Abstract
While mammalian secretory IgA (sIgA) targets mucosal pathogens for elimination, its interaction with the microbiota also enables commensal colonization and homeostasis. This paradoxical requirement in the control of pathogens versus microbiota raised the question of whether mucosal (secretory) immunoglobulins (sIgs) evolved primarily to protect mucosal surfaces from pathogens or to maintain microbiome homeostasis. To address this central question, we used a primitive vertebrate species (rainbow trout) in which we temporarily depleted its mucosal Ig (sIgT). Fish devoid of sIgT became highly susceptible to a mucosal parasite and failed to develop compensatory IgM responses against it. IgT depletion induced also a profound dysbiosis marked by the loss of sIgT-coated beneficial taxa, expansion of pathobionts, tissue damage, and inflammation. Restitution of sIgT levels in IgT-depleted fish led to a reversal of microbial translocation and tissue damage, as well as to restoration of microbiome homeostasis. Our findings indicate that specialization of sIgs in pathogen and microbiota control occurred concurrently early in evolution, thus revealing primordially conserved principles under which primitive and modern sIgs operate in the control of microbes at mucosal surfaces.
One sentence summary:
Mucosal immunoglobulins of primitive and modern vertebrates evolved to control pathogens and preserve microbiota homeostasis
Introduction
Mucosal surfaces of vertebrates are inhabited by incredibly dense and complex populations of commensal microbiota, and are also continuously exposed to a myriad of pathogens. Mucosal (secretory) immunoglobulins (sIgs) constitute the main humoral adaptive immune molecules contributing to the homeostasis and defense of mucosal surfaces (1, 2). In mammals, the central mucosal immunoglobulin is secretory IgA (sIgA) (1). Intriguingly, sIgA plays two seemingly paradoxical functions: on the one hand, sIgA targets mucosal pathogens for their neutralization and clearance (1, 3-5), while on the other hand, sIgA interaction with the microbiota enables commensal colonization, persistence and homeostasis (6-8). Moreover, sIgA contributes to, but is not essential for, protection of mucosal surfaces against pathogens, as evidenced by the very modest increased susceptibility to respiratory and intestinal infections observed in patients with selective sIgA deficiency (9). In contrast, recent studies have shown that sIgA is indispensable for the preservation of microbiota homeostasis, since profound microbiome changes have been reported in IgA-deficient mice and humans (10-15). Such changes are likely due to the involvement of IgA in microbiota colonization (6) as well as to the coating of both beneficial bacteria as well as pathobionts by sIgA (8, 11, 16). These paradoxical requirements for sIgA in the control of pathogens versus microbiota raise the question of whether sIgs initially evolved to protect mucosal surfaces from pathogens, or arose instead to establish and maintain homeostatic relationships with microbiota. Moreover, whether those roles were independently acquired during the evolution of sIgA in higher vertebrates or represent instead convergently evolved functions common to all vertebrate mucosal Igs, remains to be determined.
Whereas the role of innate immunity in mucosal host-microbial pathogenic or mutualistic interactions has often benefited from studies in invertebrates and lower vertebrates (17, 18), the study of sIg function in such interactions has almost exclusively been limited to mammalian sIgA (1, 8). Since teleost fish represent the oldest living bony vertebrates with an adaptive immune system similar to that of mammals, fish represent a unique model to understand critical aspects of immunoglobulin evolution and function (19-21). Teleosts have three bona fide immunoglobulin isotypes (i.e., IgM, IgD and IgT), have B and T lymphocytes, and respond to pathogens by inducing antigen-specific antibody (i.e., IgM and IgT) responses (21, 22). However, in contrast to warm-blooded vertebrates, fish lack organized lymphoid structures (i.e., lymph nodes, Peyer’s patches) and B cells in fish do not undergo isotype class-switching (22). Up until recently it was thought that bony fish were devoid of specialized mucosal immunoglobulins. Breaking this paradigm, we identified teleost IgT as the most ancient antibody class specialized in mucosal immunity (23). We found that IgT is expressed by a unique subset of IgT+ B cells that do not express IgD or IgM, and analogously to IgA, IgT exists in the serum as a monomer and in a polymeric form in mucosal secretions (23). Importantly, we determined that in contrast to sIgM or sIgD, sIgT was highly induced at mucosal surfaces by pathogens (23-25). Critically, we also showed that sIgT is the predominant Ig isotype coating a large portion of the fish microbiota (23-25).
To interrogate whether early sIgs emerged primarily to control pathogens or to establish homeostatic relationships with the microbiota, here we developed a novel teleost fish model in which we were able to selectively deplete IgT in adult animals. In addition to representing the only non-mammalian model system devoid of its main sIg, our IgT-depleted fish also embody the only vertebrate model in which a sIg can be temporarily depleted during adulthood, rather than permanently from birth (i.e., all existing mammalian models lacking sIgA). The effects of IgT depletion were analyzed on the gill mucosa, a bona fide mucosal surface where we have previously demonstrated the local induction of pathogen-specific IgT responses (24) that is also known to harbor a rich and diverse microbiota (26). Overall, our findings demonstrate that sIgT is required for the control of mucosal pathogens as well as for the preservation of microbiota homeostasis. Thus, our results indicate that pathogen and microbiota control at mucosal sites are guided by primordially conserved principles that emerged early in vertebrate evolution, and that are inherent to both primitive and modern sIgs.
Results
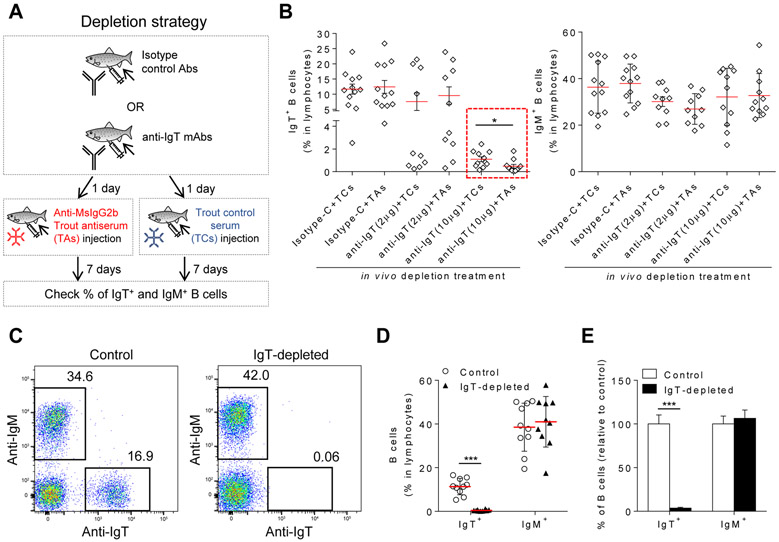
Strategy to generate IgT-depleted fish
We previously generated an anti-trout IgT monoclonal antibody (mAb) (23) (a mouse IgG2b mAb) that was exploited here to deplete IgT+ B cells in vivo. The anti-trout IgT mAb was used in conjunction with an anti-mouse IgG2b trout antiserum (TAs) generated by immunizing fish with purified mouse IgG2b (fig. S1A, upper panel). The specificity of TAs was assessed by ELISA (fig. S1B) and Western blotting (fig. S1C, lane 3). Moreover, flow cytometric analysis using mouse anti-trout IgM mAb revealed that the TAs specifically recognized the mouse anti-trout IgT mAb on IgT+ B cells (fig. S1, D and E), but trout control serum (TCs) did not (fig. S1F). We then tested the capacity of the anti-IgT mAb to deplete IgT+ B cells in vivo, in combination with the TAs or TCs (Fig. 1A). At a dose of 10 μg/fish, the anti-IgT mAb in combination with the TCs or the TAs, depleted 90.6±5.8% and 96.1±4.1% of blood IgT+ B cells respectively, indicating that addition of TAs had a minor but significant positive effect when compared to the addition of TCs in the overall depletion efficiency (Fig. 1B, left panel, red dotted box). In contrast, IgT+ B cells were not depleted in fish treated with 10 μg/fish of isotype IgG2b control antibody (Isotype-C) either in combination with TCs or TAs (Fig. 1B, left panel). Importantly, none of the treatments affected the % of IgM+ B cells (Fig. 1B, right panel), supporting the specificity of IgT+ B cell depletion. For the in vivo studies performed thereafter, we used a dose of 25 μg/fish of anti-IgT mAb or its respective isotype control, followed by injection 24 hrs later with 30 μl of a pooled TAs. Using these conditions, the % of IgT+ B cell depletion achieved in blood was consistently over 95%, while the % of IgM+ B cells was not altered (Fig. 1, C to E). In conclusion, we developed a strategy to selectively and consistently deplete IgT+ B cells that did not alter the % of IgM+ B cells.
Fig. 1. Development of a trout IgT+ B cell depletion model.
(A) Scheme of the strategy used to deplete trout IgT+ B cells. Fish were injected with either isotype control Abs (mouse IgG2b) or with anti-trout IgT mAbs. A day after, half of the fish from both groups were injected with anti-mouse (Ms) IgG2b trout antiserum (TAs) and the other half with trout control serum (TCs). Seven days later, the % of IgT+ and IgM+ B cells from blood leukocytes of all fish was analyzed by flow cytometry. (B) Percentage of IgT+ (left panel) or IgM+ B cells (right panel) in blood leukocytes from trout injected with isotype control (Isotype-C) antibodies (10 μg/fish) or with mouse anti-trout IgT mAbs (2 μg/fish or 10 μg/fish). Isotype-C- and anti-trout IgT-treated groups were thereafter injected with TAs or TCs (n = 10-12 fish per group) as depicted in (A). The groups injected with anti-IgT mAb (10 μg) are shown in red dotted box. (C) Flow cytometry of blood leukocytes from control (left) and IgT+ B cell depleted (right) fish. Numbers adjacent to outlined areas indicate percentage of IgM+ cells (top left) or IgT+ cells (bottom right) in the lymphocyte population. (D) The percentage of IgT+ or IgM+ B cells in the lymphocyte population of control or IgT-depleted fish (n = 10 fish per group). (E) The % of IgT+ and IgM+ B cells in the IgT-depleted group relative to that of control fish (mean ± s.e.m.; n = 10 fish). Fish in (C to E) were injected with either 25 μg of anti-trout IgT mAb (IgT-depleted) or 25 μg of the isotype control antibody (control) followed by injection with TAs. Each symbol (B and D) represents an individual fish; small horizontal red lines (B and D) indicate the mean (± s.e.m.). Data are pooled from two independent experiments (B) or representative of two independent experiments (C to E). Statistical analysis was performed by unpaired Student’s t-test. *P < 0.05, ***P < 0.001.
IgT-B cell and IgT depletion persisted for several weeks in all analyzed organs and fluids respectively
To evaluate the degree and duration of IgT+ B cell depletion in both systemic and mucosal lymphoid organs, as well as its effect on serum IgT and gill mucus sIgT levels, we performed a time course study. When compared to control levels, IgT+ B cells were depleted to a high degree (87.7-97.6%) in all tested tissues from day 1 up until day 28 post-depletion treatment (Fig. 2, A to C, left panels). Thereafter, IgT+ B cell numbers started to recover and by day 56 post-depletion treatment they were similar to levels in control fish. For all three tissues tested, the % of IgM+ B cells remained unchanged throughout the treatment, indicating a lack of a compensatory response by IgM+ B cells (Fig. 2, A to C, right panels). The absence of IgT+ B cells in depleted but not control animals was confirmed by immunofluorescence microscopy studies (Fig. 2, D to F). While the depletion of IgT+ B cells occurred quickly (1-3 days), the % of IgT depletion from serum (Fig. 3A) and gill mucus (Fig. 3D) did not become significant until day 14 post-depletion treatment (48.8-72.6% depletion compared to controls). The % of IgT depletion reached its peak 21-28 days post-depletion treatment (78.2-92.3%), with IgT levels being completely restored by day 56. In contrast to IgT, serum and gill mucus IgM and IgD levels remained unchanged at all times post-depletion treatment, confirming a lack of IgM and IgD compensatory responses (Fig. 3, B, C, E and F). Importantly, no mortalities were observed throughout the 56-day period in either fish group (fig. S2). In conclusion, the IgT+ B cell depletion treatment induced a near complete depletion of IgT+ B cells and IgT immunoglobulin in all lymphoid tissues and fluids analyzed respectively, and did not induce measurable compensatory responses from other immunoglobulin isotypes.
Fig. 2. Analysis of IgT+ and IgM+ B cells in blood, head kidney and gill upon IgT+ B cell depletion treatment.
(A to C) Percentage of IgT+ (left) and IgM+ B cells (right) within the lymphocyte population of blood (A), head kidney (B) and gill (C) leukocytes in control and IgT-depleted fish (n = 10-12 fish per group). Data are representative of at least three independent experiments (mean ± s.e.m.). Statistical analysis was performed by unpaired Student’s t-test. **P < 0.01, ***P < 0.001. (D to F) Immunofluorescence microscopy of trout blood leukocytes (D), head kidney tissue (E) and gill tissue (F), stained for IgT (green) and IgM (red), from control (left) and IgT-depleted (right) fish. Nuclei are stained with DAPI (blue). Isotype-matched control antibody staining in fig. S2. Scale bar, 50 μm. Data are representative of at least three independent experiments.
Fig. 3. IgT, IgM and IgD concentration in serum and gill mucus upon IgT+ B cell depletion treatment.
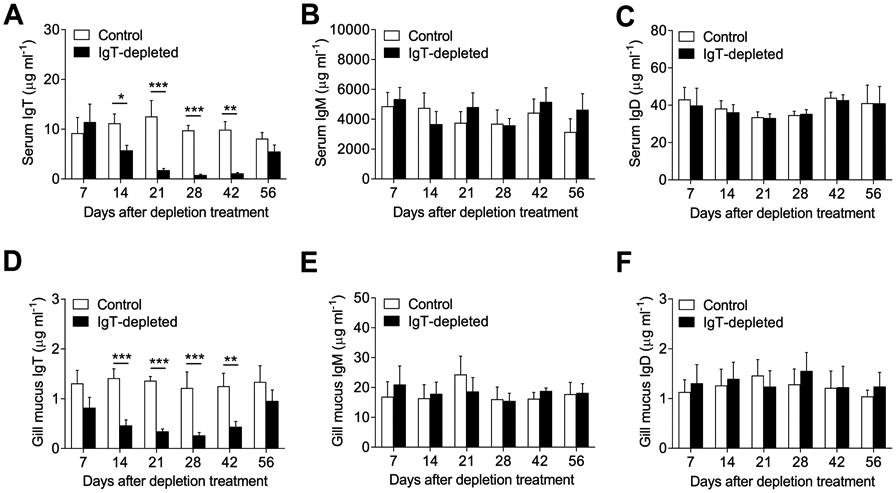
(A to C) Concentration of IgT (A), IgM (B) and IgD (C) in serum of control and IgT-depleted fish. (D to F) Concentration of sIgT (D), sIgM (E) and sIgD (F) in gill mucus of control and IgT-depleted fish. n = 10-12 fish per group. Data are representative of at least three independent experiments (mean ± s.e.m.). Statistical analysis was performed by unpaired Student’s t-test. *P < 0.05, **P < 0.01 and ***P < 0.001.
IgT depletion significantly increased the susceptibility of fish to Ich infection
We have previously shown that trout infected with the mucosal parasite Ichthyophthirius multifiliis (Ich) generate strong local parasite-specific sIgT responses in the gill mucosa, while parasite-specific IgM responses are primarily detected in the serum (24). To determine if sIgT is critical for protecting the gill epithelium against this parasite, we first generated immune fish (i.e., fish exposed to three successive rounds of Ich infection [Fig. 4A]). Immune fish were then divided into two groups; one group underwent IgT-depletion treatment (as shown in Fig. 1A), while the other (non-depleted group) was injected instead with isotype control antibodies and TAs (Fig. 4A). After 14 days, both groups were re-exposed to the parasite and thereafter, Ig responses, pathogen load, and fish mortalities were recorded (Fig. 4A). While non-depleted immune fish contained significant parasite-specific sIgT titers in the gill mucus, as expected (Fig. 4B), the IgT-depleted immune group did not (Fig. 4C). In contrast, IgT-depletion did not have any effect on the low parasite-specific IgM titers in the mucus (Fig. 4, B and C) or the high parasite-specific IgM titers in the serum (fig. S3, A and B). Critically, the parasite load increased dramatically in the IgT-depleted immune group when compared to that of non-depleted immune fish (Fig. 4, D and E), which was consistent with the higher expression of the Ich 18S rRNA gene in the gills of IgT-depleted immune fish (Fig. 4F). In addition, upon challenge with Ich, the IgT-depleted immune group had a significantly higher cumulative mortality when compared to that of non-depleted immune fish (Fig. 4G). Overall, these results indicate that sIgT is critical for parasite control at the gill mucosa.
Fig. 4. IgT depletion significantly increases pathogen load and fish mortalities upon Ich infection.
(A) Scheme of the experimental strategy. Immune fish (fish which survived monthly infections by bath with ~1000 theronts per fish, during a three month period) were injected with either isotype control Abs (non-depleted immune fish) or anti-IgT mAb (IgT-depleted immune fish). A day after, fish were injected with TAs. At 14 days post-TAs injection, the non-depleted and IgT-depleted groups were infected by bath with Ich (~1000 theronts per fish), and at 21, 10 and 30 days post-infection, different groups of fish were analyzed for Ig responses, pathogen load and mortalities, respectively. (B and C) Specific Ig responses against the parasite were measured by pull-down assays in which we assessed the IgT-, IgM- and IgD-specific binding to Ich in dilutions of gill mucus from non-depleted (B) and IgT-depleted fish (C). Results are presented as relative values to those of uninfected naive fish (n = 10-12 fish per group). (D) Representative stereomicroscope images of gill arches infected with Ich from non-depleted (left) and IgT-depleted fish (right). White arrowheads point to Ich trophonts. (E) Parasites numbers on gills from non-depleted and IgT-depleted fish upon infection with Ich (n = 17 fish per group). Each symbol represents an individual fish; small horizontal red lines indicate the mean. (F) Real-time PCR analysis of Ich 18S rRNA gene in gills from non-depleted and IgT-depleted fish. DNA abundance was normalized to that of non-depleted fish, which is set as 1 (n = 10 fish per group). (G) Percentage survival of non-depleted and IgT-depleted fish infected with Ich. Data (B to F) are representative of three independent experiments (mean ± s.e.m.) or two independent experiments (G). Statistical analysis was performed by unpaired Student’s t-test (B, C, E and F), or log-rank (Mantel-Cox) test (G). *P < 0.05, **P < 0.01 and ***P < 0.001.
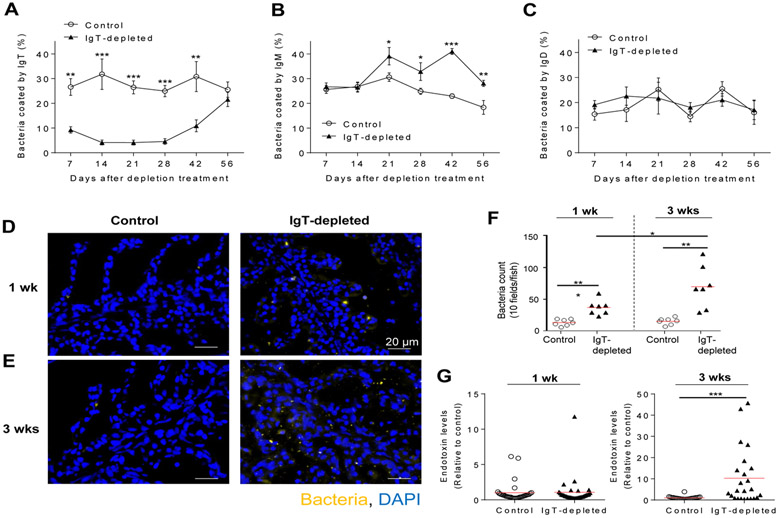
Decreased sIgT-coating of microbiota correlates with bacterial translocation into the gill tissue
Mammalian sIgA coats and homeostatically regulates microbiota at mucosal surfaces (8). We have previously shown that sIgT is the predominant sIg coating the gill microbiota (24). Here, upon IgT depletion, we observed a remarkable reduction (65.3%) in the percentage of sIgT-coated microbiota by day 7 when compared to control fish (Fig. 5A). This decrease in sIgT-coating was maximal between days 14 and 28 post-depletion treatment when coating was reduced to 81.7-86.9% of control levels (Fig. 5A). After day 28, the percentage of microbiota coated by sIgT started to recover, and were fully restored by day 56 (Fig. 5A), thus mirroring the presence of sIgT in gill mucus of IgT-depleted fish (Fig. 3D). Interestingly, the percentage of sIgM-coated bacteria increased significantly from day 21 post-depletion when compared to that of control fish, and it remained significantly elevated throughout the rest of the sampling period (Fig. 5B). In contrast, sIgD coating levels in the control group were not different from those found in IgT-depleted fish (Fig. 5C). Notably, in the absence of sIgT-coating, significant levels of bacteria translocated from the mucus layer across the gill epithelium, especially by week 3 post-depletion (Fig. 5, D to F, fig. S4). Moreover, translocated bacteria entered the systemic circulation, as evidenced by significant increases in LPS levels in the serum of a substantial number of IgT-depleted fish at week 3 post-depletion treatment (Fig. 5G, right). These results demonstrate a critical role of sIgT in containing microbiota within the mucosal areas and in avoiding its systemic dissemination.
Fig. 5. IgT-depletion leads to a major decrease of sIgT coating of the gill microbiota and to their translocation across the gill epithelium.
(A to C) Percentage of gill bacteria coated with sIgT (A), sIgM (B) or sIgD (C) at days 7, 14, 21, 28, 42 and 56 after depletion treatment (n = 10-12 fish per group). Data are representative of at least two independent experiments (mean ± s.e.m.). (D and E) Detection of bacteria by fluorescence in situ hybridization in gill cryosections from control (left) and IgT-depleted fish (right) at 1 week (D) and 3 weeks (E) post depletion treatment. Eubacteria were detected with Cy5-EUB338 oligoprobe (yellow). Nuclei were stained with DAPI (blue). Scale bar, 20 μm. (F) Translocated bacteria were quantified in gill cryosections of control and IgT-depleted fish (10 microscopy fields per fish, n = 7 per group). (G) Endotoxin levels were measured by LAL chromogenic endpoint assay in serum from control and IgT-depleted fish (n=25 per group) at 1 week (left) and 3 weeks (right) post-depletion treatment. Endotoxin levels in IgT-depleted fish were normalized to those in control fish, which were set as 1. Each symbol (F and G) represents an individual fish; small horizontal red lines (F and G) indicate the mean. Data are representative of two independent experiments (D to E) or pooled from two independent experiments (F and G). Statistical analysis was performed by unpaired Student’s t-test. *P < 0.05 and ***P < 0.001.
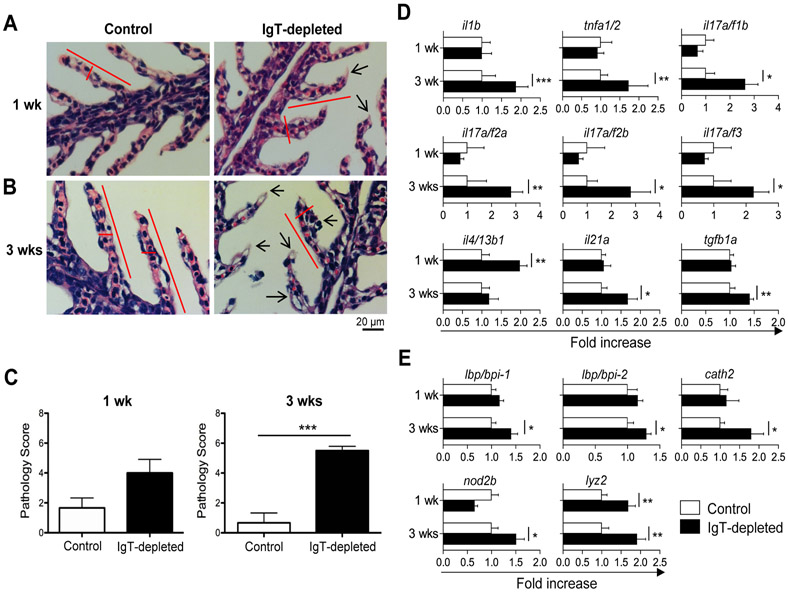
The gills of IgT-depleted fish show signs of tissue damage and inflammation
We next assessed if the translocation of microbiota into the gill tissue in IgT-depleted fish promoted tissue damage and/or inflammation. While moderate changes in gill histology were observed at 1 week post-depletion, significant signs of tissue damage were detected at 3 weeks, as evidenced by wider and shorter secondary lamellae with edema and signs of inflammation (Fig. 6, A to C). In addition, the expression of several key pro-inflammatory cytokines was increased, mostly 3 weeks post-depletion (Fig. 6D). Specifically, we detected a 1.67-2.81 fold increase relative to control fish for il-1β, tnfα ½, il17 a/f1b, il17 a/f2a, il-17 a/f2b, il-17 a/f3, il-17a/f1b and il-21a transcripts. The only pro-inflammatory gene upregulated at week 1 post-depletion was il4/13b1 (i.e., 1.98 fold increase), while the only mildly upregulated anti-inflammatory gene (1.41 fold increase) was tgfβ1a at 21 days post-depletion. Translocation and dysbiosis of microbiota at mammalian mucosal surfaces have often been associated with increases in antimicrobial peptide (AMP) gene expression (27). Similarly, at 3 weeks post-depletion we observed moderate increases (1.29-1.91 fold relative to control fish) in transcript levels of two LPS binding proteins (lbp/bpi1 and lbp/bpi2), the AMP cathelicidin (cath2), the antimicrobial molecule lysozyme (lyz), and nucleotide binding oligomerization domain 2 (nod2b), an intracellular bacterial pattern recognition receptor. Other cytokines and AMP genes remained unchanged throughout the depletion period (fig. S5). To evaluate whether the observed impact on tissue damage and homeostasis was due to a potential impairment in TLR signaling in IgT-depleted fish, we injected control and IgT-depleted animals with three agonists of TLR signaling, including lipoteichoic acid (LTA), flagellin, and resiquimod (R848). We observed no differences in the mRNA expression levels of il-1β, il-6, il-8 and tnfα ½ in gills between control and IgT-depleted fish upon treatment with any of three TLR agonists (fig. S6), thus suggesting that TLR responses remain intact in IgT-depleted trout. Together, our results underscore the importance of sIgT in the maintenance of mucosal barrier integrity, and suggest that in the absence of sIgT the loss of tissue integrity is not related to a defect in TLR signaling.
Fig. 6. IgT-depletion leads to damage and inflammation of the gill tissue.
(A and B) Hematoxylin-eosin staining of gill paraffin sections in control (left) and IgT-depleted (right) fish at 1 week (A) and 3 weeks (B) post-depletion treatment. (C) Pathology score of gill tissue in control and IgT-depleted fish at 1 week (left) and 3 weeks (right) post-depletion treatment (mean ± s.e.m.; n = 6 fish per group). (D and E) Real-time PCR analysis of cytokine genes (D) and antimicrobial peptide genes (E) in gill tissue from control and IgT-depleted fish. Expression levels in IgT-depleted fish were normalized to those in control fish, which were set as 1 (mean ± s.e.m.; n = 8 fish per group). Data are representative of two independent experiments. Statistical analysis was performed by unpaired Student’s t-test. *P < 0.05, **P < 0.01 and ***P < 0.001.
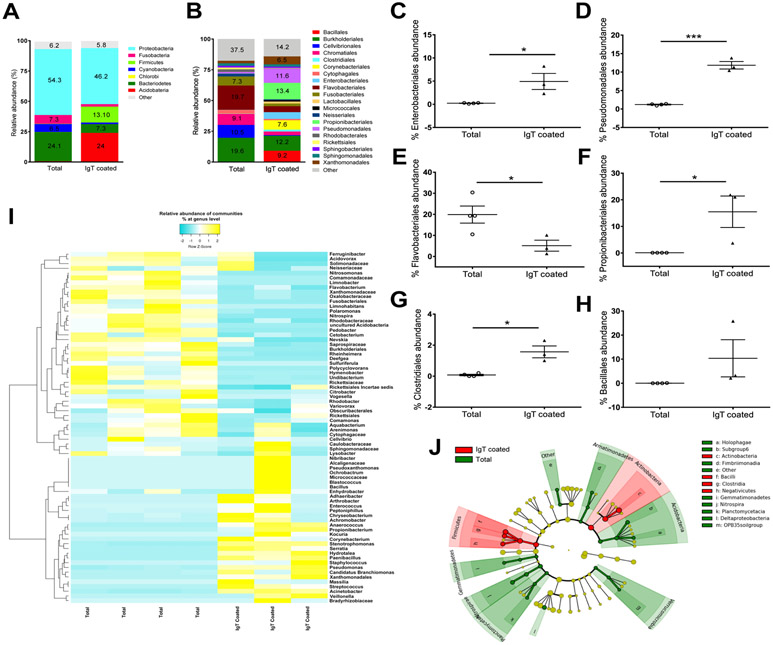
sIgT targets taxonomically distinct subsets of bacteria
In order to identify which bacterial taxa are coated by mammalian sIgA, recent substantial inroads have been made using the powerful IgA-seq technique that enables the sorting and taxa identification of sIgA-coated bacteria (16). As a first step to understand the involvement of sIgT in microbiota homeostasis upon sIgT depletion, here we adapted the IgA-seq technique to identify the trout gill bacteria taxa coated by sIgT (IgT-seq). To this end, we sorted sIgT+ microbiota from healthy fish and subjected it to 16S rDNA Illumina sequencing. IgT-seq revealed that sIgT coated a well-defined subset of bacteria (~300 different Operational Taxonomic Units [OTUs]) which accounted for approximately 25% of all the microbial diversity present in the gills (fig. S7A and table S1). The sIgT-targeted bacterial community was composed mostly of anaerobes and facultative anaerobes and was highly enriched in Actinobacteria and Firmicutes (24% and 13%, respectively) compared to the total bacterial community (0.62% and 0.23%, respectively) (Fig. 7A and fig. S7B). Bacteroidetes, Cyanobacteria and Fusobacteria, in turn, were more abundant in the total bacterial community (24.1%, 6.4% and 7.3%, respectively) compared to the sIgT-coated bacterial community (7.3%, 1.3% and 2.2%, respectively) (Fig. 7A and fig. S7B). Proteobacteria, the most common phylum in the trout gill microbiome (26), was the predominant phylum in both groups, accounting for 54.3% of the diversity in the total bacterial community and 46.2% in the sIgT-coated community (Fig. 7A and fig. S7B). At the order level, sIgT coated potentially pathogenic bacteria (Fig. 7B and table S2). Examples included members of the orders Enterobacteriales (5.5% sIgT-coated vs. 0.12% total bacteria) and Pseudomonadales (11.5% sIgT-coated vs. 1.3% total bacteria) (Fig. 7, B to D). In contrast, the order Flavobacteriales, which contains several well-known fish pathogens, was significantly enriched in the total bacteria (19.7%) compared to the sIgT-coated community (4.8%) (Fig. 7, B and E). The sIgT-coated microbial community was also enriched in potentially beneficial taxa known to produce short-chain fatty acids (SCFAs), including Propionibacteriales (13.36% sIgT-coated vs. 0.1% total bacteria) and Clostridiales (1.4% sIgT-coated vs. 0.14% total bacteria) (Fig. 7, B, F and G). There was also an enrichment trend in other SCFA-producing bacteria, including members of the order Bacillales (9.2% sIgT-coated vs. 0.03% total bacteria) (Fig. 7, B and H) and the order Lactobacillales (1.6% sIgT coated vs. <0.1% total bacteria) (table S2), although these changes did not reach statistical significance. The top 75 differentially abundant OTUs further showed that subsets of bacteria that are preferentially targeted by sIgT in gills include both beneficial bacteria such as Propionibacterium sp., Corynebacterium sp. (28) and Paenibacillus sp. (29) as well as disease-driving taxa such as Candidatus Branchiomonas, Acinetobacter sp., Serratia sp., Veillonela sp., Streptococcus sp. and Stenotrophomonas sp. (30, 31) (Fig. 7I). Linear discriminant analysis (LDA) effect size (LEfSe) method showed that the sIgT-coated community is enriched in 16 taxonomic features including Firmicutes, Bacilli, Clostridiales, Lactobacillales, Enterobacteriales and Pseudomonadales (Fig. 7J and fig. S7C). These results indicate that sIgT coats a broad but well-defined range of bacteria with both beneficial and pathogenic characteristics.
Fig. 7. sIgT coats specific subset of gill microbiota with beneficial and pathogenic characteristics.
Total bacteria and sIgT-coated bacteria from the gill mucosa were FACS sorted and high-throughput 16S rDNA sequencing of the V1-V3 region was performed (IgT-seq). (A) Mean relative abundance at the phylum level of the total gill bacterial community (Total) and the sIgT-coated bacterial community (IgT Coated). (B) Mean relative abundance at the order level of the total gill bacterial community (Total) and the sIgT-coated bacterial community (IgT Coated). (C to H) Relative abundance (%) of Enterobacteriales (C), Pseudomonadales (D), Flavobacteriales (E), Proprionibacteriales (F), Clostridiales (G) and Bacillales (H) in the total and sIgT-coated gill bacterial community. Data were analyzed in Qiime 1.8. Differential abundances were determined by unpaired Student’s t-test. *P < 0.05, ***P < 0.001. (I) Heatmap of the top 75 OTUs with differential abundances in the total (n = 4 pools, 4 fish per pool) and sIgT-coated (n = 3 pools, 4 fish per pool) gill bacterial communities. (P < 0.05). The heatmap was generated using the online free tool Heatmapper (78) using average linkage as a clustering method followed by the Spearman rank correlation for distance measurement. (J) Cladogram representation of LEfSe analysis showing bacterial taxa that were significantly associated with the total bacterial community (green) or the sIgT-coated bacterial community (red) (P < 0.05). n=3-4 pools, 4 fish per pool. Results are representative of two independent experiments.
IgT-depletion induces a profound dysbiosis in the gill
We next asked if the observed translocation of bacteria in IgT-depleted fish (Fig. 5, D and E) reflected a defect in maintenance of microbiota homeostasis. 16S rDNA sequencing of gill tissue of control and IgT-depleted trout showed changes in the microbial community with kinetics that mirrored those of tissue damage and bacterial translocation. Although no significant changes in Shannon diversity index (a metric that weights the numbers of species by their relative evenness data) were detected 1 week after depletion (fig. S8A), the gill bacterial community composition of IgT-depleted animals already showed some minor differences compared to that of control fish by this time point. At the phylum level (fig. S8B), we observed a decrease in the overall abundance of Proteobacteria as a result of IgT depletion (65% in controls vs. 33.7% in IgT-depleted) as well as increases in Bacteroidetes abundance (8.4% in controls vs. 19.5% in IgT-depleted) suggesting that sIgT may be critical for Proteobacteria colonization at least in the initial stages of dysbiosis. A total of 7 OTUs had significantly different abundances in both experimental groups (table S3). For instance, we observed significant decreases in the abundance of Caulobacteriales (3.6% control vs. 0.2% IgT-depleted) and Sphingomonadales (14.2% control and 5% IgT-depleted) and moderate but not significant increases in Flavobacteriales (8.7% control and 16.1% IgT-depleted) (fig. S8C and table S4). Additionally, Flavobacterium sp. and Deefgea sp. abundance increased in IgT-depleted trout while it decreased in the family Moraxellaceae (fig. S8D). Abundance changes at the genus level resulting from week 1 post-depletion treatment can be found in table S5. LEfSe analysis and cladogram representation highlighted that 12 taxa were more abundant in the control group than in the IgT-depleted group including Comamonadaceae, Caulobacteriales, Actinomycales and Corynebacteriales (fig. S8, E and F). Conversely these analyses also identified three taxa that were more abundant in the IgT-depleted group than in control fish, including Pseudomonadaceae, Cyanobacteria and Betaproteobacteria (fig. S8, E and F). One week post “treatment” (i.e. IgT depletion) was however not a significant predictor of gill microbial community composition by analysis of similarities (ANOSIM) with P=0.763. These results indicated that IgT depletion has moderate effects on the microbial community composition of the gill early on during the depletion treatment likely due to the incomplete depletion of mucus sIgT at that time point.
At 3 weeks post-depletion, the Shannon diversity index of the IgT-depleted fish gill microbiome was significantly lower (P=0.026) compared to that of the control group (Fig. 8A) indicating that fewer OTUs contributed to a higher proportion of the overall bacterial abundance in the community in the IgT-depleted group compared to controls. At the phylum level, the gill microbial community composition of control samples was dominated by Fusobacteria (29.6%) followed by Proteobacteria (21%) and Bacteroidetes (16%) (Fig. 8B). In response to IgT depletion, the community shifted and became dominated by Bacteroidetes (34%), followed by Proteobacteria (26%) and Fusobacteria (18.5%) (Fig. 8B). Importantly, the abundance of Firmicutes (shown to be enriched in the sIgT-coated fraction) dropped from 3.5% in controls to 0.75% in IgT-depleted animals (Fig. 8B). As a result of this shift, the Bacteroidetes to Firmicutes ratio increased ~10-fold in IgT-depleted animals. At the order level, IgT depletion resulted in significant reductions in the abundance of Actinomycetales (12.3% control vs. 2.5% IgT-depleted) and beneficial SCFA producers such as Bacillales (2.8% control vs. 0.16% IgT-depleted) (Fig. 8C and table S6). On the other hand, other orders were found at higher abundances. As examples, significant expansions of the orders Flavobacteriales (7.9% control vs. 35.3% IgT-depleted) and Neisseriales (1.1% control vs. 4.3% IgT-depleted) were observed in IgT-depleted gills (Fig. 8, C and D, table S6). Overall, a total of 39 OTUs showed significantly different abundances between both experimental groups indicating a global state of dysbiosis in the gills 3 weeks after IgT depletion (Fig. 8E and table S7) (ANOSIM P=0.028). As examples of abundance changes at the genus level, significant decreases in beneficial taxa such as Propionibacterium sp. (7.6% control vs. 1% IgT-depleted) and Pseudomonas sp. (1.3% control vs. 0.3% IgT-depleted) were observed (Fig. 8, E and F, and table S8). LEfSe analysis determined the features more likely to define control and IgT-depleted gill microbial communities, and highlighted that in the absence of sIgT, Flavobacteriales and Neisseriales expand at the expense of losses in Firmicutes, Pseudomonales, Bacillales, Pasterurellales and Propionibacteriaceae (Fig. 8, G and H). Combined, these data indicate that sIgT depletion has profound and complex effects on the microbial community composition in the gill of rainbow trout. Critically, sIgT deficiency triggered a state of dysbiosis characterized by loss of potential beneficial SCFA-producing bacteria and expansion of pathobionts, underscoring the importance of sIgT in maintaining colonization of beneficial microbes and controlling disease-driving bacteria.
Fig. 8. IgT depletion results in significant gill dysbiosis 3 weeks post-depletion.
The microbial community composition of control and IgT-depleted gills (n = 5-6 per group) was determined by high-throughput 16S rDNA sequencing 3 weeks after IgT depletion treatment. (A) Shannon-diversity index (mean ± s.e.m.) of the control and IgT-depleted gill microbial communities. (B) Mean relative abundance at the phylum level of the control and IgT-depleted gill bacterial community. (C) Mean relative abundance at the order level of the control and IgT-depleted gill bacterial community. (D and F) Relative abundance (%) of Flavobacteriales (D, top) and Neisseriales (D, bottom), Propionibacterium sp. (F, top) and Pseudomonas sp. (F, bottom) in the control and IgT-depleted gill bacterial community at 3 weeks post-depletion. Data were analyzed in Qiime 1.8. Differential abundances were determined by unpaired Student’s t-test. *P < 0.05, **P < 0.01. (E) Heatmap of the top 25 OTUs with significantly different abundance in control and IgT-depleted gills 3 weeks post-depletion. The full list of the 39 differentially abundant OTUs can be found in table S7. Each column represents one individual fish. (G) Bar chart of the log-transformed LDA score of bacterial taxa found to be significantly associated with control and IgT-depleted 3 week-post depletion showing the presence of 15 taxonomical features with higher abundance in controls (red) and 5 taxonomical features with greater abundance in IgT-depleted gills (green) by LEfSe (P < 0.05). (H) Cladogram representation of LDA analysis in (G) showing the phylogenetic relationships among the bacterial taxa found to be significantly associated with control (red) or IgT-depleted gills (green) by LEfSe. Results are representative of two independent experiments.
IgT recovery is sufficient to restore tissue and microbiome homeostasis
In our model, gill IgT+ B cells recover almost completely by 8 weeks post depletion treatment (Fig. 2). This enabled us to ask whether sIgT recovery is sufficient to restore tissue and microbiome homeostasis in trout gills. To this end, we sampled fish at 13 weeks post-depletion treatment to ensure that IgT recovery was complete. At that time point, the % of IgT+ B cells had recovered to the levels of control fish both in gills and head kidney (fig. S9, A and B) and this recovery paralleled that of IgT protein levels both in serum and gill mucus (fig. S9, C and F). Meanwhile, the % of IgM+ B cells remained unchanged in these tissues (fig. S9, A and B). Importantly, the levels of microbiota coated by sIgT were also fully restored (fig. S9I). Interestingly, the increased levels of sIgM-coated bacteria observed in IgT-depleted fish from weeks 3-8 post-depletion (Fig. 5B) had faded away and were comparable to levels observed in control fish (fig. S9J). As previously observed for week 8 post-depletion (Fig. 5C), levels of microbiota coated by sIgD remained unchanged (fig. S9K). Restoration of IgT levels after 13 weeks post-depletion treatment was sufficient to revert bacterial translocation levels to those of control fish (fig. S9L), as reflected also by the absence of systemic LPS in the IgT-recovered fish (fig. S8M). Importantly, absence of translocated bacteria correlated also with a complete reversal of tissue damage in the gills (fig. S9N). These data indicate that the deleterious effects in tissue homeostasis induced by the lack of sIgT are reversed upon the recovery of physiological sIgT levels in the IgT-depleted group after 13 weeks of the IgT depletion treatment.
With regards to the microbiome composition, by week 13 post-depletion there were no significant differences in the Shannon diversity index of the gill microbial community of control and IgT-depleted fish (P=0.235) (fig. S10A). The lack of dysbiosis was reflected both at the phylum level where no significant differences were observed between both groups (fig. S10B and table S9), as well as at the order level where only three OTUs were significantly different between both groups, albeit with marginal significance (p-value=0.048), including Staphylococcus sp., Cetabacterium sp. and an unassigned OTU (fig. S10C and tables S10 and S11). Thus, by week 13 post-depletion the gill microbial community composition of IgT-depleted fish was not significantly different from that of control animals (ANOSIM P=0.584). Combined, the microbiome results indicate that restoring sIgT levels is sufficient to reestablish the balance in the microbial ecosystem of the gills.
Discussion
Mucosal Igs (sIgs) emerged in both primitive and modern vertebrates in response to common selective pressures and physiological demands imposed by pathogens and commensals at mucosal surfaces. However, the function of sIgs in vertebrates in limiting and regulating microbes at mucosal sites is not well understood (1, 8, 32). In that regard, mammalian sIgA appears to have antagonistic roles in the control of mucosal microbes as sIgA is important for mucosal pathogen clearance (1, 3-5), while at the same time, it is required for microbiota colonization and homeostasis (6-8). Because the selective forces that have shaped fish and mammalian sIgs are similar, we reasoned that the study of a primitive sIg could shed light on whether sIgs arose to fight mucosal pathogens and/or to enable the establishment of a healthy microbiota. This fascinating question was addressed here by developing a new teleost fish model in which its mucosal sIg was selectively depleted during adulthood.
To determine the contributions of sIgT in pathogen control and microbiota homeostasis, we designed a strategy to selectively deplete IgT+ B cells in rainbow trout, a teleost species. This strategy proved highly effective in depleting most of the IgT and IgT+ B cells from all tested fluids and lymphoid organs respectively, for over a 4-week period. It is worth pointing out that extended B cell depletions spanning several weeks have also been reported in mice and humans with the use of a single antibody treatment (33, 34). Naive IgA-deficient mice, and patients with selective sIgA deficiency (sIgAdef) show compensatory increased levels of total IgM and IgG in serum and mucus (9, 35, 36). In contrast, here we show that IgM or IgD in serum and mucus did not increase upon IgT depletion, thus indicating a lack of compensatory responses by other fish Ig isotypes. In conclusion, our IgT-depletion model represents the only vertebrate in vivo model that allows for the temporal and selective depletion of a specialized mucosal immunoglobulin during adulthood. In contrast, all existing IgA-deficient mammalian models lack sIgA from birth (9, 35). This fundamental difference between our fish and the current mammalian models allows us to elucidate the function of a sIg isotype in an animal where both a wild-type mucosal immune system and a homeostatic microbiota have co-existed and matured together prior to the sIg depletion.
Our previous findings in fish have shown a marked compartmentalization of pathogen-specific IgT and IgM responses in mucosal and systemic areas respectively, suggesting an important role for sIgT in the control of mucosal pathogens (23-25). Supporting this hypothesis, here we show that depletion of IgT dramatically increases the susceptibility of fish to a mucosal parasite. Interestingly, we did not observe a compensatory IgM response in IgT-depleted fish. In contrast, IgA-deficient mice have been reported to develop stronger IgM titers than do their littermate controls in several infection models (35, 37-39). Similarly, respiratory infections in sIgAdef patients are more recurrent when these individuals lack sIgM in their lungs (36). Moreover, sIgAdef patients with high levels of IgM in saliva have shown a lower infection incidence (40). As mammalian IgA arises from class switch recombination, it is tempting to propose that the potential compensatory response of IgM in mammals is derived from IgM+ B cells with the same antibody specificity, but which have not yet class switched. In that regard, we have previously reported that the germline repertoire between fish IgT and IgM is very different (41), and class-switch recombination does not exist in these species (22), thus providing a potential explanation for the inability of IgM to compensate for pathogen-specific IgT responses in fish.
The symbiosis between microbiota and the mucosal surfaces of all metazoans is one of the most ancient and successful relationships found in nature. Our previous work has shown that similar to sIgA in mammals, sIgT is the prevalent antibody isotype coating the microbiota of fish (23-25). As expected, IgT depletion led to a dramatic decrease in the sIgT coating of gill microbiota. Interestingly, we observed an increase in the % of IgM-coated microbiota from week 3 through week 6 post-depletion treatment, which coincides with the time points at which sIgT concentration was at its lowest in the gill mucus of IgT-depleted fish. This increase in sIgM-coated microbiota could be explained by several processes. First, the possibility exists for an sIgM compensatory mechanism similar to what has recently been reported in human sIgAdef patients, in which sIgM partially compensates for the lack of sIgA by coating microbiota in a much less targeted and less specific manner (11, 15). Accordingly, in the absence of sIgT, compensatory sIgM would then coat microbiota subsets that would otherwise be coated by sIgT. Alternatively, or concurrently, it is possible that some of the bacterial taxa that expand during dysbiosis in IgT-depleted fish are those found initially coated by sIgM in control individuals, which would increase in abundance upon IgT depletion. Further work is warranted to analyze all the aforementioned possibilities. In addition to most gill-associated bacteria losing their sIgT coating by 3 weeks post-depletion, large numbers of bacteria translocated within the gill lamellae, and significant amounts of LPS were found in blood, thus indicating that the potential sIgM-mediated compensatory mechanism postulated above is not sufficient to overcome the deleterious effects of dysbiosis. Microbiota translocation in IgT-depleted fish suggests that sIgT is required for microbiota colonization, anchoring and containment of sIgT-coated microbiota within the mucosal layer of the gills, as suggested also for sIgA at mammalian mucosal surfaces (6, 8). Alternatively, as sIgA has also been shown to regulate microbiota composition by promoting microbiota symbiosis (7), it is likely that dysbiosis induced in the absence of sIgT may allow the outgrowth of microbiota that can then translocate across the mucosal epithelium and reach systemic sites, as has been observed also in microbiota of murine models devoid of sIgA (14, 42-44). Intriguingly, sIgAdef patients have no increase in LPS serum levels, suggesting the involvement of IgA-independent mechanisms implicated in the containment of uncoated or dysbiotic bacteria in humans (11). It is conceivable, however, that translocation of non-Gram negative bacteria occurs in these patients, a possibility that warrants further investigation. In our teleost model, the significant bacterial translocation observed at week 3 post-depletion correlated with the presence of significant tissue damage and upregulation of critical pro-inflammatory cytokines and AMPs. The latter strongly suggests that the invasion of microbiota into the gill tissue results in inflammatory and anti-microbial responses in the affected area, similar to what has also been described in murine models lacking sIgA (11 , 43). Interestingly, our studies with TLR agonists imply that the observed tissue damage in sIgT-depleted fish was not due to a defect in TLR signaling, and thus, further studies are warranted to understand the mechanisms involved in the loss of tissue integrity in the absence of sIgT.
While a large percentage of microbiota are coated by sIgA in mammals, the functional significance of such coating is poorly understood (8, 16). Recently the newly developed IgA-seq technique has enabled the identification of IgA-coated taxa (16). Although these studies are in their infancy and have only been performed in mammals, they have revealed that despite of the polyreactive nature of sIgA, the sIgA-coated microbiota fraction contains a large but well-defined range of bacteria with distinct taxonomies, while entire taxa are devoid of IgA coating (11, 16, 45). Here we developed IgT-seq for our fish model, and found that similar to what has been described for sIgA, sIgT targets a broad but distinct repertoire of bacterial taxa belonging to seven different phyla which represent ~25% of all OTUs found in the gills. Comparable to mammalian sIgA (8, 13), sIgT preferentially targeted Proteobacteria (the most abundant phylum in fish external microbiomes (26)), followed by Actinobacteria and specific groups of Firmicutes. In addition, like sIgA (8, 16, 45, 46), sIgT preferentially coated a number of beneficial bacteria, including several SCFA-producing taxa such as the orders Lactobacillales and Clostridiales, which have anti-inflammatory properties (47, 48) and support antibody production in mice (49, 50). Our findings suggest that SCFA-producing taxa may play a conserved role across vertebrates in supporting tissue barrier integrity and sIg production. Further studies are warranted to explore this hypothesis. We also found that the sIgT-coated microbiota fraction was significantly enriched in several opportunistic or pathogenic bacteria, akin to the colitogenic bacteria and pathobionts with inflammatory properties that are highly coated by mammalian sIgA (8, 16). Overall, our IgT-Seq data support the notion that mucosal immunoglobulins evolved to target well-defined taxa of both beneficial and pathogenic bacteria that form part of a healthy vertebrate microbiome under homeostatic conditions.
The IgT-seq data demonstrating that sIgT coated a distinct set of microbial taxa led us to hypothesize a critical role for sIgT in the preservation of microbiota homeostasis. In support, we found that the gill microbiome shifted upon sIgT depletion in a time-dependent fashion that mirrored sIgT levels. The most profound shifts in the microbial community composition were detected by week 3 post-depletion, when mucus sIgT levels were negligible. The observed dysbiosis was also associated with significant tissue damage, inflammation and microbial translocation. The microbiome changes were characterized by significant abundance shifts in a large number of OTUs, decreased Shannon diversity, and a ~10-fold decrease in the Firmicutes to Bacteriodetes ratio in IgT-depleted animals. Interestingly, large decreases in the Firmicutes to Bacteroidetes ratios in mammals are also characteristic of dysbiotic states (51, 52). Similar to sIgAdef patients and murine IgA KO models (11, 15), sIgT deficiency triggered losses of typically beneficial resident taxa, including Bacillales (known to secrete large amounts of antimicrobial compounds (53)) and Propionibacteriaceae (known to produce SCFAs (54)) which in our IgT-Seq dataset were identified as preferentially coated by sIgT. In agreement with a very recent study in mice demonstrating the previously unrecognized ability of certain commensals to use sIgA for mucosal colonization (6), our current findings suggest that sIgT coating of bacteria likely mediates bacterial colonization under homeostatic conditions. Further studies are warranted to test this hypothesis. Importantly, IgT depletion also resulted in significant expansions of pathogenic taxa including the members of the orders Flavobacteriales and Neisseriales. Interestingly, these subsets were not preferentially coated by sIgT according to IgT-Seq, indicating that expansions of pathobionts upon IgT depletion occur via indirect mechanisms. For instance, the absence of sIgT may prevent certain microbiota taxa from colonizing or being contained in the mucosa, thus allowing space and resources for the outgrowth of non-sIgT coated microbiota (i.e., pathobionts). Similarly, expansions of pathobionts, including segmented filamentous bacteria (SFB) and Clostridia, occur in sIgA-deficient mammals (10, 12), although in general, these bacterial taxa appear to be coated by sIgA (16, 55-57). Overall our findings demonstrate the previously unrecognized requirement of a primitive mucosal immunoglobulin in the maintenance of microbiota homeostasis, and parallel those reported in mammals lacking sIgA which show also significant alterations in the composition of their microbiomes.
While we see important commonalities in the way bacterial communities shift between our IgT-depleted fish and the current mammalian models lacking sIgA, it is worth pointing out two significant differences. First, Shannon diversity doesn’t change significantly in sIgAdef patients (11, 15), while we find a significant decrease of that index in sIgT-depleted fish. Second, in mammalian models lacking sIgA, the pathobionts expanding are generally those found coated by sIgA in control individuals (11), whereas in IgT-depleted fish, the expanding pathobionts are generally found not coated with sIgT in control fish. These discrepancies likely arise from the fact that sIgA-deficient mammalian models lack sIgA from birth, which prevents the initial colonization of many taxa, whereas our fish model results in the selective and temporary depletion of sIgT during adulthood. Thus, in our fish model, microbiota diversity is optimal under homeostatic conditions (i.e., in the presence of sIgT), but when a perturbation is introduced (i.e., temporal depletion of sIgT), the existing communities significantly shift in their abundance as reflected by the decreased Shannon diversity. We propose that our model provides a more physiologically relevant scenario in which to test the involvement of sIg in host-microbial interactions, and thus, it may portray a more accurate picture of what may follow in conditions in which a mucosal Ig is temporarily reduced or depleted in a mucosal surface (e.g., reductions of IgA levels upon stress or antibiotic treatment) (58, 59). In this respect, our recovery experiments significantly contribute to our understanding of immune-mediated regulation of microbial community stability. Microbiota recovery following a perturbation depends on the type of perturbation (i.e. agent or condition, duration, intensity) as well as the stability and resilience of the dysbiotic microbial community (60, 61). Our study shows that restoring sIgT levels is sufficient to restitute a normal homeostatic gill microbial community in trout. This finding supports the notion that adaptive immunity promotes stability of microbiomes at mucosal barriers (62-64) and that this is a well-conserved mechanism across all vertebrate species that contain specialized mucosal immunoglobulins. In addition, previous studies have shown that the immune system reestablishes a healthy microbiome by suppressing harmful microbial species (62-64). Thus, the restitution of a healthy microbiota upon sIgT-depletion may be explained by several mechanisms. On the one hand, it is possible that increasing sIgT levels during the recovery period results in a density-dependent regulation of harmful bacteria that leads to a dampening of positive feedback loops in the community, as previously suggested (60). On the other hand, restoration of sIgT levels probably re-enables the colonization of sIgT-dependent microbiota as well as the control (through immune exclusion) of potential pathobionts whose growth and containment within luminal areas is controlled by sIgT coating. The aforementioned potential mechanisms responsible for restoration of microbiome composition and homeostasis by sIgT warrant further studies. It is worth pointing that recovery of microbiome homeostasis was accompanied by restoration of mucosal tissue integrity. Since recovery of sIgT levels in IgT-depleted fish led to a reversal in bacterial tissue translocation and systemic LPS, it is plausible to infer that the tissue damage and inflammation induced at the peak of the IgT-depletion period was due to the significant dysbiosis and translocation of microbiota. Thus, sIgT is critical to both maintaining microbiome and mucosal tissue homeostasis.
In conclusion, our data supports the notion that specialization of sIgs in protection of mucosal sites from pathogens and preservation of microbiota homeostasis occurred concurrently after the emergence of bony fish. Importantly, these data also imply that both sIg functions emerged early in evolution rather than representing functions recently acquired by sIgA and IgX in tetrapods. Our findings support further the notion that IgT and IgA are phylogenetically distant immunoglobulins that specialized in mucosal immune responses through convergent evolution (23) and reveal the existence of primordially conserved principles by which mucosal immunoglobulins of primitive and modern vertebrates control both pathogens and microbiota. On one hand, sIgT and sIgA are required to control mucosal pathogens, albeit the role of sIgA in that area appears to have become less essential throughout evolutionary time, likely due to the advent of class switch recombination (CSR) (not present in fish) which enabled IgM and sIgA to share their antigen-binding sites. On the other hand, sIgT and sIgA have also evolved to be indispensable in preservation of microbiota homeostasis at mucosal sites, and the number of functional commonalities between the two Igs in this area is remarkable. Most relevantly, both Igs have evolved to coat broad but exquisitely distinct subsets of beneficial and pathogenic microbial taxa. As a consequence, in the absence of sIgT or sIgA, the microbiome undergoes profound dysbiosis that is characterized by losses of beneficial taxa, large increases in the Bacteroidetes to Firmicutes ratio, and an expansion of pathobionts. Moreover, sIgT- or sIgA-mediated dysbiosis is associated with increased microbial translocation, mucosal tissue damage, and inflammation. In addition to revealing these primordial principles, our studies offer a unique model and unbiased phylogenetic dimension to the field, thus providing the potential to uncover previously undiscovered mechanisms of microbial control and regulation by mucosal Igs.
Materials and Methods
Study Design
The objective of this study was to evaluate the involvement of a primitive fish mucosal immunoglobulin (sIgT) in the control of mucosal pathogens and the maintenance of microbiota homeostasis. To this end we first developed a fish model lacking IgT. This in vivo model of IgT depletion was performed in rainbow trout with the injection of anti-rainbow trout IgT mAbs and rainbow trout antiserum against mice IgG. Using the newly generated IgT-depleted fish, we performed parasite (Ich) infections to evaluate the specific contribution of sIgT in protecting fish against this mucosal pathogen. We also evaluated the effect of IgT depletion on mucosal tissue homeostasis and microbiome composition.
Fish Maintenance.
2-3 g rainbow trout (Oncorhynchus mykiss) were obtained from Troutlodge (Summer, WA) and were maintained as previously described (23). Fish were acclimatized for 2 weeks at 15 °C in an aerated recirculating aquarium with internal biofilters and fed daily with dry pellets at 1% biomass/day. The fish used are of the May strain and are not genetically inbred. As indicated by Troutlodge, the May strain is an outbred strain, and it is also a closed population in which no new genetic material has been introduced for 9 generations. This strain was initiated in the year 2000.All animal procedures were approved by the Institutional Animal Care and Use Committees of the University of Pennsylvania.
Production of trout antiserum to mouse IgG2b (TAs).
Fish (~50-100 g) were first immunized with 25 μg of mouse IgG2b (Biolegend) emulsified in Freund’s complete adjuvant (Sigma) by intraperitoneal injection. Thereafter, fish were injected monthly with the same amount of antigen emulsified in Freund’s incomplete adjuvant (Sigma) at least two times. Antisera from immunized fish (TAs) were obtained two weeks after the last immunization. Trout control sera (TCs) were collected from fish immunized with adjuvant alone following the same immunization scheme described above. Harvested sera was sterilized with Millex-GV syringe filters (Millipore) and kept in −80 °C until further use. The specificity of TAs was evaluated by Western blot analysis. In brief, mouse IgG2b (0.1 μg) was resolved on 4-15% Mini-PROTEAN TGX Gels (BioRad) under nonreducing condition. The gel was transferred onto Sequi-Blot PVDF membranes (Bio-Rad). The membrane was blocked with 8% skim milk and incubated with TAs or TCs at a dilution of 1:10,000, followed by incubation with biotinylated mouse anti-trout IgM mAb, and then incubation with HRP-conjugated streptavidin (Thermo Fisher Scientific). Immunoreactive bands were visualized using the HyGLO Chemiluminescent HRP Ab Detection Reagent (Denville Scientific Products). TAs IgM titers against mouse IgG2b were determined by ELISA as described previously (23). TAs was only used for depletion purposes when titers were above 1:204,800.
Depletion of IgT+ B cells and IgT immunoglobulin from tissues and body fluids respectively.
To initially evaluate the IgT+ B cell depletion effect of anti-trout IgT mAbs and TAs in vivo, fish (~2-3 g) were intraperitoneally injected with two different doses (2 or 10 μg) of mouse anti-trout IgT mAbs (clone 41.8; IgG2b) (23) or 10 μg of mouse IgG2b as isotype control antibody (Biolegend). After 24 hours, half of the fish from each group were further injected with 30 μl of TAs (titer ≥ 1:204,800), whereas the other half were injected with TCs. Seven days after antibody injection, blood leukocytes were obtained and the IgT+ and IgM+ B cell populations were stained and evaluated by flow cytometry as described in the Supplementary Methods. After confirmation that effective IgT+ B cell depletion was achieved with the treatment of 10 μg of anti-trout IgT mAbs and TAs, we used for the rest of experimental procedure 2.5 times the amount of the anti-trout IgT mAb (25 μg/fish) to ensure a consistent and high level of IgT+ B cell depletion in all fish. A time course of IgT+ B cell depletion from blood, head kidney and gill were thereafter performed using the 25 μg/fish dose of anti-trout IgT mAb or its corresponding isotype control, in combination with the subsequent injection of TAs in all groups. Concentration of IgT, IgM and IgD in the serum and gill mucus of these fish were measured as described in the Supplementary Methods.
In vivo treatment of IgT-depleted and control fish with TLR agonists
To investigate whether TLR signaling remains intact in IgT-depleted trout, individual control and IgT-depleted fish (at 3 weeks after depletion treatment) were injected intraperitoneally with PBS or lipoteichoic acid (LTA) from Bacillus subtilis (Sigma, 10 μg/g fish), resiquimod (R848, Sigma, 10 μg/g fish), or flagellin from Salmonella typhimurium (InvivoGen, 1 μg/g fish) (n = 7 fish per group). Six hours post injection, fish were sacrificed and the gills were collected for total RNA extraction. As readouts of TLR activation we measured transcript levels of selected fish cytokines including il1β, il6, il8 and tnfα ½ using real-time PCR (as described in the Supplementary Methods).
IgT-seq and microbiome sequencing from control and IgT-depleted fish.
IgT-seq allows for the identification of taxa-specific microbiota coated by sIgT. This technique was adapted here for fish from the recently reported IgA-seq procedure (16). Gill microbiota were collected as described in Supplementary Methods from healthy fish (2-3 g). To sort sIgT-coated bacteria, we prepared four different pools of microbiota, each pool containing microbiota from 4 different fish. Bacteria from each pool were stained with biotinylated anti-trout IgT mAb for 30 min at 4 °C. Stained bacteria were detected with Brilliant Violet 421-conjugated streptavidin (Biolegend). A portion of the whole bacterial suspension from each pool was stored as the pre-sort sample while 500,000 sIgT+ bacteria were sorted per pool, using a BD FACSAria II flow cytometer (BD Biosciences). Bacteria were preserved in a non-toxic sucrose lysis buffer (SLB) (73) at −80 °C until further use. For total (pre-sort sample) gill bacteria microbiome analysis, we used 4 bacterial pools, and for the sIgT-coated bacteria microbiome analysis, we used 3 bacterial pools. Results are representative of two independent experiments.
To compare the gill microbiome of control versus IgT-depleted fish, gill tissue from 5-6 fish per group was collected 1, 3 and 13 weeks after IgT-depletion and control treatment (described above). Tissues were individually preserved in SLB as previously described (73).
To extract DNA from bacteria and gill tissue, we followed the cetyltrimethylammonium bromide (CTAB) buffer method as previously described (74). When using tissue, this was placed in TissueLyser II (Qiagen) and homogenized using sterile 3-mm tungsten carbide beads (Qiagen). Purified DNA obtained from bacteria or gill tissue was then resuspended in 30 μl of DNase- and RNase-free molecular biology grade water. Sample DNA concentration and purity were measured in a NanoDrop ND 1000 (Thermo Scientific). Negative controls consisting of SLB only and positive controls consisting of a known mock bacterial community of 7 bacterial species were included in each sequencing run as we have previously reported (75). Bacterial community composition was determined as by next generation sequencing of the prokaryotic 16S rRNA (74). Briefly, total genomic DNA for each sample was diluted 1 in 10 or 1 in 100 in RNase-free water and amplified in triplicate using Illumina adapter fused primers that target V1-V3 variable regions of the prokaryotic 16S rRNA sequences. Gene specific primer sequences used were: 28F 5’-GAGTTTGATCNTGGCTCAG-3’ and 519R, 5’-GTNTTACNGCGGCKGCTG-3’ (where N = any nucleotide, and K = T or G). The amplification was carried out with initial activation of the enzyme (5PRIME HotMaster Taq DNA Polymerase, Quanta Bio) at 94 °C for 90 sec followed by 33 cycles of the following: 94 °C for 30 sec, annealing at 52 °C for 30 sec, and 72 °C for 90 sec, and a 7 min extension cycle at 72 °C with a final holding temperature of 4 °C. PCR amplicons were purified using Axygen AxyPrep Mag PCR Clean-up Kit (Thermo Fisher Scientific) per manufacturer’s instructions. Samples were then indexed by ligating index barcode to Illumina adapters onto the PCR amplicon using the Nextera XT Index Kit v2 Set A (Illumina). DNA concentrations in each sample were quantified, pooled and adjusted to a DNA concentration of 200 ng/μl. Pooled samples were purified again using the Axygen AxyPrep Mag PCR Clean-up Kit and sequenced in an Illumina MiSeq platform at the Translational Science Center at University of New Mexico Health Sciences Center.
Microbiome sequence analysis and statistics.
Sequence data was analyzed using Quantitative Insights Into Microbial Ecology (QIIME 1.9) pipeline (76) within the web-based platform Galaxy at the University of New Mexico (77). Operational Taxonomic Units (OTUs) were selected by open reference picking using sumaclust method. OTUs were aligned in SILVA 16S/18S database with a 97% identity level. In order to generate rarefaction curves and assess sampling depth, rarefaction analysis was performed in QIIME using several alpha diversity metrics (PD_whole_tree, chao1, and observed_otus). Core diversity analysis was run on the remaining samples with a normalized sampling depth of 5,800 sequences for whole tissue sequencing and 11,000 sequences for IgT-Seq samples. Alpha diversity metrics including were obtained in QIIME. Non-phylogenetic and phylogenetic beta-diversity analyses were performed in QIIME using the Bray-Curtis metric or the unweighted and weighted UniFrac, respectively. Principal coordinate analysis and taxonomic summaries were produced in QIIME to compare the bacterial community in all experimental groups. Statistical analyses (ANOSIM) were performed in QIIME comparing IgT-depletion treatment with its corresponding age-matched isotype control. Differential abundance analysis was performed by unpaired Student’s t-test in GraphPad Prism version 6. Differences were considered statistically significant when P < 0.05.
Ich parasite isolation and infection.
Isolation of Ichthyophthirius multifiliis parasite (Ich) was performed as previously reported by us (24, 25). Fish (n = 80) were exposed monthly to ~1000 theronts/fish over a 3-month period to obtain fish survivors from Ich infection (immune fish). Mock-infected (uninfected) fish were exposed to the same tank water, but without the parasite. One month after the last exposure, survivor and uninfected fish were divided each into two groups, one group was treated for IgT-depletion (IgT-depleted fish) with anti-IgT mAbs, as described above, while the other group was treated with isotype control Abs (non-depleted fish). Fourteen days after the depletion treatment, non-depleted and IgT-depleted groups were infected with the same dose of parasites. At 10 and 21 days post-infection, fish were sacrificed with an overdose of tricaine methanesulfonate (MS-222, Syndel), and gill tissue samples and fluids (serum and gill mucus) were collected as described in Supplementary Methods, to measure pathogen loads and pathogen-specific antibody titers, respectively. Experiments were repeated two or three times.
Pathogen load, binding of IgT, IgM and IgD to Ich, and fish mortalities.
Pathogen load was obtained by counting the number of the Ich parasites (trophonts) on one side of gill tissue in a double-blind fashion by two independent researchers using a stereomicroscope. Images of the gill tissue were acquired by using an Olympus SZX12 stereomicroscope and DP21 camera (Olympus). The other side of gill tissue of the same fish was used for DNA extraction with DNeasy Blood & Tissue Kit (Qiagen) to measure Ich burden by real-time PCR as described in Supplementary Methods. Primer sequences are shown in table S12.
To assess whether non-depleted and IgT-depleted fish infected with Ich parasites contained pathogen-specific immunoglobulins, we measured the capacity of IgT, IgM and IgD from their serum and gill mucus to bind to Ich using pull-down assays as previously described by us(24). In short, parasites (~100 tomonts) were preincubated with a solution of 0.5% BSA in PBS (pH 7.2) for 2 h at 4 °C. Thereafter, parasites were incubated with diluted gill mucus or serum obtained from the different fish groups for 2 h at 4 °C in a 300 μl volume. Dilutions were made with PBS containing 0.5% BSA (pH 7.2). After incubation, the tomonts were washed with PBS, and bound proteins were eluted with Laemmli Sample Buffer (Bio-Rad) and boiled for 5 min at 95 °C. The eluted material was resolved on 4-15% Mini-PROTEAN TGX Gels (BioRad) under nonreducing conditions, and the presence of IgT, IgM or IgD was detected by Western blotting using the anti-trout IgT, IgM or IgD antibodies as described in Supplementary Methods section. IgT-, IgM- and IgD-specific binding to Ich in dilutions of gill mucus and serum were evaluated by densitometric analysis of immunoblots and presented as relative values to those of uninfected (non-immune) control fish. Fish (25 fish per group) mortalities were recorded daily for 30 days post Ich infection.
Statistical analysis.
The sample sizes were determined based on preliminary studies. Thus, the sample sizes for each in vivo experiment were determined by power analyses to ensure a statistical power of 80-100% depending on the readout. The sample size and number of independent experiments are indicated in the figure captions. Fish were sampled from experimental tanks in a randomized manner, and experiments were repeated at least twice as described throughout the paper. Fish were not repeatedly sampled, but sacrificed at each time point. Histological examination of H&E stains to determine pathology scores in gills and parasite counting for pathogen load in gills were performed blindly by two independent researchers. No data were excluded. An unpaired Student’s t-test and Mantel-Cox test was performed in Prism (GraphPad) for analysis of differences between groups. P values of 0.05 or less were considered statistically significant.
Supplementary Material
Fig. S1. Generation of trout antiserum to mouse IgG2b.
Fig. S2. IgT-depletion does not affect the survival rate of IgT-depleted fish.
Fig. S3. Ich-specific immunoglobulin responses in the serum from non-depleted and IgT-depleted fish after infection.
Fig. S4. Control probe staining of trout gill cryosections.
Fig. S5. Cytokines and antimicrobial peptides with unchanged expression of gene transcripts in gill tissue from control and IgT-depleted fish.
Fig. S6. Cytokine expression in gill tissue from control and IgT-depleted fish upon in vivo treatment with TLR agonists.
Fig. S7. sIgT coats specific subset of bacteria with beneficial and pathogenic characteristics.
Fig. S8. IgT depletion results in mild gill dysbiosis 1 week post-depletion.
Fig. S9. Levels of IgT+ B cells, IgT protein, sIgT-microbiota coating, gill microbial translocation and tissue damage of IgT-depleted fish revert to those of control fish at 13 weeks post-depletion treatment.
Fig. S10. IgT recovery at 13 weeks post-depletion treatment is sufficient to restore microbiome composition.
Table S1. Alpha diversity metrics in gill sIgT-coated bacteria.
Table S2. Bacterial community composition of trout gill sIgT coated bacteria at the order level (in %). Each column represents one individual.
Table S3. Significant OTUs 1 week post IgT depletion in trout gill.
Table S4. Bacterial community composition in control and IgT-depleted trout gill at the order level (in %) 1 week post-depletion. Each column represents one individual.
Table S5. Bacterial community composition in control and IgT-depleted trout gill at the genus level (in %) 1 week post-depletion. Each column represents one individual.
Table S6. Bacterial community composition in control and IgT-depleted trout gill at the order level (in %) 3 weeks post-depletion. Each column represents one individual.
Table S7. Significant OTUs 3 weeks post IgT depletion in trout gill.
Table S8. Bacterial community composition in control and IgT-depleted trout gill at the genus level (in %) 3 weeks post-depletion. Each column represents one individual.
Table S9: Bacterial community composition in control and IgT-depleted trout gill at the Phylum level (in %) 13 weeks post-depletion. Each column represents one individual.
Table S10. Significant OTUs 13 weeks post IgT depletion in trout gill.
Table S11. Bacterial community composition in control and IgT-depleted trout gill at the genus level (in %) 13 weeks post-depletion. Each column represents one individual.
Table S12. Primer sequences used for real-time PCR.
Data file S1. Raw data file (Excel spreadsheet).
Acknowledgements:
We thank Dr. Darrel Dinwiddie for sharing the Illumina sequencer and to Dr. Lijing Bu for help with microbiome bioinformatics analysis. The anti-rainbow trout IgD mAb was provided by the U.S. Veterinary Immune Reagent Network, which was funded by the USDA NIFA #2010-65121-20649 award.
Funding: This work was supported by the National Science Foundation Grant NSF-IOS-1457282 to J.O.S., the US Department of Agriculture Grant USDA-NIFA-2016-09400 to J.O.S., the National Institutes of Health Grant NIH 2R01GM085207-09 to J.O.S. and I.S., the National Institutes of Health Grant NIH P20GM103452 to I.S., the National Natural Science Foundation of China 31879045 to Z.X., Japan Society for the Promotion of Science (JSPS) KAKENHI JP19K21158 to F.T., JSPS Overseas Research Fellowships to F.T. and Y.S., and a grant from the University of New Mexico's Initiative for Maximizing Student Development (IMSD) Program to T.J.C.S.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. The 16S rRNA sequencing data was deposited at the NCBI Sequence Read Archive (SRA) with an accession number of Bioproject PRJNA601439.
References and Notes
- 1.Woof JM, Mestecky J, Mucosal immunoglobulins. Immunological reviews 206, 64–82 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Macpherson AJ, Yilmaz B, Limenitakis JP, Ganal-Vonarburg SC, IgA Function in Relation to the Intestinal Microbiota. Annual review of immunology 36, 359–381 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Dann SM, Manthey CF, Le C, Miyamoto Y, Gima L, Abrahim A, Cao AT, Hanson EM, Kolls JK, Raz E, Cong Y, Eckmann L, IL-17A promotes protective IgA responses and expression of other potential effectors against the lumen-dwelling enteric parasite Giardia. Experimental parasitology 156, 68–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renegar KB, Small PA Jr., Boykins LG, Wright PF, Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract Journal of immunology (Baltimore, Md. : 1950) 173, 1978–1986 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Sun K, Johansen FE, Eckmann L, Metzger DW, An important role for polymeric Ig receptor-mediated transport of IgA in protection against Streptococcus pneumoniae nasopharyngeal carriage Journal of immunology (Baltimore, Md. : 1950) 173, 4576–4581 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, Mazmanian SK, Gut microbiota utilize immunoglobulin A for mucosal colonization Science (New York, N.Y.) 360, 795–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, Kuwahara T, Tsuruyama T, Yamada S, Matsuura M, Nakase H, Peterson DA, Fagarasan S, Suzuki K, IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med 215, 2019–2034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunker JJ, Bendelac A, IgA Responses to Microbiota. Immunity 49, 211–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yel L, Selective IgA deficiency. Journal of clinical immunology 30, 10–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T, Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora Science (New York, N.Y.) 298, 1424–1427 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, Pons N, Le Chatelier E, Levenez F, Kennedy S, Galleron N, de Barros JP, Malphettes M, Galicier L, Boutboul D, Mathian A, Miyara M, Oksenhendler E, Amoura Z, Dore J, Fieschi C, Ehrlich SD, Larsen M, Gorochov G, Microbial ecology perturbation in human IgA deficiency. Sci Transl Med 10, (2018). [DOI] [PubMed] [Google Scholar]
- 12.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S, Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proceedings of the National Academy of Sciences of the United States of America 101, 1981–1986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, Hooper LV, Yarovinsky F, Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut microbes 5, 28–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richmond BW, Brucker RM, Han W, Du RH, Zhang Y, Cheng DS, Gleaves L, Abdolrasulnia R, Polosukhina D, Clark PE, Bordenstein SR, Blackwell TS, Polosukhin VV, Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nature communications 7, 11240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catanzaro JR, Strauss JD, Bielecka A, Porto AF, Lobo FM, Urban A, Schofield WB, Palm N, IgA-deficient humans exhibit gut microbiota dysbiosis despite production of compensatory IgM. bioRxiv, 446724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA, Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas AE, The Drosophila model for microbiome research. Lab animal 47, 157–164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dishaw LJ, Flores-Torres JA, Mueller MG, Karrer CR, Skapura DP, Melillo D, Zucchetti I, De Santis R, Pinto MR, Litman GW, A Basal chordate model for studies of gut microbial immune interactions. Frontiers in immunology 3, 96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper MD, The early history of B cells. Nature reviews. Immunology 15, 191–197 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Sunyer JO, Fishing for mammalian paradigms in the teleost immune system. Nature immunology 14, 320–326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salinas I, Zhang YA, Sunyer JO, Mucosal immunoglobulins and B cells of teleost fish. Developmental and comparative immunology 35, 1346–1365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flajnik MF, A cold-blooded view of adaptive immunity. Nature reviews. Immunology 18, 438–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO, IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nature immunology 11, 827–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Takizawa F, Parra D, Gomez D, von Gersdorff Jorgensen L, LaPatra SE, Sunyer JO, Mucosal immunoglobulins at respiratory surfaces mark an ancient association that predates the emergence of tetrapods. Nature communications 7, 10728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, Parra D, Gomez D, Salinas I, Zhang YA, von Gersdorff Jorgensen L, Heinecke RD, Buchmann K, LaPatra S, Sunyer JO, Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proceedings of the National Academy of Sciences of the United States of America 110, 13097–13102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowrey L, Woodhams DC, Tacchi L, Salinas I, Topographical Mapping of the Rainbow Trout (Oncorhynchus mykiss) Microbiome Reveals a Diverse Bacterial Community with Antifungal Properties in the Skin. Applied and environmental microbiology 81, 6915–6925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostaff MJ, Stange EF, Wehkamp J, Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO molecular medicine 5, 1465–1483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP, Staphylococcus aureus Shifts toward Commensalism in Response to Corynebacterium Species. Frontiers in microbiology 7, 1230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravi AV, Musthafa KS, Jegathammbal G, Kathiresan K, Pandian SK, Screening and evaluation of probiotics as a biocontrol agent against pathogenic Vibrios in marine aquaculture. Letters in applied microbiology 45, 219–223 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Kozińska A, Paździor E, Pękala A, Niemczuk W, Acinetobacter johnsonii and Acinetobacter lwoffii-the emerging fish pathogens. Bull Vet Inst Pulawy 58, 193–199 (2014). [Google Scholar]
- 31.Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, Ibba I, Lai MA, Orru S, Blois S, Loizedda AL, Griffin JL, Usai P, Caboni P, Atzori L, Manzin A, Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Scientific reports 7, 9523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lycke NY, Bemark M, The regulation of gut mucosal IgA B-cell responses: recent developments. Mucosal immunology 10, 1361–1374 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Uchida J, Hamaguchi Y, Oliver JA, Ravetch JV, Poe JC, Haas KM, Tedder TF, The innate mononuclear phagocyte network depletes B lymphocytes through Fc receptor-dependent mechanisms during anti-CD20 antibody immunotherapy. J Exp Med 199, 1659–1669 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Looney RJ, Anolik JH, Campbell D, Felgar RE, Young F, Arend LJ, Sloand JA, Rosenblatt J, Sanz I, B cell depletion as a novel treatment for systemic lupus erythematosus: a phase I/II dose-escalation trial of rituximab. Arthritis Rheum 50, 2580–2589 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Harriman GR, Bogue M, Rogers P, Finegold M, Pacheco S, Bradley A, Zhang Y, Mbawuike IN, Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes Journal of immunology (Baltimore, Md. : 1950) 162, 2521–2529 (1999). [PubMed] [Google Scholar]
- 36.Brandtzaeg P, Karlsson G, Hansson G, Petruson B, Bjorkander J, Hanson LA, The clinical condition of IgA-deficient patients is related to the proportion of IgD- and IgM-producing cells in their nasal mucosa. Clinical and experimental immunology 67, 626–636 (1987). [PMC free article] [PubMed] [Google Scholar]
- 37.Furuya Y, Kirimanjeswara GS, Roberts S, Racine R, Wilson-Welder J, Sanfilippo AM, Salmon SL, Metzger DW, Defective anti-polysaccharide IgG vaccine responses in IgA deficient mice. Vaccine 35, 4997–5005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murthy AK, Sharma J, Coalson JJ, Zhong G, Arulanandam BP, Chlamydia trachomatis pulmonary infection induces greater inflammatory pathology in immunoglobulin A deficient mice. Cellular immunology 230, 56–64 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez A, Tjarnlund A, Ivanji J, Singh M, Garcia I, Williams A, Marsh PD, Troye-Blomberg M, Fernandez C, Role of IgA in the defense against respiratory infections IgA deficient mice exhibited increased susceptibility to intranasal infection with Mycobacterium bovis BCG. Vaccine 23, 2565–2572 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Mellander L, Bjorkander J, Carlsson B, Hanson LA, Secretory antibodies in IgA-deficient and immunosuppressed individuals. Journal of clinical immunology 6, 284–291 (1986). [DOI] [PubMed] [Google Scholar]
- 41.Castro R, Jouneau L, Pham HP, Bouchez O, Giudicelli V, Lefranc MP, Quillet E, Benmansour A, Cazals F, Six A, Fillatreau S, Sunyer O, Boudinot P, Teleost fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS pathogens 9, e1003098 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reikvam DH, Derrien M, Islam R, Erofeev A, Grcic V, Sandvik A, Gaustad P, Meza-Zepeda LA, Jahnsen FL, Smidt H, Johansen FE, Epithelial-microbial crosstalk in polymeric Ig receptor deficient mice. European journal of immunology 42, 2959–2970 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, Orandle M, Mayer L, Macpherson AJ, McCoy KD, Fraser-Liggett C, Matzinger P, Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med 17, 1585–1593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rigoni R, Fontana E, Guglielmetti S, Fosso B, D’Erchia AM, Maina V, Taverniti V, Castiello MC, Mantero S, Pacchiana G, Musio S, Pedotti R, Selmi C, Mora JR, Pesole G, Vezzoni P, Poliani PL, Grassi F, Villa A, Cassani B, Intestinal microbiota sustains inflammation and autoimmunity induced by hypomorphic RAG defects. J Exp Med 213, 355–375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, Ishizuka IE, Dent AL, Wilson PC, Jabri B, Antonopoulos DA, Bendelac A, Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 43, 541–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, Warner BB, Gordon JI, Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534, 263–266 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tedelind S, Westberg F, Kjerrulf M, Vidal A, Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World j gastroenterol 13, 2826–2832 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vieira EL, Leonel AJ, Sad AP, Beltrao NR, Costa TF, Ferreira TM, Gomes-Santos AC, Faria AM, Peluzio MC, Cara DC, Alvarez-Leite JI, Oral administration of sodium butyrate attenuates inflammation and mucosal lesion in experimental acute ulcerative colitis. The Journal of nutritional biochemistry 23, 430–436 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Kim M, Qie Y, Park J, Kim CH, Gut Microbial Metabolites Fuel Host Antibody Responses. Cell host & microbe 20, 202–214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu W, Sun M, Chen F, Cao AT, Liu H, Zhao Y, Huang X, Xiao Y, Yao S, Zhao Q, Liu Z, Cong Y, Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal immunology 10, 946–956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamboli CP, Neut C, Desreumaux P, Colombel JF, Dysbiosis in inflammatory bowel disease. Gut 53, 1–4 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S, Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60, 631–637 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Ilinskaya ON, Ulyanova VV, Yarullina DR, Gataullin IG, Secretome of Intestinal Bacilli: A Natural Guard against Pathologies. Frontiers in microbiology 8, 1666 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jan G, Belzacq AS, Haouzi D, Rouault A, Metivier D, Kroemer G, Brenner C, Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell death and differentiation 9, 179–188 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Moens F, De Vuyst L, Inulin-type fructan degradation capacity of Clostridium cluster IV and XIVa butyrate-producing colon bacteria and their associated metabolic outcomes. Beneficial microbes 8, 473–490 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Fernández J, Redondo-Blanco S, Gutierrez-del-Rio I, Miguélez EM, Villar CJ, Lombo F, Colon microbiota fermentation of dietary prebiotics towards short-chain fatty acids and their roles as anti-inflammatory and antitumour agents: A review. J Funct Foods 25, 511–522 (2016). [Google Scholar]
- 57.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F, From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Jarillo-Luna A, Rivera-Aguilar V, Garfias HR, Lara-Padilla E, Kormanovsky A, Campos-Rodriguez R, Effect of repeated restraint stress on the levels of intestinal IgA in mice. Psychoneuroendocrinology 32, 681–692 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Fransen F, Zagato E, Mazzini E, Fosso B, Manzari C, El Aidy S, Chiavelli A, D’Erchia AM, Sethi MK, Pabst O, Marzano M, Moretti S, Romani L, Penna G, Pesole G, Rescigno M, BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity. Immunity 43, 527–540 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Coyte KZ, Schluter J, Foster KR, The ecology of the microbiome: Networks, competition, and stability Science (New York, N.Y.) 350, 663–666 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P, The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15, 630–638 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI, Host-bacterial mutualism in the human intestine Science (New York, N.Y.) 307, 1915–1920 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Lee YK, Mazmanian SK, Has the microbiota played a critical role in the evolution of the adaptive immune system? Science (New York, N.Y.) 330, 1768–1773 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazmanian SK, Round JL, Kasper DL, A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Takizawa F, Magadan S, Parra D, Xu Z, Korytar T, Boudinot P, Sunyer JO, Novel Teleost CD4-Bearing Cell Populations Provide Insights into the Evolutionary Origins and Primordial Roles of CD4+ Lymphocytes and CD4+ Macrophages Journal of immunology (Baltimore, Md. : 1950) 196, 4522–4535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeLuca D, Wilson M, Warr GW, Lymphocyte heterogeneity in the trout, Salmo gairdneri, defined with monoclonal antibodies to IgM. European journal of immunology 13, 546–551 (1983). [DOI] [PubMed] [Google Scholar]
- 67.Ramirez-Gomez F, Greene W, Rego K, Hansen JD, Costa G, Kataria P, Bromage ES, Discovery and characterization of secretory IgD in rainbow trout: secretory IgD is produced through a novel splicing mechanism Journal of immunology (Baltimore, Md. : 1950) 188, 1341–1349 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Tacchi L, Musharrafieh R, Larragoite ET, Crossey K, Erhardt EB, Martin SAM, LaPatra SE, Salinas I, Nasal immunity is an ancient arm of the mucosal immune system of vertebrates. Nature communications 5, 5205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roth RI, Levin FC, Levin J, Optimization of detection of bacterial endotoxin in plasma with the Limulus test. The Journal of laboratory and clinical medicine 116, 153–161 (1990). [PubMed] [Google Scholar]
- 70.Zilberg D, Munday B, Pathology of experimental amoebic gill disease in Atlantic salmon, Salmo salar L., and the effect of pre‐maintenance of fish in sea water on the infection. J Fish Dis 23, 401–407 (2000). [Google Scholar]
- 71.Ringo E, Salinas I, Olsen RE, Nyhaug A, Myklebust R, Mayhew TM, Histological changes in intestine of Atlantic salmon (Salmo salar L.) following in vitro exposure to pathogenic and probiotic bacterial strains. Cell and tissue research 328, 109–116 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Baxter EJ, Sturt MM, Ruane NM, Doyle TK, McAllen R, Harman L, Rodger HD, Gill damage to Atlantic salmon (Salmo salar) caused by the common jellyfish (Aurelia aurita) under experimental challenge. PloS one 6, e18529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mitchell KR, Takacs-Vesbach CD, A comparison of methods for total community DNA preservation and extraction from various thermal environments. Journal of industrial microbiology & biotechnology 35, 1139–1147 (2008). [DOI] [PubMed] [Google Scholar]
- 74.Reid KM, Patel S, Robinson AJ, Bu L, Jarungsriapisit J, Moore LJ, Salinas I, Salmonid alphavirus infection causes skin dysbiosis in Atlantic salmon (Salmo salar L.) post-smolts. PloS one 12, e0172856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown RM, Wiens GD, Salinas I, Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish & shellfish immunology 86, 497–506 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R, QIIME allows analysis of high-throughput community sequencing data. Nature methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A, Galaxy: a platform for interactive large-scale genome analysis. Genome research 15, 1451–1455 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS, Heatmapper: web-enabled heat mapping for all. Nucleic acids research 44, W147–153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Generation of trout antiserum to mouse IgG2b.
Fig. S2. IgT-depletion does not affect the survival rate of IgT-depleted fish.
Fig. S3. Ich-specific immunoglobulin responses in the serum from non-depleted and IgT-depleted fish after infection.
Fig. S4. Control probe staining of trout gill cryosections.
Fig. S5. Cytokines and antimicrobial peptides with unchanged expression of gene transcripts in gill tissue from control and IgT-depleted fish.
Fig. S6. Cytokine expression in gill tissue from control and IgT-depleted fish upon in vivo treatment with TLR agonists.
Fig. S7. sIgT coats specific subset of bacteria with beneficial and pathogenic characteristics.
Fig. S8. IgT depletion results in mild gill dysbiosis 1 week post-depletion.
Fig. S9. Levels of IgT+ B cells, IgT protein, sIgT-microbiota coating, gill microbial translocation and tissue damage of IgT-depleted fish revert to those of control fish at 13 weeks post-depletion treatment.
Fig. S10. IgT recovery at 13 weeks post-depletion treatment is sufficient to restore microbiome composition.
Table S1. Alpha diversity metrics in gill sIgT-coated bacteria.
Table S2. Bacterial community composition of trout gill sIgT coated bacteria at the order level (in %). Each column represents one individual.
Table S3. Significant OTUs 1 week post IgT depletion in trout gill.
Table S4. Bacterial community composition in control and IgT-depleted trout gill at the order level (in %) 1 week post-depletion. Each column represents one individual.
Table S5. Bacterial community composition in control and IgT-depleted trout gill at the genus level (in %) 1 week post-depletion. Each column represents one individual.
Table S6. Bacterial community composition in control and IgT-depleted trout gill at the order level (in %) 3 weeks post-depletion. Each column represents one individual.
Table S7. Significant OTUs 3 weeks post IgT depletion in trout gill.
Table S8. Bacterial community composition in control and IgT-depleted trout gill at the genus level (in %) 3 weeks post-depletion. Each column represents one individual.
Table S9: Bacterial community composition in control and IgT-depleted trout gill at the Phylum level (in %) 13 weeks post-depletion. Each column represents one individual.
Table S10. Significant OTUs 13 weeks post IgT depletion in trout gill.
Table S11. Bacterial community composition in control and IgT-depleted trout gill at the genus level (in %) 13 weeks post-depletion. Each column represents one individual.
Table S12. Primer sequences used for real-time PCR.
Data file S1. Raw data file (Excel spreadsheet).