Abstract
Glaucoma is the leading cause of irreversible blindness worldwide. The molecular etiology of glaucoma is complex and unclear. At present, there are few drugs available for glaucoma treatment. The aim of the present study was to perform a systematic analysis of glaucoma candidate drugs/chemicals based on glaucoma genes, including genetic factors and differentially expressed (DE) genes. In total, 401 genes from the genetic databases and 1656 genes from the DE gene analysis were included in further analyses. In terms of glaucoma-related genetic factors, 54 pathways were significantly enriched (FDR < 0.05), and 96 pathways for DE genes were significantly enriched (FDR < 0.05). A search of the PheWAS database for diseases associated with glaucoma-related genes returned 1,289 diseases, and a search for diseases associated with DE glaucoma-related genes returned 1,356 diseases. Cardiovascular diseases, neurodegenerative diseases, cancer, and ophthalmic diseases were highly related to glaucoma genes. A search of the DGIdb, KEGG, and CLUE databases revealed a set of drugs/chemicals targeting glaucoma genes. A subsequent analysis of the electronic medical records (EMRs) of 136,128 patients treated in Sichuan Provincial People’s Hospital for candidate drug usage and the onset of glaucoma revealed nine candidate drugs. Among these drugs, individuals treated with nicardipine had the lowest incidence of glaucoma. Taken together with the information from the drug databases, the 40 most likely candidate drugs for glaucoma treatment were highlighted. Based on these findings, we concluded that the molecular mechanism of glaucoma is complex and may be a reflection of systemic diseases. A set of ready-to-use candidate drugs targeting glaucoma genes may be developed for glaucoma clinical drug treatments. Our results provide a systematic interpretation of glaucoma genes, interactions with other systemic diseases, and candidate drugs/chemicals.
Subject terms: Data mining, Data processing
Introduction
Glaucoma is a set of progressive optic neuropathies1 and the leading cause of irreversible blindness worldwide2. Glaucoma is characterized by a loss of retinal ganglion cells and consequent visual field loss. The two most common forms of glaucoma are primary open-angle glaucoma (POAG) and primary angle-closure glaucoma3. The main known risk factors for glaucoma include high intraocular pressure (IOP), older age, African race, high myopia4, a high vertical cup/intervertebral disc ratio5, and a reduction in the optic disk area and central corneal thickness6. Epidemiological studies have shown that the prevalence of glaucoma is expected to reach 76 million by 2020 and 118 million globally by 2040 due to population aging7. The mechanism underlying the development of glaucoma is not fully understood.
Glaucoma is a complex hereditary disease. Mutations in the OPTN, MYOC, and WDR36 genes have been identified as the causes of POAG8. Thus far, 14 genome-wide association studies have identified 97 single-nucleotide polymorphisms near 75 genes associated with glaucoma in the GWAS catalog, including ABCA19, CAV1/CAV210, TMCO111, CDKN2B-AS112, SIX1/SIX613, GAS714, and ATOH715.
The treatment of glaucoma includes drug use and surgery16. Antiglaucoma drugs reduce IOP mainly by reducing aqueous humor production and promoting aqueous humor discharge17. At present, there are four kinds of drugs for glaucoma treatment: β-receptor blockers, prostaglandins, α-2 agonists and carbonic anhydrase inhibitors. Laser peripheral iridectomy (LPI) is used in glaucoma with anterior chamber angle occlusion and occlusion18. Selective laser trabeculoplasty (SLT)19 can be used as primary or auxiliary treatment for primary open-angle glaucoma or early- and late-stage glaucoma after LPI. Microinvasive glaucoma surgery (MIGS) reduces IOP and reduces dependence on glaucoma medications20.
It can cost billions of dollars to develop a new drug and often takes several years. Drug reuse is a strategy for the identification of new uses of drugs for approval or research beyond the scope of the original medical indications21. This new phase of genomics, which is increasingly referred to as precision medicine, has sparked a new chapter in the relationship between genomics and drug development22. Compared with the development of new drugs for specific indications, drug reuse has several advantages23, including safety and less investment24. For example, thalidomide, developed in 1957, was originally used as a sedative25. Later, it was found to be effective in patients with intermediate thalassemia26 and multiple xanthogranuloma in adults27. Given the high failure rate and high costs of new drug development, the reuse of “old” drugs to treat human diseases is becoming an attractive proposition. Some trials for the reuse of “old” drugs in glaucoma are underway. e.g., nicotinamide. Thus far, there has been no systematic analysis of drug reuse for glaucoma.
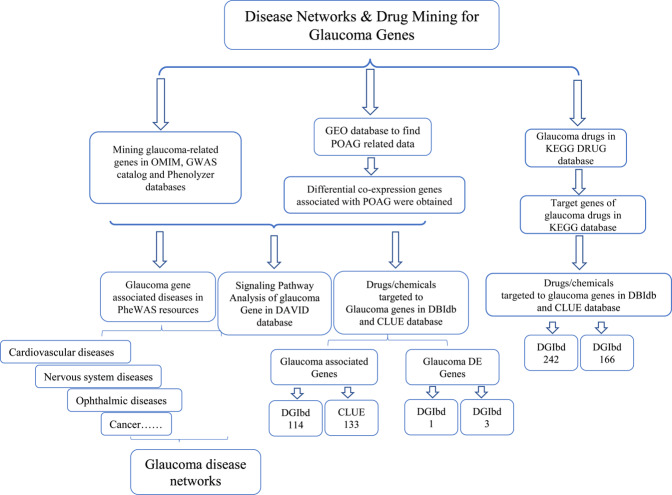
Given recent progress in genomics, it is now possible to rapidly identify and interpret genetic variations underlying a single disease in a single patient, thereby aiding individualized (tailored) drug therapy22. The aim of the present study was to provide new information for candidate drug development for glaucoma. The study design is shown in Fig. 1.
Figure 1.
The overall design of this study.
Methods
Gene mining for glaucoma
Information on glaucoma-associated/mutated genes was obtained from the GWAS Catalog28, OMIM29, Phenolyzer30, and published papers14. We searched the GEO database for human glaucoma-related tissues, optic papillary astrocytes, and the trabecular meshwork. In all the datasets, a large amount of data were available only for human optic nerve head astrocytes (GSE9963)31 and the trabecular meshwork (GSE27276)32. Lukas TJ et al. reported 1,204 differentially expressed genes in the optic nerve head of POAG31. Liu Y et al. reported 495 differentially expressed genes in the trabecular meshwork of POAG32. The genetic factors and DE genes were combined and considered glaucoma genes. In total, 1656 genes were identified for further analysis.
Glaucoma pathway analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID)33 is a bioinformatics data resource composed of an integrated biological knowledge base and analysis tools, which are used to extract meaningful biological information from a large number of gene and protein collections. The database collects and integrates a variety of gene identifiers. We used the DAVID for glaucoma pathway analysis.
Mining gene interactions
Cytoscape can be used in conjunction with large databases of protein–protein, protein–DNA, and genetic interactions to develop biomolecular interaction networks34. ClueGO is a user Cytoscape plug-in that analyzes interrelations of terms and functional groups in networks35. We used ClueGO embedded in Cytoscape 3.6 for the gene interaction analysis and selected only pathways with p values ≤ 0.05.
Disease association analysis
Phenome-wide association studies analyze many phenotypes compared to a single genetic variant (or other attribute). Such studies were originally based on electronic medical record (EMR) data from the EMR linked to the Vanderbilt DNA Biobank, BioVU. However, they can also be applied to other rich phenotype sets36. In the present study, the Phenome-wide Association Studies (PheWAS) database was used for glaucoma-related disease analysis.
Mining drugs/chemicals for glaucoma genes
The CLUE database, Drug-Gene Interaction database (DGIdb), and KEGG database were used for drug/chemical discovery. CLUE (LINCS) L100037 is a complete gene expression database with information on over 20,000 small-molecule compounds, gene overexpression, and gene knockouts. P values of each drug/chemical were calculated by the chi-squared test. An FDR < 0.05 was used for candidate drug/chemical filtering in this study. The DGIdb was set up by the University of Washington in 2013 to collect drug targets38. The updated version of the database contains 15 different gene–drug interaction sources, including DrugBank, TTD, and PharmGKB. The database provides detailed information on drugs and drug targets. This database generated drugs for each searched gene. Multigene target treatments are used for diseases such as cancers39 and cardiovascular disease40. Therefore, we calculated the p values of drugs targeted to the glaucoma gene list. Drugs with lower p values gain higher probabilities. The KEGG is a database resource that integrates genomic, biological, and functional information41. KEGG DRUG is a comprehensive drug information resource for approved drugs in Japan, the U.S., and Europe. It contains information on chemical structures and/or chemical components, therapeutic targets, metabolizing enzymes, and other molecular network information. We used KEGG DRUG to generate drugs for glaucoma disease and then generated genes for those drugs. After the genes were generated, we searched the drugs targeting these genes in CLUE and DGIdb.
Electronic medical records (EMRs)
To obtain information about the candidate drugs and glaucoma, we searched the EMR data of Sichuan Provincial People’s Hospital from August 2015 to August 2018 (N = 136,128) for the usage of the candidate drugs and the onset of glaucoma. For each drug usage, the number of total patients and the proportions of glaucoma patients were calculated. The study was approved by the institutional ethics committee of Sichuan Provincial People’s Hospital and was conducted according to the Declaration of Helsinki principles. Informed consent was obtained from the participants.
Results
Mining glaucoma genes
The search included mutation/association genes, as well as IOP genes for glaucoma genetic factors42–44. The search identified 159 genes in the OMIM database, 144 genes in the GWAS catalog (before 06/20/2017), and 86 genes in the Phenolyzer database. In total, 401 genes were obtained for further analysis as glaucoma genetic factors. In addition, 1,024 DE genes and 495 DE genes were identified in the POAG individuals in the GSE9963 and GSE27276 datasets, respectively. In total, 1,656 genes from the DE gene analysis corresponding to glaucoma were included in subsequent analysis.
Gene pathways for glaucoma
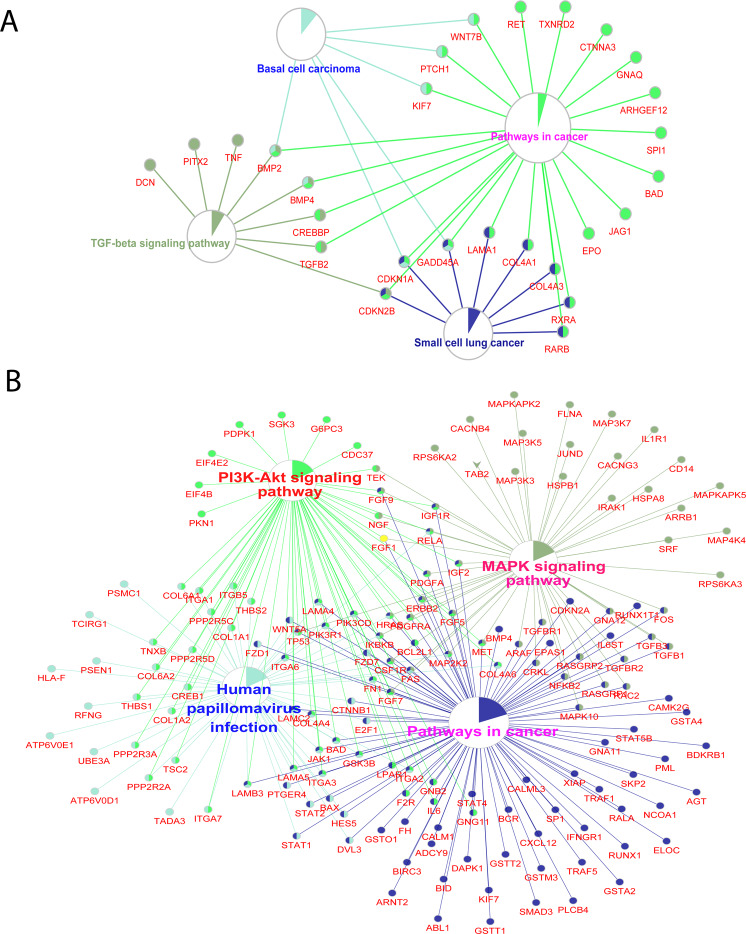
Among the 401 glaucoma genetic factors, 13 pathways were significantly enriched (FDR < 0.05), including pathways in cancer, focal adhesion, amoebiasis, the PI3K-Akt signaling pathway, and the TGF-beta signaling pathway (Table S1A). We used ClueGO35, a Cytoscape plug-in, for biological interpretation of the genetically associated genes for glaucoma (Fig. 2A).
Figure 2.
Gene interrelated graph of glaucoma genes. (A), Connected graph of genetic factors of glaucoma. (B), Connected graph of DE genes of glaucoma.
For 1,656 glaucoma DE genes, 53 pathways were significantly enriched (FDR < 0.05) (Table S1B), including pathways of Alzheimer’s disease (AD), non-alcoholic fatty liver disease, hypertrophic cardiomyopathy (HCM), arrhythmogenic right ventricular cardiomyopathy (ARVC), and dilated cardiomyopathy (Table 1). Using ClueGO, the interactions connected several pathways; the neurological degenerative diseases were tightly connected together (Fig. 2B).
Table 1.
Highly enriched pathways in glaucoma.
| KEGG_PATHWAY | Genes | P-Value | FDR (<0.05) |
|---|---|---|---|
| hsa04932: Nonalcoholic fatty liver disease (NAFLD) | BID, UQCRC2, NDUFB8, CYC1, TGFB1, NDUFS7, NDUFS5, MAP3K5, NDUFS8, FAS, PIK3R1, NDUFA4, NDUFA2, IL6, NDUFA4L2, RELA, PIK3CD, ADIPOR2, MAPK10, SDHA, SDHB, EIF2S1, GSK3B, BAX, COX6A2, IKBKB | 0.014658 | 3.42E-02 |
| hsa05410: Hypertrophic cardiomyopathy (HCM) | IL6, TNNC1, ITGA1, LMNA, TGFB3, ITGA2, ITGB5, CACNG3, ITGA3, CACNB4, TPM4, TGFB1, ITGA6, DMD, ITGA7, SGCA | 0.015053 | 3.42E-02 |
| hsa05414: Dilated cardiomyopathy | TNNC1, ITGA1, LMNA, TGFB3, ITGA2, ITGB5, CACNG3, ITGA3, CACNB4, TPM4, TGFB1, ITGA6, ADCY9, DMD, ITGA7, SGCA | 0.028256 | 4.73E-02 |
| hsa05010: Alzheimer’s disease | UQCRC2, ATP5D, BID, NDUFA4, NDUFA2, ADAM10, NDUFB8, NDUFA4L2, CYC1, ATP5G1, BAD, ATP5G3, NDUFS7, SDHA, SDHB, NDUFS5, PLCB4, PSEN1, CALML3, GSK3B, NDUFS8, COX6A2, ATP5C1, ADAM17, PSENEN, FAS, CALM1 | 0.028736 | 4.73E-02 |
| hsa05412: Arrhythmogenic right ventricular cardiomyopathy (ARVC) | ITGA6, DMD, ITGA7, LMNA, ITGA1, ITGB5, GJA1, ITGA2, CACNG3, ITGA3, CDH2, CACNB4, SGCA, CTNNB1 | 0.021189 | 3.96E-02 |
Glaucoma-related diseases
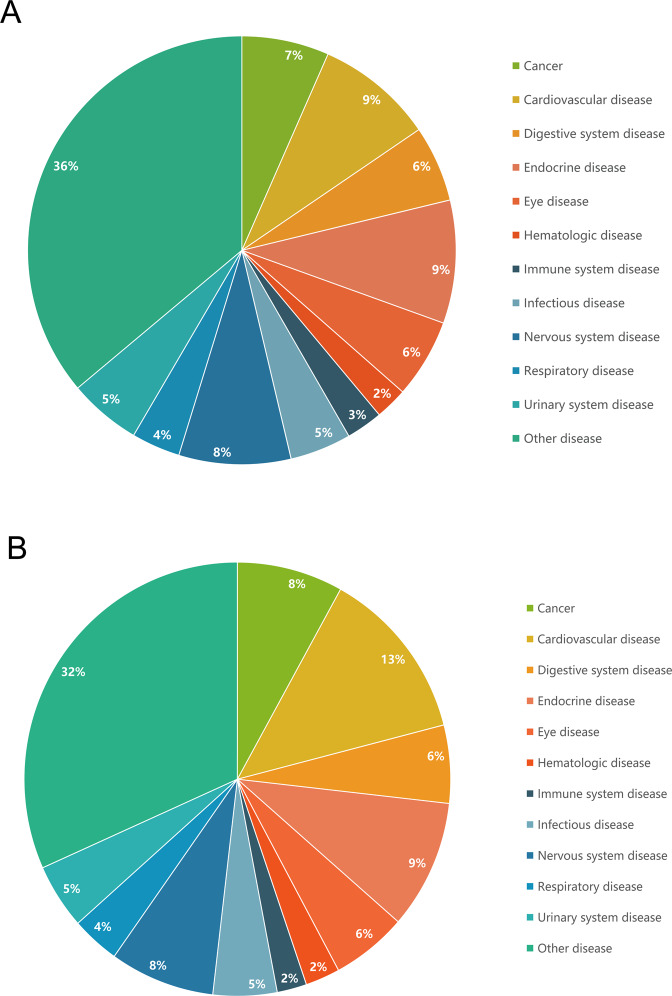
The search of the PheWAS database identified 1,289 diseases related to glaucoma genetic factors (Table S2). Pathways related to cardiovascular disease (13%), endocrine disease (9%), nervous system disease (8%), and eye disease (6%) were enriched in glaucoma-related genes (Fig. 3A). The database search revealed 1,357 diseases related to DE glaucoma genes (Table S3). The disease spectrum for these DE genes was very similar to the disease spectrum of glaucoma genetic factors (Fig. 3B). Cardiovascular disease (11%), endocrine disease (11%), nervous system disease (9%), and eye diseases (6%) were enriched in glaucoma DE genes.
Figure 3.
PheWAS diseases of glaucoma genes. (A), The percentage of diseases obtained from the PheWAS database of glaucoma genetic factors. (B), The percentage of diseases obtained from the PheWAS database of glaucoma DE genes.
Drug discovery for glaucoma genes
The search of the DGIdb identified 114 drugs with effects on glaucoma-related genes with an FDR < 0.05 (Table S4). The above results indicate that retinoic acid, cyclosporin, collagenase clostridium histolyticum, and taprazole have smaller P values. Of these candidate drugs, dorzolamide is used for antiglaucoma treatment45. The top 50 drugs are presented in Table 2.
Table 2.
The top 50 candidate drugs in the DGIdb targeted glaucoma genetic factors.
| No | Drug | P-value | FDR < 0.05 |
|---|---|---|---|
| 1 | Tretinoin | 2.06E-017 | 1.03E-015 |
| 2 | Ocriplasmin | 5.76E-12 | 6.1E-011 |
| 3 | Collagenase clostridium histolyticum | 2.44E-12 | 9.6E-011 |
| 4 | Talarozole | 2.87E-07 | 3.58E-06 |
| 5 | Bevacizumab | 1.46E-06 | 1.08E-04 |
| 6 | Tromethamine | 2.21E-05 | 1.98E-03 |
| 7 | Tipifarnib | 8.72E-05 | 4.27E-03 |
| 8 | Pumosetrag | 8.72E-05 | 4.27E-03 |
| 9 | Rg3487 | 8.72E-05 | 4.27E-03 |
| 10 | Granisetron | 8.72E-05 | 4.27E-03 |
| 11 | Mofarotene | 8.72E-05 | 4.27E-03 |
| 12 | Dorzolamide | 2.55E-04 | 9.18E-03 |
| 13 | Dolasetron | 2.55E-04 | 9.18E-03 |
| 14 | Adapalene | 2.55E-04 | 9.18E-03 |
| 15 | Alitretinoin | 2.55E-04 | 9.18E-03 |
| 16 | Pemetrexed disodium | 6.04E-04 | 2.04E-02 |
| 17 | Cerivastatin | 1.23E-03 | 3.31E-02 |
| 18 | Ascorbate | 1.12E-03 | 3.31E-02 |
| 19 | Chembl1188853 | 1.23E-03 | 3.31E-02 |
| 20 | Acitretin | 1.23E-03 | 3.31E-02 |
| 21 | Triamcinolone | 5.70E-03 | 3.74E-02 |
| 22 | Bupivacaine | 3.69E-03 | 3.74E-02 |
| 23 | Irinotecan | 5.27E-03 | 3.74E-02 |
| 24 | Denufosol tetrasodium | 7.48E-03 | 3.74E-02 |
| 25 | Emixustat hydrochloride | 7.48E-03 | 3.74E-02 |
| 26 | Tafamidis meglumine | 7.48E-03 | 3.74E-02 |
| 27 | Chembl1232343 | 7.48E-03 | 3.74E-02 |
| 28 | Chembl503075 | 7.48E-03 | 3.74E-02 |
| 29 | Chembl472925 | 7.48E-03 | 3.74E-02 |
| 30 | Chembl574602 | 7.48E-03 | 3.74E-02 |
| 31 | Chembl2064657 | 7.48E-03 | 3.74E-02 |
| 32 | Chembl475346 | 7.48E-03 | 3.74E-02 |
| 33 | Dexniguldipine | 7.48E-03 | 3.74E-02 |
| 34 | Chembl89093 | 7.48E-03 | 3.74E-02 |
| 35 | Chembl340868 | 7.48E-03 | 3.74E-02 |
| 36 | Chembl317382 | 7.48E-03 | 3.74E-02 |
| 37 | Chembl329722 | 7.48E-03 | 3.74E-02 |
| 38 | Chembl321845 | 7.48E-03 | 3.74E-02 |
| 39 | Chembl322524 | 7.48E-03 | 3.74E-02 |
| 40 | Chembl108705 | 7.48E-03 | 3.74E-02 |
| 41 | Chembl118044 | 7.48E-03 | 3.74E-02 |
| 42 | Chembl130499 | 7.48E-03 | 3.74E-02 |
| 43 | Chembl71053 | 7.48E-03 | 3.74E-02 |
| 44 | Chembl1208337 | 7.48E-03 | 3.74E-02 |
| 45 | Mifamurtide sodium hydrate | 7.48E-03 | 3.74E-02 |
| 46 | Celecoxib | 6.67E-03 | 3.74E-02 |
| 47 | Nms-1116354 | 7.48E-03 | 3.74E-02 |
| 48 | Rxdx-103 | 7.48E-03 | 3.74E-02 |
| 49 | Bms-863233 (chembl3544943) | 7.48E-03 | 3.74E-02 |
| 50 | Chembl179583 | 7.48E-03 | 3.74E-02 |
The search of the KEGG DRUG database returned 24 drugs for glaucoma. Then, we identified 13 target genes for the 24 drugs from the KEGG database. A search of the DGIdb revealed drugs targeting these genes (Table S5). The top 100 candidate drugs with antiglaucoma or therapeutic effects on glaucoma are listed in Table 3. Acetylcholinesterase inhibitors, alpha-adrenergic receptor antagonists, antihypertensive drugs, and antipsychotic drugs had higher frequencies.
Table 3.
Top 100 candidate drugs in the DGIdb targeted to KEGG DRUG-targeted genes.
| No | Drug | P values | FDR < 0.05 | No | Drug | P values | FDR < 0.05 |
|---|---|---|---|---|---|---|---|
| 1 | Choline | 0 | 0 | 51 | Mivacurium | 0 | 0 |
| 2 | Gallamine triethiodide | 0 | 0 | 52 | Oxyphencyclimine | 0 | 0 |
| 3 | Demecarium | 0 | 0 | 53 | Darifenacin | 0 | 0 |
| 4 | Physostigmine | 0 | 0 | 54 | Tridihexethyl | 0 | 0 |
| 5 | Ambenonium | 0 | 0 | 55 | Benzquinamide | 0 | 0 |
| 6 | Tubocurarine | 0 | 0 | 56 | Brompheniramine | 0 | 0 |
| 7 | Decamethonium | 0 | 0 | 57 | Tolterodine | 0 | 0 |
| 8 | Echothiophate | 0 | 0 | 58 | Pilocarpine | 0 | 0 |
| 9 | Neostigmine methylsulfate | 0 | 0 | 59 | Pipecuronium | 0 | 0 |
| 10 | Hexafluorenium bromide | 0 | 0 | 60 | Fesoterodine | 0 | 0 |
| 11 | Pyridostigmine bromide | 0 | 0 | 61 | Aclidinium | 0 | 0 |
| 12 | Tacrine hydrochloride | 0 | 0 | 62 | Umeclidinium | 0 | 0 |
| 13 | Rivastigmine | 0 | 0 | 63 | Acetylcholine | 0 | 0 |
| 14 | Donepezil | 0 | 0 | 64 | Bethanechol | 0 | 0 |
| 15 | Dipivefrin | 0 | 0 | 65 | Xanomeline | 0 | 0 |
| 16 | Aripiprazole | 0 | 0 | 66 | Amitriptyline | 0 | 0 |
| 17 | Olanzapine | 0 | 0 | 67 | Benztropine mesylate | 0 | 0 |
| 18 | Loxapine | 0 | 0 | 68 | Atropine | 0 | 0 |
| 19 | Promazine | 0 | 0 | 69 | Biperiden (chembl1101) | 0 | 0 |
| 20 | Propiomazine | 0 | 0 | 70 | Ipratropium | 0 | 0 |
| 21 | Carvedilol | 0 | 0 | 71 | Oxybutynin | 0 | 0 |
| 22 | Dronedarone | 0 | 0 | 72 | Propantheline | 0 | 0 |
| 23 | Epinephrine bitartrate | 0 | 0 | 73 | Solifenacin | 0 | 0 |
| 24 | Phenylephrine | 0 | 0 | 74 | Clidinium | 0 | 0 |
| 25 | Doxazosin | 0 | 0 | 75 | Tiotropium (chembl1900528) | 0 | 0 |
| 26 | Terazosin | 0 | 0 | 76 | Norepinephrine | 0 | 0 |
| 27 | Tamsulosin | 0 | 0 | 77 | Cabergoline | 0 | 0 |
| 28 | Fenoldopam | 0 | 0 | 78 | Clozapine | 0 | 0 |
| 29 | Bethanidine | 0 | 0 | 79 | Levomepromazine | 0 | 0 |
| 30 | Labetalol | 0 | 0 | 80 | Droxidopa | 0 | 0 |
| 31 | Metoprolol succinate | 0 | 0 | 81 | Noradrenaline | 0 | 0 |
| 32 | Isoetharine | 0 | 0 | 82 | Hydroxyamphetamine hydrobromide | 0 | 0 |
| 33 | Propranolol | 0 | 0 | 83 | Batefenterol | 0 | 0 |
| 34 | Pirbuterol | 0 | 0 | 84 | Epinephrine | 0 | 0 |
| 35 | Betaxolol | 0 | 0 | 85 | Galantamine hydrobromide | 6.83E-07 | 5.96E-06 |
| 36 | Dobutamine | 0 | 0 | 86 | Edrophonium chloride | 1.33E-06 | 5.96E-06 |
| 37 | Oxprenolol hydrochloride | 0 | 0 | 87 | Promethazine | 5.29E-07 | 5.96E-06 |
| 38 | Metipranolol hydrochloride | 0 | 0 | 88 | Thiethylperazine | 5.80E-07 | 5.96E-06 |
| 39 | Penbutolol sulfate | 0 | 0 | 89 | Doxepin | 6.35E-07 | 5.96E-06 |
| 40 | Sotalol hydrochloride | 0 | 0 | 90 | Neostigmine | 2.43E-06 | 1.18E-05 |
| 41 | Timolol maleate | 0 | 0 | 91 | Isoflurophate | 2.72E-06 | 1.66E-05 |
| 42 | Nebivolol hydrochloride | 0 | 0 | 92 | Malathion | 3.36E-06 | 1.66E-05 |
| 43 | Levobunolol hydrochloride | 0 | 0 | 93 | Labetalol hydrochloride | 2.66E-06 | 1.66E-05 |
| 44 | Isoproterenol hydrochloride | 0 | 0 | 94 | Mephentermine sulfate | 2.82E-06 | 1.66E-05 |
| 45 | Esmolol hydrochloride | 0 | 0 | 95 | Carvedilol phosphate | 3.36E-06 | 1.66E-05 |
| 46 | Propafenone hydrochloride | 0 | 0 | 96 | Dipivefrin hydrochloride | 3.46E-06 | 1.66E-05 |
| 47 | Desipramine | 0 | 0 | 97 | Phenserine | 7.43E-06 | 3.82E-05 |
| 48 | Isoproterenol | 0 | 0 | 98 | Itopride | 1.07E-05 | 5.95E-05 |
| 49 | Carteolol | 0 | 0 | 99 | Dapiprazole | 1.34E-05 | 6.96E-05 |
| 50 | Glycopyrrolate bromide | 0 | 0 | 100 | Risperidone | 1.50E-05 | 7.95E-05 |
The search of the CLUE database for glaucoma genetic factors returned 133 drugs/chemicals with an FDR < 0.05 (Table S6). The top 50 candidate drugs/chemicals for glaucoma are listed in Table 4. Most of the drugs identified in the search were antitumor, antihypertension, and analgesic drugs. In the CLUE database, 3 candidate gene-matched drugs/chemicals with an FDR < 0.05 were returned by searching for glaucoma DE genes (Table 5).
Table 4.
Top 50 candidate drugs/chemicals in the CLUE database targeted to glaucoma genetic factors.
| Order | Drug | FDR < 0.05 | Order | Drug | FDR < 0.05 |
|---|---|---|---|---|---|
| 1 | Alitretinoin | 0 | 26 | MRS-1220 | 8E-08 |
| 2 | Acitretin | 0 | 27 | BFL-50481 | 8E-08 |
| 3 | Linifanib | 0 | 28 | Apoptosis-activator-Ii | 8E-08 |
| 4 | Catechin | 0 | 29 | Flavanone | 8E-08 |
| 5 | Pyrazinamide | 0 | 30 | Sa-792709 | 8E-08 |
| 6 | 10h-Phenothiazin-10-Yl)(P-tolyl)methanone | 0 | 31 | Carbenoxolone | 2.1E-07 |
| 7 | Importazole | 0 | 32 | Hydroxycholesterol | 3.49E-06 |
| 8 | Meptazinol | 0 | 33 | Deoxycholic acid | 3.49E-06 |
| 9 | Le-135 | 0 | 34 | Withaferin-A | 3.49E-06 |
| 10 | Carvedilol | 1E-08 | 35 | Valdecoxib | 3.49E-06 |
| 11 | Retinol | 3E-08 | 36 | Fostamatinib | 3.49E-06 |
| 12 | Pyridine-2-Aldoxime | 8E-08 | 37 | Rita | 3.49E-06 |
| 13 | Mestinon | 8E-08 | 38 | TTNPB | 3.49E-06 |
| 14 | Edrophonium | 8E-08 | 39 | Psb-11 | 3.49E-06 |
| 15 | Tolcapone | 8E-08 | 40 | Probucol | 3.49E-06 |
| 16 | Tamibarotene | 8E-08 | 41 | Pp-1 | 3.49E-06 |
| 17 | Tak-715 | 8E-08 | 42 | Gw-9662 | 3.49E-06 |
| 18 | BCI-Hydrochloride | 8E-08 | 43 | Eriodictyol | 3.49E-06 |
| 19 | Celastrol | 8E-08 | 44 | Umbelliferone | 3.49E-06 |
| 20 | Physostigmine | 8E-08 | 45 | Rofecoxib | 3.49E-06 |
| 21 | AZD-7762 | 8E-08 | 46 | Paxilline | 3.49E-06 |
| 22 | Tacrine | 8E-08 | 47 | PSB-1115 | 3.49E-06 |
| 23 | CAY-10470 | 8E-08 | 48 | Gw-3965 | 3.49E-06 |
| 24 | Cytarabine | 8E-08 | 49 | Flufenamic acid | 6.44E-06 |
| 25 | Ac-55649 | 8E-08 | 50 | Caffeine | 2.44E-05 |
Table 5.
Candidate chemicals targeted to DE genes in the CLUE database.
| Name | P values | FDR < 0.05 |
|---|---|---|
| Pentoxifylline | 3.29E-03 | 4.28E-02 |
| Parthenolide | 2.90E-04 | 3.77E-03 |
| W-13 | 2.90E-04 | 3.77E-03 |
For the 13 KEGG glaucoma drug-targeted genes, 166 drugs/chemicals were enriched in the CLUE database with an FDR < 0.05 (Table S7), including carteolol, betaxolol and latanoprost. Among these, propranolol derivatives were the most enriched chemical. The US FDA approved a new glaucoma drug, netarsudil/latanoprost (Rocklatan) (Aerie Pharmaceuticals, the US), in March 2019. The drug consisted of an ophthalmic solution (0.02%/0.005%) (Table S7). The top 25 selected candidate drugs/chemicals for glaucoma are listed in Table 6.
Table 6.
Top 25 drugs in the CLUE database for KEGG DRUG targeted genes.
| Order | Drug | FDR < 0.05 |
|---|---|---|
| 1 | Diphemanil | 4.31E-05 |
| 2 | Ethoprop | 4.31E-05 |
| 3 | Velnacrine | 4.31E-05 |
| 4 | Isoxsuprine | 4.31E-05 |
| 5 | Propentofylline | 4.31E-05 |
| 6 | Practolol | 4.31E-05 |
| 7 | Zamifenacin | 4.31E-05 |
| 8 | Harpagoside | 4.31E-05 |
| 9 | J-104129 | 4.31E-05 |
| 10 | Meptazinol | 4.31E-05 |
| 11 | Buphenine | 4.31E-05 |
| 12 | Procaterol | 4.31E-05 |
| 13 | Ritodrine | 4.31E-05 |
| 14 | Salmeterol | 4.31E-05 |
| 15 | 10H-Phenothiazin-10-Yl)(P-tolyl)methanone | 4.31E-05 |
| 16 | BRD-K66896231 | 4.31E-05 |
| 17 | Huperzine-A | 4.31E-05 |
| 18 | Esmolol | 4.31E-05 |
| 19 | Latanoprost | 4.31E-05 |
| 20 | Orciprenaline | 4.31E-05 |
| 21 | Terbutaline | 4.31E-05 |
| 22 | Bisoprolol | 4.31E-05 |
| 23 | Clebopride | 4.31E-05 |
| 24 | Desoxypeganine | 4.31E-05 |
| 25 | Donepezil | 4.31E-05 |
The analysis of 136,128 EMR histories revealed nine candidate drugs of all the mined glaucoma drugs mentioned above, which were used in Sichuan Provincial People’s Hospital from August 2015 to August 2018 (Table 7). Of 435 patients treated with cytarabine (242 of whom were older than 40 years), none of the patients had glaucoma. The prevalence of glaucoma was 0.11% in theophylline-treated patients (N = 4,594), 0% in nicardipine-treated patients (N = 564), 0.058% in celecoxib-treated patients (N = 1,488), and 0.035% in nicardipine-treated patients (N = 564). The incidence of glaucoma was significantly lower in these drug-use cohorts than in healthy individuals (1% in those aged older than 40 years). Thus, these drugs may have antiglaucoma effects. Among 1,293 hospitalized AD patients, 48 (3.8%) patients had glaucoma. This was significantly higher than the prevalence rate of glaucoma in the healthy population, suggesting that the incidence of glaucoma may be elevated in individuals with AD (P = 7.99E-05 assuming 1000 samples, OR = 3.9). Finally, by P-value and FDR ranking, we selected 40 drugs/chemicals most likely to prevent or treat glaucoma, and we selected 40 most likely drugs/chemicals for the prevention or treatment of glaucoma (Table 8).
Table 7.
Candidate drug usage and the onset of glaucoma in Sichuan Provincial People’s Hospital in 136,128 electronic medical records (EMRs) from August 2015 to August 2018.
| Drug | Repetitions | Effect | Cases | Glaucoma Patients | Glaucoma Prevalence | Cases (age > 40) | Glaucoma Patients (age > 40) | Pa | ORb |
|---|---|---|---|---|---|---|---|---|---|
| Cytarabine | 6 | Anticancer, antimetabolism, antiviral, DNA polymerase inhibitor | 435 | 0 | 0 | 242 | 0 | 4.30E-03 | 0 |
| Caffeine | 6 | Doping (central), adenosine receptor antagonist, phosphodiesterase inhibitor | 3 | 0 | 0 | 0 | 0 | \ | \ |
| Dipyridamole | 6 | Vasodilators (coronary arteries), platelet aggregation inhibitors, phosphodiesterase inhibitors | 46 | 0 | 0 | 15 | 0 | 4.30E-03 | 0 |
| Paclitaxel | 5 | Antineoplastic, tubulin depolymerization inhibitor | 634 | 0 | 0 | 573 | 0 | 4.30E-03 | 0 |
| Dasatinib | 5 | Antineoplastic, tyrosine kinase inhibitors | 6 | 0 | 0 | 2 | 0 | 4.30E-03 | 0 |
| Celecoxib | 5 | Analgesic, anti-inflammatory, COX-2 inhibitors | 1719 | 1 | 5.82E-04 | 1488 | 1 | 1.74E-03 | 0.067 |
| Theophylline | 6 | Bronchodilator, phosphodiesterase inhibitor | 4594 | 5 | 1.09E-03 | 4397 | 5 | 7.60E-06 | 0.113 |
| Aspirin | 5 | Analgesic, anti-inflammatory, antipyretic, antirheumatic, antiplatelet, COX inhibitors | 5358 | 16 | 2.99E-03 | 5197 | 13 | 9.80E-04 | 0.248 |
| Nicardipine | 6 | Antihypertensive, vasodilators, calcium channel blockers | 588 | 2 | 3.40E-03 | 564 | 2 | 8.30E-02 | 0.35 |
We found 9 drugs used in this hospital. a, P values for each drug assuming we had 1000 participants who used the drugs and 1000 participants in the healthy population age > 40 when the investigated samples were less than 1000. b, OR: odds ratio.
Table 8.
Forty most likely candidate drugs for glaucoma.
| Name | Medical uses | FDR |
|---|---|---|
| Alitretinoin | Chronic hand eczema (1) | 2.41E-03 |
| Vinblastine | Interference with tubulin (2) | 6.76E-05 |
| Paclitaxel | Non-small-cell lung cancer (3) | 2.41E-03 |
| Meptazinol | Analgesia (4) | 4.31E-05 |
| Caffeine | Neurodegenerative diseases (5) | 6.00E-07 |
| Auranofin | Rheumatoid arthritis (6) | 0 |
| Cilomilast | Asthma and chronic obstructive pulmonary disease (7) | 5.71E-04 |
| Dipyridamole | Antithrombosis (8) | 5.71E-04 |
| Loperamide | Acute infectious diarrhea (9) | 1.23E-03 |
| Ketotifen | Allergic diseases (10), chronic urticaria (11) | 2.41E-03 |
| Tipifarnib | Prevents hypoxia-induced pulmonary hypertension (12), tumor (13) | 0 |
| Pentoxifylline | Venous leg ulcer (14) | 0 |
| Aspirin | Antitumor (15), pain (16) | 0 |
| Sulfasalazine | Rheumatoid arthritis (17) | 4.00E-07 |
| Docetaxel | Breast cancer (18) | 2.27E-04 |
| Tretinoin | Acne vulgaris, keratosis pilaris, acute promyelocytic and leukemia (19) | 2.41E-03 |
| Celecoxib | Osteoarthritis, rheumatoid arthritis and ankylosing spondylitis (20) | 3.74E-02 |
| Retinol | Reduces symptoms of skin aging (21) | 3E-08 |
| Carvedilol | Systemic hypertension and myocardial dysfunction (22) | 1E-08 |
| Pyrazinamide | Tuberculosis (23) | 0 |
| Acitretin | Pediatric psoriasis (24) | 0 |
| Linifanib | Cancer (25) | 0 |
| Cytarabine | Acute myeloid leukemia (26) | 8E-08 |
| Sulpiride | Schizophrenia (27) | 1.41E-04 |
| Ocriplasmin | Symptomatic vitreomacular adhesion (28) | 0 |
| Bevacizumab | Recurrent glioblastoma (29) | 1.08E-04 |
| Dexamethasone | Acute spinal cord injury (30) | 0 |
| Cyclosporine | Transplant rejection (31) | 1.454-04 |
| Everolimus | Tumors (32) | 4.31E-02 |
| Fluorouracil | Tumors (33) | 2.23E-04 |
| Tamoxifen | Breast cancer (34) | 1.08E-04 |
| Etretinate | Psoriasis and many other skin diseases (35) | 0 |
| Trametinib | Melanoma (36) | 2.94E-04 |
| Selumetinib | Advanced/metastatic non-small-cell lung cancer (37) | 3.75E-04 |
| Sirolimus | Prevention of transplant rejection (38) | 1.45E-04 |
| Dasatinib | Chronic myeloid leukemia (39) | 5.98E-04 |
| Gemcitabine | Cancer (40) | 2.94E-04 |
| Cetuximab | Cancer (41) | 2.51E-04 |
| Arsenic trioxide | Acute promyelocytic leukemia (42) | 5.43E-04 |
| Labetalol | Hypertension (43) | 0 |
Discussion
Glaucoma is a set of disorders that cause damage to the optic nerve and worsen over time. Pathological ocular hypertension, race, a family history of glaucoma, vasospasms, and peripheral vascular disease are common contributing factors46. Current drug treatments for glaucoma are mainly directed toward lowering IOP. Previous research reported that various pathways, including focal adhesion, extracellular matrix–receptor interaction, cancer, and the PI3K-Akt pathway, were significantly related to IOP43, pointing to its complex etiology. Current glaucoma medications reduce IOP by reducing the production of fluid in the eye or increasing its outflow.
In the present study, the pathway analysis identified several nervous system disorders, including AD, that were strongly associated with glaucoma. Glaucoma is part of a set of age-related neurodegenerative diseases47. AD is the most common neurodegenerative disease among elderly individuals. Previous studies have demonstrated that AD and glaucoma share several biological characteristics48,49. Signaling pathways such as dilated cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy (ARVC), and hypertrophic cardiomyopathy (HCM) are enriched, suggesting that glaucoma may be associated with signaling pathways in cardiovascular disease.
In the present study, we presented an enhanced and updated perspective on glaucoma-related genes, associated diseases, and drugs targeting these diseases (Fig. 1). The analysis of diseases linked to glaucoma genes showed that cardiovascular diseases were most closely associated with glaucoma50. Previous research pointed to an abnormal hemorheological pattern in glaucoma patients, with increased plasma viscosity leading to the hypoperfusion of the ophthalmic artery, which can potentially aggravate optic nerve injury51. Other research has suggested that peripheral vascular endothelial dysfunction may be related to the progression of glaucoma52. High blood viscosity plays a role in the occurrence of glaucoma53. A previous study showed that when the shear rate of retinitis decreased and the blood viscosity increased, the low perfusion of retinal blood flow can lead to local ischemia and aggravate vision atrophy54. The hemodynamics of the ophthalmic artery and central retinal artery are correlated with POAG55.
From a drug discovery standpoint, the identification of glaucoma genes provides valuable information to reveal its causative mechanisms and drugs targeting this disorder. In the CLUE database, carbonic anhydrase inhibitors acetazolamide and dipivefrin had antiglaucoma effects56. The results of the present study suggested that anticancer drugs, analgesics, and antihypertension drugs may have potential in the prevention and treatment of glaucoma. Potential candidate drugs identified in the search of the CLUE database included propranolol derivatives, which are nonselective beta-1 and beta-2 adrenergic receptor blockers. Propranolol derivatives have similar pharmacological effects to those of currently used glaucoma drugs and have potential for drug development as antiglaucoma agents. In the DGIdb, celecoxib, paclitaxel, and cyclosporine appeared frequently, suggesting that antineoplastic drugs may represent a new direction for glaucoma drug screening57.
Screening of the KEGG database, DGIdb database, and CLUE database identified α1/β adrenergic receptor antagonists. This antagonist can reduce cyclic adenosine phosphate in ciliary epithelial cells. Alpha 1/beta adrenergic receptor antagonists might have potential in reducing not only the generation of aqueous humor but also the outflow of aqueous humor through the trabecular meshwork. Carvedilol, a nonselective beta-adrenergic receptor blocker (beta 1 and beta 2) and alpha-adrenergic receptor blocker (alpha 1), is currently used for the treatment of hypertension58.
This study has two limitations. First, although we discovered many candidate drugs for glaucoma and provided gene–drug pair information in the supplementary data, it was based on the mixed forms of glaucoma (most genes are involved in POAG). Because different glaucoma types are very different diseases and should have different gene involvement and drug targets, the readers should refer to the gene of a subtype of glaucoma to find the proper candidate drugs in the supplementary data. Second, although we performed statistical analysis of significance, further experiments are still needed for the verification of the treatment of glaucoma.
In summary, we investigated genetic factors and DE genes in glaucoma. We interpreted the pathways of these glaucoma genes and systematically investigated diseases related to glaucoma genes. In this study, we screened the top drug candidates for glaucoma, such as tretinoin, ocriplasmin, collagenase clostridium histolyticum, Talarozole and bevacizumab. Tretinoin is also known as all-trans retinoic acid. Talarozole is a systemic all-trans retinoic acid metabolism blocking agent that increases intracellular levels of endogenous all-trans retinoic acid. Tretinoin is an intermediate product of vitamin A metabolism in the body. Vitamin A is known for its function in the retina with importance for rhodopsin visual phototransduction, and it protects against free radicals, i.e., it acts as an antioxidant59. Regarding dietary intake of retinol equivalents, two large studies reported a protective effect on POAG59–61. Ocriplasmin is a recombinant protease with activity against fibronectin and laminin, components of the vitreoretinal interface, and may lower IOP by degrading vitreous or connective tissue. McClintock et al. reported the case of a glaucoma patient who received a single intravitreal injection of 125 µg ocriplasmin for vitreomacular traction in the right eye. Its final visual acuity was 20/50 + , and IOP was 18 mmHg at 16 weeks after surgery, with IOP reduction and serous choroidal effusion after ocriplasmin injection62. Collagenase clostridium histolyticum is an enzyme produced by the bacterium Clostridium histolyticum that dismantles collagen. The collagen matrix is the main structure of the trabecular meshwork, which plays an important role in high-tension glaucoma63. Collagenase clostridium histolyticum had a drug effect that may lower IOP by degrading adhesive collagens in the hole of the trabecular meshwork. Bevacizumab is a monoclonal antibody developed against vascular endothelial growth factor (VEGF). It is used for neovascular glaucoma64 and for reducing glaucoma surgical scars65. Of these drug candidates, we still need more mechanistic studies in the future. Subsequently, we mined drugs/chemicals targeting glaucoma genes. In addition, we analyzed the usage of candidate drugs and the onset of glaucoma in clinical EMRs. Finally, we selected the 40 most likely candidate drugs for the prevention and treatment of glaucoma. The results provide a systematic interpretation of glaucoma-related genes, diseases, and candidate drugs. Our research provides comprehensive data that can enrich the understanding of glaucoma and potential glaucoma drugs.
URLs
GWAS Catalog:https://www.ebi.ac.uk/gwas/;
Online Mendelian Inheritance in Man® (OMIM®): https://www.omim.org/;
GEO: https://www.ncbi.nlm.nih.gov/geo/;
DAVID: Bioinformatics Resources: https://david.ncifcrf.gov/;
PheWAS: https://phewascatalog.org/;
CLUE: https://clue.io;
DGIdb: www.dgidb.org.
Supplementary information
Acknowledgements
This research project was supported by the National Natural Science Foundation of China (81970839 (L.H.), 81670895 (L.H.) and 81300802 (L.H.)) and the Department of Science and Technology of Sichuan Province, China (2017JQ0024 (L.H.), 2016HH0072 (L.H.), 2015JQO057 (L.H.) and 2013JY0195 (L.H.)).
Author contributions
L.H. designed the study. H.W., Y.D., and L.W. performed the statistical analysis. H.W. and L.H. wrote the manuscript. All of the authors critically revised and provided final approval for this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-66350-w.
References
- 1.Weinreb RN, Khaw PT. Primary open-angle glaucoma. Lancet. 2004;363:1711–1720. doi: 10.1016/S0140-6736(04)16257-0. [DOI] [PubMed] [Google Scholar]
- 2.He S, Stankowska DL, Ellis DZ, Krishnamoorthy RR, Yorio T. Targets of Neuroprotection in Glaucoma. J Ocul Pharmacol Ther. 2018;34:85–106. doi: 10.1089/jop.2017.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta D, Chen PP. Glaucoma. American family physician. 2016;93:668–674. [PubMed] [Google Scholar]
- 4.Weinreb RN, et al. Primary open-angle glaucoma. Nat Rev Dis Primers. 2016;2:16067. doi: 10.1038/nrdp.2016.67. [DOI] [PubMed] [Google Scholar]
- 5.Dias DT, et al. Eyes with Suspicious Appearance of the Optic Disc and Normal Intraocular Pressure: Using Clinical and Epidemiological Characteristics to Differentiate Those with and without Glaucoma. PLoS One. 2016;11:e0158983. doi: 10.1371/journal.pone.0158983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sng CC, Ang M, Barton K. Central corneal thickness in glaucoma. Curr Opin Ophthalmol. 2017;28:120–126. doi: 10.1097/ICU.0000000000000335. [DOI] [PubMed] [Google Scholar]
- 7.Tham YC, et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Yao YH, et al. A recurrent G367R mutation in MYOC associated with juvenile open angle glaucoma in a large Chinese family. Int J Ophthalmol. 2018;11:369–374. doi: 10.18240/ijo.2018.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nature genetics. 2014;46:1115–1119. doi: 10.1038/ng.3078. [DOI] [PubMed] [Google Scholar]
- 10.Nunes HF, et al. Investigation of CAV1/CAV2 rs4236601 and CDKN2B-AS1 rs2157719 in primary open-angle glaucoma patients from Brazil. Ophthalmic Genet. 2018;39:194–199. doi: 10.1080/13816810.2017.1393830. [DOI] [PubMed] [Google Scholar]
- 11.Zanon-Moreno, V. et al. A Multi-Locus Genetic Risk Score for Primary Open-Angle Glaucoma (POAG) Variants Is Associated with POAG Risk in a Mediterranean Population: Inverse Correlations with Plasma Vitamin C and E Concentrations. Int J Mol Sci18 (2017). [DOI] [PMC free article] [PubMed]
- 12.Chidlow G, et al. Ocular expression and distribution of products of the POAG-associated chromosome 9p21 gene region. PLoS One. 2013;8:e75067. doi: 10.1371/journal.pone.0075067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sang J, et al. Association of three single nucleotide polymorphisms at the SIX1-SIX6 locus with primary open angle glaucoma in the Chinese population. Sci China Life Sci. 2016;59:694–699. doi: 10.1007/s11427-016-5073-y. [DOI] [PubMed] [Google Scholar]
- 14.Wiggs JL, Pasquale LR. Genetics of glaucoma. Hum Mol Genet. 2017;26:R21–R27. doi: 10.1093/hmg/ddx184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao D, et al. CDKN2B polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados, West Indies. PLoS One. 2012;7:e39278. doi: 10.1371/journal.pone.0039278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebmann JM, Lee JK. Current therapeutic options and treatments in development for the management of primary open-angle glaucoma. Am J Manag Care. 2017;23:S279–S292. [PubMed] [Google Scholar]
- 17.Cheema A, Chang RT, Shrivastava A, Singh K. Update on the Medical Treatment of Primary Open-Angle Glaucoma. Asia Pac. J Ophthalmol (Phila) 2016;5:51–58. doi: 10.1097/APO.0000000000000181. [DOI] [PubMed] [Google Scholar]
- 18.Chen YY, et al. Long-term intraocular pressure fluctuation of primary angle closure disease following laser peripheral iridotomy/iridoplasty. Chin Med J (Engl) 2011;124:3066–3069. [PubMed] [Google Scholar]
- 19.McAlinden C. Selective laser trabeculoplasty (SLT) vs other treatment modalities for glaucoma: systematic review. Eye (Lond) 2014;28:249–258. doi: 10.1038/eye.2013.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saheb H. & Ahmed, II. Micro-invasive glaucoma surgery: current perspectives and future directions. Curr Opin Ophthalmol. 2012;23:96–104. doi: 10.1097/ICU.0b013e32834ff1e7. [DOI] [PubMed] [Google Scholar]
- 21.Pushpakom, S. et al. Drug repurposing: progress, challenges and recommendations. Nature reviews. Drug discovery (2018). [DOI] [PubMed]
- 22.Dugger SA, Platt A, Goldstein DB. Drug development in the era of precision medicine. Nature reviews. Drug discovery. 2018;17:183–196. doi: 10.1038/nrd.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakravarti T. The Association of Socioeconomic Status with Severity of Glaucoma and the Impacts of Both Factors on the Costs of Glaucoma Medications: A Cross-Sectional Study in West Bengal, India. J Ocul Pharmacol Ther. 2018;34:442–451. doi: 10.1089/jop.2017.0135. [DOI] [PubMed] [Google Scholar]
- 24.Pammolli F, Magazzini L, Riccaboni M. The productivity crisis in pharmaceutical R&D. Nature reviews. Drug discovery. 2011;10:428–438. doi: 10.1038/nrd3405. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, Ando H, Handa H. Teratogenic effects of thalidomide: molecular mechanisms. Cell Mol Life Sci. 2011;68:1569–1579. doi: 10.1007/s00018-010-0619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, et al. Thalidomide has a significant effect in patients with thalassemia intermedia. Hematology. 2018;23:50–54. doi: 10.1080/10245332.2017.1354427. [DOI] [PubMed] [Google Scholar]
- 27.Debois D, Marot L, Andre M, Dachelet C. Thalidomide as an effective treatment for adult multiple xanthogranuloma. JAAD Case Rep. 2018;4:896–898. doi: 10.1016/j.jdcr.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes B. Overview of Statistical Methods for Genome-Wide Association Studies (GWAS) Methods Mol Biol. 2013;1019:149–169. doi: 10.1007/978-1-62703-447-0_6. [DOI] [PubMed] [Google Scholar]
- 29.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic acids research. 2015;43:D789–798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nature methods. 2015;12:841–843. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukas TJ, et al. Susceptibility to glaucoma: differential comparison of the astrocyte transcriptome from glaucomatous African American and Caucasian American donors. Genome biology. 2008;9:R111. doi: 10.1186/gb-2008-9-7-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, et al. Gene expression profile in human trabecular meshwork from patients with primary open-angle glaucoma. Investigative ophthalmology & visual science. 2013;54:6382–6389. doi: 10.1167/iovs.13-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang DW, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research. 2007;35:W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan Q, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic acids research. 2014;42:W449–460. doi: 10.1093/nar/gku476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cotto KC, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic acids research. 2018;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang C, Lin X, Zhao Z. Applications of CRISPR/Cas9 Technology in the Treatment of Lung Cancer. Trends Mol Med. 2019;25:1039–1049. doi: 10.1016/j.molmed.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Khodadi, E. Platelet Function in Cardiovascular Disease: Activation of Molecules and Activation by Molecules. Cardiovasc Toxicol (2019). [DOI] [PubMed]
- 41.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khawaja AP, et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet. 2018;50:778–782. doi: 10.1038/s41588-018-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang L, et al. Genome-wide analysis identified 17 new loci influencing intraocular pressure in Chinese population. Science China. Life sciences. 2019;62:153–164. doi: 10.1007/s11427-018-9430-2. [DOI] [PubMed] [Google Scholar]
- 44.MacGregor S, et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nature genetics. 2018;50:1067–1071. doi: 10.1038/s41588-018-0176-y. [DOI] [PubMed] [Google Scholar]
- 45.Fechtner RD, Realini T. Fixed combinations of topical glaucoma medications. Curr Opin Ophthalmol. 2004;15:132–135. doi: 10.1097/00055735-200404000-00013. [DOI] [PubMed] [Google Scholar]
- 46.Manalastas PIC, et al. Reproducibility of Optical Coherence Tomography Angiography Macular and Optic Nerve Head Vascular Density in Glaucoma and Healthy Eyes. J Glaucoma. 2017;26:851–859. doi: 10.1097/IJG.0000000000000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson LN. Glaucoma as a Neurodegenerative Disease: Why We Must ‘Look for the Protein’. R I Med J (2013) 2016;99:18–21. [PubMed] [Google Scholar]
- 48.Lin IC, et al. Glaucoma, Alzheimer’s disease, and Parkinson’s disease: an 8-year population-based follow-up study. PLoS One. 2014;9:e108938. doi: 10.1371/journal.pone.0108938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mancino R, et al. Glaucoma and Alzheimer Disease: One Age-Related Neurodegenerative Disease of the Brain. Curr Neuropharmacol. 2018;16:971–977. doi: 10.2174/1570159X16666171206144045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aggarwal M, Aggarwal B, Rao J. Integrative Medicine for Cardiovascular Disease and Prevention. Med Clin North Am. 2017;101:895–923. doi: 10.1016/j.mcna.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Binggeli T, Schoetzau A, Konieczka K. In glaucoma patients, low blood pressure is accompanied by vascular dysregulation. EPMA J. 2018;9:387–391. doi: 10.1007/s13167-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu CH, et al. Association Between Peripheral Vascular Endothelial Function and Progression of Open-Angle Glaucoma. Medicine (Baltimore) 2016;95:e3055. doi: 10.1097/MD.0000000000003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klaver JH, Greve EL, Goslinga H, Geijssen HC, Heuvelmans JH. Blood and plasma viscosity measurements in patients with glaucoma. Br J Ophthalmol. 1985;69:765–770. doi: 10.1136/bjo.69.10.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trope GE, Salinas RG, Glynn M. Blood viscosity in primary open-angle glaucoma. Can J Ophthalmol. 1987;22:202–204. [PubMed] [Google Scholar]
- 55.Liu X, Ge J, Zhou W, Lin Y, Cai X. Hemodynamics of ophthalmic artery and central retinal artery and correlation with other factors in patients with primary open angle glaucoma. Yan ke xue bao = Eye science. 1998;14:138–144. [PubMed] [Google Scholar]
- 56.Lichter PR, Musch DC, Medzihradsky F, Standardi CL. Intraocular pressure effects of carbonic anhydrase inhibitors in primary open-angle glaucoma. Am J Ophthalmol. 1989;107:11–17. doi: 10.1016/0002-9394(89)90807-6. [DOI] [PubMed] [Google Scholar]
- 57.Kondkar AA, et al. Association of increased levels of plasma tumor necrosis factor alpha with primary open-angle glaucoma. Clin Ophthalmol. 2018;12:701–706. doi: 10.2147/OPTH.S162999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alves JM, Prado LD, Rocha HV. Evaluation and correlation of the physicochemical properties of carvedilol. Pharm Dev Technol. 2016;21:856–866. doi: 10.3109/10837450.2015.1073740. [DOI] [PubMed] [Google Scholar]
- 59.Ramdas, W. D., Schouten, J. & Webers, C. A. B. The Effect of Vitamins on Glaucoma: A Systematic Review and Meta-Analysis. Nutrients10 (2018). [DOI] [PMC free article] [PubMed]
- 60.Ramdas WD, et al. Nutrient intake and risk of open-angle glaucoma: the Rotterdam Study. Eur J Epidemiol. 2012;27:385–393. doi: 10.1007/s10654-012-9672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giaconi JA, et al. The association of consumption of fruits/vegetables with decreased risk of glaucoma among older African-American women in the study of osteoporotic fractures. Am J Ophthalmol. 2012;154:635–644. doi: 10.1016/j.ajo.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McClintock M, MacCumber MW. Lowered intraocular pressure in a glaucoma patient after intravitreal injection of ocriplasmin. Clin Ophthalmol. 2015;9:1995–1998. doi: 10.2147/OPTH.S85509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Filla MS, Dimeo KD, Tong T, Peters DM. Disruption of fibronectin matrix affects type IV collagen, fibrillin and laminin deposition into extracellular matrix of human trabecular meshwork (HTM) cells. Exp Eye Res. 2017;165:7–19. doi: 10.1016/j.exer.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olmos LC, et al. Long-term outcomes of neovascular glaucoma treated with and without intravitreal bevacizumab. Eye (Lond) 2016;30:463–472. doi: 10.1038/eye.2015.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, et al. Inhibition of vascular endothelial growth factor reduces scar formation after glaucoma filtration surgery. Invest Ophthalmol Vis Sci. 2009;50:5217–5225. doi: 10.1167/iovs.08-2662. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.