Abstract
Many obligate intracellular apicomplexan parasites have adapted a distinct invasion mechanism involving a close interaction between the parasite ligands and the sialic acid (SA) receptor. We found that sialic acid binding protein-1 (SABP1), localized on the outer membrane of the zoonotic parasite Toxoplasma gondii, readily binds to sialic acid on the host cell surface. The binding was sensitive to neuraminidase treatment. Cells preincubated with recombinant SABP1 protein resisted parasite invasion in vitro. The parasite lost its invasion capacity and animal infectivity after the SABP1 gene was deleted, whereas complementation of the SABP1 gene restored the virulence of the knockout strain. These data establish the critical role of SABP1 in the invasion process of T. gondii. The previously uncharacterized protein, SABP1, facilitated T. gondii attachment and invasion via sialic acid receptors.
Keywords: Toxoplasma gondii, sialic acid, receptor, ligand, attachment, invasion
A Toxoplasma gondii surface protein TgSABP1 was identified as a ligand of sialic acid receptor. Recombinant SABP1 and SABP1-specific antibodies blocked T. gondii invasion into host cells. Parasites with TgSABP1 gene deletion lost their invasion capability.
Apicomplexan parasites cause a number of severe diseases, including malaria and toxoplasmosis. Toxoplasma gondii, the causative agent of toxoplasmosis, can infect all warm-blooded mammals, including humans [1]. Invasion into a host cell is a key step for T. gondii proliferation and dissemination throughout host tissues [2]. It is initiated by the attachment of parasites that utilize gliding motility driven by glidesomes to slide over the cell surface seeking a suitable site for invasion [3]. A critical stage during invasion is the formation of an intimate interaction between the parasites and their host cells, which requires the participation of interacting proteins secreted by the parasites. For example, T. gondii actively releases proteins, like those of the RON family, from the rhoptry neck. They interact with micronemal proteins such as apical membrane antigen-1 (AMA1), which adheres to the surface of the host cell and forms the moving junction structure [4, 5]. The moving junction moves from the apex to the cell membrane, enabling parasites to invade cells [6]. However, the pantropic characteristic of T. gondii suggests that many uncharacterized proteins on the parasite surface, or discharged from the invasion-associated organelles, are critical for cell recognition and invasion.
Sialic acids (SAs) are a large family of monosaccharide derivatives containing 9 carbon atoms. The SAs of glycolipids and glycoproteins on the cell surface are receptors for influenza viruses [7, 8]. SA exposed on the surface of macrophages greatly facilitates phagocytosis of Trypanosoma cruzi [9, 10]. SA is also an essential receptor for the merozoites of Plasmodium falciparum to recognize host erythrocytes before invasion [11, 12]. Erythrocyte-binding antigen-175 (EBA-175) is the best characterized protein of the erythrocyte-binding ligand family of P. falciparum [13, 14], which triggers the invasion process by binding to erythrocytic SA [15]. T. gondii appears to attach to host cells through interaction with SA prior to invading host cells [16, 17]. The micronemal proteins of T. gondii, TgMIC1 and TgMIC13, have been characterized as SA ligands [18, 19]. However, other T. gondii molecules, especially those expressed on the tachyzoite surface and initiating host cell interaction, are poorly characterized.
In this study, an SA-binding protein, SABP1, on the T. gondii surface was identified and characterized. SABP1 readily bound to SA in vitro and its binding capacity to mammalian cells was eliminated by neuraminidase treatment. T. gondii parasites with SABP1 gene deletion lost their ability to attach and invade cells and had no pathogenicity to mice. This defect could be overcome by gene complementation. Our data suggest a critical role of SABP1 in host cell recognition and invasion by T. gondii.
MATERIALS AND METHODS
Ethical Statement
The experiments using mice, rats, and rabbits were approved by the Ethics Committee on Animal Experiments of the Laboratory Animal Center of Shenyang Agricultural University, China.
Toxoplasma gondii Culture
T. gondii tachyzoites were cultured in a confluent monolayer of Vero cells and incubated at 37°C with 5% CO2.
Sialic Acid-Binding Proteome of T. gondii
To obtain the SA-binding proteome of T. gondii, purified tachyzoites were suspended in phosphate-buffered saline (PBS) containing protease inhibitors and centrifuged at 12 000 rpm for 10 minutes. The supernatants of parasite lysates were incubated separately with SA-agarose (Vector laboratories) and agarose beads (Vector laboratories) in triplicate at 4°C overnight. The eluted proteins were subjected to liquid chromatography-sequential window acquisition of all theoretical fragment ion spectra (LC-SWATH) analysis. Proteins with an abundance ratio of SA-agarose/agarose ≥ 2 were selected as potential SA-binding proteins.
Molecular Cloning and Expression of Recombinant SABP1 Proteins
The gene coding for SABP1 (gene ID, TGGT1_225940) was obtained from T. gondii strain RH tachyzoites using polymerase chain reaction (PCR) with gene-specific primers (Supplementary Table 6). The amplicons were cloned into pDEST-15 and pDEST-17 gateway cloning vectors (Invitrogen). Two recombinant plasmids were expressed in E. coli BL21 (DE3) as previously described [20, 21]. Glutathione S-transferase (GST)–SABP1 and His-SABP1 were purified by affinity purification.
Generation of SABP1-Specific Antibodies
SABP1-specifc polyclonal antibodies were obtained by immunization of rabbits and rats with His-SABP1 fusions. The rabbits and rats were subcutaneously immunized 4 times in Freund’s adjuvants. Rabbit–anti-SABP1 IgG was purified from rabbit sera using protein A Sepharose 4 Fast Flow (GE Healthcare).
Expression and Immunofluorescence Analysis of SABP1 in T. gondii
To examine the expression of the native SABP1 in T. gondii, T. gondii lysate was dissolved in 5 × sodium dodecyl sulfate (SDS) loading buffer (Beyotime), run on a 10% SDS- polyacrylamide gel electrophoresis (PAGE) gel and transferred to a polyvinylidene fluoride membrane (Millipore). The membrane was blocked with 5% defatted milk at 37°C for 1 hour and incubated in PBS with Tween 20 (PBST) containing SABP1-specific IgG at 4°C for 8–10 hours. After 3 washes with PBST, the membrane was incubated with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG; EASYBIO) for 1 hour. The membrane was then washed with PBST 4 times and developed with a 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) alkaline phosphatase color development kit (Beyotime).
To confirm the localization of the SABP1 protein, purified T. gondii tachyzoites were fixed in 4% paraformaldehyde (PFA) and permeabilized with 0.25% Triton X-100 (or without permeabilization to determine the precise localization of TgSABP1) as previously described [22]. The slides were blocked with 5% defatted milk at 37°C for 30 minutes and a rabbit anti-SAG1 antibody and a rat anti-SABP1 sera were incubated as primary antibody. The slides were then washed 5 times with PBS and incubated with secondary antibodies. Following 3 washes with PBS, the slides were stained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) before being examined with a confocal laser scanning microscope (Leica SP8).
Sialic Acid Binding and Inhibition Assays In Vitro
GST-SABP1 and GST proteins were incubated separately with SA-agarose and agarose at 4°C for 2 hours. The beads were washed 3 times with PBS and centrifuged at 500g for 5 minutes between each wash. The pellets were boiled with 5 × SDS loading buffer followed by western blot analysis.
For the SA-binding inhibition assay, GST-SABP1 proteins interacted with SA reagent (Adhoc Intertek) at a concentration of 0.001–100 mg/mL at 37°C for 1 hour before incubation with SA-agarose. The agarose beads were then centrifuged at 500g and washed with PBS. The inhibitory effect was assessed by western blot.
Cell Adhesion and Invasion Inhibition Assay With Recombinant SABP1
GST-SABP1 fusion protein and GST protein were incubated with Vero and dendritic cells as previously described [23]. The cell pellets were washed 3 times with PBS and mixed with 5 × SDS loading buffer (Beyotime), and analyzed by western blot. For the immunofluorescence assay (IFA), Vero and dendritic cells on coverslips were incubated with GST-SABP1 and GST protein, respectively. After 3 washes with PBS, each coverslip was fixed with 4% PFA and detected with an anti-GST monoclonal antibody.
The invasion inhibition assay was conducted as previously described [24]. Briefly, GST-SABP1 (0, 0.1, 0.5, 1, 2, and 5 μM) was preincubated with semiconfluent Vero cells for 60 minutes. GST protein was used as a negative control. The cells were then washed with PBS and cocultured with RH tachyzoites for 4 hours before 3 washes with PBS to remove uninvaded parasites. The invaded parasites were further cultured for 12 hours and stained with acridine orange. The number of cells and parasites were counted with a fluorescence microscope (Leica DM4B).
Effect of Neuraminidase Treatment on Cell Binding of SABP1 Protein
Vero and dendritic cells were pretreated with neuraminidase (0, 0.5, 1, 2, 5, and 10 U/mL; Biotopped) at 37°C and incubated with GST-SABP1 fusion protein and GST, respectively. Subsequently, 50% of the cells were mixed with 5 × SDS loading buffer followed by SDS-PAGE and binding of GST-SABP1 fusion protein to the cells was detected by western blot. For IFA detection, the cells were blocked with 3% bovine serum albumin (BSA) for 30 minutes and detected with an anti-GST monoclonal antibody. The mean fluorescence intensity of host cells was determined with a FACSAria III flow cytometer (BD Biosciences).
Invasion inhibition Assay With TgSABP1-Specific Antibodies In Vitro
The ability of TgSABP1-specific antibodies to inhibit T. gondii invasion was tested as previously described [25]. RH tachyzoites were preincubated with different concentrations of TgSABP1-specific IgG (0, 10, 25, 50, 100, 150, and 250 μg/mL) at 37°C for 60 minutes prior to adding them to Vero cell monolayers. An IgG from unimmunized rabbits was used as a negative control. After 4 hours, each slide was washed with PBS buffer and the invaded parasites were cultivated for 12 hours and then stained with acridine orange. The number of parasites was counted.
Generation of SABP1 Gene Knockout and Complemented Strains Using CRISPR/CAS9 System
We used the CRISPR/Cas9 system to generate an SABP1 gene knockout strain as previously described [26]. Briefly, a total of 10 μg linear dihydrofolate reductase (DHFR) cassette flanked by 5′ and 3′ SABP1 sequences was transfected together with 50 μg pSAG1-CAS9-U6-sgSABP1 plasmid into 2 × 107 RH tachyzoites and selected by pyrimethamine (Sigma) for 3−5 days. By a limiting-dilution method, single positive knockout clones (termed ΔSABP1) were verified by PCR with specific primers (Supplementary Table 2), IFA, and western blot analysis.
To reintroduce the SABP1 gene back into the genome of ΔSABP1 parasites, the pSAG1-CAS9-U6-sgUPRT plasmid was transfected with an amplicon containing the coding sequence of TgSABP1 flanked by the 5′ and 3′ untranslated region of the uracil phosphoribosyltransferase (UPRT) gene. Under selection with 10 μM fluorodeoxyribose (FUDR) (Sigma) for 3−5 days, PCR-positive clones (ΔSABP1-C) were verified by western blot and IFA.
Phenotypic Characterization of ΔSABP1 and ΔSABP1-C Parasites
The invasion efficiency of the wild-type RH strain, ΔSABP1, and ΔSABP1-C parasites was compared in red/green assays as previously described [27]. Briefly, the parasites were inoculated on cells for 1 hour at 37°C, then washed with PBS 3 times and fixed with 4% PFA for 15 minutes. A rat anti-SAG1 and a rabbit anti-ROP9 was used to stain extracellular or intracellular parasites. An attachment assay using cytochalasin D (Sigma) was performed as described [27]; parasites were incubated with Endo buffer (44.7 mM K2SO4, 10 mM MgSO4, 106 mM sucrose, 5 mM glucose, 20 mM Tris, 0.35% wt/vol BSA, pH 8.2) containing 1 μM cytochalasin D for 10 minutes at room temperature and inoculated onto cells at 37°C for 15 minutes. The medium was then replaced by prewarmed 10% DMEM containing 1 μM cytochalasin D and incubated for another 20 minutes before an immunofluorescence assay. Twelve fields were randomly counted for each slide. Analyses of plaque formation and gliding motility of the parasites were also assessed as previously described [28].
Pathogenicity of the SABP1 Gene Knocked-Out and Complemented Parasites in Mice
Briefly, 80 Balb/C female mice were divided into 8 groups and intraperitoneally inoculated with 50, 500, 5 × 103, or 5 × 105 of either wild-type or ΔSABP1 parasites. For the test of SABP1-complemented parasites, 50 ΔSABP1-C parasites were similarly inoculated and then monitored for 28 days.
RESULTS
Sialic Acid-Binding Proteome of T. gondii
SA-agarose (SA conjugated to agarose beads) was used to select high-affinity proteins from parasites lysates; unlabeled agarose was used as a control to minimize the effect of nonspecific binding proteins. The SA-binding proteome of T. gondii was acquired by LC-MS/MS analysis of the affinity-purified proteins. In total, 224 proteins were obtained and quantified by LC-SWATH. Only 59 proteins showed high affinity to SA (Supplementary Table 2), and these were categorized according to cellular localization and biological process (Supplementary Figure 1) in gene ontology annotations [29]. Apart from a few proteins such as RON8 (a rhoptry-derived protein), which was associated with parasite invasion, most of the SA-binding proteome comprised proteins with unknown function (Supplementary Figure 1). To determine the function of the SA-binding proteins, a protein termed sialic acid binding protein 1 (SABP1, TGGT1_225940), which shared a similar amino acid sequence domain with the conserved MAR domain (SA-binding domain) of MIC1 protein (Supplementary Figure 2), was selected for further investigation.
SABP1 Was Localized on the Surface of T. gondii Tachyzoites
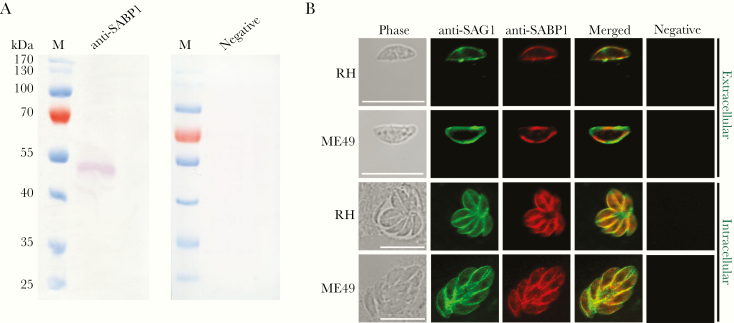
To localize the expression of native SABP1 in T. gondii, His-tagged SABP1 fusion protein was generated (Supplementary Figure 3A) and immunized into animals to produce SABP1-specific antibodies. The native SABP1, with a molecular weight of approximately 50 kDa in T. gondii was specifically detected by western blot (Figure 1A). The localization of TgSABP1 on T. gondii was further determined using an immunofluorescence assay. The fluorescence signal of TgSABP1 overlapped with TgSAG1 and was observed on the surface of T. gondii tachyzoites of both intracellular and extracellular parasites (Figure 1B), and the fluorescence was not affected by permeabilization (Supplementary Figure 3B). The data demonstrated that TgSABP1 was localized on the outer membrane of T. gondii.
Figure 1.
Toxoplasma gondii sialic acid binding protein-1 (TgSABP1) expressed in T. gondii is localized on the outer membrane of tachyzoites. A, Western blot analysis of native SABP1 expression in T. gondii detected by SABP1-specific antibodies. Sera of a healthy rat was used as the negative control. B, The localization of TgSABP1 (red) overlapped with surface antigen-1 (SAG1, green) was detected on the surface of both intracellular and extracellular parasites (without permeabilization) of T. gondii (RH and ME49 strains). Sera of a healthy rat was used as the negative control. Scale bar, 10 μm.
SABP1 Specifically Interacted With Sialic Acid In Vitro
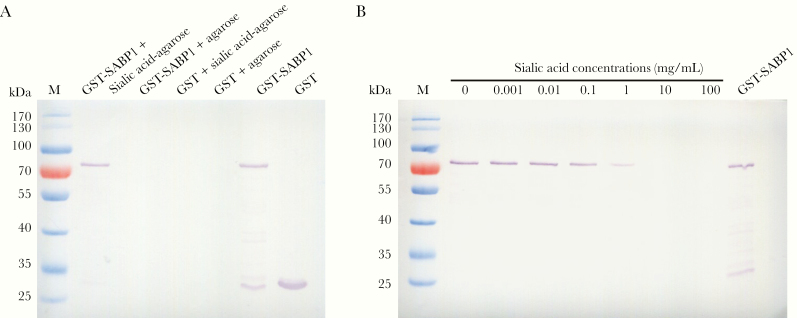
To evaluate and confirm the SA-binding property of SABP1, GST-tagged SABP1 fusion protein was generated (Supplementary Figure 3A). GST-SABP1 specifically bound to SA-agarose but not to the agarose alone. GST did not bind to SA-agarose (Figure 2A). The specific binding of SABP1 to SA was also verified in an inhibition assay. The binding of GST-SABP1 to the SA-agarose beads was blocked by SA in a concentration-dependent manner (Figure 2B), which confirmed the specific interaction between SABP1 and SA.
Figure 2.
Glutathione S-transferase–sialic acid binding protein-1 (GST-SABP1) specifically bound to sialic acid in vitro. A, Evaluation of the binding specificity between SABP1 and sialic acid by western blot assay. GST-SABP1 bound to the sialic acid-agarose beads but not to the agarose. GST did not bind to sialic acid-agarose beads. B, Binding of GST-SABP1 to the sialic acid-agarose beads was inhibited by preincubation of the recombinant protein with sialic acid.
Recombinant SABP1 Specifically Bound to Vero and Dendritic Cells and Inhibited Parasite Invasion
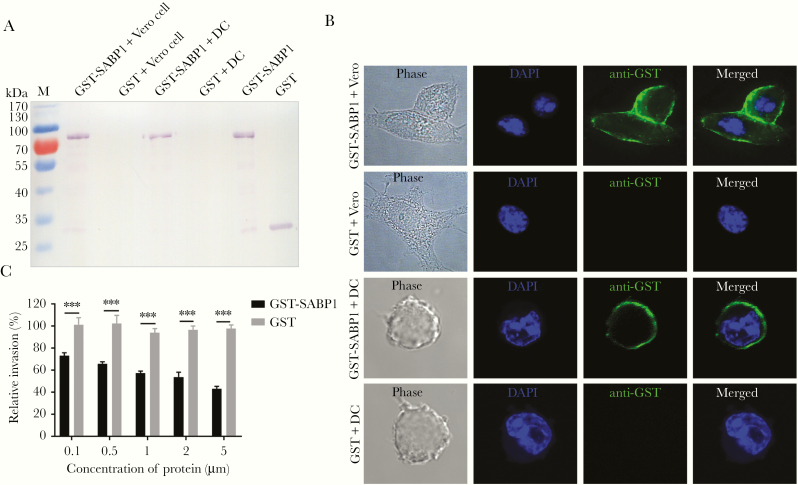
The binding properties of TgSABP1 with host cells were studied by incubating GST-SABP1 with both Vero and dendritic cells followed by western blot and immunofluorescence analysis. GST-SABP1 showed specific binding to both Vero and dendritic cells, and the GST control protein did not bind to the cells (Figure 3A and 3B).
Figure 3.
Recombinant sialic acid binding protein-1 (SABP1) bound to both Vero and dendritic cells in vitro and inhibited Toxoplasma gondii invasion. A, Western blot analysis of cell adhesion in vitro. Glutathione S-transferase (GST)-SABP1 fusion protein and GST protein were incubated with Vero and dendritic cells (DC). GST-SABP1 bound to the cells but the GST protein did not. B, Indirect immunofluorescence assay showed a specific interaction between GST-SABP1 fusion protein and host cells (green). C, Preincubation of GST-SABP1 fusion protein with host cells significantly inhibited parasite invasion. Error bars indicate the mean ± SD (n = 3). *** P < .001. Abbreviation: DAPI, 4′,6-diamidino-2-phenylindole.
We showed that SABP1 was a ligand participating in T. gondii invasion using an invasion inhibition assay. Parasite invasion of host cells was impaired by GST-SABP1 protein in a concentration-dependent manner, while GST protein had no inhibitory effect on T. gondii invasion (Figure 3C). This suggests that the host cell recognition or interaction of T. gondii tachyzoites was blocked by GST-SABP1 protein. These data indicate that TgSABP1 is a parasite ligand essential for T. gondii-SA interaction during parasite invasion.
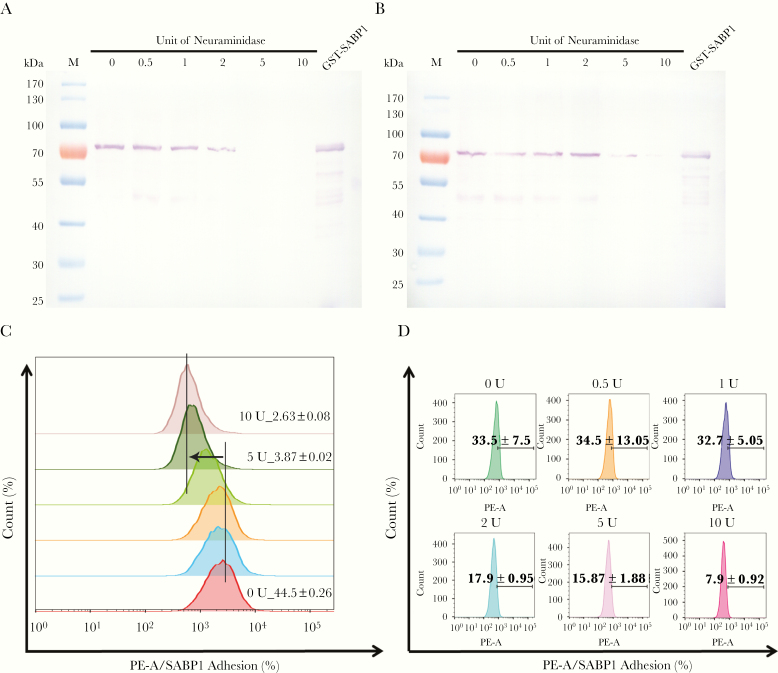
To determine whether the binding of GST-SABP1 to Vero and dendritic cells was via specific interaction with the cell surface SA receptor, both Vero and dendritic cells were pretreated with neuraminidase before incubation with the GST-SABP1 protein. The binding of GST-SABP1 protein to Vero cells disappeared after enzymatic treatment when the concentration of neuraminidase reached 5 U/mL (Figure 4A). Similarly, the binding of GST-SABP1 protein to dendritic cells was abolished when the enzyme concentration reached 10 U/mL (Figure 4B and Supplementary Figure 4). The binding of GST-SABP1 protein to both Vero and dendritic cells was analyzed with flow cytometry; the data (Figure 4C and 4D and Supplementary Figure 4) confirmed that SA is a specific receptor for TgSABP1.
Figure 4.
Binding of glutathione S-transferase–sialic acid binding protein-1 (GST-SABP1) to host cells was sensitive to neuraminidase treatment. Western blot analysis showing the effect of neuraminidase treatment on the binding of GST-SABP1 to Vero (A) and dendritic cells (B). Vero and dendritic cells were treated with neuraminidase before incubation with GST-ABP1 protein. C and D, Binding of GST-SABP1 to Vero and dendritic cells was gradually abolished with increased neuraminidase concentration; GST-SABP1 protein levels detected on (C) Vero and (D) dendritic cells surface. PE-A: the red fluorescence signal detected in PE channel by flow cytometer (PE: phycoerythrin, A: area).
SABP1-Specific Antibodies Inhibited T. gondii Invasion
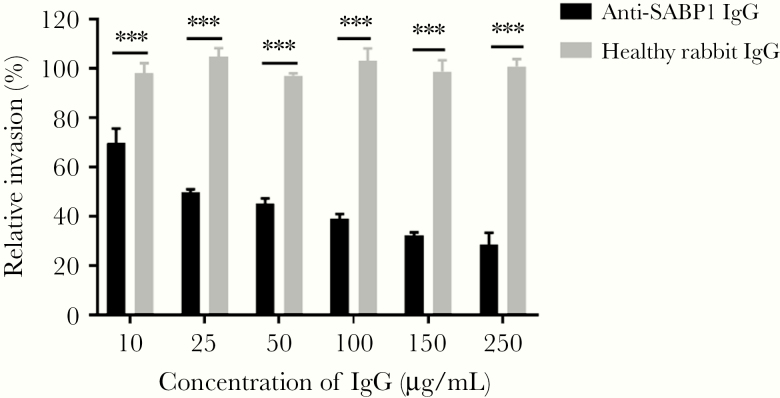
To determine whether TgSABP1 exerted a role in T. gondii invasion, an invasion inhibition assay was performed. The invasion efficiency of T. gondii was significantly inhibited by SABP1-specific antibodies in a concentration-dependent manner (Figure 5), while the control antibodies had no inhibitory effect on the parasites. The data indicate that SABP1 is a ligand associated with the invasion of T. gondii into host cells.
Figure 5.
Toxoplasma gondii sialic acid binding protein-1 (TgSABP1)-specific antibodies inhibited T. gondii invasion into host cells. Preincubation of the tachyzoites with concentration of SABP1-specific immunoglobulin G (IgG) from 10 to 250 μg/mL significantly inhibited parasite invasion. Error bars represent mean ± SD (n = 3). *** P < .001.
Deletion of the Gene Encoding TgSABP1 Diminished Host Cell Recognition/Attachment and Invasion by T. gondii
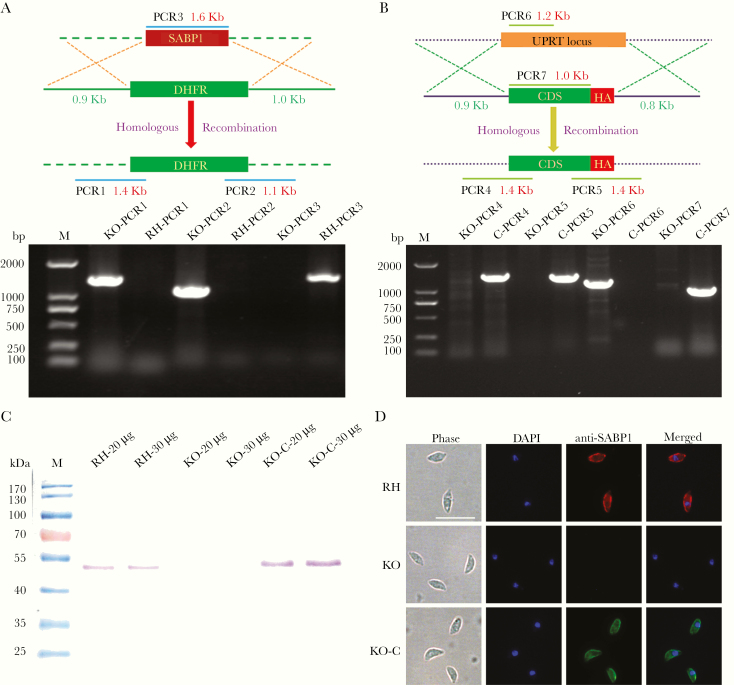
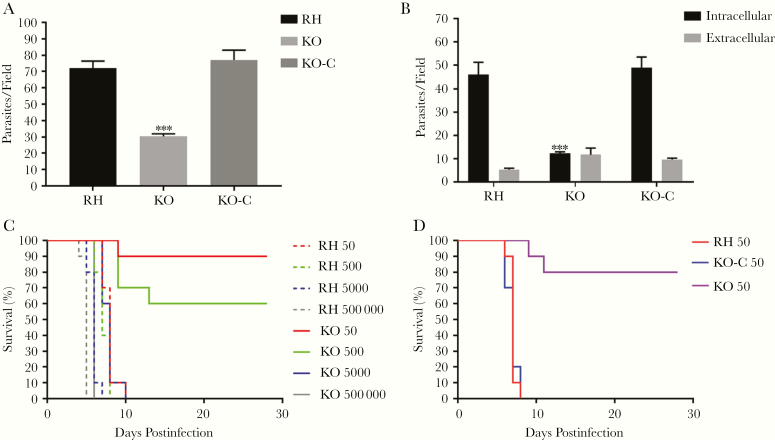
To confirm that TgSABP1 is essential for parasite invasion, the SABP1 gene of T. gondii was deleted using the CRISPR/Cas9 system. The knockout strain (ΔSABP1/KO) was cloned and verified using PCR with gene-specific primers, western blot, and IFA (Figure 6). The cell recognition, attachment, and invasion ability of ΔSABP1 strain was severely diminished in vitro compared to the wild-type strain (Figure 7). The gene encoding SABP1 linked with an HA tag was reintroduced into the genome of the ΔSABP1 strain at the UPRT locus (Figure 6) and a clonal strain (ΔSABP1-C/KO-C) was obtained (Figure 6). The expression of SABP1 in the ΔSABP1-C strain was confirmed by western blot and IFA with SABP1-specific antibodies (Figure 6). Consequently, the recognition/attachment and invasion efficiency of ΔSABP1-C to HFF cells was completely restored (Figure 7). Plaque assays performed on the ΔSABP1 parasites indicated a defect in parasite invasion (Supplementary Figure 5). Further phenotypic analysis on the ΔSABP1 strain revealed that SABP1 depletion did not affect parasite gliding motility (Supplementary Figure 5).
Figure 6.
CRISPR/Cas9 strategy used to generate sialic acid binding protein-1 gene (SABP1) knockout (KO) and gene complemented strain. A, Verification of single clonal SABP1-deficient parasite by PCR. The positive KO-PCR1, PCR2 (DNA of ΔSABP1 parasites was used as the template) and negative RH-PCR3 (DNA of RH parasites was used as the template) indicate that the SABP1 gene was replaced by the DHFR cassette. B, SABP1 gene was reintroduced into the genome of the KO strain. PCR of ΔSABP1-C clonal strains (C-PCR4 +, C-PCR5 +, C-PCR7 +, and C-PCR6 −) demonstrated that SABP1 gene was completely inserted at the UPRT locus of the KO genome. C, The expression of SABP1 in RH, ΔSABP1, and ΔSABP1-C parasites analyzed by western blot (20 and 30 μg lysates of each strain) with the SABP1-specific antibody to demonstrate the depletion and complementation of TgSABP1 gene. D, Immunofluorescence assays with SABP1-specific antibodies illustrate that the protein is expressed on the surface of wild-type RH strain (red) and ΔSABP1-C parasites (green), but was not detected on ΔSABP1 parasites. Scale bar, 10 μm. Abbreviation: DAPI, 4′,6-diamidino-2-phenylindole.
Figure 7.
Deletion of the gene encoding Toxoplasma gondii sialic acid binding protein-1 (TgSABP1) reduced the recognition/attachment and invasion of T. gondii. Parasite attachment (A) and invasion efficiency (B) were significantly weakened in SABP1-deficient parasites (KO) compared to wild-type and ΔSABP1-C (KO-C) parasites. Rat anti-SAG1 and rabbit anti-ROP9 was used to stain extracellular or intracellular parasites. Error bars represent the mean ± SD (n = 3). *** P < .001. C and D, Survival of mice challenged with 50, 500, 5 × 103, and 5×105 wild-type RH, ΔSABP1 (C) and infected with 50 wild-type RH, ΔSABP1, and ΔSABP1-C parasites (D). All mice were monitored for 28 days.
Infectivity of ΔSABP1 Parasites Was Attenuated
The infectivity of the SABP1-depleted strain was also verified in an animal infection experiment. The pathogenicity of ΔSABP1 strain was severely reduced in mice at doses of 50 and 500 tachyzoites and, importantly, the SABP1 gene complemented strain fully rescued the virulence (Figure 7). These data suggest that TgSABP1 is an important molecule associated with host cell recognition or attachment by T. gondii.
DISCUSSION
The first step for all apicomplexan parasites to infect and proliferate in mammals is recognition and adhesion to the host cell. In this process, proteins on the parasite surface play a critical role. T. gondii, which almost infects any nucleated cells, has a variety of surface proteins, including those from the SAG families [30] and other uncharacterized proteins. After attachment of tachyzoites to the host cell surface, proteins, like RON2 and AMA1, are sequentially released from invasion-associated organelles such as the microneme and rhoptry. These proteins mediate junction formation of the tachyzoite with the host cell membrane and progressive movement into the cell. Compared to the parasite ligands, which have been relatively well characterized, the documented host cell receptors for T. gondii invasion only include a few glycan molecules, such as SA [31].
SA is widely distributed on the cell surface of all vertebrates and commonly occurs as a component of oligosaccharide chains of glycolipids, glycoproteins, and mucins, and occupies terminal position within these molecules on the cell surfaces [32, 33]. SA is a major determinant of T. gondii invasion into a host cell [18, 34]. TgMIC1 and TgMIC13, two micronemal proteins of T. gondii, specifically bind sialylated oligosaccharides and are characterized as SA-binding ligands involved in the SA-dependent invasion pathway of T. gondii [18, 19]. Therefore, exposure of parasite-derived invasion-associated lectins to SAs on host cells might be of primary importance in T. gondii invasion.
Previous studies on P. falciparum indicated that the parasite-derived proteins involved in host cell recognition could be diverse and localized in different organelles before release [35]. Therefore, with a similar approach, we generated an SA-binding proteome and proteins with an unknown function were prioritized for more detailed analysis. SABP1 was one of the proteins selected and was found to contain a considerable similarity in amino sequence with the MAR domains (SA-binding domain) of both NcMIC1 and TgMIC1 (Supplementary Figure 2), indicating SABP1 could potentially bind to SA. The molecular weight of both native TgSABP1 and His-SABP1 recombinant protein recognized by the specific antibody was approximately 50 kDa, which is larger than its predicted molecule weight. This is likely due to the amino acid content or the highly hydrophilic nature of TgSABP1, which binds more SDS contributing to its slower migration in the gel.
SABP1 is clearly a membrane-bound protein because it was labeled on the surface of both Triton-treated and untreated parasites, and deletion of the SABP1-coding gene caused the loss of its expression on the parasite’s surface. The protein was also observed on the surface of intracellular tachyzoites, likely because it needs to be synthesized and located at the surface for the next round of invasion after egress. Further, recombinant GST-SABP1 showed specific binding to SA and host cells in vitro (Figure 2 and Figure 3). Also, the invasion efficiency of the parasite to the host cells preincubated with GST-SABP1 fusion protein was significantly inhibited (Figure 3C), indicating that recognition of the native SABP1 by host cells was blocked by GST-SABP1. Additionally, the interaction of GST-SABP1 with host cells was sensitive to neuraminidase treatment (Figure 4). These data indicate that the SABP1-SA interaction is important for the invasion process. A significant reduction in invasion efficiency occurred after incubation of SABP1-specific antibody with tachyzoites (Figure 5), indicating the importance of SABP1 in the invasion of T. gondii into host cells.
The role of SABP1 in the invasion process was further supported by gene deletion and complementation using the CRISPR/Cas9 system [36]. In mice, the SABP1-deficient strain was less virulent than wild-type parasites, and the infectivity was restored when the SABP1 gene was reintroduced into the knockout parasites (Figure 7). These data support the conclusion that SABP1 is a vital parasite ligand associated with parasite recognition or attachment to host cells.
In summary, the data has increased our understanding of the ligand-receptor interaction between T. gondii and its hosts. The identification of TgSABP1, a novel SA-binding protein of T. gondii, may aid in the development of methods for prevention and treatment of toxoplasmosis.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We appreciate very much the support of Professor Shen Bang, Huazhong Agricultural University, for supplying the CRISPR/Cas9:sgUPRT plasmid. Also, we thank Professor Suo Xun, China Agricultural University, for technological assistance.
Financial support. This work was supported by the National Key Research and Development Program of China (grant number 2017YFD0500400); the National Natural Science Foundation of China (grant number 31672546, 31972702); and the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (grant number 2019-I2M-5–042).
Potential conflicts of interest. Q. C. was responsible for conceptualization, writing, and editing. M. X. and N. Y. curated data and M. X. and D. W. carried out analysis. M. X., X. S., Y. F., R. C., and X. W. were responsible for methodology. Q. C., N. Y., and N. J. acquired funding. N. J. carried out project administration. M. X. wrote the first draft. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Dubey JP. Toxoplasmosis of animals and humans, 2nd ed. Beltsville, MA: CRC Press, 2016. [Google Scholar]
- 2. Sibley LD. Toxoplasma gondii: perfecting an intracellular life style. Traffic 2003; 4:581–6. [DOI] [PubMed] [Google Scholar]
- 3. Soldati D. Molecular dissection of host cell invasion by the apicomplexans: the glideosome. Parasite 2008; 15:197–205. [DOI] [PubMed] [Google Scholar]
- 4. Besteiro S, Dubremetz JF, Lebrun M. The moving junction of apicomplexan parasites: a key structure for invasion. Cell Microbiol 2011; 13:797–805. [DOI] [PubMed] [Google Scholar]
- 5. Lamarque M, Besteiro S, Papoin J, et al. . The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog 2011; 7:e1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol 1997; 73:114–23. [PubMed] [Google Scholar]
- 7. Ng PS, Böhm R, Hartley-Tassell LE, et al. . Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat Commun 2014; 5:5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin SC, Huang MH, Tsou PC, Huang LM, Chong P, Wu SC. Recombinant trimeric HA protein immunogenicity of H5N1 avian influenza viruses and their combined use with inactivated or adenovirus vaccines. PLoS One 2011; 6:e20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Araujo-Jorge T. The biology of Trypanosoma cruzi-macrophage interaction. Mem Inst Oswaldo Cruz 1989; 84:441–62. [DOI] [PubMed] [Google Scholar]
- 10. Araujo-Jorge T, Souza W. Effect of carbohydrates, periodate and enzymes in the process of endocytosis of Trypanosoma cruzi by macrophages. Acta Trop 1984; 41:17–28. [PubMed] [Google Scholar]
- 11. Friedman MJ, Blankenberg T, Sensabaugh G Tenforde TS. Recognition and invasion of human erythrocytes by malarial parasites: contribution of sialoglycoproteins to attachment and host specificity. J Cell Biol 1984; 98:1672–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ord RL, Rodriguez M, Yamasaki T, Takeo S, Tsuboi T, Lobo CA. Targeting sialic acid dependent and independent pathways of invasion in Plasmodium falciparum. PLoS One 2012; 7:e30251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sim BKL. EBA-175: an erythrocyte-binding ligand of Plasmodium falciparum. Parasitol Today 1995; 11:213–7. [DOI] [PubMed] [Google Scholar]
- 14. Chowdhury P, Sen S, Datta Kanjilal SD, Sengupta S. Genetic structure of two erythrocyte binding antigens of Plasmodium falciparum reveals a contrasting pattern of selection. Infect Genet Evol 2017; 57:64–74. [DOI] [PubMed] [Google Scholar]
- 15. Orlandi PA, Klotz FW, Haynes JD. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2- 3)Gal- sequences of glycophorin A. J Cell Biol 1992; 116:901–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monteiro VG, Pacheco Soares C, de Souza W. Host cell surface sialic acid residues are involved on the process of penetration of Toxoplasma gondii into mammalian cells. FEMS Microbiol Lett 1998; 164:323–7. [DOI] [PubMed] [Google Scholar]
- 17. Carruthers VB, Håkansson S, Giddings OK, Sibley LD. Toxoplasma gondii uses sulfated proteoglycans for substrate and host cell attachment. Infect Immun 2000; 68:4005–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blumenschein TM, Friedrich N, Childs RA, et al. . Atomic resolution insight into host cell recognition by Toxoplasma gondii. EMBO J 2007; 26:2808–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friedrich N, Santos JM, Liu Y, et al. . Members of a novel protein family containing microneme adhesive repeat domains act as sialic acid-binding lectins during host cell invasion by apicomplexan parasites. J Biol Chem 2010; 285:2064–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moll K, Ahuja S, Chene A, Bejarano MT, Chen Q. Optimized expression of Plasmodium falciparum erythrocyte membrane protein I domains in Escherichia coli. Malar J 2005; 3:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 1990; 185:60–89. [DOI] [PubMed] [Google Scholar]
- 22. Erler H, Ren B, Gupta N, Beitz E. The intracellular parasite Toxoplasma gondii harbors three druggable FNT-type formate and L-lactate transporters in the plasma membrane. J Biol Chem 2018; 293:17622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baum J, Chen L, Healer J, et al. . Reticulocyte-binding protein homologue 5—an essential adhesin involved in invasion of human erythrocytes by Plasmodium falciparum. Int J Parasitol 2009; 39:371–80. [DOI] [PubMed] [Google Scholar]
- 24. Zhang D, Jiang N, Chen Q. ROP9, MIC3, and SAG2 are heparin-binding proteins in Toxoplasma gondii and involved in host cell attachment and invasion. Acta Trop 2019; 192:22–9. [DOI] [PubMed] [Google Scholar]
- 25. Zhao X, Chang Z, Tu Z, et al. . PfRON3 is an erythrocyte-binding protein and a potential blood-stage vaccine candidate antigen. Malar J 2014; 13:490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen B, Brown KM, Lee TD, Sibley LD. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio 2014; 5:e01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Straub KW, Peng ED, Hajagos BE, Tyler JS, Bradley PJ. The moving junction protein RON8 facilitates firm attachment and host cell invasion in Toxoplasma gondii. PLoS Pathog 2011; 7:e1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Graindorge A, Frénal K, Jacot D, Salamun J, Marq JB, Soldati-Favre D. The conoid associated motor MyoH is indispensable for Toxoplasma gondii entry and exit from host cells. PLoS Pathog 2016; 12:e1005388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Jiang N, Lu H, et al. . Proteomic analysis of Plasmodium falciparum schizonts reveals heparin-binding merozoite proteins. J Proteome Res 2013; 12:2185–93. [DOI] [PubMed] [Google Scholar]
- 30. Lekutis C, Ferguson DJ, Grigg ME, Camps M, Boothroyd JC. Surface antigens of Toxoplasma gondii: variations on a theme. Int J Parasitol 2001; 31:1285–92. [DOI] [PubMed] [Google Scholar]
- 31. Kato K. How does Toxoplama gondii invade host cells? J Vet Med Sci 2018; 80:1702–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Severi E, Hood D, Thomas G. Sialic acid utilization by bacterial pathogens. Microbiology 2007; 153:2817–22. [DOI] [PubMed] [Google Scholar]
- 33. Wang B, Brand-Miller J. The role and potential of sialic acid in human nutrition. Eur J Clin Nutr 2003; 57:1351–69. [DOI] [PubMed] [Google Scholar]
- 34. Baba M, Sato M, Kitoh K, Takashima Y. The distribution pattern of α2,3- and α2,6-linked sialic acids affects host cell preference in Toxoplasma gondii. Exp Parasitol 2015; 155:74–81. [DOI] [PubMed] [Google Scholar]
- 35. Wright GJ, Rayner JC. Plasmodium falciparum erythrocyte invasion: combining function with immune evasion. PLoS Pathog 2014; 10:e1003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.