Abstract
Cancer metastases are a challenge for cancer treatment due to their organ specificity and pathophysiological complexity. Engineering 3D in vitro models capable of replicating native cancer dissemination can significantly improve our understanding of cancer biology and can help to guide the development of more effective treatments. In order to better mimic the behavior of native cancer, a triculture metastatic model has been created using a stereolithography printing technique with optimized inks for investigating the invasion of breast cancer (BrCa) cells into vascularized bone tissue. The printed system allows us to study transendothelial migration and the colony-forming behavior of metastatic BrCa cells. The key steps of BrCa cell progression, including expansion, migration, and colonization have been continuously monitored, and the interactions between cancer cells, vascular cells, and bone cells have been systematically investigated. The study results demonstrate that the 3D printed tissue construct by incorporating multiple cells and various favorable ink matrices, provides a suitable model to study the interaction between these cells in a complex vascular microenvironment. As such, our 3D printed tricultured model may serve as a valuable tool for studying metastatic breast cancer progression in bone.
Keywords: 3D printing, optimized inks, cancer metastasis, vascularized bone, cancer model
ToC figure
To better study cancer metastasis, a 3D printed in vitro vascularized cancer model is created by integrating a tumor, endothelialized vessels, and nano- bone matrix using various inks to demonstrate the multiple interactions between breast cancer cells and the vascularized bone environment. It is expected that the model will be notably valuable for future cancer research and new therapeutic discovery.
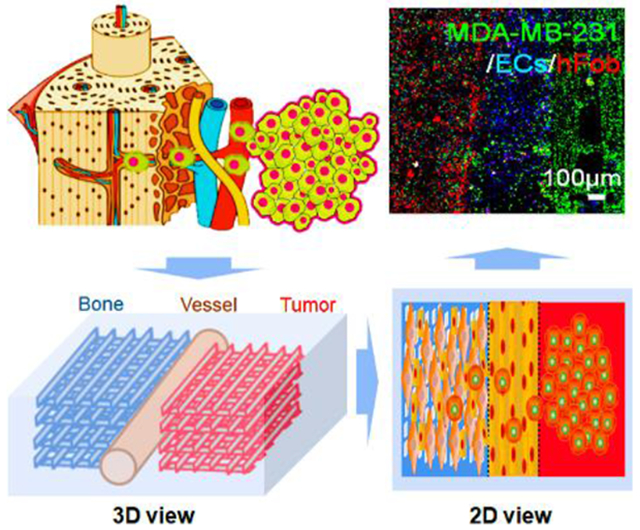
1. Introduction
Cancer metastasis is responsible for more than 90% of cancer-related mortality and represents a major challenge to effective therapy [1, 2]. Cancer development involves many complex biological mechanisms, and therefore, understanding the underlying biology is an important first step for the development of effective therapies [2, 3]. Clinical evidence has shown that breast cancer often metastasizes to the bones, liver, lungs, and brain [2, 4]. The process of metastasis has proven to be difficult to study in individual patients, and several experimental models have been developed to create simpler and more predictable systems for identifying the important role of biological, chemical, and physical factors involved in cancer pathophysiology [2, 4]. However, conventional 2D culture approaches do not accurately replicate the characteristics of the native cancer microenvironment, and animal models are plagued by several fundamental limitations, such as a lack of similarity to the human tissue environment and immune system, general variability, the complexity xenograft procedures, and high overall expense [2, 5, 6]. To better recapitulate the specificity and complexity of living human tissues, engineered 3D tissue matrices have been developed as an alternative model to provide a more highly controllable microenvironment for studying cancer development [6–9]. The developed 3D culture platforms enable cancer cells to display heterogeneous growth, resistance to drug treatment, and have metastatic behavior [9–11]. However, existing engineered 3D systems often cannot provide a complex physiological environment to simulate the diverse features of different tissues in one platform [8–10]. Therefore, it is difficult to utilize traditional manufacturing techniques to create a physiologically relevant metastatic model. As such, 3D printing offers a novel engineering technique to address these complexities by precisely controlling the spatial arrangement of living cells and their supporting matrices for complex tissue fabrication in vitro [12, 13]. 3D printing facilitates the customized design and high-throughput fabrication through predesigned computer-aided designs (CAD) to target the repair and regeneration of different tissues/organs, such as bone, cartilage, nerve, and myocardium [14, 15], in complex tissue models.
Metastatic breast cancer (BrCa) cells have a high propensity to invade the stroma of large bones. It has been demonstrated that 70% of advanced breast cancer patients have skeletal metastases, leading to severe pain, bone fractures, and often mortality [16, 17]. Recent studies have demonstrated that many 3D printed cancer models more closely mimic the natural behavior of cancer and bone cells, both phenotypically and genotypically in their response to anticancer drugs, than 2D cultured systems [18, 19, 20]. Our previous studies have shown that the presence of osteoblasts or mesenchymal stem cells promoted morphological changes, migratory behavior and drug resistance of BrCa cells in a physiologically-relevant 3D matrix [19–21]. These findings suggested that 3D printed tissue can serve as a promising model to study the BrCa bone metastasis. However, these studies primarily focused on the final stage of BrCa metastasis to investigate the direct interaction between BrCa cells and bone cells. To better understand metastatic BrCa cell behavior, in vitro models need to take into account cell migration from the primary site to a distant secondary site [2, 4], and the role that the vascular environment and associated endothelial cells play in transendothelial migration [22, 23]. Compared to current vascularized models that involve the use of microfluidic systems [24], it is expected that a 3D printed vascularized tissue model will have an advantage in precisely controlling the ink matrix composition, spatial distribution, and architectural complexity of the metastatic niche.
In the present study, we developed a novel in vitro localized cancer model using stereolithography (SL) 3D printing (beam-scanning, or laser direct writing technique) that allows the study of BrCa cell metastasis from its migration through an endothelialized vessel to its final colonization in bone by incorporating various biocomponents and photocurable inks into a multifunctional 3D tissue matrix with suitable mechanical strength and cell distribution. The localized model here allowed the complicated metastatic process was simplified to a particular area where some typical features of the “tumor-vessel-bone” metastatic niche were replicated, i.e., the BrCa cells detach from first tumor tissue, migrate through the vessel, and invasive into bone tissue for secondary tumor formation. Additionally, less invasive (or non-invasive) MCF-7 cells were compared to the more invasive and metastatic MDA-MB-231 cell line to evaluate the printed 3D cancer bone model with respect to the interplay between osteoblasts and BrCa cells and the effect of vascular environment.
2. Results and Discussion
2.1. Ink Optimization and 3D Printing of Cancer Bone Model
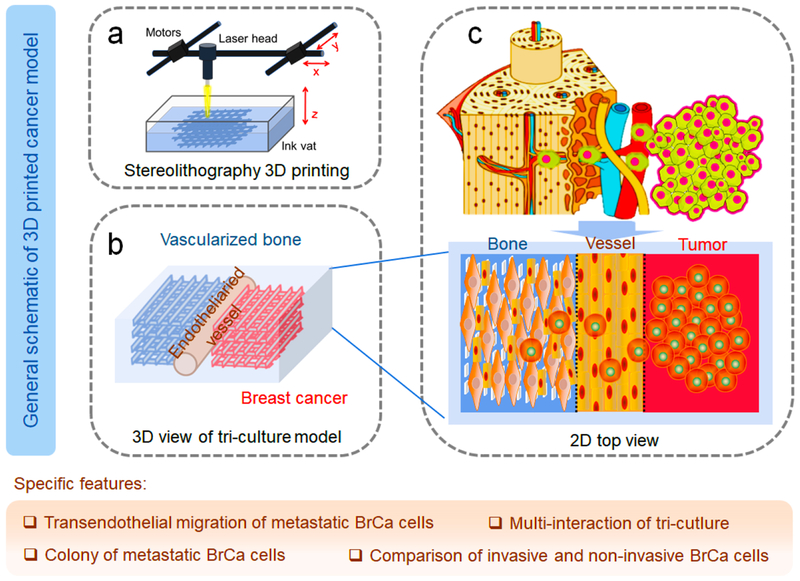
To replicate the BrCa cell metastasis into vascularized bone, a localized cancer metastatic niche was created with vascularized bone, an endothelium channel, and BrCa cells. As mentioned above, this model specifically focused on transendothelial migration and colonization in the bone tissue region to explore cell-cell interactions between BrCa cells and osteoblasts (hFob), and to elucidate the influence of endothelial cells on this process as well as the metastatic diversity of two BrCa cell lines. The conceptual design of the metastatic model is illustrated in Schematic 1, and is comprised of three neighboring tissue regions consisting of microvascularized bone, endothelial vessels, and BrCa tissue.
Schematic 1.
Conceptual design of the 3D printed BrCa bone metastatic models. (a) Beam-scanning SL 3D printing of BrCa bone model. (b) Schematic 3D view of the triculture model. (c) Schematic of the in vivo invasion of MDA-MB-231 cells into bone, and 2D view of our triculture model. The localized niche has three neighboring regions, consisting of microvascularized bone, endothelial vessels, and BrCa cells. The specific features of the model are also highlighted in the tan shaded area.
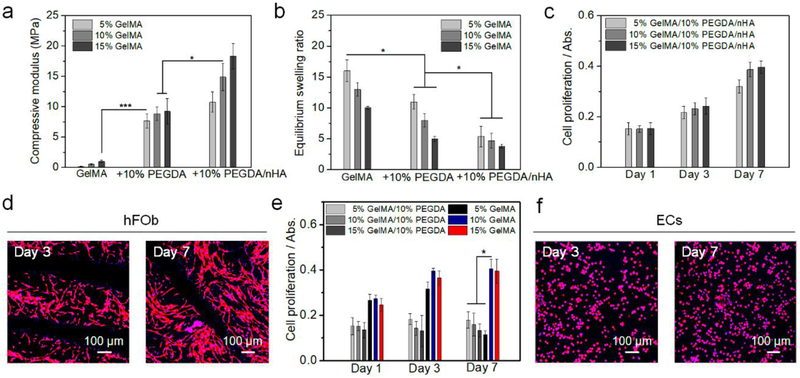
In order to replicate the human bone environment, a nanostructured bone ECM was printed utilizing ink I, which consisted of a photocrosslinkable gelatin-based ink (GelMA), PEGDA ink, and nHA. The major mineral component of bone is nanocrystalline hydroxyapatite (Ca10(PO4)6(OH)2), which constitutes an average 50~60 wt% of the bone matrix [25]. Thus, we selected a 50 wt% nHA (vs. the dry weight of GelMA and PEGDA) for printing mineralized bone scaffolds. Additionally, the organic matrix makes up ∼20 wt% of native bone and is primarily comprised of collagen [26]. Gelatin is partially denatured collagen and is less immunogenic compared to the collagen so that the biochemical properties of gelatin and its derivative (GelMA) are similar to the bone organic component. The photocrosslinkable PEGDA ink is nominally immunogenic and generally non-toxic after it is fully crosslinked, and has been widely used in biomedical research. The PEGDA solution was mixed with GelMA to decrease the swelling volume, and to increase the mechanical modulus and structural stability of the printed hydrogel. Thus, we obtained different bone constructs by varying the ratio of GelMA and PEGDA inks. Mechanical testing indicated that the matrices printed with the ink with a higher concentration of PEGDA or GelMA had a higher compressive modulus (Figure 1a). The compressive modulus of the printed matrices with nHA was greater than the corresponding matrices without nHA. Although the mechanical moduli of the hydrogel-based scaffolds were still lower than that of the native bone, the nHA/gelatin-based scaffold provided both a high surface modulus and a physiologically active component for bone cell growth. The swelling behavior of a hydrogel-based biomaterial can ultimately affect its surface properties, structural stability, and bulk structural features (Figure 1b). The swelling rate of the hydrogel with a lower concentration of PEGDA or GelMA was greater than the high concentration samples, and the swelling rate of the pure hydrogels was higher than their nHA containing counterparts. After the optimization of the mechanical modulus, swelling behavior, and hFob proliferation results (Figure 1c), inks containing 10 % GelMA, 10 % PEGDA, and nHA (weight vs. the volume of PBS in the ink) were selected to print all bone tissue constructs. After 3 days of culture, it was observed that the hFob cells reached their highest proliferation rate and assumed their natural spindle-like morphology on the optimized samples (Figure 1d).
Figure 1.
Characterization and optimization of inks. (a) The compressive moduli of different matrices. The mean ± standard deviation., n≥6 with triplicates, *P <0.05, ***P <0.001. (b) Equilibrium swelling ratios of different matrices. The mean ± standard deviation, n≥6 with triplicates, *P <0.05. (c) The proliferation of hFob on different matrices for 1, 3, and 7 days. The mean ± standard deviation, n≥6 with triplicates. (d) hFob spreading morphology on printed 10% GelMA/10% PEGDA/nHA matrix after 3 and 7 days of culture. (e) The proliferation of ECs on different matrices for 1, 3, and 7 days. The mean ± standard deviation, n≥6 with triplicates, *P <0.05. (f) Morphology of ECs on printed 10% GelMA matrix after 3 and 7 days of culture.
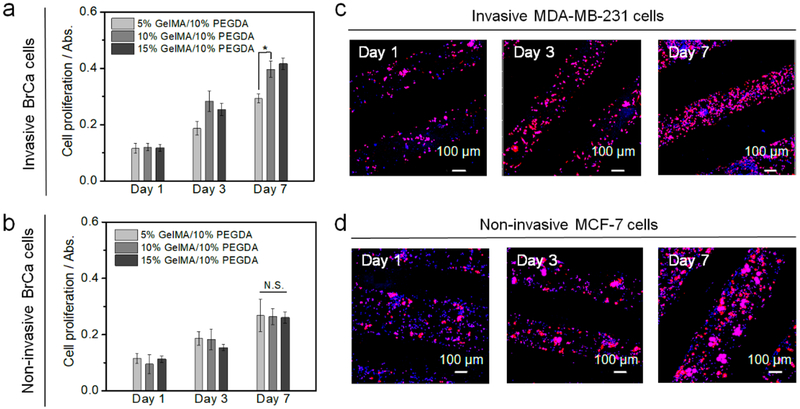
Additionally, an endothelialized vessel was created to provide a vascular environment for BrCa metastasis. Therein, the BrCa metastatic process, involves the cancer cell intravasation through the endothelial barrier, and final extravasation to the bone region. By changing the ratio of GelMA and PEGDA inks, we found the ECs growing within the pure 10% and 15% wt GelMA vessel exhibited higher metabolic activity when compared to the other groups (Figures 1e and f). It is assumed that the bioactivity of pure GelMA is better than the mixture and the pure GelMA of low concentration has a softer surface, which is better for EC adhesion and growth. Notably, pure 5% wt GelMA exhibited a decrease in EC proliferation on day 3 and day 7, which was likely due in part to the weak structural stability of the printed gel matrix with this concentration. Finally, BrCa cells were cultured within the printed 3D hydrogel network to mimic the cancerous tissue environment and recapitulate the initiation of cancer spreading. In our study, two typical osteotropic metastatic BrCa cell lines, MDA-MB-231, and MCF-7 with different phenotypes and metastatic potential, were compared to evaluate the efficiency of the vascularized model. Specifically, MDA-MB-231 cells have been used in a metastatic model because of their high propensity to metastasize to the bone. Conversely, MCF-7 cells have a lower metastatic capacity, are estrogen and progesterone receptor-positive and, are human epidermal growth factor receptor 2 (HER2)-negative (HER2-) as compared to the triple negative MDA-MB-231 cells [27]. We found that MDA-MB-231 cells exhibited higher proliferation on 10% GelMA/10% PEGDA scaffold when compared to other inks, and MCF-7 cells also had an excellent proliferation (Figures 2a and b). Confocal imaging showed that MDA-MB-231 cells displayed endothelial-like morphology and stellate projections, while the MCF-7 cells had more clustered structures (Figures 2c and d).
Figure 2.
Optimization of BrCa matrices. (a) The proliferation of invasive MDA-MB-231 cells on different matrices for 1, 3, and 7 days. The mean ± standard deviation, n≥6 with triplicates, *P <0.05. (b) The proliferation of non-invasive MCF-7 cells on different matrices for 1, 3, and 7 days. The mean ± standard deviation, n≥6 with triplicates, N.S. indicates no significance. (c) Morphology of invasive MD-MB-231 cells on printed 10% GelMA/10% PEGDA matrix for 1, 3, and 7 days. (d) Morphology of non-invasive MCF-7 cells on printed 10% GelMA/10% PEGDA matrix for 1, 3, and 7 days.
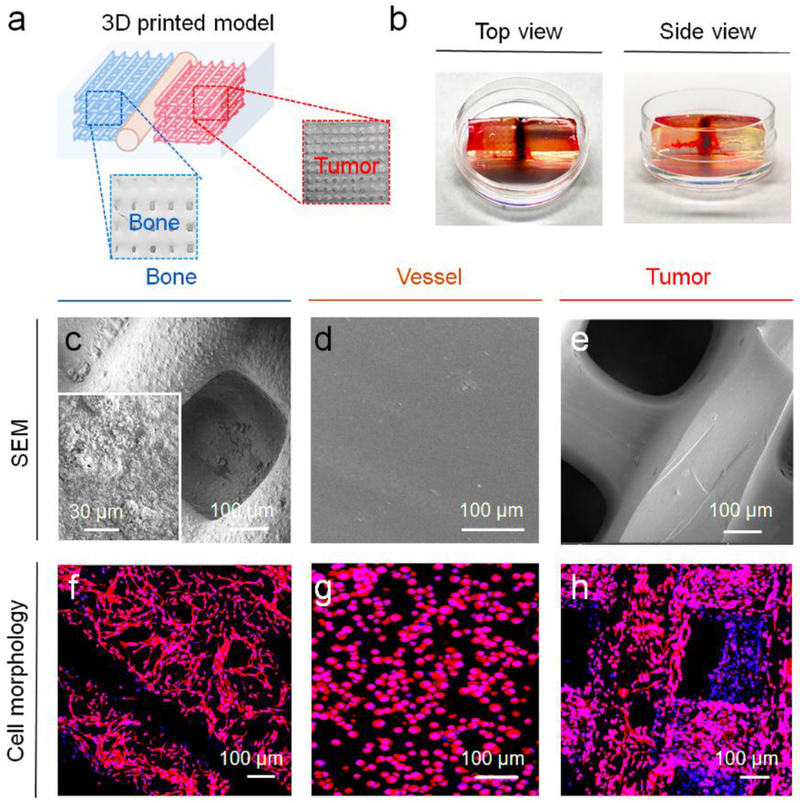
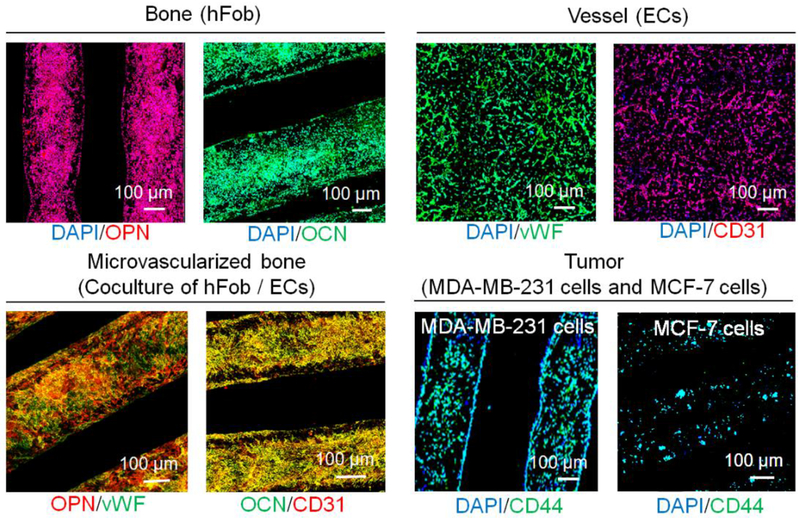
Utilizing the CAD design, our cancer model was printed using optimized inks with an SL printer as is shown in Figures 3a and b. The CAD model was illustrated in Figure S1, Supporting Information. In our previous bone model study, we found that printed tissue matrices with small square patterns have higher surface areas and higher porosities when compared to other printed patterns [21]. Therefore, the small square printing pattern was used for the current study. Our entire model is able to be directly printed, however, in order to facilitate the seeding of different cell lines in the different tissue regions of the model, a typical “two-step” method was used to fabricate the cancer model. First, the bone matrix and the tumor matrix (8 mm × 8 mm × 1 mm) were printed with four layers using different inks, respectively. Then, they were mounted in 10% wt GelMA ink and the vessel with a diameter of 500 μm was printed with 10% wt GelMA ink between the bone matrix and the tumor matrix. Computational analysis results showed each printed matrix (bone and tumor) had ~55 mm3 of total volume, ~60 mm2 of surface area, and ~25 pores/mm3 of porosity. The matrix morphologies of different regions were characterized by SEM (Figures 3c–e). The enlarged images showed that the bone matrix had a mineralized nanosurface with considerable roughness, while the cancer matrix and vessel matrix exhibited smoother surfaces. For the cell seeding, the hFob (or hFob/ECs), and BrCa cells (MDA-MB-231 and MCF-7) were seeded onto the bone matrix and the tumor matrix, respectively. After matrix mounting and vessel printing, the ECs in excess were injected into the vessel channel for the formation of the endothelialized lumen. In order to minimally affect the cell migration and medium exchange, the thin, soft hydrogel was printed between the bone and tumor to fabricate the vessel channel. Figures 3f–h illustrates the morphological characteristics of the cells on the different matrices with F-actin staining, and the cellular phenotypes of different regions are identified with immunostaining after 7 days of culture (Figure 4). hFob and ECs were stained with the osteogenic markers OCN and OPN and the angiogenic markers CD31 and vWF. The images illustrated that favorable microvascularized bone formation occurred on the 3D printed bone matrix. Although ECs had a lower proliferation rate on the bone matrix, the depreciated growth rate did not affect the vascularized bone formation in the coculture. Cell adhesion molecule (CD44) expression is upregulated in many cancers and contributes to the regulation of metastasis [28]. Metastatic MDA-MB-231 cells exhibited higher expression of CD44 compared to non-metastatic MCF-7 cells (Figure 4), and endothelial cells in the vessel region exhibited positive expression of the endothelial markers vWF and CD31 (endothelial cell adhesion molecule).
Figure 3.
3D printing of BrCa bone model. (a) Schematic of printed BrCa model and photo images of different regions. (b) Photo images of 3D printed BrCa model with top view and side view (the thickness of the model construct is ~ 3 mm). (c) SEM image of the bone matrix region. (d) SEM image of the vessel matrix region. (e) SEM image of the BrCa (tumor) matrix region. (f) Morphology of hFob on the bone matrix region. (g) Morphology of ECs on the vessel matrix region. (h) Morphology of BrCa (MDA-MB-231) cells on the tumor matrix region.
Figure 4.
Immunofluorescent images of different regions for printed BrCa bone model after 7 days of culture with DAPI staining. Bone: osteogenic OPN and OCN staining of hFob. Vessel: angiogenic vWF and CD31 staining of ECs. Vascularized bone: OPN/vWF and OCN/CD31 dual staining. Tumor: cancer CD44 staining of BrCa MDA-MB-231 and MCF-7 cells.
2.2. BrCa Migration and Proliferation Study of 3D Printed Vascularized Bone model
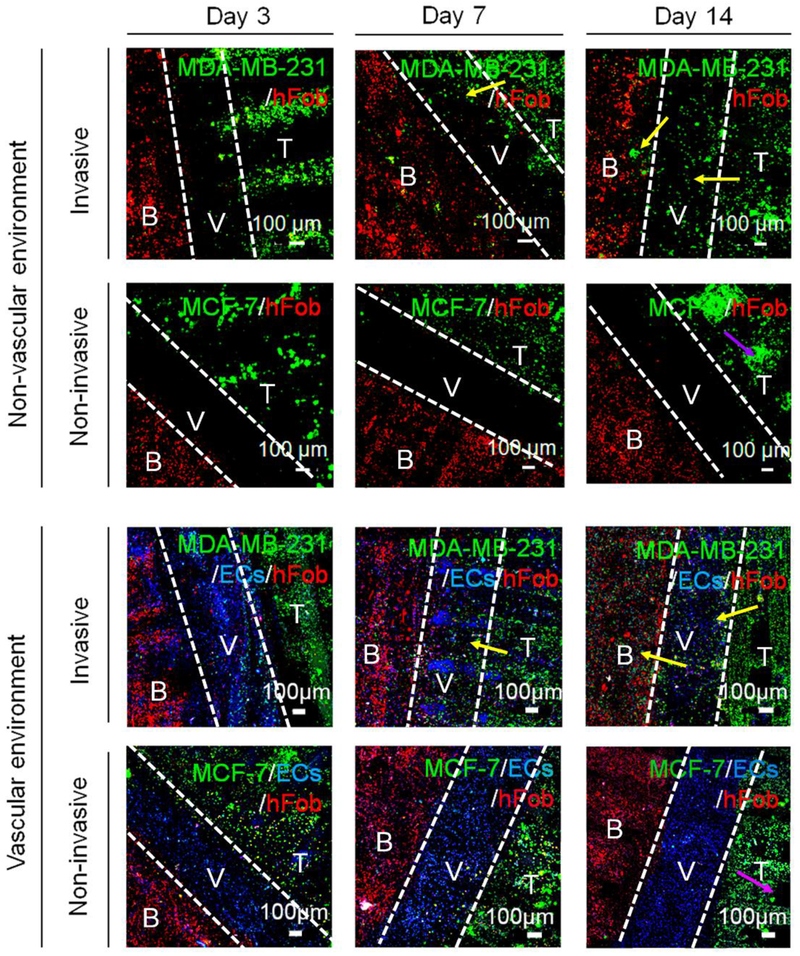
To visualize the cell distribution and BrCa cell progression in the 3D matrices, cells were prelabeled with fluorescent dyes, wherein hFob cells were colored red, ECs were colored blue, and BrCa cells were colored green. As shown in Figure 5, clusters of cancer cells, bone cells, and vascular cells were precisely placed in our designed model through the utilization of 3D printing approach. The fluorescent data were also quantified by Image J software, and the results were shown in Figure S2, Supporting Information. The various cell types within the different matrix regions demonstrated increased proliferation and expansion with time. Importantly, by comparing the cell distribution at each time point, different invasive behaviors between MDA-MB-231 cells and MCF-7 cells were observed. MDA-MB-231 cells showed distinct migration to the bone matrix over a 7-day culture period, while MCF-7 cells exhibited less invasive behavior. Next, we investigated whether the vascular environment generated by EC seeding could affect BrCa migratory behavior, since the most critical step in metastasis is the intravasation and extravasation process of cancer cells at the vessel site. The result reveals that the growth of BrCa cells was accelerated by the presence of vascular cells, and the vascular environment also increased the migration of MDA-MB-231 cells over 14 days. Compared to monoculture without ECs, the more invasive MDA-MB-231 cells migrated towards the ECs, whereas, the non-invasive phenotype MCF-7 cells were weakly affected. It was also noticed that ECs exhibited increased proliferation prior to 3 days when cocultured with MDA-MB-231 cells or MCF-7 cells, which can likely be attributed to the secretion of angiogenic factors by the BrCa cells [29], by marked endothelial and cancer cell expansion [23, 30]. When cocultured with non-invasive MCF-7 cells, ECs population were maintained at a stable level, demonstrating the important role of the vascular environment during oncogenesis for BrCa directional migration [31]. Additionally, when MDA-MB-231 cells colonized the bone matrix, the proliferation rate of hFob cells significantly decreased in comparison to monoculture, in contrast, the proliferation rate of the MDA-MB-231 cells was increased, suggesting that cytokines secreted by the osteoblasts promote BrCa cell growth [30].
Figure 5.
Development of BrCa (non-invasive MCF-7 and invasive MDA-MB-231 cells) metastasis toward bone over 14 days of the culture period. Cell tracker imaging was conducted to monitor the BrCa invasive process, including BrCa growth, transendothelial migration, and colonization. B: bone tissue, V: Vessel, T: tumor tissue. The white dotted lines in each image are used to divide the different tissue regions. The yellow arrows indicate the migration of invasive BrCa cells. The purple arrows indicate the in situ expansion of non-invasive BrCa cells.
The cellular growth rates between models with and without vascular cells were also quantified by the WST-8 assay (Figure 6a and b). In order to avoid an inaccurate assessment of potential BrCa invasion into other regions, we designed three built-in chambers located at each end of the different matrix regions where culture medium was allowed to exchange between these chambers. Our indirect co-culture system can measure different cell proliferation rates with alterations in the conditioned media, but not the direct crosstalk between BrCa cells, ECs, and hFob. Unlike the fluorescent imaging results, the proliferation of ECs at 7 days remained constant, suggesting that when MDA-MB-231 cells do not directly invade the vessel region, as there is no highly competitive growth advantage [30]. However, EC growth in coculture was much slower than that in monoculture, suggesting that the secretion of factors from BrCa cells inhibited the proliferation of ECs [32]. Compared to monoculture, both MDA-MB-231 and MCF-7 cells in co-culture exhibited increased proliferation, whereas, the proliferation of hFob was decreased. After 2 weeks of culture, a more significant difference in the proliferation of each cell type was observed, where the effect of MDA-MB-231 cells on the growth of ECs was much higher than that of MCF-7 cells. These phenomena were observed in separated chambers, and therefore, it is difficult to distinguish the real influence of osteoblasts when BrCa directly colonize the bone matrix.
Figure 6.
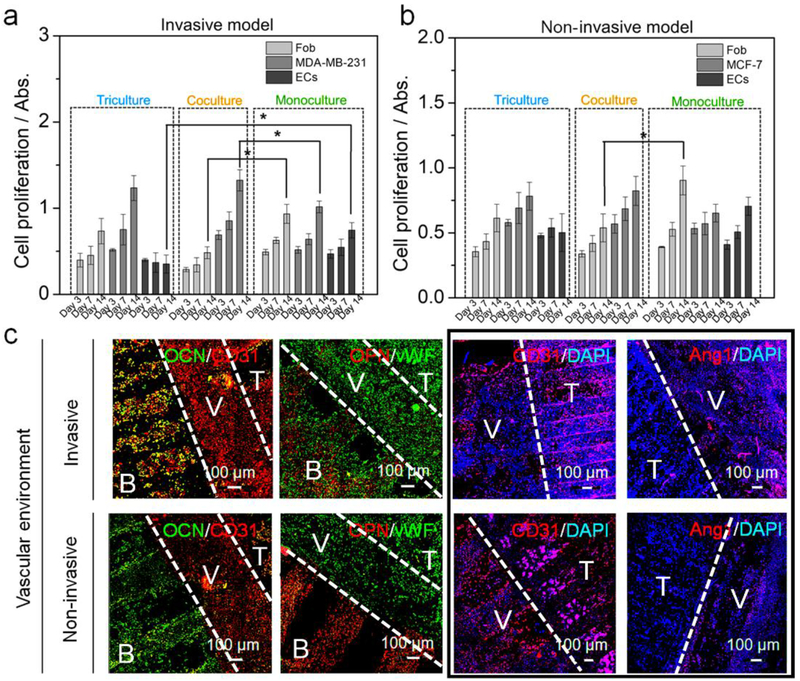
Colonization of BrCa toward bone. (a) The proliferation of hFob, ECs, MDA-MB-231 cells in monoculture, coculture, and triculture with ECs conditions for 14 days. The mean ± standard deviation, n≥6 with triplicates, *P <0.05. (b) The proliferation of hFob, ECs, MCF-7 cells in monoculture, coculture, and triculture conditions for 14 days. The mean ± standard deviation, n≥6 with triplicates, *P <0.05. (c) Immunofluorescent images of hFob and BrCa (MDA-MB-231 and MCF-7 cells) in a vascular environment with DAPI staining after 14 days of culture. CD31 and vWF staining were used to identify both EC and BrCa cells. Osteogenesis of hFob was characterized by OCN and OPN staining. Combining CD31 and Ang1 was used to distinguish the BrCa cells and ECs. B: bone tissue, V: Vessel, T: tumor tissue. The white dotted lines in each image are used to divide the different tissue regions. The four images in the black frame are shown to compare the marker expression of ECs and BrCa cells in the vessel and tumor regions.
2.3. Immunostaining and Gene Analysis of BrCa on 3D Printed Vascularized Bone Model
To highlight the advantage of our 3D printed cancer model in replicating the local vascularized bone environment, we investigated BrCa colonization of the vascularized bone using immunostaining. Figure 6c shows the expression of cell markers of MDA-MB-231 and MCF-7 cells after 2 weeks in culture. Compared to their initial distribution on the 3D printed cancer matrix, BrCa cells exhibited increased expansion that occupied the entire matrix space. As the angiogenic markers CD31 and vWF were expressed both the ECs and the BrCa cells, both the vessel region and the tumor region were positive for these markers. However, a significant difference was observed between the two BrCa cell types, where MDA-MB-231 cells significantly colonized the vascularized bone matrix, whereas, the MCF-7 cells exhibited limited expansion and migration. With the invasive MDA-MB-231 cell line, the fluorescent intensity of the osteogenic markers OPN and OCN was relatively lower when compared to that of the non-invasive counterpart. The comparison of CD31 and Ang1 (EC specific marker) expression indicates vessel maturation and cancer progression in the invasive model and in the non-invasive model [33]. These results confirm that our cancer model is able to mimic in vivo BrCa behavior and can be used to distinguish between different invasive potentials.
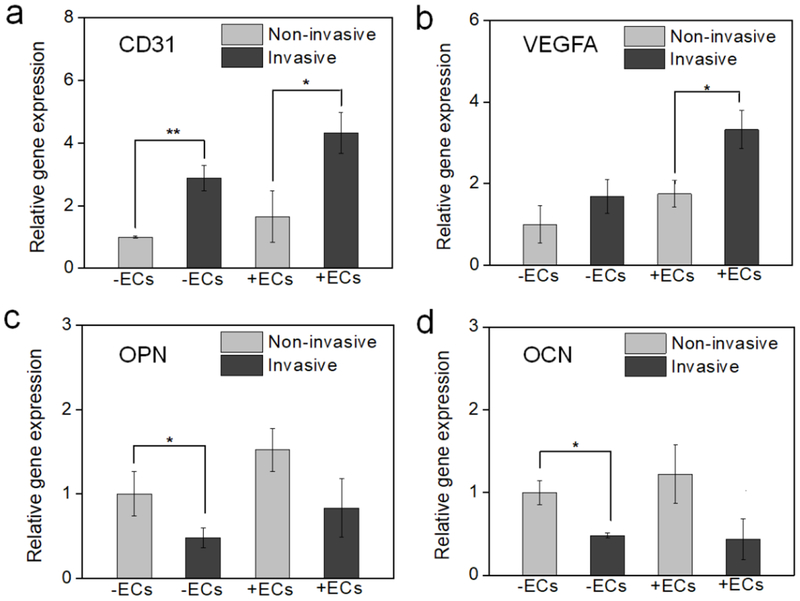
The gene expression in relation to cancer angiogenesis and osteogenesis was also analyzed after 2 weeks of culture (Figure 7). After BrCa cells colonized the bone matrix, CD31 was up-regulated, whereas, OPN and OCN expression were down-regulated. This suggested that the invasion of MDA-MB-231 cells into the bone matrix had a significantly greater effect compared to the MCF-7 cell line [34]. A similar phenomenon was observed in the vascularized bone model co-cultured with ECs and in the bone model without ECs. In the presence of ECs, the changes in gene expression of VEGFA were more remarkable. Compared to the previous proliferation results, this difference may possibly be explained by the inhibitory effect of BrCa cells on osteogenesis. Conversely, BrCa cell growth was promoted by the presence of the vascular environment in the bone matrix. These results indicate that our 3D printed cell-laden nano-construct effectively mimics a natural vascularized bone matrix and can provide a preferable microenvironment for BrCa migration and colonization [17].
Figure 7.
Gene expression of BrCa bone model (a) CD31, (b) VEGFA, (c) OPN, and (d) OCN using RT-PCR analysis to study the effect of non-invasive (MCF-7)/invasive (MDA-MB-231) BrCa cells, and vascular/non-vascular environment. The mean ± standard deviation, n≥6 with triplicates, *P <0.05, **P <0.01.
Compared to previous BrCa bone models, the model presented here possesses several important advantages: (1) inclusion of an endothelialized compartment for exploring EC function during metastasis, (2) inclusion of an osteoblast-laden nanostructured mineral bone matrix to study the interaction osteogenesis and oncogenesis, (3) allowance of cancer cells to contact adjacent endothelial tissue in order to provide insight into the role of transendothelial migration in cancer progression, and (4) use of 3D SL printing to achieve the accurate placement of multiple inks for the fabrication of complex “cancer-vessel-tissue” niches in vitro. Although the current study also has some limitations, i.e., the capillaries sprouted from endothelialized vessel were not observed both in tumor and bone regions during 2 weeks of culture; the BrCa metastasis was mainly involved in the transendothelial migration on surface of endothelialized channel without migrating through microvessels; and osteoclasts were not included in the bone region to investigate the osteolysis. In the future, these features will be included in our design for further improving the cancer model.
3. Conclusion
In summary, an engineered BrCa metastatic model consisting of a nanostructured bone matrix and cancer cells separated by an endothelialized vascular channel was developed using 3D SL printing in order to recapitulate the pathophysiology of cancer metastasis and to advance our understanding of the interplay between osteoblasts, BrCa cells, and the vascular environment. By combining the advantages of control over spatial distribution and physiological features, our 3D printed cancer model provides an approach to mimic transendothelial migration and colonization of cancer cells, which will be invaluable for the screening of novel anticancer drugs as well as the development of patient-specific diagnostics and therapeutics.
4. Experimental Section
Ink Synthesis and Preparation:
Nanohydroxyapatites (nHA) were synthesized using a hydrothermal treatment method as described in our previous study [19]. Briefly, 0.6 M ammonium phosphate was added to water and adjusted to a pH of 10 with ammonium hydroxide. 1 M calcium nitrate was slowly titrated into the above mixture to obtain an HA precipitate and then heated at 200°C for 20 hours in a Teflon liner to yield nHA rods. The resulting nHA had a grain size of ∼25 nm in width and 50−100 nm in length. In addition, gelatin methacrylate (GelMA) was synthesized as a photocurable ink for SL printing [35]. Specifically, 1% (v/v) methacrylic anhydride was added dropwise to the gelatin (Type A, Sigma, 10% in PBS (w/w)) solution while stirring, and then the mixture was reacted for 3 hours at 50 °C. The GelMA solution was dialyzed against deionized water for 7 days at 50 °C. The dialyzed GelMA solution was lyophilized and stored at room temperature. Before use, a GelMA polymer solution was prepared by dissolving the freeze-dried GelMA and the photoinitiator (Irgacure 2959) (0.5 w/v%) in PBS (0.01 M). The 3D printable inks were prepared by mixing the 5, 10, and 15 wt% GelMA with/without 10 wt% polyethylene glycol diacrylate (PEGDA, Mn = 700, Sigma) or 5, 10, 15 wt% nHA, in order to obtain the 3D printed scaffolds with different mechanical and biochemical properties. The compressive moduli of the samples (8 mm diameter and 5 mm thickness) were determined using the standard flat compression platen on a universal mechanical testing system (MTS). The test speed was controlled to 2 mm/min, and Young’s moduli were calculated from the linear regions of the stress-strain curves.
Cell Culture:
Osteoblasts (human fetal osteoblasts, hFOB) were obtained from the American Type Culture Collection (ATCC) (CRL-11372) and cultured in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F12 (DMEM-F12) supplemented with Fetal Bovine Serum (FBS) (10%, v/v), penicillin–streptomycin (1%, v/v), and L-glutamine (1%, v/v)). BrCa cell line (MDA-MB-231 and MCF-7) was obtained from the ATCC (HTB-26 and HTB-22) and cultured in DMEM supplemented with 10% FBS and 1% penicillin–streptomycin. Human umbilical vein endothelial cells (ECs, ThermoFisher Scientific) were cultured in endothelial cell growth medium (Cell Application). All cells were incubated at 37 °C, 5% CO2, and 95% relative humidity.
Cell Proliferation and Morphology:
Cell proliferation on the printed tissue matrices was quantitatively investigated using a CCK-8 kit (Dojindo). After the predetermined period, the incubation medium was changed with WST-8 solution (10% v/v in medium). After 3 h of incubation, the absorbance values of the supernatant solution were measured at 450 nm on a photometric plate reader (Thermo Scientific). The cells’ cytoskeleton was identified with double staining of actin (red) using Texas Red labeled phalloidin and nuclei (blue) using 4, 6-diamidino-2-phenylindole dihydrochloride (DAPI) (Invitrogen). Cells were fixed in 10% formalin for 15 min, permeabilized in 0.1% Triton X-100, and blocked with 1% bovine serum albumin (BSA, Sigma-Aldrich). Cells were then incubated with phalloidin for 30 min and DAPI for 5 min. Samples were observed and imaged using sing a Zeiss 710 confocal microscope.
3D Printing of Cancer Bone Model and Characterization:
3D “cancer-vessel-bone” model was designed by CAD and printed with a tabletop beam-scanning (or laser direct writing) SL 3D printer. The printer was developed in our lab based on the existing rapid prototyping platform (Printrbot®), which includes a temperature-controllable stage, an X–Y–Z tool head for motion, and an optic-coupled solid-state ultraviolet (UV) laser fiber [12, 15, 20, 35, 36]. The parameters were set as follows: ~190 μm laser beam with 355 nm wavelength, 20 μJ at 15 kHz intensity output of emitted UV, and 5 mm/s printing speed. The 3D printed model consists of three chambers: microvascularized bone, endothelialized vessel, and cancer tumor. In order to easily seed the different cell lines in the different regions, a “two-step” method was used to fabricated the cancer model. The microvascularized bone chamber and the cancer chamber were printed using nHA doped GelMA/PEGDA ink and GelMA/PEGDA, respectively. After that, these two chambers were placed and mounted in the GelMA ink, and then the vessel channel chamber was printed in the middle region using GelMA ink to separate the bone and tumor chamber (a microneedle-based subtractive technique was performed to obtain a lumen structure in the hydrogel). Representative CAD models and Slic3r configurations of the 3D model were used to analyze and calculate all structural parameters, including the scaffold volume, surface area, and porosity. The morphologies of the 3D printed models were assessed by scanning electron microscopy (SEM) (Zeiss Nvision 40FIB). Samples were sputter-coated with a ∼10 nm layer of gold prior to SEM imaging.
BrCa Migration and Proliferation Study of 3D Printed Vascularized Bone model:
The cell-laden 3D BrCa bone construct was fabricated by a multiple cell seeding process. To visually monitor the cell behaviors of hFob, ECs, and BrCa (MDA-MB-231 or MCF-7) cells on the scaffolds, the cells were pre-labeled with CellTracker™ Green CMFDA Dye, CellTracker™ Blue CMAC Dye, CellTracker™ Orange CMRA Dye (Molecular Probes). hFob and ECs with a ratio of 1:1 (104 /mL) were co-cultured in the 3D vascularized bone scaffolds, and BrCa cells (MDA-MB-231 or MCF-7 cells) were cultured in the cancerous scaffolds. After the printing of the vessel between bone and tumor, ECs (104 /mL) were cultured in the 3D vessel channel. The coculture was conducted in mixed media (OM, BM, and EGM with a ratio of 1:1:1). To observe the migration, growth, and colonization of BrCa within the vascularized bone, after 1, 3, and 7 days of co-culture, the samples were imaged under a confocal microscope. The level of fluorescence was quantified to obtain BrCa area% in the bone region by Image J analysis software. Six visible areas were randomly selected for quantifying statistical analysis on each sample; there were three samples in each group. To investigate the cell interactions among hFob, ECs, BrCa (MDA-MB-231 or MCF-7) cells, the three chambers of the bone models were completely separated in a customized PDMS dish, and the medium with secreted cytokines was only allowed for the exchange. The metabolic activity was evaluated by a CCK-8 assay. The absorbance values were measured at 570 nm on a photometric plate reader (Thermo Scientific).
Immunostaining and Gene Analysis of BrCa on 3D Printed Vascularized Bone Model:
After 2 weeks of co-culture on the 3D printed model, the samples were fixed with 10% formalin for 15 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes, and blocked with 1% BSA for 1 hour. Then the samples were incubated with primary antibodies overnight and colored with secondary antibodies for 2 hours. The following primary antibodies were purchased from Abcam and used for staining: sheep polyclonal anti-von Willebrand factor (vWF), mouse monoclonal anti-CD31, rabbit monoclonal anti-CD44, mouse monoclonal anti-Angiopoietin 1 (Ang1), rabbit polyclonal anti-osteocalcin (OCN), and mouse monoclonal anti-osteopontin (OPN). The following secondary antibodies were purchased from ThermoFisher Scientific and used: donkey anti-sheep IgG-H&L Alexa Fluor®488, goat anti-rabbit IgG-H&L Alexa Fluor®488, and goat anti-mouse IgG-H&L Alexa Fluor®594. Finally, the cell nucleus was counterstained with DAPI and imaged using a confocal microscope.
After 2 weeks, the gene expression profiles associated with osteogenesis, angiogenesis, and invasion were characterized by a real-time quantitative reverse transcription polymerase chain reaction (RT-PCR) assay. The isolation of total RNA was performed on the samples using Trizol reagent. Total RNA purity and concentration were determined using a microplate reader (OD 260/280 nm within 1.8-2.0). cDNA was synthesized using a real-time RT-PCR kit (PrimeScript RT Master Mix, Takara) according to the manufacturer’s instructions. The quantitative PCR analysis was performed in triplicate per sample using SYBR qPCR kit (SYBR Premix Ex Taq II (Tli RNase H Plus), Takara) and CFX384 Real-Time System (Bio-Rad, USA). Relative quantification of gene expression was analyzed using the standard 2-(ΔΔCt) method, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the endogenous housekeeping gene. Primer sequences are as follows: GAPDH, forward 5’-GGAGCGAGATCCCTCCAAA-3’, and reverse 5’-GGCTCCCCCCTGCAAA-3’; CD31, forward 5’-GAGTCCTGCTGACCCTTCTG-3’ and reverse 5’-CACTCCTTCCACCAACACCT-3’; VEGFA: forward, 5′-AAGAAATCCCGTCCCTGTGG-3′ and reverse, 5′-GCAACGCGAGTCTGTGTTTT-3′; OCN, forward 5’-GACTGTGACGAGTTGGCTGA-3’, and reverse 5’-CTGGAGAGGAGAACTGG-3’; OPN, forward 5’-CATCACCTGTGCCATACCAG-3’, and reverse 5’-GCCACAGCATCTGGGTATTT-3’.
Statistical Analysis:
For quantitative experiments, data are presented as the mean ± standard deviation; sample size is greater than or equal to 6 unless otherwise indicated, and all experiments were replicated at least three times. One-way analysis of variance (ANOVA) followed by the posthoc Tukey’s test was used to calculate statistical significance using OriginPro Software, and ***p < 0.001, **p < 0.01, *p < 0.05 was defined as statistical significance in all analyses.
Supplementary Material
Acknowledgements
We would like to thank the NIH Director’s New Innovator Award 1DP2EB020549-01 for financial support.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Haitao Cui, Department of Mechanical and Aerospace Engineering, The George Washington University, 3590 Science and Engineering Hall, 800 22nd Street NW, Washington, DC 20052, USA.
Timothy Esworthy, Department of Mechanical and Aerospace Engineering, The George Washington University, 3590 Science and Engineering Hall, 800 22nd Street NW, Washington, DC 20052, USA.
Xuan Zhou, Department of Mechanical and Aerospace Engineering, The George Washington University, 3590 Science and Engineering Hall, 800 22nd Street NW, Washington, DC 20052, USA.
Sung Yun Hann, Department of Mechanical and Aerospace Engineering, The George Washington University, 3590 Science and Engineering Hall, 800 22nd Street NW, Washington, DC 20052, USA.
Robert I. Glazer, Department of Oncology, and Lombardi Comprehensive Cancer Center, Georgetown University Medical Center, Washington, DC 20007, USA
Rong Li, Department of Biochemistry & Molecular Medicine, The George Washington University, Washington, DC 20052, USA.
Lijie Grace Zhang, Departments of Electrical and Computer Engineering, Department of Biomedical Engineering, Department of Medicine, The George Washington University, Washington, DC 20052, USA; Department of Mechanical and Aerospace Engineering, The George Washington University, 3590 Science and Engineering Hall, 800 22nd Street NW, Washington, DC 20052, USA.
References
- [1].Malandrino A, Kamm RD, Moeendarbary E, ACS Biomater. Sci. Eng 2018, 4, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alemany-Ribes M, Semino CE, Adv. Drug Deliv. Rev 2014, 79–80, 40. [DOI] [PubMed] [Google Scholar]
- [3].Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, Thompson SK, Zollo M, Spano D, Dhawan P, Sliva D, Subbarayan PR, Sarkar M, Honoki K, Fujii H, Georgakilas AG, Amedei A, Niccolai E, Amin A, Ashraf SS, Ye L, Helferich WG, Yang X, Boosani CS, Guha G, Ciriolo MR, Aquilano K, Chen S, Azmi AS, Keith WN, Bilsland A, Bhakta D, Halicka D, Nowsheen S, Pantano F, Santini D, Semin. Cancer Biol 2015, 35 Suppl, S244. [DOI] [PubMed] [Google Scholar]
- [4].Gupta GP, Massague J, Cell 2006, 127, 679. [DOI] [PubMed] [Google Scholar]
- [5].Ferreira LP, Gaspar VM, Mano JF, Acta Biomater. 2018. [DOI] [PubMed] [Google Scholar]
- [6].Peela N, Truong D, Saini H, Chu H, Mashaghi S, Ham SL, Singh S, Tavana H, Mosadegh B, Nikkhah M, Biomaterials 2017, 133, 176. [DOI] [PubMed] [Google Scholar]
- [7].Guiro K, Arinzeh TL, Breast Cancer (Auckl) 2015, 9, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rijal G, Li W, Biomaterials 2016, 81, 135. [DOI] [PubMed] [Google Scholar]
- [9].Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M, van der Kuip H, Consortium IP, Biotechnol. J 2014, 9, 1115. [DOI] [PubMed] [Google Scholar]
- [10].Asghar W, El Assal R, Shafiee H, Pitteri S, Paulmurugan R, Demirci U, Mater. Today (Kidlington) 2015, 18, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Herrmann D, Conway JR, Vennin C, Magenau A, Hughes WE, Morton JP, Timpson P, Carcinogenesis 2014, 35, 1671. [DOI] [PubMed] [Google Scholar]
- [12].Cui H, Nowicki M, Fisher JP, Zhang LG, Adv. Healthcare Mater 2017, 6, 1601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cui H, Miao S, Esworthy T, Zhou X, Lee SJ, Liu C, Yu ZX, Fisher JP, Mohiuddin M, Zhang LG, Adv. Drug Deliv. Rev 2018, 132, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cui H, Zhu W, Holmes B, Zhang LG, Adv. Sci 2016, 3, 1600058; [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhou X, Nowicki M, Cui HT, Zhu W, Fang XQ, Miao SD, Lee SJ, Keidar M, Zhang LJG, Carbon 2017, 116, 615; [Google Scholar]; Cui H, Miao S, Esworthy T, Lee S, Zhou X, Hann SY, Webster TJ, Harris BT, Zhang L, Nano Res 2019, 12, 1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhu W, Cui H, Boualam B, Masood F, Flynn E, Rao RD, Zhang ZY, Zhang LG, Nanotechnology 2018, 29, 185101; [DOI] [PubMed] [Google Scholar]; Zhou X, Cui H, Nowicki M, Miao S, Lee SJ, Masood F, Harris BT, Zhang LG, ACS Appl. Mater. Interfaces 2018, 10, 8993; [DOI] [PubMed] [Google Scholar]; Miao S, Cui H, Nowicki M, Xia L, Zhou X, Lee SJ, Zhu W, Sarkar K, Zhang Z, Zhang LG, Adv. Biosyst 2018, 2, 1800101; [DOI] [PMC free article] [PubMed] [Google Scholar]; Miao S, Cui H, Nowicki M, Lee SJ, Almeida J, Zhou X, Zhu W, Yao X, Masood F, Plesniak MW, Mohiuddin M, Zhang LG, Biofabrication 2018, 10, 035007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shupp AB, Kolb AD, Mukhopadhyay D, Bussard KM, Cancers (Basel) 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Villasante A, Marturano-Kruik A, Vunjak-Novakovic G, Biomaterials 2014, 35, 5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S, Trends Biotechnol. 2015, 33, 504; [DOI] [PubMed] [Google Scholar]; Meng F, Meyer CM, Joung D, Vallera DA, McAlpine MC, Panoskaltsis-Mortari A, Adv. Mater 2019, e1806899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou X, Zhu W, Nowicki M, Miao S, Cui H, Holmes B, Glazer RI, Zhang LG, ACS Appl. Mater. Interfaces 2016, 8, 30017. [DOI] [PubMed] [Google Scholar]
- [20].Zhu W, Castro NJ, Cui H, Zhou X, Boualam B, McGrane R, Glazer RI, Zhang LG, Nanotechnology 2016, 27, 315103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhu W, Holmes B, Glazer RI, Zhang LG, Nanomedicine 2016, 12, 69. [DOI] [PubMed] [Google Scholar]
- [22].Mastro AM, Vogler EA, Cancer Res. 2009, 69, 4097. [DOI] [PubMed] [Google Scholar]
- [23].Butler JM, Kobayashi H, Rafii S, Nat. Rev. Cancer 2010, 10, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Blaha L, Zhang C, Cabodi M, Wong JY, Biofabrication 2017, 9, 045001; [DOI] [PMC free article] [PubMed] [Google Scholar]; Bhatia SN, Ingber DE, Nat. Biotechnol. 2014, 32, 760. [DOI] [PubMed] [Google Scholar]
- [25].Allen MR, Burr DB, in Basic and Applied Bone Biology, 2019, 85; [Google Scholar]; JafariNasabian P, Inglis JE, Reilly W, Kelly OJ, Ilich JZ, J. Endocrinol 2017, 234, R37; [DOI] [PubMed] [Google Scholar]; Olszta MJ, Cheng X, Jee SS, Kumar R, Kim Y-Y, Kaufman MJ, Douglas EP, Gower LB, Mater. Sci. Eng. R Rep 2007, 58, 77. [Google Scholar]
- [26].Fuchs RK, Thompson WR, Warden SJ, in Bone Repair Biomaterials (Second Edition), (Eds: Pawelec KM, Planell JA), Woodhead Publishing, 2019, 15. [Google Scholar]
- [27].Holliday DL, Speirs V, Breast Cancer Res. 2011, 13, 215; [DOI] [PMC free article] [PubMed] [Google Scholar]; Curtin P, Youm H, Salih E, Biomaterials 2012, 33, 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R Jr., Badve S, Nakshatri H, Breast Cancer Res. 2006, 8, R59; [DOI] [PMC free article] [PubMed] [Google Scholar]; Hiraga T, Ito S, Nakamura H, Cancer Res. 2013, 73, 4112. [DOI] [PubMed] [Google Scholar]
- [29].Alvarez-Garcia V, Gonzalez A, Alonso-Gonzalez C, Martinez-Campa C, Cos S, J. Pineal. Res 2013, 54, 373. [DOI] [PubMed] [Google Scholar]
- [30].Szot CS, Buchanan CF, Freeman JW, Rylander MN, Tissue Eng., Part C 2013, 19, 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Khodarev NN, Cell Sci J. 2003, 116, 1013; [DOI] [PubMed] [Google Scholar]; Shoval H, Karsch-Bluman A, Brill-Karniely Y, Stern T, Zamir G, Hubert A, Benny O, Sci. Rep 2017, 7, 10428; [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang W, Xu J, Fang H, Tang L, Chen W, Sun Q, Zhang Q, Yang F, Sun Z, Cao L, Wang Y, Guan X, FASEB J. 2018, 32, 276. [DOI] [PubMed] [Google Scholar]
- [32].Peyri N, Berard M, Fauvel-Lafeve F, Trochon V, Arbeille B, Lu H, Legrand C, Crepin M, Anticancer Res. 2009, 29, 2347. [PubMed] [Google Scholar]
- [33].Hendrix MJ, Seftor EA, Kirschmann DA, Seftor RE, Breast Cancer Res. 2000, 2, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Marturano-Kruik A, Villasante A, Yaeger K, Ambati SR, Chramiec A, Raimondi MT, Vunjak-Novakovic G, Biomaterials 2018, 150, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cui H, Zhu W, Nowicki M, Zhou X, Khademhosseini A, Zhang LG, Adv. Healthcare Mater 2016, 5, 2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou X, Castro NJ, Zhu W, Cui H, Aliabouzar M, Sarkar K, Zhang LG, Sci. Rep 2016, 6, 32876; [DOI] [PMC free article] [PubMed] [Google Scholar]; Miao S, Zhu W, Castro NJ, Nowicki M, Zhou X, Cui H, Fisher JP, Zhang LG, Sci. Rep 2016, 6, 27226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.