Qiu et al. present a novel deep learning strategy to generate high-resolution visualizations of Alzheimer’s disease risk in humans that are highly interpretable and can accurately predict Alzheimer’s disease status. They then test the model by comparing its performance to that of neurologists and neuropathological data.
Keywords: dementia, biomarkers, Alzheimer’s disease, structural MRI, neurodegeneration
Abstract
Alzheimer’s disease is the primary cause of dementia worldwide, with an increasing morbidity burden that may outstrip diagnosis and management capacity as the population ages. Current methods integrate patient history, neuropsychological testing and MRI to identify likely cases, yet effective practices remain variably applied and lacking in sensitivity and specificity. Here we report an interpretable deep learning strategy that delineates unique Alzheimer’s disease signatures from multimodal inputs of MRI, age, gender, and Mini-Mental State Examination score. Our framework linked a fully convolutional network, which constructs high resolution maps of disease probability from local brain structure to a multilayer perceptron and generates precise, intuitive visualization of individual Alzheimer’s disease risk en route to accurate diagnosis. The model was trained using clinically diagnosed Alzheimer’s disease and cognitively normal subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset (n = 417) and validated on three independent cohorts: the Australian Imaging, Biomarker and Lifestyle Flagship Study of Ageing (AIBL) (n = 382), the Framingham Heart Study (n = 102), and the National Alzheimer’s Coordinating Center (NACC) (n = 582). Performance of the model that used the multimodal inputs was consistent across datasets, with mean area under curve values of 0.996, 0.974, 0.876 and 0.954 for the ADNI study, AIBL, Framingham Heart Study and NACC datasets, respectively. Moreover, our approach exceeded the diagnostic performance of a multi-institutional team of practicing neurologists (n = 11), and high-risk cerebral regions predicted by the model closely tracked post-mortem histopathological findings. This framework provides a clinically adaptable strategy for using routinely available imaging techniques such as MRI to generate nuanced neuroimaging signatures for Alzheimer’s disease diagnosis, as well as a generalizable approach for linking deep learning to pathophysiological processes in human disease.
Introduction
Millions worldwide continue to suffer from Alzheimer’s disease (Scheltens et al., 2016), while attempts to develop effective disease-modifying treatments remain stalled. Though tremendous progress has been made towards detecting Alzheimer’s disease pathology using CSF biomarkers (Frisoni et al., 2010; Jack et al., 2013; Harper et al., 2014), as well as PET amyloid (Nordberg, 2004; Bohnen et al., 2012), and tau imaging (Mattsson et al., 2019; Ossenkoppele et al., 2019), these modalities often remain limited to research contexts. Instead, current standards of diagnosis depend on highly skilled neurologists to conduct an examination that includes inquiry of patient history, an objective cognitive assessment such as bedside Mini-Mental State Examination (MMSE) or neuropsychological testing (McKhann et al., 2011), and a structural MRI to rule in findings suggestive of Alzheimer’s disease (Frisoni et al., 2010). Clinicopathological studies suggest the diagnostic sensitivity of clinicians ranges between 70.9% and 87.3% and specificity between 44.3% and 70.8% (Beach et al., 2012). While MRIs reveal characteristic cerebral changes noted in Alzheimer’s disease such as hippocampal and parietal lobe atrophy (Whitwell et al., 2012), these characteristics are considered to lack specificity for imaging-based Alzheimer’s disease diagnosis (van de Pol et al., 2006; Barkhof et al., 2007; Raji et al., 2009; Frisoni et al., 2010). Given this relatively imprecise diagnostic landscape, as well as the invasive nature of CSF and PET diagnostics and a paucity of clinicians with sufficient Alzheimer’s disease diagnostic expertise, advanced machine learning paradigms such as deep learning (LeCun et al., 2015; Hinton, 2018; Topol, 2019), offer ways to derive high accuracy predictions from MRI data collected within the bounds of neurology practice.
Recent studies have demonstrated the application of deep learning approaches such as convolutional neural networks (CNNs) for MRI and multimodal data-based classification of cognitive status (Qiu et al., 2018). Despite the promising results, these models have yet to achieve full integration into clinical practice for several reasons. First, there is a lack of external validation of deep learning algorithms since most models are trained and tested on a single cohort. Second, there is a growing notion in the biomedical community that deep learning models are ‘black-box’ algorithms (Castelvecchi, 2016). In other words, although deep learning models demonstrate high accuracy classification across a broad spectrum of disease, they neither elucidate the underlying diagnostic decisions nor indicate the input features associated with the output predictions. Lastly, given the uncertain onset and heterogeneity of symptoms seen in Alzheimer’s disease, a computerized individual-level characterization of Alzheimer’s disease remains unresolved. Considering these factors, we surmise that the clinical potential of deep learning is attenuated by a lack of external validation of single cohort-driven models, and an increasing use of opaque decision-making frameworks. Thus, overcoming these challenges is not only crucial to harness the potential of deep learning algorithms to improve patient care, but to also pave the way for explainable evidence-based machine learning in the medical imaging community. To address these limitations, we developed a novel deep learning framework that links a fully convolutional network (FCN) to a traditional multilayer perceptron (MLP) to generate high-resolution visualizations of Alzheimer’s disease risk that can then be used for accurate predictions of Alzheimer’s disease status (Fig. 1). Four distinct datasets were chosen for model development and validation: Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset, Australian Imaging, Biomarker and Lifestyle Flagship Study of Ageing (AIBL), Framingham Heart Study (FHS), and National Alzheimer’s Coordinating Center (NACC) (Table 1 and Supplementary Fig. 1). Association of model predictions with neuropathological findings along with a head-to-head comparison of the model performance with a team of neurologists underscored the validity of the deep learning framework.
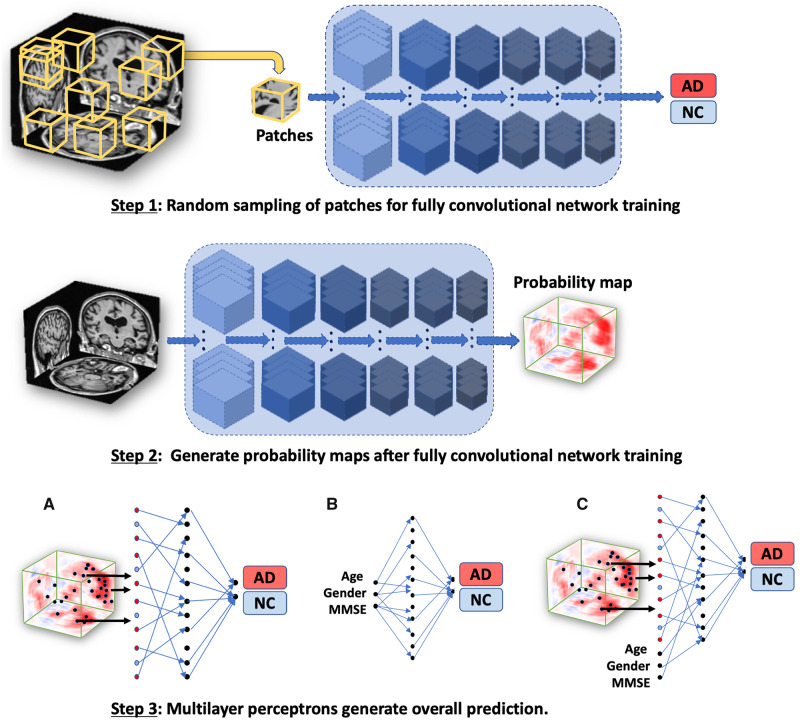
Figure 1.
Schematic of the deep learning framework. The FCN model was developed using a patch-based strategy in which randomly selected samples (sub-volumes of size 47 × 47 × 47 voxels) of T1-weighted full MRI volumes were passed to the model for training (Step 1). The corresponding Alzheimer’s disease status of the individual served as the output for the classification model. Given that the operation of FCNs is independent of input data size, the model led to the generation of participant-specific disease probability maps of the brain (Step 2). Selected voxels of high-risk from the disease probability maps were then passed to the MLP for binary classification of disease status (Model A in Step 3; MRI model). As a further control, we used only the non-imaging features including age, gender and MMSE and developed an MLP model to classify individuals with Alzheimer’s disease and the ones with normal cognition (Model B in Step 3; non-imaging model). We also developed another model that integrated multimodal input data including the selected voxels of high-risk disease probability maps alongside age, gender and MMSE score to perform binary classification of Alzheimer’s disease status (Model C in Step 3; Fusion model). AD = Alzheimer’s disease; NC = normal cognition.
Table 1.
Study population and characteristics
| Dataset | ADNI |
AIBL |
FHS |
NACC |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic | NC | AD | P-value | NC | AD | P-value | NC | AD | P-value | NC | AD | P-value |
| (n = 229) | (n = 188) | (n = 320) | (n = 62) | (n = 73) | (n = 29) | (n = 356) | (n = 209) | |||||
| Age, years, median [range] | 76 [60, 90] | 76 [55, 91] | 0.4185 | 72 [60, 92] | 73 [55, 93] | 0.5395 | 73 [57, 100] | 81 [67, 94] | <0.0001 | 74 [56, 94] | 77 [55, 95] | 0.0332 |
| Education, years, median [range] | 16 [6, 20] | 16 [4, 20] | <0.0001 | NAa | NAa | NA | 14 [8, 25] | 13 [5, 25] | 0.3835 | 16b [0, 22] | 14.5b [2, 24] | 0.8363 |
| Gender, male (%) | 119 (51.96) | 101 (53.72) | 0.7677 | 144 (45.00) | 24 (38.71) | 0.40 | 37 (50.68) | 12 (41.38) | 0.51 | 126 (35.39) | 95 (45.45) | 0.0203 |
| MMSE, median [range] | 29 [25, 30] | 23.5 [18, 28] | <0.0001 | 29 [25, 30] | 21 [6, 28] | <0.0001 | 29c [22, 30] | 25c [10, 29] | <0.0001 | 29c [20, 30] | 22c [0, 30] | <0.0001 |
| APOE4, positive (%) | 61 (26.65) | 124 (65.97) | <0.0001 | 11 (3.44) | 12 (19.35) | <0.0001 | 13 (17.81) | 11d (40.74) | 0.035 | 102 (28.65) | 112 (53.59) | <0.0001 |
Four independent datasets were used for this study including: the ADNI dataset, the AIBL, the FHS, and the NACC. The ADNI dataset was randomly split in the ratio of 3:1:1, where 60% of it was used for model training, 20% of the data were used for internal validation and the rest was used for internal testing. The best performing model on the validation dataset was selected for making predictions on the ADNI test data as well as on the AIBL, FHS and NACC datasets, which served as external test datasets for model validation. All the MRI scans considered for this study were performed on individuals within ±6 months from the date of clinical diagnosis. AD = Alzheimer’s disease; NA = not available; NC = normal cognition.
Years of education not available for all AIBL study participants.
Years of education not available for some study participants.
MMSE scores not available for some subjects in the study cohort within 6 months of diagnosis.
APOE4 (genetic) information not available for some subjects in the study cohort.
Materials and methods
Study participants and data collection
Data from ADNI, AIBL, FHS, and NACC cohorts were used in the study (Table 1 and Supplementary Fig. 1). ADNI is a longitudinal multicentre study designed to develop clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of Alzheimer’s disease (Petersen et al., 2010). AIBL, launched in 2006, is the largest study of its kind in Australia and aims to discover biomarkers, cognitive characteristics, and lifestyle factors that influence the development of symptomatic Alzheimer’s disease (Ellis et al., 2010). The FHS is a longitudinal community cohort study and has collected a broad spectrum of clinical data from three generations (Massaro et al., 2004). Since 1976, the FHS expanded to evaluate factors contributing to cognitive decline, dementia, and Alzheimer’s disease. Finally, the NACC, established in 1999, maintains a large relational database of standardized clinical and neuropathological research data collected from Alzheimer’s disease centres across the USA (Beekly et al., 2004).
Model training, internal validation and testing were performed on the ADNI dataset. Following training and internal testing on the ADNI data, we validated the predictions on AIBL, FHS, and NACC. The criterion for selection included individuals aged ≥55 years, with 1.5 T, T1-weighted MRI scans taken within ±6 months from the date of clinically confirmed diagnosis of Alzheimer’s disease or normal cognition (Supplementary Fig. 1). We excluded cases including Alzheimer’s disease with mixed dementia, non-Alzheimer’s disease dementias, history of severe traumatic brain injury, severe depression, stroke, and brain tumours, as well as incident major systemic illnesses. Note that this inclusion and exclusion criterion was adapted from the baseline recruitment protocol developed by the ADNI study (Petersen et al., 2010), and to maintain consistency, the same criterion was applied to other cohorts as applicable. This led to the selection of 417 individuals from the ADNI cohort, 382 individuals from AIBL, 102 FHS participants, and 565 individuals from the NACC cohort. If an individual had multiple MRI scans taken within the time window, then we selected the scan closest to the date of clinical diagnosis. For most of these selected cases, age, gender and MMSE score were available.
Algorithm development
An FCN was designed to input a registered volumetric MRI scan of size 181 × 217 × 181 voxels and output the Alzheimer’s disease class probability at every location. We used a novel, computationally efficient patch-wise training strategy to train the FCN model (Fig. 1). This process involved random sampling of 3000 volumetric patches of size 47 × 47 × 47 voxels from each training subject’s MRI scan and used this information to predict the output of interest (Supplementary Fig. 2). The size of the patches was the same as the receptive field of the FCN.
The FCN consists of six convolutional blocks (Supplementary Table 1). The first four convolutional blocks consist of a 3D convolutional layer followed by the following operations: 3D max pooling, 3D batch-normalization, Leaky Relu and Dropout. The last two convolutional layers function as dense layers in terms of the classification task and these two layers play a key role in boosting model efficiency (Shelhamer et al., 2017). The network was trained de novo with random initialization of the weights. We used the Adam optimizer with a 0.0001 learning rate and a mini-batch size of 10. During the training process, the model was saved when it achieved the lowest error on the ADNI validation dataset. After FCN training, single volumetric MRI scans were forwarded to obtain complete arrays of disease probabilities that we refer to as disease probability maps. Once trained, the process of obtaining disease probability maps from test cases took ∼1 s on an NVIDIA GTX Titan GPU.
The FCN was trained by repeated application to cuboidal patches of voxels randomly-sampled from a full volume of sequential MRI slices. Because the convolutions decrease the size of the input across successive layers of the network, the size of each patch was selected such that the shape of the final output from each patch was equal to 2 × 1 × 1 × 1 (Supplementary Table 1); i.e. the application of the FCN to each patch during training produced a list of two scalar values. These values can be converted to respective Alzheimer’s disease and normal cognition probabilities by application of a softmax function, and the greater of the two probabilities was then used for classification of disease status. In this way, the model was trained to infer local patterns of cerebral structure that suggested an overall disease state.
After generating disease probability maps for all subjects, an MLP model framework was developed to perform binary classification to predict Alzheimer’s disease status by selecting Alzheimer’s disease probability values from the disease probability maps. This selection was based on observation of the overall performance of the FCN classifier as estimated using the Matthew’s correlation coefficient values on the ADNI training data. Specifically, we selected disease probability map voxels from 200 fixed locations that were indicated to have high Matthew’s correlation coefficient values (Supplementary Table 2). The features extracted from these locations served as input to the MLP model that performed binary classification of Alzheimer’s disease status (MRI model in Fig. 1, Step 3). Two additional MLP models were developed where one model used age, gender and MMSE score values as input to predict Alzheimer’s disease status (non-imaging model in Fig. 1, Step 3), and the other MLP took the 200 features along with age, gender and MMSE score as input to predict Alzheimer’s disease status (Fusion model in Fig. 1, Step 3). All the MLP models comprised a single hidden layer and an output layer (Supplementary Table 3). The MLP models also included non-linear operators such as ReLu and Dropout.
Image registration, intensity normalization and volumetric MRI segmentation
The MRI scans from all the datasets were obtained in NIFTI format. We used the MNI152 template (ICBM 2009c Nonlinear Symmetric template, McGill University, Canada) to register all the scans. We used the FLIRT tool available within the FSL package (Wellcome Center, University of Oxford, UK), to align the scans with respect to the MNI152 template. A careful manual review of the registered images revealed that the automatic registration was done reasonably well on a large majority of the ADNI, AIBL and NACC cases. For cases that were not registered well (mainly within FHS), we performed affine transformations to perform manual registration using known regions as landmarks. Given that there may not be a registration method that would work for all MRI scans, our two-step process resulted in a reasonable set of registered images.
After image registration, we normalized intensities of all the voxels [mean = 0 and standard deviation (SD) = 1]. We then adjusted the intensity of these voxels and other outliers by clipping them to the range: [−1, 2.5], where any voxel with intensity lower than −1 was assigned a value of −1, and a voxel with intensity higher than 2.5 was assigned a value of 2.5. We then performed background removal where all the voxels from background regions outside of the skull were set to −1 to ensure uniform background intensity.
Cortical and subcortical structures from volumetric MRI scans of 11 individuals from the FHS cohort, with brain autopsies, were segmented using FreeSurfer (Fischl, 2012). In-built functions such as ‘recon-all’, ‘mri_annotation2label’, ‘tkregister2’, ‘mri_label2vol’, ‘mri_convert’ and ‘mris_calc’ were used to obtain the segmented structures.
Neuropathological validation
We validated the FCN model’s ability to identify regions of high Alzheimer’s disease risk by overlapping the predicted brain regions with post-mortem findings. Eleven individuals from the FHS dataset had histopathological evaluations of autopsied brains, and four individuals out of the 11 had confirmed Alzheimer’s disease. A blinded assessment to all demographic and clinical information was conducted during the neuropathological evaluation. Detailed descriptions of the neuropathological evaluation have been previously reported (Au et al., 2012). For this study, we examined the density of neurofibrillary tangles, diffuse senile, neuritic or compacted senile plaques, from paraffin-embedded sections extracted within the cortical and subcortical regions. The sections were stained using Bielschowsky silver stain. Immunocytochemistry was performed for phosphorylated tau protein (Innogenetics, AT8, 1:2000) and amyloid-β protein (Dako, 6F-3D, 1:500, pretreated in 90% formic acid for 2 min). The maximum density of neurofibrillary tangles per 200× field was assessed semi-quantitatively and scores ranging from 1 to 4 were assigned (1+: 1 neurofibrillary tangle/field; 2+: 2–5 neurofibrillary tangles/field; 3+: 6–9 neurofibrillary tangles/field; and 4+: ≥10 neurofibrillary tangles/field). Similarly, diffuse senile, and neuritic or compacted senile plaques were examined in a 100× microscopic field and rated separately with scores ranging between 1 and 4 (1+: 1–9 plaques/field; 2+: 10–19/field, 3+: 20–32/field, and 4+: >32/field). The final determinations were made by averaging the count in three microscopic fields. The density of neurofibrillary tangles, diffuse senile, and neuritic or compacted senile plaques in each brain region were qualitatively compared with the model’s Alzheimer’s disease probability in that region.
Neurologist-level validation
Nine US board-certified practicing neurologists and two non-US practicing neurologists (all referred to as neurologists) were asked to provide a diagnostic impression (Alzheimer’s disease versus normal cognition) of 80 randomly selected cases from the ADNI dataset that were not used for model training. For each case, the neurologists were provided with full volumetric, T1-weighted MRI scan, subject’s age, gender and their MMSE score for evaluation. The same parameters were used for training the model (Fusion model in Fig. 1). To obtain estimates of how the deep learning model compared to an average neurologist, the characteristics of neurologist performance were averaged across the neurologists who individually evaluated each test case. More details on the neurologist approach to the ratings can be found in the Supplementary material.
Convolutional neural network model development
A 3D CNN was created to perform classification of Alzheimer’s disease and normal cognition cases and its results compared with the FCN model. The CNN model was trained, validated and tested on the same split of data that were used for the FCN model. To facilitate direct comparison with the FCN model, one CNN model was developed using the MRI data alone, as well as an additional MLP that included the CNN model-derived features along with age, gender and MMSE score. Similar to the FCN-MLP model, we merged the CNN-based imaging features (i.e. feature vector after the first dense layer of the CNN) and non-imaging features for MLP training.
The CNN model consisted of four convolutional layers followed by two dense layers (Supplementary Fig. 3 and Supplementary Table 4). Each convolution layer was followed by ReLu activations. Max-pooling layers between the convolution blocks were used to down-sample the feature maps. Batch normalization, Leaky ReLu, and dropout were applied after each convolutional layer. Dropout and Leaky ReLu were applied on the feature vectors of the dense layers. Softmax was used on the final dense layer. The CNN model was trained from scratch with the same optimizer and loss function as the FCN model. We used a learning rate of 0.0001 and mini-batch size of six. The CNN model with the best performance on the ADNI validation dataset was used to predict Alzheimer’s disease status on the test datasets.
Random forests model
Derived MRI measures (n = 117), available from the MRI SPM voxel-based morphometry analysis table in the ADNI dataset were used as inputs to construct a random forests (RF) classifier to predict the Alzheimer’s disease status. The random forests model construction was repeated 10 times using different random seeds, and the average model performance was reported.
Performance metrics
The models were constructed on the ADNI data, which was randomly divided into three groups for training, validation and testing, respectively. The models were built on each training and validation split, and the performance on the test datasets (ADNI test, AIBL, FHS and NACC) were evaluated, and this process was repeated five times. Performance was presented as mean and standard deviation over the model runs. The scans from the ADNI testing dataset were used for the head-to-head comparison with the neurologists.
We generated sensitivity-specificity and precision-recall curves based on model predictions on the ADNI test data as well as on the other independent datasets (AIBL, FHS and NACC). For each sensitivity-specificity and precision-recall curve, we also computed the area under curve (AUC) values. Additionally, we computed sensitivity, specificity, F1-score and Matthews correlation coefficient on each set of model predictions. The F1-score considers both precision and recall of a test and is defined as:
| (1) |
Here, TP denotes true positive values, and FP and FN denote false-positive and false-negative cases, respectively. Matthew’s correlation coefficient (MCC) is a balanced measure of quality for dataset classes of different sizes of a binary classifier and defined as follows:
| (2) |
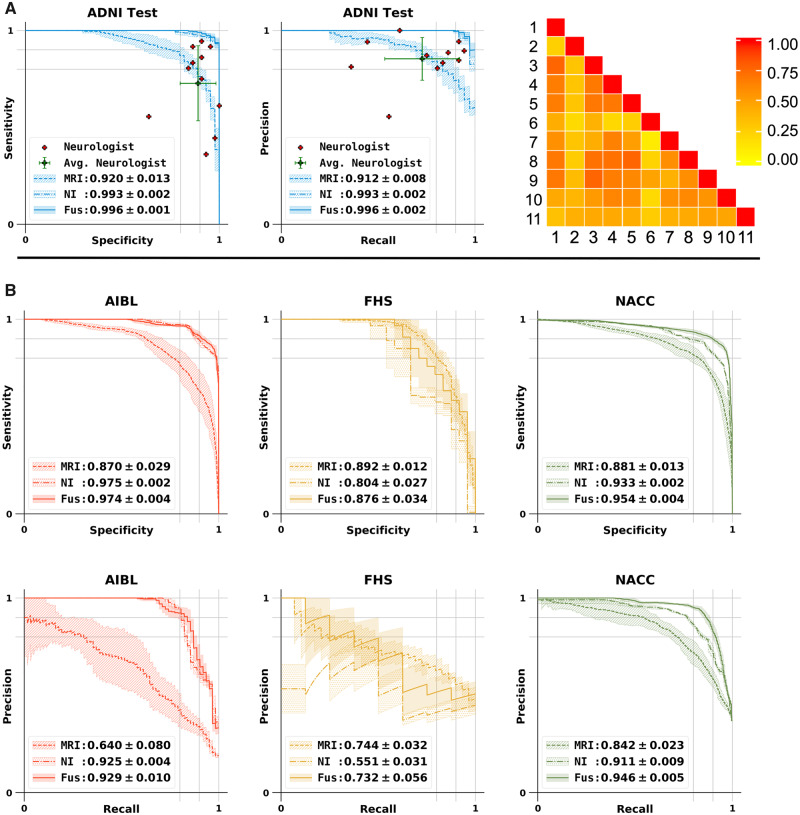
The TN denotes true negative values. We also calculated inter-annotator agreement using Cohen’s kappa (κ), as the ratio of the number of times two annotators agreed on a diagnosis. The κ-statistic measures inter-rater agreement for categorical items. A κ-score of 1 indicates perfect agreement between the annotators. Average pairwise κ was computed that provided an overall measure of agreement between the neurologists.
Statistical analysis
To assess the overall significant levels of differences between normal cognition and Alzheimer’s disease groups, two-sample t-test and the χ2 test were used for continuous and categorical variables, respectively. The FCN model’s ability to identify regions of high Alzheimer’s disease risk was evaluated by overlapping the disease probability maps with post-mortem histopathological findings. A subset of 11 individuals from the FHS study sample had undergone brain autopsy and were used for the analysis. In these participants, the locations and frequencies of amyloid-β and tau pathologies, semi-quantitatively reported by neuropathologists, were associated with high-Alzheimer’s disease risk regions. The Spearman’s rank correlation coefficient test was used to determine the strength and direction (negative or positive) of the relationship between these regional Alzheimer’s disease probabilities and pathology scores.
Data availability
Python scripts and sample data are made available on GitHub (https://github.com/vkola-lab/brain2020).
Results
Our deep learning pipeline can link an FCN to an MLP to predict Alzheimer’s disease status directly from MRI data or from a combination of MRI data and readily available non-imaging data (Fig. 1). The FCN portion of the framework generated high-resolution visualizations of overall Alzheimer’s disease risk in individuals as a function of local cerebral morphology. We refer to these visualizations as disease probability maps. The MLP then used the disease probability maps directly (MRI model in Fig. 1), or a set of non-imaging features such as age, gender and MMSE score (non-imaging model in Fig. 1), or a multimodal input data comprising disease probability maps, MMSE score, age and gender (fusion model in Fig. 1), to accurately predict Alzheimer’s disease status across four independent cohorts (Table 1). We chose these known Alzheimer’s disease risk factors because they can be easily obtained by non-Alzheimer’s disease specialists. The FCN was trained to predict disease probability from randomly selected patches (sub-volumes) of pixels sampled from the full MRI volume (Fig. 1 and Supplementary Table 1). Given that this type of network accepts input of arbitrary size, application of the sub-volumetrically trained FCN could then be used to construct high resolution disease probability maps without the need to redundantly decompose full-sized test images.
Rapid processing of individual MRI volumes generated volumetric distributions of local Alzheimer’s disease probabilities in the brains of affected and unaffected individuals, respectively (Fig. 2). To assess the anatomical consistency of Alzheimer’s disease-suggestive morphology hot spots derived from these distributions, population-wide maps of Matthew’s correlation coefficient were constructed. This mapping enabled identification of areas from which correct predictions of disease status were most frequently derived (Fig. 3), thus acting as a means to demonstrate structures most affected by neuropathological changes in Alzheimer’s disease.
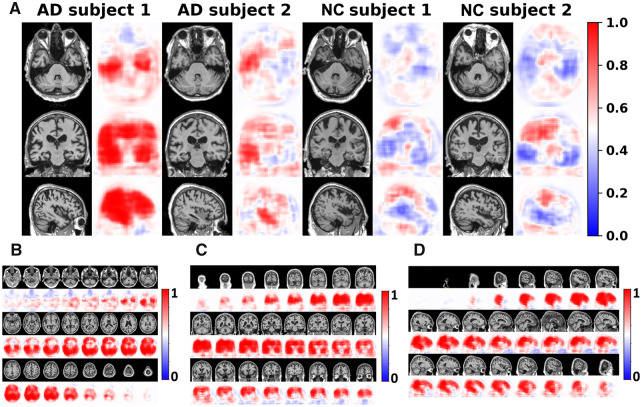
Figure 2.
Subject-specific disease probability maps. (A) Disease probability maps generated by the FCN model highlight high-risk brain regions that are associated with Alzheimer’s disease pathology. Individual cases are shown where the blue colour indicates low-risk and red indicates high-risk of Alzheimer’s disease. The first two individuals were clinically confirmed to have normal cognition whereas the other two individuals had clinical diagnosis of Alzheimer’s disease. (B–D) Axial, coronal and sagittal stacks of disease probability maps from a single subject with clinically confirmed Alzheimer’s disease are shown. All imaging planes were used to construct 3D disease probability maps. Red colour indicates locally inferred probability of Alzheimer’s disease >0.5, whereas blue indicates <0.5. AD = Alzheimer’s disease; NC = normal cognition.
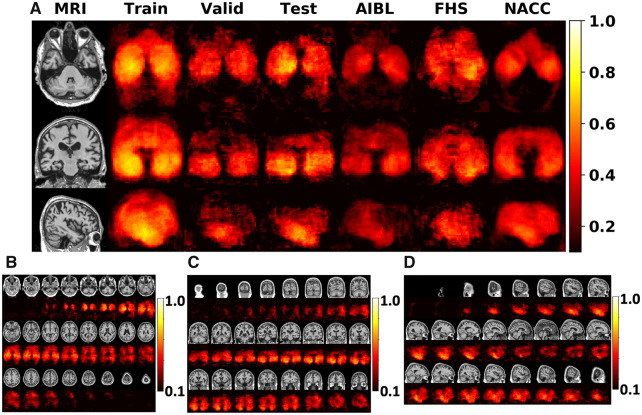
Figure 3.
Summary of the FCN model performance. (A) Voxel-wise maps of Matthew’s correlation coefficient (MCC) were computed independently across all the datasets to demonstrate predictive performance derived from all regions within the brain. (B–D) Axial, coronal and sagittal stacks of the MCC maps at each cross-section from a single subject, are shown. These maps were generated by averaging the MCC values on the ADNI test data.
As confirmation, average regional probabilities extracted from selected segmented brain regions (Fig. 4), were highly associated with Alzheimer’s disease positive findings reported in post-mortem neuropathology examinations. Specifically, these regions correlated with the locations and numerical frequency of amyloid-β and tau pathologies reported in available autopsy reports from the FHS dataset (n = 11) (Supplementary Table 5). Post-mortem data indicated that, in addition to predicting higher region-specific Alzheimer’s disease probabilities in individuals with disease compared to those without, proteinopathies were more frequent in cerebral regions implicated by the model in Alzheimer’s disease (Fig. 4). Model-predicted regions of high Alzheimer’s disease risk overlapped with the segmented regions that were indicated to have high localized deposition of amyloid-β and tau. Additionally, predicted Alzheimer’s disease risk within these zones increased with pathology scores. Given that these post-mortem findings are definitive in terms of confirming Alzheimer’s disease, these physical findings grounded our computational predictions in biological evidence.
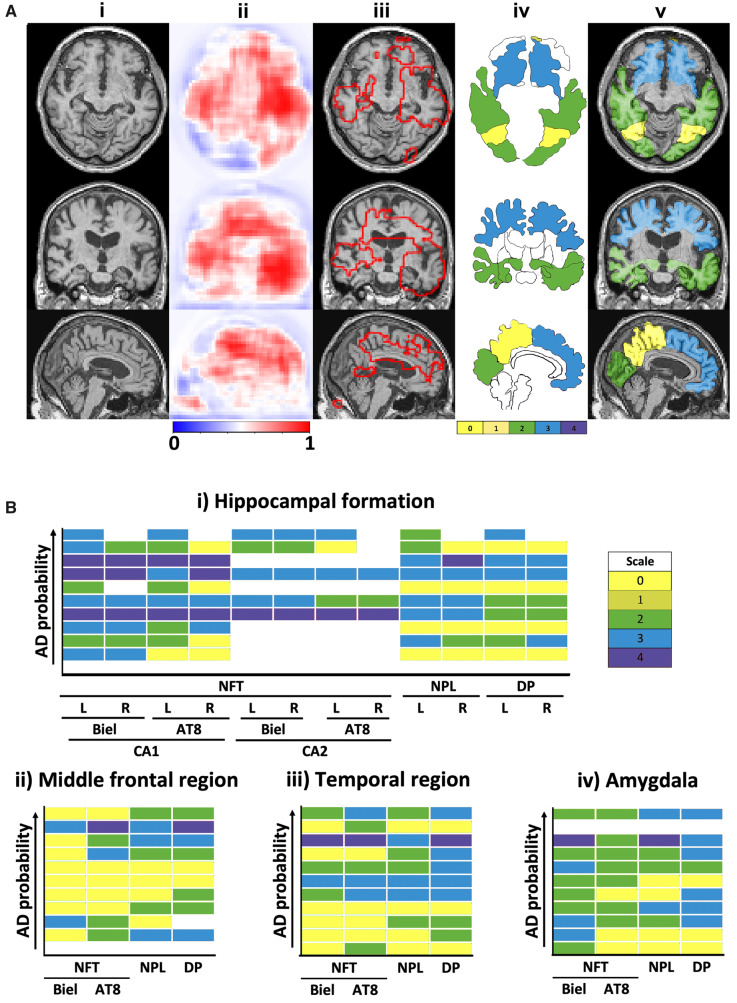
Figure 4.
Correlation of model findings with neuropathology. (A) Overlap of model predicted regions of high Alzheimer’s disease risk with post-mortem findings of Alzheimer’s disease pathology in a single subject. This subject had clinically confirmed Alzheimer’s disease with affected regions including the bilateral asymmetrical temporal lobes and the right-side hippocampus, the cingulate cortex, the corpus callosum, part of the parietal lobe and the frontal lobe. The first column (i) shows MRI slices in three different planes followed by a column (ii), which shows corresponding model predicted disease probability maps. A cut-off value of 0.7 was chosen to delineate the regions of high Alzheimer’s disease risk and overlapped with the MRI scan in the next column (iii). The next column (iv), depicts a segmented mask of cortical and subcortical structures of the brain obtained from FreeSurfer (Fischl, 2012). A sequential colour-coding scheme denotes different levels of pathology ranging from green (0, low) to pale red (4, high). The final column (v), shows the overlay of the magnetic resonance scan, disease probability maps of high Alzheimer’s disease risk and the colour-coded regions based on pathology grade. (B) We then qualitatively assessed trends of neuropathological findings from the FHS dataset (n = 11). The same colour-coding scheme as described above was used to represent the pathology grade (0–4) in the heat maps. The boxes coloured in ‘white’ in the heat maps indicate missing data. Using the Spearman’s Rank correlation coefficient test, an increasing Alzheimer’s disease probability risk was associated with a higher grade of amyloid-β and tau accumulation, in the hippocampal formation, the middle frontal region, the amygdala and the temporal region, respectively. Biel = Bielschowsky stain; L = left; R = right.
Furthermore, disease probability maps provided an information-dense feature that yielded sensitive and specific binary predictions of Alzheimer’s disease status when passed independently to the MLP portion of the framework (MRI model in Fig. 5A and B). An MLP trained using just the non-imaging features such as age, gender and MMSE score also was predictive of Alzheimer’s disease status (non-imaging model in Fig. 5A and B). Model performance was further improved by expanding the MLP input to include disease probability maps, gender, age, and MMSE score (fusion model in Fig. 5A and B). When other non-imaging features such as APOE status were included, model performance slightly improved (Supplementary Fig. 4 and Supplementary Table 6). Given the proportionality between age and global cerebral atrophy (van de Pol et al., 2006; Raji et al., 2009), addition of non-imaging variables at the MLP stage also allowed us to control for the natural progression of cerebral morphological changes over the lifespan.
Figure 5.
Performance of the MLP model for Alzheimer’s disease classification and model comparison with neurologists. (A) Sensitivity-specificity and precision-recall curves showing the sensitivity, the true positive rate, versus specificity, the true negative rate, calculated on the ADNI test set. Individual neurologist performance is indicated by the red plus symbol and averaged neurologist performance along with the error bars is indicated by the green plus symbol on both the sensitivity-specificity and precision-recall curves on the ADNI test data. Visual description of pairwise Cohen’s kappa (κ), which denotes the inter-operator agreement between all the 11 neurologists is also shown. (B) Sensitivity-specificity and PR curves calculated on the AIBL, FHS and NACC datasets, respectively. For all cases, model A indicates the performance of the MLP model that used MRI data as the sole input, model B is the MLP model with non-imaging features as input and model C indicates the MLP model that used MRI data along with age, gender and MMSE values as the inputs for binary classification.
We also compared performance of the deep learning models against an international group of clinical neurologists recruited to provide impressions of disease status from a randomly sampled cohort of ADNI participants whose MRI, MMSE score, age and gender were provided. The performance of the neurologists (Fig. 5A), indicated variability across different clinical practices, with a moderate inter-rater agreement as assessed by pairwise kappa (κ) scoring (Fig. 5A; average κ = 0.493 ± 0.16). Interestingly, we noted that the deep learning model that was based on MRI data alone (MRI model; accuracy: 0.834 ± 0.020; Table 2), outperformed the average neurologist (accuracy: 0.823 ± 0.094; Supplementary Table 7). When age, gender and MMSE information were added to the model, then the performance increased significantly (fusion model; accuracy: 0.968 ± 0.014; Table 2).
Table 2.
Performance of the deep learning models
| Accuracy | Sensitivity | Specificity | F1-score | MCC | |
|---|---|---|---|---|---|
| MRI | |||||
| ADNI test | 0.834 ± 0.020 | 0.767 ± 0.036 | 0.889 ± 0.030 | 0.806 ± 0.024 | 0.666 ± 0.042 |
| AIBL | 0.870 ± 0.022 | 0.594 ± 0.119 | 0.924 ± 0.025 | 0.593 ± 0.088 | 0.520 ± 0.095 |
| FHS | 0.766 ± 0.064 | 0.901 ± 0.096 | 0.712 ± 0.123 | 0.692 ± 0.044 | 0.571 ± 0.056 |
| NACC | 0.818 ± 0.033 | 0.764 ± 0.031 | 0.849 ± 0.052 | 0.757 ± 0.033 | 0.613 ± 0.059 |
| Non-imaging | |||||
| ADNI test | 0.957 ± 0.010 | 0.924 ± 0.019 | 0.983 ± 0.032 | 0.951 ± 0.010 | 0.915 ± 0.020 |
| AIBL | 0.915 ± 0.022 | 0.872 ± 0.037 | 0.923 ± 0.034 | 0.772 ± 0.035 | 0.731 ± 0.035 |
| FHS | 0.760 ± 0.042 | 0.517 ± 0.043 | 0.842 ± 0.068 | 0.512 ± 0.026 | 0.367 ± 0.053 |
| NACC | 0.854 ± 0.021 | 0.881 ± 0.013 | 0.838 ± 0.041 | 0.817 ± 0.019 | 0.703 ± 0.033 |
| Fusion | |||||
| ADNI test | 0.968 ± 0.014 | 0.957 ± 0.014 | 0.977 ± 0.031 | 0.965 ± 0.014 | 0.937 ± 0.026 |
| AIBL | 0.932 ± 0.031 | 0.877 ± 0.032 | 0.943 ± 0.042 | 0.814 ± 0.054 | 0.780 ± 0.059 |
| FHS | 0.792 ± 0.039 | 0.742 ± 0.185 | 0.808 ± 0.082 | 0.633 ± 0.076 | 0.517 ± 0.098 |
| NACC | 0.852 ± 0.037 | 0.924 ± 0.025 | 0.810 ± 0.068 | 0.824 ± 0.032 | 0.714 ± 0.053 |
Three models were constructed for explicit performance comparison. The MRI model predicted Alzheimer’s disease status based upon imaging features derived from the patch-wise trained FCN. The non-imaging model consisted of an MLP that processed non-imaging clinical variables (age, gender, MMSE). The fusion model appended the clinical variables used by the MRI model to the MLP portion of the non-imaging model in order to form a multimodal imaging/non-imaging input. Accuracy, sensitivity, specificity, F1-score, and Matthew’s correlation coefficient (MCC) are demonstrated for each. The fusion model was found to outperform the other models in nearly all metrics in each of the four datasets. Of interest, however, we noted that the performance of the MRI model and the non-imaging model still displayed higher specificity and sensitivity than many of the human neurologists, all of whom used the full suite of available data sources to arrive at an impression.
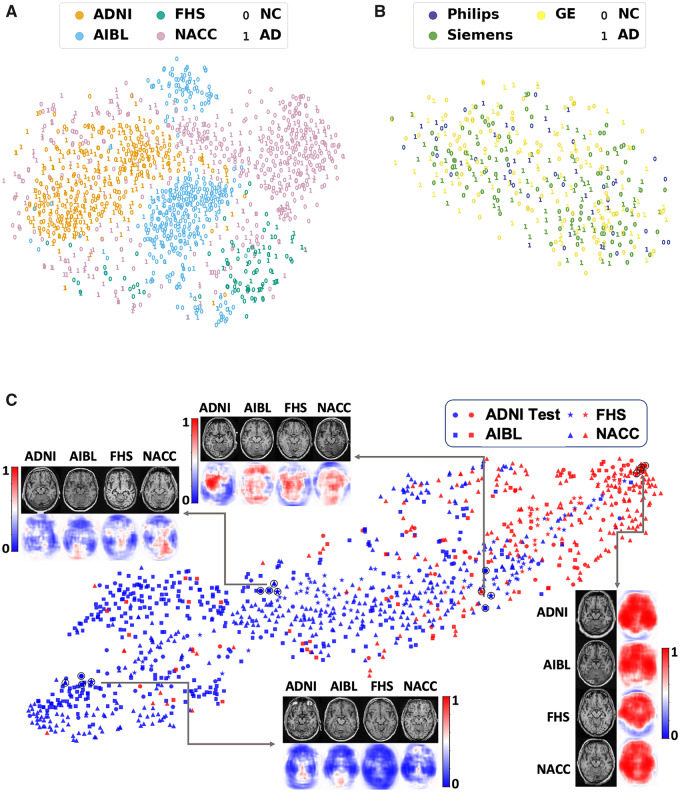
Consistent, high classification performance of the deep learning model across the external datasets was confirmed using other metrics (Table 2). We performed t-distributed stochastic neighbour embedding (t-SNE) (van der Maaten and Hinton, 2008), on the volumetric MRI scans using the intensity values as inputs from all the four datasets. The t-SNE method takes high-dimensional data and creates a low-dimensional representation of those data, so that it can be easily visualized. While the t-SNE plot resulted in site-specific clustering of the scans (Fig. 6A), intra-site distribution of cases revealed no clear differentiation between Alzheimer’s disease and normal cognition cases. This observation underscores a rationale for utilizing a supervised learning strategy to predict Alzheimer’s disease status using MRI scan data alone. We believe this is a strength of our study because despite site-specific differences, the FCN model was able to generalize well on the external datasets. We then used scanner-specific info from the ADNI cohort and generated another t-SNE visualization, which also revealed no discernible clustering of Alzheimer’s disease or normal cognition cases (Fig. 6B). This implies that any potential scanner-specific differences may not have influenced the model training process. Further, we examined the model performance visually by respective clustering of Alzheimer’s disease and normal cognition cases in a t-SNE, which used features before the final hidden layer of the MLP (Fig. 6C).
Figure 6.
Visualization of data. (A) Voxel-level MRI intensity values from all four datasets (ADNI, AIBL, FHS and NACC) were used as inputs and a two-dimensional plot was generated using t-SNE, a method for visualizing high-dimensional data. The colour in the plot represents the site and the digit ‘0’ was used to present cases who had normal cognition (NC) and the digit ‘1’ was used to show cases who had confirmed Alzheimer’s disease (AD). (B) This t-SNE plot was generated only on using the ADNI dataset, where the colour was used to represent the scanner. The digit ‘0’ was used for normal cognition cases and ‘1’ for Alzheimer’s disease cases. (C) FCN-based outputs that served as input features to the MLP model were embedded in a two-dimensional plot generated using t-SNE for the two classes (Alzheimer’s disease and normal cognition). The colour (blue versus red) was used to distinguish normal cognition from Alzheimer’s disease cases, whereas a unique symbol shape was used to represent individuals derived from the same cohort. Several individual cases that were clinically confirmed to have Alzheimer’s disease or normal cognition are also shown (indicated as a black circle overlying the respective datapoint). The plot also indicates co-localization of subjects in the feature space based on the disease state and not on the dataset of origin.
It is worth noting that our strategy represents a significant increase in computational efficiency over a traditional CNN approach to the same task (Step 1 in Fig. 1 versus Supplementary Fig. 5). Given fixed dense layer dimensions, generation of disease probability maps from traditional CNNs requires not only sub-volumetric training, but also sub-volumetric application to full-sized MRI volumes (Supplementary Table 8 versus Table 2), obligating repeated computations in order to calculate local probabilities of disease status. By circumventing this rigidity, our approach readily generates disease probability maps (Fig. 1, Step 2), which can be integrated with multimodal clinical data for Alzheimer’s disease diagnosis (Fig. 1, Step 3). As such, this work extends recently reported efforts to abstract visual representations of disease risk directly from medical images (Coudray et al., 2018), and also represents an application of FCNs to disease classification tasks as opposed to semantic segmentation (Shelhamer et al., 2017). Additionally, the FCN model performed at the same level as a traditional CNN model with fully connected layers in predicting Alzheimer’s disease status, and this result was consistent across all the datasets (Supplementary Fig. 5 and Supplementary Table 8). Of note, the FCN model outperformed a traditional machine learning model that was constructed using derived MRI features (Supplementary Fig. 6 and Supplementary Table 9).
Discussion
Our deep learning framework links a fully convolutional network to a multilayer perceptron and generates high resolution disease probability maps for neurologist-level diagnostic accuracy of Alzheimer’s disease status. The intuitive local probabilities outputted by our model are readily interpretable, thus contributing to the growing movement towards explainable artificial intelligence in medicine, and deriving an individualized phenotype of insidious disease from conventional diagnostic tools. Indeed, the disease probability maps provide a means for tracking conspicuous brain regions implicated in Alzheimer’s disease during diagnosis. We then aggregated disease probability maps across the entire cohort to demonstrate population-level differences in neuroanatomical risk mapping of Alzheimer’s disease and normal cognition cases. Critically, by the standards of several different metrics, our model displayed good predictive performance, yielding high and consistent values on all the test datasets. Such consistency between cohorts featuring broad variance in MRI protocol, geographic location, and recruitment criteria, suggests a strong degree of generalizability. Thus, these findings demonstrate innovation at the nexus of medicine and computing, simultaneously contributing new insights to the field of computer vision while also expanding the scope of biomedical applications of neural networks.
Disease probability maps were created by element-wise application of a softmax function to the final array of activations generated by the FCN. This step enabled the conversion of abstract tensor encodings of neuroanatomical information to probability arrays demonstrating the likelihood of Alzheimer’s disease at different locations in the brain given their local geometry. Alternatively put, the model develops a granular conceptualization of Alzheimer’s disease-suggestive morphologies throughout the brain, and then uses this learning information in test cases to assess the probability of Alzheimer’s disease-related pathophysiological processes occurring at each region. The simple presentation of these probabilities as a coherent colour map displayed alongside traditional neuroimaging thus allows a point-by-point prediction of where disease-related changes are likely to be present (Fig. 4). Recent work has also demonstrated effective differentiation of Alzheimer’s disease and normal cognition cases using a patch-based sampling algorithm (Lu et al., 2018), but is limited by simultaneous reliance on MRI and fluorodeoxyglucose PET as well as a model whose inputs are computed as scalar averages of intensities from multi-voxel cerebral loci. Furthermore, we believe that the broader notion of disease process mapping with deep learning has the potential to be applied in many fields of medicine. The simple presentation of disease risk as a coherent colour map overlaid on traditional imaging modalities aids interpretability. This is in contrast to saliency mapping strategies that highlight certain pixels based only on their utility to the internal functioning of a network (Shelhamer et al., 2017), as well as methods that highlight penultimate-layer activation values (Lu et al., 2018). Consequently, informative anatomical information is abstracted and lost. Our work builds upon such advances by requiring just a single imaging modality en route to mapping an array of raw pixel values to a disease probability map that isomorphically preserves neuroanatomical information.
While traditional deep neural networks such as a CNN with fully connected layers require an input of fixed size, FCNs are capable of acting on inputs of arbitrary size. This is potentially useful in datasets where heterogeneously-sized scans can be processed without training separate classifiers for scans of each size. Moreover, FCNs can efficiently process volumetric scans, because their fully convolutional nature allows them to evaluate multiple patches simultaneously. This does not imply that the FCNs enforce global structure into the individual patch-level predictions. Rather, the generated disease probability maps lead to a contiguous volumetric interpretation denoting high probability regions of Alzheimer’s disease risk.
Certainly, limitations to the current study must be acknowledged. We considered a case-control population in which two subpopulations were chosen in advance that were either cognitively normal or have the diagnosis (Alzheimer’s disease). This scenario is not exactly representative of the standard clinical decision-making process faced by the neurologist. Patients often present with a set of symptoms and results from standard neurological testing that are indicative of a spectrum of neurodegenerative disease as opposed to a binary scenario. Therefore, our method is not directly applicable in its current state but serves as a first step towards building a more comprehensive framework to characterize multiple aetiologies of neurodegeneration. Of note, the non-imaging data-based models performed better on AIBL and NACC data, while the MRI-based model performed better on the FHS data. As such, the MMSE value was a key element in the study criteria for ADNI, AIBL and NACC, and this may explain why the non-imaging data-based model performed better on these datasets. Because the FHS is a community cohort, it served as a relatively unbiased dataset for model validation. Despite this study selection, our FCN model can associate MRI changes with regional neuropathology, and provides compelling evidence that the use of an imaging biomarker alone can accurately assess Alzheimer’s disease status. We acknowledge that the CNN model with fully connected layers used in this study is based on a specific architecture and one could design CNN models that may even outperform the FCN models, at least in terms of test accuracy. Nonetheless, the ability for the FCN model to generate interpretable disease probability maps makes it more appealing than using CNN models with fully connected layers for predicting Alzheimer’s disease status.
Our approach has significant translational potential beyond Alzheimer’s disease diagnosis. Indeed, the tissue-level changes predicted by our model suggest the prospect of directly highlighting areas of pathophysiology across a spectrum of disease. It may be of interest in future studies to determine whether the well-defined pattern of high-risk findings from the currently presented framework may follow regions of interest from PET scans. In such cases, our model may aid in non-invasive monitoring of Alzheimer’s disease development.
In conclusion, our deep learning framework was able to obtain high accuracy Alzheimer’s disease classification signatures from MRI data, and our model was validated against data from independent cohorts, neuropathological findings and expert-driven assessment. If confirmed in clinical settings, this approach has the potential to expand the scope of neuroimaging techniques for disease detection and management. Further validation could lead to improved care and outcomes compared with current neurological assessment, as the search for disease-modifying therapies continues.
Supplementary Material
Acknowledgements
The authors thank the ADNI, AIBL, FHS and NACC investigators for providing access to the data.
Glossary
- ADNI =
Alzheimer’s Disease Neuroimaging Initiative
- AIBL =
Australian Imaging, Biomarker and Lifestyle Flagship Study of Ageing
- CNN =
convolutional neural network
- FCN =
fully convolutional network
- FHS =
Framingham Heart Study
- MLP =
multilayer perceptron
- MMSE =
Mini-Mental State Examination
- NACC =
National Alzheimer’s Coordinating Center
- t-SNE =
t-distributed stochastic neighbour embedding
Funding
This project was supported in part by the National Center for Advancing Translational Sciences, National Institutes of Health, through BU-CTSI Grant (1UL1TR001430), a Scientist Development Grant (17SDG33670323) from the American Heart Association, and a Hariri Research Award from the Hariri Institute for Computing and Computational Science & Engineering at Boston University, Framingham Heart Study’s National Heart, Lung and Blood Institute contract (N01-HC-25195; HHSN268201500001I) and NIH grants (R56-AG062109, AG008122, R01-AG016495, and R01-AG033040). Additional support was provided by Boston University’s Affinity Research Collaboratives program and Boston University Alzheimer’s Disease Center (P30-AG013846).
Competing interests
The authors report no competing interests.
References
- Au R, Seshadri S, Knox K, Beiser A, Himali JJ, Cabral HJ, et al. The Framingham Brain Donation Program: neuropathology along the cognitive continuum. Curr Alzheimer Res 2012; 9: 673–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhof F, Polvikoski TM, van Straaten EC, Kalaria RN, Sulkava R, Aronen HJ, et al. The significance of medial temporal lobe atrophy: a postmortem MRI study in the very old. Neurology 2007; 69: 1521–7. [DOI] [PubMed] [Google Scholar]
- Beach TG, Monsell SE, Phillips LE, Kukull W.. Accuracy of the clinical diagnosis of Alzheimer disease at National Institute on Aging Alzheimer Disease Centers, 2005-2010. J Neuropathol Exp Neurol 2012; 71: 266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekly DL, Ramos EM, van Belle G, Deitrich W, Clark AD, Jacka ME, et al. The National Alzheimer’s Coordinating Center (NACC) Database: an Alzheimer disease database. Alzheimer Dis Assoc Disord 2004; 18: 270–7. [PubMed] [Google Scholar]
- Bohnen NI, Djang DS, Herholz K, Anzai Y, Minoshima S.. Effectiveness and safety of 18F-FDG PET in the evaluation of dementia: a review of the recent literature. J Nucl Med 2012; 53: 59–71. [DOI] [PubMed] [Google Scholar]
- Castelvecchi D. Can we open the black box of AI? Nature 2016; 538: 20–3. [DOI] [PubMed] [Google Scholar]
- Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyo D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med 2018; 24: 1559–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis KA, Rowe CC, Villemagne VL, Martins RN, Masters CL, Salvado O, et al. Addressing population aging and Alzheimer’s disease through the Australian imaging biomarkers and lifestyle study: collaboration with the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement 2010; 6: 291–6. [DOI] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. Neuroimage 2012; 62: 774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR Jr, Scheltens P, Thompson PM.. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol 2010; 6: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L, Barkhof F, Scheltens P, Schott JM, Fox NC.. An algorithmic approach to structural imaging in dementia. J Neurol Neurosurg Psychiatry 2014; 85: 692–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton G. Deep learning-a technology with the potential to transform health care. JAMA 2018; 320: 1101–2. [DOI] [PubMed] [Google Scholar]
- Jack CR Jr, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013; 12: 207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G.. Deep learning. Nature 2015; 521: 436–44. [DOI] [PubMed] [Google Scholar]
- Lu D, Popuri K, Ding GW, Balachandar R, Beg MF.. Alzheimer’s disease neuroimaging I. Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural MR and FDG-PET images. Sci Rep 2018; 8: 5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaro JM, D’Agostino RB Sr, Sullivan LM, Beiser A, DeCarli C, Au R, et al. Managing and analysing data from a large-scale study on Framingham Offspring relating brain structure to cognitive function. Stat Med 2004; 23: 351–67. [DOI] [PubMed] [Google Scholar]
- Mattsson N, Insel PS, Donohue M, Jogi J, Ossenkoppele R, Olsson T, et al. Predicting diagnosis and cognition with (18)F-AV-1451 tau PET and structural MRI in Alzheimer’s disease. Alzheimers Dement 2019; 15: 570–80. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011; 7: 263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg A. PET imaging of amyloid in Alzheimer’s disease. Lancet Neurol 2004; 3: 519–27. [DOI] [PubMed] [Google Scholar]
- Ossenkoppele R, Smith R, Ohlsson T, Strandberg O, Mattsson N, Insel PS, et al. Associations between tau, Abeta, and cortical thickness with cognition in Alzheimer disease. Neurology 2019; 92: e601–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 2010; 74: 201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Chang GH, Panagia M, Gopal DM, Au R, Kolachalama VB.. Fusion of deep learning models of MRI scans, Mini-Mental State Examination, and logical memory test enhances diagnosis of mild cognitive impairment. Alzheimer's Dementia 2018; 10: 737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji CA, Lopez OL, Kuller LH, Carmichael OT, Becker JT.. Age, Alzheimer disease, and brain structure. Neurology 2009; 73: 1899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, et al. Alzheimer’s disease. Lancet 2016; 388: 505–17. [DOI] [PubMed] [Google Scholar]
- Shelhamer E, Long J, Darrell T. . Convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 2017; 39: 640–51. [DOI] [PubMed] [Google Scholar]
- Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019; 25: 44–56. [DOI] [PubMed] [Google Scholar]
- van de Pol LA, Hensel A, Barkhof F, Gertz HJ, Scheltens P, van der Flier WM.. Hippocampal atrophy in Alzheimer disease: age matters. Neurology 2006; 66: 236–8. [DOI] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G.. Visualizing data using t-SNE. Mach Learn 2008; 9: 2579–605. [Google Scholar]
- Whitwell JL, Dickson DW, Murray ME, Weigand SD, Tosakulwong N, Senjem ML, et al. Neuroimaging correlates of pathologically defined subtypes of Alzheimer’s disease: a case-control study. Lancet Neurol 2012; 11: 868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Python scripts and sample data are made available on GitHub (https://github.com/vkola-lab/brain2020).