Abstract
Herpes simplex keratitis (HSK) is the most common cause of corneal blindness in developed nations, caused by primary or recurrent herpes simplex virus 1 (HSV-1) infection of the cornea. Latent infection of HSV-1, especially in the trigeminal ganglion (TG), causes recurrence of HSV-1 infection. As antiviral treatment is not effective on latent HSV-1, to test the possibility of inhibiting HSV-1 by SpCas9 (Streptococcus pyogenes Cas9) or SaCas9 (Staphylococcus aureus Cas9), ICP0 and ICP4, two important genes required for HSV-1 replication and reactivation, were chosen as targets. In Vero cells, SpCas9 and SaCas9 targeting ICP0 or ICP4 can effectively inhibit the proliferation of HSV-1 without affecting cell viability. No significant guide RNA (gRNA)-dependent off-targets were observed in the human genome by digenome sequencing and deep sequencing verification. Adeno-associated virus 1 (AAV1)-mediated delivery of SaCas9 inhibits HSV-1 replication by targeting ICP4 in mouse primary TG neuronal cells. SpCas9 and SaCas9 are able to inhibit HSV-1 infection in Vero cells and mouse TG neuronal cultures with high efficiency and good biosafety. AAV1-mediated delivery of SaCas9 shows great potential in treating HSK and inhibiting HSV-1 in TG neurons. Further investigations may be needed to test the inhibition of latent infections, which may result in the development of novel methods for treating viral diseases.
Keywords: SaCas9, AAV, HSV-1, HSK, trigeminal ganglion neuron
Graphical Abstract
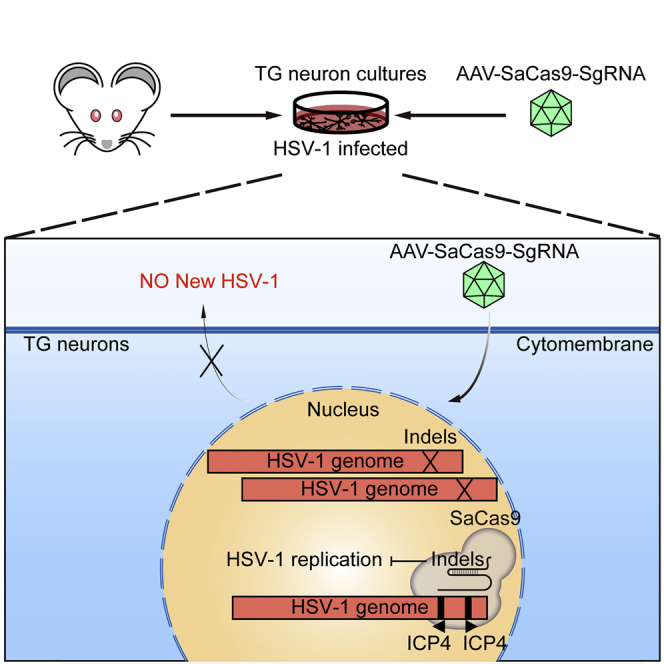
There is no effective treatment that can eliminate persistent HSV-1 within trigeminal ganglion (TG) neurons. Even though the meganucleases and CRISPR-SpCas9 system can interrupt the HSV-1 genome, they could not be packaged in an AAV system for in vivo therapy. In this study, the authors show the potential use of a single AAV1-mediated delivery system of ICP4-targeting SaCas9 to inhibit HSV-1 in TG neurons.
Introduction
Herpes simplex virus 1 (HSV-1) and HSV-2 are two major double-stranded DNA (dsDNA) simplexviruses infecting humans. According to the World Health Organization (WHO), around 67% of the global population under the age of 50 has HSV-1 infection,1 and 11% of the global population aged 15–49 has HSV-2 infection.2 Most infections are asymptomatic. Typical symptomatic infections are marked by multiple small blisters, fever blisters, or small ulcers. After primary infection, active HSVs are cleared by the immune system. However, the remaining HSVs migrate to neuron cell bodies, in which they become latent in the ganglion without active replication.
A severe type of HSV-1 infection is herpes simplex keratitis (HSK). HSK is caused by primary or recurrent HSV-1 infection in the cornea. There are about 40,000 new cases of HSK each year, and it is the most common cause of cornea-derived blindness in developed nations.3 Primary infections of HSV-1 in the cornea, oral cavity, or other regions travel from sensory nerves or hematogenously spread to trigeminal ganglion (TG) and become latent in the ganglion. When triggered by certain stimuli, HSV-1 is reactivated and transported to cornea, which induces a severe immune response and HSV-1-related lesions in the cornea with blindness as the end result.3
Acyclovir, valacyclovir, trifluridine, and vidarabine4, 5, 6 are nucleoside analogs that act as viral DNA synthesis inhibitors targeting viral DNA polymerase or viral thymidine kinase.7 These drugs have been shown to be useful to shorten the duration or reduce the incidence of primary or recurrent HSV-1 infections.8,9 However, there is no effective treatment that can eliminate persistent HSV-1 that becomes latent within TG, resulting in symptom recurrence after treatment.10 More importantly, mutation or deficiency in viral thymidine kinase leading to resistance to acyclovir further emphasizes the need for new and effective therapies.11,12
The emergence of engineered endonucleases such as meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas system have been shown to interrupt the quiescent HSV-1 genome as well as other DNA viruses. Meganucleases can inhibit HSV-1 replication and disrupt latent HSV-1 both in vitro and in vivo to some degree.4,13,14 Successful editing of the HSV-1 genome in gE, thymidine kinase, ICP0, UL23, UL29, UL52, and UL8 by Streptococcus pyogenes Cas9 (SpCas9, 4,101 bp) was shown recently.15, 16, 17, 18, 19 However, the inhibitory effect of SpCas9 targeting latency-related genes in TG, which limits the potential application of the CRISPR-Cas system in eliminating HSV-1 in TG, was not estimated. The large size of SpCas9 also can become a setback for adeno-associated virus (AAV)-mediated delivery in TG neurons, whereas Staphylococcus aureus Cas9 (SaCas9, 3,156 bp) can be packaged in AAV vectors for genome editing in vivo.20
ICP0 and ICP4 are two important immediate-early proteins involved in HSV-1 gene expression, viral replication, and reactivation,21, 22, 23 which may be suitable targets for inactivating HSV-1 in TG neurons. The shorter SaCas9 shows significant advantage in AAV packaging than SpCas9 and may be a better choice for inhibiting HSV-1 in TG neurons. It is not clear whether SaCas9 can inhibit HSV-1 infection and replication by targeting ICP0 or ICP4 in TG neurons, and the biosafety and off-target effects of SaCas9 in inhibiting HSV-1 remain unknown. The application of SaCas9 to inhibit HSV-1 may be a novel potential method for treating HSK and HSV-1 acute or latent infection and for previously “incurable” viral diseases.
Results
Effectively Editing HSV-1 ICP0 and ICP4 Loci by SpCas9 and SaCas9
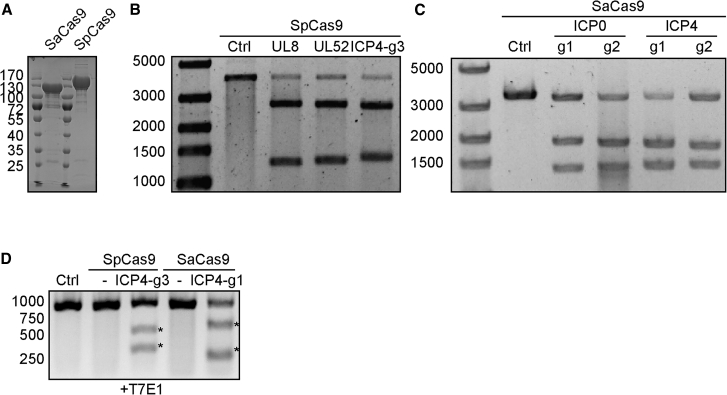
As reported by previous studies, ICP0 and ICP4 are two important immediate-early proteins for HSV-1 gene expression and viral replication.21,22 To test whether disrupting either ICP0 or ICP4 loci by SpCas9 or SaCas9 will impair HSV-1 replication, we designed six guide RNA (gRNA) targets for SpCas9 (SpICP0 g1–g3 targeting ICP0, SpICP4 g1–g3 targeting ICP4) and four gRNA targets for SaCas9 (SaICP0-g1 and SaICP0-g2 targeting ICP0, SaICP4-g1 and SaICP4-g2 targeting ICP4) (Figure S1). As positive controls, SpCas9/gRNAs for UL8 and UL52 were used for comparison in our experiment.18 We first expressed and purified SpCas9 and SaCas9 from E. coli (Figure 1A). Cleavage capabilities of gRNAs were verified by in vitro digestion. By incubating the cleavage templates carrying the targeted sites with purified SpCas9 or SaCas9 protein and gRNAs, cleavage activity was confirmed by gel electrophoresis (Figures 1B and 1C; Figure S2). All gRNAs showed high activity of in vitro digestion of targeted sites with purified SpCas9 or SaCas9, as indicated by the generation of clear cleavage bands (Figures 1B and 1C; Figure S2). To further confirm the gRNA activity in human cells, SpCas9/gRNA and SaCas9/gRNA targeting ICP4 were transfected into HEK293T cells followed by infection with HSV-1 (MOI of 1). Indel formation was clearly shown by a T7E1 assay and Sanger sequencing in the ICP4 locus, indicating effective editing of targeting sites (Figure 1D; Figure S3). The above data suggest that SpCas9 or SaCas9 with gRNAs can cleave HSV-1 ICP0 and ICP4 loci in vitro and in HEK293T cells.
Figure 1.
Validation of HSV-1 Targeting SpCas9 and SaCas9 gRNAs
(A) Coomassie staining of purified SpCas9 and SaCas9. (B) In vitro cleavage of UL8, UL52, and ICP4 targeted sites by SpCas9 with indicated gRNA. (C) In vitro cleavage of ICP4 targeted sites by SaCas9 with indicated gRNA. (D) T7E1 assay showing cleavage of HSV-1 genome in SpCas9- or SaCas9-expressing cells. Generation of indels are indicated by an asterisk. Ctrl, control.
SpCas9/gRNA- or SaCas9/gRNA-Overexpressing Cells Resist HSV-1 Infection
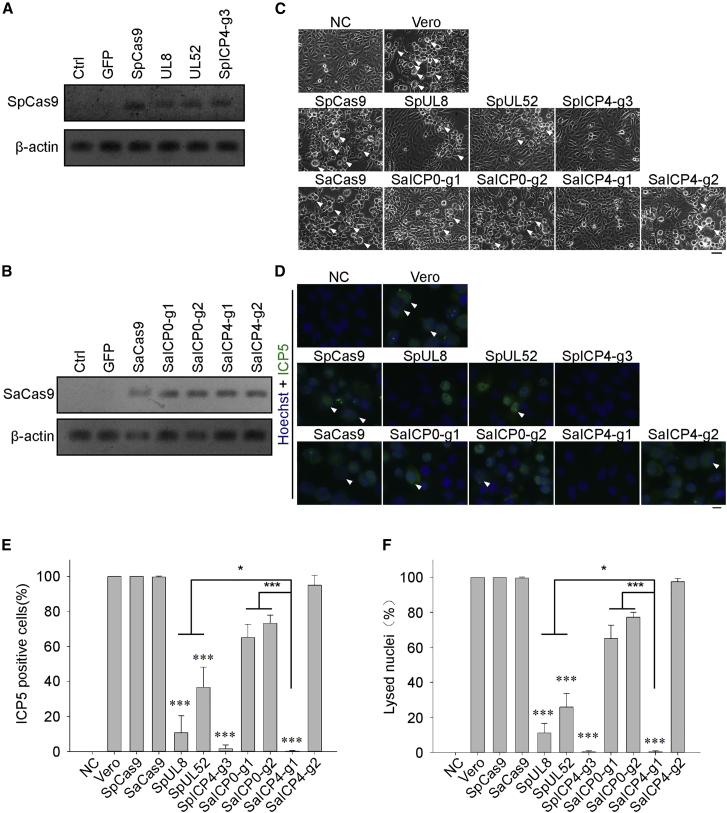
As it was observed that expression of SpCas9 or SaCas9 with gRNAs can lead to genome modification in the HSV-1 genome, we suspected that expression of the CRISPR-Cas9 system could defend against HSV-1 infection. To address this question, we used non-human primate Vero cell lines, which are known to be a suitable model for studying HSV-1 infection and replication, for stably expressing SpCas9 or SaCas9 with the indicated gRNAs. Expression of SpCas9 or SaCas9 was confirmed by RT-PCR (Figures 2A and 2B). When wild-type (WT) Vero cells were challenged with HSV-1 (MOI of 0.1) for 2 days and checked for signs of HSV-1 infection, all cells became round and detached from the plate bottom, which are known as cytopathic effects (CPEs) (Figure 2C, upper panel). As expected, expression of SpCas9 or SaCas9 alone did not offer resistance to HSV-1 in Vero cells, while cells bearing SpUL8 or SpUL52 gRNA showed mild signs of infection (Figure 2C). Surprisingly, SpICP4-g3 and SaICP4-g1 did not show any symptoms of HSV-1 infection, while ICP0-targeting SpCas9 or SaCas9 gRNAs showed minimal CPE formation (Figure 2C; Figure S4A), demonstrating the protective role of the CRISPR system against HSV-1 infection. To further confirm the infectivity of HSV-1 in Cas9/gRNA-expressing cells, we used ICP5, one of the major capsid proteins, to visualize HSV-1-infected cells. Using immunofluorescence, clear reduction of ICP5-positive cells was found in SpUL8 (10.73% ± 7.90%) and SpUL52 (36.68% ± 9.34%) cells compared to WT Vero cells (100%) (Figures 2D and 2E). Almost no ICP5 signal was detected in SpICP4-g3 (1.58% ± 1.78%) and SaICP4-g1 (0.18% ± 0.25%) in Vero cells (Figures 2D and 2E). Ring-shaped lysed nuclei were observed as a late symptom in the HSV-1 lytic stage, indicating a severe CPE. The percentage of lysed nuclei was significantly lower in SpICP4-g3 (0.31% ± 0.44%) and SaICP4-g1 (0.32% ± 0.45%) (Figure 2F). In summary, the above data showed that expression of SpICP4-g3 (1.58% ± 2.18%) and SaICP4-g1 (0.18% ± 0.31%) can reduce HSV-1 infectivity in Vero cells, suggesting that ICP4 may be a better target to inhibit HSV-1 compared to ICP0 (65.06% ± 7.60% and 73.21% ± 4.76%, p < 0.05), UL8 (10.73% ± 9.68%, p < 0.05), and UL52 (36.681% ± 11.44%, p < 0.05).
Figure 2.
Expression of SpCas9/gRNA or SaCas9/gRNA Inhibits HSV-1 Infection
(A) Expression analysis of SpCas9 mRNA in Vero cell lines by RT-PCR. (B) Expression analysis of SaCas9 mRNA in Vero cell lines by RT-PCR. (C) Morphology of HSV-1-infected SpCas9 or SaCas9 cell lines 3 days after infection with HSV-1 (MOI of 0.1). White arrows indicate the cytopathic effect (CPE). Scale bar, 30 μm. (D) Immunostaining of ICP5 in HSV-1-infected SpCas9 or SaCas9 cell lines 3 days after infection with HSV-1 (MOI of 0.1). Hoechst 33342 was used to visualize the nuclei. White arrows indicate ring-shaped lysed nuclei. Scale bar, 10 μm. (E) Statistical analysis of ICP5-positive cells in (D). More than 300 cells were collected from three independent experiments. (F) Statistical analysis of ring-shaped lysed nuclei in (D). More than 300 cells were collected from three independent experiments. Significance was determined by a one-way ANOVA. ∗∗∗p < 0.001.
Expression of SpCas9/gRNA or SaCas9/gRNA Compromised HSV-1 Replication
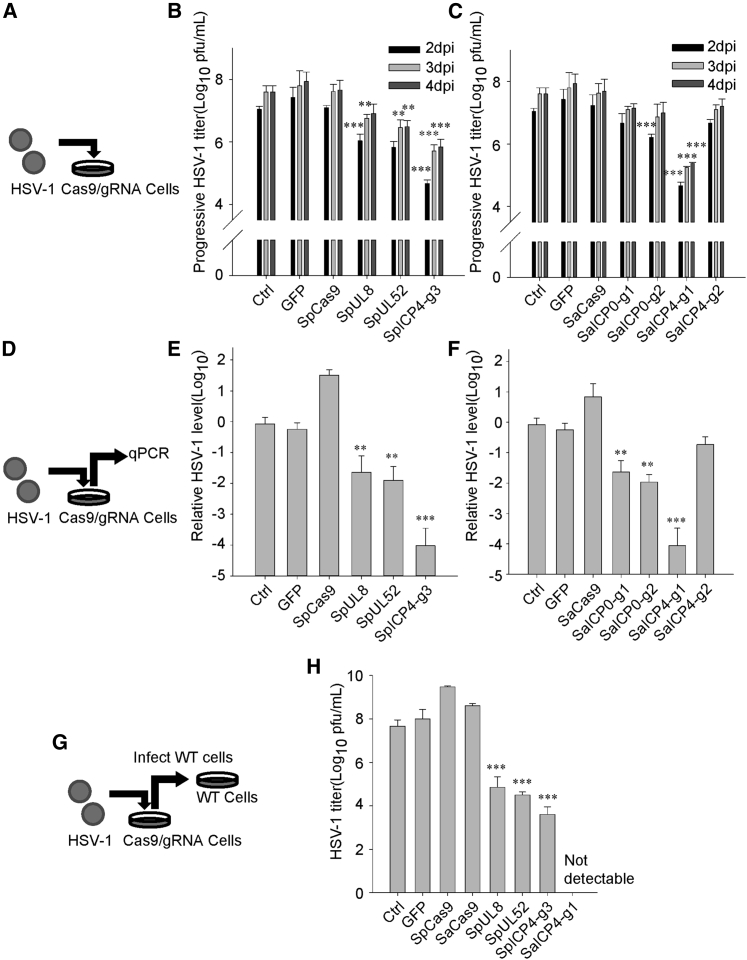
As HSV-1 infectivity was reduced in SaCas9/gRNA- or SaCas9/gRNA-expressing Vero cells (Figure 2), it is possible that replication of HSV-1 was compromised due to disruption of the HSV-1 genome. Infection of WT and Cas9/gRNA-expressing Vero cells with HSV-1 (Figure 3A) resulted in dramatically decreased plaque formation in SpICP4-g3 and SaICP4-g1 (105 plaque-forming units [PFU]/mL versus 107 PFU/mL) cells from 2 days post-infection (dpi) to 4 dpi (Figures 3B and 3C; Figure S4B). To compare the differences of HSV-1 replication in WT and Cas9/gRNA-expressing Vero cells, medium was discarded and refreshed after infecting with HSV-1 for 1 h. Furthermore, we collected the daughter virus-containing supernatant and measured HSV-1 production in HSV-1-infected cells (Figure 3D). qPCR quantification showed an almost 1,000-fold reduction of HSV-1 was found in supernatants from SpICP4-g3- or SaICP4-g1-expressing cells (Figures 3E and 3F; Figure S4C), reflecting impaired HSV-1 replication. To examine the production of active HSV-1, we saw a similar reduction in viral titer in indicated supernatants using the 50% tissue culture infective dose (TCID50) method (control, 107.7 PFU/mL; SpICP4-g3,103.7 PFU/mL; SaICP4-g1, not detectable) (Figures 3G and 3H). Expression of ICP4 is important for HSV-1 replication and its entrance into the lytic cycle.24 Disruption of the transcription factor ICP4 may likely have greater impact than ICP0 on HSV-1 replication (Figure 3; Figure S4). These data suggest that our gRNAs targeting ICP4 significantly reduced HSV-1 replication by editing the ICP4 locus, which demonstrated the ability of CRISPR-Cas9 to inhibit HSV-1 infection.
Figure 3.
Expression of SpCas9/gRNA or SaCas9/gRNA Inhibits HSV-1 Replication
(A) Schedule of the experimental process in (B) and (C). HSV-1 was used to infect Cas9 stably expressing Vero cells. (B) Control and SpCas9/gRNA-expressing Vero cells were infected with gradient-diluted HSV-1. Infection progress on each day after infection was determined by the Karber method. (C) Control and SaCas9/gRNA-expressing Vero cells were infected with gradient-diluted HSV-1. Infection progress on each day after infection was determined by the Karber method. (D) Schedule of the experimental process in (E) and (F). HSV-1 (MOI of 0.1) was used to infect Cas9 stably expressing Vero cells. Culture medium containing HSV-1 was collected 3 days post-infection (dpi) and then qPCR was performed to measure HSV-1 genomes. (E) qPCR analysis of HSV-1 genomes in the culture medium of control and SpCas9-expressing Vero cells infected with HSV-1 (MOI of 0.1) at 72 h. (F) qPCR analysis of relative HSV-1 genome copy in the culture medium of control and SaCas9-expressing Vero cells infected with HSV-1 (MOI of 0.1) at 72 h. Data were collected from three independent experiments. Error bars represent standard deviation (SD). Significance was determined by a one-way ANOVA. ∗∗p < 0.01. (G) Schedule of the experimental process in (H). HSV-1 (MOI of 0.1) was used to infect Cas9 stably expressing Vero cells. Culture medium containing HSV-1 was collected at 3 dpi. Viral titer in the medium was measured by the standard Karber method. (H) Titer of HSV-1 in the culture medium of control, SpCas9/gRNA-, or SaCas9/gRNA-expressing Vero cells infected with HSV-1 (MOI of 0.1) at 72 h. Titer was determined by infecting WT Vero cells and calculated by the Karber method on the fifth day after infection. Data were collected from three independent experiments. Error bars represent SD. Significance was determined by a one-way ANOVA. ∗∗p < 0.01, ∗∗∗p < 0.001.
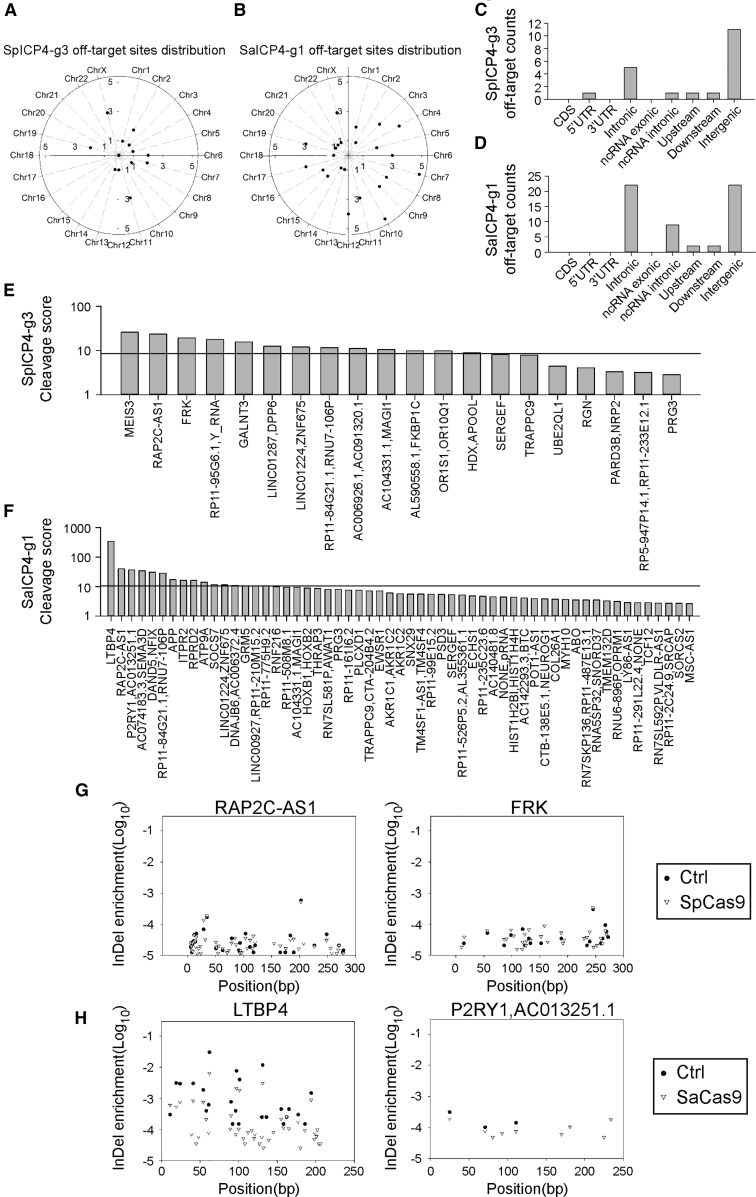
Off-target Analysis of SpCas9 or SaCas9 with ICP4 gRNAs
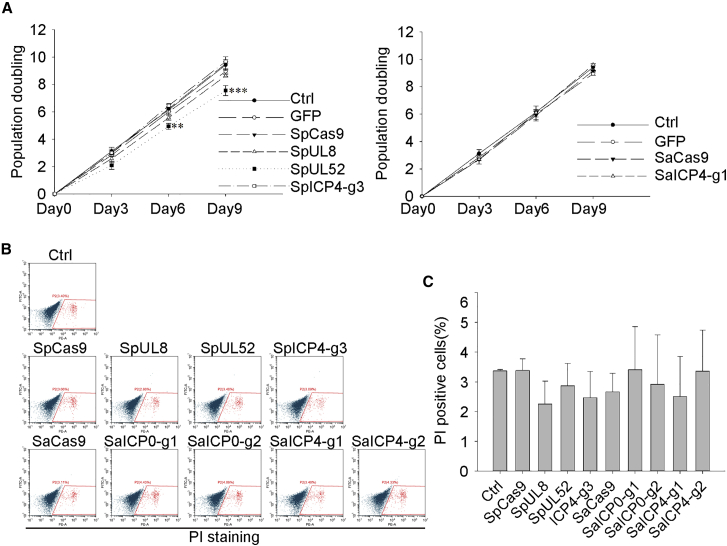
Off-target effects are one of the greatest concerns in the application of the CRISPR-Cas9 system. Except for SpUL52, no significant changes in cell proliferation or viability were observed in nearly all SpCas9/gRNA- or SaCas9/gRNA-expressing cells (Figures 4A–4C). Considering the potential clinical usage of the anti-HSV-1 gRNAs, we preformed digenome sequencing, a method for higher sensitivity, putative off-target site detection in the whole human genome,25,26 to detect any off-target effects in the ICP4-targeted gRNAs. Digenome sequencing revealed that only a few high-confidence, potential off-target sites of SpICP4-g3 (20 sites) were identified, while more were found in the SaICP4-g1-treated sample (57 sites) in the whole genome (Figures 5A and 5B). Most of the putative off-target sites were in non-coding regions, including introns (SpICP4-g3, 5 sites; SaICP4-g1, 22 sites) and intergenic regions (SpICP4-g3, 11 sites; SaICP4-g1, 22 sites) (Figures 5C and 5D). Most of the putative off-target sites found in SpICP4-g3 and SaICP4-g1 had low cleavage scores (SpICP4-g3, ranging from 25.86 to 2.84; SaICP4-g1, ranging from 39.61 to 2.6) (Figures 5E and 5F; Table S1). Considering that the average cleavage score in control samples was around 10 (Figures 5E and 5F, black line), those bearing scores less than 10 were less likely to be true off-target sites. However, the score in LTBP4 was obviously higher than the others (338.58 versus 39.61), suggesting a higher off-target potential in this site (Figure 5F). To further confirm whether the off-target effects can be detected in living cells, HEK293T cells were transfected with SpICP4-g3 or SaICP4-g1 to examine the high-score off-target sites. These sites were then amplified and sent for deep sequencing. Deep sequencing results showed that these sites had no significant enrichment of indels compared to the negative control (Figures 5G and 5H; Figures S5 and S6). Collectively, the two best performing gRNAs did not show detectable off-target effects in the human genome, demonstrating safety in further application of these gRNAs.
Figure 4.
Normal Cell Viability in SpCas9/gRNA- and SaCas9/gRNA-Expressing Cells
(A) Growth curve of control, SpCas9/gRNA-, and SaCas9/gRNA-expressing Vero cells. Error bars represent SD. Significance was determined by a one-way ANOVA. ∗∗p < 0.01, ∗∗∗p < 0.001. (B) Fluorescence-activated cell sorting (FACS) analysis of SpCas9/gRNA- and SaCas9/gRNA-expressing Vero cells. Cells were stained with propidium iodide (PI) for measurement of dead cells. (C) Statistical analysis of PI-positive cells in (B). Data were collected from three independent experiments. Significance was determined by one-way ANOVA.
Figure 5.
Digenome Sequencing-Based Off-target Analysis of ICP4-Targeted SpCas9 and SaCas9 gRNAs
(A) Distribution of off-target sites of SpICP4-g3 discovered by digenome sequencing in the whole genome. (B) Distribution of off-target sites of SaICP4-g3 discovered by digenome sequencing in the whole genome. (C) Distribution of off-target sites of SpICP4-g3 discovered by digenome sequencing in different genomic elements. (D) Distribution of off-target sites of SaICP4-g1 discovered by digenome sequencing in different genomic elements. (E) Off-target scores of SpICP4-g3 in each putative off-target site. Black line indicates average score of the control sample. (F) Off-target scores of the SaICP4-g1 gRNA in each putative off-target site. Black line indicates average score of the control sample. (G) Example of deep sequencing confirmation of SpICP4-g3 off-target sites in HEK293T cells. Frequency of indels at a certain position was calculated as the “indel enrichment” score. Scores of control and sample were plotted to identify Cas9 cleavage. (H) Example of deep sequencing confirmation of SaICP4-g1 off-target sites in HEK293T cells. Frequency of indels in a certain position was calculated as the indel enrichment score. Scores of control and sample were plotted to identify Cas9 cleavage.
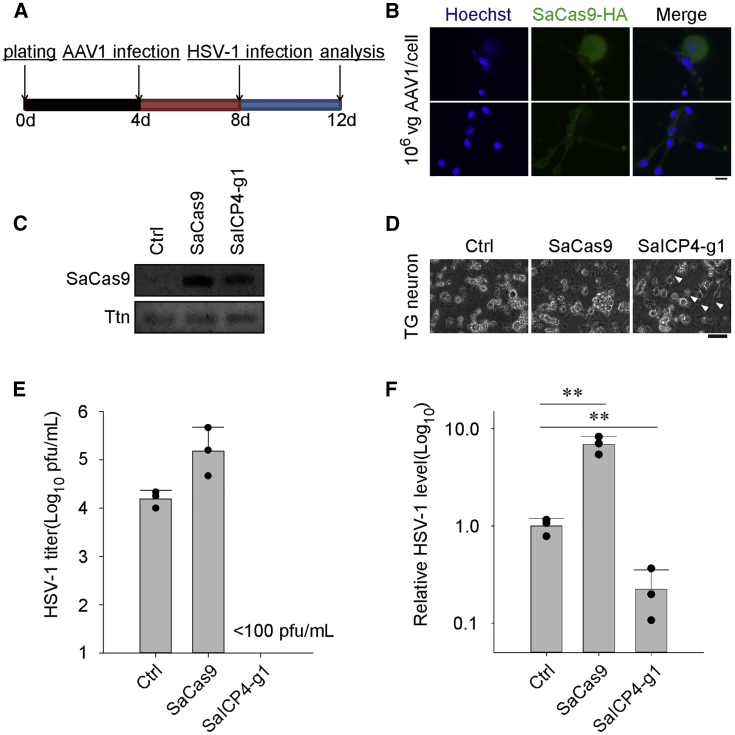
Suppression of HSV-1 Infection in Cultured TG Neuronal Cells
New treatments for latent infection of HSV-1 require efficient transduction of effectors, such as meganucleases and TG neurons.4 AAV has shown great promise for gene delivery in vivo as well as in TG neurons.27, 28, 29, 30, 31 Due to the packaging limit of AAV (<4,500 bp), SaCas9 (3,156 bp) is more suitable for AAV-mediated delivery than SpCas9 (4,101 bp), which leaves no spaces for other regulatory elements. In vivo genome editing and excision of HIV provirus are possible by delivery of SaCas9 by AAV.20,32 To test whether SaCas9 can be used to eliminate HSV-1 in TG neurons, we established in vitro mouse primary TG neuronal cultures for further experiments. Cells in TG were isolated and maintained as previously described.33 TG neuronal cells were first cultured for 4 days and then infected with AAV1 for SaCas9/gRNA expression and finally challenged with HSV-1 for another 4 days (Figure 6A). Successful delivery of SaCas9 was confirmed by both immunofluorescent microscopy and RT-PCR (Figures 6B and 6C). After challenging with HSV-1, neuronal features were greatly lost in control TG neuronal cultures compared to SaICP4-g1-expressing cells (Figure 6D). We measured HSV-1 levels in all TG neuronal cells. As predicted, HSV-1 replication was greatly compromised in cells expressing SaICP4-g1 (control, 104 PFU/mL; SaICP4-g1, <100 PFU/mL), as no detectable HSV-1 was found by qPCR (Figure 6E). Furthermore, DNA was extracted to quantify the HSV-1 genomes in TG neuronal cells. Consistent with the HSV-1 titer, an almost 10-fold decrease of HSV-1 was observed in SaICP4-g1 cells (Figure 6F). Taken together, these data indicate that AAV1-mediated delivery of SaCas9 together with ICP4-targeting gRNA may suppress HSV-1 replication in TG neuronal cultures, suggesting this may be a promising treatment for inhibition of HSV-1 in TG neurons.
Figure 6.
Expression of SaCas9/gRNA Inhibits HSV-1 Replication in Cultured Trigeminal Ganglion (TG) Neuronal Cells
(A) Schedule of the experimental process. (B) Immunostaining of SaCas9-HA in TG neurons infected with AAV (MOI of 106). Hoechst 33342 marks the nuclei. SaCas9-HA was visualized through immunostaining with hemagglutinin (HA) antibody. Scale bar, 10 μm. (C) Expression analysis of SaCas9 mRNA in TG neurons by RT-PCR. Ttn served as the endogenous control. (D) Morphology of HSV-1-infected TG neuronal cells 4 dpi at and MOI of 0.1. Arrows indicate neuronal morphology remaining in the culture. Scale bar, 30 μm. (E) Titer of HSV-1 in the culture medium of control or SaCas9-expressing TG neuronal cells infected with HSV-1 (MOI of 0.1) at 96 h. Titer was determined by infecting WT Vero cells and calculated by the Karber method on the fifth day after infection. Data were collected from three independent experiments. Error bars represent SD. (F) qPCR quantification of HSV-1 in TG neuronal cultures infected with HSV-1 (MOI of 0.1) at 96 h. Data were collected from three independent experiments. Error bars represent SD. Significance was determined by one-way ANOVA. ∗∗p < 0.01.
Discussion
Latent infection of HSV-1 is considered incurable due to the quiescent existence of HSV-1 genomes in cell bodies, especially in the TG neurons, in which HSV-1 hides from the immune system.21 In this study, we attempted to inhibit HSV-1 replication through gene-editing technology, which permits direct abrogation of the HSV-1 genome. We showed that both SpCas9 and SaCas9 can cleave ICP0 and ICP4 in vitro and in cell models. Expression of SpCas9 or SaCas9 together with ICP0- or ICP4-targeted gRNAs can inhibit HSV-1 infection and replication in Vero cells without affecting cell viability. We further identified a new target site on the ICP4 locus to inhibit HSV-1 replication by SaCas9. Digenome sequencing combined with deep sequencing confirmation revealed that both selected ICP4-targeting gRNAs showed high biosafety without gRNA-dependent off-target effects in the human genome. Additionally, we demonstrated for the first time that AAV1 delivery of SaCas9 can inhibit HSV-1 replication in cultured primary TG neurons. In this study, we showed that the CRISPR-Cas9 system can effectively inhibit HSV-1 replication in Vero cells or TG neurons. Furthermore, our data suggest a novel method for inactivating HSV-1 in HSK by AAV1-mediated SaCas9 delivery, which may be useful for targeting HSV-1 in TG neurons. To date, gene therapy has shown great promise in treating previously incurable diseases. In our study, we showed the potential application of AAV1-mediated SaCas9 delivery for inhibiting HSV-1 replication by targeting the ICP4 locus. In-depth research on whether SaCas9 can inhibit HSV-1 recurrence, eliminate quiescent HSV-1, and treat primary or recurrent HSK remains to be conducted.
Four distinct regions can be identified in the structure of HSV-1, i.e., the dsDNA genome, capsid, tegument, and envelope.7 ICP0 and ICP4 are two components that can be found in the HSV-1 tegument.34 Further investigation on the function of ICP0 and ICP4 reveal that they work as transcription activators for the transition from latency to reactivation.34 The E3 ubiquitin ligase ICP0 is important for initiating early and late gene transcription and anti-host defense.35 ICP4 acts as a transcription activator by promoting viral replication and controlling early- and late-related gene transcription, such as latency-associated promoters.24,36,37 Previous attempts have been made to inhibit HSV-1 replication by editing UL19, UL30, ICP0, UL7, UL23, UL24, UL8, and UL52,4,18,19,27,38 all of which show a certain degree of reduction of HSV-1 infectivity in cultured cells. Importantly, we found that a single gRNA targeting ICP4 can effectively inhibit HSV-1. A single gRNA targeting the ICP4 locus with SpCas9 or SaCas9 effectively compromised HSV-1 replication in culture cells. Since ICP0 and ICP4 can be upregulated by stress-induced transcription factors in the cells,39,40 it is considerable that the major activator ICP4 may be a more suitable target for inhibiting HSV-1. We identified no putative gRNA-dependent off-target effects of SpCas9 and SaCas9 targeting the ICP4 locus compared to control. This indicates that ICP4 might be a better target for attenuating HSV-1 infection in terms of effectivity and biosafety. However, not all of the tested ICP0 or ICP4 gRNAs were effective in inhibiting HSV-1, which suggests that careful selection and testing of the gRNA are of great importance in further applications.
It had been shown that the combination of different gRNAs targeting different loci would enhance the inhibition effect in other studies.17,18 A combination of UL8 and UL52 SpCas9/gRNA had been shown previously, which can greatly enhance the HSV-1 inhibition efficiency (nearly no detectable HSV-1 production).18 However, the presence of two gRNAs may likely double the probability of off-targets. One of the advantages of using ICP4 as a target is that ICP4 is the repeat region on the HSV-1 genome. Each HSV-1 genome has two repeats of ICP4, which means the SaCas9/gRNA would have two cut sites with one sequence accounting for off-targets, compared to UL8 and UL52 bearing only a single digestion site in the HSV-1 genome (Figure S1C). Also, we have shown that a single SaCas9/gRNA targeting ICP4 would effectively inhibit HSV-1 production without harnessing dual gRNAs.
AAV-mediated gene therapy is greatly described in preclinical tests, and AAV drugs are widely accepted in clinical settings as well.41,42 Current AAV therapy mainly focuses on delivery of functional genes to treat loss-of-function diseases.43,44 However, infectious diseases, especially those that have become latent, require disrupting the viral genome or gene regulation to inhibit viral reactivation. Recently, it was reported that meganucleases targeting HSV-1 genome can effectively inhibit HSV-1 replication and disrupt the latent HSV-1 genome in mice models.4 Due to the large size of SpCas9, it is difficult to package both the SpCas9 and gRNA in a single AAV, even though it has been shown that SpCas9 can edit and inhibit HSV-1 in cell models.15,18,19,27 With this consideration, smaller Cas9 orthologs may be better choices than SpCas9. SaCas9 is much smaller than SpCas9, and it has been confirmed that AAV-mediated delivery of SaCas9 achieves effective gene editing in vivo.20 It has been reported that SaCas9 is also capable of inhibiting HIV and hepatitis B virus (HBV).32,45,46 In the present study, we showed that HSV-1 can be inhibited by SaCas9 through single AAV1 delivery strategy in TG neurons, which suggests that a potential therapy using AAV1 in combination with SaCas9 to attenuate HSV-1 infection and interrupt the HSV-1 genome in the TG neuron could prevent HSV-1 recurrence or treat HSK. Further investigation may require in-depth study on the effect of AAV-mediated delivery of SaCas9 to inhibit HSV-1 reactivation in latent infection in animal models, and may contribute to the development of novel methods for treating viral diseases, especially those caused by recurrence of latent infections.
Materials and Methods
Mice
Mice were housed under standard conditions (22°C ± 1°C) in a specific pathogen-free animal facility with a 14-h light/10-h dark cycle in Sun Yat-sen University. The Institutional Animal Care and Use Committee of Sun Yat-sen University (P.R. China) approved all experimental protocols concerning the handling of mice.
Expression and Purification of SpCas9 and SaCas9
BL21 Star (DE3) E. coli cells were transformed with pET28a-His-SpCas9 or pET28a-His-SaCas9. A single clone was cultured in 5 mL of Luria-Bertani (LB) broth with 50 μg/mL kanamycin at 37°C overnight. The cells were inoculated into 1 L of LB broth at 37°C and grown to an optical density 600 (OD600) of 0.6. The cells were incubated at 4°C for 1 h and added with 0.5 mM IPTG (isopropyl β-d-thiogalactoside) to induce protein expression at 18°C for 14–16 h. Induced cells were collected by centrifugation at 5,000 × g for 10 min and lysed by sonication (2 s pulse-on, 5 s pulse-off for 6 min total) in 10 mL of lysis buffer (50 mM tris(hydroxymethyl)aminomethane [Tris]-HCl [pH 8.0], 0.5 M NaCl, 5% glycerol, and 20 mM imidazole; Sigma-Aldrich). The lysate supernatant was poured into a column with Ni Sepharose 6 FF (fast flow) (GE Healthcare) and washed with about 40 mL of lysis buffer. Protein was eluted with elution buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 5% glycerol, 270 mM imidazole) and further purified by a 1-mL SP Sepharose cation exchange column (GE Healthcare) with 20 mL of low-salt buffer (50 mM Tris-HCl [pH 8.0], 0.1 M NaCl, 5% glycerol) and washed. The Cas9 protein was eluted by elution buffer (50 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 5% glycerol) for gel filtration chromatography. The final protein was concentrated to 300 μL and filtered through a 0.22-μm polyvinylidene fluoride membrane (Millipore) and stored at 4°C.
In Vitro Cleavage of dsDNA Target
Synthetic target DNA was cloned to pGSI by Igebio (the sequence of pGSI is shown in the Supplemental Information). Plasmid was incubated with FastDigest ScaI (Thermo Scientific) overnight and purified with a PCR cleanup kit (QIAGEN). 100 ng of linearized DNA was used for Cas9 digestion in a reaction volume of 10 μL containing 1 μL of 10× NEBuffer 3, 300 nM Cas9, and 900 nM gRNA for 1 h at 37°C, after removal of gRNA and protein using RNase A (Takara) and Proteinase K (Thermo Scientific).
Plasmid Construction
Oligonucleotides corresponding to HSV-1 targets were annealed and ligated to lentiCRISPR v2, SaCas9 lenti V2, or pX601 as previously described.44,45 gRNA sequences for different targets were as follows: SpICP0-g1, 5′-GCCCCCCCTCGAGAGGACGG-3′; SpICP0-g2, 5′-AGGCGGAGTCGTCGTCATGG-3′; SpICP0-g3, 5′-ACCAGCTTGGCGTTGCACAG-3′; SpICP4-g1, 5′-CGGCCCTCATGTTTGACCCG-3′; SpICP4-g2, 5′-GGGGGTCTTCGCGCGCGTGG-3′; SpICP4-g3, 5′-GAAGGAGCTGCTGTTGCGCG -3′; SaICP0-g1, 5′-GGCTGCTGGGGCCGCAGGGC-3′; SaICP0-g2, 5′-GATAGTGGGCGTGACGCCCA-3′; SaICP4-g1, 5′-CACGTCCCCGGGGACCACGC-3′; and SaICP4-g2, 5′-CACCCCGGACGCGGAGGCCA-3′.
Cell Lines and Virus
293T cells came from ATCC. Vero cells and HSV-1 virus were gifts from J.C. 293T cells and Vero cells were cultured in DMEM (HyClone, SH30022.01B) containing penicillin/streptomycin (Invitrogen, 10378016) and 10% fetal bovine serum (FBS) (HyClone, SH30084.03).
Establishment of Cas9-gRNA Co-expression Cell Lines
HEK293T cells were seeded in six-well plates and transfected at about 80% confluency with Cas9-gRNA co-expressive plasmid lentiCRISPR v2 and lentivirus packaging vectors pMD2.G and pSPAX2. Vero cells were infected with the lentivirus produced form HEK293T cells supplemented with 8 μg/mL Polybrene. Two days after infection, Vero cells were selected in culture media with puromycin (2.5 μg/mL) for at least 5 days.
HSV-1 Titer Assay
5,000 Vero cells were planted in 96-well plates per well and infected by gradient dilution of HSV-1 after 48 h. The percentage of herpesvirus-infected wells was monitored 2 days to 5 days after infection. The HSV-1 titer from the origin sample was calculated by Karber’s method.
HSV-1 Produced from Cas9-gRNA Co-expression Cells
Cas9-gRNA co-expression Vero cells were seeded in six-well plates and infected with HSV-1 (MOI of 0.1) at approximately 85% confluency for 1 h before refreshing culturing medium. The cells were cultured for 72 h. Then, the supernatant was gradient diluted and infected WT Vero cells in 96-well plates for titer determination by Karber’s method.
HSV-1 Genome Copy Number Detection
We performed quantitative real-time PCR (GoTaq qPCR master mix; Promega, Madison, WI, USA, A6002) using the supernatant collected from HSV-1-infected cells at 72 h (for those collected from TG neuronal cultures, detected at 96 h) to quantify the relative level of HSV-1 genomic DNA in HSV-1-infected wells at an MOI of 0.1. The reaction mixture was heated at 95°C for 10 min, then 40 cycles of amplification at 95°C for 15 s and 60°C for 60 s. Data were analyzed by the StepOnePlus system (Applied Biosystems). PCR primers were as follows: HSV, forward, 5′-TTCTCGTTCCTCACTGCCTCCC-3′, reverse, 5′-CGTCTGGACCAACCGCCACAC-3′; Ttn, forward, 5′-GACACCACAAGGTGCAAAGTC-3′, reverse, 5′-CCCACTGTTCTTGACCGTATCT-3′.
Immunofluorescence Assay
Vero cells were infected with HSV-1 (MOI of 1). 48 h after infection, the cells were washed with PBS twice and fixed in 4% paraformaldehyde in PBS for 30 min at 4°C. Then, samples were permeabilized by 0.5% Triton X-100, 20 mM HEPES, 50 mM NaCl, 3 mM MgCl2, and 300 mM sucrose in water and incubated with 5% goat serum in PBS. Mouse anti-ICP5 (1:200, Abcam, anti-HSV1 + HSV2 ICP5 major capsid protein antibody [3B6; ab6508]) antibody was used for immunostaining for 2 h at room temperature. Alexa Fluor 555-conjugated donkey anti-mouse immunoglobulin G (IgG) (1:2,000, Life Technologies) antibody containing 10 μg/mL Hoechst 33342 in PBS (Invitrogen, H3570) was used to visualize the primary antibody and label the nuclei.
Cell Viability Assay
Vero cells were harvested in a 1.5-mL Eppendorf (EP) tube from six-well plates after 48 h and planted using 0.25% trypsin. The samples were centrifuged for 3 min at 1,200 rpm. The pellets were washed with 1 mL of PBS once and mixed with 1 mL of propidium iodide (PI) staining reagent (Beyotime). The samples were incubated for 30 min at room temperature in the dark and analyzed by fluorescence-activated cell sorting (FACS).
Digenome Sequencing
Human genomic DNA was purified from HEK292T cells using a DNeasy Blood & Tissue Kit (QIAGEN). About 8 μg of genomic DNA was used for Cas9 digestion in a reaction volume of 400 μL, which contained 40 μL of 10× NEBuffer 3, 300 nM Cas9, and 900 nM gRNA for 8 h at 37°C. After removal of gRNA and protein using RNase A and Proteinase K, genomic DNA was purified again with a DNeasy Blood & Tissue Kit (QIAGEN). The digenome sequencing sample was prepared and sequenced as previously described.25,26 Digenome sequencing to discover in vitro cleavage sites was performed using a program at https://github.com/snugel/digenome-toolkit. The off-target score was calculated accordingly.25,26
Deep Sequencing
According to the analysis results of digenome sequencing, the off-target sites were amplified with a KOD PCR kit (Toyobo) and barcode-containing primers to generate a deep sequencing library. Barcode-containing products were then sequenced by paired-end 150 HiSeq 2000 (Illumina). Indel formation was analyzed as previously described.25,26
Culture of TG Neuronal Cells
Neuronal cells in TG were isolated from female C57BL/6 mice (8 weeks old) and cultured as previously reported with modification.4 After removing the brain, ganglia were cut and cleaned by Hanks’ balanced salt solution (HBSS) (130 mM NaCl, 5 mM KCl, 2 mM KH2PO4, 10 mM glucose, and 10 mM HEPES [pH 7.2]), then chopped and incubated with papain and then collagenase II/dispase II. TG neurons were separated by Percoll gradients by centrifugation. Dissociated cells were seeded in 12-well plates and cultured with media (Neurobasal A supplemented with 2% B27, 50 ng/mL nerve growth factor, l-glutamine, and penicillin/streptomycin). TG neuronal cells were infected with AAV1 at day 4 and HSV-1 at day 8, and analyzed by qPCR and immunofluorescence (IF) at day 12.
Statistical Analysis
The statistical significances among multiple mean values were examined using one-way ANOVA with the Holm-Sidak test for data with equal variance and normal distribution (SigmaPlot 12.5). Otherwise, ANOVA on ranks with the Kruskal-Wallis test was used (SigmaPlot 12.5). Arcsine transformation was performed before testing of percentage data. Data were considered significant at p < 0.05 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Data Accessibility
Supporting information is available from the Wiley Online Library or from the author.
Author Contributions
J.H. and Y.C. designed the experiments. Y.C., S.Z., P.L., Q. Zhang, M.L., Q. Zhao, J.R., and J.C. performed and analyzed the experiments. J.H., Y.L., and Z.S. supervised the research. Y.C. and J.H. wrote the paper. All authors discussed the results and commented on the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key R&D Program of China (2017YFC1001901), the National Natural Science Foundation of China (31671540 and 31971365), the Natural Science Foundation of Guangdong Province (2017A030313491), the Guangdong Special Support Program (2019BT02Y276) and the Guangzhou Science and Technology Project (201803010020).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.05.011.
Contributor Information
Junjiu Huang, Email: hjunjiu@mail.sysu.edu.cn.
Yizhi Liu, Email: liuyizh@mail.sysu.edu.cn.
Zhou Songyang, Email: songyanz@mail.sysu.edu.cn.
Supplemental Information
References
- 1.Looker K.J., Magaret A.S., May M.T., Turner K.M., Vickerman P., Gottlieb S.L., Newman L.M. Global and regional estimates of prevalent and incident herpes simplex virus type 1 infections in 2012. PLoS ONE. 2015;10:e0140765. doi: 10.1371/journal.pone.0140765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Looker K.J., Magaret A.S., Turner K.M., Vickerman P., Gottlieb S.L., Newman L.M. Global estimates of prevalent and incident herpes simplex virus type 2 infections in 2012. PLoS ONE. 2015;10:e114989. doi: 10.1371/journal.pone.0114989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farooq A.V., Shukla D. Herpes simplex epithelial and stromal keratitis: an epidemiologic update. Surv. Ophthalmol. 2012;57:448–462. doi: 10.1016/j.survophthal.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubert M., Madden E.A., Loprieno M., DeSilva Feelixge H.S., Stensland L., Huang M.L., Greninger A.L., Roychoudhury P., Niyonzima N., Nguyen T. In vivo disruption of latent HSV by designer endonuclease therapy. JCI Insight. 2016;1:e88468. doi: 10.1172/jci.insight.88468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsatsos M., MacGregor C., Athanasiadis I., Moschos M.M., Hossain P., Anderson D. Herpes simplex virus keratitis: an update of the pathogenesis and current treatment with oral and topical antiviral agents. Clin. Exp. Ophthalmol. 2016;44:824–837. doi: 10.1111/ceo.12785. [DOI] [PubMed] [Google Scholar]
- 6.Wilhelmus K.R. Antiviral treatment and other therapeutic interventions for herpes simplex virus epithelial keratitis. Cochrane Database Syst. Rev. 2015;1:CD002898. doi: 10.1002/14651858.CD002898.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X., Che Y., Li Q. HSV-1 tegument protein and the development of its genome editing technology. Virol. J. 2016;13:108. doi: 10.1186/s12985-016-0563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudesh S., Laibson P.R. The impact of the herpetic eye disease studies on the management of herpes simplex virus ocular infections. Curr. Opin. Ophthalmol. 1999;10:230–233. doi: 10.1097/00055735-199908000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Miserocchi E., Modorati G., Galli L., Rama P. Efficacy of valacyclovir vs acyclovir for the prevention of recurrent herpes simplex virus eye disease: a pilot study. Am. J. Ophthalmol. 2007;144:547–551. doi: 10.1016/j.ajo.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Wald A., Corey L., Cone R., Hobson A., Davis G., Zeh J. Frequent genital herpes simplex virus 2 shedding in immunocompetent women. Effect of acyclovir treatment. J. Clin. Invest. 1997;99:1092–1097. doi: 10.1172/JCI119237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan D., Kaye S.B., Hopkins M., Kirwan R., Hart I.J., Coen D.M. Common and new acyclovir resistant herpes simplex virus-1 mutants causing bilateral recurrent herpetic keratitis in an immunocompetent patient. J. Infect. Dis. 2014;209:345–349. doi: 10.1093/infdis/jit437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voigt S., Hofmann J., Edelmann A., Sauerbrei A., Kühl J.S. Brincidofovir clearance of acyclovir-resistant herpes simplex virus-1 and adenovirus infection after stem cell transplantation. Transpl. Infect. 2016;18:791–794. doi: 10.1111/tid.12582. [DOI] [PubMed] [Google Scholar]
- 13.Grosse S., Huot N., Mahiet C., Arnould S., Barradeau S., Clerre D.L., Chion-Sotinel I., Jacqmarcq C., Chapellier B., Ergani A. Meganuclease-mediated inhibition of HSV1 infection in cultured cells. Mol. Ther. 2011;19:694–702. doi: 10.1038/mt.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aubert M., Boyle N.M., Stone D., Stensland L., Huang M.L., Magaret A.S., Galetto R., Rawlings D.J., Scharenberg A.M., Jerome K.R. In vitro inactivation of latent HSV by targeted mutagenesis using an HSV-specific homing endonuclease. Mol. Ther. Nucleic Acids. 2014;3:e146. doi: 10.1038/mtna.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suenaga T., Kohyama M., Hirayasu K., Arase H. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol. Immunol. 2014;58:513–522. doi: 10.1111/1348-0421.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell T.A., Stefanovic T., Tscharke D.C. Engineering herpes simplex viruses by infection-transfection methods including recombination site targeting by CRISPR/Cas9 nucleases. J. Virol. Methods. 2015;213:18–25. doi: 10.1016/j.jviromet.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Roehm P.C., Shekarabi M., Wollebo H.S., Bellizzi A., He L., Salkind J., Khalili K. Inhibition of HSV-1 replication by gene editing strategy. Sci. Rep. 2016;6:23146. doi: 10.1038/srep23146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Diemen F.R., Kruse E.M., Hooykaas M.J., Bruggeling C.E., Schürch A.C., van Ham P.M., Imhof S.M., Nijhuis M., Wiertz E.J., Lebbink R.J. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Bi Y., Xiao H., Sun L., Ren Y., Li Y., Chen C., Cun W. CRISPR-Cas9 system-driven site-specific selection pressure on Herpes simplex virus genomes. Virus Res. 2018;244:286–295. doi: 10.1016/j.virusres.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umbach J.L., Kramer M.F., Jurak I., Karnowski H.W., Coen D.M., Cullen B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagglund R., Roizman B. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 2004;78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinnoji R.C., Bedadala G.R., George B., Holland T.C., Hill J.M., Hsia S.C. Repressor element-1 silencing transcription factor/neuronal restrictive silencer factor (REST/NRSF) can regulate HSV-1 immediate-early transcription via histone modification. Virol. J. 2007;4:56. doi: 10.1186/1743-422X-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grondin B., DeLuca N. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J. Virol. 2000;74:11504–11510. doi: 10.1128/jvi.74.24.11504-11510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D., Kim S., Kim S., Park J., Kim J.S. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex digenome-seq. Genome Res. 2016;26:406–415. doi: 10.1101/gr.199588.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.-I., Kim J.-S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 27.Lin C., Li H., Hao M., Xiong D., Luo Y., Huang C., Yuan Q., Zhang J., Xia N. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci. Rep. 2016;6:34531. doi: 10.1038/srep34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goater J., Müller R., Kollias G., Firestein G.S., Sanz I., O’Keefe R.J., Schwarz E.M. Empirical advantages of adeno associated viral vectors in vivo gene therapy for arthritis. J. Rheumatol. 2000;27:983–989. [PubMed] [Google Scholar]
- 29.Ross C.J., Twisk J., Bakker A.C., Miao F., Verbart D., Rip J., Godbey T., Dijkhuizen P., Hermens W.T., Kastelein J.J. Correction of feline lipoprotein lipase deficiency with adeno-associated virus serotype 1-mediated gene transfer of the lipoprotein lipase S447X beneficial mutation. Hum. Gene Ther. 2006;17:487–499. doi: 10.1089/hum.2006.17.487. [DOI] [PubMed] [Google Scholar]
- 30.Bockstael O., Foust K.D., Kaspar B., Tenenbaum L. Recombinant AAV delivery to the central nervous system. Methods Mol. Biol. 2011;807:159–177. doi: 10.1007/978-1-61779-370-7_7. [DOI] [PubMed] [Google Scholar]
- 31.Dang C.H., Aubert M., De Silva Feelixge H.S., Diem K., Loprieno M.A., Roychoudhury P., Stone D., Jerome K.R. In vivo dynamics of AAV-mediated gene delivery to sensory neurons of the trigeminal ganglia. Sci. Rep. 2017;7:927. doi: 10.1038/s41598-017-01004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin C., Zhang T., Qu X., Zhang Y., Putatunda R., Xiao X., Li F., Weidong Xiao W., Zhao H., Dai S. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol. Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malin S.A., Davis B.M., Molliver D.C. Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat. Protoc. 2007;2:152–160. doi: 10.1038/nprot.2006.461. [DOI] [PubMed] [Google Scholar]
- 34.Loret S., Guay G., Lippé R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith M.C., Boutell C., Davido D.J. HSV-1 ICP0: paving the way for viral replication. Future Virol. 2011;6:421–429. doi: 10.2217/fvl.11.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perron H., Suh M., Lalande B., Gratacap B., Laurent A., Stoebner P., Seigneurin J.M. Herpes simplex virus ICP0 and ICP4 immediate early proteins strongly enhance expression of a retrovirus harboured by a leptomeningeal cell line from a patient with multiple sclerosis. J. Gen. Virol. 1993;74:65–72. doi: 10.1099/0022-1317-74-1-65. [DOI] [PubMed] [Google Scholar]
- 37.Halford W.P., Kemp C.D., Isler J.A., Davido D.J., Schaffer P.A. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 2001;75:6143–6153. doi: 10.1128/JVI.75.13.6143-6153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X., Fan S., Zhou J., Zhang Y., Che Y., Cai H., Wang L., Guo L., Liu L., Li Q. The mutated tegument protein UL7 attenuates the virulence of herpes simplex virus 1 by reducing the modulation of α-4 gene transcription. Virol. J. 2016;13:152. doi: 10.1186/s12985-016-0600-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinani D., Cordes E., Workman A., Thunuguntia P., Jones C. Stress-induced cellular transcription factors expressed in trigeminal ganglionic neurons stimulate the herpes simplex virus 1 ICP0 promoter. J. Virol. 2013;87:13042–13047. doi: 10.1128/JVI.02476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.El-Mayet F.S., Sawant L., Thunuguntla P., Jones C. Combinatorial effects of the glucocorticoid receptor and Krüppel-like transcription factor 15 on bovine herpesvirus 1 transcription and productive infection. J. Virol. 2017;91:e00904-17. doi: 10.1128/JVI.00904-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu W., Wu Z. In vivo applications of CRISPR-based genome editing in the retina. Front. Cell Dev. Biol. 2018;6:53. doi: 10.3389/fcell.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J.H., Wang J.H., Chen J., Li F., Edwards T.L., Hewitt A.W., Liu G.S. Gene therapy for visual loss: opportunities and concerns. Prog. Retin. Eye Res. 2019;68:31–53. doi: 10.1016/j.preteyeres.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Burnight E.R., Giacalone J.C., Cooke J.A., Thompson J.R., Bohrer L.R., Chirco K.R., Drack A.V., Fingert J.H., Worthington K.S., Wiley L.A. CRISPR-Cas9 genome engineering: treating inherited retinal degeneration. Prog. Retin. Eye Res. 2018;65:28–49. doi: 10.1016/j.preteyeres.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiCarlo J.E., Mahajan V.B., Tsang S.H. Gene therapy and genome surgery in the retina. J. Clin. Invest. 2018;128:2177–2188. doi: 10.1172/JCI120429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaminski R., Bella R., Yin C., Otte J., Ferrante P., Gendelman H.E., Li H., Booze R., Gordon J., Hu W., Khalili K. Excision of HIV-1 DNA by gene editing: a proof-of-concept in vivo study. Gene Ther. 2016;23:690–695. doi: 10.1038/gt.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott T., Moyo B., Nicholson S., Maepa M.B., Watashi K., Ely A., Weinberg M.S., Arbuthnot P. ssAAVs containing cassettes encoding SaCas9 and guides targeting hepatitis B virus inactivate replication of the virus in cultured cells. Sci. Rep. 2017;7:7401. doi: 10.1038/s41598-017-07642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting information is available from the Wiley Online Library or from the author.