Abstract
Methylation of the N6 position of adenosine (m6A) is a widespread RNA modification that is critical for various physiological and pathological processes. Although this modification was also found in the RNA of several viruses almost 40 years ago, its biological functions during viral infection have been elucidated recently. Here, we investigated the effects of viral and host RNA methylation during porcine epidemic diarrhea virus (PEDV) infection. The results demonstrated that the m6A modification was abundant in the PEDV genome and the host methyltransferases METTL3 and METTL14 and demethylase FTO were involved in the regulation of viral replication. The knockdown of the methyltransferases increased PEDV replication while silencing the demethylase decreased PEDV output. Moreover, the proteins of the YTHDF family regulated the PEDV replication by affecting the stability of m6A-modified viral RNA. In particular, PEDV infection could trigger an increasement of m6A in host RNA and decrease the expression of FTO. The m6A modification sites in mRNAs and target genes were also altered during PEDV infection. Additionally, part of the host responses to PEDV infection was controlled by m6A modification, which could be reversed by the expression of FTO. Taken together, our results identified the role of m6A modification in PEDV replication and interactions with the host.
Keywords: N6-methyladenosine, Porcine epidemic diarrhea virus, Coronavirus, Anti-Viral mechanism, Posttranscriptional modification
Abbreviations
- m6A
N6-methyladenosine
- PEDV
porcine epidemic diarrhea virus
- METTL3
methyltransferase-like protein 3
- METTL14
methyltransferase-like protein 14
- WTAP
Wilms Tumor 1 associated protein
- RBM15
RNA binding motif protein 15
- ZC3H13
zinc finger CCCH domain-containing protein 13
- FTO
fat mass and obesity-associated protein
- ALKBH5
AlkB homolog 5
- YTHDF
YTH N6-methyladenosine RNA binding protein
- hm6A
N6-hydroxymethyladenosine
- f6A
N6-formyladenosine
- HIV-1
human immunodeficiency virus 1
- PED
Porcine epidemic diarrhea
- UTR
untranslated region
- ORFs
open reading frames
- CPE
cytopathic effect
- TEM
transmission electron microscope
- ncRNA
noncoding RNA
- cds
coding sequence
- GO
Gene Ontology
- BP
biological process
- MF
Molecule function
1. Introduction
Epigenetic regulation refers to heritable changes in gene regulation that do not result from changes in the DNA/RNA sequence. Epigenetic modifications include DNA methylation, histone modification, genomic imprinting, maternal effects, RNA methylation, etc. Among these modifications, N6-methyladenosine (m6A) methylation is characterized by its dynamic regulation and abundance in mRNA. M6A was first discovered in the 1970s in a purified poly (A) RNA fraction (Desrosiers et al., 1974). Later, it was found to play critical roles in various processes of mRNA metabolism, including transcription, decay, degeneration, and translation (Zhao et al., 2017). In 2010, antibody-based immune precipitation followed by high-throughput sequencing combined with the identification of the m6A complex facilitated the studies of m6A modification (Dominissini et al., 2012; Jia et al., 2011; Zheng et al., 2013). Since then, novel biological functions of m6A have been widely explored.
M6A modification is added by methyltransferases (writers) or removed by demethylases (erasers) respectively, and exerts its function by m6A binding proteins (readers). In mammalian cells, methyltransferases are a complex mainly consisting of methyltransferase-like protein 3 (METTL3) and METTL14 (Bokar et al., 1994, 1997; Liu et al., 2014; Ping et al., 2014). Other associated proteins have also been identified, such as Wilms Tumor 1 associated protein (WTAP) (Ping et al., 2014), Virilizer (KIAA1429) (Schwartz et al., 2014; Horiuchi et al., 2013), RNA binding motif protein 15 (RBM15) (Patil et al., 2016), and zinc finger CCCH domain-containing protein 13 (ZC3H13) (Knuckles et al., 2018; Wen et al., 2018). However, The METTL3, a catalytic subunit, and METTL14, an RNA-binding platform, are recognized as key factors (Liu et al., 2014). Two demethylases, fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), are responsible for the removal of m6A. The m6A demethylase activity of FTO was first discovered in 2011 (Jia et al., 2011). It can oxidize m6A to Adenine (A) and generate N6-hydroxymethyladenosine (hm6A) as an intermediate product and N6-formyladenosine (f6A) as a further oxidized product (Gerken et al., 2007; Jia et al., 2008). The second demethylase, ALKBH5, was identified in 2013 (Zheng et al., 2013). However, ALKBH5 may function in a specific sequence or structural context. In addition, the function of m6A modification on target mRNAs is mediated by specific reader proteins. Members of the YTH family have been shown to specifically bind to m6A-containing precursor RNAs. The main m6A readers, YTHDF1, YTHDF2, and YTHDF3, are located in the cytoplasm. YTHDF1 has been demonstrated to interact with methylated mRNA near the stop codon and enhances its translational efficiency (Wang et al., 2015). YTHDF2 can reduce the stability of m6A modified RNA (Wang et al., 2014; Du et al., 2016). YTHDF3 has been shown to cooperate with YTHDF1 and YTHDF2 (Li et al., 2017). The combination of methyltransferases, demethylases and m6A-binding proteins contributed to the dynamic regulation of this modification. Its flexibility highly correlates with the metabolism of RNA and plays critical roles in transcription, decay, degeneration and translation. Almost all biological processes related to RNAs have been identified to be affected by m6A modification.
The replication of all the viruses requires RNA except the Prions. The involvement of m6A in viral infection was discovered in the 1970s. M6A was found to be present on viral RNAs, for instance, the influenza virus and even several DNA viruses (Krug et al., 1976; Sommer et al., 1976; Moss et al., 1977; Furuichi et al., 1975). However, the specific roles of m6A in viral replication remain unclear until 2016, when transcriptome-wide profiling of m6A was made possible through antibody-based immunoprecipitation followed by high-throughput sequencing. The replication of human immunodeficiency virus 1 (HIV-1), RNA viruses within the family Flaviviridae, and influenza virus were first reported to be affected by m6A (Tirumuru et al., 2016; Kennedy et al., 2016; Gokhale et al., 2016; Lichinchi et al., 2016a, 2016b). However, there is much still unknown regarding the functions of m6A during virus infection, e.g. whether m6A is present in the RNA of other viruses, and the dynamic m6A modification of host RNA during viral infection.
Porcine epidemic diarrhea (PED) is a devastating enteric disease in pigs that can cause mortality of up to 100% in piglets younger than one week old. Since its re-emergence in 2010, this disease has rapidly spread worldwide and caused substantial economic losses to the global swine industry (Jung and Saif, 2015; Davies, 2015; Wang et al., 2016). Porcine epidemic diarrhea virus (PEDV), the causative agent of PED, is an RNA virus belonging to the Alphacornavirus genus within the Coronaviridae family. The genome of PEDV is approximately 28 kb in size with a 5′ untranslated region (UTR), a 3’ UTR, and at least seven open reading frames (ORFs) (Kocherhans et al., 2001). PEDV has been recognized as one of the largest RNA viruses and has a complicated genomic structure. Therefore, we attempted to investigate whether m6A modification exists within the PEDV genome and its regulation of viral replication.
In this study, the m6A ratio of the PEDV genome was detected by m6A RNA methylation quantification. Antibody-based immune precipitation followed by high-throughput sequencing was used to map the specific m6A peaks within the PEDV genome. In addition, the biological functions of key factors related to m6A methylation on PEDV replication were elucidated. Finally, the dynamic m6A modification of host RNA in PEDV infected cells was also analyzed. We believe our studies will facilitate the understanding of m6A modification in viral RNA and its regulation in host RNA during viral infection.
2. Materials and methods
2.1. Cell culture and virus infection
Vero cells and LLC-PK1 cells were cultured in DMEM (Sigma, Germany) and MEM-199 (Sigma) supplemented with 10% fetal bovine serum (Gemini, USA) and 100 units/ml penicillin (Sigma) in an incubator (5% CO2, at 37 °C). The PEDV LJX01/GS/2014 strain was isolated and stored in our laboratory.
2.2. PEDV titration
Vero cells were seeded into 96-well plates (approximately 5 × 104 cells per well) for PEDV titration with the collected supernatant. The collected supernatant was first serially diluted 10-fold and then 100 μl of each dilution was inoculated into four wells of the 96-well plate. The plates were incubated at 37 °C in 5% CO2 until 2 days post infection. Titers were calculated from the supernatant dilution that caused cytopathologic effects in half of the cultures (TCID50/mL) based on the Reed-Muench method.
2.3. PEDV purification
Cells were seeded into 10-cm dishes and inoculated with PEDV at a MOI = 0.1. The supernatant was collected 36 h post-infection (hpi) and then centrifuged at 2000 × g for 15 min to remove cellular debris. After centrifugation at 100000 × g for 2 h at 4 °C, the pellets were resuspended in PBS and purified by sucrose density gradient centrifugation. The sucrose cushion was 20%, 40%, and 50%. PEDV particles were between 40% and 50% sucrose cushions.
2.4. Electron microscopy
Purified PEDV particles were diluted with PBS and spotted onto Formvar-coated grids. Ten minutes later, the PBS was removed. The grids were directly negatively stained with phosphotungstic acid for 10 min at room temperature. A Hitachi electron microscope (HT7700) was used at 80 kV for viral particle observation.
2.5. M6A-seq
High-throughput sequencing of PEDV methylation was carried out by m6A-seq following a previously described protocol (Dominissini et al., 2013). In brief, rRNA was removed from the total RNA by the RiboMinus Eukaryote System v2 (Thermo, USA). Then, the RNA was fragmented using Ambion RNA Fragmentation Reagents (Thermo, USA) and mixed with 25 μg of affinity purified anti-m6A polyclonal antibody (Synaptic Systems, Germany) at 4 °C for 2 h. Sequencing libraries were prepared with eluted RNA, as well as input RNA, using the TruSeq RNA sequencing (RNA-seq) kit (Illumina, USA). Sequencing was carried out on Illumina HiSeq 2000 according to the manufacturer's instructions and was performed by Novel Bioinformatics Company.
2.6. Quantification of RNA m6A level
For the quantification of the host RNA m6A levels, the cells were harvested at the indicated time points and prepared for RNA extraction. The genomic RNA of PEDV was extracted from purified viral particles by the RNeasy Mini Kit (Qiagen, Germany). The EpiQuick m6A RNA Methylation Quantification Kit (EpiGentek, USA) was used in the following quantification. Briefly, RNA was bound to strip wells using an RNA high-binding solution. Specific capture N6-methyladenosine antibody and detection antibody were then incubated with bound RNAs. The detection signal was enhanced and detected at a wavelength of 450 nm. The amount of m6A was proportional to the OD intensity measured. Three replicate samples were used to ensure that the generated signal was validated. Finally, the percentage of m6A was calculated according to the formula provided.
2.7. RNA interference
For the knockdown of genes involved in the process of methylation, siRNAs (200 nM) were transfected into Vero cells or LLC-PK1 cells using X-tremeGENE siRNA Transfection reagent (Roche, Switzerland) according to the manufacturer's instructions. Cells were harvested for western blot analysis. All siRNAs were synthesized by RiboBio Company. The target sequences of the siRNAs are listed in Table 1 .
Table 1.
The target sequences of siRNAs.
| Gene name | Species | Target Sequence |
|---|---|---|
| FTO | Sus scrofa | GCACCTACAAGTACCTGAA |
| METTL3 | Sus scrofa | CTGAACCAACAATCTACTA |
| METTL14 | Sus scrofa | AGAGACAGATGAAGACAAA |
| YTHDF1 | Sus scrofa | CTCCGCCCATAAAGCATAA |
| YTHDF2 | Sus scrofa | CAAGGAAACAAAGTGCAAA |
| YTHDF3 | Sus scrofa | GGGAGAGAAATAGAAACAA |
| FTO | Chlorocebus sabaeus | TGACGATGTGGACAATGGT |
| METTL3 | Chlorocebus sabaeus | GCAGTTCCTGAATTAGCTA |
| METTL14 | Chlorocebus sabaeus | CCTCCTCCCAAATCTAAAT |
| YTHDF1 | Chlorocebus sabaeus | CTACCTTACTGGACAGTCA |
| YTHDF2 | Chlorocebus sabaeus | GCCCAATAATGCGTATACT |
| YTHDF3 | Chlorocebus sabaeus | GGTCAACATGGATTTAACT |
2.8. Immunoprecipitation and RNA
Cells were transfected with plasmids carrying MYC-YTHDF1-3 or an empty vector (EV). The cells were then infected with PEDV at MOI = 0.1 24 h post-transfection. When CPE appeared, the cells were UV-cross-linked and lysed in NP40 buffer. The supernatant of the lysate was then mixed with anti-Myc antibodies and protein G beads (Genscript, China) and rotated for 2 h at 4 °C. After incubation, the beads were washed 3 times with NP40 buffer. Coimmunoprecipitated RNA was isolated from the immunoprecipitates using TRIzol (Takara, Japan) and RNeasy columns (Qiagen) with an on-column DNase I treatment (Sigma, USA) and eluted with RNase-free water. Equal volumes of RNA were used as a template for first-strand cDNA synthesis, according to the manufacturer's instructions.
2.9. RNA stability assay
Cells were transfected with siRNAs (200 nM) and/or treated with actinomycin D (4 μg/ml) (Solarbio, China) in DMSO. Cells were then infected with PEDV at a MOI = 0.1. The total RNA was collected at the indicated time points and reverse-transcribed into cDNA. The mRNA stability was measured by analysis of the relative expression at 0, 3 h, and 6 h after PEDV infection. The primers for target gene detection were as follows: PEDV-N forward: 5′-GATACTTTGGCCTCTTGTGT-3′, reverse: 5′- CACAACCGAATGCTATTGACG-3’; porcine-IL-8 forward: 5′-CTGAGAGTGATTGAGAGT-3′, reverse: 5′-TTACTGCTGTTGTTGTTG-3’; African green monkey-IL-8 forward: 5′- TGGACCACACTGCGTCAATA-3′, reverse: 5′-ACAACCCTAGACACCCATGGTA-3’.
2.10. Construction of pseudovirus and stable cell lines
To construct stable cell lines with Mettl3/Mettl14 knockdown for PEDV production, duplexes of synthesized oligonucleotides containing target sequences were inserted into the lentivirus vector pLVX-shRNA2. The plasmids (21 μg) were then transfected into HEK293T cells with the packaging plasmids psPAX2 (14 μg) and pMD 2.0G (7 μg). At 48 h post transfection, the culture supernatant was concentrated by Amicon Ultra-15 centrifugal filters. The concentrated pseudovirus was transduced into LLC-PK1 cells for further studies. For the construction of LLC-PK1 cells with FTO over-expressing, the FTO gene was inserted into the pFUGW vector using SfiI to generate the pFUGW-FTO plasmid. The packing and transduction procedures were the same as mentioned above. The target sequences and primers for lentivirus construction were listed in Table 2 .
Table 2.
The target sequences and primers for lentivirus construction of porcine genes.
| Gene name | Sequence | Note |
|---|---|---|
| METTL3 | GCATTTGGATCTACGGAATCC | Target |
| METTL14 | GCATTGGTGCCGTGTTAAATA | Target |
| FTO | F: ACAGGCCATTACGGCCATGAACGAGGAGGCCATCTGCA R: TACGGCCGAGGCGGCCCTAAAATAAATTTAAAGATGGT |
Primer |
2.11. Real-time PCR analysis
To assess the number of PEDV genomes binding to YTHDF proteins, real-time PCR was used for the quantitation assay. Real-time PCR was performed in a Bio-Rad CFX96 system with TransStart Probe qPCR SuperMix (Transgen, China). Briefly, the reactions were incubated at 94 °C for 30 s, followed by 40 cycles at 94 °C for 5 s and 60 °C for 30 s. The primers (forward: 5′-GATACTTTGGCCTCTTGTGT-3′, reverse: 5′- CACAACCGAATGCTATTGACG-3′) and probe (5′-FAM-TTCAGCATCCTTATGGCTTGCATC-TAMRA-3′) were derived from our previous study (Huang et al., 2019). For the detection of host gene expression levels, real-time qPCR was performed with a unique aptamer qPCR SYBR green master mix (Novogen, China). Briefly, the reactions were incubated at 95 °C for 5 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 30 s. All reactions were run in triplicate. The probe and primer sets are listed in Table 3 . The ΔΔCt method was used to measure the expression level of target genes.
Table 3.
The primers used for m6A real-time PCR analysis.
| Gene Name | Gene Discription | Sequence |
|---|---|---|
| CDC42 | cell division cycle 42 | F: AAAAGGGGAAAGCCGATGCT R: TTCAGCTTTCTGTGCCTGGT |
| CTTN | cortactin | F: AGACCGACAGGACAAGTGTG R: CCTGCCTCCGAAACCAGTCTT |
| MYH | A/G-specific adenine DNA glycosylase | F: CAAGCTGGCCAAGGAGAAGA R: TTGGCGAGGCTCTTGGATTT |
| NRAS | NRAS proto-oncogene | F: GCGCAAGTCAGAGAAGAGGT R: AGGATCCCTCTCAGTCGCAT |
| YBX3 | Y-box binding protein 3 | F: CCCCTATAACTATCGGCGGC R: AGCAGGGTTGTCAGATGGTG |
| CDK4 | cyclin dependent kinase 4 | F: GGCCCCGAGATGTGTCTCTA R: CCATCTCAGGTACCACCGAC |
| CDKN1A | cyclin dependent kinase inhibitor 1A | F: CACAGGCACCATGTCAGAGT R: TCGAAGTTCCATCGCTCTCG |
| GADD45B | growth arrest and DNA damage inducible beta | F: GGGAGCAGGGGCTGAATTTG R: CTGTAAGCCTCCCATCTCTCTT |
| LOC100737977 | tumor necrosis factor receptor superfamily member 10B-like | F: GAACATCTACTGGAACCGGCA R: CTCAAGCATTCTGTGGGGTCT |
| MDM2 | MDM2 proto-oncogene | F: TCCAGCACATCTGTGAGTGAAA R: TTTCTCTTCCTGTAGCTCCTGC |
| PMAIP1 | phorbol-12-myristate-13-acetate-induced protein 1 | F: CCTCTACTGTTGGGGCCTCT R: AAACAACTGGGGCAAACGAC |
| RRM2 | ribonucleotide reductase regulatory subunit M2 | F: CCTTCGGAGCAGAGAGTGAAAG R: TGTCTGCCACGAACTCGAT |
| ATF2 | activating transcription factor 2 | F: CTCCTGGGGTGGTTGGTAAA R: TGCTGCCAGGAATCCCTTAAC |
| DAXX | death domain associated protein | F: GTTTCTGAGGGGGTGTCGAG R: ACGTCGACATCCGGTCTTTC |
| DUSP6 | dual specificity phosphatase 6 | F: AGTGCAACAGACTCCGATGG R: TGGGGGTGACGTTCAAGATG |
| HSP70 | heat shock protein 70 | F: TGATCAACGACGGGGACAAG R: TCTTGGTCAGCACCATCGAC |
| DUSP10 | dual specificity phosphatase 10 | F: AAGAGCCACATCCAAGGAGC R: GACAGTGATCTTGCCCTGCT |
| MAP2K5 | mitogen-activated protein kinase kinase 5 | F: CTTGGAAGAATTGCAGTGGCG R: AGCTTGACCTGTCCCCTTGT |
| MAPK8IP3 | mitogen-activated protein kinase 8 interacting protein 3 | F: CGAGTTCGAAGATGCCTTGG R: GGGCGTTGTACTCCTTCTTCA |
| MAP3K4 | mitogen-activated protein kinase kinase kinase 4 | F: ATGGGCACTGTTTTGGGCAT R: GTCTTCAGGCTCGTTGCCTT |
| MYC | MYC proto-oncogene, bHLH transcription factor | F: AGAACTGCTTGCTGGCCATT R: CAAGACCCAGCCAAGGTTGT |
| PAK | p21-activated kinase | F: GGAACACCAGCACTGCATAC R: TGGTCGTGGGGCAATAACAG |
| RAC | AKT serine/threonine kinase 1 | F: TCGCTGTCTGGATGCGAAAC R: CAGATCTGCCTGAGTGGGAT |
| SIX4 | SIX homeobox 4 | F: ATTTCCAGGGCTGATACCCAG R: CACTACAGAGCCTCCGTCC |
| SRF | serum response factor | F: CCCCGCTCAGACCCTACCA R: CTGGTAGGTGAGGTCTGTCTCT |
| TAB2 | TAK1-associated binding protein 2 | F: CTCTGCCACATACCTCAGCCC R: AAGACTGTGAGCTACCAGACGA |
| TGFB3 | transforming growth factor beta 3 | F: GCTGGCTCTGAGAATCACTGT R: TATCCCAAATCCCATGGCCC |
| TOK2 | TOK2 potassium channel | F: TTCCTTGTGTGCAGACCCCT R: GGACATGCTTGGCACTGAAT |
2.12. Statistical analysis
The differences between matched groups were examined for statistical significance using Student's t-test. An unadjusted P value of less than 0.05 was considered to be significant, and a P value of less than 0.01 was considered to be highly significant. NS means not significant.
3. Results
3.1. The PEDV genome contains m6A modifications
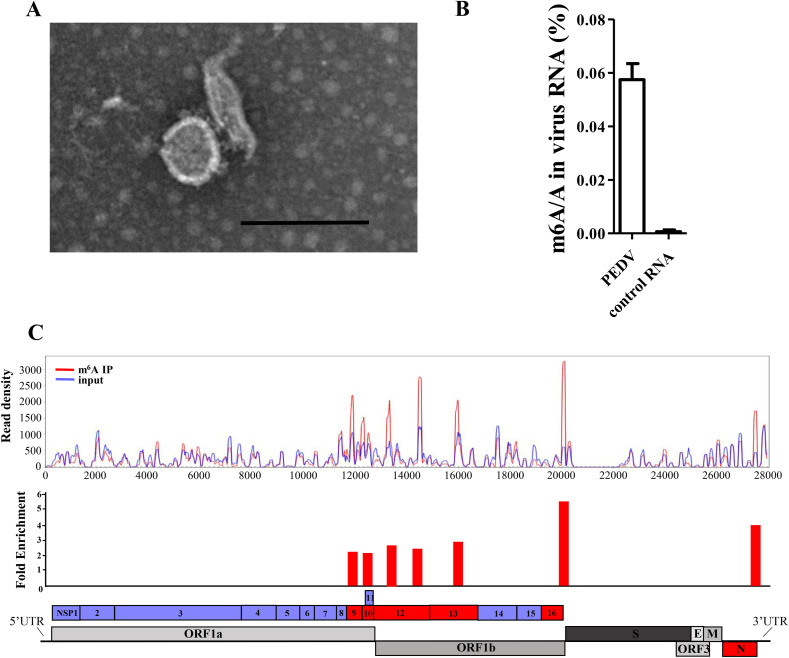
Although m6A was proven to be present in the RNA of several viruses, there have been no reports about its existence in the genomic RNA of PEDV. To investigate the presence of m6A in PEDV RNA, purified PEDV particles were prepared for viral RNA extraction. To do so, the LLC-PK1 cells were infected with the PEDV LJX01/GS/2014 strain. When a significant cytopathic effect (CPE) was observed, the supernatant was collected and then subjected to sucrose density gradient centrifugation for PEDV purification. The purified viral particles were observed with a transmission electron microscope (TEM). The results showed that the collected particles exhibited classical morphology of coronavirus and were of high purity (Fig. 1 A). After the extraction of viral RNA, we first quantified the m6A level of the PEDV RNA genome with an m6A RNA Methylation Quantification Kit. The result demonstrated that the m6A ratio of PEDV RNA was approximately 0.05–0.06% of the total adenosines (Fig. 1B).
Fig. 1.
The PEDV genome was modified by m6A methylation.
The PEDV genome was extracted from purified viral particles and used for m6A quantification and m6A-seq. A) Purified PEDV particles were observed by transmissible electron microscope analysis. Scale bar, 200 nm. B) The m6A level of PEDV RNA was quantified by ELISA. The synthetic scrambled RNA was employed as a negative control. C) Map of m6A reads on the PEDV genome. The red line represents m6A-seq, and the blue line indicates input RNA-seq. The red bars of the PEDV genome indicate m6A peaks identified in duplicate experiments by MeRIPPeR analysis (p < 0.05). The blue bars indicate the fragment of the PEDV genome without m6A modification.
To further map the m6A peaks within PEDV RNA, we performed immunoprecipitation with viral RNA. Viral RNA was fragmented into approximately 200 bp fragments and then conserved as input control or immunoprecipitated with m6A-specific antibodies. The RNA was then eluted and analyzed by high-throughput RNA sequencing. The reads were mapped to identify the regions of the PEDV genome enriched in m6A. Seven m6A peaks were identified in PEDV genomic RNA, mainly in the genes that can be translated into nonstructural proteins (Fig. 1C). The specific information is listed in Table 4 .
Table 4.
The peaks identified in the PEDV genome.
| Peak Position | p-value | Enrichment | GENE |
|---|---|---|---|
| 11835-11913 | 1.55E-15 | 2.073908274 | NSP9 |
| 12311-12405 | 2.22E-16 | 2.000712104 | NSP10 |
| 13309-13413 | 0 | 2.544230775 | NSP12 |
| 14449-14557 | 6.66E-16 | 2.261206607 | NSP12 |
| 15921-16103 | 6.62E-07 | 2.696092108 | NSP13 |
| 20081-20125 | 0 | 5.255663421 | NSP16 |
| 27452-27551 | 0 | 3.787781048 | N |
Together, the results above confirmed that the genomic RNA of PEDV contained m6A modifications at multiple sites within the viral genome.
3.2. The m6A modification regulates PEDV replication in different cell lines
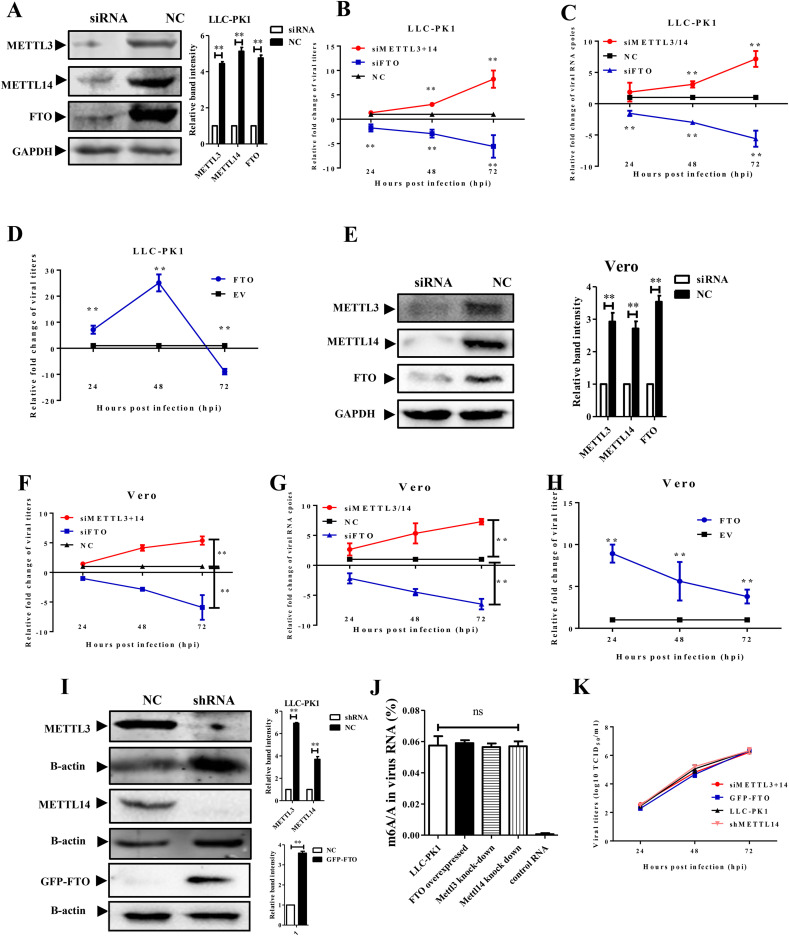
Since the PEDV genomic RNA was modified by m6A, we next attempted to determine whether this modification could regulate PEDV replication. The siRNAs against porcine FTO, METTL3, and METTL14 were synthesized and transfected into LLC-PK1 cells. At 48 h post transfection, the cells were collected for western blotting analysis. The results demonstrated that these molecules were efficient in the knockdown of the corresponding genes (Fig. 2 A). The cells were also infected with MOI = 0.01 PEDV at 24 h after transfection. The supernatant was collected at 24, 48, and 72 hpi and used for virus titration and real-time PCR analysis. We found that the METTL3 and METTL14 knockdown facilitated PEDV replication, while the FTO knockdown resulted in a significant decrease of viral titers at 24, 48, and 72 hpi. To further verify whether this effect may reduce the viral RNA replication or impact the infectivity of the virus, viral RNA copy numbers were then measured by real-time RT-qPCR analysis. The results showed that the METTL3/14 knockdown increased the PEDV RNA copy numbers at 48 and 72 hpi (Fig. 2C). The knockdown of FTO also decreased PEDV copies at all three time points. These results were consistent with the viral titers and suggested m6A modification directly reduced viral RNA production. Myc-tagged FTO gene was then overexpressed in LLC-PK1 cells to test whether it had a similar effect on PEDV growth. The viral titers of PEDV were significantly increased by the overexpressed exogenous FTO except at 72 hpi (Fig. 2D). The overexpression of FTO significantly accelerated the PEDV replication and resulted in cell apoptosis and a decrease of viral loads at 72 hpi while the viral titers of the control group remained increase at this time point. These data showed that the m6A modification inhibited PEDV replication in LLC-PK1 cells.
Fig. 2.
PEDV replication was regulated by m6A modification.
The siRNAs against porcine METTL3/METTL14/FTO were transfected into different cells and the cells were infected with PEDV at a MOI = 0.1. The supernatants were collected at different time points for viral titration. A) Western blotting analysis of LLC-PK1 cells transfected with siRNAs and scrambled siRNA as the negative control (NC) at 48 hpi. B) The relative fold change of viral titers in LLC-PK1 cells transfected with siRNAs or NC at different time points. C) The relative fold change of viral RNA copies of PEDV in LLC-PK1 cells at different time points. D) The relative fold change of viral titers in LLC-PK1 cells overexpressing FTO at different time points. E) Western blotting analysis of Vero cells transfected with siRNAs or NC at 48 hpi. F) The relative fold change of viral titers in Vero cells transfected with siRNAs or NC at different time points. G) The relative fold change of viral RNA copies of PEDV in Vero cells at different time points. H) The relative fold change of viral titers in Vero cells overexpressing FTO at different time points. I) Western blotting analysis of constructed cell lines with METTL3/METTL14 knockdown and with GFP-FTO overexpressed. J) The m6A level of PEDV RNA produced in different cell lines was quantified by ELISA. The synthetic scrambled RNA was taken as a negative control. K) The growth curve of PEDV derived from different cell lines. *P < 0.05, **P < 0.01, ns means not significant.
Vero is another cell line commonly used for PEDV isolation and propagation. This cell line was derived from the African green monkey kidney epithelium. M6A modification can be varied among different cell lines. Therefore, we also used this cell line to test the effect of m6A on PEDV replication. The siRNAs molecules against green monkey FTO, METTL3, and METTL14 were synthesized and later proved to be efficient in gene knockdown (Fig. 2E). We next infected Vero cells with PEDV and collected the supernatant at different time points. Similar to the results obtained from LLC-PK1 cells, the silence of FTO delayed PEDV growth, while knockdown of METTL3 and METTL14 accelerated PEDV replication at 24, 48, and 72 hpi (Fig. 2F). The viral RNA copy numbers also exhibited significant differences at different time points (Fig. 2G). The overexpression of exogenous FTO accelerated PEDV replication compared to that in cells transfected with an empty vector (Fig. 2H). Compared with the knockdown results, overexpression of FTO was more efficient in accelerating PEDV replication.
The results above also raised other scientific concerns. What is the m6A level of PEDV genomic RNA without the help of METTL3/14 or FTO? Does m6A modification influences the PEDV production or its infectivity? To answer these questions, different cell lines with METTL3/14 knockdown or FTO over-expression were constructed using a pseudovirus vector delivery system. We used the LLC-PK1 cells as the parent cell lines. The western blotting analysis showed the target proteins were successfully knocked-down or overexpressed (Fig. 2I). PEDV was inoculated to these cell lines and then collected for m6A quantification and titration. Although the production of viral particles differs in different cells, the ELISA result of m6A level exhibited no significant difference in their genomic RNA (Fig. 2J), suggested that there might be other proteins involving in this modification. We then titrated the viruses derived from the above cell lines. The growth curves demonstrated similar patterns between different groups, indicated that the virus m6A might influence the PEDV production but not its infectivity (Fig. 2K).
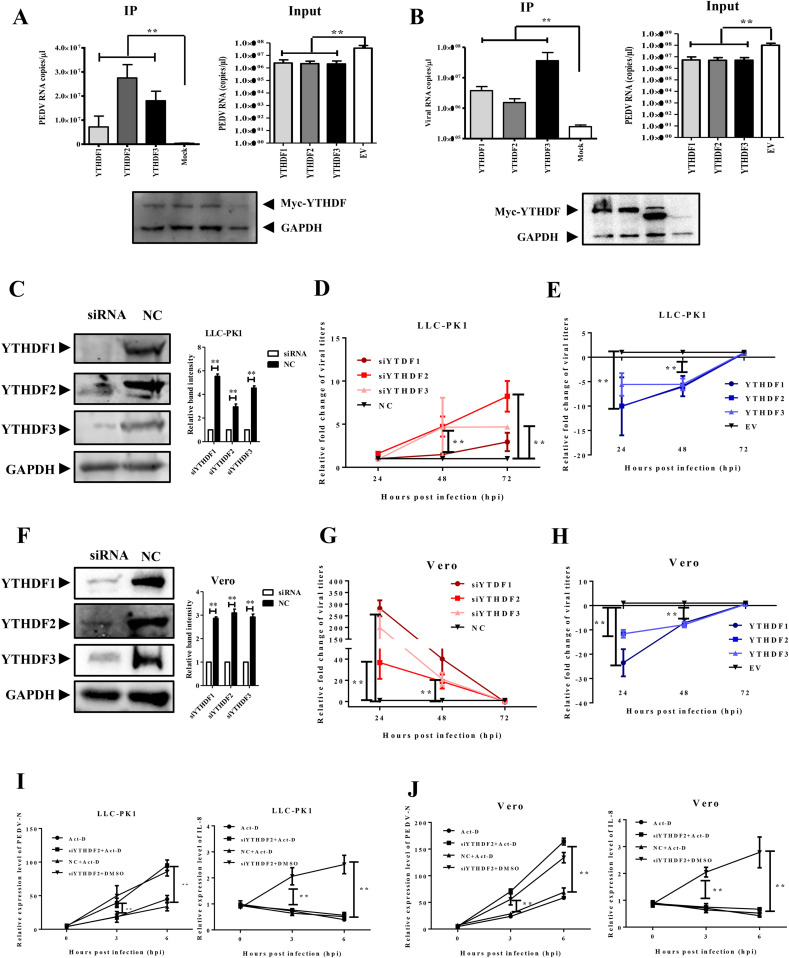
3.3. YTHDF1-3 proteins bind to PEDV RNA and negatively regulate PEDV replication
The YTHDF proteins are the m6A readers, which play a role in the RNA-binding process. Given that m6A regulates PEDV infection, we next tested whether YTHDF1-3 could bind to the PEDV genome and contribute to PEDV replication. We transfected Myc-tagged porcine YTHDF1/YTHDF2/YTHDF3 or empty vectors into LLC-PK1 cells and then infected the cells with MOI = 0.1 PEDV at 24 h post transfection. The PEDV genome binding activity to YTHDF proteins was analyzed by real-time PCR (Fig. 3 A). The results showed that PEDV replicated in all groups. The three YTHDF proteins had a much higher affinity to the PEDV genome than the control. The YTHDF2 exhibited a superior affinity than the other two proteins. YTHDF1/YTHDF2/YTHDF3 of green monkey or empty vectors were also transfected into Vero cells. The results also showed that YTHDF1-3 could bind to PEDV RNA but with different affinities compared to porcine proteins. YTHDF3 had the highest affinity in Vero cells (Fig. 3B). The siRNA molecules against both porcine and green monkey YTHDF proteins were synthesized. Western blot analysis showed that these molecules could efficiently knock down the endogenous genes (Fig. 3C and F). We then determined the virus titers in the cells. The knockdown of YTHDF proteins led to an increase in virus titers at 48 and 72 hpi in LLC-PK1 cells except YTHDF3 (Fig. 3D) and at 24 and 48 hpi in Vero cells (Fig. 3G). The depletion of YTHDF exerted the most significant upregulation of viral production at 24 hpi in Vero cells and at 72 hpi in LLC-PK1 cells. These results suggested YTHDFs might be critical for PEDV replication in Vero cells at early stage. Besides that, YTHDF2 played more important roles in LLC-PK1 cells than that in Vero cells, suggested that the function of YTHDF proteins may vary in different cell lines. In addition, the plasmids carrying YTHDF genes were transfected into both cell lines to observe their function on PEDV replication. The results illustrated that the overexpression of YTHDF proteins dramatically delayed the PEDV replication. The viral titers in YTHDF-transfected groups still increased while the EV group dropped at 72 hpi (Fig. 3E and H). Taken together, these data suggested that the YTHDF proteins could regulate PEDV infection.
Fig. 3.
YTHDF 1–3 proteins negatively regulated PEDV replication.
Cells were transfected with Myc-YTHDF1-3 and infected with PEDV at a MOI = 0.1. The cells were then lysed and used for RNA-binding analysis 24 hpi. A) Real-time PCR analysis of PEDV genome binding to YTHDF1-3 proteins in LLC-PK1 cells. EV represents the empty vector. B) Real-time PCR analysis of PEDV genome binding to YTHDF1-3 proteins in Vero cells. C) Western blotting analysis of LLC-PK1 cells transfected with siRNAs and NC at 48 hpi. D) The relative fold change of viral titers in LLC-PK1 cells transfected with siRNAs or NC at different time points. E) The relative fold change of viral titers in LLC-PK1 cells transfected with myc-YTHDFs or EV at different time points. F) Western blotting analysis of Vero cells transfected with siRNAs or NC at 48 hpi. G) The relative fold change of viral titers in Vero cells transfected with siRNAs or NC at different time points. H) The relative fold change of viral titers in Vero cells transfected with myc-YTHDFs or EV at different time points. I) Effect of YTHDF2 knockdown combined with Actinomycin D (Act-D) treatment on the expression and stability of mRNA in LLC-PK1 cells. J) Effect of YTHDF2 knockdown combined with Act-D treatment on the expression and stability of mRNA in Vero cells. *P < 0.05, **P < 0.01.
YTHDF2 reduces the stability of m6A modified RNA (Wang et al., 2014; Du et al., 2016). To test whether YTHDF2 decrease PEDV replication by the degradation of viral RNA, we employed Actinomycin D combined with YTHDF2 siRNA for further studies. Actinomycin D is an inhibitor of DNA transcription and replication but doesn't affect the function of RNA viruses (Koba and Konopa, 2005). PEDV N was selected as the target gene to verify the RNA stability because it was modified by m6A. The Vero and LLC-PK1 cells were transfected with YTHDF2 siRNA or scrambled siRNA, treated with Actinomycin D (4 mg/ml) or DMSO, and then infected with PEDV at a MOI = 0.1. At 0, 3, and 6 hpi, the total RNA was extracted for RT-qPCR analysis. The results demonstrated that YTHDF2 siRNA increased the PEDV RNA copy numbers while Actinomycin D didn't affect PEDV replication in both Vero and LLC-PK1 cells (Fig. 3I and J). We also evaluated the IL-8 expression because the IL-8 mRNA was not modified by m6A but upregulated by PEDV infection (Xu et al., 2013a, 2013b). The results illustrated that the IL-8 mRNA level was upregulated by siYTHDF2 transfection and PEDV infection but slightly downregulated by Actinomycin D treatment (Fig. 3I and J). These results suggested that YTHDF2 protein could reduce the stability of m6A modified viral RNA.
3.4. Host m6A RNA methylation is enhanced by PEDV infection
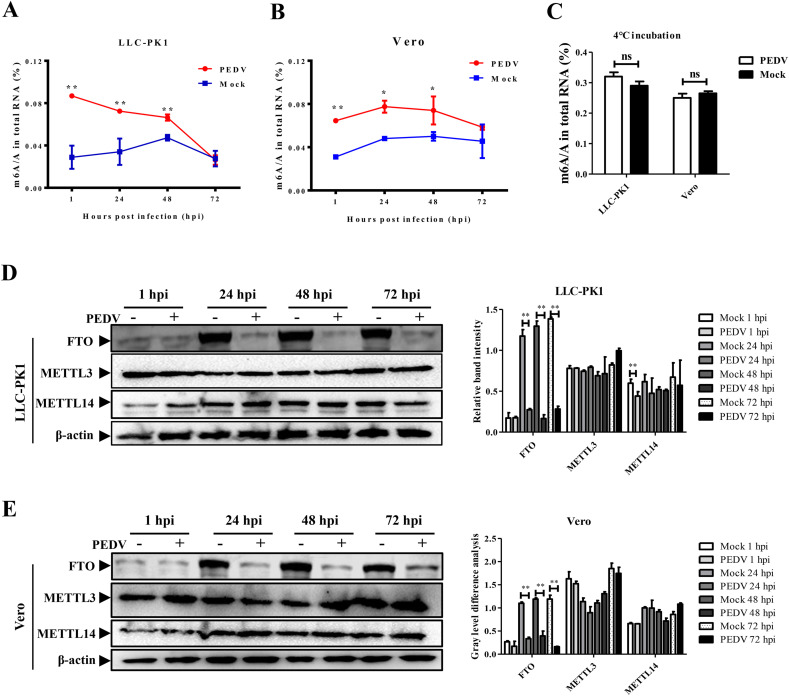
M6A modifications exist in both viral and host RNAs. Since we have demonstrated that m6A could inhibit PEDV infection, we next investigated its effect on host RNAs. Both LLC-PK1 and Vero cells were infected with PEDV and harvested for total RNA and proteins at different time points. Using the m6A RNA Methylation Quantification Kit, we first detected the m6A ratio of total RNA. It was interesting to find that PEDV infection could trigger a drastic increase in the m6A ratio in PEDV-infected cells compared to mock infection (Fig. 4 A and B). The initial modification percentages of mock cells were nearly the same in LLC-PK1 and Vero cells at approximately 0.03%. After PEDV infection, the m6A ratio immediately climbed to approximately 0.09% in LLC-PK1 cells and over 0.06% in Vero cells at 1 hpi. Although it decreased with the course of viral replication, the m6A ratio of infected cells was higher than that of mock cells. At 72 hpi in LLC-PK1 cells or 48 hpi in Vero cells, the PEDV infection had disrupted most of the cells, and the curve of the ratio flattened out thereafter. This enhancement of m6A modification occurred as early as 1 hpi, suggested that it might closely correlated with PEDV entry. To prove this, cells were incubated with or without PEDV at 4 °C and then subject to RNA extraction and ELISA analysis. The results showed that there were no significant differences between the infection group and mock group (Fig. 4C), further indicated that PEDV infection enhanced host m6A RNA methylation.
Fig. 4.
PEDV infection enhanced the m6A modification in different cells and influenced the expression of FTO
A) The m6A modification level of total RNA was enhanced by PEDV infection in LLC-PK1 cells. B) The m6A modification level of total RNA was enhanced by PEDV infection in Vero cells. C) The cells were incubated with or without PEDV at 4 °C for 1 h and then subject to m6A level quantification by ELISA. D) PEDV infection in LLC-PK1 cells influenced the expression of demethylase. The histogram shows the relative expression of proteins at different time points. E) PEDV infection in Vero cells influenced the expression of demethylase. The histogram shows the relative expression of proteins at different time points. Ns means not significant, *P < 0.05, **P < 0.01, ns means not significant.
We then detected the expression pattern of m6A methyltransferases and demethylases in LLC-PK1 cells to determine their relationship with the curve of the m6A ratio. The western blotting analysis demonstrated that the FTO expression was easily influenced by different factors. The expression level of FTO was pretty low at 1 hpi due to the suffering of trypsin treatment when cell passaging. However, it was increased in the mock groups at 24, 48 and 72 hpi while and remained low expression level at infection groups. In comparison, the expression of the methyltransferases METTL3 and METTL14was not changed at 24, 48 and 72 hpi (Fig. 4D). In Vero cells, the expression levels of FTO were lower in the infection groups compared to mock groups at 24, 48, and 72 hpi. The expression of the methyltransferases remained stable (Fig. 4E). Both host RNA quantification of and western blotting analysis showed that PEDV infection could enhance the m6A ratio in host RNA, which suggested that the host may restrict PEDV replication by m6A modification though different mechanisms.
3.5. The topology of the m6A RNA methylome during PEDV infection
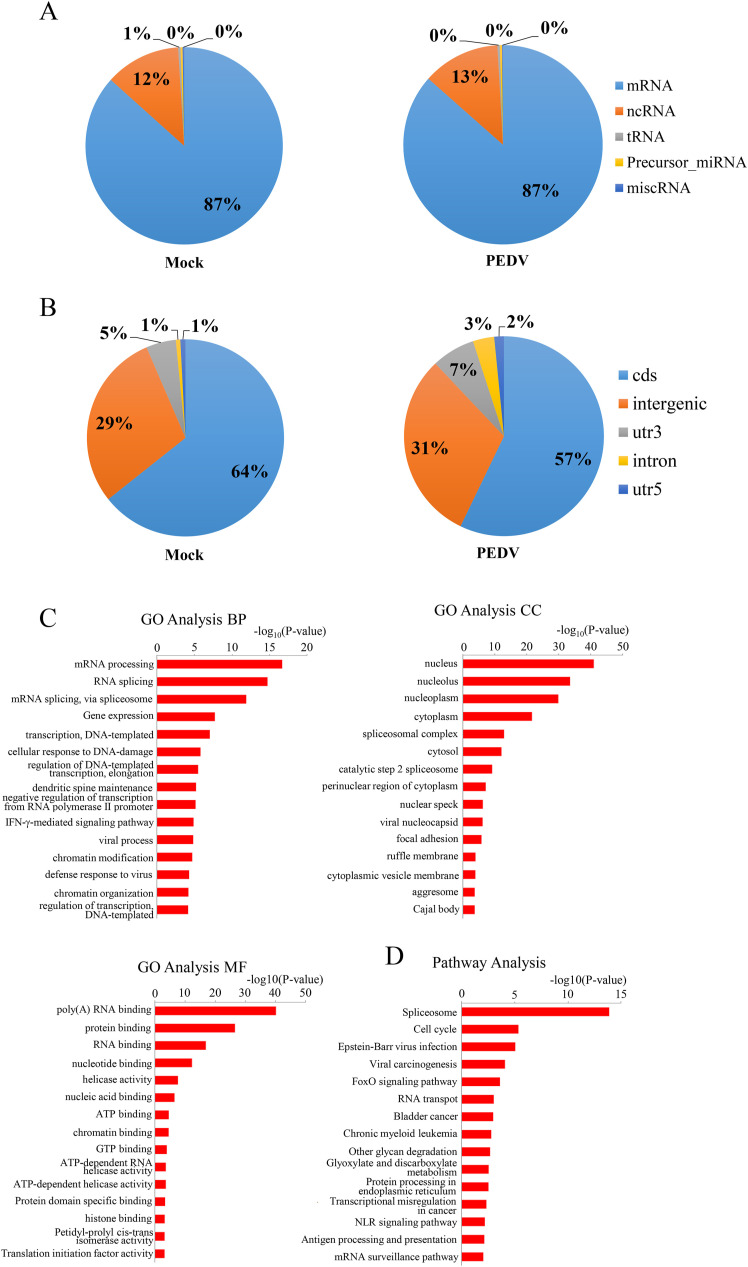
In addition to the m6A quantification of host RNA, we also performed m6A-seq to determine the dynamics of host RNA methylation after PEDV infection. The distribution of m6A on host transcripts was not significantly changed during viral infection. Of the total transcripts with m6A modification, mRNA accounted for approximately 87% in both groups. The percentage of methylated noncoding RNA (ncRNA) was 12% in the mock group and 13% in the PEDV-infected group. The m6A modification was rarely identified in tRNA, misc RNA, and precursor miRNA (Fig. 5 A). However, the detailed analysis further demonstrated that the distribution of m6A on gene structures in PEDV-infected cells was different from that in mock cells. PEDV infection increased the distribution of m6A on the untranslated regions (UTRs) and decreased its distribution on the coding sequence (cds). In mock cells, 64% of m6A was located in the cds, while in PEDV-infected cells, the percentage dropped to 57%. In contrast, the percentage of m6A distribution on the UTR had dropped by approximately 6% due to PEDV infection (Fig. 5B). Next, functional analysis of m6A-modified mRNAs was carried out. Gene Ontology (GO) and KEGG databases were used in this process. GO annotation analysis revealed that most m6A-modified mRNAs were involved in the process of metabolism. The mRNA processing and RNA splicing were the top two processes belonging to the biological process (BP) ontology of the GO analysis. Molecule function (MF) showed that most transcripts functioned in RNA or protein binding. In addition, the nucleus was the main location of the proteins translated from m6A-modified mRNAs (Fig. 5C). All these results demonstrated that m6A modification was a key player in the regulation of RNA processes. KEGG pathway analysis showed that the spliceosome and cell cycle were the top two pathways in which m6A-modified genes were involved. Several pathways related to virus infection and host immune response were also identified in this analysis, which suggested that the m6A modification also affected virus infection and host defenses (Fig. 5D).
Fig. 5.
PEDV infection influences RNA methylation of host cell transcripts
A) Distribution of m6A peaks in different types of host RNA transcripts. B) Distribution of m6A peaks in different structures of host mRNA. The m6A-modified mRNAs were searched in the C) GO database for functional significance and in the D) KEGG pathway database for pathway analysis.
3.6. Validation of m6A modification to gene expression
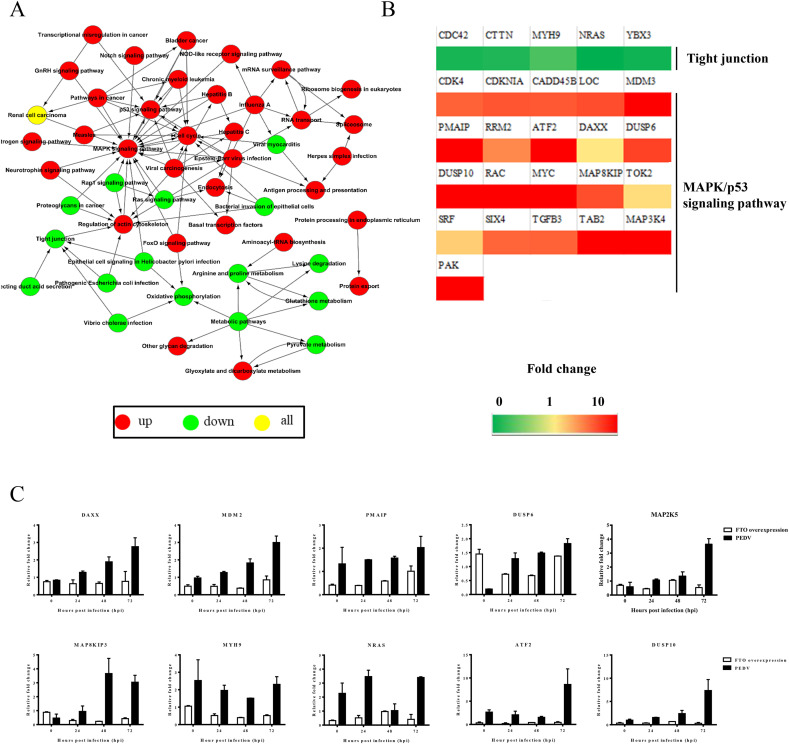
The results above showed that many genes had changes in their m6A modification during PEDV infection. According to our statistical analysis, 647 genes exhibited m6A modification, and 67 genes lost this modification with a p-value<0.05. The differently modified genes were then clustered by the pathways that they were involved in (Fig. 6 A). In this network, the MAPK signaling pathway was identified as the hotpot of upregulated genes, while the tight junction pathway was identified as the hotpot of downregulated genes. We then selected 26 genes that were differentially modified during PEDV infection and were involved in the above pathways for further analysis. First, real-time PCR was performed to confirm the dynamic modification of m6A in these fragmented RNAs prepared for m6A-seq. The primers were designed according to m6A-modified sequences obtained from m6A-seq (Table 3). As no genes have been found to be stably modified by m6A, no internal control was set in the analysis but equal amounts of RNA were used for reverse-transcription. The results showed that all these genes had changed their modification during PEDV infection, except DAXX, SRF, and TOK2 (Fig. 6B).
Fig. 6.
M6A modification regulates the host response to PEDV infection
A) The pathway network analysis of different signaling pathways in which the m6A-modified genes were involved. B) Real-time PCR verification of m6A-modified genes involved in the tight junction and MAPK/p53 signaling pathway. C) Real-time PCR analysis of selected m6A-modified genes in LLC-PK1 cells. The expression levels of all genes were normalized to GAPDH levels (internal control). The 2−ΔΔCt method was used to calculate the relative gene expression data. All experiments were performed in triplicate.
Next, we attempted to determine how these candidate genes respond to PEDV infection and whether demethylase can reverse their influence. LLC-PK1 cells were divided into three groups: untreated cells, cells infected with PEDV, cells overexpressing FTO, and infected with PEDV. The gene expression level in cells infected with PEDV was compared with that in the other two groups. The real-time PCR analysis showed that most genes had changed expression levels during PEDV infection. However, the expressionpattern of these genes was reversed after the FTO transfection. For example, DAXX was upregulated during PEDV infection, while the expression of FTO decreased its transcription (Fig. 6C). Other genes had similar expression patterns. These results indicated that PEDV infection could change the distribution of m6A in mRNA. The distribution in most transcripts was upregulated, and demethylase could reverse the expression level of these genes. This result correlated with the previous hypothesis (Hoernes et al., 2016). The dynamic methylation changes in host RNA implied that the enhancement of m6A modification during PEDV infection might be a regulation driven by host immunity.
4. Discussion
M6A modification has been shown to regulate the replication of several viruses. However, the specific role and mechanism still need further exploration. In this study, m6A modification was identified to be present in PEDV genomic RNA. We then quantified and mapped seven internal m6A peaks in PEDV RNA. Further studies showed that m6A modification reduced PEDV replication in different cell lines. The knockdown of METTL3 and METTL14 accelerated PEDV replication, while knockdown of FTO suppressed PEDV replication. The replication of viral RNA was also influenced. Subsequently, we investigated the role of m6A in host response during PEDV infection in LLC-PK1 and Vero cells. The ratio of m6A modification increased while the expression level of FTO decreased after PEDV infection. High-throughput sequencing revealed that there were many genes differently modified by m6A during PEDV infection. Real-time PCR analysis also proved these results. The overexpression of FTO could reverse the host response. Our findings demonstrated that m6A modification represents an important component in host regulation of virus replication.
Modifications in biological molecules have been recognized as an important regulatory measure for various life processes. The modification of RNAs can alter their biochemical structure and cell biology function. To date, 163 posttranscriptional modifications of RNA have been discovered to introduce functional diversity (Boccaletto et al., 2018). The m6A modification is the most abundant internal modification in eukaryotic RNAs. After its discovery in the 1970s (Desrosiers et al., 1974), researchers attempted to determine whether this modification also existed within viral RNA. With the development of the isotope-labeling method and other detection methods, m6A was found in the RNA of several viruses, including RNA viruses, DNA viruses, and retroviruses (Krug et al., 1976; Sommer et al., 1976; Moss et al., 1977; Furuichi et al., 1975). However, sequencing techniques and poor understanding of the regulation process have restricted further exploration. In 2011, 2013, the discovery of demethylases combined with the previous identification of a methyltransferase complex and RNA-binding proteins highly accelerated the studies of m6A (Jia et al., 2011; Zheng et al., 2013; Bokar et al., 1994; Liu et al., 2014). In 2012, antibody-based immunoprecipitation followed by high-throughput sequencing enabled the transcriptome-wide profiling of m6A (Dominissini et al., 2012). These developments promoted the studies on m6A in viral replication.
HIV-1 was the first virus with m6A peaks mapped within its genomic RNA. In 2016, three groups investigated the role of m6A during HIV-1 infection (Tirumuru et al., 2016; Kennedy et al., 2016; Lichinchi et al., 2016a). Two studies showed that m6A promoted the replication of HIV, while the other reported the opposite result. Later, the replications of the Zika virus, hepatitis C virus, and members of the Flaviviridae family were found to be negatively regulated by m6A (Gokhale et al., 2016; Lichinchi et al., 2016a). For the influenza virus and enterovirus 71, it was demonstrated that m6A contributed to their replication (Hao et al., 2019; Courtney et al., 2017). In our study, m6A inhibited and overexpression of FTO facilitated PEDV growth. All of these studies suggest that m6A modification could play different roles in the regulation of viral replication. Various factors may account for this. First, m6A contributes not only to the regulation of viral RNA but also to host RNAs. Both viral and host factors may influence the replication of viruses (Gokhale and Horner, 2017). The output of viral replication is determined by the balance of interaction effects between viral and host factors. Therefore, it is reasonable that the final effects of m6A are different among different viruses. Second, viruses have hypervariable regions among their RNAs, which also exhibit different characteristics (Prentoe and Bukh, 2018; Smith, 1999). The presence of m6A may also be divergent for different strains, which may have different effects. Third, different cell lines usually are originated from different species and organisms and have different features and regulate viral growth by different pathways. Finally, the reversible process of m6A may have different effects on viral replication. The time points selected for analysis may also affect the results.
The presence of m6A also varies among different kinds of viruses. Although the number of identified m6A peaks varied among the studies of HIV, the studies agreed on the presence of m6A at the 3ʹ end of HIV-1 gRNA (Tirumuru et al., 2016; Kennedy et al., 2016; Lichinchi et al., 2016a). NS5B and NS3 were the main regions in which m6A peaks were present among viruses within the Flaviviridae family (Gokhale et al., 2016; Lichinchi et al., 2016a). For influenza virus and enterovirus 71, m6A were peaked are located in the genes encoding structural proteins (Hao et al., 2019; Courtney et al., 2017), while our results showed that the m6A peaks were mainly presented in ORF1b, which encodes nonstructural proteins. As m6A peaks are present in different regions of viral genomic RNA, the specific mechanism of m6A in regulating viral replication may vary between different viruses. For HCV, mutation of m6A sites within the E1 region can increase HCV RNA binding to the Core protein (Gokhale et al., 2016). The expression of HIV-1 gag and p24 was reduced by the knockdown of METTL3 and METTL14, which suggested that m6A might facilitate the translation of viral proteins (Tirumuru et al., 2016). Although m6A has been found to affect most posttranscriptional steps in gene expression, such as mRNA stability, splicing, translational efficacy and pri-microRNA processing (Zheng et al., 2013; Wang et al., 2015; Alarcon et al., 2015; Ozkurede et al., 2019), the regulatory mechanism has not been clearly elucidated and needs to be explored further.
With a better understanding of the effects of m6A on viral replication, the identification of specific sites modified by m6A requires additional research. The m6A-seq technique employed in the above studies was based on captured nucleotides of 100–200 bp length. Therefore, the clusters of m6A within 200 nucleotides cannot be precisely located by sequencing. The identification of Tth DNA polymerase I solved this problem. This DNA polymerase was previously shown to act as reverse transcriptase in the presence of Mn2+. Later, it was found that Tth DNA polymerase I could discriminate m6A from adenosine (A) in RNA. It was selective by up to 18-fold for the incorporation of thymidine opposite unmodified A over m6A (Harcourt et al., 2013). The identification of two m6A modification sites in the EV71 genome benefited from the use of this polymerase. The adenines at 3055 and 4555 in the EV71 genome were proven to be m6A-modified. Mutations at these two sites resulted in decreased virus replication (Hao et al., 2019). However, this method needs to incorporate radiolabeled probes which have limited its use, such as for our study. The locations of m6A modification in RNAs at nucleotide resolution will no doubt improve the understanding of their function.
It was of great interest to us to find that PEDV infection increases the m6A ratio in host RNAs. KEGG analysis also showed that there were more genes with increased m6A modification. Most genes were upregulated during PEDV infection, while the overexpression of FTO downregulated their expression. These results suggested that m6A also functioned in the host response to PEDV infection. This finding correlated with previous studies showing that reversible m6A could regulate gene expression (Fu et al., 2014). This phenotype was also reported recently in HIV and VSV (Lichinchi et al., 2016b; Tirumuru and Wu, 2019; Liu et al., 2019). The infection of both viruses can enhance m6A modification in host RNA. It is interesting to further identify whether virus infection always enhances m6A modification and potential reasons. Besides, we found that the expression of FTO was altered by virus infection. FTO, as the key component of demethylases, was decreased during PEDV infection. It is not clear whether the infection of other viruses can drive similar changes. These results suggested that m6A modification could be an important mechanism in regulating gene expression, especially for the host response to viral infection.
To our knowledge, this is the first evidence to report the presence of m6A in coronaviruses and its regulation of both virus and host responses. We presented global m6A profiling of PEDV and found that this modification can inhibit PEDV infection. Furthermore, we found that the m6A modification in host RNAs was also drastically changed during PEDV infection. Our study suggests that m6A modification may represent a novel and conserved target for antiviral mechanisms. We believe that the results from this study will facilitate the understanding of the role of m6A and broaden our knowledge on virus-host interactions at the RNA level.
Author contribution
GL and JC conceived the project, designed the experiments, analyzed the data, wrote and edited the manuscript. JC, LJ, ZW, LW, QC, YC performed the experiments. All authors approved the final manuscript.
CRediT authorship contribution statement
Jianing Chen: Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Project administration, Funding acquisition. Li Jin: Investigation. Zemei Wang: Investigation. Liyuan Wang: Investigation. Qingbo Chen: Investigation. Yaru Cui: Investigation. Guangliang Liu: Conceptualization, Methodology, Validation, Resources, Data curation, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31702209, 31572498, 31972689), and by the National Key R&D Program of China (2016YFD0500103).
References
- Alarcon C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A., Helm M., Bujnicki J.M. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–d307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA (New York, N.Y.) 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Courtney D.G., Kennedy E.M., Dumm R.E., Bogerd H.P., Tsai K., Heaton N.S., Cullen B.R. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe. 2017;22:377–386. doi: 10.1016/j.chom.2017.08.004. e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P.R. The dilemma of rare events: porcine epidemic diarrhea virus in North America. Prev. Vet. Med. 2015;122:235–241. doi: 10.1016/j.prevetmed.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Dominissini D., Rechavi G., He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A.J., Stavnezer E., Bishop J.M. Blocked, methylated 5'-terminal sequence in avian sarcoma virus RNA. Nature. 1975;257:618–620. doi: 10.1038/257618a0. [DOI] [PubMed] [Google Scholar]
- Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S., McDonough M.A., Cunliffe S., McNeill L.A., Galvanovskis J., Rorsman P., Robins P., Prieur X., Coll A.P., Ma M., Jovanovic Z., Farooqi I.S., Sedgwick B., Barroso I., Lindahl T., Ponting C.P., Ashcroft F.M., O'Rahilly S., Schofield C.J. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science (New York, N.Y.) 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale N.S., Horner S.M. vol. 13. 2017. (RNA Modifications Go Viral). e1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale N.S., McIntyre A.B.R., McFadden M.J., Roder A.E., Kennedy E.M., Gandara J.A., Hopcraft S.E., Quicke K.M., Vazquez C., Willer J., Ilkayeva O.R., Law B.A., Holley C.L., Garcia-Blanco M.A., Evans M.J., Suthar M.S., Bradrick S.S., Mason C.E., Horner S.M. N6-Methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016;20:654–665. doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Hao S., Chen H., Chen Z., Zhang Y., Wang J., Wang H., Zhang B., Qiu J., Deng F., Guan W. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 2019;47:362–374. doi: 10.1093/nar/gky1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt E.M., Ehrenschwender T., Batista P.J., Chang H.Y., Kool E.T. Identification of a selective polymerase enables detection of N(6)-methyladenosine in RNA. J. Am. Chem. Soc. 2013;135:19079–19082. doi: 10.1021/ja4105792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoernes T.P., Huttenhofer A., Erlacher M.D. mRNA modifications: dynamic regulators of gene expression? RNA Biol. 2016;13:760–765. doi: 10.1080/15476286.2016.1203504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chen J., Yao G., Guo Q., Wang J., Liu G. vol. 103. 2019. pp. 4943–4952. (A TaqMan-Probe-Based Multiplex Real-Time RT-qPCR for Simultaneous Detection of Porcine Enteric Coronaviruses). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Yang C.G., Yang S., Jian X., Yi C., Zhou Z., He C. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. vol. 204. Veterinary journal; London, England: 2015. Porcine Epidemic Diarrhea Virus Infection: Etiology, Epidemiology, Pathogenesis and Immunoprophylaxis; pp. 134–143. 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E.M., Bogerd H.P., Kornepati A.V., Kang D., Ghoshal D., Marshall J.B., Poling B.C., Tsai K., Gokhale N.S., Horner S.M., Cullen B.R. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P., Lence T., Haussmann I.U., Jacob D., Kreim N., Carl S.H., Masiello I., Hares T., Villasenor R., Hess D., Andrade-Navarro M.A., Biggiogera M., Helm M., Soller M., Buhler M., Roignant J.Y. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Gene Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koba M., Konopa J. [Actinomycin D and its mechanisms of action] Postępy Higieny Medycyny Doświadczalnej. 2005;59:290–298. [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Gene. 2001;23:137–144. doi: 10.1023/A:1011831902219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R.M., Morgan M.A., Shatkin A.J. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J. Virol. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Chen Y.S., Ping X.L., Yang X., Xiao W., Yang Y., Sun H.Y., Zhu Q., Baidya P., Wang X., Bhattarai D.P., Zhao Y.L., Sun B.F., Yang Y.G. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G., Zhao B.S., Wu Y., Lu Z., Qin Y., He C., Rana T.M. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016;20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G., Gao S., Saletore Y., Gonzalez G.M., Bansal V., Wang Y., Mason C.E., Rana T.M. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.11. 16011-16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., Dai Q., Chen W., He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., You Y., Lu Z., Yang J., Li P., Liu L., Xu H., Niu Y., Cao X. N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science (New York, N.Y.) 2019;365:1171–1176. doi: 10.1126/science.aax4468. [DOI] [PubMed] [Google Scholar]
- Moss B., Gershowitz A., Stringer J.R., Holland L.E., Wagner E.K. 5'-Terminal and internal methylated nucleosides in herpes simplex virus type 1 mRNA. J. Virol. 1977;23:234–239. doi: 10.1128/jvi.23.2.234-239.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkurede U., Kala R., Johnson C., Shen Z., Miller R.A., Garcia G.G. Cap-independent mRNA translation is upregulated in long-lived endocrine mutant mice. J. Mol. Endocrinol. 2019;63:123–138. doi: 10.1530/JME-19-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S., Zhao X., Li A., Yang Y., Dahal U., Lou X.M., Liu X., Huang J., Yuan W.P., Zhu X.F., Cheng T., Zhao Y.L., Wang X., Rendtlew Danielsen J.M., Liu F., Yang Y.G. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentoe J., Bukh J. Hypervariable region 1 in envelope protein 2 of hepatitis C virus: a linchpin in neutralizing antibody evasion and viral entry. Front. Immunol. 2018;9:2146. doi: 10.3389/fimmu.2018.02146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D., Sanjana N.E., Freinkman E., Pacold M.E., Satija R., Mikkelsen T.S., Hacohen N., Zhang F., Carr S.A., Lander E.S., Regev A. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.B. Evolution of the hypervariable region of hepatitis C virus. J. Viral Hepat. 1999;6(Suppl. 1):41–46. doi: 10.1046/j.1365-2893.1999.00010.x. [DOI] [PubMed] [Google Scholar]
- Sommer S., Salditt-Georgieff M., Bachenheimer S., Darnell J.E., Furuichi Y., Morgan M., Shatkin A.J. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res. 1976;3:749–765. doi: 10.1093/nar/3.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N., Wu L. HIV-1 envelope proteins up-regulate N (6)-methyladenosine levels of cellular RNA independently of viral replication. J. Biol. Chem. 2019;294:3249–3260. doi: 10.1074/jbc.RA118.005608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N., Zhao B.S., Lu W., Lu Z., He C., Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife. 2016;5 doi: 10.7554/eLife.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., Ren B., Pan T., He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Lv R., Ma H., Shen H., He C., Wang J., Jiao F., Liu H., Yang P., Tan L., Lan F., Shi Y.G., He C., Shi Y., Diao J. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038. doi: 10.1016/j.molcel.2018.02.015. e1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Huang Y., Dong J., Liang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013;164:212–221. doi: 10.1016/j.vetmic.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhang H., Zhang Q., Dong J., Liang Y., Huang Y., Liu H.J., Tong D. Porcine epidemic diarrhea virus E protein causes endoplasmic reticulum stress and up-regulates interleukin-8 expression. Virol. J. 2013;10:26. doi: 10.1186/1743-422X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H., Lu Z., Bosmans R.P., Dai Q., Hao Y.J., Yang X., Zhao W.M., Tong W.M., Wang X.J., Bogdan F., Furu K., Fu Y., Jia G., Zhao X., Liu J., Krokan H.E., Klungland A., Yang Y.G., He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]