochemically (IHC) detected, and the routine clinico-pathological prognostic features of breast cancer were collected. For both cohorts of breast cancer patients, a prognostic model was assembled based on the optimal combination of biomarkers of the metaphase-anaphase transition. The prognostic value of the models was evaluated in comparison with the clinically applied routine prognosticators of breast cancer.
All patients were treated with surgical resection or mastectomy with axillary evacuation, radiation and/or adjuvant treatment with anti-estrogenic or cytostatic drugs depending on the patients’ age, hormone receptor and lymph node (N) status according to the international guidelines for breast cancer treatment at the time of diagnosis [19]. No pre-operative adjuvant treatments were administered. Complete clinical data was collected from pathology reports and patient files and registered applying the criteria presented by WHO [20] and St. Gallen International Expert Consensus [21]. Intrinsic subtypes were approximated by immunohistochemistry according to international guidelines [22]. Causes of death were obtained from autopsy reports, death certificates and from the national cancer registry (Statistics Finland, Helsinki, Finland).
Tissue materials
Tissue materials were prepared according to standard histology practice, i.e. fixed in buffered formalin (pH 7.0) and embedded into paraffin blocks. Tissue micro arrays (TMAs) were prepared using the representative tumor area of each patient. The TMAs included two tissue cores with diameters 0.6 mm (cohort I) or 1 mm (cohort II) from each tumor.
IHC methods
Immunohistochemistry was performed on sections of TMAs cut at 3 μm. Immunohistochemical stainings for Securin, Separase, Cdc20, Pttg1IP, SA2 subunit of Cohesin and CyclinB1 were performed as previously described ([9, 13, 14, 16, 17], Additional file 1). IHC for detecting Cdk1 was performed on an automated immunostaining platform Discovery XT (Roche Diagnostics/Ventana Medical Systems, Tucson, AZ). Deparaffinization, epitope retrieval, and antibody incubation were performed before detection with OmniMap DAB Detection Kit (Roche/Ventana). IHC for Ki-67, estrogen (ER) and progesterone (PR) receptors and HER2, and HER2-amplification with in situ hybridization (ISH) followed standard protocols.
Interpretation of IHC
Immunoexpressions for Securin, Separase, Cdc20, Pttg1IP, SA2, Cdk1 and CyclinB1 were observed as combinations of nuclear and cytoplasmic staining and registered as average fractions (%) of positively staining cancer cells [13, 14, 16, 23]. In each case, the number of immunopositive cells was calculated in sets of one hundred cancer cells (minimum 100 and maximum 3 × 100 cancer cells evaluated) and registered as an average fraction (%) of immunopositivity for each patient. Interpretations for IHC of Ki-67, ER, PR, and IHC and ISH for HER2 followed previous literature and generally accepted international guidelines [22, 24]. All IHC interpretations were performed by experienced histopathologists (HR, PK).
Statistical analysis
The cutpoints for immunoexpressions of the studied biomarkers were set based on previous literature, histopathological observations and statistical analyses involving the mean, median and univariate prognostic values of each parameter [9, 13, 14, 16, 17]. In prognostic analyses, Kaplan-Meier estimates were performed to demonstrate the cumulative percentages of breast cancer specific mortality. Cox’s proportional hazard models were used to test associations between disease outcome, biomarker expressions and clinical prognostic features, i.e. tumor size, axillary lymph node status, histological and intrinsic classifications and histological grade. The risk of breast cancer death associated with the studied proteins and the routine prognosticators was quantitated as hazard ratio (HR) with 95% confidence interval (CI). P-values < 0.05 were considered statistically significant. The computations were performed with SAS for Windows, Version 9.3 (I-III) and 9.4 (IV) (SAS Institute, Cary, NC, USA). Kaplan-Meier survival plots were generated using R 2.15.0.
Results
In IHC of the invasive breast carcinomas, Securin was detected as predominantly nuclear but occasionally, showed both nuclear and/or cytoplasmic immunoreaction. Separase showed two distinct and apparently mutually excluding expression patterns observed in the nucleus or in the cytoplasm of cancer cells. Nuclear immunoexpression was observed for Cdc20, SA2, Cdk1 and CyclinB1 whereas Pttg1IP showed cytoplasmic expression only. Table 2 summarizes the fractions of immunopositive breast carcinomas among all breast cancer subtypes (cohort I) and in subgroups divided according to tumor size, nodal status, histological grade and survival.
Table 2.
Fraction (%) of carcinomas immunopositive for the studied proteins in the whole material and in subgroups
| n | All | N- | N+ | T1 | T2–3 | low grade | high grade | alive | DOD | |
|---|---|---|---|---|---|---|---|---|---|---|
| Securin | 604 | 34 | 28 | 52 | 19 | 36 | 28 | 52 | 19 | 42 |
| Separase | 401 | 31 | 18 | 32 | 15 | 19 | 17 | 57 | 18 | 34 |
| Cdc20 | 387 | 5 | 3 | 7 | 3 | 6 | 4 | 11 | 4 | 8 |
| Pttg1IP | 420 | 56 | 59 | 53 | 65 | 51 | 66 | 28 | 60 | 51 |
| SA2 | 470 | 28 | 32 | 22 | 29 | 26 | 28 | 26 | 33 | 19 |
| Cdk1 | 384 | 23 | 9 | 24 | 8 | 24 | 12 | 26 | 6 | 26 |
| CyclinB1 | 455 | 57 | 53 | 61 | 49 | 60 | 55 | 66 | 51 | 67 |
N- node-negative, N+ node-positive, T1 tumor size < 2 cm, T2–3 tumor size ≥2 cm, low grade = grades 1–2, high grade grade 3, DOD dead of disease
The prognostic impacts of the studied regulators of metaphase-anaphase transition were first analyzed for all breast cancer subtypes (cohort I, n = 781) with Cox’s proportional hazard model. Among all studied biomarkers, statistically significant prognostic value in univariate analyses was observed for Securin (HR 2.1, p < 0.0001, CI 1.6–2.8), nuclear Separase (HR 2.0, p < 0.0004, CI 1.4–3.0), Cdk1 (HR 2.5, p < 0.0001, CI 1.7–3.6) and CyclinB1 (HR 2.0, p = 0.04, CI 1.0–2.0).
In the next phase, these biomarkers were further tested in combinations in order to assemble a prognostic model producing the most significant prognostic impact among all breast cancer subtypes. The optimal model for detecting favorable outcome of disease was determined as the combination of low expressions for Securin (< 10% of cancer cells), Separase (< 1% of cancer cells) and Cdk1 (< 10% of cancer cells). This model was a significant indicator of survival of disease while the opposite detected patients in risk of breast cancer death (HR 8.4, p < 0.0001). The Kaplan-Meier curves demonstrate the survival difference among all patients and in subgroups with N+ and N- disease (Fig. 1). Concluding from the survival analyses, favorable outcome of the model indicated that the majority (> 75%) of patients were alive 18.4 years after primary diagnosis while unfavorable outcome of the model suggested that one quarter (25%) of the patients were already dead of breast cancer after 2.5 years of diagnosis. Among the subgroup of N- patients, no cancer-related deaths were observed among patients exhibiting favorable outcome of the model. Instead, the unfavorable outcome suggested cancer mortality for every fourth patient within 5.3 years from diagnosis. Correspondingly, the majority of N+ patients with favorable and unfavorable outcome of the model were alive after 17.6 and 2.0 years from the primary diagnosis, respectively.
Fig. 1.
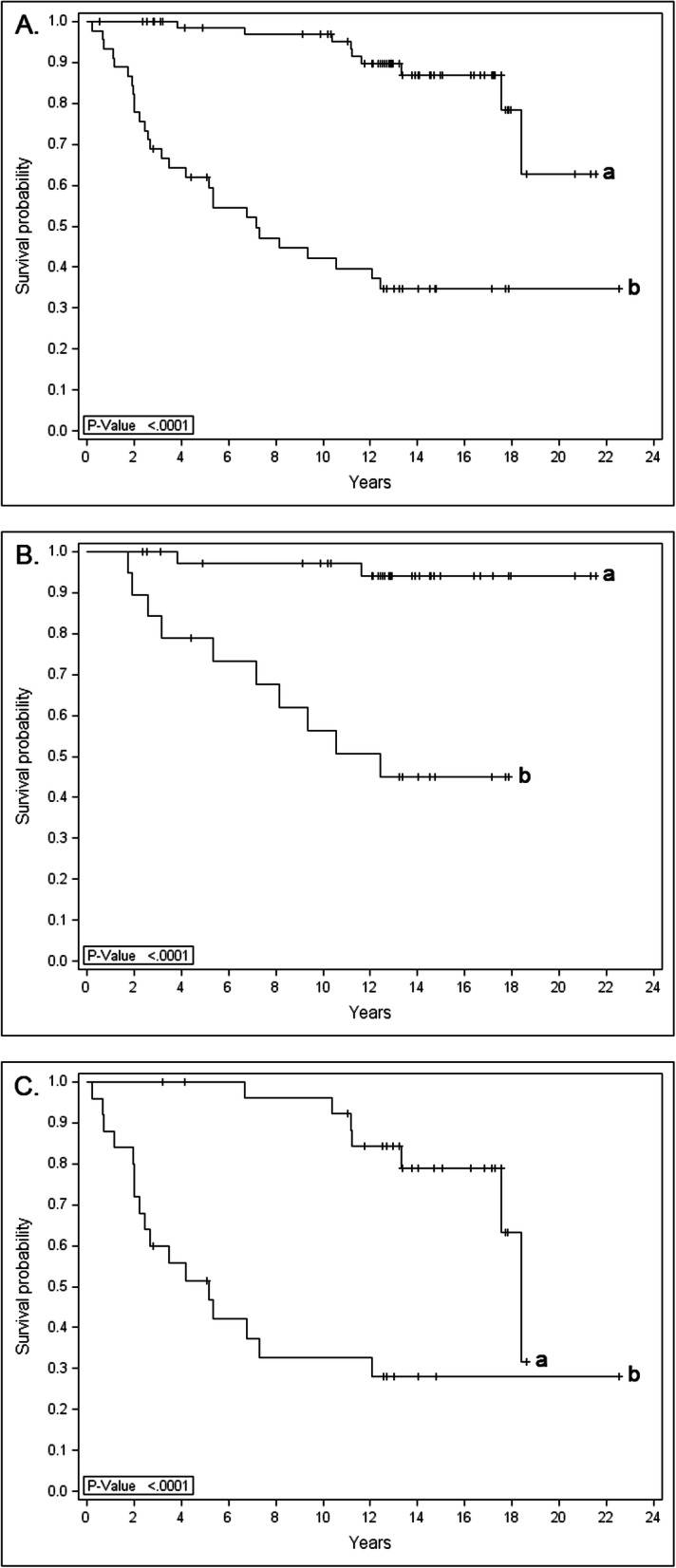
Kaplan-Meier curves show the survival difference between favorable (curve a: low immunoexpressions for Securin, Separase and Cdk1) vs unfavorable (curve b: high expressions of Securin, Separase and Cdk1) outcome of the prognostic model for all breast carcinomas (a) and for subgroups with N+ (b) and N- (c) disease (cohort I, n = 781)
Finally, in multivariable analyses of all breast cancer subtypes (Table 3), the designed model was compared with the established clinical prognosticators of breast cancer, i.e. tumor size, nodal status, intrinsic classification and histological grade for superior prognostic power in predicting the risk of breast cancer mortality. In the whole material, significant prognostic impact was observed for axillary lymph node status along with the designed model. Independent prognostic value was detected for the model also among N+ and N- patients. Tumor size (diameter < 2 cm vs ≥2 cm), intrinsic classification or histological grade (1–2 vs 3) were not detected with independent prognostic value in any of the performed analyses.
Table 3.
Multivariate analyses involving the prognostic modela with nodal status, tumor size, intrinsic classification and histological grade
| HRb | p | CI | |
|---|---|---|---|
| All patients (n = 781) | |||
| Model | 8.4 | < 0.0001 | 3.4–20.7 |
| Nodal status | 4.3 | < 0.0001 | 2.6–7.0 |
| Tumor size | ns. | ||
| Tumor grade | ns. | ||
| Intrinsic classification | ns. | ||
| N+ patients (n = 350) | |||
| Model | 6.5 | 0.0003 | 2.3–17.9 |
| Tumor size | ns. | ||
| Tumor grade | ns. | ||
| Intrinsic classification | ns. | ||
| N- patients (n = 431) | |||
| Model | 19.5 | 0.006 | 2.3–163.8 |
| Tumor size | ns. | ||
| Tumor grade | ns. | ||
| Intrinsic classification | ns. | ||
(≥1% of cancer cells) and Cdk1 (≥10% of cancer cells)
aHigh expression for Securin (≥10% of cancer cells), Separase
bThe hazard ratio of breast cancer death
ns not statistically significant
As the cohort I contained a relatively old patient material, a more recent patient material (cohort II) was collected to evaluate the prognostic impact of the regulators of metaphase-anaphase transition in luminal (n = 208) and triple-negative breast carcinomas (n = 146). As the result of luminal breast carcinomas, Securin (HR 1.1, p = 0.02, CI 1.0–1.2) and nuclear Separase (HR 5.7, p = 0.002, CI 1.9–17.2) – but not Cdk1 remained the most powerful predictors of cancer mortality, along with nodal status (HR 4.9, p = 0.003, CI 1.7–13.7). Also as combined into a model, Securin and Separase showed significant prognostic impact (p = 0.0006). Similar trends for the prognostic model were observed also in separate analyses of N+ (n = 42) and N- (n = 166) patients although the associations were not statistically significant in the small patient groups.
In TNBC (n = 146), no statistically significant prognostic value could be detected for any of the immunohistochemically studied biomarkers or clinicopathological features.
Discussion
In the present study, prognostic models involving regulators of the metaphase-anaphase transition of the cell cycle were introduced for predicting survival of breast cancer patients. The model was assembled based on data from a total of 1135 breast cancer patients with complete clinical information and up to 22-year follow-up.
The results show that combining high immunoexpressions for Securin, Separase and Cdk1 comprises a promising prognostic model indicating 8.4-fold increased risk of breast cancer death (p < 0.0001). In luminal breast carcinomas, the combination of Securin and Separase resulted in independent prognostic impact. In all analyses, the prognostic value of the combination of Securin and Separase with or without Cdk1 outperformed the impact of tumor size and histological grade whereas axillary lymph node status remained a strong and independent prognosticator in all analyses. At highest, the introduced prognostic model predicted 19.5-fold increased risk of breast cancer death among N- (p = 0.006) and 6.5-fold increased risk of mortality among N+ breast carcinomas (p = 0.0003). This suggests that the model may provide additional prognostic information to nodal status in treatment decisions of breast cancer patients. Among N- disease, the model may provide information to identify patients with aggressive course of disease, potentially in need of adjuvant treatments. In N+, the model could provide evidence for withholding chemotherapy from patients with favorable outcome.
TNBC is an aggressive subtype of breast cancer resulting in high mortality. It is also a therapeutically challenging subgroup as the lack of estrogen and progesterone receptors limits the treatment options available. The ongoing intense research has not yet revealed promising novel prognostic biomarkers or treatment targets for TNBC [2]. The present study did not reveal a prognostic impact for the routine clinicopathological parameters or for the studied biomarkers in TNBC. This may be due to the small cohort size as well as the heterogeneous nature of TNBC comprising several molecular subtypes [2].
The value of the introduced model lies in the pivotal role of the studied biomarkers in cancer progression. Loss of control of the cell cycle is a hallmark of malignancy resulting in aneuploidy and genomic instability [25]. Cell cycle checkpoints, including the mitotic checkpoint SAC, are the major cell cycle control mechanism and specifically deregulated in cancer cells. The presently studied regulators of metaphase-anaphase transition are involved in ensuring the fidelity of chromosome segregation. In previous literature, the studied biomarkers have been shown with prognostic impact in breast carcinoma as well as in other malignancies [14, 16, 17, 26–33]. In addition, numerous strategies have been proposed for the design of cell cycle-selective therapies in cancer, including targeting the metaphase-anaphase transition [8]. In all, the introduced model appears to identify biological drivers that could add to the conventional histopathological evaluation on the proliferative behavior of the tumor.
The reliability of the introduced prognostic model is increased by repeated statistical analysis of the studied biomarkers. The first approach applied for all breast cancer subtypes (cohort I) was based on testing combinations of biomarkers for their independent prognostic impact in comparison to each other, the clinical prognostic features and disease survival. In the second approach applied for analysis luminal and triple-negative breast carcinomas (cohort II), the optimal combination of biomarkers was extracted together with the clinical prognostic markers in sequential multivariate analyses of breast cancer-specific survival. Each approach was independently applied on separate patient materials originating from two Finnish breast cancer centers. As a conclusion, irrespective of the patient material or statistical procedure, the combination of Securin and Separase showed the most significant prognostic impact among all breast carcinomas and among luminal breast cancer.
From the practical point of view, immunohistochemical detection of Securin and Separase comprises a biology driven, cost-effective and reliable prognostic method (Fig. 2). By comparison, Ki-67 - the established proliferation marker of cancer diagnostics - has been criticized for high variability across individual pathologist and institutions, as well as for poor prognostic value [34, 35]. In the literature, numerous prognostic models have been introduced for breast cancer but only a few of them have shown impact exceeding that of the routine clinical prognosticators [36–38].
Fig. 2.

The combination of immunoexpressions for Securin and Separase indicate favorable (Securin a and Separase b) vs unfavorable (Securin c and Separase d) outcome of breast cancer
Conclusions
In our results from a total of 1135 breast cancer patients with complete clinical information and up to 22-year follow-up, high immunoexpression for the combination of Securin and Separase comprises a powerful prognostic tool to identify patients in risk of breast cancer death. In our scenario, the proposed model may facilitate personalized clinical decision suggesting less aggressive therapy for patients associated with low risk of mortality and indicating benefits from adjuvant therapy for a subgroup of patients with aggressive disease. Despite the accumulating data on multiparametric prognostic models, the clinical judgement remains the key determinant on selecting between the different treatment schemes.
Supplementary information
Additional file 1. Details of IHC for detecting Securin, Separase, Cdc20, Pttg1IP, SA2, Cdk1 and CyclinB1 and routine clinical biomarkers of breast cancer ER, PR, Ki-67 and HER2. The datasheet contains the details of the immunohistochemical methods used in this study.
Acknowledgements
The authors wish to thank Mrs. Sinikka Collanus for preparing the immunohistochemical stainings.
Abbreviations
- APC/C
Anaphase-Promoting Complex / Cyclosome
- Cdc20
Cell division cycle 20
- Cdk1
Cyclin-Dependent Kinase protein 1
- CI
Confidence interval
- CyclinB1
G2/mitotic-specific cyclin-B1
- ER
Estrogen receptor
- ESPL1
Extra Spindle Pole Bodies Like protein 1
- HER2
Human epidermal growth factor 2
- HR
Hazard ratio
- IHC
Immunohistochemistry
- ISH
In situ hybridization
- N +
Lymph node positive
- N-
Lymph node negative
- PBF
Pituitary tumor-transforming gene 1 binding factor
- PR
Progesterone receptor
- Pttg1
Pituitary tumor-transforming gene 1
- Pttg1IP
Pituitary tumor-transforming gene 1 interacting protein
- SA2
Stromal antigen 2
- SAC
Spindle Assembly Checkpoint
- TMA
Tissue microarray
- TNBC
Triple negative breast cancer
Authors’ contributions
HR participated in the design, histological and immunohistochemical analyses and writing of the manuscript, EL performed the statistical analyses, SK and LK selected and provided the patient specimen from Auria biobank, TK selected and provided the patient specimen from the Biobank of Central Finland, KT participated in analyzing the data and writing the manuscript, PK supervised the study and prepration of the manuscript. All authors read and approved the final manuscript.
Funding
This study has been financially supported by the Cancer Society of Finland, Turku University Hospital and Finska Läkaresällskapet foundation.
The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The data that support the findings of this study are available from Auria Biobank (www.auriabiobank.fi), Turku University Hospital, Turku, and Biobank of Central Finland, Jyväskylä, Finland but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the Biobanks stated above.
Ethics approval and consent to participate
The research was performed in accordance with the ethical standards of institutional and national research committees approved by the Regional Ethical Review Boards of Turku University Hospital and Auria Biobank, Turku, Finland, Central Hospital of Central Finland, Jyväskylä, Finland, and Finnish Cancer Registry, Cancer Society of Finland, Helsinki, Finland (permit numbers 6/2002, AB15–9859 and TK-53-716-16). All research procedures were performed in accordance with the 1964 Helsinki declaration and its amendments (www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects) and the Finnish Biobank Act (688/2012, Ministry of Social Affairs and Health, Helsinki, Finland). The patients have been included in the study according to the Finnish Biobank Act 688/2012, Ministry of Social Affiars and Health (https://www.finlex.fi/en/laki/kaannokset/2012/20120688). According to the Act, the national biobank organization, after ethical pre-evaluation, allows collection, storage and processing of patient specimen and information for medical research.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12885-020-07045-3.
References
- 1.Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee A, Djamgoz MBA. Triple negative breast cancer: emerging therapeutic modalities and novel combination therapies. Cancer Treat Rev. 2018;62:110–122. doi: 10.1016/j.ctrv.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Rakha EA, Soria D, Green AR, Lemetre C, Powe DG, Nolan CC, et al. Nottingham prognostic index plus (NPI+): a modern clinical decision making tool in breast cancer. Br J Cancer. 2014;110:1688–1697. doi: 10.1038/bjc.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):26–35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Filipits M, Nielsen TO, Rudas M, Greil R, Stöger H, Jakesz R, et al. The PAM50 risk-of-recurrence score predicts risk for late distant recurrence after endocrine therapy in postmenopausal women with endocrine-responsive early breast cancer. Clin Cancer Res. 2014;20:1298–1305. doi: 10.1158/1078-0432.CCR-13-1845. [DOI] [PubMed] [Google Scholar]
- 6.Potapova T, Gorbsky GJ. The consequences of chromosome segregation errors in mitosis and meiosis. Biology. 2017;6:E12. doi: 10.3390/biology6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol. 2015;25:R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez-Brauer C, Thu KL, Mason JM, Blaser H, Bray MR, Mak TW. Targeting mitosis in cancer: emerging strategies. Mol Cell. 2015;60:524–536. doi: 10.1016/j.molcel.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Talvinen K, Tuikkala J, Nevalainen O, Rantanen A, Hirsimäki P, Sundström J, et al. Proliferation marker securin identifies favourable outcome in invasive ductal breast cancer. Br J Cancer. 2008;99:335–340. doi: 10.1038/sj.bjc.6604475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grizzi F, Di Biccari S, Fiamengo B, Štifter S, Colombo P. Pituitary tumor-transforming gene 1 is expressed in primary ductal breast carcinoma, lymph node infiltration, and distant metastases. Dis Markers. 2013;35:267–272. doi: 10.1155/2013/912304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solbach C, Roller M, Peters S, Nicoletti M, Kaufman M, Knecht R. Pituitary tumor-transforming gene (PTTG): a novel target for anti-tumor therapy. Anticancer Res. 2005;25:121–125. [PubMed] [Google Scholar]
- 12.Smith VE, Franklyn JA, McCabe CJ. Pituitary tumor-transforming gene and its binding factor in endocrine cancer. Expert Rev Mol Med. 2010;12:e38. doi: 10.1017/S1462399410001699. [DOI] [PubMed] [Google Scholar]
- 13.Karra H, Repo H, Ahonen I, Löyttyniemi E, Pitkänen R, Lintunen M, et al. Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer. 2014;110:2905–2913. doi: 10.1038/bjc.2014.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gurvits N, Löyttyniemi E, Nykänen M, Kuopio T, Kronqvist P, Talvinen K. Separase is a marker for prognosis and mitotic activity in breast cancer. Br J Cancer. 2017;117:1383–1391. doi: 10.1038/bjc.2017.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J, Lu M, Cui Q, Zhang D, Kong D, Liao X, et al. Overexpression of ASPM, CDC20, and TTK confer a poorer prognosis in breast cancer identified by gene co-expression network analysis. Front Oncol. 2019;9:e310. doi: 10.3389/fonc.2019.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Repo H, Löyttyniemi E, Nykänen M, Lintunen M, Karra H, Pitkänen R, et al. The expression of cohesin subunit SA2 predicts breast cancer survival. Appl Immunohistochem Mol Morphol. 2016;24:615–621. doi: 10.1097/PAI.0000000000000240. [DOI] [PubMed] [Google Scholar]
- 17.Repo H, Gurvits N, Löyttyniemi E, Nykänen M, Lintunen M, Karra H, et al. PTTG1-interacting protein (PTTG1IP/PBF) predicts breast cancer survival. BMC Cancer. 2017;17:705–712. doi: 10.1186/s12885-017-3694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang N, Pati D. Biology and insights into the role of cohesin protease separase in human malignancies. Biol Rev Camb Philos Soc. 2017;92:2070–2083. doi: 10.1111/brv.12321. [DOI] [PubMed] [Google Scholar]
- 19.Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, et al. Thresholds for therapies: highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2009. Ann Oncol .2009;20:1319–1329. [DOI] [PMC free article] [PubMed]
- 20.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ, editors. WHO classification of Tumours of the breast. 4. Lyon: IARC; 2012. pp. 10–11. [Google Scholar]
- 21.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, et al. Tailoring therapies - improving the management of early breast cancer: St Gallen international expert consensus on the primary therapy of early breast Cancer. Ann Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. Goldhirsch, EP, Winer AS, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer. Ann Oncol. 2013;24:2206–2223. [DOI] [PMC free article] [PubMed]
- 23.Suzuki T, Urano T, Miki Y, Moriya T, Akahira J, Ishida T, et al. Nuclear cyclin B1 in human breast carcinoma as a potent prognostic factor. Cancer Sci. 2007;98:644–651. doi: 10.1111/j.1349-7006.2007.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241–256. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamenz J, Hauf S. Time to split up: dynamics of chromosome separation. Trends Cell Biol. 2017;27:42–54. doi: 10.1016/j.tcb.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Tong Y, Zhao W, Zhou C, Wawrowsky K, Melmed S. PTTG1 attenuates drug-induced cellular senescence. PLoS One. 2011;6:e23754. doi: 10.1371/journal.pone.0023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer R, Fofanov V, Panigrahi A, Merchant F, Zhang N, Pati D. Overexpression and mislocalization of the chromosomal segregation protein separase in multiple human cancers. Clin Cancer Res. 2009;15:2703–2710. doi: 10.1158/1078-0432.CCR-08-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi R, Sun Q, Sun J, Wang X, Xia W, Dong G, et al. Cell division cycle 20 overexpression predicts poor prognosis for patients with lung adenocarcinoma. Tumour Biol. 2017;39:1–10. doi: 10.1177/1010428317692233. [DOI] [PubMed] [Google Scholar]
- 29.Ding ZY, Wu HR, Zhang JM, Huang GR, Ji DD. Expression characteristics of CDC20 in gastric cancer and its correlation with poor prognosis. Int J Clin Exp Pathol. 2014;7:722–727. [PMC free article] [PubMed] [Google Scholar]
- 30.Read ML, Fong JC, Modasia B, Fletcher A, Imruetaicharoenchoke W, Thompson RJ, et al. Elevated PTTG and PBF predicts poor patient outcome and modulates DNA damage response genes in thyroid cancer. Oncogene. 2017;36:5296–5308. doi: 10.1038/onc.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Cho H, Shin HY, Chung JY, Kang ES, Lee EJ, et al. Accumulation of cytoplasmic Cdk1 is associated with cancer growth and survival rate in epithelial ovarian cancer. Oncotarget. 2016;7:49481–49497. doi: 10.18632/oncotarget.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung WW, Lin YM, Wu PR, Yen HH, Lai HW, Su TC, et al. High nuclear/cytoplasmic ratio of Cdk1 expression predicts poor prognosis in colorectal cancer patients. BMC Cancer. 2014;14:951–957. doi: 10.1186/1471-2407-14-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun X, Zhangyuan G, Shi L, Wang Y, Sun B, Ding Q. Prognostic and clinicopathological significance of cyclin B expression in patients with breast cancer: a meta-analysis. Medicine. 2017;96:e6860. doi: 10.1097/MD.0000000000006860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11:174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 35.Penault-Llorca F, Radosevic-Robin N. Ki67 assessment in breast cancer: an update. Pathology. 2017;49:166–171. doi: 10.1016/j.pathol.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Phung MT, Tin Tin S, Elwood JM. Prognostic models for breast cancer: a systematic review. BMC Cancer. 2019;19:230–247. doi: 10.1186/s12885-019-5442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green AR, Soria D, Stephen J, Powers DG, Nolan CC, Kunkler I, et al. Nottingham prognostic index plus: validation of a clinical decision making tool in breast cancer in an independent series. J Pathol Clin Res. 2016;2:32–40. doi: 10.1002/cjp2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albergaria A, Ricardo S, Milanezi F, Carneiro V, Amendoeira I, Vieira D, et al. Nottingham prognostic index in triple-negative breast cancer: a reliable prognostic tool? BMC Cancer. 2011;11:299–306. doi: 10.1186/1471-2407-11-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Details of IHC for detecting Securin, Separase, Cdc20, Pttg1IP, SA2, Cdk1 and CyclinB1 and routine clinical biomarkers of breast cancer ER, PR, Ki-67 and HER2. The datasheet contains the details of the immunohistochemical methods used in this study.
Data Availability Statement
The data that support the findings of this study are available from Auria Biobank (www.auriabiobank.fi), Turku University Hospital, Turku, and Biobank of Central Finland, Jyväskylä, Finland but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the Biobanks stated above.


