Abstract
Tuber mustard, which is the raw material of Fuling pickle, is a crop with great economic value. However, during growth and development, tuber mustard is frequently attacked by the pathogen Plasmodiophora brassicae and frequently experiences salinity stress. Jasmonic acid (JA) is a hormone related to plant resistance to biotic and abiotic stress. Jasmonate ZIM domain proteins (JAZs) are crucial components of the JA signaling pathway and play important roles in plant responses to biotic and abiotic stress. To date, no information is available about the characteristics of the JAZ family genes in tuber mustard. Here, 38 BjJAZ genes were identified in the whole genome of tuber mustard. The BjJAZ genes are located on 17 of 18 chromosomes in the tuber mustard genome. The gene structures and protein motifs of the BjJAZ genes are conserved between tuber mustard and Arabidopsis. The results of qRT-PCR analysis showed that BjuA030800 was specifically expressed in root, and BjuA007483 was specifically expressed in leaf. In addition, 13 BjJAZ genes were transiently induced by P. brassicae at 12 h, and 7 BjJAZ genes were induced by salt stress from 12 to 24 h. These results provide valuable information for further studies on the role of BjJAZ genes in the regulation of plant growth and development and in the response to biotic and abiotic stress.
Introduction
Tuber mustard is the raw material of Fuling pickle, which has great economic value. As a Cruciferae plant species, tuber mustard is frequently attacked by soil pathogens, such as Plasmodiophora brassicae. Discovering the mechanism by which tuber mustard responds to pathogens is important for improving the production of this crop. Jasmonic acid (JA) is a plant hormone that is responsible for plant resistance to biotic and abiotic stress. Studying the roles of JA signaling-related genes in the regulation of plant growth and development will shed light on the molecular mechanism underlying the resistance of tuber mustard to pathogens such as P. brassicae.
JA is an endogenous growth-regulating substance found in higher plants. Methyl ester JA (MeJA) and isoleucine-conjugated JA (JA-Ile) are derivatives of JA and have similar functions to JA. Collectively, these hormones are known as jasmonates. Jasmonates are involved in many biological processes, such as plant growth and development [1–3] and plant responses to herbivore damage [4]. JAs also play roles in plant responses to biotic stress, such as the response of Brassica. napus to P. brassicae [5], and plant responses to abiotic stress, such as the salinity tolerance of wheat [6]. JA functions through a cascade of molecular signal transductions, and the Coronatine Insensitive1(COI1) and jasmonate ZIM domain (JAZ) proteins are the coreceptors of JA-Ile [7]. The interaction between COI1 and JAZ proteins activated by JA-Ile leads to the degradation of JAZ proteins by the 26S proteasome, which eventually activates the function of some JA-responsive transcription factors (TFs), such as MYCs, MYBs, NACs, ERFs and WRKYs [8–9], resulting in JA-related responses.
JAZ proteins, which contain a ZIM domain (TIFY) at the N-terminus, are repressors of the JA signaling pathway and exhibit homo- and heteromeric interactions [10]. JAZ proteins play roles in plant responses to biotic and abiotic stress. It has been reported that overexpression of TaJAZ1 led to enhanced resistance against powdery mildew in bread wheat [11]. In rice, OsJAZ8 was reported to be involved in the induction of monoterpene linalool, which conferred resistance to rice bacterial blight [12]. Some PtJAZ genes, including PtJAZ3, PtJAZ6, PtJAZ9 and PtJAZ15, were found to be induced by leaf rust pathogen infection in poplar, suggesting their potential roles in plant responses to abiotic stresses [13]. In Arabidopsis, jaz10 mutants exhibit enhanced susceptibility to the pathogen Pseudomonas syringae DC3000 [14]. These studies suggest that JAZ proteins play crucial roles in plant resistance to biotic stress. In addition, JAZ proteins are widely involved in plant tolerance to salt stress. Overexpression of GhJAZ2 resulted in enhanced sensitivity to salt stress in transgenic cotton [15]. Overexpression of OsJAZ8 in rice led to improved salt tolerance in transgenic seedlings [16].
JAZ proteins also exist in tuber mustard; however, the functions of JAZ family genes in this crop are still unknown. Here, we identified JAZ family members in tuber mustard and analyzed their gene and protein characteristics. We also investigated the expression patterns of the BjJAZ genes during plant growth and development and their expression profiles in the roots of tuber mustard seedlings under pathogen and salinity treatments. This study will be useful for functional studies of JAZs in tuber mustard.
Materials and methods
Plant materials and growth conditions
To evaluate the gene expression patterns in different tissues, seedlings of the tuber mustard cultivar ‘Yong An XiaoYe’ were planted in a field for six months and grown to the reproductive stage, and the roots, stems, leaves, flowers and pods were sampled for qRT-PCR analysis. For stress treatments, seeds of the tuber mustard cultivar ‘Yong An XiaoYe’ were sown in plant pots in vermiculite and turfy soil (2:1) medium, and the culture temperature was 22°C with a 16/8 h light/dark regime. For the salt-stress treatments, two-week-old seedlings were irrigated with a nutrient solution with or without 200 mM NaCl for 0, 6, 12, 24 and 48 h, and the roots were sampled for qRT-PCR analysis. For pathogen treatment, two-week-old seedlings were inoculated with a 5-mL resting spore suspension of P. brassicae (OD600 = 0.07) for 0, 12, 24, 36 and 72 h, and the roots were sampled for qRT-PCR analysis.
Identification of BjJAZ proteins in tuber mustard and bioinformatic analysis
The JAZ protein sequences of A. thaliana were obtained from the Arabidopsis information resource website (https://www.arabidopsis.org/), the gene ID and protein sequences of these AtJAZ genes are listed in S1 File. A BLASTP analysis was performed using the AtJAZ proteins as the query sequences to search the putative JAZ proteins in the Brassica. juncea genome database (chromosome V1.5, http://brassicadb.org/brad/), the parameters were set as Evalue < 1e-10, identity > 70%, query coverage > 95%, and the other parameters were defaulted. The Pfam database [17] (Pfam; http://pfam.xfam.org/)was used to analyze the conserved domains of all the selected nonredundant sequences, and the proteins that contained both the TIFY domain and Jas domain were defined as belonging to the JAZ family. For gene location analysis, the region of each BjJAZ genomic sequence was downloaded from the B. juncea genome database, and the location of each BjJAZ gene was marked in the tuber mustard chromosome using MapDraw V2.1 software [18]. The coding sequence and corresponding genomic sequences of BjJAZ were downloaded from the B. juncea genome database, and the exon-intron structure diagram was drawn using online software for the GSDS2.0 server (http://gsds.cbi.pku.edu.cn/). The phylogenetic analysis was performed using MEGA5 software with the neighbor-joining method [19], the bootstrap value was used for 1000 replicates, and the “p-distance model” and “Pairwise deletion” were used for “Model/Method” and “Gaps/Missing” data treatment, respectively. The amino acid isoelectric point (pI) and molecular weight (MW) were predicted with ExPASy software (https://web.expasy.org/compute_pi/), and protein motifs were analyzed with the NCBI-CDD (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The diagram of the protein motifs was drawn by ExPASy (https://prosite.expasy.org/prosite.html), and the sequence logo for the TIFY domain and Jas domain was generated with WebLogo 3 (http://weblogo.berkeley.edu/). The comparison and annotation of orthologous gene clusters of the JAZ genes among tuber mustard and Arabidopsis were analyzed using the online software OrthoVenn2 under default parameters [20]. The duplication events were analyzed by performing BLASTP for proteins against each other, which were defined when both the identity and query coverage was > 80% between partner sequences [21,22]. Tandem duplication genes were defined according to the methods of Holub, where a chromosomal region within 200 kb containing two or more genes is defined as a tandem duplication event [23]. The online software MEME (Version 5.1.1; http://meme-suite.org/tools/meme) was used to elucidate conserved motifs of BjJAZs, the maximum number of motifs was 9, and the other parameters were defaulted [24].
RNA extraction and qRT-PCR analysis
Total RNA was extracted from different plant materials using TRIzol reagent (Tiangen Biotech (Beijing) Co., Ltd., China) following the manufacturer’s instructions, and the concentration and quality was detected using Nanodrop One (Thermo Fisher Scientific, USA) and 1% agarose gel electrophoresis. The total RNA samples were treated with DNase I (Invitrogen Biotech Co., Ltd., USA) to remove contaminating genomic DNA. First-strand cDNA was synthesized from 500 ng RNA in 20 μL volume using a FastQuant RT Kit (Tiangen Biotech (Beijing) Co., Ltd., China) following the manufacturer’s instructions. qRT-PCR was performed using SuperReal PreMix Plus (SYBR Green; Tiangen Biotech (Beijing) Co., Ltd., China). For qRT-PCR analysis, each reaction was performed in 10 μL volume containing 0.3 μL diluted (50x) cDNA sample as template, 5 μL SuperReal PreMix Plus, 0.5 μL (10 μmol/L) each of forward and reverse primers, and 3.7 μL ddH2O. The prepared samples were subjected to the following conditions: 95°C for 15 min, followed by 40 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 30 s. The fluorescence was determined observing the last step of each cycle, and the Melting Curves program was added in the last step. LightCycler 480 II (Roche, Germany) was used for amplification detection. Each of three biological and technical replicates was examined for each stress treatment. The 18S rRNA gene (BjuA046942) was used as the internal reference gene for qRT-PCR, and the gene-specific primers are listed in S1 Table.
Statistical analysis
The qRT-PCR gene relative expression levels were calculated using the 2−ΔΔCt method and the data were analyzed using SigmaPlot 10.0 (Systat Software Inc., USA) and SPSS 16.0 software (IBM Corporation, USA). The averages and standard deviations of all results were calculated using SPSS 16.0 software, and one-way ANOVA followed by the LSD test was used for difference significance analysis. Significant differences are designated as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Results
Identification of JAZ family genes in tuber mustard
To identify the BjJAZ genes in the tuber mustard genome, protein sequence BLAST searches were performed in the Brassica Database using the Arabidopsis JAZ protein sequences as the queries. The protein sequences in the tuber mustard genome with more than 70% identity to AtJAZ proteins were selected as the candidate homologs of JAZs, and the JAZ proteins were further identified by gene structure and protein domain conservation analyses. A total of 38 nonredundant BjJAZ genes were identified in the genome of tuber mustard, and the identification was supported by conserved functional domain and multiple sequence alignment analyses. The sequences of the predicted BjJAZ proteins and gene IDs are listed in S2 File. The gene and protein characteristics of the identified JAZ genes were investigated by bioinformatic analysis. The lengths of the BjJAZ genomic sequences ranged from 458 bp to 2929 bp, and the lengths of the protein-coding sequences (CDSs) ranged from 369 bp to 1065 bp. The predicted lengths of the proteins encoded by the BjJAZ genes varied from 122 amino acids to 354 amino acids, with the corresponding molecular weights (MW) ranging from 13.74 kDa to 37.50 kDa, and the theoretical isoelectric (pI) values ranged from 5.20 to 10.33 (Table 1).
Table 1. The BjJAZ family genes in tuber mustard.
| Sequence ID | Locus | Genomics (bp) | CDS (bp) | Protein (aa) | pI | MW (kD) |
|---|---|---|---|---|---|---|
| BjuB032431 | B03 | 1403 | 822 | 273 | 9.49 | 29.82 |
| BjuB031487 | B03 | 1411 | 825 | 274 | 9.49 | 30.09 |
| BjuA025820 | A06 | 1296 | 762 | 253 | 9.73 | 27.15 |
| BjuB043409 | B03 | 1047 | 756 | 251 | 9.22 | 27.33 |
| BjuA030800 | A08 | 1121 | 780 | 259 | 9.49 | 28.31 |
| BjuB021388 | B06 | 1185 | 687 | 228 | 9.01 | 25.11 |
| BjuA027037 | A07 | 1170 | 675 | 224 | 9.18 | 24.49 |
| BjuA029428 | A07 | 1150 | 738 | 245 | 9.20 | 26.63 |
| BjuB029798 | B03 | 1213 | 759 | 252 | 9.36 | 27.68 |
| BjuA005572 | A02 | 1199 | 723 | 240 | 9.23 | 26.07 |
| BjuB011370 | B05 | 1246 | 738 | 245 | 9.01 | 26.72 |
| BjuA045157 | A01 | 2929 | 1062 | 353 | 9.49 | 37.44 |
| BjuB007213 | B07 | 2676 | 1065 | 354 | 9.53 | 37.50 |
| BjuA046021 | A05 | 2574 | 1008 | 335 | 9.51 | 35.87 |
| BjuB025543 | B01 | 2602 | 1020 | 339 | 9.47 | 36.17 |
| BjuB043343 | B03 | 1269 | 786 | 261 | 9.14 | 29.18 |
| BjuA022138 | A06 | 1287 | 771 | 256 | 9.28 | 28.75 |
| BjuB029203 | B04 | 1231 | 771 | 256 | 10.33 | 28.96 |
| BjuA030507 | A08 | 1286 | 813 | 270 | 8.32 | 30.03 |
| BjuA001107 | A09 | 1137 | 798 | 265 | 9.71 | 29.88 |
| BjuB030035 | B03 | 1886 | 684 | 227 | 9.21 | 25.45 |
| BjuB010656 | B05 | 1338 | 819 | 272 | 8.99 | 30.33 |
| BjuA007483 | A02 | 1320 | 822 | 273 | 8.95 | 30.18 |
| BjuB026559 | B01 | 458 | 369 | 122 | 9.27 | 13.74 |
| BjuB032915 | B03 | 586 | 396 | 131 | 9.69 | 15.04 |
| BjuB029529 | B04 | 568 | 396 | 131 | 9.23 | 15.07 |
| BjuA034780 | A09 | 579 | 402 | 133 | 9.47 | 15.39 |
| BjuA007387 | A02 | 2078 | 822 | 273 | 9.82 | 29.27 |
| BjuA027135 | A07 | 1133 | 639 | 212 | 9.17 | 23.50 |
| BjuB030369 | B03 | 1935 | 834 | 277 | 9.85 | 29.88 |
| BjuA027422 | A07 | 2148 | 804 | 267 | 9.76 | 28.81 |
| BjuB035964 | B02 | 1605 | 591 | 196 | 9.90 | 21.87 |
| BjuA041687 | A03 | 1486 | 588 | 195 | 9.95 | 21.79 |
| BjuB014771 | B08 | 1673 | 588 | 195 | 9.80 | 21.79 |
| BjuA022588 | A06 | 1341 | 666 | 221 | 5.20 | 23.10 |
| BjuO008948 | Contig | 1108 | 633 | 210 | 8.93 | 22.45 |
| BjuA047148 | A10 | 1079 | 612 | 203 | 6.92 | 21.43 |
| BjuA001950 | A05 | 549 | 372 | 123 | 9.90 | 14.01 |
Phylogenetic analysis of BjJAZs
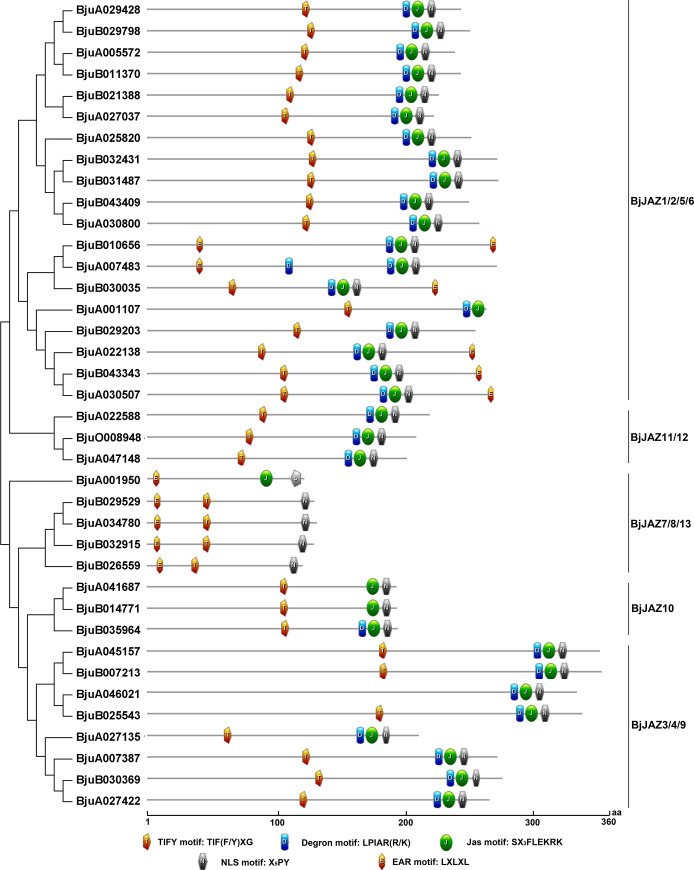
To characterize the evolutionary relationships among the JAZ genes in tuber mustard and Arabidopsis, a phylogenetic analysis was performed using MEGA 5 software, and the BjJAZ and AtJAZ protein sequences were used for the analysis. As shown in the neighbor-joining phylogenetic tree, these JAZs were assigned tofive separate clades: JAZ1/2/5/6, JAZ11/12, JAZ7/8/13, JAZ10, and JAZ3/4/9 (Fig 1), and the details are listed in Table 1. We also analyzed the orthologous relationships among the BjJAZ and AtJAZ genes using the online software OrthoVenn2, and 13 clusters were identified, of those, 11 orthologous clusters were found shared between tuber mustard and Arabidopsis, expect for 2 single-copy gene clusters (S3 File).
Fig 1. Phylogenetic analysis of JAZs in Arabidopsis thaliana and tuber mustard.
The protein sequence of each gene was used for the alignment, and the phylogenetic neighbor-joining tree was constructed using MEGA5 phylogenetic analysis software.
Chromosomal locations of BjJAZs in the genome of tuber mustard
Thirty-seven of the thirty-eight BjJAZ genes were anchored on 18 tuber mustard chromosomes except for chromosome A04 (Fig 2), while BjuO008948 could not be anchored in the tuber mustard genome. Chromosome B03 had the highest density of BjJAZ genes (8 members), and only one BjJAZ gene could be found in each of 8 chromosomes (A01, A03, A05, A10, B02, B06, B07 and B08). Four BjJAZ genes were located on chromosome A07, and 3 BjJAZ genes were located on chromosomes A02 and A06. Each of the chromosomes A08, A09, B04 and B05 contained 2 BjJAZ genes (Fig 2). We also analyzed the tandem duplication genes in the whole genome of tuber mustard, and 2588 tandem duplication pairs were identified (S4 File). However, no tandem duplication pairs were found in 38 BjJAZ genes.
Fig 2. Gene locations of BjJAZs in the genome of tuber mustard.
Thirty-seven of thirty-eight identified JAZ homologous genes were mapped to 17 out of 18 chromosomes. The chromosome name is at the top of each bar. The scale of the chromosome is in millions of bases (Mb).
Exon/intron structure analysis of BjJAZs
The exon/intron structure of each BjJAZ gene was revealed by comparing the CDS with the corresponding genomic sequences. The number of exons in the BjJAZ genes varied from 2 (BjuA001107) to 7 (BjJAZ3/4/9 group genes, except for BjuA027135). In BjJAZ1/2/5/6 group, each of the 6 genes in sub-Clade I contained 5 exons with the intron phase pattern “1,2,1,2”, and each of the 5 genes in sub-Clade II contained 4 exons with the intron phase pattern “1,2,1”. Seven of the eight sub-Clade III genes contained 4 exons with intron phase pattern “1,2,2”, the exception was BjuA001107, which contained 3 exons; all of the BjJAZ11/12 and BjJAZ10 group genes contained 5 exons, with the intron phase pattern “2,0,1,2” and “1,2,1,2”, respectively; all of the BjJAZ7/8/13 group genes contained 3 exons with the intron phase pattern “2,0”, except for BjuA001107 (Fig 3). We also analyzed the intron phase patterns in the TIFY and Jas domains. The results showed that a phase “2” intron inserted in the Jas domain was highly conserved at the 18th amino acid. The BjJAZ3/4/9, BjJAZ7/8 and BjJAZ11/12 family genes were inserted by a phase “0” intron in the TIYF domain, and no intron insertions were found for the other BjJAZ genes (except for BjuA022138, which exhibited a phase “2” insertion) (Fig 3). Collectively, most of the genes in the same phylogenetic group (or subclade) exhibited the same exon/intron numbers and intron phase patterns.
Fig 3. The exon/intron structures of the BjJAZ genes.
The intron phase was defined as the position of the introns within or between codons. Phase 0: the intron was between codons; phase 1: the intron was between the first and second bases of a codon; phase 2: the intron was between the second and third bases of a codon.
Protein motif analysis of BjJAZs
The TIFY domain was previously known as the ZIM domain, which is crucial for interaction with an adapter protein called NINJA. NINJA contains an ethylene-responsive element binding factor associated amphiphilic repression (EAR) motif to recruit TOPLESS. The secondary protein domain prediction results showed that each of the BjJAZ proteins except for BjuA001950 contained one TIFY domain (S1 Fig). Another important domain of the JAZ protein is called the Jas domain (also known as the CCT_2 domain), which is responsible for the COI1-JAZ and MYC2-JAZ interactions. Most of the BjJAZ proteins contained a Jas domain, however, the Jas domain could not be found in BjuB026559 and BjuA001950, and a truncated Jas domain was found in the C-terminus of BjuA001107 (S1 Fig). Several motifs in the TIFY and Jas domains are crucial for the function of JAZ proteins, including the TIFY motif in the TIFY domain; Degron motif, Jas motif, and NLS motif in the Jas domain; and the EAR motif in some JAZ family proteins. The TIFY motif TIF(F/Y)XG is the key component of the TIFY domain, playing important roles in JAZ-NINJA and JAZ-JAZ protein interactions. All the BjJAZ proteins contained a TIFY motif except for BjuA001950 and BjuA046021 (Fig 4). Although no canonical TIFY motif existed in BjuA046021, the TIFY domain was predicted in NCBI-CDD for this protein. The degron motif LPIAR(R/K) is crucial for the COI1-JA-Ile-JAZ complex formation. All the BjJAZ1/2/5/6, BjJAZ3/4/9, and BjJAZ11/12 group proteins contained the degron motif; in contrast, all the BjJAZ7/8/13 group proteins lacked this motif. Interestingly, for the BjJAZ10 group, the degron motif existed in BjuB035964 but not in BjuA041687 and BjuB014771 (Fig 4), and the divergence indicated the different mechanisms by which these BjJAZ10 proteins function in the JA signaling pathway. The Jas motif (SX3FLEKRK) is involved in the binding of JAZ to downstream transcription factors. Except for four proteins in BjJAZ7/8/13 group (BjuB026559, BjuB032915, BjuB029529 and BjuA034780), all the other BjJAZ proteins contained this motif. The EAR motif (LxLxL) is related to the interaction with TOPLESS proteins and the resultant repression of JA signaling. The EAR motif existed in 6 protiens of BjJAZ1/2/5/6 group and all of the proteins of BjJAZ7/8/13 group; in particular, there were two EAR motifs in BjuB010656 (Fig 4). Together, the protein motifs were conserved in the same group (except for BjJAZ1/2/5/6); most of the proteins in BjJAZ3/4/9, BjJAZ11/12 and BjJAZ10 groups shared the same motifs. We also analyzed the conservation of the TIFY domain and Jas domain of BjJAZ proteins in tuber mustard and Arabidopsis. The results showed that the tuber mustard TIFY domain and Jas domain were well conserved and very similar to those of Arabidopsis (Fig 5). The consensus core region of the TIFY motif and Jas motif were conserved between tuber mustard and Arabidopsis, and the variation in their flanking regions was also very similar between the two species (Fig 5). Furthermore, to better understand the sequence characteristics of the BjJAZ genes, conserved motifs were predicted using MEME based on 38 BjJAZ protein sequences, and 9 motifs (motif 1–9) were identified (S2 Fig). Each of the BjJAZ proteins contained 2 (BjJAZ7/8/13, BjJAZ10 and BjJAZ11/12 proteins) to 9 (part of proteins in BjJAZ3/4/9 group) conserved motifs. The sequence of motif 1 includes the core sequence of Jas motif, and this motif was found in all the BjJAZ proteins (except for BjuA001107); motif 2 includes the core sequence of TIFY motif, and all the BjJAZ proteins contained this motif. Besides that, the distribution of the other motifs was conserved among BjJAZ7/8/13, BjJAZ10 and BjJAZ11/12 groups, but not the other groups (S2 Fig).
Fig 4. Protein motif prediction for BjJAZs.
Five conserved motifs were predicted, and the diagram of the protein motifs was drawn by ExPASy.
Fig 5.
Consensus sequences of the TIFY domain (A) and Jas domain (B) in Arabidopsis and tuber mustard. The sequence logos for the TIFY domain and Jas domain were generated with WebLogo 3.
Analysis of transcriptional expression patterns of BjJAZs in different tissues
To investigate the gene expression patterns of BjJAZs, different tissues (root, stem, leaf, flower and pod) of tuber mustard were collected and qRT-PCR assays were performed to detect the expression levels of each BjJAZ gene in different organs. The results showed that the genes BjuB043409, BjuA022138, BjuB029203, BjuB026559, BjuB035964, BjuA041687, BjuB014771 and BjuA001950 were expressed at low levels in most organs, while BjuA005572, BjuB010656, BjuB032915, BjuB029529, BjuA027422, BjuA022588, BjuO008948 were expressed at high levels in roots; BjuB029798, BjuA005572, BjuB011370, BjuB043343, BjuB010656, BjuB030369, BjuA027422 were highly expressed in stems; and BjuB029798, BjuA005572, BjuB043343, BjuB010656, BjuB029529, BjuA034780, BjuB030369, BjuA027422 were highly expressed in leaves. All the BjJAZ genes except for BjuA005572, BjuB043343, BjuB010656 and BjuA027422 were expressed at low levels in flowers (except for BjuB043343 and BjuB010656) and pods (except for BjuA005572 and BjuA027422). BjuA030800, BjuA022588 and BjuA047148 were expressed more strongly in the roots than in the other organs. BjuA027037, BjuB011370 and BjuB030035 showed higher expression levels in stems than in the other organs (Fig 6). BjuB043343 and BjuA007483 showed relatively higher expression levels in leaves than in the other organs (Fig 6). The results suggested that these BjJAZ genes with tissue-specific expression might play roles in the regulation of the relative organs growth and development.
Fig 6. Transcriptional expression patterns of BjJAZs in different tissues of tuber mustard.
The expression patterns of BjJAZ genes were analyzed by qRT-PCR. The Bj18S rRNA gene was used as the internal control. The boxes display the gene expression levels using the 2−ΔΔCT method, and different colors represent the different expression levels of each gene.
Analysis of transcriptional expression patterns of BjJAZs under Plasmodiophora brassicae treatment
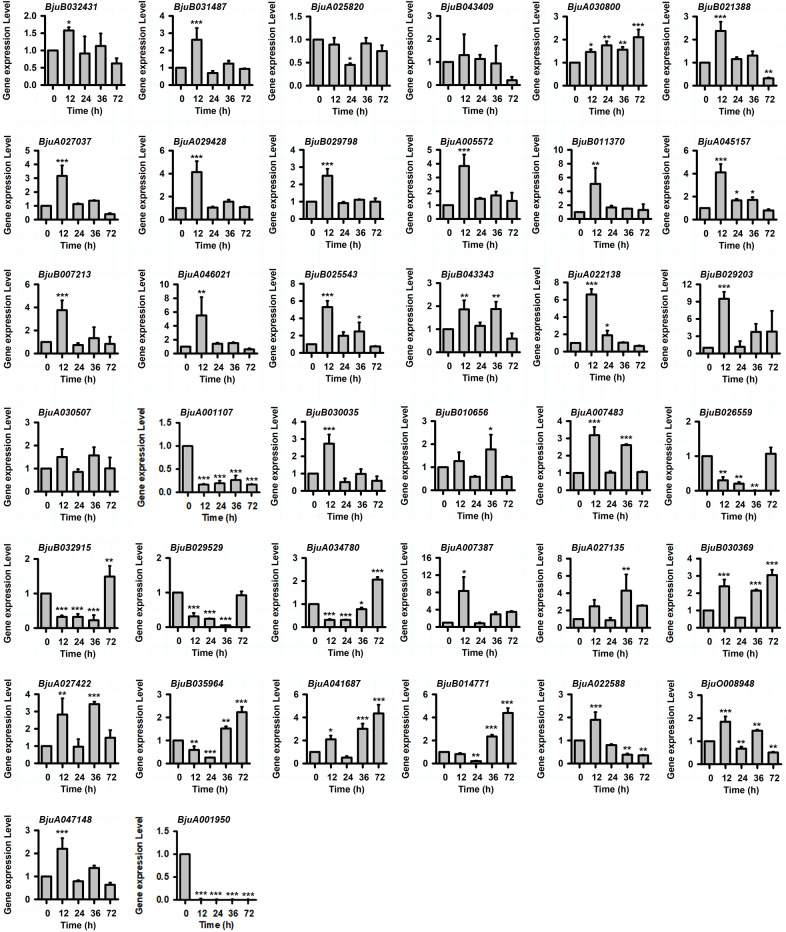
P. brassicae, the main pathogen of tuber mustard, is a severe pathogen that frequently attacks tuber mustard seedlings and causes substantial production losses. To investigate the functions of the BjJAZ genes in plant resistance to the pathogen P. brassicae, the transcriptional expression levels of these genes were assessed in the tuber mustard roots treated with P. brassicae. The results showed that BjuB031487, BjuB021388, BjuA027037, BjuA029428, BjuB029798, BjuA005572, BjuA045157, BjuB007213, BjuB025543, BjuA022138, BjuB030035, BjuA022588 and BjuA047148 were upregulated at 12 h after P. brassicae treatment compared to the control, and then the expression of these genes decreased (Fig 7). BjuA007483 was mainly induced by P. brassicae at 12 h and 36 h, while all three BjJAZ10 group genes were mainly induced at 36 and 72 h (Fig 7). In contrast, the expression levels of BjuB026559, BjuB032915, BjuB029529 and BjuA034780 were significantly decreased after 12, 24, and 36 h of P. brassicae treatment, and the expression of these genes decreased at 72 h. BjuA001107 and BjuA001950 were significantly repressed by P. brassicae from 12 to 72 h. These results indicate that these genes might be involved in the tuber mustard response to P. brassicae stress.
Fig 7. Expression patterns of BjJAZ genes under treatment with the pathogen Plasmodiophora brassicae.
The expression patterns of BjJAZ homologous genes were analyzed by qRT-PCR. The Bj18S rRNA gene was used as the internal control. Three independent biological repeats were performed, and all data points are the means of three biological replicates ± standard errors (SEs). Significant differences: *, p < 0.05.; **, p < 0.01.; ***, and p < 0.001.
Analysis of transcriptional expression patterns of BjJAZs under salt-stress treatment
Tuber mustard is frequently attacked by salt stress during plant growth in the field. To identify the BjJAZ genes involved in the plant response to salt stress, 2‐week‐old seedlings were treated with 200 mM NaCl for 0, 6, 12, 24 and 48 h, and the expression levels of the BjJAZ family genes were evaluated by qRT-PCR. The results showed that nearly no BjJAZ gene was downregulated by salt-stress treatment; in contrast, the expression levels of some BjJAZ genes increased at different time points after 200 mM NaCl treatment. The genes BjuB032431, BjuB007213 and BjuB043343 were continuously induced from 6 to 24 h; BjuB031487, BjuB011370, BjuA030507, BjuB010656, BjuA007483, BjuB032915 and BjuB035964 were mainly upregulated at 12 and 24 h. Some genes were mainly induced by salt stress at 24 h after treatment, such as BjuA029428, BjuB029798, BjuA005572, BjuB025543, BjuB029529, BjuA034780 and BjuA027422 (Fig 8). In contrast, the expression level of BjuA001107 was significantly decreased at 12 to 24 h, and the expression of BjuA001950 was decreased at 24 h. The results indicate that these genes might play roles in the tuber mustard response to salt stress.
Fig 8. Expression patterns of BjJAZ genes under salt stress treatment.
The expression patterns of BjJAZ homologous genes were analyzed by qRT-PCR. Bj18S rRNA gene was used as the internal control. Three independent biological repeats were performed, and all data points were the means of three biological replicates ± standard error (SE). Significant differences: *, p < 0.05.; **, p < 0.01.; and ***, p < 0.001.
Discussion
Tuber mustard is the raw material of Fuling pickle, which has great economic value. The mechanisms involved in the regulation of tuber mustard growth and development and responses to biotic and abiotic stress remain to be further investigated. Although it has been reported that some JAZ genes play roles in plant growth, development and responses to abiotic stress [25,26], the identities and functions of tuber mustard JAZ genes remain unknown. Here, we identified the BjJAZ genes in the whole genome of tuber mustard and investigated their expression patterns during growth and development and their possible roles in plant responses to salt and pathogen stress.
According to the results of gene sequence identity and protein domain conservation analyses, 38 BjJAZ genes were identified in the genome of tuber mustard, and the number of BjJAZs is more than double that in Arabidopsis (13), rice (15) and Brachypodium distachyon (15) [27–29]. The reason might be that tuber mustard is an allotetraploid species formed by hybridization between the diploid ancestors of Brassica rapa and Brassica nigra with subsequent genome duplication [30]; genome duplication is also found in moso bamboo, the genome of which contains 18 PeJAZ genes [31]. Gene duplication was also found among JAZ genes in apple, the genome of which contained 18 MdJAZ genes [32]. In a previous study, the origin and evolution of TIFY transcription factors were investigated using 14 genomes from all kingdoms of life, and the results indicated that whole genome duplication and tandem duplication contributed to the expansion of TIFY family genes [33]. However, no tandem duplication pairs were found in 38 BjJAZ genes. Whole genome duplication might be the main factors resulting in the expansion of BjJAZ family genes.
Exon/intron structural divergence plays important roles in the evolution of multiple gene families. Most of the BjJAZ genes have the same number of exons comparing with the relative JAZ genes in Arabidopsis. However, the divergence of exon number was also found among BjJAZ and AtJAZ genes. For example, although five BjJAZ genes (BjuA025820, BjuB032431, BjuB031487, BjuB043409, BjuA030800) and AtJAZ1 formed a single orthologous cluster (S3 File), they had different number of exons: 4 exons were found in the five BjJAZ genes, but only 2 in AtJAZ1 (Fig 3). The exon/intron gene structure reflected the diversification and evolution of these genes through gain/loss and insertion/deletions, and the diversity of exon/intron structure between different groups may be related to their functional evolution. JAZ family proteins are defined by the highly conserved TIFY domain and Jas domain. However, the recently identified JAZ13 lacks a TIFY domain. It was reported that the Arabidopsis JAZ13 protein together with its orthologs in Brassicaceae species contained a “NAFXXG” motif in place of the highly conserved “TIF(F/Y)XG” motif [28]. Similar to the JAZ13 protein in Arabidopsis, BjuA001950 also contains a divergent “NAFYSG” sequence instead of a canonical TIFY motif (S1 Fig). In Arabidopsis, the EAR motif specifically exists in JAZ5, JAZ6, JAZ7 and JAZ8 [34]. In tuber mustard, EAR motifs were also found insome proteins of BjJAZ1/2/5/6 and BjJAZ7/8/13 group (Fig 4). Taken together, the results show that the common motifs are shared by tuber mustard and Arabidopsis, suggesting the conserved evolutionary processes between the two species.
Gene expression patterns often have a connection with gene function, and some BjJAZ genes with tissue-specific expression were identified in tuber mustard, which suggested their roles in specific tissues. In wheat, TaJAZ5 was observably highly expressed in leaf tissues, and TaJAZ6 was specifically expressed in the roots [35]. In contrast, BjuA030800 and BjuA022588 were expressed more strongly in the roots than in the other organs, suggesting the different roles of these genes in the two species (Fig 6).
P. brassicae is the main pathogen during tuber mustard growth and development, and a number of BjJAZ genes are induced by this pathogen, suggesting the common roles of JA signaling in plant responses to this pathogen. The roles of JAZ genes in plant resistance to pathogens have been reported in other crops. For example, transgenic lines overexpressing apple MdJAZ2 were less susceptible to P. syringae pv. tomato DC3000 than wild-type seedlings of Arabidopsis [36]. In Arabidopsis, all AtJAZ genes, except for AtJAZ3, AtJAZ4, AtJAZ11 and AtJAZ12, were induced by infection with the pathogen P. syringae DC3000 from 24 to 48 h [14]. In accordance with this finding, most BjJAZ genes were induced by treatment with the pathogen P. brassicae (Fig 7), suggesting similar roles in the plant response to pathogen attacks. However, four BjJAZ genes (BjuB026559, BjuB032915, BjuB029529, BjuA034780) were significantly repressed by treatment with this pathogen from 12 to 36 h (Fig 7), indicating their different roles in the tuber mustard response to P. brassicae. In rice, although the expression level of OsJAZ8 was stable after the seedlings were treated with the pathogen X. oryzae pv. oryzae (Xoo), the overexpression of Jas domain-truncated OsJAZ8 confirmed that this gene negatively regulated JA-induced resistance to Xoo in rice [37]. In tuber mustard, BjuA034780 and BjuB029529 were identified as co-orthologs of AtJAZ8 (S3 File), and they were significantly repressed by the pathogen P. brassicae, which merits further study.
Salinity is the main abiotic stress factor for tuber mustard in the planting area. According to the expression patterns of BjJAZ genes under salt-stress treatment, the salt stress responsive BjJAZ genes might play roles in tuber mustard tolerance to salinity. Similarly, in Gossypium hirsutum, 14 of 30 GhJAZ genes were significantly induced by salt-stress treatment from 3 to 6 h [15]. In Glycine soja, the transcriptional level of GsJAZ2 increased from 1 to 6 h of 200 mM NaCl treatment in the roots, and overexpression of GsJAZ2 enhanced the salt tolerance of transgenic Arabidopsis seedlings [38]. In addition, overexpression of the apple MdJAZ2 gene also improved plant tolerance to NaCl stress treatment in transgenic Arabidopsis [36]. In tuber mustard, three AtJAZ2 orthologous genes (BjuA029428, BjuA005572, BjuB011370) were significantly induced by 200 mM NaCl (Fig 8, S3 File), suggesting similar roles for these genes in plant tolerance to salt stress. In wheat, several TaJAZ genes, such as TaJAZ1, TaJAZ2, TaJAZ5, TaJAZ9 and TaJAZ10, were induced by high-salinity treatment [35]. The qRT-PCR results also showed that the expression of a number of BjJAZ1/2/5/6 and BjJAZ10 group genes was induced by salt treatment, indicating that the functions of the JAZ genes in the plant response to salinity are probably conserved between the two species. In moss, the expression level of PnJAZ1 was increased after NaCl treatment, and both the transgenic lines of Arabidopsis and Physcomitrella overexpressing PnJAZ1 showed increased tolerance to salt stress [39], suggesting the conserved function of JAZ1 in the plant response to salt stress in lower and higher plant species. Some divergent expression patterns of JAZ genes were also found in other species. For example, in Solanum lycopersicum, SlJAZ3 and SlJAZ10 were consistently induced by salt treatment in leaves but downregulated in roots [40]. The different expression patterns of JAZ-orthologous genes in different species indicated the complex roles of JAZ proteins in the regulation of JA signaling. Further study of the detailed functions of biotic and abiotic stress-responsive BjJAZ genes will shed light on the mechanism of resistance of tuber mustard to these forms of stress.
Supporting information
(DOCX)
(TIF)
(TIF)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work is supported by the National Natural Science Foundation of China (Program Nos. 31701451, 31770407 and 31800311), Zhaoming Cai, Diandong Wang and Jingjing Liao; the Chongqing Basic Research and Frontier Exploration Project (Program No.cstc2019jcyj-msxmX0285), Zhaoming Cai; the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN201901406), Zhaoming Cai. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carvalhais LC, Schenk PM, Dennis PG. Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 2017, 37, 42–47. 10.1016/j.mib.2017.03.009 [DOI] [PubMed] [Google Scholar]
- 2.Huang H, Liu B, Liu L, Song S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. 10.1093/jxb/erw495 [DOI] [PubMed] [Google Scholar]
- 3.Niwa T, Suzuki T, Takebayashi Y, Ishiguro R, Higashiyama T, et al. Jasmonic acid facilitates flower opening and floral organ development through the upregulated expression of SlMYB21 transcription factor in tomato. Biosci. Biotechnol. Biochem. 2018, 82, 292–303. 10.1080/09168451.2017.1422107 [DOI] [PubMed] [Google Scholar]
- 4.Qin J, Wu M, Liu H, Gao Y, Ren A. Endophyte infection and methyl jasmonate treatment increased the resistance of Achnatherum sibiricum to insect herbivores independently. Toxins 2018, 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu L, Yang H, Ren L, Chen W, Liu L, et al. Jasmonic acid-mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Front. Plant Sci. 2018, 9, 750 10.3389/fpls.2018.00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Dong W, Zhang N, Ai X, Wang M, et al. A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 2014, 164, 1068–1076. 10.1104/pp.113.227595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. 10.1038/nature09430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. 10.1038/nature05960 [DOI] [PubMed] [Google Scholar]
- 9.Ruan J, Zhou Y, Zhou M, Yan J, Khurshid M, et al. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, E2479 10.3390/ijms20102479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. 10.1111/j.1365-313X.2009.03852.x [DOI] [PubMed] [Google Scholar]
- 11.Jing Y, Liu J, Liu P, Ming D, Sun J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 2019, 9, 5691 10.1038/s41598-019-42177-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi S, Hosokawa-Shinonaga Y, Tamaoki D, Yamada S, Akimitsu K, et al. Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 2014, 37, 451–461. 10.1111/pce.12169 [DOI] [PubMed] [Google Scholar]
- 13.Xia W, Yu H, Cao P, Luo J, Wang N. Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. Front. Plant Sci. 2017, 8, 493 10.3389/fpls.2017.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demianski AJ, Chung KM, Kunkel BN. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol. Plant Pathol. 2012, 13, 46–57. 10.1111/j.1364-3703.2011.00727.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H, Chen L, Li J, Hu M, Ullah A, et al. The JASMONATE ZIM-Domain gene family mediates JA signaling and stress response in cotton. Plant Cell Physiol. 2017, 58, 2139–2154. 10.1093/pcp/pcx148 [DOI] [PubMed] [Google Scholar]
- 16.Peethambaran PK, Glenz R, Höninger S, Shahinul Islam SM, Hummel S, et al. Salt-inducible expression of OsJAZ8 improves resilience against salt-stress. BMC Plant Biol. 2018, 18, 311 10.1186/s12870-018-1521-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finn RD, Alex B, Jody C, Penelope C, Eberhardt RY, et al. Pfam: the protein families database. Nucleic Acids Res. 2014, 42, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu R, Meng J. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 2003, 25, 317–321. [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu L, Dong Z, Fang L, Luo Y, Wei Z, et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. 10.1093/nar/gkz333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, et al. Basic local alignment search tool (BLAST). J. Mol. Bio. 1990, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 22.Kong X, Lv W, Jiang S, Zhang D, Cai G, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics. 2013, 14, 433–433. 10.1186/1471-2164-14-433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. 10.1038/35080508 [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL, Bodén M, Buske FA, Frith M, Grant CE, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song S, Qi T, Huang H, Ren Q, Wu D, et al. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011, 23, 1000–1013. 10.1105/tpc.111.083089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS. Jasmonate ZIM-Domain (JAZ) protein regulates host and nonhost pathogen-induced cell death in tomato and Nicotiana benthamiana. PLoS ONE 2013, 8, e75728 10.1371/journal.pone.0075728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye H, Du H, Tang N, Li X, Xiong L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. 10.1007/s11103-009-9524-8 [DOI] [PubMed] [Google Scholar]
- 28.Thireault C, Shyu C, Yoshida Y, St Aubin B, Campos ML, et al. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. 10.1111/tpj.12841 [DOI] [PubMed] [Google Scholar]
- 29.Zhang L, You J, Chan Z. Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 2015, 128, 995–1005. 10.1007/s10265-015-0755-2 [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Liu D, Wang X, Ji C, Cheng F, et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1234. 10.1038/ng.3657 [DOI] [PubMed] [Google Scholar]
- 31.Huang Z, Jin S, Guo H, Zhong X, He J, et al. Genome-wide identification and characterization of TIFY family genes in Moso Bamboo (Phyllostachys edulis) and expression profiling analysis under dehydration and cold stresses. PeerJ 2016, 4, e2620 10.7717/peerj.2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Yin X, Wang H, Li J, Guo C, et al. Genome-wide identification and analysis of the apple (Malus×domestica Borkh.) TIFY gene family. Tree Genet. Genomes. 2015, 11, 808. [Google Scholar]
- 33.Bai Y, Meng Y, Huang D, Qi Y, Chen M. Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 2011, 98, 0–136. [DOI] [PubMed] [Google Scholar]
- 34.Garrido-Bigotes A, Valenzuela-Riffo F, Figueroa CR. Evolutionary analysis of JAZ proteins in plants: an approach in search of the ancestral sequence. Int. J. Mol. Sci. 2019, 20, E5060 10.3390/ijms20205060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Qiao L, Bai J, Wang P, Duan W, et al. Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.). BMC Genomics 2017, 18, 152 10.1186/s12864-017-3582-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An X, Hao Y, Li E, Xu K, Cheng C. Functional identification of apple MdJAZ2 in Arabidopsis with reduced JA-sensitivity and increased stress tolerance. Plant Cell Rep. 2016, 36, 1–11. 10.1007/s00299-016-2073-0 [DOI] [PubMed] [Google Scholar]
- 37.Yamada S, Kano A, Tamaoki D, Miyamoto A, Shishido H, et al. Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 2012, 53, 2060–2072. 10.1093/pcp/pcs145 [DOI] [PubMed] [Google Scholar]
- 38.Zhu D, Cai H, Luo X, Bai X, Deyholos MK, et al. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012, 426, 273–279. 10.1016/j.bbrc.2012.08.086 [DOI] [PubMed] [Google Scholar]
- 39.Liu S, Zhang P, Li C, Xia G. The moss jasmonate ZIM-domain protein PnJAZ1 confers salinity tolerance via crosstalk with the abscisic acid signalling pathway. Plant Sci. 2019, 280, 1–11. 10.1016/j.plantsci.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 40.Chini A, Ben-Romdhane W, Hassairi A, Aboul-Soud MAM. Identification of TIFY/JAZ family genes in Solanum lycopersicum and their regulation in response to abiotic stresses. PLoS ONE 2017, 12, e0177381 10.1371/journal.pone.0177381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
(TIF)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.