Abstract
We investigated gene expression profiles of the corpus luteum (CL) at the time of maternal recognition to evaluate the functional changes of the CL during early pregnancy in cows and help improve reproductive efficiency and avoid defective fetuses. Microarray analyses using a 15 K bovine oligo DNA microarray detected 30 differentially expressed genes and 266 differentially expressed genes (e.g., PPARD and CYP21A2) in the CL on pregnancy days 15 (P15) and 18 (P18), respectively, compared with the CL on day 15 (NP15) of non-pregnancy (n = 4 for each group). PPARD expression was the highest while the CYP21A2 expression was the lowest in P15 and P18 compared with that of NP15. These microarray results were validated by quantitative real-time PCR analysis. The addition of interferon-τ and supernatants derived from homogenized fetal trophoblast increased ISG15 and MX1 expressions in the cultured luteal tissue (P < 0.01), but did not affect PPARD and CYP21A2 expressions. PPARD expression in the luteal tissue was stimulated (P < 0.05) by GW0742, known as a selective PPARD agonist, and PPARD ligands (i.e., arachidonic, linoleic and linolenic acids). In contrast, CYP21A2 mRNA expression was not affected by both agonist and ligands. The concentration of prostaglandin (PG) E2 and PGF2α decreased after GW0742 stimulation and increased after arachidonic acid stimulation (P < 0.05). The addition of GW0742 and arachidonic acid increased progesterone (P4) concentration. Collectively, these findings suggest that high expression levels of PPARD and low expression levels of CYP21A2 in the CL during early pregnancy may support P4 production by bovine luteal cells.
Keywords: Corpus luteum, Cow, Maternal recognition, Microarray analysis, Unsaturated fatty acid
The corpus luteum (CL) is a transient ovarian organ formed by follicle cells following ovulation. Progesterone (P4), the primary product of the CL, is necessary for the establishment and maintenance of pregnancy. If pregnancy does not occur, the CL degenerates. On the other hand, when pregnancy is established, the CL lifespan is prolonged, and the CL continues to produce P4 during the gestation period in cows [1]. In all mammals, the CL is required during the early phase of pregnancy. The CL is required for the entire gestation period in some species (e.g., cows, pigs, goats and dogs) [2], but not for other species (e.g., primates, sheep) as luteal P4 secretion can be replaced by placental P4 secretion [3]. These findings suggest that the physiological roles of the pregnant CL differ based on the species, and that normal CL function is crucial for maintaining pregnancy in cows.
Interferon-τ (IFNT), a pregnancy recognition hormone in ruminants, is produced by the mononuclear trophectoderm cells of conceptuses at a critical time to prevent the regression of CL. IFNT binds to interferon receptors (IFNαR1 and IFNαR2)[4] and inhibits prostaglandin (PG) F2α production by endometrium of ruminants [5]. A previous study demonstrated that the infusion of IFNT into the uterine vein inhibited spontaneous luteolysis in ewes [6]. The concentration of bioactive IFNT has been found to be higher in uterine venous blood than in arterial blood on day 15 of pregnancy in ewes [7]. Interferon-stimulated gene 15 ubiquitin-like modifier (ISG15), a marker gene of IFN response, was up-regulated in the CL during the maternal recognition period in pregnant cows [8, 9] and pregnant ewes [7]. Furthermore, the number of neutrophils and interleukin-8 expression increased in the bovine CL during early pregnancy (day 16 after artificial insemination) and increased after exposure to IFNT in vitro [10]. Thus, IFNT may influence not only the uterine environment but also the CL function in cows via local circulation or peripheral circulation. Understanding the role of CL function during pregnancy may help to determine a way for the improvement of the reproductive efficiency and reduction of the number of defective fetuses. Therefore, in the present study we evaluated the functional changes of the CL during early pregnancy in cows by assessing the global gene expression profiles of the CL at the time of maternal recognition.
Materials and Methods
Collection of bovine CL
Bovine ovaries containing CL were obtained from Japanese Black cows at the institute ranch within 10–30 min of exsanguination. For microarray analysis and immunohistochemistry, tissue samples were collected from cows on days 15 and 18 after artificial insemination (n = 4 animals/stage). The day of artificial insemination was designated as day 1. The uterine horn ipsilateral of the CL was obtained and immediately cut open to see the endometrium. The presence or absence of fetal trophoblast was assessed macroscopically to determine whether the cows were pregnant or not. The CLs were immediately separated from the ovaries, cut into small pieces (< 0.5 cm3), submerged in RNAlater (Qiagen GmbH, Hilden, Germany) or in 10% neutral formalin, and stored until later use.
Supernatants derived from the homogenized fetal trophoblast on day 18 of pregnancy (FMP) were collected by a previously described method [11]. Briefly, the fetal trophoblast was transferred into 2.5 ml of chilled homogenized buffer (300 mM sucrose, 25 mM Tris-HCl, 2 mM EDTA, pH 7.4) containing a proteinase inhibitor tablet (cOmplete Ultra tablet EDTA-free, Roche Diagnostics, Tokyo, Japan), and was homogenized in an ice bath with a rotor-stator homogenizer (TissueRuptor; Qiagen) using three 30 s bursts at maximum speed with 20 sec intervals of cooling between each burst. The homogenate was subsequently centrifuged at 23,500 × g for 30 min at 4˚C. The supernatant was collected, and the total protein concentration was measured using the commercial protein assay kit (DC Protein Assay Kit, #500-0111JA, Bio-Rad Laboratories, Tokyo, Japan).
All procedures for animal experiments were performed in accordance with guidelines approved by the Animal Ethics Committee of the National Institute of Agrobiological Sciences (#H18-036-3).
Microarray analysis
A custom-made 15 K bovine oligo DNA microarray (Agilent Technologies, Palo Alto, CA, USA) was used for the microarray analysis and performed according to methods described by previous reports [12, 13]. After verifying the quality of the RNA with a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technology, Wilmington, DE, USA) and an Experion RNA StdSens kit (#700-7104JA, Bio-Rad Laboratories), a one-color microarray analysis was performed. The RNA integrity was confirmed, and all samples had a A260/280 ratio and an RNA integrity number greater than 1.8 and 8.2, respectively. The microarray data from each sample were imported into the GeneSpring 12 (Agilent Technologies) software to use its normalization algorithm and for the detection of the candidate gene. Normalization was performed by dividing each measurement of each array by the median of all measurements in that array (per chip normalization). The Gene Expression Omnibus (GEO, available online: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) accession numbers are as follows: GPL9284 for the platform, GSM3683318 to GSM3683329 for the samples, and GSE128706 for the series.
Real-time PCR
Total RNA isolation and subsequent reverse transcription and real-time PCR steps were performed as per a previously described method [14]. The primers encoding the bovine sequences were chosen using an online software package (http://primer3.ut.ee/) and synthesized as listed in Table 1. The primer length (18–22 bp) and GC contents of each primer (50 to 60%) were selected to avoid primer dimer formation.
Table 1. Primers used in real-time PCR.
| Gene | Sequence (5'-3') | GenBank accession no. | bp | |
|---|---|---|---|---|
| PPARD | For | ACTCACTTCCTTCCAGCAGC | NM_001083636 | 111 |
| Rev | TATTGAGGCTGCCACACGAG | |||
| CYP21A2 | For | GCTCAGAAACCTCCACCTCC | NM_174639 | 148 |
| Rev | GTTCAGCACCACCACCTCTT | |||
| ISG15 | For | GCAGACCAGTTCTGGCTGTCT | NM_174366 | 58 |
| Rev | CCAGCGGGTGCTCATCAT | |||
| MX1 | For | GAGGTGGACCCCCAAGGA | NM_173940 | 58 |
| Rev | CCACCAGATCGGGCTTTGT | |||
| MX2 | For | GGGCAGCGGAATCATCAC | NM_173941 | 55 |
| Rev | CTCCCGCTTTGTCAGTTTCAG | |||
| OAS1 | For | CCAAGTCAAACAAGCCATCGA | NM_178108 | 62 |
| Rev | CACATCGGAAACACCTCTCCTT | |||
| IFNαR1 | For | GGTCCAAAAAATACAGGCAAGC | NM_174552 | 101 |
| Rev | CCAATGACAGGCCTTTCAAGA | |||
| IFNαR2 | For | TCAGCGATTGGACCACTGAA | NM_174553 | 74 |
| Rev | GGCGCAACATACGAAATACCA | |||
| RN18S1 | For | AACGGCTACCACATCCAAGG | DQ222453 | 115 |
| Rev | ATTCCAATTACAGGGCCCCG | |||
For, forward; Rev, reverse.
Gene expression was measured by real-time PCR using the Mx3000P Real-time PCR analyzing system (Agilent Technologies) and a QuantiFast SYBR Green PCR kit (#204054, Qiagen) starting with 600 ng of reverse-transcribed total RNA. The expression of 18S ribosomal RNA (RN18S1) was used as an internal control. PCR was performed under the following conditions: (first step) 95˚C for 5 min, 45 cycles of 95˚C for 15 sec, 60˚C for 30 sec and (second step) 95˚C for 60 sec, and 60˚C for 30 sec. The reaction was then held at 25˚C. Each PCR was followed by determination of the melting curve to ensure single-product amplification. As standard curves, serial dilutions of appropriate cDNA were used for gene quantification. The obtained data were normalized on the basis of RN18S1 mRNA content. Use of the Mx3000P Real-time PCR analyzing system at elevated temperatures resulted in a reliable and sensitive quantification of the RT-PCR products with high linearity (the Pearson correlation coefficient r > 0.95). To exclude any contaminating genomic DNA, all experiments included controls that lacked the reverse transcription enzyme. As a negative control, water was used instead of RNA for the PCR to exclude possible contamination from buffers and tubes.
Immunohistochemistry
Immunohistochemistry for PPARD, CYP21A2, IFNαR1, and IFNαR2 in the bovine CLs on day 18 of pregnancy and on day 15 of non-pregnancy was performed using the automated Ventana HX System Discovery with a DabMapKit (Roche Diagnostics) as per a previously described method [15]. The 5 µm-thick sections from the paraffin-embedded luteal tissue were incubated at room temperature (20˚C) with rabbit polyclonal anti-human PPARD antibody (#ARP38765_T100, Aviva Systems Biology, San Diego, CA, USA; 1:50), rabbit polyclonal anti-human CYP21A2 antibody (#ARP60144_P050, Aviva Systems Biology; 1:25), rabbit polyclonal anti-human IFNαR1 antibody (#bs-4116R, Bioss, Woburn, MA, USA; 1:50) or rabbit polyclonal anti-human IFNαR2 antibody (#bs-7022R, Bioss; 1:50) for 12 h. The signals were detected using anti-rabbit IgG-Biotin conjugate (Sigma-Aldrich, LLC, St Louis, MO, USA; 1:100) for 1 h, and then counterstained with hematoxylin. Negative controls were assessed using normal rabbit IgG (#NBP2-24891, Novus Biologicals LLC, Littleton, CO, USA) diluted at concentrations equivalent to the primary antibodies. The sections were observed with a Leica DMRE HC microscope (Leica Microsystems, Tokyo, Japan) and photographed with a Nikon Digital Sight DS-Fi1-L2 (Nikon Instruments, Tokyo, Japan).
Luteal tissue culture
For the tissue culture study, luteal tissue samples were collected from cows on days 10–12 of the estrous cycle (n = 4 animals). The luteal tissues were obtained using a modified procedure from the one described previously [11]. The CL was washed three times with 30–50 ml sterile physiological saline supplemented with 100 IU/ml penicillin, 100 µg/ml streptomycin, and 0.1% BSA (#735078, Roche Diagnostics). The CL was sliced into approximately 0.2 mm-thick layers with a tissue slicer (#KN-822, Natsume Seisakusyo, Tokyo, Japan). The luteal strips were then cut into small square pieces (3 mm2) with sterile surgical blades. The tissues (approximately 15–25 mg wt) were pre-incubated in the Dulbecco’s Modified Eagle’s medium (DMEM; #D1152, Sigma-Aldrich) supplemented with 0.1% BSA. After pre-incubating for 4 h, the luteal tissues were placed in culture medium (DMEM/Ham’s F-12; 1:1 (v/v); #D8900, Sigma-Aldrich) supplemented with 10% (v/v) calf serum (#C6278, Sigma-Aldrich), 20 IU/ml penicillin, 20 µg/ml streptomycin, 0.05 µg/ml amphotericin B (516104, EMD Millipore, Billerica, MA, USA) and cultured at 37.5˚C in a humidified atmosphere of 5% CO2 in air. Cultured luteal tissues were further incubated in the culture medium with bovine IFNT (1.1 × 105 Units/mg), generated from the HEK293 cells as per a previously described method [16], supernatant derived from the homogenized FMP, GW0742 (a potent and highly selective PPARD agonist; #2229, Bio-Techne, Minneapolis, MN, USA), arachidonic acid (#90010, Cayman Chemical, Ann Arbor, MI, USA), linoleic acid (#L1012, Sigma-Aldrich), linolenic acid (#L2376, Sigma-Aldrich), or the bovine tumor necrosis factor-α (TNF; #2279-BT, R&D Systems, Minneapolis, MN, USA). After incubation for 18 h, the luteal tissue and supernatant were collected and stored at –80°C until use.
ELISA
The concentrations of PGE2, PGF2α and P4 in the culture media were examined using commercial ELISA kits (PGE2: #500141, PGF2α: #516011, P4: #582601; Cayman Chemical, Ann Arbor, MI, USA). To evaluate any damage in the luteal tissue during the culture period, lactate dehydrogenase (LDH) secretion in the culture media was measured using a cytotoxicity LDH assay kit-WST (#CK12, Dojindo Laboratories, Kumamoto, Japan).
Statistical analyses
Microarray data were statistically analyzed with the Student’s t-test and summarized using GeneSpring 12 (Agilent Technologies). The differences in mRNA expression in the endometrium between the non-pregnant and the pregnant groups (separately on days 15 and 18) were statistically analyzed with an unpaired Student’s t-test using the JMP 7 software (SAS Institute, Cary, NC, USA). Experimental data for real-time PCR and ELISA have been presented as the mean ± SEM. The statistical significance of differences in the abundance of mRNA expression and in protein concentrations between the control group and the treated group was analyzed using one-way ANOVA with the Dunnett’s Multiple Comparison post-hoc test with the KaleidaGraph 3.6 (Synergy Software, Reading, PA, USA) software package. A P < 0.05 was considered statistically significant.
Results
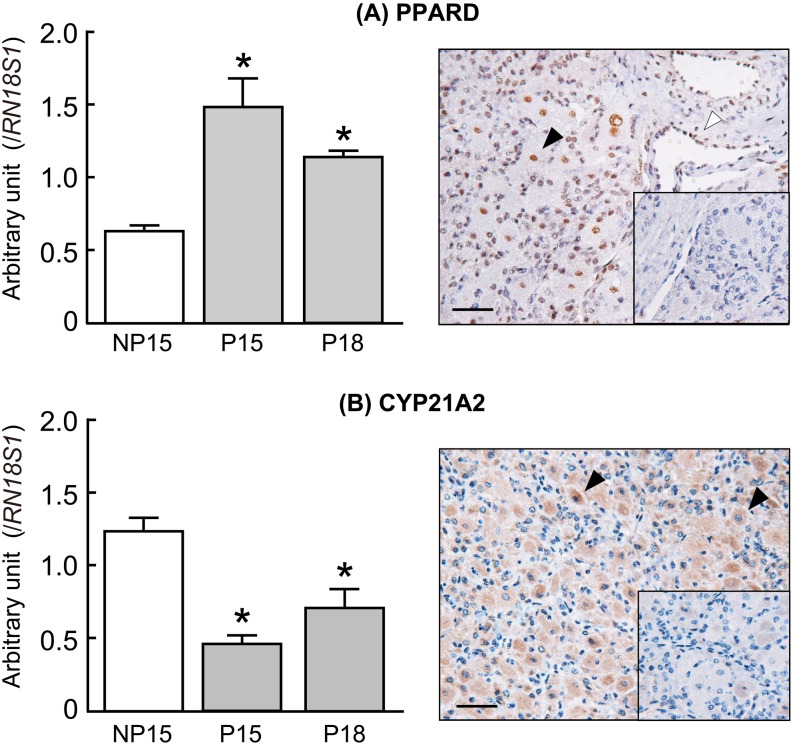
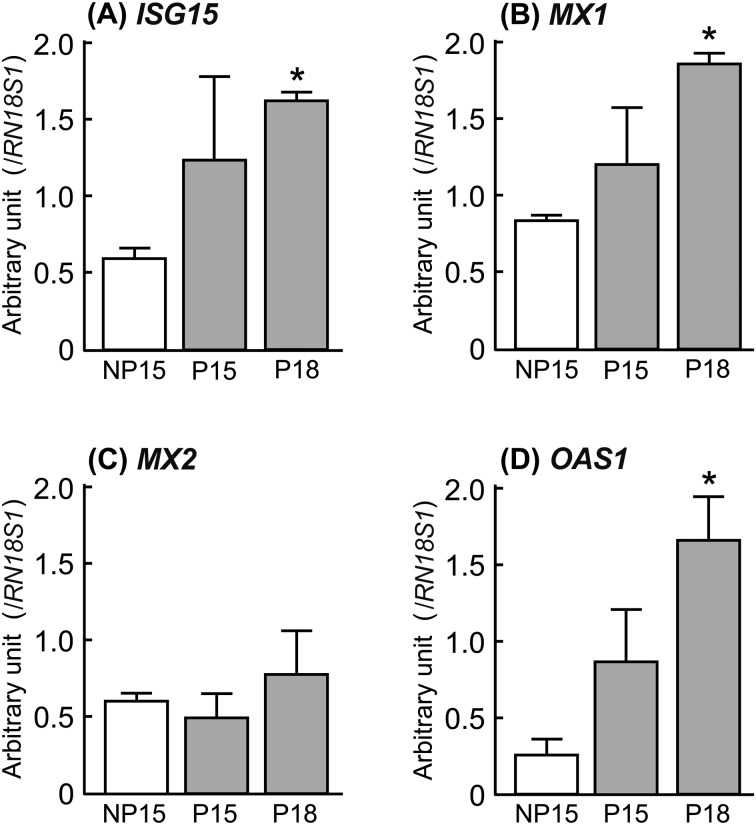
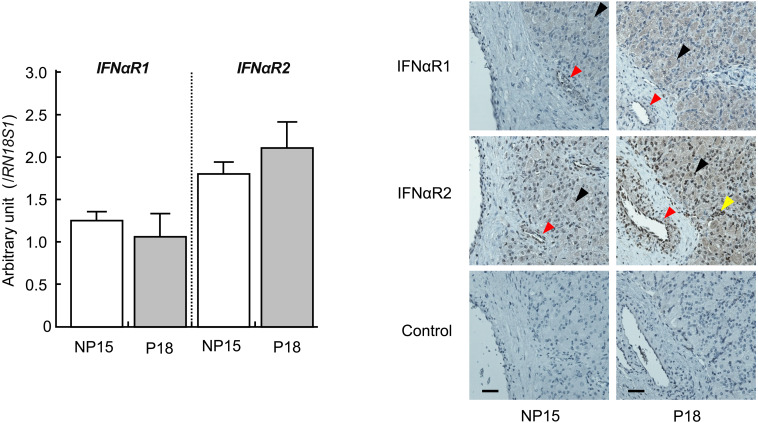
The microarray analysis detected 30 and 266 differentially expressed genes in the CL on pregnancy days 15 (P15) and 18 (P18) compared with the CL on day 15 of the non-pregnant cows (NP15; Table 2). Expression of PPARD was the highest and that of CYP21A2 was the lowest in the CL of pregnant cows (P15, P18) compared with that of the non-pregnant cows (NP15). These microarray results were validated by quantitative real-time PCR analysis (Fig. 1; P < 0.05). Immunohistochemical staining showed that PPARD was expressed in the nuclei of both the luteal and the endothelial cells and that CYP21A2 was expressed in the cytoplasm of the luteal cells (Fig. 1). In addition, transcripts of interferon-induced genes (ISG15, MX1, and OAS1) were more abundant in the CL on day 18 of pregnant cows than in the CL of non-pregnant cows (Fig. 2; P < 0.05). Although the mRNA of the type I interferon receptors (IFNαR1 and IFNαR2) was detected in the bovine CL during the estrous cycle and pregnancy, there were no significant changes in the expression of these receptors at these stages. Positive immunohistochemical staining of IFNαR1 and IFNαR2 was observed in the luteal cells, the endothelial cells, and the immune cells of the CL obtained from both pregnant and non-pregnant cows (Fig. 3).
Table 2. Top 10 genes of increased or decreased in the corpus luteum (CL) of pregnancy (P15 and P18) compared with the CL of non-pregnancy (NP15).
| Day 15 of pregnancy (30 genes) |
Day 18 of pregnancy (266 genes) |
|||||
|---|---|---|---|---|---|---|
| Gene | Accession No. | Fold | Gene | Accession No. | Fold | |
| (Increased) | (Increased) | |||||
| PPARD | NM_001083636 | 6.61 | PPARD | NM_001083636 | 15.9 | |
| MSX2 | NM_001079614 | 5.84 | CD5L | NM_001102119 | 11.9 | |
| PFN3 | NM_001077945 | 5.31 | ERCC2 | NM_001103317 | 11.8 | |
| MRPS28 | NM_001046401 | 4.63 | ISG15 | NM_174366 | 8.32 | |
| NUCB1 | NM_001075199 | 3.66 | ZNRD1 | NM_001046400 | 7.50 | |
| SSPO | NM_174706 | 3.17 | MSX2 | NM_001079614 | 5.08 | |
| TERF2IP | NM_001075412 | 3.13 | P2RY4 | AY540307 | 4.25 | |
| LAS1L | NM_001076815 | 3.11 | NOP10 | NM_001114521 | 4.04 | |
| ITGB5 | NM_174679 | 3.09 | LMO3 | NM_001046337 | 3.72 | |
| PPP2R4 | NM_001046173 | 2.96 | GJB1 | NM_174069 | 3.57 | |
| (Decreased) | (Decreased) | |||||
| CYP21A2 | NM_174639 | 5.10 | CYP21A2 | NM_174639 | 6.16 | |
| SCG2 | NM_174176 | 3.30 | CSNK1D | XM_002687933 | 4.87 | |
| EDEM3 | NM_001205853 | 2.68 | PLCG1 | NM_174425 | 4.55 | |
| PRF1 | NM_001143735 | 2.61 | FN1 | NM_001163778 | 4.30 | |
| HSD3B1 | NM_174343 | 2.60 | ANKRD50 | NM_001205949 | 4.17 | |
| UTS2R | NM_001040484 | 2.52 | HSPA1A | NM_174550 | 3.87 | |
| TPD52L1 | NM_001076033 | 2.36 | WRNIP1 | NM_001103106 | 3.85 | |
| POLR1B | NM_001100292 | 2.24 | EIF4B | NM_001035028 | 3.45 | |
| LRSAM1 | NM_001075296 | 2.17 | PPP4C | NM_001099108 | 3.41 | |
| SF3A2 | NM_001099211 | 2.10 | RPA1 | NM_001075176 | 3.34 | |
Fig. 1.
Changes in the relative amount of mRNA for (A) PPARD and (B) CYP21A2 in the bovine corpus luteum (CL) on days 15 and 18 of pregnant cows (P15 and P18, respectively) and on day 15 of non-pregnant cows (NP15). Data represent means ± SEM for four cows per stage and have been expressed as relative ratios of the mRNAs to 18S ribosomal RNA (RN18S1). Asterisks show significant differences between NP and P (* P < 0.05). Localization of PPARD and CYP21A2 in the bovine CL obtained on day 18 of pregnant cows. Intensive immunoreactivity was observed in the nuclei of luteal cells (black arrowheads) and endothelial cells (white arrowheads) for PPARD and in the cytoplasm of luteal cells (black arrowheads) for CYP21A2. No positive immunoreactivity was observed in the negative control (inserted panels). Scale bar = 50 µm.
Fig. 2.
Changes in the relative amount of mRNA for (A) ISG15, (B) MX1, (C) MX2 and (D) OAS1 in the bovine corpus luteum (CL) on days 15 and 18 of pregnant cows (P15 and P18, respectively) and on day 15 of non-pregnant cows (NP15). Data represent means ± SEM for four cows per stage and have been expressed as relative ratios of the mRNAs to 18S ribosomal RNA (RN18S1). Asterisks show significant differences between NP and P (* P < 0.05).
Fig. 3.
Changes in the relative amount of mRNA for IFNαR1 and IFNαR2 in the bovine corpus luteum (CL) on day 18 of pregnant cows (P18) and on day 15 of non-pregnant cows (NP15). Data represent means ± SEM for four cows per stage and have been expressed as relative ratios of the mRNAs to 18S ribosomal RNA (RN18S1). Localization of IFNαR1 and IFNαR2 in bovine CL obtained on P18 and NP15. Intensive immunoreactivity was observed in the luteal cells (black arrowheads), endothelial cells (red arrowheads), and immune cells (yellow arrowheads) obtained from the CL of both pregnant and nonpregnant cows. No positive immunoreactivity was observed in the negative control. Scale bar = 50 µm.
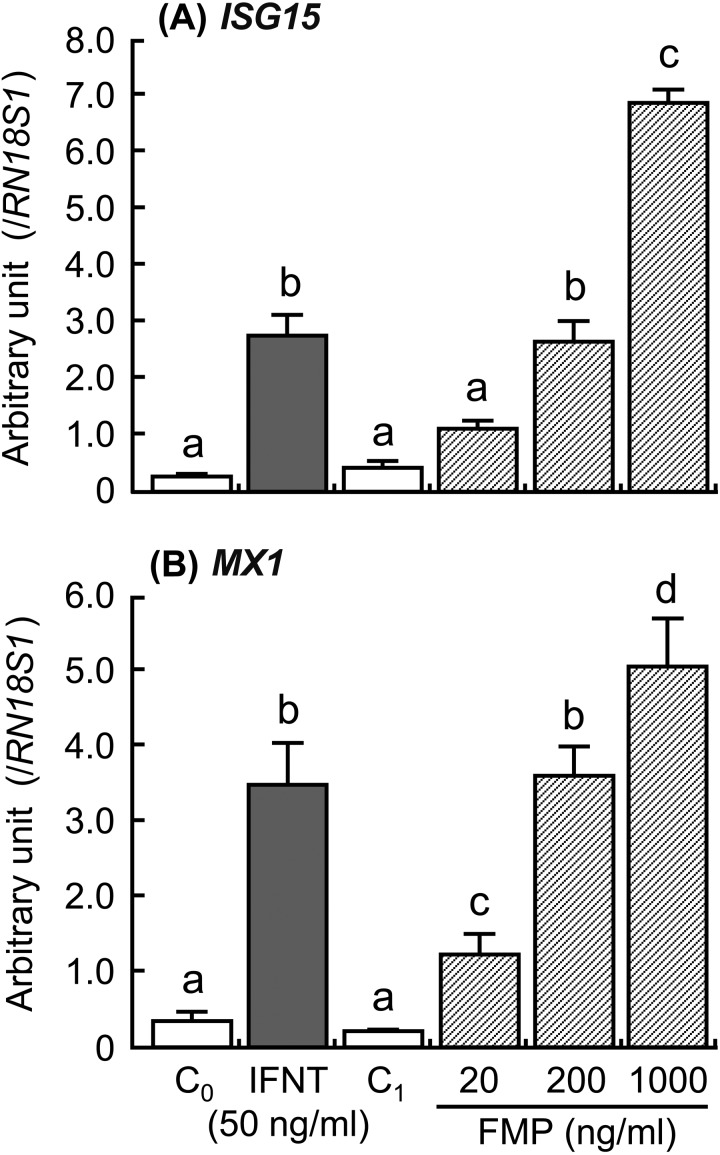
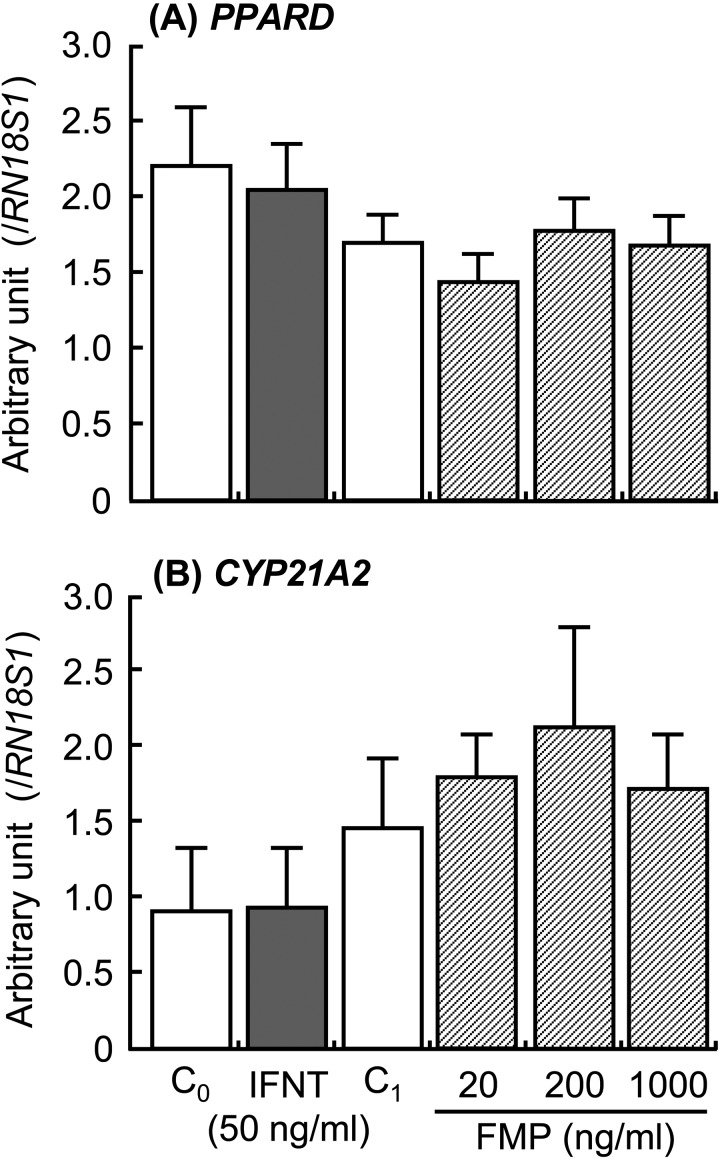
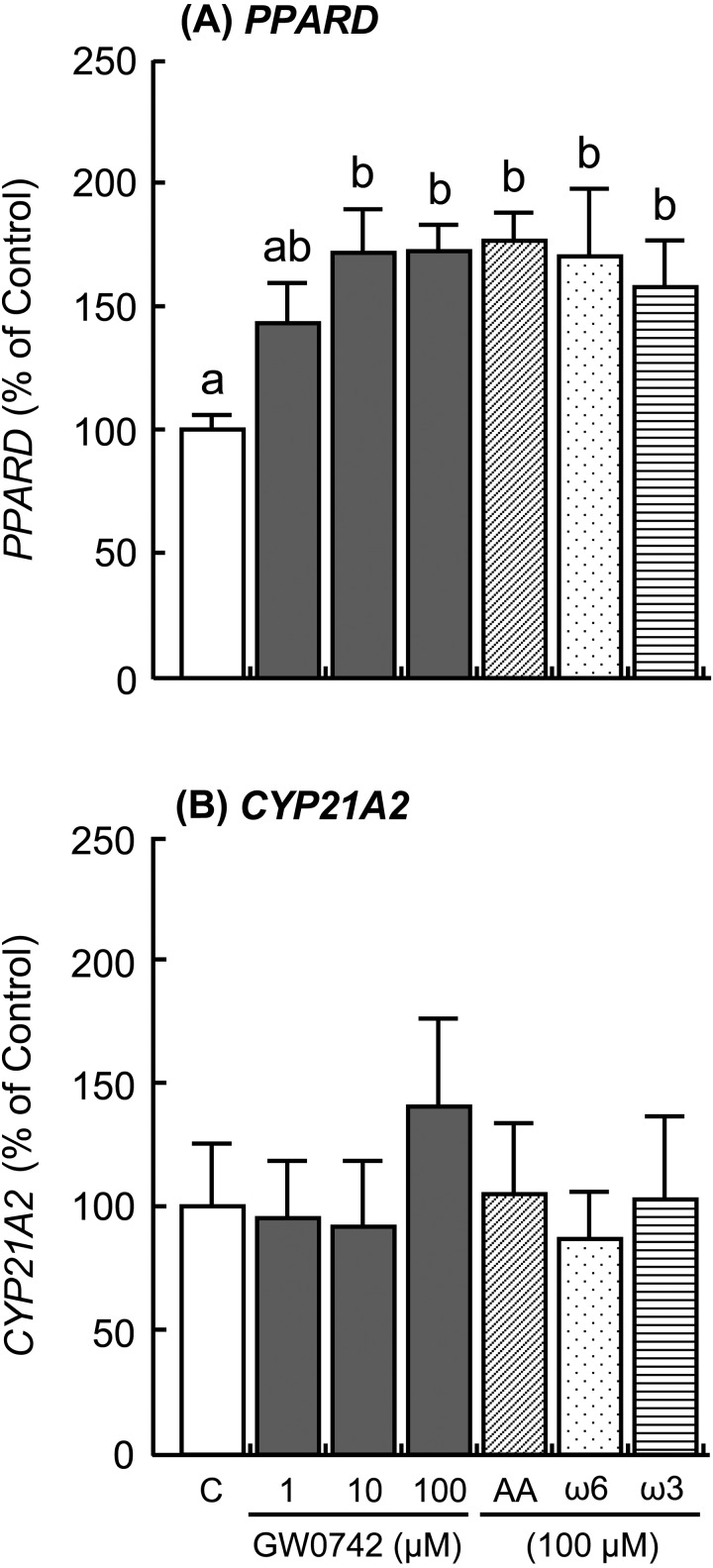
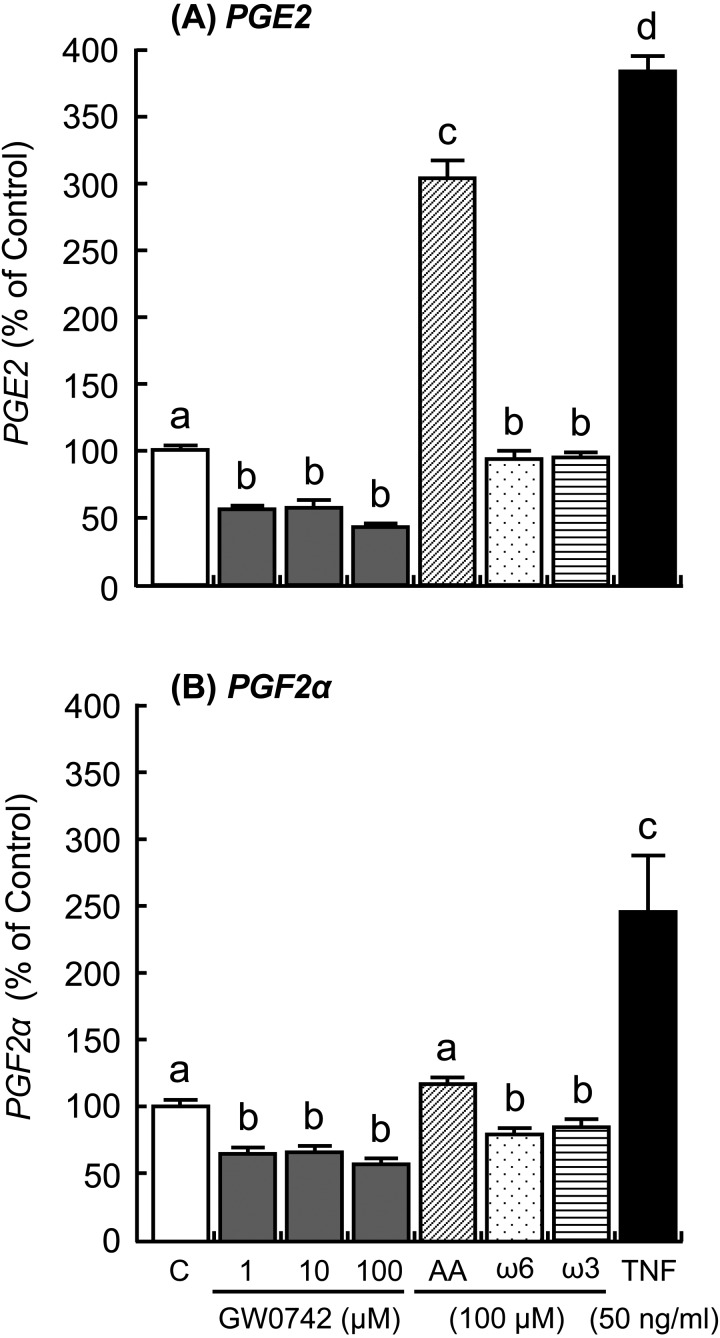
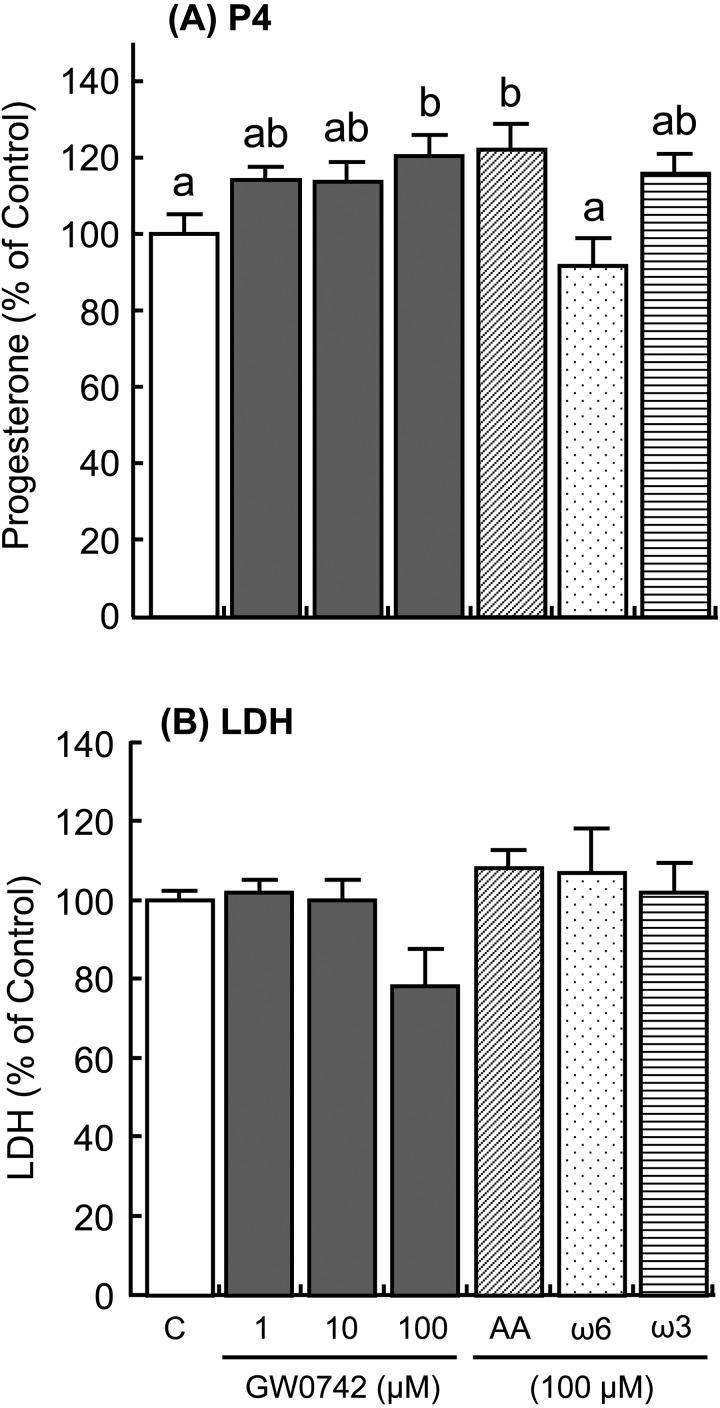
The addition of IFNT and FMP increased ISG15 and MX1 expressions in the cultured luteal tissue (Fig. 4; P < 0.01), but the addition did not affect PPARD and CYP21A2 expressions (Fig. 5). The PPARD expression in the luteal tissue was stimulated by the GW0742 drug, which is a known selective PPARD agonist (Fig. 6A; P < 0.05). Ligands of PPARD (arachidonic, linoleic, and linolenic acids) also stimulated PPARD expression (Fig. 6A; P < 0.05). In contrast, CYP21A2 mRNA expression was not affected by any of the tested substances (Fig. 6B). The concentrations of PGE2 and PGF2α decreased after stimulation with GW0742, linoleic acid (ω6) and linolenic acid (ω3), and PGE2 concentration increased after stimulation with arachidonic acid (Fig. 7; P < 0.05). TNF, which was used as a positive control, stimulated secretion of both PGE2 and PGF2α in the luteal tissue (Fig. 7; P < 0.01). Moreover, the addition of GW0742 and arachidonic acid resulted in increased P4 concentration in the luteal tissue (Fig. 8A; P < 0.05). The GW0742 and PPARD ligands did not affect LDH concentration in the tissue culture medium (Fig. 8B), suggesting that these substances may not cause serious damage to the luteal tissue.
Fig. 4.
Effects of interferon-τ (IFNT) and the supernatant derived from homogenized fetal trophoblast (FMP) on the mRNA expression of (A) ISG15 and (B) MX1 in the cultured bovine luteal tissues. Data represent means ± SEM of four cows and have been expressed as relative ratios of the mRNAs to 18S ribosomal RNA (RN18S1). C0 = culture supernatant containing 0.1%BSA-PBS; C1 = culture supernatant containing homogenized buffer. Different letters indicate significant differences (P < 0.05).
Fig. 5.
Effects of interferon-τ (IFNT) and the supernatant derived from homogenized fetal trophoblast (FMP) on the mRNA expression of (A) PPARD and (B) CYP21A2 in the cultured bovine luteal tissues. Data represent means ± SEM of four cows and have been expressed as relative ratios of the mRNAs to 18S ribosomal RNA (RN18S1). C0 = culture supernatant containing 0.1%BSA-PBS; C1 = culture supernatant containing homogenized buffer.
Fig. 6.
Effects of a selective agonist of PPARD (GW0742), arachidonic acid (AA), linoleic acid (ω6), and linolenic acid (ω3) on the mRNA expression of (A) PPARD and (B) CYP21A2 in the cultured bovine luteal tissues. Data represent means ± SEM of four cows and have been expressed as a relative percentage of the control value. C = culture supernatant containing 1% ethanol. Different letters indicate significant differences (P < 0.05).
Fig. 7.
Effects of a selective agonist of PPARD (GW0742), arachidonic acid (AA), linoleic acid (ω6), linolenic acid (ω3), and tumor necrosis factor α (TNF) on (A) PGE2 and (B) PGF2α secretion by the cultured bovine luteal tissues. Data represent means ± SEM of four cows per stage and have been expressed as a relative percentage of the control value. C = culture supernatant containing 1% ethanol. Different letters indicate significant differences (P < 0.05).
Fig. 8.
Effects of a selective agonist of PPARD (GW0742), arachidonic acid (AA), linoleic acid (ω6), and linolenic acid (ω3) on (A) progesterone (P4) and (B) lactate dehydrogenase (LDH) secretion by the cultured bovine luteal tissues. Data represent means ± SEM of four cows per stage and have been expressed as a relative percentage of the control value. C = culture supernatant containing 1% ethanol. Different letters indicate significant differences (P < 0.05).
Discussion
The results of the current study demonstrated that gene expression in bovine CL during early pregnancy differs from that in the CL during the estrous cycle. In this study, we focused on the expression of the luteal genes on P15 and P18 since IFNT production from the trophoblast drastically increases around these days [17], and the elongated conceptus is implanted into the endometrial epithelial cells on days 18–19 in cattle [18]. PPARD mRNA expression was the highest in the CL during P15 and P18, whereas the expression of CYP21A2 was the lowest, suggesting that stage-dependent changes in PPARD and CYP21A2 expression may have some roles in regulating communication in the maternal conceptus.
PPARs are nuclear transcription factors that regulate metabolic, anti-inflammatory, and tissue developmental processes. Three PPAR isotypes have been identified in mammals: PPARα, PPARβ/δ (PPARD), and PPARγ. PPARs are involved in the regulation of reproductive processes [19], i.e., the presence of PPARs has been reported in the embryos, follicular cells, luteal cells and placenta of various species [19,20,21,22,23]. In this study, PPARD mRNA expression was high in the CL on P15 and P18 compared with that on NP15, and PPARD protein was expressed in the nuclei of the luteal cells. Moreover, the PPARD agonist GW0742 inhibited secretion of both PGE2 and PGF2α by bovine luteal tissues. The role of PPARD in PG synthesis in the endometrium of cows [24] and pigs [25] has been demonstrated. Although the PPARD-ligand complex did not affect COX-2 expression in endometrial cells, it decreased PGF2α release during the mid-luteal phase of the estrous cycle and at the beginning of placental implantation [24, 26]. By contrast, GW0742 stimulated P4 production from the luteal tissues in the present study. It has been demonstrated that the PPARD-ligand complex regulates ovarian steroidogenesis [27] as well as ovarian angiogenesis and tissue remodeling [28]. The PPARD-ligand complex also induced P4 production by stimulating cholesterol uptake in the steroidogenic cells [29]. These findings may support the hypothesis that reduced PGF2α and increased P4 production during the time of maternal recognition imply PPARD involvement in CL maintenance.
To determine whether up-regulation of PPARD in the CL of early pregnancy is a pregnancy-dependent phenomenon, the effects of IFNT and supernatants derived from the homogenized fetal trophoblast (FMP) on PPARD expression were tested. Unexpectedly, both IFNT and FMP did not affect PPARD expression in the cultured luteal tissues, but they increased ISG15 and MX1 expressions. Then, since PPAR ligands include several naturally occurring long-chain polyunsaturated fatty acids, such as arachidonic, linoleic, and linolenic acids [30], the effects of these unsaturated fatty acids on PPARD expression in the cultured luteal tissues were evaluated in this study. As expected, PPARD expression in the luteal tissues was stimulated by the GW0742 as well as the arachidonic, linoleic and linolenic acids. It has been also demonstrated that docosahexaenoic acid, eicosapentaenoic acid, and eicosanoids (including leukotrienes and PGI2) are ligands to PPARs [21, 30]. Although we could not find any reports showing that the concentration of these PPARD ligands was increased in early pregnant cows, we hypothesize that unsaturated fatty acids may potentially be chosen as candidates to regulate PPARD expression in bovine CL. Further studies will be needed to clarify the regulatory mechanism of PPARD expression in the CL at the time of maternal recognition.
Dietary supplementation of fatty acids, including ω-3 (linolenic acids) and ω-6 (linoleic acids) families during the postpartum period, has the potential to increase pregnancy rates in dairy cows and alter the function of CL by improving the energy status and increasing precursors for the synthesis of reproductive hormones, such as PGs [31, 32]. In addition, it has been demonstrated that both linolenic and linoleic acids inhibit the secretion of eicosanoids in several cell types [33, 34]. In this study, both linoleic and linolenic acids inhibited the secretion of PGE2 and PGF2α in the cultured luteal tissues. A previous study also demonstrated that linolenic acids reduce the ovarian and endometrial synthesis of PGF2α by reducing the activation of PG synthase and decreasing the availability of its precursor arachidonic acid [35]. Therefore, unsaturated fatty acids, including linoleic and linolenic acids, may not only stimulate PPARD expression but may also inhibit PGE2 and PGF2α synthesis in bovine CL.
IFNT is a multifunctional cytokine that exhibits biological activity similar to that by other type-1 IFNs. It acts by binding to the cell surface receptors IFNαR1 and IFNαR2 [4]. Using immunohistochemistry, this study demonstrated that both of these receptors were present in the luteal and the endothelial cells. Moreover, IFNT stimulated the expressions of ISG15 and MX1 genes in the cultured luteal tissues. It has been previously shown that several ISGs are stimulated in the CL of ovine and bovine species during early pregnancy [7, 36], or after IFNT infusion in ewes [6]. A direct in vitro effect of IFNT on the various luteal cell types has been demonstrated in several studies [7,8,9,10, 37]. In addition, IFNT in a dose-dependent manner stimulated the expression of ISGs (ISG15 and OAS1) in the luteal tissues as well as in the endothelial cells of bovine CL [38]. Taken together, these results support the hypothesis that IFNT acts directly on bovine CL to stimulate the expression of ISGs.
Little information is available about the involvement of CYP21A2 in bovine reproduction. CYP21A2 is a cytochrome P450 with the largest number of naturally occurring mutants involved in steroidogenic hormone biosynthesis [39]. It is expressed in the adrenal cortex, plays an essential role in the biosynthesis of aldosterone and cortisol. CYP21A2 is also known to convert P4 and 17-hydroxyprogesterone (17OHP) to 11-deoxycorticosterone and 11-deoxycortisol, respectively [40, 41]. In the present study, expression of the CYP21A2 gene in the CL of early pregnancy was found to be low compared with that of non-pregnant cows. Immunohistochemical analyses revealed that the CYP21A2 protein was expressed in the cytoplasm of the luteal cells. However, CYP21A2 expression was not affected by stimulation with IFNT, FMP, arachidonic acid, linoleic acid, linolenic acid or TNF. The possible roles of CYP21A2 in regulating steroidogenesis in the CL at the time of maternal recognition will be determined in the near future.
Collectively, the results of the present study demonstrate that the number of genes whose expression changed in the CL, during the transition from the estrous cycle to pregnancy, gradually increased with the stage of gestation. This suggests that different gene expression profiles may contribute to functional changes in the bovine CL during early pregnancy, and embryonic signals, such as IFNT, may act on the CL via local circulation or peripheral circulation. Since PPARD induces P4 production by stimulating cholesterol uptake in steroidogenic cells whereas CYP21A2 enzymatically catabolizes P4 to deoxycorticosterone, high levels of PPARD expression and low levels of CYP21A2 expression in the CL of early pregnancy may support P4 production by bovine luteal cells. Moreover, PPARD expression in the CL may be up-regulated by unsaturated fatty acids but not by conceptus-derived IFNT, suggesting that these fatty acids may be involved in the establishment of pregnancy in cows.
Acknowledgments
This research was supported by the Grants-in-Aid for the Research Program on Innovative Technologies for Animal Breeding, Reproduction, and Vaccine Development (REP1001) from the Ministry of Agriculture, Forestry and Fisheries of Japan, and the JSPS KAKENHI (No. 17K08056). The authors thank Ms S Fujii, Ms H Kanahara, and the staff of the Livestock Operations Unit 1, Tsukuba Technical Support Center, NARO for their skilled technical assistance.
References
- 1.Niswender GD, Juengel JL, Silva PJ, Rollyson MK, McIntush EW. Mechanisms controlling the function and life span of the corpus luteum. Physiol Rev 2000; 80: 1–29. [DOI] [PubMed] [Google Scholar]
- 2.McDonald LE, McNutt SH, Nichols RE. On the essentiality of the bovine corpus luteum of pregnancy. Am J Vet Res 1953; 14: 539–541. [PubMed] [Google Scholar]
- 3.Henson MC. Pregnancy maintenance and the regulation of placental progesterone biosynthesis in the baboon. Hum Reprod Update 1998; 4: 389–405. [DOI] [PubMed] [Google Scholar]
- 4.Rosenfeld CS, Han CS, Alexenko AP, Spencer TE, Roberts RM. Expression of interferon receptor subunits, IFNAR1 and IFNAR2, in the ovine uterus. Biol Reprod 2002; 67: 847–853. [DOI] [PubMed] [Google Scholar]
- 5.Demmers KJ, Derecka K, Flint A. Trophoblast interferon and pregnancy. Reproduction 2001; 121: 41–49. [DOI] [PubMed] [Google Scholar]
- 6.Bott RC, Ashley RL, Henkes LE, Antoniazzi AQ, Bruemmer JE, Niswender GD, Bazer FW, Spencer TE, Smirnova NP, Anthony RV, Hansen TR. Uterine vein infusion of interferon τ (IFNT) extends luteal life span in ewes. Biol Reprod 2010; 82: 725–735. [DOI] [PubMed] [Google Scholar]
- 7.Oliveira JF, Henkes LE, Ashley RL, Purcell SH, Smirnova NP, Veeramachaneni DN, Anthony RV, Hansen TR. Expression of interferon (IFN)-stimulated genes in extrauterine tissues during early pregnancy in sheep is the consequence of endocrine IFN-τ release from the uterine vein. Endocrinology 2008; 149: 1252–1259. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Wang XL, Wan PC, Zhang LY, Wu Y, Tang DW, Zeng SM. Up-regulation of expression of interferon-stimulated gene 15 in the bovine corpus luteum during early pregnancy. J Dairy Sci 2010; 93: 1000–1011. [DOI] [PubMed] [Google Scholar]
- 9.Nitta A, Shirasuna K, Haneda S, Matsui M, Shimizu T, Matsuyama S, Kimura K, Bollwein H, Miyamoto A. Possible involvement of IFNT in lymphangiogenesis in the corpus luteum during the maternal recognition period in the cow. Reproduction 2011; 142: 879–892. [DOI] [PubMed] [Google Scholar]
- 10.Shirasuna K, Matsumoto H, Matsuyama S, Kimura K, Bollwein H, Miyamoto A. Possible role of interferon tau on the bovine corpus luteum and neutrophils during the early pregnancy. Reproduction 2015; 150: 217–225. [DOI] [PubMed] [Google Scholar]
- 11.Sakumoto R, Hayashi KG, Takahashi T. Different expression of PGE synthase, PGF receptor, TNF, Fas and oxytocin in the bovine corpus luteum of the estrous cycle and pregnancy. Reprod Biol 2014; 14: 115–121. [DOI] [PubMed] [Google Scholar]
- 12.Kizaki K, Shichijo-Kizaki A, Furusawa T, Takahashi T, Hosoe M, Hashizume K. Differential neutrophil gene expression in early bovine pregnancy. Reprod Biol Endocrinol 2013; 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakumoto R, Hayashi KG, Hosoe M, Iga K, Kizaki K, Okuda K. Gene expression profiles in the bovine corpus luteum (CL) during the estrous cycle and pregnancy: possible roles of chemokines in regulating CL function during pregnancy. J Reprod Dev 2015; 61: 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakumoto R, Komatsu T, Kasuya E, Saito T, Okuda K. Expression of mRNAs for interleukin-4, interleukin-6 and their receptors in porcine corpus luteum during the estrous cycle. Domest Anim Endocrinol 2006; 31: 246–257. [DOI] [PubMed] [Google Scholar]
- 15.Ushizawa K, Takahashi T, Hosoe M, Kizaki K, Hashizume K. Characterization and expression analysis of SOLD1, a novel member of the retrotransposon-derived Ly-6 superfamily, in bovine placental villi. PLoS One 2009; 4: e5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi T, Sakumoto R, Hayashi KG, Hosoe M, Shirai J, Hashizume K. Generation of recombinant bovine interferon tau in the human embryonic kidney cell line and its biological activity. Anim Sci J 2017; 88: 1498–1505. [DOI] [PubMed] [Google Scholar]
- 17.Ealy AD, Yang QE. Control of interferon-tau expression during early pregnancy in ruminants. Am J Reprod Immunol 2009; 61: 95–106. [DOI] [PubMed] [Google Scholar]
- 18.Bauersachs S, Ulbrich SE, Gross K, Schmidt SE, Meyer HH, Wenigerkind H, Vermehren M, Sinowatz F, Blum H, Wolf E. Embryo-induced transcriptome changes in bovine endometrium reveal species-specific and common molecular markers of uterine receptivity. Reproduction 2006; 132: 319–331. [DOI] [PubMed] [Google Scholar]
- 19.Bogacka I, Kurzynska A, Bogacki M, Chojnowska K. Peroxisome proliferator-activated receptors in the regulation of female reproductive functions. Folia Histochem Cytobiol 2015; 53: 189–200. [DOI] [PubMed] [Google Scholar]
- 20.Braissant O, Foufelle F, Scotto C, Dauça M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology 1996; 137: 354–366. [DOI] [PubMed] [Google Scholar]
- 21.Huang JC. The role of peroxisome proliferator-activated receptors in the development and physiology of gametes and preimplantation embryos. PPAR Res 2008; 2008: 732303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Chen L, Zhang X, Zhou Y, Zhang D, Huo M, Guan Y. PPARs and female reproduction: evidence from genetically manipulated mice. PPAR Res 2008; 2008: 723243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J, Jann JC, Biemann R, Koch HM, Fischer B. Effects of the environmental contaminants DEHP and TCDD on estradiol synthesis and aryl hydrocarbon receptor and peroxisome proliferator-activated receptor signalling in the human granulosa cell line KGN. Mol Hum Reprod 2014; 20: 919–928. [DOI] [PubMed] [Google Scholar]
- 24.Sheldrick EL, Derecka K, Marshall E, Chin EC, Hodges L, Wathes DC, Abayasekara DR, Flint AP. Peroxisome-proliferator-activated receptors and the control of levels of prostaglandin-endoperoxide synthase 2 by arachidonic acid in the bovine uterus. Biochem J 2007; 406: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogacka I, Bogacki M, Kurzyńska A, Chojnowska K. The involvement of peroxisome proliferator activated receptors (PPARs) in prostaglandin F2α production by porcine endometrium. Reprod Biol 2013; 13: 309–316. [DOI] [PubMed] [Google Scholar]
- 26.Kurzynska A, Chojnowska K, Bogacki M, Bogacka I. PPAR ligand association with prostaglandin F2α and E2 synthesis in the pig corpus luteum-An in vitro study. Anim Reprod Sci 2016; 172: 157–163. [DOI] [PubMed] [Google Scholar]
- 27.Kurzynska A, Bogacki M, Chojnowska K, Bogacka I. Peroxisome proliferator activated receptor ligands affect progesterone and 17β-estradiol secretion by porcine corpus luteum during early pregnancy. J Physiol Pharmacol 2014; 65: 709–717. [PubMed] [Google Scholar]
- 28.Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications—a review. Nutr J 2014; 13: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meher A, Sundrani D, Joshi S. Maternal nutrition influences angiogenesis in the placenta through peroxisome proliferator activated receptors: A novel hypothesis. Mol Reprod Dev 2015; 82: 726–734. [DOI] [PubMed] [Google Scholar]
- 30.Jump DB. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr Opin Lipidol 2002; 13: 155–164. [DOI] [PubMed] [Google Scholar]
- 31.Ambrose DJ, Kastelic JP. Dietary fatty acids and dairy cow fertility. Adv Dairy Technol 2003; 15: 35–47. [Google Scholar]
- 32.Silvestre FT, Carvalho TSM, Francisco N, Santos JEP, Staples CR, Jenkins TC, Thatcher WW. Effects of differential supplementation of fatty acids during the peripartum and breeding periods of Holstein cows: I. Uterine and metabolic responses, reproduction, and lactation. J Dairy Sci 2011; 94: 189–204. [DOI] [PubMed] [Google Scholar]
- 33.Levine L, Worth N. Eicosapentaenoic acid: its effects on arachidonic acid metabolism by cells in culture. J Allergy Clin Immunol 1984; 74: 430–436. [DOI] [PubMed] [Google Scholar]
- 34.Achard F, Gilbert M, Bénistant C, Ben Slama S, DeWitt DL, Smith WL, Lagarde M. Eicosapentaenoic and docosahexaenoic acids reduce PGH synthase 1 expression in bovine aortic endothelial cells. Biochem Biophys Res Commun 1997; 241: 513–518. [DOI] [PubMed] [Google Scholar]
- 35.Mattos R, Staples CR, Thatcher WW. Effects of dietary fatty acids on reproduction in ruminants. Rev Reprod 2000; 5: 38–45. [DOI] [PubMed] [Google Scholar]
- 36.Magata F, Shirasuna K, Strüve K, Herzog K, Shimizu T, Bollwein H, Miyamoto A. Gene expressions in the persistent corpus luteum of postpartum dairy cows: distinct profiles from the corpora lutea of the estrous cycle and pregnancy. J Reprod Dev 2012; 58: 445–452. [DOI] [PubMed] [Google Scholar]
- 37.Romero JJ, Antoniazzi AQ, Smirnova NP, Webb BT, Yu F, Davis JS, Hansen TR. Pregnancy-associated genes contribute to antiluteolytic mechanisms in ovine corpus luteum. Physiol Genomics 2013; 45: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basavaraja R, Przygrodzka E, Pawlinski B, Gajewski Z, Kaczmarek MM, Meidan R. Interferon-tau promotes luteal endothelial cell survival and inhibits specific luteolytic genes in bovine corpus luteum. Reproduction 2017; 154: 559–568. [DOI] [PubMed] [Google Scholar]
- 39.White PC, Speiser PW. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr Rev 2000; 21: 245–291. [DOI] [PubMed] [Google Scholar]
- 40.Kagawa N, Waterman MR. Evidence that an adrenal-specific nuclear protein regulates the cAMP responsiveness of the human CYP21B (P450C21) gene. J Biol Chem 1991; 266: 11199–11204. [PubMed] [Google Scholar]
- 41.Zhao B, Lei L, Kagawa N, Sundaramoorthy M, Banerjee S, Nagy LD, Guengerich FP, Waterman MR. Three-dimensional structure of steroid 21-hydroxylase (cytochrome P450 21A2) with two substrates reveals locations of disease-associated variants. J Biol Chem 2012; 287: 10613–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]