Abstract
Traditionally, phenotypic plasticity in adult somatic cells has been thought of as dedifferentiation and transdifferentiation in the context of tissue regeneration or wound healing. Although the dedifferentiation process is central to tissue repair and stemness, dedifferentiation inherently carries the risk of cancer initiation. As such, phenotypic plasticity presents a new paradigm for understanding cancer initiation, progression, and resistance to therapy. Here we discuss the general concept that, when cells exhibit plasticity, they converge on signaling processes that induce and maintain cellular dedifferentiation. Activation of these programs in turn enables the initiation and progression of carcinogenesis and underlies resistance to therapy.
Introduction
All stem cells are defined by the key properties of self-renewal (the ability to generate more of themselves) and multipotency (the ability to divide asymmetrically and generate more differentiated progeny) (reviewed in Reya et al., 2001). Two general classes of stem cells exist: embryonic stem cells (ESCs) and adult tissue stem cells. ESCs are pluripotent, and they can produce all cell types in the body. ESCs are present only during early stages of embryogenesis. Conversely, tissue stem cells have a more restricted potential, and they can produce only a limited number of cell types. However, tissue stem cells persist throughout adult life in organs that continually or periodically regenerate, such as the skin, intestine, mammary gland, and the hematopoietic system. Because of their long life, tissue stem cells have an enhanced potential to acquire the necessary oncogenic hits for tumor formation, and they are the suspected cells of origin for many cancers, including breast cancer (Visvader, 2011).
Development from a fertilized egg to a mature organism is thought to proceed in a fundamentally hierarchical manner (Marjanovic et al., 2013). Each stem cell asymmetric division produces a progressively more differentiated cell type, beginning with the zygote and ending with all of the terminally differentiated cells of the body. At the branch points of the hierarchy are stem cells and/or multipotent progenitor cells, which, during asymmetric division, generate lineage-committed progeny that no longer possess self-renewal (also termed transit amplifying cells). In most tissues, the progeny cells eventually give rise to post-mitotic, terminally differentiated cell types. The classic and best-studied example of a developmental hierarchy is the hematopoietic system (Reya et al., 2001). Long-term hematopoietic stem cells reside in the bone marrow and generate transit-amplifying progenitors and progressively more differentiated cell types, including lymphocytic and myelocytic cells. The strength of the hematopoietic paradigm has influenced the belief that solid tissues are similarly organized.
However, certain phenomena have challenged the concept of differentiation as a permanent or unidirectional process. These phenomena suggest that many ‘terminally differentiated’ cells retain the potential to change fate. Here, we use the term ‘plasticity’ to refer generally to a broad set of such phenomena including dedifferentiation (the loss of lineage commitment and reacquisition of stem cell features) and transdifferentiation (direct fate switching to another differentiated cell type) (Bonfanti et al., 2012).
Plasticity has a long history. The early literature often described dedifferentiation and transdifferentiation in the context of regeneration or wound healing. A well-described example of transdifferentiation is the regeneration of the amphibian retina by pigment epithelial cells that specifically respond to tissue damage (Okada, 1980). Similarly, as Godlewski first reported in 1928 (Godlewski, 1928) dedifferentiation of epidermal cells to generate chondrocytes and skeletal muscle cells occurs in the regenerating axolotl limb (Rose, 1947). However, generally, these observations were limited to ‘lower’ vertebrates such as amphibians, which have a capacity for tissue regeneration far exceeding that of mammals. Recently, however, it has become clear that mammalian cells can also be induced to dedifferentiate or transdifferentiate (Figure 1). Typically, investigators achieve ‘reprogramming’ of mammalian cells by introducing one or more transcription factors (TFs) into a differentiated cell type. Davis et al. performed the earliest example of this type of reprogramming with MyoD, which induced conversion to myoblasts when ectopically expressed in fibroblasts (Davis et al.,1987). Then came the seminal discovery that a combination of four transcription factors, OCT4, SOX2, KLF4, and MYC (OSKM), could ‘reprogram’ adult human or mouse fibroblasts to an embryonic stem-like state (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). The reality of induced pluripotency has led to a extensive re-evaluation of the permanence of the differentiated state. Lately, investigators have demonstrated that fibroblasts and other cell types could be transdifferentiated or “directly reprogrammed” to cardiomyocytes, neurons, and pancreatic neuroendocrine cells, among other cell types (Zhou et al., 2008; Vierbuchen et al., 2010; Szabo et al., 2010; Ieda et al., 2010; Efe et al., 2011; Kim et al., 2011, Tanabe et al., 2018).
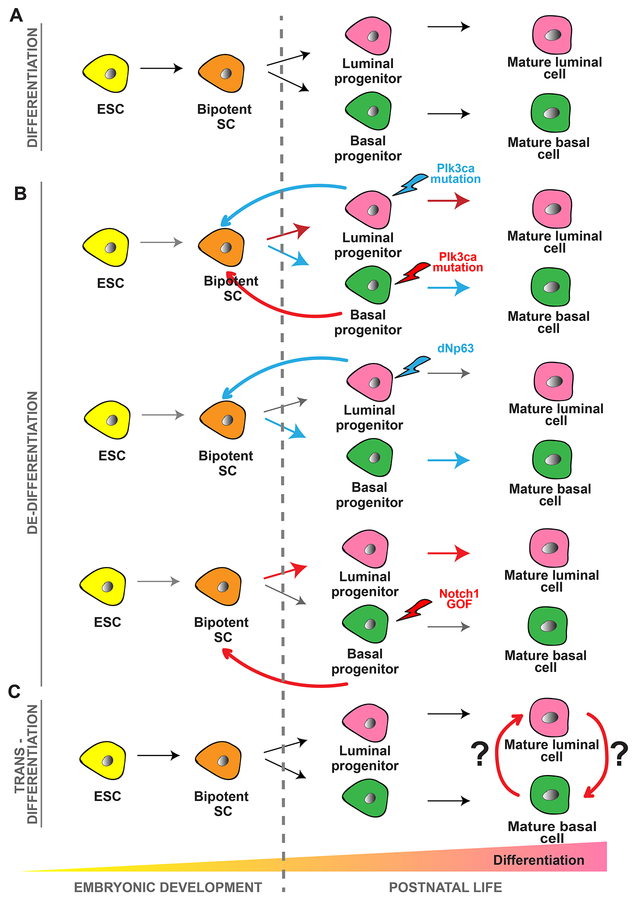
Figure 1. Types of Differentiation that Are Induced during Cellular Plasticity.
Types of epithelial differentiation and plasticity seen in the mammary gland and how it relates to more primitive states of multipotency seen during embryonic development.
All of these examples involved transient or permanent expression of one or more transcription factor in the original cell type, which appeared to transition into a different cell type without proceeding through an intermediate multipotent stage. These studies proved that differentiation states are changeable, metastable entities, and the studies showed that specific transcription factors could shift cells from one state to another.
It is useful to distinguish plasticity induced by forced expression of transcription factors, sometimes termed ‘intrinsic plasticity’, from plasticity induced by changes in the microenvironment, termed ‘extrinsic plasticity’ (Bonfanti et al., 2012; Marjanovic et al., 2013). The strongest evidence for extrinsically triggered dedifferentiation comes from recent lineage-tracing studies in diverse settings such as the lung (Tata et al., 2013) and hair follicle (Rompolas et al., 2013). Investigators have definitively mapped the fates of differentiated cells and their progeny with genetic markers following ablation of a particular cell population within the tissue. In both cases, the non-ablated, differentiated cell populations underwent facultative dedifferentiation to regenerate the ablated cells. In addition, extrinsic cues and certain pathologic states may trigger transdifferentiation. For instance, in a mouse model of calcifying atherosclerosis, adoption of an osteogenic or chondrogenic phenotype by vascular smooth muscle cells preceded calcification of the vessel intima (Speer et al., 2009). Therefore, plasticity has a regenerative function in vivo. In some of these cases, the induction or expression of certain TFs regulates the switch between hierarchy and plasticity.
Plasticity may also be triggered artificially by experimental manipulation. Ex vivo cell culture often fails to recapitulate most aspects of the tissue microenvironment, and such cell culture often results in dedifferentiation. In 2D cultures, mammary epithelial cells (MECs) stochastically acquire stem-like traits during short-term culture (Chaffer et al., 2011; Keller et al., 2012), and long-term MEC culture causes widespread epigenetic changes and the adoption of an uncommitted ectodermal stem cell phenotype (Holst et al., 2003, Keller et al., 2012; Breindel et al., 2017). However, culturing MECs within 3D matrices that recapitulate the biological and mechanical properties of in vivo tissue preserves lineage identity and functionality ex vivo (Sokol et al., 2016). Similarly, articular chondrocytes growing in monolayer culture lose the ability to express cartilage proteins, but this behavior can be reversed if the chondrocytes are grown in soft agar, which is more mechanically similar to cartilage (Benya and Shaffer, 1982). These findings underscore the importance of instructive structural inputs that determine cellular differentiation potential.
Transplanting cells from their native microenvironment to a different site in vivo can also trigger dedifferentiation or transdifferentiation because of inductive signals present in the recipient tissues. For example, Bonfanti et al. showed that thymic epithelial cells could generate hair follicle multipotent stem cells when transplanted into the inductive microenvironment of the dermis (Bonfanti et al.,2012). Again, however, the molecular signals operative in these de- or trans-differentiation processes are not clear in these and many other instances.
In this review, we discuss the role of phenotypic plasticity during cancer initiation, progression, and resistance to therapy, and we review the relevant factors that dictate the switch from hierarchy to plasticity in normal tissues and in cancer.
Plasticity and the origins of cancer
The cell of origin (also referred to as the tumor precursor cell or the tumor-initiating cell) refers to the original cell that receives the first oncogenic hits and undergoes clonal expansion in the earliest stage of tumor progression. The identity of the cell of origin can have a substantial impact on the behavior and progression of the resulting tumor because, in many cases, the characteristics of the tumor precursor cell are passed on epigenetically to the tumor cells (Gupta et al, 2005; Ince et al 2007). Conversely, the characteristics of the tumor cell of origin are not necessarily equivalent or even similar to the characteristics of the cancer stem cell (CSC) (Visvader, 2011). Moreover, although in many breast tumors the cell of origin is suspected to be a long-lived tissue stem cell, this supposition is not universally true. Even when the cell of origin is a stem cell, it is by no means guaranteed that the resulting cancer cells will resemble their original precursor or that the stem cell program will survive neoplastic transformation intact. Therefore, CSCs, tissue stem cells, and cells of origin are distinct concepts.
Identifying the cell of origin seems straightforward in principle, but identification can be quite challenging to accomplish experimentally because 1) transformation of the original precursor cell cannot usually be observed directly, and 2) the influence of the cell of origin on the tumor phenotype is not always overt. To identify the cell of origin in breast cancer, investigators have used two main approaches. The first approach involves isolating normal cell subsets by FACS and either comparing them to the tumor subtypes or using lentiviral vectors to transduce these cells ex vivo with a combination of oncogenes that will lead to tumorigenesis. Interestingly these studies revealed that the global gene expression profiles of basal-like tumors were most similar to the luminal progenitor profile in normal tissues (Lim et al., 2009). Further, transformation of luminal progenitor cells led to tumors with both luminal and basal features (Keller et al., 2012). In contrast, transformation of human cells with an EpCAMlow/CD49fhigh immunophenotype, thought to contain basal/ME, stem and/or bipotent progenitor cells, gave rise to aggressive tumors with squamous differentiation and other metaplastic features (Keller et al., 2012). These tumors were molecularly most similar to the claudin-low intrinsic subtype, which displays high expression of MaSC-associated genes and mesenchymal markers. Metaplastic breast cancer is rare in humans; therefore, these tumors may represent the rare transformation of basal/ME progenitors or stem cells (Prat and Perou, 2011).
A complementary approach is to direct conditional expression of oncogenes (or deletion of tumor suppressor genes) to specific mammary epithelial subpopulations to initiate tumorigenesis in a defined cell population. Molyneux et al. employed a mouse model in which loss of the BRCA1 tumor suppressor was targeted to either KR14-expressing basal/ME or to β-lactoglobulin (Blg)-expressing luminal cells on a p53-heterozygous background (Molyneux et al., 2010). This approach revealed that targeting BRCA1 loss to luminal cells recapitulated the basal-like phenotype of human BRCA1-associated breast tumors. KR14-driven BRCA1 loss also led to tumor formation; however, histology was that of malignant adenomyoepithelioma, which is not usually seen in BRCA1-associated human cancer.
Together, these studies enshrine progenitor cells as the likely cells of origin, but recent findings have demonstrated that plasticity is relevant to understanding the origins of tumors and their heterogeneity. Solid cancers are highly diverse, exhibiting heterogeneity both between different tumors (intertumor heterogeneity) and between cells within a single tumor (intratumor heterogeneity). It is becoming clear that tumors reactivate and/or hijack developmental differentiation programs of the tissues in which they originate as part of the mechanism by which tumor diversity is generated.
To evaluate plasticity during tumor initiation in breast cancer, investigators have used a genetic approach (Van Keymeulen et al., 2015). Investigators activated the oncogenic PIK3CA mutation, with or without p53 deletion, using K5CreER in basal cells of the mammary gland and K8CreER in luminal cells. Surprisingly, activation of PIK3CA mutation in basal cells induced the formation of luminal oestrogen receptor (ER)/progesterone receptor (PR)-positive tumors, whereas its expression in luminal cells gave rise to luminal ER+PR+ tumors or basal-like ER-PR-tumors. Interestingly, oncogenic PIK3CA mutation activated a multipotent genetic program in normally lineage-restricted populations at the early stage of tumor initiation, influencing future tumor heterogeneity (Van Keymeulen et al., 2015,). Similar observations were made in BRCA1-associated hereditary breast tissues. Recent work with mice and humans demonstrated that lineage restriction is dysregulated in preneoplastic BRCA1 cells and tissues, in which there is an overexpansion of luminal progenitor cells that fail to differentiate and aberrantly express basal epithelial cell markers (Lim et al., 2009; Molyneux et al., 2010; Proia et al., 2011). The cause of this defect appears to be aberrantly increased protein stability of the EMT TF Slug in the BRCA1 tissues. In normal tissues, Slug represses luminal differentiation in basal cells, and it is important for the mammary stem cell phenotype (Proia et al., 2011; Guo et al., 2012; Nassour et al., 2012; Phillips et al., 2014). In BRCA1 mutant tissues, however, Slug is aberrantly stabilized, and it accumulates in luminal cells, a phenomenon that likely explains why the tumors are basal-like (Proia et al., 2011) (Figure 2).
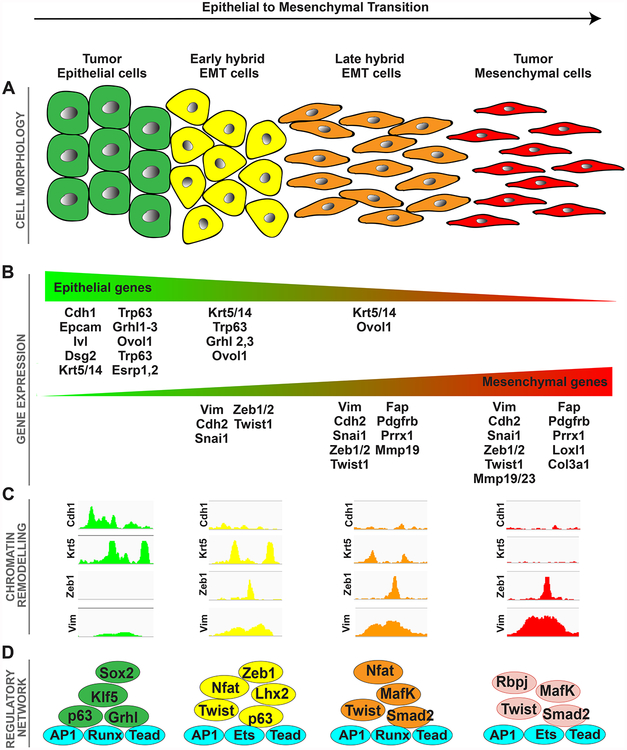
Figure 2. Tumor Transition States Occurring during EMT.
(A–D) Changes in cell morphology (A), gene expression (B), chromatin remodeling (C), and transcription factors (D) involved in the regulation of different tumor transition states occurring during EMT.
Phenotypic plasticity during tumor initiation is also driven by activation of the developmental differentiation program--the epithelial-to-mesenchymal transition (EMT) (Figure 2). EMT is the process by which cells acquire plasticity and gain the properties of stem cells. In EMT, cells of a differentiated epithelial phenotype lose apicobasal polarity, become motile, and express markers characteristic of mesenchymal cells (Thiery et al., 2009). EMT is intimately linked with an undifferentiated or stem-like state, including the capacity for extended self-renewal and the acquisition of a stem-like gene expression program (Mani et al., 2008, Morel et al., 2008). However, not all epithelial tumors activate EMT programs with the same frequency, and the dedifferentiation process that takes place leads to re-expression of primitive cell transcriptional programs and cellular metaplasia. In addition, although acquisition of metaplastic and mesenchymal traits is a prominent feature of some cancers, those traits are rarely observed in other cancers, a circumstance that may reflect intrinsic properties of their cells of origin. Recently, Latil et al. used a genetic model of skin cancer in which the oncogenic Kras mutation was activated with simultaneous deletion of p53. Combined with lineage tracing, the investigators showed that skin squamous cell carcinomas (SCCs) were derived from interfollicular epidermis (IFE). IFE (K14CreER) displayed a well differentiated phenotype, whereas skin SCC derived from hair follicle (HF) stem cells (Lgr5CreER) gave rise to tumors with a wide range of EMT, from well differentiated to totally mesenchymal or sarcoma-like tumors with increased metastatic potential (Latil et al., 2017). Interestingly, transcriptional and epigenomic profiling revealed that IFE and HF tumor-initiating cells possessed distinct chromatin landscapes and gene regulatory networks. Thus, this profiling demonstrated, for the first time, that accessibility of key epithelial and mesenchymal TF in the cancer cell of origin primes and dictates the tumor phenotype and EMT (Latil et al., 2017).
Plasticity and tumor progression/metastasis
The EMT is the mostly widely studied example of phenotypic plasticity, and its role in tumor progression and metastasis is well established. Metastasis is responsible for most cancer patient deaths (Lambert et al., 2017). When tumors spread to distant sites, life expectancy decreases significantly, and, despite important advances, treatment options are limited for patients with metastatic disease. To successfully form metastasis, tumor cells should acquire certain plasticity, thus enabling the invasion of the underlying mesenchyme, intravasation into the blood circulation, and, finally, extravasation and colonization of distant organs (Lambert et al., 2017). The hypothesis that EMT and the reverse process, mesenchymal to epithelial transition (MET), promote the invasion-metastasis cascade has been accepted for over a decade (Brabletz et al., 2018). However, recent studies have challenged the indispensability of full mesenchymal transition in the metastatic process (Fischer et al., 2015; Zheng et al., 2015). The concept of hybrid epithelial/mesenchymal phenotype has acquired increasing importance for our understanding of the EMT process and its implications for metastasis (Jolly et al., 2015; Jolly et al., 2016; Nieto et al., 2016).
Recently, investigators have identified several transition states occurring during EMT in skin squamous cell carcinoma and in mammary tumors (Pastushenko et al., 2018). The different tumor cell subpopulations associated with different EMT stages from epithelial to completely mesenchymal states, passing through intermediate hybrid states, presented similar tumor propagating cell capacity. However, the tumor cell subpopulations displayed different cell plasticity and invasiveness. Intravenous injection of different subpopulations revealed a strong increase in metastatic potential of early hybrid EMT states. The quantification of YFP+ circulating tumor cells (CTCs) confirmed this observation: the vast majority of CTCs exhibited EpCAM-CD106-CD51-CD61 phenotype that was associated with co-expression of both epithelial and mesenchymal markers. Interestingly, all tumor cells independently of their degree of EMT could revert to the epithelial state. However, the increase in metastatic capacity of the hybrid states did not correlate with the ability of tumor cells to undergo MET. Thus, other mechanisms beside MET contribute to the higher metastatic potential of these hybrid epithelial/mesenchymal populations (Pastushenko et al., 2018).
In a pancreatic cancer model, driven by PDX-cre-mediated activation of mutant KRas and p53, Zeb1 was a key factor for phenotypic plasticity, formation of precursor lesions, invasion, and, notably, metastasis. In this model, depletion of Zeb1 suppressed stemness, colonization capacity, and, particularly, phenotypic/metabolic plasticity of pancreatic tumor cells (Krebs et al., 2017). In a mouse model of breast cancer, 6% of the tumors expressed Twist1, and most of the Twist1+ cells coexpressed several other EMT TFs (Snail, Slug, Zeb2), lost ERα and luminal marker K8, and exhibited a partial EMT phenotype (ECadherin+/Vimentin+) (Xu et al., 2017). Interestingly, compared with tumors that expressed Twist1, Twist1 knockout tumor cells had largely decreased the expression of the different EMT-inducing TFs, the frequency of CTCs, and the incidence of lung metastasis (Xu et al., 2017). Snail has also been reported to have a key function in tumor growth, invasion, and metastasis in human breast cancer cell lines (Olmeda et al., 2017), mouse skin carcinoma cells lines (Olmeda et al., 2008), and gastric cancer (Shin et al., 2012), among others. Overexpression of Slug and Snail in head and neck SCC cell lines repressed miR-101, subsequently activating EZH2, and inducing EMT, migration, and invasion of cancer cells (Zheng et al., 2015).
Several lines of evidence suggest that hybrid epithelial/mesenchymal states also exist in human cancers. Tumor cells co-expressing both e-cadherin and vimentin were found in invasive breast cancer (Yamashita et al., 2015). Interestingly, the subset of tumors co-expressing these two markers exhibited the worst disease free survival (DFS) and overall survival (OS) among all breast cancer patients analyzed. We were able to detect different degrees of EMT in lung, breast, and esophagus SCC patient-derived xenografts (PDX), thus demonstrating that EMT is not a binary phenomenon in human cancers (Pastushenko et al., 2018). Computational modeling that considered mutual inhibitory loops between several miRNAs and EMT transcription drivers showed that a hybrid EMT state could potentiate the progress of developmental programs and increase metastatic potential (Jolly et al., 2015, Tian et al., 2013; Nieto et al., 2016).
The presence of tumor cells in the circulation has been associated with metastasis in multiple cancers (Aceto et al., 2015). When analysing the EMT phenotype of CTCs, most studies found an association between the presence of hybrid and mesenchymal CTCs and clinical prognosis (Yu et al., 2013; Wu et al., 2015; Hyun et al., 2016; Lecharpentier et al., 2011; Satelli et al., 2015; Zhao et al., 2017). In hepatocellular carcinoma patients, the presence of hybrid and mesenchymal CTCs correlated with more advanced clinical stages and metastasis (Boral et al., 2017). In breast cancer patients, therapy or disease progression was accompanied by an increase in mesenchymal CTCs (Yu et al., 2013). Breast cancer patients with brain metastasis also exhibited CTCs with a higher EMT score.
Despite existence of a growing body of evidence linking EMT to disease progression, recent evidence supports the notion that a partial cell-state transition without a full EMT is sufficient to drive invasive progression. For example, by upregulating expression of secreted proteases that degrade basement membrane, SMARCE1 is sufficient to drive the invasive progression of early-stage and in situ tumors (Sokol et al. 2017). SMARCE1 upregulates protease expression by forming a SWI/SNF-independent complex with the transcription factor ILF3. This association, which occurs in invasive cells that have undergone a partial EMT, directs the genomic localization of SMARCE1 to genes encoding proteases and other matrix-remodeling factors.
Plasticity, stress and resistance to therapy
The primary cause of adult cancer deaths is metastasis of epithelial tumors that are resistant to therapy. Carcinoma cells acquire both of these critical malignant traits-metastasis and drug resistance – when they undergo de-differentiation. Experimental induction of EMT or de-differentiation in cancer cell lines and mouse models is sufficient to promote invasion and metastasis (Thiery et al., 2009; Mani et al., 2008). De-differentiation is also sufficient to promote resistance to a wide spectrum of chemotherapy drugs; often, de-differentiation increases by ~10-fold the IC50 dose of a chemotherapy drug (Gupta et al., 2009; Thiery et al., 2006). Consistent with these findings in experimental models, in clinical samples, high tumor grade (Polyak et al., 2009), invasiveness (Savanger et al., 2005, Yang et al., 2009), and survival within the circulation (Tester et al., 2000) all correlate to poor response to chemotherapy (Blanco et al., 2002).
Although an increasing number of treatment options exist, in modern cancer medicine, the development of therapeutic resistance is a major challenge and the cause of treatment failure and disease recurrence. The differentiation state of a tumor is a key determinant of therapeutic resistance (Arienti et al., 2013b; Chang et al., 2011a; Haslehurst et al., 2012b; Jiang et al., 2016b; Kurrey et al., 2009; Vitali et al., 2008a; Xu et al., 2015a). Overexpression of certain transcription factors associated with EMT or metaplasia causes resistance to traditional chemotherapy such as radiation and chemotherapy drugs (Dong et al., 2016; Haslehurst et al., 2012b; Kurrey et al., 2009). Conversely, inhibition of transcription factor expression increases therapeutic efficacy of these treatments.
The downstream mechanism responsible for resistance to therapy is related to the multiple mechanisms that control target genes. Radioresistance and chemoresistance are achieved by promoting the acquisition of a de-differentiated state (Kurrey et al., 2009; DelVecchio et al., 2014) by increasing expression of stemness-related genes. This de-differentiated state causes metabolic changes that impair pro-drug activation and drug uptake (Feng et al., 2017; Feng et al., 2014; Del Vecchio et al., 2014). For example, experimental induction of Snail or Twist1 causes constitutively active Perk kinase signaling and activation of its downstream target, NRF2. Nrf2 is a master transcriptional regulator of the antioxidant response, a key mediator of therapy resistance (Feng et al., 2014; Del Vecchio et al., 2014). In addition, overexpression of Slug antagonizes cell death triggered by cancer therapies and promotes cell survival by repressing the pro-apoptotic protein PUMA (Wu et al., 2005a).
Currently, two classes of clinical interventions have been suggested that could prove useful for targeting plasticity in cancer. The first class of intervention would either block or reverse de-differentiation to prevent cancer cells from becoming metastatic and drug-resistant, for example, by neutralizing secreted factors that promote EMT or by inhibiting the expression of transcription factors that induce EMT. The second class would block a signaling pathway used by EMT cells to invade, survive in the circulation, or resist therapy. Although, in principle, both of these EMT-targeting strategies could inhibit tumor malignancy, neither on its own would be toxic to cancer cells. Because these proposed EMT-targeting therapies would lack cancer cell toxicity, the cancer cells might eventually develop resistance.
These considerations suggest that it is important to destroy cancer cells that have undergone an EMT, and not just to block or reverse EMT. Although this goal is attractive, in practice it has been difficult to find chemical compounds that selectively kill cancer cells that have undergone an EMT; on the contrary, almost invariably, such cells are highly resistant to any chemical treatment.
Plasticity and tumor stemness
In established cancers, cancer stem cells or “tumor stemness” is the ability of tumor cells to both self-renew and to produce other cell types that constitute the tumor. Activation of EMT programs has been associated not only with acquisition of mesenchymal traits, but with the expression of stem cell markers and an increased ability to form mammospheres, a property associated with mammary epithelial stem cells (Mani et al., 2008). Investigators have proposed that some properties commonly attributed to CSCs, such as invasiveness and metastatic potential, may be acquired by activation of the EMT program. Indeed, in breast cancer patients, circulating tumor cells (CTCs) commonly express EMT markers, a property that suggests EMT may enable these cells to leave the primary tumor site, intravasate into the vasculature, and travel to distant sites (Aktas et al., 2009).
Stochastic cell-state transitions may also generate cells with the properties of stem cells and/or CSCs. Recently, Chaffer et al. reported that a subpopulation of basal-like mammary epithelial cells retained the capacity to spontaneously generate stem-like cells in vitro, and the same population could generate CSC-like cells following oncogenic transformation (Chaffer et al., 2011). The transformed cells were enriched for CSC markers, and they exhibited enhanced tumorigenicity in xenotransplantation assays. Moreover, similar transitions have been observed in cultured breast cancer cell lines, in which non-CSCs isolated by FACS regenerated the CSC population at a rate that was too rapid to be explained by sorting impurities (Gupta et al., 2011). Because the in vitro tissue culture microenvironment is presumably more or less homogenous, these transitions are more likely to occur randomly instead of in a directed manner. Gupta et al. attempted to model these transitions as a Markov process, in which the cells stochastically transition between luminal-like, basal-like, and stem-like states at characteristic frequencies. Markov modeling accurately predicted the collective cell-state transition behavior of FACS-purified luminal, basal, and stem cells (Gupta et al., 2011). Markovian cell-state transitions may also occur in non-cancerous mammary cells (Phillips et al., 2014). As a caveat, investigators have not yet explored the in vivo prevalence of stochastic transitions between non-CSCs and CSCs in breast cancer. Recently, however, several groups have reported in vivo evidence of stochastic interconversion between CSCs and non-CSCs in other cancer types, including Wnt-driven intestinal tumors (Schwitalla et al., 2013).
Two major types of phenotypic plasticity exist in cancer: initiating plasticity and maintaining plasticity. Initiating plasticity is generated by the influence of the cell of origin and the specific driver mutations that occur during tumorigenesis. These two forces collaborate to generate the tumor phenotypes that are varied even within the same tissue. Conversely, maintaining plasticity is a result of genetic evolution and hierarchical and plastic interconversion between cellular phenotypes. Maintaining plasticity is also problematic from a therapeutic perspective. Plasticity significantly muddles the analysis of tumor phenotype because many common modalities used to study tumors at the genomic and molecular level (such as exome sequencing and microarrays) rely on bulk tissue, and these methods typically cannot resolve heterogeneous or rare subpopulations within a tumor. From a therapeutic standpoint, maintaining plasticity is also problematic because the presence of multiple types of cancer cells within a single tumor vastly increases the chance that a given therapy will fail to kill some of the malignant cells. Hence, great efforts have been taken to understand the origin of cellular diversity within breast and other tumors.
Molecular mechanisms underlying plasticity
Cellular differentiation states are dynamically regulated in normal cells and tissues via the activation or inactivation of specific transcriptional factors. The factors that promote cellular plasticity during development and wound healing overlap with those that generate phenotypic plasticity in cancer because both groups of factors participate in aberrant activation of developmental programs.
Activation of developmental pathways has been detected in epithelial tumors. For instance, Zeb1 and Twist1 have been shown to repress differentiation and behave as oncogenic TFs by activation of MAPK molecular pathways in melanoma (Bedogni et al., 2014). In addition, Zeb1 overexpression promoted a reversible transition to a MITF low / p75 high stem-like and tumorigenic phenotype, resulting in emergence of resistance to MAPK inhibitors in BRAF v600-mutated melanoma cell lines and tumors (Bedogni et al., 2008; Zhang et al., 2012). Expression of Notch receptors is low or undetectable in normal adult melanocytes, whereas Notch receptor expression is increased in human melanoma lesions and melanoma cell lines (Pinnix et al., 2009). Interestingly, a gradually increasing expression pattern of Notch can be observed from nevi, to primary melanoma lesions, to metastatic melanoma. Notch1 activation confers a metastatic phenotype to primary melanoma in vivo, whereas Notch4 has a crucial function in promoting cell proliferation and in regulating an aggressive phenotype of melanoma cell lines (Lin et al., 2016).
Other well-studied mechanisms of plasticity involve master transcription factors (TFs), the Snail, Zeb, and Twist families, that orchestrate transcriptional networks that drive de-differentiation. These TFs mediate sequence-specific interactions with DNA. The SNAIL family of zinc finger transcriptional repressors, of which Snail/SNAI1, Slug/SNAI2, and Smug/SNAI3 are members, are conserved among vertebrate species and have critical functions in various developmental and cellular processes. SNAIL family member functions include, but are not limited to, mesoderm formation, neural crest migration, determination of left-right asymmetry, cell migration, the regulation of cell motility, apoptosis, and cancer initiation/progression (Hemavathy et al., 2000; Inukai et al., 1999; Isaac et al., 1997; Nieto, 2002; Vega et al., 2004).
Slug and Snail both control epigenetic repression of target genes that harbor the E-box consensus CAGGTG motif recognized by the C-terminal zinc-fingers of Slug and Snail (Barrallo-Gimeno and Nieto, 2005b; Cobaleda et al., 2007b; Nieto, 2002a). The evolutionarily conserved SNAG transactivation domain, located in the N-termini of Slug and Snail, recruits epigenetic silencing complexes such as polycomb repressive complex 2 (PRC2) and corepressor Lys-specific demethylase 1 (LSD1). This coupling enables the deposition of repressive histone marks (e.g., H3K4me3) to silence the expression of Snail or Slug target genes (Chiang and Ayyanathan, 2013; Lin et al., 2010; Phillips et al., 2014; Wu et al., 2012; Choi et al., 2015; Barrallo-Gimeno and Nieto, 2005b; Cobaleda et al., 2007a; Nieto, 2002b; Nieto et al., 1994)).
The ZEB family of zinc finger proteins, of which ZEB1 and ZEB2 are members, contains two widely separated and conserved zinc-finger domain clusters with a centrally located homeodomain. This homeodomain is POU-like and does not bind DNA, so it is likely involved in protein-protein interactions. Much like the SNAIL family, the ZEB family of TFs represses transcription by an epigenetic mechanism at specific DNA sequences. The PXDLS motifs in both ZEB1 and ZEB2 recruit epigenetic silencing complexes, such as the CtBP core complex 2 and co RE1 silencing transcription factor (coREST), and this coupling enables the alteration of repressive histone marks to silence the expression of ZEB target genes.
The Twist family (Twist1 and Twist2) is composed of basic helix-loop-helix (bHLH) domain-containing transcription factors. Twist family bHLH proteins regulate expression of target genes by binding as dimers to canonical E-box responsive elements (Zhu et al, 2016; Ansieau et al., 2010). The Twist family of TFs is composed of key regulators in embryonic development and organogenesis (Zhao et al., 2017). Twist family members can act as transcriptional repressors, by recruiting histone deacetylases or inhibiting acetyl-transferases, or they can function as transcriptional activators. Twist can also regulate transcription by interacting with several TFs (MyoD, RUNX1, RUNX2, p53, NF-kB) and inhibiting or enhancing Slug gene transcription (Casas et al., 2011). Twist2 is a regulator of embryonic development, but its function in tumor initiation/metastasis/growth is not well documented (Zhu et al., 2016).
By repressing adhesion barriers, these TFs mediate the partial reprograming of epithelial cells to acquire invasive behavior (De Craene and Berx, 2013b; Lamouille et al., 2014a) and the acquisition of mesenchymal behavior by inducing matrix deposition and secretion. In addition, TF overexpression commonly correlates with tumor progression and predicts poor clinical outcomes in many cancer types (Cobaleda et al., 2007a, b; De Craene and Berx, 2013b; de Herreros et al., 2010; Lamouille et al., 2014a; Shih and Yang, 2011), thus, raising immense therapeutic interest for targeting these TFs in metastatic disease.
Zeb1, TWIST1, SNAIL, Slug or treatment with TGFb promote tumorigenicity and stemness of cancer cells. For instance, Zeb1 strongly represses the miR-200 family, whose members are potent inducers of epithelial differentiation (Wellner et al., 2009; Krebs et al., 2017) and to physically interact with Hippo pathway effector YAP (Lehmann et al., 2015), consequently increasing tumor propagating cell frequency and cell plasticity in pancreatic and colorectal cancer cells. In addition, Zeb1 promotes expression of the cancer stem cell surface marker CD44 in pancreatic and breast cancer cells (Preca et al., 2015). Knockdown of Snail or Slug in breast or pancreatic cancer cells decreased invasion, increased E-Cadherin expression, and inhibited ALDH expression, together with decreased sphere and colony forming capacity (Zhou et al., 2014; Proia et al 2011; Phillips et al., 2012). Similar observations were made in cell line-derived tumors from tongue SCC, in which overexpression of Snail was associated with EMT features and CSC-like features (Zhu et al., 2012).
The chromatin remodeling factor JMJD3 binds to and deposits the active histone mark H3K27me3 on the SNAI2 gene promoter, thereby promoting the tumor-initiating abilities of hepatocellular carcinoma cells (Tang et al., 2016). The RNA-binding protein IMP3 directly stabilizes SNAI2 transcripts, as does the deacetylase SIRT2, thereby promoting Slug protein expression and expanding TIC population in breast cancer (Samanta et al., 2016; Zhou et al., 2016). These observations suggest that activation of EMT program in cancer cells is closely related to CSC state and increased cell plasticity in many cancer types. However, these two phenomena, although closely related, are not synonymous, and some EMT-TFs promote tumor stemness independently of their affect on EMT. For example, conditional ablation of Twist1 in benign skin tumors causes increased apoptosis, reduced cell proliferation, and defective tumor maintenance and propagation independently of Twist1’s EMT function (Beck et al., 2015).
In addition, modulation of TAZ is also capable on its own of inducing plasticity and stemness in mammary epithelial cells (Cordenonsi et al., 2011; Skibinski et al., 2015). TAZ acts as molecular switch regulating luminal and basal phenotypes, and toggling of the switch is sufficient to alter differentiation state. Overexpression of TAZ causes luminal cells to adopt basal/ME features, and depletion of TAZ induces basal/ME cells to acquire luminal characteristics. The ability of TAZ to induce cellular plasticity depends on chromatin remodeling factors to effect changes in differentiation state. The SWI/SNF complex directly interacts with TAZ and is essential in mediating TAZ function (Skibinski et al 2015). Although both BRG1 and BRM retain the ability to bind to TAZ by their PPXY motifs, cellular plasticity is achieved only by BRM recruitment of TAZ to target genes and not by TAZ/BRG1 complexes. Therefore, the lack of redundancy between BRM and BRG1 may result from binding to distinct sets of cofactors or other transcription factors that provide specificity for particular promoter sequences to drive transdifferentiation. It is worth nothing that, although BRG1 does not seem to be important for TAZ-mediated transcription in mammary epithelial cells, it is possible that BRG1 regulates TAZ target genes and plasticity in other cell types.
Cellular plasticity in mammary epithelial cells can also originate from epigenetic reprogramming via a coordinated process of de novo DNA methylation by DNMT3a and gene silencing by DOT1L-mediated reduction in histone H3K79 methylation. This process causes loss of expression of both cell cycle regulators and lineage-specific genes (Breindel 2017; Hinshelwood et al., 2009). Although the temporal nature of de-differentiation is not entirely clear, this work sheds light on the epigenetic basis of cellular plasticity, knowledge that could prove useful in understanding similar instances of dedifferentiation in other systems.
Conclusions
The discovery of somatic cell plasticity in adults is an unanticipated theme of contemporary biology. The study of plasticity is gradually moving from phenomenology to a more precise identification of the mechanisms underlying dedifferentiation and transdifferentiation. Phenotypic plasticity relates directly to the cellular origins of cancer and to cancer progression and therapy response. The relevant factors that dictate the switch from hierarchy to plasticity are beginning to be identified; however, still needed is a deeper understanding about the signatures and mechanisms that drive transdifferentiation or dedifferentiation transitions. In addition, our understanding of the generation of inter- and intra-tumor diversity as a result of phenotypic plasticity is far from complete. Finally, because phenotypic plasticity may produce unexpected vulnerabilities, it is important to determine whether we could exploit such plasticity with anticancer therapies.
References
- Aceto N1, Toner M2, Maheswaran S3, Haber DA4. En Route to Metastasis: Circulating Tumor Cell Clusters and Epithelial-to-Mesenchymal Transition. (2015) Trends Cancer 1(1):44–52. [DOI] [PubMed] [Google Scholar]
- Arienti et al. , 2013b;
- Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, and Kasimir-Bauer S (2009). Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Research 11, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Lapouge G, Rorive S, Drogat B, Desaedelaere K, Delafaille S, Dubois C, Salmon I, Willekens K, Marine JC, Blanpain C.Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. (2015) Cell Stem Cell 16(1):67–79. [DOI] [PubMed] [Google Scholar]
- Bedogni et al. , 2008
- Bedogni N Pigment Cell Melanoma Res 2014; 27:162–8. [DOI] [PubMed] [Google Scholar]
- Benya PD, and Shaffer JD (1982). Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 30, 215–224. [DOI] [PubMed] [Google Scholar]
- Blanco MJ, et al. , Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene, 2002. 21(20): p. 3241–6. [DOI] [PubMed] [Google Scholar]
- Bonfanti P, Barrandon Y, and Cossu G (2012). ‘Hearts and bones’: The ups and downs of ‘plasticity’ in stem cell biology. EMBO Molecular Medicine 4, 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boral D et al. Nat Commun 2017, 8:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz T, Raghu Kalluri R, Nieto MA, Weinberg RA. (2018) Nat Rev Cancer 18 (2), 128–134. [DOI] [PubMed] [Google Scholar]
- Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, Yang J. Snail2 is an essential mediator of Twist1-induced epithelial-mesenchymal transition and metastasis. (2011) Cancer Res 71(1): 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Suc Y, Polyak K, et al. (2011). Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. U. S. A 108, 7950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang et al. , 2011a;
- Choi N, Zhang B, Zhang L, Ittmann M, and Xin L (2012). Adult Murine Prostate Basal and Luminal Cells Are Self-Sustained Lineages that Can Both Serve as Targets for Prostate Cancer Initiation. Cancer Cell 21, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui MH (2013). Insights into cancer metastasis from a clinicopathologic perspective: Epithelial-Mesenchymal Transition is not a necessary step. International Journal of Cancer 132, 1487–1495. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M1, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. (2011) Cell 147(4):759–72. [DOI] [PubMed] [Google Scholar]
- Cregan MD, Fan Y, Appelbee A, Brown ML, Klopcic B, Koppen J, Mitoulas LR, Piper KME, Choolani MA, Chong Y-, and Hartmann PE (2007). Identification of nestin-positive putative mammary stem cells in human breast milk. Cell Tissue Res 329, 129–136. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Young P, Christov K, Guzman R, Nandi S, Talamantes F, and Thordarson G (1995). Mammary phenotypic expression induced in epidermal cells by embryonic mammary mesenchyme. Acta Anat 152, 195–204. [DOI] [PubMed] [Google Scholar]
- Daniel CW, De Ome KB, Young JT, Blair PB, and Faulkin LJ Jr. (1968). The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proc. Natl. Acad. Sci. U. S. A 61, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, and Lassar AB (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. [DOI] [PubMed] [Google Scholar]
- Del Vecchio CA, Feng Y, Sokol ES, Tillman EJ, Sanduja S, Reinhardt F, Gupta PB. De-differentiation confers multidrug resistance via noncanonical PERK-Nrf2 signaling. (2014) PLoS Biol 12(9):e1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong et al. , 2016;
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, and Ding S (2011). Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat. Cell Biol 13, 215–222. [DOI] [PubMed] [Google Scholar]
- Ewald AJ, Brenot A, Duong M, Chan BS, and Werb Z (2008). Collective Epithelial Migration and Cell Rearrangements Drive Mammary Branching Morphogenesis. Developmental Cell 14, 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA, Jin DX, Reinhardt F, Ploegh HL, Wang Q, Gupta PB. (2014) Epithelial-to-mesenchymal transition activates PERK-eIF2α and sensitizes cells to endoplasmic reticulum stress. Cancer Discov 4(6):702–15. [DOI] [PubMed] [Google Scholar]
- Feng YX, Jin DX, Sokol ES, Reinhardt F, Miller DH, Gupta PB. Cancer-specific PERK signaling drives invasion and metastasis through CREB3L1. (2017) Nat Commun 8(1):1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, Schwabe RF, Vahdat LT, Altorki NK, Mittal V, Gao D. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. (2015). Nature 2015, 527:472–6; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B, and Ghila L (2010). Cell plasticity in homeostasis and regeneration. Mol. Reprod. Dev 77, 837–855. [DOI] [PubMed] [Google Scholar]
- García-Zaragoza E, Pérez-Tavarez R, Ballester A, Lafarga V, Jiménez-Reinoso A, Ramírez T, Murillas R, and Gallego MI (2012). Intraepithelial paracrine Hedgehog signaling induces the expansion of ciliated cells that express diverse progenitor cell markers in the basal epithelium of the mouse mammary gland. Dev. Biol 372, 28–44. [DOI] [PubMed] [Google Scholar]
- Gjorevski N, and Nelson CM (2010). Endogenous patterns of mechanical stress are required for branching morphogenesis. Integrative Biology 2, 424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski E Untersuchungen über Auslösung und Hemmung der Regeneration beim Axolotl Wilhelm Roux Arch. Entwicklungsmech. Organismen, 114 (1928), pp. 108–143 [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zürrer-Härdi U, Bell G, et al. (2012). Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Kuperwasser C, Brunet JP, Ramaswamy S, Kuo WL, Gray JW, Naber SP, Weinberg RA. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. (2005) Nat Genet 37(10):1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, and Lander ES (2011). Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644. [DOI] [PubMed] [Google Scholar]
- Gupta PB, et al. , Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell, 2009. 138(4): p. 645–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslehurst et al. , 2012b
- Hinshelwood RA, Melki JR, Huschtscha LI, Paul C, Song JZ, Stirzaker C, Reddel RR, and Clark SJ (2009). Aberrant de novo methylation of the p16INK4A CpG island is initiated post gene silencing in association with chromatin remodelling and mimics nucleosome positioning. Hum. Mol. Genet 18, 3098–3109. [DOI] [PubMed] [Google Scholar]
- Honeth G, Bendahl P, Ringnér M, Saal LH, Gruvberger-Saal SK, Lövgren K, Grabau D, Fernö M, Borg Å, and Hegardt C (2008). The CD44+/CD24− henotype is enriched in basal-like breast tumors. Breast Cancer Research 10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst2013.
- Hyun KA, Koo GB, Han H, Sohn J, Choi W, Kim SI, Jung HI, Kim YS. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. (2016) Oncotarget 7:24677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu J-, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, and Srivastava D (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, Iglehart JD, Weinberg RA. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. (2007) Cancer Cell 12(2):160–70. [DOI] [PubMed] [Google Scholar]
- Jiang et al. , 2016b
- Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, Onuchic JN, Levine H. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. (2015) Front Oncol 2015, 5:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly MK, Tripathi SC, Jia, Mooney SM, Celiktas M, Hanash SM, Mani SA, Pienta KJ, Ben-Jacob E, Levine H. Stability of the hybrid epithelial/mesenchymal phenotype. (2016) Oncotarget 7(19):27067–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, Smith AE, Prat A, Perou CM, Gilmore H, et al. (2012). Defining the cellular precursors to human breast cancer. Proc. Natl. Acad. Sci. U. S. A 109, 2772–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, and Ding S (2011). Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U. S. A 108, 7838–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P, Brunton VG, Pilarsky C, Winkler TH, Brabletz S, Stemmler MP, Brabletz. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. (2017) Nat Cell Biol 2017 May;19(5):518–529 [DOI] [PubMed] [Google Scholar]
- Kurrey et al. , 2009
- Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. (2017) Cell 168(4):670–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latil M, Nassar D, Beck B, Boumahdi S, Wang L, Brisebarre A, Dubois C, Nkusi E, Lenglez S, Checinska A, Vercauteren Drubbel A, Devos M, Declercq W, Yi R, Blanpain C. Cell-Type-Specific Chromatin States Differentially Prime Squamous Cell Carcinoma Tumor-Initiating Cells for Epithelial to Mesenchymal Transition. (2017) Cell Stem Cell 20(2):191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecharpentier A, Vielh, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. (2011) Br J Cancer 2011 Oct 25;105(9):1338–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q-, Zhang H, Zhao B, Zha Z-, Bai F, Pei X-, Zhao S, Xiong Y, and Guan K- (2008). TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol 28, 2426–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann W, Mossmann D, Kleemann J, Mock K, Meisinger C, Brummer T, Herr R, Brabletz S, Stemmler MP, Brabletz T. ZEB1 turns into a transcriptional activator by interacting with YAP1 in aggressive cancer types. (2016) Nat Commun 15;7:10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu M-, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, et al. (2008). Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst 100, 672–679. [DOI] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, Ding S, and Guan K- (2010). The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes and Development 24, 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat M-, Gyorki DE, Ward T, Partanen A, et al. (2009). Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med 15, 907–913. [DOI] [PubMed] [Google Scholar]
- Lin X, Sun B, Zhu D, Zhao X, Sun R, Zhang Y, Zhang D, Dong X, Gu Q, Li Y, Liu F. Notch4+ cancer stem-like cells promote the metastatic and invasive ability of melanoma. (2016). Cancer Sci 107(8):1079–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CP, Polak L, Rocha AS, Pasolli HA, Chen S-, Sharma N, Blanpain C, and Fuchs E (2012). Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 150, 136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao M-, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. (2008). The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 133, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanovic ND, Weinberg RA, and Chaffer CL (2013). Cell plasticity and heterogeneity in cancer. Clin. Chem 59, 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay F-, McCarthy A, Kendrick H, Natrajan R, MacKay A, Grigoriadis A, Tutt A, Ashworth A, Reis-Filho JS, and Smalley MJ (2010). BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7, 403–417. [DOI] [PubMed] [Google Scholar]
- Morel A-, Lièvre M, Thomas C, Hinkal G, Ansieau S, and Puisieux A (2008). Generation of breast cancer stem cells through epithelial-mesenchymal transition. Plos One 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassour M, Idoux-Gillet Y, Selmi A, Côme C, Faraldo M-M, Deugnier M-, and Savagner P. (2012). Slug Controls Stem/Progenitor Cell Growth Dynamics during Mammary Gland Morphogenesis. Plos One 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. (2016) Cell 166(1):21–45. [DOI] [PubMed] [Google Scholar]
- Olmeda D. Cancer Res. 2017.
- Olmeda D. Oncogene. 2008.
- Okada TS (1980). Cellular metaplasia or transdifferentiation as a model for retinal cell differentiation. Curr. Top. Dev. Biol 16, 349–380. [PubMed] [Google Scholar]
- Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D, Moers V, Lemaire S, De Clercq S, Minguijón E, Balsat C, Sokolow Y, Dubois C, De Cock F, Scozzaro S, Sopena F, Lanas A, D’Haene N, Salmon I5, Marine JC, Voet T, Sotiropoulou PA, Blanpain C. Identification of the tumour transition states occurring during EMT. (2018) Nature 556(7702):463–468. [DOI] [PubMed] [Google Scholar]
- Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, Skibinski A, Shirley S, Perou C, Gill G, Gupta P, and Kuperwasser C (2014) Cell-State Transitions Regulated by SLUG Are Critical for Tissue Regeneration and Tumor Initiation. Stem Cell Reports 2(5):633–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnix CC, Lee JT, Liu ZJ, McDaid R, Balint K, Beverly LJ, Brafford PA, Xiao M, Himes B, Zabierowski SE, Yashiro-Ohtani Y, Nathanson KL, Bengston A, Pollock PM, Weeraratna AT, Nickoloff BJ, Pear WS, Capobianco AJ, Herlyn M.Active Notch1 confers a transformed phenotype to primary human melanocytes. (2009) Cancer Res 69(13):5312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K and Weinberg RA, Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer, 2009. 9(4): p. 265–73. [DOI] [PubMed] [Google Scholar]
- Prat A, and Perou CM (2011). Deconstructing the molecular portraits of breast cancer. Molecular Oncology 5, 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preca BT, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, Wellner U, Hopt UT, Brummer T, Brabletz S, Brabletz T, Stemmler MP. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. (2015) Int J Cancer 137(11):2566–77. [DOI] [PubMed] [Google Scholar]
- Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, Gilmore H, Tung N, Naber SP, Schnitt S, Lander ES, and Kuperwasser C (2011). Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 8, 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, and Weissman IL (2001). Stem cells, cancer, and cancer stem cells. Nature 414, 105–111. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, and Greco V (2012). Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SM (1947). Dedifferentiation in the regenerating amphibian limb. Anat. Rec 99, 568. [PubMed] [Google Scholar]
- Šale S, Lafkas D, and Artavanis-Tsakonas S (2013). Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nat. Cell Biol 15, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S, Sun H, Goel HL, Pursell B, Chang C, Khan A, Greiner DL, Cao S, Lim E, Shultz LD, Mercurio AM. IMP3 promotes stem-like properties in triple-negative breast cancer by regulating SLUG. Oncogene 2016. March 3;35(9):1111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satelli A, Mitra A, Brownlee Z, Xia X, Bellister S, Overman MJ, Kopetz S, Ellis LM, Meng QH, Li S. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. (2015) Clin Cancer Res 21(4):899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner P, et al. , Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J Cell Physiol, 2005. 202(3): p. 858–66. [DOI] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Shin NR1, Jeong EH, Choi CI, Moon HJ, Kwon CH, Chu IS, Kim GH, Jeon TY, Kim DH, Lee JH, Park DY. Overexpression of Snail is associated with lymph node metastasis and poor prognosis in patients with gastric cancer. (2012) BMC Cancer 12:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski A, Breindel JL, Prat A, Galván P, Smith E, Rolfs A, et al. The hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Reports 2014;6(6):1059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol ES, Miller DH, Breggia A, Spencer KC, Arendt LM, Gupta PB.Growth of human breast tissues from patient cells in 3D hydrogel scaffolds. (2016) Breast Cancer Res 18(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol ES, Feng YX, Jin DX, Tizabi MD, Miller DH, Cohen MA, Sanduja S, Reinhardt F, Pandey J, Superville DA, Jaenisch R, Gupta PB. SMARCE1 is required for the invasive progression of in situ cancers. (2017) Proc Natl Acad Sci U S A 114(16):4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer MY, Yang H-, Brabb T, Leaf E, Look A, Lin W-, Frutkin A, Dichek D, and Giachelli CM (2009). Smooth muscle cells give rise to osteochondrogenic precursors and chondrocytes in calcifying arteries. Circ. Res 104, 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E, Rampalli S, Risueño RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, and Bhatia M (2010). Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S (2007). Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of Pluripotent Stem Cells from Mouse Embryonic andAdult Fibroblast Cultures by Defined Factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Ang CE, Chanda S, Olmos VH, Haag D, Levinson DF, Südhof TC, Wernig M. Transdifferentiation of human adult peripheral blood T cells into neurons. (2018) Proc Natl Acad Sci U S A Jun 4. pii: 201720273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Qi G, Tang F, Yuan S, Wang Z, Liang X, Li B, Yu S, Liu J, Huang Q, Wei Y, Zhai R, Lei B, Yu H, Tomlinson S, He S. Aberrant JMJD3 Expression Upregulates Slug to Promote Migration, Invasion, and Stem Cell-Like Behaviors in Hepatocellular Carcinoma. (2016) Cancer Res 76(22):6520–6532. [DOI] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, Medoff BD, and Rajagopal J (2013). Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester AM, et al. , MMP-9 secretion and MMP-2 activation distinguish invasive and metastatic sublines of a mouse mammary carcinoma system showing epithelial-mesenchymal transition traits. Clin Exp Metastasis, 2000. 18(7): p. 553–60. [DOI] [PubMed] [Google Scholar]
- Tian2013.
- Thiery JP and Sleeman JP, Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol, 2006. 7(2): p. 131–42. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, and Nieto MA (2009). Epithelial-Mesenchymal Transitions in Development and Disease. Cell 139, 871–890. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Lu Y, Nichols PW, Zlotnikov G, Jones PS, and Smith HS (1996). Contiguous patches of normal human mammary epithelium derived from a single stem cell: Implications for breast carcinogenesis. Cancer Res 56, 402–404. [PubMed] [Google Scholar]
- Tse GM, Tan PH, Putti TC, Lui PCW, Chaiwun B, and Law BKB (2006). Metaplastic carcinoma of the breast: A clinicopathological review. J. Clin. Pathol 59, 1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, and Blanpain C (2011). Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Lee MY, Ousset M, Brohée S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon I, Sotiriou C, Phillips WA, Blanpain C. (2015) Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525(7567):119–23. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, and Wernig M (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE (2011). Cells of origin in cancer. Nature 469, 314–322. [DOI] [PubMed] [Google Scholar]
- Vitali et al. , 2008a
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schüler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. (2009) Nat Cell Biol 11(12):1487–95. doi: 10.1038/ncb1998. Epub 2009 Nov 22. [DOI] [PubMed] [Google Scholar]
- Williams JM, and Daniel CW (1983). Mammary ductal elongation: Differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev. Biol 97, 274–290. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu S, Liu Z, Huang J, Pu X, Li J, Yang D, Deng H, Yang N, Xu J. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. (2015) PLoS One 10(4):e0123976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. PNAS. 2017.
- Xu et al. , 2015a
- Yamashita N, Tokunaga E, Inoue Y, Tanaka K, Nakashima Y, Ando K, Ogaki K, Saeki H, Oki E, Maehara Y. Clinical significance of co-expression of E-cadherin and vimentin in invasive breast cancer. J Clin Oncol (2015) 3315supple22013 [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, and McKeon F (1999). p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718. [DOI] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. (2013) Science 339(6119):580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu C-, Zha Z-, Zhao B, Yao J, Zhao S, Xiong Y, Lei Q-, and Guan K. (2009). TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J. Biol. Chem 284, 13355–13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji J-, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, and Haber DA (2009). YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol 11, 1444–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. , 2012. (BRAF melanoma)
- Zhao R et al. Oncotarget 2017, 8:9293–9302). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer.(2015) Nature 527(7579):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M et al. Oncotarget 2015, 6:6794–6810 [Google Scholar]
- Zhou W, Ni TK, Wronski A, Glass B, Skibinski A, Beck A, Kuperwasser C. The SIRT2 Deacetylase Stabilizes Slug to Control Malignancy of Basal-like Breast Cancer. (2016) Cell Rep 17(5):1302–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, and Melton DA (2008). In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455, 627–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lv R, Qi W, Wu D, Xu Y, Liu W, Mou Y, Wang L. Snail contributes to the maintenance of stem cell-like phenotype cells in human pancreatic cancer. (2014) PLoS One 9(1):e87409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LF1, Hu Y, Yang CC, Xu XH, Ning TY, Wang ZL, Ye JH, Liu LK. Snail overexpression induces an epithelial to mesenchymal transition and cancer stem cell-like properties in SCC9 cells. (2012) Lab Invest 92(5):744–52. [DOI] [PubMed] [Google Scholar]