Abstract
Background
There is an urgent need to discover biocontrol agents to control bacterial wilt. This study reports on a new lipopeptide-producing biocontrol strain FJAT-46737 and explores its lipopeptidic compounds, and this study investigates the antagonistic effects of these compounds.
Results
Based on a whole genome sequence analysis, the new strain FJAT-46737 was identified as Bacillus velezensis, and seven gene clusters responsible for the synthesis of bioactive secondary metabolites in FJAT-46737 were predicted. The antimicrobial results demonstrated that FJAT-46737 exhibited broad-spectrum antimicrobial activities in vitro against three bacteria and three fungi. Pot experiments showed that the control efficiencies for tomato bacterial wilt of the whole cultures, the 2-fold diluted supernatants and the crude lipopeptide of FJAT-46737 were 66.2%, 82.0%, and 96.2%, respectively. The above results suggested that one of the antagonistic mechanisms of FJAT-46737 was the secretion of lipopeptides consisting of iturins, fengycins and surfactins. The crude lipopeptides had significant antagonistic activities against several pathogens (including Ralstonia solanacearum, Escherichia coli and Fusarium oxysporum) and fengycins were the major antibacterial components of the lipopeptides against R. solanacearum in vitro. Furthermore, the rich organic nitrogen sources (especially yeast extracts) in the media promoted the production of fengycin and surfactin by FJAT-46737. The secretion of these two lipopeptides was related to temperature fluctuations, with the fengycin content decreasing by 96.6% and the surfactins content increasing by 59.9% from 20 °C to 40 °C. The optimal temperature for lipopeptide production by FJAT-46737 varied between 20 °C and 25 °C.
Conclusions
The B. velezensis strain FJAT-46737 and its secreted lipopeptides could be used as new sources of potential biocontrol agents against several plant pathogens, and especially the bacterial wilt pathogen R. solanacearum.
Keywords: Bacillus velezensis FJAT-46737, Lipopeptides, Antagonistic activities, Bacterial wilt, Fengycin, Culture conditions
Background
Bacterial wilt caused by Ralstonia solanacearum is a devastating disease that affects almost 250 plant species, and it seriously threaten plant growth and leads to huge losses worldwide [1, 2]. Traditional agricultural practices, such as crop rotation, field sanitation, resistant variety cultivation, and chemical bactericide application, have been widely applied to control the bacterial wilt, although they present certain shortcomings [3]. The long-term or excessive use of chemical bactericides caused pathogen resistance and has adverse effects on the environment, beneficial organisms, and human health [3]. The identification of safe, effective and alternative methods for controlling plant pathogen diseases is becoming increasingly important for improving the output and quality of agricultural products. Furthermore, biological control using antagonistic microorganisms including bacteria, yeasts, and filamentous fungi, represent a safe, effective and sustainable alternative method for fighting plant pathogens compared with chemical bactericides [4–6]. Members of the genus Bacillus have been used as effective biocontrol agents to reduce damages caused by bacterial wilt, and their derived products account for approximately half of the commercially available biopesticides [7]. However, the control of bacterial wilt is difficult and ineffective due to the high genetic variability, persistence in the environment and broad host range of R. solanacearum. Thus, the use of alternative antimicobial agents is needed [3, 8].
Studies on the biocontrol mechanisms of Bacillus agents indicated that the biocontrol effects of certain strains are primarily associated with their production of various bioactive molecules [9]. Lipopeptides belong to the most important bioactive substances and they exhibit excellent properties, such as broad-spectrum antibiotic activity, good stability, low toxicity, high biodegradability, and reduced drug resistance susceptibility [10]. Bacillus lipopeptides include three classes, namely, iturin (bacillomycin D/F/L/Lc, iturin A/C/D/E, and mycosubtilin), fengycin (fengycin A/B, and plipastatin A/B) and surfactin (halobacillin, pumilacidin and surfactin) [11–13]. All the mentioned classes share a common amphiphilic molecule structure composed of a fatty acid side chain linking to a cyclic peptide ring (Figure S1). Among them, the iturins exhibit strong antifungal activities against various types of yeast and filamentous fungi but have limited antibacterial activity; the fengycins have strong antifungal activities, especially on filamentous fungi, although their ability to inhibit bacteria has only recently been reported by Villegas-Escobar et al. [14]; and the surfactins display effective bactericidal activities and can reduce the surface tension of plant roots, which facilitates the swimming ability and biofilm formation of Bacillus strains, thereby providing protection against pathogen attacks [15].
Studies of lipopeptides have mainly focused on the control of plant fungal pathogens, such as Rhizoctonia solani [16], Pythium ultimum [4], Botrytis cinerea [17], Podosphaera fusca [17], Fusarium graminearum [18, 19], Fusarium oxysporum [19], and Sclerotinia sclerotiorum [20]. Few studies have focused on R. solanacearum bacterial pathogens. Zhu et al. [21] reported that the lipopeptide mixture (surfactin and iturin A) produced by Bacillus amyloliquefaciens XZ-173 could effectively inhibit the growth of R. solanacearum. Furthermore, the lipopeptides secreted by XZ-173 were used to manufacture lipopeptide-mineral composites to achieve 87.76% biocontrol efficacy on bacterial wilt of tomato [22]. Lipopeptides suppress plant diseases either by impeding the pathogens directly or by promoting induced systemic resistance in the host plants [17, 23, 24]. A strong correlation has been reported between defense-inducing activity and the surfactin content generated by Bacillus strains [25]. In addition, it has been reported that lipopeptides secreted by Bacillus bacteria depend on the strain it self, the culture medium components and cultivation conditions [26–28].
The present study reports on the new lipopeptide-producing biocontrol strain FJAT-46737, which was identified via a whole genome sequence analysis, and the bioactive secondary metabolite gene clusters in the strain were predicted. The suppressive effects of FJAT-46737 and its lipopeptides were evaluated in vitro and in vivo. Furthermore, the lipopeptide components were identified; and effects of the culture conditions on the antibacterial activity of the strain and its lipopeptides were investigated. This work tend to promote the application of strain FJAT-46737 and its lipopeptides as new sources of biocontrol agents against plant pathogens, especially the bacterial wilt pathogen R. solanacearum.
Results
Identification and antagonistic activities of FJAT-46737
Strain FJAT-46737 is a gram-positive and endospore-forming bacterium. Its colonies on the NA plate were flat, slightly rough, nearly circular, and light yellow (Figure S2). The phylogenetic analysis of the 16S rRNA and gyrB gene results indicated that strain FJAT-46737 belongs to the genus Bacillus and is closely related to strains B. velezensis and B. amyloliquefaciens (Figure S3, S4). The 16 S rRNA gene of FJAT-46737 exhibits 99.72, 99.71 and 99.44% similarity to the type strains Bacillus siamensis KCTC 13613T, Bacillus velezensis CR-502T and B. amyloliquefaciens DSM7T, respectively. The gyr B gene of FJAT-46737 displayed greater than 99.0% identity with the type species of B. amyloliquefaciens and B. velezensis. Because of the high degree of relatedness among B. amyloliquefaciens, B. siamensis and B. velezensis, the ANI method based on the whole-genome sequence was used to discriminate strain FJAT-46737.
The whole-genome sequence of strain FJAT-46737 contains 3,995,340 bp, and the G + C content of the chromosomal DNA was 46.5%. The ANI values between the strain FJAT-46737 and the type strains B. amyloliquefaciens DSM7T, B. siamensis KCTC 13613T, and B. velezensis KCTC 13012T were calculated as 94.16, 94.36 and 98.26%, respectively. The last one displayed ANI values of > 98%, which exceeded the recommended cut-off of 96% for species delineation. This result suggested that strain FJAT-46737 was a member of B. velezensis species.
The gene clusters of bioactive secondary metabolites in strain FJAT-46737 were analyzed using the antiSMASH method. The results have shown that strain FJAT-46737 possesses 7 gene clusters that are responsible for the synthesis of surfactin, fengycin, macrolactin, bacillaene, difficidin, bacilysin and bacillibactin (Figure S5). These findings indicate that strain FJAT-46737 presents strong antagonistic activities.
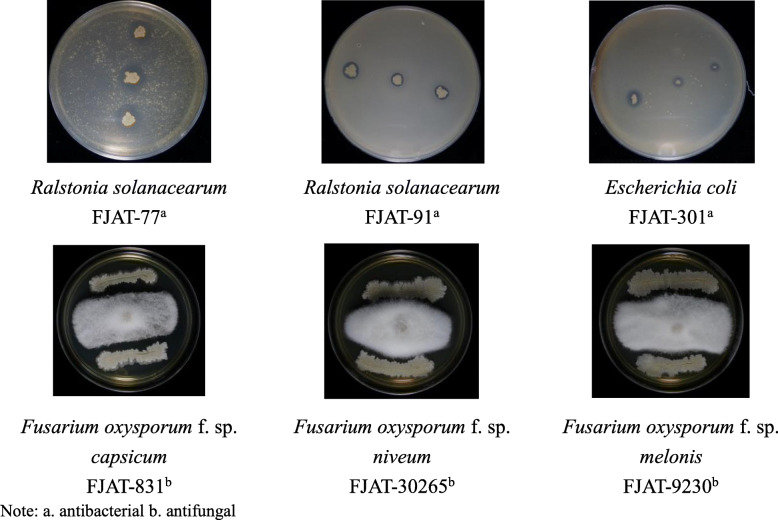
The agar disk diffusion method was used to evaluate the antagonistic activities of FJAT-46737 against several animal or plant pathogens. The results showed that this strain exhibited significant activities against gram-negative bacteria, including R. solanacearum and E. coli with the inhibition zone diamiter of 6.95–14.56 mm, and filamentous fungi, including 3 biotypes of. oxysporum (F. oxysporum f. sp. capsicum, F. oxysporum f. sp. niveum and F. oxysporum f. sp. melonis) (Fig. 1) with the inhibition rate of 65.25%–72.71%.
Fig. 1.
The spectrum of antimicrobial activity of FJAT-46737 against bacteria and filamentous fungi
Biocontrol efficacy of FJAT-46737 against tomato bacterial wilt
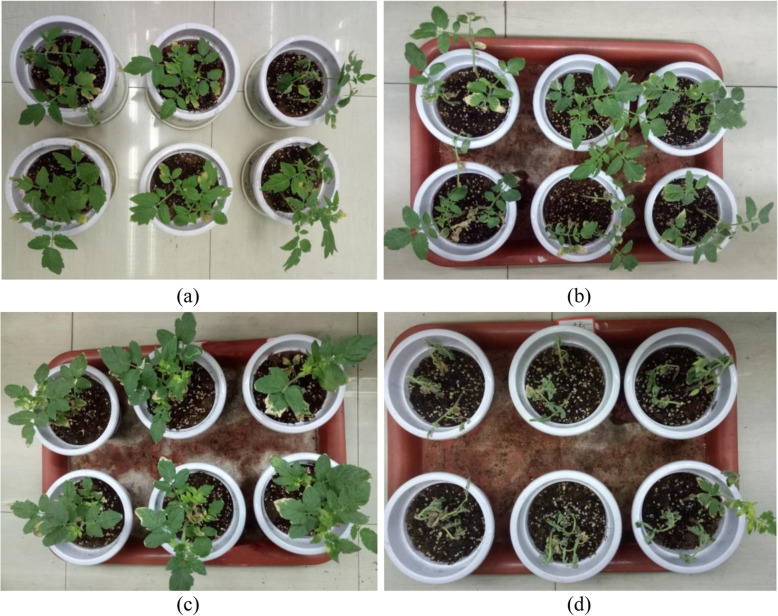
The above results showed that the B. velezensis strain FJAT-46737 had in vitro antibacterial activity against R. solanacearum. Thus, we attempted to investigate its biocontrol efficacy against tomato bacterial wilt by performing pot experiments under greenhouse conditions. First, we found that the DI of the tomato plants in the treatment group with whole cultures of FJAT-46737 (31.7%) was much lower than that of the control group (93.8%) (Fig. 2, Table 1). The biocontrol efficacy of whole cultures of FJAT-46737 against tomato bacterial wilt could reach 66.2% in the greenhouse experiments (Table 1).
Fig. 2.
Biocontrol effect of the whole cultures of FJAT-46737 (a), the culture supernatant (b), 1 mg/mL crude lipopeptides (c), and control (d) against bacterial wilt disease. Note: a. antibacterial b. antifungal
Table 1.
The biocontrol efficacies and the disease incidence in differently treatment groups of plants
| Treatment group | biocontrol efficiency | Disease incidence (DI) |
|---|---|---|
| Control group | 0% | 93.8% |
| Whole culture | 66.2% | 31.7% |
| 2-fold diluted cell-free supernatants | 82.0% | 17.5% |
| Lipopeptide (1 mg/mL) | 96.2% | 3.7% |
To clarify the biocontrol mechanisms of FJAT-46737, we subsequently evaluated the suppressive effects of its cell-free supernatants on tomato bacterial wilt. A preliminary experiment indicated that undiluted cell-free supernatant caused seedling injury while the 2-fold diluted supernatant had no injury effect. Thus, the 2-fold diluted cell-free supernatants was selected to perform the pot experiments. The results showed that an approximately 82.0% biocontrol efficiency could be achieved in the cell-free supernatant treatment group (Fig. 2, Table 1), which indicated that the biocontrol ability of FJAT-46737 could be mainly (if not entirely) attributed to its production of extracellular bioactive substances.
We further tested the biocontrol efficiency of the crude lipopeptide extracts from FJAT-46737 against tomato root infection by R. solanacearum. A preliminary experiment indicated that a high concentration (≥2.5 mg/mL) of lipopeptides led to seedling injury while 1 mg/ml lipopeptides had no injury effect. Thus, a concentration of 1 mg/mL lipopeptides was selected to perform the pot experiments. The treatment in which plantlets were soaked with the lipopeptides could significantly reduce the mortality of the tomato plants and achieve a biocontrol efficiency of 96.2% (Fig. 2, Table 1). These results suggested that one of the antagonistic mechanisms of strain FJAT-46737 is the secretion of lipopeptides.
LC-QTOF-MS/MS analyses of the lipopeptides produced by FJAT-46737
In this study, we used the LC-ESI-MS/MS method to determine the lipopeptide profile of FJAT-46737. Three classes of cyclic lipopeptides, namely, iturin (retention time, 12.8–22.3 min), fengycin (29.0–36.0 min), and surfactin (48.3–52.0 min), were detected, and the retention times, MS and MS2 spectral data and identification results for the lipopeptides from B. velezensis FJAT-46737 are summarized in Table 2. The results show that the lipopeptides consisted of C14–C16 iturin A, C13–C15 surfactins, and C16 fengycin A/B and C16 fengycin A2/B2 .
Table 2.
Identification of lipopeptides extracted from strain Bacillus velezensis FJAT-46737 through LC-QTOF-MS/MS
| Retention time (min) | MS m/z [M + H]+ /[M + Na]+ |
Identified Compounds |
|---|---|---|
| 12.833 | 1065.5a | C14Iturin A |
| 16.632 | 1079.7a | C15Iturin A |
| 17.536 | 1079.7a | C15Iturin A |
| 21.244 | 1093.6a | C16Iturin A |
| 22.284 | 1093.6a | C16Iturin A |
| 29.067 | 1449.9b | C16FengycinA2 |
| 30.423 | 1449.9b | C16FengycinA2 |
| 30.514 | 1463.9b | C16FengycinA |
| 31.056 | 1463.9b | C16FengycinA |
| 31.735 | 1449.9b | C16FengycinA2 |
| 32.413 | 1477.9b | C16FengycinB2 |
| 33.182 | 1463.9b | C16FengycinA |
| 34.132 | 1477.9b | C16FengycinB2 |
| 34.991 | 1477.9b | C16FengycinB2 |
| 35.262 | 1491.8b | C16FengycinB |
| 35.986 | 1491.8b | C16FengycinB |
| 48.376 | 1030.8a | C13Surfactin |
| 50.26 | 1044.8a | C14Surfactin |
| 50.757 | 1044.8a | C14Surfactin |
| 51.345 | 1030.8a | C13Surfactin |
| 51.978 | 1058.8a | C15Surfactin |
Note: a is [M + Na]+ and b is [M + H]+
Inhibitory spectra of the antagonistic lipopeptides
We used an agar well diffusion assay to further evaluate the antimicrobial activities of the crude lipopeptides from FJAT-46737. The results showed that the crude lipopeptides had significant activities against R. solanacearum, E. coli, and F. oxysporum in a dosage-dependent manner (Tables 3 and 4). At 48 h, the inhibition zone diameters under treatment with 10 mg/mL of the crude lipopeptides could reach up to 18.52 ± 0.73 mm, 14.55 ± 0.23 mm, and 14.57 ± 1.85 mm against the pathogens R. solanacearum FJAT-91 and FJAT-77, and E. coli FJAT-301, respectively (Table 3). The lipopeptide concentration of 0.5 mg/mL displayed antibacterial activity against R. solanacearum FJAT-91, while the concentration 0.25 mg/mL did not (Figure S6). These results implied that the crude lipopeptides of FJAT-46737 had the strongest antibacterial activity on R. solanacearum. Moreover, under 30 mg/mL of the crude lipopeptides, the inhibition zone diameters for 4 biotypes of F. oxysporum (F. oxysporum f. sp. capsicum FJAT-831, F. oxysporum f. sp. niveum FJAT-9230 and F. oxysporum f. sp. melonis FJAT-30265) were all approximately 20 mm at 72 h.
Table 3.
Antibacterial ability of lipopeptides from strain Bacillus velezensis FJAT-46747
| Diameter of inhibition zone (mm) | ||||
|---|---|---|---|---|
| indicator strains |
Ralstonia solanacearum FJAT-91 (tomato pathogens) |
Escherichia coli FJAT-301 |
Ralstonia solanacearum FJAT-77 (peanut pathogens) |
|
| concentration of crude lipopeptides (mg/mL) | 10 | 18.52 ± 0.73a | 14.55 ± 0.23a | 14.57 ± 1.85a |
| 5 | 16.04 ± 0.26b | 12.32 ± 1.14b | 13.90 ± 0.43a | |
| 2.5 | 13.87 ± 0.99b | 10.02 ± 0.01c | 11.19 ± 0.03a | |
| 1 | 12.51 ± 0.58b | weak | – | |
Note: Values were expressed as mean ± standard deviation (n = 3)
The difference letter in the same column indicated that the difference between the grades is significantly through Duncan test (p < 0.05)
Table 4.
Antifungal ability of lipopeptides from strain Bacillus velezensis FJAT-46747
| Diameter of inhibition zone (mm) | ||||
|---|---|---|---|---|
| indicator strains |
Fusarium oxysporum f. sp. capsicum FJAT-831 |
Fusarium oxysporum f. sp. melonis FJAT-9230 |
Fusarium oxysporum f. sp. niveum FJAT-30265 |
|
| concentration of crude lipopeptides (mg/mL) | 30 | 17.87 ± 0.11a | 18.77 ± 0.25a | 20.47 ± 3.08a |
| 20 | 16.62 ± 0.78a | 18.36 ± 0.44a | 17.28 ± 0.64a | |
| 10 | 13.84 ± 0.58b | 16.00 ± 0.22b | 14.51 ± 1.35a | |
Note: Values were expressed as mean ± standard deviation (n = 3)
The difference letter in the same column indicated that the difference between the grades is significantly through Duncan test (p < 0.05)
Effects of medium components and culture conditions on the antibacterial activities and contents of lipopeptides
To further improve the antagonistic activities of FJAT-46737, we analyzed the effects of the medium components and temperatures on the antibacterial activities of the cluture supernatants and crude lipopeptides. Given that the crude lipopeptides of FJAT-46737 had strong antibacterial activity on R. solanacearum, FJAT-91 was used as the indicator bacterium. The results of the antagonistic activity of the supernatant, crude lipopeptides and lipopeptide contents under different conditions are shown in Tables 5 and 6.
Table 5.
Diameter of inhibition zone of the culture supernatant and crude lipopeptides (10 mg/mL) produced by FJAT-46737 against Ralstonia solanacearum FJAT-91 and the content of lipopeptide under different culture medium
| Medium | A | B | C | D | E | F | |
|---|---|---|---|---|---|---|---|
| Diameter of inhibition zone (mm) | supernatant | – | – | 15.75 ± 0.38ab | 14.93 ± 0.77b | 16.36 ± 0.60a | 12.77 ± 0.88c |
| Lipopeptides | 13.88 ± 2.54b | – | 21.41 ± 1.60a | 19.15 ± 0.78a | 19.75 ± 0.93a | 15.67 ± 1.00b | |
| Yield of lipopeptide in the supernatant (mg/L) | iturin | 0.53 ± 0.20cd | 0.31 ± 0.09d | 0.98 ± 0.02b | 0.68 ± 0.14c | 1.99 ± 0.18a | 0.38 ± 0.02d |
| fengycin | 22.17 ± 11.94b | 9.03 ± 1.69b | 58.66 ± 9.81a | 50.37 ± 12.46a | 24.34 ± 2.75b | 9.17 ± 1.21b | |
| surfactin | 0.65 ± 0.42a | 0.55 ± 0.23a | 1.39 ± 0.06a | 1.21 ± 0.35a | 0.61 ± 0.06a | 0.59 ± 0.13a | |
| Total lipopeptides | 23.34 ± 12.54b | 10.07 ± 1.80b | 61.04 ± 9.86a | 52.27 ± 12.70a | 27.54 ± 3.90b | 10.22 ± 1.29b |
Note: Values were expressed as mean ± standard deviation (n = 3)
The difference letter in the same row indicated that the difference between the grades is significantly through Duncan test (p < 0.05). The effects of medium components on the antibacterial activities and contents of lipopeptides were carried out at 25 °C
Table 6.
Diameter of inhibition zone of the culture supernatant and crude lipopeptides (10 mg/mL) produced by FJAT-46737 against Ralstonia solanacearum FJAT-91 and the content of lipopeptide under different culture temperature
| Temperature | 20 °C | 25 °C | 30 °C | 35 °C | 40 °C | |
|---|---|---|---|---|---|---|
| Diameter of inhibition zone (mm) | supernatant | 12.36 ± 0.46b | 14.47 ± 1.06a | 12.06 ± 0.26b | – | – |
| Lipopeptides | 14.34 ± 0.90b | 17.16 ± 0.46a | 15.53 ± 1.83ab | 11.00 ± 0.49c | – | |
| Yield of lipopeptide in the supernatant (mg/L) | iturin | 0.65 ± 0.03b | 0.78 ± 0.07a | 0.86 ± 0.10a | 0.61 ± 0.07b | 0.45 ± 0.01c |
| fengycin | 55.11 ± 4.74a | 56.47 ± 6.59a | 23.62 ± 6.34b | 9.55 ± 0.27c | 1.88 ± 0.28c | |
| surfactin | 0.59 ± 0.06b | 0.83 ± 0.19b | 0.78 ± 0.13b | 1.22 ± 0.21a | 1.47 ± 0.15a | |
| Total lipopeptides | 56.30 ± 4.76a | 57.94 ± 4.76a | 25.41 ± 6.37b | 11.39 ± 0.53c | 3.72 ± 0.36c |
Note: Values were expressed as mean ± standard deviation (n = 3)
The difference letter in the same row indicated that the difference between the grades is significantly through Duncan test (p < 0.05). The effects of temperature on the antibacterial activities and contents of lipopeptides were carried out using the culture media C
Using the selected culture media A ~ F (Table S1), the supernatants and crude lipopeptides of FJAT-46737 exhibited good antibacterial activities against R. solanacearum FJAT-91, except for the culture supernatant from the medium A (Table 5). Moreover, the antibacterial activities of both the supernatants and crude lipopeptides from culture media C, D and E were stronger than those from culture media A, B and F (Table 5). These results suggested that the medium components could significantly affect the antibacterial activities of FJAT-46737.
Subsequently, the effects of temperature on the antibacterial activities of the cell-free supernatants and crude lipopeptides were determined using the culture medium C. The results showed that the culture temperature had significant effects on the antibacterial activities of the cell-free supernatants and crude lipopeptides: i) the cell-free supernatants did not exhibit antibacterial activity when the temperature was higher than 35 °C; and ii) both the cell-free supernatants and crude lipopeptides had the strongest antibacterial activities when incubated at 25 °C (Table 6).
The iturin, fengycin and surfactin contents in different culture conditions were further determined by LC-QTOF-MS and are summarized in Tables 5 and 6. The fengycins were the most abundant lipopeptide family produced by FJAT-46737. The iturin, fengycin and surfactin contents in the six culture media varied within the range of 0.31–1.99 mg/L, 9.03–58.66 mg/L and 0.55–1.39 mg/L, respectively. The contents of lipopeptides in culture media C and D were significantly higher than those in the other four media, indicating that media containing a rich organic nitrogen source might be benificial for lipopeptides production. Moreover, adding yeast extract in culture medium D significantly increased the yield of lipopeptides compared with the lack of yeast extract (in culture medium B). In culture medium C, the production of lipopeptides (61.04 ± 9.86 mg/L) was the highest compared to the other media. Moreover, the contents of the iturins, fengycins and surfactins were strongly dependent on temperature. The contents of fengycins decreased by 96.6% when the culture temperature increased from 20 °C to 40 °C while contents of surfactins increased by 59.9%. The temperature ranges of 25–30 °C, 20–25 °C and 35–40 °C were suitable for the production of iturin, fengycin and surfactin, respectively.
Subsequently, a correlation analysis was performed between the cell-free supernatant antibacterial activities and lipopeptide contents in different culture conditions (Table 7). The contents of fengycins and total lipopeptides were significantly correlated with the antibacterial activities of the cell-free supernatants of all samples (p < 0.05), whereas the contents of surfactins and iturins did not show this correlation. These results suggested that the antibacterial activities of the cell-free supernatants were mainly attributed to the secretion of the fengycins by FJAT-46737.
Table 7.
Correlation coefficients between antibacterial activities of the cultivation supernatant and the lipopeptide contents
| Pearson correlation coefficient | Kendall correlation coefficient | Spearman correlation coefficient | |
|---|---|---|---|
| Iturin | 0.570 | 0.597* | 0.740* |
| Fengycin | 0.695* | 0.559* | 0.726* |
| Surfactin | −0.100 | 0.078 | 0.033 |
| Total lipopeptide | 0.705* | 0.559* | 0.726* |
Test of significance by two-tailed
*p < 0.05
Effect of purified lipopeptides on the inhibition of R. solanacearum growth
The antibacterial activities of the purified lipopeptides against R. solanacearum were tested. The results showed that only the fraction obtained by SPE with 70% MeOH (named SPE70) exhibited antibacterial activity. Therefore, the composition of the fraction SPE70 was further determined by LC-QTOF-MS/MS (Figure S7). The results showed that the fraction SPE70 contained only fengycin, indicating that fengycin plays an important role in the growth inhibition of R. solanacearum. This result was consistent with the above correlation analysis.
Discussion
Traditional phenotypic methods and phylogenetic analysis of conserved gene are insufficient to distinguish the species of Bacillus, including B. subtilis, B. amyloliquefaciens, B. velezensis, B. siamensis, B. licheniformis, and B. pumilus, due to the close correlations between these species [29]. However, the increased availability of complete genome sequences has facilitated the discrimination of these species. The ANI value based on the complete genome sequences was calculated, and a cut-off of 96% was proposed for species delineation [29, 30]. In the present study, strain FJAT-46737 displayed 98.26% similarity to the B. velezensis type strain KCTC 13012T, which is well above of the recommended threshold of 96% for species delineation. Thus, FJAT-46737 was identified as B. velezensis.
The occurrence of bacterial wilt in crops has led to great economic losses wordwide. Chemicals are often used in controlling plant diseases, although they lead to environmental pollution and microbial pathogen resistance [3]. Many studies have shown that the use of Bacillus strains as biological control agents is as a promising and safe strategy for effective tomato bacterial wilt management. Kwon and Kim reported that B. subtilis JW-1 could lead to a > 80% reduction in bacterial wilt disease and can be used as a potential biocontrol agent of tomato bacterial wilt [31]. Xiong et al. isolated the B. amyloliquefaciens JK6 strain and found that it could effectively suppress tomato bacterial wilt with biocontrol efficacies for up to 52.9% under two greenhouse conditions [32]. B. subtilis 916 exhibited biocontrol efficiency of 55.6% on tomato bacterial wilt [33]. In the present study, we identified a new biocontrol strain B. velezensis FJAT-46737, and it exhibits broad-spectrum antimicrobial activities against gram-negative bacteria and filamentous fungi. The biocontrol efficacy of strain FJAT-46737 against tomato bacterial wilt was 66.2%, which was higher than that of many previously reported Bacillus spp. strains, such as the strains APF1 (60.3%) [34], JK6 (58.6%) [32] and 916 (55.6%) [33].
The biocontrol ability of Bacillus strains against plant pathogens has been confirmed to be achieved by the induction of host systemic resistance, antibiotic production, siderophore and lytic enzymes secretion, biofilm formation, or niches competition within the rhizosphere [35]. Bais et al. reported that B. subtilis 6051 was able to control Pseudomonas syringae root infection in Arabidopsis because of the secretion of surfactin and the formation of a biofilm on the plant roots [15]. Kwon and Kim demonstrated that B. subtilis JW-1 induced a significant bacterial wilt disease suppression effect in vivo due to the production of cyclic lipopeptides [31]. Xiong et al. reported that the secretion of surfactin by the strain JK6 played important roles in the biocontrol of tomato bacterial wilt [32]. In the present study, the biocontrol efficiency of the cell-free supernatant against tomato bacterial wilt was 82.0% while that of the lipopeptides reached up to 96.2%, indicating that one of the mechanisms of disease suppression by B. velezensis FJAT-46737 was lipopeptide secretion.
The production of lipopeptide by Bacillus species is based on the strains: some strains can coproduce two or three classes of lipopeptides, while others can yield only one class [11–13]. B. licheniformis MB01 produces surfactin only [12], B. subtilis K1 coproduces surfactins and iturins, while B. amyloliquefaciens SYBC H47 yields three types of lipopeptides, namely, bacillomycin, fengycin and surfactin [36]. In the present study, the B. velezensis strain FJAT-46737 could coproduce three types of lipopeptides: C14–C16 iturin A, C16 fengycin A/B, C16 fengycin A2/ B2, as well as C14–C15 surfactin. Furthermore, the yield of lipopeptides was significantly affected by the medium components (such as carbon and nitrogen sources, trace metals, etc.) and cultivation conditions (such as culture temperature, incubation time, rotary speed, etc.) [37]. In this study, B. velezensis FJAT-46737 could yield three types of lipopeptides in all six common culture media (A ~ F). However, the antibacterial activities of the crude lipopeptides (10 mg/mL) produced from culture media A, B or F were much weaker than that of the other three media (C, D and E). The carbon source has been considered as the primary factor influencing the lipopeptide production. For example, Li et al. found that variations in carbon sources in the culture medium changed the type of lipopeptides produced by B. licheniformis HSN221 [38]. However, the types of lipopeptides produced by FJAT-46737 were not changed if the culture medium contained glucose or not. Moreover, nitrogen sources have been reported to play an important role in the regulation of biosurfactant synthesis [39]. Our results showed that rich organic nitrogen sources in the media were beneficial for producing fengycin and surfactin. Furthermore, yeast extracts had a greater influence on the production of fengycin than iturin and surfactin, which may be related to the less sensitive response of the enzymatic complex for fengycin biosynthesis to nitrogen sources compared with that for iturin and surfactin [40].
Interestingly, opposite effects were observed for the culture temperatures on the contents of the fengycins and surfactins generated by strain FJAT-46737: with the increasing temperature, the content of fengycins decreased while that of surfactin increased. These results were not consistent with the report by Monteiro et al., who found that low temperature (15 °C) was suitable for producing surfactins and the amount of fengycin was not affected by temperature changes [37]. Fengycin and surfactin are synthesized nonribosomally by fengycin synthetases and surfactin synthetases, respectively [41]. Usually, the optimum temperature for enzyme activity is 37 °C, which can explain why the content of surfactins increases along with increase of the culture temperature. The reduction of fengycin under high temperatures could attributed to the decreased activity of fengycin synthetases. Generally, the suitable temperatures for crop growth in the field are 20 °C–30 °C. Our results indicated that 25 °C was a favorable temperature for the production of lipopeptides producing the strongest antibacterial activities, implying that strain FAJT-46737 and its lipopeptides have good ecosystem adaptability and prospects for future agricultural application. Previous studies demonstrated that surfactins display antibacterial activities while iturins show strong antifungal activities with limited antibacterial activity. The fengycin family has been considered specific against filamentous fungi, although two cases recently reported by Villegas-Escobar et al. [14] and Chen et al., [42] indicated that fengycins exhibited strong antibacterial activity against R. solanacearum in vitro. In the present study, the correlation analysis between the cell-free supernatant antibacterial activities and lipopeptide contents indicated that the antibacterial activities of cell-free supernatant were significantly positive correlated with the content of fengycins. This finding is consistent with a report showing that a significant relationship occurred between the strong antibacterial activity and production of fengycin and surfactin of Bacillus isolates [43]. In addition, only the purified fraction SPE70 exhibited antibacterial activity in vitro, and it only consisted of fengycins, indicating that the antibacterial activity of the lipopeptide mixtures could be due to the fengycins. These results further confirmed the antibacterial activity of fengycins first reported by Villegas-Escobar et al. [14] and Chen et al., [42].
Conclusions
A new B. velezensis strain FJAT-46737 with broad-spectrum antimicrobial activities was confirmed to have strong antibacterial activity against the bacterial wilt pathogen R. solanacearum by both in vivo and in vitro experiments. Moreover, suppressive effects of FJAT-46737 were associated with lipopeptide secretion, especially the content of fengycins. Therefore, FJAT-46737 and its lipopeptides have good application prospects for the biocontrol of bacterial wilt.
Methods
Strains and chemicals
Strain FJAT-46737 was isolated from a soil sample from the Huanggang Mountain area, Fujian Province, China, and preserved at the China General Microbiological Culture Collection Center (CGMCC No. 14661). The pathogenic strains R. solanacearum FJAT-91 (CGMCC No. 10692, tomato pathogen), R. solanacearum FJAT-77 (peanut pathogen), Escherichia coli FJAT-301, F. oxysporum f. sp. capsicum FJAT-831, F. oxysporum f. sp. niveum FJAT-30265 and F. oxysporum f. sp. melonis FJAT-9230 were preserved at the Agricultural Bioresources Research Institute, Fujian Academy of Agricultural Sciences, Fujian, China. All the culture media used for bacterial and fungal growth were purchased from AoBoXing Biological Technology Co., Ltd. (Beijing, China). The cultivation substrates used for the pot experiments were purchased from Xiamen Jiang Ping biological Technology Co., Ltd. (Xiamen, China). The reference standards for iturin and surfactin were produced from B. subtilis and purchased from Sigma (St. Louis, MO, USA).
Antimicrobial spectra of the strain FJAT-46737
Using the agar disk diffusion method, strain FJAT-46737 was evaluated for its in vitro potential to inhibit several animal and plant pathogens, including R. solanacearum, E. coli, F. oxysporum f. sp. capsicum, F. oxysporum f. sp. niveum and F. oxysporum f. sp. melonis. For the fungal pathogen strains, a block of mycelium with a 7 mm-diameter was placed onto the center of a sterile potato dextrose agar (PDA) plate and then cultures of strain FJAT-46737 were streaked with sterilized toothpicks at a distance of 2.5 cm away from the margins of the mycelia colony and cultivated at 28 °C for three to 7 days. The PDA plate inoculated only with the pathogenic fungi was used as control.
The antagonistic effects of strain FJAT-46737 against the bacterial pathogenic strains were investigated using a two layer plating method. Semi-solid nutrition agar (NA in Table S1 for cultivating R. solanacearum)/Luria–Bertani (LB in Table S1 for cultivating E. coli) media containing pathogenic bacterial culture suspensions were poured onto Petri plates (90 mm) coated with the corresponding solid media. After solidification, a colony of the strain FJAT-46737 was streaked on the plate, and then cocultivated at 30 °C for 2 days. The NA/LB plate inoculated only with the strain FJAT-46737 was used as control. The inhibition rate was calculated according to the method descrbied by Chen et al. [44].
Identification of strain FJAT-46737
Strain FJAT-46737 was streaked on NA agar plates and cultured at 30 °C for 48 h for investigating the colony morphology. The genomic deoxyribonucleic acid (DNA) of FJAT-46737 was extracted using the sodium dodecyl sulfate (SDS) method, detected by agarose gel electrophoresis and quantified by Qubit® 3.0. Strain FJAT-46737 was further identified based on an analysis of its 16S rRNA and partial gyrase subunit B (gyrB) gene sequences and whole-genome sequence. The 16S rRNA gene was amplified and sequenced using the universal primers 27F and 1492R. The gyrB gene was amplified using the specific primers F (5′-GAAGTCATCATGACC-3′) and R (5′-AGCAGGGTACGGAT-3′). Whole-genome sequencing was performed on an Illumina HiSeq PE150 platform at the Beijing Novogene Bioinformatics Technology Co., Ltd. The average nucleotide identity (ANI) was calculated using the OrthoANIu algorithm [45]. An ANI value cut-off of 96% is recommended for species delineation [30, 46]. The 16S rRNA gene, gyrB gene and whole-genome sequences of strain FJAT-46737 were deposited in GenBank under the accession numbers MG924092, MH470338 and CP044133, respectively. The secondary metabolite gene clusters in strain FJAT-46737 were predicted using the antibiotics Secondary Metabolite Analysis Shell (antiSMASH) tool.
Extraction and identification of the lipopeptides
FJAT-46737 was grown at 30 °C in 50 mL of culture medium (Table S1) in a shaking incubator (170 rpm). After 48 h, the culture was centrifuged at 6000 g for 5 min to remove the cells. The supernatant was adjusted to pH 2.0 with 3 mol/L HCl to obtain the crude lipopeptides. The extracted lipopeptides were dissolved in phosphate buffer and then lyophilized [42]. The obtained lipopeptide powder was dissolved in pure methanol for antimicrobial activity tests, for the qualitative and quantitative analyses and it was dissolved in water for the further pot experiments.
Qualitative and quantitative analyses of the lipopeptides were performed using liquid chromatography quadrupole time-of-flight tandem mass spectrometry (LC-QTOF-MS/MS) technology based on previous methods [42]. The structure of the lipopeptides was estimated by their accurate mass and MS/MS fragmentation patterns according to the literatures [47–51]. The content of each type lipopeptide was defined as follows: milligram of standard/liter of culture supernatant (mg/L).
Biocontrol evaluation of FJAT-46737 against tomato bacterial wilt
Strain FJAT-46737 was cultured at 30 °C for 48 h in medium E (Table S1) in a shaking incubator (170 rpm). Tomato seedlings with 3–4 emerged leaves were transplanted into pots with a diameter of 15 cm at 30 °C. After 2 days, two groups (each with thirty seedlings) were drenched with 100 mL/pot of the whole FJAT-46737 cultures (108 cfu/mL) and the double diluted culture supernatant. The lipopeptide-treated group consisted of thirty tomato seedlings that were carefully uprooted, the roots were then dipped in the crude lipopeptide solution (1 mg/mL, pH 6.5) for 1 h and then the seedlings were transplanted into pots. Each pot was drenched with 100 mL of the R. solanacearum cultures (108 cfu/mL) after 3 days. Five control groups were also included: one was infected with only the R. solanacearum cultures and the others were irrigated with only water, whole cultures, supernatant and lipopeptides. Each treatment or control group was replicated three times. The survivability of the seedlings was monitored at a regular interval (2 days). The above pot experiments were carried out from June to August 2017 in a greenhouse with the following incubation conditions: light cycle of 16-h day-8-h night; temperature of 28–32 °C; and relative humidity of 60–80%. The strain FJAT-46737, supernatant, lipopeptide and water did not cause the happen of disease in tomato plants. Thus, the disease index (di), disease incidence (DI) and biocontrol efficacy of bacterial wilt were calculated according to the method described by Xiong et al. [39].
Antimicrobial spectrum of the lipopeptides
The crude lipopeptide extracts from FJAT-46737 were dissolved in pure methanol to the final concentrations of 2.5–30 mg/mL. The antimicrobial activities of the crude lipopeptides were evaluated against eight pathogenic strains using the oxford cup method [52]. The pathogenic strains were grown at 30 °C in the corresponding liquid media for 48 h (for bacteria) /72 h (for fungi). A two layer plate containing fungal spores (105 spores/mL) or bacterial culture suspensions (106 cfu/mL) was prepared, and then four oxford cups were placed onto each plate and 150 μL of either the crude lipopeptides or cell-free supernatants was added. Methanol served as a negative control. Finally, the plates were cultivated at 30 °C for 48 h and the inhibition zone diameter was measured. All experiments were repeated three times.
Effects of cultivation parameters on the antagonistic activities of the lipopeptides
R. solanacearum FJAT-91 was selected as an indicator strain. The culture media of A, B, C, D, E and F in Table S1 and temperatures of 20, 25, 30, 35 and 40 °C were selected for cultivating strain FJAT-46737. The antagonistic activities of the lipopeptides produced under different culture conditions were determined by the aforementioned agar disk diffusion method [42].
Purification of antibacterial lipopeptides
The lipopeptides were loaded onto a solid phase extraction (SPE) C18 cartridge (6 g/60 mL) and washed with 60 mL of water and different proportions of methanol (10 to 100%). After evaporation, the obtained fraction was dissolved in water and used for the antibacterial activity test.
Statistical analysis
An analysis of variance (ANOVA) of the quantitative data was carried out using the statistical software SPSS 19.0. Duncan’s multiple range tests were performed to determine the significant differences among the data (p < 0.05).
Supplementary information
Additional file 1: Figure S1. The chemical structure of three types of cyclic lipopeptides. Figure S2. Morphology of Bacillus strain FJAT-46737. Figure S3. Phylogenetic tree based on the 16S rDNA sequences showing the position of strain FJAT-46737 (accession number: MG924092). The type strains of Bacillus sp. and representatives of some other related taxa. Scale bar represents 0.001 substitutions per nucleotide position. It is note that the strain B. amyloliquefaciens subsp. plantarum FZB42 was renamed as B. velezensis. Figure S4. Phylogenetic tree based on the gyrB sequences showing the position of strain FJAT-46737 (accession number: MH470338). The type strains of Bacillus sp. and representatives of some other related taxa. Scale bar represents 0.001 substitutions per nucleotide position. Figure S5. The prediction of gene clusters of bioactive secondary metabolites in strain FJAT-46737. Figure S6. The antibacterial photo of lipopeptide (0.1 ~ 1 mg/mL) against R. solanacearum FJAT-91. Figure S7. The full scan LC–ESI–MS chromatogram of fraction SPE70. Table S1. Culture medium components
Acknowledgements
Not applicable.
Abbreviations
- CGMCC
China General Microbiological Culture Collection Center
- PDA
Potato dextrose agar
- LB
Luria–Bertani
- NA
Nutrient agar
- SDS
Sodium dodecyl sulfate
- gyr B
Gyrase subunit B
- DNA
Deoxyribonucleic acid
- RNA
Ribonucleic acid
- ANI
Average nucleotide identity
- antiSMASH
Antibiotics Secondary Metabolite Analysis Shell
- di
Disease index
- DI
Disease incidence
- SPE
Solid phase extraction
- ANOVA
Analysis of variance
- LC-QTOF-MS/MS
Liquid chromatography coupled to quadrupole time-of-flight tandem mass spectrometry
Authors’ contributions
MC, JW, BL and YZ conceived and designed the experiments. MC, JW, YZ, RX and BL were responsible for drafting the article. MC, RX, WY, CG, and ZC were involved in the experiments preformation and data analysis. All authors approved the final version of the manuscript.
Funding
This work was funded by the Young Elite Scientists Sponsorship Program by CAST (No. 2017QNRC001); the Science and Technology Innovation Team Program (No. STIT2017-1-11), the Program (No. DEC201821206) of Fujian Academy of Agricultural Sciences. These organazations provided finance support for this study, but did not involved in the study design, data analysis, decision to publish, or in writing this manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files. The 16S rRNA, gyrB and whole genome sequences of this study have been submitted to NCBI GenBank database (accessions MG924092, MH470338 and CP044133).
Ethics approval and consent to participate
No animals, human subjects, human material, or human data are used in this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12866-020-01851-2.
References
- 1.Genin S. Research review: molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 2010;187:920–928. doi: 10.1111/j.1469-8137.2010.03397.x. [DOI] [PubMed] [Google Scholar]
- 2.Jiang G, Wei Z, Xu J, Chen HL, Zhang Y, She XM, Macho AP, Ding W, Liao DS. Bacterial wilt in China: history, current status, and future perspectives. Front Plant Sci. 2017;8:1549–1559. doi: 10.3389/fpls.2017.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuliar YAN, Koki T. Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 2015;30:1–11. doi: 10.1264/jsme2.ME14144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ongena M, Jacques P, Toure Y, Destain J, Jabrane A, Thonart P. Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl Microbiol Biotechnol. 2005;69:29–38. doi: 10.1007/s00253-005-1940-3. [DOI] [PubMed] [Google Scholar]
- 5.Maji S, Chakrabartty PK. Biocontrol of bacterial wilt of tomato caused by Ralstonia solanacearum by isolates of plant growth promoting rhizobacteria. Australian J Crop Sci. 2014;8:208–214. [Google Scholar]
- 6.Yendyo S, Ramesh GC, Pandey BR. Evaluation of Trichoderma spp., Pseudomonas fluorescens and Bacillus subtilis for biological control of Ralstonia wilt of tomato. F1000 Res. 2018;6:2028–2050. doi: 10.12688/f1000research.12448.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam HS, Yang HJ, Oh BJ, Anderson AJ, Kim YC. Biological control potential of Bacillus amyloliquefaciens KB3 isolated from the feces of Allomyrina dichotoma larvae. Plant Pathol J. 2016;32:273–280. doi: 10.5423/PPJ.NT.12.2015.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Y, Caceres-Moreno C, Jimenez-Gongora T, Wang KK, Sang YY, Lozano-Duran R, Macho AP. The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the Solanaceae family. Plant Biotech J. 2018;16:1349–1362. doi: 10.1111/pbi.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rocha DJA, Moura AB. Biological control of tomato wilt caused by Ralstonia solanacearum and Fusarium oxysporum f. sp lycopersici by rhizobacteria. Trop Plant Pathol. 2013;38:423–430. [Google Scholar]
- 10.Chowdhury SP, Uhl J, Grosch R, Alquéres S, Pittroff S, Dietel K, Schmitt-Kopplin P, Borriss R, Hartmann A. Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol Plant-Microbe Interact. 2015;28:984–995. doi: 10.1094/MPMI-03-15-0066-R. [DOI] [PubMed] [Google Scholar]
- 11.Bóka B, Manczinger L, Kecskeméti A, Chandrasekaran M, Kadaikunnan S, Alharbi NS. Ion trap mass spectrometry of surfactins produced by Bacillus subtilis SZMC 6179J reveals novel fragmentation features of cyclic lipopeptides. Rapid Commun Mass Spectrom. 2016;30:1581–1590. doi: 10.1002/rcm.7592. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Liu SA, Mou H, Ma Y, Li M, Hu X. Characterization of lipopeptide biosurfactants produced by Bacillus licheniformis MB01 from marine sediments. Front Microbiol. 2017;8:871–882. doi: 10.3389/fmicb.2017.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, Kong Q, Qin C, Chen YL, Chen YJ, Lv RH, Zhou GH. Identification of lipopeptides in Bacillus megaterium by two-step ultrafiltration and LC-ESIMS/MS. AMB Express. 2016;6:79–94. doi: 10.1186/s13568-016-0252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villegas-Escobar V, González-Jaramillo LM, Ramírez M, Moncada RN, Sierrz-Zapata L, Orduz S. Lipopeptides from Bacillus sp. EA-CB0959: active metabolites responsible for in vitro and in vivo control of Ralstonia solanacearum. Biol Control. 2018;125:20–28. [Google Scholar]
- 15.Bais HP, Fall R, Vivanco JM. Biocontrol of Bacillus subtilis against infection of arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004;134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Q, Dong W, Li S, Lu X, Wang P, Zhang X, Wang Y, Ma P. Fengycin produced by Bacillus subtilis NCD-2 plays a major role in biocontrol of cotton seeding damping-off disease. Microbiol Res. 2014;169:533–540. doi: 10.1016/j.micres.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Romero D, de Vicente A, Rakotoaly RH, Dufour SE, Veening JW, Arrebola E, Cazorla F, Kuipers OP, Paquot M, Perez-Garcia P. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol Plant-Microbe Interact. 2007;20:430–440. doi: 10.1094/MPMI-20-4-0430. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Selvaraj JN, Xing FG, Zhou L, Wang Y, Song HM, Tan XX, Sun LC, Sangare LC, Folly YME, Liu Y. Antagonistic action of Bacillus subtilis strain SG6 on Fusarium graminearum. PLoS One. 2014;9:e92486. doi: 10.1371/journal.pone.0092486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong AD, Li HP, Yuan QS, Song XS, Yao W, He WJ, Zhang JB, Liao YC. Antagonistic mechanism of iturin a and plipastatin a from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS One. 2015;10:e0116871. doi: 10.1371/journal.pone.0116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez F, Castro M, Principe A, Borioli G, Fischer S, Mori G, Jofré E. The plant-associated Bacillus amyloliquefaciens strains MEP218 and ARP23 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J Appl Microbiol. 2012;112:159–174. doi: 10.1111/j.1365-2672.2011.05182.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z, Zhang GY, Xu YC, Yang XM, Ran W, Shen QR. Bio-control and growth-promoting effects of bio-manure fermented by lipopeptide-producing bacteria. Acta Pedol Sin. 2012;49:104–112. [Google Scholar]
- 22.Wu YL. Preparation of microbial lipopeptide-mineral composites and their effects on tomato growth and against bacterial wilt of tomato. Nanjing: Nanjing Agricultural University (In Chinese); 2015.
- 23.Asaka O, Shoda M. Biocontrol of Rhizoctonia solani damping off of tomato with Bacillus subtilis RB14. Appl Environ Microbiol. 1996;62:4081–4085. doi: 10.1128/aem.62.11.4081-4085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou TT, Chen D, Li CY, Liu F, Shen QR, Shen B. Isolation and characterization of Pseudomonas brassicacearum J12 as an antagonist against Ralstonia solanacearum and identification of its antimicrobial components. Microbiol Res. 2012;167:388–394. doi: 10.1016/j.micres.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Cawoy H, Mariutto M, Henry G, Fisher C, Vasilyeva N, Thonart P, Dommes J, Ongena M. Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol Plant-Microbe Interact. 2014;27:87–100. doi: 10.1094/MPMI-09-13-0262-R. [DOI] [PubMed] [Google Scholar]
- 26.Ohno A, Aao T, Shoda M. Effect of temperature on production of lipopeptide antibiotics, iturin a and surfactin by a dual producer, Bacillus subtilis RB14, in solid-state fermentation. J Biosci Bioeng. 1995;80:517–519. [Google Scholar]
- 27.Mizumoto S, Shoda M. Medium optimization of antifungal lipopeptide, iturin a, production by Bacillus subtilis in solid-state fermentation by response surface methodology. Appl Microb Biotechnol. 2007;76:101–108. doi: 10.1007/s00253-007-0994-9. [DOI] [PubMed] [Google Scholar]
- 28.Nihorimbere V, Cawoy H, Seyer A, Brunelle A, Thonart P, Ongena M. Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneflcial strain Bacillus amyloliquefaciens S499. FEMS Microbiol Ecol. 2012;79:176–191. doi: 10.1111/j.1574-6941.2011.01208.x. [DOI] [PubMed] [Google Scholar]
- 29.Fan B, Blom J, Klenk HP, Borriss R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “Operational group B. amyloliquefaciens” within the B. subtilis species complex. Front Microbiol. 2017;8:00022. doi: 10.3389/fmicb.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc.Natl.Acad.Sci.U.S.A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon JW, Kim SD. Characterization of an antibiotic produced by Bacillus subtilis JW-1 that suppresses Ralstonia solanacearum. J Microbiol Biotechnol. 2014;24:13–18. doi: 10.4014/jmb.1308.08060. [DOI] [PubMed] [Google Scholar]
- 32.Xiong HQ, Li YT, Cai YF, Cao Y, Wang Y. Isolation of Bacillus amyloliquefaciens JK6 and identification of its lipopeptides surfactin for suppressing tomato bacterial wilt. RSC Adv. 2015;5:82042–82049. [Google Scholar]
- 33.Qiao JQ, Chen ZY, Liang XJ, Liu YF, Liu YZ. Biocontrol efficacy on tomato bacterial wilt by Bacillus subtilis Bs916. Chin J Biol Control. 2016;32:229–234. [Google Scholar]
- 34.Abdulwareth AA, Ullah KK, Zarqa N, Li B, Alisaand M, Chun-Lan Y, Xie GL. Tomato plant growth promotion and antibacterial related-mechanisms of four rhizobacterial Bacillus strains against Ralstonia solanacearum. Symbiosis. 2014;63:59–70. [Google Scholar]
- 35.Tan SY, Jiang Y, Song S, Huang JF, Ling N, Xu YC, Shen QR. Two Bacillus amyloliquefaciens strains isolated using the competitive tomato root enrichment method and their effects on suppressing Ralstonia solanacearum and promoting tomato plant growth. Crop Prot. 2013;43:134–140. [Google Scholar]
- 36.Li XH, Zhang YZ, Wei ZW, Guan ZB, Cai YJ, Liao XR. Antifungal activity of isolated Bacillus amyloliquefaciens SYBC H47 for the biocontrol of peach gummosis. PLoS One. 2016;11:1–22. doi: 10.1371/journal.pone.0162125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monteiro FP, de Medeiros FHV, Ongena M, Franzil L, de Souza PE, de Souza JR. Effect of temperature, pH and substrate composition on production of lipopeptides by Bacillus amyloliquefaciens 629. Afr J Microbiol Res. 2016;10:1506–1512. [Google Scholar]
- 38.Li YM, Haddad NIA, Yang SZ, Mu BZ. Variants of lipopeptides produced by Bacillus licheniformis HSN221 in different medium components evaluated by a rapid method ESI-MS. Int J Pept Res Ther. 2008;14:229–235. [Google Scholar]
- 39.Ghribi D, Ellouze-Chaabouni S. Enhancement of Bacillus subtilis lipopeptide biosurfactants production through optimization of medium composition and adequate control of aeration. Biotechnol Res Int. 2011;653654. 10.4061/2011/653654. [DOI] [PMC free article] [PubMed]
- 40.Akpa E, Jacques P, Wathelet B, Paquot M, Fuchs R, Budzikiewicz H, Thonart P. Influence of culture conditions on lipopeptide production by Bacillus subtilis. Appl Biochem Biotech. 2001;91-93:551–561. doi: 10.1385/abab:91-93:1-9:551. [DOI] [PubMed] [Google Scholar]
- 41.Leães FL, Velho RV, Caldas DGG, Ritter AC, Tsai SM, Brandelli A. Expression of essential genes for biosynthesis of antimicrobial peptides of Bacillus is modulated by inactivated cells of target microorganisms. Res Microbiol. 2016;167:83–89. doi: 10.1016/j.resmic.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Chen MC, Wang JP, Zhu YJ, Liu B, Yang WJ, Ruan CQ. Antibacterial activity against Ralstonia solanacearum of the lipopeptides secreted from the Bacillus amyloliquefaciens strain FJAT-2349. J Appl Microbiol. 2019;126:1519–1529. doi: 10.1111/jam.14213. [DOI] [PubMed] [Google Scholar]
- 43.Mara I, Cabrefiga J, Montesinos E. Cyclic lipopeptide biosynthetic genes and products, and inhibitory activity of plant-associated Bacillus against phytopathogenic bacteria. PLoS One. 2015;10:e0127738. doi: 10.1371/journal.pone.0127738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y, Xu YP, Zhou T, Akkaya MS, Wang LL, Li SY, Li XY. Biocontrol of Fusarium wilt disease in strawberries using bioorganic fertilizer fortifed with Bacillus licheniformis X-1 and Bacillus methylotrophicus Z-1. 3 Biotech. 2020;10:80. doi: 10.1007/s13205-020-2060-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 46.Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64(2):346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 47.Pecci Y, Rivardo F, Martinotti MG, Allegrone G. LC/ESI-MS/MS characterisation of lipopeptide biosurfactants produced by the Bacillus licheniformis V9T14 strain. J Mass Spectrom. 2010;45:772–778. doi: 10.1002/jms.1767. [DOI] [PubMed] [Google Scholar]
- 48.Pathak KV, Keharia H, Gupta K, Thakur SS, Balaram P. Lipopeptides from the banyan endophyte, Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins. J Am Soc Mass Spectrom. 2012;23:1716–1728. doi: 10.1007/s13361-012-0437-4. [DOI] [PubMed] [Google Scholar]
- 49.Pathak KV, Bose A, Keharia H. Identification and characterization of novel surfactins produced by fungal antagonist Bacillus amyloliquefaciens 6B. Biotechnol Appl Biochem. 2014;61:349–356. doi: 10.1002/bab.1174. [DOI] [PubMed] [Google Scholar]
- 50.Pathak KV, Keharia H. Identification of surfactins and iturins produced by potent fungal antagonist, Bacillus subtilis K1 isolated from aerial roots of banyan (Ficus benghalensis) tree using mass spectrometry. 3 Biotech. 2014;4:283–295. doi: 10.1007/s13205-013-0151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jemila N, Manresab A, Rabanalc F, Ben Ayed H, Hmidet N, Nasri M. Structural characterization and identification of cyclic lipopeptides produced by Bacillus methylotrophicus DCS1 strain. J Chromatogr B. 2017;1060:374–386. doi: 10.1016/j.jchromb.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 52.Shang RF, Wang GH, Xu XM, Liu SJ, Zhang C, Yi YP, Liang JP, Liu Y. Synthesis and biological evaluation of new pleuromutilin derivatives as antibacterial agents. Molecules. 2014;19:19050–19065. doi: 10.3390/molecules191119050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The chemical structure of three types of cyclic lipopeptides. Figure S2. Morphology of Bacillus strain FJAT-46737. Figure S3. Phylogenetic tree based on the 16S rDNA sequences showing the position of strain FJAT-46737 (accession number: MG924092). The type strains of Bacillus sp. and representatives of some other related taxa. Scale bar represents 0.001 substitutions per nucleotide position. It is note that the strain B. amyloliquefaciens subsp. plantarum FZB42 was renamed as B. velezensis. Figure S4. Phylogenetic tree based on the gyrB sequences showing the position of strain FJAT-46737 (accession number: MH470338). The type strains of Bacillus sp. and representatives of some other related taxa. Scale bar represents 0.001 substitutions per nucleotide position. Figure S5. The prediction of gene clusters of bioactive secondary metabolites in strain FJAT-46737. Figure S6. The antibacterial photo of lipopeptide (0.1 ~ 1 mg/mL) against R. solanacearum FJAT-91. Figure S7. The full scan LC–ESI–MS chromatogram of fraction SPE70. Table S1. Culture medium components
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. The 16S rRNA, gyrB and whole genome sequences of this study have been submitted to NCBI GenBank database (accessions MG924092, MH470338 and CP044133).