Abstract
Using cryo-electron microscopy and molecular characterization, David Sabatini and colleagues provide crucial new insights that validate and expand their model of how amino acids are sensed and signal at the lysosome to activate mechanistic target of rapamycin complex 1 (mTORC1) and cell growth-regulating processes. This work also reveals new therapeutic opportunities for mTORC1-driven diseases.
In response to changes in extra- and intra-cellular nutrient status, living organisms balance their anabolic and catabolic processes to maintain cellular homeostasis. The evolutionarily conserved Ser/Thr kinase, mTORC1, is a master regulator of this process and is regulated by a variety of nutrients including amino acids. Once activated, mTORC1 phosphorylates its downstream substrates to promote macromolecule synthesis while suppressing catabolic processes such as autophagy. mTORC1 is composed of three core components, mTOR, Raptor, and mLST8, where mLST8 binds to the mTOR catalytic site and Raptor modulates the subcellular localization of mTORC1 as well as the recruitment of downstream substrates [1]. The molecular details of this process are becoming clearer as a result of structural studies.
The lysosome is a well-established membrane-enclosed organelle that is specialized for cellular catabolism. Despite occupying a small percentage of cell volume and lipid membrane surface, there now clear evidence that it has a crucia function as a platform for regulating metabolic signaling, nutrient sensing, and quality control [2]. Specifically, lysosomes a key role in mTORC1 activation by families of Ras-like GTPases, the Rags and Rhebs, that are localized to the lysosomal surface [3]. As part of the activation process, the Rag heterodimer is recruited to the lysosomal surface-associated and nutrient-activated Ragulator complex, where RagA or RagB is GTP-loaded its associated partner, RagC or RagD, GOP-loaded via guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) such GATOR1, FLCN-FNIP, SLC38A9, and Ragulator [4]. The nucleotide state of Rag is also tightly regulated by interactions within Rag heterodimers. lntersubunit crosstalk between Rag GTPase domains, as a result of obligate heterodimerization, allows mTORC1 signaling to respond rapidly to changes in nutrient levels, and Sabatini and coworkers previously showed that GTP binding to one subunit induces GTP hydrolysis in the other subunit [5]. The activated Rags then bind to the Raptor component of mTORC1, bringing it into the proximity of lysosome-associated Rheb for activation. Maximal stimulation of mTORC1 phosphotransferase activity therefore requires not only activation of the Rag complex by amino acids and glucose, but also Rheb activation by growth factors, energy sufficiency, and/or oxygen availability [3]. How these inputs regulate mTORC1 signaling at a molecular level is becoming clearer, as highlighted in the study of Rogala et at. [6] that demonstrates how mTORC1 docks onto the lysosomal surface in response to nutrients via complex formation with Rag-Ragulator (Figure 1).
Figure 1.
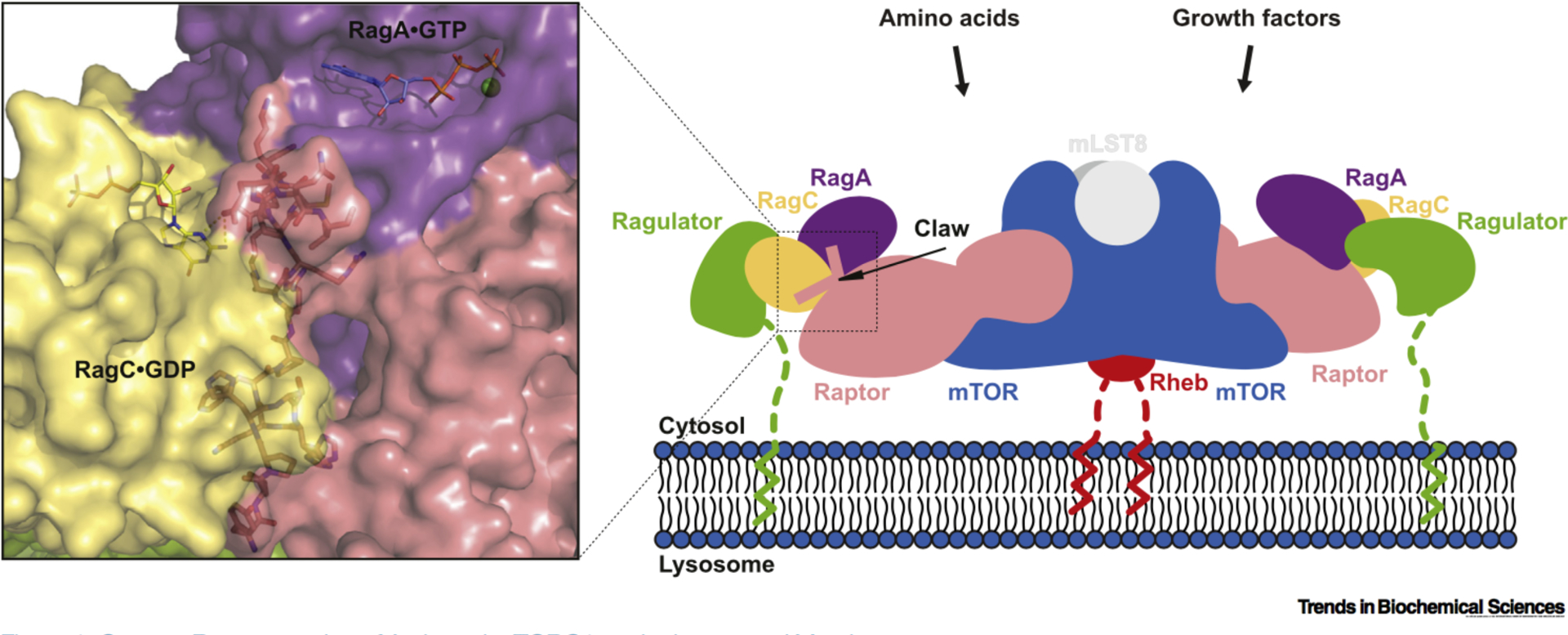
Cartoon Representation of Activated m TORC1 on the Lysosomal Membrane. mTOR kinase functions at the center of the cellular response to nutrient and growth factor availability, and controls metabolism, protein synthesis, and cell growth accordingly. Together with Raptor and mLST8, it forms the evolutionarily conserved signaling complex, mTORC1. Amino acids promote Rag GTPase–Ragulator-mediated translocation of mTORC1 to the lysosomal membrane via the myristoylated and palmitoylated 45 amino acid tail of Ragulator, enabling mTORC1 to be activated by growth factor-induced Rheb which is also localized to the membrane by a C-terminal famesyl group. The cryo-electron microscopy structure of the Raptor–Rag–Ragulator complex shows that Raptor selectively binds to the heterodimer of GTP-bound RagA and GOP-bound RagC via its own nucleotide detector, the ‘Raptor claw’, a triangular structure that threads between the GTPase domains of the Rag heterodimer (PDB 6U62). Abbreviations: mTORC1, mechanistic target of rapamycin complex 1.
Rogala et at. determined the structure of the Raptor-Rag-Ragulator supercomplex by cryo-electron microscopy, which revealed the regulatory interface between Raptor and RagA/C in molecular detail, and explains how mTORC1 discriminates between different Rag nucleotide states for translocation to the lysosome via a nutrient-sensitive interaction with Raptor. In their Raptor-Rag-Ragulator structure, Rag GTPases interact with the central region of Raptor (α-solenoid), and RagA interacts with Raptor much more extensively than does RagC. Rag binding to mTORC1 does not change its conformation, unlike the allosteric activator Rheb [7,8]. Three helices from Raptor (α24, α26, α29) form hydrogen bonds and salt bridges with the switch machinery of RagA, which agrees with the binding sites identified by hydrogen/deuterium exchange mass spectrometry (HDX-MS) analysis [8]. Mutations of Raptor residues mediating these contacts greatly reduce binding to RagA/C without affecting mTOR binding, and based on other RagA-related small GTPases, GDP binding to RagA likely causes a rearrangement of its switch machinery, thus disrupting interactions with the three Raptor helices.
In attempts to reconstitute the Raptor–Rag-Ragulator supercomplex, Rogala et at. used the RagA•GTP–RagC•GDP heterodimer obtained by taking advantage of the slow intrinsic GTPase rate of wild-type RagA and mutations (S75N, T90N) that stabilize the GOP-bound state of RagC [5]. The structure of the complex revealed a ‘Raptor claw’, a key structure corresponding to residues 916–937 of Raptor that are conserved in vertebrates and are involved in interactions with the RagA/C heterodimer. The Raptor claw plays a crucial role in the nucleotide detection mechanism, ensuring that GDP is bound to RagC (Figure 1). This triangle-shaped 22 amino acid loop, the Raptor claw, enters the space between Rag GTPases near the switch machinery of RagA and turns through 130° to exit near the nucleotide binding site of RagC. Superimposition of the available GTP-bound RagC structure (PDB 3LLU) revealed that GTP binding not only interferes with the interaction between the tip of the Raptor claw and key residues in the rigidified RagC switches, but also causes a register shift in the interswitch strands of RagC, which in turn results in a large conformational shift of the entire GTPase domain of RagC that is repositioned to avoid clashing with its the C-terminal roadblock domain (CRD).
Rogala and colleagues utilize their new Raptor–Rag–Ragulator structure [6], superimposed on the recent Rheb-bound mTORC1 structure [7], both of which contain Raptor, to propose a structural model of how the amino acid-regulated and lysosome-docked Rag–Ragulator complex (via its myristoylated and palmitoylated p18/lAMTOR1 subunit [9]) recruits mTORC1 to the lysosomal surface for activation by the farnesylated and membrane-associated Rheb (Figure 1). In this model, the Rag–Ragulator complex grabs each side of the dimeric mTORC1 to bring it closer to the lysosomal surface and, through the 45 amino acid tail of Ragulator, provides mTORC1 with an extensive search area for its activator Rheb. These new insights into the mTORC1 translocation mechanism expand our current understanding of nutrient sensing at a molecular level by revealing the importance of the tightly regulated interface between Raptor and Rag GTPases.
Dysfunctional regulation of mTORC1 is known to be associated with cancer, metabolic diseases such as diabetes, obesity, and aging, and with various inflammatory and neurological disorders, thus making this complex an attractive pharmacological target [10]. The mTORC1 allosteric inhibitors, rapamycin and rapalogs, have been extensively investigated as therapeutics. Because of their limited efficacy, however, ATP-competitive mTOR kinase inhibitors and the third-generation mTOR inhibitor ‘Rapalink’ have also been developed and characterized. These inhibitors all display limited efficacy as a result of a variety of on- and off-target effects such as suppression of various negative-feedback loops, cytotoxicity, acquired resistance, and others [10]. The current findings now lay important groundwork for the development of novel therapeutic strategies aimed at more specific inhibition of mTORC1 signaling. Guided by the discoveries of Rogala et al., the development of small molecules that target the recruitment of mTORC1 to the lysosomal surface will be an important alternative therapeutic approach in the future for the treatment of mTORC1-driven diseases.
References
- 1.Saxton AA and Sabatini DM (2017) mTOR sgnaling in growth, metabolism, and disease. Cell 169, 361–371 [DOI] [PubMed] [Google Scholar]
- 2.Lawrence RE and Zoncu R (2019) The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Bioi 21, 133–142 [DOI] [PubMed] [Google Scholar]
- 3.Sancak Y et al. (2010) Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condon KJ and Sabatini DM (2019) Nutrient regulation of mTORC1 at a glance. J. Cell Sci 132, jcs222570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen K et al. (2017) lntersubunit crosstalk in the Rag GTPase hetemdimer enables mTORC1 to respond rapidly to amino acid availability. Mol. Cell 68, 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogala KB et al. (2019) Structural basis for the docking of mTORC1 on the lysosomaj surface. Science 366, 468–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H et al. (2017) Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anandapadamanaban M et al. (2019) Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nada S et al. (2009) The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 28, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J and Guan KL (2019) mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Bioi 21, 63–71 [DOI] [PubMed] [Google Scholar]


