Abstract
Immune cells provide defense against non‐self and have recently been shown to also play key roles in diverse processes such as development, metabolism, and tumor progression. The heterogeneity of Drosophila immune cells (hemocytes) remains an open question. Using bulk RNA sequencing, we find that the hemocytes display distinct features in the embryo, a closed and rapidly developing system, compared to the larva, which is exposed to environmental and metabolic challenges. Through single‐cell RNA sequencing, we identify fourteen hemocyte clusters present in unchallenged larvae and associated with distinct processes, e.g., proliferation, phagocytosis, metabolic homeostasis, and humoral response. Finally, we characterize the changes occurring in the hemocyte clusters upon wasp infestation, which triggers the differentiation of a novel hemocyte type, the lamellocyte. This first molecular atlas of hemocytes provides insights and paves the way to study the biology of the Drosophila immune cells in physiological and pathological conditions.
Keywords: Drosophila melanogaster, immune cells, single‐cell RNA‐seq, wasp infestation
Subject Categories: Immunology, Methods & Resources
Single cell RNAseq in normal and wasp infected flies reveals distinct immune cell populations associated with different biological processes.
Introduction
The innate immune response has been the object of intense investigation in Drosophila melanogaster, as this model shows mechanisms that are conserved throughout evolution, from pattern recognition molecules to immune molecular cascades (Akira et al, 2006; Kleino & Silverman, 2014). Given the importance of innate immunity in a variety of physiological and pathological processes including tumor progression (Ratheesh et al, 2015), the current challenge is to characterize immune cell heterogeneity and identify specific hemocyte populations. This is the aim of the present work.
Three classes of hemocytes have so far been identified as follows: the plasmatocytes, the crystal cells, and the lamellocytes (Honti et al, 2014). The plasmatocytes are the most abundant cell type and are responsible for the main functions of the hemocytes: phagocytosis, secretion of extracellular matrix proteins (ECM), signaling molecules, and antimicrobial peptides (AMPs; Yasothornsrikul et al, 1997; Basset et al, 2000; Sears et al, 2003; Ferrandon et al, 2004; Baer et al, 2010; Gold & Bruckner, 2015). The crystal cells account for less than 5% of the total hemocyte population, with distinctive crystals inside them that are composed of prophenoloxidases (PPO; Rizki & Rizki, 1959). These enzymes are released in large quantity upon wounding and constitute a key component for the melanization process (Rizki & Rizki, 1959). The lamellocytes are flat and large cells that only appear upon challenge. They are considered activated immune cells (Gold & Bruckner, 2015) that arise through plasmatocyte trans‐differentiation or from a mitotic‐dedicated precursor (Anderl et al, 2016).
In the embryo, the hemocytes contribute to the clearance of apoptotic cells and the deposition of ECM‐related molecules including Peroxidasin (Pxn) and Viking (Vkg; Nelson et al, 1994; Yasothornsrikul et al, 1997). By the larval stage, the organism interacts with the external environment and responds to metabolic and oxidative stress as well as to infection‐ or injury‐related stimuli. The hemocytes must therefore adapt to these new, highly demanding, settings. In addition, while during embryogenesis, the hemocytes are highly motile and patrol the whole organism, during the larval life a large fraction of them, called resident hemocytes, colonize segmentally repeated epidermal‐muscular pockets in which cell proliferation is enhanced (Makhijani et al, 2011). Upon wounding, septic infection, or infestation by parasitic wasps, the resident hemocytes are mobilized and enter in circulation to reach the site of the immune challenge (Owusu‐Ansah & Banerjee, 2009; Dragojlovic‐Munther & Martinez‐Agosto, 2012). Thus, hemocyte localization adapts to homeostatic and challenged conditions.
We here characterize the transcriptional changes occurring during development and the different types of hemocytes present in the larva. Comparing the bulk RNA sequencing data allows us to define stage‐specific features: In the embryo, hemocytes contribute to the shaping of the tissues and are glycolytic, whereas in the larva, hemocytes show a strong phagocytic potential and a metabolic switch toward internalization of glucose and lipid and toward beta oxidation. The single‐cell RNA sequencing (scRNA‐seq) assay allows us to identify fourteen clusters of larval plasmatocytes and to assign specific molecular and cellular features, including nutrient storage, proliferative potential, antimicrobial peptide production, and phagocytosis.
Finally, as a first characterization of the immune response at the single‐cell level, we assess the transcriptional changes induced by infestation by the parasitic wasp Leptopilina boulardi, one of the most studied pathways linked to cellular immunity. The wasp lays eggs in the Drosophila larva and triggers hemocyte proliferation as well as lamellocyte differentiation (Markus et al, 2009), with subsequent encapsulation of the wasp egg and its death through the increased levels of reactive oxygen species (ROS). The scRNA‐seq assay identifies two lamellocyte populations, a mature one with a strong glycolytic signature, and a population that expresses both lamellocyte and plasmatocyte features, likely originating through trans‐differentiation (Anderl et al, 2016).
The response to wasp infestation involves the embryonic hemocytes that differentiate from the procephalic mesoderm (1st wave of hematopoiesis; Tepass et al, 1994), as well as the hemocytes that originate from the lymph gland, the site of the 2nd hematopoietic wave. While in not‐infested (NI) conditions, the lymph gland histolyses and releases hemocytes in circulation during the pupal life, upon wasp infestation (WI), it undergoes precocious histolysis so that both lymph gland and embryonic‐derived hemocytes populate the larva (Letourneau et al, 2016; Bazzi et al, 2018; Banerjee et al, 2019). Our single‐cell RNA sequencing assay identifies the same number of plasmatocyte clusters as that observed in normal conditions, strongly suggesting that the plasmatocytes from the first and second hematopoietic waves share the same features.
In sum, this work characterizes the transcriptional changes occurring during hemocyte development and the hemocyte populations present in the Drosophila larva. It also provides the molecular signature and the initial characterization of the larval hemocyte repertoire as well as numerous novel markers in NI and in WI conditions. These first bulk and single‐cell RNA‐seq data pave the way to understand the role of the immune system in development and physiology.
Results
Comparing the bulk transcriptomes from embryonic (E16) and larval (WL) hemocytes
In the embryo, insulated from most immune challenges by the eggshell, the hemocytes main functions are developmental. They clear the organism from apoptotic bodies issued from organogenesis and secrete extracellular components. In the larva, the hemocytes display new properties to respond to the microorganism‐rich environment in which they grow. To identify the changes occurring in the hemocytes during development, we compared the hemocytes’ transcriptomes from mature, stage 16 (E16) embryos and from third‐instar wandering larvae (WL).
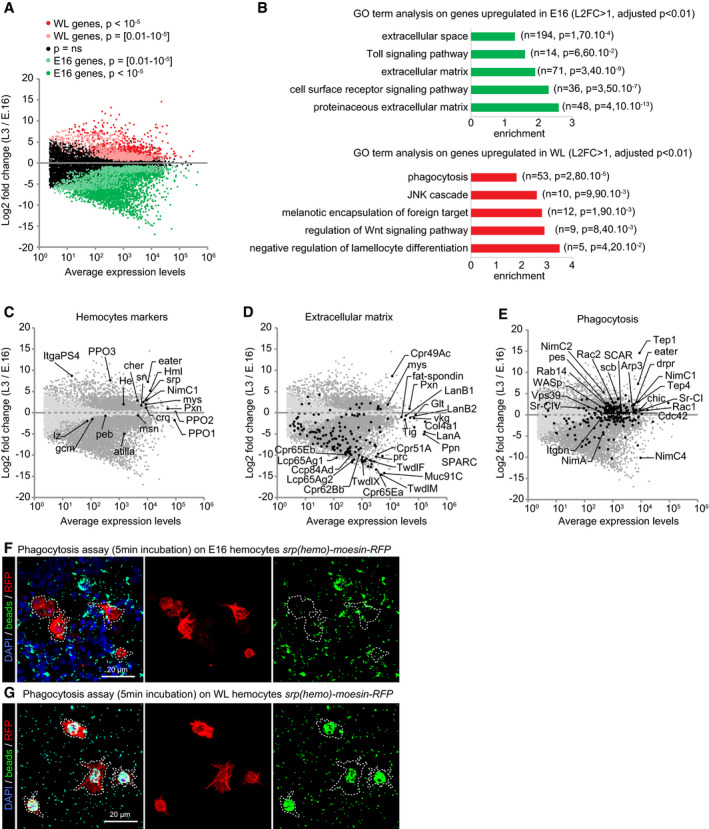
The comparison shows 3,396 genes significantly up‐regulated in E16 and 1,593 up‐regulated in WL hemocytes (Fig 1A, data in Dataset EV1). Most plasmatocyte markers such as Hemese (He; Kurucz et al, 2003), Singed (Sn; Zanet et al, 2009), Eater (Kocks et al, 2005), Hemolectin (Hml; Goto et al, 2001), Serpent (Srp; Shlyakhover et al, 2018), Nimrod C1 (NimC1, also called P1; Kurucz et al, 2007), Croquemort (Crq; Franc et al, 1996), and Pxn (Nelson et al, 1994) are strongly expressed at both stages but enriched in WL hemocytes (Fig 1C). Crystal cell markers are also present in the transcriptome: Pebbled (Peb) and Lozenge (Lz) are detected at relatively low levels, in agreement with the small number of crystal cells in the E16 and WL hemolymph (Rizki & Rizki, 1959). The crystal cell‐specific markers PPO1 and PPO2, on the other hand, are among the genes expressed at the highest levels, highlighting their key function and the sharp specialization of the crystal cells (Binggeli et al, 2014).
Figure 1. Hemocytes display distinct properties at E16 and WL stage.
-
ATranscriptome comparison of hemocytes from stage 16 (E16) embryos and wandering 3rd‐instar larvae (WL). The x‐axis is the average gene expression levels (n = 3), and the y‐axis is the log2 fold change WL/E16. P‐values are indicated with the color code.
-
BGene Ontology (GO) term enrichment analysis in E16 (green) and WL (red) hemocytes. The fold enrichments for a subset of significant GO terms are displayed; the number of genes and the P‐value of the GO term enrichment are indicated in brackets.
-
C–EScatter plots as in (A) highlighting in black subsets of known genes expressed in hemocytes (C) or genes associated with the GO term extracellular matrix (D) and phagocytosis (E).
-
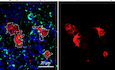
F, GPhagocytosis assay on E16 (F) and WL hemocytes (G) srp(hemo)‐moesin‐RFP. The beads (in green) are phagocytosed by the hemocytes (in red). The WL hemocytes show greater phagocytic capacity compared to the embryonic ones after 5 min of exposure. Full stacks are displayed, and the scale bars represent 20 μm.
Surprisingly, most lamellocyte markers such as myospheroid (Mys or L4; Irving et al, 2005), Misshapen (Msn; Braun et al, 1997), Cher (or L5; Rus et al, 2006), and Atilla (or L1; Honti et al, 2009) were also detected at significant levels in the hemocytes from both stages. This suggests that they are expressed at basal levels in normal hemocytes and are strongly induced in lamellocytes and/or that few lamellocytes are present in basal conditions. At last, Gcm is involved in hemocyte development in the early embryo (stages 8–10; Bernardoni et al, 1997) and is no longer expressed by E16 (Bazzi et al, 2018). Accordingly, Gcm transcripts are barely detected in E16 and WL transcriptomes (levels < 40 normalized read count). Overall, these data prove the efficiency of the experimental design to purify hemocytes.
Embryonic hemocytes express ECM components
We next carried out a GO term enrichment analysis on the genes up‐regulated in either population (|log2 fold change WL/E16| > 1, adjusted P‐value < 0.01; Dataset EV1). The E16 hemocytes display a striking enrichment for gene coding for extracellular matrix components (ECM; Fig 1B and D). Out of 162 gene coding for ECM proteins, 138 are enriched in E16 hemocytes. To confirm the expression pattern of the ECM genes, we compared these data with two in situ hybridization databases (Berkeley Drosophila Genome Project (Hammonds et al, 2013; Tomancak et al, 2002, 2007) and Fly‐FISH (Lecuyer et al, 2007; Wilk et al, 2016); Appendix Fig S1D and E). Most genes for which we could find data are specifically expressed in hemocytes in the embryo (Appendix Fig S1D and E).
The expression/secretion of few specific ECM compounds by the hemocytes during embryonic development was previously described. The integrins alphaPS1 (Mew) and Mys as well as the integrin ligand Tiggrin (Tig) are secreted by the hemocytes at the level of muscle insertion to stabilize strong attachment between the cells (Fogerty et al, 1994; Bunch et al, 1998). The laminins LanA, LanB1, LanB2, and Wb are secreted by the hemocytes for them to migrate efficiently throughout the embryo (Sanchez‐Sanchez et al, 2017). Pxn and the collagen Vkg and Col4a1 secretion by the hemocytes are essential for the condensation of the ventral nerve cord (Olofsson & Page, 2005). Finally, SPARC is produced by the hemocytes and is necessary for basal lamina assembly (Martinek et al, 2008). These 11 compounds are expressed at extremely high levels in the embryo and remain highly expressed in the larva (Fig 1D), suggesting that the role of these specific genes is preserved throughout development.
Among the remaining ECM genes enriched in E16, we distinguished a large group of ECM compounds described as constituent of the cuticle: 23 Tweedles (Twdl), 56 Cuticular Proteins (Cpr and Ccp), nine Larval Cuticle Proteins (Lcp), and nine Mucins (Muc; Fig 1D, annotated in Dataset EV1). This calls for a role of the embryonic hemocytes in cuticle deposition. We also identified 21 ECM genes strongly up‐regulated in the embryo (log2FC < −3, P‐value < 0.01, annotated in Dataset EV1). These include the two heart‐specific ECM compounds Pericardin (Prc) and Lonely heart (Loh;Maroy et al, 1988; Chavez et al, 2000; Charles, 2010), Thrombospondin (Tsp), which interacts with the integrins Mew, Mys, and If at the tendon‐muscle attachment sites (Chanana et al, 2007) and Shifted (Shf) that modulate Hedgehog diffusion (Gorfinkiel et al, 2005; not exhaustive list). This strongly calls for additional embryo‐specific pathways for the deposition of the ECM, in which future studies will elucidate.
Larval hemocytes express specific scavenger receptors
The GO terms enriched in WL compared to E16 hemocytes highlight phagocytosis and, to a lower extent, signaling pathways involved in the immune response (JNK and Wnt; Fig 1B and E, and Dataset EV1).
Among the genes involved in phagocytosis, a large panel is coding for transmembrane phagocytic receptors involved in pathogen recognition, such as the Nimrod family (Eater (Kocks et al, 2005), NimC1 (Kurucz et al, 2007) and NimC2), several scavenger receptors (Sr‐CI and Sr‐CIV (Lazzaro et al, 2004), He, Peste (Cuttell et al, 2008; Hashimoto et al, 2009) as well as the integrins Scab (alpha‐PS3) and Integrin beta‐nu (Itgbn; Nonaka et al, 2013). Noteworthy, the E16 embryonic hemocytes are specifically enriched for NimC4 (also called Simu), a receptor of the Nimrod family that is involved in the phagocytosis of apoptotic bodies (Fig 1E; Kurant et al, 2008; Roddie et al, 2019).
The WL hemocytes are also enriched for opsonins. These secreted molecules bind to the pathogens and promote their phagocytosis by the macrophages. Tep1 and Tep4 (Dostalova et al, 2017; Haller et al, 2018) are among the genes expressed at the highest levels in WL hemocytes, and Tep1 presents the strongest enrichment. Most of the genes involved in phagosome formation are also enriched at this stage: Arp3, Rac1, Rac2, SCAR, WASP, Chic, and Cdc42 (Pearson et al, 2003). Finally, genes involved in phagosome maturation (Rab14) and phagolysosome formation (Vps39) are enriched as well (Fig 1E; Garg & Wu, 2014; Jiang et al, 2014).
The scavenger receptors and the opsonins cover a large panel of pathogens (for review, see Melcarne et al, 2019), indicating an overall switch for hemocytes’ function from apoptotic body scavenging and cuticle production at embryonic stages to pathogen scavenging at the WL stage. Since it was previously shown that the hemocytes present in the embryo are able of phagocytosis (Vlisidou et al, 2009; Tan et al, 2014), we compared the phagocytic capacity of E16 and WL hemocytes upon exposing them to fluorescent beads. The results clearly show that the larval hemocytes phagocytose faster and more than the embryonic ones (Fig 1F and G).
In sum, the transcriptome analysis reveals a change in the function of the hemocytes during development, from building the ECM and the cuticle to adopting a defense profile against immune challenges.
Metabolic shift between embryonic and larval hemocytes
The properties of the immune cells are directly dependent on their metabolic state, which is constrained by their micro‐environment (reviewed in Sieow et al, 2018). We hypothesized that the hemocytes display distinct metabolic states according to the nutritional environment present in the two developmental stages, as embryos are closed systems, whereas larvae have been feeding for most of their life. To address this hypothesis, we analyzed the expression profiles of the energy metabolic pathways in E16 and WL hemocytes.
The transcriptome data comparison reveals that the larval hemocytes are most likely internalizing and metabolizing lipids through the beta oxidation pathway to generate acetyl CoA and drive the TCA cycle (Appendix Fig S2A–C). This notion is supported by the up‐regulation of genes encoding lipid‐scavenging receptors, and the down‐regulation of genes is involved in lipid biosynthetic (TAG) pathway (Appendix Fig S2A and B).
The down‐regulation of genes involved in glycolysis, mainly phosphofructokinase and pyruvate dehydrogenase (Appendix Fig S2B), implies that the larval hemocytes do not rely on this process to drive the TCA cycle. The transcriptional down‐regulation of gluconeogenic genes (phosphoenolpyruvate carboxykinase and fructose 1, six bisphosphatase) suggests the absence of gluconeogenesis in these cells. However, a significant up‐regulation of the Glut1 sugar transporter suggests active uptake of glucose by the larval hemocytes. The down‐regulation of glycolytic genes downstream of G6P and up‐regulation of genes of the pentose phosphate pathway (PPP) imply that the internalized glucose could be potentially used to generate pentose sugars for ribonucleotide synthesis and redox homeostasis through the generation of NADPH. Corroborating this observation is also the strong up‐regulation of redox homeostatic enzymes (Appendix Fig S2A).
In contrast to the larval hemocytes, the E16 hemocytes are glycolytic and rely less on oxidative metabolism (Appendix Fig S2B). This is supported by the strong up‐regulation of a key glycolytic enzyme, lactate dehydrogenase, which is essential for the conversion of pyruvate to lactate. Furthermore, these cells likely metabolize lipids at a lower level, as enzymes of the beta oxidation pathway are transcriptionally down‐regulated compared to the larval hemocytes.
Generation of single‐cell RNA‐seq datasets from NI and WI larvae
The Drosophila larva contains plasmatocytes and crystal cells that are resident or in circulation. Upon wasp infestation, the lamellocytes are produced from precursors (Anderl et al, 2016) or by plasmatocyte trans‐differentiation (Stofanko et al, 2010). To obtain a comprehensive repertoire of the hemocyte populations present in the larva, we generated two single‐cell libraries on the hemocytes from not‐infested WL (NI dataset) and from WL infested by the parasitoid wasp L. boulardi (WI dataset). The NI cells comprise the resident and the circulating embryonic‐derived hemocytes, and the WI cells include in addition the hemocytes released from the lymph gland.
The hemocytes were collected from pools of 20 female larvae. The libraries were produced using the Chromium single‐cell 3′mRNA‐seq protocol (10 × Genomics). The NI library contains 7,606 cells (mean read per cell = 37,288; median genes per cell = 959) and the WI library 8,058 cells (mean read per cell = 32,365; median genes per cell = 1,250). The libraries were merged to cluster the hemocytes presenting similar expression profiles using the Seurat toolkit (Butler et al, 2018; Stuart et al, 2019; Appendix Fig S3A). Subclustering was then applied to refine the grouping of the cells leading to the identification of 16 clusters of hemocytes (Appendix Fig S3A′ and C–D″). The identity of each cluster was assigned using the list of known markers for the crystal cells (Lz, Peb, PPO1, PPO2), for the lamellocytes (Mys, Msn, Cher, Atilla, ItgaPS4, PPO3), and for the plasmatocytes (Sn, Pxn, Hml, Eater, NimC1, Crq, He, Srp; Appendix Fig S3B). Of note, the single‐cell data show that the lamellocyte markers Mys, Msn, Cher, and Atilla detected in the bulk RNA‐seq on WL and E16 are expressed in the plasmatocytes and enriched in the lamellocytes (Appendix Fig S3B).
We identified 13 clusters of plasmatocytes and one cluster of crystal cells in both the NI and the WI larvae and two clusters of lamellocytes specifically found in the WI larvae. The name of each cluster corresponds to the name of one of the main markers or to specific biological features (Fig 2A and B). Importantly, all cells analyzed in these datasets present known hemocyte markers, which indicate a high purity of the samples.
Figure 2. Fourteen hemocyte populations can be distinguished in WL by single‐cell RNA‐seq.
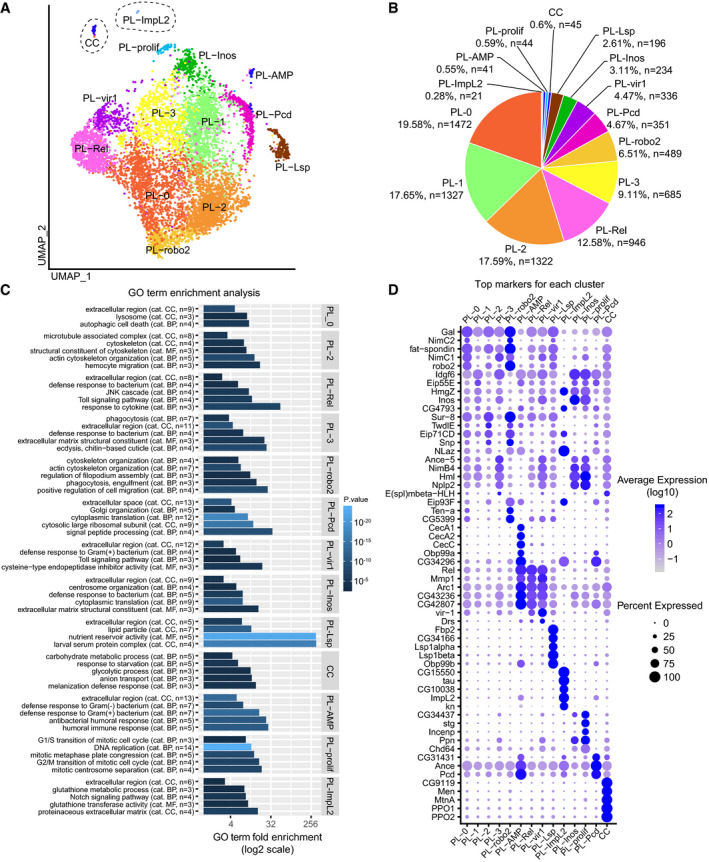
- UMAP projection representing the 14 clusters of cells identified in the hemocyte pools from OregonR WL (NI dataset).
- Number of cells and proportion of each cluster in the NI dataset.
- GO term enrichment analysis for each cluster. The x‐axis is the GO term enrichment, the color gradient (black to light blue) indicates the P‐value, and the number of genes and the GO term category (CC: cellular compound, BP: biological process, MF: molecular function) are indicated between brackets.
- Top 5 markers of each cluster. The expression levels are represented by the gradient of purple levels) and the percentage of cells with the size of the dots.
Characterization of the transcriptomic profile in normal conditions
Following the identification of the clusters, our first aim was to characterize the properties of the clusters in the NI dataset. Thus, we carried out GO term analyses on the genes enriched in each of them (Dataset EV2, Fig 2C) using DAVID (Huang da et al, 2009). In addition, to estimate whether the clusters are enriched/specifically localized in the circulating or in the resident compartments, we performed qPCR assays on hemocytes from either compartment to measure the expression levels of the strong markers of the different clusters (Fig 3A).
Figure 3. Localization of the NI hemocyte clusters.
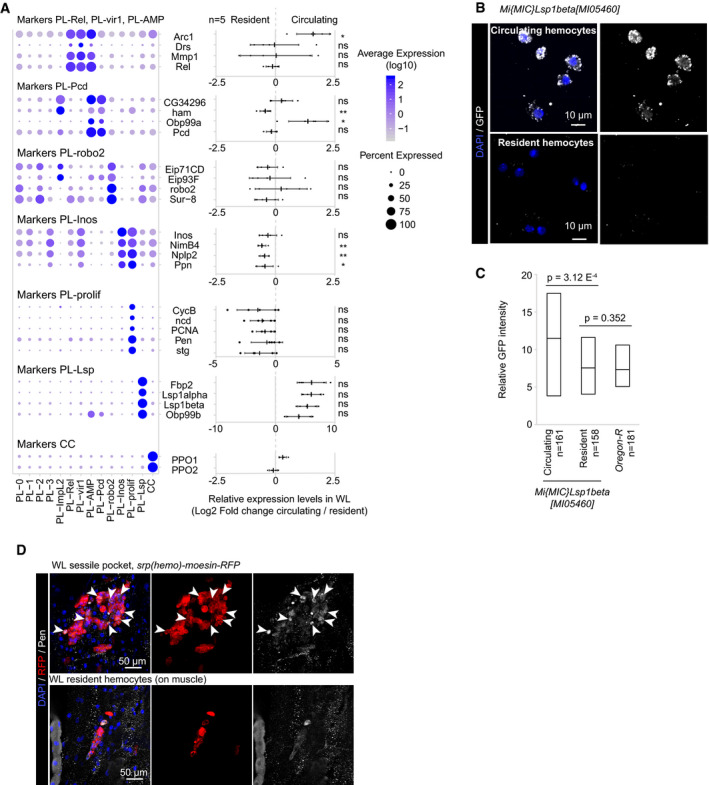
- Identification of the position (circulating/resident) of the clusters within the larva by qPCR. The left panel indicates the distribution of each marker across all clusters (as in Fig 2D), and the right panel indicates the log2 of the ratio between the expression level in the circulating versus the resident compartment. Positive values indicate an enrichment in the circulating compartment and negative values in the resident compartment (n = 5, mean ± SD is represented on the graph). The P‐values are estimated by bilateral paired Student's t‐test and indicated as follow: ns = non‐significant (> 0.05), “*” = P[0.05–0.01[, “**” = P[0.01–0.001[.
- Circulating and resident hemocytes (top and bottom panels, respectively) from Mimic‐Lsp1beta‐MI05460 WL, which express Lsp1beta tagged with GFP. The immunolabeling was done using anti‐GFP (in gray), and the nuclei were marked with DAPI (blue). Full stacks are displayed, the left panels show the overlay of DAPI and GFP, and the right panels show the GFP alone. The scale bars represent 10 μm.
- Quantification of the GFP intensity in circulating and resident hemocytes from MiMIC‐Lsp1beta‐MI05460 larvae. The OregonR value indicates the background level. The number of cells included in the analysis is displayed on the x‐axis label and is quantified from two independent preparation; P‐values were estimated after variance analysis with bilateral student test for equal variance.
- Resident hemocytes located around the oenocytes or along the muscles (top and bottom panels, respectively) from WL srp(hemo)‐moesin‐RFP that express moesin tagged with RFP in hemocytes. The immunolabeling was done using anti‐RFP (in red) and anti‐pendulin (Pen, in gray), and the nuclei were marked with DAPI (blue). Full stacks are displayed; the left panels show the overlay of DAPI, Pen, and RFP; the middle panels show RFP alone; and the right panels show Pen alone. Arrowheads in the top panels indicate cells co‐expressing RFP and Pen. The scale bars represent 50 μm.
The plasmatocyte clusters PL‐0, PL‐1, PL‐2, and PL‐3 encompass more than 60% of all the hemocytes. These clusters express most of the plasmatocyte markers (Appendix Fig S4A) but do not display strong distinct signatures (avg_logFC of the strongest markers < 0.93; Fig 2D); therefore, they can only be distinguished by combining several markers. The nine remaining plasmatocyte clusters present very specific molecular signatures in addition to the main plasmatocyte markers (Fig 2D) and are organized at the periphery of the four clusters on the graphical representation generated with the UMAP dimension reduction technique (Becht et al, 2018; Fig 2A).
PL‐Rel
The PL‐Rel cluster includes 12.6% of the total hemocyte population, with more than 100 strong markers (Dataset EV2), most of which are involved in the immune response (Fig 2C). The cluster expresses the main transcription factors of the Imd pathway (i.e., Relish, Rel) and of the Toll pathway (i.e., Dorsal, Dl), Cactus (Cact), and the secreted protein PGRP‐SA (Govind, 1999; Valanne et al, 2011; Zhai et al, 2018; Dataset EV2). In addition, it expresses proteins associated with the JNK pathway such as Jra and Puc (Martin‐Blanco et al, 1998; Zheng et al, 2017). The qPCR assay on resident and circulating hemocytes indicates that PL‐Rel hemocytes are present in both compartments (Fig 3A).
PL‐vir1
The PL‐vir1 cluster contains 4.5% of the total hemocytes and expresses the same strong markers as PL‐Rel, including Rel, Jra, and Puc (Fig 2D, Dataset EV2). Compared to PL‐Rel, however, the marker of viral infection Vir1 (Dostert et al, 2005), the protein Pastrel involved in resistance to virus infection (Magwire et al, 2012; Martins et al, 2014), the predicted peptidase Ance‐5, and the apolipoprotein Nplp2 (Rommelaere et al, 2019) are all up‐regulated. PL‐vir1 also presents the same GO terms as PL‐Rel, with respect to the defense response to bacterium and to the Toll signaling pathway (Fig 2C). The qPCR assay suggests that PL‐vir1 is present in both resident and circulating compartments.
PL‐robo2
The PL‐robo2 cluster represents 6.5% of the total hemocyte population. It does not express unique markers (Fig 2D), but displays a strong enrichment for GO terms related to migration and phagocytosis (Fig 2C). It expresses the actin‐regulatory protein enable (Tucker et al, 2011; Stedden et al, 2019) and transmembrane proteins that participate in the migration of multiple cell types. It also presents the strongest up‐regulation of the phagocytic receptors Crq and Drpr (Franc et al, 1999; Manaka et al, 2004). In addition, Crq is the main receptor involved in lipid scavenging and is a major actor of the induction of the inflammatory response to high‐fat diet initiated by the hemocytes (Woodcock et al, 2015). In line with this, PL‐robo2 is enriched in the lipid droplet‐associated protein Jabba involved in the regulation of lipid metabolism (McMillan et al, 2018). PL‐robo2 is present in both circulating and resident compartments (Fig 3A).
PL‐Pcd
The PL‐Pcd cluster contains 4.7% of the total hemocyte population and is mostly linked to translation and Golgi organization (Fig 2C): Half of the proteins present in the GO term‐enriched functions are ribosomal proteins (three RpS including Rps3 and 11 RpL), while the others are related to Golgi (Gmap, Ire‐1, Sec23; Friggi‐Grelin et al, 2006; Norum et al, 2010; Zacharogianni et al, 2011). This cluster also specifically expresses the pterin Pcd involved in amino acid metabolism, the peptidase Ance involved in proteolysis, and two uncharacterized genes CG31431 and CG34296 (Fig 2D).
The qPCR data highlight one marker (Ham) enriched in the resident compartment and one marker (Obp99a) enriched in circulation. These ambiguous results may be due to the fact that the markers are expressed in other clusters as well (PL‐AMP and PL‐ImpL2).
PL‐AMP
The PL‐AMP cluster (0.5% of the hemocytes) presents strong similarities with PL‐Rel, as they share the same markers related to the Imd pathway (Fig 2D), but distinguishes itself by the expression of the antimicrobial peptides (AMP) Cecropin A1 (CecA1), Cecropin A2 (CecA2), Cecropin C (CecC), Attacin‐A (AttA), Attacin‐B (AttB), and Attacin‐D (AttD; Fig 2D and Dataset EV2) as well as of the PL‐Pcd cluster markers Pcd, Ance, CG31431, and CG34296 (Fig 2D). The AMP are usually induced and secreted primarily by the fat body after septic wounds that trigger the Imd pathway (Govind, 1999; Zhai et al, 2018) and by a small percentage of hemocytes (Samakovlis et al, 1990; Meister et al, 1994).
The expression of CecC, CecA1, and CecA2 was not detected by qPCR in the resident nor in the circulating hemocytes. This is likely due to the low representation of these cells combined with the small size of the Cecropin transcripts (less than 400nt) that prevent the optimal design of primers and affect PCR efficiency. Therefore, we could not conclude on the localization of the PL‐AMP hemocytes in the larvae.
PL‐Inos and PL‐prolif
The clusters PL‐Inos and PL‐prolif express several common markers and represent approximately 3 and 0.6% of the total hemocyte population, respectively (Fig 2B). Initially, PL‐Inos and PL‐prolif were clustered together and were splitted upon subclustering (Appendix Fig S3A and C), which indicates that they present strong similarities and may represent two states of the same cell population.
The PL‐Inos cluster is enriched in GO terms associated with multiple functions including the response to bacteria, the ECM, cytoplasmic translation, and centrosome organization.
The PL‐prolif cluster is specifically characterized by genes involved in mitosis (Fig 2C and Dataset EV2). Klp61F, Klp67A, and Ncd, which are linked to mitotic centrosome separation (Sharp et al, 1999; Gandhi et al, 2004), as well as Ncd80 and Nuf2, which are part of the NCD80 complex, are all up‐regulated. Like their mammalian counterparts, these proteins are essential for mitotic metaphase plate congregation (Przewloka et al, 2007). Finally, PL‐prolif hemocytes express two cyclins (CycB and CycE), the nuclear protein transporter Pen and cell cycle‐related enzymes (String (Stg), the cyclin‐dependent kinases Cdk1 and Cdk2), which are linked to the G2/M and G1/S transition (Kussel & Frasch, 1995; Yuan et al, 2016).
The qPCR assays indicate that the clusters PL‐Inos and PL‐prolif are enriched in the resident compartment (Fig 3A and D). This is concordant with a previous analysis, indicating that the resident hemocytes are more proliferative than the circulating ones (Makhijani et al, 2011).
PL‐Lsp
The PL‐Lsp cluster contains approximately 3% of the total hemocyte population and is strongly associated with the GO term larval serum protein complex, nutrient reservoir activity, and lipid particle (Fig 2C and Dataset EV2). This is due to the expression of the larval serum proteins (LSP), which serve as a nutrient pool that will be used during metamorphosis (Telfer & Kunkel, 1991). The PL‐Lsp cluster also expresses the receptor responsible for the incorporation of the LSPs (i.e., Fbp1), proteins associated with lipid transport (Rfabg; Kutty et al, 1996; Massey et al, 1997; Burmester et al, 1999), and the odorant binding protein Obp99b that is considered as a storage protein (Handke et al, 2013). These proteins are usually described as secreted by the fat body, suggesting shared features and role in metabolism between this tissue and PL‐Lsp.
The qPCR on resident and circulating plasmatocytes reveals that the PL‐Lsp hemocytes are mostly in circulation (Fig 3A). This localization is supported by labeling of two independent LSP transgenic reporters (Fig 3B and C, Appendix Fig S5A–C).
PL‐ImpL2
The PL‐ImpL2 cluster comprises less than 0.3% of the hemocytes, but displays the most distinctive molecular signature (49 genes presenting an enrichment avg_logFC > 1; Dataset EV2). This includes genes involved in glutathione metabolism (GstD1, GstD3, GstE12) and specific transcription factors (Ham, Kn, Antp, Eip93F, Noc, and ElB; Fig 2C, and Appendix Fig S5D and E).
Many of these markers are usually associated with the postsignaling center (PSC) that regulates the differentiation of lamellocytes within the lymph gland (Crozatier et al, 2004; Mandal et al, 2007; Benmimoun et al, 2015). The low abundance of these cells renders them hard to track in the larva; however, the GstD reporter GstD‐LacZ (Sykiotis & Bohmann, 2008) labels subsets of WL hemocytes that may correspond to the PL‐ImpL2 cluster (Appendix Fig S5E).
CC
The CC cluster expresses the two well‐known crystal cell markers PPO1 and PPO2 (Dudzic et al, 2015). In addition, we identify new potential markers: Metallothionein A (MtnA), Malic enzyme (Men), and CG9119. Further analysis of the GO terms highlights the response to starvation (Lipin (Lpin), Mthl10) as well as enzymes essential for the biosynthesis of proteoglycans (Sugarless, sgl) and glucose homeostasis (Pfk, 6‐phosphofructo‐2‐kinase (pfrx), and Aldolase (ald); Hacker et al, 1997; Flowers et al, 2007; Ugrankar et al, 2011; Dudzic et al, 2015; Enzo et al, 2015; Sung et al, 2017; Wong et al, 2019).
Finally, GO terms related to “extracellular region” are enriched recurrently in almost every cluster (Fig 2C). Further analyses of all the genes expressed in the NI dataset and associated with the term extracellular region show that each cluster expresses different proteins (Appendix Fig S4B).
Characterization of the molecular pathways active in the clusters
To identify which regulatory networks characterize the clusters, we performed a regulon analysis using SCENIC (Aibar et al, 2017; Fig 4A). SCENIC defines the regulon as an ensemble of genes coregulated by a single transcription factor and determines the regulon(s) active in single cells in three steps. First, covariation of the expression levels of the genes is estimated in the single‐cell dataset. Then, each group of genes displaying covariation is screened for common cis‐regulatory motifs present in the group. This defines groups of genes regulated by a specific transcription factor (=regulon). Finally, the activity of each regulon is estimated in each cell of the dataset (Aibar et al, 2017). Of note, this analysis was done independently of our initial clustering based on the expression levels. The clustering of the WT NI cells based on the regulons is highly comparable to the expression‐based clustering, with a high overlap between the two clustering approaches, as estimated by the Rand index or RI (Rand, 1971; RI = 0.83; see the Materials and Methods section). This overlap highlights the robustness of our initial clustering.
Figure 4. Identification of the cluster‐specific molecular pathways and of the filiation between the NI clusters.
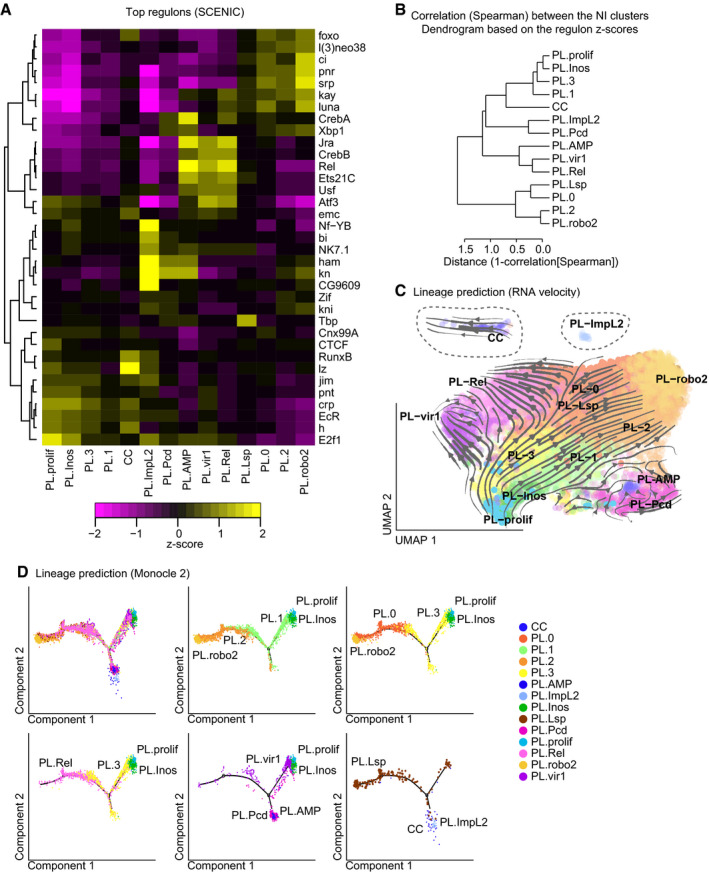
- Heatmap representing the z‐score for the top 5 regulons of each cluster determined with SCENIC. The dendrogram on the left side of the panel indicates the correlation between the regulons across the dataset. The z‐score is indicated with a gradient from magenta (z‐score < 0, the regulon is repressed) to yellow (z‐score > 0, the regulon is active).
- Dendrogram representing the distance among the clusters. The tree was built on the correlation (Spearman) calculated on the regulon matrix from the NI dataset.
- Lineage prediction using RNA velocity. The arrows and lines on the UMAP predict the “direction” taken by the cells of each cluster, based on the comparison between the levels of mRNA and pre‐mRNA. Note that the clusters CC and PL‐ImpL2 (dashed lines) have been moved from their original position to fit in the graph.
- Single‐cell trajectory reconstructed with Monocle 2 on the NI dataset. The first panel shows the overlap of all clusters, and subsequent panels show restricted number of clusters for which the RNA velocity analysis suggested a filiation.
The SCENIC analysis allowed us to associate specific regulons to each cluster (Fig 4A). Concordant with previous reports, the analysis highlighted a positive correlation between the regulon lz and the crystal cells. Lz is a Runx transcription factor essential for the differentiation of these cells (Lebestky et al, 2000). The PL‐Rel, PL‐AMP, and PL‐vir1 clusters are characterized by the regulons Jra (JNK cascade), Rel (IMD pathway), and Ets21C (which cooperates with the JNK pathway). The regulon activating transcription factor 3 (ATF3) involved in anti‐viral response in mammals (Labzin et al, 2015) is specific to PL‐vir1 and PL‐Rel, whereas the regulon CrebA regulating the secretory pathway is specific to the PL‐AMP cluster, which expresses most of the antimicrobial peptides. The PL‐prolif cluster and the closely related cluster PL‐Inos (Fig 4B) are enriched for the regulons EcR and E2F1 that are involved in the regulation of hemocyte proliferation (Sinenko et al, 2010). PL‐Lsp is associated with the Tbp regulon that is involved in the canonical transcriptional machinery. Such enrichment may indicate a higher rate of transcription, which would be concordant with the function of this cluster in producing storage proteins in preparation for pupariation. PL‐robo2 is enriched for the regulons of the GATA factors Pnr and Srp, which is known to regulate the expression of scavenger receptors in the hemocytes (Shlyakhover et al, 2018; Valanne et al, 2018). At last, PL‐ImpL2 is enriched in Ham, Kn, CG9609, and Nf‐YB. Ham was shown to limit amplifying divisions in neural stem cells (Eroglu et al, 2014). CG9609 is a zinc finger transcription factor poorly described, expressed mostly in ovaries (Robinson et al, 2013), and Nf‐YB regulates cell death and proliferation (Ly et al, 2013).
Developmental links between the different hemocyte populations in NI animals
The GO term and the regulon analyses reveal distinct functions and properties for specific hemocyte clusters. The identification of the proliferative cluster prompted us to ask whether there is a filiation among the clusters and, if so, to define their hierarchical organization. We adopted two distinct strategies to predict the hierarchy between the clusters: RNA velocity (La Manno et al, 2018) and Monocle (Trapnell et al, 2014; Qiu et al, 2017a,b).
RNA velocity compares unspliced and spliced transcripts in the single‐cell dataset, to evaluate the developmental direction of single cells and to generate a UMAP displaying the link between cells (La Manno et al, 2018). Following this, the cluster identities were appended to the RNA velocity map (Fig 4C). The map suggests that PL‐prolif/PL‐Inos is at the origin of most clusters, that PL‐0, PL‐1, PL‐2, and PL‐3 are derived from PL‐Inos, and that PL‐vir1, PL‐Rel, and PL‐robo2 are issued from PL‐3, PL‐0, and PL‐2 (Fig 4C). The comparison of the RNA velocity results with the regulons (Fig 3C) suggests the pathways involved in the acquisition of the specific properties. First, from PL‐prolif to PL‐Inos and then to PL‐1/PL‐3, we observe a gradual reduction in the regulons EcR and E2f1. Then, the JNK‐associated regulons (Jra, Ets21C) and the regulon Rel become progressively enriched starting from PL‐3 to PL‐Rel and PL‐vir1 clusters. For the PL‐robo2 branch, we observe a gradual enrichment of the regulons associated with the GATA factors Srp and Pnr and with the Hedgehog pathway (Ci) from PL‐1/PL‐3 to PL‐2/PL‐0. Concerning the remaining clusters, PL‐Lsp is scattered over the clusters PL‐0/PL‐1/PL‐2/PL‐3, suggesting that it is also issued from PL‐prolif/PL‐Inos, but the directionality is unclear. At last, no clear directionality could be drawn for PL‐AMP, PL‐Pcd, CC, and PL‐ImpL2, which suggests that their direct progenitors are not detected in our dataset (Fig 4C).
The second approach, Monocle, estimates the cell trajectories by first defining the sequences of gene expression changes required to adopt distinct cell states and then by positioning the cells on the trajectories according to their transcriptomes (Trapnell et al, 2014; Qiu et al, 2017a,b). The Monocle analysis identified trajectories in line with the branches observed with RNA velocity (Fig 4D). The PL‐prolif/PL‐Inos clusters are at one extremity, followed by PL‐1/PL‐3, then PL‐2/PL‐0, with PL‐robo2 at the other extremity. PL‐Rel follows the PL‐3 > PL‐0 axis, and PL‐vir1 follows the same direction as PL‐Inos > PL‐3.
Overall, these two distinct approaches return concordant results in terms of biological interpretation of the relationship between the clusters. We infer from these data that PL‐prolif is at the origin of PL‐0 to PL‐3, PL‐vir1, PL‐Rel, and PL‐robo2. In this model, PL‐0 to PL‐3 would represent the bulk of the plasmatocytes that may then specialize into PL‐vir1, PL‐Rel, and PL‐robo2.
Clusters’ dynamics upon wasp infestation
In response to wasp infestation, the Drosophila larva displays a strong immune reaction involving multiple organs such as the muscle, the fat body, the lymph gland, and the hemocytes (Banerjee et al, 2019). A key response is triggered by the hemocytes, with the production of the lamellocytes, a fraction of whose aggregate around the wasp egg and encapsulate it to prevent its hatching. During this process, the hemocytes present in the resident compartment and in the lymph gland are recruited through cytokine secretion by the circulating hemocytes, the fat body, and the muscles (reviewed in Banerjee et al, 2019; Kim‐Jo et al, 2019; Letourneau et al, 2016). To characterize the diversity of hemocytes generated by this systemic immune response, including the hemocytes released from the lymph gland, we produced single‐cell RNA‐seq data on the cells of the hemolymph from infested larvae.
The efficiency of the immune response of Drosophila larvae to wasp infestation critically depends on multiple parameters including the intensity of exposure (number of wasps, duration of the infestation), the genetic background of the Drosophila larvae, and the developmental stage and the temperature at which the infestation occurs (personal observations). To obtain a uniform response, we developed a protocol of mild infestation that maximizes the number of larvae containing a single wasp egg (Bazzi et al, 2018) and the survival rate of the Drosophila OregonR hosts (see Materials and Methods, Appendix Fig S6A). In these optimized conditions, we observe first the production of lamellocytes and a decrease in plasmatocytes, which suggests that the plasmatocytes trans‐differentiate into lamellocytes from the resident and circulating pool of hemocytes (24 h after infestation). Following this, the lymph glands histolyse (> 90% of the lymph glands histolysed 48 h after infestation) and the hemocytes proliferate (Appendix Fig S6B–D). By 72 h after infestation (which corresponds to the time of hemocyte collection for the scRNA‐seq), all lymph glands are histolysed and the hemolymph contains hemocytes (plasmatocytes, crystal cells, and lamellocytes) from the two hematopoietic waves (Appendix Fig S7A–F; Bazzi et al, 2018). It is important to note that the WI dataset covers only the cells that have not encapsulated the wasp egg.
Compared to the NI dataset, the single‐cell data on the WI hemolymph display two novel cell populations (LM‐1 and LM‐2) expressing the lamellocyte markers (Fig 5A and Appendix Fig S3B). The other clusters are already present in the NI dataset. PL‐Inos, PL‐prolif, PL‐vir1, and PL‐0 to PL‐3 clusters increase in cell number, and the size of the PL‐Lsp and PL‐robo2 clusters remains constant, whereas that of the CC, PL‐Pcd, and PL‐Rel clusters decreases after wasp infestation (Fig 5B).
Figure 5. Two additional hemocyte populations are produced after wasp infestation.
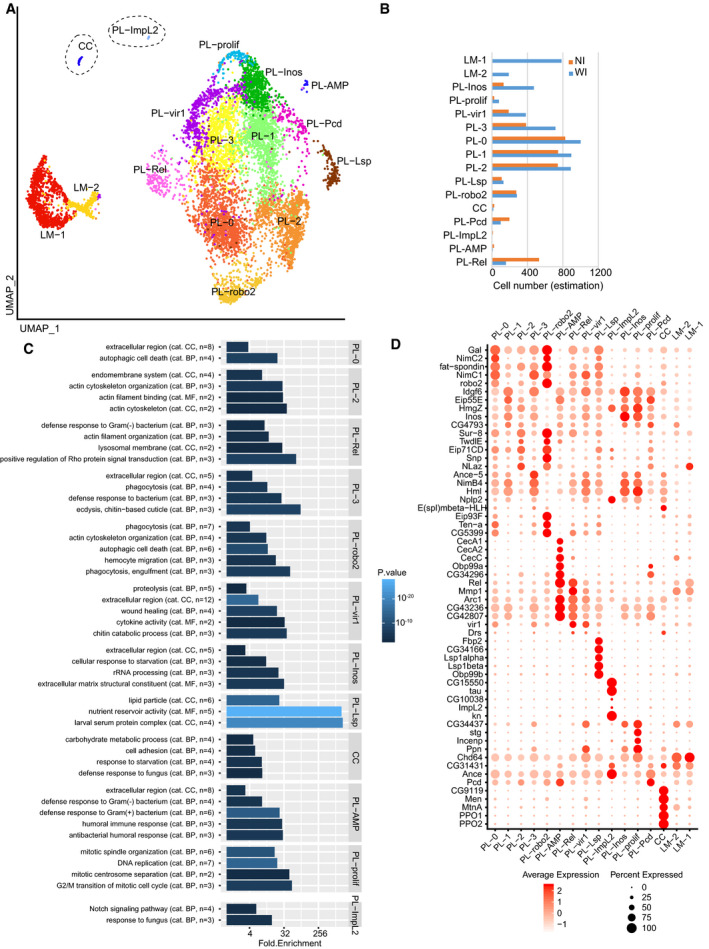
- UMAP projection representing the 16 clusters of cells identified in the WI sample. Note the presence of two additional clusters, LM‐1 and LM‐2, compared to the NI sample. The two clusters correspond to the lamellocytes.
- Estimation of the number of cells of each cluster per larva in normal condition (NI, orange) and after wasp infestation (WI, blue). For each cluster, the cell number was deduced from the total number of plasmatocytes and lamellocytes numerated in Appendix Fig S5D, and the proportion of each cluster in the single‐cell datasets.
- GO term enrichment analysis for each cluster of the WI dataset. Represented as in Fig 2C.
- Dot plot representing the expression levels (gradient of red levels) and the percentage of cells (size of the dots) that express the top 5 markers of each cluster in the WI dataset.
Comparisons between NI and WI data were carried out for each cluster but did not reveal strong transcriptomic modifications induced by the infestation. Each NI cluster is highly correlated with its WI counterpart (Appendix Fig S6E), and the markers and the GO terms remain rather similar (Fig 5C and D).
Characterization of the two lamellocyte clusters and their developmental links to the other clusters
To characterize further the two lamellocyte clusters, we compared first their transcriptomes to all the other clusters and carried out GO term enrichment analyses (Fig 6A and B). This comparison indicates an enrichment for genes involved in melanotic encapsulation, which is the primary role of the lamellocytes, a strong implication of the JNK pathway, which was previously implicated in lamellocytes production (Zettervall et al, 2004), and several GO terms related to cytoskeleton reorganization and integrin‐mediated cell adhesion, two processes that are necessary for the formation of the capsule around the wasp egg (Irving et al, 2005).
Figure 6. Characterization of the two lamellocyte clusters.
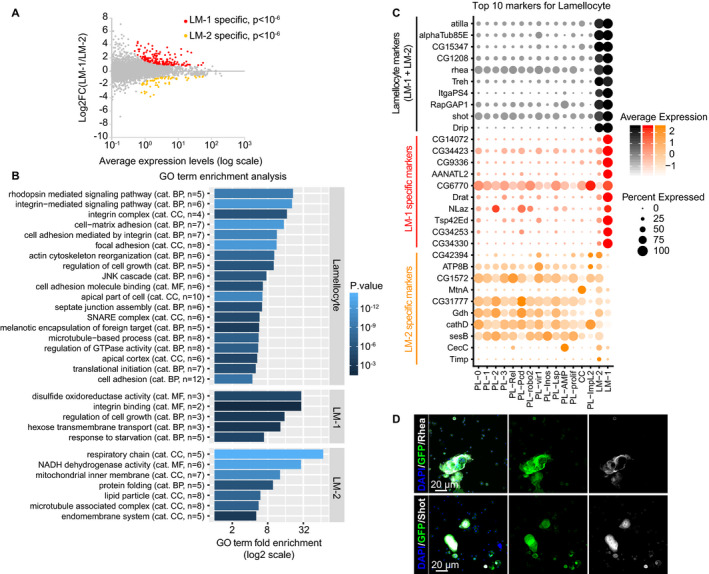
- Scatter plot comparing the transcriptome of the two lamellocyte clusters, deduced from the WI dataset. The x‐axis is the average expression levels of the genes and the y‐axis the log2 fold change (LM‐1/LM‐2). The genes significantly enriched in the LM‐1 cluster are highlighted in red (P < 10−6), those significantly enriched in the LM‐2 cluster in orange (P < 10−6).
- GO term enrichment analysis on the genes enriched in lamellocytes compared to all other clusters (top panel) and on the genes specific to LM‐1 (middle panel) or LM‐2 (lower panel). The bars represent the fold enrichment, and the color gradient indicates the P‐value of the GO term enrichment (as in Fig 2C).
- Expression levels (gradient of color) and the percentage of cells (size of the dots) for the top 10 markers of lamellocytes (in black), of markers specific to LM‐1 compared to LM‐2 (in red) and specific to LM‐2 compared to LM‐1 (in orange). Note that most markers enriched in LM‐1 compared to LM‐2 are exclusively expressed in LM‐1, while most markers enriched in LM‐2 compared to LM‐1 are also expressed in the plasmatocyte and crystal cell clusters.
- Hemocytes from WL OregonR infested by wasp. The immunolabeling was done with antibodies targeting the new lamellocyte markers identified in this study Rhea (in gray, top panels) and Shot (in gray, lower panels). Phalloidin‐FITC (in green) labels the actin filament particularly abundant in lamellocytes (Tokusumi et al, 2009), and the nuclei were marked with DAPI (blue). Full stacks are displayed; the left panels show the overlay of DAPI, FITC, and the lamellocyte markers; the middle panels show the FITC alone; and the right panels show the lamellocyte markers alone. The scale bars represent 20 μm.
We then compared the LM‐1 and LM‐2 clusters and found 157 genes up‐regulated in LM‐1 and 58 genes up‐regulated in LM‐2 (Fig 6A). Only few GO terms are enriched specifically in one or the other cluster: The LM‐1‐specific genes are involved in integrin processing, while the LM‐2‐specific genes are involved in cytoskeleton and mitochondrial processes (Fig 6B). The analysis of the top 10 markers for each population shows that the markers common to LM‐1 and LM‐2 include known and novel lamellocyte‐specific markers (including Atilla, ItgaPS4, Rhea, Shot; in black in Fig 6C and D; listed in Dataset EV6). The markers enriched in LM‐1 are expressed at low levels in LM‐2 and in the other hemocyte clusters, whereas the markers enriched in LM‐2 are also expressed in most hemocyte clusters (Fig 6C). This suggests that the LM‐1 cluster represents the mature lamellocytes and LM‐2 represents cells at a plasmatocyte/lamellocyte intermediate state.
To investigate the link between the two lamellocyte populations and the plasmatocytes, we carried out RNA velocity and Monocle analyses. The two analyses place LM‐2 as intermediate between the plasmatocytes and LM‐1, and also suggest that the main cluster producing the lamellocytes is PL‐vir1. Otherwise, the same major branches from PL‐prolif/PL‐Inos to PL‐robo2, PL‐Rel, and PL‐vir1 as those identified in the NI larvae are observed (Fig 7A and B).
Figure 7. Identification of the filiation between the WI clusters.
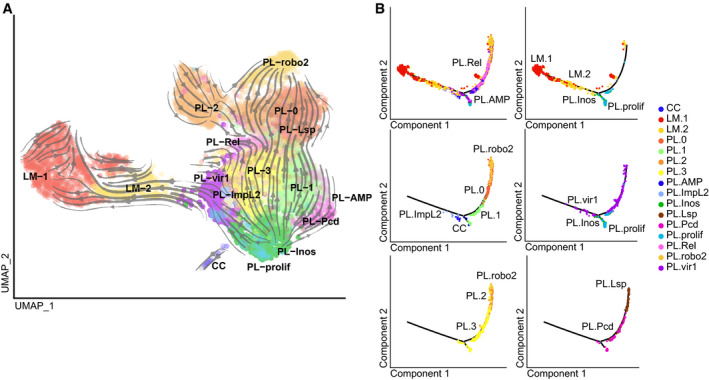
- Lineage prediction for the clusters from the WI sample using RNA velocity, as in Fig 4C.
- Single‐cell trajectory reconstructed with Monocle 2 on the WI dataset. The panels show restricted number of clusters for which the RNA velocity analysis suggested a filiation.
Markers’ dynamics upon WI
Considering the strong immune response induced by the WI, we expected a very robust modification of the transcriptional landscape of most hemocytes; however, the transcriptome of the plasmatocyte clusters remains overall similar in WI compared to NI (see above and Appendix Fig S6E). To rule out that this observation is due to the low depth of the scRNA‐seq, we used qPCR to quantify the main clusters’ markers in NI and WI hemocytes from WL (Fig 8A). In agreement with the scRNA‐seq data, the large majority of the NI plasmatocyte markers maintain the same expression profile upon WI. As expected, the lamellocyte markers are strongly up‐regulated in WI (Fig 8A). In addition to the lamellocyte markers, the following markers display significant up‐regulation: NimB4 (marker of PL‐Inos), CecC (PL‐AMP), Hml and Sr‐C1 (PL‐prolif), and Lz (CC). This increase can reflect a modification of the size of their affiliated clusters, which is the case for PL‐Inos and PL‐prolif (Fig 5B). It also reflects the molecular response to the WI, which is likely the case for CC and PL‐AMP whose size remains low upon WI.
Figure 8. Timing of the production of the different clusters in the hemolymph of WI larvae.
-
AExpression levels of the cluster markers in the hemolymph of WI larvae compared to NI larvae. The left panel indicates the distribution of each marker across all clusters (as in Fig 2D), and the graph on the right panel indicates the log2 of the ratio between the expression level in WI versus NI. Positive values indicate an up‐regulation upon wasp infestation and negative values a down‐regulation (n = 3, pool of 10 larvae per replicate, mean ± SD is represented on the graph). The P‐value are estimated by bilateral Student's t‐test and indicated as follow: ns = non‐significant (> 0.05), “*” = P[0.05–0.01[, “**” = P[0.01–0.001[.
-
B, CExpression levels of the cluster markers increasing in WI compared to NI according to (A) during the progression of the immune response to wasp infestation. Collections were carried out at t0 (time of infestation, L2), 24 h (early L3), 48 h (mid L3), and 72 h (WL) after wasp infestation (n = 3). The timeline is on the x‐axis and the normalized expression levels on the y‐axis. The expression levels were normalized to the levels at t0. The dotted lines indicate t 0 relative level (=1). The markers that do not display strong variability during the timeline are presented in (B), and the ones showing a strong up‐regulation are in (C).
-
DExpression levels during the infestation timeline as in (B, C) for the markers of proliferation.
-
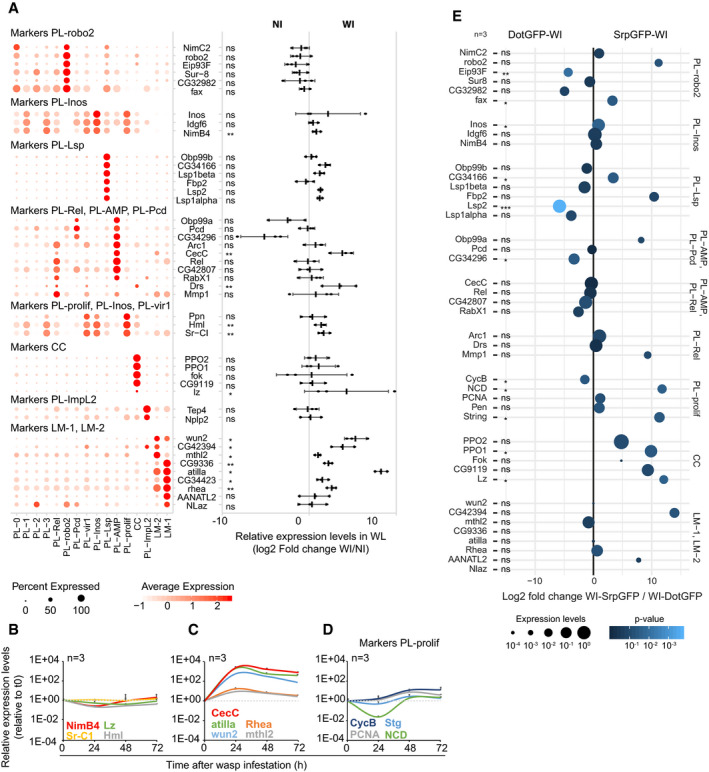
EWeighted dot plot representing the enrichment (log2 fold change) of the markers in hemocytes originating from the lymph gland (DotGFP‐WI) compared to embryonic hemocytes (srpGFP‐WI) from WI larva. The lymph gland hemocytes and embryonic hemocytes were traced using the lymph gland driver DotGal4 and the embryonic driver Srp(hemo)Gal4, respectively, combined with a lineage tracing construct. The relative expression levels are indicated by the size of the dots and the P‐value of the log2 fold change with the gradient dark blue (not significant) to light blue (P < 10−3). The P‐value is estimated by bilateral Student's t‐test and indicated as follow: ns = non‐significant (> 0.05), “*” = P[0.05–0.01[, “**” = P[0.01–0.001[, “***” = P < 0.001].
To characterize the progression of the immune response to the infestation, we collected the hemocytes 24, 48 and 72 h after WI and quantified the expression levels of specific markers by qPCR (Fig 8B–D). CecC and the lamellocyte markers undergo strong up‐regulation in the 24 h following the WI. Within this timeframe, the lymph gland is still intact (Appendix Fig S6C), indicating that this early response is produced by the hemocytes of embryonic origin. The high expression levels of the markers are then maintained until 72 h, suggesting that they are also expressed by the hemocytes released from the lymph gland (Fig 8C). The other markers (i.e., NimB4, Sr‐C1, Lz, and Hml) do not display a strong modulation during the response (Fig 8B). We also characterized the expression levels of the PL‐prolif markers CycB, PCNA, Stg, and NCD. The four markers display stable expression at 24 h and up‐regulation at 48 h, which is maintained at 72 h after WI (Fig 8D). This suggests that the number of dividing cells stays constant at 24 h and increases later on. These data are concordant with the hemocyte counts at the different stages of infestation (Appendix Fig S6D). Within 24 h of infestation, the number of hemocytes remains the same, with lamellocytes being produced upon plasmatocyte trans‐differentiation. In the following time points, there is an increase in plasmatocyte number, which can be explained by the combination of the release of hemocytes from the lymph gland and increased proliferation (Fig 8D and Appendix Fig S6D).
Finally, to determine the origin of the hemocytes, we analyzed the expression levels of the main clusters’ markers in the hemocytes originating from the lymph gland or from the embryo in WI conditions (Fig 8E). The hemocytes were traced using the lymph gland‐specific driver DotGal4 or the embryonic hemocyte driver Srp(hemo)Gal4 combined with two lineage tracing transgenes (gtrace, see Materials and Methods section and Appendix Fig S7B, D and F). The hemocytes were sorted by FACS before quantification by qPCR. Of note, the filtration steps necessary for the FACS sorting removed most lamellocytes from the samples, which explains the low levels of lamellocyte markers in these data (personal observation). The analysis reveals enrichment for the markers of PL‐prolif and CC in the embryonic‐derived hemocytes. All other clusters display markers in the hemocytes from both origins. This suggests that the lymph gland produces hemocytes highly similar to the embryonic‐derived hemocytes upon WI. It also suggests that the lymph gland releases only few crystal cells and proliferating cells in the hemolymph after WI.
Overall, this analysis indicates that the hemocytes from the lymph gland express most markers found in embryonic‐derived hemocytes (in the NI dataset). Thus, at the present level of resolution of the scRNA‐seq data, we cannot identify markers specific to the origin (i.e., embryonic or lymph gland) of the hemocytes.
Metabolic properties of the clusters
To observe the potential metabolic differences among the clusters, we analyzed the expression profile of the main actors of energy metabolism across the NI and WI datasets (Appendix Fig S8) in the same fashion as it was done on the bulk transcriptomes (Appendix Fig S2). Although the analyses carried out on the single cell are not as precise as the ones on the bulk transcriptome, most plasmatocyte clusters display metabolic markers in line with the observations made with the bulk RNA‐seq. We can hence use these data to draw first conclusions.
In the larva, the hemocytes import and metabolize lipids to drive the TCA cycle. Lipid‐scavenging receptors and the genes of the fatty acid degradation pathway are expressed in most of them. The three exceptions are PL‐AMP, PL‐Pcd, and PL‐ImpL2 that express low levels of lipid‐scavenging receptors and fatty acid degradation genes as well as low levels of TCA genes, suggesting a lower metabolism for these clusters (Appendix Fig S8 left panel).
The crystal cells distinguish themselves by the expression of the glucose transporters (Sut1 and Glut1) and genes involved in gluconeogenesis (fbp, Ald) and lipid biosynthesis (Lpin), suggesting a metabolism involving glucose and lipid uptake (Appendix Fig S8 left panel).
The wasp infestation enhances the expression levels of the lipid‐scavenging receptors and of the fatty acid degradation genes in the plasmatocytes, including in the clusters PL‐Pcd and PL‐AMP, which suggests a stronger metabolic activity (Appendix Fig S8 right panel). We also noticed increased levels of the glucose transporter Glut1 and of the genes involved in glycolysis in PL‐robo2 hemocytes, suggesting that the infestation induces a diversification of the energy source for this cluster (Appendix Fig S8 right panel). Finally, the lamellocyte clusters display a strong expression of the glucose transporter sut1. In addition, LM‐1 displays a prominent glycolysis pathway, which may indicate that lamellocyte metabolism relies mostly on glucose (Appendix Fig S8 right panel).
The wasp infestation induces a strong metabolic shift in the larvae where the resources are deviated from development toward the immune response (Rauw, 2012). This shift, mediated by the hemocytes through the production of extracellular adenosine, increases considerably the sugar levels in the hemolymph (Bajgar et al, 2015). Our data suggest that this additional sugar is mostly used by the lamellocytes and by the PL‐robo2 clusters to respond to the wasp infestation, while the other hemocyte clusters keep using lipids.
Discussion
The immune cells provide the first line of defense against the non‐self, and accumulating evidence strongly suggests that their function exceeds the immune response. Due to their ability to communicate with the other organs and tissues, immune cells provide ideal sensors for the internal state and homeostasis during development and ontogeny. This raises the issue of immune cell heterogeneity; that is, can we identify subpopulations with specific potentials? The understanding of immune cell biology heavily relies on the thorough characterization of these cells as well as on the identification of specific markers and subpopulations. This work provides the first atlas of the Drosophila hemocytes, by specifically focusing on those that originate from the first hematopoietic wave. We show that these hemocytes undergo a molecular and metabolic shift during development. We show the existence of distinct hemocyte populations and identify a large panel of novel markers specific to the different populations. Monitoring the larval response against the wasp L. boulardi reveals the hemocyte behavior upon challenge and defines intermediate and mature lamellocyte populations. Finally, we use multiple bioinformatics tools to predict a temporal progression among the different hemocyte clusters in control and in challenged conditions.
The developmental shift between the embryonic and the larval stages
Immune cells are considered as static components of our defense system; however, these cells constantly interact with the ever‐changing environment. In addition, the cells that are born in the early embryo experience the extensive rearrangements that occur during development, including tissue and organ formation. We here show that the Drosophila hemocytes undergo a significant transcriptional shift that fully complies with the requirements of the embryonic and larval stages.
The highly migratory hemocytes present in the differentiated embryo display a strong developmental role: They allow tissue reshaping by secreting several constituents of the extracellular matrix and by engulfing dead cells through specific scavenger receptors such as NimC4. They display high levels of gluconeogenesis and TAG synthesis, processes that provide adequate levels of glucose and fatty acid for tissue/organ development. They secrete cuticle proteins. The larval hemocytes, on the other hand, express high levels of transcripts that are linked to the immune response, in accordance with the exposure to pathogens occurring after hatching, and are highly phagocytic. Moreover, they express fewer molecules associated with the extracellular matrix as compared to those observed in the embryo. Finally, they strongly express the molecular pathways that release stored energy (beta oxidation, TCA cycle), most likely in preparation for the metamorphosis and to help building the adult tissues.
Different types of hemocytes in wild‐type larvae
The single‐cell analysis on the NI animals reveals the presence of 14 different hemocyte clusters, based on the profile of gene expression, on the enrichment in specific GO terms and regulons as well as on the in vivo validation. Indeed, a number of clusters are identified by a single regulon (Tbp for PL‐Lsp, lz for CC, E2f1 for PL‐prolif) or by a specific combination of regulons in the case of related clusters (e.g., PL‐vir1, PL‐Rel, and PL‐Amp). These data provide us with a battery of novel‐specific markers and will make it possible to generate more targeted genetic tools. Excitingly, we can already define distinct features and functions of the different clusters.
The PL‐Rel cluster likely provides a cellular reservoir for a specific immune response, and the closely related PL‐AMP hemocytes seem more specifically dedicated to the humoral response, whereas PL‐vir1 hemocytes seem associated with the anti‐viral response. These three clusters share GO terms and regulons associated with immune functions, suggesting that they respond to a variety of challenges. These data suggest that specific hemocyte subtypes target different pathogens. This does not necessarily mean that only these hemocytes will respond to the infection, as these cells may just respond faster than others to a specific challenge.
The circulating PL‐Lsp hemocytes represent the nutrient reservoir that stores amino acids and has a role in homeostasis. The PL‐Lsp and PL‐AMP hemocytes are associated with the major roles of the fat body, the metabolic homeostasis, and the humoral immune response, suggesting that they contribute to the fat body–hemocyte axis acting in physiological and pathological conditions. This axis is bidirectional. For example, (i) the small secreted peptide Edin produced in the fat body controls the number of plasmatocytes in circulation upon wasp infestation; (ii) the hemocyte expression of the Spaetzle ligand controls the activity of Toll signaling in the fat body and affects the response to infection (Shia et al, 2009) as well as tumor growth (Parisi et al, 2014); and (iii) the metabolically induced production of the NimB5 protein from the fat body adjusts the number of hemocytes to the physiological state of the larva (preprint: Ramond et al, 2019).
The PL‐robo2 hemocytes are associated with phagocytosis and may serve as a sensor of lipid levels in the hemolymph (Woodcock et al, 2015). This cluster shares features with the large PL‐0 and PL‐2 clusters that are mildly enriched for the regulon related to the phagocytic abilities (srp), in agreement with the finding that the vast majority of the larval hemocytes is phagocytic (Fig 1). The PL‐0, PL‐2, PL‐1, and PL‐3 clusters, which do not display strong specific molecular features, may serve different purposes, perhaps less efficiently than the more specialized hemocytes, or may express enhanced potentials in response to specific challenges.
The unexpected finding that a cluster expresses a high rate of ribosomal protein suggests that either translation is higher in PL‐Pcd hemocytes compared to other clusters or that the ribosomal proteins high in this cluster act on the immune response in an uncanonical fashion. Notably, studies in mammals indicate that ribosomal protein like RPS3 selectively modulates the target genes of NF‐κB, the orthologue of Rel (Zhou et al, 2015).
The data on the small CC cluster validate the role of these cells in melanization and reveal a distinct metabolism as they seem to use glucose as energy source, whereas the plasmatocytes mostly use lipids.
The PL‐prolif cells, mostly localized in the resident compartment, likely provide the pool of mitotic precursors for most of the hemocyte clusters identified in the larva and PL‐Inos the immature progenitors, respectively.
The PL‐ImpL2 cluster defines PSC‐like cells present outside of the lymph gland in a still unknown location. These hemocytes are likely set aside in the embryo, in line with the findings that they are significantly enriched for transcripts that are specific to the E16 hemocytes (bulk transcriptome analyses, unpublished data) and that they do not seem to originate from the PL‐prolif cluster.
The identification of different populations of specialized plasmatocytes in the Drosophila larva prompts us to draw parallels with the mammalian immune cells. The closest relative to plasmatocytes is the monocytes and the macrophages (Wood & Martin, 2017). Monocytes are equipped with Toll‐like receptors, scavenger receptors, and their main function is to patrol as well as remove microorganisms, lipids, and dying cells via phagocytosis. Upon inflammation, they infiltrate specific tissues and differentiate into macrophages. The macrophages keep phagocytosing, induce an inflammatory response by releasing cytokines, and participate to repair of the tissue (Yang et al, 2014). The scRNA‐seq assay reveals that hemocytes express markers such as Integrin alphaPS2 (If), EcR/Hr96, Lamp1, Rgh, Tfc/lectin‐46Cb/CG34033, and Lz, which are the Drosophila orthologues of CD11b, PPARγ, CD68, Dectin, CD207, and RUNX, respectively. In mammals, these proteins are responsible for migration, adhesion, phagocytosis, differentiation from monocytes to macrophages, and pathogen recognition (Ramprasad et al, 1996; Voon et al, 2015; Podolnikova et al, 2016; Daley et al, 2017; Heming et al, 2018).
Single‐cell analyses of the kind we performed on the fly hemocytes bring about the important question of whether the differences observed between clusters are stable or transient; in other words, do the clusters represent different and stable identities or do they reflect the presence of transient, unstable, states? Based on our data, we speculate that some clusters represent specific populations (the LSP producing plasmatocytes, the crystal cells, etc.), because they display very unique features. The Prolif and the Inos plasmatocytes, on the other hand, share a number of markers and may represent states of the same cluster. Plasmatocyte clusters 0–3, which can only be resolved using a combination of markers, may also represent developmental states, in line with the trajectories identified by the bioinformatic analyses. In this regard, investigating the similarities and differences to the adult hemocytes will shed light on some of these issues.
The larval response to wasp infestation
The single‐cell analysis upon WI reveals the reduced representation of some clusters such as PL‐Rel and the expanded representation of “early” clusters (e.g., PL‐prolif, PL‐Inos). Thus, specific hemocyte clusters may preferentially survive/proliferate upon challenge. The majority of the clusters, however, remain equally represented in the two conditions, and the correlation between the average transcriptomes in NI and WI conditions reveals strong similarity between most of the identified clusters (Pearson = 0.97). This implies that the hemocytes produced by the 1st and the 2nd hematopoietic waves share major features.
Two new populations of cells appear, LM‐1 and LM‐2, the second one representing an intermediate state characterized by the co‐expression of lamellocyte and plasmatocyte genes. Interestingly, LM‐2 also expresses a specific identity that is linked to energy supply (e.g., respiratory chain, NADH activity), whereas LM‐1 cells are mostly devoted to encapsulation.
With respect to the mode of lamellocyte production, the bioinformatics predictions (RNA velocity and Monocle) could support the hypothesis of a dedicated precursor, the lamelloblast (Anderl et al, 2016). The PL‐prolif cluster seems to rapidly branch out with one arm giving rise to lamellocytes, partly associated with the PL‐vir1 cluster, and the other arm giving rise to the other plasmatocyte clusters. In this model, the 1st hematopoietic wave would produce lamellocytes through trans‐differentiation (the expression of LM‐2 markers already increases in the first 24 h after infestation), whereas the 2nd wave would do it (also) through the mitotically active lamelloblast. At the level of resolution provided by the scRNA‐seq assay, we may have lost the lamelloblast cluster.
In sum, the different clusters identified by the scRNA‐seq assay exhibit distinct features, which can be now tested functionally using the newly identified markers and the associated genetic tools that are publically available (Gal4 drivers, RNAi and overexpressing transgenes, mutations). Future technological refinements may enhance the depth of the analyses, as the current scRNA‐seq assays only allow for the identification of a subset of genes for each cluster, the most expressed ones. As an example, the larval hemocytes do not all phagocytose with the same efficiency, but we cannot allocate the different potentials to specific clusters (Appendix Fig S5F). Nevertheless, our data on the bulk and single‐cell transcriptomes of the Drosophila hemocytes provide a powerful framework to understand the role of immune cells in physiological and pathological conditions.
Materials and Methods
Fly strains and genetics
All flies were raised on standard media at 25°C. For the bulk sequencing, the hemocytes from stage 16 (E16) embryos were collected from srp(hemoGal4/+;UAS‐RFP/+) animals obtained upon crossing srp(hemo)Gal4 (gift from K. Brückner; Bruckner et al, 2004) and UAS‐RFP flies (RRID:BDSC_8547). The wandering L3 (WL) hemocytes were collected from staged HmlΔRFP/+ animals upon crossing HmlΔRFP (Makhijani et al, 2011) with OregonR flies (108–117 h After Egg Laying, h AEL).
For the single‐cell sequencing, OregonR flies were used as the wild‐type (WT) strain for all the experiments and for the single‐cell RNA sequencing.
Validation of the single‐cell data involved the following stocks: srp(hemo)‐moesin‐RFP [stock D2244 on chr 2, gift from D. Siekhaus (Gyoergy et al, 2018)], Mimic‐Lsp1beta‐MI05460 (RRID:BDSC_40782), Lsp2‐Gal4 (RRID:BDSC_6357), the lineage tracing line UAS‐FLP,Ubi‐p63E(FRT.STOP)Stinger (RRID:BDSC_28282) that was combined with UAS‐FLP; act5c‐FRT,y+,FRT‐Gal4,UASmCD8GFP [gift from I. Ando (Honti et al, 2010)], and Dot‐Gal4 (RRID:BDSC _6903) and GstD‐LacZ [gift from D. Bohmann (Sykiotis & Bohmann, 2008)].
FACS sorting of embryonic and larval hemocytes
Staged egg laying was carried out to produce E16 embryos as follows. The cross to produce srp(hemo)Gal4/+;UAS‐RFP/+ embryos (with at least 100 females) was transferred to egg laying cages on a yeasted apple juice agar at 25°C. After a pre‐lay period of 30 min, the agar plates containing yeast were replaced with fresh plates and flies were left to lay for 3 h at 25°C. Agar plates were then removed, and the embryos were raised for 11 h and 40 min at 25°C until they reached stage 16. Embryos were then isolated from the medium and washed on a 100‐μm mesh. The collected embryos were transferred into a cold solution of phosphate‐buffered saline (PBS) in a Dounce Homogeniser on ice. The embryos were dissociated using the large clearance pestle than the small clearance pestle, and then, the cells were filtered through a 70‐μm filter to prepare them for FACS sorting. The cells were sorted using FACS Aria II (BD Biosciences) at 4°C in three independent biological replicates. Live cells were first selected based on the forward scatter and side scatter, and only single cells were taken into account. OregonR cells were used as a negative control to set the gate for the sorting of RFP‐positive cells only (Appendix Fig S1). RFP‐positive hemocytes were collected in 1 ml of TRI reagent (MRC) for RNA extraction.
For the wandering L3, HmlΔRFP/+ hemocytes, staged lay of 3 h, were carried out at 25°C to prevent overcrowding of the vials (between 50 and 100 embryos per vial) and wandering larvae were collected 108–117 h AEL, bled in cold PBS containing PTU (Sigma‐Aldrich P7629) to prevent hemocyte melanization (Lerner & Fitzpatrick, 1950), filtered through a 70‐μm filter to isolate them, and sorted by FACS as described for the embryonic hemocytes.
The purity of the sorted populations was assessed prior to the collection of the sample for RNA extraction by carrying out a post‐sort step. The FACS sorter was set up to produce hemocyte pools displaying at least 80% of purity on the post‐sort analysis.
RNA extraction and bulk RNA sequencing
The sorted cells were homogenized then left at room temperature (RT) for 5 min to ensure complete dissociation of nucleoprotein complexes. 0.2 ml of chloroform was added to each sample followed by centrifugation at 12,000 g for 15 min at 4°C. The upper aqueous phase containing the RNA was collected and transferred to a fresh autoclaved tube. 0.5 ml of 2‐propanol was added, and the samples were incubated for 5–10 min at RT. The RNA was precipitated by centrifugation, washed with 1 ml of 75% ethanol, precipitated again, and air‐dried. 20 μl of RNase‐free water was added to each sample before incubation at 55°C for 15 min. Single‐end mRNA‐seq libraries were prepared using the SMARTer (Takara) Low Input RNA Kit for Illumina sequencing. All samples were sequenced in 50‐length single read. At least 40 × 106 reads were produced for each replicate (Appendix Fig S1).
Analysis of bulk RNA‐seq data
The data analysis was performed using the GalaxEast platform, the Galaxy instance of east of France (http://www.galaxeast.fr/, RRID:SCR_006281; Afgan et al, 2018). First, summary statistics was computed on the raw FastQ Illumina files of the dataset using the quality control tool for high‐throughput sequence data FastQC (Babraham Bioinformatics, RRID:SCR_014583). FastQ Illumina files were converted to FastQ Sanger using the FastQ Groomer tool after assessing the quality of the sequencing. The FastQ Sanger files were then mapped onto the D. melanogaster reference genome Dm6 using TopHat (RRID:SCR_013035; Trapnell et al, 2009). As for the expression levels, the analysis of differential gene expression was based on the number of reads per annotated gene. This was done by using Htseq‐Count (RRID:SCR_011867; Anders et al, 2015), and the comparison and normalization of the data between the different cell types were done in Deseq2 (Appendix Fig S1; RRID:SCR_015687; Anders & Huber, 2010). The gene ontology studies presented in Fig 1B and Dataset EV1 were done using the Database for Annotation, Visualisation and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/,RRID:SCR_001881; Huang da et al, 2009) for the identification of biological processes.
The metabolic pathway analysis (Appendix Fig S2) was done as follows. Genes that showed a fold change ≥ 2 with an adjusted P‐value of less than 0.05 were considered for gene set enrichment analysis. Gene ontology and KEGG pathway enrichment analysis of the differentially expressed genes were done in ShinyGO v0.60 webserver (preprint: Ge & Jung, 2018). Genes associated with metabolic pathways considered in this study were retrieved from the KEGG database (http://www.genome.jp/, RRID:SCR_012773; Kanehisa & Goto, 2000). The log2FC values of the metabolic genes (q < 0.05) in the hemocytes were then plotted using R (version 3.4.0; R Core Team, 2017). The corresponding expression data for these genes in the “Embryo_16‐18 hr” and “larva_L3_puffstage_7‐9” developmental stages from modENCODE database (RRID:SCR_006206; Graveley et al, 2011) were downloaded using the webtool DGET (Hu et al, 2017; https://www.flyrnai.org/tools/dget/web/). These data were used to calculate the fold change and are represented as bar‐plots. For genes with paralogs, the paralog with highest fold change has been considered for the analysis. The details of all the genes (including all the paralogs) from these pathways are present in Table EV1, and the genes represented in the bar‐plots (Appendix Fig S2) are highlighted in yellow.
Phagocytosis assay
Hemocytes from E16 embryos and wandering larvae underwent the phagocytosis assay with latex beads. Briefly, srp(hemo)‐moesin‐RFP flies were staged for 3 h at 25°C, and then, the embryos were incubated at 25°C for 12 h in order to reach stage 16. Then, the embryos were collected and dechorionated in 25% bleach for 5 min at RT. Upon that the embryos were washed, homogenized with Dounce Homogeniser in Schneider medium complemented with 10% fetal calf serum (FCS), 0.5% penicillin, 0.5% streptomycin (PS), and few crystals of N‐phenylthiourea ≥ 98% (PTU) (Sigma‐Aldrich P7629) to prevent hemocyte melanization (Lerner & Fitzpatrick, 1950), and filtered with a 70‐μm filter. Twenty third‐instar larvae were bled in Schneider medium. Hemocytes for both stages were treated at the same time with latex beads 0.50 μm (Polysciences Inc., cat 17152) for 5 and 20 min, cytospinned at 700 rpm for 3 min, fixed for 10 min in 4% paraformaldehyde/PBS at RT, incubated for 30 min with DAPI to label nuclei (Sigma‐Aldrich) (diluted to 10–3 g/l in blocking reagent) and phalloidin Cy3 (only for the WL3 hemocytes due to low moesin‐RFP signal), and then mounted in Aqua‐Poly/Mount (Polysciences, Inc.). The slides were analyzed by confocal microscopy (Leica Spinning Disk) using identical settings.
For the phagocytosis assay on NimC1/P1‐negative hemocytes, 20 third‐instar larvae were bled in Schneider medium supplemented with PTU and were treated with latex beads 0.50 μm diluted 1/500 for 2, 5, or 10 min. Cells were then fixed and labeled with rabbit anti‐Srp(Bazzi et al, 2018) and mouse anti‐P1(Vilmos et al, 2004). We then used the secondary antibodies Cy5 goat anti‐mouse IgG (Jackson ImmunoResearch Labs Cat# 115‐177‐003, RRID:AB_2338719) and Cy3 goat anti‐rabbit IgG (Jackson ImmunoResearch Labs Cat# 111‐165‐144, RRID:AB_2338006) and DAPI. Images were acquired using Leica Spinning Disk microscope. Images produced were analyzed in Fiji (RRID:SCR_002285; Schindelin et al, 2012).
Imaging
The images produced for this paper were acquired on a Leica Spinning Disk from the Imaging center of the IGBMC (http://ici.igbmc.fr/). The acquisition step was 0.5 μm with a 40 × magnification. For the quantifications, three or more fields per sample were used with more than 50 cells in total. The intensity of latex beads or protein levels was measured with the Imaris software (version 9.5).
Hemocyte immunolabeling
Ten 3rd‐instar larvae per sample were bled in Schneider medium complemented with 10% fetal calf serum (FCS), 0.5% penicillin, 0.5% streptomycin (PS), and few crystals of N‐phenylthiourea ≥ 98% (PTU). For the collection of circulating hemocytes, the hemolymph was gently allowed to exit, while resident hemocytes were scraped and/or jabbed off the carcass in a second well as described in Petraki et al (2015). For the infested larvae, there was no separation of circulating from resident hemocytes and we used five larvae per sample. The cells were cytospinned at 700 rpm for 3 min; then, the samples were fixed for 10 min in 4% paraformaldehyde/PBS at RT, incubated with blocking reagent (Roche) for 1 h at RT, incubated overnight at 4°C with primary antibodies diluted in blocking reagent, washed three times for 10 min with PTX (PBS, 0.1% Triton X‐100), incubated for 1 h with secondary antibodies, washed twice for 10 min with PTX, incubated for 30 min with DAPI and phalloidin GFP, and then mounted Aqua‐Poly/Mount (Polysciences, Inc.). The slides were analyzed by confocal microscopy (Leica Spinning Disk) using identical settings between control and infested samples. The following combination of primary antibodies was used to determine the fraction of lamellocytes: mouse anti‐Relish [1:40; supernatant from the Developmental Studies Hybridoma Bank (DSHB Cat# anti‐Relish‐C 21F3, RRID:AB_1553772)], mouse anti‐Shot (1:40; supernatant, DSHB Cat# anti‐Shot mAbRod1, RRID:AB_528467), mouse anti‐Talin (Rhea) (1:40; DSHB Cat# Talin A22A, RRID:AB_10660289) mouse anti‐Talin (Rhea) (1:40,DSHB Cat# Talin E16B, RRID:AB_10683995), and rat anti‐Elav (1:200; DSHB Cat# Rat‐Elav‐7E8A10 anti‐elav, RRID:AB_528218). The secondary antibodies, Cy3 donkey anti‐mouse IgG (Jackson ImmunoResearch Labs Cat# 715‐165‐151, RRID:AB_2315777), Cy3 goat anti‐rat IgG (Jackson ImmunoResearch Labs Cat# 112‐165‐167, RRID:AB_2338251), Cy5 goat anti‐mouse IgG (Jackson ImmunoResearch Labs Cat# 115‐177‐003, RRID:AB_2338719), and Cy5‐AffiniPure goat anti‐rat IgG (H+L) (Jackson ImmunoResearch Labs Cat# 112‐175‐167, RRID:AB_2338264), were used at 1:500.
Quantitative PCR
For the comparison between resident and circulating hemocytes, 20 3rd‐instar larvae per sample were bled on ice‐cold PBS and the circulating hemocytes were separated from the resident ones as described above. The cells were then centrifuged at 300 g, 4°C, and RNA isolation was performed with the RNeasy Mini Kit (Qiagen) by following the manufacture's protocol. The DNase treatment was performed with the TURBO DNA‐free Kit (Invitrogen), and the reverse transcription (RT) was done by using the Super‐Script IV (Invitrogen) with random primers. The cycle program is used for the RT 65°C for 10 min, 55°C for 20 min, 80°C for 10 min. The qPCR we used is FastStart Essential DNA Green Master (Roche). The primers are listed in Dataset EV5. The P‐values and statistical test used are indicated in Table EV1.
For the quantitative PCR done on UAS‐FLP/+;srp(hemo)Gal4/gtrace‐mCD8‐GFP;gtrace‐nls‐GFP/+ and UAS‐FLP/+;DotGal4/gtrace‐mCD8‐GFP;gtrace‐nls‐GFP/+, 30 wandering third‐instar larvae were collected per replicate, the larvae were bled in PBS supplemented with PTU, and the cells were filtered through a 70‐μm filter to prepare them for FACS sorting. The cells were sorted using FACS Aria II (BD Biosciences) at 4°C in three independent biological replicates. Live cells were first selected based on the forward scatter and side scatter, and only single cells were taken into account. UAS‐FLP/+;gtrace‐mCD8‐GFP/+;gtrace‐nls‐GFP/+ cells were used as a negative control to set the gate for the sorting of GFP‐positive cells only. GFP‐positive hemocytes were collected, and the RNA isolation and qPCR were done as mentioned above.
Single‐cell sample preparation, sequencing, and analysis
OregonR females were used for the generation of the single‐cell data. For the NI sample, 20 female larvae were collected at the wandering L3 stage (108–117 h AEL at 25°C) and bled in Schneider medium complemented with PTU on ice. Both circulating and resident pools of hemocytes were collected as described in Petraki et al (2015). Briefly, the larval cuticle was first punctured to release the circulating hemocytes and then scrapped with fine forceps to release the resident hemocytes. The efficiency of the method was assessed with the HmlΔRFP strain by inspecting visually the larval carcass for remaining hemocytes.
For the WI sample, staged larvae were infested at the L2 stage (48–56 h AEL) for 2 h at 20°C with 20 female wasps (L. boulardi) per 100 Drosophila larvae. Following infestation, the larvae were raised at 25°C until the wandering L3 stage. Of note, the wasp infestation induces a developmental delay, as infested larvae reach the wandering stage at ~ 120 h AEL. In an effort to obtain reproducible results, the infestation conditions were optimized so that the majority of the larvae carry a single wasp egg (Bazzi et al, 2018). Twenty female larvae were processed as described above for the NI condition.
After bleeding, the hemolymph from NI and WI larvae was filtered on a 100‐μm mesh to remove cell aggregates and wasp eggs. The cell viability was assessed with trypan blue (over 90%), and cell concentration was estimated with a hemocytometer. 10,000 cells of each condition were used to prepare the 3′mRNA‐seq libraries with the Chromium Single Cell 3′ Reagent Kits v2 (10× Genomics). The libraries were then sequenced on the sequencer Illumina Hiseq 4000, on two lanes using paired sequencing of 2 × 100nt. The raw data were analyzed using Cell Ranger v3.0.1 (pipeline from 10× Genomics) and mapped to the Drosophila genome assembly BDGP6_ens95. Cells with low unique molecular identifiers (< 200) were removed.
Clustering the single‐cell data
The single‐cell data were further analyzed using the R based toolkit Seurat v3 (RRID:SCR_016341; Butler et al, 2018; Stuart et al, 2019). Following this, both NI and WI datasets were combined following the standard workflow for data integration (Stuart et al, 2019). Briefly, first the datasets were normalized using log‐normalization and the variable features (set up to 2,000 with the variance stabilizing transformation method) were determined for each dataset individually. Then, the anchors common between the two datasets were determined (dimensionality set up to 50) and the two datasets were corrected for batch effect before integration. The integrated data were clustered using PCA and visualized with UMAP. The optimal number of dimensions for the generation of the UMAP was determined using the tools DimHeatmap and Elbowplots, and the clustering was done on 20 dimensions with a resolution of 0.55. These parameters returned 14 clusters. Further clustering was then attempted on each cluster separately (manual curation). If the manual curation returned more than 10 distinct markers (|avg_logFC| > 1, ROC analysis returning AUC > 0.75), the cluster was subsequently subdivided. This protocol led to the redefinition of the PL‐Rel and PL‐Inos clusters (see Appendix Fig S3).
The GO term enrichment analysis was carried out based on gene expression levels in NI condition to identify the main features of the clusters. The number of lamellocytes being negligible in the NI dataset (eight cells), they were excluded from this analysis. The genes enriched in each cluster (log2FC > 0.25, adjusted P‐value < 0.01, determined with FindAllMarkers from Seurat, Dataset EV2) were analyzed using DAVID. The list of cluster‐specific genes was compared to the list of genes expressed in the whole dataset. The whole GO term results are available in Dataset EV3, and for each hemocyte cluster, the hemocyte‐related GO terms displaying the strongest enrichment are presented in Fig 2C.
RNA velocity analysis
To visualize the ongoing transcriptional changes in single cell, we adopted the approach described in La Manno et al (2018); i.e., we calculated the “velocity” of each cell in the high‐dimensional gene expression space. We started by generating the loom file, containing spliced and unspliced reads with velocyto, version 0.17.17 (http://velocyto.org/velocyto.py; La Manno et al, 2018). As input to velocyto, we gave the “run10x” option (specific for 10 × data), the CellRanger output, and Ensembl annotation v95 (ftp://ftp.ensembl.org/pub/release-95/gtf/drosophila_melanogaster/). Our NI dataset includes 4.2% intronic sequences, on which velocyto analysis was based. The representation of the data was done with the package scVelo (https://scvelo.readthedocs.io/; preprint: Bergen et al, 2019) that implements UMAP representations.
Monocle graph reconstruction
To complement the RNA velocity analysis, we run also Monocle 2 (Trapnell et al, 2014; Qiu et al, 2017a,b). We use the R version 3.5.1. The CellDataSet has been built using “expressionFamily=negbinomial.size()”. We adopted “DDRTree” method for dimensionality reduction, and a maximum number of components equals 2.
Regulon analysis
To identify the regulons involved in the hematopoietic system, we ran Single‐Cell regulatory Network Inference and Clustering (SCENIC, RRID:SCR_017247; Aibar et al, 2017) through its Python implementation pySCENIC, version 0.9.19 (https://pyscenic.readthedocs.io/en/latest/). The source code was downloaded from the GitHub repository https://github.com/aertslab/pySCENIC.git. The supplemental files necessary to run SCENIC were obtained from https://resources-mirror.aertslab.org/cistarget/. For the analysis, we chose the motifs version 8 (http://cistarget/motif2tf/motifs-v8-nr.flybase-m0.001-o0.0.tbl) and the regulatory elements within 5 kb upstream the TSS and the transcript introns (http://cistarget/databases/drosophila_melanogaster/dm6/flybase_r6.02/mc8nr/gene_based/dm6-5kb-upstream-full-tx-11species.mc8nr.feather). Finally, to identify the most significant regulons showing a different activity among clusters, we performed a Wilcoxon rank‐sum test (Mann & Whitney, 1947), between the AUC scores given by SCENIC in a specific cluster versus all the rest of the clusters.
Comparison of regulon‐based and gene‐based clusters
We performed clustering based on the regulon AUC scores per cell, by adopting the “Louvain” algorithm with a resolution equal to 1.3 in Scanpy (Wolf et al, 2018) version 1.4.4, in order to obtain the same number of clusters (=16) than in the previous gene‐based analysis. To compare the results from gene‐based and the regulon‐based clustering, we calculated the Rand index (Rand, 1971), obtaining a value of 0.83. Rand index (RI) = 0 means no overlaps between the clusters; RI = 1 means perfect overlap.
Author contributions
Conceptualization, PBC and AG; Methodology, PBC, AG, RS, AP, CD, NM, AR, TM, and NH; Investigation, PBC, AG, RS, AP, CD, NM, AR, TM, and NH; Writing—Original Draft, PBC, AG, RS, and AP; Writing—Review and Editing, PBC, AG, RS, and AP; Funding Acquisition, AG, NM, and TM; Resources, AG; Supervision, PBC and AG.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Table EV1
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Review Process File
Acknowledgements
We thank the Imaging Center of the IGBMC for technical assistance. This study was supported by the grant ANR‐10‐LABX‐0030‐INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d'Avenir ANR‐10‐IDEX‐0002‐02. The sequencing was performed by the GenomEast platform, a member of the “France Génomique” consortium (ANR‐10‐INBS‐0009). We thank C. Thibault‐Carpentier, V. Alunni, and C. Keime for handling the single‐cell library preparation, the sequencing, and the initial analysis on Cell Ranger. We thank I. Ando, K. Bruckner, M. Crozatier, M. Meister, D. Siekhaus, and D. Bohmann for providing fly and wasp stocks. In addition, stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and antibodies obtained from the Developmental Studies Hybridoma Bank created by the NICHD of the NIH and maintained at The University of Iowa (Department of Biology, Iowa City, IA 52242) were used in this study. We thank Dasaradhi Palakodeti for useful discussion on transcriptome analysis. This work was supported by INSERM, CNRS, UDS, Ligue Régionale contre le Cancer, Hôpital de Strasbourg, ARC, CEFIPRA, ANR grants, and by the CNRS/University LIA Calim. T. Mukherjee and Nivedita Hariharan are funded by the Council for Scientific and Industrial Research fellowship. A. Pavlidaki is an IGBMC International PhD Programme fellow supported by LabEx INRT funds. The IGBMC was also supported by a French state fund through the ANR labex.
EMBO Journal (2020) 39: e104486
See also: V Hartenstein (June 2020)
Contributor Information
Pierre B Cattenoz, Email: cattenoz@igbmc.fr.
Angela Giangrande, Email: angela@igbmc.fr.
Data availability
The datasets produced in this study are available in the following databases:
The scRNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐8698.
The RNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐8702.
References
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA et al (2018) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46: W537–W544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S, Gonzalez‐Blas CB, Moerman T, Huynh‐Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J et al (2017) SCENIC: single‐cell regulatory network inference and clustering. Nat Methods 14: 1083–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124: 783–801 [DOI] [PubMed] [Google Scholar]
- Anderl I, Vesala L, Ihalainen TO, Vanha‐Aho LM, Ando I, Ramet M, Hultmark D (2016) Transdifferentiation and proliferation in two distinct hemocyte lineages in Drosophila melanogaster larvae after wasp infection. PLoS Pathog 12: e1005746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics (Oxford, England) 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer MM, Bilstein A, Caussinus E, Csiszar A, Affolter M, Leptin M (2010) The role of apoptosis in shaping the tracheal system in the Drosophila embryo. Mech Dev 127: 28–35 [DOI] [PubMed] [Google Scholar]
- Bajgar A, Kucerova K, Jonatova L, Tomcala A, Schneedorferova I, Okrouhlik J, Dolezal T (2015) Extracellular adenosine mediates a systemic metabolic switch during immune response. PLoS Biol 13: e1002135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee U, Girard JR, Goins LM, Spratford CM (2019) Drosophila as a genetic model for hematopoiesis. Genetics 211: 367–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, Lemaitre B (2000) The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci USA 97: 3376–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi W, Cattenoz PB, Delaporte C, Dasari V, Sakr R, Yuasa Y, Giangrande A (2018) Embryonic hematopoiesis modulates the inflammatory response and larval hematopoiesis in Drosophila . Elife 7: e34890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW (2018) Dimensionality reduction for visualizing single‐cell data using UMAP. Nat Biotechnol 37: 38–44 [DOI] [PubMed] [Google Scholar]
- Benmimoun B, Polesello C, Haenlin M, Waltzer L (2015) The EBF transcription factor Collier directly promotes Drosophila blood cell progenitor maintenance independently of the niche. Proc Natl Acad Sci USA 112: 9052–9057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ (2019) Generalizing RNA velocity to transient cell states through dynamical modeling. bioRxiv 10.1101/820936 [PREPRINT] [DOI] [PubMed] [Google Scholar]
- Bernardoni R, Vivancos V, Giangrande A (1997) glide/gcm is expressed and required in the scavenger cell lineage. Dev Biol 191: 118–130 [DOI] [PubMed] [Google Scholar]
- Binggeli O, Neyen C, Poidevin M, Lemaitre B (2014) Prophenoloxidase activation is required for survival to microbial infections in Drosophila . PLoS Pathog 10: e1004067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A, Lemaitre B, Lanot R, Zachary D, Meister M (1997) Drosophila immunity: analysis of larval hemocytes by P‐element‐mediated enhancer trap. Genetics 147: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner K, Kockel L, Duchek P, Luque CM, Rorth P, Perrimon N (2004) The PDGF/VEGF receptor controls blood cell survival in Drosophila . Dev Cell 7: 73–84 [DOI] [PubMed] [Google Scholar]
- Bunch TA, Graner MW, Fessler LI, Fessler JH, Schneider KD, Kerschen A, Choy LP, Burgess BW, Brower DL (1998) The PS2 integrin ligand tiggrin is required for proper muscle function in Drosophila . Development 125: 1679–1689 [DOI] [PubMed] [Google Scholar]
- Burmester T, Antoniewski C, Lepesant JA (1999) Ecdysone‐regulation of synthesis and processing of fat body protein 1, the larval serum protein receptor of Drosophila melanogaster . Eur J Biochem 262: 49–55 [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanana B, Graf R, Koledachkina T, Pflanz R, Vorbruggen G (2007) AlphaPS2 integrin‐mediated muscle attachment in Drosophila requires the ECM protein Thrombospondin. Mech Dev 124: 463–475 [DOI] [PubMed] [Google Scholar]
- Charles JP (2010) The regulation of expression of insect cuticle protein genes. Insect Biochem Mol Biol 40: 205–213 [DOI] [PubMed] [Google Scholar]
- Chavez VM, Marques G, Delbecque JP, Kobayashi K, Hollingsworth M, Burr J, Natzle JE, O'Connor MB (2000) The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 127: 4115–4126 [DOI] [PubMed] [Google Scholar]
- Crozatier M, Ubeda JM, Vincent A, Meister M (2004) Cellular immune response to parasitization in Drosophila requires the EBF orthologue collier. PLoS Biol 2: E196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttell L, Vaughan A, Silva E, Escaron CJ, Lavine M, Van Goethem E, Eid JP, Quirin M, Franc NC (2008) Undertaker, a Drosophila junctophilin, links draper‐mediated phagocytosis and calcium homeostasis. Cell 135: 524–534 [DOI] [PubMed] [Google Scholar]
- Daley D, Mani VR, Mohan N, Akkad N, Ochi A, Heindel DW, Lee KB, Zambirinis CP, Pandian GSB, Savadkar S et al (2017) Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat Med 23: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostalova A, Rommelaere S, Poidevin M, Lemaitre B (2017) Thioester‐containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps. BMC Biol 15: 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C, Jouanguy E, Irving P, Troxler L, Galiana‐Arnoux D, Hetru C, Hoffmann JA, Imler JL (2005) The Jak‐STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila . Nat Immunol 6: 946–953 [DOI] [PubMed] [Google Scholar]
- Dragojlovic‐Munther M, Martinez‐Agosto JA (2012) Multifaceted roles of PTEN and TSC orchestrate growth and differentiation of Drosophila blood progenitors. Development 139: 3752–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudzic JP, Kondo S, Ueda R, Bergman CM, Lemaitre B (2015) Drosophila innate immunity: regional and functional specialization of prophenoloxidases. BMC Biol 13: 81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzo E, Santinon G, Pocaterra A, Aragona M, Bresolin S, Forcato M, Grifoni D, Pession A, Zanconato F, Guzzo G et al (2015) Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J 34: 1349–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu E, Burkard TR, Jiang Y, Saini N, Homem CCF, Reichert H, Knoblich JA (2014) SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell 156: 1259–1273 [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Imler JL, Hoffmann JA (2004) Sensing infection in Drosophila: toll and beyond. Semin Immunol 16: 43–53 [DOI] [PubMed] [Google Scholar]
- Flowers JM, Sezgin E, Kumagai S, Duvernell DD, Matzkin LM, Schmidt PS, Eanes WF (2007) Adaptive evolution of metabolic pathways in Drosophila . Mol Biol Evol 24: 1347–1354 [DOI] [PubMed] [Google Scholar]
- Fogerty FJ, Fessler LI, Bunch TA, Yaron Y, Parker CG, Nelson RE, Brower DL, Gullberg D, Fessler JH (1994) Tiggrin, a novel Drosophila extracellular matrix protein that functions as a ligand for Drosophila alpha PS2 beta PS integrins. Development 120: 1747–1758 [DOI] [PubMed] [Google Scholar]
- Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA (1996) Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4: 431–443 [DOI] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RA, White K (1999) Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila . Science 284: 1991–1994 [DOI] [PubMed] [Google Scholar]
- Friggi‐Grelin F, Rabouille C, Therond P (2006) The cis‐Golgi Drosophila GMAP has a role in anterograde transport and Golgi organization in vivo, similar to its mammalian ortholog in tissue culture cells. Eur J Cell Biol 85: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A (2004) The Drosophila kinesin‐like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell 15: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Wu LP (2014) Drosophila Rab14 mediates phagocytosis in the immune response to Staphylococcus aureus . Cell Microbiol 16: 296–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SX, Jung D (2018) ShinyGO: a graphical enrichment tool for animals and plants. bioRxiv 10.1101/315150 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold KS, Bruckner K (2015) Macrophages and cellular immunity in Drosophila melanogaster . Semin Immunol 27: 357–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Sierra J, Callejo A, Ibanez C, Guerrero I (2005) The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid‐modified Hedgehog. Dev Cell 8: 241–253 [DOI] [PubMed] [Google Scholar]
- Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, Kadowaki T, Beck K, Kitagawa Y (2001) A Drosophila haemocyte‐specific protein, hemolectin, similar to human von Willebrand factor. Biochem J 359: 99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind S (1999) Control of development and immunity by rel transcription factors in Drosophila . Oncogene 18: 6875–6887 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW et al (2011) The developmental transcriptome of Drosophila melanogaster . Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyoergy A, Roblek M, Ratheesh A, Valoskova K, Belyaeva V, Wachner S, Matsubayashi Y, Sanchez‐Sanchez BJ, Stramer B, Siekhaus DE (2018) Tools allowing independent visualization and genetic manipulation of Drosophila melanogaster macrophages and surrounding tissues. G3 (Bethesda) 8: 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker U, Lin X, Perrimon N (1997) The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development 124: 3565–3573 [DOI] [PubMed] [Google Scholar]
- Haller S, Franchet A, Hakkim A, Chen J, Drenkard E, Yu S, Schirmeier S, Li Z, Martins N, Ausubel FM et al (2018) Quorum‐sensing regulator RhlR but not its autoinducer RhlI enables Pseudomonas to evade opsonization. EMBO Rep 19: e44880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds AS, Bristow CA, Fisher WW, Weiszmann R, Wu S, Hartenstein V, Kellis M, Yu B, Frise E, Celniker SE (2013) Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol 14: R140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handke B, Poernbacher I, Goetze S, Ahrens CH, Omasits U, Marty F, Simigdala N, Meyer I, Wollscheid B, Brunner E et al (2013) The hemolymph proteome of fed and starved Drosophila larvae. PLoS One 8: e67208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Tabuchi Y, Sakurai K, Kutsuna M, Kurokawa K, Awasaki T, Sekimizu K, Nakanishi Y, Shiratsuchi A (2009) Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. J Immunol 183: 7451–7460 [DOI] [PubMed] [Google Scholar]
- Heming M, Gran S, Jauch S‐L, Fischer‐Riepe L, Russo A, Klotz L, Hermann S, Schäfers M, Roth J, Barczyk‐Kahlert K (2018) Peroxisome proliferator‐activated receptor‐γ modulates the response of macrophages to lipopolysaccharide and glucocorticoids. Front Immunol 9: 893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honti V, Kurucz E, Csordas G, Laurinyecz B, Markus R, Ando I (2009) In vivo detection of lamellocytes in Drosophila melanogaster . Immunol Lett 126: 83–84 [DOI] [PubMed] [Google Scholar]
- Honti V, Csordas G, Markus R, Kurucz E, Jankovics F, Ando I (2010) Cell lineage tracing reveals the plasticity of the hemocyte lineages and of the hematopoietic compartments in Drosophila melanogaster . Mol Immunol 47: 1997–2004 [DOI] [PubMed] [Google Scholar]
- Honti V, Csordas G, Kurucz E, Markus R, Ando I (2014) The cell‐mediated immunity of Drosophila melanogaster: hemocyte lineages, immune compartments, microanatomy and regulation. Dev Comp Immunol 42: 47–56 [DOI] [PubMed] [Google Scholar]
- Hu Y, Comjean A, Perrimon N, Mohr SE (2017) The Drosophila Gene Expression Tool (DGET) for expression analyses. BMC Bioinformatics 18: 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, Hoffmann JA, Hetru C, Meister M (2005) New insights into Drosophila larval haemocyte functions through genome‐wide analysis. Cell Microbiol 7: 335–350 [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N (2014) The HOPS complex mediates autophagosome‐lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 25: 1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim‐Jo C, Gatti JL, Poirie M (2019) Drosophila cellular immunity against parasitoid wasps: a complex and time‐dependent process. Front Physiol 10: 603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleino A, Silverman N (2014) The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol 42: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM et al (2005) Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila . Cell 123: 335–346 [DOI] [PubMed] [Google Scholar]
- Kurant E, Axelrod S, Leaman D, Gaul U (2008) Six‐microns‐under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell 133: 498–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz E, Zettervall CJ, Sinka R, Vilmos P, Pivarcsi A, Ekengren S, Hegedus Z, Ando I, Hultmark D (2003) Hemese, a hemocyte‐specific transmembrane protein, affects the cellular immune response in Drosophila . Proc Natl Acad Sci USA 100: 2622–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz E, Markus R, Zsamboki J, Folkl‐Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E et al (2007) Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol 17: 649–654 [DOI] [PubMed] [Google Scholar]
- Kussel P, Frasch M (1995) Pendulin, a Drosophila protein with cell cycle‐dependent nuclear localization, is required for normal cell proliferation. J Cell Biol 129: 1491–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty RK, Kutty G, Kambadur R, Duncan T, Koonin EV, Rodriguez IR, Odenwald WF, Wiggert B (1996) Molecular characterization and developmental expression of a retinoid‐ and fatty acid‐binding glycoprotein from Drosophila. A putative lipophorin. J Biol Chem 271: 20641–20649 [DOI] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A et al (2018) RNA velocity of single cells. Nature 560: 494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labzin LI, Schmidt SV, Masters SL, Beyer M, Krebs W, Klee K, Stahl R, Lutjohann D, Schultze JL, Latz E et al (2015) ATF3 is a key regulator of macrophage IFN responses. J Immunol 195: 4446–4455 [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Sceurman BK, Clark AG (2004) Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303: 1873–1876 [DOI] [PubMed] [Google Scholar]
- Lebestky T, Chang T, Hartenstein V, Banerjee U (2000) Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 288: 146–149 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Lerner AB, Fitzpatrick TB (1950) Biochemistry of melanin formation. Physiol Rev 30: 91–126 [DOI] [PubMed] [Google Scholar]
- Letourneau M, Lapraz F, Sharma A, Vanzo N, Waltzer L, Crozatier M (2016) Drosophila hematopoiesis under normal conditions and in response to immune stress. FEBS Lett 590: 4034–4051 [DOI] [PubMed] [Google Scholar]
- Ly LL, Suyari O, Yoshioka Y, Tue NT, Yoshida H, Yamaguchi M (2013) dNF‐YB plays dual roles in cell death and cell differentiation during Drosophila eye development. Gene 520: 106–118 [DOI] [PubMed] [Google Scholar]
- Magwire MM, Fabian DK, Schweyen H, Cao C, Longdon B, Bayer F, Jiggins FM (2012) Genome‐wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster . PLoS Genet 8: e1003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhijani K, Alexander B, Tanaka T, Rulifson E, Bruckner K (2011) The peripheral nervous system supports blood cell homing and survival in the Drosophila larva. Development 138: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaka J, Kuraishi T, Shiratsuchi A, Nakai Y, Higashida H, Henson P, Nakanishi Y (2004) Draper‐mediated and phosphatidylserine‐independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem 279: 48466–48476 [DOI] [PubMed] [Google Scholar]
- Mandal L, Martinez‐Agosto JA, Evans CJ, Hartenstein V, Banerjee U (2007) A Hedgehog‐ and Antennapedia‐dependent niche maintains Drosophila haematopoietic precursors. Nature 446: 320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18: 50–60 [Google Scholar]
- Markus R, Laurinyecz B, Kurucz E, Honti V, Bajusz I, Sipos B, Somogyi K, Kronhamn J, Hultmark D, Ando I (2009) Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster . Proc Natl Acad Sci USA 106: 4805–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroy P, Kaufmann G, Dubendorfer A (1988) Embryonic ecdysteroids of Drosophila‐melanogaster . J Insect Physiol 34: 633–637 [Google Scholar]
- Martin‐Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez‐Arias A (1998) puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila . Genes Dev 12: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek N, Shahab J, Saathoff M, Ringuette M (2008) Haemocyte‐derived SPARC is required for collagen‐IV‐dependent stability of basal laminae in Drosophila embryos. J Cell Sci 121: 1671–1680 [DOI] [PubMed] [Google Scholar]
- Martins NE, Faria VG, Nolte V, Schlotterer C, Teixeira L, Sucena E, Magalhaes S (2014) Host adaptation to viruses relies on few genes with different cross‐resistance properties. Proc Natl Acad Sci USA 111: 5938–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey HC Jr, Kejzlarova‐Lepesant J, Willis RL, Castleberry AB, Benes H (1997) The Drosophila Lsp‐1 beta gene. A structural and phylogenetic analysis. Eur J Biochem 245: 199–207 [DOI] [PubMed] [Google Scholar]
- McMillan EA, Longo SM, Smith MD, Broskin S, Lin B, Singh NK, Strochlic TI (2018) The protein kinase CK2 substrate Jabba modulates lipid metabolism during Drosophila oogenesis. J Biol Chem 293: 2990–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister M, Braun A, Kappler C, Reichhart JM, Hoffmann JA (1994) Insect immunity. A transgenic analysis in Drosophila defines several functional domains in the diptericin promoter. EMBO J 13: 5958–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcarne C, Lemaitre B, Kurant E (2019) Phagocytosis in Drosophila: from molecules and cellular machinery to physiology. Insect Biochem Mol Biol 109: 1–12 [DOI] [PubMed] [Google Scholar]
- Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olson PF, Parker CG, Fessler JH (1994) Peroxidasin: a novel enzyme‐matrix protein of Drosophila development. EMBO J 13: 3438–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Nagaosa K, Mori T, Shiratsuchi A, Nakanishi Y (2013) Integrin alphaPS3/betanu‐mediated phagocytosis of apoptotic cells and bacteria in Drosophila . J Biol Chem 288: 10374–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norum M, Tang E, Chavoshi T, Schwarz H, Linke D, Uv A, Moussian B (2010) Trafficking through COPII stabilises cell polarity and drives secretion during Drosophila epidermal differentiation. PLoS One 5: e10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson B, Page DT (2005) Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev Biol 279: 233–243 [DOI] [PubMed] [Google Scholar]
- Owusu‐Ansah E, Banerjee U (2009) Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461: 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F, Stefanatos RK, Strathdee K, Yu Y, Vidal M (2014) Transformed epithelia trigger non‐tissue‐autonomous tumor suppressor response by adipocytes via activation of Toll and Eiger/TNF signaling. Cell Rep 6: 855–867 [DOI] [PubMed] [Google Scholar]
- Pearson AM, Baksa K, Ramet M, Protas M, McKee M, Brown D, Ezekowitz RA (2003) Identification of cytoskeletal regulatory proteins required for efficient phagocytosis in Drosophila . Microbes Infect 5: 815–824 [DOI] [PubMed] [Google Scholar]
- Petraki S, Alexander B, Bruckner K (2015) Assaying blood cell populations of the Drosophila melanogaster larva. J Vis Exp e52733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolnikova NP, Kushchayeva YS, Wu Y, Faust J, Ugarova TP (2016) The role of integrins αMβ2 (Mac‐1, CD11b/CD18) and αDβ2 (CD11d/CD18) in macrophage fusion. Am J Pathol 186: 2105–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Zhang W, Costa P, Archambault V, D'Avino PP, Lilley KS, Laue ED, McAinsh AD, Glover DM (2007) Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster . PLoS One 2: e478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C (2017a) Single‐cell mRNA quantification and differential analysis with census. Nat Methods 14: 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C (2017b) Reversed graph embedding resolves complex single‐cell trajectories. Nat Methods 14: 979‐982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2017) R: a language and environment for statistical computing [Google Scholar]
- Ramond E, Petrignani B, Dudzic JP, Boquete J‐P, Poidevin M, Kondo S, Lemaitre B (2019) Metabolic adjustment of Drosophila hemocyte number and sessility by an adipokine. bioRxiv 10.1101/648626 [PREPRINT] [DOI] [Google Scholar]
- Ramprasad MP, Terpstra V, Kondratenko N, Quehenberger O, Steinberg D (1996) Cell surface expression of mouse macrosialin and human CD68 and their role as macrophage receptors for oxidized low density lipoprotein. Proc Natl Acad Sci USA 93: 14833–14838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand WM (1971) Objective criteria for the evaluation of clustering methods. J Am Stat Assoc 66: 846–850 [Google Scholar]
- Ratheesh A, Belyaeva V, Siekhaus DE (2015) Drosophila immune cell migration and adhesion during embryonic development and larval immune responses. Curr Opin Cell Biol 36: 71–79 [DOI] [PubMed] [Google Scholar]
- Rauw WM (2012) Immune response from a resource allocation perspective. Front Genet 3: 267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki MT, Rizki RM (1959) Functional significance of the crystal cells in the larva of Drosophila melanogaster . J Biophys Biochem Cytol 5: 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SW, Herzyk P, Dow JA, Leader DP (2013) FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster . Nucleic Acids Res 41: D744–D750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddie HG, Armitage EL, Coates JA, Johnston SA, Evans IR (2019) Simu‐dependent clearance of dying cells regulates macrophage function and inflammation resolution. PLoS Biol 17: e2006741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere S, Boquete JP, Piton J, Kondo S, Lemaitre B (2019) The exchangeable apolipoprotein Nplp2 sustains lipid flow and heat acclimation in Drosophila . Cell Rep 27: 886–899 e6 [DOI] [PubMed] [Google Scholar]
- Rus F, Kurucz E, Markus R, Sinenko SA, Laurinyecz B, Pataki C, Gausz J, Hegedus Z, Udvardy A, Hultmark D et al (2006) Expression pattern of Filamin‐240 in Drosophila blood cells. Gene Expr Patterns 6: 928–934 [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Kimbrell DA, Kylsten P, Engstrom A, Hultmark D (1990) The immune response in Drosophila: pattern of cecropin expression and biological activity. EMBO J 9: 2969–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Sanchez BJ, Urbano JM, Comber K, Dragu A, Wood W, Stramer B, Martin‐Bermudo MD (2017) Drosophila embryonic hemocytes produce laminins to strengthen migratory response. Cell Rep 21: 1461–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears HC, Kennedy CJ, Garrity PA (2003) Macrophage‐mediated corpse engulfment is required for normal Drosophila CNS morphogenesis. Development 130: 3557–3565 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM (1999) Antagonistic microtubule‐sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol 1: 51–54 [DOI] [PubMed] [Google Scholar]
- Shia AK, Glittenberg M, Thompson G, Weber AN, Reichhart JM, Ligoxygakis P (2009) Toll‐dependent antimicrobial responses in Drosophila larval fat body require Spatzle secreted by haemocytes. J Cell Sci 122: 4505–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlyakhover E, Shklyar B, Hakim‐Mishnaevski K, Levy‐Adam F, Kurant E (2018) Drosophila GATA factor serpent establishes phagocytic ability of embryonic macrophages. Front Immunol 9: 266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieow JL, Gun SY, Wong SC (2018) The sweet surrender: how myeloid cell metabolic plasticity shapes the tumor microenvironment. Front Cell Dev Biol 6: 168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinenko SA, Hung T, Moroz T, Tran QM, Sidhu S, Cheney MD, Speck NA, Banerjee U (2010) Genetic manipulation of AML1‐ETO‐induced expansion of hematopoietic precursors in a Drosophila model. Blood 116: 4612–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedden CG, Menegas W, Zajac AL, Williams AM, Cheng S, Ozkan E, Horne‐Badovinac S (2019) Planar‐polarized semaphorin‐5c and plexin A promote the collective migration of epithelial cells in Drosophila . Curr Biol 29: 908–920 e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stofanko M, Kwon SY, Badenhorst P (2010) Lineage tracing of lamellocytes demonstrates Drosophila macrophage plasticity. PLoS One 5: e14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive integration of single‐cell data. Cell 177: 1888–1902.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung EJ, Ryuda M, Matsumoto H, Uryu O, Ochiai M, Cook ME, Yi NY, Wang H, Putney JW, Bird GS et al (2017) Cytokine signaling through Drosophila Mthl10 ties lifespan to environmental stress. Proc Natl Acad Sci USA 114: 13786–13791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila . Dev Cell 14: 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KL, Vlisidou I, Wood W (2014) Ecdysone mediates the development of immunity in the Drosophila embryo. Curr Biol 24: 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer WH, Kunkel JG (1991) The function and evolution of insect storage hexamers. Annu Rev Entomol 36: 205–228 [DOI] [PubMed] [Google Scholar]
- Tepass U, Fessler LI, Aziz A, Hartenstein V (1994) Embryonic origin of hemocytes and their relationship to cell‐death in Drosophila . Development 120: 1829–1837 [DOI] [PubMed] [Google Scholar]
- Tokusumi T, Sorrentino RP, Russell M, Ferrarese R, Govind S, Schulz RA (2009) Characterization of a lamellocyte transcriptional enhancer located within the misshapen gene of Drosophila melanogaster . PLoS One 4: e6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE et al (2002) Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol 3: RESEARCH0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM (2007) Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol 8: R145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics (Oxford, England) 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL (2014) The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK, Evans IR, Wood W (2011) Ena drives invasive macrophage migration in Drosophila embryos. Dis Model Mech 4: 126–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrankar R, Liu Y, Provaznik J, Schmitt S, Lehmann M (2011) Lipin is a central regulator of adipose tissue development and function in Drosophila melanogaster . Mol Cell Biol 31: 1646–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanne S, Wang JH, Ramet M (2011) The Drosophila toll signaling pathway. J Immunol 186: 649–656 [DOI] [PubMed] [Google Scholar]
- Valanne S, Vesala L, Ramet M (2018) Commentary: Drosophila GATA Factor serpent establishes phagocytic ability of embryonic macrophages. Front Immunol 9: 1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilmos P, Nagy I, Kurucz E, Hultmark D, Gateff E, Ando I (2004) A rapid rosetting method for separation of hemocyte sub‐populations of Drosophila melanogaster . Dev Comp Immunol 28: 555–563 [DOI] [PubMed] [Google Scholar]
- Vlisidou I, Dowling AJ, Evans IR, Waterfield N, ffrench‐Constant RH, Wood W (2009) Drosophila embryos as model systems for monitoring bacterial infection in real time. PLoS Pathog 5: e1000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon DC‐C, Hor YT, Ito Y (2015) The RUNX complex: reaching beyond haematopoiesis into immunity. Immunology 146: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R, Hu J, Blotsky D, Krause HM (2016) Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev 30: 594–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, Theis FJ (2018) SCANPY: large‐scale single‐cell gene expression data analysis. Genome Biol 19: 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KKL, Liao JZ, Verheyen EM (2019) A positive feedback loop between Myc and aerobic glycolysis sustains tumor growth in a Drosophila tumor model. Elife 8: e46315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W, Martin P (2017) Macrophage functions in tissue patterning and disease: new insights from the fly. Dev Cell 40: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock KJ, Kierdorf K, Pouchelon CA, Vivancos V, Dionne MS, Geissmann F (2015) Macrophage‐derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid‐rich diet. Immunity 42: 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang L, Yu C, Yang X‐F, Wang H (2014) Monocyte and macrophage differentiation: circulation inflammatory monocyte as biomarker for inflammatory diseases. Biomark Res 2: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasothornsrikul S, Davis WJ, Cramer G, Kimbrell DA, Dearolf CR (1997) viking: Identification and characterization of a second type IV collagen in Drosophila . Gene 198: 17–25 [DOI] [PubMed] [Google Scholar]
- Yuan K, Seller CA, Shermoen AW, O'Farrell PH (2016) Timing the Drosophila mid‐blastula transition: a cell cycle‐centered view. Trends Genet 32: 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharogianni M, Kondylis V, Tang Y, Farhan H, Xanthakis D, Fuchs F, Boutros M, Rabouille C (2011) ERK7 is a negative regulator of protein secretion in response to amino‐acid starvation by modulating Sec16 membrane association. EMBO J 30: 3684–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J, Stramer B, Millard T, Martin P, Payre F, Plaza S (2009) Fascin is required for blood cell migration during Drosophila embryogenesis. Development 136: 2557–2565 [DOI] [PubMed] [Google Scholar]
- Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, Hultmark D (2004) A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci USA 101: 14192–14197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Huang X, Yin Y (2018) Beyond immunity: the Imd pathway as a coordinator of host defense, organismal physiology and behavior. Dev Comp Immunol 83: 51–59 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Ma A, Yuan L, Gao N, Feng Q, Franc NC, Xiao H (2017) Apoptotic cell clearance in Drosophila melanogaster . Front Immunol 8: 1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao WJ, Liao JM, Liao P, Lu H (2015) Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol 7: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Table EV1
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Review Process File
Data Availability Statement
The datasets produced in this study are available in the following databases:
The scRNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐8698.
The RNA‐seq data have been deposited in the ArrayExpress database at EMBL‐EBI (www.ebi.ac.uk/arrayexpress) under accession number E‐MTAB‐8702.


