See Doyle et al. (doi:10.1093/brain/awaa149) for a scientific commentary on this article.
Lowering TMEM106B levels has been proposed as a disease-modifying strategy in FTD and other neurodegenerative diseases. However, using two independent Tmem106b deficiency mouse models, Zhou et al. reveal a key role for TMEM106B in myelination, with potential consequences for its use as a therapeutic target.
Keywords: TMEM106B, myelin, cuprizone, oligodendrocytes, lysosome trafficking
Abstract
Genetic variants that define two distinct haplotypes at the TMEM106B locus have been implicated in multiple neurodegenerative diseases and in healthy brain ageing. In frontotemporal dementia (FTD), the high expressing TMEM106B risk haplotype was shown to increase susceptibility for FTD with TDP-43 inclusions (FTD-TDP) and to modify disease penetrance in progranulin mutation carriers (FTD-GRN). To elucidate the biological function of TMEM106B and determine whether lowering TMEM106B may be a viable therapeutic strategy, we performed brain transcriptomic analyses in 8-month-old animals from our recently developed Tmem106b−/− mouse model. We included 10 Tmem106b+/+ (wild-type), 10 Tmem106b+/− and 10 Tmem106−/− mice. The most differentially expressed genes (153 downregulated and 60 upregulated) were identified between Tmem106b−/− and wild-type animals, with an enrichment for genes implicated in myelination-related cellular processes including axon ensheathment and oligodendrocyte differentiation. Co-expression analysis also revealed that the most downregulated group of correlated genes was enriched for myelination-related processes. We further detected a significant loss of OLIG2-positive cells in the corpus callosum of Tmem106b−/− mice, which was present already in young animals (21 days) and persisted until old age (23 months), without worsening. Quantitative polymerase chain reaction revealed a reduction of differentiated but not undifferentiated oligodendrocytes cellular markers. While no obvious changes in myelin were observed at the ultrastructure levels in unchallenged animals, treatment with cuprizone revealed that Tmem106b−/− mice are more susceptible to cuprizone-induced demyelination and have a reduced capacity to remyelinate, a finding which we were able to replicate in a newly generated Tmem106b CRISPR/cas9 knock-out mouse model. Finally, using a TMEM106B HeLa knock-out cell line and primary cultured oligodendrocytes, we determined that loss of TMEM106B leads to abnormalities in the distribution of lysosomes and PLP1. Together these findings reveal an important function for TMEM106B in myelination with possible consequences for therapeutic strategies aimed at lowering TMEM106B levels.
See Doyle et al. (doi:10.1093/brain/awaa149) for a scientific commentary on this article.
Introduction
Frontotemporal dementia (FTD) is the second most frequent early-onset dementia after Alzheimer’s disease. It accounts for 20–25% of pre-senile dementias (Ratnavalli et al., 2002; Neary et al., 2005). A large subset of FTD cases are pathologically characterized by abnormal inclusions of TAR DNA binding protein 43 (TDP-43) (FTD-TDP) (Mackenzie et al., 2009) and haploinsufficiency of the progranulin protein (PGRN) due to mutations in the progranulin gene (GRN) is one of the leading causes of FTD-TDP (Baker et al., 2006; Cruts et al., 2006; Gass et al., 2006).
In 2010, a genome-wide association study identified transmembrane protein 106B (TMEM106B) as a genetic risk factor for FTD-TDP (Van Deerlin et al., 2010). Subsequent studies established TMEM106B as a major modifier of disease risk, with the strongest association in FTD with GRN mutations (FTD-GRN) (Finch et al., 2011). In particular, individuals with GRN mutations who also carry a TMEM106B ‘protective’ haplotype have ∼50% lower odds of developing FTD symptoms as compared to carriers of the ‘risk’ haplotype (Finch et al., 2011; Pottier et al., 2018). The same TMEM106B haplotypes have been associated with important gene expression changes during ageing and have been implicated in other neurodegenerative diseases including Alzheimer’s disease and hippocampal sclerosis (Amador-Ortiz et al., 2007; Rutherford et al., 2012; Murray et al., 2014; Rhinn and Abeliovich, 2017; Li et al., 2020), highlighting the importance of TMEM106B.
While the presence of ‘protective’ and ‘risk’ haplotypes at the TMEM106B locus has clearly been established, it has been challenging to pinpoint the functional variant(s) responsible for the risk modifying effect. Within the TMEM106B locus, one coding variant [rs3173615, encoding p.T185S (T185S)] and several non-coding variants in strong linkage disequilibrium have been reported to be associated with disease risk in FTD-TDP patients in general and FTD-GRN carriers in particular (Van Deerlin et al., 2010; Finch et al., 2011; Pottier et al., 2018). Whether this coding variant has any effect on the TMEM106B protein function remains unclear and alternative candidates, such as the non-coding variant rs1990620, have been suggested to influence TMEM106B expression (Gallagher et al., 2017). Nevertheless, most studies agree that the ‘risk’ haplotype is associated with higher protein levels of TMEM106B as compared to the ‘protective’ haplotype (Van Deerlin et al., 2010; Nicholson et al., 2013; Gallagher et al., 2017). TMEM106B is a type II transmembrane protein with its main intracellular localization at lysosomes (Chen-Plotkin et al., 2012; Lang et al., 2012; Brady et al., 2013). Overexpression of TMEM106B in vitro leads to lysosomal dysfunctions, including enlarged lysosomal size, reduced lysosomal pH, and deficits of endolysosomal trafficking (Chen-Plotkin et al., 2012; Lang et al., 2012; Brady et al., 2013; Schwenk et al., 2014; Stagi et al., 2014). Furthermore, animal experiments have shown TMEM106B levels are significantly increased in Grn−/− mouse brains, and overexpression of TMEM106B further exacerbated FTD pathologies (increased lipofuscin and lysosome dysfunction) in Grn−/− brains at older ages (Zhou et al., 2017b). In line with these findings, a recent study demonstrated that a complete loss of TMEM106B ameliorates both the lysosomal and FTD-related phenotypes in Grn−/− mice (Klein et al., 2017). Thus, lowering TMEM106B levels might serve as an attractive strategy for the therapeutic treatment of FTD-GRN; however, there is limited knowledge about the normal function of TMEM106B in vivo. In this study, we investigated TMEM106B function in vivo by using Tmem106b−/− mice.
Materials and methods
Mouse lines
The original C57BL/6N Tmem106b−/− mice were generated as described (Nicholson et al., 2018). The Tmem106b CRISPR knockout mouse model was generated by CRISPR/Cas9 technology using guide RNA: 5′-TTAAACAACATAACTAACAT-3′ in The Jackson Laboratory. Genotyping was performed on genomic DNA using PCR primers: 5′-TTCCCAGTAAATGAGATATAGTTCCA-3′ (Forward); 5′-GATTTTCAGAGGGGAACCAG-3′ (Reverse), and sequencing using the forward primer. The ages of the mice are described in each experiment. Male and female mice were used and the sex of the mice in each experiment was matched between conditions or genotypes. All mice were housed in the animal facility at Mayo Clinic Jacksonville and animal procedures were approved by the Institutional Animal Care and Use Committee at Mayo Clinic.
RNA sequencing and bioinformatic data analysis
Brains from 30 mice were harvested from 8-month-old Tmem106b mice [10 Tmem106b+/+ (wild-type), 10 Tmem106b+/− and 10 Tmem106b−/−, five males and five females per genotype]. All animals were 8 months old at harvest; however, for each genotype, mice were derived from two cohorts: six mice were harvested at the first time point and four mice were harvested at a later time point. RNA extraction from the cerebral hemispheres of all 30 brains was performed in one experiment using the RNeasy® Plus Mini Kit (#74136; Qiagen) as in our previous study (Nicholson et al., 2018). After measuring RNA integrity number (RIN) with a 2100 Bioanalyzer using the RNA Nano Chip (Agilent), mRNA samples were sequenced at Mayo Clinic’s Genome Analysis Core using Illumina HiSeq 2000. Reads were mapped to the mouse genome mm10. Raw exon and gene read counts, along with sequencing QC, were generated using the in-house-developed RNA-Seq analytic pipeline: MAP-RSeq version 2.1.0 (Kalari et al., 2014). Conditional quantile normalization (CQN) was applied to raw gene counts to correct for gene length, library size, GC bias and technical variations, and to obtain similar quantile-by-quantile distributions of gene expression levels across samples (Hansen et al., 2012). Based on the bimodal distribution of the CQN-normalized and log2-transformed reads per kilobase per million (RPKM) gene expression values, genes with an average log2 RPKM < 1 across all genotypes were considered not expressed and were excluded from further analysis. After filtering, 21 209 genes remained.
Differential gene expression analyses were performed using Partek Genomics Suite (Partek Inc.). Gene expression between Tmem106b−/−, Tmem106b+/− and wild-type mice were compared using ANOVA models, while correcting for RIN, sex, and cohort (time of harvest). The Benjamini–Hochberg procedure (BH step-up procedure) was performed to adjust for multiple testing and control for false discovery rate. Differentially expressed genes (DEGs) were defined by the threshold of BH step-up P-values <0.05. DEGs with |fold change| ≥ 1.2 were selected for the pathway analysis by using MetaCore pathway analysis (Thomson Reuters; Version 6.25).
To identify groups of genes correlated with Tmem106b genotype, we performed weighted gene co-expression network analysis (WGCNA) (https://labs.genetics.ucla.edu/horvath/CoexpressionNetwork/Rpackages/WGCNA/) using residual expression values after corrections for RIN, sex, and cohort. A signed hybrid co-expression network was built using the soft thresholding power of 10, minimum module size of 40 and minimum height for merging modules at 0.4. Each module was summarized by the first principal component of the scaled module expression profile (module eigengene). To assess the correlation of the module eigengenes and Tmem106b genotype, we defined the wild-type genotype as 0, and Tmem106b−/− genotype as 1. Modules that differed significantly between Tmem106b−/− and wild-type were annotated using R function GO enrichment analysis. Volcano plots and heat maps were generated using MATLAB (R2015a).
Quantitative PCR
Brain RNA (500 ng) was reverse transcribed using the SuperScript™ III complementary DNA (cDNA) synthesis kit with an equal ratio of random hexamers and Oligo dT primers (Thermo Fisher Scientific). Real-time quantitative PCRs (qPCRs) were conducted using either TaqMan® gene expression or SYBR green assays (Applied Biosystems). For TaqMan® gene expression assays, the following probes were used: Fa2h (Mm00626259_m1), Mal (Mm01339780_m1), Opalin (Mm00463365_m1), Enapp6 (Mm00624107_m1), Plin4 (Mm00491061_m1), and Gapdh (Mm99999915_g1). All probes were purchased from Life Technologies. For the SYBR® green assay, the following primers were used: Cnp: forward 5′-AGAGACCTCCAGGTGTGCTG-3′, reverse 5′-TCTCTTCACCACCTCCTGCT-3′; Sox10: forward 5′-AACGCCTTCATGGTGTGG-3′, reverse 5′-CAACCTCCAGAGCTTGCCTA-3′; Plp: forward 5′-CAGAGGCCAACATCAAGCTC-3′, reverse 5′-CATAGGTGATGCCCACAAACT-3′; Mbp: forward 5′-TCACACACGAGAACTACCCATT-3′, reverse 5′-CGAGGTGTCACAATGTTCTTG-3′; Mog: forward 5′-CTCCATCGGACTTTTGATCC-3′, reverse 5′-CAGATGATCAAGGCAACCAG-3′; Nkx2.2: forward 5′-ACCGAGGGCCTCCAATACT-3′, reverse 5′-TTGTCATTGTCCGGTGACTC-3′; Pdgfr: forward 5′-GAGGAACAGACACAGCTCACA-3′, reverse 5′-CGATGGTCTCGTCCTCTCTC-3′; Ng2/Cspg4: forward 5′-AGGATGCCTCCAGGTCAGA-3′, reverse 5′-AGCTGTCGGAGACAGTGAGC-3′; and Gapdh: forward 5′-GTCGGTGTGAACGGATTTG-3′, reverse 5′-TCAATGAAGGGGTCGTTGAT-3′. Results were analysed with the QuantStudioTM Real-time PCR Software and the comparative CT method. Data were expressed as 2−ΔΔCT for the experimental gene of interest normalized to the housekeeping gene (Gapdh) and presented as fold-change relative to control.
Cell culture and CRISPR-mediated genome editing
Primary neuron-glia mixed cultures were prepared as in our previous study (Zhou et al., 2015). Briefly, brains of newborn pups from wild-type and Tmem106b−/− mice were collected and cortices were further dissected and digested in trypsin (Gibco) for 15 min at 37°C. DNase (Sigma-Aldrich) was added to a final concentration of 0.05 mg/ml for 5 min or until no DNA clumps were visible. After a gentle trituration, the single cells were collected and seeded on poly-d-lysine-coated (Sigma-Aldrich) glass coverslips (12-mm diameter) in Dulbecco’s modified Eagle medium (DMEM)/F12 medium containing 10% foetal calf serum (Gibco), 10% horse serum (Gibco) and 1% penicillin/streptomycin (Invitrogen) at a density of 80 000 cells/coverslip. Cells were cultured at 37°C in a humidified atmosphere and 5% CO2 and the medium was refreshed every 2 days. The cells were maintained for 7–10 days to obtain mature neuron-glia mixed cultures.
HeLa cells (ATCC) were maintained in Eagle’s minimum essential medium (EMEM) (ATCC) supplemented with 10% foetal bovine serum (Gibco) and 1% penicillin-streptomycin (Invitrogen) in a humidified incubator at 37°C and 5% CO2. To generate the TMEM106B−/− cell line, HeLa cells were transfected with Cas9 and guide RNA targeted to human TMEM106B [oligos with 5′-CACCGGAGTCACATCTGAAAACATG-3′ and 5′-AAACCATGTTTTCAGATGTGACTCC-3′ were ligated to pX459 (#62988, Addgene)] using Lipofectamine™ 2000 Transfection Reagent (Invitrogen) as in our previous study (Nicholson et al., 2018). Two days after transfection, single cells were seeded in 96-well plates by flow cytometry (Attune NxT analytical flow cytometer; Thermo Fisher Scientific) and single clones were selected by sequencing using the following primers: 5′-CACGACGTTGTAAAACGACAGGACCTGTTGAGACTGTG-3′ and 5′-GGATAACAATTTCACACAGGTTGACTGAAGGTAACTGATATG-3′.
Immunohistochemistry and quantitative analysis
Mice were perfused with 4% paraformaldehyde (PFA). Brains were dissected and fixed in 4% PFA in phosphate-buffered saline (PBS) for 24 h at 4°C. After dehydration with 70% ethanol, brain tissues were embedded in paraffin and cut into 5-μm sections. For staining, paraffin sections were deparaffinized with xylene and rehydrated in a series of ethanol washes. For immunohistochemistry using rabbit anti-OLIG2 (1:200, ab109186; Abcam), antigen retrieval was performed by steaming slides for 30 min with distilled water before blocking in 0.03% hydrogen peroxide. The immunostaining of sections was done using a Dako Autostainer and EnVision+ HRP system (Dako) per manufacturer’s instructions. After dehydration with a series of alcohols and xylene, slides were mounted using Cytoseal XYL (Thermo Fisher Scientific). Stained images were scanned with an Aperio ScanScope AT2 Slide Scanner (Leica Biosystems). For OLIG2 positive cell counting, the corpus callosum was annotated as the region of interest with ImageScope software (v12.1.0.5029; Leica). A custom-designed algorithm was applied to detect the number of OLIG2-positive nuclei per area (mm2) as in a previous study (Chew et al., 2015) by a blinded investigator.
Immunofluorescence staining and quantitative analysis
Wild-type HeLa cells and TMEM106B CRISPR knockout HeLa cells were plated on coverslips the day prior to transfection. Cells were fixed with 4% PFA in PBS 24 h post-transfection with PLP1 (OHu25924; GenScript) or GFP-tagged MOG [a gift from Dr Markus Reindl (Addgene plasmid #126463)]. After washing three times with PBS, coverslips were blocked with Odyssey Blocking Buffer (LI-COR Biosciences) containing 0.1% saponin (Sigma) for 1 h at room temperature. Cells were then incubated with the following primary antibodies: rabbit anti-TMEM106B (1:100) (Dr F. Hu, Cornell University, Ithaca, USA) (Brady et al., 2013), rabbit anti-PLP1 (1:250, ab28486; Abcam), and mouse anti-LAMP1 (1:250, #555798, BD Biosciences) in Odyssey Blocking Buffer overnight at 4°C. Cells were washed with PBS, followed by incubation with a 1:2000 mixture of Hoechst and secondary antibody (donkey anti-rabbit/mouse IgG Alexa Fluor® 488 and 594) at room temperature for 2 h. Cells were washed three times with PBS and mounted with Fluoromount G® (Southern Biotech) and imaged with a Zeiss LSM 700 laser scanning confocal microscope. Similarly, the cells from primary neuron-glia mixed cultures were fixed with 4% PFA in PBS at DIV (day in vitro) 7 or 10 and stained with rat-PLP1 (1:100) (clone aa3, Drs C. Linington and K. Muecklisch, University of Glasgow, Glasgow, UK) (Yamamura et al., 1991) and goat anti-mouse PSAP (1:250) (Dr Y. Sun, University of Cincinnati College of Medicine, Cincinnati, USA) (Sun et al., 2005).
For quantification of lysosome distribution, images of cells were randomly captured using a 63× objective. Images were then analysed in ImageJ (NIH) as previously described (Li et al., 2016) with some modification. Briefly, the whole-cell and nuclear areas were selected by the Freehand Selection tool and the perinuclear regions (defined as 5 μm around the nuclear membrane) were selected using the Enlarge tool. Intensities of LAMP1 were measured for the whole cell (Itotal), nucleus (Inuclear), and perinuclear areas (Iperinuclear). The percentage of perinuclear distributed lysosomes (Pperinuclear) was calculated as:
| (1) |
To quantify lysosomal PLP1 and MOG (ILyso) levels, the regions of interest for lysosomal areas were selected based on LAMP1 signals, the regions of interest were then applied to PLP1 channel or MOG and the fluorescence intensity was measured as in our previous study (Zhou et al., 2017a). To quantify PLP1 levels in the plasma membrane region (defined as a region 1 μm within the outline of the whole cell), the region of the whole cell and the region of whole cell without the plasma membrane region were selected by Freehand Selection and Enlarge tools. The intensities of PLP1 were measured for the selected whole cell (Itotal), and whole-cell without plasma membrane (Iw-pm). The percentage of lysosomal PLP1 or MOG (PLyso) and plasma membrane region PLP1 or MOG (PPM) were calculated, respectively as:
| (2) |
| (3) |
For the HeLa cell experiments, the measurements were made on images from three independent experiments (n = 20 per experiment) for all groups. For the primary neuron-glia mixed cultures, the measurements were made on images from 12 primary oligodendrocytes and results were confirmed by at least two additional independent experiments.
For immunofluorescence staining of MBP, paraffin-embedded mouse brain sections were deparaffinized as described above. Antigen retrieval was performed by microwaving in citrate buffer (pH 6.0) for 18 min. Slides were stained with rat anti-MBP (1:500, MCA409S; Bio-Rad). Images were acquired on an Aperio ScanScope FL Scanner (Leica Biosystems)
Western blotting
Brain tissues were homogenized with radioimmunoprecipitation assay (RIPA) buffer (Boston BioProducts) supplemented with protein inhibitors at a ratio of 1:20 (1 mg tissue : 20 μl RIPA). The supernatant was collected after centrifugation at 14 000 rpm for 15 min. Protein concentration was measured using the BCA assay (Thermo Fisher Scientific). Protein samples were denatured, separated, and transferred as a previous study (Zhou et al., 2017a). Primary antibodies included: rabbit anti-TMEM106B [a gift from Dr F. Hu (Brady et al., 2013), 1:1000], mouse anti-CNP (1:2000, 11-5B, MAB326; Millipore), rat anti-MBP (1:5000, MCA409S; Bio-Rad), mouse anti-GAPDH (H86504M; Meridian Life Sciences). Bands of western blots were quantified using ImageJ (NIH).
Electron microscopy
Mice were transcardially perfused with the electron microscopy (EM) fixation buffer (2% glutaraldehyde, 2% PFA in 0.1 M PBS). Brains were then dissected and further fixed in EM fixation buffer for 24 h at 4°C. The corpus callosum was dissected from the fixed brains and placed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4 overnight. After PBS washing, tissue was post-fixed in 1% OsO4, en bloc, washed three times in pure water, stained with 1% uranyl acetate and 50% ethanol, dehydrated in 70%, 80%, 95% and 100% ethanols and propylene oxide, infiltrated and embedded in Epon® 812 (Polysciences). Ultrathin sections were cut from the Epon® 812 embedded samples using a Leica Ultramicrotome, counterstained with uranyl acetate and lead citrate, and imaged with a Philips 208S electron microscope. To calculate the G-ratios of myelinated fibres, the diameter of the axon and the diameter of the axon with the myelin sheath were measured using ImageJ (NIH). The G-ratio of myelinated fibres was calculated as the ratio of the diameter of the axon to the diameter of the axon with the myelin sheath. Measurements were made on electron micrographs from three pairs of mice (n = 35 axons per mouse) in all experiments.
Luxol fast blue and toluidine blue staining
For Luxol fast blue staining, paraffin-embedded mouse brain sections were deparaffinized and hydrated as described above. Slides were incubated in Luxol fast blue solution (l0% Luxol fast blue dissolved in 95% ethanol and 5% acetic acid) overnight at room temperature. Stained slides were washed once in 95% ethanol and distilled water. Washed slides were incubated in a saturated lithium carbonate solution for ∼3 min and then washed once in distilled water. Stained brain slides were dehydrated again in ethanol and xylene and mounted with permanent mounting media. Images were captured using an Aperio ScanScope AT2 Slide Scanner (Leica Biosystems). The intensities of the Luxol fast blue staining (in blue colour) were measured with ImageJ (NIH).
For toluidine blue staining, mice were perfused with EM fixation buffer. Sciatic nerves were dissected and further fixed in EM fixation buffer overnight at 4°C. Fixed nerves were embedded and cut into 5 μm sections. Sciatic nerve tissues were incubated in 95% ethanol for 30 s and stained in 0.5% toluidine blue solution for 1 h at room temperature. Slides were then dehydrated and mounted as above. Stained sciatic nerves were imaged using an Aperio ScanScope AT2 Slide Scanner (Leica Biosystems). The G-ratios of the myelinated fibres were measured as described above using ImageJ. Measurements were made on images from three or four mice per group (n = 50 axons per mouse) in all experiments.
Statistical analysis
In all experiments, data were expressed as mean ± standard eror of the mean (SEM). One-way ANOVA followed by Bonferroni’s multiple comparison test was used to test for statistical significance between multiple groups. Student’s t-test was used to compare two groups. All statistical analyses were performed using the GraphPad Prism5 software (GraphPad Software, San Diego, CA). P-values <0.05 were considered statistically significant.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary material. The raw RNAseq data is available at Gene expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE145314) with the accession number: GSE145314.
Results
Transcriptomic analyses reveal global myelination deficits in Tmem106b−/− mouse brain
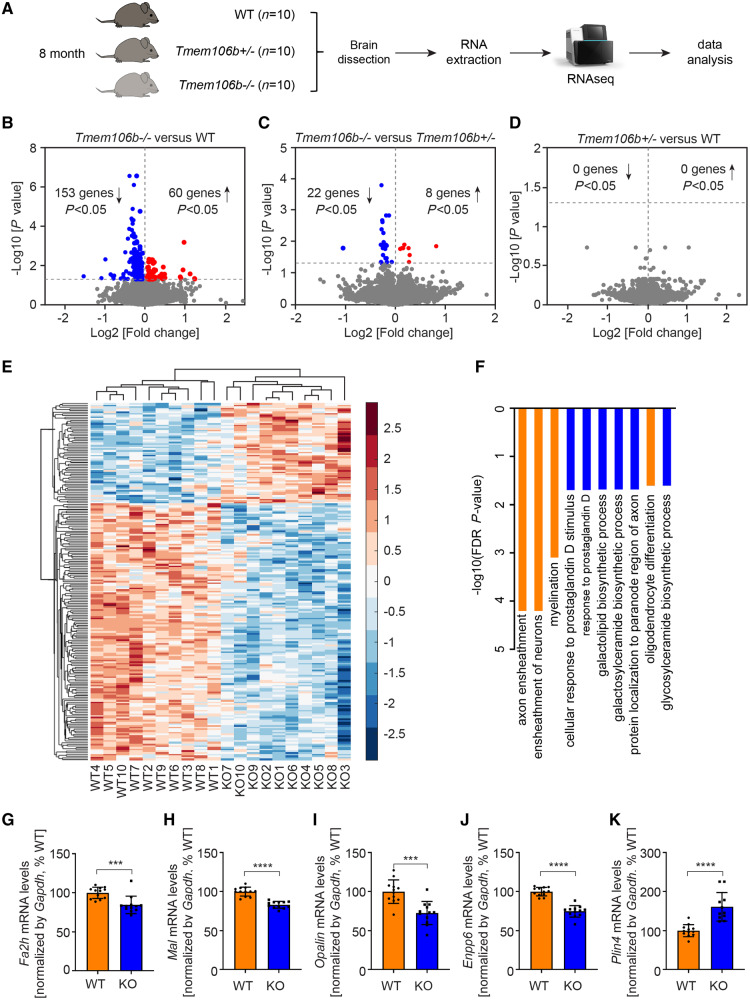
To understand the normal function of TMEM106B in vivo, we performed a large scale RNA sequencing study in our previously generated Tmem106b−/− mice (Nicholson et al., 2018). We included 10 Tmem106b+/+ (wild-type, 8 months), 10 Tmem106b+/− (8 months) and 10 Tmem106b−/− (8 months) mice with equal number of males and females (Fig. 1A). After gene normalization and filtering (log2 RPKM > 1), we identified 21 209 genes expressed across all genotypes for inclusion in the differential gene expression study. We identified 213 DEGs (153 downregulated and 60 upregulated) between Tmem106b−/− and wild-type (Fig. 1B, E and Supplementary Table 1), and 30 DEGs (22 downregulated and eight upregulated) between Tmem106b−/− and Tmem106b+/− (Fig. 1C and Supplementary Table 2) (step-up P-values < 0.05). No DEGs were identified when we compared Tmem106b+/− with wild-type animals (Fig. 1D). Notably, among the 30 DEGs identified between Tmem106b−/− and Tmem106b+/− mice, 25 overlapped with the DEGs identified when comparing Tmem106b−/− and wild-type mice (Supplementary Fig. 1A).
Figure 1.
Transcriptomics analysis reveals global changes in myelination pathways in TMEM106B deficient mice. (A) Diagram showing experimental procedures of transcriptomic analysis with Tmem106b+/+ (wild-type, WT), Tmem106b+/− and Tmem106b−/− mouse brains. (B–D) Volcano plots show significant DEGs between Tmem106b−/− and wild-type mice (B), Tmem106b−/− and Tmem106b+/− (C), and Tmem106b+/− and wild-type mice (D). Cut-off step-up P < 0.05. Red and blue colours represent upregulated and downregulated genes in Tmem106b−/− mice, respectively. (E) Hierarchical clustering of 213 DEGs between Tmem106b−/− and wild-type mice, step-up P < 0.05. Red and blue colours represent upregulated and downregulated genes in Tmem106b−/− mice, respectively. (F) Top 10 cellular processes identified by GO enrichment analysis from the DEGs between Tmem106b−/− and wild-type mice. Myelination-related cellular processes are highlighted in orange. (G–K) Quantitative PCR validation of selected down- (G–J) and upregulated (K) DEGs using 8-month-old brains from wild-type and Tmem106b−/− (KO) mice. Graphs represent the mean ± SEM. Data were analysed by Student’s t-test (n = 12 per group). ***P < 0.001, ****P < 0.0001.
Given that the comparison between Tmem106b−/− and wild-type mice showed the most DEGs, we performed GO (gene ontology) analysis based on the 213 DEGs identified. Interestingly, myelination-related cellular processes including axon ensheathment, ensheathment of neurons, myelination, and oligodendrocyte differentiation were identified as significantly enriched pathways (Fig. 1F). Moreover, processes involved in the biosynthesis of myelin abundant sphingolipids: galacolipid, galactosylceramide, and glycosylceramide biosynthetic processes (Chrast et al., 2011), also stood out as the top significantly enriched cellular processes (Fig. 1F). Further analysis revealed that almost 20% (37/213) of the DEGs were oligodendrocyte-enriched or myelination-related genes (Supplementary Fig. 1A) and all of these genes (except Gfap) were downregulated in Tmem106b−/− mice (Supplementary Fig. 1B). We subsequently carried out a WGCNA to identify highly correlated gene clusters/modules that were either up- or downregulated in a genotype-specific manner (Supplementary Fig. 2A). We identified 27 modules of which nine were significantly correlated with Tmem106b genotype: five upregulated and four downregulated modules (Supplementary Fig. 2A). Enrichment analyses revealed that the most significantly downregulated module (red module) was enriched for myelination-related processes [axon ensheathment Bonferoni P = 3.71 × 10−5; myelination Bonferoni P = 0.00018 (Supplementary Fig. 2B)], whereas translation-related processes (cytosolic ribosome Bonferoni P = 7.10 × 10−7 and ribosomal subunit Bonferoni P = 4.33 × 10−5) were enriched in the most significantly upregulated (turquoise) module (Supplementary Fig. 2C). We validated several of the most down- (Fa2h, Mal, Opalin, Enpp6) and upregulated (Plin4) DEGs by qPCR in 8-month-old mouse brains (Fig. 1G–K). Additional qPCR analyses across different ages revealed that the transcriptional changes in these genes were detectable at 3 months and did not appear to change further with age (Supplementary Fig. 3). Taken together, these results suggest that the loss of TMEM106B affects myelination in vivo.
Reduced number of oligodendrocytes but no obvious myelination deficits in Tmem106b−/− mouse brain
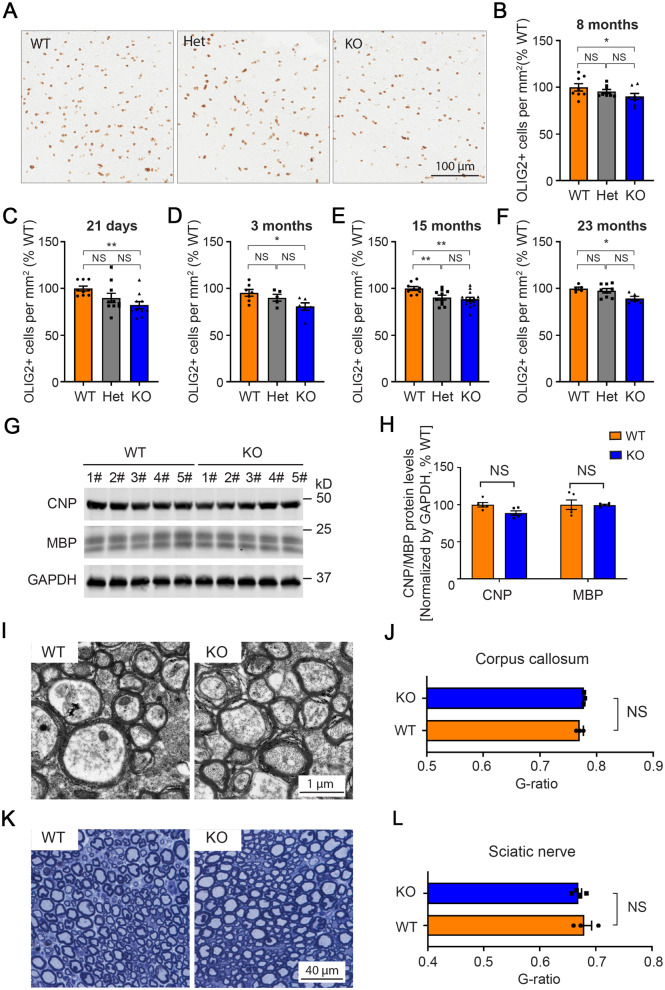
To investigate myelination in TMEM106B-deficient mice, we examined oligodendrocyte cell numbers in the corpus callosum of 8-month-old wild-type, Tmem106b+/−, and Tmem106b−/− mouse brains by immunohistochemistry staining for OLIG2 (Fig. 2A). We found a significant reduction of OLIG2-positive cells in Tmem106b−/− mice compared to wild-type mice (P < 0.05) (Fig. 2B). While the reduction in OLIG2-positive cells appeared to be dose-dependent, comparisons of wild-type with Tmem106b+/− and Tmem106b+/− with Tmem106b−/− mice did not reach significance (Fig. 2B). To examine whether ageing could enhance the loss of OLIG2-positive cells in a genotype-dependent manner, we quantified the number of OLIG2-positive cells in both 15- and 23-month-old mice and found that the relative loss of OLIG2-positive cells did not appear to increase with age (Fig. 2E and F). We also examined OLIG2 counts in younger mice (Fig. 2C and D) and determined that the reduction in OLIG2 counts in the TMEM106B-deficient mice were present at postnatal Day 21 (Fig. 2C). These results suggest that the reduction in oligodendrocytes in TMEM106B-deficient mice is not an ageing, but rather a developmental deficit. We also examined the OLIG2-positive cells in the cortex, a brain region with less density of oligodendrocytes. While OLIG2 cells also appeared to be reduced in the cortex of Tmem106b−/− mice, there was no statistically significant reduction of OLIG2-positive cells when compared to wild-type mice (Supplementary Fig. 4), possibly suggesting different regional vulnerabilities to the loss of TMEM106B.
Figure 2.
Assessment of the myelin in Tmem106b−/− mice. (A) Representative images of OLIG2 immunostaining in corpus callosum of 8-month-old Tmem106b+/+ (wild-type, WT), Tmem106b+/− (Het), and Tmem106b−/− knockout (KO) mouse brains. (B–F) Quantification of OLIG2-positive cells in the corpus callosum of the wild-type, Het, and KO mouse brains at indicated ages (n = 5–14 per group). (G) Western blots show protein levels of CNP, MBP, and GAPDH in Tmem106b+/+ (WT) and Tmem106b−/− (KO) adult mice. (H) Quantification of the blots in g (n = 5 per group). (I) Representative EM images of myelinated fibres in corpus callosum from wild-type and KO adult mice. (J) Measurement of the G-ratios in myelinated fibres in G (n = 3 per group). (K) Representative toluidine blue-stained sections of sciatic nerves from wild-type and KO adult mice. (L) Measurement of the G-ratios in myelinated fibres in g (n = 3–4 per group). Graphs represent the mean ± SEM. Data were analysed by Student’s t-test. NS = not significant, *P < 0.05, **P < 0.01.
We next examined the protein levels of myelin-associated proteins by western blot and determined that both CNP and MBP are only mildly reduced in Tmem106b−/− (P > 0.05) as compared to wild-type mice (Fig. 2G and H). We also investigated the fine structures of myelinated nerve fibres in both the central and peripheral nervous systems. No obvious difference in the thickness of the myelin was detected in either the corpus callosum or sciatic nerve between wild-type and Tmem106b−/− mice (Fig. 2I–L).
Tmem106b −/− mice are more susceptible to cuprizone-induced demyelination and have a reduced capacity to remyelinate
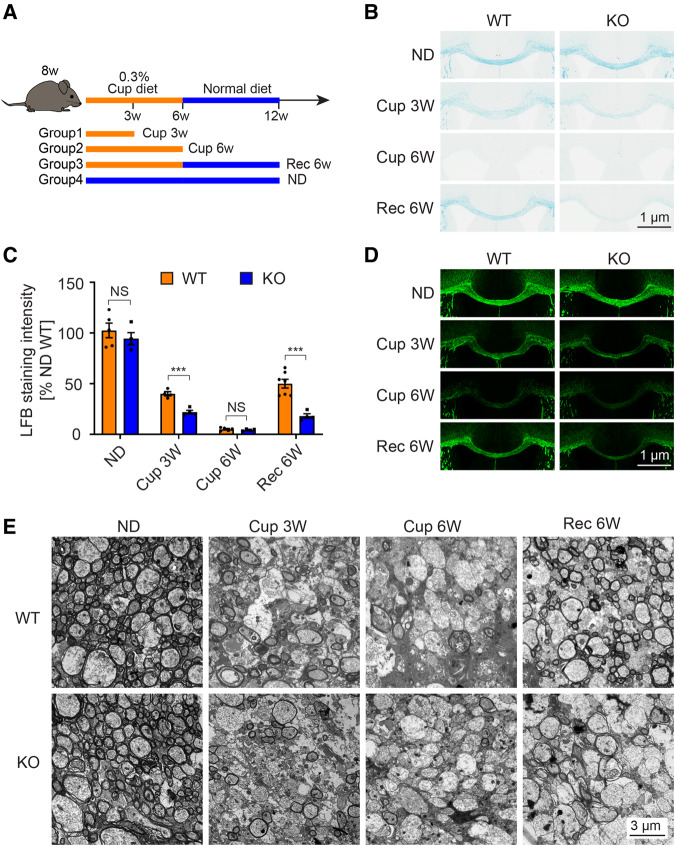
To investigate the role of TMEM106B in myelination, we challenged our Tmem106b−/− mice with cuprizone, a widely used toxin to study myelination in the corpus callosum (Skripuletz et al., 2011). To this end, mice were fed a cuprizone-containing diet and harvested at different time points (Fig. 3A). In wild-type mice, as previously reported (Kang et al., 2012; Guo et al., 2018), we observed rapid demyelination in the corpus callosum within 6 weeks of cuprizone treatment, which was followed by remyelination when the mice were switched back to a normal diet for 6 weeks (Fig. 3B). Also consistent with our western blot results (Fig. 2G and H), untreated mice showed no significant difference on Luxol fast blue stains of myelin in the corpus callosum between wild-type and Tmem106b−/− mice that were fed with a normal diet for 12 weeks (Fig. 3B and C). On the other hand, after 3 weeks of cuprizone treatment, Tmem106b−/− mice showed significantly reduced myelin (∼50% of wild-type, P < 0.001) as compared to wild-type mice (Fig. 3B and C). After 6 weeks of cuprizone treatment, this effect was no longer obvious. There was, however, significantly less regeneration of myelin (∼40% of wild-type, P < 0.001) in Tmem106b−/− mice compared to wild-type mice after 6 weeks of recovery on a normal diet. (Fig. 3B and C). MBP immunofluorescence staining (Fig. 3D) and electronic microscopy analysis of myelin in the corpus callosum (Fig. 3E) showed results comparable to the Luxol fast blue staining. Consistent with a previous study (Vega-Riquer et al., 2019), we also observed cuprizone treatment induced robust microglial activation in the corpus callosum (Supplementary Fig. 5A and B) and Tmem106b−/− mice showed a significant increase of microglia activation compared to wild-type mice (Supplementary Fig. 5A and B).
Figure 3.
Increased demyelination and reduced remyelination in Tmem106b−/− mice in a cuprizone-induced de- and remyelination mouse model. (A) Diagram shows experimental groups and procedures in the cuprizone-induced demyelination mouse model. (B) Representative images of Luxol fast blue (LFB) staining of corpus callosum from wild-type (WT) and Tmem106b−/− (KO) mice treated with indicated diets: Cup 3W = 3 weeks of cuprizone diet; Cup 6W = 6 weeks of cuprizone diet; ND = normal diet; Rec 6W = 6 weeks of cuprizone diet plus 6 weeks of normal diet. (C) Quantification of Luxol fast blue stained in B. (D) Representative images of MBP immunofluorescence staining of corpus callosum from the mice in B. (E) Representative EM images of corpus callosum from wild-type and KO mice treated as described in B. Graph represents the mean ± SEM. Data were analysed by Student’s t-test (n = 4–7 per group). NS = not significant, ***P < 0.001.
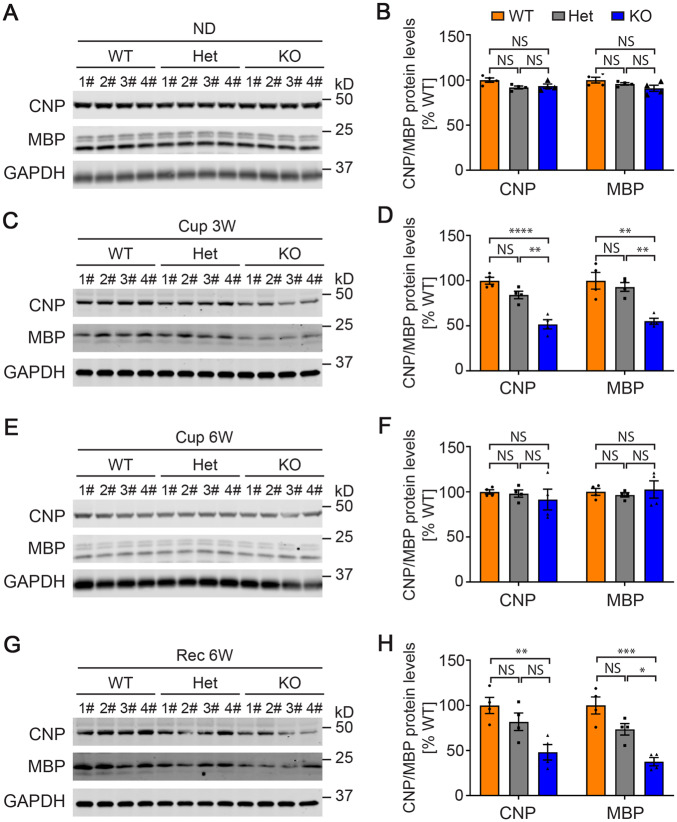
To verify these findings, we generated a second cohort of cuprizone-treated mice in which we also included Tmem106b+/− mice. We again observed significantly less residual and regenerated myelin-associated proteins (CNP and MBP) in Tmem106−/− mice compared to wild-type mice after 3 weeks of cuprizone treatment (P < 0.0001 for CNP, P < 0.01 for MBP) (Fig. 4C and D) and after 6 weeks of treatment followed by 6 weeks of recovery (P < 0.01 for CNP, P < 0.001 for MBP) (Fig. 4G and H). There was no significant difference between Tmem106b−/− and wild-type mice when fed a normal diet for 12 weeks (Fig. 4A and B) or after 6 weeks of cuprizone treatment (Fig. 4E and F). In addition, we observed dose-dependent effects of TMEM106B on both myelin loss in response to cuprizone treatment (Fig. 4C and D) and in the ability to increase myelin-associated proteins during recovery (Fig. 4G and H).
Figure 4.
Myelination-related protein levels in cuprizone diet-treated wild-type and TMEM106B-deficient mice. (A, C, E and G) Western blots show protein levels of CNP, MBP, and GAPDH in Tmem106b+/+ (wild-type, WT), Tmem106b+/− (heterozygous, Het), and Tmem106b−/− (KO) mice treated with normal diet (ND, A), 3 weeks of cuprizone diet (Cup 3W, C), 6 weeks of cuprizone diet (Cup 6W, E), and 6 weeks of cuprizone diet plus 6 weeks of normal diet (Rec 6W, G), respectively. (B, D, F and H) Quantification of the blots in A, C, E and G, respectively. Graphs represent the mean ± SEM. Data were analysed by Student’s t-test (n = 4). NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We previously identified a residual N-terminal fragment of TMEM106B in our Tmem106b−/− mouse model (Nicholson et al., 2018). Since it is unknown whether this residual N-terminal fragment of TMEM106B has a biological function, it raises the concern that the myelination deficits found in this knockout model might be due to this undefined TMEM106B fragment, rather than the loss of normal TMEM106B function. To exclude this possibility, we generated a new Tmem106b−/− mouse line by using CRISPR/Cas9 technology. In this new Tmem106b CRISPR knockout (KO) mouse, the CRISPR editing introduced a 5-bp deletion, which causes an early stop codon (Supplementary Fig. 6A and B). Unlike our previous line, we did not detect any residual TMEM106B fragment, nor did we detect any full-length TMEM106B by western blot (Supplementary Fig. 6C–F). Using this newly established Tmem106b CRISPR KO mouse line, we repeated the cuprizone experiments and verified the findings obtained in the initial TMEM106B deficient model. Specifically, Tmem106b CRISPR KO had a more rapid demyelination after 3 weeks of treatment and a reduced capacity to remyelinate after 6 weeks of treatment followed by 6 weeks of recovery based on assessment of Luxol fast blue stained sections (Supplementary Fig. 7A). These findings were confirmed by the quantitative analysis of CNP and MBP protein levels by western blot (Supplementary Fig. 7B–I). Taken together, these data suggest that loss of TMEM106B not only increases the susceptibility to the cuprizone-induced demyelination but also results in deficits in remyelination during recovery.
Loss of TMEM106B leads to a reduction in differentiated oligodendrocytes and PLP1 localization deficits
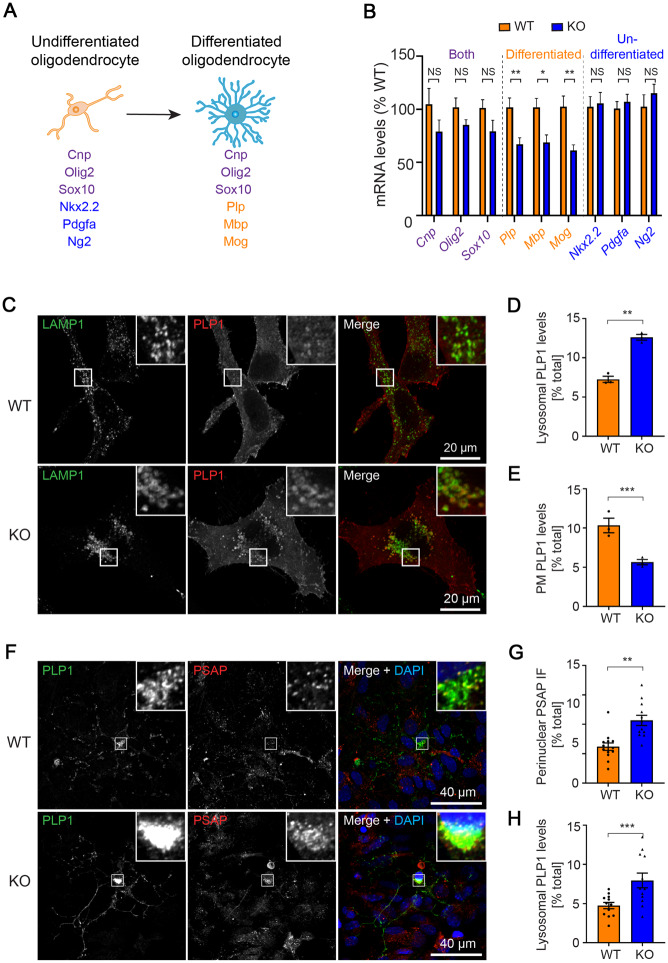
OLIG2 is a general oligodendrocyte marker expressed in both undifferentiated (oligodendrocyte progenitor cells and pre-oligodendrocytes) and differentiated (immature and mature oligodendrocytes) oligodendrocytes (Trapp et al., 1997). To determine whether the reduction in OLIG2-positive cells observed in the TMEM106B-deficient mice (Fig. 2A–F) was the result of a loss of undifferentiated oligodendrocytes, differentiated oligodendrocytes or both, we measured the mRNA levels of different oligodendrocyte cell markers by qPCR in 3-month-old wild-type and Tmem106b−/− mice (Fig. 5A and B). Expression levels of molecular markers present in both undifferentiated and differentiated oligodendrocytes including Olig2, Cnp, and Sox10, were reduced in Tmem106b−/− mice compared to wild-type mice (Fig. 5B), consistent with the results from OLIG2 counting. Interestingly, only the cell markers specific for differentiated oligodendrocytes (Plp1, Mbp, Mog), but not those unique to undifferentiated oligodendrocytes (Nkx2.2, Pdgfa, Ng2), were reduced to a significant extent in Tmem106b−/− as compared to wild-type mice (Fig. 5B). This suggests that the loss of oligodendrocytes in Tmem106b−/− mice is likely due to a reduction in differentiated oligodendrocytes.
Figure 5.
Reduction in differentiated oligodendrocytes and impaired PLP distribution in TMEM106B-deficient cells. (A) Diagram shows cellular markers of undifferentiated and differentiated oligodendrocytes. (B) Quantitative PCR shows the relative mRNA levels of immature and mature oligodendrocyte cellular markers in 3-month-old Tmem106b−/− (KO) mice compared to Tmem106b+/+ (wild-type, WT) (n = 6 per group). (C) Representative immunofluorescent images of LAMP1 and PLP1 from wild-type (WT) HeLa and TMEM106B CRISPR KO (KO) HeLa cells 24 h after PLP1 transfection. (D and E) Quantification of lysosomal (D) and plasma membrane (E) localized PLP1 in C (n = 3). (F) Representative immunofluorescent images of PSAP, PLP1, and DAPI from wild-type and Tmem106b−/− (KO) mouse primary neuron-glial mixed cultures. (G and H) Quantification of perinuclear-distributed PSAP+ vesicles (G) and lysosomal localized PLP1 (H) in F (n = 12). Graphs in B, D, E, G and H represent the mean ± SEM. Data were analysed by Student’s t-test. NS = not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
During oligodendrocyte maturation, the fate (either survival or degeneration) of an oligodendrocyte is largely determined by myelin formation, which in turn relies on proper trafficking of myelin protein to the plasma membrane (Trapp et al., 1997; Gow et al., 1998). PLP1 and MOG are two main myelin proteins in mature oligodendrocytes (Trajkovic et al., 2006) with distinct trafficking pathways. The plasma membrane targeting of PLP1 is mainly dependent on the lysosomal trafficking whereas MOG is mainly dependent on the trafficking of recycling endosomes (Winterstein et al., 2008; Baron and Hoekstra, 2010). To investigate the role of TMEM106B on PLP1 and MOG trafficking, we generated a TMEM106B−/− HeLa cell line by using CRISPR/Cas9 technology (Supplementary Fig. 8). Consistent with a previous study (Schwenk et al., 2014), immunofluorescence staining in these cells revealed that a complete loss of TMEM106B leads to significant accumulation/clustering of lysosomes, as visualized by LAMP1 staining, in the perinuclear region of the cells (Supplementary Fig. 9A and B). Moreover, when we transiently overexpressed PLP1 in these cells, we found PLP1 accumulated in lysosomes in TMEM106B−/− cells as evidenced by a significant increase of PLP1 in LAMP1+ compartments, and a significant decrease of PLP1 at the plasma membrane region in the TMEM106B−/− cells as compared to wild-type HeLa cells (Fig. 5C–E). In contrast, loss of TMEM106B failed to change the distribution of MOG (Supplementary Fig. 9C–E). By using primary neuron-glia mixed cultures, we further confirmed that loss of TMEM106B also results in significant clustering of lysosomes, as visualized by prosaposin (PSAP, another lysosomal resident protein) staining, in the perinuclear region of primary oligodendrocytes (Fig. 5F and G). Moreover, similar to the finding in TMEM106B−/− Hela cells, accumulated PLP1 was also found in the Tmem106b−/− primary oligodendrocytes (Fig. 5F and H). Taken together, these results suggest that loss of TMEM106B mainly leads to distribution deficits of the lysosomal-dependent myelin protein PLP1, but not to lysosomal-independent proteins such as MOG.
Discussion
While initially reported in relation to FTD, recent studies have revealed a broader impact of TMEM106B variants on brain ageing, including a profound effect on gene expression in aged human brain (Rhinn and Abeliovich, 2017; Ren et al., 2018; Li et al., 2020), and an association with neuronal proportion in brain tissue derived from neurodegenerative disease patients and elderly, cognitively healthy, controls (Li et al., 2020). Accumulating evidence suggests that the TMEM106B haplotype that confers disease risk (and reduced neuronal number) is associated with increased levels of TMEM106B (Van Deerlin et al., 2010; Busch et al., 2013). Reducing TMEM106B levels, therefore, seems to be an attractive therapeutic or disease-modifying approach, especially in patients with FTD-GRN, where the strongest genetic association is observed (Pottier et al., 2018). In this study, we characterized our previously generated TMEM106B-deficient mouse model to determine the potential risks of such a treatment strategy and to further characterize the normal function of TMEM106B.
Using brain transcriptomics, we found enrichment for differently expressed genes implicated in myelination-related processes, with a significant reduction in the expression of these genes in Tmem106b−/− as compared to wild-type or Tmem106b+/− mice. Genes involved in the biosynthesis of galactolipids, such as galactosylceramide, which are important components of the myelin sheath (Hoshi et al., 1973), were also reduced in Tmem106b−/− mouse brain. Co-expression analysis confirmed these findings and revealed a module enriched for genes implicated in exon ensheathment and myelination as the most correlated with the Tmem106b genotype. The observed gene expression changes appeared to result from a significant loss of oligodendrocytes in Tmem106b−/− compared to wild-type mice, which we quantified using immunohistochemistry in the corpus callosum. The cell loss was already present in animals at 21 days of age and did not appear to worsen with age (studied until 23 months). Interestingly, the effect of TMEM106B levels on the number of oligodendrocytes appeared dose-dependent at most time-points. The significant effect on myelination-related gene expression and oligodendrocyte cell loss in Tmem106b−/− suggested an important role for TMEM106B in myelination; however, at the ultrastructural levels, we did not detect obvious changes in myelin and only a mild and non-significant reduction of myelin-associated proteins was detected. This prompted us to challenge our model with cuprizone, a commonly used toxin which leads to the selective loss of oligodendrocytes and microglia accumulation and extensive areas of demyelination and gliosis in the corpus callosum (Vega-Riquer et al., 2019). Cuprizone treatment clearly showed that loss of TMEM106B increased the susceptibility towards myelin loss and decreased the capability to regenerate myelin, a finding which we validated using a newly generated Tmem106b CRISPR KO mouse. The addition of this new model excludes the involvement of the small, residual TMEM106B N-terminal fragment detected in the original knockout model (Nicholson et al., 2018) in these mechanisms, and reaffirms loss TMEM106B function as the primary driver of the myelination deficit.
The proper function of lysosomes including their trafficking is essential for normal myelination as demonstrated by the frequent occurrence of myelination deficits in lysosomal storage diseases, including Niemann-Pick disease, Gaucher disease and metachromatic leukodystrophy (Folkerth, 1999; Faust et al., 2010; Boustany, 2013). PLP1, which is the most abundant myelin-specific protein in oligodendrocytes, is largely localized at lysosomes (Shen et al., 2016) and its function is dependent on intact lysosomal function. Specifically, following its biosynthesis in the rough endoplasmic reticulum (ER), PLP1 is transported to the Golgi and plasma membrane, from where it is internalized through endocytosis and stored in late endosomes and lysosomes in the absence of neuronal signals. During myelination, PLP1 is targeted to the plasma membrane through lysosomal exocytosis (Trajkovic et al., 2006; Feldmann et al., 2011). Since the loss of TMEM106B has previously been reported to cause impairments in lysosomal acidification (Klein et al., 2017) and lysosomal trafficking (Schwenk et al., 2014), we hypothesized a possible effect on PLP1 upon TMEM106B loss. Indeed, using a TMEM106B CRISPR KO HeLa cell line and primary cultured oligodendrocytes, we confirmed that the loss of TMEM106B leads to an accumulation of lysosomes in the perinuclear region (Schwenk et al., 2014) and we observed that this apparent defect in lysosomal trafficking resulted in the accumulation of lysosomal PLP1 and a reduction in PLP1 at the plasma membrane area. In line with our expectations, the localization of MOG, an important myelin protein which traffics independent of lysosomes, was not altered. Given the limitation that we only used fixed cells in this study, which only reflect the condition at a single point in time, dynamic studies of PLP1 trafficking using sophisticated live imaging techniques will be needed to confirm these findings. Notably, increased TMEM106B has been recently associated with increased lysosomal exocytosis (Kundu et al., 2018) and impaired lysosomal exocytosis has been shown to directly lead to plasma membrane trafficking deficits of PLP1 (Shen et al., 2016). Thus, it is possible that loss of TMEM106B might reduce lysosomal exocytosis thereby further impairing the PLP1 trafficking from lysosomes to the plasma membrane. Whether this is the case in Tmem106b−/− model also needs further investigation. Importantly, the trafficking of PLP1 from lysosomes to the plasma membrane is a key process during myelination and controls oligodendrocyte cell death (Trapp et al., 1997). We observed significant oligodendrocyte loss as early as postnatal Day 21 in our Tmem106b−/− mouse brain (Fig. 2C). The transcriptional analysis of the oligodendrocyte differentiation markers (Fig. 5B) showed a specific loss of differentiated oligodendrocytes, suggesting that cell loss may happen during myelination. In sum, these findings suggest that TMEM106B may influence myelination through control of lysosomal trafficking. When disrupted, it may induce PLP1 trafficking deficits and associated oligodendrocyte cell loss.
Our identification of an important role for TMEM106B in myelination is also relevant in relation to a recently reported recurrent de novo mutation in TMEM106B (p.D252N) in patients with hypomyelinating leukodystrophy (Simons et al., 2017). In fibroblasts derived from one p.D252N-patient carrier, normal levels of TMEM106B were reported; however, lysosomal acidification was impaired and the activity of specific lysosomal enzymes was reduced (Ito et al., 2018). The effect of this mutation on PLP1 trafficking remains to be studied but our findings suggest that loss of normal TMEM106 function could contribute to the phenotype in these patients.
Within the CNS, TMEM106B is expressed in many cell types including neurons, astrocytes, oligodendrocytes, and microglia (Zhang et al., 2014). Our transcriptomic analysis found a significant increase in Gfap expression in Tmem106b−/− when compared with wild-type mice (Supplementary Table 1 and Supplementary Fig. 1B), which we confirmed by GFAP immunohistochemical analysis (data not shown), suggesting that the loss of TMEM106B could activate astrocytes. In Tmem106b−/− mouse brains, we further observed a significant increase in the expression of Trem2, Tyrobp, Lyz2, Plek, C1qb and C4b which are part of the TYROBP network of genes associated with microglial neuroinflammation (Supplementary Fig. 10A and B) (Klein et al., 2017; Takahashi et al., 2017). The fact that most of these genes were also increased in progranulin-deficient mice (Lui et al., 2016; Klein et al., 2017; Takahashi et al., 2017) may suggest that the loss of Tmem106b might exacerbate Grn−/− pathologies through the inflammatory response. As such, TMEM106B might also play an important role in the regulation of astrocyte and microglia activation, in addition to myelination.
While it is known that TMEM106B plays an important role in lysosomes (Nicholson and Rademakers, 2016), we did not find general transcriptional changes in terms of lysosomal-related genes. Loss of TMEM106B has been reported to ameliorate the phenotypes of progranulin-deficient mice partially by correcting lysosomal deficits of Grn−/− mice (Klein et al., 2017). However, we did not observe any suppression of the top upregulated lysosomal genes reported in Grn−/− mice in our Tmem106b−/− mice (Supplementary Fig. 11). In contrast, several important lysosomal enzymes such cathpesin B (Ctsb), cathpesin S (Ctss), and cathpesin D (Ctsd) (P < 0.05) were found to be increased in Tmem106b−/− as compared to wild-type mice (Supplementary Fig. 11) suggesting loss of TMEM106B does not normalize the lysosomal changes in Grn−/− mice, at least at the transcriptional level.
Together our analyses of Tmem106b−/− mice raise important concerns in relation to the lowering of TMEM106B levels as a therapeutic strategy in FTD-GRN, or age-associated neurodegenerative diseases in general. While a partial reduction of TMEM106B could be considered, we note that a non-significant trend towards reduced oligodendrocytes number and altered myelination and remyelination was observed in our cuprizone-induced toxicity model in Tmem106b+/− animals (Figs 2A–F and 4). Furthermore, a partial loss of TMEM106B did not correct lysosomal abnormalities and FTD related phenotypes due to progranulin haploinsufficiency (Arrant et al., 2018), although a complete loss of TMEM106B has been shown to rescue lysosomal and FTD-related phenotypes in progranulin-deficient mice (Klein et al., 2017). As the field continues to focus on TMEM106B as a possible therapeutic target, it will be important to take these findings into consideration; however, until we fully understand the biological consequence of the TMEM106B haplotypes discussions on therapies remain hypothetical. It also remains possible that the myelination phenotype we observed is mainly developmental in origin which would leave antisense oligonucleotide-like treatments in adults as a viable strategy (Clayton et al., 2018). Finally, one should also consider that it remains possible that the coding variant p.T185S induces a functional difference between the TMEM106B risk and protective haplotype on top of the change in protein levels and that this functional change is responsible for the protective effects observed in people (Pottier et al., 2018). Future studies should interrogate this.
Supplementary Material
Acknowledgements
The authors thank Dr Katja Muecklisch and Dr Christopher Linington for providing antibodies against mouse PLP1, Dr Ying Sun for providing antibody against mouse PSAP, and Dr Fenghua Hu for providing antibody against TMEM106B.
Funding
The research leading to these results has received funding by the National Institutes of Health (NIH) grant R35 NS097261 and the Bluefield Project to Cure FTD. The generation of the CRISP/Cas9 Tmem106b knock-out mouse model at The Jackson Laboratory was supported by The Center for Precision Genetics, NIH U54 OD020351 and partially supported by the Jackson Laboratory’s Genetic Engineering Technologies Scientific Service. X.Z. is supported by a research fellowship from The Bluefield Project to Cure FTD and a Developmental grant from the Mayo Clinic ADRC (NIH P30 AG062677).
Competing interests
The authors report no competing interests.
Glossary
- DEG =
differentially expressed gene
- FTD =
frontotemporal dementia
References
- Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, et al. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer’s disease. Ann Neurol 2007; 61: 435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrant AE, Nicholson AM, Zhou X, Rademakers R, Roberson ED.. Partial Tmem106b reduction does not correct abnormalities due to progranulin haploinsufficiency. Mol Neurodegener 2018; 13: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006; 442: 916–9. [DOI] [PubMed] [Google Scholar]
- Baron W, Hoekstra D.. On the biogenesis of myelin membranes: sorting, trafficking and cell polarity. FEBS Lett 2010; 584: 1760–70. [DOI] [PubMed] [Google Scholar]
- Boustany RM. Lysosomal storage diseases–the horizon expands. Nat Rev Neurol 2013; 9: 583–98. [DOI] [PubMed] [Google Scholar]
- Brady OA, Zheng Y, Murphy K, Huang M, Hu F.. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum Mol Genet 2013; 22: 685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch JI, Martinez-Lage M, Ashbridge E, Grossman M, Van Deerlin VM, Hu F, et al. Expression of TMEM106B, the frontotemporal lobar degeneration-associated protein, in normal and diseased human brain. Acta Neuropathol Commun 2013; 1: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Unger TL, Gallagher MD, Bill E, Kwong LK, Volpicelli-Daley L, et al. TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J Neurosci 2012; 32: 11213–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 2015; 348: 1151–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrast R, Saher G, Nave KA, Verheijen MH.. Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J Lipid Res 2011; 52: 419–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton EL, Milioto C, Muralidharan B, Norona FE, Edgar JR, Soriano A, et al. Frontotemporal dementia causative CHMP2B impairs neuronal endolysosomal traffic-rescue by TMEM106B knockdown. Brain 2018; 141: 3428–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006; 442: 920–4. [DOI] [PubMed] [Google Scholar]
- Faust PL, Kaye EM, Powers JM.. Myelin lesions associated with lysosomal and peroxisomal disorders. Expert Rev Neurother 2010; 10: 1449–66. [DOI] [PubMed] [Google Scholar]
- Feldmann A, Amphornrat J, Schonherr M, Winterstein C, Mobius W, Ruhwedel T, et al. Transport of the major myelin proteolipid protein is directed by VAMP3 and VAMP7. J Neurosci 2011; 31: 5659–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology 2011; 76: 467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkerth RD. Abnormalities of developing white matter in lysosomal storage diseases. J Neuropathol Exp Neurol 1999; 58: 887–902. [DOI] [PubMed] [Google Scholar]
- Gallagher MD, Posavi M, Huang P, Unger TL, Berlyand Y, Gruenewald AL, et al. A dementia-associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am J Hum Genet 2017; 101: 643–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet 2006; 15: 2988–3001. [DOI] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Lazzarini RA.. Disrupted proteolipid protein trafficking results in oligodendrocyte apoptosis in an animal model of Pelizaeus-Merzbacher disease. J Cell Biol 1998; 140: 925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Suo N, Cui X, Yuan Q, Xie X.. Vitamin C promotes oligodendrocytes generation and remyelination. Glia 2018; 66: 1302–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, Wu Z.. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 2012; 13: 204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi M, Williams M, Kishimoto Y.. Characterization of brain cerebrosides at early stages of development in the rat. J Neurochem 1973; 21: 709–12. [DOI] [PubMed] [Google Scholar]
- Ito Y, Hartley T, Baird S, Venkateswaran S, Simons C, Wolf NI, et al. Lysosomal dysfunction in TMEM106B hypomyelinating leukodystrophy. Neurol Genet 2018; 4: e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA, et al. MAP-RSeq: mayo analysis pipeline for RNA sequencing. BMC Bioinformatics 2014; 15: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Z, Liu L, Spangler R, Spear C, Wang C, Gulen MF, et al. IL-17-induced Act1-mediated signaling is critical for cuprizone-induced demyelination. J Neurosci 2012; 32: 8284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ZA, Takahashi H, Ma M, Stagi M, Zhou M, Lam TT, et al. Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 2017; 95: 281–96 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu ST, Grzeskowiak CL, Fradette JJ, Gibson LA, Rodriguez LB, Creighton CJ, et al. TMEM106B drives lung cancer metastasis by inducing TFEB-dependent lysosome synthesis and secretion of cathepsins. Nat Commun 2018; 9: 2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang CM, Fellerer K, Schwenk BM, Kuhn PH, Kremmer E, Edbauer D, et al. Membrane orientation and subcellular localization of transmembrane protein 106B (TMEM106B), a major risk factor for frontotemporal lobar degeneration. J Biol Chem 2012; 287: 19355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rydzewski N, Hider A, Zhang X, Yang J, Wang W, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol 2016; 18: 404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Farias FHG, Dube U, Del-Aguila JL, Mihindukulasuriya KA, Fernandez MV, et al. The TMEM106B FTLD-protective variant, rs1990621, is also associated with increased neuronal proportion. Acta Neuropathol 2020; 139: 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell 2016; 165: 921–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol 2009; 117: 15–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray ME, Cannon A, Graff-Radford NR, Liesinger AM, Rutherford NJ, Ross OA, et al. Differential clinicopathologic and genetic features of late-onset amnestic dementias. Acta Neuropathol 2014; 128: 411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary D, Snowden J, Mann D.. Frontotemporal dementia. Lancet Neurol 2005; 4: 771–80. [DOI] [PubMed] [Google Scholar]
- Nicholson AM, Finch NA, Wojtas A, Baker MC, Perkerson RB 3rd, Castanedes-Casey M, et al. TMEM106B p.T185S regulates TMEM106B protein levels: implications for frontotemporal dementia. J Neurochem 2013; 126: 781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson AM, Rademakers R.. What we know about TMEM106B in neurodegeneration. Acta Neuropathol 2016; 132: 639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson AM, Zhou X, Perkerson RB, Parsons TM, Chew J, Brooks M, et al. Loss of Tmem106b is unable to ameliorate frontotemporal dementia-like phenotypes in an AAV mouse model of C9ORF72-repeat induced toxicity. Acta Neuropathol Commun 2018; 6: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier C, Zhou X, Perkerson RB 3rd, Baker M, Jenkins GD, Serie DJ, et al. Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study. Lancet Neurol 2018; 17: 548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnavalli E, Brayne C, Dawson K, Hodges JR.. The prevalence of frontotemporal dementia. Neurology 2002; 58: 1615–21. [DOI] [PubMed] [Google Scholar]
- Ren Y, van Blitterswijk M, Allen M, Carrasquillo MM, Reddy JS, Wang X, et al. TMEM106B haplotypes have distinct gene expression patterns in aged brain. Mol Neurodegener 2018; 13: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn H, Abeliovich A.. Differential aging analysis in human cerebral cortex identifies variants in TMEM106B and GRN that regulate aging phenotypes. Cell Syst 2017; 4: 404–15.e5. [DOI] [PubMed] [Google Scholar]
- Rutherford NJ, Carrasquillo MM, Li M, Bisceglio G, Menke J, Josephs KA, et al. TMEM106B risk variant is implicated in the pathologic presentation of Alzheimer disease. Neurology 2012; 79: 717–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk BM, Lang CM, Hogl S, Tahirovic S, Orozco D, Rentzsch K, et al. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J 2014; 33: 450–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YT, Gu Y, Su WF, Zhong JF, Jin ZH, Gu XS, et al. Rab27b is involved in lysosomal exocytosis and proteolipid protein trafficking in oligodendrocytes. Neurosci Bull 2016; 32: 331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C, Dyment D, Bent SJ, Crawford J, D’Hooghe M, Kohlschutter A, et al. A recurrent de novo mutation in TMEM106B causes hypomyelinating leukodystrophy. Brain 2017; 140: 3105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripuletz T, Gudi V, Hackstette D, Stangel M.. De- and remyelination in the CNS white and grey matter induced by cuprizone: the old, the new, and the unexpected. Histol Histopathol 2011; 26: 1585–97. [DOI] [PubMed] [Google Scholar]
- Stagi M, Klein ZA, Gould TJ, Bewersdorf J, Strittmatter SM.. Lysosome size, motility and stress response regulated by fronto-temporal dementia modifier TMEM106B. Mol Cell Neurosci 2014; 61: 226–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Quinn B, Witte DP, Grabowski GA.. Gaucher disease mouse models: point mutations at the acid beta-glucosidase locus combined with low-level prosaposin expression lead to disease variants. J Lipid Res 2005; 46: 2102–13. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Klein ZA, Bhagat SM, Kaufman AC, Kostylev MA, Ikezu T, et al. Opposing effects of progranulin deficiency on amyloid and tau pathologies via microglial TYROBP network. Acta Neuropathol 2017; 133: 785–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Dhaunchak AS, Goncalves JT, Wenzel D, Schneider A, Bunt G, et al. Neuron to glia signaling triggers myelin membrane exocytosis from endosomal storage sites. J Cell Biol 2006; 172: 937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD, Nishiyama A, Cheng D, Macklin W.. Differentiation and death of premyelinating oligodendrocytes in developing rodent brain. J Cell Biol 1997; 137: 459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet 2010; 42: 234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Riquer JM, Mendez-Victoriano G, Morales-Luckie RA, Gonzalez-Perez O.. Five decades of cuprizone, an updated model to replicate demyelinating diseases. Curr Neuropharmacol 2019; 17: 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterstein C, Trotter J, Kramer-Albers EM.. Distinct endocytic recycling of myelin proteins promotes oligodendroglial membrane remodeling. J Cell Sci 2008; 121: 834–42. [DOI] [PubMed] [Google Scholar]
- Yamamura T, Konola JT, Wekerle H, Lees MB.. Monoclonal antibodies against myelin proteolipid protein: identification and characterization of two major determinants. J Neurochem 1991; 57: 1671–80. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun L, Bracko O, Choi JW, Jia Y, Nana AL, et al. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations. Nat Commun 2017. a; 8: 15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun L, Brady OA, Murphy KA, Hu F.. Elevated TMEM106B levels exaggerate lipofuscin accumulation and lysosomal dysfunction in aged mice with progranulin deficiency. Acta Neuropathol Commun 2017. b; 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zoller T, Krieglstein K, Spittau B.. TGFbeta1 inhibits IFNgamma-mediated microglia activation and protects mDA neurons from IFNgamma-driven neurotoxicity. J Neurochem 2015; 134: 125–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its Supplementary material. The raw RNAseq data is available at Gene expression Omnibus (GEO) repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE145314) with the accession number: GSE145314.