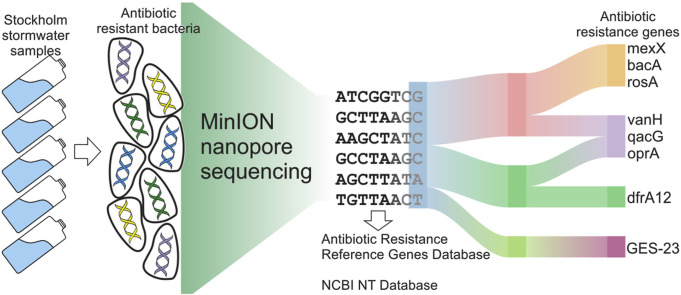
Abstract
Discharge of urban stormwater containing organic matter, heavy metals and sometime human feces, to the natural aquatic reservoirs without any treatment is not only an environmental problem. It can lead to prevalence of antibiotic resistant bacteria in stormwater systems and transmission of antibiotic resistance genes to the environment. We performed antibiotic resistome identification and virus detection in stormwater samples from Stockholm, using publicly available metagenomic sequencing MinION data. A MinION platform offers low-cost, precise environmental metagenomics analysis. 37 groups of antibiotic resistant bacteria (ARB), 11 resistance types with 26 resistance mechanisms – antibiotic resistance genes (ARGs) giving tolerance to the aminoglycoside, beta-lactams, fosmidomycin, MLS, multidrug and vancomycin were identified using ARGpore pipeline. The majority of the identified bacteria species were related to the natural environment such as soil and were not dangerous to human. Alarmingly, human pathogenic bacteria carrying resistance to antibiotics currently used against them (Bordetella resistant to macrolides and multidrug resistant Propionibacterium avidum) were also found in the samples. Most abundant viruses identified belonged to Caudovirales and Herpesvirales and they were not carrying ARGs. Unlike the virome, resistome and ARB were not unique for stormwater sampling points. This results underline the need for extensive monitoring of the microbial community structure in the urban stormwater systems to assess antimicrobial resistance spread.
Keywords: MinION, Metagenomics, ARG, ARB, Stormwater
Graphical abstract
1. Introduction
The urban population density and number of citizens, size of areas modified for urban needs and scale of the intrusion of human-made structures in the landscape are growing over time and that all together accelerates the spread and creation of new antibiotic resistance mechanisms. We already know that physicochemical parameters, presence of antibiotics, antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARGs) in the runoff from the cities can have negative impact on water bodies. Despite that, the quality of water and the risk of infection are usually investigated only in municipal sewage systems and in wastewater treatment plants’ (WWTPs) effluents while urban runoff water is often neglected. As Hamilton et al. (2020) reported only 15 quantitative studies focused on ARGs and ARB were published so far. Even though only few studies investigated dissemination of ARB and ARGs in urban runoffs, their results are alarming (Almakki et al., 2019).
Anthropogenic pressure together with storms changing hydro regime and overflow from WWTPs and agricultural area (waste lagoons, manure-treated fields, animal feeding lots) create a new hot-spot and at the same time vehicle for creation and dissemination of ARGs and ARB. The low concentration of antibiotics stimulate creation of ARGs and their spread via horizontal gene transfer (HGT). That is also important as potential route of ARGs transfer from environmental bacteria to pathogens (Almakki et al., 2019). Selection of resistance is highly probable in stormwater due to fecal contamination as well as a presence of heavy metals, the other aspect is absorption of antibiotics and other medicines at organic particles (Almakki et al., 2019). Then during rain out the settled matter can get to the sewer system and to WWTPs. Such flush within stormwater can also increase amount of sediments and partially remove biofilm in the sewage network. This phenomenon is frequent in WWTPs with combined sewer systems during stormwater events, resulting in overflows and direct transfer of untreated wastewater to the receiving aquatic environment (Eggen and Vogelsang, 2015). Global climate changes and dramatic weather phenomena including severe storms have noticeable impact on the pool of ARGs and ARB (Almakki et al., 2019).
The water network in the cities and premise plumbing contain lots of bacteria autochthonous to natural waters, mostly representatives of phyla Acidobacteria, Actinobacteria, Bacteroidetes and Proteobacteria (Wang et al., 2013). Opportunistic human pathogens called in this case opportunistic premise plumbing pathogens (OPPPs) including Legionella pneumophila, Pseudomonas aeruginosa, and non-tuberculous mycobacteria (inherently resistant to disinfectants) are also abundant in the system. However, peculiar milieu in the pipes favors bacterial groups’ colonization, proliferation and spread via the network. OPPPs are additionally adjusted to grow in premise pipes, growth in biofilms saves them not only from being consumed by Protozoa, but also from biocides and thermal shocks (Falkinham et al., 2015).
Kwak et al. (2015) observed increasing frequencies of antibiotic resistance in untreated Stockholm wastewater (UW) during 1 year study of resistance patterns of Escherichia coli. Such results may reflect increasing fecal carriage of ARB in the Swedish urban population and the antimicrobial resistance (AMR) spread. Resistance to one of the 10 antibiotics (ampicillin, cefotaxime, ceftazidime, chloramphenicol, ciprofloxacin, gentamicin, nalidixic acid, cefpodoxime, tetracycline, trimethoprim) were detected in 34% of the UW isolates and 2.4% isolates were resistant to all of 3 beta-lactam antibiotics included in the study. 2.9% from the UW isolates were suspected extended-spectrum beta-lactamase-producing E. coli (ESBL-EC) (Kwak et al., 2015). In presented study we considered all bacteria in urban stormwater, not only connected to fecal contamination, which can have a source in leakages in wastewater system using long-read metagenomic sequencing data. Metagenomics can show ARB and ARGs co-occurrence, connect their abundances in the same (waste)water samples and be a powerful tool helping investigation of HGT as well as the antimicrobial resistance (AMR) dynamics. All without biases caused by primers use or cultivation methods. Although high throughput sequencing methods became cheaper and more available, nowadays trend in metagenomics is to gather more information in higher quality owing to longer read length offered by techniques like nanopore (i.e. MinION; Oxford Nanopore Technologies) and PacBio (Pacific Biosciences) sequencing. We believe that potential of these techniques still has not been fully revealed. In particular no study, to our knowledge, has considered metagenomics using MinION sequencing to gather information about the AMR in stormwater. The common strategies for AMR stormwater investigation are bacterial culturing joined antibiotic resistance testing, screening for specific ARGS with quantitative polymerase chain reaction (qPCR), obtaining ARGs relative abundance next-generation sequencing (both environmental DNA and cultured isolates can be the input), metagenomic using short read sequencing (Hamilton et al., 2020).
We therefore analyzed ARB and ARGs in Stockholm stormwater samples and investigated whether MinION will provide new information regarding AMR. Our analysis revealed various ARB carrying multiple antibiotic resistance mechanisms suggesting need for large-scale metagenomic studies and microbial monitoring of urban stormwater in the face of growing antibiotic resistance.
2. Materials and methods
2.1. Metagenomic sequencing data
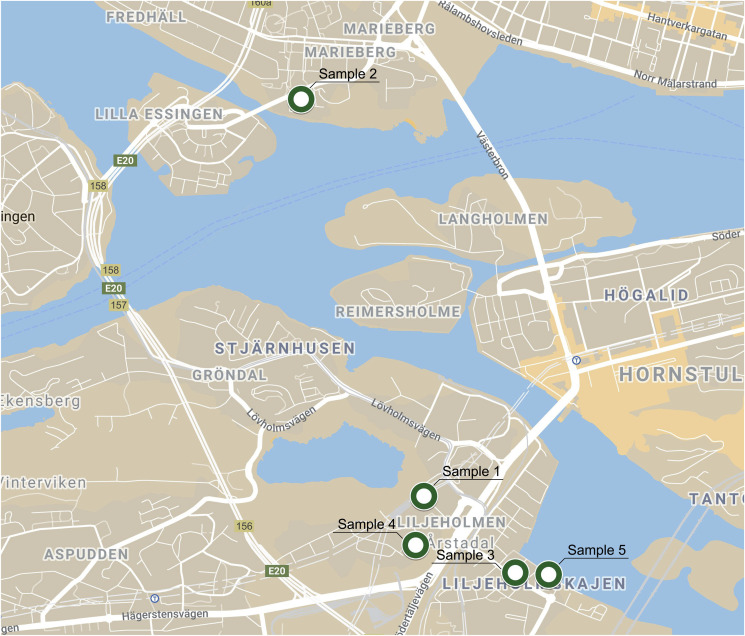
We performed ARB and ARG identification and virus detection in stormwater samples using publicly available metagenomic MinION sequencing data from Sequence Read Archive (SRA), accession number PRJEB20562 (Hu et al., 2018). Authors have taken total of 73 samples from stormwater street manholes not linked in the stormwater system in the city of Stockholm, except for the sample named ‘Bromma’ in supplementary material originating from WWTP. E. coli culturing was performed to assess potential contamination resulting from misconnections of the sanitary sewer pipes (wastewater from kitchens, toilets, and bathrooms) connected to the stormwater system. From all stormwater samples, five were randomly selected for MinION sequencing, two with high (>242,000 MPN per 100 ml, those were Sample 2 and Sample 4) and three with low (<100 MPN, Sample 1, Sample 3, Sample 5) E. coli culturing counts and sufficient DNA amount for MinION sequencing. Total of 5 samples, coming from two different sampling areas and 5 different points (Samples 1, 3, 4, 5 comes from Liljeholmen, according to Hu et al. (2018): C8, C13, C16, C21 respectively; Sample 2 is from Kungsholmen, C12 in Hu et al. (2018); geographic locations are shown in Fig. 1 ) The map was modified manually using images from Google Maps Map of Stockholm, 2017. Swedish urban stormwater systems were sequenced using Oxford Nanopore MinION long-read sequencing technology. FASTQ files deposited to SRA were converted to FASTA files using SRA toolkit (Leinonen et al., 2011).
Fig. 1.
Map showing the sampling locations.
The procedure of DNA extraction and sequencing was described in details in Hu et al. (2018). Briefly, after filtration through 0.22 μm pore-size polyethersulfone membranes the genomic DNA was extracted with the PowerWater® DNA Isolation kit (MO BIO Laboratories Inc.). Later the DNA was sheared and purified before the Nanopore library preparation using the MinION PCR barcoding kit DEV-MAP004 (Oxford Nanopore Technologies; Oxford, UK). The sequencing was performed with ONT protocol and the kit SQK-NSK007 (version R9; Oxford Nanopore Technologies) using 75.0 μl of the library and the 48-h sequencing protocol (NC_48Hr_Sequencing_Run_Flo_MIN106_SQK_LSK208.py) on the MinKNOW control software (version 1.3.25) a base-calling was done on the Metrichor software (version 1.125) and 2D Base-calling plus barcoding program (for FLO-MIN106: “2D Base-calling plus Barcoding for FLO-MIN106 250 bp”).
2.2. Resistome investigation
ARGpore (Xia et al., 2017) was used to investigate antibiotic resistance in stormwater. The tool was design to find AMR genes and their hosts in metagenomic data utilizing BLAST, HMMER, UBLAST using databases derived from ARDB and CARD. ARGpore uses Resistome prediction Algorithm (FASTA sequence was searched against nt-version SARG database (v1.0); valid alignment with >80% similarity over 70% alignment length were kept for further filtering of overlap regions and if two hit regions on the same read overlapped for > 50% alignment length, only the one with longest ARG hit were kept). To identify the host of ARGs ARGpore uses another algorithm - Taxa Algorithm. The Nanopore 2D reads are aligned to MetaPhlAn 2 database of taxon-defining marker genes (Segata et al., 2012). MetaPhlAn (Metagenomic Phylogenetic Analysis) maps reads against a set of clade-specific marker sequences, it compares each metagenomic read from a sample to the marker catalog to identify high-confidence matches (Segata et al., 2012). Only the best alignment (with the highest bit score) showing similarity higher than 80% over more than 70% of the marker gene length is kept for taxonomy classification. The generated output is the list of ARG-containing nanopore reads with taxa annotated (arg.w.taxa.tab). Additional classification of species was made manually based on MiDAS 2.0 (McIlroy et al., 2017).
2.3. Viruses investigation
Virus detection in sequenced samples was done using Kraken tool (Wood and Salzberg, 2014). Kraken assigns taxonomic labels to metagenomics DNA reads by examining the k-mers within a read and then querying a selected database with those k-mers. Kraken was run on the Galaxy platform using default parameters and the viruses database (Galaxy Version 1.2.4). Classification results were summarized using Kraken-report tool (usegalaxy.org).
PPR-Meta ver. 1.1 was used to identify metagenomic sequences as phages, chromosomes or plasmids (Fang et al., 2019). It uses a novel neural network architecture named as the Bi-path Convolutional Neural Network for 3-class classification of sequences based on comparison to prokaryote chromosomes, prokaryote plasmids, and phages from NCBI genome database. The virtual machine version of PPR-Meta was run through the executable file with default parameters. FASTA files containing metagenomic sequences were split in three parts (representing phages, chromosomes and plasmid sequences) based on PPR-Meta classification using seqinr package in R and further analyzed separately using ARGpore as described above.
2.4. Sankey diagram
Sankey Diagram was created using package ggvis in R (Chang and Wickham, 2018). Results of ARGpore analysis for all samples were plotted together with genus levels, resistance types and resistance genes displayed on the diagram nodes.
3. Results
3.1. Main resistance types and ARB harboring them
In total 11 resistance types with 26 resistance mechanisms were found (details are given in Table 1 ). Those were antibiotic inactivation of aminoglycoside, beta-lactam, chloramphenicol, rifamycin (8 such mechanisms were found); antibiotic target alteration of bacitracin, MLS, vancomycin (5); antibiotic target replacement of beta-lactam and trimethoprim (2), antibiotic efflux towards fosmidomycin, multidrug and puromycin (11).
Table 1.
Resistance types, mechanisms and prevalence among the sequenced genomes/plasmids/phages in investigated stormwater samples assembled and supplemented by CARD data (Jia et al., 2017).
| resistance type | resistance mechanisms | prevalence among the sequenced genomes and plasmids available at NCBI for 82 important pathogens |
PPR-Meta prediction of resistance mechanisms presence among samples analyzed in this study (numbered from 1 to 5) on different genetic elements |
||||
|---|---|---|---|---|---|---|---|
| chromosomal | plasmid | chromosomal | plasmid | phages | |||
| aminoglycoside | aac (3)-X | antibiotic inactivation | ND | ND | 1 | ||
| aac (6′)-I | antibiotic inactivation | – | – | 1, 3 | 2 | ||
| aph (3′)-I | antibiotic inactivation | + | + | 4 | 2 | ||
| aph (3′)-IIb | antibiotic inactivation | + | – | 2 | 5 | ||
| bacitracin | bacA | antibiotic target alteration | + | only S. enterica (0.18%) | 2 | 4 | 4 |
| beta-lactam | class A | antibiotic inactivation | ND | ND | 2 | 3 | |
| GES-23 | antibiotic inactivation | ND | ND | 3,4 | |||
| mecI | antibiotic target replacement | + | – | 2 | |||
| chloramphenicol | chloramphenicol | antibiotic inactivation | ND | ND | 3,5 | ||
| fosmidomycin | rosA | antibiotic efflux | ND | ND | 2 | ||
| macrolide-lincosamide-streptogramin | ermO | antibiotic target alteration | ND | ND | 4 | 3, 4 | |
| multidrug | abeS | antibiotic efflux | + | – | 5 | 2 | |
| major facilitator superfamily transporter | antibiotic efflux | ND | ND | 3, 5 | 3, 4 | 2 | |
| mexE | antibiotic efflux | + | – | 4, 5 | 3 | ||
| mexX | antibiotic efflux | – | – | 2, 3, 4 | 3, 4 | 1, 3 | |
| ompR | antibiotic efflux | ND | ND | 3 | |||
| opcM | antibiotic efflux | – | – | 2, 4 | |||
| oprA | antibiotic efflux | + | – | 3 | |||
| oprN | antibiotic efflux | + | – | 4 | 3, 4 | ||
| qacG | antibiotic efflux | ND | ND | 2 | |||
| puromycin | puromycin | antibiotic efflux | ND | ND | 3 | ||
| rifamycin | ADP-ribosylating | antibiotic inactivation | ND | ND | 3 | ||
| trimethoprim | dfrA12 | antibiotic target replacement | + | + | 2 | ||
| vancomycin | vanH | antibiotic target alteration | ND | ND | 2 | ||
| vanR | antibiotic target alteration | ND | ND | 2 | |||
| vanS | antibiotic target alteration | ND | ND | 2 | |||
The ARGs were present not only on chromosomes and plasmids but also on phages. Most of them were present on chromosomes. However, some of the ARGs in our study were found only on mobile genetic elements (MGEs), like aac (6′)-I, aac (3)-X, qacG and vanR. The last three were found only on plasmids.
Seven main resistance types along with species of ARB carrying them are presented in Table 2 . Ten of the found ARB are usually encountered in activated sludge in WWTPs. One identified group of ARB is biofilm forming pathogen previously noted as responsible for prosthetic hip joint infections (Wildeman et al., 2016). Interestingly, identical ARB with the same resistance was found in two pairs of samples: 2 and 3; 4 and 5, independently of number of reads achieved in sequencing and the sampling point 2 (Kungsholmen) was not located next to the other sampling area (Liljeholmen). Sphingomonas sp. resistant to aminoglycoside were found in samples 1, 4 and 5. Limnohabitans sp. were found both in sampling point 2–5, but bacteria in samples 2 and 3, presented resistance toward aminoglycoside while in 4 and 5 to beta-lactams.
Table 2.
Antibiotic resistant bacteria (ARB) identified in stormwater.
| Resistance type/ARB in sewage sample | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| aminoglycoside | Polymorphum gilvum; Rhizobium etli; Sphingomonas sp.∗ | Azospirillum lipoferum; Acidovorax delafieldii∗; Limnohabitans sp.; Rubrivivax benzoatilyticus | Sphingomonas sp.∗; Leifsonia xyli | ||
| beta-lactam | Bacteroides stercoris; Anaerostipes hadrus∗ | Limnohabitans sp. | |||
| fosmidomycin | Acinetobacter tandoii∗ | Mycobacterium parascrofulaceum⋆ | |||
| MLS | Alicycliphilus denitrificans∗; Leptothrix cholodnii∗; Pseudomonas sp.∗; Sphingomonas sp.∗; Sandarakinorhabdus limnophila | ||||
| multidrug | Pseudomonas sp.∗ | Acinetobacter tandoii∗; Propionibacterium avidum✫ | |||
| unclassified | Slackia sp.∗; Coprococcus sp.∗; Eubacterium eligens; Prochlorococcus sp. | ||||
| vancomycin | Eubacterium eligens; Thermoanaerobacter wiegelii | ||||
| number of reads | 56,220 | 122,506 | 44,337 | 69,316 | 31,756 |
Description: MLS: macrolide-lincosamide-streptogramin, species encountered in wastewater treatment plants are marked with∗ (based on MiDAS 2.0), opportunistic pathogen are marked with⋆ and pathogens are marked with✫.
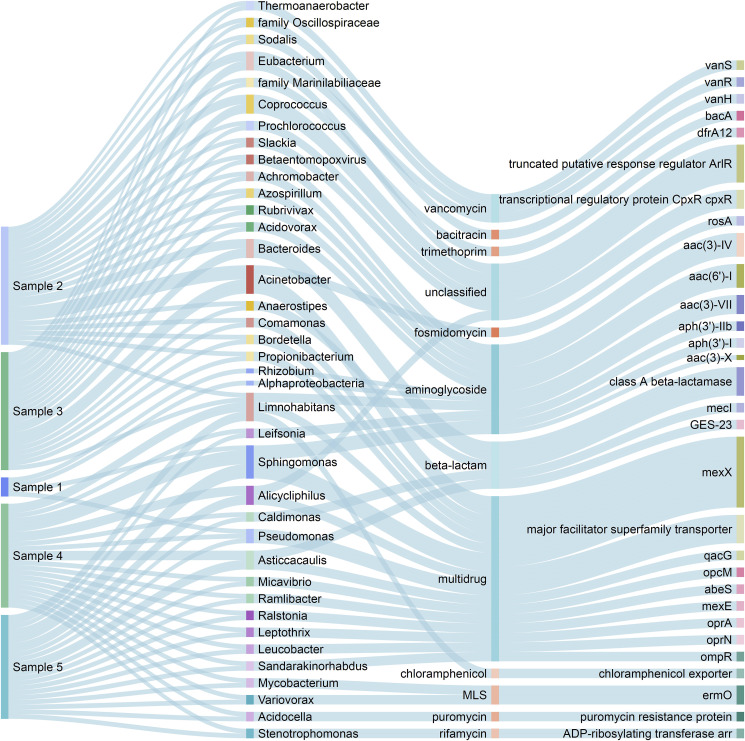
All found ARB identified to genus level (with except for the unclassified representatives of Marinilabiliaceae and Oscillospiraceae) are presented in Fig. 2 together with carried genes and resistance type. Also Betaentomopoxviruses carrying resistance towards aminoglycoside were encountered.
Fig. 2.
Sankey Diagram showing results of ARGpore classification analysis. The nodes represent genus levels of bacteria with antibiotic resistance mechanisms and genes found in the metagenomic sequencing data from Samples 1–5.
3.2. Viruses
Caudovirales, Herpesvirales, Polydnaviridae were present in all tested samples. Pandoravirus dulcis, Pandoravirus salinus and Baculoviridae were not found only in sample 2. Micromonas pusilla virus 12 T were in samples 1 and 3. Poxviridae in samples 2, 3, 4; uncultured phage crAssphage in sample 3. The Coronaviridae responsible for COVID-19 pandemics were not found in the samples. The order Nidovirales contains this family and was noted only in Sample 4 with the Ball python nidovirus.
Caudovirales and Herpesvirales were the most abundant orders consisting 20%–88% of all viruses in the sample. Classification sometimes even to species level and the viral abundance can be found in Krona diagram (Supplementary Figure 1) (Ondov et al., 2011).
4. Discussion
4.1. Resistance mechanisms
The most widespread resistance mechanism according to Zhang et al. (2009), is the target bypass, which is inaccessibility of the antibiotics to their target enzyme by mutational changes or loss on the enzyme gene (Huovinen et al., 1995; Happi et al., 2005). Surprisingly, such ARGs were not noticed in this study. We showed that in Stockholm’s stormwater most common resistance mechanisms were based on antibiotic efflux (11 out of 26), contrary to the findings of Zhang et al. (2009) for environmental ARB in the water. However, those results tie well with previous study by Nesme et al. (2014). Wherein they investigated environmental metagenomes. Mentioned study showed that bacteria with the efflux pumps were able to revoke the dosing of antibiotics used in veterinary and human healthcare namely beta-lactam, vancomycin, and tetracycline.
Others presented resistance determinants in bacterial genome have broader spectrum and are more connected to long environmental pressure than just a defense against human-produced antibiotics. Martínez (2017) remarks environmental profits like new functions obtained with ARGs as essential to keep being fixed and preserved in genome. The efflux pumps are good example of exaptation in AMR, which is organism’s procuring a new function that did not exist or differs from its original function which had been derived by evolution.
From our results on ARB having unique resistant phenotype and efflux mechanism being the most frequent one in the study, it is clear that environmental organisms may achieve resistance to antibiotics as exaptation. The efflux pumps are able to discard molecules from cytoplasm through membrane and wall of the cell. Such machinery does not target defined particles, it removes whole variety of metabolic products. What is more, in case of overexpression or mutation in pump, even antibiotics, biocides, heavy metals and pesticides can be ejected by the efflux system at the same time giving resistance to the host (Alonso et al., 1999; Baker-Austin et al., 2006; Buffet-Bataillon et al., 2016). Often it is also the reason for co-resistance to antibiotics and heavy metals in bacteria (Piddock, 2006).
Mentioned evolutionary trait was noted for the beta-lactamases. The enzymes are involved in cell wall biosynthesis as well as hydrolyzing both natural and man-made antibiotics of the beta-lactam family evolved from the same ancestor (Meroueh et al., 2003).
We found 4 other systems: (1) antibiotic inactivation through direct or indirect deactivation of antibiotic molecule (Wright, 2005); (2) modification of drug targeted structures including modification of the action sites of antibiotics (Lambert, 2005); (3) antibiotic target alteration and (4) antibiotic target replacement. It is noteworthy that the resistance to certain antibiotic may be associated with different ARGs based on more than one mechanism.
Prevalence of the identified mechanisms exclusively in chromosomes was noted in 50%, and in 17% both in chromosomes and plasmids. For most (54%) of the found ARGs such classification is still not available (Table 1).
4.2. ARB in stormwater
4.2.1. Environmental ARB
In line with previous studies many of the encountered ARB are associated with different environments: (1) WWTPs: half of the ARB species in this study (marked with ∗ in Table 2); (2) freshwater: Sandarakinorhabdus limnophila and Limnohabitans sp.; (3) lake water: Ramlibacter and Variovorax (Hahn et al., 2010); (4) soil habitat and plant growth, for example: Comamonas, Achromobacter, Azospirillum, Rhizobium, Xanthomonadaceae, Acidovorax (phytopathogen), Leifsonia xyli (sugar cane pathogen), Ralstonia (phytopathogen); (5) extreme environments: Asticcacaulis (deserts) and Caldimonas (hot springs); (6) endosymbiosis in insects: Sodalis, which is also natural hosts for Betaentomopoxvirus (present in our tested samples and carrying ARGs).
A similar pattern of results for Variovorax was obtained by Roberts (2011), where owing to efflux pumps it was resistant to tetracyclines (tetA and tetL). Another bacteria, Asticcacaulis excentricus were previously recorded as resistant to beta-lactams (Kanehisa and Goto, 2000), in our study we also noted in the genome the presence of genes coding protein from major facilitator superfamily able to give resistance to these and many other drugs.
Limnohabitans sp. was noted in 80% of samples, but the resistance presented by this species varied between locations (aminoglycoside in 2, beta-lactams in 3) suggesting that ARGs were obtained in effect of local environmental pressure.
4.2.2. ARB that can be transferred from/to human body environment
Most of the found taxa are not able to survive in the human body. Other like Propionibacterium are human commensals or constitute human gut microbiota like Bacteroides stercoris and Eubacterium eligens.
Another species, Mycobacterium parascrofulaceum is an opportunistic pathogen, like many other nontuberculous mycobacterial species (Tortoli et al., 2005). Even though M. parascrofulaceum have rosA gene, cationic antimicrobial peptides (CAMPs) efflux/K+ antiporter, it is not additional danger when it comes to spread. Described subunit typical for Mycobacteria. M. tuberculosis is not affected by fosmidomycin due to a lack of its uptake, not thanks to rosA. As it was described by Brow and Parish (Brown and Parish, 2008) mycobacteria are naturally resistant at the cell level, not connected to efflux pump. Usually Mycobacteria are resistant to antibiotic owing to cell wall rich in long-chain fatty acids covalently linked to the arabinogalactan-peptidoglycan layer. Moreover, the porins, which are specific protein channels allowing hydrophilic molecules to enter the cell via diffusion, are rare in portrayed taxa (Trias et al., 1992).
Our analysis found evidence for the presence of pathogenic ARB in stormwater. It is Bordetella causing ‘whooping cough’ respiratory diseases, which can develop to vomiting, convulsions, coma and death (Mattoo and Cherry, 2005). Roberts (2011) reported resistant Bordetella with tetA and tetC genes. Macrolides are recommended for ‘whooping cough’ treatment (erythromycin, clarithromycin, or azithromycin (Altunaiji et al., 2007)), therefore it is disturbing that in investigated samples Bordetella were carrying mexX giving resistance also to this group of drugs.
Propionibacteria are members of the human skin microbiota, but are also opportunistic pathogens responsible for prosthetic hip joint infections (Achermann et al. 2014, 2018; Wildeman et al., 2016). Lately, Achermann et al. (2018) reported large series of periprosthetic joint infections in Sweden caused by P. avidum. 11 out of 12 tested strains were susceptible to clindamycin, levofloxacin, and rifampin. The authors also used whole genome sequencing and found EPS-encoding island, probably obtained by HGT (flanked by transfer RNA genes). They connected the capacity to initiate biofilms on medical implants with potentially key virulence trait of P. avidum. However, the authors did not manage to point out a specific risk factor for the increasing number of P. avidum periprosthetic joint infections in recent years. Interestingly, we found resistant representatives of P. avidum in stormwater in the samples from year 2013, which was within the period (1997–2015) of Achermann et al. (2018) investigation. What is more, the mexX mechanisms characterizing P. avidum in our study gives resistance towards drugs tested in Achermann et al. (2018) study: penman (penicillin), lincosamide (clindamycin), fluoroquinolones (ciprofloxacin and levofloxacin) and cephalosporin (cefuroxime), but not to lincosamide (clindamycin) and rapamycin (rifampin) for which the lowest minimal inhibitory concentrations were noted in the mentioned study (Achermann et al., 2018 – Table 3 in original work).
Risk assessment is essential to unravel the relation between human exposure to ARB potentially infecting humans and environmental ARGs (Pruden et al., 2018). We identified the pathogens carrying ARGs, the question of human exposition to bacteria from urban stormwater stays open.
4.3. Comparison of ARGs in stormwater and wastewater
These basic findings are consistent with research showing ARGs occurrence in stormwater. We found genes giving tolerance to the aminoglycoside, beta-lactams, fosmidomycin, MLS, multidrug and vancomycin. Some of them may be the source of spread of AMR to those drugs to WWTPs with stormwater influent. The result of our analysis was compared with the first broad-spectrum metagenomic investigation of WWTP’s resistome Yang et al. (2014). He found the same ARGs types giving resistance toward drugs listed above and 12 more. Those additional groups were acridine, acriflavine, bacitracin, bicyclomycin, chloramphenicol, fosfomycin, polymyxin, quinolone, sulfonamide, tetracycline and trimethoprim.
While only ARGs giving resistance towards sulfonamides (sul 1), tetracyclines (tetM), beta-lactamases (bla OXA58, bla TEM, bla OXA-48, bla CTX-M-32, bla KPC-3) and colistins (mcr-1) were monitored and reported by Cacace et al. (2019) with qPCR in the European WWTPs. Unfortunately, Sweden being in focus in our article was not included in the survey. The resistance to aminoglycoside (aacA, aadA, strA, strB, aphA) (Ferreira Da Silva et al., 2007; Moura et al., 2012); macrolides (ermA, B, F) (Agga et al., 2015; Marti et al., 2013; Szczepanowski et al., 2009); glycopeptides (vancomycin, vanA) (Araújo et al., 2010; Morris et al., 2012; Rosenberg Goldstein et al., 2014); trimethoprim (dfr) (Ferreira da Silva et al., 2007; Moura et al., 2012; Schwartz et al., 2006) and quinolones (accA6-ib-cr, qrnA, qrnB, qrnS) (Agga et al., 2015; Figueira et al., 2011a; Szczepanowski et al., 2009) were also noted in different studies.
Our study suffers from the limitations associated with a lack of data regarding HGT and sources of ARGs in various environmental compartments.
The ARGs found in stormwater may be considered worth investigating in WWTP. Additional analysis of ARB carrying the genes may show if there was a potential transfer of resistance from one bacterial group to another. Also similarities in the sequences responsible for resistance can indicate the route of ARGs spread.
4.4. Viruses and potential HGT in stormwater
Others confirmed that CrAssphage can be a marker of human fecal contamination (Karkman et al., 2019), which implicates the sampling point 3 to be contaminated with human feces through sewer pipe damage. However, we do not see the typical fecal ARB composition which is also identical in the sample 2, where we did not find crAssphage.
Caudovirales order constitute a majority in phages. The Siphoviridae phage is noted in highest abundance in many habitats, also WWTPs (Nigro et al., 2017; Parmar et al., 2018; Wang et al., 2018). Siphoviridae constituted 3–25% of all viruses and 30–57% of Caudovirales in tested samples (Supplementary material).
Order Caudovirales was the most abundant in the hospital wastewater investigated by Subirats et al. (2016) and in aquaculture wastewater by Colombo et al. (2016), both studies focused on AMR.
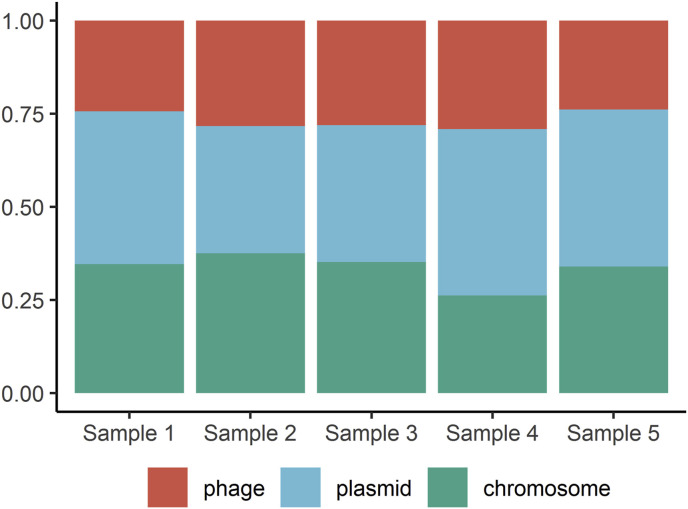
The relative abundance of DNA from chromosome, plasmid and phage DNA was determined for each sample (Fig. 3 ) using prediction algorithm, indicating highest contribution of plasmid in samples 1, 4, 5, with 40%, 44%, 42% respectively. Phages were most abundant in samples 2, 3 and 4 (with 28%, 28%, 29% respectively). High percentage of MGEs and phages may be associated with frequent HGT. Especially that 3 ARGs were found exclusively on plasmids. The resistance mechanisms aac (6′)-I was present both on plasmids and phages sequences. Six other ARGs were also identified on phages contigs without confirmation of their presence in other contigs on chromosomes (Table 1). One can assume that they could have been transferred by those phages.
Fig. 3.
Percentages of phages, chromosomes, and plasmids in the stormwater samples. The sequences of phages, chromosomes, and plasmids in metagenomic data obtained by long-read sequencing were predicted using PPR-Meta and the sequence percentages of phages, chromosomes, and plasmids were calculated.
In the Subirats et al. (2016) hospital wastewater study the relative abundance of ARGs in the phages was increased by half in comparison to the bacterial DNA fraction. As the phages are the most abundant biological entities around the world and they are able to transfer genes among bacterial hosts, the transduction should be carefully investigated in AMR studies. The transposons, integrative conjugative elements, plasmids and bacteriophages and other MGEs may be involved in HGT. Especially phages are directly involved in pickup and recombination of foreign DNA by bacteria (Marti et al., 2013). To verify the frequency of the HGT and confirm which ARGs were obtained this way, additional experiments performed on live microbes are required.
4.5. Comparison between sampling points
Resistome and ARB were not unique for sampling points in the study. However, the virome was individual for every point. The ARB in sampling points varied between locations and at the same time were similar for pairs of sampling points. Pseudomonas were in samples 1, 4 and 5. Contrary to the findings of Yeom et al. (2017) on soil microbiome, we did not find Pseudomonas spp. Presenting strong resistance to sulfathiazole.
Samples 4 and 5 contain same groups of bacteria. Also set of ARB in sample 2 and 3 were the same, additionally they contained resistant Betaentomopoxvirus (family Poxviridae).
The sampling points were not connected by the stormwater system, coming from two different sampling areas. The similarity in ARB composition is hard to explain by geographic location (Fig. 1) as samples 3, 4, 5 were gathered in Liljeholmen, while sample 2 in the island area Kungsholmen. The limitations of the present studies using sequencing data from repository naturally include limitations connected to metadata absence. We see the results of some factors influencing the ARB composition, but we are unable neither to indicate which factor those may be nor to prove their differentiating effects. However, we acknowledge that there are considerable discussions among researchers concerning hydrological conditions affecting the AMR (Hamilton et al., 2020). The hydrological and meteorological variables could have transported stressing factor, bacteria carrying ARGs prone to HGT or even ARB. Unfortunately we cannot asses given the influence as those factors were neglected during original study providing the sequencing data.
Sphingomonas sp. Resistant to aminoglycoside were found in 2 out of 3 sampling point - 1 and 3. Sphingomonadaceae were studied by Vaz-Moreira et al. (2011) as isolates from a drinking water, treatment plant, tap water, cup fillers for dental chairs and a water demineralization filter. Unlike bacterium from Swedish stormwater, none of them presented resistance to aminoglycoside (Vaz-Moreira et al., 2011). We speculate that this might be due to extraordinary habitat in stormwater system and urban factors driving AMR. However, one of the species S. paucimobilis has been previously reported resistant toward penicillin and first generation cephalosporin, third generation cephalosporin and fluoroquinolone and susceptible to tetracycline, chloramphenicol, aminoglycosides, carbapenems and trimethoprim and sulphamethoxazole (Krishna et al., 2011). Therefore, the revealed resistance mechanisms are not unique to stormwater Sphingomonadaceae.
As postulated by Hamilton et al. (2020), the measurement of AMR stressors in stormwater environments would be very helpful (e.g. heavy metals and antibiotics concentrations). The possibilities of drawing conclusions is confined here due to lack of context.
4.6. Opportunities connected to MinION sequencing
The results demonstrated in this chapter match state of the art methods. ARGs detection based on sequencing can help to avoid ARG primers limitation or to design primers adjusted to environmental variety of ARGs. AMR studies based on high-throughput sequencing is already popular in environmental studies on the distribution of ARGs in various ecosystems (Freilich et al., 2010; Li et al., 2015). One limitation is found in this case, most of the sequencing techniques (e.g., Illumina, 454) could hardly achieve the real-time resistome profiling which is required to guide the resistance control measures. The genes acquired by HGT including bacterial virulence and antibiotic resistance genes, have an inherent repetitive nature resulting in numerous gaps in the short-read assemblies. Due to those obstacles, the assembles of the short reads are also fragmented due to their nature. It hinders the identification of the ARB carrying found ARGs.
The long-reads provided by third generation sequencing techniques like nanopore i.e. MinION contribute in high quality assembles of viral and bacterial genomes. The longer the read, the bigger chance of finding a matching sequence to the reference database (Huson et al., 2018). MinION sequencing was already used by Xia et al. (2017) for multiple antibiotic resistant coliform bacteria detection in wastewater. It was a sufficient tool for quick monitoring and parallel phylogenetic identification. It is also helpful in monitoring genomic divergence obtained by HGT. Due to the inherent repetitiveness or flanking of genome parts acquired by HGT the short read sequencing (like MiSeq, Illumina), would leave numerous gaps in the in assembles of resistance and pathogenicity islands (Ashton et al., 2015).
4.7. Limitations of the study
The ultimate goal of this study was to verify if the genomic DNA MinION sequencing can be used in metagenomic identification of bacterial and viral AMR in stormwater using publicly available data and open source bioinformatics tools. The data from the repository were used in the study of E. coli in stormwater, thus many of the metadata regarding many aspects that would be useful in our study were not collected. Issues connected to the lack of metadata in stormwater studies have been lately described in review by Hamilton et al. (2020), some of them were mentioned in chapter 4.5. The authors advocates for filing the well-defined knowledge gaps regarding (1) characterization of background conditions; (2) survey of AMR stressors in stormwater environments; (3) survey of hydrological conditions; (4) global agreement on needed defined sampling targets. Our study has contribution in the latest gap. We presented spectrum of ARB and ARGs occurring in stormwater in European capital, the region neglected in studies listed in Hamilton et al. (2020). In their review only European representative was Italy, other studies were performed in USA and China.
Other limitations are connected to the MinION sequencing technique. It is still not perfect and often characterized by high error rate. However, it according to Loman et al. (2015) it has no significant impact the ARG/ARB prediction. There are plenty of bioinformatic algorithms addressing issues with error correction and de novo assembly of nanopore long reads (Loman et al., 2015). First of all MinION sequencing can have a low accuracy (typically 80–90%), but errors distribution is relatively random. Therefore, during identification of most ARGs based on BLASTN alignments some degree of sequence mismatches in reads should not badly impact the overall identification. Kamathewatta et al. (2019) proved this thesis in their research by angling alignments between individual MinION presumably including ARGs and corresponding sequences from the ResFinder database.
We used ARGpore, as the problem of high error rate for ARGs was already explained by authors of the tool in their paper (Xia et al., 2017). Despite the quite high error rate (assess for their study as 9% by Phred Score and 15.4% by mapping to Illumina assembly), reliable ARG survey was possible using MinION sequences as their length (on average > 800 bp) was almost equal to the whole length of reference sequences in SARG database (1131 bp on average).
5. Conclusions
The study concludes how to process long read metagenomes in search of ARB connected with defined resistance type. Coexistence of environmental and pathogenic bacteria in the same niches in stormwater and detailed investigation regarding horizontal gene transfer of extracellular ARGs between those groups should be continued. At this point the possibility of identifying not only pathogens but also resistance encoding genetic material that may be integrated by pathogens is crucial. MinION sequencing and work frame presented in the study allowed not only identification of ARGs (resistance mechanism), but also ARB carrying them. This assumption might be addressed in future studies on more samples and comparison between short and long read sequencing.
Overall, our findings indicate that ARB in Swedish stormwater are organisms typical for WWTPs, associated with soil, lake and freshwater, plant growth as well as insects’ endosymbionts. Alarmingly Bordetella causing ‘whooping cough’ resistant to macrolides was identified in two samples. To our knowledge, this is the first report of source of Propionibacterium avidum carrying mexX that may be connected to large series of periprosthetic joint infection in Sweden.
Resistome and ARB were not unique for stormwater sampling points, but the virome was. Caudovirales, Herpesvirales, Polydnaviridae were present in stormwater, in most samples Betaentomopoxviruses (family Poxviridae) carrying resistance towards aminoglycoside was noted.
CRediT authorship contribution statement
Maciej Białasek: Formal analysis, Visualization, Writing - review & editing. Aleksandra Miłobędzka: Conceptualization, Formal analysis, Supervision, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors, Maciej Białasek and Aleksandra Miłobędzka declare no conflict of interest.
Acknowledgements
We would like to thank Yue Hu, Anders Andersson and their co-authors for open data sharing as well as providing additional information regarding sampling points in private communication. The researcher working on this project was supported by the European Structural and Investment 671 Funds, OP RDE-funded project ’ChemJets’ (No. CZ.02.2.69/0.0/0.0/16_027/0008351).
Handling Editor: Jian-Ying Hu
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.chemosphere.2020.127392.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Achermann Y., Goldstein E.J., Coenye T., Shirtliff M.E. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin. Microbiol. Rev. 2014;2014(27):419–440. doi: 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achermann Y., Liu J., Zbinden R., Zingg P.O., Anagnostopoulos A., Barnard E. Propionibacterium avidum: a virulent pathogen causing hip periprosthetic joint infection. Clin. Infect. Dis. 2018;2018(66):54–63. doi: 10.1093/cid/cix665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agga G.E., Arthur T.M., Durso L.M., Harhay D.M., Schmidt J.W. Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PloS One. 2015;10 doi: 10.1371/journal.pone.0132586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almakki A., Jumas-Bilak E., Marchandin H., Licznar-Fajardo P. Science of the total environment antibiotic resistance in urban runoff. Sci. Total Environ. 2019;667:64–76. doi: 10.1016/j.scitotenv.2019.02.183. [DOI] [PubMed] [Google Scholar]
- Alonso A., Rojo F., Martínez J.L. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environ. Microbiol. 1999;1:421–430. doi: 10.1046/j.1462-2920.1999.00052.x. [DOI] [PubMed] [Google Scholar]
- Altunaiji S., Kukuruzovic R., Curtis N., Massie J. Antibiotics for whooping cough (pertussis) Cochrane Database Syst. Rev. 2007;3 doi: 10.1002/14651858.CD004404.pub3.PMID17636756. CD004404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo C., Torres C., Silva N., Carneiro C., Gonçalves A., Radhouani H., Correia S., da Costa P.M., Paccheco R., Zarazaga M., Ruiz-Larrea F., Poeta P., Igrejas G. Vancomycin- resistant enterococci from Portuguese wastewater treatment plants. J. Basic Microbiol. 2010;50:605–609. doi: 10.1002/jobm.201000102. [DOI] [PubMed] [Google Scholar]
- Ashton P.M., Nair S., Dallman T., Rubino S., Rabsch W., Mwaigwisya S. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. 2015. 33. [DOI] [PubMed]
- Baker-Austin C., Wright M.S., Stepanauskas R., McArthur J.V. Co-selection of anti- biotic and metal resistance. Trends Microbiol. 2006;14:176–182. doi: 10.1016/j.tim.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Brown A.C., Parish T. Dxr is essential in Mycobacterium tuberculosis and fosmidomycin resistance is due to a lack of uptake. BMC Microbiol. 2008;8:1–9. doi: 10.1186/1471-2180-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffet-Bataillon S., Tattevin P., Maillard J.-Y., Bonnaure-Mallet M., Jolivet-Gougeon A. Efflux pump induction by quaternary ammonium compounds and fluoroquinolone resistance in bacteria. Future Microbiol. 2016;11:81–92. doi: 10.2217/fmb.15.131. [DOI] [PubMed] [Google Scholar]
- Cacace D., Fatta-Kassinos D., Manaia C.M., Cytryn E., Kreuzinger N., Rizzo L., Karaolia P., Schwartz T., Alexander J., Merlin C., Garelick H., Schmitt Daisy, de Vries, Carsten U Schwermer, Sureyya Meric, Can Burak Ozkal, Marie H., Pons N., Kneis D., Berendonk T.U. Water Research; 2019. Antibiotic Resistance Genes in Treated Wastewater and in the Receiving Water Bodies: A Pan-European Survey of Urban Settings. [DOI] [PubMed] [Google Scholar]
- Chang W., Wickham H. 2018. Ggvis: Interactive Grammar of Graphics. [Google Scholar]
- Tortoli E., Chianura L., Fabbro L., Mariottini A., Martín-Casabona N., Mazzarelli G. Infections due to the newly described species Mycobacterium parascrofulaceum. J. Clin. Microbiol. 2005;43:4286–4287. doi: 10.1128/JCM.43.8.4286-4287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S., Arioli S., Guglielmetti S., Lunelli F., Mora D. Virome-associated antibiotic-resistance genes in an experimental aquaculture facility. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Ecol. 2016;ume 92(3) doi: 10.1093/femsec/fiw003. March 2016, fiw003. [DOI] [PubMed] [Google Scholar]
- Eggen T., Vogelsang C. Vol. 67. Elsevier; 2015. Occurrence and fate of pharmaceuticals and personal care products in wastewater. Compr. Anal. Chem. [DOI] [Google Scholar]
- Falkinham J.O., Hilborn E.D., Arduino M.J., Pruden A., Edwards M.A. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environ. Health Perspect. 2015;123:749–758. doi: 10.1289/ehp.1408692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Tan J., Wu S., Li M., Xu C., Xie Z., Zhu H. PPR-Meta: a tool for identifying phages and plasmids from metagenomic fragments using deep learning. GigaScience. 2019;8(6):giz066. doi: 10.1093/gigascience/giz066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira da Silva M., Vaz-Moreira I., Gonzalez-Pajuelo M., Nunes O.C., Manaia C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban waste water treatment plant. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Ecol. 2007;60:166e176. doi: 10.1111/j.1574-6941.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- Figueira V., Serra E., Manaia C.M. Differential patterns of antimicrobial resistance in population subsets of Escherichia coli isolated from waste- and surface waters. Sci. Total Environ. 2011;409:1017–1023. doi: 10.1016/j.scitotenv.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Freilich S., Kreimer A., Meilijson I., Gophna U., Sharan R., Ruppin E. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010;38:3857–3868. doi: 10.1093/nar/gkq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M.W., Kasalický V., Jezbera J., Brandt U., Jezberova J., Šimek K. Limnohabitans curvus gen. nov., sp. nov., a planktonic bacterium isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 2010;2010(60):1358–1365. doi: 10.1099/ijs.0.013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton K.A., Garner E., Joshi S., Ahmed W., Ashbolt N., Medema G., Pruden A. Antimicrobial resistant microorganisms and their genetic determinants in stormwater: a systematic review. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.02.012. [DOI] [Google Scholar]
- Happi C.T., Gbotosho G.O., Folarin O.A., Akinboye D.O., Yusuf B.O., Ebong O.O., Sowunmi A., Kyle D.E., Milhous W., Wirth D.T., Oduola A.M.J. Polymorphisms in Plasmodium falciparum dhfr and dhps genes and age related in vivo sulfaxine-pyrimethamine resistance in malaria-infected patients from Nigeria. Acta Trop. 2005;95:183–193. doi: 10.1016/j.actatropica.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Hu Y., Ndegwa N., Alneberg J., Johansson S., Logue J., Huss M. Stationary and portable sequencing-based approaches for tracing wastewater contamination in urban stormwater systems. Sci. Rep. 2018;8:11970. doi: 10.1038/s41598-018-29920-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huovinen P., Sundstrom L., Swedberg G., Skold O. Trimethoprim and sulfonamide resistance. Antimicrob. Agents Chemother. 1995;39:279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Albrecht B., Bagci C., Bessarab I., Gorska A., Jolic D. MEGAN-LR: new algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biol. Direct. 2018;13(1):6. doi: 10.1186/s13062-018-0208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K. Card 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamathewatta K., Bushell R., Young N., Stevenson M., Billman-Jacobe H., Browning G. Exploration of antibiotic resistance risks in a veterinary teaching hospital with Oxford Nanopore long read sequencing. PloS One. 2019;14 doi: 10.1371/journal.pone.0217600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkman A., Pärnänen K., Larsson D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019;10 doi: 10.1038/s41467-018-07992-3. Article number: 80 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Ciraj A.M., Bairy I., Shobha K.L. Sphingomonas paucimobilis urinary tract infection in a renal transplant recipient: a rare case. Int. J. Med. Publ. Health. 2011;1(No. 1):47–49. 2011. [Google Scholar]
- Kwak Y.-K., Colque P., Byfors S., Giske C.G., Möllby R., Kühn I. Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: does it reflect the resistance trends in the society? Int. J. Antimicrob. Agents. 2015;45:25–32. doi: 10.1016/j.ijantimicag.2014.09.016. [DOI] [PubMed] [Google Scholar]
- Lambert P.A. Bacterial resistance to antibiotics: modified target sites. Adv. Drug Deliv. Rev. 2005;57:1471–1485. doi: 10.1016/j.addr.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Leinonen R., Sugawara H., Shumway M. The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Yang Y., Ma L., Ju F., Guo F., Tiedje J.M. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015;9:2490–2502. doi: 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loman N.J., Quick J., Simpson J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 2015;12:733–735. doi: 10.1038/nmeth.3444. [DOI] [PubMed] [Google Scholar]
- Map of Stockholm (Map Data © Google). Available at: https://www.google.se/maps/place/Sztokholm,+Szwecja/@59.3258414,17.7018721,10z/data=!3m1!4b1!4m5!3m4!1s0x465f763119640bcb:0xa80d27d3679d7766!8m2!3d59.3293235!4d18.0685808.
- Marti E., Jofre J., Balcazar J.L. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PloS One. 2013;8 doi: 10.1371/journal.pone.0078906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez J.L. Ecology and evolution of chromosomal gene transfer between environmental microorganisms and pathogens. Microbiol. Spectr. 2017;6(1):1–16. doi: 10.1128/microbiolspec.MTBP-0006-8862016. [DOI] [PubMed] [Google Scholar]
- Mattoo S., Cherry J.D. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 2005;18:326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy S.J., Kirkegaard R.H., McIlroy B., Nierychlo M., Kristensen J.M., Karst S.M., Albertsen M., Nielsen P.H. 2017. MiDAS 2.0: an ecosystem-specific taxonomy and online database for the organisms of wastewater treatment systems expanded for anaerobic digester groups. Database 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroueh S.O., Minasov G., Lee W., Shoichet B.K., Mobashery S. Structural aspects for evolution of beta-lactamases from penicillin-binding proteins. J. Am. Chem. Soc. 2003;125:9612–9618. doi: 10.1021/ja034861u. [DOI] [PubMed] [Google Scholar]
- Morris D., Galvin S., Boyle F., Hickey P., Mulligan M., Cormican M. Enterococcus faecium of the vanA genotype in rural drinking water, effluent, and the aqueous environment. Appl. Environ. Microbiol. 2012;78:596–598. doi: 10.1128/AEM.06636-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A., Pereira C., Henriques I., Correia A. Novel gene cassettes and integrons in antibiotic- resistant bacteria isolated from urban wastewaters. Res. Microbiol. 2012;163:92–100. doi: 10.1016/j.resmic.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Nesme J., Cecillon S., Delmont T.O., Monier J.M., Vogel T.M., Simonet P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014;24(2014):1096–1100. doi: 10.1016/j.cub.2014.03.036. [DOI] [PubMed] [Google Scholar]
- Nigro O.D., Jungbluth S.P., Lin H.T., Hsieh C.C., Miranda J.A., Schvarcz C.R., Rappe M.S., Steward G.F. Viruses in the oceanic basement. mBio. 2017;2017(8) doi: 10.1128/mBio.02129-16. e02129-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinf. 2011;12(1):385. doi: 10.1186/1471-2105-12-385. 2011 Sep. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K., Dafale N., Pal R., Tikariha H., Purohit H. An insight into phage diversity at environmental habitats using comparative metagenomics approach. Curr. Microbiol. 2018;2018(75):132–141. doi: 10.1007/s00284-017-1357-0. [DOI] [PubMed] [Google Scholar]
- Piddock L.J.V. Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Pruden A., Alvarez P.J.J., Ashbolt N., Bischel H., Alcalde R.E., Capiro N.L., Riquelme M.V. An environmental science and engineering framework for combating antimicrobial resistance an environmental science and engineering framework for combating antimicrobial resistance, (December) 2018. [DOI]
- Roberts M.C. Antimicrobial Resistance in the Environment. 2011. Mechanisms of bacterial antibiotic resistance and lessons learned from environmental tetracycline-resistant bacteria; pp. 93–121. [DOI] [Google Scholar]
- Rosenberg Goldstein R.E., Micallef S.A., Gibbs S.G., George A., Claye E., Sapkota A., Joseph S.W., Sapkota A.R. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 2014;466(67):404–411. doi: 10.1016/j.scitotenv.2013.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subirats J., Sanchez-Melsio A., Borrego C.M., Balcazar J.L., Simonet P. Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int. J. Antimicrob. Agents. 2016;48(2):163–167. doi: 10.1016/j.ijantimicag.2016.04.028. 2016. [DOI] [PubMed] [Google Scholar]
- Szczepanowski R., Linke B., Krahn I., Gartemann K., Gu T., Eichler W., Pu A., Schlu A. Detection of 140 clinically relevant antibiotic- resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology. 2009;155:2306–2319. doi: 10.1099/mic.0.028233-0. [DOI] [PubMed] [Google Scholar]
- Trias J., Jarlier V., Benz R. Porins in the cell wall of mycobacteria. Science. 1992;1992(258):1479–1481. doi: 10.1126/science.1279810. [DOI] [PubMed] [Google Scholar]
- Vaz-Moreira I., Nunes O., Manaia C. Diversity and antibiotic resistance patterns of Sphingomonadaceae isolates from drinking water. Appl. Environ. Microbiol. 2011;77(16):5697–5706. doi: 10.1128/AEM.00579-11. 2011 Published by American Society for Microbiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Pryor M.A., Edwards M.A., Falkinham J.O., Pruden A. Effect of GAC pre- treatment and disinfectant on microbial community structure and opportunistic pathogen occurrence. Water Res. 2013;47:5760–5772. doi: 10.1016/j.watres.2013.06.052. [DOI] [PubMed] [Google Scholar]
- Wang Y., Jiang X., Liu L., Li B., Zhang T. High-resolution temporal and spatial patterns of Virome in wastewater treatment systems. Environ. Sci. Technol. 2018;2018(52):10337–10346. doi: 10.1021/acs.est.8b03446. [DOI] [PubMed] [Google Scholar]
- Wildeman P., Brüggemann H., Scholz C.F.P., Leimbach A., Söderquist B. Propionibacterium avidum as an etiological agent of prosthetic hip joint infection. PloS One. 2016;11(6) doi: 10.1371/journal.pone.0158164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.D. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 2005;57:1451–1470. doi: 10.1016/j.addr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Xia Y., Li A.D., Deng Y., Jiang X.T., Li L.G., Zhang T. MinION Nanopore sequencing enables correlation between resistome phenotype and genotype of coliform bacteria in municipal sewage. Front. Microbiol. 2017;8(OCT):1–13. doi: 10.3389/fmicb.2017.02105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li B., Zou S., Fang H.H.P., Zhang T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014;62:97–106. doi: 10.1016/j.watres.2014.05.019. [DOI] [PubMed] [Google Scholar]
- Yeom J.R., Su Y., Kim C.G. Quantification of residual antibiotics in cow manure being spread over agricultural land and assessment of their behavioral effects on antibiotic resistant bacteria. Chemosphere. 2017;182:771–780. doi: 10.1016/j.chemosphere.2017.05.084. [DOI] [PubMed] [Google Scholar]
- Zhang X.X., Zhang T., Fang H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009;82(3):397–414. doi: 10.1007/s00253-008-1829-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.