Abstract
Intraspecific trait variation (ITV), based on available genetic diversity, is one of the major means plant populations can respond to environmental variability. The study of functional trait variation and diversity has become popular in ecological research, for example, as a proxy for plant performance influencing fitness. Up to now, it is unclear which aspects of intraspecific functional trait variation (iFDCV) can be attributed to the environment or genetics under natural conditions. Here, we examined 260 individuals from 13 locations of the rare (semi‐)dry calcareous grassland species Trifolium montanum L. in terms of iFDCV, within‐habitat heterogeneity, and genetic diversity. The iFDCV was assessed by measuring functional traits (releasing height, biomass, leaf area, specific leaf area, leaf dry matter content, Fv/Fm, performance index, stomatal pore surface, and stomatal pore area index). Abiotic within‐habitat heterogeneity was derived from altitude, slope exposure, slope, leaf area index, soil depth, and further soil factors. Based on microsatellites, we calculated expected heterozygosity (He) because it best‐explained, among other indices, iFDCV. We performed multiple linear regression models quantifying relationships among iFDCV, abiotic within‐habitat heterogeneity and genetic diversity, and also between separate functional traits and abiotic within‐habitat heterogeneity or genetic diversity. We found that abiotic within‐habitat heterogeneity influenced iFDCV twice as strong compared to genetic diversity. Both aspects together explained 77% of variation in iFDCV ( = .77, F 2, 10 = 21.66, p < .001). The majority of functional traits (releasing height, biomass, specific leaf area, leaf dry matter content, Fv/Fm, and performance index) were related to abiotic habitat conditions indicating responses to environmental heterogeneity. In contrast, only morphology‐related functional traits (releasing height, biomass, and leaf area) were related to genetics. Our results suggest that both within‐habitat heterogeneity and genetic diversity affect iFDCV and are thus crucial to consider when aiming to understand or predict changes of plant species performance under changing environmental conditions.
Keywords: (semi‐)dry grasslands, environmental heterogeneity, functional traits, intraspecific functional trait variation (iFDCV), mountain clover, population genetics
Up to now, it is unclear which aspect of intraspecific functional trait variation (ITV) can be attributed to the environment or to genetics under natural and present environmental conditions. Here, we used data from an extensive field study (260 individuals from 13 locations) on the (semi-)dry calcareous grassland species Trifolium montanum (mountain clover). We demonstrated that abiotic within-habitat heterogeneity had twice as much impact as genetic diversity on ITV explaining together 77% of variation.

1. INTRODUCTION
Functional traits are morphological, (eco‐)physiological, and also reproductive traits that impact an individual's growth, reproduction, and survival and thus influence plant fitness indirectly but also directly (Nock, Vogt, & Beisner, 2016; Violle et al., 2007). Gained from direct measurements or databases (e.g., TRY, Kattge et al., 2011; Kattge et al., 2020), they have been used to investigate responses of populations, species, communities, and ecosystems to the environment (e.g., land use, climate; Bernhardt‐Römermann, Gray, et al., 2011; Bucher et al., 2016; Díaz & Cabido, 2001; Gratani, 2014; König et al., 2018; Nicotra et al., 2010; Römermann, Bernhardt‐Römermann, Kleyer, & Poschlod, 2009; Violle et al., 2007).
Intraspecific trait variation (ITV) depends on the available phenotypic trait plasticity of individuals within a population. Phenotypic plasticity, that is, the phenotypic variation expressed by a single genotype under different environmental conditions (Hufford & Mazer, 2003; Nicotra et al., 2010; Sultan, 2000), might be one of the most important mechanisms for plants in reacting to environmental changes (e.g., land use, climate change; Agrawal, 2001; Arnold, Kruuk, & Nicotra, 2019; Gratani, 2014; Via et al., 1995). In general, environment and genetics can generate ITV (de Bello et al., 2011; Violle et al., 2012). The complex relationships between population‐based ITV of functional traits and environmental heterogeneity of the habitats where traits of populations have been investigated, however, have not yet received much attention.
Genetic diversity is considered to be fundamental for population fitness and evolutionary processes, and influences adaptive potential of a species with respect to environmental changes, competitors or pathogens (Arnold et al., 2019; Boulding, 2008; Chevin & Hoffmann, 2017; Karbstein, Tomasello, & Prinz, 2019; Lande, 2009; Nicotra et al., 2010; Reed & Frankham, 2003). Relationships between genetic diversity and “traditional” fitness parameters (e.g., number of flowers, seeds and fruits, and seed weight) or life‐history traits have been investigated intensely both within and across species (Freeland, Kirk, & Petersen, 2011; Leimu, Mutikainen, Koricheva, & Fischer, 2006; Reed & Frankham, 2003; Reisch & Bernhardt‐Römermann, 2014). In some model plants, functional trait plasticity related to morphology (e.g., plant height), ecophysiology (e.g., water‐use efficiency), and life history (e.g., flowering time and seed traits) was found to be under genetic control (Ackerly et al., 2000; Hughes, Soppe, & Albani, 2019; Locascio, Lucchin, & Varotto, 2009; Thornsberry et al., 2001). Moreover, studies also indicate phenotypic and genetic connections, and also, more explicitly, correlations between phenotypic traits and genetic variation (Csilléry et al., 2020; Karbstein et al., 2019; Waitt & Levin, 1998). Waitt and Levin (1998) impressively showed positive correlations between the phenotypic variation of morphology‐related functional traits and genetic variation in several species from different plant families. Nevertheless, trait variation, if it is for example entirely plastic, does not necessarily coincide with genetic variation (see also Chevin & Hoffmann, 2017). Relationships between intraspecific functional trait variation and genetic diversity at population‐level and under natural environmental conditions remain poorly understood.
Considerable functional differences may provide improved resource partitioning due to differences in niche exploitation and/or a more flexible response to environmental changes (see Bucher et al., 2016; MacArthur & Wilson, 1967; Schweiger et al., 2018; Simpson, 1949; Violle et al., 2012). Within populations, increased ITV based on within‐habitat heterogeneity should be able to increase adaptability with positive consequences for growth, reproduction, and survival. Environmental heterogeneity within habitats may thus lead to an increased number of different functional phenotypes and thus enhances ITV. Moreover, habitat heterogeneity is expected to influence the genotypic range of variation within a habitat: Variable environments can exert different selective pressures generating genetic heterogeneity (Gratani, 2014; Linhardt & Grant, 1996; Sakaguchi et al., 2019). Nevertheless, within a habitat of a population, environmental differences are usually lower and gene flow more frequently (for example due to missing geographical barriers) than between habitats across larger scales. Within‐habitat heterogeneity might also enhance the occurrence of different genotypes due to different resource exploitation possibilities, increasing genetic variation (see, e.g., Agashe & Bolnick, 2010; Reusch, Ehlers, Hämmerli, & Worm, 2005). Therefore, within‐habitat heterogeneity affects ITV directly and genetic diversity indirectly. However, ITV, within‐habitat heterogeneity, and genetic diversity may interact in complex ways under natural environmental conditions. For example, plasticity of traits (generating ITV) is able to influence the selective effect of within‐habitat heterogeneity on genetic diversity, whereas connectivity and dispersal (gene flow) among habitats can affect selection on genetic diversity (Ghalambor, McKay, Carroll, & Reznick, 2007; Linhardt & Grant, 1996; Reisch & Schmid, 2019; Vellend & Geber, 2005).
In this study, we aim to investigate the relative effects of abiotic within‐habitat heterogeneity and genetic diversity on intraspecific trait variation (ITV). Investigations are based on 260 individuals from 13 Central European populations of the rare (semi‐)dry calcareous grassland species Trifolium montanum L. (mountain clover; Figure 1). We addressed the following questions: Is ITV related to abiotic within‐habitat heterogeneity and/or genetic diversity? If so, to what extent is ITV explained by either aspect? Which functional traits are related to abiotic within‐habitat heterogeneity and/or genetic diversity?
FIGURE 1.

Trifolium montanum in different habitats. (a) Location Bottendorf (Bo): Small T. montanum individuals grow on continental‐dry grasslands. (b) Location Jena‐Wogau (Wo): T. montanum individuals inhabit semi‐dry Bromus erectus grasslands along the forest margin. Mountain clover is characterized by denticulate leaflets with silky abaxial leaf surfaces (see a and b). Image source: Karbstein (2016)
2. MATERIALS AND METHODS
2.1. Model species
Trifolium montanum L. (Fabaceae) is a perennial herb of extensively used, calcareous grasslands distributed in Europe and Western Russia (Figures 1 and 2; GBIF Secretariat, 2017; Hahn, Kettle, Ghazoul, Hennig, & Pluess, 2013; Schleuning & Matthies, 2008; Schleuning, Niggemann, Becker, & Matthies, 2009). Populations usually inhabit (semi‐)dry grasslands (Jäger, 2011), but they also occur along with shrub and forest margins. In Central Europe, the species is quite rare because of degradation and fragmentation of (semi‐)dry grasslands (Garve, 2004; Schleuning & Matthies, 2008; Schleuning et al., 2009). Trifolium montanum is diploid with 2n = 16 (Rice et al., 2015).
FIGURE 2.
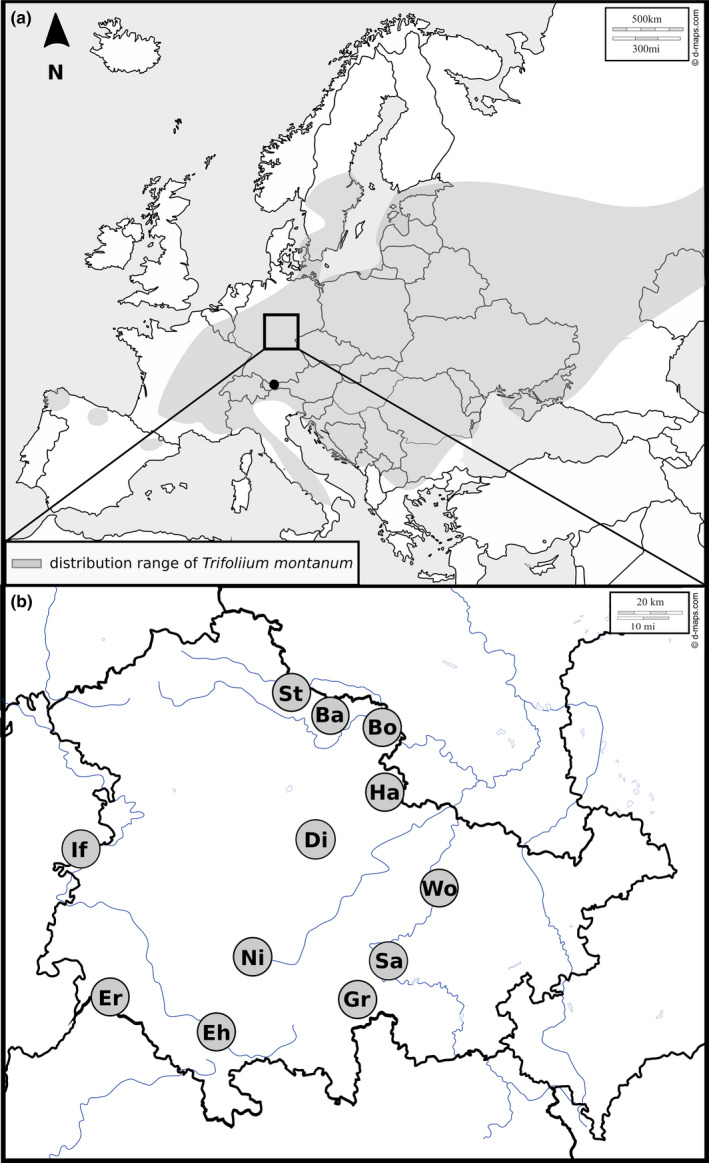
(a) Distribution range of Trifolium montanum in Europe (light gray) according to Meusel and Jäger (1998). The black square indicates the sampling area in Central Germany. The black dot represents the sampling location in Austria (“KW”). (b) Sampling scheme of the present study in Central Germany (see Table 1 for abbreviations and detailed information). Black circles represent sampling locations. Within circles, location abbreviations are given. Black lines indicate borders of the German Federal States (focus on Thuringia). Basic geographical maps were downloaded from d‐maps.com
2.2. Study locations and sampling
We focused on 13 locations in total, from which 12 are situated in Germany and, to cover a larger range of environmental conditions, one in Austria (Table 1). Trifolium montanum populations at the 13 different locations are independent of each other: The average distance of mating events in this species is quite low (10 m), and thus, the majority of mating events occur on small distances (pollen dispersal up to a distance of 324 m possible; Matter, Kettle, Ghazoul, Hahn, & Pluess, 2013). This indicates a reduced potential for pollen‐mediated long‐distance dispersal. For two Trifolium species, the estimated long‐distance dispersal via seeds is also estimated to be only six to 10 m (Vittoz & Engler, 2007). We estimated an average distance among study locations of ca. 133 km (80 km without the distant location KW) and standard deviation of 124 km (41 km without the distant location KW, see Table S3 for details and the applied “geosphere” r package vers. 1.5‐5 (Hijmans, 2016) using the Vincenty ellipsoid method). Distances are thus too large for direct pollen exchange or seed dispersal. There is only the possibility for direct gene flow between locations Bo and Ha, and Ba and St. However, locations are separated by large agrarian areas (particularly Ha), forests, and roads, minimizing the probability of pollen exchange or seed‐mediated long‐distance dispersal and thus the potential of gene flow. Trifolium montanum is consumed by grazing animals, but Bo was grazed by sheep and goats, St by cattle, and the management of Ha is unknown, suggesting rather very local grazer movements instead of habitat connection via transhumance.
TABLE 1.
Location, date of sampling, latitude (lat. (N), longitude (long. (E.), and mean coefficients (with standard errors in brackets) for variation of intraspecific functional trait variation (iFDCV), abiotic within‐habitat heterogeneity (HD), and mean genetic diversity (GD; He) of 13 Trifolium montanum populations
| Location | Date | Lat. (N) | Long. (E) | iFDCV | HD | GD |
|---|---|---|---|---|---|---|
| Riezlern (KW) | 17.07.2015 | 47.361,036 | 10.173,825 | 0.173 (±0.044) | 0.143 (±0.044) | 0.597 (±0.083) |
| Bottendorf (Bo) | 22.05.2016 | 51.316,042 | 11.396,525 | 0.228 (±0.058) | 0.303 (±0.091) | 0.612 (±0.072) |
| Hardisleben (Ha) | 25.05.2016 | 51.162,917 | 11.446,789 | 0.205 (±0.055) | 0.235 (±0.056) | 0.630 (±0.098) |
| Jena‐Wogau (Wo) | 29.05.2016 | 50.924,306 | 11.665,083 | 0.210 (±0.054) | 0.257 (±0.064) | 0.654 (±0.080) |
| Bad Frankenhausen (Ba) | 31.05.2016 | 51.367,267 | 11.103,056 | 0.196 (±0.048) | 0.244 (±0.054) | 0.666 (±0.060) |
| Steinthaleben (St) | 05.06.2016 | 51.409,550 | 11.004,850 | 0.265 (±0.076) | 0.357 (±0.102) | 0.686 (±0.060) |
| Saalfeld (Sa) | 08.06.2016 | 50.631,003 | 11.383,729 | 0.242 (±0.061) | 0.325 (±0.107) | 0.637 (±0.073) |
| Ifta (If) | 12.06.2016 | 51.086,633 | 10.148,017 | 0.184 (±0.044) | 0.150 (±0.042) | 0.678 (±0.067) |
| Niederwillingen (Ni) | 15.06.2016 | 50.776,294 | 11.027,711 | 0.204 (±0.049) | 0.169 (±0.053) | 0.661 (±0.085) |
| Dielsdorf (Di) | 19.06.2016 | 51.095,233 | 11.188,406 | 0.202 (±0.056) | 0.295 (±0.111) | 0.647 (±0.072) |
| Erbenhausen (Er) | 23.06.2016 | 50.565,556 | 10.157,383 | 0.224 (±0.043) | 0.346 (±0.097) | 0.658 (±0.078) |
| Großneundorf (Gr) | 28.06.2016 | 50.532,456 | 11.294,961 | 0.149 (±0.036) | 0.151 (±0.044) | 0.570 (±0.090) |
| Ehrenberg (Eh) | 29.06.2016 | 50.478,583 | 10.665,786 | 0.153 (±0.031) | 0.193 (±0.064) | 0.595 (±0.084) |
From each location, we collected 20 individuals of a T. montanum population, totaling 260 individuals. We attempted to distribute individual sampling points equally within a habitat. Fieldwork was done in July 2015 (Austria) and from May to June 2016 (Germany).
2.3. Functional traits—measurements and ecological meaning
As traits change with the season (Bucher et al., 2019; Römermann, Bucher, Hahn, & Bernhardt‐Römermann, 2016), we started sampling in lowlands and finished sampling in higher altitudes (compare also Tautenhahn, Grün‐Wenzel, Jung, Higgins, & Römermann, 2019). To ensure the comparability of functional traits among populations, we only sampled flowering and early fruiting individuals to control for phenology (Römermann et al., 2016). All functional traits were measured on 20 different individuals per population, and all leaf functional traits were measured on two leaves per individual.
In the field, we measured Fv/Fm and PI on absorption basis after 30 min by high intensity focused LED (3,500 µmol/m2 * s−1 intensity and wavelength peak 627 nm) with Pocket PEA, preparing leaves according to the manufacturer's instructions (Hansatech Instruments Ltd., King´s Lynn, England; Strasser, Srivastava, & Tsimilli‐Michael, 2000; Strasser, Tsimilli‐Michael, & Srivastava, 2004). The ratio of variable fluorescence to maximal fluorescence (Fv/Fm) is related to the efficiency of PS II electron transport and indicates abiotic and/or biotic stress due to photoinhibition (Butler & Kitajima, 1975; Griffin, Epstein, & Boelman, 2013; Maxwell & Johnson, 2000; Paillotin, 1976). The performance index (PI) represents the photosynthetic performance of a chlorophyll molecule, the vitality of the plant, and its ability to resist constraints from outside (Bucher, Bernhardt–Römermann, & Römermann, 2018; Clark, Landolt, Bucher, & Strasser, 2000; Strasser et al., 2000). We also determined releasing height (RH) as the shortest distance between the ground and the highest flower head [m] (Cornelissen et al., 2003), and cut total fresh aboveground biomass directly above taproot.
In the laboratory, we weighed total dry aboveground biomass (AGB) per individual [g]. We determined the weight of two fresh leaves per individual, conducted leaf scans, and calculated leaf area (LA) using the “LeafTraits” R package vers. 1.0 (Bernhardt‐Römermann, unpubl. data). RH and AGB are related to competitive ability (Chen et al., 2011; Cornelissen et al., 2003; Gaudet & Keddy, 1988; Moles et al., 2009). Leaf area has important consequences for light interception, carbon exchange, and water balance (Díaz et al., 2016; Farquhar, Buckley, & Miller, 2002; Givnish, 1987). Leaf area forms an allometric complex together with RH and AGB due to anatomical and architectural consequences, representing morphology‐related functional traits (Ackerly & Donoghue, 1998; Bartelink, 1997; Cornelissen et al., 2003).
Subsequently, leaves were oven‐dried and weighed again [mg]. We calculated individual mean values for SLA (Cornelissen et al., 2003; Pérez‐Harguindeguy et al., 2013) as the ratio of one‐sided fresh LA [mm2] and its oven‐dry mass [mg], and LDMC (Pérez‐Harguindeguy et al., 2013) as the ratio of oven‐dry mass of a leaf [mg] and its water‐saturated fresh mass [g]. Specific leaf area (SLA) tends to be positively correlated with potential relative growth rate, but negatively with leaf dry matter content (LDMC) that represents leaf longevity/robustness (Cornelissen et al., 2003; Pérez‐Harguindeguy et al., 2013; Römermann et al., 2016).
We took stomata imprints using nail polish from two leaves per individual. Utilizing an Olympus CH40 microscope, we counted stomata density at 200× magnification and measured guard cell length and width at 400× magnification. We determined individual mean values of four measurements for stomata density and of eight measurements for stomata length and width measurements of the abaxial leaf surface. We also calculated stomatal pore surface (SPS) as guard cell length [µm] * guard cell width [µm * π * 4–1 (Balasooriya et al., 2009) and stomatal pore area index (SPI) as the product of (guard cell length)2 [mm2] * stomatal density [1/mm2] (Sack, Cowan, Jaikumar, & Holbrook, 2003). SPS characterizes stomata size, SPI indicates stomatal conductance, and both traits are known to change along abiotic environmental gradients (Bucher et al., 2016, 2017; Woodward, Lake, & Quick, 2002).
Selected functional traits cover a broad range of plant trait space (see, e.g., Díaz et al., 2016; Gratani, 2014), and many of them are already known to respond to environmental conditions (see, e.g., Cornelissen et al., 2003; Nicotra et al., 2010; Pérez‐Harguindeguy et al., 2013).
2.4. Assessment of habitat characteristics
We characterized each location with a maximum of five environmental replicate measurements. The replicates (grid cell 2 m × 2 m) were equally distributed within the local range of each population (20 to 4,600 m2, unpubl. data). Therefore, the distance between replicates differed according to habitat sizes. For one location (Eh, see Tables 1 and 2), we reduced the number of replicates to four due to the limited habitat size (~20 m2). In total, we analyzed n = 64 records.
TABLE 2.
Coefficients of variation (CV) of particular functional traits (n = 260 individuals), abiotic factors (n = 64 replicates), and population genetic indices (n = 255 individuals) based on nine microsatellite markers (Matter et al., 2012) of 13 Trifolium montanum populations. F v/F m and PI measurements are missing at the location Riezlern (KW). Due to data completeness and comparability of iFDCV among populations, we approximated these values by linear regressions
| Population | iFDCV | Nind | CVRH | CVAGB | CVLA | CVSLA | CVLDMC | CVFv/Fm | CVPI | CVSPS | CVSPI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (KW) | 20 | 0.161 | 0.429 | 0.276 | 0.096 | 0.050 | 0.011 | 0.275 | 0.105 | 0.157 | |
| (Bo) | 20 | 0.233 | 0.427 | 0.305 | 0.097 | 0.060 | 0.023 | 0.542 | 0.144 | 0.225 | |
| (Ha) | 20 | 0.199 | 0.455 | 0.490 | 0.101 | 0.049 | 0.011 | 0.212 | 0.147 | 0.178 | |
| (Wo) | 20 | 0.183 | 0.493 | 0.319 | 0.098 | 0.102 | 0.011 | 0.413 | 0.100 | 0.171 | |
| (Ba) | 20 | 0.232 | 0.470 | 0.278 | 0.094 | 0.068 | 0.015 | 0.326 | 0.115 | 0.170 | |
| (St) | 20 | 0.361 | 0.696 | 0.496 | 0.132 | 0.067 | 0.019 | 0.349 | 0.090 | 0.176 | |
| (Sa) | 20 | 0.324 | 0.585 | 0.380 | 0.108 | 0.070 | 0.015 | 0.334 | 0.120 | 0.238 | |
| (If) | 20 | 0.167 | 0.400 | 0.311 | 0.087 | 0.050 | 0.012 | 0.310 | 0.121 | 0.196 | |
| (Ni) | 20 | 0.180 | 0.407 | 0.452 | 0.128 | 0.064 | 0.010 | 0.228 | 0.141 | 0.223 | |
| (Di) | 20 | 0.172 | 0.457 | 0.312 | 0.121 | 0.062 | 0.021 | 0.458 | 0.079 | 0.135 | |
| (Er) | 20 | 0.280 | 0.381 | 0.373 | 0.174 | 0.104 | 0.011 | 0.337 | 0.130 | 0.230 | |
| (Gr) | 20 | 0.138 | 0.350 | 0.258 | 0.091 | 0.056 | 0.008 | 0.210 | 0.090 | 0.137 | |
| (Eh) | 20 | 0.156 | 0.307 | 0.203 | 0.088 | 0.074 | 0.014 | 0.262 | 0.121 | 0.151 |
| HD | Nrep | CValtitude | CVslope exposure | CVslope | CVLAI | CVsoil depth | CVCECpot | CVpH | CVN | CVP | CVK | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (KW) | 5 | 0.001 | 0.000 | 0.000 | 0.282 | 0.294 | 0.038 | 0.091 | 0.129 | 0.349 | 0.248 | |
| (Bo) | 5 | 0.013 | 0.895 | 0.520 | 0.587 | 0.350 | 0.051 | 0.030 | 0.206 | 0.125 | 0.251 | |
| (Ha) | 5 | 0.022 | 0.373 | 0.288 | 0.496 | 0.322 | 0.016 | 0.164 | 0.102 | 0.465 | 0.100 | |
| (Wo) | 5 | 0.021 | 0.566 | 0.508 | 0.381 | 0.223 | 0.018 | 0.041 | 0.172 | 0.421 | 0.217 | |
| (Ba) | 5 | 0.018 | 0.000 | 0.315 | 0.537 | 0.408 | 0.149 | 0.146 | 0.298 | 0.227 | 0.345 | |
| (St) | 5 | 0.011 | 1.044 | 0.684 | 0.437 | 0.251 | 0.028 | 0.048 | 0.428 | 0.413 | 0.226 | |
| (Sa) | 5 | 0.013 | 0.722 | 0.974 | 0.621 | 0.368 | 0.030 | 0.007 | 0.096 | 0.203 | 0.218 | |
| (If) | 5 | 0.011 | 0.141 | 0.276 | 0.400 | 0.274 | 0.012 | 0.007 | 0.097 | 0.086 | 0.193 | |
| (Ni) | 5 | 0.012 | 0.000 | 0.516 | 0.316 | 0.281 | 0.117 | 0.007 | 0.084 | 0.132 | 0.229 | |
| (Di) | 5 | 0.012 | 0.000 | 1.129 | 0.342 | 0.424 | 0.028 | 0.048 | 0.090 | 0.406 | 0.471 | |
| (Er) | 5 | 0.009 | 0.639 | 0.572 | 0.509 | 0.438 | 0.030 | 0.012 | 0.156 | 0.889 | 0.210 | |
| (Gr) | 5 | 0.009 | 0.124 | 0.233 | 0.447 | 0.266 | 0.036 | 0.024 | 0.044 | 0.124 | 0.204 | |
| (Eh) | 4 | 0.006 | 0.000 | 0.389 | 0.552 | 0.334 | 0.029 | 0.007 | 0.071 | 0.365 | 0.177 |
| Population | GD | Nind | NA | PAP | He | Ho | I |
|---|---|---|---|---|---|---|---|
| (KW) | 20 | 52 | 5.77 | 0.597 | 0.533 | 1.251 | |
| (Bo) | 20 | 63 | 1.59 | 0.612 | 0.604 | 1.343 | |
| (Ha) | 20 | 71 | 5.63 | 0.630 | 0.594 | 1.450 | |
| (Wo) | 20 | 68 | 4.41 | 0.654 | 0.654 | 1.460 | |
| (Ba) | 19 | 53 | 0.00 | 0.666 | 0.560 | 1.384 | |
| (St) | 19 | 63 | 1.59 | 0.686 | 0.662 | 1.472 | |
| (Sa) | 20 | 56 | 3.57 | 0.637 | 0.622 | 1.347 | |
| (If) | 18 | 59 | 1.69 | 0.678 | 0.667 | 1.465 | |
| (Ni) | 20 | 64 | 4.69 | 0.661 | 0.630 | 1.473 | |
| (Di) | 20 | 56 | 0.00 | 0.647 | 0.607 | 1.369 | |
| (Er) | 20 | 59 | 5.08 | 0.658 | 0.690 | 1.419 | |
| (Gr) | 19 | 49 | 0.00 | 0.570 | 0.531 | 1.185 | |
| (Eh) | 20 | 52 | 3.85 | 0.595 | 0.575 | 1.241 |
Abbreviations: AGB, total dry aboveground biomass; CECpot, soil potential cation‐exchange capacity; F v/F m, ratio of variable to maximal fluorescence; He, expected heterozygosity, Ho, observed heterozygosity; I, Shannon's diversity index; K, plant‐available soil potassium content; LA, leaf area; LAI, leaf area index; LDMC, leaf dry matter content; N, soil nitrogen content; NA, allelic richness; Nind, number of evaluated individuals; Nrep, number of replicates; P, plant‐available soil phosphor content; PAp, private allelic richness; pH, soil reaction; PI, performance index; RH, releasing height; SLA, specific leaf area; soil potassium content; SPI, stomatal pore area index; SPS, stomatal pore surface.
Per replicate, we assessed GPS coordinates and altitude [m.a.s.l.] (eTrex 30, Garmin GmbH, Garching, Germany), slope exposure [°] and slope [°] (TruPulse 200/B Laser Rangefinder, Laser Technology Inc., Lincoln, USA). In addition, LAI (leaf area index; LAI‐2200 Plant Canopy Analyzer, LI‐COR Inc., Lincoln, USA) and soil depth [cm] were measured five and ten times, respectively. We also took five random soil samples per replicate (2 m × 2 m, 4 m2), which were mixed and dried. Potential cation‐exchange capacities [cmol/kg] were determined as a measure of all potentially exchangeable cations in total (CECpot), for sodium (CECNa), potassium (CECK), calcium (CECCa), and magnesium (CECMg). Soil reaction was obtained by using soil suspension mixed with deionized water (pHH2O, pH) and potassium chloride (pHKCl). Furthermore, soil contents were analyzed concerning organic carbon (Corg) [%], lime (Canorg × 8.3 = CaCO3) [%], nitrogen (N) [%], plant‐available phosphor (P) [mg/100 g], and plant‐available potassium (K) [mg/100 g]. All soil analyses were conducted in the soil laboratory of the Thüringer Landesanstalt für Landwirtschaft und Ländlichen Raum (TLLLR) following standardized protocols. Mean annual temperature (Ta) and mean annual precipitation (Pa) were interpolated for study locations using ArcMap (vers. 10.5; ESRI Inc., Redlands, USA) and data from WorldClim 1.4 global climate database from 1960 to 1990 (Hijmans, Cameron, Parra, Jones, & Jarvis, 2005; www.worldclim.org).
2.5. Population genetics and laboratory work
We isolated DNA of sampled individuals from approximately 20–25 mg dry leaf material using a modified CTAB protocol (Doyle & Doyle, 1987; Saghai‐Maroof, Soliman, Jorgensen, & Allard, 1984; modified by adding 1% PVP to CTAB buffer). Nine microsatellite loci were applied to quantify genetic diversity of T. montanum populations (Matter, Määttänen, Kettle, Ghazoul, & Pluess, 2012). We conducted PCRs with labeled primers (IRD 700/IRD 800) in 10 µl reaction volumes containing 1× Dream Taq buffer (Thermo Fisher Scientific Inc., Waltham, USA), 0.2 mM dNTPs, 0.1 µM (IRD 700)/0.2 µM (IRD 800) of tailed forward primer, 0.1 µM (IRD 700)/0.2 µM (IRD 800) reverse primer, 0.025 U * µl−1 Dream Taq polymerase (Thermo Fisher Scientific Inc., Waltham, USA), and 1.5 µl of undiluted template DNA. PCR cycling was performed using primer‐specific annealing temperatures (ta): 65°C (ats002 and ats032), 51°C (ats006), 58°C (ats029), 56°C (Tm10 and Tm12), 55°C (Tm21 and Tm24), and 61°C (Tm16). We carried out locus‐specific touchdown programs to increase PCR specificity (Korbie & Mattick, 2008). Programs comprised (a) initial denaturation 95°C/15 min; (b) 11× [denaturation 95°C/30 s, primer‐specific ta in touchdown (ta + 5°C) to (ta) to (ta − 5°C)/ 45 s, extension 72°C/45 s]; (c) 9× (IRD 700) and 15× (IRD 800) [denaturation 95°C/30 s, primer‐specific ta/45 s, extension 72°C/45 s]; 15× (d) [denaturation 95°C/30 s, annealing 53°C/45 s, extension 72°C/45 s]; and (e) final extension 72°C/30 min. All PCRs were stored at 4°C. We combined differently labeled PCR products for electrophoretic analyses of fragment lengths using a LI‐COR Long Readir 4200 (Global Edition IR2 DNA Sequencer, LI‐COR Inc., Lincoln, USA).
2.6. Data analyses
All statistical analyses (except genetic diversity calculations) were performed with R vers. 3.6.0 (R Core Team, 2019). We calculated means for numerical variables, medians for the ordinal variable slope exposure, and coefficients of variation (CV) for diversity variables as the ratio of standard deviation to mean. To evaluate the multiple linear regression model, we exceptionally used the adjusted coefficient of determination () instead of the coefficient of variation (R 2). This has the benefit of avoiding model overfitting in R 2 calculation (see Crawley, 2007).
2.6.1. Intraspecific functional trait variation (iFDCV)
To detect erroneous entries (errors in measurement) in functional traits, we excluded all records (seven trait measurements in total) from the dataset with a distance of >4 standard deviations from the mean of all individuals (compare Díaz et al., 2016; Kattge et al., 2011). We deleted Fv/Fm outliers and the respective PI values (identical source, Pocket PEA). We also checked collinearity among traits, that is, when two or more traits were highly correlated (r > ~│.7│; Dormann et al., 2013). We assessed correlations among functional traits with Spearman's rank coefficient (rSP, cor.test()) due to non‐normal distribution of data. The r package “corrgram” vers. 1.13 (Wright, 2018) was used to visualize correlations. As correlation coefficient values (rSp) were below ~│.7│, we did not exclude particular functional traits (see Figure S1).
ITV (or functional diversity, “FD”) can be calculated in different manners. Functional trait dissimilarity among or within species is used to calculate FD via trait distance matrices and dendrograms (Petchey & Gaston, 2006; Tilman, 2001; see individual‐based FD (iFD) calculations by Cianciaruso, Batalha, Gaston, & Petchey, 2009 across species and Wood, McKinney, & Loftin, 2017 within species). Hypervolumes can also be applied to capture trait space and variation. However, axes (traits) sometimes have to be reduced to follow recommendations about the ratio between observations and number of dimensions and orthogonality among traits (e.g., to four axes, see Benavides, Scherer‐Lorenzen, & Valladares, 2019). The “iFDCV” method used herein is a trait‐by‐trait approach simply incorporating individual trait variation based on population‐wise coefficients of variation (CVs) within a species (see, e.g., Helm et al., 2019). We ensured the absence of collinearity among functional traits (see above), and thus, there was no further need to do axes (trait) reductions like in hypervolume approaches. Our concept focuses on the functional variation within the population of a species, instead of functional differences. Each trait contributes independently and with different weight to the index because standardized trait CVs have different value ranges (variation), and, for example, traits with larger variation contribute stronger to iFDCV than traits with a smaller variation. Therefore, our population‐wise trait‐by‐trait approach (iFDCV) is appropriate for studying environmental or genetic effects on ITV.
We assessed population‐wise iFDCV as the mean CV of RH (CVRH), AGB (CVAGB), LA (CVLA), SLA (CVSLA), LDMC (CVLDMC), F v/F m (CVFv/Fm), PI (CVPI), SPS (CVSPS), and SPI (CVSPI; Tables 1 and 2; see also Helm et al., 2019 for CV functional trait calculation).
2.6.2. Within‐habitat heterogeneity (HD)
We tested for correlations among environmental factors with Spearman's rank coefficient (r SP, cor.test()) due to non‐normal distribution of data. The r package “corrgram” vers. 1.13 (Wright, 2018) was used to visualize correlations. As explained above for functional traits, we checked for collinearity (r > ~│.7│; Dormann et al., 2013) and excluded the factors CECK, CECCa, pHKCl, Corg, CaCO3, Ta, Pa (r SP > .7), and CECMg (r SP = .50), and one variable due to an almost complete absence of variation (CECNa; see Figure S2 for correlations). Afterward, we calculated abiotic within‐habitat heterogeneity (HD) as location‐wise mean CV of altitude (CValtitude), slope exposure (CVslope exposure), slope (CVslope), leaf area index (CVLAI), soil depth (CVsoil depth), potential soil cation‐exchange capacity (CVCECpot), pH (CVpH), soil nitrogen content (CVN), soil phosphor content (CVP), and soil potassium content (CVK; see Tables 1 and 2).
2.6.3. Genetic diversity (GD)
We scored microsatellite fragments with an internal size standard. We proved the scoring procedure at least three times and removed ambiguous results. Analyses were conducted for all individuals characterized by at least four microsatellite loci resulting in a final sample size of n = 255 individuals (see Table 2). Mean loci coverage was 90% per individual, that is, in mean, 90% of loci were present in an individual. To ensure that sampled populations represent the same genetic line of the species, we calculated individual‐ and population‐wise distance matrices based on Nei´s genetic distance (Nei, 1978) and conducted principal coordinate analyses (PCoAs) in GenAlEx vers. 6.503 (Peakall & Smouse, 2006, 2012). Moreover, we performed analyses in STRUCTURE vers. 2.3.4 (Pritchard, Stephens, & Donnelly, 2000) setting an admixture model (with correlated allele frequencies), burn‐in to 5,000, MCMC to 50,000, and K to one to 13 (10 replicates per K). The optimal K was determined by STRUCTURE HARVESTER (Earl & vonHoldt, 2012) using the Evanno method. We merged the replicates of each optimal K (K = 2 and K = 9) with CLUMPP vers. 1.1.2 (Jakobsson & Rosenberg, 2007) and plotted results with DISTRUCT vers. 1.1 (Rosenberg, 2004). PCoA and STRUCTURE results showed that all populations have high levels of admixture and the geographically distant population KW does not belong to a separate genetic lineage of T. montanum (see Figures S3–S5). Moreover, relationships among iFDCV, HD, and GD in KW fit those observed among Central German populations (see Table 1 and Figures 3, 4, 5). To examine the effect of genetic diversity on iFDCV, we were careful to sample T. montanum populations of different sizes (about 50–20,000 individuals, Karbstein et al., unpubl. data). Population size of T. montanum was positively related to genetic diversity (Karbstein et al., unpubl. data), and varying genetic diversity is needed to examine its effect on iFDCV.
FIGURE 3.
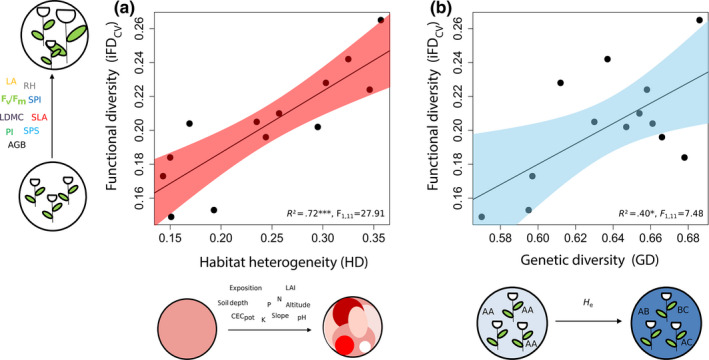
Significant positive relationships between iFDCV and (a) abiotic within‐habitat heterogeneity and (b) genetic diversity (He) including 13 Trifolium montanum populations (n = 255 to 260 individuals) of Central Europe. Confidence intervals (95%) are drawn. See Table 2 for abbreviations. Significance levels: ***p < .001 and *p < .05
FIGURE 4.
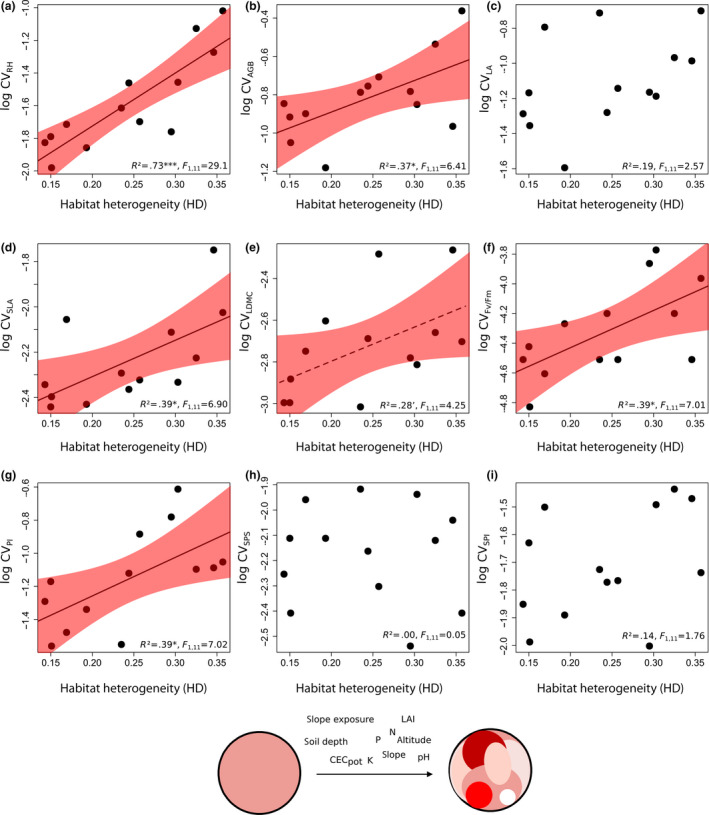
Relationships between coefficient of variation of particular traits (CVtraits) and abiotic within‐habitat heterogeneity (HD) in 13 Trifolium montanum populations (n = 260 individuals) of Central Europe. 95% confidence intervals are drawn for all (marginal) significant relationships. Dotted regression lines represent only marginal significant relationships (0.05 < p < .1). See Table S1 for detailed model statistics, and Table 2 for abbreviations. Significance levels: ***p < .001, *p < .05 and ‘=.1 > p > .05
FIGURE 5.
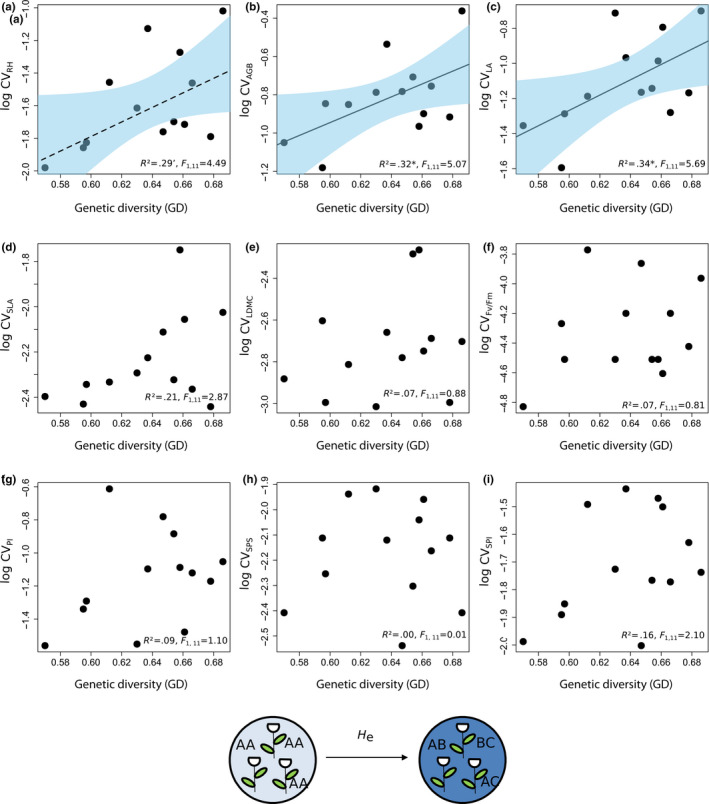
Relationships between coefficient of variation of particular traits (CVtraits) and genetic diversity (GD, He) in 13 Trifolium montanum populations (n = 255 individuals) of Central Europe. 95%‐confidence intervals are drawn for all (marginal) significant relationships. Dotted regression lines represent only marginal significant relationships (.05 < p < .1). See Table S1 for detailed model statistics, and Table 2 for abbreviations. Significance levels: *p < .05 and ‘=.1 > p> .05
We calculated population‐wise multilocus allelic richness (NA) and mean values of private allelic richness (PAp), observed (Ho) and expected heterozygosity (He), and Shannon´s diversity index (I), using GenAlEx vers. 6.503 (Hardy, 1908; Nei, 1973, 1973, 1978; Peakall & Smouse, 2006, 2012; Shannon, 1948; Weinberg, 1908; see Tables 1 and 2).
2.6.4. Saturation of iFDCV, HD and GD
To examine whether iFDCV, HD, and GD (see Statistical modeling section) are saturated within populations/habitats, we used an r script (Karbstein, 2020) that randomly chose one to 20, one to five and one to 18 (18 individuals genotyped per population at a minimum) samples and calculated iFDCV, HD and GD, respectively with 100 iterations per step. Mean iFDCV, HD and GD values were plotted against sample size for each population/habitat. We used r functions implemented in the packages “dbplyr” vers. 1.4.2 (Wickham & Ruiz, 2019) to randomly sample populations/habitats and “adegenet” vers. 2.1.1 (Jombart et al., 2018) to calculate expected heterozygosity (He).
2.6.5. Statistical modeling
To assess whether iFDCV is related to within‐habitat heterogeneity and genetic diversity (and to assess which specific genetic diversity index best explains iFDCV), we employed a multiple linear regression model with iFDCV as the dependent variable, and HD and genetic diversity indices NA, PAp, Ho, He, and I as explaining, independent variables. To fulfill statistical assumptions, we standardized independent variables to zero mean for unit variance (z‐transformation). We conducted linear regression model simplification with the standard backward selection approach by always excluding the least significant variable (p > .1) until the final minimal adequate model was attained (see Crawley, 2015). Next, we carried out an ANOVA implemented in the R function aov() and additionally calculated the Akaike information criterion (AIC) with AIC() for model comparison to justify each simplification step. We checked our final model using the R function plot(), and it visually fulfilled the assumptions for normality, homoscedasticity, and linearity. The final multiple linear regression model contained HD and expected heterozygosity (He) as explaining, independent variables (see Table S1). He is widely used as genetic diversity index, and it less depends on population history (e.g., bottlenecks) compared to the other indices (Freeland et al., 2011; Kalinowski, 2004; Szczecińska, Sramko, Wołosz, & Sawicki, 2016). Therefore, we used population‐wise multilocus mean values of expected heterozygosity (He) as genetic diversity (GD; see also section Discussion, Relationships between intraspecific functional trait variation and genetic diversity).
To illustrate separate relationships between iFDCV and HD or GD (see Figure 3), we executed two linear regression models and plotted regression results. Additionally, we conducted a linear regression model to assess whether HD is associated with GD. GD was handled as dependent and HD as the independent variable.
To assess which functional traits were related to HD and/or GD (see Table S1), we performed linear regression models with log‐transformed trait variation as dependent variable and HD or GD as independent variable. We log‐transformed the dependent model variable to achieve normality and/or linearity, and checked normality with the Shapiro–Wilcox test. Model assumptions were again visually examined as described above.
To test for spatial autocorrelation among populations/habitats (e.g., closer populations with more similar genetic diversity and more similar iFDCV) within linear regression models, we calculated Moran's I (Moran, 1950) values using the R function correlog() function included in the r package “ncf” vers. 1.2‐6 (Bjornstad & Cai, 2018) based on the model residuals. Moran's I values mostly resided around ≤±1. Permutation of two‐sided p values per distance class (with 1,000 resamples under the null distribution) comprised only six to 11% significant p values on average with different increment settings across linear regression models indicating only very weak spatial autocorrelation. Hence, we did not consider models accounting for spatial autocorrelation.
To support the interpretation of results found between iFDCV and HD, we also examined correlations among particular functional traits and abiotic environmental factors chosen to calculate HD. We added a value of 1 to all CVs and logarithmized traits and environmental factor CVs (CVs have to be >1) to achieve normal distribution. We used the rcorr() function within the r package “hmisc” vers. 4.2‐0 (Harrell, 2019) to calculate a correlation matrix based on Pearson's rank correlation coefficient. We carried out the corrplot() function implemented in the r package “corrplot” vers. 0.84 (Taiyun & Simko, 2017) to visualize the correlation matrix only considering p values below .1.
3. RESULTS
3.1. Relationships among iFDCV, HD, and GD
The results of the multiple linear regression model demonstrated that iFDCV was positively and significantly related to abiotic within‐habitat heterogeneity (HD) and genetic diversity (He, GD), accounting for 67.42% (p < .001) and 32.58% (p < .05) of explained variation, respectively (see Table S1). Together both aspects significantly explained 77% of iFDCV ( = .77, F 2, 10 = 21.66, p < .001).
Individual relationships between iFDCV and HD, and iFDCV and GD were also statistically significant (Figure 3): The iFDCV formed a well‐explained positive linear relationship with HD (R 2 = .72, F 1, 11 = 27.91, p < .001) whereas a linear, though weaker, relationship was found between iFDCV and GD (R 2 = .40, F 1, 11 = 7.48, p < .05).
HD and GD of T. montanum populations were not significantly related in our study (R 2 = .18, F 1, 11 = 2.37, p = .15). We observed saturation of all diversity variables. For iFDCV, HD and GD, the curves began to saturate between 5–10, 4–5, and 10–15 samples, respectively (see Figures S6–S8).
3.2. Relationships of functional traits with environmental factors and genetic diversity
Most functional traits were significantly positively related to HD and some to GD (Figures 4 and 5; see Table S1 for detailed statistics). We observed a higher number of (marginally) significant relationships between functional traits (CVRH, CVAGB, CVSLA, CVLDMC, CVFv/Fm, and CVPI) and HD than between functional traits (CVRH, CVAGB and CVLA) and GD. CVRH is strongest related to HD (R 2 = .73), followed by CVSLA (R 2 = .39), CVFv/Fm (R 2 = .39), CVPI (R 2 = .39), CVAGB (R 2 = .37), and CVLDMC (R 2 = .28). With GD, CVLA (R 2 = .34), CVAGB (R 2 = .32), and CVRH (R 2 = .29) exhibited significant relationships of similar strength.
We found (marginally) significant positive correlations between functional traits and abiotic environmental factors chosen to calculate HD (Figure 6, see Table S2): Some trait CVs are positively correlated to CVslope exposure (CVRH, CVAGB, CVLA, and CVSPI) and CVslope (CVRH, CVFv/Fm and CVPI). Many traits were positively correlated to CV of soil nutrients, that is, CVN (CVRH and CVAGB), CVP (CVSLA and CVLDMC), and CVK (CVFv/Fm and CVPI). No significant trait CV correlations were found to CValtitude, CVLAI, CVsoil depth, CVCECpot, and CVpH. Correlations between trait CVs and environmental CVs revealed widely positive coefficients (~71%).
FIGURE 6.
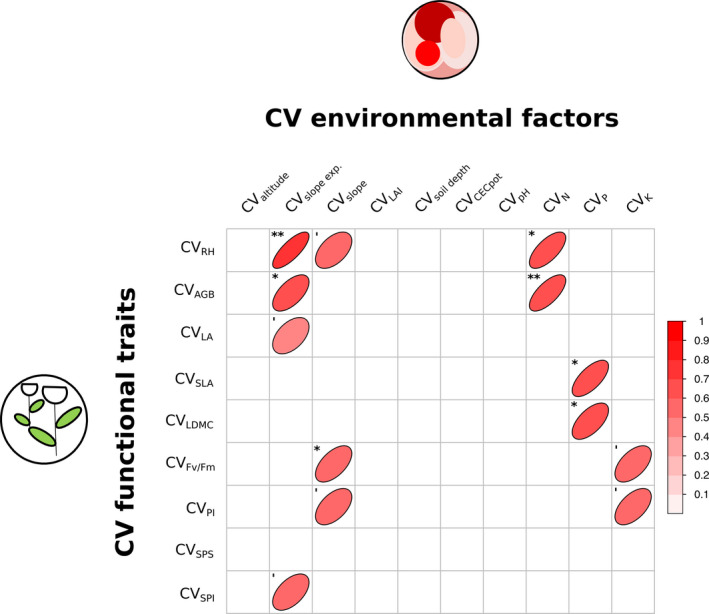
Visualized correlation matrix based on Pearson correlation coefficients between variation of particular traits (CVtrait) and particular abiotic environmental factors (CVfactor) in 13 Trifolium montanum populations (n = 260 individuals) of Central Europe. We only illustrated (marginal) significant results (see Results). Width of an ellipse reflects the correlation coefficient, that is, the higher a correlation coefficient (in positive and negative direction), the narrower the ellipse. See Table 2 for abbreviations, and Table S2 for statistics. Significance levels: **=p < .01, *=p < .05 and ‘=0.1 > p > .05
4. DISCUSSION
Connecting intraspecific functional trait variation with genetic and environmental variation is an important ecological challenge. This study showed that population‐wise intraspecific functional trait variation (iFDCV) of T. montanum can be attributed to a high extent (77%) to both abiotic within‐habitat heterogeneity and population genetic diversity under natural environmental conditions (Figure 3). Interestingly, within‐habitat heterogeneity statistically affected iFDCV considerably stronger than genetic diversity (Figures 4 and 5).
4.1. Relationships between intraspecific functional trait variation and within‐habitat heterogeneity
Variation of morphology‐related functional traits RH, AGB, and LA was mainly correlated to variation of habitat slope exposure and slope, whereas variation of (eco‐)physiology‐related traits (SLA, LDMC, Fv/Fm, PI, SPS, and PCI) was predominantly correlated to variation of slope characteristics and soil factors (Figure 6). Within a habitat, different slopes and slope exposures, influencing soil humidity, may have led to an increase or reduction in height, biomass, and leaf area of T. montanum. For example, drought stress is known to limit nutrient uptake and thus photosynthesis and plant growth (Farooq, Wahid, Fujita, & Basra, 2009; Jaleel et al., 2009; Pérez‐Harguindeguy et al., 2013). Also, RH and AGB were positively associated with soil nitrogen content (N), and N‐deficient soils in T. montanum habitats probably constrained height and biomass accumulation of individuals as well (see, e.g., Ågren, Wetterstedt, & Billberger, 2012). (Positive) associations between plant height or biomass to soil properties and particularly N are in line with literature (Cornelissen et al., 2003; Razaq, Zhang, & Shen, 2017; Reich & Hobbie, 2013). In contrast, variation of SLA and LDMC was correlated to soil phosphorous content (P). Phosphor regulates protein biosynthesis and development of new plant tissue (see Kerkhoff, Fagan, Elser, & Enquist, 2006), and P‐deficient soils may thus impact relative growth rate and leaf robustness. SLA and LDMC respond also to soil properties (Cornelissen et al., 2003). P has been shown to form only rare and weak positive intraspecific relationships with SLA and particularly in herbaceous species when N is in abundant supply (wild rice; Dwyer, Hobbs, & Mayfield, 2014; Sims, Pastor, & Dewey, 2012). Moreover, soil potassium content (K), besides slope affecting soil humidity (see above), influenced performance and vitality (PI and F v/F m). High soil potassium content was mainly found in CaCO3‐rich soils, which T. montanum prefers (Jäger, 2011). Due to physiological constraints, low K and CaCO3 conditions may decrease the performance/vitality of T. montanum individuals while high K and CaCO3 may increase it. Studies already revealed that F v/F m and PI respond positively to increased calcium supply due to stabilization of chlorophyll and an increase of photosystem II activity (Coffea arabica; Ramalho, Rebelo, Santos, Antunes, & Nunes, 1995; Zea mays and Solanum lycopersicum cultivars; Kalaji et al., 2014). In general, several functional traits may respond simultaneously to environmental heterogeneity in T. montanum habitats. Intraspecific functional trait variation captured by iFDCV is probably the response of populations to increased abiotic and biotic environmental differences within their habitats (see also Ghalambor et al., 2007; Nicotra et al., 2010, 2015): as shown, iFDCV was positively correlated to within‐habitat heterogeneity, suggesting that the more environmentally variable a habitat, the higher the intraspecific functional trait variation in T. montanum populations. Results are line with literature showing associations between functional trait values and diversity, and environmental conditions within and across species (e.g., Albert et al., 2010; Bernhardt‐Römermann, Gray, et al., 2011; Bucher et al., 2016; Díaz & Cabido, 2001; Gratani, 2014; Karbstein et al., 2019; König et al., 2018; Violle et al., 2007). Intraspecific functional differences are known to facilitate a more flexible response to varying abiotic conditions (see, e.g., Bucher et al., 2016 for elevational gradients; Karbstein et al., 2019 for small‐scale habitat differences). Thus, iFDCV likely affects population growth and reproduction with positive consequences for survival and fitness (see also Nock et al., 2016; Violle et al., 2007).
4.2. Relationships between intraspecific functional trait variation and genetic diversity
Genetic diversity, in terms of microsatellite variation, was positively related to intraspecific functional trait variation (iFDCV; Figures 3b and 5). This observation is also in line with literature indicating positive relationships between trait variation/plant fitness (iFDCV, plant fitness, see above) and genetic diversity within species (e.g., Leimu et al., 2006; Waitt & Levin, 1998). Microsatellite markers are frequently applied to capture population genetic diversity (e.g., Matter et al., 2013; Matter et al., 2012; Prinz, Weising, & Hensen, 2009). They are widely distributed throughout genomes, and while regulatory functions in gene expression are known, these markers are presumed to predominantly occur in non‐coding regions and thus to be under neutral selection (see also Ellegren, 2004; Li, Korol, Fahima, Beiles, & Nevo, 2002; Vieira, Santini, Diniz, & Munhoz, 2016). The weak relationship of iFDCV with genetic diversity may be explained by the neutrality of applied microsatellite markers in relation to selected functional traits. However, our intention was not to explain functional traits with particular microsatellites but to assess whether iFDCV and/or particular trait variation coincide with genetic diversity.
It is likely that genetic diversity limits and influences the range of iFDCV. Mutation and recombination events create genetic variation and thus novel functional trait variation within a population. Natural selection probably acts on the genetic basis of trait variation, which in turn probably affects the range of iFDCV. After modeling, expected heterozygosity (He) best‐explained iFDCV. He is based on allelic structure, represents genotype and allele frequencies, and is less sensitive to population history (Freeland et al., 2011; Kalinowski, 2004; Szczecińska et al., 2016). Heterozygosity within individuals and populations probably influenced iFDCV because it enhances the reaction norm and adaptability and thus affects intraspecific trait variation (Boulding, 2008; Freeland et al., 2011; Reed & Frankham, 2003). Interestingly, only the variation of morphology‐related traits was associated with genetic diversity in T. montanum (see also Waitt & Levin, 1998). In contrast, both variation of morphology‐ and (eco‐)physiology‐related traits was linked to within‐habitat heterogeneity. (Eco‐)physiology‐related traits (gas exchange and photosynthesis) tend to have a higher heritability (trait variation due to genetic variation) compared to morphology‐related traits (morphology and vegetative performance, Geber & Griffen, 2003), and should thus be more sensitive to genetic variation . However, an explanation might be that heterozygosity effects (see, e.g., Boulding, 2008; Freeland et al., 2011; Reed & Frankham, 2003) are stronger pronounced in morphology‐related traits leading to similar trait variation and genetic variation based on microsatellites.
However, a positive relationship between genetic diversity and iFDCV may be strengthened by the self‐incompatible nature of T. montanum (see, e.g., Leimu et al., 2006; Reed & Frankham, 2003; Schleuning et al., 2009). Observations between classical fitness parameters and genetic diversity of self‐incompatible species are frequent and can be explained by pollinator limitation in fragmented populations with a low density of flowering individuals (Leimu et al., 2006; Schleuning et al., 2009). This process enhances the loss of genetic diversity in smaller populations, and it extends the range of genetic variation between small and big populations, probably also affecting the range of IFDCV. Thus, relationships between genetic diversity and iFDCV, which influences plant fitness directly and indirectly (Nock et al., 2016; Violle et al., 2007), might be strengthened in T. montanum.
4.3. Differentiated view on relationships among intraspecific functional trait variation, within‐habitat heterogeneity, and genetic diversity
Relations between iFDCV, within‐habitat heterogeneity and genetic diversity (Figure 7) are reported from several species and discussed in literature cited above. Under natural environmental conditions, within‐habitat heterogeneity and genetic diversity probably act on iFDCV in complex ways. For example, several factors may influence whether environmental variation within a habitat promotes the occurrence of different genotypes. Although a positive relationship between within‐habitat heterogeneity and population genetic diversity was expected from literature (particularly for self‐incompatible, outcrossing species, like T. montanum, with moderate to high population genetic diversity), we did not observe a significant effect. Several reasons can explain this result. Large phenotypic plasticity of traits can enable individuals to inhabit different environmental niches potentially shielding them from natural selection (e.g., Ghalambor et al., 2007). Thus, there would have been no need to select for higher functional trait variation, weakening the relationship between within‐habitat heterogeneity and population genetic diversity. Moreover, some close T. montanum populations are potentially connected (or were at least in the past) due to sheep and goat grazing (transhumance). Connectivity would have allowed for gene flow among them (Linhardt & Grant, 1996; Reisch & Schmid, 2019; Vellend & Geber, 2005) superseding (partially) local genotypes and altering population genetic diversity and adaptation to local environmental habitat conditions. However, some T. montanum populations in lowly/highly variable habitats are characterized by low/high genetic diversity (and low/high iFDCV) indicating an association between environment and genetics (and trait variation) within habitats of a species (see also Gram & Sork, 2001; Huenneke, 1991; Linhardt & Grant, 1996). Varying resource exploitation of different genotypes may explain the observed pattern (see, e.g., Agashe & Bolnick, 2010; Reusch et al., 2005). Moreover, within‐habitat heterogeneity could be underestimated in large T. montanum habitats (e.g., Er, If, Bo, St) characterized by large population sizes (Karbstein et al., in prep.), genetic diversity, and iFDCV. More environmental replicates would have potentially led to higher within‐habitat heterogeneity estimates additionally strengthening positive relationships among iFDCV, within‐habitat heterogeneity, and genetic diversity.
FIGURE 7.
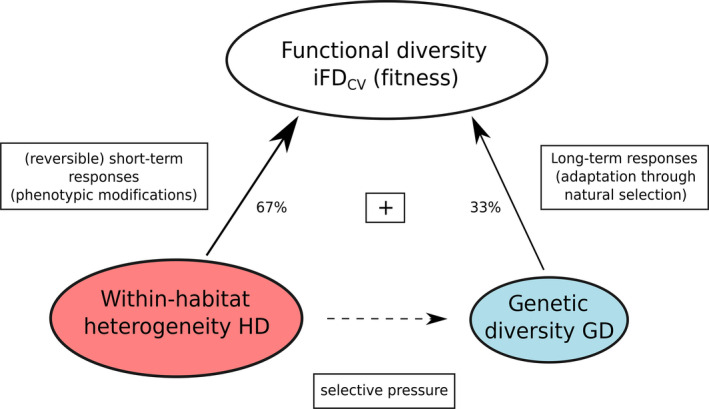
A conceptual model of relationships among intraspecific trait variation (functional diversity; iFDCV), abiotic within‐habitat heterogeneity (HD), and genetic diversity (GD) in T. montanum. HD influenced iFDCV twice as much as GD symbolized by different circle sizes and arrow strengths. Results and percents are extracted from the multiple linear regression model ( = .77, F 2, 10 = 21.66, p < .001). HD can lead to (reversible) short‐term responses, that is, to modification of functional trait expression (phenotypic modifications, variation). In contrast, GD is controlled by selection and is a prerequisite for adaptation through natural selection. iFDCV thus also depends on the available genetic variation within a population. Habitat heterogeneity and genetic diversity are not significantly related in this study (R = .10, F 1, 11 = 2.37, p = .15; dashed line)
Saturation of diversity variables (iFDCV, HD, and GD) is an important feature. Unsaturated variables can bias relationships, and can lead to false results and conclusions. In T. montanum, at least five to 10 samples were sufficient to saturate iFDCV within populations (see also Bastias et al., 2017). Within‐habitat heterogeneity was saturated by three to five samples per habitat, and genetic diversity reached the plateau at 10 to 15 samples per population, not biasing our regression models.
In addition, epigenetic processes, like DNA methylation or activation of transposable elements, in response to environmental variation can also alter phenotypic plasticity (Nicotra et al., 2015; Weinhold, 2006) and thus ITV potentially explaining a particular amount of unexplained variation in regression models. Our primary goal was not to separate the environmental component from the genetic one but to understand the relative importance of both environment and genetics on iFDCV, and particularly, how populations react under natural environmental conditions. To clearly separate the environment from the genetic impact on iFDCV, common garden experiments under controlled environmental conditions are necessary. Moreover, genomic data will provide more insights into genetic variation of populations, and in investigating relationships between functional trait variation and genetic variation. Connecting intraspecific functional trait, environmental, and genetic variation remains still an important ecological challenge.
4.4. Impact on biodiversity research
Trait variation is probably of major importance to plants short‐term adjustment on (rapid) environmental changes (see also Arnold et al., 2019; Gratani, 2014). Genetic diversity influences the range of trait plasticity and thus trait variation within a population, which can be advantageous for short‐term responses (e.g., land use abandonment) by offering genetic variants that are fitter under novel environmental conditions. Moreover, in the long term (e.g., considering anthropogenic climate change), genetic diversity offers variation for natural selection to act and thus allows for adaptation to novel habitat conditions.
Environmental habitat aspects and population genetics should be considered in biodiversity research dealing with intraspecific functional trait variation at population, community, and ecosystem level. Consideration of these aspects can prevent bias and misinterpretation of trait variation analyses, for example, comparing trait variation between sites where trait differences cannot be attributed to environment or genetics. Habitat features are directly extractable from field measurements as shown in this study or potentially from databases with a high spatial resolution (e.g., WorldClim; Hijmans et al., 2005). If genetic features of a species cannot be investigated due to a lack of suitable markers, population size can be used as a cautious proxy of genetic diversity (see Leimu et al., 2006).
Our study demonstrates the potential of deriving intraspecific functional trait variation based on environmental and genetic aspects (or its proxies) and provides empirical evidence to encourage the incorporation of intraspecific functional trait variation into interspecific comparisons (see also Albert et al., 2010; de Bello et al., 2011; Violle et al., 2012). Directly measured species‐specific intraspecific functional trait variation, but also values from databases provide the possibility for a better understanding of community and ecosystem responses to environmental changes and a more realistic estimation of ecosystem functioning.
CONFLICT OF INTEREST
None.
AUTHORS' CONTRIBUTION
Kevin Karbstein: Conceptualization (supporting); Investigation (equal); Methodology (equal); Software (equal); Visualization (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Kathleen Prinz: Conceptualization (lead); Methodology (equal); Project administration (equal); Supervision (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Frank Hellwig: Conceptualization (lead); Project administration (equal); Writing‐original draft (supporting); Writing‐review & editing (supporting). Christine Römermann: Conceptualization (lead); Formal analysis (equal); Investigation (equal); Methodology (equal); Project administration (equal); Supervision (equal); Writing‐original draft (equal); Writing‐review & editing (equal).
The study was designed by K.P., F.H., and C.R. K.K. sampled and analyzed the data Statistical data analyses were supported by K.P. and C.R. K.K. wrote the manuscript with equal contributions from K.P., F.H., and C.R.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
The Institute of Ecology and Evolution (Friedrich‐Schiller‐University, Jena, Germany) financially supported this research. We acknowledge the lower nature conservation authorities (“UNB”) in Arnstadt, Bad Salzungen, Jena, Rudolstadt, and Sondershausen for permitting access to conservation areas, and the Thüringer Landesanstalt für Landwirtschaft und Ländlichen Raum (TLLLR) for preparing and conducting soil analyses. We also thank Janin Nauman, Annika Lambert, and Andreas Gerth for technical assistance, and Isabell Schellin, Manuela Karbstein, Ronald Karbstein, Marco Schmidt, Viola Schellin, Andreas Schellin, and Solveig Franziska Bucher for support during demanding fieldwork. Claudia Pätzold, Ladislav Hodac, and Emma Jardine kindly provided comments, improvement suggestions, and language editing on previous versions of the manuscript.
Karbstein K, Prinz K, Hellwig F, Römermann C. Plant intraspecific functional trait variation is related to within‐habitat heterogeneity and genetic diversity in Trifolium montanum L. Ecol Evol. 2020;10:5015–5033. 10.1002/ece3.6255
DATA AVAILABILITY STATEMENT
Code availability. R scripts used in analyses are available from the corresponding author on request. The specific R script to calculate diversity saturation of iFDCV, HD, and GD is available on Github (https://github.com/KK260/saturation‐of‐diversity‐variables).
Data availability. Basic data supporting the findings of this study are available within the manuscript. Functional trait, environmental, and genetic data will be made available upon publication on Dryad data repository (https://doi.org/10.5061/dryad.n02v6wwtd). Functional trait data will additionally be deposited on TRY database (www.try‐db.org) upon publication.
REFERENCES
- Ackerly, D. D. , & Donoghue, M. J. (1998). Leaf size, sapling allometry, and Corner’s rules: Phylogeny and correlated evolution in maples (Acer). The American Naturalist, 152(6), 767–791. 10.1086/286208 [DOI] [PubMed] [Google Scholar]
- Ackerly, D. D. , Dudley, S. A. , Sultan, S. E. , Schmitt, H. , Coleman, J. S. , Linder, C. R. , … Lechowicz, M. J. (2000). The evolution of plant ecophysiological traits: Recent advances and future directions. BioScience, 50(11), 979–995. 10.1641/0006-3568(2000)050[0979:TEOPET]2.0.CO;2 [DOI] [Google Scholar]
- Agashe, D. , & Bolnick, D. I. (2010). Intraspecific genetic variation and competition interact to influence niche expansion. Proceedings of the Royal Society B: Biological Sciences, 277(1696), 2915–2924. 10.1098/rspb.2010.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, A. A. (2001). Phenotypic plasticity in the interactions and evolution of species. Science, 294(5541), 321–326. 10.1126/science.1060701 [DOI] [PubMed] [Google Scholar]
- Ågren, G. I. , Wetterstedt, J. Å. M. , & Billberger, M. F. K. (2012). Nutrient limitation on terrestrial plant growth ‐ modeling the interaction between nitrogen and phosphorus. New Phytologist, 194(4), 953–960. 10.1111/j.1469-8137.2012.04116.x [DOI] [PubMed] [Google Scholar]
- Albert, C. H. , Thuiller, W. , Yoccoz, N. G. , Douzet, R. , Aubert, S. , & Lavorel, S. (2010). A multi‐trait approach reveals the structure and the relative importance of intra‐ vs. interspecific variability in plant traits. Functional Ecology, 24(6), 1192–1201. 10.1111/j.1365-2435.2010.01727.x [DOI] [Google Scholar]
- Arnold, P. A. , Kruuk, L. E. B. , & Nicotra, A. B. (2019). How to analyse plant phenotypic plasticity in response to a changing climate. New Phytologist, 222(3), 1235–1241. 10.1111/nph.15656 [DOI] [PubMed] [Google Scholar]
- Balasooriya, B. L. W. K. , Samson, R. , Mbikwa, F. , Vitharana, U. W. A. , Boeckx, P. , & Van Meirvenne, M. (2009). Biomonitoring of urban habitat quality by anatomical and chemical leaf characteristics. Environmental and Experimental Botany, 65(2–3), 386–394. 10.1016/j.envexpbot.2008.11.009 [DOI] [Google Scholar]
- Bartelink, H. (1997). Allometric relationships for biomass and leaf area of beech (Fagus sylvatica L.). Annales Des Sciences Forestieres, 54(31), 39–50. 10.1051/forest:19970104 [DOI] [Google Scholar]
- Bastias, C. C. , Fortunel, C. , Valladares, F. , Baraloto, C. , Benavides, R. , Cornwell, W. , … Kraft, N. J. B. (2017). Intraspecific leaf trait variability along a boreal‐to‐tropical community diversity gradient. PLoS ONE, 12(2), 1–16. 10.1371/journal.pone.0172495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavides, R. , Scherer‐Lorenzen, M. , & Valladares, F. (2019). The functional trait space of tree species is influenced by the species richness of the canopy and the type of forest. Oikos, 128, 1435–1445. 10.1111/oik.06348 [DOI] [Google Scholar]
- Bernhardt‐Römermann, M. , Gray, A. , Vanbergen, A. J. , Bergès, L. , Bohner, A. , Brooker, R. W. , … Stadler, J. (2011). Functional traits and local environment predict vegetation responses to disturbance: A pan‐European multi‐site experiment. Journal of Ecology, 99(3), 777–787. 10.1111/j.1365-2745.2011.01794.x [DOI] [Google Scholar]
- Bjornstad, O. N. , & Cai, J. (2018). ncf: spatial covariance functions.
- Boulding, E. G. (2008). Genetic diversity, adaptive potential, and population viability in changing environments In Carroll S. P., & Fox C. W. (Eds.), Conservation biology: Evolution in action (pp. 201–222). Oxford, UK: Oxford University Press. [Google Scholar]
- Bucher, S. F. , Auerswald, K. , Grün‐Wenzel, C. , Higgins, S. I. , Garcia Jorge, J. , & Römermann, C. (2017). Stomatal traits relate to habitat preferences of herbaceous species in a temperate climate. Flora: Morphology, Distribution, Functional Ecology of Plants, 229, 107–115. 10.1016/j.flora.2017.02.011 [DOI] [Google Scholar]
- Bucher, S. F. , Auerswald, K. , Tautenhahn, S. , Geiger, A. , Otto, J. , Müller, A. , & Römermann, C. (2016). Inter‐ and intraspecific variation in stomatal pore area index along elevational gradients and its relation to leaf functional traits. Plant Ecology, 217(3), 229–240. 10.1007/s11258-016-0564-2 [DOI] [Google Scholar]
- Bucher, S. F. , Bernhardt‐Römermann, M. , & Römermann, C. (2018). Chlorophyll fluorescence and gas exchange measurements in field research: An ecological case study. Photosynthetica, 56(4), 1161–1170. 10.1007/s11099-018-0809-5 [DOI] [Google Scholar]
- Bucher, S. F. , Feiler, R. , Buchner, O. , Neuner, G. , Rosbakh, S. , Leiterer, M. , & Römermann, C. (2019). Temporal and spatial trade‐offs between resistance and performance traits in herbaceous plant species. Environmental and Experimental Botany, 157, 187–196. 10.1016/j.envexpbot.2018.10.015 [DOI] [Google Scholar]
- Butler, W. L. , & Kitajima, M. (1975). Fluorescence quenching in photosystem II of chloroplasts. BBA ‐ Bioenergetics, 376(1), 116–125. 10.1016/0005-2728(75)90210-8 [DOI] [PubMed] [Google Scholar]
- Chen, X. , Visser, E. J. W. , de Kroon, H. , Pierik, R. , Voesenek, L. A. C. J. , & Huber, H. (2011). Fitness consequences of natural variation in flooding‐induced shoot elongation in Rumex palustris . New Phytologist, 190(2), 409–420. 10.1111/j.1469-8137.2010.03639.x [DOI] [PubMed] [Google Scholar]
- Chevin, L. M. , & Hoffmann, A. A. (2017). Evolution of phenotypic plasticity in extreme environments. Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1723), 10.1098/rstb.2016.0138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciaruso, M. V. , Batalha, M. A. , Gaston, K. J. , & Petchey, O. L. (2009). Including intraspecific variability in functional diversity. Ecology, 90(1), 81–89. 10.1890/07-1864.1 [DOI] [PubMed] [Google Scholar]
- Clark, A. J. , Landolt, W. , Bucher, J. B. , & Strasser, R. J. (2000). Beech (Fagus sylvatica) response to ozone exposure assessed with a chlorophyll a fluorescence performance index. Environmental Pollution, 109(3), 501–507. 10.1016/S0269-7491(00)00053-1 [DOI] [PubMed] [Google Scholar]
- R Core Team . (2019). R: A language and environment for statistical computing. Vienna, Austria: Foundation for Statistical Computing; Retrieved from http://www.r‐project.org [Google Scholar]
- Cornelissen, J. H. C. , Lavorel, S. , Garnier, E. , Díaz, S. , Buchmann, N. , Gurvich, D. E. , … Poorter, H. (2003). A handbook of protocols for standardised and easy measurement of plant functional traits worldwide. Australian Journal of Botany, 51(4), 335 10.1071/BT02124 [DOI] [Google Scholar]
- Crawley, M. J. (2007). The R book. Chichester, UK: John Wiley & Sons, Ltd; 10.1002/9780470515075 [DOI] [Google Scholar]
- Crawley, M. J. (2015). Statistics: An introduction using R (2nd edition). Chichester, UK: John Wiley & Sons. [Google Scholar]
- Csilléry, K. , Ovaskainen, O. , Sperisen, C. , Buchmann, N. , Widmer, A. , & Gugerli, F. (2020). Adaptation to local climate in multi‐trait space: Evidence from silver fir (Abies alba Mill.) populations across a heterogeneous environment. Heredity, 124(1), 77–92. 10.1038/s41437-019-0240-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bello, F. , Lavorel, S. , Albert, C. H. , Thuiller, W. , Grigulis, K. , Dolezal, J. , … Lepš, J. (2011). Quantifying the relevance of intraspecific trait variability for functional diversity. Methods in Ecology and Evolution, 2(2), 163–174. 10.1111/j.2041-210X.2010.00071.x [DOI] [Google Scholar]
- Díaz, S. , & Cabido, M. (2001). Vive la différence: Plant functional diversity matters to ecosystem processes. Trends in Ecology & Evolution, 16(11), 646–655. 10.1016/S0169-5347(01)02283-2 [DOI] [Google Scholar]
- Díaz, S. , Kattge, J. , Cornelissen, J. H. C. , Wright, I. J. , Lavorel, S. , Dray, S. , … Gorné, L. D. (2016). The global spectrum of plant form and function. Nature, 529(7585), 167–171. 10.1038/nature16489 [DOI] [PubMed] [Google Scholar]
- Dormann, C. F. , Elith, J. , Bacher, S. , Buchmann, C. , Carl, G. , Carré, G. , … Lautenbach, S. (2013). Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography, 36(1), 027–046. 10.1111/j.1600-0587.2012.07348.x [DOI] [Google Scholar]
- Doyle, J. , & Doyle, J. L. (1987). Genomic plant DNA preparation from fresh tissue‐CTAB method. Phytochemical Bulletin, 19(11), 11–15. [Google Scholar]
- Dwyer, J. , Hobbs, R. , & Mayfield, M. (2014). Specific leaf area responses to environmental gradients through space and time. Ecology, 95(2), 399–410. [DOI] [PubMed] [Google Scholar]
- Earl, D. A. , & vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Ellegren, H. (2004). Microsatellites: Simple sequences with complex evolution. Nature Reviews Genetics, 5(6), 435–445. 10.1038/nrg1348 [DOI] [PubMed] [Google Scholar]
- Farooq, M. , Wahid, A. , Fujita, K. D. , & Basra, S. M. A. (2009). Plant drought stress: Effects, mechanisms and management. Agronomy for Sustainable Development, 29, 185–212. 10.1051/agro [DOI] [Google Scholar]
- Farquhar, G. D. , Buckley, T. N. , & Miller, J. M. (2002). Optimal stomatal control in relation to leaf area and nitrogen content. Silva Fennica, 36(3), 625–637. 10.14214/sf.530 [DOI] [Google Scholar]
- Freeland, J. R. , Kirk, H. , & Petersen, S. D. (2011). Molecular ecology (2nd edition). Hoboken, NJ: John Wiley & Sons; 10.1002/9780470979365 [DOI] [Google Scholar]
- Garve, E. (2004). Rote Liste und Florenliste der Farn‐und Blütenpflanzen in Niedersachsen und Bremen. Hannover, Germany: Niedersächsisches Landesamt für Ökologie‐Fachbehörde für Naturschutz. [Google Scholar]
- Gaudet, C. L. , & Keddy, P. A. (1988). A comparative approach to predicting competitive ability from plant traits. Nature, 334(6179), 242–243. 10.1038/334242a0 [DOI] [Google Scholar]
- Geber, M. A. , & Griffen, L. R. (2003). Inheritance and natural selection on functional traits. International Journal of Plant Sciences, 164(S3), S21–S42. 10.1086/368233 [DOI] [Google Scholar]
- Ghalambor, C. K. , McKay, J. K. , Carroll, S. P. , & Reznick, D. N. (2007). Adaptive versus non‐adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Functional Ecology, 21(3), 394–407. 10.1111/j.1365-2435.2007.01283.x [DOI] [Google Scholar]
- Givnish, T. J. (1987). Comparative studies of leaf form: Assessing the relative roles of selective pressures and phylogenetic constraints. New Phytologist, 106, 131–160. 10.1111/j.1469-8137.1987.tb04687.x [DOI] [Google Scholar]
- Gram, W. K. , & Sork, V. L. (2001). Association between environmental and genetic heterogeneity in forest tree populations. Ecology, 82(7), 2012–2021. 10.2307/2680065 [DOI] [Google Scholar]
- Gratani, L. (2014). Plant phenotypic plasticity in response to environmental factors. Advances in Botany, 2014, 1–17. 10.1155/2014/208747 [DOI] [Google Scholar]
- Griffin, K. L. , Epstein, D. J. , & Boelman, N. T. (2013). Hill slope variations in chlorophyll fluorescence indices and leaf traits in a small arctic watershed. Arctic, Antarctic, and Alpine Research, 45(1), 39–49. 10.1657/1938-4246-45.1.39 [DOI] [Google Scholar]
- Hahn, T. , Kettle, C. J. , Ghazoul, J. , Hennig, E. I. , & Pluess, A. R. (2013). Landscape composition has limited impact on local genetic structure in mountain clover, Trifolium Montanum L. Journal of Heredity, 104(6), 842–852. 10.1093/jhered/est058 [DOI] [PubMed] [Google Scholar]
- Hardy, G. H. (1908). Mendelian proportions in a mixed population In Peters J. A. (Ed.), Classic papers in genetics (pp. 60–62). Englewood Cliffs, NJ: Prentice‐Hall Biological Science Series; 10.1126/science.28.706.49 [DOI] [Google Scholar]
- Harrell, F. E. (2019). Hmisc ‐ Harrell Miscellaneous. Retrieved fromhttps://github.com/harrelfe/Hmisc
- Helm, J. , Dutoit, T. , Saatkamp, A. , Bucher, S. F. , Leiterer, M. , & Römermann, C. (2019). Recovery of Mediterranean steppe vegetation after cultivation: Legacy effects on plant composition, soil properties and functional traits. Applied Vegetation Science, 22(1), 71–84. 10.1111/avsc.12415 [DOI] [Google Scholar]
- Hijmans, R. J. (2016). geosphere: Spherical trigonometry. Retrieved fromhttps://CRAN.R‐project.org/package=geosphere
- Hijmans, R. J. , Cameron, S. E. , Parra, J. L. , Jones, P. G. , & Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology, 25(15), 1965–1978. 10.1002/joc.1276 [DOI] [Google Scholar]
- Huenneke, L. F. (1991). Ecological implications of variation in plant populations In Falk D. A., & Holsinger K. E. (Eds.), Genetics and conservation of rare plants (pp. 31–44). New York, NY: Oxford University Press. [Google Scholar]
- Hufford, K. M. , & Mazer, S. J. (2003). Plant ecotypes: Genetic differentiation in the age of ecological restoration. Trends in Ecology and Evolution, 18(3), 147–155. 10.1016/S0169-5347(03)00002-8 [DOI] [Google Scholar]
- Hughes, P. W. , Soppe, W. J. J. , & Albani, M. C. (2019). Seed traits are pleiotropically regulated by the flowering time gene PERPETUAL FLOWERING 1 (PEP1) in the perennial Arabis alpina . Molecular Ecology, 28(5), 1183–1201. 10.1111/mec.15034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger, E. (2011). Trifolium montanum L(20th edition). Heidelberg, Germany: Springer Spektrum. [Google Scholar]
- Jakobsson, M. , & Rosenberg, N. A. (2007). CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23(14), 1801–1806. 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Jaleel, C. A. , Manivannan, P. , Wahid, A. , Farooq, M. , Al‐Juburi, H. J. , Somasundaram, R. , & Panneerselvam, R. (2009). Drought stress in plants: A review on morphological characteristics and pigments composition. International Journal of Agriculture & Biology, 11, 100–105. [Google Scholar]
- Jombart, T. , Kamvar, Z. N. , Collins, C. , Lustrik, R. , Beugin, M.‐P. , Knaus, B. J. , & Jombart, M. T. (2018). adegenet: A R package for the multivariate analysis of genetic markers. Retrieved fromhttps://github.com/thibautjombart/adegenet [DOI] [PubMed]
- Kalinowski, S. T. (2004). Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conservation Genetics, 5(4), 539–543. 10.1023/B:COGE.0000041021.91777.1a [DOI] [Google Scholar]
- Karbstein, K. (2020). Saturation‐of‐diversity‐variables. Github Repositoryhttps://github.com/KK260/saturation‐of‐diversity‐variables
- Karbstein, K. , Tomasello, S. , & Prinz, K. (2019). Desert‐like badlands and surrounding (semi‐)dry grasslands of Central Germany promote small‐scale phenotypic and genetic differentiation in Thymus praecox . Ecology and Evolution, 9, 14066–14084. 10.1002/ece3.5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattge, J. , Bönisch, G. , Díaz, S. , Lavorel, S. , Prentice, I. C. , Leadley, P. , … Wirth, C. (2020). TRY plant trait database ‐ enhanced coverage and open access. Global Change Biology, 26(1), 119–188. 10.1111/gcb.14904 [DOI] [PubMed] [Google Scholar]
- Kattge, J. , Díaz, S. , Lavorel, S. , Prentice, I. C. , Leadley, P. , Bönisch, G. , … Wirth, C. (2011). TRY ‐ a global database of plant traits. Global Change Biology, 17(9), 2905–2935. 10.1111/j.1365-2486.2011.02451.x [DOI] [Google Scholar]
- Kerkhoff, A. J. , Fagan, W. F. , Elser, J. J. , & Enquist, B. J. (2006). Phylogenetic and growth form variation in the scaling of nitrogen and phosphorus in the seed plants. The American Naturalist, 168(4), E103–E122. 10.1086/507879 [DOI] [PubMed] [Google Scholar]
- König, P. , Tautenhahn, S. , Cornelissen, J. C. , Kattge, J. , Bönisch, G. , & Römermann, C. (2018). Advances in flowering phenology across the Northern Hemisphere are explained by functional traits. Global Ecology and Biogeography, 27(3), 310–321. 10.1111/geb.12696 [DOI] [Google Scholar]
- Korbie, D. J. , & Mattick, J. S. (2008). Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nature Protocols, 3(9), 1452–1456. 10.1038/nprot.2008.133 [DOI] [PubMed] [Google Scholar]
- Lande, R. (2009). Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. Journal of Evolutionary Biology, 22(7), 1435–1446. 10.1111/j.1420-9101.2009.01754.x [DOI] [PubMed] [Google Scholar]
- Leimu, R. , Mutikainen, P. , Koricheva, J. , & Fischer, M. (2006). How general are positive relationships between plant population size, fitness and genetic variation? Journal of Ecology, 94(5), 942–952. 10.1111/j.1365-2745.2006.01150.x [DOI] [Google Scholar]
- Li, Y. C. , Korol, A. B. , Fahima, T. , Beiles, A. , & Nevo, E. (2002). Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Molecular Ecology, 11(12), 2453–2465. 10.1046/j.1365-294X.2002.01643.x [DOI] [PubMed] [Google Scholar]
- Linhardt, Y. B. , & Grant, M. C. (1996). Evolutionary significance of local genetic differentiation in plants. Annual Review of Ecology and Systematics, 27, 237–277. 10.1146/annurev.soc.29.010202.100030 [DOI] [Google Scholar]
- Locascio, A. , Lucchin, M. , & Varotto, S. (2009). Characterization of a MADS FLOWERING LOCUS C‐LIKE (MFL) sequence in Cichorium intybus: A comparative study of CiMFL and AtFLC reveals homologies and divergences in gene function. New Phytologist, 182(3), 630–643. 10.1111/j.1469-8137.2009.02791.x [DOI] [PubMed] [Google Scholar]
- MacArthur, R. , & Wilson, E. (1967). The theory of island biogeography. Princeton, NJ: Princeton University Press. [Google Scholar]
- Matter, P. , Kettle, C. J. , Ghazoul, J. , Hahn, T. , & Pluess, A. R. (2013). Evaluating contemporary pollen dispersal in two common grassland species Ranunculus bulbosus L. (Ranunculaceae) and Trifolium montanum L. (Fabaceae) using an experimental approach. Plant Biology, 15(3), 583–592. 10.1111/j.1438-8677.2012.00667.x [DOI] [PubMed] [Google Scholar]
- Matter, P. , Määttänen, K. , Kettle, C. J. , Ghazoul, J. , & Pluess, A. R. (2012). Eleven microsatellite markers for the mountain clover Trifolium montanum (Fabaceae). American Journal of Botany, 99(11), 447–449. 10.3732/ajb.1200102 [DOI] [PubMed] [Google Scholar]
- Maxwell, K. , & Johnson, G. N. (2000). Chlorophyll fluorescence – A practical guide. Journal of Experimental Botany, 51(345), 659–668. 10.1093/jxb/51.345.659 [DOI] [PubMed] [Google Scholar]
- Meusel, H. , & Jäger, E. (1998). Vergleichende Chorologie der Zentraleuropäischen Flora‐Band I, II, III. Jena, DE: Gustav Fischer Verlag. [Google Scholar]
- Moles, A. T. , Warton, D. I. , Warman, L. , Swenson, N. G. , Laffan, S. W. , Zanne, A. E. , … Leishman, M. R. (2009). Global patterns in plant height. Journal of Ecology, 97(5), 923–932. 10.1111/j.1365-2745.2009.01526.x [DOI] [Google Scholar]
- Moran, P. A. P. (1950). Notes on continuous stochastic phenomena. Biometrika Trust, 37(1–2), 17–23. 10.2307/2332142 [DOI] [PubMed] [Google Scholar]
- Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proc Nat Acad Sci USA, 70(12), 3321–3323. 10.1073/pnas.70.12.3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89(3), 583–590. 10.3390/ijms15010277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra, A. B. , Atkin, O. K. , Bonser, S. P. , Davidson, A. M. , Finnegan, E. J. , Mathesius, U. , … van Kleunen, M. (2010). Plant phenotypic plasticity in a changing climate. Trends in Plant Science, 15(12), 684–692. 10.1016/j.tplants.2010.09.008 [DOI] [PubMed] [Google Scholar]
- Nicotra, A. B. , Segal, D. L. , Hoyle, G. L. , Schrey, A. W. , Verhoeven, K. J. F. , & Richards, C. L. (2015). Adaptive plasticity and epigenetic variation in response to warming in an Alpine plant. Ecology and Evolution, 5(3), 634–647. 10.1002/ece3.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock, C. A. , Vogt, R. J. , & Beisner, B. E. (2016). Functional traits. ELS, February, 1–8. 10.1002/9780470015902.a0026282 [DOI] [Google Scholar]
- Paillotin, G. (1976). Movement of excitations in the photosynthetic domains of photosystem II. Journal of Theoretical Biology, 58(1), 237–252. 10.1016/0022-5193(76)90150-8 [DOI] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6(1), 288–295. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research – An update. Bioinformatics, 28(19), 2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Harguindeguy, N. , Díaz, S. , Garnier, E. , Lavorel, S. , Poorter, H. , Jaureguiberry, P. , … Cornelissen, J. H. C. (2013). New handbook for standardised measurement of plant functional traits worldwide. Australian Journal of Botany, 61(3), 167–234. 10.1071/BT12225 [DOI] [Google Scholar]
- Petchey, O. L. , & Gaston, K. J. (2006). Functional diversity: Back to basics and looking forward. Ecology Letters, 9(6), 741–758. 10.1111/j.1461-0248.2006.00924.x [DOI] [PubMed] [Google Scholar]
- Prinz, K. , Weising, K. , & Hensen, I. (2009). Genetic structure of coastal and inland populations of the annual halophyte Suaeda maritima (L.) dumort. in Central Europe, inferred from amplified fragment length polymorphism markers. Plant Biology, 11(6), 812–820. 10.1111/j.1438-8677.2008.00178.x [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho, J. C. , Rebelo, M. C. , Santos, M. E. , Antunes, M. L. , & Nunes, M. A. (1995). Effects of calcium deficiency on Coffea arabica. Nutrient changes and correlation of calcium levels with some photosynthetic parameters. Plant and Soil, 172(1), 87–96. 10.1007/BF00020862 [DOI] [Google Scholar]
- Razaq, M. , Zhang, P. , Shen, H.‐L. , & Salahuddin, (2017). Influence of nitrogen and phosphorous on the growth and root morphology of Acer mono . PLoS ONE, 12(2), e0171321 10.1371/journal.pone.0171321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, D. H. , & Frankham, R. (2003). Correlation between fitness and genetic diversity. Conservation Biology, 17(1), 230–237. 10.1046/j.1523-1739.2003.01236.x [DOI] [Google Scholar]
- Reich, P. B. , & Hobbie, S. E. (2013). Decade‐long soil nitrogen constraint on the CO2 fertilization of plant biomass. Nature Climate Change, 3(3), 278–282. 10.1038/nclimate1694 [DOI] [Google Scholar]
- Reisch, C. , & Bernhardt‐Römermann, M. (2014). The impact of study design and life history traits on genetic variation of plants determined with AFLPs. Plant Ecology, 215(12), 1493–1511. 10.1007/s11258-014-0409-9 [DOI] [Google Scholar]
- Reisch, C. , & Schmid, C. (2019). Species and genetic diversity are not congruent in fragmented dry grasslands. Ecology and Evolution, 9(1), 664–671. 10.1002/ece3.4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch, T. B. , Ehlers, A. , Hämmerli, A. , & Worm, B. (2005). Ecosystem recovery after climatic extremes enhanced by genotypic diversity. Proceedings of the National Academy of Sciences of the United States of America, 102(8), 2826–2831. 10.1073/pnas.0500008102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, A. , Glick, L. , Abadi, S. , Einhorn, M. , Kopelman, N. M. , Salman‐Minkov, A. , … Mayrose, I. (2015). The Chromosome Counts Database (CCDB) – A community resource of plant chromosome numbers. New Phytologist, 206(1), 19–26. 10.1111/nph.13191 [DOI] [PubMed] [Google Scholar]
- Römermann, C. , Bernhardt‐Römermann, M. , Kleyer, M. , & Poschlod, P. (2009). Substitutes for grazing in semi‐natural grasslands – Do mowing or mulching represent valuable alternatives to maintain vegetation structure? Journal of Vegetation Science, 20(6), 1086–1098. [Google Scholar]
- Römermann, C. , Bucher, S. F. , Hahn, M. , & Bernhardt‐Römermann, M. (2016). Plant functional traits ‐ fixed facts or variable depending on the season? Folia Geobotanica, 51(2), 143–159. 10.1007/s12224-016-9250-3 [DOI] [Google Scholar]
- Rosenberg, N. A. (2004). DISTRUCT: A program for the graphical display of population structure. Molecular Ecology Notes, 4(1), 137–138. 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- Sack, L. , Cowan, P. D. , Jaikumar, N. , & Holbrook, N. M. (2003). The ‘hydrology’ of leaves: Co‐ordination of structure and function in temperate woody species. Plant, Cell and Environment, 26(8), 1343–1356. 10.1046/j.0016-8025.2003.01058.x [DOI] [Google Scholar]
- Saghai‐Maroof, M. A. , Soliman, K. M. , Jorgensen, R. A. , & Allard, R. W. (1984). Ribosomal DNA spacer‐length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences of the United States of America, 81(24), 8014–8018. 10.1073/pnas.81.24.8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi, S. , Horie, K. , Ishikawa, N. , Nishio, S. , Worth, J. R. , Fukushima, K. , … Ito, M. (2019). Maintenance of soil ecotypes of Solidago virgaurea in close parapatry via divergent flowering time and selection against immigrants. Journal of Ecology, 107(1), 418–435. 10.1111/1365-2745.13034 [DOI] [Google Scholar]
- Schleuning, M. , & Matthies, D. (2008). Habitat change and plant demography: Assessing the extinction risk of a formerly common grassland perennial. Conservation Biology, 23(1), 174–183. 10.1111/j.1523-1739.2008.01054.x [DOI] [PubMed] [Google Scholar]
- Schleuning, M. , Niggemann, M. , Becker, U. , & Matthies, D. (2009). Negative effects of habitat degradation and fragmentation on the declining grassland plant Trifolium montanum . Basic and Applied Ecology, 10(1), 61–69. 10.1016/j.baae.2007.12.002 [DOI] [Google Scholar]
- Schweiger, A. K. , Cavender‐Bares, J. , Townsend, P. A. , Hobbie, S. E. , Madritch, M. D. , Wang, R. , … Gamon, J. A. (2018). Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nature Ecology and Evolution, 2(6), 976–982. 10.1038/s41559-018-0551-1 [DOI] [PubMed] [Google Scholar]
- GBIF Secretariat . (2017). Trifolium montanum L. GBIF Backbone Taxonomy. Checklist Dataset. Retrieved from 10.15468/39omei [DOI]
- Shannon, C. E. (1948). A mathematical theory of communication. The Bell System Technical Journal, 27, 379–423. 10.1145/584091.584093 [DOI] [Google Scholar]
- Simpson, E. H. (1949). Measurement of diversity. Nature, 163(4148), 688 10.1038/163688a0 [DOI] [Google Scholar]
- Sims, L. , Pastor, J. , Lee, T. , & Dewey, B. (2012). Nitrogen, phosphorus and light effects on growth and allocation of biomass and nutrients in wild rice. Oecologia, 170(1), 65–76. 10.1007/s00442-012-2296-x [DOI] [PubMed] [Google Scholar]
- Strasser, R. J. , Srivastava, A. , & Tsimilli‐Michael, M. (2000). The fluorescence transient as a tool to characterize and screen photosynthetic samples In Yunus M., Pathre U., & Mohanty P. (Eds.), Probing photosynthesis: Mechanisms, regulation and adaptation (pp. 445–483). London, UK and New York, NY: Taylor and Francis. [Google Scholar]
- Strasser, R. J. , Tsimilli‐Michael, M. , & Srivastava, A. (2004). Analysis of the chlorophyll a fluorescence transient In Papageorgiou G. C. & Govindjee (Eds.), Chlorophyll a fluorescence (2nd edition, pp. 321–362). Dordrecht, NL: Springer; 10.1007/978-1-4020-3218-9_12 [DOI] [Google Scholar]
- Sultan, S. E. (2000). Phenotypic plasticity for plant development, function and life history. Trends in Plant Science, 5, 537–542. 10.1016/S1360-1385(00)01797-0 [DOI] [PubMed] [Google Scholar]
- Szczecińska, M. , Sramko, G. , Wołosz, K. , & Sawicki, J. (2016). Genetic diversity and population structure of the rare and endangered plant species Pulsatilla patens (L.) Mill in East Central Europe. PLoS ONE, 11(3), 1–24. 10.1371/journal.pone.0151730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiyun, W. , & Simko, V. (2017). corrplot ‐ visualization of a correlation matrix. Retrieved fromhttps://github.com/taiyun/corrplot
- Tautenhahn, S. , Grün‐Wenzel, C. , Jung, M. , Higgins, S. , & Römermann, C. (2019). On the relevance of intraspecific trait variability ‐ A synthesis of 56 dry grassland sites across Europe. Flora, 254, 161–172. 10.1016/j.flora.2019.03.002 [DOI] [Google Scholar]
- Thornsberry, J. M. , Goodman, M. M. , Doebley, J. , Kresovich, S. , Nielsen, D. , & Buckler, E. S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nature Genetics, 28(3), 286–289. 10.1038/90135 [DOI] [PubMed] [Google Scholar]
- Tilman, D. (2001). Functional diversity. Encyclopedia of Biodiversity, 2, 587‐596. 10.1016/B978-0-12-384719-5.00061-7 [DOI] [Google Scholar]
- Vellend, M. , & Geber, M. A. (2005). Connections between species diversity and genetic diversity. Ecology Letters, 8(7), 767–781. 10.1111/j.1461-0248.2005.00775.x [DOI] [Google Scholar]
- Via, S. , Gomulkiewicz, R. , De Jong, G. , Scheiner, S. M. , Schlichting, C. D. , & Van Tienderen, P. H. (1995). Adaptive phenotypic plasticity: Consensus and controversy. Trends in Ecology & Evolution, 10(5), 212–217. 10.1016/S0169-5347(00)89061-8 [DOI] [PubMed] [Google Scholar]
- Vieira, M. L. C. , Santini, L. , Diniz, A. L. , & Munhoz, C. D. F. (2016). Microsatellite markers: What they mean and why they are so useful. Genetics and Molecular Biology, 39(3), 312–328. 10.1590/1678-4685-GMB-2016-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violle, C. , Enquist, B. J. , McGill, B. J. , Jiang, L. , Albert, C. H. , Hulshof, C. , … Messier, J. (2012). The return of the variance: Intraspecific variability in community ecology. Trends in Ecology & Evolution, 27(4), 244–252. 10.1016/j.tree.2011.11.014 [DOI] [PubMed] [Google Scholar]
- Violle, C. , Navas, M. , Vile, D. , Kazakou, E. , Fortunel, C. , Hummel, I. , & Garnier, E. (2007). Let the concept of trait be functional!. Oikos, 116(5), 882–892. 10.1111/j.2007.0030-1299.15559.x [DOI] [Google Scholar]
- Vittoz, P. , & Engler, R. (2007). Seed dispersal distances: A typology based on dispersal modes and plant traits. Botanica Helvetica, 117, 109–124. [Google Scholar]
- Waitt, D. E. , & Levin, D. A. (1998). Genetic and phenotypic correlations in plants: A botanical test of Cheverud’s conjecture. Heredity, 80(3), 310–319. 10.1046/j.1365-2540.1998.00298.x [DOI] [Google Scholar]
- Weinberg, W. (1908). Über Vererbungsgesetze beim Menschen. Zeitschrift Für Induktive Abstammungs‐ Und Vererbungslehre, 1(1), 377–392. 10.1007/BF01990605 [DOI] [Google Scholar]
- Weinhold, B. (2006). Epigenetics: The science of change. Environmental Health Perspectives, 114(3), 160–167. 10.1289/ehp.114-a160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. , & Ruiz, E. (2019). A ‘dplyr’ back end for databases.
- Wood, C. M. , McKinney, S. T. , & Loftin, C. S. (2017). Intraspecific functional diversity of common species enhances community stability. Ecology and Evolution, 7(5), 1553–1560. 10.1002/ece3.2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, F. I. , Lake, J. A. , & Quick, W. P. (2002). Stomatal development and CO2: Ecological consequences. New Phytologist, 153(3), 477–484. 10.1046/j.0028-646X.2001.00338.x [DOI] [PubMed] [Google Scholar]
- Wright, K. (2018). corrgram: Plot a correlogram. Retrieved from https://cran.r‐project.org [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Code availability. R scripts used in analyses are available from the corresponding author on request. The specific R script to calculate diversity saturation of iFDCV, HD, and GD is available on Github (https://github.com/KK260/saturation‐of‐diversity‐variables).
Data availability. Basic data supporting the findings of this study are available within the manuscript. Functional trait, environmental, and genetic data will be made available upon publication on Dryad data repository (https://doi.org/10.5061/dryad.n02v6wwtd). Functional trait data will additionally be deposited on TRY database (www.try‐db.org) upon publication.


