Abstract
Background
The aim of this study was to investigate the expression of tumor-derived exosomal RNA eIF4E (exo-eIF4E) in non-small cell lung cancer (NSCLC) and its correlation with prognosis.
Material/Methods
The Cancer Genome Atlas (TCGA) data was exacted to investigate the role of tissue eIF4E in NSCLC. We enrolled 99 NSCLC patients and 40 healthy volunteers with corresponding serum samples in this study. The levels of exo-eIF4E in the peripheral blood of each group were tested by quantitative polymerase chain reaction (PCR). The chi-squared test and the log-rank test were applied to analyze the correlation between the expression levels of exo-eIF4E and the patients’ clinical-pathological data, including the overall survival.
Results
TCGA data showed that increased eIF4E in NSCLC tissues was associated with late-stage disease (P=0.0497) and inferior overall survival (P=0.017). The expression of exo-eIF4E in the serum of the NSCLC group was significantly higher than that in healthy individuals (P<0.001). Furthermore, advanced TNM stage (P=0.003), distant metastasis (P=0.008), and serum positive cytokeratin fragment 19 (CYFRA21-1) (P=0.023) are more likely present in NSCLC patients with higher exo-eIF4E expression. Moreover, the multivariate combined with univariate analyses verified exo-eIF4E as an independent prognostic factor for shorter overall survival (P=0.01) and progression-free survival (P=0.005). Shorter overall survival (P=0.0005) and inferior progression-free survival (P=0.0017) are more likely present in NSCLC patients with higher exo-eIF4E.
Conclusions
Tumor-derived exo-eIF4E in serum can be a practical tool to predict the prognosis of NSCLC.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; Eukaryotic Initiation Factor-4E; Exosome Multienzyme Ribonuclease Complex; Prognosis
Background
Lung cancer is a malignant tumor with the highest morbidity and mortality in the world [1]. Non-small cell lung cancer (NSCLC) is its most crucial type [2]. Most cases of NSCLC have different changes in tumor suppressor genes and proto-oncogenes caused by smoking, domestic gas inhalation, and occupational exposure [2]. The activation of proto-oncogenes and the disorder of the body’s immune system eventually lead to abnormal proliferation and apoptosis of pulmonary epithelial cells, resulting in NSCLC [3]. Changes in useful markers are related to the treatment effect and different prognosis of patients with NSCLC, guiding the choice of the treatment strategy. Molecular targeted therapies have improved the survival of NSCLC subgroups divided by detection of genomic aberrations. Patient cases with exon 19 (deletions) or exon 21 (Leu858Arg) positive mutations in NSCLC will benefit from treatment with epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor (TKI) [4]. Progression-free survival and objective response rate increase in patients with anaplastic lymphoma kinase (ALK) rearrangements receiving TKIs [5]. Therefore, novel biomarkers for predicting NSCLC prognosis, especially disease-free survival, might be a promising strategy to intervene in the development and relapse of NSCLC.
Exosomes are cystic vesicles of 30 to 100 nm in size and contain specific proteins or RNAs secreted by host cells [6]. These exosome-derived RNAs or proteins can be targeted to the recipient cells by fusion with the cell membrane of the recipient cell, and the specific biological effect on the receptor cells can be exerted through a non-contact manner [7]. Exosomal-derived regulatory signals can not only mediate the malignant behavior of tumor cells but also affect interstitial angiogenesis [8]. Moreover, biomarkers in circulating exosomes can improve the accuracy of early NSCLC diagnosis [9]. The downward trend of specific microRNAs (miRNAs) in exosomes can also indicate partial remission in immunotherapy [10]. The eukaryotic translation initiation factor 4G2 (eIF4E) acts as a “scaffold protein” that interferes with the translation of cap-dependent and non-dependent proteins in eukaryotic. Thus, the proliferation, apoptosis, cell cycle, migration, invasion, and angiogenesis of tumor cells will be affected, which will ultimately affect the occurrence and development of tumors [11–13]. However, at present, the expression of eIF4E in serum exosomes and its associations with disease characteristics in patients with NSCLC are not clear.
This study examined the expression of exosomal eIF4E (exo-eIF4E) in the serum of NSCLC patients and healthy individuals and analyzed the correlation between exo-eIF4E expression and prognosis. We focused on eIF4E RNA in circulating exosomes to explore its value in prognostic judgment in NSCLC patients.
Material and Methods
Extraction and analysis of The Cancer Genome Atlas (TCGA) data
The Cancer Genome Atlas (TCGA) data were extracted and analyzed at the Gene Expression Profiling Interactive Analysis (GEPIA) website (http://gepia.cancer-pku.cn/). eIF4E as a keyword was searched in GEPIA to extract data of the comparison of eIF4E between NSCLC and adjacent normal tissues, and the relation between eIF4E tissue expression and clinical features (including TNM stage, overall survival, and disease-free survival).
Patient enrollment and blood samples
A total of 99 NSCLC patients (59 males and 40 females) between March and October 2017 were selected. All patients with complete clinical information were diagnosed according to the histological biopsy. In addition, 40 healthy individuals were enrolled. Individuals with cancer or other diseases were excluded. All the participants gave their written informed consent. The Ethical Committee of the Yantai Yuhuangding Hospital approved this study.
Whole blood samples (3 mL) were collected in a coagulation tube and were coagulated at 37°C for 30 minutes. The serum and blood cells were separated by centrifugation at 2000 g for 10 minutes. The collected serum was centrifuged at 10 000 g for 30 minutes to obtain a supernatant further. After being treated with a 0.22 μm filter (Millipore, Billerica, MA, USA), serum was stored in a cryopreservation tube at −80°C for further analysis.
Exosomes extraction and identification
According to the manufacturer’s instruction, we used a Total Serum Exosome Isolation Kit (Thermo, Carlsbad, CA, USA) to extract exosomes from the stored serum. Briefly, 1 mL stored serum was supplemented with 200 μL exosome isolation reagent. After being blended mildly, the mixtures were incubated at 4°C for 30 minutes. Following a 10 000 g centrifugation for 10 minutes, the exosome pellet was collected at the bottom of the tubes. Phosphate-buffered saline (200 μL) was used to resuspend the exosome pellet.
Formvar solution (0.125%) and exosome pellet (10 uL) were mixed to fix the exosome pellet. After being stained using uranyl acetate, the exosome pellet was photographed using a JCM-7000 TEMSCAN microscope (JEOL, Tokyo, Japan). After a calibration via standardized dilutions, a NanoSight NS300 Instrument (NanoSight Ltd., Amesbury, UK) was used to investigate the volume distribution of the nanoparticle-based on the instruction. Besides, several specific markers (CD9, CD63, and tumor susceptibility gene 101-TSG101), and endoplasmic reticulum (calnexin) [14] were evaluated by western blot to verify the exosome element.
Western blotting assay
Radioimmunoprecipitation assay (RIPA) buffer (Solarbio, China) was applied to extract total proteins, supplementing with 1% phosphorylation and protease inhibitors (Thermo Fisher Scientific, USA). According to the manufacturer’s protocol, the concentration of the protein samples was tested by the bicinchoninic acid (BCA) protein assay kit (Tiangen, China). After denatured at 96°C for 10 minutes, 9% SDS-PAGE (sodium dodecyl sulphate-polyacrylamide gel electrophoresis) (Solarbio, China) used to divide the target proteins. The PVDF (polyvinylidene fluoride membrane) (Millipore, USA) was used for transfer. After incubation with 5% non-fat milk for a blockade of non-specific signals, PVDF membranes were incubated with primary antibodies against CD9 (1: 1000), CD63 (1: 2000), TSG101 (1: 3000), calnexin (1: 2000) (Cell Signaling Technology, USA) overnight at 4°C. Then the PVDF membrane was dealt with horseradish peroxidase (HRP) conjugated secondary antibody (1: 5000, Cell Signaling Technology, USA). The protein blots were photographed using a western imaging system (General Electric Company, USA). The density of bands was quantified by ImageJ software (Bio-Rad, Hercules, CA, USA).
Total RNA extraction and quantitative analysis
Total RNA of tissue and cell line was extracted using RNAiso Plus (TAKARA, Beijing, China) according to the instruction. The extracted RNA was synthesized to cDNA by the PrimeScript™ RT reagent Kit (TAKARA, Beijing, China). Quantitative polymerase chain reaction (qPCR) was done using SYBR® Green Realtime PCR Master Mix (TOYOBO, Shanghai, China) on the Applied Biosystems Veriti Thermal Cycler (Thermo Fisher Scientific, USA). The melting curve and amplification plot are shown in Supplementary Figure 1. The quantitation of the target RNA expression was assessed using the endogenous control by the 2−ΔΔCt method (β-actin as an internal control). The primers of eIF4E are as follows:
-
forward 5′-GAAACCACCCCTACTCCTAA TCC-3′;
reverse 5′-AGAGTGCCCATCTGTTC TGTA-3′.
Qubit Flex Fluorometer (Thermo Fisher Scientific, USA) was used to evaluate the quality of the prepared RNA and cDNA was measured.
Statistical analysis
All data were presented as the mean±standard deviation. The Student’s t-test made a comparison between the means from 2 groups. The optimal cutoff was calculated as the value that maximized the sum of sensitivity and specificity receiver the operating characteristic curves were applied to determine the optimal cutoff of serum exosome eIF4 for predicting NSCLC prognosis. A comparison of the predicting efficiency between serum exosome eIF4 and the other biomarkers was made using the areas under these curves. The univariate and the multivariate Cox proportional hazards regression were applied to identify the independent predicting factors for overall survival. Survival analysis was described using the Kaplan-Meier method, followed by comparison using the log-rank test. Every experiment was finished at least 3 independent times. All statistical analyses were carried out by SPSS 22.0 software (International Business Machines Corporation, USA). Statistical significance was defined as P<0.05.
Results
eIF4E expression in NSCLC tissues was highly related to advanced stage and inferior overall survival shown by TCGA data
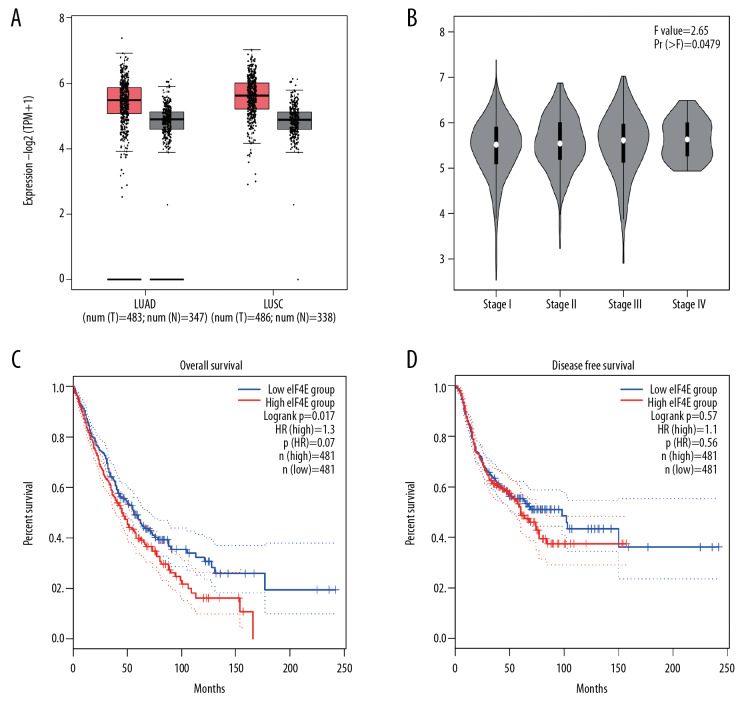
Tissue expression data of eIF4E in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) was acquired from the TCGA database. We found eIF4E inclined notably in cancerous tissues relative to normal tissues (Figure 1A, both P<0.01). Patients with advanced TNM stages exhibited mildly increased eIF4E expression than those with early stages (Figure 1B, P=0.0497). Besides, a higher expression of eIF4E was associated with inferior overall survival (Figure 1C, P=0.017), but not disease-free survival (Figure 1D, P=0.57). Altogether, our results indicated that eIF4E is upregulated in NSCLC tissues, and it demonstrates an oncogenic role in NSCLC.
Figure 1.
EIF4E expression in NSCLC tissues is highly related to advanced stage and inferior overall survival shown by TCGA data. (A) Expression of eIF4E in LUAD, LUSC tissues, and normal tissues. (B) Tissue expression of eIF4E in different stages of NSCLC patients. (C) Overall survival of NSCLC patients with high and low eIF4E expression. (D) Disease-free survival of NSCLC patients with high and low eIF4E expression. NSCLC – non-small cell lung cancer; TCGA – The Cancer Genome Atlas; LUAD – lung adenocarcinoma; LUSC – lung squamous cell carcinoma.
Identification of exosomes from serum
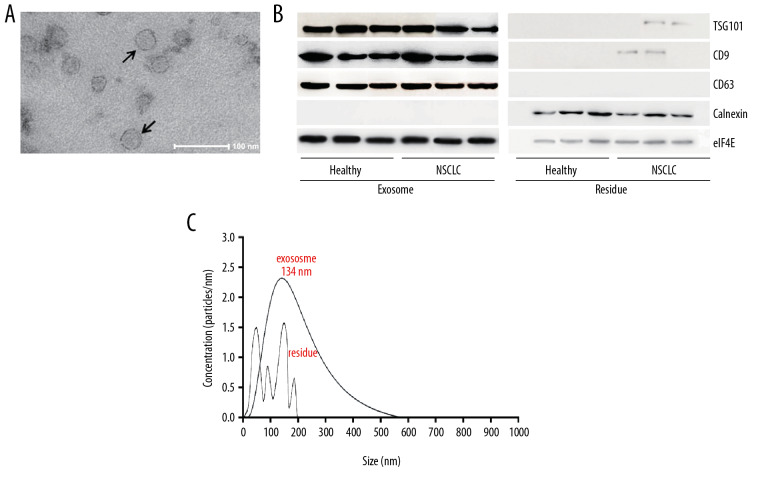
As shown in Figure 2A, the microbubbles pointed by the arrows are exosomes. Western blot assay was used to verify the positive expression of TSG101, CD9, CD63, eIF4E, and negative expression of calnexin in exosomes from both NSCLC and healthy individuals (compared to the residue, Figure 2B). The exosomes have an average diameter of 134 nm (Figure 2C).
Figure 2.
Identification of exosomes from serum. (A) The micromorphology of exosomes from serum using an electron microscope. (B) Western blot assay showed the positive TSG101, CD9, and CD63 expression and negative calnexin expression in exosomes from serum in NSCLC and healthy individuals. (C) Nanoparticle tracking analysis showed a diameter distribution of exosome. ** P<0.01, * P<0.05. NSCLC – non-small cell lung cancer.
Exo-eIF4E in NSCLC was strongly relative to disease progression
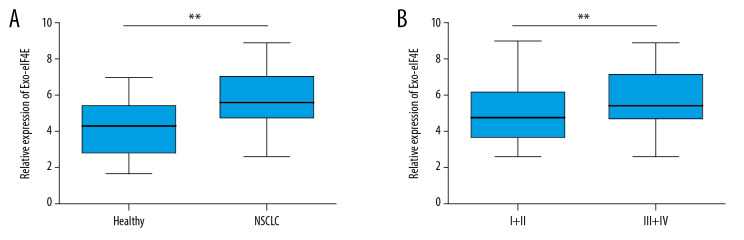
We tested the exo-eIF4E RNA levels in both the NSCLC patients and the healthy volunteers to investigate the vital impact of exo-eIF4E in the tumorigenesis of NSCLC. The data show that the exo-eIF4E levels are more likely to increase in NSCLC patients than om the controls (Figure 3A, P<0.001). Moreover, late-stage NSCLC patients exhibit higher exo-eIF4E in serum than patients with early-stage disease (III+IV versus I+II, Figure 3B, P=0.0048). These findings suggest that exo-eIF4E is a helpful biomarker in NSCLC development.
Figure 3.
Exo-eIF4E in NSCLC is strongly relative to disease progression. (A) qPCR showed a comparison of exosome eIF4E levels between NSCLC patients (n = 99) and healthy individuals (n=40). (B) qPCR showed the comparison of exosome eIF4E levels between NSCLC patients with TNM I+II stage (n=39) and III+IV stage (n=60). ** P<0.01, * P<0.05. exo-eIF4E – eoxsomal eIF4E; NSCLC – non-small cell lung cancer; qPCR – quantitative polymerase chain reaction.
Exo-eIF4E might be a prognostic factor for NSCLC patients
We studied the correlations between clinical-pathological features and the exo-eIF4E levels in serum. According to the serum levels of exo-eIF4E, we divided the NSCLC patients into 2 groups based on exo-eIF4E levels: high ≥6.23; low <6.23). We observed no significant associations between exo-eIF4E expression and age, sex, tumor size, smoking status, histology, lymph node metastasis, or serum carcinoembryonic antigen (CEA) (Table 1, all P>0.05). However, a high level of exo-eIF4E was found to be associated with the advanced TNM stage (Table 1, P=0.003), distant metastasis (Table 1, P=0.008), and serum positive cytokeratin fragment 19 (CYFRA21-1) (Table 1, P=0.023), all of which are significant prognostic factors for NSCLC. Altogether, our data indicated that exos-eIF4E might predict the prognosis of NSCLC patients.
Table 1.
Correlations between expression levels of serum exo-eIF4E and clinical characteristics (n=99).
| n | Serum exo-eIF4E level | P-value | ||
|---|---|---|---|---|
| Low (n=50) | High (n=49) | |||
| Age (years) | 0.265 | |||
| <65 | 53 | 24 | 29 | |
| ≥65 | 46 | 26 | 20 | |
| Sex | 0.461 | |||
| Male | 59 | 28 | 31 | |
| Female | 40 | 22 | 18 | |
| Smoking status | 0.774 | |||
| Smoker | 39 | 19 | 20 | |
| Nonsmoker | 60 | 31 | 29 | |
| Size of tumor | 0.056 | |||
| <4 cm | 50 | 30 | 20 | |
| ≥4 cm | 49 | 20 | 29 | |
| Histology | 0.355 | |||
| Adenocarcinoma | 43 | 24 | 19 | |
| Squamous carcinoma | 56 | 26 | 30 | |
| TNM stage | 0.003* | |||
| I–II | 39 | 27 | 12 | |
| III–IV | 60 | 23 | 37 | |
| Lymph node metastasis | 0.481 | |||
| Negative | 35 | 16 | 19 | |
| Positive | 64 | 34 | 30 | |
| Distant metastasis | 0.008* | |||
| Negative | 67 | 40 | 27 | |
| Positive | 32 | 10 | 22 | |
| CYFRA21-1 (ng/mL) | 0.023* | |||
| <3.3 | 32 | 23 | 9 | |
| ≥3.3 | 67 | 37 | 40 | |
| CEA (ug/mL) | 0.776 | |||
| <5 | 33 | 16 | 17 | |
| ≥5 | 66 | 34 | 32 | |
exo-eIF4E – exosomal eIF4E; CYFRA21-1 – cytokeratin fragment 19; CEA – carcinoembryonic antigen.
Exo-eIF4E, an independent prognostic factor for overall survival and disease progression of NSCLC
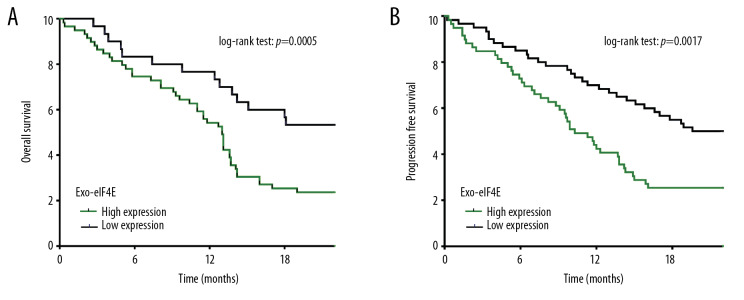
Shown by multivariate statistics, advance TNM stage (III+IV, P=0.003) and high level of exo-eIF4E (P=0.01, hazard ratio [HR]=1.744, 95% confidence interval [CI]=1.040–3.093) were independent predictors for cancer-relative death in NSCLC (Table 2). Moreover, advance TNM stage (III+IV, P=0.001), positive CYFRA21-1 (P=0.023) and high level of exo-eIF4E (P=0.005, HR=1.478, 95% CI=1.134–2.972) in NSCLC can independently predict the disease progression (Table 3). Based on Kaplan-Meier analysis, a higher expression level of exo-eIF4E (74.6%, 54.2%, and 25.4% versus. 83.3%, 76.7%, and 53.3%) was associated with inferior overall survival at 6, 12, and 18 months, respectively (Figure 4A, P=0.0005). NSCLC patients with low exo-eIF4E levels (85.0%, 70.0%, and 55.0%) exhibited notably decreased rates of disease progression than NSCLC patients with high exo-eIF4E levels (73.3%, 43.3%, and 25.0%) at 6, 12, and 18 months, respectively (Figure 4B, P=0.0017).
Table 2.
Independent risk factors for overall survival in NSCLC patients (n=99).
| Variable | Univariate | Multivariate analysis | ||
|---|---|---|---|---|
| P-value | P-value | Hazard ratio (95% CI) | ||
| Size of tumor (cm) | ≥4 vs. <4 | 0.003 | ||
| TNM stage | I–II vs. III–IV | 0.001 | 0.003 | 1.845 (1.176–3.313) |
| Lymph node metastasis | Negative vs. positive | 0.008 | ||
| CYFRA21-1 | <3.3 vs. ≥3.3 | 0.005 | ||
| Exo-eIF4E | <6.23 vs. ≥6.23 | 0.003 | 0.01 | 1.744 (1.040–3.093) |
NSCLC – non-small cell lung cancer; CI – confidence interval; CYFRA21-1 – cytokeratin fragment 19; Exo-eIF4E – eoxsomal eIF4E.
Table 3.
Independent risk factors for disease-free survival in NSCLC patients (n=99).
| Variable | Univariate | Multivariate analysis | ||
|---|---|---|---|---|
| P-value | P-value | Hazard ratio (95% CI) | ||
| Size of tumor (cm) | ≥4 vs. <4 | 0.002 | ||
| TNM stage | I–II vs. III–IV | 0.001 | 0.001 | 1.538 (1.299–2.984) |
| Lymph node metastasis | Negative vs. positive | 0.003 | ||
| CYFRA21-1 | <3.3 vs. ≥3.3 | 0.005 | 0.023 | 1.328 (1. 436–3.182) |
| CEA | <5 vs. ≥5 | 0.003 | ||
| Exo-eIF4E | <6.23 vs. ≥6.23 | 0.001 | 0.005 | 1.478 (1.134–2.972) |
NSCLC – non-small cell lung cancer; CI – confidence interval; CYFRA21-1 – cytokeratin fragment 19; CEA – carcinoembryonic antigen; Exo-eIF4E – eoxsomal eIF4E.
Figure 4.
Exo-eIF4E, an independent prognostic factor for overall survival and disease progression of NSCLC. (A) Overall survival of high (n=49) and low (n=50) exo-eIF4E levels in NSCLC patients. (B) Progression-free survival of high (n=49) and low (n=50) exo-eIF4E levels in NSCLC patients. ** P<0.01, * P<0.05. Exo-eIF4E – eoxsomal eIF4E; NSCLC – non-small cell lung cancer.
Discussion
The survival rate for NSCLC patients undergoing surgery or chemotherapy within 5 years has reached 20%, and unfortunately, the relapse rates has remained high for the past 2 decades [15]. Despite extensive research on the molecular mechanisms of drug resistance, there is still a lack of an effective intervention strategy. Various molecular drugs have been designed to inhibit the proliferation, apoptosis, invasion, and metastasis and angiogenesis of NSCLC cells targeting the signal transduction process. Therefore, novel sensitive markers are needed for the treatment and diagnosis of NSCLC.
We studied the levels of exo-eIF4E in the serum of patients with NSCLC to analyze the correlation between exo-eIF4E expression and the development of NSCLC (including chemotherapy and overall survival). Previously, researchers have shown that eIF4E is involved in different processes of lung cancer, but its role is controversial. eIF4E can promote epithelial-mesenchymal transition in NSCLC and induce metastasis [16]. The secretion of human bone marrow mesenchymal stem cells can reduce its invasiveness by inhibiting eIF4E in NSCLC [17], further supporting the vital role of eIF4E in NSCLC metastasis. However, it has been reported that high expression of eIF4E is associated with docetaxel sensitivity in NSCLC patients [18]. Inhibition of eIF4E by miR-141 promotes resistance of NSCLC cells to docetaxel [18]. In this study, we found that exo-eIF4E expression was different in NSCLC patients and healthy individuals (Figure 2). The expression of exo-eIF4E in circulating exosomes was related to the TNM stage, distant metastasis, and serum positive CYFRA21-1 (Table 1). Besides, our study further demonstrated the value of exo-eIF4E in predicting the prognosis of NSCLC (Figure 3).
Exosomes are specialized vesicles containing proteins or RNA secreted by cells [19]. These proteins or RNAs are “injected” into target cells by cell membrane fusion of target cells, which make various biological effects be exerted by regulating genes or proteins in cells, including anti-oxidative stress and regulation of tumor cell invasiveness [20]. The stability in the systemic circulation and tissue-specific uptake make exosomes promising targets for new therapies. Different exosomes are involved in promoting cancer growth, recurrence after treatment, metastasis, and immune cell regulation [21,22]. Alpha-smooth muscle actin in exosomes from NSCLC cells not only boosts cancer cell proliferation but restrain fibroblasts apoptosis, resulting in the development of NSCLC [21]. Exosomes-derived miR-214 has been shown to reduce the chemotherapy response of NSCLC cells to gefitinib [22].
Besides, studies have shown that RNA in exosomes is more stable than other free RNA and therefore has potential as a diagnostic marker for the disease [23]. These exosomal characteristics represent the molecular characteristics of the cells, making liquid biopsy possible. Using tissue biopsy as a standard, the peripheral blood exosome T790M mutation detection, a biomarker for sensitivity to chemotherapy basing on third-generation tyrosine kinase inhibitors, achieved significant accuracy (sensitivity and specificity: 92% and 89%, respectively) [24]. Exosome-derived long noncoding RNA growth arrest-specific transcript 5 (GAS5) can effectively predict the diagnosis of NSCLC with the assistance of CEA (AUC=0.929) [25]. Exosome-derived YKT6 in NSCLC can not only regulate exosome release but also predict the prognosis of NSCLC patients after surgery [26]. Exosome mRNA has its unique advantages in liquid biopsy. Exosome derived mRNA can increase the detection sensitivity of activating EGFR mutations and EGFR T790M mutations to 98% and 90% (82% and 84% for ctDNA only), respectively [27]. Pleural effusion-derived exosome mRNA expression profiles can distinguish benign lung disease from lung adenocarcinoma, especially lipocalin-2 (LCN2), with the highest AUC (0.9916) [28]. These suggest that exosomal mRNA is a potential screening biomarker for NSCLC.
It has been proven that eIF4E acts as an oncogene in the development of NSCLC. Various eIF4 components have a different potential impact on the tumorigenesis of NSCLC. Compared to adjacent normal tissues, the histological level of eIF4G1 in cancer tissues is increased [29]. However, highly expressed eIF4A2 is closely related to better overall and disease-free survival in NSCLC [30]. Initially, researchers found that high eIF4E was more likely present in NSCLC with an inferior prognosis, while high cyclin D1 was not [31]. Yoshizawa et al. showed that increased p-eIF4E was an independent predictor for NSCLC prognosis using multivariate analysis [32]. Compared with eIF4E non-phosphorylated patients, phosphorylated ones had significantly inferior overall survival [32]. The expression of phosphorylated eIF4E is sharply higher in patients with early-stage (T1) compared with patients with advanced-stage (T3) in NSCLC and gastrointestinal malignancies [33]. However, few studies investigate the role of exosome-derived eIF4E in the development of NSCLC, let alone its prediction for NSCLC prognosis. Compared with traditional biopsies of tissue eIF4E, the ubiquitous presence of exosomes in the blood makes it easier to monitor the dynamic changes.
However, some limitations must be considered in understanding our results. First, the sample size was insufficient and no external validation was performed using data from independent biological samples. Second, the relationships between serum exosomes-derived eIF4E and more molecular pathological features was not analyzed, and its mechanism involved in the pathogenesis of NSCLC was not explored. Therefore, large-scale, prospective, extensive clinical trials are needed to validate our findings.
Conclusions
This study demonstrates that the exosome derived eIF4E in serum can be used as a useful predictor for NSCLC survival.
Supplementary Data
The representing amplification plot (A) and melting curve (B) in quantitative analysis of exosome derived eIF4E.
Acknowledgment
Thanks to Beijing Genecast Biotechnology Co. (Beijing, P.R. China) for technical support in molecular biology experiments.
Footnotes
Data availability statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Balata H, Fong KM, Hendriks LE, et al. Prevention and early detection for NSCLC: Advances in Thoracic Oncology 2018. J Thorac Oncol. 2019;14:1513–27. doi: 10.1016/j.jtho.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Tsao AS, Scagliotti GV, Bunn PA, Jr, et al. Scientific advances in lung cancer 2015. J Thorac Oncol. 2016;11:613–38. doi: 10.1016/j.jtho.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–77. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 6.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–89. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, Park S, Kim H, et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett. 2020;475:2–13. doi: 10.1016/j.canlet.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Yu L, Lin X, et al. Combination of serum miRNAs with serum exosomal miRNAs in early diagnosis for non-small-cell lung cancer. Cancer Manag Res. 2020;12:485–95. doi: 10.2147/CMAR.S232383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng XX, Yu RY, Wu X, et al. Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J Immunother Cancer. 2020;8(1) doi: 10.1136/jitc-2019-000376. pii: e000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggero D, Montanaro L, Ma L, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–86. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 12.Coleman LJ, Peter MB, Teall TJ, et al. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer. 2009;100:1393–99. doi: 10.1038/sj.bjc.6605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang XM, Yu XN, Huang RZ, et al. Prognostic significance of eukaryotic initiation factor 4E in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2016;142:2309–17. doi: 10.1007/s00432-016-2232-2. [DOI] [PubMed] [Google Scholar]
- 14.Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012;(59):e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 16.Attar-Schneider O, Drucker L, Gottfried M. Migration and epithelial-to-mesenchymal transition of lung cancer can be targeted via translation initiation factors eIF4E and eIF4GI. Lab Invest. 2016;96:1004–15. doi: 10.1038/labinvest.2016.77. [DOI] [PubMed] [Google Scholar]
- 17.Attar-Schneider O, Zismanov V, Drucker L, Gottfried M. Secretome of human bone marrow mesenchymal stem cells: An emerging player in lung cancer progression and mechanisms of translation initiation. Tumour Biol. 2016;37:4755–65. doi: 10.1007/s13277-015-4304-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang D, Ma J, Ji X, et al. miR-141 regulation of EIF4E expression affects docetaxel chemoresistance of non-small cell lung cancer. Oncol Rep. 2017;37:608–16. doi: 10.3892/or.2016.5214. [DOI] [PubMed] [Google Scholar]
- 19.Jia Y, Chen Y, Wang Q, et al. Exosome: Emerging biomarker in breast cancer. Oncotarget. 2017;8:41717–33. doi: 10.18632/oncotarget.16684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dini L, Tacconi S, Carata E, et al. Microvesicles and exosomes in metabolic diseases and inflammation. Cytokine Growth Factor Rev. 2020;51:27–39. doi: 10.1016/j.cytogfr.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Ding Z, Luo Q, Xu W. Cancer cell-derived exosomes promote cell proliferation and inhibit cell apoptosis of both normal lung fibroblasts and non-small cell lung cancer cell through delivering alpha-smooth muscle actin. Am J Transl Res. 2019;11:1711–23. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Li M, Hu C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem Biophys Res Commun. 2018;507:457–64. doi: 10.1016/j.bbrc.2018.11.061. [DOI] [PubMed] [Google Scholar]
- 23.Rana S, Zoller M. Exosome target cell selection and the importance of exosomal tetraspanins: A hypothesis. Biochem Soc Trans. 2011;39:559–62. doi: 10.1042/BST0390559. [DOI] [PubMed] [Google Scholar]
- 24.Castellanos-Rizaldos E, Grimm DG, Tadigotla V, et al. Exosome-based detection of EGFR T790M in plasma from non-small cell lung cancer patients. Clin Cancer Res. 2018;24:2944–50. doi: 10.1158/1078-0432.CCR-17-3369. [DOI] [PubMed] [Google Scholar]
- 25.Li C, Lv Y, Shao C, et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721–27. doi: 10.1002/jcp.28678. [DOI] [PubMed] [Google Scholar]
- 26.Ruiz-Martinez M, Navarro A, Marrades RM, et al. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget. 2016;7:51515–24. doi: 10.18632/oncotarget.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018;29:700–6. doi: 10.1093/annonc/mdx765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hydbring P, De Petris L, Zhang Y, et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer. 2018;124:45–52. doi: 10.1016/j.lungcan.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Cao Y, Wei M, Li B, et al. Functional role of eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) in NSCLC. Oncotarget. 2016;7:24242–51. doi: 10.18632/oncotarget.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaoyan X, Juanjuan Y, Yalan T, et al. Downregulation of EIF4A2 in non-small-cell lung cancer associates with poor prognosis. Clin Lung Cancer. 2013;14:658–65. doi: 10.1016/j.cllc.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Khoury T, Alrawi S, Ramnath N, et al. Eukaryotic initiation factor-4E and cyclin D1 expression associated with patient survival in lung cancer. Clin Lung Cancer. 2009;10:58–66. doi: 10.3816/CLC.2009.n.009. [DOI] [PubMed] [Google Scholar]
- 32.Yoshizawa A, Fukuoka J, Shimizu S, et al. Overexpression of phospho-eIF4E is associated with survival through AKT pathway in non-small cell lung cancer. Clin Cancer Res. 2010;16:240–48. doi: 10.1158/1078-0432.CCR-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan S, Ramalingam SS, Kauh J, et al. Phosphorylated eukaryotic translation initiation factor 4 (eIF4E) is elevated in human cancer tissues. Cancer Biol Ther. 2009;8:1463–69. doi: 10.4161/cbt.8.15.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The representing amplification plot (A) and melting curve (B) in quantitative analysis of exosome derived eIF4E.