Abstract
Background
Circular RNAs (circRNAs) are frequently aberrantly expressed in non-small cell lung cancer (NSCLC) and are considered to exert a pivotal role in the occurrence and development of NSCLC via targeting and negatively regulating microRNAs (miRNAs). We aimed to investigate the role of hsa_circ_0109320 in the proliferation, invasion and apoptosis of NSCLC, and explore its underlying molecular mechanism.
Material/Methods
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis was performed to determine the circ_0109320 and miR-595 expression in tissues or cells. Western blot analysis was conducted to examine the cleaved caspase-3, Bax, Bcl-2, and E2F7 protein expression. Transwell detection was used to evaluate the invasion level of NSCLC cell lines.
Results
The results of present study indicated that circ_0109320 expression in NSCLC patients was upregulated significantly in tumor tissues compared with tissues adjacent to carcinoma. Upregulated circ_0109320 level was significantly associated with TNM stages as well as lymph node metastasis of NSCLC. Moreover, downregulation of circ_0109320 attenuated proliferation and invasion while promoting apoptosis in NSCLC cells. We further confirmed that circ_0109320 could sponge miR-595 to upregulate E2F7 expression. Silencing of miR-595 or overexpression of E2F2 could partially reversed the inhibitory role of circ_0109320 knockdown in NSCLC cells. These data provided evidence that the suppression of circ_0109320 attenuates NSCLC cell proliferation and invasion and enhances apoptosis through the miR-595/E2F7 pathway.
Conclusions
Circ_0109320/miR-595/E2F2 axis may exert a pivotal role in the pathological mechanism of NSCLC progression, and it has potential application in the future treatment of NSCLC.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; E2F2 Transcription Factor; MicroRNAs; RNA, Untranslated
Background
Lung cancer is a worldwide malignant disease with high mortality, which seriously threatens human health and quality of life [1,2]. Currently, lung cancer is still the leading cancer disease in the world [3]. Among all types of lung cancer, the 5-year survival rate after radical resection of non-small cell lung cancer (NSCLC) is still unsatisfactory, mainly due to the rapid proliferation of tumor cells in NSCLC [4]. Therefore, strengthening the research on the mechanism of proliferation and invasion of NSCLC cells is conducive to its treatment and will have a positive significance for improving the survival rate of patients.
With the rapid development of modern molecular detection technology, NSCLC has been detected and proven to have a variety of tumor-driving genes [3,5]. The occurrence and progression of tumors are closely related to the disorder of tumor-related gene function [6]. Previous studies have found that microRNAs (miRNAs) can regulate the expression of downstream tumor-related genes, and affect the occurrence, development, and metastasis of tumors, including NSCLC [7,8]. The research on the function and potential molecular mechanism of miRNAs in tumors has been a hot topic in last decade.
In recent years, the discovery of the role of non-coding circular RNAs (circRNAs) on the sponge of miRNAs has made circRNA research a new international research hotspot [9,10]. CircRNAs are new type of non-coding RNAs that differ from linear RNAs such as miRNAs. They have a closed ring structure and are widely expressed in eukaryotic transcriptomes [11]. Studies have proven that circRNAs play important roles in many tumor diseases. For example, hsa_circRNA_100290 regulates oral squamous cell carcinoma cell growth through acting as a competing endogenous RNA (ceRNA) for miR-378a [12]. CircPAPPA inhibition attenuates the proliferation and invasion of trophoblast cells via targeting the miR-384/STAT3 pathway [13]. Although some progress has been made in the formation process and mechanism of circRNAs, the role of circRNAs in NSCLC and its molecular mechanism are still not fully clear.
In our study, we combined the results of pre-screening of circRNAs chips [14], and verification of the high expression of circ_0109320 in NSCLC in vivo and in vitro. Upregulated circ_0109320 expression level was associated with TNM stages as well as lymph node metastasis of NSCLC. In addition, circ_0109320 exerts oncogenic roles through the miR-595/E2F7 pathway in NSCLC. Collectively, our study revealed a novel oncogenic circRNA in the progression of NSCLC.
Material and Methods
Patient selection
A total of 57 tumor tissues and corresponding non-tumor normal specimens were collected from NSCLC patients who were undergoing surgery in the Second Hospital of Lianyungang between July 2016 and July 2018. The inclusion criteria were that patients met the diagnostic criteria of NSCLC and surgical resection was conducted in these patients. Patients with malignant tumors of other organs were excluded. The project was authorized by the Ethics Committee of Lianyungang Second Hospital. All patients read the informed consent and signed it to participate in the project.
Cell culture
Normal human airway epithelial cells (16HBE) and NSCLC cell lines (H358, H1299, H1581, and A549) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). NSCLC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, MA, USA) and were grown with 5% CO2 in a humidified atmosphere at 37°C.
Cell transfection
H358 and A549 cells were transfected with 50 nM circ_0109320 knockdown and overexpression plasmids (Genechem, Shanghai, China), or with 50 nM overexpression plasmids E2F2 or miR-595 inhibitor together (GeneChem, Shanghai, China) utilizing Lipofectamine® 3000 (Thermo Fisher Scientific, MA, USA) for 48 hours.
Cell Counting Kit-8 (CCK-8) assay
The proliferation of A549 and H358 cells was determine using Cell Counting Kit-8 (CCK-8) assay kits (Beyotime, Nanjing, China). Cells were digested and seeded at 1×104 cells/well in 96-well plates and were cultured for 24 to 96 hours. Then, CCK-8 assay was conducted at 0, 24, 48, 72, and 96 hours to measure and analyze the optical density values at 490 nm.
TUNEL
Apoptosis in tumor cells was detected by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) via an In situ Cell Death Detection kit (Roche, Germany). The kit was used according to the manufacturer’s protocol.
Cell invasion assay
A549 and H358 cells were grown in DMEM containing 10% FBS. When growing to 80% to 90% density, the cells were digested and suspended using trypsin and 1% FBS. Next, pretreated cell suspension was added to each well for 36 hours. After 24 hours of culture, the invasive cells were processed by 4% paraformaldehyde-fixed and dyed with 0.5% crystal violet. The number of invasive cells was counted and photographed in six randomly selected microscopic fields.
Dual-luciferase reporter assay
Potential miRNAs targeting circ_0109320 sequences were predicted using the circInteractome database (https://circinteractome.nia.nih.gov/index.html). TargetScan (http://www.targetscan.org/) was used to analyze the potential targets of miR-595. Wild type (WT) and mutant (Mut) 3′ UTR of E2F2 or circ_0109320 were separately constructed into the vectors (pGL4.10, Promega, Madison, WI, USA) and co-transfected with miR-595 or NC mimics into 2 cell lines using Lipofectamine 3000. Double luciferase reporter assay system (Solabio Technology, Beijing, China) was performed to detect luciferase activity at 48 hours post-transfection.
Immunoblotting assay
Proteins from the NSCLC cells were extracted by lysis buffer (Beyotime, Nanjing, China). After examining the concentration of protein, loading buffer was added into the protein sample and boiled for 10 minutes. Protein (30 μg each lane) was subjected to 10% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and transferred to PVDF (Polyvinylidene fluoride) membranes. After blocked with 5% bovine serum albumin (BSA) for 2 hours, the membranes were cultured with antibodies against Bax, cleaved caspase-3, Bcl-2, E2F2, and GAPDH at 4°C overnight (all antibodies; Abcam, Cambridge, MA, USA). After washing for 5 minutes, the membrane was cultured for 2 hours at room temperature with a 1: 5000 dilution of secondary antibody buffer (Beyotime, Nanjing, China). The bands were detected with a chemiluminescence kit (Beyotime, Nanjing, China.) and analyzed using ImageJ version.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from NSCLC tissues and cells was extracted using TRIzol (Invitrogen, CA, USA). SYBR Green RT-PCR SuperMix UDG reagents (Invitrogen, CA, USA) and the MX3000P system (Stratagene, CA, USA) were used to amplify cDNA. Different from the commonly used convergent primers, divergent primers were designed for RT-qPCR analysis of circ_0109320. U6 was used as the internal control of miR-595, and 18S was used as the control of circ_0109320 and E2F7. Specific primers used in our study are shown in Table 1.
Table 1.
Sequences of primers for PCR.
| Genes | Forward primer | Reverse primer |
|---|---|---|
| hsa_circ_0109320 | ATGCTTTGCCACATTCTTCACA | TAAGATAATTCACACTGGAGAGA |
| 18S | CTTTCGTACGGTACGCCT | GCCAAGAGGGGCTTAGT |
| U6 | TTGCTTCGTGTTTCTCAGT | AGTCCGGTTTTCCCCGAC |
| MiR-1228 | GATTACGCGCTGCCCGATA | GTTGCCGACTAGGTCCAA |
| MiR-1243 | ATGCCTCGTTCATCAAGAA | TAGTCGGCCTAACACATCG |
| MiR-330-3p | GCCTCGCCCCACTTGCAG | TGCCTCGGGAACCCGCAGG |
| MiR-483-3p | TGGTTGATCCGGCAATAC | TCCCGTAGCTGTGGGTCTTC |
| MiR-583 | GGCGTATGGCCCAGTGTG | TAGGCTACTTCCTCCCCTAG |
| MiR-651 | CAGTCTTGGACTGGACTT | GTTGACCCGATTGGTTGTAC |
| MiR-595 | ACGGTCCCTTGGAGTTTTC | AATGGCTCACCCAAACTGC |
| E2F7 | GGAAAGGCAACAGCAAACTCT | TGGGAGAGCACCAAGAGTAGAAGA |
PCR – polymerase chain reaction.
Statistical analysis
SPSS 22.0 (Chicago, IL, USA) were utilized for data analyses. The data were presented using Student’s t-test or ANOVA. Univariate and multivariate, and further Kaplan-Meier analysis were used for analysis and statistics of clinical data. All the experiments in present study were conducted 3 times independently. P<0.05 was considered statistically different.
Results
Expression of circ_0109320 is increased in NSCLC specimens and cells
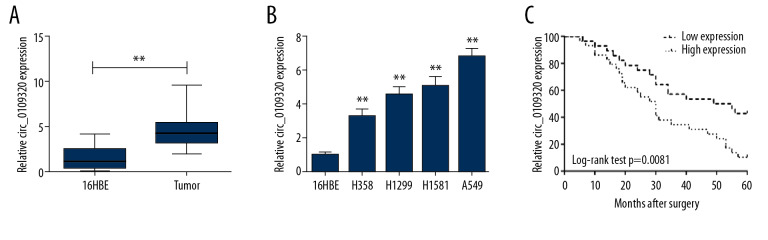
We firstly examined the expression level of circ_0109320 in tumor tissues compared with tissues adjacent to carcinoma. We confirmed that the circ_0109320 level was elevated significantly in the NSCLC specimens (Figure 1A). Similarly, compared with 16HBE, circ_0109320 was upregulated in NSCLC cells (Figure 1B). In further study, we recruited 57 NSCLC patients and divided them into low (n=28) and high (n=29) circ_0109320 expression group according to the mean value of circ_0109320 expression. Kaplan-Meier analysis indicated that high circ_0109320 expression was related to a worse overall survival (Figure 1C). Moreover, upregulated circ_0109320 expression level was associated with TNM subsets as well as lymph node metastasis. However, other features, including gender, age, smoking status and differentiation, were not related to circ_0109320 expression, as shown in Table 2. Univariate and further multivariate analysis confirmed that circ_0109320 was an independent prognostic variable in NSCLC (Table 3). These results announced that circ_0109320 might play a vital role in the NSCLC progression.
Figure 1.
Circ_0109320 is increased in NSCLC specimens and cells. (A) Circ_0109320 expression was elevated significantly in the NSCLC specimens. (B) Circ_0109320 expression was upregulated in NSCLC cell lines. (C) The association between circ_0109320 and overall survival in NSCLC. ** P<0.01. NSCLC – non-small cell lung cancer.
Table 2.
Circ_0109320 expression and clinical characteristics of NSCLC patients.
| Characteristics | N | circ_0109320 | P | |
|---|---|---|---|---|
| High | Low | |||
| Gender | 0.707 | |||
| Male | 40 | 21 | 19 | |
| Female | 17 | 8 | 9 | |
| Age (years) | 0.689 | |||
| <60 | 29 | 14 | 15 | |
| ≥60 | 28 | 15 | 13 | |
| Smoking status | 0.872 | |||
| No | 34 | 17 | 17 | |
| Yes | 23 | 12 | 11 | |
| Lymph node metastasis | 0.043 | |||
| No | 21 | 7 | 14 | |
| Yes | 36 | 22 | 14 | |
| Differentiation | 0.234 | |||
| High | 23 | 11 | 12 | |
| Low | 34 | 18 | 16 | |
| TNM subsets | 0.012 | |||
| I–II stage | 27 | 9 | 18 | |
| III–IV stage | 30 | 20 | 10 | |
NSCLC – non-small cell lung cancer.
Table 3.
Analysis of prognostic factors for overall survival rate in patients with NSCLC.
| Index | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Overall survival rate | ||||||
| Gender | 0.836 | 0.432–1.732 | 0.673 | |||
| Age, years (≥60 vs. <60) | 1.276 | 0.713–2.268 | 0.368 | |||
| Smoking status (yes vs. no) | 1.103 | 0.679–2.457 | 0.703 | |||
| Lymph node metastasis (yes vs. no) | 1.569 | 0.784–3.187 | 0.069 | |||
| Differentiation (low vs. high) | 1.335 | 0.748–2.342 | 0.422 | |||
| TNM subsets (III–IV stage vs. I–II stage) | 2.589 | 1.399–4.762 | 0.003 | 2.012 | 1.198–4.013 | 0.012 |
| Circ_0109320 expression (high vs. low) | 2.698 | 1.435–4.909 | 0.001 | 2.132 | 1.212–4.168 | 0.004 |
NSCLC – non-small cell lung cancer; HR – hazard ratio; CI – confidence interval.
Circ_0109320 exerts oncogenic roles in NSCLC cells
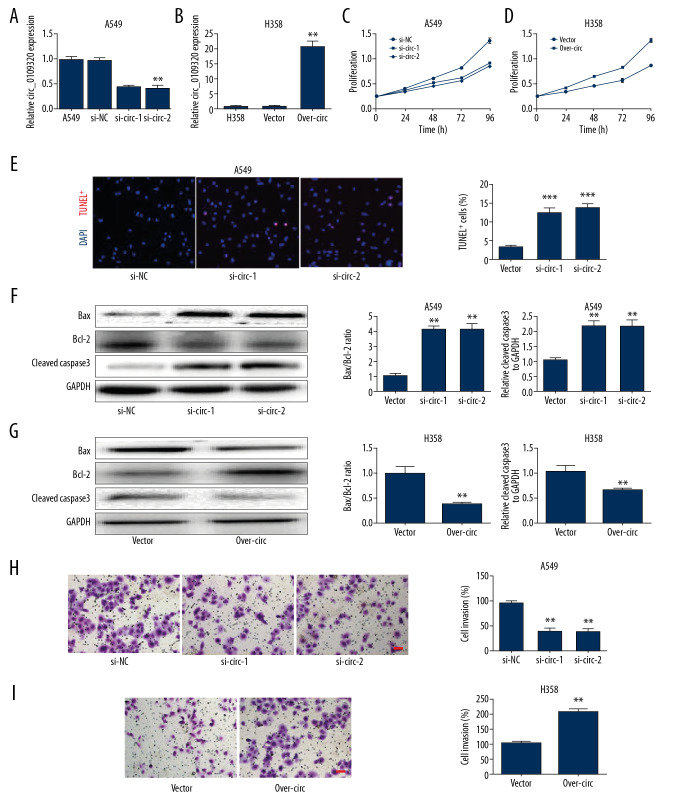
Then, in order to verify the oncogenic function of circ_0109320 in NSCLC, we knocked out the expression of circ_0109320 in A549 cells (which had the supreme expression), and overexpressed the circ_0109320 expression in H358 cells (which had the lowest expression) (Figure 2A, 2B). Afterwards, functional experiments including CCK-8 assay, TUNEL staining, western blot analysis, and Transwell assays were conducted to determine the cell proliferation, apoptosis, and invasion, respectively. The results of TUNEL staining showed that knockdown of circ_0109320 could increase the proportion of TUNEL positive cells and promote the apoptosis of tumor cells (Figure 2E). The data indicated that silencing of circ_0109320 could inhibit cell proliferation, increase cell apoptosis and downregulate cell invasion significantly. (Figures 2C, 2F, 2H). However, excessive circ_0109320 in H358 cells would promote the cell viability, attenuate the apoptosis and enhance the invasion (Figure 2D, 2G, 2I). These results suggested that circ_0109320 might exert oncogenic roles in NSCLC cells.
Figure 2.
Circ_0109320 exerts oncogenic roles in NSCLC cells. (A, B). Relative circ_0109320 expression in A549 and H358 cells after transfection. (C, D) CCK-8 assays was used to detect the proliferation. (E) TUNEL staining was used to detect the effect of knockdown of circ_0109320 on tumor cell apoptosis. (F, G). Apoptosis was evaluated using western blot. (H, I). Transwell assays were applied for exploring cell invasion. * P<0.05, ** P<0.01. NSCLC – non-small cell lung cancer; CCK-8 – Cell Counting Kit-8; TUNEL – terminal deoxynucleotidyl transferase dUTP nick end labeling.
Circ_0109320 sponges miR-595 to upregulate the E2F7 expression
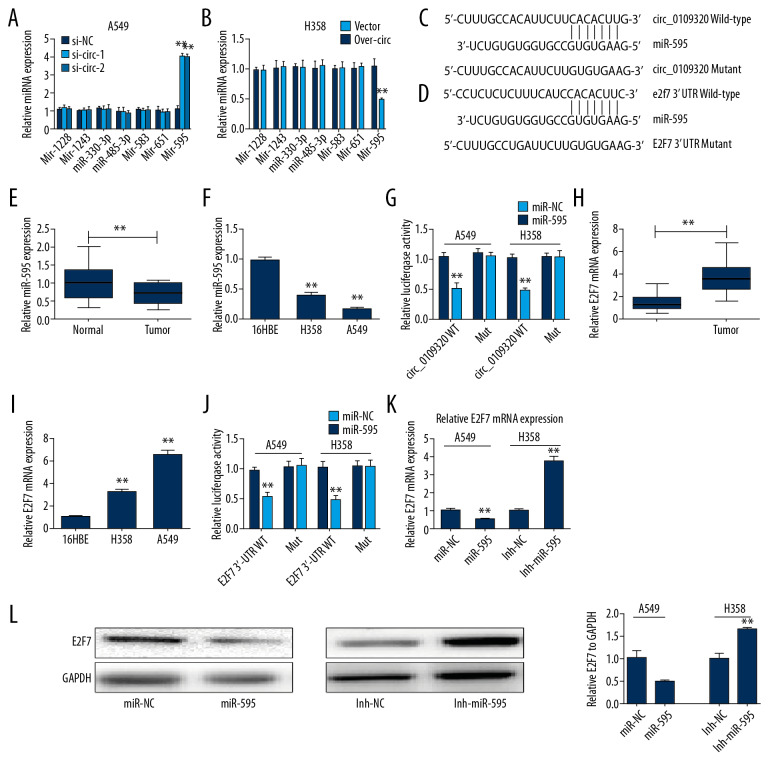
To further explore the possible mechanism of circ_0109320 in NSCLC, several potential miRNAs that could be sponged with circ_0109320 were predicted. Potential miRNAs targeting circ_0109320 sequences were predicted using the circInteractome database (https://circinteractome.nia.nih.gov/index.html). Through the analysis of biological information, we selected the top 10 miRNAs that have the most potential binding with circ_0109320. Among these miRNAs, 7 miRNAs play important regulatory role in tumor proliferation, invasion, and apoptosis. Therefore, we selected these 7 miRNAs for further screening through experimental verification. The RT-qPCR analysis results confirmed that miR-595 could be negative regulated by circ_0109320 (Figure 3A, 3B). The potential blinding site of miR-595 with circ_0109320 are shown in Figure 3C. Interestingly, miR-595 expression was decreased obviously in NSCLC specimens and cell lines, which was contrary to the circ_0109320 expression level (Figure 3E, 3F). To evaluate the binding ability between circ_0109320 and miR-595, dual-luciferase reporter experiment was taken. The data indicated miR-595 overexpression could obviously attenuate the luciferase intensity driven by circ_0109320 WT instead of circ_0109320 Mut in the A549 and H 358 cell lines (Figure 3G). Subsequently, we predicted that E2F7 was the potential downstream target of miR-595 via bioinformatics analysis. The potential binding site of miR-595 in the 3′-UTR of E2F7 is shown in (Figure 3D). Oppositely, E2F7 mRNA expression was significantly upregulated in tumor tissues and cell lines (Figure 3H, 3I). Dual-luciferase reporter experiment indicated that co-transfection of miR-595 mimics with E2F7 3′-UTR WT, instead of E2F7 3′-UTR Mut, obviously alleviate the luciferase activity in the 2 cell lines (Figure 3J). Thus, the mRNA and protein expression levels of E2F7 were significantly inhibited in the miR-595 overexpression group in A549 cells. On the contrary, in H358 cells, the expression of E2F7 was enhanced after silencing of miR-595 (Figure 3K, 3L).
Figure 3.
Circ_0109320 upregulated the E2F7 expression via sponging miR-595. (A, B). RT-qPCR analysis confirmed that miR-595 could be negative regulated by circ_0109320. (C) Potential blinding site of miR-595 with circ_0109320. (D) Potential binding site of miR-595 in the 3′-UTR of E2F7. (E, F) MiR-595 expression was decreased obviously in NSCLC specimens and cell lines. (G) The miR-595 and circ_0109320 blinding ability was determined by dual-luciferase reporter assay. (H, I) E2F7 mRNA expression was significantly upregulated in NSCLC tissues and cell lines. (J) The miR-595 and E2F7 3′-UTR blinding ability was determined by dual-luciferase reporter assay. (K, L) The E2F7 mRNA and protein expression was negatively correlated with the miR-595 expression. ** P<0.01. RT-qPCR – reverse transcription-quantitative polymerase chain reaction; NSCLC – non-small cell lung cancer.
Circ_0109320 exerts oncogenic roles through the miR-595/E2F7 pathway in NSCLC
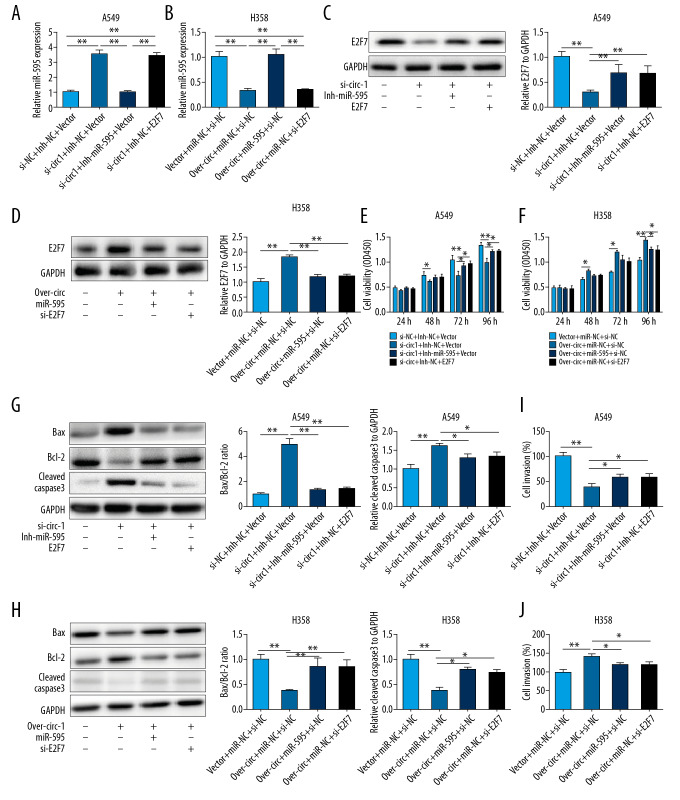
To evaluate whether the miR-595/E2F7 pathway was involved in circ_0109320 exerting oncogenic roles in NSCLC, functional experiments were conducted. RT-qPCR assay demonstrated that miR-595 expression was significantly increased after silencing of circ_0109320. However, cotransfecting with miR-595 inhibitor could partly restore the miR-595 expression in A549 cells (Figure 4A). Whereas, in H358 cells, overexpressed circ_0109320 obviously decreased miR-595 expression while miR-595 mimics would partly rescue the change (Figure 4B). Western blot analysis claimed that E2F7 expression was significantly decreased after knocking out circ_0109320. However, after cotransfected with miR-595 inhibitor, the E2F7 expression could be partly increased in A549 cells (Figure 4C). For H358 cell line, overexpression of circ_0109320 significantly elevated the E2F7 expression while miR-595 mimics could partially renovate the change as the si-E2F7 did (Figure 4D). Furthermore, knocking down miR-595 or upregulating E2F7 could in part alleviate the inhibitory effect of si-circ_0109320 on NSCLC cells (Figures 4E, 4G, 4I). On the contrary, both miR-595 mimic and si-E2F7 vectors could partially restore the carcinogenic function due to the overexpression of circ_0109320 (Figures 4F, 4H, 4J).
Figure 4.
Circ_0109320 exerts oncogenic roles through the miR-595/E2F7 pathway in NSCLC. (A, B) RT-qPCR was used to determine the miR-595 expression after transfection. (C, D) Western blot was carried out to evaluate the E2F7 protein expression after transfection. (E, F) CCK-8 assays was used to detect the cell viability after transfection. (G, H) Apoptosis after transfection was evaluated using western blot. (I, J) Transwell assays were applied for exploring cell invasion after transfection. * P<0.05, ** P<0.01. NSCLC – non-small cell lung cancer; RT-qPCR – reverse transcription-quantitative polymerase chain reaction; CCK-8 – Cell Counting Kit-8.
Discussion
Chemotherapy is the main treatment for non-small cell lung cancer (NSCLC) [15]. The standard platinum-containing 2-drug regimen for first-line treatment has reached the bottleneck stage, and its side effects are also large, so it is difficult to make further breakthroughs in the efficacy [15,16]. In the past decade, miRNAs and lncRNAs have been the focus of research [17,18]. With the discovery of circRNAs, more and more research has begun to focus on the role of circRNAs in human diseases.
CircRNAs, as a class of stable closed circular non-coding RNA, are abundant in eukaryotic transcriptome [10]. Most of the circRNAs are composed of exon sequences, which are conservative in different species, and have expression specificity at different stages of development [19]. CircRNAs can compete bind with miRNAs and play important roles in many cancer diseases, including NSCLC [20,21]. In our study, we confirmed that circ_0109320 expression was significant increase in NSCLC tissues and cell lines. Besides, upregulated circ_0109320 expression level was associated with TNM stages as well as lymph node metastasis, but not with gender, age, smoking, and differentiation grade. These data suggest that circ_0109320 may have significant tumorigenic effect in NSCLC.
Numerous studies have shown that circRNAs can play a spongy role in miRNAs in different physiological and pathological processes, and can competently bind to miRNAs, thereby negatively regulating the expression of target gene [21,22]. MiRNAs, as a group of non-coding small RNAs composed of 19–22 nucleotides, have been widely proven to play important roles in NSCLC [8]. In our study, we confirmed that circ_0109320 exerts oncogenic roles through negatively regulating miR-595 in NSCLC. Previous studies have shown that miR-595 plays an important role in many tumors [23,24], but its role in NSCLC is still unclear. Our study demonstrated that miR-595 also plays an important regulatory role in NSCLC. In addition, we further proved that E2F7 was the downstream target of miR-595. As a novel mammalian E2F family member, the role of E2F7 in cancers, including NSCLC, has been identified [25]. Ma et al. revealed that overexpression of miR-302 promotes self-renewal capability and cell cycle entry of liver cancer stem cells by inhibition of E2F7 expression [26]. Similarly, using bioinformatics tools, Wang et al. found that E2F2 was closely related to the overall survival rate of NSCLC [27]. Consistent with previous findings, we found upregulation of E2F7 could partially block the inhibitory effect of si-circ_0109320 on NSCLC cells. These data demonstrate that circ_0109320 exerts oncogenic roles through the miR-595/E2F7 pathway in NSCLC.
Conclusions
Our study demonstrated that circ_0109320 exerts a critical role in NSCLC oncogenesis. Moreover, the present study revealed that si-circ_0109320 might have potential application in the future treatment of NSCLC. However, further in vivo experiments should be carried out to demonstrate that circ_0109320 is mediated in the progress of NSCLC to verify the current results.
Footnotes
Conflict and interest
None.
Source of support: This study was supported by Lianyungang Health Family Planning Science and Technology Project (No. 201718)
References
- 1.Skaug K, Eide GE, Gulsvik A. What is the minimum number of patients for quality control of lung cancer management in Norway? Clin Respir J. 2016;10:707–13. doi: 10.1111/crj.12274. [DOI] [PubMed] [Google Scholar]
- 2.Argyri E, Tsimplaki E, Marketos C, et al. Investigating the role of human papillomavirus in lung cancer. Papillomavirus Res. 2017;3:7–10. doi: 10.1016/j.pvr.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasapoglu US, Arinc S, Gungor S, et al. Prognostic factors affecting survival in non-small cell lung carcinoma patients with malignant pleural effusions. Clin Respir J. 2016;10:791–99. doi: 10.1111/crj.12292. [DOI] [PubMed] [Google Scholar]
- 4.Faris NR, Smeltzer MP, Lu F, et al. Evolution in the surgical care of patients with non-small cell lung cancer in the mid-south quality of surgical resection cohort. Semin Thorac Cardiovasc Surg. 2017;29:91–101. doi: 10.1053/j.semtcvs.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishio M, Hida T, Atagi S, et al. Multicentre phase II study of nivolumab in Japanese patients with advanced or recurrent non-squamous non-small cell lung cancer. ESMO Open. 2016;1:e000108. doi: 10.1136/esmoopen-2016-000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatt VR, D’Souza SP, Smith LM, et al. Epidermal growth factor receptor mutational status and brain metastases in non-small-cell lung cancer. J Glob Oncol. 2017;3:208–17. doi: 10.1200/JGO.2016.003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Wang Y, Wang J. MicroRNA-584 inhibits cell proliferation and invasion in non-small cell lung cancer by directly targeting MTDH. Exp Ther Med. 2018;15:2203–11. doi: 10.3892/etm.2017.5624. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Yang Z, He J, Gao P, et al. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget. 2017;8:113558–70. doi: 10.18632/oncotarget.23060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kou P, Zhang C, Lin J, Wang H. Circular RNA hsa_circ_0078602 may have potential as a prognostic biomarker for patients with hepatocellular carcinoma. Oncol Lett. 2019;17:2091–98. doi: 10.3892/ol.2018.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Zhang C, Zhang B, et al. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. 2019;17:388–97. doi: 10.3892/ol.2018.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Ding W, Tariq MA, et al. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–69. doi: 10.7150/thno.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Yu J, Tian H, et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol. 2019;234:19130–40. doi: 10.1002/jcp.28692. [DOI] [PubMed] [Google Scholar]
- 13.Zhou W, Wang H, Yang J, et al. Down-regulated circPAPPA suppresses the proliferation and invasion of trophoblast cells via the miR-384/STAT3 pathway. Biosci Rep. 2019;39(9) doi: 10.1042/BSR20191965. pii: BSR20191965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu YT, Han XH, Xing PY, et al. Circular RNA profiling identified as a biomarker for predicting the efficacy of Gefitinib therapy for non-small cell lung cancer. J Thorac Dis. 2019;11:1779–87. doi: 10.21037/jtd.2019.05.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng H, Li XJ, Wang XJ, et al. A meta-analysis of adjuvant EGFR-TKIs for patients with resected non-small cell lung cancer. Lung Cancer. 2019;137:7–13. doi: 10.1016/j.lungcan.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Hopkins AM, Shahnam A, Zhang S, et al. Prognostic model of survival outcomes in non-small cell lung cancer patients initiated on afatinib: Pooled analysis of clinical trial data. Cancer Biol Med. 2019;16:341–49. doi: 10.20892/j.issn.2095-3941.2018.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui J, Mo J, Luo M, et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:12400–9. [PMC free article] [PubMed] [Google Scholar]
- 18.Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Glazar P, Papavasileiou P, Rajewsky N. CircBase: A database for circular RNAs. RNA. 2014;20:1666–70. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas LF, Saetrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30:2243–46. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Tan S, Liu WR, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18:134. doi: 10.1186/s12943-019-1061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513:207–12. doi: 10.1016/j.bbrc.2019.03.213. [DOI] [PubMed] [Google Scholar]
- 23.Hao Y, Zhang S, Sun S, et al. MiR-595 targeting regulation of SOX7 expression promoted cell proliferation of human glioblastoma. Biomed Pharmacother. 2016;80:121–26. doi: 10.1016/j.biopha.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Tian S, Zhang M, Chen X, et al. MicroRNA-595 sensitizes ovarian cancer cells to cisplatin by targeting ABCB1. Oncotarget. 2016;7:87091–99. doi: 10.18632/oncotarget.13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitxelena J, Apraiz A, Vallejo-Rodriguez J, et al. An E2F7-dependent transcriptional program modulates DNA damage repair and genomic stability. Nucleic Acids Res. 2019;47:7716–17. doi: 10.1093/nar/gkz587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SI, Jeon MH, Kim JS, et al. The miR-302 cluster transcriptionally regulated by POUV, SOX and STAT5B controls the undifferentiated state through the post-transcriptional repression of PBX3 and E2F7 in early chick development. Mol Reprod Dev. 2014;81:1103–14. doi: 10.1002/mrd.22429. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Yin H, Zhang L, et al. The construction and analysis of the aberrant lncRNA-miRNA-mRNA network in non-small cell lung cancer. J Thorac Dis. 2019;11:1772–78. doi: 10.21037/jtd.2019.05.69. [DOI] [PMC free article] [PubMed] [Google Scholar]