Abstract
Renal fibrosis is associated with the reduction in the functional renal parenchyma and in most cases progresses to end-stage kidney failure, a devastating condition that requires lifelong dialysis or kidney transplantation. However, due to the extreme complexity in the pathogenesis of renal fibrosis and our limited knowledge, therapeutic options for renal fibrosis in the clinical setting are still scarce and often ineffective. Hence, further studies on the molecular mechanisms underlying renal fibrosis are compellingly needed. Multiple miRNAs have demonstrated to participate in kidney diseases in a TGF-β dependent or independent manner, but there is very little known about miR-155-5p on renal fibrosis. In the present study, we firstly explored the expression level and functions of miR-155-5p in the setting of renal fibrosis. Our research revealed that miR-155-5p is highly expressed in kidney tissues from patients and unilateral ureteral obstruction (UUO) rat models, and miR-155-5p knockdown significantly blocks renal fibrosis both in vivo and in vitro. In mechanism, our data demonstrate that miR-155-5p promotes renal fibrosis by increasing the phosphorylated activation of STAT3 via targeting SOCS1/6. Altogether, our findings highlight a miR-155-5p/SOCS/STAT3 axis in the pathogenesis of renal fibrosis, which may provide promising therapeutic targets for clinical prevention of this disease.
1. Introduction
Renal fibrosis, characterized by tubulointerstitial fibrosis and glomerulosclerosis, is almost the final pathological process of chronic kidney disease (CKD) and ultimately causes kidney failure and organ death in most cases [1–4]. In histological features, renal fibrosis manifests the excessive accumulation and deposition of extracellular matrix components, which is always companied by various pathological alterations in cells including epithelial-to-mesenchymal transition, fibroblast activation, immune cell infiltration, and apoptosis [1–3]. In recent epidemiological studies, in the United States, over 30 million people already have been diagnosed as CKD, and around 50% of people are projected to develop the disease in their lifetime [5]. Currently, due to the complexity in the pathogenesis of renal fibrosis, the effective intervention for this disease is still lacking. Hence, further studies on the molecular mechanisms underlying renal fibrosis are compellingly needed.
Previously, tremendous evidence has revealed that TGF-β is substantially upregulated in renal fibrosis, and overexpression of active TGF-β is capable of inducing renal fibrosis [4, 6]. In contrast, blocking TGF-β using a neutralizing antibody, antisense oligonucleotides, inhibitors, or genetic deletion of receptors effectively alleviates the degree of renal fibrosis both in vivo and in vitro [7]. In the progression of renal fibrosis, TGF-β1 signaling promotes the deposition of the extracellular matrix via activating transcriptional factors either Smad (such as Smad2/3/4) or STAT (such as STAT3) family [6]. Kuratsune and colleagues reported that a significantly elevated activation of STAT3 is observed in the renal tubular interstitial cells of the kidneys with unilateral ureteral obstruction (UUO) [8]. Moreover, Pang and colleagues demonstrated that S3I-201, a widely used STAT3 inhibitor, remarkably suppresses the expression of fibronectin, α-SMA (alpha-smooth muscle actin), and collagen I proteins and effectively alleviates UUO-induced renal interstitial fibrosis of animals [9]. All the previous evidence indicates that STAT3 plays a critical role in the pathogenesis of renal fibrosis. However, why the expression of STAT3 is upregulated in fibrotic kidney tissues remains unclear.
MicroRNAs (miRNAs), a kind of well-investigated small endogenous noncoding RNAs with 20-22 nucleotides in length, are ultimately assembled into the RNA-induced silencing complex (RISC) to bind the 3′UTR of target mRNAs for the posttranscriptional gene silencing by mRNA degradation or translation inhibition [10, 11]. Extensive miRNAs have been demonstrated to participate in kidney development, hemostasis, and especially kidney fibrosis in a TGF-β-dependent or TGF-β-independent manner, such as miR-21, miR-23a, miR-27, miR-34a-5p, miR-93, miR-132, miR-135, and miR-146 which are reported highly expressed in renal fibrotic tissues and contribute to the fibrogenesis of renal; while miR-23b, miR-29, miR-30, miR-34a, miR-192, miR-200, and miR-433 are described downregulated and revealed an inhibitory effect on kidney fibrosis [12–18].
miR-155-5p, a typical multifunctional miRNA, is first identified in B-cells and is processed from an exon of a noncoding RNA transcribed from the B-cell Integration Cluster (BIC) located on the chromosome [19–21]. miR-155-5p is ubiquitously expressed in various tissues and plays crucial roles both in physiological and pathological processes including hematopoietic lineage differentiation, immunity, inflammation, cancer, and cardiovascular diseases [21–23]. In the kidney, miR-155-5p was reported abnormally expressed in renal cancer, end-stage renal disease, and renal transplantation undergoing acute cellular rejection [24–30]. More importantly, miR-155-5p was observed to be upregulated in fibrotic lung tissues, and hypoxia-induced miRNA-155-5p promotes the fibrosis of proximal tubule cells in vitro [31–35], implying that it may participate in the pathology of renal fibrosis. However, whether and how miR-155-5p involves renal fibrosis remain unknown.
In this study, we firstly explored the expression level and functions of miR-155-5p in renal fibrosis patients and unilateral ureteral obstruction (UUO) rat models. Noticeably, due to the capability of generating progressive renal fibrosis, UUO has become one of the most popular models for chronic kidney disease research in vivo [36]. We observed that miR-155-5p was highly expressed in the kidney tissues from renal disease patients and UUO rats. To further investigate the underlying mechanisms by which miR-155-5p involved in the pathogenesis of renal fibrosis, we identified SOCS1/6 as its putative targets using bioinformatics tools, and these results were experimentally validated in NRK-49F and HK-2 cells and clinical specimens. Finally, our work revealed that miR-155-5p promotes renal fibrosis by increasing the phosphorylated activation of STAT3 both in vitro and in vivo. Altogether, our findings highlight a miR-155-5p/SOCS/STAT3 axis in the pathogenesis of renal fibrosis, which may provide promising therapeutic targets for clinical prevention or treatment of this disease.
2. Materials and Methods
2.1. Patients
In this study, 6 patients (4 males and 2 females; mean age = 51.3 ± 4.7 years old) with tubulointerstitial fibrosis (TIF) disease were collected from the Affiliated Changzhou No.2 People's Hospital of Nanjing Medical University. Six healthy donors (ND; 3 males and 3 females; mean age = 49.6 ± 10.1 years old) who died from traffic accidents were enrolled as normal controls with the following exclusion criteria: with other renal diseases, 20 years of age, inability to provide informed consent, presence of active infection, and pregnancy. The kidney tissue was harvested, and RNA samples and protein samples were extracted using the Qiagen RNeasy Mini Kit and RIPA buffer, respectively. All experimental protocols and procedures involved in the present study were approved by the Ethics Committee of the Southeast University, and the study complied with the declaration of Helsinki. The written informed consent was obtained from all participants for the publication of this article.
2.2. Unilateral Ureteral Obstruction (UUO) Rat Models
All animal experiments were performed following the protocols approved by the Southeast University institutional committee for the care and use of laboratory animals and compliance with local authorities (2018-2404). Adult male rats (250-300 g body weight) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China). All animals were housed in sterile and filter-capped cages in the present study. The surgery of UUO was carried out as previously described [37]. In brief, animals were anesthetized and a midline incision was made in the abdominal wall, the left ureter was dissected out and ligated with 4.0 silk at two points along its length, and the kidneys were harvested at the seventh day after surgery.
2.3. Cell Culture, Transfection, and miR-155-5p Knockdown
HEK293 cell line and kidney cell lines NRK-49F and HK-2 were maintained in DMEM medium plus with 10% heat-inactivated fetal bovine serum and 1% Penicillin-Streptomycin and kept in culture with 37°C and 5% CO2 condition. For plasmid transfection, the cells were seeded in the 6-well plates and transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the instructions. For miR-155-5p knockdown, shRNA-expressing lentivirus was packaged in HEK293 cells by cotransfected vectors and packaging plasmids; 48 hours later, the virus was harvested and subjected to infect cells. The miR-155-5p knockdown cells were obtained by selecting with puromycin.
2.4. Luciferase Reporter Assay
HEK293 cells were transfected with the indicated reporter constructs containing either the wild-type or mutated miR-155-5p binding sequences in the 3′UTRs of SOCS1, SOCS5, and SOCS6 and control Renilla luciferase reporter and miR-155-5p or NC. Luciferase activity was determined by using a dual luciferase assay kit (Promega, Madison, WI, USA) 48 hours after transfection, and firefly luciferase activity was measured and normalized with cotransfected Renilla luciferase activity.
2.5. RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from human renal disease patients, rat kidney tissues, or cultured cells by TRIzol reagent or Qiagen RNeasy Mini Kit according to the manufacturer's instruction. An equal amount RNA was subjected to reverse the transcription for cDNAs using the Takara PrimeScript RT-PCR Kit (Takara, Dalian, China). Real-time PCR was carried out using the TB Green Fast qPCR Mix (Takara, Dalian, China). Quantification was done using ΔΔCt values, and the reference gene GAPDH was used as the internal control.
For the detection of mature miR-155-5p, the One-Step PrimeScript miRNA cDNA Synthesis Kit (Takara, Dalian, China) was used for RNA reverse transcription. The relative expression of miRNA was calculated using the Ct method, and the expression of U6 small nuclear RNA (snRNA) was used as an internal control. The primers used in the study were listed as follows: SOCS1 forward: 5′-CAC GCA CTT CCG CAC ATT C-3′; SOCS1 reverse: 5′-TAA GGG CGA AAA AGC AGT TCC-3′; SOCS5 forward: 5′-GTG CCA CAG AAA TCC CTC AAA-3′; SOCS5 reverse: 5′-TCT CTT CGT GCA AGT CTT GTT C-3′; SOCS6 forward: 5′-ATC ACG GAG CTA TTG TCT GGA-3′; SOCS6 reverse: 5′-CTG ACT CTC ATC CTC GGG GA-3′; GAPDH forward: 5′-ATC ACT GCC ACC CAG AAG AC-3′; GAPDH reverse: 5′-ATG AGG TCC ACC ACC CTG TT-3′; miR-155-5p forward: 5′-CTT AAT GCT AAT CGT GAT AGG GGT-3′; U6 forward: 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′; and U6 reverse: 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′. Noticeably, the reverse primer of miR-155-5p is the universal primer from the company.
2.6. Immunoblotting (IB)
Cells or kidney tissues were lysed in RIPA buffer and quantitated using a BCA kit (Abcam, Cambridge, MA, USA). Subsequently, the protein samples were separated by SDA-PAGE and then transferred to PVDF (polyvinylidene difluoride) membranes (Millipore, Billerica, MA, USA). Then, the membranes were blocked with 5% BSA and blotted with primary antibodies overnight. The next day, the immunoreactive bands were exposed using the enhanced chemiluminescence (ECL). β-Actin (or β-tubulin) was used as internal control. Antibodies against human SOCS1 (#3950), STAT3 (#12640), phospho-STAT3 (Tyr705) (#9145), α-smooth muscle actin (#19245), β-tubulin (#2128) were purchased from Cell Signaling Technology (Cell Signaling Technology, USA); antibodies of SOCS5 (ab97283), SOCS6 (ab197335), fibronectin (ab2413), collagen I (ab34710), and β-actin (ab8227) were purchased from Abcam (Abcam, Cambridge, MA, USA).
2.7. Hematoxylin and Eosin (H&E) Staining and Immunohistochemistry (IHC)
For histological analysis, rats or renal disease patient's kidney tissues were collected and embedded in paraffin, and 4 μm tissue sections were subjected to hematoxylin and eosin staining. The interstitial injury was scored in a blinded manner according to the widely used semiquantitative scale for renal tubular injury: no injury (0), mild: less than 25% (1), moderate: less than 50% (2), severe: less than 75%, and (3) very severe: more than 75% (4). Immunohistochemical analysis was applied in paraffin sections that were deparaffinized in xylene and rehydrated in a gradient of ethanol and distilled water. Antigen retrieval was finished with a microwave-based technique. Tissue sections were then blocked with 5% BSA and subjected to primary antibodies collagen I and α-SMA. Isotype-matched rabbit immunoglobulin G was used as negative controls, and the nuclei were counterstained with hematoxylin.
2.8. In Situ Hybridization (ISH)
For in situ hybridizations, specific 5′FITC-labeled antisense-locked nucleic acid oligonucleotides for mmu-miR-155 and a scrambled probe as negative control were obtained commercially. Paraffin-embedded kidney tissue slides were deparaffinization and deproteinization and subsequently were prehybridized with hybridization buffer. The hybridization was carried out overnight with the FITC-anti-sense microRNA probe at 45°C. Signals were visualized and detected under the fluorescence microscope.
2.9. Bioinformatics Analysis and Statistics Analysis
Potential miR-155-5p targets were predicted with in-house bioinformatics tools MicroCible and MicroTopTable. In this study, all the data were expressed as mean ± SEM, and the duplication numbers were specified in the figure legends. Statistical analysis was carried out using the software of GraphPad Prism7 (San Diego, CA, USA). For the comparison of two samples, the student's t-test was used after F-test to compare their variances; for the comparison of multiple samples, the one-way ANOVA test was used; in the linear regression for correlation analysis, R2 was calculated for the fit goodness. p value < 0.05 was considered statistically significant.
3. Results
3.1. miR-155-5p Is Highly Expressed in the Fibrotic Kidney Tissues
To investigate the roles of miR-155-5p in renal fibrosis, we first determined the expression level of miR-155-5p in the kidney tissues from the patients diagnosed as tubulointerstitial fibrosis (TIF). Intriguingly, we observed that, compared to normal donors, the miR-155-5p expression was significantly increased in the kidney tissues from patients (Figure 1(a)). To further confirm our observation, we generated a rat renal fibrosis model using UUO surgery, a widely used animal model for chronic kidney disease. Seven days after UUO surgery, we observed that the expressions of fibronectin and α-SMA were dramatically increased determined by IB (Figure 1(b)), indicating the animal models were built successfully. Consistent with the results of human patients, we also found that the miR-155-5p expression was dramatically upregulated (days 5-7) in the kidney tissues of UUO rets and highly correlated to the surgery course (Figure 1(c)). Furthermore, we visualized the expression pattern and the altered level of miR-155-5p in kidney sections using in situ hybridization. Our results revealed that the miR-155-5p was mainly expressed in renal tubular epithelial cells and the expression level gradually increased with the time course and α-SMA expression (Figures 1(d) and 1(e)). Besides, we observed that the miR-155-5p expression level was tightly correlated to the interstitial injury score of UUO rats (Figure 1(e)). Altogether, our observations indicated that miR-155-5p expression is increased in the fibrotic kidney tissues both from human patients and UUO animal models.
Figure 1.
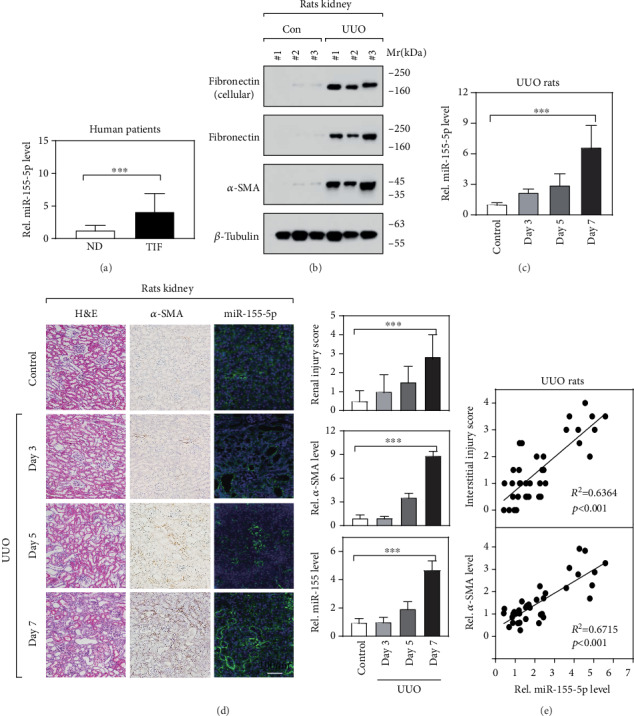
miR-155-5p is highly expressed in the fibrotic kidney tissues. (a) RT-qPCR showed the expression level of miR-155-5p in the kidney tissues from normal donors (ND, n = 6) and tubulointerstitial fibrosis (TIF) patients (n = 6). ∗∗∗p < 0.001 by t-test. (b) Immunoblot determined the protein levels of fibronectin and α-SMA in the kidney tissues. UUO: unilateral ureteral obstruction surgery; Mr: molecular weight; KDa: kilo Dalton. (c) RT-qPCR determined miR-155-5p expression in the kidney from UUO rats at different time points (after UUO surgery). n = 12, ∗∗∗p < 0.001 by t-test. (d) Histological analysis determined renal fibrosis injury (H&E staining), α-SMA (IHC), and miR-155-5p (ISH) expression in kidney sections. The optical density value of IHC and ISH was obtained using the ImageJ software, and all the values were normalized by the control group. n = 6, ∗∗∗p < 0.001 by one-way ANOVA. (e) The correlation between miR-155-5p expression and renal injury score/α-SMA expression in UUO rats. R: Pearson's correlation coefficient, ∗∗∗p < 0.001.
3.2. miR-155-5p Is Essential for Renal Fibrosis
To further explore the roles of miR-155-5p in the renal fibrogenesis, we constructed three miR-155-5p knockdown cell lines in NRK-49F using the lentivirus-mediated method. As shown in Figure 2(a), miR-155-5p expression was effectively suppressed in these NRK-49F cell lines, determined by RT-qPCR. Intriguingly, the reduction in the expression of fibronectin and α-SMA was observed in miR-155-5p knockdown cell lines (Figures 2(b) and 2(c)), suggesting that the knockdown of miR-155-5p alleviates renal fibrosis. Furthermore, we silenced miR-155-5p in UUO rat renal tissues using an adeno-associated virus type 2- (AAV2-) mediated shRNA-expressing system. The RT-qPCR results showed that more than 50% of miR-155-5p expression was inhibited when compared to the control shRNA-expressing tissues (Figure 2(d)). In miR-155-5p-silenced kidney tissues, a significant decrease in collagen I and α-SMA protein level was also observed (Figure 2(e)), indicating that the degree of renal fibrosis is alleviated. Consistently, immunostaining results also showed a dramatic downregulation of collagen I and α-SMA on protein level (Figures 2(f) and 2(g)). Collectively, our data indicate an essential function of miR-155-5p on renal fibrosis both in vitro and in vivo.
Figure 2.
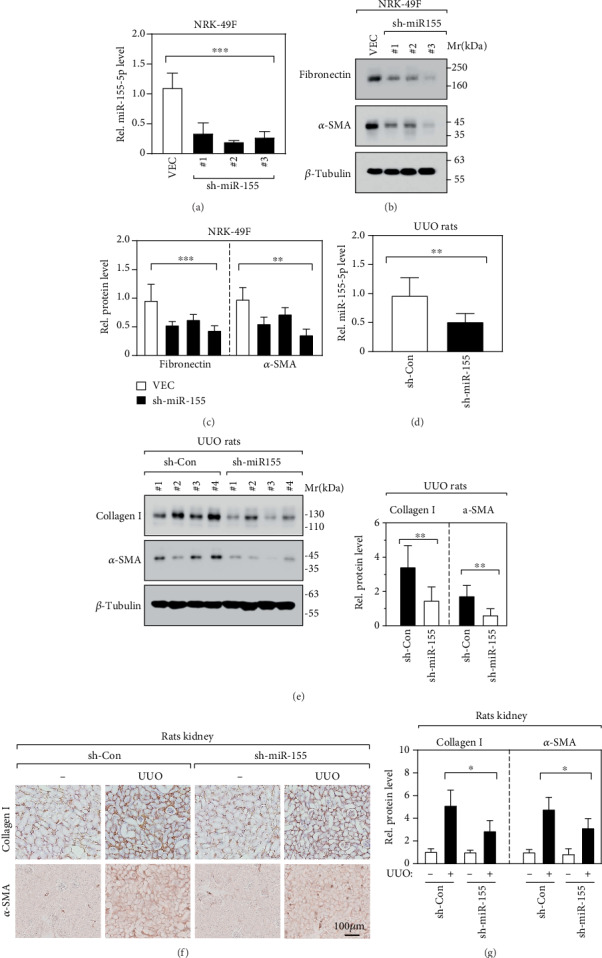
miR-155-5p is essential for renal fibrosis. (a) RT-qPCR determined miR-155-5p expression level in NRK-48F cell lines. VEC: empty vector. n = 6, ∗∗∗p < 0.001 by one-way ANOVA. (b, c) Fibronectin and α-SMA protein levels were determined by immunoblot in NRK-48F cell lines (b). The relative quantification of protein bands was carried out (c). n = 6, ∗∗p < 0.01 and ∗∗∗p < 0.001 by one-way ANOVA. (d) RT-qPCR revealed the miR-155-5p expression level in UUO rat renal tissues. n = 6, ∗∗p < 0.01 by t-test. (e) Immunoblot and its relative quantification represent the expression of collagen I and α-SMA in kidney tissues. n = 6, ∗∗p < 0.01 by t-test. (f, g) IHC (f) and its relative quantification (g) represent the expression of collagen I and α-SMA in kidney tissue sections. n = 6, ∗p < 0.05 by t-test.
3.3. SOCS1/6 Is the Direct Targets of miR-155-5p
To determine the exact mechanism by which miR-155-5p is implicated in renal fibrosis, we used the bioinformatics method to predict the potential binding sequences in target genes. We found that genes SOCS1/5/6 were potentially the direct targets of miR-155-5p, indicated by the complementary sequences between their mRNA and miR-155-5p (Figure 3(a)). Subsequently, we designed luciferase reporter constructs containing either the wild-type or mutated miR-155-5p binding sequences in SOCS1/5/6 to experimentally validate the bindings with miR-155-5p. Intriguingly, we observed that target 1 (in SOCS1), target 3 (in SOCS5), and target 4 (in SOCS6) were specifically bonded by miR-155-5p (Figure 3(b)). Furthermore, we found that ectopic miR-155-5p remarkably downregulated the mRNA level of SOCS1/6 expression but not SOCS5 in HK-2 and NRK-49 cells (Figures 3(c) and 3(d)), suggesting that only SOCS1/6 is a direct target of miR-155-5p. Meanwhile, the ectopic miR-155-5p also significantly reduced the protein level of SOCS1 and SOCS6 in HK-2 and NRK-49 cells (Figure 3(e)). Besides, we determined the expression of SOCS1 and SOCS6 in TIF patients and observed a significant decrease in SOCS1 and SOCS6 protein levels when compared to ND patients (Figure 3(f)). Altogether, miR-155-5p directly targets the mRNA of SOCS1/6 both in vitro and in vivo.
Figure 3.
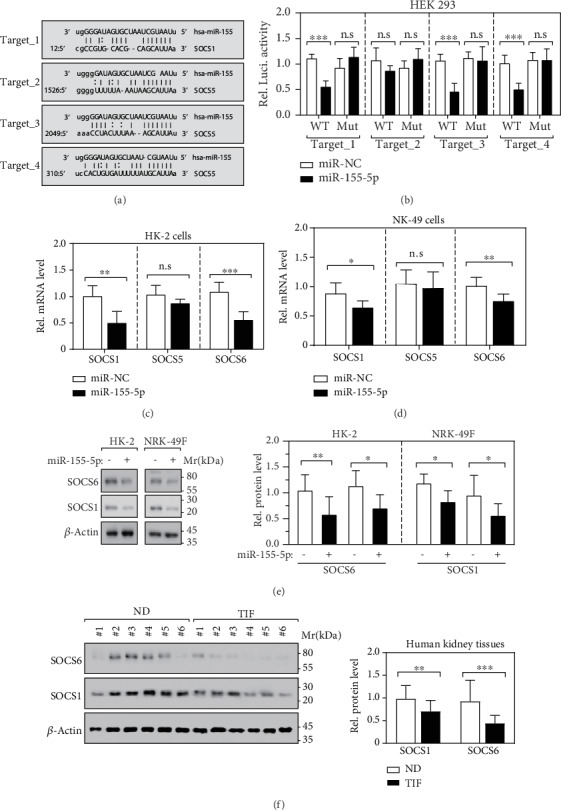
SOCS1/6 is the direct target of miR-155-5p. (a) SOCS1/5/6 are potential miR-155-5p targets predicted by in-house bioinformatics tools MicroCible and MicroTopTable. (b) Luciferase reporter assay determined the direct binding between SOCS1/5/6 and miR-155-5p, respectively. n = 6; n.s.: no significance, ∗∗∗p < 0.001 by t-test. (c) RT-qPCR determined the expression of SOCS1/5/6 in miR-155-5p overexpressed HK-2 cells. n = 6; n.s.: no significance, ∗∗p < 0.01 and ∗∗∗p < 0.001 by t-test. (d) RT-qPCR determined the expression of SOCS1/5/6 in miR-155-5p overexpressed NRK-49 cells. n = 6; n.s.: no significance, ∗p < 0.05 and ∗∗p < 0.01 by t-test. (e) Immunoblot and its relative quantification represent the expression of collagen I and α-SMA in HK-2 and NRK-49 cells. n = 3; ∗p < 0.05 and ∗∗p < 0.01 by t-test. (f) Immunoblot and its relative quantification represent the expression of SOCS1/6 in human kidney tissues. n = 12; ∗∗p < 0.01 and ∗∗∗p < 0.001 by t-test.
3.4. miR-155-5p Is Involved in Renal Fibrosis via Regulating the Phosphorylation of STAT3
Previously, extensive studies have demonstrated that SOCS proteins are involved in the inflammatory response in the progress of renal fibrosis by negatively regulating the JAK-STAT signal cascade [38–40]. Mechanistically, SOCS proteins suppress the JAK tyrosine kinase activity via their kinase inhibitory regions (KIR) and then inhibit STAT protein phosphorylation and activation [40]. In our experiments, we observed that miR-155-5p directly targets SOCS1/6 to regulate their protein level both in vitro and in vivo. Hence, we speculated that miR-155-5p may influence the phosphorylation level of STAT3 at Y705. Indeed, we observed dramatic elevation in the Y705 phosphorylation level of STAT3 in kidney tissues of UUO rats (Figures 4(a)–4(d)). More importantly, knockdown of miR-155-5p expression in the kidney significantly alleviated the increase of STAT3 phosphorylation induced by UUO surgery (Figures 4(c) and 4(d)). These observations support our speculation mentioned above. Meanwhile, we also found that the increased protein level of collagen I induced by UUO surgery was partially reversed by knockdown miR-155-5p (Figures 4(c) and 4(d)), implying that STAT3 may mediate renal fibrosis. Therefore, we knocked down STAT3 expression in NRK-49F cells to test whether it is required for miR-155-5p's biological functions in vitro. Our results revealed that the elevated expression of fibronectin and α-SMA induced by ectopic expression of miR-155-5p in NRK-49F cells was effectively eliminated by loss function of STAT3 (Figures 4(e) and 4(f)), suggesting that miR-155-5p is involved in renal fibrosis via regulating the phosphorylation of STAT3.
Figure 4.
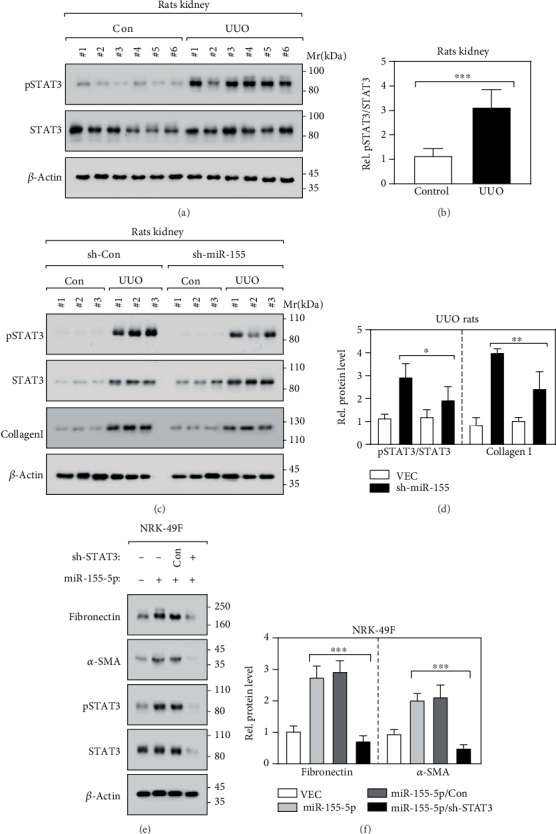
miR-155-5p is involved in renal fibrosis via regulating the phosphorylation of STAT3 A-B. Immunoblot (a) and its relative quantification (b) revealed the STAT3 phosphorylation at Y705 in kidney tissues. n = 12; ∗∗∗p < 0.001 by t-test. (c, d) Immunoblot (c) and its relative quantification (d) revealed the indicated protein levels in kidney tissues. n = 9; ∗p < 0.05 and ∗∗p < 0.01 by t-test. (e, f) Immunoblot (e) and its relative quantification (f) revealed the indicated protein levels in NRK-49 cells. n = 6; ∗∗∗p < 0.01 by t-test.
4. Discussion
The pathogenesis of renal fibrosis is a progressive process that ultimately leads to end-stage renal failure, which is regulated by various signaling including miRNAs [1–3]. Multiple miRNAs were observed functioning on renal fibrosis through regulating numerous cellular activities including epithelial-mesenchymal transition (EMT), fibroblast activation, inflammatory reaction, and apoptosis [12–15, 28–30, 41–43]. For instance, studies have demonstrated that TGF-β/Smad3 promotes renal fibrosis by inducing the expression of miR-21, miR-433, and miR-192, which in turn suppressed PTEN, Smad7, ZEB1/2, and AZIN1, respectively [12–14, 16, 30, 41, 44]. Besides, TGF-β/Smad3 is also revealed to suppress HDAC4 and E-Cadherin expression to accelerate renal fibrosis by miR-29 and miR-200, respectively [17, 43]. Furthermore, miR-192, miR-433, and miR-21 were reported playing a pathological role in renal fibrosis through a feed-forward loop to amplify TGF-β signaling [14]. As a typical multifunctional miRNA, miR-155-5p is reported widely expressed in various tissues and plays a crucial role in various physiological and pathological processes [21–23]. In the kidney, miR-155-5p was reported abnormally expressed in renal cancer, end-stage renal disease, and renal transplantation undergoing acute cellular rejection [24–30], suggesting the essential effects of miR-155-5p on renal pathology. Various studies implied the tumor-promoting function of miR-155-5p on renal cancer [22, 26, 27, 45]. It is reported that miR-155-5p silencing inhibits proliferation and migration by upregulating BACH1 in renal cancer cells [46]. Besides, miR-155-5p is revealed capable of increasing the proliferation and invasion of renal cancer cells by targeting E2F2 [26]. More importantly, it is suggested that miR-155-5p may implicate in regulating renal fibrosis. For instance, in 2015, Shenping Xie and colleagues found that in HK-2 cells, HIF-1α-induced miR-155-5p promoted the fibrosis of proximal tubule cells by regulating both TGF-β1 and the process of EMT under hypoxia [35]. In the present study, we firstly explored the expression level and functions of miR-155-5p in renal fibrosis patients and unilateral ureteral obstruction (UUO) rat models. Our work demonstrated the profibrotic function of miR-155-5p on renal fibrosis in vitro and UUO rat models. We observed that miR-155-5p was highly expressed in the kidney tissues from renal disease patients and UUO rats. Further study revealed that knockdown miR-155-5p can significantly block renal fibrogenesis in vitro and in vivo. Our work again highlights the significance of miR-155-5p on renal fibrosis.
SOCS1 and SOCS6 are members of SOCS (suppressor of cytokine signaling) family proteins that includes 8 members, SOCS1-7 and CIS, which negatively regulate JAK/STAT pathway activation by suppressing JAK and STAT phosphorylation and affect multiple cellular activations that respond to LIF, TPO, IL-6, etc. [40]. The JAK/STAT pathway was determined as crucial mediators in renal fibrosis, via regulating EMT, apoptosis, and inflammatory response [47]. STAT3 activation was observed in rat tubular epithelial cells and myofibroblasts after UUO treatment [8], and STAT3 inhibitor S3I-201 significantly attenuates renal interstitial fibroblast activation and interstitial fibrosis [9], suggested as the profibrotic role of JAK/STAT in renal injury and fibrosis. But, early studies did not clarify the that the mechanism underlying JAK/STAT activation implicates in kidney dysfunction. Consistent with other studies, we observed a significant increase in the STAT3 activation in renal fibrosis, and miR-155-5p is capable of activating STAT3 both in vitro and in vivo. Our work showed that UUO treatment dramatically increases STAT3 phosphorylation, which was remarkably restored by miR-155-5p knocking down. Furthermore, renal fibrosis induced by miR-155-5p overexpression is blocked by STAT3 deficiency. Collectively, these data demonstrated that miR-155-5p regulates STAT3 activation and STAT3 mediated the profibrotic function of miR-155-5p on renal injury. In consideration of miR-155-5p directly targeting SOCS1 and SOCS6, there are logical reasons to assume that the molecular mechanism by which miR-155-5p regulates STAT3 phosphorylation is via targeting SOCS1 and SOCS6.
In conclusion, our work uncovered a profibrotic effect of miR-155-5p on renal injury and illustrated a potential mechanism that miR-155-5p regulating STAT3 activation by directly targeting SOCS1 and SOCS6. Our findings may provide a deep insight into renal fibrosis and a new therapeutic target for clinical prevention and treatment of renal fibrosis.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Wanfen Zhang and Xiaoping Li contributed equally to this work.
Supplementary Materials
The statistic test results of Figure 3, indicating the significance of each panel.
References
- 1.Boor P., Ostendorf T., Floege J. Renal fibrosis: novel insights into mechanisms and therapeutic targets. Nature Reviews Nephrology. 2010;6:643–656. doi: 10.1038/nrneph.2010.120. [DOI] [PubMed] [Google Scholar]
- 2.Eddy A. A. Molecular basis of renal fibrosis. Pediatric Nephrology. 2000;15(3-4):290–301. doi: 10.1007/s004670000461. [DOI] [PubMed] [Google Scholar]
- 3.Min H. C. Renal fibrosis. Korean Journal of Pediatrics. 2010;53(7):735–740. doi: 10.3345/kjp.2010.53.7.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanwar Y. S. TGF-β and renal fibrosis. American Journal of Pathology. 2012;181(4):1147–1150. doi: 10.1016/j.ajpath.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.New J. P., Middleton R. J., Klebe B., et al. Assessing the prevalence, monitoring and management of chronic kidney disease in patients with diabetes compared with those without diabetes in general practice. Diabetic Medicine. 2007;24(4):364–369. doi: 10.1111/j.1464-5491.2007.02075.x. [DOI] [PubMed] [Google Scholar]
- 6.Chung A. C. K., Lan H. Y. Molecular mechanisms of TGF-β signaling in renal fibrosis. Current Pathobiology Reports. 2013;1(4):291–299. doi: 10.1007/s40139-013-0027-6. [DOI] [Google Scholar]
- 7.Zhang Y., Meng X. M., Huang X. R., Lan H. Y. The preventive and therapeutic implication for renal fibrosis by targetting TGF-β/Smad 3 signaling. Clinical Science. 2018;132(13):1403–1415. doi: 10.1042/CS20180243. [DOI] [PubMed] [Google Scholar]
- 8.Kuratsune M., Masaki T., Hirai T., et al. Signal transducer and activator of transcription 3 involvement in the development of renal interstitial fibrosis after unilateral ureteral obstruction. Nephrology. 2007;12(6):565–571. doi: 10.1111/j.1440-1797.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 9.Pang M., Ma L., Gong R., et al. A novel STAT3 inhibitor, S3I-201, attenuates renal interstitial fibroblast activation and interstitial fibrosis in obstructive nephropathy. Kidney International. 2010;78(3):257–268. doi: 10.1038/ki.2010.154. [DOI] [PubMed] [Google Scholar]
- 10.Pratt A. J., Macrae I. J. The RNA-induced silencing complex: a versatile gene-silencing machine. Journal of Biological Chemistry. 2009;284(27):17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paroo Z., Liu Q., Wang X. Biochemical mechanisms of the RNA-induced silencing complex. Cell Research. 2007;17(3):187–194. doi: 10.1038/sj.cr.7310148. [DOI] [PubMed] [Google Scholar]
- 12.Loboda A., Sobczak M., Jozkowicz A., Dulak J. TGF-β1/Smads and miR-21 in Renal Fibrosis and Inflammation. Mediators of Inflammation. 2016;2016(1):12. doi: 10.1155/2016/8319283.8319283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chau B. N., Xin C., Hartner J., et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Science Translational Medicine. 2012;4(121, article 121ra118) doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung A. C. K., Huang X. R., Meng X., Lan H. Y. miR-192 mediates TGF-β/Smad3-Driven renal fibrosis. Journal of the American Society of Nephrology. 2010;21(8):1317–1325. doi: 10.1681/ASN.2010020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin W., Chung A. C. K., Huang X. R., et al. TGF-β/Smad 3 signaling promotes renal fibrosis by inhibiting mi R-29. Journal of the American Society of Nephrology. 2011;22(8):1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong X., Chung A. C. K., Chen H. Y., et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56(3):663–674. doi: 10.1007/s00125-012-2804-x. [DOI] [PubMed] [Google Scholar]
- 17.Sene L. B., Rizzi V. H. G., Gontijo J. A. R., Boer P. A. Gestational low-protein intake enhances whole-kidney miR-192 and miR-200 family expression and epithelial-to-mesenchymal transition in rat adult male offspring. Journal of Experimental Biology. 2018;221(10, article jeb171694) doi: 10.1242/jeb.171694. [DOI] [PubMed] [Google Scholar]
- 18.Fan Y., Chen H., Huang Z., Zheng H., Zhou J. Emerging role of miRNAs in renal fibrosis. RNA Biology. 2020;17(1):1–12. doi: 10.1080/15476286.2019.1667215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ademokun A., Turner M. Regulation of B-cell differentiation by micro RNAs and RNA-binding proteins. Biochem Soc Trans. 2008;36(6):1191–1193. doi: 10.1042/BST0361191. [DOI] [PubMed] [Google Scholar]
- 20.Yin Q., Wang X., Mcbride J., Fewell C., Flemington E. B-cell receptor activation induces BIC/mi R-155 expression through a conserved AP-1 element. Journal of Biological Chemistry. 2008;283(5):2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mashima R. Physiological roles of mi R-155. Immunology. 2015;145(3):323–333. doi: 10.1111/imm.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faraoni I., Antonetti F. R., Cardone J., Bonmassar E. miR-155 gene: A typical multifunctional microRNA. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2009;1792(6):497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Vigorito E., Kohlhaas S., Lu D., Leyland R. miR-155: an ancient regulator of the immune system. Immunological Reviews. 2013;253(1):146–157. doi: 10.1111/imr.12057. [DOI] [PubMed] [Google Scholar]
- 24.Chen S., Shan J., Niu W., et al. Micro RNA-155 inhibitor as a potential therapeutic strategy for the treatment of acute kidney injury (AKI): a nanomedicine perspective. Rsc Advances. 2018;8(29):15890–15896. doi: 10.1039/c7ra13440a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Zheng Z. J., Jia Y. J., Yang Y. L., Xue Y. M. Role of p53/miR-155-5p/sirt1 loop in renal tubular injury of diabetic kidney disease. Journal of Translational Medicine. 2018;16(1):p. 146. doi: 10.1186/s12967-018-1486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y., Ma X., Yao Y., et al. miR-155 regulates the proliferation and invasion of clear cell renal cell carcinoma cells by targeting E2F2. Oncotarget. 2016;7(15):20324–20337. doi: 10.18632/oncotarget.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji H., Tian D., Zhang B., Zhang Y., Yan D., Wu S. Overexpression of miR-155 in clear-cell renal cell carcinoma and its oncogenic effect through targeting FOXO3a. Experimental and Therapeutic Medicine. 2017;13(5):2286–2292. doi: 10.3892/etm.2017.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G., Kwan B. C. H., Lai F. M. M., Chow K. M., Li P. K. T., Szeto C. C. Elevated levels of miR-146a and miR-155 in kidney biopsy and urine from patients with IgA nephropathy. Disease Markers. 2011;30(4):171–179. doi: 10.1155/2011/304852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Peng W., Shen X., Huang Y., Ouyang X., Dai Y. Circulating levels of inflammation-associated miR-155 and endothelial-enriched miR-126 in patients with end-stage renal disease. Brazilian Journal of Medical and Biological Research. 2012;45(12):1308–1314. doi: 10.1590/S0100-879X2012007500165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zununi Vahed S., Poursadegh Zonouzi A., Ghanbarian H., et al. Differential expression of circulating miR-21, miR-142-3p and miR-155 in renal transplant recipients with impaired graft function. International Urology and Nephrology. 2017;49(9):1681–1689. doi: 10.1007/s11255-017-1602-2. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharyya S., Kumar P., Tsuchiya M., Bhattacharyya A., Biswas R. Regulation of miR-155 biogenesis in cystic fibrosis lung epithelial cells: antagonistic role of two mRNA-destabilizing proteins, KSRP and TTP. Biochemical and Biophysical Research Communications. 2013;433(4):484–488. doi: 10.1016/j.bbrc.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 32.Christmann R. B., Wooten A., Sampaio-Barros P., et al. miR-155 in the progression of lung fibrosis in systemic sclerosis. Arthritis Research & Therapy. 2016;18(1):p. 155. doi: 10.1186/s13075-016-1054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasoo M. Regulation of liver X receptors by microRNA-155 in pulmonary fibrosis. University of Glasgow; 2013. [Google Scholar]
- 34.Tsuchiya M., Kalurupalle S., Kumar P., et al. RPTOR, a novel target of miR-155, elicits a fibrotic phenotype of cystic fibrosis lung epithelium by upregulating CTGF. RNA Biology. 2016;13(9):837–847. doi: 10.1080/15476286.2016.1197484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie S., Chen H., Li F., Wang S., Guo J. Hypoxia-induced microRNA-155 promotes fibrosis in proximal tubule cells. Molecular Medicine Reports. 2015;11(6):p. 4555. doi: 10.3892/mmr.2015.3327. [DOI] [PubMed] [Google Scholar]
- 36.Chevalier R. L., Forbes M. S., Thornhill B. A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney International. 2009;75(11):1145–1152. doi: 10.1038/ki.2009.86. [DOI] [PubMed] [Google Scholar]
- 37.Li A., Zhang X., Shu M., et al. Arctigenin suppresses renal interstitial fibrosis in a rat model of obstructive nephropathy. Phytomedicine. 2017;30:28–41. doi: 10.1016/j.phymed.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Chikuma S., Kanamori M., Mise-Omata S., Yoshimura A. Suppressors of cytokine signaling: potential immune checkpoint molecules for cancer immunotherapy. Cancer Sci. 2017;108(4):574–580. doi: 10.1111/cas.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y., Xu W. D., Peng H., Pan H. F., Ye D. Q. SOCS signaling in autoimmune diseases: molecular mechanisms and therapeutic implications. Eur J Immunol. 2014;44(5):1265–1275. doi: 10.1002/eji.201344369. [DOI] [PubMed] [Google Scholar]
- 40.Croker B. A., Kiu H., Nicholson S. E. SOCS regulation of the JAK/STAT signalling pathway. Seminars in Cell & Developmental Biology. 2008;19(4):414–422. doi: 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glowacki F., Savary G., Gnemmi V., et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS One. 2013;8(2, article e58014) doi: 10.1371/journal.pone.0058014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho J., Kreidberg J. A. MicroRNAs in renal development. Pediatric Nephrology. 2013;28(2):219–225. doi: 10.1007/s00467-012-2204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B., Komers R., Carew R., et al. Suppression of microRNA-29 expression by TGF-β1 promotes collagen expression and renal fibrosis. Journal of the American Society of Nephrology. 2012;23(2):252–265. doi: 10.1681/asn.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li R., Chung A. C. K., Dong Y., Yang W., Zhong X., Lan H. Y. The microRNA miR-433 promotes renal fibrosis by amplifying the TGF-β/Smad3-Azin1 pathway. Kidney International. 2013;84(6):1129–1144. doi: 10.1038/ki.2013.272. [DOI] [PubMed] [Google Scholar]
- 45.Pang W., Su J., Wang Y., et al. Pancreatic cancer-secreted miR-155 implicates in the conversion from normal fibroblasts to cancer-associated fibroblasts. Cancer Sci. 2015;106(10):1362–1369. doi: 10.1111/cas.12747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S., Chen T., Zhong Z., Wang Y., Li Y., Zhao X. MicroRNA-155 silencing inhibits proliferation and migration and induces apoptosis by upregulating BACH1 in renal cancer cells. Molecular Medicine Reports. 2012;5(4):949–954. doi: 10.3892/mmr.2012.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chuang P. Y., He J. C. JAK/STAT signaling in renal diseases. Kidney International. 2010;78(3):231–234. doi: 10.1038/ki.2010.158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The statistic test results of Figure 3, indicating the significance of each panel.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


