Abstract
Polyamines are important for regulating biofilms and the exopolysaccharide of the biofilm matrix of Bacillus subtilis. Understanding how enzymes can regulate polyamine concentrations is critical for learning more about how these processes occur in diverse bacteria. Here, we describe the structure and function of another member of the spermidine/spermine acetyltransferases (SSAT) found in Bacilli. The SpeG enzyme from B. thuringiensis (BtSpeG) binds polyamines in its allosteric site and adopts a dodecameric oligomeric state similar to other SpeG enzymes from Gram-negative bacteria. Our kinetic results show the catalytic efficiency of BtSpeG was greater than any previously characterized SpeG to date, and in contrast to other SpeG proteins it exhibited very similar kinetic properties toward both spermine and spermidine. Similar to the SpeG enzyme from E. coli, BtSpeG was able to acetylate spermidine on the N1 and N8 positions. The turnover of BtSpeG toward spermine and spermidine was also two to three orders of magnitude greater than any other Bacilli SSAT enzyme that has been previously characterized. SpeG proteins from Bacilli, including B. cereus, B. thuringiensis and B. anthracis share nearly identical sequences and therefore our results likely provide insight into the structure/function relationship across multiple Bacillus species.
Keywords: acetyltransferase, SpeG, dodecamer, x-ray crystallography, polyamine, Bacillus
Introduction
Polyamines are small aliphatic cations that play major roles in growth, development, and pathogenesis and are found across all domains of life [1–4]. In prokaryotes such as Shigella, the polyamines spermine and spermidine increase bacterial survival under oxidative stress [5], while in Escherichia coli, increased levels of spermidine can be toxic [6]. Polyamines can also act as signalling molecules to regulate biofilms in Vibrio cholerae through NspS/MbaA [7]. Regulation of intracellular polyamine concentrations is largely attributed to spermidine/spermine N-acetyltransferases (SSAT), which can partially neutralize polyamine charge through the catalytic transfer of an acetyl group from acetyl co-enzyme A (AcCoA) to a primary amine on the polyamine. These enzymes belong to the Gcn5-related N-acetyltransferase (GNAT) superfamily, and while they share a conserved GNAT fold they exhibit structural differences.
Many SSAT enzymes have been discovered in bacteria, including BltD, PaiA, and SpeG. All of these enzymes have been shown to acetylate polyamines, but currently SpeG is the only SSAT enzyme with an allosteric site that binds polyamines [8]. Moreover, the SpeG enzyme from Gram-negative bacteria is the only SSAT to form a unique dodecameric assembly, which is unusual for polyamine acetyltransferases and other members of the GNAT superfamily. It has also been shown that spermine binding to V. cholerae SpeG can alter oligomerisation and tighten the overall dodecameric structure of the enzyme [9]. In addition to its catabolic function, SpeG has been implicated in a variety of additional roles in bacteria. For example, the E. coli SpeG enzyme is critical for acetylating polyamines under stressful conditions [6,10] and it can regulate the transcription of the small RNA rprA by interacting with the transcription factor RcsB [11]. SpeG can also regulate a variety of genes in Salmonella enterica, including those important for type I and III secretion systems, flagellar biosynthesis, and histidine biosynthesis, among others [12]. Interestingly, the SpeG enzyme is not conserved in all bacteria but in certain cases it can be acquired. For example, most strains of the Gram-positive bacterium Staphylococcus aureus do not have the speG gene, but the methicillin resistant S. aureus USA 300 strain has recently acquired it via an Arginine Catabolic Mobile Element (ACME) [13]. The acquisition of this gene appears to be important for its pathogenicity [14,15], and the gene encodes a functional protein capable of acetylating polyamines [16].
With the exception of the recently characterized S. aureus SpeG enzyme [16], most of what is known about SpeG enzymes stems from studies with Gram-negative bacteria. On the other hand, much of the knowledge that exists in regards to other polyamine acetyltransferases (e.g. BltD and PaiA) is from Gram-positive B. subtilis.Bacilli are rod-shaped, spore forming bacteria that include opportunistic and pathogenic species such as B. cereus (food-born pathogen [17,18]), B. thuringiensis (biological insecticide [19,20]) and B. anthracis (air-born pathogen [21]). They are also normal residents of the soil and gastrointestinal tract of animals (e.g. B. subtilis). These bacterial species are closely related and have been reported to perform horizontal gene transfer [22]. Since biofilms are critical for the life of many Bacilli and it has been shown that spermidine is required for B. subtilis biofilms [23,24] and can regulate transcription of exopolysaccharides important for the B. subtilis biofilm matrix [25], we chose to study the SpeG enzyme from Bacillus. To our knowledge, B. subtilis does not contain a SpeG homolog in its genome, but other Bacillus species such as B. cereus, B. anthracis, and B. thuringiensis do and share very high sequence similarity (≥97%; Supplemental Materials Figure 1). These three bacteria are considered to be the same species [26] and are only differentiated by the plasmids they retain. Therefore, it is highly likely SpeG proteins from these organisms will be structurally and functionally similar and provide insight into how this protein functions across these pathogens. Here, we specifically focused on structurally and functionally characterizing the SpeG enzyme from B. thuringiensis (BtSpeG) since its sequence was available at the time we began the study. Ultimately, the motivation for our study was to compliment and expand the current knowledge of structural and functional properties of SSAT enzymes in Gram-positive bacteria.
Materials and Methods
Materials
Spermine tetrahydrochloride, spermidine trihydrochloride, agmatine sulfate, cadaverine dihydrochloride, putrescine dihydrochloride, and acetyl-coenzyme A trilithium salt, and dansyl chloride were purchased from Millipore Sigma. N1-acetyl spermidine and N8- acetylspermidine were purchased from Santa Cruz Biotechnology.
Cloning
The speG gene from Bacillus thuringiensis (NCBI Accession: WP_000076212; UniProt ID: A0A0G3E2X5) was codon optimised, synthesised, and cloned into the pMCSG21 vector (Genescript, Piscataway, NJ) using the SspI restriction site [27]. This vector encodes an N- terminal 6xHis-tag and TEV protease cleavage site for tag removal prior to the start codon of the plasmid and is spectinomycin resistant.
Recombinant Protein Expression
For protein crystallization
The plasmid was transformed into E. coli BL21 pLysS competent cells using the heat-shock method followed by recombinant protein expression using auto-induction methods [28]. After 30 h of incubation at room temperature, cells were harvested by centrifugation and resuspended in 50 mM phosphate lysis buffer pH 8.0 containing 300 mM NaCl and 20 mM imidazole. Cells were further lysed by two repetitive freeze-thaw cycles at −20°C and addition of 40 mg of lysozyme and 0.5 mg of DNAse.
For enzyme kinetics assays
The plasmid was transformed into E. coli BL21 (DE3) competent cells, which were prepared using the Zymo Research Mix & Go E. coli Transformation Kit. A single colony was used to make a glycerol stock and it was stored at - 80°C until ready to use. Cells were grown, heterologously expressed, and harvested using the procedure described [29] except that 50 μg/mL of spectinomycin was added instead of ampicillin in all cultures.
Protein Purification and Crystallisation
The cell debris was pelleted by centrifugation at 12,000 rpm for 30 min and filtered by passing through a Millipore 0.45 μm syringe filter. The cleared lysate was then loaded onto a pre-equilibrated 5 ml Ni2+ column (HisTrap HP, GE Healthcare). Following a 10-column volume wash, the target protein was eluted using an imidazole gradient over five column volumes in elution buffer containing 50 mM phosphate buffer pH 8.0, 300 mM NaCl and 500 mM imidazole. Eluted protein fractions were combined and treated for 12 h with TEV protease and further purified by size-exclusion chromatography (Superdex 200pg 26/600 column, GE Healthcare) using 50mM Tris-buffered saline pH 8.0, 125 mM NaCl. Eluted fractions containing the target protein were collected and concentrated using a 10 kDa MWCO Amicon ultrafiltration device (Millipore) to 8 mg/ml and stored at −80°C. Diffraction quality crystals were formed using the hanging drop vapour diffusion method, with each coverslip containing 1.5 μL of protein and 1.5 μL of reservoir solution suspended over 300 μL of reservoir solution in 48-well plates. For ligand-bound protein, BtSpeG was mixed with a 10-molar excess of spermine and AcCoA than the protein concentration prior to screening for crystals. The reservoir solution was comprised of either 0.1M Tris pH 7.5, 7% PEG 4000 and 0.2 M lithium sulfate for ligand-free protein crystals, or 30% ethanol for ligand-bound crystals. Crystals were preserved in a cryoprotectant containing reservoir solution incorporating 20% glycerol before being flash-cooled at 100 K in liquid nitrogen.
Protein Purification for Enzyme Kinetics Assays
Protein was purified using nickel affinity chromatography and the polyhistidine tag was cleaved as described [30] (Supplemental Materials Figure 2) since the polyhistidine tag can affect enzymatic activity of SpeG homologs from other species (eg Vibrio cholerae SpeG homolog ). After TEV cleavage, a buffer exchange was performed (10 mM Tris-HCl pH 8.3 and 500 mM NaCl) because beta-mercaptoethanol interferes with the enzyme kinetics assay. Protein was concentrated to 6 mg/ml using 10K MWCO Amicon Ultra centrifugation devices (Millipore) and stored in small aliquots at −80°C until ready to use. The protein concentration was calculated using Beer’s Law after measuring the absorbance spectrophotometrically at A280nm with a Nanodrop Spectrophotometer (Thermo Fisher Scientific). The concentration was calculated based on the molecular weight of the monomer without affinity tag and TEV cleavage site (20.5 kDa) and extinction coefficient of 23,380 M−1cm-1.
Data Collection, Structure Determination and Refinement
X-ray diffraction data were collected using the MX2 crystallography beamline (Eiger X 16M detector) at the Australian Synchrotron [31]. A dataset with a total of 360° of data was collected, resulting in 3600 images from 0.1° oscillations. The Eiger data was converted to cbf files using the eiger2cbf [32], indexed and integrated in iMosflm [33], and scaled and merged in AIMLESS [34]. Data was phased by molecular replacement using Phaser [35] and the E. coil SpeG protein structure was used as a search model (PDB ID: 3WR7). Model rebuilding was performed using Coot [36–38] and refined using Refmac5 [39–41] and Phenix [42]. Validation of the structures was performed in Phenix. The DALI server was used to search for structural homologues [43]. Structures of the ligand-free and ligand-bound BtSpeG protein were deposited into the Protein Data Bank with accession codes 6VFM and 6VFN, respectively.
Steady-State Enzyme Kinetics
We assayed the BtSpeG enzyme for activity toward spermine and spermidine in clear polystyrene 96-well microplates using a discontinuous colorimetric assay [44]. Substrate saturation curves were performed toward spermine and spermidine using the following procedure. A 100X enzyme stock solution was prepared in 4 mM polyamine, 90 mM Bicine pH 7.8, 100 mM NaCl, and 0.1% BSA. Enzyme was added to buffered polyamine solutions and plated at 40 μL per well. The reactions were initated with 10μL of AcCoA and allowed to proceed for 5 min at 37 °C. Each 50 μL reaction contained a final concentration of 70 mM Bicine pH 7.8, 20 mM NaCl, 0.02% BSA, 1 mM AcCoA, varying concentrations of polyamine (0–4 mM), and enzyme. The enzyme concentration in each reaction was 4 nM for spermine and 8.5 nM for spermidine. Fifty microliters of a solution containing 6 M guanidine hydrochloride and 0.1 M Tris-HCl pH 8.0 was used to stop the reactions and then 200 μl of Ellman’s reagent solution (0.2 mM DTNB, 0.1 M Tris-HCl pH 8.0, and 1 mM EDTA) was added. The absorbance was measured at 415 nm. At least two replicates were performed for each experiment. A calibration curve was constructed using L-cysteine as a sulfhydryl standard. Data were fitted to the Michaelis Menten equation using non-linear regression in GraphPad Prism 8.1. We also screened the BtSpeG enzyme for activity toward additional polyamine substrates, including cadaverine, putrescine, and agmatine. These reactions contained 4 mM polyamine, 1 mM AcCoA, 50 mM Bicine 7.8, and 20 mM NaCl, and the reaction was initiated with 0.3 μM enzyme.
Polyamine derivatization and analysis by LC/MS
Polyamines were derivatized based on the procedure outlined by Tapuhi et al. [45]. 25 μL of the reaction was removed and mixed with an equal volume of 1 M guanidine hydrochloride to stop the reaction. Next, 200 μL of 0.2 M sodium tetraborate (pH 9.3) was added followed by 250 μL of 1.5 mg/mL dansyl chloride solution in acetonitrile. The samples were vortexed and incubated for 30 min at 60°C, briefly centrifuged and the supernatant was transferred to HPLC vials. The samples were then analysed using an Agilent 1290II UPLC system connected to Agilent 6530 Q-TOF mass spectrometer. The chromatographic separation was performed using a Phenomenex Kinetex F5 core-shell column (2.6 μm particle size, 100 Å pore size, 3.0 × 50 mm). Buffer A contained water with 0.1% v/v trifluoroacetic acid and buffer B contained acetonitrile with 0.1% v/v trifluoroacetic acid (both of HPLC grade purchased from Thermo-Fisher Scientific). The percentage of buffer B in the mobile phase was varied linearly from 2 to 98% over 6.6 min following a 0.2 min initial hold at 2% buffer B with the total analysis time of 8.5 min. A total of 5 μL of standards or samples were injected using an autosampler. The absorbance at 293 nm was monitored using a UV diode- array detector. The mass spectrometer was operated in a positive ionization, single-stage TOF mode and was equipped with a Dual AJS electrospray ionization source.
Results and Discussion
Structure of ligand-free SpeG from B. thuringiensis
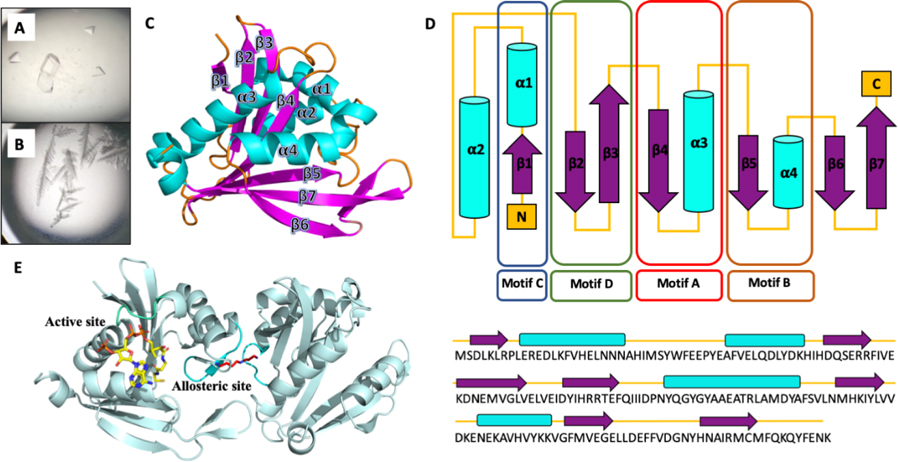
BtSpeG was heterologously expressed in E. coli and purified to greater than 95% purity (see Methods for details). Diamond shaped protein crystals of BtSpeG (ligand-free) grew in a solution containing 0.1M Tris pH 7.5, 7% PEG 4000 and 0.2 M lithium sulfate after 2 days, and diffracted to 2.7 Å resolution (Figure 1). The diffraction data were indexed in the space group P63 2 2, and two molecules were placed in the asymmetric unit (Figure 1). Following rebuilding and refinement, a final model was obtained with Rcryst and Rfree of 0.23 and 0.27, respectively and good stereochemistry (Table 1).
Figure 1.
A, Diamond shaped SpeG crystals in ligand-free form were obtained in Tris pH 7.5, PEG 4000, and 0.2 M lithium sulfate. B, SpeG crystals grown in the presence of spermine and 30% ethanol produced small irregular crystals. Small single crystals could be mounted in the loop. C, Tertiary structure of BtSpeG monomer in cartoon format. Secondary structures including β-strands, α-helices, and connecting loops colored in purple, cyan and orange respectively. The BtSpeG protomer is comprised of a 7-stranded anti-parallel sheet, with the exception of parallel strands β5 and β6, wrapped around 4-α -helices. D, The secondary structure elements represent the N-acetyltransferase sequence motifs C (β1-α1), D (β2—β3), A (β4-α3) and B (β5-α4) highlighted in colored boxes in blue, green, red and brown, respectively. The least conserved secondary structure elements Motif C, β-strand 6 and β7 and helix α2, can be absent in some GNAT proteins. Topology of BtSpeG and associated primary sequence is presented in cartoon form. E, A dimer was present in the asymmetric unit of BtSpeG in ligand-free form. A spermine molecule positioned in the allosteric site is shown in stick mode (red), and modelled AcCoA molecule (yellow) is shown in the active site (yellow).
Table 1.
Data collection and refinement statistics BtSpeG. Statistics for the highest-resolution shell are shown in parentheses.
| BtSpeG |
BtSpeG with spermine |
|
|---|---|---|
| Resolution range | 8.86 – 2.80 (2.80 – 2.67) | 29.87 – 2.5 (2.59 – 2.5) |
| Space group | P 63 2 2 | P 1 21 1 |
| Unit cell | 159.72 159.72 106.46 90 90 120 | 100.65 125.60 101.02 90 106.50 90 |
| Total reflections | 589520 | 488797 |
| Unique reflections | 23127 (3001) | 83267 (4568) |
| Multiplicity | 25.5 (27.1) | 5.9 (6) |
| Completeness (%) | 99.00 (99.56) | 99.92 (100.00) |
| Mean I/sigma(I) | 3.2 | 1.9 |
| Wilson B-factor | 42.09 | 39.53 |
| R-merge | 0.315 (1.476) | 0.114 (0.901) |
| R-meas | 0.325 (1.529) | 0.139 (1.106) |
| R-pim | 0.089 (0.411) | 0.079 (0.633) |
| CC1/2 | 0.998 (0.734) | 0.995 (0.662) |
| Reflections in refinement | 23031 (2261) | 83235 (8299) |
| Reflections for R-free | 1170 (106) | 4214 (370) |
| R-work | 0.2748 (0.3997) | 0.2259 (0.3008) |
| R-free | 0.2962 (0.4301) | 0.2522 (0.3377) |
| Non-hydrogen atoms | 2842 | 16935 |
| macromolecules | 2842 | 16767 |
| ligands | - | 168 |
| Protein residues | 334 | 2004 |
| RMS(bonds) | 0.004 | 0.003 |
| RMS(angles) | 0.66 | 0.57 |
| Ramachandran | ||
| favored | 97.27 | 97.07 |
| allowed (%) | 2.73 | 2.93 |
| outliers (%) | 0.00 | 0.00 |
| Rotomer outliers (%) | 0.33 | 0.45 |
| Clashscore | 3.24 | 6.51 |
| Average B-factor | 45.32 | 42.93 |
| macromolecules | 45.32 | 42.98 |
| ligands | - | 37.39 |
The two protomers in the asymmetric unit were highly similar, exhibiting an r.m.s.d of 0.13 Å, and we therefore limit discussion to molecule A in our model. The SpeG protomer is comprised of seven β-strands with two (β4 and β5) forming a V-shape groove, characteristic of GNAT structures (Figure. 1). The β-strands are arranged as two-interconnected β-sheets in a mixed arrangement. These sheets are surrounded by 4 α-helices, with α1 and α2 on one side, and α3 and α4 on the opposite side. Motif C, the least conserved motif and absent in certain GNATs, is present at the N-terminus and is comprised of α1 and β1 [46]. Motif D, comprised of β2 and β3, interacts with motif C and forms a rigid domain that represents the first half of the SpeG protomer. β4 and α3 comprise the well characterized motif A involved in acetyl-CoA binding and harbors the highly conserved residues QGYGYA typical of GNATs and the R/Q-G-X-G-X-G/A motif [47,48,55]. The V-like shape formed by a β-bulge on β4 and β5 is another distinctive characteristic of GNAT proteins [46,49]. The last motif, motif B is also highly conserved among different SpeG proteins and is comprised of β5 and α4, with the latter forming extensive interactions with AcCoA.
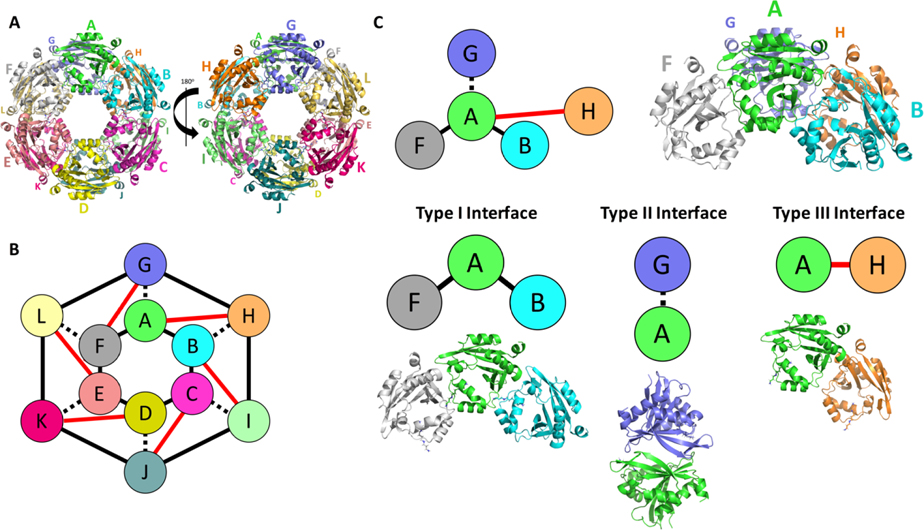
To expand our understanding of interactions and possible biological assembly of SpeG protomers we used PISA [50] software to analyse the BtSpeG structure. The analysis predicted a biological unit comprised of a dodecameric assembly (Figure 2), which has been previously observed in other SpeG homologs. In the BtSpeG assembly, each protomer interacts with its two other protomers located on one hexameric ring (type-I interface) (Figure 2). The strongest interaction was formed between monomers of the opposite hexamer (type-II interface). A minor interface also exists where a protomer binds with the adjacent protomer to the type-II interface. These interfaces are summarised in Table 2
Figure 2. Dodecamer of BtSpeG produces three binding interfaces.
A, Biological unit of BtSpeG is composed of a dimer of hexamers with D6 symmetry. Larger letters correspond to the monomers on the top face of the hexamer and the smaller letters correspond to monomers on the bottom face of the hexamer. Three interfaces mediate assembly of the dodecameric biological unit with two hexamers stacked on top of each other with D6 symmetry. B, schematic representation of dodecameric assembly of BtSpeG. C, Schematic representation of the four monomers that produce the 3 binding interfaces involved in biological unit assembly with schematic representation. The type I interface is between neighboring monomers of the same hexamer (A/B) and (A/ F). The type II interface is formed between monomers (A/ G) of opposing hexamers. The type III interface is formed between diagonal adjacent partners of the opposing hexamers.
Table 2.
Interface details for BtSpeG. Interface surface area, number of hydrogen bonds and salt bridges at the interfaces I/II/III. Detailed bond information can be found in the Supplementary section.
| Interaction
Interface | |||
|---|---|---|---|
| Type I | Type II | Type III | |
| Hydrogen bonds | 6 | 12 | 2 |
| Salt bridges | 7–8 | 12 | 0 |
| Buried surface area Å2 | 680 | 895 | 312 |
Structure of SpeG with spermine
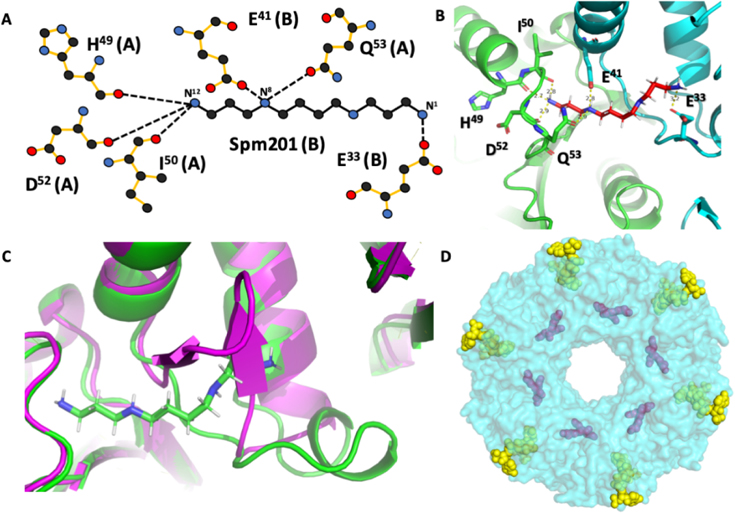
To better understand how BtSpeG recognizes its substrates and whether or not it undergoes any possible conformational changes to accommodate ligand binding, we also determined its structure by co-crystallizing the BtSpeG protein in the presence of spermine and acetyl-CoA. Crystals formed in 30% ethanol diffracted to a resolution of 2.6 Å and were indexed in the space group P2121 (Table 1). In this space group, all twelve molecules comprising the dodecameric biological unit were present in the asymmetric unit (Figure 2). Each protomer was highly similar in structure, with the greatest r.m.s.d of 0.1 Å between any monomer. Comparison with the ligand free protomer also revealed a highly similar protomer with an r.m.s.d of 0.614 Å. The greatest differences were observed in the loop region from residues 25–38 to accommodate ligand binding (Figure 3). Using PISA [50] we also compared the interfaces between the ligand-free and ligand-bound structures. We observed similar interactions between these two structures, including all types of interfaces analysed in the ligand-free structure (Supplemental Materials Table 1). In the BtSpeG liganded structure, there were twelve individual spermine molecules observed in the dodecamer within the allosteric sites of the protein. The backbone oxygens of residues His49, Ile50 and Asp52 of one chain form hydrogen bonds with N12 of spermine. Gln53 of the same chain, and Glu41 of another chain hydrogen bond with N8. Gln33 of the same chain as Glu41 binds with N1 of spermine (Fig. 3). These residues have been previously established to be important for polyamine interactions in this site [8], with the exception of Gln53. The fact that both Glu41 and Gln53 form hydrogen bonds with the N10 of spermine is a newly observed interaction between polyamine and the allosteric site of SpeG.
Figure 3. Spermine interactions with BtSpeG.
A, Schematic representation of detailed interaction between spermine and BtSpeG dimer. Hydrogen bonds are represented by dotted lines. B, graphical representation of bond formed between spermine and two monomers of BtSpeG molecule, monomer A in green and B in cyan. Spermine binds in the allosteric site between two monomers that form a type I interface of the structure. C, Superimposition of flexible loop region residues 25–38 of BtSpeG in ligand-bound (green) and ligand-free (magenta) state. Movement of the flexible loop is necessary in order to accommodate the spermine molecule in the allosteric site to avoid a clash. D, Dodecameric assembly of BtSpeG biological unit (cyan) with twelve spermine molecules in the allosteric site (purple) and putative active site with twelve AcCoA molecules (yellow and green).
Although we co-crystallized the protein in the presence of both reactants of the enzymatic reaction (AcCoA and spermine) no density for AcCoA/CoA or spermine/acetyl-spermine was observed in the BtSpeG liganded structure. In addition to crystallising BtSpeG with spermine, we also attempted crystallisation with spermidine. However, crystals formed in the presence of spermidine did not reveal any density that could support ligand binding. It is common to not observe all ligands in structures even when they are included in the cocrystallization solution. In some cases this can be attributed to lower binding affinities of different ligands or protein conformational changes that must occur to accommodate specific ligands. Moreover, during catalysis the enzyme would favour release of products unless the concentrations of these in solution were significantly higher than the Kd for these compounds in the active site. It also may be possible that AcCoA/CoA binding was not observed due to crystallographic packing that interferes with the putative AcCoA/CoA binding site.
Comparison with other SpeG homologs
Whilst we observed significant variation in the crystallographic asymmetric units between ligand-bound and ligand-free crystals, it is highly likely that the dodecameric structure we observed in the spermine bound form represents the biological assembly. This structure was also observed in the ligand-free form of the enzyme by expansion of the asymmetric unit, and related SpeG homologs also exhibit a similar dodecameric structure. To compare BtSpeG against other closely related structures, we performed a DALI search to investigate the most structurally similar proteins. The greatest structural similarity was observed for SpeG proteins from Yersinia pestis (PDB code 5WIF), V. cholerae (PDB code 4JJX), Coxiella burnetti (PDB code 3TTH) and E. coli (PDB code 3WR7) with sequence identity of 68% and r.m.s.d. of 0.413, 57% and 0.542, 64% and 0.583, and 71% and 0.690, respectively. Interfaces within our structure and predicted biological assemblies compared to other SpeG proteins were similar (Supplemental Materials Figure 3). Additionally, the AcCoA binding site was structurally conserved within these proteins and the only significant difference between apo and liganded structures was the positioning of the allosteric loop (residues 25–38 BtSpeG; Figure 3). This is a common characteristic of SpeG to accommodate the polyamine in the allosteric site, as described previously [8].
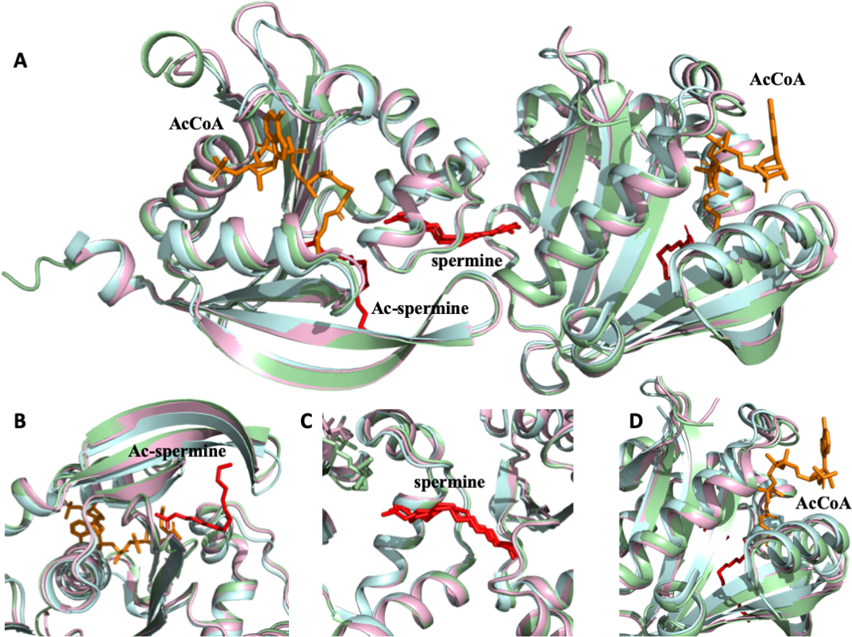
To elucidate whether any additional conformational changes occurred in the BtSpeG structure compared to other previously crystallized SpeG homologs, the BtSpeG protein was compared to the V. cholerae (VcSpeG) enzyme since many structures in different liganded states have been determined. Specifically, the VcSpeG structure (PDB code 4R87) that contains both non-acetylated spermine in the allosteric site and CoA in the active site, and the VcSpeG structure (PDB code 6E1X) with acetylated spermine in the active site and non-acetylated in the allosteric site were compared (Figure 4). No significant structural variation was been observed in any region of the molecule, and the allosteric and active sites of all three SpeG molecules showed highly conserved morphology.
Figure 4. Structural alignment of BtSpeG (6VFN), VcSPeG (4R87) and VcSpeG (6E1X).
A, Structural alignment of GNAT dimer of BtSpeG (6VFN) in light cyan, VcSpeG (4R87) in light green and VcSpeG (6E1X) in light magenta revealed no significant structural differences. B, Enlarged view of active site alignment with visible acetylated spermine (ac-seprmine) in red from VcSpeG (6E1X) shows no significant differences. C, Zoomed in view of allosteric site with spermine molecule (red) of three SpeG protomers shows nearly identical configuration. D, Close view of highly conserved active site with a CoA molecule (orange) shows no significant structural differences.
BtSpeG kinetic activity
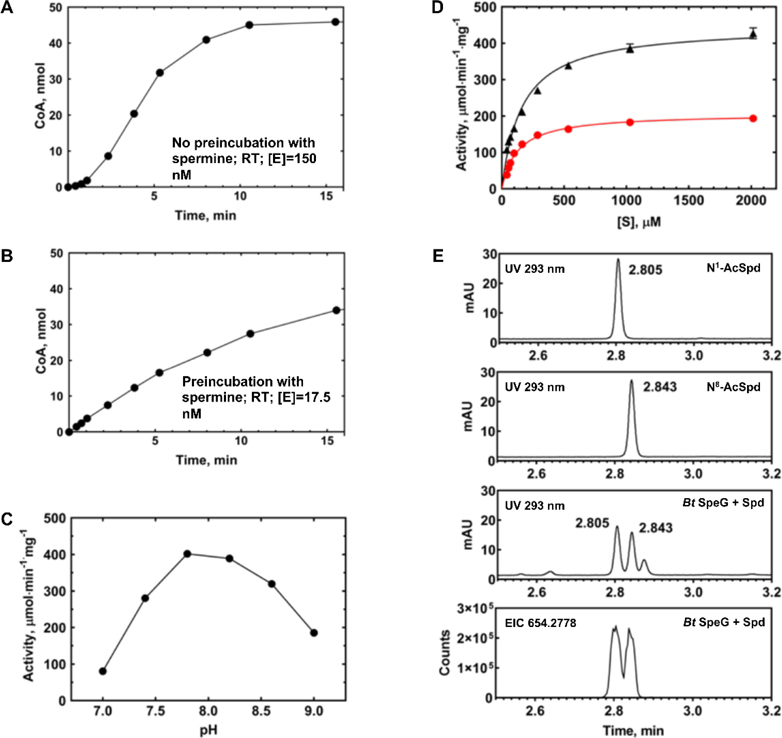
We investigated the kinetic activity of the BtSpeG enzyme toward spermine and spermidine since previously characterized SpeG enzymes acetylate them. During our preliminary experiments we observed a hysteretic behaviour of the BtSpeG enzyme. For example, when we initiated the reaction with enzyme there was a significant lag present at the beginning of time course experiments (Figure 5A). However, when we preincubated the enzyme with polyamine prior to initiating the reaction, the lag was removed and the reaction progress curve was linear at the beginning of the time course (Figure 5B). By pre-incubating BtSpeG with polyamine, we both removed the lag and used less enzyme in the reaction by one order of magnitude compared to no preincubation. While the cause of the lag was not further investigated, we suspect a relatively slow conformational change upon binding polyamine may be a contributing factor. For example, other SpeG enzymes from Gram-negative bacteria have been reported to form several oligomeric states [8] and the slow transition between these states upon polyamine binding may be the source of the hysteretic behaviour observed. Additional experiments are currently underway in our laboratory to further investigate this matter.
Figure 5. Kinetic characterization of BtSpeG.
A, Reaction time course at RT, no preincubation of enzyme with spermine. BtSpeG enzyme concentration was 150 nM and the reaction contained 70 mM Bicine pH 7.8, 20 mM NaCl, 4 mM spermine, and 1 mM AcCoA. B, Reaction time course at RT, with preincubation of enzyme with spermine. BtSpeG enzyme concentration was 17.5 nM. C, Activity of BtSpeG pre-incubated with spermine as a function of pH of Bicine buffer. Each reaction contained 70 mM Bicine buffer at different pH values, 20 mM NaCl, 4 mM spermine, and 1 mM AcCoA and reactions were performed for 5 min at 37°C. Enzyme was preincubated with spermine. D, Substrate saturation curves of BtSpeG toward spermine (black triangles) and spermidine (red circles). Enzyme was preincubated with either spermine or spermidine depending upon which polyamine was being assayed. 4 nM enzyme was used for spermine saturation curves and 8.5 nM enzyme was used for spermidine saturation curves. All reactions were performed in duplicate and the average and standard deviation of at least two biological replicates are shown. E, Identification of products of enzymatic acetylation of spermidine by LC/MS. Products of the 5 min reaction containing 11.8 nM BtSpeG, 4 mM spermidine, and 1 mM AcCoA at 22 deg C (Bt SpeG +spd) are compared to 200 uM solutions of N1 and N8 acetylspermidine standards. The bottom trace is the ion chromatogram for the fully dansylated acetylspermidine extracted using a 20 ppm window.
To further characterize the properties of the BtSpeG enzyme, we screened it for activity from pH 7–9 in Bicine buffer. We determined the optimal pH for measuring BtSpeG enzymatic activity was 7.8 (Figure 5C). Next, we performed substrate saturation curves toward spermine and spermidine at this pH. We found the turnover of the BtSpeG enzyme was higher by approximately 2-fold toward spermine compared to spermidine, but the catalytic efficiency for both polyamines was the same order of magnitude (Figure 5D; Table 3). The apparent affinity of the enzyme for both polyamines was quite similar, and to our knowledge no other SpeG enzyme has exhibited this characteristic. All previously characterized SpeG enzymes show a significantly higher (by one order of magnitude) apparent affinity for spermine compared to spermidine. To determine whether BtSpeG could acetylate other polyamines, we also screened it for activity towards agmatine, putrescine, and cadaverine. Under the described reaction conditions, we found BtSpeG could acetylate agmatine (5.6 μmol/min*mg) and cadaverine (2.4 μmol/min*mg), but did not acetylate putrescine. This activity was two orders of magnitude lower than that observed for both spermine and spermidine, which indicates spermine and spermidine are the preferential substrates of BtSpeG (Figure 5D).
Table 3.
BtSpeG kinetic parameters calculated from substrate saturations curves for different polyamines. All reactions were performed for 5 min at 37°C and enzyme was preincubated with polyamine prior to performing kinetics. Parameters were calculated from fitting the Michaelis Menten equation to data as described in Materials and Methods.
| Substrate | KM (mM) | kcat (s−1) | kcat/KM (M−1s−1) | ||
|---|---|---|---|---|---|
| Sperminea | 0.162 | ± 0.011 | 153 | ± 3 | 9.46×105 |
| Spermidinea | 0.127 | ± 0.008 | 70.6 | ± 1.2 | 5.57×105 |
Data were collected from 0–4 mM polyamine but only data from 0–2 mM are plotted and fitted because curves reached complete saturation at concentrations of polyamines less than 4 mM. No substrate inhibition was observed at higher concentrations of polyamine.
Unlike spermine, primary amino groups of spermidine are non-equivalent (aminopropyl and aminobutyl) which leaves room for the possibility that SSAT enzymes could exhibit a preference for either N1 or N8 positions of spermidine or acetylate them at different rates. Therefore, we tested whether the BtSpeG enzyme could acetylate one or both positions of spermidine using LC/MS. We found the enzyme formed both N1 and N8 acetylspermidine in nearly equimolar ratios under the described reaction conditions (Figure 5). From these data, it appears the enzyme exhibits no preference in aminopropyl or aminobutyl acetylation of spermidine and is in line with other previously reported studies on the SpeG enzyme from E. coli [10,51]. However, it is possible that the rates of formation of acetylated products at different substrate concentrations may differ and will require further studies to elucidate whether this is indeed the case.
Comparison of BtSpeG kinetic activity with other SpeG acetyltransferases
SpeG enzymes from Gram-negative bacteria have been investigated for decades, but their homologs in Gram-positive bacteria have only recently been explored. For example, the only other Gram-positive bacterium SpeG that has been kinetically characterized is from Staphylococcus aureus USA300 [16]. This enzyme had a similar apparent affinity (Km) for spermine (0.295 mM) compared to BtSpeG (0.162 mM), but S. aureus SpeG showed a one order of magnitude lower apparent affinity for spermidine (1.33 mM) compared to BtSpeG (0.127 mM). Additionally, the turnover number was one to two orders of magnitude lower for S. aureus SpeG (2.48 s−1 for spermidine and 3.52 s−1 for spermine) than BtSpeG (153 s−1 for spermine and 70.6 s−1 for spermidine). Not all bacteria are able to produce or uptake the same polyamines, and polyamine concentrations can vary across different species. In B. megaterium, spermidine was shown to be the predominant polyamine during all stages of growth and its concentration increased 2–2.5-fold when cells were dormant or in stationary phase compared to during log phase [52]. This was also true for other Bacilli, including B. cereus T and B. subtilis SB-133. It was also shown that B. megaterium could uptake high concentrations of exogenous polyamines spermine and spermidine, and spermine could completely replace spermidine during sporulation. Therefore, the higher apparent affinity of BtSpeG for spermidine compared to other SpeG enzymes may be due to the fact that spermidine is the predominant polyamine in this organism.
Redundancy of polyamine acetyltransferases in Bacilli
Interestingly, there are three different types of SSATs that have now been described in Bacilli: SpeG, BltD, and PaiA [53,54]. The sequence similarity between SpeG, BltD, and PaiA proteins from different Bacilli is very low, and SpeG is the only one that has an allosteric site that binds polyamines. Each of these proteins acetylates both spermine and spermidine, but with different efficiencies. For example, PaiA acetylated spermine, spermidine, and aminopropylcadaverine, but its catalytic efficiency for spermine was one order of magnitude higher than the other two polyamines and still one order of magnitude lower than that observed for BtSpeG. The apparent affinity (Km) of PaiA for these polyamines was 0.076 and 0.323 mM for spermine and spermidine, respectively but the turnover was significantly impaired (two to three orders of magnitude lower) compared to BtSpeG. Similar to PaiA, BltD acetylated spermine and spermidine and exhibited a higher apparent affinity for spermine than spermidine (0.067 and 0.200 mM, respectively). The apparent Vmax was one order of magnitude lower for spermine and two orders of magnitude lower for spermidine compared to PaiA.
Conclusions
Our results show that the SpeG enzyme from the Gram-positive bacterium Bacillus adopts a similar dodecameric structure to those of previously characterized SpeG enzymes from Gram-negative bacteria. Moreover, we have shown that the structure and kinetic activity of the Bacillus SpeG enzyme deviates significantly from other Bacillus SSAT enzymes that have been characterized previously. The catalytic efficiency of BtSpeG for both spermine and spermidine is much higher when compared to other Bacillus polyamine acetyltransferases PaiA and BltD. The fact that BtSpeG has an allosteric site that binds polyamines and it exhibits structural differences compared to PaiA (no structure of BltD has been determined) may contribute to its overall ability to more efficiently acetylate polyamines and regulate various cellular processes in some Bacilli.
Supplementary Material
Acknowledgements
This research was possible through the use of the MX2 beamline at the Australian Synchrotron (ANSTO). Research reported in this work was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM133506 (to MLK). Van Le was supported by a NIH MBRS-RISE fellowship (R25-GM059298). We also thank Ee Qing Lim, Ee Qi Lim, and Imani Warren for their technical assistance at SFSU. A.S. is an employee of Gilead Sciences. We sincerely thank Anthony Michael for his helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Pegg AE, Functions of Polyamines in Mammals, J Biol Chem 291 (2016) 14904–14912. 10.1074/jbc.R116.731661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pegg AE, McCann PP, Polyamine metabolism and function, Am J Physiol 243 (1982) C212–221. 10.1152/ajpcell.1982.243.5.C212. [DOI] [PubMed] [Google Scholar]
- [3].Pegg AE, Spermidine/spermine-N(1)-acetyltransferase: a key metabolic regulator, Am J Physiol Endocrinol Metab 294 (2008) E995–1010. 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
- [4].Michael AJ, Polyamine function in archaea and bacteria, J Biol Chem 293 (2018) 18693–18701. 10.1074/jbc.TM118.005670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barbagallo M, Di Martino ML, Marcocci L, Pietrangeli P, De Carolis E, Casalino M, Colonna B, Prosseda G, A new piece of the Shigella Pathogenicity puzzle: spermidine accumulation by silencing of the speG gene [corrected], PLoS One 6 (2011) e27226 10.1371/journal.pone.0027226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Limsuwun K, Jones PG, Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli, J Bacteriol 182 (2000) 5373–5380. 10.1128/jb.182.19.5373-5380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Karatan E, Duncan TR, Watnick PI, NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine, J Bacteriol 187 (2005) 7434–7443. 10.1128/jb.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Filippova EV, Kuhn ML, Osipiuk J, Kiryukhina O, Joachimiak A, Ballicora MA, Anderson WF, A novel polyamine allosteric site of SpeG from Vibrio cholerae is revealed by its dodecameric structure, J Mol Biol 427 (2015) 1316–1334. 10.1016/j.jmb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Filippova EV, Weigand S, Osipiuk J, Kiryukhina O, Joachimiak A, Anderson WF, 2015. Substrate-induced allosteric change in the quaternary structure of the spermidine N-acetyltransferase SpeG.. J. Mol. Biol 427 (22) 3538–3553. doi: 10.1016/j.jmb.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carper SW, Willis DG, Manning KA, Gerner EW, Spermidine acetylation in response to a variety of stresses in Escherichia coli, J Biol Chem 266 (1991) 12439–12441. [PubMed] [Google Scholar]
- [11].Hu LI, Filippova EV, Dang J, Pshenychnyi S, Ruan J, Kiryukhina O, Anderson WF, Kuhn ML, Wolfe AJ, The spermidine acetyltransferase SpeG regulates transcription of the small RNA rprA, PLoS One 13 (2018) e0207563 10.1371/journal.pone.0207563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fang SB, Huang CJ, Huang CH, Wang KC, Chang NW, Pan HY, Fang HW, Huang MT, Chen CK, speG Is Required for Intracellular Replication of Salmonella in Various Human Cells and Affects Its Polyamine Metabolism and Global Transcriptomes, Front Microbiol 8 (2017) 2245 10.3389/fmicb.2017.02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Joshi GS, Spontak JS, Klapper DG, Richardson AR, Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines, Mol Microbiol 82 (2011) 9–20. 10.1111/j.1365-2958.2011.07809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR, Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus, Cell Host Microbe 13 (2013) 100–107. 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Planet PJ, LaRussa SJ, Dana A, Smith H, Xu A, Ryan C, Uhlemann AC, Boundy S, Goldberg J, Narechania A, Kulkarni R, Ratner AJ, Geoghegan JA, Kolokotronis SO, Prince A, Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin, mBio 4 (2013) e00889–00813. 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li B, Maezato Y, Kim SH, Kurihara S, Liang J, Michael AJ, Polyamine-independent growth and biofilm formation, and functional spermidine/spermine N-acetyltransferases in Staphylococcus aureus and Enterococcus faecalis, Mol Microbiol 111 (2019) 159–175. 10.1111/mmi.14145. [DOI] [PubMed] [Google Scholar]
- [17].Nguyen AT, Tallent SM, Screening food for Bacillus cereus toxins using whole genome sequencing, Food Microbiol 78 (2019) 164–170. 10.1016/j.fm.2018.10.008. [DOI] [PubMed] [Google Scholar]
- [18].Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM, Foodborne illness acquired in the United States--major pathogens, Emerg Infect Dis 17 (2011) 7–15. 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hernandez E, Ramisse F, Ducoureau JP, Cruel T, Cavallo JD, Bacillus thuringiensis subsp. konkukian (serotype H34) superinfection: case report and experimental evidence of pathogenicity in immunosuppressed mice, J Clin Microbiol 36 (1998) 2138–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ibrahim MA, Griko N, Junker M, Bulla LA, Bacillus thuringiensis: a genomics and proteomics perspective, Bioeng Bugs 1 (2010) 31–50. 10.4161/bbug.1.1.10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Moayeri M, Leppla SH, Vrentas C, Pomerantsev AP, Liu S, Anthrax Pathogenesis, Annu Rev Microbiol 69 (2015) 185–208. 10.1146/annurev-micro-091014-104523. [DOI] [PubMed] [Google Scholar]
- [22].Kolsto AB, Tourasse NJ, Okstad OA, What sets Bacillus anthracis apart from other Bacillus species?, Annu Rev Microbiol 63 (2009) 451–476. 10.1146/annurev.micro.091208.073255. [DOI] [PubMed] [Google Scholar]
- [23].Hobley L, Kim SH, Maezato Y, Wyllie S, Fairlamb AH, Stanley-Wall NR, Michael AJ, Norspermidine is not a self-produced trigger for biofilm disassembly, Cell 156 (2014) 844–854. 10.1016/j.cell.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ, Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation, J Biol Chem 285 (2010) 39224–39238. 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hobley L, Li B, Wood JL, Kim SH, Naidoo J, Ferreira AS, Khomutov M, Khomutov A, Stanley-Wall NR, Michael AJ, Spermidine promotes Bacillus subtilis biofilm formation by activating expression of the matrix regulator slrR, J Biol Chem 292 (2017) 12041–12053. 10.1074/jbc.M117.789644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolsto AB, Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis--one species on the basis of genetic evidence, Appl Environ Microbiol 66 (2000) 2627–2630. 10.1128/aem.66.6.2627-2630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khandokar YB, Londhe A, Patil S, Forwood JK, Expression, purification and crystallization of acetyl-CoA hydrolase from Neisseria meningitidis, Acta Crystallogr Sect F Struct Biol Cryst Commun 69 (2013) 1303–1306. 10.1107/s1744309113028042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Studier FW, Protein production by auto-induction in high density shaking cultures, Protein Expr Purif 41 (2005) 207–234. [DOI] [PubMed] [Google Scholar]
- [29].Czub MP, Zhang B, Chiarelli MP, Majorek KA, Joe L, Porebski PJ, Revilla A, Wu W, Becker DP, Minor W, Kuhn ML, A Gcn5-Related N-Acetyltransferase (GNAT) Capable of Acetylating Polymyxin B and Colistin Antibiotics in Vitro, Biochemistry 57 (2018) 7011–7020. 10.1021/acs.biochem.8b00946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Majorek KA, Kuhn ML, Chruszcz M, Anderson WF, Minor W, Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa, J Biol Chem 288 (2013) 30223–30235. 10.1074/jbc.M113.501353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Aragão D, Aishima J, Cherukuvada H, Clarken R, Clift M, Cowieson NP, Ericsson DJ, Gee CL, Macedo S, Mudie N, Panjikar S, Price JR, Riboldi-Tunnicliffe A, Rostan R, Williamson R, Caradoc-Davies TT, MX2: a high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron, Journal of synchrotron radiation 25 (2018) 885–891. 10.1107/S1600577518003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nakane T, eiger2cbf, https://github.com/biochem-fan/eiger2cbf.
- [33].Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW, iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM, Acta Crystallographica Section D: Biological Crystallography 67 (2011) 271–281. 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Evans P, Scaling and assessment of data quality, Acta Crystallogr D Biol Crystallogr 62 (2006) 72–82. 10.1107/s0907444905036693. [DOI] [PubMed] [Google Scholar]
- [35].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, Journal of Applied Crystallography 40 (2007) 658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA, REFMAC5 for the refinement of macromolecular crystal structures, Acta Crystallographica Section D: Biological Crystallography 67 (2011) 355–367. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Emsley P, Cowtan K, Coot: model-building tools for molecular graphics, Acta Crystallographica Section D 60 (2004) 2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- [38].Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallographica Section D: Biological Crystallography 66 (2010) 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kovalevskiy O, Nicholls RA, Long F, Carlon A, Murshudov GN, Overview of refinement procedures within REFMAC5: utilizing data from different sources, Acta Crystallogr D Struct Biol 74 (2018) 215–227. 10.1107/s2059798318000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA, REFMAC5 for the refinement of macromolecular crystal structures, Acta Crystallogr D Biol Crystallogr 67 (2011) 355–367. 10.1107/s0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Murshudov GN, Vagin AA, Dodson EJ, Refinement of macromolecular structures by the maximum-likelihood method, Acta Crystallogr D Biol Crystallogr 53 (1997) 240–255. 10.1107/s0907444996012255. [DOI] [PubMed] [Google Scholar]
- [42].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD, Towards automated crystallographic structure refinement with phenix.refine, Acta Crystallogr D Biol Crystallogr 68 (2012) 352–367. 10.1107/s0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Holm L, Laakso LM, Dali server update, Nucleic Acids Research 44 (2016) W351–W355. 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kuhn ML, Majorek KA, Minor W, Anderson WF, Broad-substrate screen as a tool to identify substrates for bacterial Gcn5-related N-acetyltransferases with unknown substrate specificity, Protein Sci 22 (2013) 222–230. 10.1002/pro.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tapuhi Y, Schmidt DE, Lindner W, Karger BL, Dansylation of amino acids for high- performance liquid chromatography analysis, Anal Biochem 115 (1981) 123–129. 10.1016/0003-2697(81)90534-0. [DOI] [PubMed] [Google Scholar]
- [46].Favrot L, Blanchard JS, Vergnolle O, Bacterial GCN5-Related N-Acetyltransferases: From Resistance to Regulation, Biochemistry 55 (2016) 989–1002. 10.1021/acs.biochem.5b01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dyda F, Klein DC, Hickman AB, GCN5-related N-acetyltransferases: a structural overview, Annu Rev Biophys Biomol Struct 29 (2000) 81–103. 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Neuwald AF, Landsman D, GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein, Trends Biochem Sci 22 (1997) 154–155. 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- [49].Richardson JS, Getzoff ED, Richardson DC, The beta bulge: a common small unit of nonrepetitive protein structure, Proc Natl Acad Sci U S A 75 (1978) 2574–2578. 10.1073/pnas.75.6.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Krissinel E, Henrick K, Inference of macromolecular assemblies from crystalline state, J Mol Biol 372 (2007) 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- [51].Fukuchi J, Kashiwagi K, Takio K, Igarashi K, Properties and structure of spermidine acetyltransferase in Escherichia coli, J Biol Chem 269 (1994) 22581–22585. [PubMed] [Google Scholar]
- [52].Setlow P, Polyamine levels during growth, sporulation, and spore germination of Bacillus megaterium, J Bacteriol 117 (1974) 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Woolridge DP, Martinez JD, Stringer DE, Gerner EW, Characterization of a novel spermidine/spermine acetyltransferase, BltD, from Bacillus subtilis, Biochem J 340 ( Pt 3) (1999) 753–758. [PMC free article] [PubMed] [Google Scholar]
- [54].Forouhar F, Lee IS, Vujcic J, Vujcic S, Shen J, Vorobiev SM, Xiao R, Acton TB, Montelione GT, Porter CW, Tong L, Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase, J Biol Chem 280 (2005) 40328–40336. 10.1074/jbc.M505332200. [DOI] [PubMed] [Google Scholar]
- [55].Neuwald AF, Green P Detecting patterns in protein sequences J. Mol. Biol, 239 (1994), pp. 698–712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.