Abstract
Objective
Breast cancer has become the most common malignancy among women worldwide; therefore, novel diagnostic and prognostic markers and therapeutic targets are urgently required. NF-κB signaling plays a pivotal role in enhancing breast cancer malignant phenotypes, especially cancer invasion and metastasis, which is the main cause of death in cancer patients. TNIP2, an important inhibitor of the NF-κB pathway, is known to involve a negative feedback loop of the NF-κB signaling cascade and to regulate tumor aggressiveness in various cancer types. However, the mRNA level of TNIP2 is barely altered in breast cancer; thus, the mechanism that regulates TNIP2 in breast cancer still needs to be elucidated.
Methods
We analyzed the expression and prognosis of miR-423 in a TCGA BRCA miRNA cohort and in clinical specimens. We detected the invasive capacity through a Matrigel-coated Transwell penetration assay, a three-dimensional (3D) spheroid invasion assay and a wound healing assay. Then, we applied luciferase assays, real-time PCR assays and Western blotting to further study the mechanism.
Results
In this study, analysis of the TCGA BRCA miRNA cohort and clinical specimens demonstrated that miR-423 was upregulated in human breast cancers and was positively correlated with clinical stage, poor overall survival and metastasis classification. Moreover, the invasiveness of breast cancer cells was enhanced by ectopic expression of miR-423 and inhibited by miR-423 downregulation. Mechanistically, upregulation of miR-423 led to activation of the NF-κB signaling pathway and elevated expression of snail and twist, while repression of miR-423 inhibited this pathway. Furthermore, the results indicated that TNIP2 is a target gene of miR-423, and suppression of TNIP2 resulted in increased invasiveness in miR-423-silenced cells.
Conclusion
Our results suggest that miR-423 is a crucial factor that enhances breast cancer cell invasion through the NF-κB signaling pathway and shed light on miR-423 as a promising prognostic and therapeutic marker for metastatic breast cancer.
Keywords: breast cancer, miR-423, TNIP2, NF-κB signaling pathway
Introduction
Breast cancer is the most common malignancy among women and the second leading cause of cancer-related death worldwide, accounting for approximately 22% of all new cancer cases and 13.7% of cancer-related deaths in females.1 In recent decades, the incidence of breast cancer has risen in many developing countries, including China.2,3 There is a high likelihood for the occurrence of metastasis in breast cancer, which is the main cause of death in breast cancer patients.4 Metastasis has been demonstrated as the late stage of progression of breast cancer and involves multiple steps, including invasion. The 5-year survival rate is reduced from 85% in patients with early-stage disease to 23% in patients with late-stage disease.5 Currently, treatments for metastatic late-stage breast cancer are mainly palliative and inefficient due to poor knowledge of the mechanisms of metastasis.6 Therefore, it is of great clinical value to find new biomarkers for diagnosis of early-stage cancers and to identify molecular targets for new therapies. This can be accomplished by the expansion and utilization of our understanding of the molecular mechanisms involved in metastasis.
Previous studies have shown that the NF-κB signaling pathway plays a major role in the malignant biological functions of breast cancer, including metastasis, apoptosis and angiogenesis, which always exhibit a highly sustained state of activation in a variety of tumors, such as breast and lung cancers.7–9 When the NF-κB signaling pathway is in the resting state, the NF-κB/Rel family protein binds to its inhibitory factor IκB and remains in the cytoplasm.10,11 In response to an external signal, the zinc finger protein A20, which is a ubiquitin editing enzyme that controls ubiquitination and deubiquitination of target proteins, binds to members of the TNIP family, including TNIP1 (TNFAIP3 interacting protein 1), TNIP2 (TNFAIP3 interacting protein 2), and TNIP3 (TNFAIP3 interacting protein 3); subsequently the complex promotes ubiquitination and proteasomal degradation of proteins such as TRAF2, RIP and NEMO, leading to phosphorylation of IκB, and as a result, suppresses NF-κB activation.12,13 However, in breast cancers, the mechanisms that regulate the factors in the negative feedback loop of the NF-κB signaling pathway require further delineation.
MicroRNAs (miRNAs) are a class of small, noncoding, single-stranded RNAs that inhibit gene expression by binding to the 3ʹ-untranslated region (UTR) of target mRNAs at the posttranscriptional level.14,15 It has been reported that miRNAs are involved in various aspects of cancer cell biology, including proliferation, apoptosis, and metastasis.16–18 Though miRNAs occupy only 20–25 nucleotides of their target sequence, miRNAs regulate a majority of gene expression; therefore, miRNA dysregulation can trigger tumor progression.19 It is widely accepted that miRNAs can function as oncogenes or tumor suppressors by targeting the 3ʹ-UTRs of matched genes.20,21 In tumors, miR-423-5p has been demonstrated to contribute to the development of malignant phenotypes and temozolomide resistance in glioblastoma and promote autophagy in hepatocellular carcinoma cells.22,23 Furthermore, plasma miR-423-5p level has the potential to serve as a novel biomarker for colorectal cancer detection, particularly in the early stage.24 miR-423-5p was observed to be a marker for responsiveness to sorafenib therapy in hepatocellular carcinoma, since 75% of patients with increased plasma miR-423-5p levels achieved partial remission or stable disease after 6 months from the beginning of therapy.23 These results demonstrated the potential role of miR-423-5p in the diagnosis and therapy of cancer. However, the expression and role of miR-423-5p in breast cancer remains to be determined, and is therefore the aim of the present study.
In the present study, we provided evidence for a novel mechanistic link between miR-423 and the oncogenic NF-κB signaling pathway in breast cancer. We found that miR-423 was significantly upregulated in breast cancer and correlated with clinical stage and poor prognosis. Overexpression of miR-423 promoted breast cancer cell invasion in vitro, while downregulation of miR-423 levels inhibited invasion. Furthermore, we demonstrated that miR-423 enhanced the NF-κB signaling activation pathway by directly targeting TNIP2, the vital negative regulator of the NF-κB pathway. Taken together, our results indicated that miR-423 functions as an oncomiR in breast cancer and may represent an important target for clinical intervention in metastatic breast cancer.
Materials and Methods
Cell Culture
Normal human breast epithelial cells (NBECs) were obtained from Clonetics-Biowhittaker (Walkersville, MD, USA) and cultured in KSFM medium (Clonetics-Biowhittaker). The breast cancer cell lines MCF-10A, MDA-MB-468, T47D, MCF-7, SKBR3, Bcap37 and MDA-MB-231 were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were maintained in D-Medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) and 1% penicillin/streptomycin (Invitrogen) at 37°C in a humidified atmosphere containing 5% CO2. These cells were authenticated by short tandem repeat PCR fingerprinting at the Forensic Medicine Laboratory of Sun Yat-Sen University (Guangzhou, China) after experiments were finished.
Patient Tissue Specimens
A group of 157 paraffin-embedded archived breast cancer samples and 2 normal mammary gland samples were collected from the Sun Yat-sen University Cancer Center (SYSUCC) from 2010 to 2012. The clinical and pathological classifications and stages were determined according to the American Joint Committee on Cancer (AJCC) criteria . The histologic grade of each sample was determined according to the Elston-Ellis modification of the Scarff-Bloom-Richardson (SBR) system. To use these clinical materials for research purposes, prior consent from the patients and approval from the Institutional Research Ethics Committee were obtained. The clinical information for the patient samples is summarized in Tables 1 and 2.
Table 1.
Clinicopathologic Features of miR-423 Expression in TCGA Breast Cancer Cohort
| Variable | Low miR-423 Expression (n = 446) | High miR-423 Expression (n = 447) | P-value |
|---|---|---|---|
| Age, y, mean ± SEM | 57.77 ± 0.57 | 59.08 ± 0.58 | 0.11 |
| TNM Stage | |||
| I | 131 (29.37%) | 26 (5.82%) | < 0.01 |
| II | 265 (59.42%) | 255 (57.05%) | |
| III | 48 (10.76%) | 151 (33.78%) | |
| IV | 2 (0.45%) | 15 (3.35%) | |
| Tumor Size | |||
| T1 | 164 (36.77%) | 69 (15.44%) | < 0.01 |
| T2 | 241 (54.04%) | 286 (63.98%) | |
| T3 | 36 (8.07%) | 64 (14.32%) | |
| T4 | 5 (1.12%) | 28 (6.26%) | |
| Nodes Pathologic | |||
| N0 | 266 (59.64%) | 176 (39.37%) | < 0.01 |
| N>0 | 180 (40.36%) | 271 (60.63%) | |
| Metastasis Pathologic | |||
| M0 | 444 (99.55%) | 431(96.42%) | < 0.01 |
| M1 | 2 (0.45%) | 16 (3.58%) |
Note: Values are n (%) unless otherwise indicated.
Table 2.
Clinicopathologic Features of miR-423 Expression in SYSUCC Cohort
| Variable | Low miR-423 expression (n = 79) | High miR-423 expression (n = 78) | P-value |
|---|---|---|---|
| Age, y, mean ± SEM | 53.08 ± 1.26 | 54.68 ± 1.13 | 0.35 |
| TNM Stage | |||
| I | 19 (24.05%) | 9 (11.54%) | < 0.01 |
| II | 42 (53.16%) | 30 (38.46%) | |
| III | 17 (21.52%) | 32 (41.03%) | |
| IV | 1 (1.27%) | 7 (8.97%) | |
| Tumor Size | |||
| T1/2 | 64 (81.01%) | 51 (65.38%) | < 0.01 |
| T3/4 | 15 (18.99%) | 27 (34.62%) | |
| Nodes Pathologic | |||
| N0 | 46 (58.23%) | 29 (37.18%) | 0.02 |
| N>0 | 33 (41.77%) | 49 (62.82%) | |
| Metastasis Pathologic | |||
| M0 | 78 (98.73%) | 71 (91.03%) | 0.03 |
| M1 | 1 (1.27%) | 7 (8.97%) |
Note: Values are n (%) unless otherwise indicated.
Plasmids and Transfection
The TNIP2 3ʹ-UTR was PCR amplified from NBEC RNA and cloned into the SacI/XmaI restriction sites of the pGL3-basic luciferase reporter (Promega, Madison, WI, USA) and pGFP-C3 (Clontech, Mountain View, CA, USA) plasmids. The miR-423 mimics, negative control, and anti-miR-423 inhibitor were purchased from RiboBio (Guangzhou, Guangdong, China). pNF-κB-luc and control plasmids (Clontech, Mountain View, CA, USA) were used to quantitatively examine NF-κB activity. Transfection of plasmids was performed using the Lipofectamine 2000 reagent (Invitrogen).
Western Blot Analysis
Western blot analysis was performed according to standard methods using anti-TNIP2, anti-Snail, and anti-twist antibodies. Anti-α-tubulin monoclonal antibody was also applied (Sigma, St. Louis, MO, USA) as a loading control.
RNA Extraction and Real-Time Quantitative PCR
Total RNA from tissues or cells was extracted using TRIzol (Life Technologies) according to the manufacturer’s instructions. Then, cDNA was synthesized from 5 ng of total RNA using the TaqMan miRNA or mRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). The expression levels of miR-423 were quantified using the miRNA-specific TaqMan MiRNA Assay Kit (Applied Biosystems). Expression levels were defined based on the threshold cycle (Ct), and relative expression levels were normalized to U6 small nuclear RNA using the formula 2−[(Ct of miR−423)-(Ct of U6)]. For mRNA analysis, ChamQ SYBR qPCR Master Mix (Vazyme Biotech, China) was used to quantify gene expression. The primers used were as follows: SNAI1 sense (5ʹ-ACGAGGTGTGACTAACTATG-3ʹ) and antisense (5ʹ-CGACAAGTGACAGCCATT-3ʹ), and TWIST1 sense (5ʹ-ACCATCCTCACACCTCTG-3ʹ) and antisense (5ʹ-GATTGGCACGACCTCTTG-3ʹ). Error bars represent the mean ± sd of 3 independent experiments.
Three-Dimensional Spheroid Invasion Assay
Cells (1 × 104) were trypsinized and seeded in 24-well plates coated with Matrigel (2%; BD Biosciences), and the medium was changed every second day. Microscope images were taken at 2-day intervals for 8 days.
Transwell Matrix Penetration Assay
Cells (1 × 104) were plated into the upper chamber of BioCoat Invasion Chambers (BD, Bedford, MA) containing a polycarbonate Transwell filter coated with Matrigel and incubated at 37°C for 22 h. Cells that remained inside the upper chamber were removed using cotton swabs. Cells that had invaded the lower membrane surface were fixed in 1% paraformaldehyde, stained with hematoxylin, and counted (ten random fields per well at 100× magnification). Cell counts are expressed as the mean number of cells per field of view. Three independent experiments were performed, and the data are presented as the mean ± sd.
Wound Healing Assay
Treated MCF-7 SKBR3 and MDA-MB-231 cells were seeded in 6-well plates and cultured to 80% confluence. Thereafter, small linear wounds were created creating a scratch with a disinfected Eppendorf tip. After washing the cell debris with FBS-free medium, images of the wound were captured under a microscope after 24 and 48 h to assess the distance remaining.
TCGA Data Analysis
Level 3 miRNA-Seq data of breast cancers were downloaded from the TCGA breast cancer data set portal (https://portal.gdc.cancer.gov/). Among these breast tumor and adjacent normal sample cohorts, miR-423 (miR-423-5p) expression data were extracted, and paired t-tests were performed to evaluate significant differences between breast tumor and adjacent normal samples.
Statistical Analysis
All statistical analyses were performed using the SPSS 20.0 statistical software package. Sample size was determined by power analysis to achieve a minimum effect size of 0.5 with a P value of <0.05. The patients were divided into two groups according to whether they exhibited a high (>the median) or low (≤the median) miR-423 expression level. Survival curves were analyzed by the Kaplan–Meier method, and a Log rank test was used to assess significance. Univariate and multivariate survival analyses were performed using Cox regression analysis. Comparisons between groups were performed using Student’s t-test, and multiple analyses between groups were performed using ANOVA with Dunnett’s t-test. All error bars represent the mean ± sd derived from three independent experiments. Data analysis was performed by two independent investigators who were blinded to the sample groups. P values <0.05 were considered statistically significant.
Results
miR-423 Is Upregulated in Human Breast Cancer and Correlates with Poor Prognosis
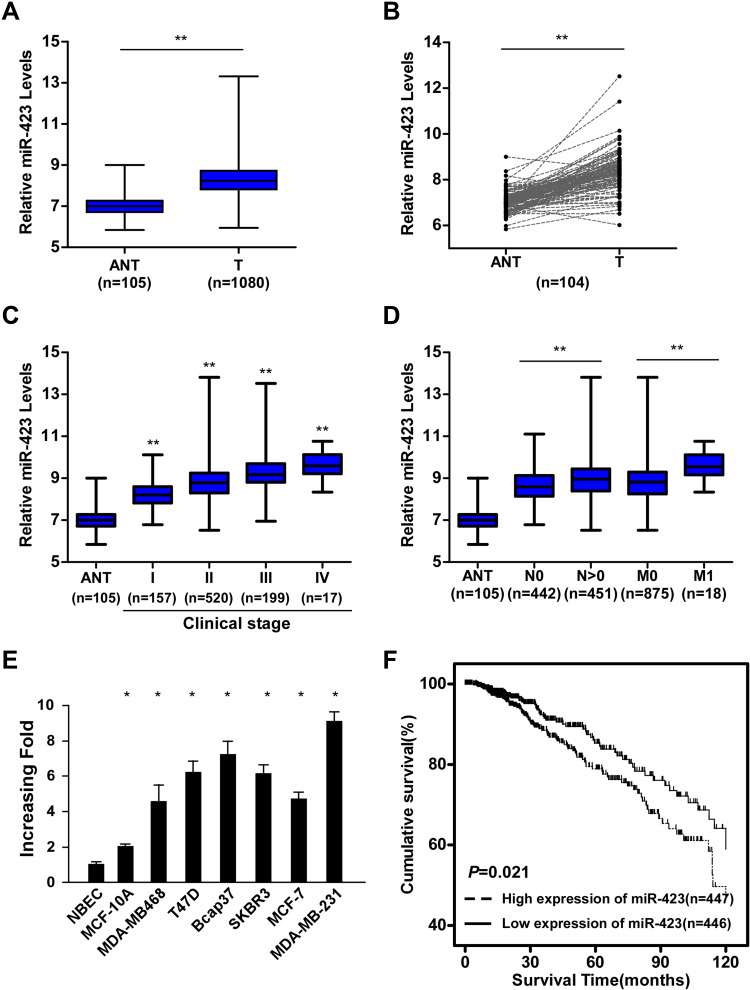
By analyzing the miRNA sequencing datasets from The Cancer Genome Atlas (TCGA) BRCA cohort, we found that miR-423 was significantly upregulated in human breast cancer tissues compared to adjacent nontumor tissues (column and paired analyses, P<0.01, respectively, Figure 1A and B). Furthermore, analyses of TCGA BRCA samples from patients with available clinical data showed that expression of miR-423 was upregulated with progressed clinical stage, and a similar trend was also obtained in the Sun Yat-sen University Cancer Center (SYSUCC) cohort (P<0.01, Figures 1C and Supplementary Figure 1A). More specifically, analyses of TCGA BRCA and SYSUCC cohorts indicated that miR-423 levels were higher in patients with tumor lymphoid node metastasis or distant metastasis (P<0.01, Figures 1D and Supplementary Figure 1B). Moreover, upregulated miR-423 expression was observed in metastatic breast cancer cell lines compared with a normal breast epithelial cell (NBEC) and MCF-10A cell lines (Figure 1E). Kaplan–Meier analysis and the Log rank test indicated that, stratified by a median cutoff of miR-423 expression, breast cancer patients with high miR-423 expression had shorter OS than those expressing low levels of miR-423 in the TCGA BRCA and SYSUCC cohorts (Log rank test, P = 0.021 and P < 0.01, respectively; Figures 1F and Supplementary Figure 1C). These data suggest that miR-423 expression is upregulated in breast cancer tissues and cell lines and that a high level of miR-423 is associated with poor prognosis and tumor metastasis in breast cancer patients.
Figure 1.
miR-423 is upregulated in human breast cancer and correlates with poor prognosis. (A) Analysis of miR-423 expressions in TCGA BRCA cohort data. (B) Analysis of miR-423 expressions between human breast cancer tissues and matched adjacent non-tumor tissues in TCGA BRCA cohort data (number of pairs = 104). (C) Analysis of miR-423 expression in different clinical stages of TCGA BRCA cohort data. The boundaries of the boxes represent the lower and upper quartiles, the lines within the boxes and whiskers denote the median and outer limits. (D) Analysis of miR-423 expression in different N (node metastasis) and M (distant metastasis) classifications of TCGA BRCA cohort data. The boundaries of the boxes represent the lower and upper quartiles, the lines within the boxes and whiskers denote the median and outer limits. (E) Real-time PCR analysis of miR-423 expression in breast cancer cell lines and normal breast epithelial cells (NBECs). Transcript levels were normalized to U6 expression. (F) Kaplan–Meier analysis of the correlation between the miR-423 level and the overall survival (OS) of breast cancer patients in TCGA BRCA cohort. Bars represent the mean ± SD of three independent experiments; **P < 0.01, *P < 0.05.
miR-423 Increases the Invasiveness of Breast Cancer Cells
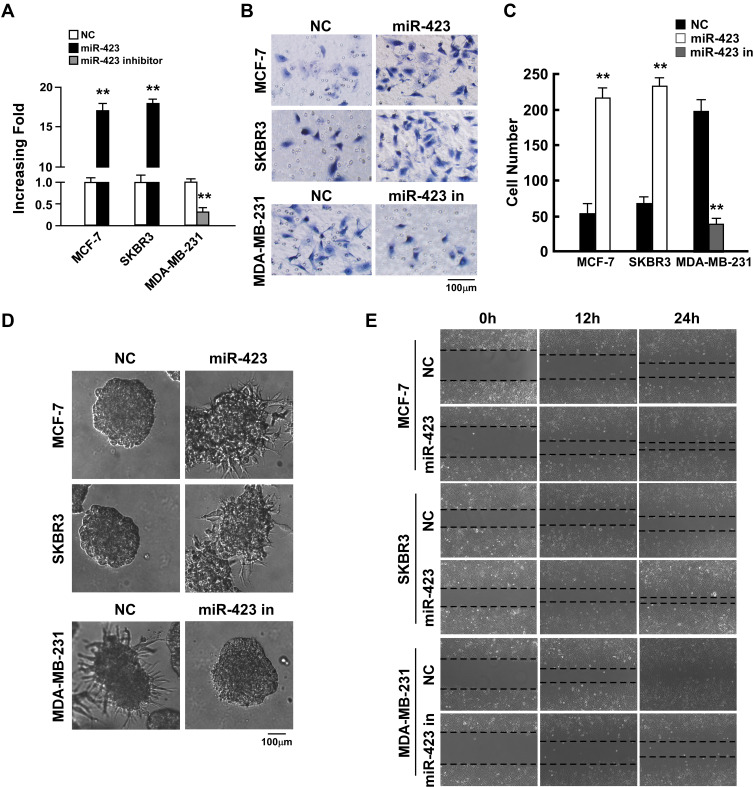
To investigate the biological function of miR-423 in breast cancer, we established MCF-7 and SKBR3 breast cancer cell lines with ectopically overexpressed miR-423 (Figure 2A) and measured the effect of overexpressed miR-423 on cell invasion. The invasive capacity of cells was examined using the Matrigel-coated Transwell penetration assay. In both tested cell lines, overexpression of miR-423 resulted in an increased number of cells that penetrated the gel-membrane barrier (Figure 2B upper panel and C, P<0.01). Furthermore, in a three-dimensional (3D) spheroid invasion assay, cells that overexpressed miR-423 had more outward projections than control cells (Figure 2D, upper panel) and displayed an altered morphology that is typical of highly invasive cells. Moreover, cell migration (measured using a wound healing assay) was increased in MCF-7 and SKBR3 cells that overexpressed miR-423 compared to control cells (Figure 2E, upper panel). These results demonstrated that miR-423 upregulation promotes the invasion of breast cancer cells.
Figure 2.
miR-423 promotes invasiveness of breast cancer cells. (A) Real-time PCR analysis of miR-423 expression in indicated cells. (B) Transwell assays performed with indicated cells, scale bar, 100 μm. (C) Relative quantification of cells in transwell assays with indicated cells. (D) 3D growth capacity showed with indicated cells. (E) Time course of wound healing assays performed with indicated cells. Bars represent the mean ± SD of three independent experiments; **P < 0.01.
We next transfected a miR-423 inhibitor into highly invasive MDA-MB-231 breast cancer cells, which led to significant repression of miR-423 (Figure 2A; P<0.01). Consistent with our previous findings, the Matrigel-coated Transwell penetration assay indicated that cells transfected with the miR-423 inhibitor were less likely to pass through the gel-membrane barrier than negative control cells (Figure 2B lower panel and C, P<0.01). Furthermore, 3D culture in Matrigel and wound healing assays demonstrated that the invasive ability of MDA-MB-231 cells was significantly decreased following transfection with the miR-423 inhibitor (Figure 2D and E, lower panel). Taken together, our data suggest that miR-423 plays an important role in controlling the invasiveness of breast cancer cells.
miR-423 Directly Targets TNIP2 in Breast Cancer Cells
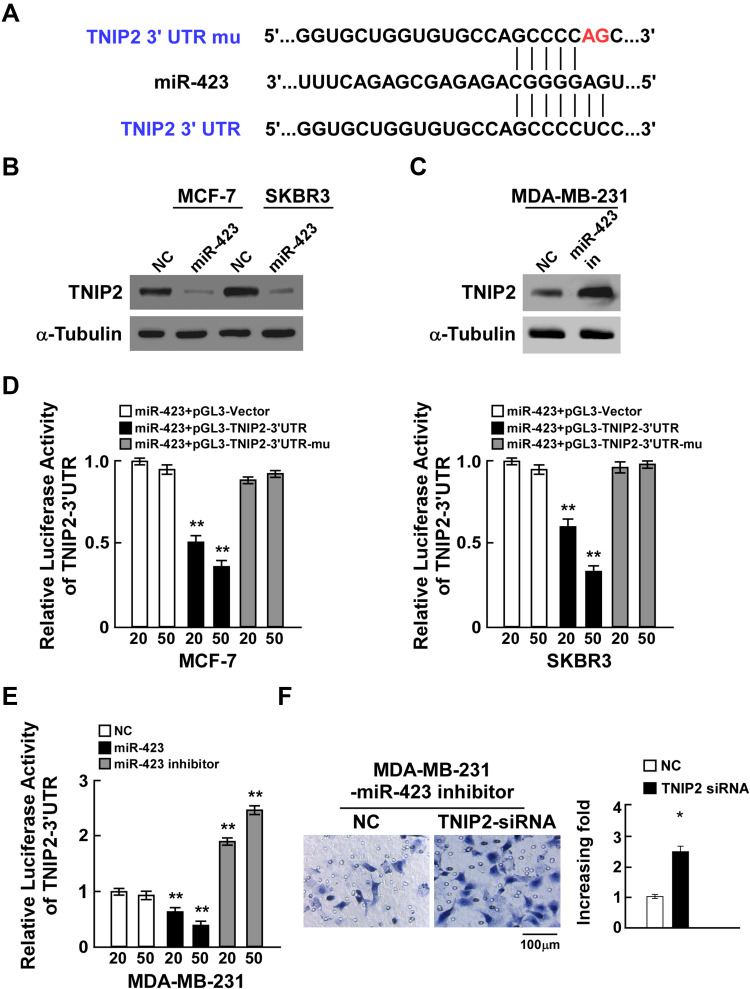
To further elucidate the underlying mechanism by which miR-423 promotes breast cancer invasiveness, TargetScan prediction was used and showed that TNIP2 was one of the conserved targets of miR-423 (Figure 3A). Notably, TNIP2 mRNA levels were barely altered in breast cancer compared to normal adjacent tissues, which suggested that the expression of TNIP2 might be regulated in a posttranscriptional manner (Supplementary Figure 2A and B). In light of the aforementioned clues, we observed that ectopic miR-423 expression resulted in decreased TNIP2 protein levels in MCF-7 and SKBR3 cells, while repression of miR-423 in MDA-MB-231 cells increased TNIP2 protein levels, supporting our identification of TNIP2 as a potential miR-423 target gene (Figure 3B and C). To confirm that miR-423 inhibition of TNIP2 is mediated by binding to the 3ʹ-UTR, we cloned the TNIP2 3ʹ-UTR into luciferase reporter plasmids and investigated the effects of miR-423 overexpression and inhibition on reporter activity. miR-423 transfection reduced the luciferase activity of the TNIP2 3ʹ-UTR luciferase reporter in MCF-7 and SKBR3 breast cancer cells in a dose-dependent manner (Figure 3D). The repression of luciferase reporter activity was abrogated when point mutations were generated in the miR-423 seed region of the TNIP2 3ʹ-UTR (Figure 3D). Moreover, transfection of a miR-423 inhibitor increased the basal activity of the TNIP2 3ʹ-UTR reporter in MDA-MB-231 cancer cells in a dose-dependent manner (Figure 3E). Furthermore, cell invasion was significantly enhanced in MDA-MB-231 cells that were cotransfected with miR-423 inhibitor and TNIP2 siRNA compared to cells that were transfected with miR-423 inhibitor alone (Figure 3F). Taken together, these results showed that TNIP2 is a direct target of miR-423 in breast cancer cells.
Figure 3.
miR-423 directly targets TNIP2 in breast cancer cells. (A) Sequence of the TNIP2 3ʹ-UTR shows the miR-423 binding seed region and mutation of the TNIP2 3ʹ-UTR seed region. (B) TNIP2 protein expression in MCF-7 and SKBR3 cells transfected with miR-423 or negative control (NC). α-Tubulin was used as loading control. (C) TNIP2 protein expression in MDA-MB231 cells transfected with miR-423 inhibitor or negative control (NC). (D) Relative luciferase activity of MCF-7 and SKBR3 cells co-transfected with increasing amounts of miR-423 mimic oligonucleotides (20 and 50 nM), and the pGL3 control reporter, pGL3-TNIP2-3ʹUTR reporter, or pGL3-TNIP2-3ʹUTR-mu reporter. (E) Relative luciferase activity of MDA-MB-231 cells transfected with increasing amounts of miR-423 mimic oligonucleotides (20 and 50 nM) or miR-423 inhibitor oligonucleotides (20 and 50 nM). (F) Transwell assay of indicated cells. Bars represent the mean ± SD of three independent experiments; **P < 0.01,*P < 0.05.
miR-423 Activates the NF-κB Pathway Through Regulation of TNIP2
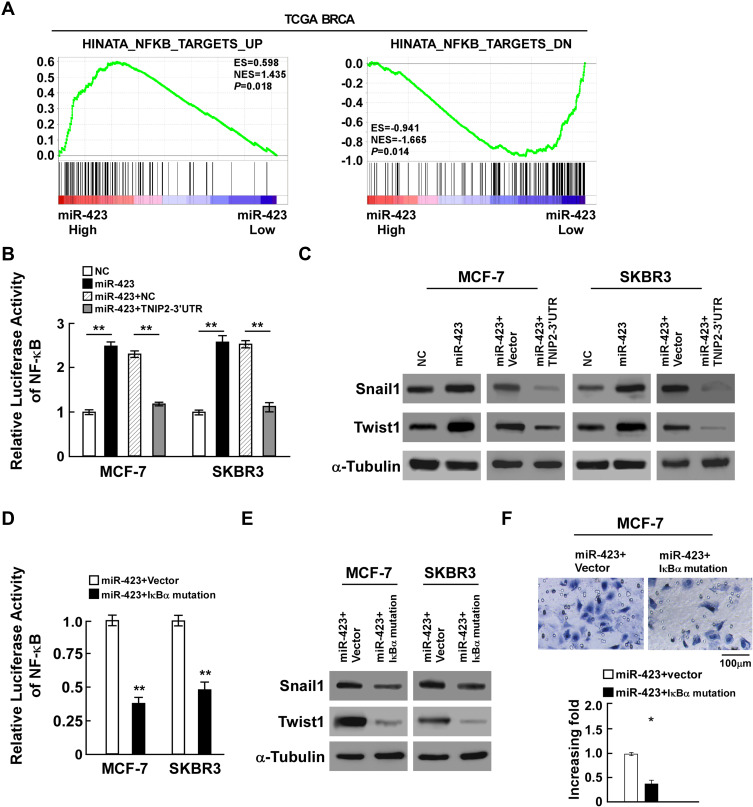
Since TNIP2 is an important negative regulator of the NF-κB pathway, to further investigate the role of miR-423 in breast cancer malignancy, gene set enrichment analysis (GSEA) of the TCGA BRCA cohort was performed and showed that higher miR-423 levels were positively correlated with the NF-κB-induced gene signature and inversely correlated with the NF-κB-suppressed gene signature (Figure 4A). In accordance with the results, luciferase reporter assays proved that overexpressed miR-423 significantly enhanced the activity of the NF-κB luciferase reporter activity, whereas the luciferase activity in cells cotransfected with miR-423 and TNIP2 3ʹ-UTR was lower than that in cells transfected with miR-423 alone (Figure 4B). Consistently, we also observed that upregulation of miR-423 substantially increased the protein and mRNA expression levels of the NF-κB downstream genes snail1 and twist1, whereas coexpression of miR-423 and the TNIP2 3ʹ-UTR resulted in decreased expression of these genes (Figures 4C and Supplementary Figure 3A). To further validate whether miR-423 promoted the invasiveness of breast cancer cells by the NF-κB pathway, a plasmid of mutated IκBα was employed to block the activation of the NF-κB pathway. In miR-423-overexpressing cells, the NF-κB luciferase activity of cells transfected with mutated IκBα was repressed (Figure 4D), and the expression of snail1 and twist1 was also downregulated (Figures 4E and Supplementary Figure 3B). In addition, transwell assays indicated that the invasiveness of MCF-7 cells was inhibited in miR-423-overexpressing cells after transfection with mutated IκBα (Figure 4F). Furthermore, after transfecting TNIP2 ORF in miR-423 overexpressed MCF-7 cells, we found that the TNIP2 expression was increased (Supplemental Figure 4A), and that NF-κB activity was suppressed compared to control (Supplemental Figure 4B), indicating that TNIP2 down-regulation was essential for miR-423 enhanced NF-κB activation. In conclusion, our data suggest that miR-423 activates the NF-κB pathway through regulation of TNIP2.
Figure 4.
miR-423 activates the NF-κB pathway through regulation of TNIP2. (A) GSEA plot showing miR-423 expression was positively correlated with NF-κB-induced gene signature and AKT-activated gene signature, and inversely correlated with NF-κB-suppressed gene signature in TCGA BRCA cohorts. (B) Luciferase activity of NF-κB activity in indicated cells. (C) Western blot analysis of snail1 and twist1 expression in indicated cells, α-Tubulin was used as loading control. (D) Luciferase activity of NF-κB activity in miR-423 overexpressing cells with transfected the plasmids of vector or IκBα mutation. (E) Western blot analysis of snail and twist expression in miR-423 overexpressing cells with transfected the plasmids of vector or IκBα mutation, α-Tubulin was used as loading control. (F) Transwell assays showed the invasiveness of MCF-7 cells was inhibited in miR-423-overexpressing cells after transfected with IκBα mutation. Bars represent the mean ± SD of three independent experiments; **P < 0.01,*P < 0.05.
Discussion
In recent decades, it has been generally recognized that TNIP family proteins play an important role in regulating NF-κB pathway activity during cancer progression.25–27 TNIP2 (also known as ABIN2) was initially discovered in a yeast two-hybrid screen as the binding partner of A20, a negative regulator of NF-κB signaling.28 Overexpression of TNIP2 was reported to inhibit TNFα-induced NF-κB activation and contribute to suppressing cell proliferation in human hepatocellular carcinoma.29 Dong et al reported that A20, TNIP-1/2, and CARD11 mutations have prognostic value in gastrointestinal diffuse large B-cell lymphoma, suggesting that TNIP2 is associated with carcinogenesis.30 Currently, the regulation of TNIP proteins has been emphasized in multiple studies. Zhou et al reported that miR-1180 targets TNIP2 to promote tumor progression during the development of hepatocellular carcinoma.31 In addition, miR-let-7a was shown to regulate USP35 expression, which then inhibited NF-κB activation by deubiquitination and stabilization of TNIP2 protein.32 However, the regulation of TNIP2 in breast cancer has not yet been clearly elucidated. In breast cancers, NF-κB constitutive activation is extensively observed, whereas the mRNA level of the NF-κB negative regulator TNIP2 is barely altered compared to normal tissues, which indicates that TNIP2 may be regulated in a posttranscriptional manner. Our current study determined that the microRNA miR-423 is upregulated in breast cancers and suppresses TNIP2 expression by directly targeting its 3ʹ-UTR. These findings expand our knowledge of TNIP2 regulation in cancers.
Presently, the early diagnosis and treatment of early breast cancer has greatly improved the five-year survival rate for patients with early stage breast cancer (survival rate of approximately 90%). However, the 5-year survival rate for patients with advanced breast cancer is only approximately 23% after tumor metastasis.33 The discovery of effective diagnostic and prognostic biomarkers and therapeutic methods is urgently required for the diagnosis and treatment of breast cancer. Numerous studies have shown that miRNAs may be considered valuable diagnostic and prognostic markers for cancer.34–36 In this study, we found that miR-423 is significantly upregulated in breast cancers. Additionally, miR-423 expression levels are correlated with breast cancer progression and overall patient survival, indicating that upregulated miR-423 may contribute to the development and progression of breast cancer and may be considered a potential diagnostic and prognostic biomarker.
Tumor metastasis is an important biological feature of malignant tumors and is one of the most important causes of cancer-related death.37 Multiple signaling pathways are involved in regulating the metastasis of cancers, including the NF-κB signaling pathway, which further activates its downstream genes, such as the metastasis-related transcription factors snail and twist.38,39 Large-scale bioinformatics analyses indicated that upregulated miR-423 expression is correlated with NF-κB signaling pathway-altered transcriptional signatures and the metastasis classification of breast cancers. Furthermore, we determined that miR-423 promotes invasion of breast cancer cells by enhancing NF-κB signaling pathway activation and elevating its downstream gene expression. Thus, our findings unveil a novel mechanism that promotes and sustains NF-κB constitutive activation in breast cancers, which further suggests that miR-423 should be considered as a novel target in the therapy of metastasis in breast cancer.
Previously, it was reported that miR-423 was upregulated in some types of cancers, including lung cancer,40 prostate cancer,41 head and neck squamous cell carcinoma.42 However some reports indicated that miR-423 served as tumor suppressor, such as colon cancer,43 ovarian.44 It seemed that the role of miR-423 was quite different dependent on the type of cancers. In breast cancer, miR-423 was reported to be upregulated in breast cancer tissues compared to adjacent normal tissues.45 However, the clinic significance of miR-423 in breast cancer was not clear. In this study, we found that overexpression of miR-423 was correlated with poor prognosis in breast cancer. Besides, in breast cancer, analysis of single nucleotide polymorphisms (SNP) demonstrated that CC genotype in pre-miR-423 reduced the risk of breast cancer.46 And the mutation type, pre-miR-423-12A, obviously correlated with clinicopathologic variables and promoted proliferation.47 Deep sequencing preliminarily indicated that patients with metastasis showed increased expression of miR-423.48 Consistently, in lung adenocarcinoma miR-423-5p was found to be a marker of bone metastasis.49 However, the biological role of miR-423 was still unclear. We found that miR-423 promoted breast cancer cell invasion and further investigated the detailed mechanisms. In the previous studies, miR-423 was found to target MYBL2 in lung adenocarcinoma,40 GRIM-19 in prostate cancer,41 NACC1 in ovarian cancer.44 Our results revealed that miR-423 promoted NF-κB signaling pathway via downregulating TNIP2, and increased the expression of Snail and Twist, providing new insights for the regulation of NF-κB pathway. In the future, we will focus our work on investigate whether miR-423 promoted bone metastasis in breast cancer.
In conclusion, the key findings of the current study provide new insights into the important role of miR-423 overexpression in the activation of the NF-κB signaling pathway and the promotion of breast cancer metastasis by directly targeting TNIP2, the vital negative regulator of the NF-κB pathway. Therefore, our results demonstrate that miR-423 functions as an oncomiR in breast cancer and reveal a novel mechanism for NF-κB activation in breast cancer, which suggests that miR-423 is a potential diagnostic marker and promising therapeutic target for breast cancer, especially in NF-κB-driven breast cancer metastasis.
Acknowledgments
We are grateful to Jian Qi for technical assistance. Ting Dai, Xiaohui Zhao, and Yun Li are co-first authors for this study.
Funding Statement
This work was supported by the Provincial Natural Science Foundation of Guangdong (Grant Nos. 2019A1515011201, 2018A030313560, 2020A1515010009), the Education Department of Guangdong Province (Nos. 2019KTSCX137, 2016KTSCX111), National Natural Science Foundation of China (Nos. 81501049, 81502103), Guangzhou Municipal Science and Technology Project (Grant No. 201904010067), the Medical Scientific Research Foundation of Guangdong Province, China (Grant No. A2018354), and Academic Project of Guangzhou Medical University (B185004136).
Ethical Approval and Consent to Participate
All the experiments were approved and monitored by Medical Ethics Committee of The First Affiliated Hospital of Sun Yat-sen University (20141214579). For each patient a written informed consent was obtained.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Porter P. “Westernizing” women’s risks? Breast cancer in lower-income countries. N Engl J Med. 2008;358:213–216. doi: 10.1056/NEJMp0708307 [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krop IE, Kim SB, González-Martín A, et al. TH3RESA study collaborators. Trastuzumab emtansine versus treatment of physician’s choice for pretreated HER2-positive advanced breast cancer (TH3RESA): a randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:689–699. doi: 10.1016/S1470-2045(14)70178-0 [DOI] [PubMed] [Google Scholar]
- 5.Lorusso G, Rüegg C. New insights into the mechanisms of organ-specific breast cancer metastasis. Semin Cancer Biol. 2012;22:226–233. doi: 10.1016/j.semcancer.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 6.Lee JH, Nan A. Combination drug delivery approaches in metastatic breast cancer. J Drug Deliv. 2012;2012:915375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karin M, Greten FR. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703 [DOI] [PubMed] [Google Scholar]
- 8.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- 9.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber MA, Azoitei N, Baumann B, et al. NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;14:569–581. doi: 10.1172/JCI200421358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870 [DOI] [PubMed] [Google Scholar]
- 12.Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune Responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225 [DOI] [PubMed] [Google Scholar]
- 13.Sakai Y, Uchida K, Nakayama H. A20 and ABIN-3 possibly promote regression of trehalose 6,6ʹ-dimycolate (TDM)-induced granuloma by interacting with an NF-kappa B signaling protein, TAK-1. Inflamm Res. 2012;61:245–253. doi: 10.1007/s00011-011-0406-6 [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 15.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruvkun G. Clarifications on miRNA and cancer. Science. 2006;311:36–37. doi: 10.1126/science.311.5757.36d [DOI] [PubMed] [Google Scholar]
- 17.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godlewski J, Nowicki MO, Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629 [DOI] [PubMed] [Google Scholar]
- 19.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 20.Rutnam ZJ, Yang BB. The non-coding 3ʹ UTR of CD44 induces metastasis by regulating extracellular matrix functions. J Cell Sci. 2012;125:2075–2085. doi: 10.1242/jcs.100818 [DOI] [PubMed] [Google Scholar]
- 21.Jiang S, Zhang HW, Lu MH, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250 [DOI] [PubMed] [Google Scholar]
- 22.Li S, Zeng A, Hu Q, Yan W, Liu Y, You Y. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017;19:55–65. doi: 10.1093/neuonc/now129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiuso P, Potenza N, Lombardi A, et al. MicroRNA-423-5p promotes autophagy in cancer cells and is increased in serum from hepatocarcinoma patients treated with sorafenib. Mol Ther Nucleic Acids. 2015;4:e233. doi: 10.1038/mtna.2015.8 [DOI] [PubMed] [Google Scholar]
- 24.Fang Z, Tang J, Bai Y, et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. 2015;34:86. doi: 10.1186/s13046-015-0198-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banks CA, Boanca G, Lee ZT, et al. TNIP2 is a hub protein in the NF-κB network with both protein and RNA mediated interactions. Mol Cell Proteomics. 2016;15:3435–3449. doi: 10.1074/mcp.M116.060509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang L, Verstrepen L, Heyninck K, et al. ABINs inhibit EGF receptor-mediated NF-κB activation and growth of EGF receptor-overexpressing tumour cells. Oncogene. 2008;27:6131–6140. doi: 10.1038/onc.2008.208 [DOI] [PubMed] [Google Scholar]
- 27.Van Huffel S, Delaei F, Heyninck K, De Valck D, Beyaert R. Identification of a novel A20-binding inhibitor of nuclear Factor-κB activation termed ABIN-2. J Biol Chem. 2001;276:30216–30223. doi: 10.1074/jbc.M100048200 [DOI] [PubMed] [Google Scholar]
- 28.Tan G, Wu L, Tan J, et al. MiR-1180 promotes apoptotic resistance to human hepatocellular carcinoma via activation of NF-κB signaling pathway. Sci Rep. 2016;6:22328. doi: 10.1038/srep22328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong G, Chanudet E, Zeng N, et al. A20, ABIN-1/2, and CARD11 mutations and their prognostic value in gastrointestinal diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:1440–1451. doi: 10.1158/1078-0432.CCR-10-1859 [DOI] [PubMed] [Google Scholar]
- 30.Zhou X, Zhu HQ, Ma CQ, et al. MiR-1180 promoted the proliferation of hepatocellular carcinoma cells by repressing TNIP2 expression. Biomed Pharmacother. 2016;79:315–320. doi: 10.1016/j.biopha.2016.02.025 [DOI] [PubMed] [Google Scholar]
- 31.Liu C, Wang L, Chen W, et al. USP35 activated by miR let-7a inhibits cell proliferation and NF-κB activation through stabilization of ABIN-2. Oncotarget. 2015;6:27891–27906. doi: 10.18632/oncotarget.4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gluck S. The prevention and management of distant metastases in women with breast cancer. Cancer Invest. 2007;25:6–13. doi: 10.1080/07357900701226974 [DOI] [PubMed] [Google Scholar]
- 33.Tu CC, Cheng LH, Hsu HH, et al. Activation of snail and EMT-like signaling via the IKKαβ/NF-κB pathway in apicidin-resistant HA22T hepatocellular carcinoma cells. Chin J Physiol. 2013;56:326–333. doi: 10.4077/CJP.2013.BAB158 [DOI] [PubMed] [Google Scholar]
- 34.Kim JH, Park S, Chung H, Oh S. Wnt5a attenuates the pathogenic effects of the Wnt/β-catenin pathway in human retinal pigment epithelial cells via down-regulating β-catenin and snail. BMB Rep. 2015;48:525–530. doi: 10.5483/BMBRep.2015.48.9.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Klymkowsky MW. Unexpected functional redundancy between Twist and Slug (Snail2) and their feedback regulation of NF-κB via Nodal and Cerberus. Dev Biol. 2009;331:340–349. doi: 10.1016/j.ydbio.2009.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan Z, Zheng H, Liu X, et al. MicroRNA-1229 overexpression promotes cell proliferation and tumorigenicity and activates Wnt/β-catenin signaling in breast cancer. Oncotarget. 2016;7:24076–24087. doi: 10.18632/oncotarget.8119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang L, Yu L, Zhang X, et al. miR-892b silencing activates NF-kappaB and promotes aggressiveness in breast cancer. Cancer Res. 2016;76:1101–1111. doi: 10.1158/0008-5472.CAN-15-1770 [DOI] [PubMed] [Google Scholar]
- 38.Jiang L, Lin C, Song L, et al. MicroRNA-30e* promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-κB/IκBα negative feedback loop. J Clin Invest. 2012;122:33–47. doi: 10.1172/JCI58849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weidner N, Semple J, Welch WR, et al. Tumor angiogenesis and metastasis–correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101 [DOI] [PubMed] [Google Scholar]
- 40.Wang R, Li G, Zhuang G, Sun S, Song Z. Overexpression of microRNA-423-3p indicates poor prognosis and promotes cell proliferation, migration, and invasion of lung cancer. Diagn Pathol. 2019;14:53. doi: 10.1186/s13000-019-0831-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin H, Lin T, Lin J, et al. Inhibition of miR-423-5p suppressed prostate cancer through targeting GRIM-19. Gene. 2019;88:93–97. doi: 10.1016/j.gene.2018.11.021 [DOI] [PubMed] [Google Scholar]
- 42.Hui AB, Lenarduzzi M, Krushel T, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166 [DOI] [PubMed] [Google Scholar]
- 43.Jia W, Yu T, An Q, Cao X, Pan H. MicroRNA-423-5p inhibits colon cancer growth by promoting caspase-dependent apoptosis. Exp Ther Med. 2018;16:1225–1231. doi: 10.3892/etm.2018.6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang X, Zeng X, Huang Y, et al. miR-423-5p serves as a diagnostic indicator and inhibits the proliferation and invasion of ovarian cancer. Exp Ther Med. 2018;15:4723–4730. doi: 10.3892/etm.2018.6015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun X, Huang T, Zhang C, et al. Long non-coding RNA LINC00968 reduces cell proliferation and migration and angiogenesis in breast cancer through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell Cycle. 2019;18:1908–1924. doi: 10.1080/15384101.2019.1632641 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Smith RA, Jedlinski DJ, Gabrovska PN, Weinstein SR, Haupt L, Griffiths LR. A genetic variant located in miR-423 is associated with reduced breast cancer risk. Cancer Genomics Proteomics. 2012;9:115–118. [PubMed] [Google Scholar]
- 47.Zhao H, Gao A, Zhang Z, et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumour Biol. 2015;36:4763–4771. doi: 10.1007/s13277-015-3126-7 [DOI] [PubMed] [Google Scholar]
- 48.Farazi TA, Horlings HM, Ten Hoeve JJ, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–4453. doi: 10.1158/0008-5472.CAN-11-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun G, Ding X, Bi N, et al. Molecular predictors of brain metastasis-related microRNAs in lung adenocarcinoma. PLoS Genet. 2019;15:e1007888. doi: 10.1371/journal.pgen.1007888 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]