Abstract
Ribonuclease 6 (RNase 6) is one of eight catalytically active human pancreatic-type RNases that belong to a superfamily of rapidly evolving enzymes. Like some of its human homologs, RNase 6 exhibits host defense properties such as antiviral and antibacterial activities. Recently solved crystal structures of this enzyme in its nucleotide-free form show conservation of the prototypical kidney-shaped fold preserved among vertebrate RNases, in addition to revealing the presence of a unique secondary active site. In this study, we determine the structural and conformational properties experienced by RNase 6 upon binding to substrate and product analogs. We present the first crystal structures of RNase 6 bound to a nucleotide ligand (adenosine 5’-monophosphate), in addition to RNase 6 bound to phosphate ions. While the enzyme preserves B2 subsite ligand preferences, our results show lack of typical B2 subsite interactions normally observed in homologous ligand-bound RNases. Comparison of the dynamical properties of RNase 6 in its apo, substrate-, and product-bound states highlight unique dynamical properties experienced on time scales ranging from nano- to milliseconds. Overall, our results confirm the specific evolutionary adaptation of RNase 6 relative to its unique catalytic and biological activities.
Keywords: Ribonuclease 6, RNase K6, nuclear magnetic resonance, conformational exchange, Carr-Purcell-Meiboom-Gill, computational simulation, crystal structure, ligand binding
Introduction
Human ribonuclease 6 (RNase 6, also referred to as RNase K6) is one of the eight catalytically active pancreatic-type RNases identified in humans1. Like other canonical members of this superfamily, this enzyme primarily catalyzes the endonucleolytic cleavage of RNA with strict conservation of catalytic triad residues His15/Lys38/His122 and distinct phosphate/nucleotide binding subsites2. RNase 6 was detected in various human tissues, including the lung, heart, placenta, and kidneys3. The occurrence of RNase 6 in monocytes and neutrophils suggested its role in host defense, a hypothesis that was subsequently confirmed by identification of antimicrobial activity against uropathogenic bacterial strains4, 5. The mechanism of action of this antimicrobial activity was shown to be similar to that determined for human RNase 3, which involves the agglutination and bacterial membrane leakage of Gram-negative bacteria6. N-terminal residues Trp1 and Ile13 in RNase 6 were shown to be essential for preservation of antimicrobial activity6. Recent studies also revealed that this enzyme is involved in the inhibition of HIV, confirming its role as an antiviral agent7.
Catalytic activity comparison of RNase superfamily members illustrates that RNase 6 is 10 times faster than human RNase 3 and >200-fold slower than bovine RNase A at cleaving a model dinucleotide RNA substrate (UpA)8. Recent studies reporting crystal structures of RNase 6 also revealed the presence of a unique secondary active center clustered around residues His36, His39, and Lys878, 9. Mutations in the primary and secondary active sites highlighted the role played by the secondary site in the catalytic mechanism of RNase 69. Crystal structures further illustrated higher overall atomic flexibility for residues 1–5 (N-terminus), 17–21 (loop 1), 34–39 (loop 2), and 85–90 (loop 7), which were inferred from the B-factors. The flexibility of the N-terminal region was proposed to promote anchoring of Trp1 to the bacterial surface to facilitate destabilization of the bacterial membrane9.
Ligand binding induces long-range conformational rearrangements in enzymes and was previously shown to alter the conformational exchange profiles of select RNases, including many human homologs10. Here, we characterize and compare the structural and dynamical properties of the ligand-free and ligand-bound states of RNase 6. We present the first holo crystal structure of human RNase 6—i.e. bound to mononucleotide product analog 5’-AMP—in addition to the crystal structure of phosphate-bound RNase 6. Using a combination of NMR relaxation experiments and computational simulations, we also probe the effect of substrate and product analog binding on the structural and dynamical properties of RNase 6 on the nanosecond to millisecond timescales. Our results illustrate the distinctively unique conformational exchange properties experienced by RNase 6 relative to closely related RNase homologs. These observations indicate the unique evolutionary adaptations that may be associated with the specific catalytic and biological functions of RNase 6 in human.
Materials and Methods
Protein expression and purification
The Escherichia coli codon-optimized human sequence of RNase 6 was inserted into NdeI/HindIII-digested expression vector pJexpress414 (ATUM, Newark, CA, USA). The DNA construct was transformed into E. coli BL21(DE3) cells for protein expression. The recombinant enzyme was expressed for 4.5 hours at 37 °C and observed within recovered inclusion bodies. The protein was properly refolded and purified according to a previous protocol11. Protein concentration was determined by UV-Vis.
Protein crystallization and data collection
The pure, wild-type RNase 6 enzyme was dissolved in milliQ water to a final concentration of 20 mg/mL (1.31 mM). Crystallization was carried out using the sitting drop method, using a reservoir solution of 70 μL containing 0.2 M sodium phosphate buffer at pH 7.0 with 20% (w/v) polyethylene glycol (PEG) 3350. Each well contained 1 μL of protein with a similar volume of buffer. The plate was sealed with Crystal Clear tape and incubated in a polystyrene box at room temperature in a non-vibrating environment. RNase 6 crystals took 5–7 days to grow to completion. A cryo-protectant was prepared by supplementing the crystallization solution reservoir with 25% glycerol. Diffraction data for the ligand-free RNase 6 structure were collected at the LS-CAT Sector 21 of the Argonne National Laboratory (IL, USA).
For the ligand-bound crystal structure, the pure RNase 6 and 5’-AMP ligand were dissolved in milliQ water to final concentrations of 15 mg/mL (0.98 mM) and 21.8 mM, respectively. The mixture was incubated at 4°C for 2 hours before being plated. The reservoir solution consisted of 35 μL of 0.2 M sodium malonate buffer with 20% (w/v) PEG 3350 (Solution 2–12, PACT premier kit MD1–29, Molecular Dimensions). The crystallization well was filled with 1 μL of the RNase 6/5’AMP mixture and 1 μL of the reservoir solution. The plate was sealed with Crystal Clear tape and incubated in a polystyrene box at room temperature in a non-vibrating environment. Crystals started to appear after 14 days, but were allowed to grow for 2 months. They were then carefully washed with paratone for data collection. Diffraction data were collected on a Rigaku rotating copper anode X-ray generator.
Data processing and structure refinement
For the free RNase 6 structure, data processing and scaling were carried out using HKL200012. Molecular replacement using PDB number 1GQV (human RNase 2), a structural homolog of RNase 6, was performed with CCP4 Phaser MR13, 14. The model was improved by alternate cycles of refinement and manual rebuilding using refmac515, 16 and Coot17. For the ligand-bound RNase 6 crystal data, molecular replacement was carried out using Phenix18 using the crystal structure of RNase 6 in the presence of sulphate anions (PDB 4X09). Multiple refinement cycles were performed using Phenix18 and Coot17. The 3D structures were validated using MolProbity4 (http://molprobity.biochem.duke.edu/). Bond lengths and angles were evaluated by Ramachandran scoring. Final data collection and refinement statistics are listed in Table 1.
Table 1.
Data Collection and Refinement statistics of apo and 5’-AMP bound RNase 6
| Data Collection | RNase 6 | RNase 6/5’-AMP |
|---|---|---|
| Space group | P22121 | P212121 |
| Unit cell (Å) | 27.8, 38.7, 108.5 | 27.6, 38.8, 97.9 |
| Resolution (Å) | 26.5 – 1.5 (1.55 – 1.5) | 36.1 – 2.59 (2.71–2.59) |
| Rmeas | 4.8 (28.3) | 4.6 (8.3) |
| I / σI | 14.5 (8.1) | 17.2 (10.4) |
| Completeness (%) | 99.1 (98.9) | 98.6 (97.4) |
| Redundancy | 3.1 (2.4) | 2.4 (2.0) |
| Refinement | ||
| Resolution (Å) | 26.5 – 1.5 | 36.1 – 2.6 |
| No. reflections | 19372 | 3537 |
| Rwork / Rfree | 17.3/22.8 | 22.0/25.1 |
| Number of Atoms | ||
| Protein | 1868 | 1023 |
| Water | 108 | 27 |
| Phosphate | 15 | - |
| AMP | - | 32.3 |
| B-factors (Å2) | ||
| Protein | 16.8 | 19.0 |
| Water | 30.1 | 17.3 |
| Phosphate | 44.7 | - |
| AMP | - | 23 |
| RMSD | ||
| Bond lengths (Å) | 0.006 | 0.003 |
| Bond angles (°) | 1.048 | 0.703 |
| Molprobity scores | 1.5 | 1.43 |
| Ramachandran favored (%) | 100 | 99.2 |
| Ramachandran allowed (%) | 0 | 0.8 |
| PDB ID | 6MV6 | 6MV7 |
Values in () are for the last resolution shell.
NMR spectroscopy
15N- or {15N, 13C}-labeled protein samples were analyzed on Varian (Agilent) 500 and 800 MHz NMR spectrometers equipped with a triple-resonance cold probe. All experiments were recorded at 298 K on samples containing 160–600 μM enzyme in 15 mM sodium acetate buffer, pH 5.0 or 5.3, with 10% D2O. All spectra were processed using NMRPipe19, and CcpNmr Analysis20. Two- and three-dimensional {1H-15N}-HSQC, HNCACB and CBCA(CO)NH spectra were recorded to perform backbone resonance assignments of the protein. Titration experiments were performed using 5’-AMP or deoxyribopentanucleotide d(ACACA) dissolved in the NMR buffer. {1H-15N}-HSQC spectra were recorded in the presence of 0, 0.174, 0.393, 0.691, 1.31, 2.71, 6.0, 12.0 and 18.0 molar equivalents of 5’-AMP, or 0, 0.84, 1.68, 2.52 and 3.36 molar equivalents of d(ACACA), as further additions of d(ACACA) led to precipitation in the NMR tube. Compounded chemical shifts (Δδ) were calculated using equation 1.
| [1] |
The dissociation constant (Kd) was determined by calculating a shared value for all residues that displayed compounded chemical shifts greater than 0.05 ppm. The Kd corresponds to the ligand concentration at which 50% of the chemical shift variations are observed. While we observed saturation curves for the titration of d(ACACA), we were unable to fit the data using the estimated ligand ratios. This suggested a potential scaling inaccuracy for the initial bulk concentration of d(ACACA). We therefore applied a scaling factor to equation 2 21 for each of the [L]/[P] instances, and fitted the data using a shared value for all datasets, providing the ligand ratios listed above:
| [2] |
Backbone amide 15N-Carr-Purcell-Meiboom-Gill (15N-CPMG) relaxation dispersion experiments were recorded on the apo, 5’-AMP-, and d(ACACA)-bound enzyme forms. Interleaved spectra were recorded with τCPMG inter-refocusing pulse delays of 0.625, 0.714 (x2), 1.000, 1.125, 1.667, 2.000, 2.500 (x2), 3.333, 5.000 and 10.000 ms, using a total relaxation period of 40.0 ms. Relaxation parameters were extracted by fitting the 800 MHz CPMG relaxation data to the fast exchange CPMG equation22 using GraphPad Prism 6. The apo protein was also submitted to a 15N-Chemical Exchange Saturation Transfer (15N-CEST) experiment23 using a 15 Hz B1 irradiation field on the 800 MHz magnet at 25°C. A set of 114 2D datasets was acquired, corresponding to 15N offsets interspaced by 0.25 ppm (~20 Hz) spanning the whole resonance array of the analyzed protein. ZZ exchange experiments for apo RNase 6 were run at 800 MHz and 25°C, using mixing times of 20, 300 and 500 ms24.
Microscale thermophoresis (MST)
RNase 6 and RNase A (BioBasic, Canada) were labeled using the RED-NHS labeling kit (Nanotemper Technologies, Germany) in accordance with the manufacturer’s protocol. Unreacted dye was removed using the provided purification columns. Binding affinity assays between these labeled proteins and ligands 5’-AMP and d(ACACA) were performed in 20 mM MES (2-(N-morpholino)ethanesulfonic acid) buffer, 150 mM NaCl, 20mM L-Arginine at pH 6.4, supplemented with 0.05% (v/v) Tween 20. A series of 16 1:1 dilutions (protein:ligand) were prepared to determine Kd values, using labeled proteins at a final concentration of 5 nM and ligands at concentrations ranging from 2 μM to 75 mM for 5’-AMP, and from 3 nM to 100 μM for d(ACACA). Samples were loaded into standard Monolith NT.115 capillaries and microscale thermophoresis was measured using a Monolith NT.115 Pico instrument (Nanotemper Technologies, Germany) at ambient temperature. Data of 3 independently pipetted experiments were analyzed with the MO.Affinity software, version 2.3 (Nanotemper Technologies, Germany) using MST-on time between 5 s and 10 s to determine Kd values.
System preparation for computational simulations
Crystal structure coordinates of RNase 6 determined in this study (PDB 6MV6) were used for the simulations of the apo, 5’-AMP, and ACAC bound states of RNase 6. The ACAC-bound form represents the substrate bound state and is used to characterize the conformational changes associated with substrate binding. The 5’-AMP bound form of RNase 6 represents the product bound state. The coordinates of the tetranucleotide ligand were obtained through superposition of the three dimensional structure of RNase 6 with that of RNase A bound to DNA ligand d(ATAA) (PDB 1RCN). Protonation states of histidine residues in the ligand-bound state were determined based on pKa values, in addition to structural interactions of histidines with other residues of the protein and the ligand. System preparation and MD simulations were performed using AMBER v14 and the ff14SB force field25. Two independent simulations, each 500 ns long, were performed for the apo and ligand-bound RNase enzymes for a total of 1 μs for each state. Simulations were performed under constant energy (NVE) conditions using a 2 fs timestep. The enzyme (enzyme-ligand) systems were solvated using the SPC(E) water model in a rectangular box with periodic boundary conditions. The systems were neutralized through the addition of counter ions (Cl−). The pre-processing steps were followed by equilibration steps, as described previously26. All simulations were performed using the GPU-enabled version of AMBER’s simulation engine pmemd. A Particle Mesh Ewald (PME) cut-off of 8 Å was used, as suggested for GPU-enabled simulations27.
Computational analyses
All-atom root mean square fluctuations (RMSFs) were computed using the cpptraj program28 by analyzing all conformational snapshots collected during the MD simulations for the apo and ligand bound states of RNase 6. All conformations were first aligned to a common structure to remove rotations and translations over the course of the simulation. The top ten modes were used to identify global motions on the slower timescales of the MD simulations. The dynamic cross-correlation maps (DCCMs) were generated using Gnuplot v5.0 [http://www.gnuplot.info/].
Results
Three-dimensional structure of RNase 6 in the free and 5’-AMP bound states
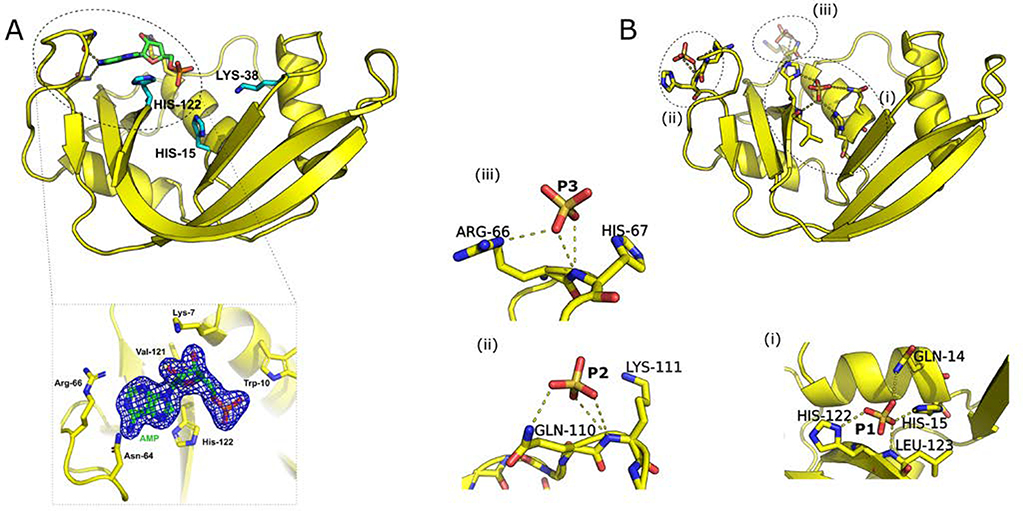
We determined the first crystal structure of holo RNase 6, i.e. bound to mononucleotide ligand 5’-AMP (PDB 6MV7). The crystal was resolved to 2.6 Å (Figure 1A) and belonged to the P212121 space group (Table 1). The overall structure of RNase 6 showed a kidney-shaped fold, preserving the prototypical fold observed for other structurally resolved members of this family. The quality of the omit map (Figure 1) enabled the unambiguous placement of the 5’-AMP ligand in the RNase 6 structure. The position of the ligand in the B2 nucleotide binding subsite was similar to that observed for other members of this superfamily, including bovine RNase A2,29. We observed polar contacts between the purine base and loop 4 residues Asn64 and Asn68, while the ribose moiety showed interactions with Lys7 of α-helix 1. In addition, water-mediated interactions were observed with residues Lys63, Asn64 in loop 4, and Asp124 of the C-terminal β-strand. Other RNase homologs (including human RNases 2, 3, and bovine RNase A) show interactions between the phosphate group of the ligand in the B2 subsite with active-site residues. For instance, phosphate interacts with residues Gln11, His12, and Phe120 in RNase A. In contrast, RNase 6 does not display any hydrogen bonding or electrostatic interactions between the phosphate group of 5’-AMP and residues in the vicinity of the catalytic triad.
Figure 1.
Crystal structures of (A) 5’-AMP bound, and (B) apo RNase 6 with bound phosphate ions. Catalytic residues His15, Lys38, and His122 are represented as cyan sticks on the top left panel. Details of ligand interactions (represented as sticks) and phosphate ions (labeled P1-P3) in the phosphate-bound state are shown using dashed lines in panels below each structure. The omit map for the 5’-AMP ligand is shown as a blue mesh structure with neighboring residues below panel A.
In addition to the 5’-AMP bound state, we determined the crystal structure of RNase 6 in the presence of phosphate ions (PDB 6MV6). The final refined model contained 127 residues and 108 water molecules. The structure was resolved to 1.5 Å and belongs to the orthorhombic crystal system and the P22121 space group (Table 1). The phosphate-bound structure of RNase 6 displayed a root mean square deviation of 0.3 Å and 0.54 Å with the sulfate and phosphate bound crystal structures reported previously for this enzyme8, 9. Interactions of the three phosphate ions in RNase 6 are shown in Figure 1B. All phosphates displayed interactions similar to the recently reported phosphate bound structure of RNase 6:9 P1 showed interactions with the catalytic His15 and His122, in addition to Gln14 and Leu123 (Figure 1B(i)), while P2 displayed interactions with residues Arg66 and His67 of loop 4 (Figure 1B(ii)). The third phosphate ion (P3, Figure 1B(iii)) displayed interactions with residues Gln110 and Lys111 of loop 7. However, our structures do not show phosphate binding interactions with residues of loop 2 reported by Prats-Ejarque et al.8, 9. Crystal structures of both phosphate and 5’-AMP bound RNase 6 showed large B-factors for residues in the N-terminal region and loop 1 (residues 17–21), in addition to loop 6 (residues 85–91), suggesting large atomic fluctuations in these regions (Figure S1).
NMR assignments and population of RNase 6 conformers in solution
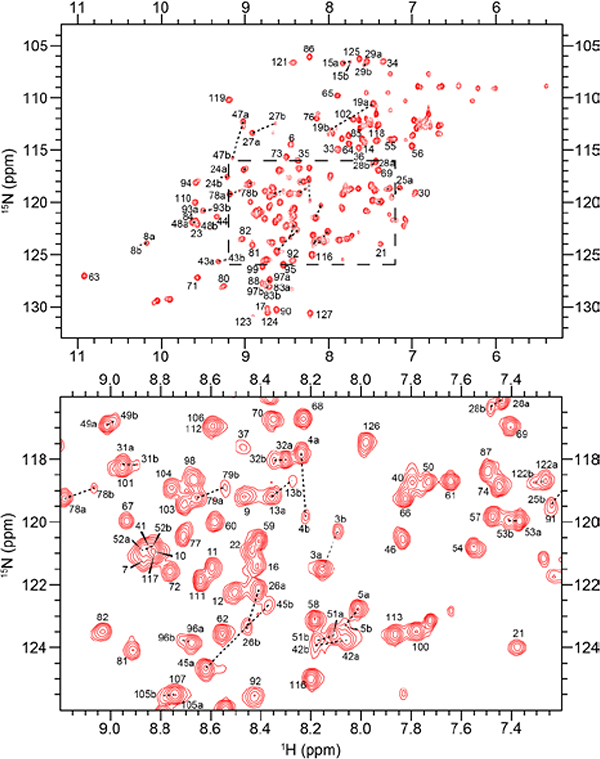
The {1H-15N}-Heteronuclear Single Quantum Coherence (HSQC) spectrum of RNase 6 showed nicely dispersed resonances, corresponding to a well-folded protein (Figure 2). Resonance assignments were performed using a combination of two-dimensional ({1H-15N}-HSQC) and three-dimensional (HNCACB and CBCA(CO)NH) experiments. We assigned 114 out of an expected total of 116 residues, excluding proline residues and the N-terminal Trp1, corresponding to 98.28% of non-proline residues. Resonance assignments were achieved for 95.28% of the Cα and 94.4% of non-glycine Cβ peaks, respectively. Unassigned residues were localized to loop 2 and correspond to Lys38 and His39. Line-broadening observed for residues belonging to the primary and secondary catalytic residues, respectively, indicated exchange of these residues on an intermediate timescale. An interesting feature of this assignment is the consistent and reproducible presence of two conformers in all prepared samples of this protein, which could not be separated by standard purification methods. We successfully assigned both the major and the minor conformers, and determined that only 70 atoms out of the 470 assigned peaks were split between the two conformers. By calculating the occupancy of each peak from a pair, we determined the population of the major (minor) conformation to be 76.93% (23.07%). With the exceptions of His122, Ala105 and six residues of α-helix 3, the majority of double-peaked atoms were located in the V2 β-sheet (Figure S2). Chemical shifts of both conformers, including the peak occupancy for each atom, was deposited in the Biological Magnetic Resonance Bank, under accession number 27659.
Figure 2.
{1H-15N}-HSQC spectrum of 15N labeled apo RNase 6 in a buffer containing 15 mM sodium acetate at pH 5.0 and 298 K. A close-up view of the central region of the spectrum is shown in the panel below. Resonances differing between the major and minor conformers are linked by dashed lines.
Effect of ligand binding to RNase 6
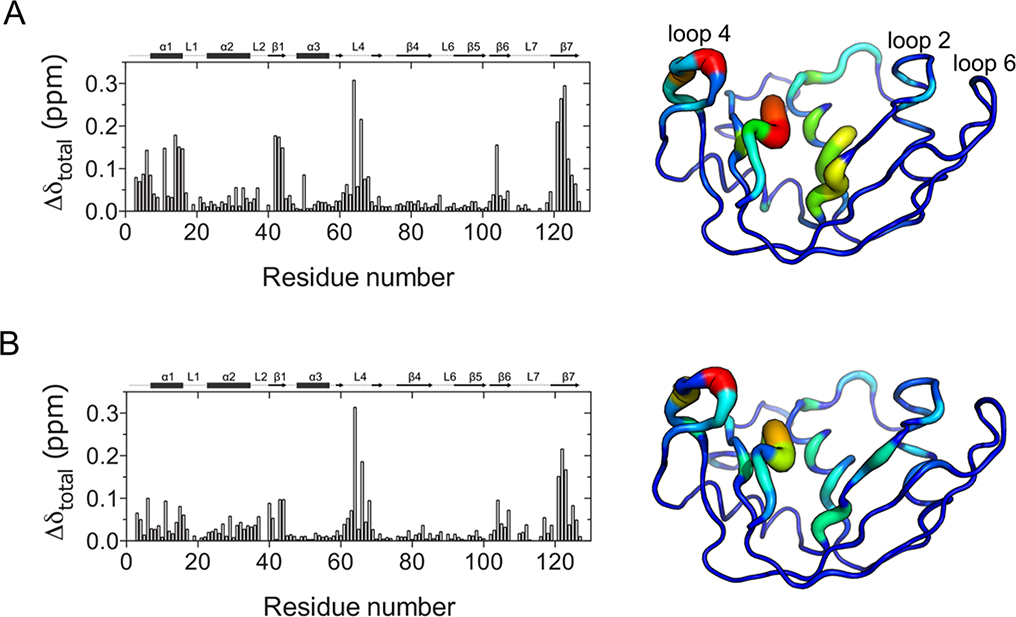
To identify residues perturbed by ligand binding, we performed NMR titrations on 15N-labeled RNase 6 using two ligands, 5’-AMP and d(ACACA), mimicking single-nucleotide product and deoxypentanucleotide substrate analogs, respectively. Figure 3 shows the mapping of compounded chemical shift changes experienced upon binding of 5’-AMP and d(ACACA) to RNase 6 on the three-dimensional structure of RNase 6 using a tube representation. Our results show that binding of both ligands induce large changes in loop 4 (residues 62–66 and 68) and the C-terminal β-strand (residues 121–126). Additional changes were observed in the N-terminal region (residues 3–20), residues 43–45 (β1 strand), and a number of loop 2 residues. Recent studies revealed a secondary active center in RNase 6 involved in its ribonucleolytic activity and located near residues His36, His39, and Lys878, 9. Our results show that binding of both ligands induced small yet notable changes near loop 2. We note that residues near the secondary active center (including His39) showed significant NMR line broadening, and thus ligand binding effects on these residues could not be determined.
Figure 3.
Effect of ligand binding to RNase 6. Compounded chemical shifts (Δδ) experienced upon binding of (A) 5’-AMP (single nucleotide product analog), and (B) d(ACACA) (deoxypentanucleotide substrate analog) to RNase 6 are plotted as a function of residue number. Δδ are calculated as the difference between the ligand-bound state (at the highest protein:ligand ratio) and the apo form using equation Δδ=((ΔδNH2 + (0.2 ΔδN)2)/2)½.42 Compounded chemical shifts are mapped on the 3D structure of RNase 6 to the right of the plots, with the blue and red ends of the spectrum corresponding to 0 and 0.3 ppm respectively. Thickness and blue-to-red spectrum coloring of the tube are used to represent chemical shift changes. Thicker tubes colored in the red end of the spectrum correspond to larger chemical shift changes, while thinner tubes colored in the blue end of the spectrum correspond to smaller chemical shift changes.
To determine the binding affinity of the 5’-AMP and d(ACACA) ligands to RNase 6, we calculated the dissociation constants (Kd) by fitting compounded chemical shift as a function of ligand concentration for each of the residues displaying Δδ > 0.05 ppm using equation 121. By calculating a shared value for all affected residues exhibiting straight resonance displacements typical of a fast exchange regime (Figure S6), we obtained a measured Kd = 154 ± 5 μM for 5’-AMP binding to RNase 6, similar to ITC values previously reported for RNase A with the same ligand (105 ± 1 μM30 and 124 ± 4 μM31). These values suggest similar properties for 5’-AMP binding to the B2 purine subsite of RNase 6 and RNase A. In contrast, Kd for the d(ACACA) analog was measured at 8 ± 2 μM, indicating strong affinity for this longer substrate analog. Since ligand concentrations required for NMR titration experiments are much higher than the calculated binding affinity of d(ACACA), in addition to the fact that simplified treatment of spectral titrations can be deceptive when only linear peak shifts are analyzed32, we also used microscale thermophoresis (MST) to measure binding affinities between RNase 6 and ligands 5’-AMP and d(ACACA) (Figure S7). Using this technique, we measured a Kd = 12 ± 10 μM for substrate analog d(ACACA), confirming the low micromolar affinity observed by NMR. However, MST experiments could not adequately reproduce the NMR and ITC binding affinities measured with product analog 5’-AMP, instead yielding a much lower affinity toward RNase 6 (Kd = 8.76 ± 4.2 mM). Although MST can overcome many shortcomings of many other biophysical techniques (namely with respect to protein sample concentration), it requires fluorescent labeling of the target protein. This labeling is performed using a fluorescent dye carrying a reactive NHS-ester group that primarily modifies amines on lysine residues33. Since Lys38 lies in the vicinity of the 5’-AMP binding sub-site in RNase 6, we believe that chemically modifying this lysine residue negatively impacts binding affinity of this mononucleotide product analog. In contrast, additional sub-site interactions conferred by the longer d(ACACA) substrate most likely overcome this detrimental effect observed with the single nucleotide. These results suggest that additional enzyme-ligand subsite interactions decrease binding entropy and contribute to increased binding affinity.
Conformational exchange experienced by RNase 6
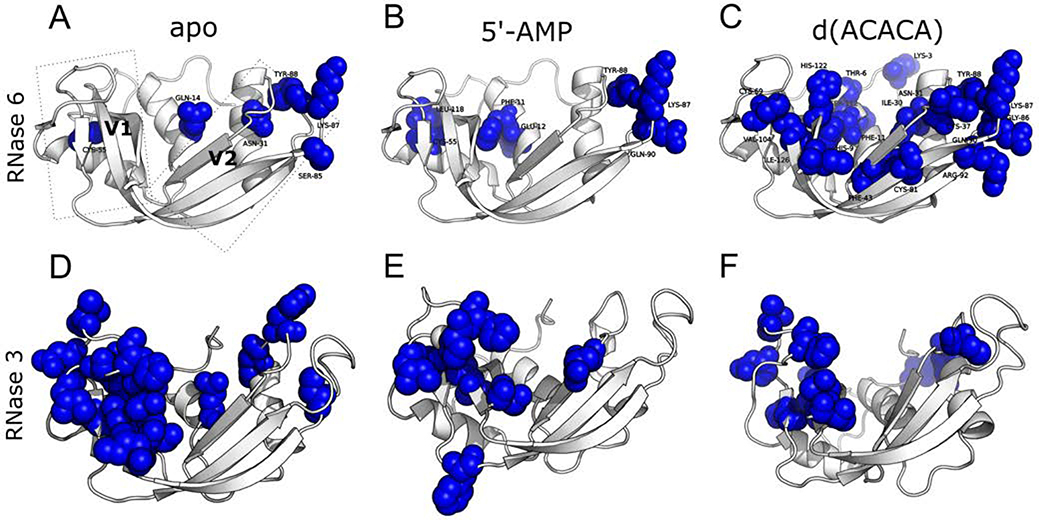
The role of conformational motions have been shown to be essential for enzyme function in a variety of systems34–36, including bovine RNase A37–40. Recent studies further revealed the conservation of dynamical patterns across homologous RNases grouped into phylogenetic subfamilies that share similar biological functions41. NMR relaxation dispersion and/or 15N-Chemical Exchange Saturation Transfer (15N-CEST) experiments performed on free, ligand-bound, and/or working enzyme states provide atomic-scale dynamic information on a per-residue basis, illuminating residues that experience conformational exchange on a timescale that correlates with enzyme function. These methodologies thus offer means to identify and allocate potential motional and/or allosteric network interactions experienced by an enzyme in its apo state and/or subjected to distinct functional perturbations (ligand binding, mutation, etc.)43. Here, we performed 15N-CPMG relaxation dispersion experiments to probe the overall conformational exchange experienced by RNase 6 on the catalytically relevant millisecond timescale for the apo and ligand-bound states (Figure 4A–C). Few residues exhibited conformational exchange on the millisecond timescale in the apo state (Figure 4A). These include residues primarily localized in the V2 domain (such as Gln14, Asn31) and residues within loop 6 (Ser85, Lys87, Tyr88). Interestingly, line-broadened residues Lys38 and His39 are also located in the vicinity of the exchanging residues of loop 6, suggesting a potential coupling of millisecond timescale motion between these exchanging residues and the two residues of the primary and secondary catalytic sites.
Figure 4.
Millisecond timescale conformational exchange experienced by RNase 6 and RNase 3 in the free and ligand-bound states. 15N-CPMG relaxation dispersion experiments were performed on the apo (A, D), 5’-AMP- (B, E) and d(ACACA)-bound (C, F) states of 15N-labeled RNase 6 and RNase 3. Residues identified as undergoing conformational exchange on the millisecond timescale (ΔR2,eff (1/τcp) > 1.5 s−1, as previously described44) are depicted using a space-filling representation. Representative relaxation dispersion curves for residues in the apo and ligand-bound states are shown in Figure S5.
Binding of the product analog 5’-AMP resulted in a modest gain of conformational exchange for Phe11, Glu12, Gln90, and Leu118, while a decrease in exchange was observed for Gln14 and Ser85 relative to the apo state (Figure 4B). However, binding of the substrate analog d(ACACA) led to a dramatic increase in the number and location of residues experiencing millisecond timescale exchange. These residues are located within and beyond the active site, and include the catalytic His122, α-helix 1 residues (Thr6, His9, Phe11), and residues from loop 2 (Cys37), loop 4 (Cys69), loop 6 (Gly86, Lys87, Tyr88), and loop 7 (Ser112) (Figure 4C). Interestingly, Lys87, identified as forming part of the secondary active center8, 9, experiences conformational exchange in the ligand-free and ligand-bound states, and was observed to display one of the highest B-factors outside the N-terminal tail in both crystal structures (Figure S1). In fact, the electron density of this side-chain could not be resolved, further highlighting the flexibility of this residue. A comparison of RNase 6 (Figure 4A–C) with RNase 3 (Figure 4D–F) and RNase 2 (Figure S3) showed the distinctly different localization of residues exhibiting conformational exchange. While RNases 2 and 3 predominantly show conformational exchange in the V1 domain, RNase 6 displays conformational exchange in the V2 domain.
Fast timescale motions experienced by RNase 6
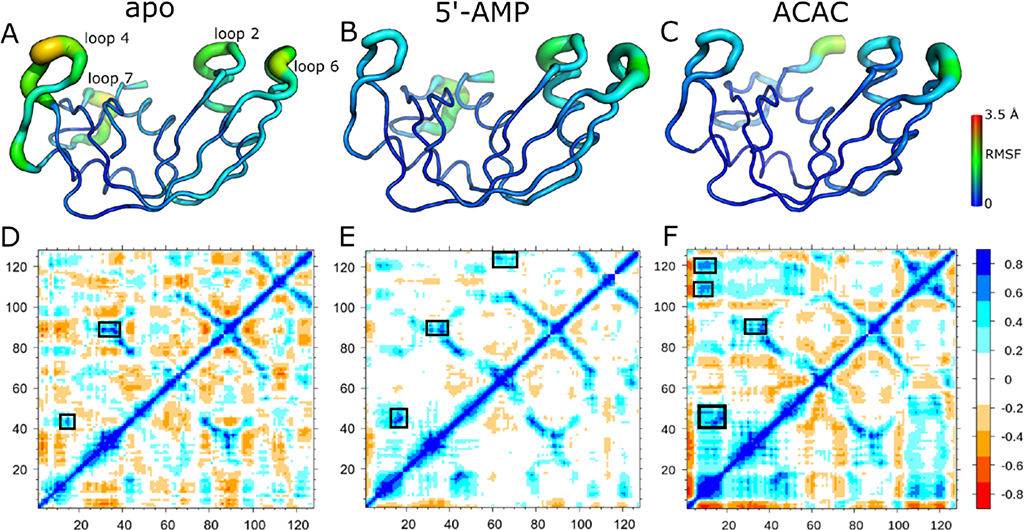
To complement the slow timescale conformational motions observed by NMR and to further characterize potential interactions involved in ligand binding and stabilization in this enzyme, we also performed nanosecond-to-microsecond timescale MD simulations of RNase 6 in its free and bound states (Figures 5 and S4). Although fast enzyme turnovers prevent experimental NMR investigation of conformational exchange using ribonucleotide substrates (hence the use of a non-hydrolyzable d(ACACA) analog), MD simulations allow for dynamical characterization using real RNA substrates. As a result, we modeled the RNase 6-ACAC complex based on a previously crystallized tetranucleotide-bound RNase A structure45. We characterized the dynamical properties of RNase 6 using root mean square fluctuations (RMSFs), which represent a measure of the average displacement of an atom relative to a reference structure. Previous studies demonstrated that the slowest ten modes (RMSF10) account for nearly 80% of the fluctuations observed in proteins, and allowed characterization of slower dynamics by removing faster stochastic motions in proteins46. A comparison of the top ten modes of the apo and ligand-bound RNase 6 showed notable changes upon ligand binding. The apo form of the enzyme showed the largest fluctuations for residues in loop 4 (residues 61–68) and loop 7 (residues 108–118) (Figure 5A). Binding of both substrate and product ligands led to reduced dynamics in loop 4 (Figure 5B–C). Substrate binding further led to reduced motions for residues in loop 7 (Figure 5C).
Figure 5.
Dynamical properties of RNase 6 on the nanosecond timescale. (A-C) Root mean square fluctuations (RMSFs) of the ten slowest modes are shown for apo (A), 5’-AMP (B), and ACAC (C) bound states of RNase 6. (D-F) Dynamical cross-correlation maps for apo (D), 5’-AMP (E), and ACAC (F) are shown with regions exhibiting relatively large (anti-)correlated motions highlighted using boxes. Positive (blue end of the spectrum) and negative (red end of the spectrum) values respectively correspond to correlated and anti-correlated motions observed between different regions of the protein. Residues of the N-terminal region, which showed large RMSFs (see Figure S4), are hidden for clarity.
To characterize the degree of correlation between the dynamics observed in the different regions of the enzyme, we constructed the dynamical cross-correlation maps (DCCMs) of RNase 6 in the free and ligand-bound states (Figure 5 D–F). Correlated motions (positive values) between two residues corresponds to motions in the same direction while anticorrelated motions (negative values) between residues indicates motions in opposite directions. The apo form of RNase 6 (Figure 5D) showed transient anticorrelated motions throughout the protein. Correlated motions were observed between residues near the catalytic His15 and residues 41–43, corresponding to the conserved (CKXX)NTF signature motif47. Additional correlated motions were observed between residues around loop 6 (residues 86–91) and loop 2 (residues 36–39), a region that encompasses residues forming the secondary active center8, 9. Interestingly, these correlated motions between residues around loop 6 and loop 2 were also observed upon binding of the two ligands (Figure 5E–F). Overall, binding of 5’-AMP led to fewer (anti)correlated motions in RNase 6. The ACAC-bound state showed transient correlated motions between different regions of the protein. In addition to correlated motions near loops 2 and 6, correlated motions were also observed between residues of α-helix 1 (residues 6–16), residues near loop 7 (105–110), and residues near the catalytic His122.
To characterize the changes in atomistic interactions upon ligand binding, we computed the average hydrogen bonding occupancy between the enzyme and ligand molecules. Hydrogen bonding occupancy was defined as the fraction of structures (in percentage) of the MD trajectory that display interactions within a distance less than 3.3 Å and an angle of 135° between the donor and acceptor atoms. Our results show that substrate ACAC is stable in the binding pocket of RNase 6 throughout the simulation, consistent with the higher affinity (smaller Kd) determined experimentally with ligand d(ACACA). High occupancies were observed between the adenine base in the B2 subsite and residues Gln14 (86% occupancy), Asn68 (85%), Trp10 (80%), while the cytidine in the B1 subsite showed moderate interactions with Leu123 (47%) and Thr43 (21%) of RNase 648. In contrast to the substrate, 5’-AMP did not exhibit stable interactions with residues of the B2 subsite and exited the binding pocket after 10 ns, suggesting a relatively lower affinity for this ligand, consistent with experimental observations. Interestingly, binding of both ligands led to changes in the intramolecular hydrogen bonding patterns in RNase 6. A comparison of the hydrogen bonding interactions of loop 4 residues in the apo and ligand-bound state showed formation of new intramolecular interactions between Cys62 and Arg65, and loss of interactions between Asn64 and Asp124 upon binding of both ligands.
Discussion
In this study, we characterized the structural and dynamical properties of human RNase 6 in the ligand-free and ligand-bound states. We report the crystal structure of phosphate-bound RNase 6 and the first crystal structure of a ligand-bound RNase 6, i.e. in the presence of mononucleotide product analog 5’-AMP. Our observations show that while RNase 6 preserves nucleotide preference in the B2 subsite and displays binding affinity in the same range as that reported for bovine RNase A, the interaction pattern of the product analog is distinct from that observed in other RNases (Table S1). The crystal structure of RNase 6 in the presence of phosphate ions is very similar to the sulphate and phosphate bound structures previously reported by Prats-Ejarque et al. (RMSD of 0.3 Å and 0.54 Å)8, 9. However, our structure does not show interactions of phosphate ions with residues of the recently identified secondary active center8, 9, nor do our NMR titrations identify residues from this secondary active center as affected by binding of either ligand. The reason for such lack of phosphate binding interaction in this region remains unclear, and may perhaps be attributed to differences in crystallization conditions and/or large motions observed for residues involved in binding with phosphate ions in this region (Figure 4). Further experiments are necessary to identify the factors contributing to this difference. However, the lack of binding to the secondary active site might be explained by its activity towards longer substrates identified by Prats-Ejarque et al.8. Indeed, given the position of this secondary catalytic triad on the surface of the enzyme rather than in a cleft or groove, it could be possible that this additional nucleotide-binding subsite cannot properly position and stabilize short ligands in this position. Thus, binding to the primary active site groove, along with the stabilizing interactions in the B1, B2, B-1, p-2, and p-3 subsites8, might be necessary for proper binding and positioning of a multiple-nucleotide ligand in the secondary active site, which would also explain why our short ligands did not affect residues of the secondary active site.
NMR characterization of RNase 6 revealed the presence of two distinct conformers of the apo enzyme in solution, corresponding to 77% and 23% of the population, respectively. Interestingly, residues exhibiting minor populations were predominantly localized in the V2 domain of RNase 6 (Figure S1). The presence of two separate peaks for a single residue on a {1H-15N}-HSQC is often due to slow exchange between two conformations occurring on the millisecond-to-second timescale. However, characterization of slower timescale motions using ZZ-exchange and 15N-CEST NMR experiments showed no detectable changes for any of the double-peaked residues observed on the {1H-15N}-HSQC. This does not necessarily suggest an absence of exchange, but rather that if these conformations are exchanging between themselves, they do so on a slower timescale. Alternatively, the required in vitro glutathione refolding protocol that was found to be critical to obtain stable folded samples of all human and bovine pancreatic-type RNases of this superfamily11, 40, 41, 49 could have reproducibly generated two subtly distinct yet stable RNase 6 folds that we were unable to separate using standard chromatography procedures. Analogously, we note that human RNase 5 (angiogenin) also displays two peaks for multiple residues on its {1H-15N}-HSQC50, possibly due to a similar phenomenon than the one we observe in RNase 6.
The role of conformational motions occurring over functionally relevant timescales to preserve optimal catalytic activity has been reported for a number of enzyme systems, including for the bovine RNase A structural homolog37–40. Recent studies also showed that conserved dynamical traits within three-dimensional regions of structurally similar RNases are evolutionarily conserved to preserve functional integrity44, while conservation of global dynamical patterns was shown to cluster into phylogenetic subfamilies41. Our results illustrate the unique conformational exchange properties experienced by RNase 6 in its free form, which primarily exhibits conformational exchange in the V2 domain (Figure 4A). This is in contrast to the most closely related human RNase 2 (Figure S3) and RNase 3 (Figure 4D), which primarily display conformational exchange in the V1 domain41, 51. Binding of the substrate analog led to major changes on the millisecond exchange profile experienced by RNase 6, suggesting a binding mechanism distinct from other related RNases. This is in contrast with human RNases 2 and 3, which displayed subtle changes while preserving the localization of residues exhibiting conformational exchange to the V1 domain (Figures 4 and S3). Millisecond timescale motions were observed in the vicinity of the primary and secondary active sites (Figure 4A–C). In addition to regions important for catalytic activity, RNase 6 residues involved in membrane binding and antimicrobial properties (such as Trp1)6 also exhibited significant motions within this region in the apo and ligand-bound states (Figures S1 and S4). These observations highlight a potential impact of the conformational dynamics in these regions on the biochemical and biological functions of RNase 6. Further experiments are necessary to characterize the potential impact of the unique structural, dynamical, and functional features of RNase 6 relative to its numerous structural homologs found in human and vertebrates.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tara Sprules of the Québec/Eastern Canada High Field NMR Facility (McGill University) for her technical assistance.
Funding Sources
This work was supported in part by a grant from NIGMS/NIH under award number R01GM105978 (to N.D. and P.K.A.) and a Discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) under award number RGPIN-2016-05557 (to N.D.). The authors also acknowledge the support from scholarship, fellowship and salary award programs, including the NSERC Canada Graduate Scholarships - Doctoral Program (D.N.B.), the Postdoctoral Fellowship of the Fondation Armand-Frappier (C.N.), the Postdoctoral Research Scholarship of the Fonds de recherche du Québec - Nature et technologies (FRQNT) (J.G.) and the Research Scholar Junior 2 Career award (number 32743) of Fonds de Recherche Québec - Santé (FRQS) (N.D.). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. XSEDE computing allocation were awarded to P.K.A. (MCB-180199 and MCB-190044). No competing financial interests have been declared.
Supporting Information: RNase interactions with ligands, B-factor values for free and ligand-bound RNase 6, Structural mapping of RNase 6 residues exhibiting two {1H-15N}-HSQC resonances, Conformational exchange experienced by free and ligand-bound RNase 6 and RNase 2, RMSFs of apo and ligand-bound RNase 6, Representative relaxation dispersion curves for free and ligand-bound RNase 6, Representative NMR titration curves for free and ligand-bound RNase 6, Representative MST graphs for binding of ligands to RNase 6.
ABBREVIATIONS
- RNase
Ribonuclease
- 5’-AMP
Adenosine 5’-monophosphate
- RNA
Ribonucleic acid
- NMR
Nuclear Magnetic Resonance
- CPMG
Carr-Purcell-Meiboom-Gill
- RMSF
Root Mean Square Fluctuation
- MST
Microscale Thermophoresis
Footnotes
Accession Codes
PDB entries 6MV6 and 6MV7.
REFERENCES
- [1].Sorrentino S (2010) The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins, FEBS Lett 584, 2194–2200. [DOI] [PubMed] [Google Scholar]
- [2].Boix E, Blanco JA, Nogues MV, and Moussaoui M (2013) Nucleotide binding architecture for secreted cytotoxic endoribonucleases, Biochimie 95, 1087–1097. [DOI] [PubMed] [Google Scholar]
- [3].Lu L, Li J, Moussaoui M, and Boix E (2018) Immune modulation by human secreted RNases at the extracellular space, Front Immunol 9, 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Becknell B, Eichler TE, Beceiro S, Li B, Easterling RS, Carpenter AR, James CL, McHugh KM, Hains DS, Partida-Sanchez S, and Spencer JD (2015) Ribonucleases 6 and 7 have antimicrobial function in the human and murine urinary tract, Kidney Int 87, 151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Koczera P, Martin L, Marx G, and Schuerholz T (2016) The ribonuclease A superfamily in humans: canonical RNases as the buttress of innate immunity, Int J Mol Sci 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pulido D, Arranz-Trullen J, Prats-Ejarque G, Velazquez D, Torrent M, Moussaoui M, and Boix E (2016) Insights into the antimicrobial mechanism of action of human RNase6: structural determinants for bacterial cell agglutination and membrane permeation, Int J Mol Sci 17, 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Christensen-Quick A, Lafferty M, Sun L, Marchionni L, DeVico A, and Garzino-Demo A (2016) Human Th17 cells lack HIV-inhibitory RNases and are highly permissive to productive HIV infection, J Virol 90, 7833–7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Prats-Ejarque G, Arranz-Trullen J, Blanco JA, Pulido D, Nogues MV, Moussaoui M, and Boix E (2016) The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement, Biochem J 473, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Prats-Ejarque G, Blanco JA, Salazar VA, Nogues VM, Moussaoui M, and Boix E (2019) Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates, Biochim Biophys Acta Gen Subj 1863, 105–117. [DOI] [PubMed] [Google Scholar]
- [10].Narayanan C, Bernard DN, Bafna K, Gagne D, Agarwal PK, and Doucet N (2018) Ligand-induced variations in structural and dynamical properties within an enzyme superfamily, Front Mol Biosci 5, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Boix E (2001) Eosinophil cationic protein, Methods Enzymol 341, 287–305. [DOI] [PubMed] [Google Scholar]
- [12].Otwinowski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode, Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- [13].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software, J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS (2011) Overview of the CCP4 suite and current developments, Acta Crystallogr D Biol Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, and Murshudov GN (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use, Acta Crystallogr D Biol Crystallogr 60, 2184–2195. [DOI] [PubMed] [Google Scholar]
- [16].Murshudov GN, Vagin AA, and Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method, Acta Crystallogr D Biol Crystallogr 53, 240–255. [DOI] [PubMed] [Google Scholar]
- [17].Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot, Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- [20].Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, and Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline, Proteins 59, 687–696. [DOI] [PubMed] [Google Scholar]
- [21].Williamson MP (2013) Using chemical shift perturbation to characterise ligand binding, Prog Nucl Magn Reson Spectrosc 73, 1–16. [DOI] [PubMed] [Google Scholar]
- [22].Lisi GP, and Loria JP (2016) Using NMR spectroscopy to elucidate the role of molecular motions in enzyme function, Prog Nucl Magn Reson Spectrosc 92–93, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vallurupalli P, Bouvignies G, and Kay LE (2012) Studying “invisible” excited protein states in slow exchange with a major state conformation, J Am Chem Soc 134, 8148–8161. [DOI] [PubMed] [Google Scholar]
- [24].Farrow NA, Zhang O, Forman-Kay JD, and Kay LE (1994) A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium, J Biomol NMR 4, 727–734. [DOI] [PubMed] [Google Scholar]
- [25].Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham I, T.E., Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej B, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, and Kollman PA (2014) AMBER 14, University of California, San Francisco. [Google Scholar]
- [26].Ramanathan A, and Agarwal PK (2011) Evolutionarily conserved linkage between enzyme fold, flexibility, and catalysis, PLoS Biol 9, e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, and Walker RC (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald, J Chem Theory Comput 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- [28].Roe DR, and Cheatham TE 3rd. (2013) PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data, J Chem Theory Comput 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- [29].delCardayre SB, and Raines RT (1994) Structural determinants of enzymatic processivity, Biochemistry 33, 6031–6037. [DOI] [PubMed] [Google Scholar]
- [30].Gagné D, French RL, Narayanan C, Simonovic M, Agarwal PK, and Doucet N (2015) Perturbation of the conformational dynamics of an active-site loop alters enzyme activity, Structure 23, 2256–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Doucet N, Jayasundera TB, Simonovic M, and Loria JP (2010) The crystal structure of ribonuclease A in complex with thymidine-3’-monophosphate provides further insight into ligand binding, Proteins 78, 2459–2468. [DOI] [PubMed] [Google Scholar]
- [32].Kovrigin EL (2012) NMR line shapes and multi-state binding equilibria, J Biomol NMR 53, 257–270.22610542 [Google Scholar]
- [33].Selvaraj N, Kedage V, and Hollenhorst PC (2015) Comparison of MAPK specificity across the ETS transcription factor family identifies a high-affinity ERK interaction required for ERG function in prostate cells, Cell Commun Signal 13, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guo J, and Zhou HX (2016) Protein Allostery and conformational dynamics, Chem Rev 116, 6503–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lisi GP, and Loria JP (2017) Allostery in enzyme catalysis, Curr Opin Struct Biol 47, 123–130. [DOI] [PubMed] [Google Scholar]
- [36].Narayanan C, Bernard DN, and Doucet N (2016) Role of conformational motions in enzyme function: selected methodologies and case studies, Catalysts 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gagné D, and Doucet N (2013) Structural and functional importance of local and global conformational fluctuations in the RNase A superfamily, FEBS J 280, 5596–5607. [DOI] [PubMed] [Google Scholar]
- [38].Watt ED, Shimada H, Kovrigin EL, and Loria JP (2007) The mechanism of rate-limiting motions in enzyme function, Proc Natl Acad Sci U S A 104, 11981–11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Doucet N, Khirich G, Kovrigin EL, and Loria JP (2011) Alteration of hydrogen bonding in the vicinity of histidine 48 disrupts millisecond motions in RNase A, Biochemistry 50, 1723–1730. [DOI] [PubMed] [Google Scholar]
- [40].Doucet N, Watt ED, and Loria JP (2009) The flexibility of a distant loop modulates active site motion and product release in ribonuclease A, Biochemistry 48, 7160–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Narayanan C, Bernard DN, Bafna K, Gagne D, Chennubhotla CS, Doucet N, and Agarwal PK (2018) Conservation of dynamics associated with biological function in an enzyme superfamily, Structure 26, 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Grzesiek S, Stahl SJ, Wingfield PT, and Bax A (1996) The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR, Biochemistry 35, 10256–10261. [DOI] [PubMed] [Google Scholar]
- [43].Sekhar A, and Kay LE (2019) An NMR view of protein dynamics in health and disease, Annu Rev Biophys 48, 297–319. [DOI] [PubMed] [Google Scholar]
- [44].Narayanan C, Gagne D, Reynolds KA, and Doucet N (2017) Conserved amino acid networks modulate discrete functional properties in an enzyme superfamily, Sci Rep 7, 3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fontecilla-Camps JC, de Llorens R, le Du MH, and Cuchillo CM (1994) Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition, J Biol Chem 269, 21526–21531. [DOI] [PubMed] [Google Scholar]
- [46].Ramanathan A, and Agarwal PK (2009) Computational identification of slow conformational fluctuations in proteins, J Phys Chem B 113, 16669–16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Rosenberg HF, Zhang J, Liao YD, and Dyer KD (2001) Rapid diversification of RNase A superfamily ribonucleases from the bullfrog, Rana catesbeiana, J Mol Evol 53, 31–38. [DOI] [PubMed] [Google Scholar]
- [48].Bafna K, Narayanan C, Chennubhotla SC, Doucet N, and Agarwal PK (2019) Nucleotide substrate binding characterization in human pancreatic-type ribonucleases, PLoS One 14, e0220037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].delCardayre SB, Ribo M, Yokel EM, Quirk DJ, Rutter WJ, and Raines RT (1995) Engineering ribonuclease A: production, purification and characterization of wild-type enzyme and mutants at Gln11, Protein Eng 8, 261–273. [DOI] [PubMed] [Google Scholar]
- [50].Lequin O, Thuring H, Robin M, and Lallemand JY (1997) Three-dimensional solution structure of human angiogenin determined by 1H,15N-NMR spectroscopy--characterization of histidine protonation states and pKa values, Eur J Biochem 250, 712–726. [DOI] [PubMed] [Google Scholar]
- [51].Gagné D, Charest LA, Morin S, Kovrigin EL, and Doucet N (2012) Conservation of flexible residue clusters among structural and functional enzyme homologues, J Biol Chem 287, 44289–44300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.