Abstract
Liver hepatocellular carcinoma (LIHC) is one of the most common malignant tumors in the world with an increasing number of fatalities. Identification of novel prognosis biomarker for LIHC may improve treatment and therefore patient outcomes. The availability of public gene expression profiling data offers the opportunity to discover prognosis biomarkers for LIHC. We developed an online consensus survival analysis tool named OSlihc using gene expression profiling and long-term follow-up data to identify new prognosis biomarkers. OSlihc consists of 637 cases from four independent cohorts. As a risk assessment tool, OSlihc generates the Kaplan–Meier survival plot with hazard ratio (HR) and p value to evaluate the prognostic value of a gene of interest. To test the reliability of OSlihc, we analyzed 65 previous reported prognostic biomarkers in OSlihc and showed that all of which have significant prognostic values. Furthermore, we identified four novel potential prognostic biomarkers (ATG9A, WIPI1, CXCL1, and CSNK2A2) for LIHC, the elevated expression of which predict the unfavorable survival outcomes. These genes (ATG9A, WIPI1, CXCL1, and CSNK2A2) may be potentially new biomarkers to identify at-risk LIHC patients when further validated. By OSlihc, users can evaluate the prognostic abilities of genes of their interest, which provides a platform for researchers to identify prognostic biomarkers to further develop targeted therapy strategies for LIHC patients. OSlihc is public and free to the users at http://bioinfo.henu.edu.cn/LIHC/LIHCList.jsp.
Keywords: liver hepatocellular carcinoma, prognosis biomarker, survival analysis, gene expression profiling, survival outcome
Introduction
As one of the most common tumors, hepatocellular carcinoma (HCC) also known as liver hepatocellular carcinoma (LIHC), is a leading cause of death of cancer globally (Yu et al., 2010; Pinato et al., 2014; Cristea et al., 2015). A number of staging and grading systems have been commonly used for LIHC, including TNM, Barcelona Clinic Liver Cancer (BCLC), albumin-bilirubin (ALBI) grading, and Okuda system (Selcuk, 2017). For LIHC patients, surgical excision is an optimal treatment (Mullath and Krishna, 2019). However, the majority of LIHC patients have the risks of recurrence and metastasis, leading to a much lower 5 years survival (Buonaguro et al., 2015). To improve patients’ quality of life, treatment should be tailored according to more accurate predicted disease outcome (Pinato et al., 2014). So far, some biomarkers have been identified to provide insights of disease outcomes. For example, α-fetoprotein (AFP) and des-gamma-carboxy-prothrombin (DCP), PCNA and Ki-67 are the most frequently used biomarkers for LIHC (King et al., 1998; Lee et al., 2013; Lin et al., 2015; Lee et al., 2016; Ma et al., 2016; Yao et al., 2016). However, due to the high heterogeneity of LIHC, more prognosis biomarkers for LIHC need to be discovered.
In the present study, we developed an easy-to-use web server named OSlihc, to provide an online platform to evaluate potential prognostic biomarkers for LIHC in multiple independent cohorts. In addition, we have validated the prognostic ability of 65 previously reported biomarkers using this web tool, and have identified four novel potential prognostic biomarkers for LIHC, including ATG9A, WIPI1, CXCL1, and CSNK2A2. By using OSlihc, researchers and clinicians can expediently evaluate the prognostic value of the genes of their interests.
Methods
Data Collection and Processing
Gene expression profiling data and clinical follow-up information of hepatocellular carcinoma were collected from TCGA (The Cancer Genome Atlas) and GEO (Gene Expression Omnibus) database. For TCGA dataset, gene expression profiling (RNA-seq, level-3, HiSeqV2) and follow-up data (361 cases) were downloaded in 2018 (Table 1). For GEO datasets, the keywords, including “hepatocellular carcinoma”, “gene expression” and “survival” were used to search in GEO database. Then the datasets containing mRNA expression and survival data with at least 50 cases were included. As a result, three GEO datasets (GSE76427, GSE20140, and GSE27150) with 115, 80, and 81 clinical LIHC cases, respectively (Table 1), were collected.
Table 1.
Summary of datasets in OSlihc.
| Dataset | Sample size | Data type | Platform | No. of death | Median of OS (months) | Survival terms |
|---|---|---|---|---|---|---|
| TCGA | 361 | RNA-seq | Illumina HiSeqV2 | 129 | 20.03 | OS, DSS, DFI, PFI |
| GSE76427 | 115 | cDNA array | GPL10558 | 23 | 13.92 | OS, RFS |
| GSE20140 | 80 | cDNA array | GPL5474 | 32 | 96.15 | OS |
| GSE27150 | 81 | cDNA array | GPL13128 | 28 | 39 | OS |
| Total | 637 | 212 | 22.43 |
Five clinical survival terms, including OS (overall survival), DSS (disease-specific survival), RFS (relapse free survival), DFI (disease-free interval), and PFI (progression-free interval) (Liu et al., 2018), were used in OSlihc web server (Table 1). The clinicopathologic characteristics of LIHC patients in OSlihc database are summarized in Tables S1 and S2.
Development of OSlihc
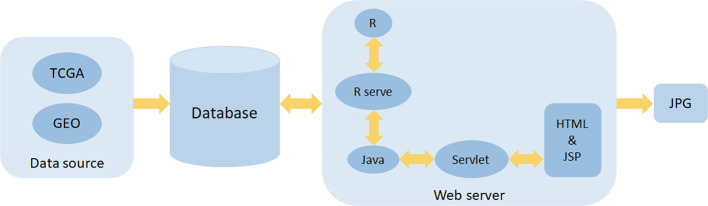
OSlihc includes two major assemblies: data storage and analysis. The OSlihc web server is hosted in a tomcat 7.0 server in Windows 2008 system, the web interface is developed by HTML and JSP languages, the controlling part is developed by Servlet, and the database is handled by SQL Server which stores and manages the gene expression profiling and clinical data. The Servlet acts as a connecting layer between the frontend and the backend to provide interaction to users. Moreover, OSlihc web server is operated by R and Java, which manipulates the request from users and then returns the analysis results to users, the R package “Rserve” acts as a connecting layer between R and Java. The JDBC package serves as the connection middleware between R, Java, and SQL. In addition, the R package “survival” generates Kaplan–Meier survival curves and calculates hazard ratio with 95% confidence intervals and p value. System architecture flow diagram is presented in Figure 1, as previous described (Wang et al., 2019a; Wang et al., 2019b; Xie et al., 2019; Zhang et al., 2019). OSlihc is available at http://bioinfo.henu.edu.cn/LIHC/LIHCList.jsp.
Figure 1.
Flowchart of web server architecture.
Validation of Prognostic Biomarkers in OSlihc
To assess the performance and reliability of prognostic analysis of OSlihc web server, previously reported prognostic biomarkers for LIHC were searched in PubMed using the keywords “hepatocellular carcinoma”, “survival”, “prognosis”, and “biomarker”. As a result, we collected 67 papers with 65 reported prognostic biomarkers. The prognostic capabilities of these reported prognostic genes were evaluated by plotting the survival curve in OSlihc. We input gene symbols of the 65 reported prognostic biomarkers into the “Gene symbol” box individually, and then clicked the “Kaplan–Meier plot” button to obtain the survival curve plot. We listed “clinical survival terms”, “cut-off”, “case”, “p value”, “HR”, “95%CI”, “detection level”, and “validation”, and compared the “prognostic outcome” between “In OSlihc” and “In reference” when the reported prognostic gene exhibited the higher expression level.
Identification of Novel Potential Prognostic Biomarkers in OSlihc
To identify prognostic biomarker candidates for LIHC, we genome-widely evaluated the prognostic values of human genes for each dataset using Cox regression analysis. Genes significantly related to prognosis were selected from each dataset (cox p value < 0.05). Then these genes were overlapped among the four datasets, as a result, four genes were identified as potential prognostic biomarkers for they were significantly related to prognosis in three datasets, including ATG9A, WIPI1, CXCL1, and CSNK2A2.
Results
Clinicopathologic Characteristics of LIHC Patients in OSlihc
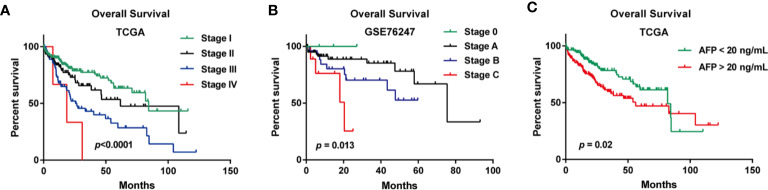
In the TCGA cohort, the median age of a total of 361 LIHC patients was 61. When examining the disease stages of LIHC patients, stage I patients accounts for 46% of all the LIHC patients (n=167), stage II accounts for 23% (n=82), stage III accounts for 23% (n=84), and stage IV accounts for 1% (n=4) (Table S1). In the GSE76427 cohort, the median age of a total of 115 patients was 64, and the distribution of BCLC stage was that stage A accounts for 64% (n=74), stage B accounts for 24% (n=28), stage C accounts for 8% (n=9), and stage 0 accounts for 3% (n=4) (Table S2). In the GSE20140 cohort, microvascular invasion was present in 21% (n=17) of LIHC patients, and absent in 56% (n=45) of LIHC patients (Table S2). The clinicopathologic information of GSE27150 dataset were unavailable. The median OS time is 22.43 months of all LIHC patients in OSlihc (Table 1). The summaries of clinicopathologic characteristics of each cohort were presented in Tables S1 and S2. The Kaplan–Meier plots for LIHC patients in OSlihc stratified by different pathologic stage and BCLC stage were presented in Figure 2. The pathologic stage and BCLC stage were significantly associated with OS (p<0.0001 and p=0.013, respectively) (Figures 2A, B). And the serum AFP level was also significantly associated with OS (p=0.02) (Figure 2C).
Figure 2.
Survival analysis of clinical characteristics of the patients included in OSlihc. (A) Pathologic stage, (B) BCLC stage, (C) Serum AFP level.
Application of OSlihc
OSlihc has the ability of drawing a forest plot for all the cohorts to highlight the prognostic significance of interested genes (cutoff: upper 50% vs. lower 50%). In OSlihc, “Gene symbol”, “Data Source”, “Survival”, and “Split patients by” are the four main parameters to evaluate the prognostic value of a gene of interest in LIHC (Figure 3A). In general, the official gene symbol is required in the “Gene symbol” box by the users. A red warning message would be shown if the input was not an official gene symbol (Figure 3B). Drop-down menu of “Data source” provides five options for users to take each of the four individual cohorts (TCGA, GSE76427, GSE20140, and GSE27150 datasets) or a combined cohort pooling all four above mentioned cohorts into one for survival analysis (Figure 3C). The combined cohort denotes that each cohort was individually stratified into subgroups by choosing a cutoff value for gene expression level, which were then pooled together for prognosis analysis. As a result, users have the choice to assess the prognostic value of a candidate gene in a single or the combined cohort according to the needs. In OSlihc, five survival terms could be generated, including OS, DSS, RFS, DFI, and PFI. OS could be measured in all the cohorts and in combined cohort, while RFS could be calculated only in GSE76427, DSS, DFI, and PFI could be analyzed only in the TCGA cohort (Figures 3D, E). “Split patients by” option could categorize LIHC patients into 2–4 subgroups by the expression level of the inputted gene (such as Upper or Lower 25%, 30%, 50%), users can select the different cutoff values (Figure 3F).
Figure 3.
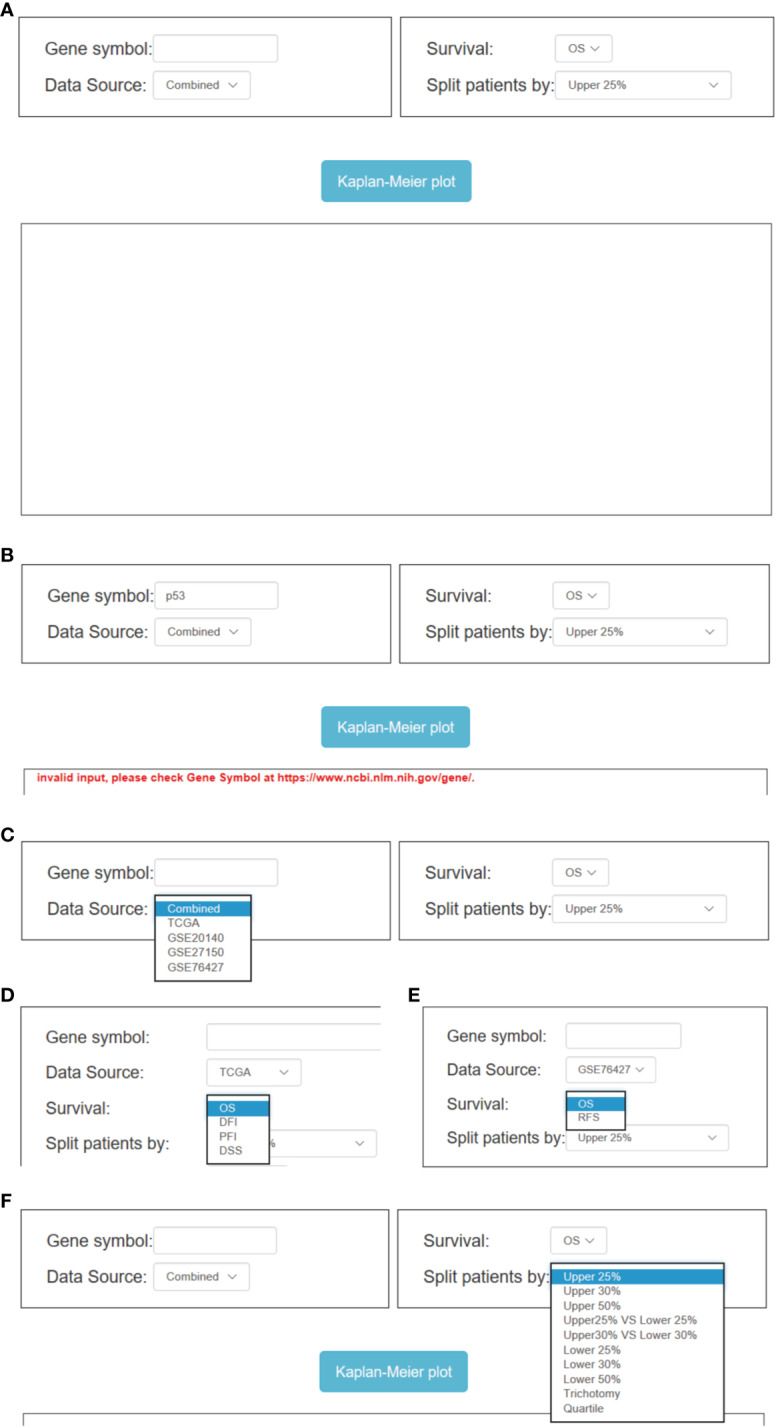
Overview of OSlihc. (A) Screenshot of OSlihc main interface. (B–F) Input and output interface of OSlihc.
In addition, additional multiple optional parameters could also be set as confounding clinical factors to sub-categorize LIHC patients for users to choose, including “Gender”, “BCLC Stage”, “TNM”, “Grade”, “Alcohol”, “Race”, etc. (Figure S1). Take “BCLC Stage” (on the interface of GSE76427 cohort) for example, users can select BCLC Stage (All, A, B, C, or 0) of LIHC from the drop-down menu box in OSlihc to evaluate the BCLC Stage-specific prognostic value of putative genes (Figure S1C). When all the options have been set, user could click the “Kaplan–Meier plot” button, OSlihc will receive the user’s request and return the prognosis analysis results for the inputted gene to users on the output web page, which graphically displays the Kaplan–Meier survival curve with p value and HR (with 95% confidence interval).
Validation of Previously Reported LIHC Prognostic Biomarkers in OSlihc
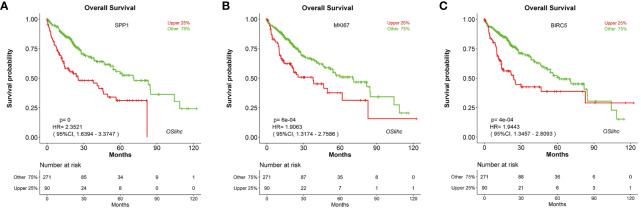
In order to assess the reliability and capability of OSlihc web server in performing prognosis analysis, we collected 65 previously reported prognostic biomarkers at protein, mRNA or serum levels from 67 literatures. CDK1, HDAC2, RORA, FOXO3, and PCNA were among these 65 genes to be evaluated in OSlihc. The analysis results showed that all the 65 previously reported prognostic biomarkers have been demonstrated significant prognostic abilities (not only OS, but also DSS, DFI, and PFI) in OSlihc web server (Table 2 and Table S3). As previously described (Ouyang et al., 2016), SPP1, MKI67, and BIRC5 genes are significantly correlated with survival (OS, DSS, DFI, and PFI) in OSlihc (Table 2, Figure 4, and Figure S2). Patients with higher SPP1, MKI67, or BIRC5 expression have shorter survival (p<0.0001, p=6e-04 or 4e-04, respectively), while patients with lower SPP1, MKI67, or BIRC5 expression exhibited longer survival (Table 2 and Figure 4). When compared with normal tissue, SPP1, MKI67, or BIRC5 expression was significantly increased in LIHC tissues (Figure S3).
Table 2.
Validation of previously published prognostic biomarkers for LIHC in OSlihc.
| Gene symbol | Biomarker name | Clinical survival terms | In OSlihc | In reference | Prognostic outcome (higher expression) | Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cut-off | p value | HR | 95%CI | Case | Cut-off | p value | case | Detection level | Validation | |||||
| SPP1 | Osteopontin (OPN) | OS | Upper 25% | 0 | 2.3521 | 1.6394-3.3747 | 361 | / | <0.05 | 110 | serum | Yes, serum biochemical assay | worse | (Yang et al., 2008; Jin et al., 2014; Qin, 2014) |
| DSS | 0.0126 | 1.8422 | 1.1403-2.9763 | / | ||||||||||
| DFI | 0.0183 | 1.5 | 1.071-2.1008 | DFS: p < 0.05 | ||||||||||
| PFI | 0.0078 | 1.5611 | 1.1242-2.1679 | / | ||||||||||
| BIRC5 | Survivin | OS | Upper 25% | 4e-04 | 1.9443 | 1.3457-2.8093 | 361 | upper n = 31/lower n = 41 | / | 72 | protein | Yes, IHC assay | worse | (Fields et al., 2004) |
| DSS | 2e-04 | 2.4062 | 1.5199-3.8095 | / | ||||||||||
| DFI | 9e-04 | 1.753 | 1.2585-2.4419 | DFS: 0.0098 | ||||||||||
| PFI | 0.0012 | 1.7165 | 1.2375-2.3809 | / | ||||||||||
| MKI67 | Ki-67 | OS | Upper 25% | 6e-04 | 1.9063 | 1.3174-2.7586 | 361 | upper n = 47/lower n = 0 | 0.0009 | 67 | protein (Ki-67 labeling index >10% indicated poor prognosis) | Yes, IHC assay | worse | (King et al., 1998) |
| DSS | 0.0011 | 2.1783 | 1.3664-3.4729 | / | ||||||||||
| DFI | 0.002 | 1.6821 | 1.2089-2.3404 | DFS: 0.02 | ||||||||||
| PFI | 0.0027 | 1.6475 | 1.1891-2.2827 | / | ||||||||||
Figure 4.
Validation of the top three high-frequency reported biomarkers in OSlihc. Kaplan–Meier plots for (A) SPP1, (B) MKI67, and (C) BIRC5 in terms of OS. p-value, confidence interval (95%CI) and number at risk are as shown. The y-axis represents survival rate and the x-axis represents survival time (months). p = 0 denotes p < 0.0001.
Identification of Novel Potential LIHC Prognostic Biomarkers in OSlihc
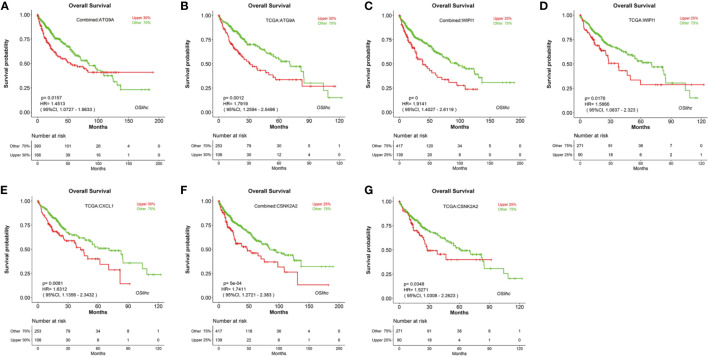
To identify new prognostic biomarker candidates for LIHC, we genome-widely evaluated the prognostic values of human genes using Cox regression analysis, and identified four genes which show significant association with survival in OSlihc, and these four genes including ATG9A (Tang et al., 2013), WIPI1 (D’Arcangelo et al., 2018), CXCL1 (Zou et al., 2014; le Rolle et al., 2015; Nakashima et al., 2015; Wang et al., 2016; Cabrero-de Las Heras and Martinez-Balibrea, 2018; Zhuo et al., 2018), and CSNK2A2 have not been reported to exhibit the prognostic values in LIHC up to now and were subject to the prognosis analysis in OSlihc (Table S4). Notably, all these four genes exhibited good performance in predicting LIHC patient outcome (Table 3 and Figure 5). Moreover, we found that patients with higher ATG9A, WIPI1, CXCL1, or CSNK2A2 expression have the worst survival rate (Figure 5), indicating that the elevated expression of all these potential predictors could predict the unfavorable prognosis.
Table 3.
Evaluation of novel potential prognostic biomarkers for LIHC in OSlihc.
| Gene symbol | In OSlihc | case | ||||
|---|---|---|---|---|---|---|
| cut-off | survival terms | p value | HR | 95%CI | ||
| ATG9A | upper 30% | OS | 0.0157 | 1.4513 | 1.0727–1.9633 | 556 |
| OS | 0.0012 | 1.7919 | 1.2594–2.5496 | 361 | ||
| DSS | 0.0246 | 1.6859 | 1.0693–2.6581 | |||
| DFI | no significance | |||||
| PFI | no significance | |||||
| WIPI1 | upper 25% | OS | <0.0001 | 1.9141 | 1.4027–2.6119 | 556 |
| OS | 0.0176 | 1.5866 | 1.0837–2.323 | 361 | ||
| DSS | no significance | |||||
| DFI | no significance | |||||
| PFI | no significance | |||||
| CXCL1 | upper 30% | OS | no significance | 636 | ||
| OS | 0.0081 | 1.6312 | 1.1356–2.3432 | 361 | ||
| DSS | 0.0454 | 1.6077 | 1.0098–2.5595 | |||
| DFI | no significance | |||||
| PFI | no significance | |||||
| CSNK2A2 | upper 25% | OS | 5e-04 | 1.7411 | 1.2721–2.383 | 556 |
| OS | 0.0348 | 1.5271 | 1.0308–2.2623 | 361 | ||
| DSS | 0.0197 | 1.7965 | 1.0979–2.9396 | |||
| DFI | 0.0208 | 1.4965 | 1.0631–2.1065 | |||
| PFI | 0.0254 | 1.4692 | 1.0485–2.0586 | |||
Figure 5.
Evaluation of the prognostic values of potential biomarkers in OSlihc. Kaplan–Meier plots for high (red) and low (green) ATG9A, WIPI1, CXCL1, or CSNK2A2-expression cohort in combined dataset (A, C, F) and TCGA dataset (B, D, E, G). p-value, confidence interval (95%CI) and number at risk are as shown. The y-axis represents survival rate and the x-axis represents survival time (months).
Discussion
LIHC is a leading malignant tumor worldwide with high metastasis and recurrence rate. Due to the complex molecular heterogeneity and poor prognosis of LIHC, it is urgent to develop biomarkers for LIHC prognosis. In the present study, we established a web server OSlihc using gene expression profiling and long-term clinical follow-up data. This prognosis analysis web server aims to assess the association between candidate gene and survival by an easy and interactive way. In current study, we have validated the association between survival and 65 previously published biomarkers for LIHC in our OSlihc server. As examples, three frequently-used biomarkers in LIHC, Osteopontin (OPN) (encoded by SPP1 gene), Survivin (encoded by BIRC5 gene) and Ki-67 (encoded by MKI67 gene), showed prognostic abilities in OSlihc, and the elevated expression of these genes predicted the unfavorable prognosis, consistent with previous reports (King et al., 1998; Fields et al., 2004; Yang et al., 2008; Jin et al., 2014; Qin, 2014). These results demonstrated the reliability and capability of OSlihc web server in prognosis analysis.
The limitation of OSlihc is that only 637 LIHC clinical cases are currently available in OSlihc. When new datasets with profiling and clinical follow-up data become available, we will update the repository of OSlihc server to improve the performance.
In conclusion, we developed an online prognosis analysis tool named OSlihc, which provides a platform for researchers to discover the new prognostic biomarkers and may offer opportunities to develop novel targeted strategies for LIHC, conducing to clinical translation of potential biomarkers.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and GEO database (https://www.ncbi.nlm.nih.gov/gds/?term=).
Author Contributions
YA, QW, and XG developed the server, performed the evaluation of novel prognostic biomarkers and drafted the paper. GZ, LZ, and FS performed the validation of previous reported biomarkers. HL, YL, and YP collected LIHC datasets. WZ and SJ contributed to data analysis and paper revision. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by National Natural Science Foundation of China (No.81602362), Supporting grants of Henan University (No.2015YBZR048, No.B2015151, No.2019YLXKJC04), Yellow River Scholar Program (No.H2016012), and Program for Innovative Talents of Science and Technology in Henan Province (No. 18HASTIT048), Program for Science and Technology Development in Henan Province (No.162102310391, No.172102210187), Program for Scientific and Technological Research of Henan Education Department (No.14B520022), Program for Young Key Teacher of Henan Province (No.2016GGJS-214), Kaifeng Science and Technology Major Project (No.18ZD008), Supporting grant of Bioinformatics Center of Henan University (No.2018YLJC01), and Innovation Project for College Students of Henan University (No.2019101904). All the funding sources have roles in study design, data collection, data analysis, and paper writing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.00875/full#supplementary-material
Screenshot of OSlihc subfield interface. (A) TCGA, (B) GSE20140, (C) GSE76427 dataset (using BCLC stage as an example).
Validation of the top three high-frequency reported biomarkers in OSlihc. Kaplan–Meier plots for (A–C) SPP1, (D–F) MKI67, and (G–I) BIRC5 in terms of DSS, DFI and PFI.
Gene expression analysis of SPP1 (A), MKI67 (B) and BIRC5 (C) at GEPIA with comparison between tumor and normal tissues.
Clinicopathologic Characteristics of LIHC patients in TCGA dataset.
Clinicopathologic Characteristics of LIHC patients in GEO datasets.
Validation of previously published predictors for LIHC survival in OSlihc.
Identification of novel potential prognostic biomarkers for LIHC in OSlihc.
References
- Buonaguro L., Tagliamonte M., Petrizzo A., Damiano E., Tornesello M. L., Buonaguro F. M. (2015). Cellular prognostic markers in hepatocellular carcinoma. Future Oncol. 11 (11), 1591–1598. 10.2217/fon.15.39 [DOI] [PubMed] [Google Scholar]
- Cabrero-de Las Heras S., Martinez-Balibrea E. (2018). CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J. Gastroenterol. 24 (42), 4738–4749. 10.3748/wjg.v24.i42.4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea C. G., Gheonea I. A., Sandulescu L. D., Gheonea D. I., Ciurea T., Purcarea M. R. (2015). Considerations regarding current diagnosis and prognosis of hepatocellular carcinoma. J. Med. Life 8 (2), 120–128. [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo D., Giampietri C., Muscio M., Scatozza F., Facchiano F., Facchiano A. (2018). WIPI1, BAG1, and PEX3 Autophagy-Related Genes Are Relevant Melanoma Markers. Oxid. Med. Cell Longev. 2018, 1471682. 10.1155/2018/1471682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields A. C., Cotsonis G., Sexton D., Santoianni R., Cohen C. (2004). Survivin expression in hepatocellular carcinoma: correlation with proliferation, prognostic parameters, and outcome. Mod. Pathol. 17 (11), 1378–1385. 10.1038/modpathol.3800203 [DOI] [PubMed] [Google Scholar]
- Jin Y., Chen J. N., Feng Z. Y., Zhang Z. G., Fan W. Z., Wang Y., et al. (2014). OPN and alphavbeta3 expression are predictors of disease severity and worse prognosis in hepatocellular carcinoma. PloS One 9 (2), e87930. 10.1371/journal.pone.0087930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K. L., Hwang J. J., Chau G. Y., Tsay S. H., Chi C. W., Lee T. G., et al. (1998). Ki-67 expression as a prognostic marker in patients with hepatocellular carcinoma. J. Gastroenterol. Hepatol. 13 (3), 273–279. 10.1111/j.1440-1746.1998.01555.x [DOI] [PubMed] [Google Scholar]
- le Rolle A. F., Chiu T. K., Fara M., Shia J., Zeng Z., Weiser M. R., et al. (2015). The prognostic significance of CXCL1 hypersecretion by human colorectal cancer epithelia and myofibroblasts. J. Transl. Med. 13, 199. 10.1186/s12967-015-0555-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Kim S. U., Kim D. Y., Ahn S. H., Lee K. H., Lee D. Y., et al. (2013). Prognostic value of alpha-fetoprotein and des-gamma-carboxy prothrombin responses in patients with hepatocellular carcinoma treated with transarterial chemoembolization. BMC Cancer 13, 5. 10.1186/1471-2407-13-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Rhim H., Kim Y. S., Kang T. W., Song K. D. (2016). Post-ablation des-gamma-carboxy prothrombin level predicts prognosis in hepatitis B-related hepatocellular carcinoma. Liver 36 (4), 580–587. 10.1111/liv.12991 [DOI] [PubMed] [Google Scholar]
- Lin J. C., Wu Y. C., Wu C. C., Shih P. Y., Wang W. Y., Chien Y. C. (2015). DNA methylation markers and serum alpha-fetoprotein level are prognostic factors in hepatocellular carcinoma. Ann. hepatology. 14 (4), 494–504. 10.1016/S1665-2681(19)31171-8 [DOI] [PubMed] [Google Scholar]
- Liu J., Lichtenberg T., Hoadley K. A., Poisson L. M., Lazar A. J., Cherniack A. D., et al. (2018). An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 173 (2), 400–16 e11. 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Yang J., Li J., Song J. (2016). The clinical utility of the proliferating cell nuclear antigen expression in patients with hepatocellular carcinoma. Tumour Biol. 37 (6), 7405–7412. 10.1007/s13277-015-4582-9 [DOI] [PubMed] [Google Scholar]
- Mullath A., Krishna M. (2019). Hepatocellular carcinoma - time to take the ticket. World J. Gastrointest. Surg. 11 (6), 287–295. 10.4240/wjgs.v11.i6.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima M., Matsui Y., Kobayashi T., Saito R., Hatahira S., Kawakami K., et al. (2015). Urine CXCL1 as a biomarker for tumor detection and outcome prediction in bladder cancer. Cancer Biomark. 15 (4), 357–364. 10.3233/CBM-150472 [DOI] [PubMed] [Google Scholar]
- Ouyang J., Sun Y., Li W., Zhang W., Wang D., Liu X., et al. (2016). dbPHCC: a database of prognostic biomarkers for hepatocellular carcinoma that provides online prognostic modeling. Biochim. Biophys. Acta 1860 (11 Pt B), 2688–2695. 10.1016/j.bbagen.2016.02.017 [DOI] [PubMed] [Google Scholar]
- Pinato D. J., Pirisi M., Maslen L., Sharma R. (2014). Tissue biomarkers of prognostic significance in hepatocellular carcinoma. Adv. Anat. Pathol. 21 (4), 270–284. 10.1097/PAP.0000000000000029 [DOI] [PubMed] [Google Scholar]
- Qin L. (2014). Osteopontin is a promoter for hepatocellular carcinoma metastasis: a summary of 10 years of studies. Front. Med. 8 (1), 24–32. 10.1007/s11684-014-0312-8 [DOI] [PubMed] [Google Scholar]
- Selcuk H. (2017). Prognostic Factors and Staging Systems in Hepatocellular Carcinoma. Exp. Clin. Transplant. 15 (Suppl 2), 45–49. 10.6002/ect.TOND16.L11 [DOI] [PubMed] [Google Scholar]
- Tang J. Y., Hsi E., Huang Y. C., Hsu N. C., Chen Y. K., Chu P. Y., et al. (2013). ATG9A overexpression is associated with disease recurrence and poor survival in patients with oral squamous cell carcinoma. Virchows Arch. 463 (6), 737–742. 10.1007/s00428-013-1482-5 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang C., Xu J., Wu H., Peng J., Cai S., et al. (2016). CXCL1 gene silencing inhibits HGC803 cell migration and invasion and acts as an independent prognostic factor for poor survival in gastric cancer. Mol. Med. Rep. 14 (5), 4673–4679. 10.3892/mmr.2016.5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wang F., Lv J., Xin J., Xie L., Zhu W., et al. (2019. a). Interactive online consensus survival tool for esophageal squamous cell carcinoma prognosis analysis. Oncol. Lett. 18 (2), 1199–1206. 10.3892/ol.2019.10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Xie L., Dang Y., Sun X., Xie T., Guo J., et al. (2019. b). OSlms: A Web Server to Evaluate the Prognostic Value of Genes in Leiomyosarcoma. Front. Oncol. 9, 190. 10.3389/fonc.2019.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Wang Q., Dang Y., Ge L., Sun X., Li N., et al. (2019). OSkirc: a web tool for identifying prognostic biomarkers in kidney renal clear cell carcinoma. Future Oncol. 15 (27), 3103–3110. 10.2217/fon-2019-0296 [DOI] [PubMed] [Google Scholar]
- Yang G. H., Fan J., Xu Y., Qiu S. J., Yang X. R., Shi G. M., et al. (2008). Osteopontin combined with CD44, a novel prognostic biomarker for patients with hepatocellular carcinoma undergoing curative resection. Oncologist 13 (11), 1155–1165. 10.1634/theoncologist.2008-0081 [DOI] [PubMed] [Google Scholar]
- Yao M., Zhao J., Lu F. (2016). Alpha-fetoprotein still is a valuable diagnostic and prognosis predicting biomarker in hepatitis B virus infection-related hepatocellular carcinoma. Oncotarget 7 (4), 3702–3708. 10.18632/oncotarget.6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Li D., Zhang H., Xue H., Pan C., Zhao S., et al. (2010). Resveratrol Inhibits Invasion and Metastasis of Hepatocellular Carcinoma Cells. J. Anim. Vet. Adv. 9 (24), 3117–3124. 10.3923/javaa.2010.3117.3124 [DOI] [Google Scholar]
- Zhang G., Wang Q., Yang M., Yuan Q., Dang Y., Sun X., et al. (2019). OSblca: A Web Server for Investigating Prognostic Biomarkers of Bladder Cancer Patients. Front. Oncol. 9, 466. 10.3389/fonc.2019.00466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo C., Wu X., Li J., Hu D., Jian J., Chen C., et al. (2018). Chemokine (C-X-C motif) ligand 1 is associated with tumor progression and poor prognosis in patients with colorectal cancer. Biosci. Rep. 38 (4), BSR20180580. 10.1042/BSR20180580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou A., Lambert D., Yeh H., Yasukawa K., Behbod F., Fan F., et al. (2014). Elevated CXCL1 expression in breast cancer stroma predicts poor prognosis and is inversely associated with expression of TGF-beta signaling proteins. BMC Cancer 14, 781. 10.1186/1471-2407-14-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Screenshot of OSlihc subfield interface. (A) TCGA, (B) GSE20140, (C) GSE76427 dataset (using BCLC stage as an example).
Validation of the top three high-frequency reported biomarkers in OSlihc. Kaplan–Meier plots for (A–C) SPP1, (D–F) MKI67, and (G–I) BIRC5 in terms of DSS, DFI and PFI.
Gene expression analysis of SPP1 (A), MKI67 (B) and BIRC5 (C) at GEPIA with comparison between tumor and normal tissues.
Clinicopathologic Characteristics of LIHC patients in TCGA dataset.
Clinicopathologic Characteristics of LIHC patients in GEO datasets.
Validation of previously published predictors for LIHC survival in OSlihc.
Identification of novel potential prognostic biomarkers for LIHC in OSlihc.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and GEO database (https://www.ncbi.nlm.nih.gov/gds/?term=).