Abstract
Viral defective interfering particles (DIPs) were intensely studied several decades ago but research waned leaving open many critical questions. New technologies and other advances led to a resurgence in DIP studies for negative-strand RNA viruses. While DIPs have long been recognized, their exact contribution to the outcome of acute or persistent viral infections has remained elusive. Recent studies have identified defective viral genomes (DVGs) in human infections, including respiratory syncytial virus and influenza, and growing evidence indicates that DVGs influence disease severity and may contribute to viral persistence. Further, several studies have advanced our understanding of key viral and host factors that regulate DIP formation and activity. Here we review these discoveries and highlight key questions moving forward.
An Interfering Activity in Viral Cultures
In the mid-20th century, von Magnus discovered an interference phenomenon while culturing influenza virus [1]. Specifically, following high multiplicity infections, large quantities of ‘incomplete’ influenza virus particles were produced that interfered with the production of standard infectious virus particles (see Glossary) [1]. In the ensuing years, interfering particles were found to be produced by several RNA viruses including Rift Valley fever virus [2], vesicular stomatitis virus (VSV) [3], lymphocytic choriomeningitis virus (LCMV) [4], and Sendai virus (SeV) [5]. Importantly, it was shown that these interfering particles mediate homologous interference, but not heterologous interference [6]. For example, interfering particles from VSV did not interfere with the heterologous encephalomyocarditis virus, demonstrating that this interference phenomenon was not simply caused by a soluble antiviral molecule, such as interferon [7]. Recognizing this growing body of literature, Alice Huang and David Baltimore proposed that these particles may be a critical determinant of infection outcome for many viruses and coined the term defective interfering particles (DIPs) in a 1970 review [6]. In the next decade and a half, a flurry of activity significantly advanced this field of research. Seminal findings included the demonstration that DIPs correlated with the establishment of persistent in vitro infections, that DIPs could be isolated from experimentally infected animals, and that purified DIPs administered separately from infectious virus could protect animals from infectious virus-induced disease or death [8]. Despite these advances, evidence supporting a role for DIPs during the course of naturally acquired infections remained elusive. Investigations were hindered by the inability to prevent DIP-forming viruses from making DIPs during the course of in vivo infections and the lack of next-generation sequencing and reverse genetics technologies. Only more recently, armed with these tools, has the field begun to provide strong evidence to suggest that DIPs can influence the outcome of natural infections. Further, we now have a deeper understanding of the mechanisms by which DIPs interfere with standard infectious virus propagation and virus–host interactions that regulate DIP production. While DIPs have been described for a wide range of viruses, including DNA viruses, retroviruses, and positive-strand RNA viruses, here we focus on recent discoveries concerning DIPs produced by enveloped, negative-strand RNA viruses. Recent reviews on DIPs by other groups have covered topics not addressed here and readers may find those articles a useful complement [9–12].
Properties of DIPs
DIPs are virus particles released from cells that are biochemically and morphologically similar to standard virus particles but, in most instances, have been shown to harbor deletions in their genomes [13,14]. DIPs containing such defective viral genomes (DVGs) can often be readily separated by centrifugation, as with VSV DIPs, which are physically shorter than the distinct, bullet-shaped standard virus particles [15]. However, DIPs from viruses not known to contain large deletions in the genomes of their DIPs, like arenaviruses, cannot be separated from standard virus particles by gradient centrifugation [16]. Since DIPs contain DVGs, they are incapable of expressing the full complement of viral proteins that standard virus particles with full-length genomes express [14]. However, the ability to be replicated is critical for a DVG’s capacity to interfere with the production of standard virus [17]. While there is significant heterogeneity in the size, type, and location of deletions in DVGs, they must retain the replication initiation and termination elements that allow them to be replicated by the viral polymerase [17].
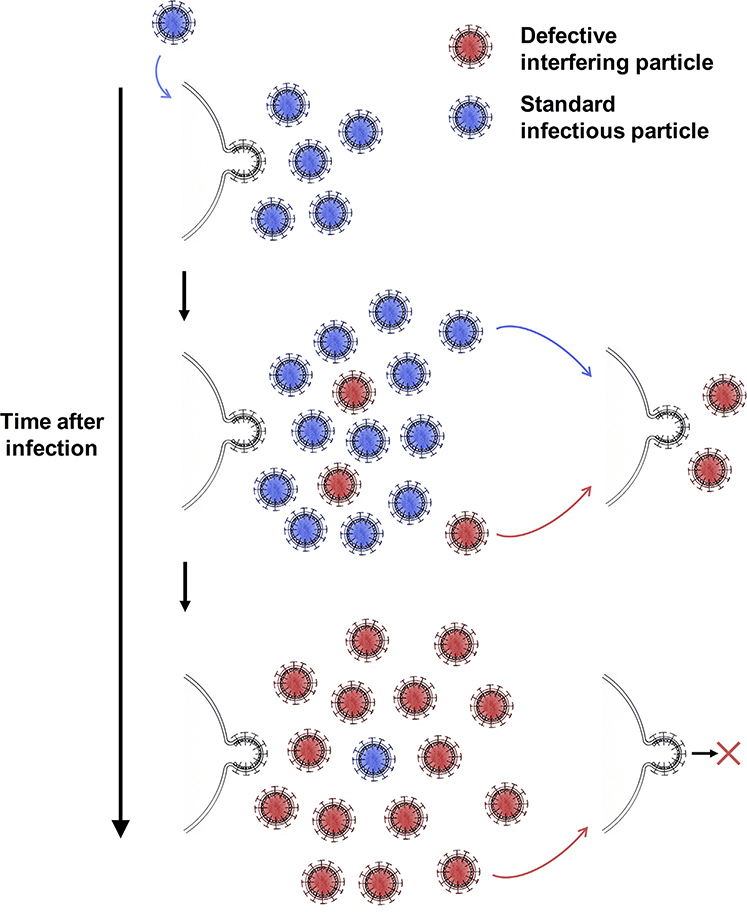
DIPs that package DVGs interfere with the production of standard virus through a highly specific, genome-mediated mechanism. This homologous interference is exquisitely specific: even different serotypes of the same virus species only exhibit partial homologous interference [7]. Various studies have demonstrated that DIPs do not interfere with standard virus by simply binding all the viral receptors on the surface of cells. Rather, DIPs interfere within the cell, as DIP-mediated interference can still occur if DIPs are added to a culture after standard virus [7,18]. RNA virus genome replication is fundamentally an error-prone process and the several classes of DVGs that exist are thought to arise as aberrant replication products produced when the viral polymerase, without release of the nascent strand, jumps to a distal location on either the template strand or the nascent strand itself, yielding an incomplete genome (reviewed in [17]). Viruses possess an intrinsic ability to generate DIPs, as even stocks of clonally pure standard virus, such as plaque-purified virus, will generate DIPs in the normal course of propagation [19]. A DVG, once formed, can be replicated by the viral polymerase and packaged into virus particles, forming DIPs, which can then enter new cells. If DIPs and standard virus particles infect the same cell, the standard virus will act as a ‘helper’ virus and DIP production will predominate (Figure 1). This replicative advantage of DIPs was originally thought to be mediated by the shorter length of DVGs [6,17], but increased promoter strength [20–23] and packaging efficiency [24,25] in certain DVGs are more likely mechanisms. DIPs, by definition, are unable to self-replicate, meaning that if a cell is only infected by DIPs, no virus particles will be produced [6].
Figure 1. A Model for the Generation of Defective Interfering Particles and the Outcomes of Infection of a Cell with Standard Virus Particles, Defective Interfering Particles, or Both.
A host cell infected with a standard virus particle will initially produce primarily standard infectious particles. Later, following the de novo generation of defective viral genomes, defective interfering particles will be produced. As defective genomes have a replicative advantage over standard genomes, they will begin to outnumber standard virus genomes. This will result in the cell producing primarily defective interfering particles. Once a defective interfering particle is released, it can infect other cells. A cell that is infected only by a defective interfering particle will presumably not produce defective interfering or standard virus particles. However, if an uninfected cell is infected with a standard infectious particle and a defective interfering particle, the defective interfering particle will block the production of new infectious particles. However, additional defective interfering particles may be produced in this setting.
The molecular basis by which some DIPs interfere with standard infectious virus propagation is not known. Notably, a substantial body of work has accumulated on arenavirus DIPs. For example, LCMV, which exhibits the majority of properties that define DIPs, does not produce classic DI genomes containing large internal deletions within open reading frames. Sanger sequencing efforts in the 1990s revealed that small deletions (2–40 nucleotides) in the 3′ and 5′ untranslated regions of the LCMV genome accumulate during acute and persistent infection (presumably at the same time DIPs accumulate), both in vitro and in vivo [26,27]. However, it is currently unknown whether these candidate DVGs can interfere with standard virus replication. Thus, there is an opportunity to apply next-generation sequencing and reverse genetics to map candidate DVGs and test whether they are truly the cause of interference by LCMV DIPs or whether a genome-independent mechanism may be at work [28].
Technological Advances Enabling Progress in DIP Research
A notable challenge in the study of DIPs has been the limited sensitivity of assays to either measure them directly or alternatively to measure their interfering activity. The interference caused by DIPs in a virus culture can be observed with some viruses when titering by plaque assay, particularly for viruses that produce high levels of DIPs, like LCMV (Figure 2) [28]. In wells inoculated with a high concentration of virus, little or no cytopathic effect (CPE; i.e., plaque formation) is observed. Only after the sample is diluted sufficiently (presumably so that infectious virus particles can enter a cell without an accompanying DIP) can CPE and countable plaques be discerned. While qualitatively useful, more precise assays are required to accurately quantitate DIPs. Historically, DIPs were quantified indirectly by combining the sample of interest containing DIPs and a constant amount of standard virus and inoculating this mixture onto cells followed by measurement of a reduction in standard virus yield (i.e., release of standard virus into supernatant) [3] or plaque formation [16]. These assays generally lacked great sensitivity, which limited the study of DIPs, particularly in vivo, where titers were often low [14,29,30]. However, for high DIP producing viruses, they still represent a valuable tool.
Figure 2. DIP-Mediated Interference with Infectious Virus Propagation Can Be Visualized by Plaque Assay.
Pictured is a representative plaque assay performed on a laboratory stock of LCMV strain Armstrong 53b. The highest concentration of infectious virus was added to the far left well and the wells to the right received successive serial 10-fold dilutions of the initial inoculum. Each well was stained with crystal violet and the image converted to gray scale (i.e., black staining indicates intact areas of the Vero E6 monolayer, while white areas/foci represent LCMV plaques). Starting from the left, note that despite receiving the most infectious virus, the first three dilutions show no or few plaques. This is presumably because all of the available cells were infected with DIPs that blocked propagation of any standard infectious particles that also infected those cells. Only by the fourth and fifth serial dilutions are individual plaques visible, presumably because DIPs were sufficiently diluted to allow standard infectious particles to productively infect cells in the absence of DIPs and thus create plaques. Abbreviations: DIPs, defective interfering particles; LCMV, lymphocytic choriomeningitis virus.
Several major technological advances since the late 1980s have enabled investigators to break new ground in the characterization of DIPs. The advent of PCR occurred in the mid-1980s [31], but a PCR assay to selectively amplify certain DVGs was not described until 1992 [32]. Such DVG-specific PCR assays have been subsequently employed to directly detect DVGs in human infections of measles virus (MeV) [33] and respiratory syncytial virus (RSV) [34]. DVG-specific PCR assays, however, require at least some sequence information about the DVGs themselves and often cannot detect the full range of DVGs that are present in a sample [35]. Modern next-generation sequencing technologies are changing this landscape and recently have been used to characterize DVGs found in several paramyxoviruses [35–40], influenza viruses [38,41–43], VSV [44], and flock house virus [45,46]. Next-generation sequencing has also permitted characterization of DVGs from clinical isolates of influenza patients [41,43,47], ultimately leading to some of the most compelling evidence that DVGs may influence the outcome of natural infections [41]. While these modern approaches for measuring DVGs are highly sensitive, they do not directly measure the interfering capacity of these genomes. It remains important for the field to demonstrate that presumptive DVGs uncovered by PCR and/or sequencing are (i) defective for permitting the full viral life cycle to be completed and (ii) able to interfere with standard virus propagation, which could be measured by classic assays such as the yield reduction assay described earlier. This validation of candidate DVGs is critical in cases where accumulation of these DVGs is being correlated with a particular outcome of infection.
In addition to sensitive detection of DVGs at the cell population level, technological advances have aided the analysis of DVGs at the single-cell level. High multiplicity infections can result in cell protection, less efficient standard virus production, and enhanced DIP production [48]. The theory behind this holds that during high multiplicity infections, a large proportion of cells will be infected with both infectious viruses and DIPs. As DIPs can inhibit standard virus production with single-hit kinetics, a single DIP infecting a cell may be enough to completely inhibit standard virus production in that cell and protect it from death [49–51]. Conversely, in low multiplicity infections, cells may be infected with DIPs only, standard virus only, or both. While this model has existed since Huang and Baltimore first proposed it [6], only recently has substantial evidence supporting the heterogeneous nature of infection in a population of cells begun to accumulate. Fluorescent reporter viruses and RNA fluorescence in situ hybridization coupled with fluorescence microscopy or flow cytometry have been used to monitor the spread of DIPs and standard virus during infection at the single cell level [52–54]. These technologies have provided mechanistic insight into how DIP-infected cells, despite the ability of some DVGs to potently activate the innate antiviral response, are able to selectively elicit a prosurvival pathway and escape death [54]. Furthermore, these technologies have shown that SeV DVGs fail to interact with host trafficking proteins and consequently, little or no DIPs are produced from cells with high levels of DVGs [53]. Collectively, this recent work has allowed a deeper mechanistic understanding of the dynamic interplay between DIPs and standard virus.
A significant barrier to determining the role of DIPs during viral infections has been the difficulty in separating the function of standard virus and DIPs. Viruses have a natural propensity to produce DIPs, meaning that even when starting with clonally pure standard infectious virus, DIPs will eventually be made. Several approaches have been used to circumvent this problem to the extent possible. Functionally pure DIPs can be obtained by irradiation with UV light for viruses with DIPs that are less sensitive to UV light relative to standard virus, like arenaviruses and influenza A virus (IAV) [16,55]. Differential centrifugation can be used for viruses like VSV in which the DIPs were different enough in size compared with standard virus [15]. DIP-enriched virus preparations can also be derived from persistently infected cells or from the acute phase of infection in cells that were inoculated at a high multiplicity of infection [15]. The ability to add DIPs of a certain genome type to cultures or to manipulate elements of the viral genome in order to selectively control DIP production was a feat that required reverse genetics systems. As an example, VSV DVGs were the first to be successfully generated from cloned cDNA and packaged into ‘infectious’ DIPs [56,57]. The development of reverse genetics systems for other negative-strand viruses [58] provided tools to help determine elements of viruses that contribute to the production of DIPs as will be discussed in the following sections.
Regulation of DIP Production by Viral and Host Factors
While DIPs accumulate during a wide range of viral infections, both viral and host factors can regulate their formation (see Table 1, Key Table, for a summary). Accumulation of DIPs is presumably a combined result of the de novo generation of new DVGs from the wild-type parental genome followed by the subsequent replication and incorporation of these DVGs into new virus particles. Replication of RNA virus genomes is inherently an error-prone process. Accordingly, de novo generation of DVGs is often described as a result of this error-prone process. However, recent evidence has demonstrated that viruses can reproducibly produce DVGs with defined sequences, suggesting that DVG generation can be directed and is not exclusively a product of random replication errors [59]. Identification of specific sequences in the genome of RSV that regulate the formation of copy-back DVGs has enabled the engineering of viruses that do not produce specific DVGs [59].
Table 1.
Viral and Host Factors That Regulate the Formation of DIPs
| Virus | Genomic or protein level | Host or viral factor | De novo generation, replication, or packaging | Proposed mechanism ofaction | Refs |
|---|---|---|---|---|---|
| Lymphocytic choriomeningitis virus | Protein | Viral: Z matrix protein | Unknown | The PPXY late domain in the viral Z protein promotes DIP production | [28] |
| Lymphocytic choriomeningitis virus | Not applicable (n/a) | Host: ESCRT, host kinases, Nedd4 family E3 ubiquitin ligases | Unknown | Thecellular ESCRT pathway, host kinase-driven phosphorylation of the viral matrix protein, and certain Nedd4 family E3 ubiquitin ligases promote DIP production | [28,78,79] |
| Respiratory syncytial virus | Genomic | Viral | De novo | Nucleotide composition at specific sites in the genomic RNA increases the rate at which the viral polymerase generates copy-back DVGs | [59] |
| Influenza A virus | Unkown | Viral | Unknown | A mutation in the PA subunit in the polymerase results in a significant increase in DIPs. Function could be at the genomic level as the mutation in PA results in increased levels of DVGs with mutations in genome segment containing PA. The mutation could also affect the ability of PA to stay bound to the RNA template which would increase the chances of aberrant RNA replication | [74] |
| Influenza A virus | Protein | Viral | De novo | Mutations in NS2 increase generation of DIPs specifically containing deletions in PA gene | [69] |
| Vesicular stomatitis virus | n/a | Host | De novo | Human chromosome 16 in human–mouse somatic cell hybrids suppressed de novo generation of DVGs | [75] |
| Rabies virus | Genomic | Virus | Replication | Promoter strength of an engineered copy-back DVG permitted strongly biased amplification of the DVG over full-length genomes | [65] |
| Influenza A virus | Genomic | Viral | n/a packaging | IAV DVGs are packaged into virus particles more efficiently than their cognate full-length genomes | [24,66,67] |
| Influenza A virus | Genomic | Viral | Replication | A specific type of IAV DVGs has a mutation in its replication promoter that substantially increases the rate of replication relative to wild-type genomes | [61] |
| Vesicular stomatitis virus, Sendai virus | Genomic | Viral | Replication | Complementary ends and a lack of a transcriptional promoter allow 5′ copy-back and snap-back DVGs to replicate at a higher rate than full-length genomes | [61–64] |
| Influenza A virus | Likely Protein | Viral | Unknown | A mutation (D529N) in the PA subunit of the polymerase inhibited the production of DVGs | [41] |
| Influenza A virus | Likely Protein | Viral | Unknown | Mutations in M1 and M2 increase the production of DVGs | [41,73] |
| Sendai virus, measles virus | Likely protein | Viral | Unknown | The C protein suppresses DVG production | [36,70,71] |
| Sendai virus | Protein | Viral | De novo | A point mutation in the N protein increases production of DVGs, possibly by the lower density of viral nucleocapsids | [68] |
| Influenza A virus | Protein | Viral | Unknown | The NS gene segment from a highly pathogenic H5N1 IAV induces robust production of DIPs compared with the NS gene segment from H1N1 | [70] |
A DVG, once generated, typically will replicate at a much higher rate than full-length genomes. Huang and Baltimore originally proposed that this replicative advantage was a product of the shorter length of DVGs [6]; however, evidence has accumulated showing that factors other than the length of the genome may also contribute to this increase in the replicative capacity of DVGs. For example, a full-length, but defective interfering genome found in IAV can replicate more efficiently due to its increased promoter strength [60]. In addition, evidence has demonstrated that copy-back DVGs possess a significant replicative advantage by way of their terminal complementarity and, for 5′ copy-back DVGs, the lack of a transcriptional promoter [61–65]. Beyond de novo generation and replication, other factors may also influence the accumulation of DIPs. For example, IAV DVGs can be packaged more efficiently than their cognate full-length genomes [24,66,67].
Several viral proteins have been shown to regulate the de novo generation of DVGs. For example, specific mutations in the IAV NS2 protein and the SeV N protein increase DVG production [68,69] while the C protein of both SeV and MeV suppress DVG synthesis [36,70,71]. In addition, the IAV NS protein from the highly pathogenic H5N1 virus strongly drives the production of DIPs while NS from H1N1 does not [72]. Further, the IAV proteins M1 and M2 and the PA subunit of the polymerase also appear to regulate DIP production [41,73,74]. The production of DIPs is not only regulated by viral factors, but also by the cellular host. It has been shown that certain cell types produce little or no DIPs [14]. Interestingly, the de novo generation of DVGs is restricted in certain cell types [75–77]. However, the underlying mechanism(s) for these restrictions are largely unknown.
Our group recently identified both viral and host factors that regulate the formation of LCMV DIPs. Specifically, the PPXY late domain encoded by the LCMV matrix protein and the cellular ESCRT (endosomal sorting complexes required for transport) pathway, which is recruited by this late domain, are required for DIP production, but not standard infectious virus particles [28]. Further, certain host Nedd4 family E3 ubiquitin ligases, which bind to PPXY late domains, also specifically promote DIP production [78]. Last, phosphorylation of the LCMV matrix protein at multiple sites, including the PPXY late domain, appears to drive DIP formation, suggesting that arenaviruses can dynamically adjust DIP production in response to external environmental factors [28,79]. Collectively, these findings suggest that divergent pathways and regulatory mechanisms exist for the formation of standard particles versus DIPs in the setting of arenavirus infection.
DIPs in Natural Infections
Determining the role of DIPs during natural infections has been a major challenge and evidence that they can dictate infection outcome has only slowly emerged. The interference mediated by DIPs was first associated with ‘incomplete’ virus in the 1950s using experimental infections with Rift Valley fever virus and IAV in mice [2,80]. However, it took nearly two decades before the presence of DIPs from VSV [81], rabies virus [81], and LCMV [82] was confirmed in experimental infections in mice. In addition to their production in vivo, exogenous DIPs have been shown to reduce the pathology or delay or prevent death associated with homologous virus challenge in animals [8]. Arguably, some of the earliest studies of DIPs in natural infections were with LCMV, which is carried in nature by the common house mouse [82,83]. In addition, during a lethal outbreak of IAV H5N2 in chickens in the 1980s, it was found that the virulent strain failed to produce detectable DVGs while an avirulent strain did, suggesting that DIPs were a virulence factor [84,85]. DVGs have also been found in human infections, notably in brain tissue of humans with subacute sclerosing panencephalitis (SSPE) associated with MeV [86] and more recently in patients with IAV [41,43,47] and RSV [34] infections.
Evidence has also emerged that DIPs are not only present during natural infections, but can also help determine the course of disease. A clinical isolate of IAV H1N1 from a fatal case was identified that produced few DVGs while an isolate from a mild case of disease produced high levels of DVGs [41]. The specific genetic differences between the isolates that controlled DVG levels were identified, making it possible to recover recombinant viruses with genetic control over DVG production [41]. The high levels of DVGs produced by the ‘mild’ virus elicited an innate immune response in mice that helped control viremia and prevented neutrophil invasion and excessive inflammation that are associated with more severe disease [41]. In addition, in other IAV clinical isolates, increased levels of DVGs were associated with mild disease whereas clinical isolates from fatal cases had low levels of DVGs [41]. Similarly, detection of DVGs in the respiratory tract of children infected with RSV correlated with the expression of antiviral genes [34]. This work builds on in vitro studies showing that VSV DIPs activate the interferon response [87] and more recent studies demonstrating the critical role DVGs have in stimulating the innate immune response during several acute viral infections (for more a more detailed review of the immune response to DVGs, see [88]). DVGs are recognized by pattern recognition receptors, including retinoic acidinducible gene 1 (RIG-I), resulting in the production of interferon and other proinflammatory cytokines as well as the maturation of dendritic cells [9,89]. Specifically, copy-back or snapback DVGs for the nonsegmented, negative-strand RNA viruses or short deletion mutants in IAV have been implicated in activating antiviral innate immunity [14,38]. Indeed, the DVGs of these viruses, rather than standard virus genomes, are the primary activators of the interferon response [88]. The innate immune response activated by DVGs, at least in some cases, confers protection to the cells containing them by specifically activating a prosurvival pathway [54]. While this cellsparing activity initiated by the DVGs was first shown in 1977 for VSV [87], it is only recently that the signaling pathway through which interferon activation by DVGs results in cell sparing has been determined [54]. Collectively, DIPs appear to confer protection to their host by inhibiting standard virus production and/or activating the innate immune response resulting in a broad range of downstream consequences, but additional mechanisms may be at work.
DIPs in Persistent Infections
Viruses that establish persistent infections in their hosts must balance their own replication with the potential damage it may cause. Some non-retroviral RNA viruses are capable of establishing and maintaining persistent infections even though they cannot integrate their genomes into host chromosomes, like retroviruses, or maintain their genomes as episomes, like DNA viruses [90–92]. Huang and Baltimore originally proposed that DIPs may be a critical factor during persistent viral infections [6]. DIPs have been studied in the context of persistent infections in vitro with several viruses and are characterized by cyclical infections in which the levels of DIPs and standard virus rise and fall in alternating patterns [14,48]. There is significantly less known about DIPs and persistent infections in vivo. LCMV infection in mice is a particularly relevant model as LCMV is normally maintained in nature through persistent, asymptomatic infections in Mus musculus [93]. DIPs have been found in persistent LCMV infections in mice and are suspected to function during these infections [82], though numerous other factors are likely involved [94]. It has been particularly difficult to determine how DIPs dictate the course of persistent infections given the intrinsic ability of standard virus to generate DIPs, resulting in a mixture of DIPs and standard viruses during infection. In the case of LCMV, we have now engineered viruses that no longer produce DIPs (but still make normal levels of standard virus particles) [28,78], which opens the door to exploring their impact on the establishment and maintenance of an asymptomatic, persistent infection.
There has also been substantial interest in whether DIPs contribute to viral persistence in human infections including in Ebola virus and MeV infections, which are typically acute in humans, but are increasingly suspected to cause persistent infections. DIPs have been found in the persistent manifestation of MeV infection in the brain, SSPE, and it has been suggested that DIPs may play a role in this disease by helping to establish or maintain persist infection which can lead to SSPE [33,92]. For Ebola virus, during the unprecedented outbreak in 2014, some survivors harbored active infections long after resolution of acute disease [95,96], which may facilitate sexual transmission [97], or development of neurological disease [98]. It has been suggested that DIPs may have a role in this persistent state [99], as Ebola virus DIPs have been detected during in vitro infections [100], but to date it is unclear whether these patients harbor significant levels of DIPs [101].
Concluding Remarks
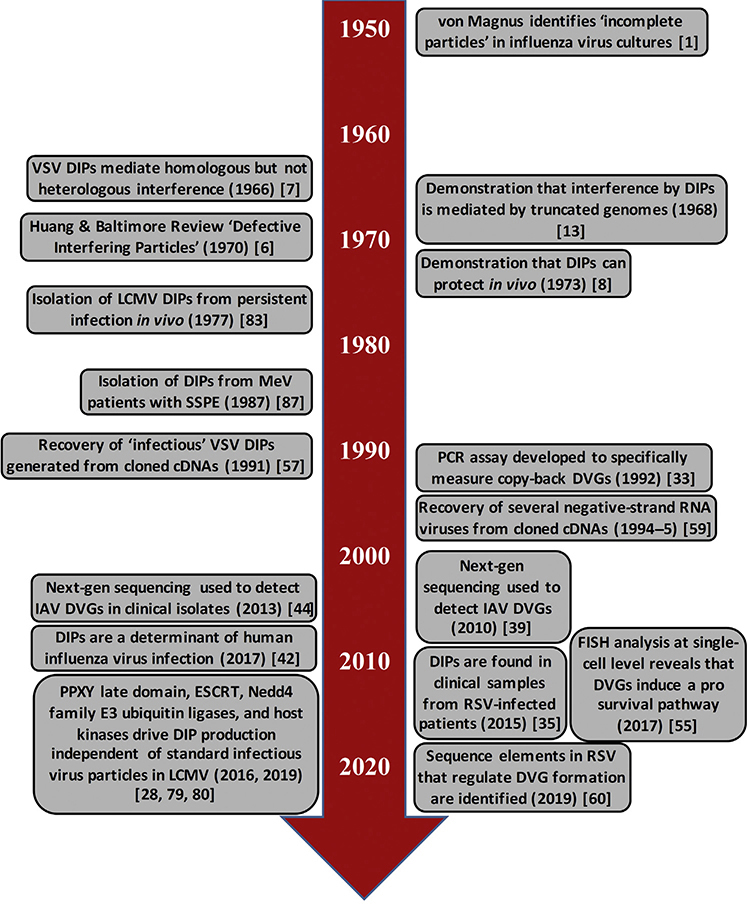
The collective understanding of this interfering phenomenon from ‘incomplete’ virus has come a long way in the past 70 years (see Figure 3 for a timeline of notable discoveries) but several key questions remain to be answered (see Outstanding Questions). For example, it remains to be determined whether DIPs function in a broader range of natural infections and the various mechanisms underlying their function. At present, little is known about viral and host factors that regulate the production of DIPs, but progress can certainly be made in this area. An improved understanding of these factors could provide strategies for transforming virulent viruses into live-attenuated vaccines by enhancing their capacity to produce DIPs. The mechanisms underlying persistent infections could also potentially be advanced if the tools that have been used to study DIPs in acute viral infections were applied. Finally, many important viral pathogens are zoonotic and are carried by their hosts persistently (e.g., orthohantaviruses and mammarenaviruses). Understanding the role that DIPs may have in the maintenance of these viruses in nature will be critical for understanding virus–host ecology and transmission dynamics.
Figure 3. Timeline of Notable Defective Interfering Particle Discoveries. See also [1,6–8,13,28,32,34,38,41,43,54,56,58,59,78,79,82,86].
Abbreviations: DIPs, defective interfering particles; DVGs, defective viral genomes; FISH, fluorescence in situ hybridization; IAV, influenza A virus; LCMV, lymphocytic choriomeningitis virus; MeV, measles virus; RSV, respiratory syncytial virus; SSPE, subacute sclerosing panencephalitis; VSV, vesicular stomatitis virus.
Outstanding Questions.
Are DIPs a general virulence factor for many viruses in the setting of human infection? If so, could detection and/or quantitation of DIPs inform treatment decisions?
What role do DIPs play in the setting of persistent viral infection of sylvatic reservoir species (i.e., mammarenavirus or orthohantavirus infection of rodents)? Are they critical for protection of host fitness and thus viral maintenance in nature? What is their role during persistent infection in humans (i.e., Ebola or Lassa viruses)?
What is the molecular basis of interference employed by DIPs produced by mammarenaviruses and orthohantaviruses? Is it DVG independent? If not, what classes of DVGs are produced and by what molecular mechanism?
How important are DIPs for the development and durability of protective immunity, particularly antiviral memory B and T cells?
What are the different viral and host factors that regulate DIP production for different viruses? How do these factors mechanistically drive or repress DIP production?
Can mechanistic knowledge of the virus and host factors involved in DIP production be harnessed to provide better treatment for acutely ill individuals (e.g., is DIP production a legitimate antiviral target?)?
How can we genetically modify viruses or manipulate host factors to inhibit or enhance DIP production? Can this knowledge be used to engineer virulent viruses into live attenuated vaccines by directing them to produce higher levels of DIPs?
How can we improve the sensitivity of assays to measure DIPs or their activity?
Many candidate DVGs are being identified using next-generation sequencing platforms. Are they able to mediate interference against standard infectious particles? While this question is difficult to answer, more direct testing is needed to move beyond correlative associations of a particular DIP/DVG signature and a particular disease state (i.e., enhanced or improved disease severity).
Highlights.
New technologies including reverse genetics, PCR, and next-generation sequencing have led to a resurgence in DIP research.
DVG synthesis was presumed to occur randomly due to the error-prone nature of viral polymerases. Recent work suggests that DVG generation can be a highly directed process.
Several novel viral and host factors required for DIP production have been discovered.
For certain viruses, DIP production appears to be a highly regulated and dynamic process. Importantly, it is now possible to engineer viruses that no longer create DIPs but still produce normal quantities of standard infectious virus particles.
DIPs have long been hypothesized to reduce virulence of a particular virus for its host. New evidence strongly suggests DIPs derived by several viruses including influenza and respiratory syncytial virus are protective in humans.
Acknowledgments
We thank the National Institutes of Health (NIH) for the following grant support: T32 AI055402 (C.M.Z.), T32 HL076122 (C.M.Z.), R21 AI088059 (J.W.B.), and P30GM118228 (Immunobiology and Infectious Disease COBRE award) (J.W.B.).
Glossary
- Defective interfering particles (DIPs)
DIPs are biochemically similar to standard infectious virus particles, but differ because in most examples they package DVGs. DIPs are defective because they package a faulty genome and thus cannot complete the viral life cycle without help from standard infectious virus particles. DIPs are interfering because they block the propagation of standard infectious virus particles (in the setting where a host cell is co-infected with both a DIP and a standard infectious virus particle)
- Defective viral genomes (DVGs)
viral genomes that arise through viral polymerase-driven mutation or recombination. They lack critical regions of the wild-type genome required for successful completion of the viral life cycle but retain the elements required for their replication by the viral polymerase. DVGs are the molecular basis for DIP-mediated interference with standard infectious virus particles. Specifically, DVGs outcompete wild-type genomes for access to viral genome replication machinery
- Heterologous interference
a phenomenon whereby DIPs produced by a specific virus species can interfere with the propagation of standard infectious virus particles of a different virus species (i.e., LCMV DIPs interfering with Lassa virus standard virus particles)
- Homologous interference
a phenomenon whereby DIPs produced by a specific virus species can interfere with the propagation of standard infectious virus particles produced by that same species of virus (i.e., LCMV DIPs interfering with LCMV standard virus particles)
- Standard infectious virus particles
virus particles that package a wild-type viral genome. These virus particles can enter a host cell, replicate the viral genome, express the full complement of viral proteins, and successfully complete the viral life cycle by producing new infectious virions
References
- 1.von Magnus P (1954) Incomplete forms of influenza virus In Advances in Virus Research (Smith KM and Lauffer MA, eds), pp. 59–79, Academic Press; [DOI] [PubMed] [Google Scholar]
- 2.Mims CA (1956) Rift Valley fever virus in mice. IV. Incomplete virus; its production and properties. Br. J. Exp. Pathol 37, 129–143 [PMC free article] [PubMed] [Google Scholar]
- 3.Bellett AJ and Cooper PD (1959) Some properties of the transmissible interfering component of vesicular stomatitis virus preparations. J. Gen. Microbiol 21, 498–509 [DOI] [PubMed] [Google Scholar]
- 4.Lehmann-Grube F et al. (1969) A persistent and inapparent infection of L cells with the virus of lymphocytic choriomeningitis. J. Gen. Virol 5, 63–81 [DOI] [PubMed] [Google Scholar]
- 5.Kingsbury DW et al. (1970) Properties of incomplete Sendai virions and subgenomic viral RNAs. Virology 42, 857–871 [DOI] [PubMed] [Google Scholar]
- 6.Huang AS and Baltimore D (1970) Defective viral particles and viral disease processes. Nature 226, 325–327 [DOI] [PubMed] [Google Scholar]
- 7.Huang AS and Wagner RR (1966) Defective T particles of vesicular stomatitis virus. Virology 30, 173–181 [DOI] [PubMed] [Google Scholar]
- 8.Barrett ADT and Dimmock NJ (1986) Defective interfering viruses and infections of animals In Current Topics in Microbiology and Immunology (Clarke A et al., eds), pp. 55–84, Springer; [DOI] [PubMed] [Google Scholar]
- 9.Vignuzzi M and López CB (2019) Defective viral genomes are key drivers of the virus–host interaction. Nat. Microbiol 4, 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genoyer E and López CB (2019) The impact of defective viruses on infection and immunity. Ann. Rev. Virol 6, 547–566 [DOI] [PubMed] [Google Scholar]
- 11.Yang Y et al. (2019) The antiviral and antitumor effects of defective interfering particles/genomes and their mechanisms. Front. Microbiol 10, 1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rezelj VV et al. (2018) The defective component of viral populations. Curr. Opin. Virol 33, 74–80 [DOI] [PubMed] [Google Scholar]
- 13.Duesberg PH (1968) The RNA of influenza virus. Proc. Natl. Acad. Sci 59, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland JJ et al. (1980) Defective interfering RNA viruses and the host-cell response In Comprehensive Virology: Vol. 16: Virus-Host Interactions: Viral Invasion, Persistence, and Disease (Fraenkael-Conrat H and Wagner RR, eds), pp. 137–192, Springer [Google Scholar]
- 15.Huang AS et al. (1966) Defective T particles of vesicular stomatitis virus. Virology 30, 161–172 [DOI] [PubMed] [Google Scholar]
- 16.Welsh RM et al. (1972) Properties of defective lymphocytic choriomeningitis virus. J. Gen. Virol 17, 355–359 [DOI] [PubMed] [Google Scholar]
- 17.Lazzarini RA et al. (1981) The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26, 145–154 [DOI] [PubMed] [Google Scholar]
- 18.Dutko FJ and Pfau CJ (1978) Arenavirus defective interfering particles mask the cell-killing potential of standard virus. J. Gen. Virol 38, 195–208 [DOI] [PubMed] [Google Scholar]
- 19.Stampfer M et al. (1971) Absence of interference during high-multiplicity infection by clonally purified vesicular stomatitis virus. J. Virol 7, 409–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao DD and Huang AS (1982) Interference among defective interfering particles of vesicular stomatitis virus. J. Virol 41, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meier E et al. (1984) Sites of copy choice replication involved in generation of vesicular stomatitis virus defective-interfering particle RNAs. J. Virol 51, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattnaik AK et al. (1995) The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology 206, 760–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T and Pattnaik AK (1997) Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology 232, 248–259 [DOI] [PubMed] [Google Scholar]
- 24.Duhaut SD and McCauley JW (1996) Defective RNAs inhibit the assembly of influenza virus genome segments in a segment-specific manner. Virology 216, 326–337 [DOI] [PubMed] [Google Scholar]
- 25.Odagiri T and Tashiro M (1997) Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J. Virol 71, 2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer BJ and Southern PJ (1997) A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J. Virol 71, 6757–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer BJ and Southern PJ (1994) Sequence heterogeneity in the termini of lymphocytic choriomeningitis virus genomic and antigenomic RNAs. J. Virol 68, 7659–7664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziegler CM et al. (2016) The lymphocytic choriomeningitis virus matrix protein PPXY late domain drives the production of defective interfering particles. PLoS Pathog. 12, e1005501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welsh RM and Oldstone MB (1977) Inhibition of immunologic injury of cultured cells infected with lymphocytic choriomeningitis virus: role of defective interfering virus in regulating viral antigenic expression. J. Exp. Med 145, 1449–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux L and Waldvogel FA (1983) Defective interfering particles of Sendai virus modulate HN expression at the surface of infected BHK cells. Virology 130, 91–104 [DOI] [PubMed] [Google Scholar]
- 31.Saiki RK et al. (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239, 487. [DOI] [PubMed] [Google Scholar]
- 32.Calain P et al. (1992) Molecular cloning of natural paramyxovirus copy-back defective interfering RNAs and their expression from DNA. Virology 191, 62–71 [DOI] [PubMed] [Google Scholar]
- 33.Sidhu MS et al. (1994) Defective measles virus in human subacute sclerosing panencephalitis brain. Virology 202, 631–641 [DOI] [PubMed] [Google Scholar]
- 34.Sun Y et al. (2015) Immunostimulatory defective viral genomes from respiratory syncytial virus promote a strong innate antiviral response during infection in mice and humans. PLoS Pathog. 11, e1005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosma TJ et al. (2019) Identification and quantification of defective virus genomes in high throughput sequencing data using DVG-profiler, a novel post-sequence alignment processing algorithm. PLOS ONE 14, e0216944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sánchez-Aparicio MT et al. (2017) Loss of Sendai virus C protein leads to accumulation of RIG-I immunostimulatory defective interfering RNA. J. Gen. Virol 98, 1282–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Killip MJ et al. (2013) Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J. Virol 87, 4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baum A et al. (2010) Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci 107, 16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beauclair G et al. (2018) DI-tector: defective interfering viral genomes’ detector for next-generation sequencing data. RNA 24, 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Hoogen BG et al. (2014) Excessive production and extreme editing of human metapneumovirus defective interfering RNA is associated with type I IFN induction. J. Gen. Virol 95, 1625–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasilijevic J et al. (2017) Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients. PLoS Pathog. 13, e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alnaji FG et al. (2019) Sequencing framework for the sensitive detection and precise mapping of defective interfering particle-associated deletions across influenza A and B viruses. J. Virol 93 e00354–00319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saira K et al. (2013) Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J. Virol 87, 8064–8074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timm C et al. (2014) Quantitative characterization of defective virus emergence by deep sequencing. J. Virol 88, 2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Routh A et al. (2012) Nucleotide-resolution profiling of RNA recombination in the encapsidated genome of a eukaryotic RNA virus by next-generation sequencing. J. Mol. Biol 424, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaworski E and Routh A (2017) Parallel ClickSeq and nanopore sequencing elucidates the rapid evolution of defective-interfering RNAs in Flock House virus. PLoS Pathog. 13, e1006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lui W-Y et al. (2019) SMRT sequencing revealed the diversity and characteristics of defective interfering RNAs in influenza A (H7N9) virus infection. Emerg. Microbes Infect 8, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang AS (1973) Defective interfering viruses. Annu. Rev. Microbiol 27, 101–117 [DOI] [PubMed] [Google Scholar]
- 49.Sekellick MJ and Marcus PI (1980) Viral interference by defective particles of vesicular stomatitis virus measured in individual cells. Virology 104, 247–252 [DOI] [PubMed] [Google Scholar]
- 50.Popescu M et al. (1976) Homologous interference of lymphocytic choriomeningitis virus: detection and measurement of interference focus-forming units. J. Virol 20, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stauffer Thompson KA et al. (2009) Multiple-hit inhibition of infection by defective interfering particles. J. Gen. Virol 90, 888–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baltes A et al. (2017) Inhibition of infection spread by cotransmitted defective interfering particles. PLOS ONE 12, e0184029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genoyer E and López CB (2019) Defective viral genomes alter how Sendai virus interacts with cellular trafficking machinery, leading to heterogeneity in the production of viral particles among infected cells. J. Virol 93 e01579–01518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J et al. (2017) Replication defective viral genomes exploit a cellular pro-survival mechanism to establish paramyxovirus persistence. Nat. Commun 8, 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nayak DP et al. (1978) Homologous interference mediated by defective interfering influenza virus derived from a temperature-sensitive mutant of influenza virus. J. Virol 28, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pattnaik AK and Wertz GW (1991) Cells that express all five proteins of vesicular stomatitis virus from cloned cDNAs support replication, assembly, and budding of defective interfering particles. Proc. Natl. Acad. Sci 88, 1379–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pattnaik AK et al. (1992) Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 58.Neumann G et al. (2002) A decade after the generation of a negative-sense RNA virus from cloned cDNA – what have we learned? J. Gen. Virol 83, 2635–2662 [DOI] [PubMed] [Google Scholar]
- 59.Sun Y et al. (2019) A specific sequence in the genome of respiratory syncytial virus regulates the generation of copy-back defective viral genomes. PLoS Pathog. 15, e1007707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kupke SY et al. (2019) A novel type of influenza A virusderived defective interfering particle with nucleotide substitutions in its genome. J. Virol 93 e01786–01718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolakofsky D (1979) Studies on the generation and amplification of Sendai virus defective-interfering genomes. Virology 93, 589–593 [DOI] [PubMed] [Google Scholar]
- 62.Leppert M and Kolakofsky D (1980) Effect of defective interfering particles on plus- and minus- strand leader RNAs in vesicular stomatitis virus-infected cells. J. Virol 35, 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wertz GW et al. (1994) Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc. Natl. Acad. Sci 91, 8587–8591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whelan SPJ and Wertz GW (1997) Defective interfering particles of vesicular stomatitis virus: functions of the genomic termini. Semin. Virol 8, 131–139 [Google Scholar]
- 65.Finke S and Conzelmann K-K (1999) Virus promoters determine interference by defective RNAs: selective amplification of mini-RNA vectors and rescue from cDNA by a 3′ copy-back ambisense rabies virus. J. Virol 73, 3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duhaut SD and Dimmock NJ (2002) Defective segment 1 RNAs that interfere with production of infectious influenza A virus require at least 150 nucleotides of 5′ sequence: evidence from a plasmid-driven system. J. Gen. Virol 83, 403–411 [DOI] [PubMed] [Google Scholar]
- 67.Meng B et al. (2017) Unexpected complexity in the interference activity of a cloned influenza defective interfering RNA. Virol. J. 14, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida A et al. (2018) A single amino acid substitution within the paramyxovirus Sendai virus nucleoprotein is a critical determinant for production of interferon-beta-inducing copybacktype defective interfering genomes. J. Virol 92 e02094–02017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Odagiri T and Tobita K (1990) Mutation in NS2, a nonstructural protein of influenza A virus, extragenically causes aberrant replication and expression of the PA gene and leads to generation of defective interfering particles. Proc. Natl. Acad. Sci 87, 5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pfaller CK et al. (2014) Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J. Virol 88, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfaller CK et al. (2015) Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J. Virol 89, 7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ngunjiri JM et al. (2012) Influenza virus subpopulations: exchange of lethal H5N1 virus NS for H1N1 virus NS triggers de novo generation of defective-interfering particles and enhances interferon-inducing particle efficiency. J. Interf. Cytokine Res 33, 99–107 [DOI] [PubMed] [Google Scholar]
- 73.Pérez-Cidoncha M et al. (2014) An unbiased genetic screen reveals the polygenic nature of the influenza virus anti-interferon response. J. Virol 88, 4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fodor E et al. (2003) A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase promotes the generation of defective interfering RNAs. J. Virol 77, 5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang CY et al. (1981) Suppression of vesicular stomatitis virus defective interfering particle generation by a function (s) associated with human chromosome 16. J. Virol 40, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kingsbury DW and Portner A (1970) On the genesis of incomplete Sendai virions. Virology 42, 872–879 [DOI] [PubMed] [Google Scholar]
- 77.Whistler T et al. (1996) Generation of defective interfering particles by two vaccine strains of measles virus. Virology 220, 480–484 [DOI] [PubMed] [Google Scholar]
- 78.Ziegler CM et al. (2019) NEDD4 family ubiquitin ligases associate with LCMV Z’s PPXY domain and are required for virus budding, but not via direct ubiquitination of Z. PLoS Pathog. 15, e1008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ziegler CM et al. (2016) A novel phosphoserine motif in the LCMV matrix protein Z regulates the release of infectious virus and defective interfering particles. J. Gen. Virol 97, 2084–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernkopf H (1950) Study of infectivity and hemagglutination of influenza virus in deembryonated eggs. J. Immunol 65, 571. [PubMed] [Google Scholar]
- 81.Holland JJ and Villarreal LP (1975) Purification of defective interfering T particles of vesicular stomatitis and rabies viruses generated in vivo in brains of newborn mice. Virology 67, 438–449 [DOI] [PubMed] [Google Scholar]
- 82.Popescu M and Lehmann-Grube F (1977) Defective interfering particles in mice infected with lymphocytic choriomeningitis virus. Virology 77, 78–83 [DOI] [PubMed] [Google Scholar]
- 83.Buchmeier MJ et al. (1980) The virology and immunobiology of lymphocytic choriomeningitis virus infection In Advances in Immunology (Dixon FJ and Kunkel HG, eds), pp. 275–331, Academic Press; [DOI] [PubMed] [Google Scholar]
- 84.Bean WJ et al. (1985) Characterization of virulent and avirulent A/chicken/Pennsylvania/83 influenza A viruses: potential role of defective interfering RNAs in nature. J. Virol 54, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chambers TM and Webster RG (1987) Defective interfering virus associated with A/Chicken/Pennsylvania/83 influenza virus. J. Virol 61, 1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cattaneo R et al. (1987) Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marcus PI and Sekellick MJ (1977) Defective interfering particles with covalently linked [±]RNA induce interferon. Nature 266, 815–819 [DOI] [PubMed] [Google Scholar]
- 88.Manzoni TB and López CB (2018) Defective (interfering) viral genomes re-explored: impact on antiviral immunity and virus persistence. Futur. Virol 13, 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yount JS et al. (2006) A novel role for viral-defective interfering particles in enhancing dendritic cell maturation. J. Immunol 177, 4503–4513 [DOI] [PubMed] [Google Scholar]
- 90.Perrault J (1981) Origin and replication of defective interfering particles. Curr. Top. Microbiol. Immunol 93, 151–207 [DOI] [PubMed] [Google Scholar]
- 91.Boldogh I et al. (1996) Persistent viral infections In Medical Microbiology (Baron S, ed.), University of Texas Medical Branch at Galveston; [PubMed] [Google Scholar]
- 92.Randall RE and Griffin DE (2017) Within host RNA virus persistence: mechanisms and consequences. Curr. Opin. Virol 23, 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salazar-Bravo J et al. (2002) Mammalian reservoirs of arenaviruses In Arenaviruses I (Oldstone MA, ed.), pp. 25–63, Springer; [DOI] [PubMed] [Google Scholar]
- 94.King BR et al. (2018) Visualization of arenavirus RNA species in individual cells by single-molecule fluorescence in situ hybridization suggests a model of cyclical infection and clearance during persistence. J. Virol 92 e02241–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deen GF et al. (2015) Ebola RNA persistence in semen of Ebola virus disease survivors – final report. N. Engl. J. Med 377, 1428–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Varkey JB et al. (2015) Persistence of Ebola virus in ocular fluid during convalescence. N. Engl. J. Med 372, 2423–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mate SE et al. (2015) Molecular evidence of sexual transmission of Ebola virus. N. Engl. J. Med 373, 2448–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jacobs M et al. (2016) Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 388, 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Calain P et al. (2016) Defective interfering genomes and Ebola virus persistence. Lancet 388, 659–660 [DOI] [PubMed] [Google Scholar]
- 100.Calain P et al. (1999) Ebola virus defective interfering particles and persistent infection. Virology 262, 114–128 [DOI] [PubMed] [Google Scholar]
- 101.Whitmer SLM et al. (2018) Active Ebola virus replication and heterogeneous evolutionary rates in EVD survivors. Cell Rep. 22, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]