Abstract
Objective
To determine the susceptibility of the endometrium to infection by—and thereby potential damage from—SARS-CoV-2.
Design
Analysis of SARS-Cov-2 infection-related gene expression from endometrial transcriptomic data sets.
Setting
Infertility research department affiliated with a public hospital.
Patient(s)
Gene expression data from five studies in 112 patients with normal endometrium collected throughout the menstrual cycle.
Intervention(s)
None.
Main Outcome Measure(s)
Gene expression and correlation between viral infectivity genes and age throughout the menstrual cycle.
Result(s)
Gene expression was high for TMPRSS4, CTSL, CTSB, FURIN, MX1, and BSG; medium for TMPRSS2; and low for ACE2. ACE2, TMPRSS4, CTSB, CTSL, and MX1 expression increased toward the window of implantation. TMPRSS4 expression was positively correlated with ACE2, CTSB, CTSL, MX1, and FURIN during several cycle phases; TMPRSS2 was not statistically significantly altered across the cycle. ACE2, TMPRSS4, CTSB, CTSL, BSG, and MX1 expression increased with age, especially in early phases of the cycle.
Conclusion(s)
Endometrial tissue is likely safe from SARS-CoV-2 cell entry based on ACE2 and TMPRSS2 expression, but susceptibility increases with age. Further, TMPRSS4, along with BSG-mediated viral entry into cells, could imply a susceptible environment for SARS-CoV-2 entry via different mechanisms. Additional studies are warranted to determine the true risk of endometrial infection by SARS-CoV-2 and implications for fertility treatments.
Key Words: ACE2, coronavirus, COVID-19, endometrial transcriptomics, SARS-CoV-2
Abstract
Evaluación del riesgo de infección por SARS-CoV-2 en el endometrio: expresión génica relacionada con la infección viral a lo largo del ciclo menstrual.
Objetivo
Determinar la susceptibilidad del endometrio a la infección -y el consiguiente daño potencial ocasionado- por el SARS-CoV2.
Diseño
Análisis del conjunto de datos de transcriptómica endometrial de expresión génica relacionados con la infección por el SARS-CoV2.
Entorno
Departamento de investigación en infertilidad afiliado a un hospital público.
Paciente(s)
Datos de expresión génica de cinco estudios en 112 pacientes con endometrio normal, recogidas a lo largo del ciclo menstrual.
Intervención(es)
Ninguna.
Medida del resultado principal
Expresión génica y correlación entre los genes de infectividad viral y la edad a lo largo del ciclo menstrual.
Resultado(s)
La expresión génica fue alta para TMPRSS4, CTSL, CTSB, FURIN, MX1 y BSG; media para rTMPRSS2 y baja para ACE2. La expresión de ACE2, TMPRSS4, CTSB, CTS y dMX1 se incrementó hacia la ventana de implantación. La expresión de TMPRSS4 se correlacionó de manera positiva con ACE2, CTSB, CTSL, MX1 y FURIN durante varias fases del ciclo; TMPRSS2 no estaba alterado de manera estadísticamente significativa a lo largo del ciclo. La expresión de ACE2, TMPRSS4, CTSB, CTSL, BSG y MX1 se incrementó con la edad, especialmente en las fases tempranas del ciclo.
Conclusión(es)
El tejido endometrial está probablemente a salvo de la entrada celular por el SARS-CoV2 basándonos en la expresión de ACE2 y TMPRSS2, pero la susceptibilidad se incrementa con la edad. Además, la entrada del virus al interior de las células mediado por TMPRSS4 junto con BSG podría implicar una susceptibilidad ambiental para la entrada del SARS-CoV2 a través de diferentes mecanismos. Se recomiendan más estudios para determinar el verdadero riesgo de infección endometrial por SARS-CoV2 y sus implicaciones para los tratamientos de fertilidad.
Discuss: You can discuss this article with its authors and other readers at https://www.fertstertdialog.com/users/16110-fertility-and-sterility/posts/30632
The rapid international spread of novel coronavirus disease 2019 (COVID-19) (1) has resulted in 5,103,006 confirmed cases and 333,401deaths worldwide as of May 23, 2020 (2). COVID-19 is caused by SARS-coronavirus 2 (SARS-CoV-2) infection in the lower respiratory tract (3), but the mechanisms underlying infection remain poorly understood (4). The rapid spread of SARS-CoV-2, genetically closely related to severe acute respiratory syndrome coronavirus (SARS-CoV-1) (5), has resulted in a global health emergency with long-term consequences on economics markets, nutritional habits, and mental and physical well-being (5, 6, 7, 8, 9).
As information is still emerging about the health consequences of COVID-19, assisted reproductive treatments (ARTs) have been delayed due to fear of the unknown impact of SARS-Cov-2 on fertility (10, 11). Fertility is compromised by age, and the longer treatments are delayed, the less likely that successful outcomes will be achieved (12). Furthermore, effects of SARS-CoV-2 infection increase in severity with host age (13, 14, 15), meaning that women of more advanced reproductive age undergoing ART could be at higher risk of infection. It is therefore critical to determine exactly how the virus affects reproductive physiology. Although the studies to date have been discordant, vertical transmission during pregnancy seems to occur infrequently, ranging from 0 to 11% (11, 16, 17, 18). To date there is no information about how infection affects early implantation and development.
The ability of SARS-CoV-2 to damage tissue is determined by its capacity to enter and infect cells in that tissue (4). The SARS-CoV-2 entry point on the cell is angiotensin-converting enzyme 2 (ACE2) (19, 20), which plays a key role in the renin-angiotensin system, cleaving angiotensin II to angiotensin 1–7. The system is disrupted after SARS-CoV-2 gains cell entry and down-regulates the expression of ACE2, leading to up-regulation of the proinflammatory response by angiotensin II (21, 22, 23). ACE2 exhibits moderately increased expression with age (24), which could explain disease severity in older people. Another path of entry, using Basigin (BSG) as receptor instead of ACE2, has been proposed (25).
To enter the cell, SARS-CoV-2 uses its spike S protein to bind to ACE2, which leads to fusion with the cell membrane and endocytosis (4, 20, 26). TMPRSS2, a transmembrane protease, cleaves the S protein (27). Cleavage is necessary for the virus to bind to ACE2 and spread through the infected host (19). Other proteases are under investigation as possible implications in SARS-CoV-2 infectivity related to S protein cleaving. TMPRSS4 increased virus infectivity on its own, at least in gut epithelial cells (28), while cathepsins B and L (CTSB and CTSL, respectively) had residual cleaving activity of viral S protein in TMPRSS2- cells (19). FURIN, another protease predicted to cleave S protein, presents alongside ACE2 in epithelial layers of several oral mucosal tissues (29, 30). MX dynamin-like GTPase 1 (MX1) regulates neutrophil infiltration, favoring infection through protein S modification by neutrophil elastase (31).
The clinical presentation of COVID-19 ranges from mild respiratory symptoms to severe progressive pneumonia, gastrointestinal symptoms, fecal shedding, multiorgan failure, and even death (32, 33), but few studies have focused on the virus’s effect on fertility and damage to reproductive tissues, or on concerns regarding the use of reproductive treatments. Leydig and Sertoli cells in the testis (34, 35, 36, 37), oocytes (38), and ovarian tissue (37) are likely to experience damage due to their medium-high expression of the ACE2 receptor. The endometrium is crucial for human reproduction and embryo implantation, but studies of the effect of SARS-CoV-2 infection on menstrual cycle progression have not been performed. Delineating the virus’s impact on the tissue is important for determining risk to ART, given that a healthy endometrium is needed for embryo implantation and growth.
The endometrium is a complex tissue subjected to a cycle of cell death and renewal approximately every 28 days (39). Numerous transcriptomic studies have sought to understand gene expression changes throughout the menstrual cycle (40), and most of these data sets are available in public repositories, such as the Gene Expression Omnibus (GEO) database (41). According to the Human Protein Atlas (HPA) (42), ACE transcript is in low abundance in endometrium and not present as protein. According to HPA expression levels, TMPRSS4 and FURIN RNA expression is low, and protein levels are medium, whereas CTSB, MX1, and BSG have medium RNA expression and high protein expression (43). However, there is little information on how the virus could affect endometrial receptivity and embryo implantation.
We analyzed the impact of SARS-CoV-2 infection on the endometrium by measuring endometrial ACE2, TMPRSS2, TMPRSS4, CTSB, CTSL, FURIN, MX1, and BSG gene expression. Transcriptomic data sets available across the phases of endometrial progression were used to evaluate molecularly the risk of SARS-CoV-2 infection during the COVID-19 pandemic.
Materials and methods
Search and selection of SARS-CoV-2 infectivity-related proteins
A thorough literature search identified proteins related to the SARS-CoV-2 disease-causing mechanism, including genes related to cell entry and genes with implications in health and fertility, among others. The search for relevant genes associated with viral infectivity was made chronologically up to May 10, 2020. The keywords searched in PubMed (44) included all possible combinations between “SARS-CoV-2,” “COVID-19,” “coronavirus,” “cell entry mechanisms,” “long-term implications,” and “fertility.”
Search and selection of endometrial transcriptomic data sets
Public transcriptomic data sets were used to analyze expression of SARS-CoV-2 infectivity-related genes throughout the menstrual cycle. Endometrial transcriptomic experiments for control patients (without any known endometrial pathology) were systematically searched in the GEO database (41) with no restrictions on publication date or language and according to preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines (45). Keywords were: uter∗ OR endometr∗, filtered by “homo sapiens.” Experiments were selected if
-
•
RNA was extracted directly from human endometrial biopsies.
-
•
Endometrial biopsies were collected at different times during the menstrual cycle.
-
•
Cycle phase at the time of biopsy was available for all samples.
-
•
Endometrial gene expression was evaluated by microarray or RNA sequencing using Affymetrix, Illumina, or Agilent platforms.
-
•
Raw gene expression data were freely available to download from GEO.
Preprocessing and integrative analysis
Transcriptomic raw data were downloaded from the GEO database and processed according to the standards of the technology used (microarrays or RNA-seq) using limma R-package (version 3.34.9) (46). Expression data from each experiment were annotated with biomaRt (version 2.30.0) (47), log transformed, and quantile normalized using limma (46). Principal components analysis (PCA) was done using the prcomp() function, and the scores were displayed using the ggplot2 R-package (48) to look for possible batch effects and were corrected if using linear models. Outliers were deleted from posterior analysis if there was a distinct transcriptional behavior according to PCA.
To integrate the included endometrial transcriptomic experiments, several steps were followed as recommended by Tajti et al. (49): after being independently normalized, selected studies were joined in a unique data set, and batch effects were corrected using linear models (limma R package) (43). A relative expression value of low, medium, and high expression were established. The thresholds respectively correspond to 1% to 10%, 11% to 50%, and 51% to 100% categories of gene expression values of the entire integrated data set.
Pairwise differential expression analysis between experiments was performed using limma to identify genes with expression differences. P values were corrected using false discovery rates (FDR) (50), and genes differentially expressed between experiments were removed from the analysis (FDR <.05).
Differential gene expression throughout the menstrual cycle
An analysis of variance (ANOVA) was performed for each selected gene related to SARS-CoV-2 infectivity to assess which genes showed statistically significant differences between endometrial phases. Analysis of variance was followed by a pairwise t-test to determine statistically significant differences between phases. Fold changes in gene expression from phase to phase were also calculated. The mean expression and confidence intervals for each gene in each phase were represented using ggplot2 R-package (version 3.0.0) (45).
Coexpression of infectivity genes and the effect of age
Pearson’s correlation (51) was used to assess coexpression between each pair of selected proteins related to SARS-CoV-2 infectivity and to analyze the effect of age (from 23 to 50 years) on the expression of each protein. All programming and statistical tests (t-test, Pearson’s correlation, and analysis of variance) were implemented in the R environment (version 3.4.4, 2018-03-15) (52).
Results
Menstrual cycle clock in integrated endometrial data sets
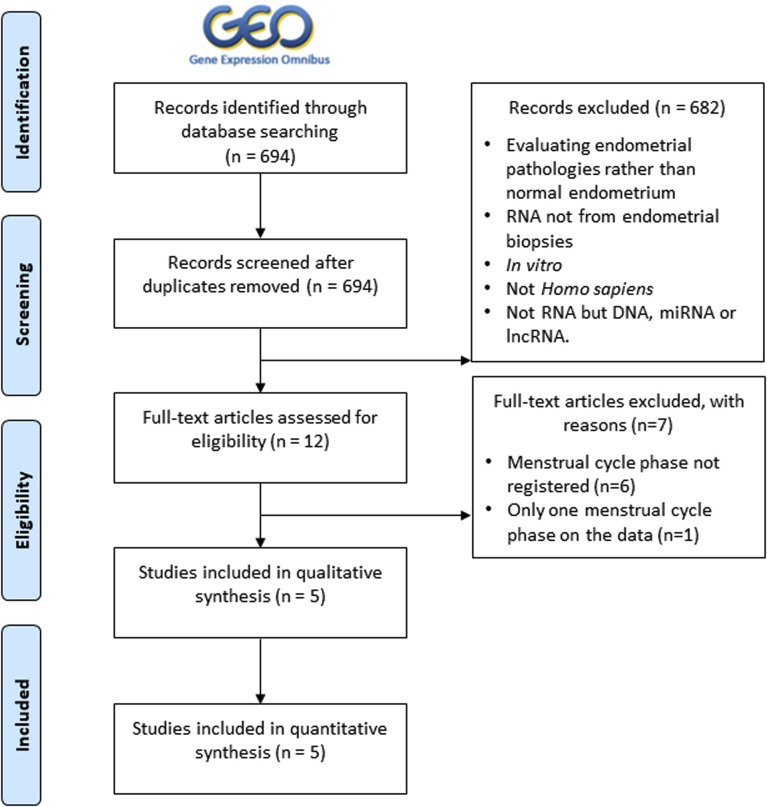
From a total of 694 studies retrieved from GEO, our search identified five unique studies that evaluated endometrial gene expression in women with normal endometrium, comprising 112 samples (Table 1 ; see Supplemental Fig. 1, available online, for detailed filtering steps). All studies were analyzed separately for correction of possible batch effects, and three samples were excluded from analysis (details in Supplemental Fig. 2, available online). A unique combined data set with a population of 109 patients was obtained, comprising 29 samples in the proliferative phase, 29 in the early secretory phase, 43 in the medium secretory phase, and eight in the late secretory phase (Table 1).
Table 1.
Characterization of endometrial transcriptomic data sets with GEO data sets identifier, experiment name given for this study, cycle type, method of cycle phase dating, population from which samples were collected, age, transcriptomic platform used to measure gene expression, number of genes of each data set, number of samples in each data set, number of samples per cycle phase, and publication in which data were initially employed.
| GEO ID | Experiment name | Cycle type | Cycle phase dating method | Population | Age | Platform | No. genes | No. samples | No. samples per cycle phase | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| GSE4888 | Talbi 2006 | Normo-ovulatory; regular (24–35 d); 3 mo since last hormone treatment | Noyes et al. (39) reviewed by four pathologists | Caucasian (n = 17); black (n = 6); Asian (n = 1); other (n = 2) | 23–50 | hgu133plus2 Affymetrix | 19,361 | 27 | PF (n = 6); ESE (n = 4); MSE (n = 9); LSE (n = 8) | (53) |
| GSE29981 | Bradley 2010 | Regular | Days from LH peak: PF (LH-14–LH-1), ESE (LH+1–LH+4), MSE (LH+6–LH+7) | Collected in Belgium | 20–39 | hgu133plus2 Affymetrix | 19,361 | 19 | PF (n = 10); ESE (n = 6); MSE (n = 3) | — |
| GSE98386 | Altmäe 2017 | Natural cycle | Days from LH peak. We classified it in ESE = LH+2; MSE = LH+8 | Collected in Estonia | — | Illumina HiSeq 2500 | 16,426 | 38 | ESE (n = 19); MSE (n = 19) | (54) |
| GSE86491 | Sigurgeirsson 2017 | Regular; 3 mo since last hormone treatment | Urinary LH ovulation predictor kit for MSE-LSE, days after the start of the subsequent menstruation for PF. Both confirmed by a gynecologic pathologist through histopathologic examination. | Collected in Iceland | 24–30 | Illumina HiSeq 2500 | 15,939 | 14 | PF (n = 7); MSE (n = 7) | (55) |
| GSE119209 | Kelleher 2017 | — | — | Collected in U.S. | — | Illumina HiSeq 2500 | 17,934 | 11 | PF (n = 6); MSE (n = 5) | — |
Note: Last row indicates the total number of samples accounted and genes in common between all data sets for all menstrual cycle phases (53, 54, 55). ESE = early secretory; GEO = Gene Expression Omnibus; LH = luteinizing hormone; LSE = late secretory; MSE = medium secretory; PF = proliferative phase.
The 109 samples were grouped depending on the experiment rather than by the menstrual cycle phase (Fig. 1 A, left). To make data sets comparable after the integration of all samples, this batch effect was removed (Fig. 1A, right). The resulting gene expression samples showed a behavior based on menstrual cycle phases rather than study nature (Fig. 1B).
Figure 1.
Endometrial data set integration and menstrual cycle clock. (A) Data set experiment effect and correction. Each point of the principal component analysis (PCA) plots represents endometrial gene expression of one sample and is colored by the endometrial transcriptomic data sets to which it belongs. Principal component 1 (PC1) and principal component 2 (PC2) explain the percentage of variability due to these components for each PCA. Integration of the transcriptomic studies showed a clear batch effect by the nature of each experiment (left PCA plot). After correction (right PCA plot), all genes in common between data sets were retained, amounting to a total of 13,437 genes, as no differentially expressed genes were detected between experiments. (B) Menstrual cycle effect. The samples of the integrated data sets are colored by the menstrual cycle phase to show how they are grouped by phases of the menstrual cycle. ESE = early secretory endometrium; LSE = late secretory endometrium; MSE = midsecretory endometrium; PF = proliferative phase.
Viral infectivity genes show a different expression landscape across the menstrual cycle
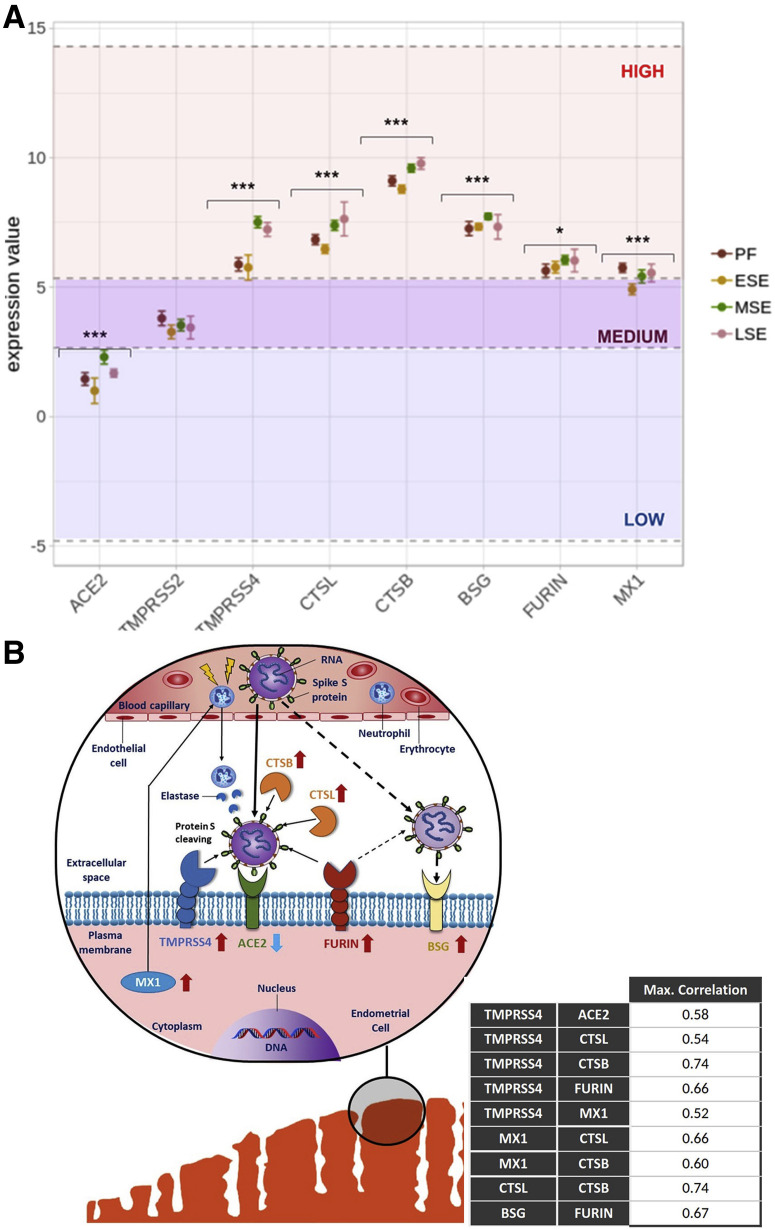
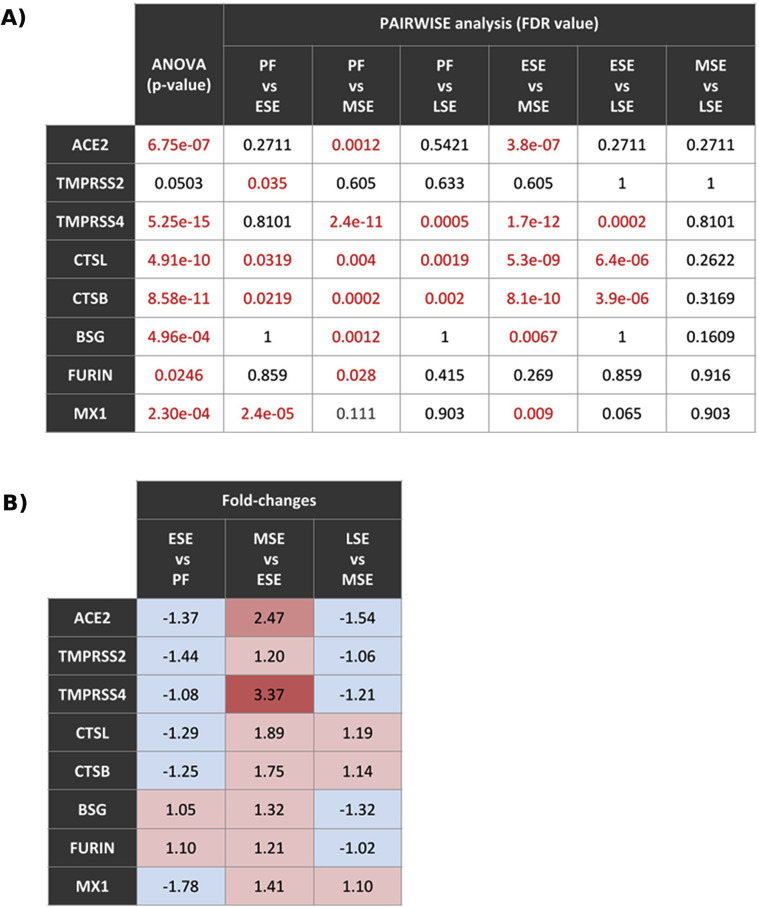
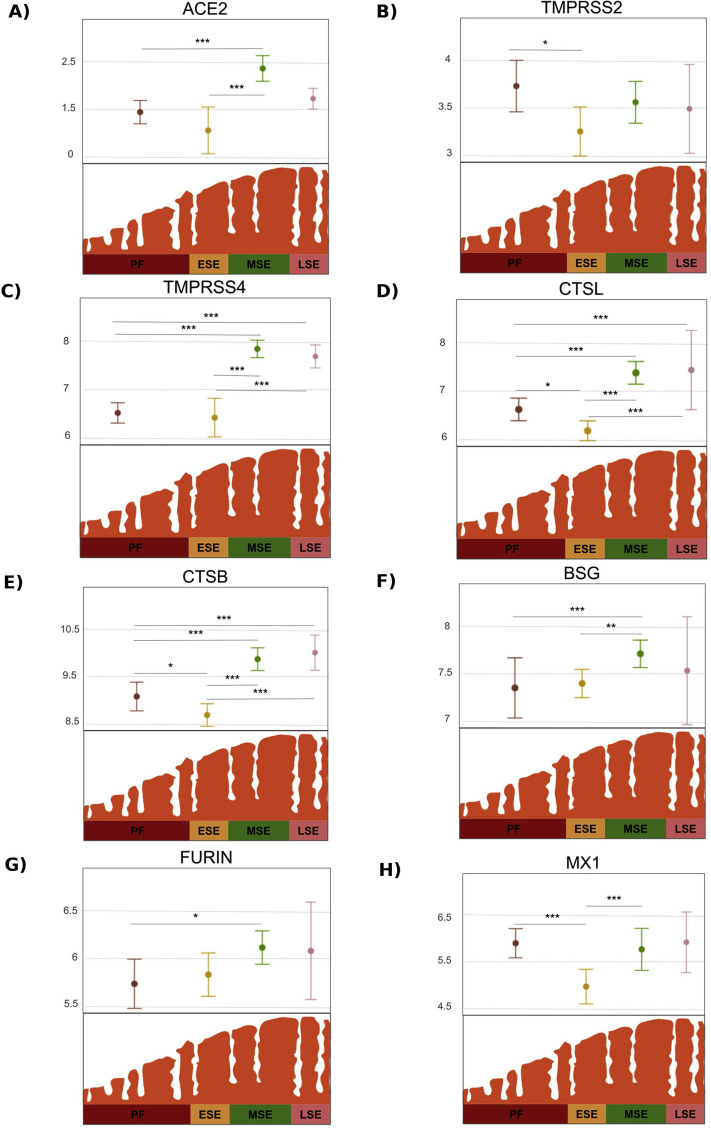
The gene expression landscapes for each viral gene across the cycle are shown in Figure 2 B (56). TMPRSS4, CTSL, CTSB, FURIN, MX1, and BSG were highly expressed throughout the cycle; TMPRSS2 was moderately expressed, and ACE2 was low (Fig. 2A). Gene expression of viral proteins depended on menstrual cycle phase. Calculated P values and adjusted P values are presented in Supplemental Fig. 3A (available online). All genes except TMPRSS2 (P=.053) had statistically significant changes in expression across the menstrual cycle (Fig. 2A). Specifically, the genes most affected by menstrual cycle progression (P<.0001) were ACE2, which increased expression from early secretory to midsecretory; TMPRSS4, whose expression increased from proliferative to midsecretory and from early secretory to midsecretory (P<.0001); CTSL and CTSB, which increased from early secretory to midsecretory (P<.0001); TMPRSS2 with decreased expression from proliferative to early secretory (P<.05); and BSG and FURIN, which increased from proliferative to midsecretory (P<.01) (Fig. 2A). All genes, including TMPRSS2, showed increased expression from early secretory to midsecretory, as indicated in Supplemental Figure 3B. Detailed expression changes across the menstrual cycle are provided for each gene in Supplemental Figure 4 (available online).
Figure 2.
Gene expression of viral infection-related genes throughout the menstrual cycle. (A) Landscape of expression changes. Genes were located depending on their relative expression against the whole set. Low, medium, and high expression thresholds correspond to 1% to 10%, 11% to 50%, and 51% to 100% categories of gene expression values of the entire integrated data set, respectively. Analysis of variance results for overall change of expression during cycle are shown for each gene. ∗P<.05; ∗∗∗P<.0001. (B) Molecular scheme of SARS-CoV-2 endometrial infection. ACE2, TMPRSS4, FURIN, and BSG are shown in plasma membrane of an endometrial cell (lower left figure). CTSL and CTSB are represented outside the cell. MX1 is shown in cytoplasm. Expression of viral genes in comparison to whole transcriptomic set is represented as arrows next to their names: up = highly expressed; down = lowly expressed. Viral genes are positioned in their schematized cell locations as stablished by GeneCards database > Localization section (release 4.14) (56). Only maximum confidence levels (5 and 4) for compartments-derived cell locations were used. Proteins were grouped considering the highest coactivation values between pairs of viral genes during the menstrual cycle, which are shown in the lower right table in the figure. Discontinuous arrow shows less evidence according to our results, given that only FURIN showed high activation with BSG and that further studies are needed to understand BSG-related mechanisms of SARS-CoV-2 entry. ESE = early secretory endometrium; LSE = late secretory endometrium; MSE = midsecretory endometrium; PF = proliferative phase.
Correlations between genes showed activations and repressions between them throughout the menstrual cycle (Supplemental Table 1, available online). ACE2 and TMPRSS4 were positively correlated in the early secretory phase (0.58). A weak correlation (activation) was found for ACE2 with TMPRSS4 and CTSL in the window of implantation (0.17 and 0.25, respectively). Coactivations were also detected in the proliferative phase between CTSB and TMPRSS4; the early secretory phase between FURIN and BSG; the midsecretory phase between CTSB with CTSL, and MX1 with CTSB, CTSL, and TMPRSS4; and the late secretory phase between TMPRSS4 and CTSB, FURIN, CTSB, and CTSL. Based on our results, high coactivation values were prioritized to build a molecular scheme of SARS-CoV-2 plausible infection of the endometrium based on two mechanisms of entry: one via ACE2 receptor and the other via BSG receptor. This scheme is shown in Figure 2B.
In this model, TMPRSS4, which is highly expressed in all phases of the menstrual cycle, especially during the window of implantation, is the protease responsible for protein S cleaving, giving SARS-CoV-2 the capacity to bind to ACE2 and infect cells even if ACE2 is lowly expressed. The model adds CTSL, CTSB, and FURIN as novel actors in the SARS-CoV-2 mechanism of cell entry. These proteases are highly expressed and coactivate with TMPRSS4. They could also help with the cleaving of protein S in different sites, thereby increasing infectivity. We also included MX1, which regulates neutrophil activity and is highly expressed, bringing neutrophils into the tissue with elastases to help cleave S protein. Finally, BSG, the alternative receptor for SARS-CoV-2, also showed high expression, which would favor infectivity in this model, and activation with FURIN, which would favor infectivity in this model.
Age influences the expression of viral genes throughout the menstrual cycle
For this analysis, only samples from the study Talbi et al. (53) were used because it was the only study that provided the age for each sample (in a range of 23–50 years old; n = 27). ACE2 showed increased expression with age in the proliferative, early secretory, and midsecretory phases (0.4, 0.73, 0.44, respectively; Fig. 3 A). Most correlations by age were stronger in early phases of the menstrual cycle (proliferative and early secretory) than in subsequent endometrial stages (Fig. 3B). TMPRSS4 and MX1 were highly correlated with age in the early secretory phase; TMPRSS4, CTSL, and CTSB had positive correlation in the proliferative phase; and TMPRSS4 and BSG had positive correlation in the late secretory phase. In addition, high negative correlations were detected for BSG in the early secretory phase and for FURIN in the midsecretory phase.
Figure 3.
Impact of age on viral-related infectivity gene expression throughout the menstrual cycle. (A) Effect of age on ACE2 expression. Gene expression is represented for ACE2 in each phase of the cycle according to the age of the sample analyzed. The range of age from patients involved in this study was 23 to 50 years. (B) Effect of age on viral gene expression. Pearson correlation R2 values are shown for each gene studied through of the phases of the menstrual cycle. Gray scale represents the magnitude of the correlation of increase or decrease in expression with age. High values are colored darker, and low values are colored lighter. ESE = early secretory endometrium; LSE = late secretory endometrium; MSE = midsecretory endometrium; PF = proliferative phase.
Discussion
It is important to understand the consequences of SARS-CoV-2 infection as the virus continues to spread worldwide. Fertility, specifically embryo implantation, is impacted by changes in endometrial gene expression throughout the menstrual cycle. Between menstrual cycle phases, gene expression varies to accompany physiologic changes (57). The differences in gene expression from one phase to another create a changing landscape for viral infection, and viral proteins themselves alter their expression throughout the menstrual cycle, setting the stage for a cyclical landscape of viral infectivity risk.
ACE2, the receptor far for SARS-CoV-2 cell entry, showed low expression in the endometrium in our study, as reported previously in the Human Protein Atlas (HPA) (42). ACE2 is also reduced in critical tissues such as lungs (24); however, we cannot suggest that low levels of ACE2 expression imply no effect of the virus on the tissue. Further studies are needed to elucidate the effect of decreased ACE2 in the endometrium. It is interesting that ACE2 increased (fold change = 2.47) from the early secretory to midsecretory phases, implying an increase in ACE2 in the window of implantation and a high risk of viral infectivity at this stage of the menstrual cycle.
Several possible mechanisms of SARS-CoV-2 cell entry have been proposed (19, 28). TMPRSS2, the most-reported protease involved with SARS-CoV-2 infectivity alongside ACE2, had medium endometrial expression in our study. However, there was no correlation between TMPRSS2, ACE2, and the rest of the genes studied. These results imply that the endometrium should be safe against SARS-CoV-2 infectivity mediated by TMPRSS2, though the expression of other proteases associated with S protein cleavage show a different landscape than TMPRSS2. TMPRSS4 statistically significantly changes its expression throughout the menstrual cycle, showing a statistically significant increase in the midsecretory phase. This protein also showed an interesting correlation with other genes, including an increase alongside ACE2 in the early secretory phase. Although TMPRSS2 is implicated in cell entry, the fact that S proteins could be targeted by TMPRSS4 and that TMPRSS4 increased infectivity in gut epithelial cells (26, 28) could mean a vulnerability of the endometrium to infection by SARS-CoV-2 mediated by TMPRSS4. Furthermore, TMPRSS4 up-regulation was correlated with CTSL and CTSB in all phases except the early secretory phase. Cell infection by SARS-CoV-2 was observed in TMRPSS2- cell lines expressing both CTSL and CTSB, so these proteins may have a residual function of cleaving viral S protein (19).
We propose that a synergy of CTSL and CTSB with TMPRSS4 through most of the cycle favors SARS-CoV-2 infectivity in the endometrium. FURIN, another protease predicted to cleave protein S (37, 58, 59), also showed a positive correlation with TMPRSS4 in the late secretory phase; with both targeting S protein, higher infectivity may occur. Finally, TMPRSS4 was positively correlated with MX1 in the midsecretory phase. MX1 actively participates in SARS-CoV-2 infection by attracting neutrophils to infected tissue and employing their elastases to cleave protein S along with TMPRSS2 (31); if TMPRSS4 participates alongside MX1, a more severe infection could occur during the window of implantation. TMPRSS4, CTSB, CTSL, FURIN, and MX1 showed statistically significant expression changes throughout the cycle. Protein expression of TMPRSS4, CTSB, FURIN, and MX1 (42) supports a susceptible environment for infection in the early secretory and midsecretory phases. Future studies are needed to fully understand the mechanism of SARS-CoV-2 cell entry and the roles of TMPRSS4, CTSB, CTSL, FURIN, and MX1 in the endometrium.
BSG, the alternative receptor for SARS-CoV-2 entry (25), showed high expression with statistically significant changes between phases and strong activation with FURIN through most of the cycle; however, its repression was correlated with ACE2 and TMPRSS2. Our results may indicate that only FURIN could be involved in S viral protein cleavage, if necessary, for SARS-CoV-2 binding to BSG, though other unknown proteases could also be involved. Further studies are needed to provide insight on BSG function in SARS-CoV-2 infection in the endometrium.
We also investigated the effect of age on viral gene expression throughout the menstrual cycle. In a recent study in lungs, ACE2 expression was detected as highly variable among individuals ranging from 0.17 transcripts per million to 14.5 transcripts per million in the same demographic group and highlighted a skewness of expression that statistically significantly increased with age (24). Our results showed a positive correlation between age and ACE2 from the proliferative to midsecretory phases, especially in the early secretory phase, meaning that the endometrium in older women (to age 50) could be more susceptible to SARS-CoV-2 infection. TMPRSS2 decreased with age, particularly in the midsecretory and late secretory phases, which may mean that cell-mediated entry of the virus is lowered with increased age. However, TMPRSS4 increased in expression from the proliferative to early secretory phases similar to CTSB, CTSL, and MX1. These results, in conjunction with ACE2 expression changes, highlighted the same behavior related to age as in the lungs (24), implying a higher risk of endometrial infection in older women, especially in the early phases of the menstrual cycle, as well as potential implantation problems. However, our results are limited to only one study with 27 patients, and there are no prospective studies showing the real clinical implications.
ACE2 plays an important role in endometrial changes because it cleaves angiotensin II and impacts spiral artery vasoconstriction, posterior proliferation, and renewal of tissue (60, 61, 62). ACE2 down-regulation by SARS-CoV-2 could alter the endometrial balance (63); however, given that the landscape of the tissue is modified with each menstrual cycle renewal, the consequences of SARS-CoV-2 infection could be reset from cycle to cycle.
Because our study integrates data sets from GEO, this approach is limited by the study design. This also means that results depend on the experimental transcriptomic procedures of prior reported results and the sample cohort included in each study. However, the inclusion criteria for the selected studies were very careful, and uniform and raw data were preprocessed to control undesired effects. Likewise, the virus differentially affects individuals due to each person’s genetic profile (31, 64). Further prospective research is needed to determine the mechanisms of SARS-CoV-2 infection in the endometrium and how it could affect the fertility of each patient.
Conclusion
Although TMPRSS2 expression implies a safe environment against infection, the roles of proteases and neutrophil regulating proteins TMPRSS4, CTSB, CTSL, FURIN, and MX1 could imply susceptibility of infection, especially during the early secretory and midsecretory phases. Additionally, the high expression of BSG could imply an alternative mechanism independent of ACE2 for endometrial viral infection. Our findings show that viral gene expression increases with age, suggesting that the endometrium of older women undergoing ART is at higher risk of viral infection. A careful approach is advised, with precautions implemented when resuming ART.
Acknowledgments
The authors thank the IVI-RMA IVI Foundation and the University of Valencia for their research support.
Footnotes
I.H.-C. has nothing to disclose. P.S.-L. has nothing to disclose. A.D.-P. has nothing to disclose. A.P. has nothing to disclose. P.D.-G. has nothing to disclose.
I.H.-C. and P.S.-L. should be considered similar in author order.
Supported by the IVI-RMA IVI Foundation, Valencia, Spain (2005-FIVI-043-PD), and the ACIF/2019/148 predoctoral program fellowship from the Conselleria de Educacion Investigacion Cultura y Deporte, Generalitat de Valencia, Spain (to I.H.-C.), and an FPU/15/01398 predoctoral program fellowship from the Ministry of Science, Innovation and Universities, Government of Spain (to A.D.P.).
Supplementary data
Relation of activation and repression among viral genes for each cycle phase. Pearson correlation R2 values are shown for each pair of genes through endometrial progression. Grayscale represents the magnitude of activation or repression between viral genes. High values are colored darker, and lower are colored lighter.
Supplemental Figure 1.
Flowchart of the selection of transcriptomic studies evaluating the endometrium of control patients (without any known endometrial pathology). The selection of suitable individual transcriptomic studies at Gene Expression Omnibus (GEO) and the number of individual studies excluded and remaining after each filtering step are shown (n, number of studies; N, sample size; lncRNA, long noncoding RNA).
Supplemental Figure 2.
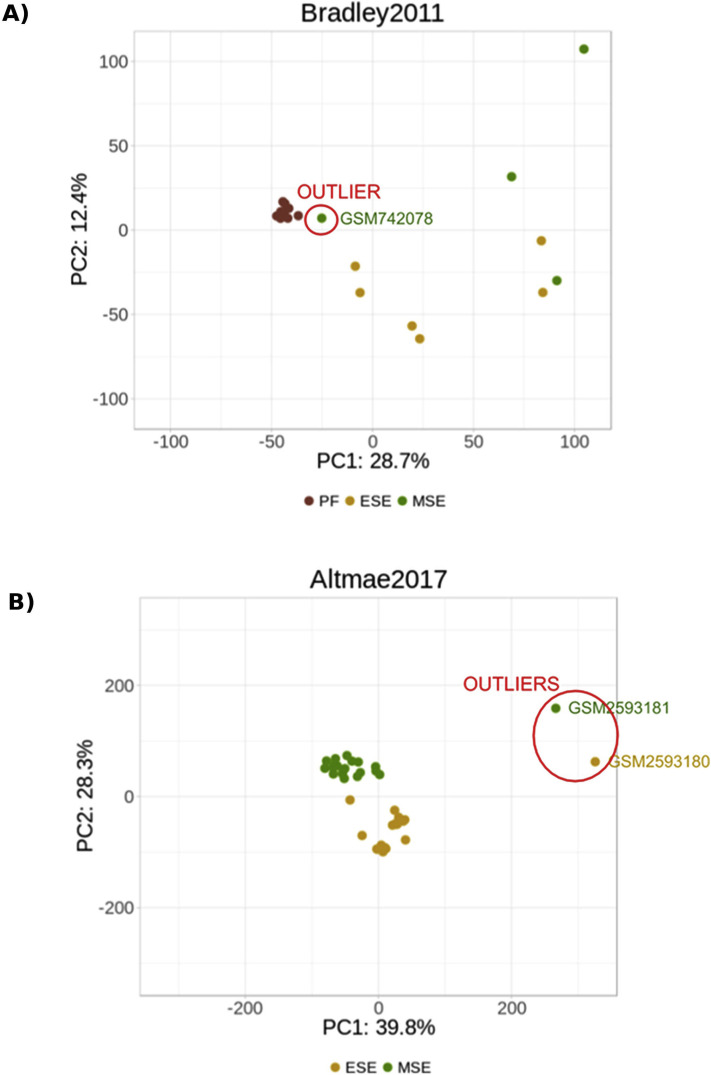
Excluded samples from endometrial data sets. (A) An early secretory sample in the Bradley 2011 data set was grouped to proliferative samples at the transcriptomic level due to mistaken labeling, as progesterone levels for that sample were the same as the samples from the proliferative phase. Given the doubt, this sample was removed from further analysis. (B) After correcting a batch effect due to different sequencing protocols in the data set of Altmäe et al. (63), two outliers were detected due to different transcriptomic behaviors and consequently removed. ESE = early secretory endometrium; LSE = late secretory endometrium; MSE = midsecretory endometrium; PF = proliferative phase.
Supplemental Figure 3.
Statistical tests for expression changes in viral genes throughout the menstrual cycle. (A) Analysis of variance P values and pairwise t-test P adjusted values are shown for each gene through the phases of the menstrual cycle. Pairwise t-test P values were adjusted by false discovery rate (FDR). Statistically significant values (FDR <0.05) are represented in red. (B) Fold change expression calculated for each gene from each phase to the phase immediately following in endometrial progression. Proliferative to early secretory, early secretory to midsecretory, and midsecretory to late secretory increases or decreases of expression are shown for each gene. ESE = early secretory endometrium; LSE = late secretory endometrium; MSE = midsecretory endometrium; PF = proliferative phase.
Supplemental Figure 4.
Expression changes across menstrual cycle phases. Gene expression distributions of each viral infectivity related gene are shown for each phase of the menstrual cycle in figures A to H. Statistically significant pairwise t-test results evaluating expression changes between menstrual cycle stages is indicated as ∗P<.1; ∗∗P<.01; ∗∗∗P<.001. ESE = early secretory; LSE, late secretory; MSE = medium secretory; PF = proliferative phase.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Novel coronavirus (n2019): situation report 124. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200523-covid-19-sitrep-124.pdf Available at:
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Fang L. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler MJ, Barrientos RM. The impact of nutrition on COVID-19 susceptibility and long-term consequences. Brain Behav Immunol. Published online April 18, 2020. Available at: 10.1016/j.bbi.2020.04.040. [DOI] [PMC free article] [PubMed]
- 8.Xiong T.Y., Redwood S., Prendergast B., Chen M. Coronaviruses and the cardiovascular system: acute and long-term implications. Eur Heart J. 2020;41:1798–1800. doi: 10.1093/eurheartj/ehaa231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorillo A., Gorwood P. The consequences of the COVID-19 pandemic on mental health and implications for clinical practice. Eur Psychiatry. 2020;63:e32. doi: 10.1192/j.eurpsy.2020.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Requena A., Cruz M., Vergara V., Prados N., Galliano D., Pellicer A. A picture of the covid-19 impact on IVIRMA fertility treatment clinics in Spain and Italy. Reprod Biomed Online. 2020;41:1–5. doi: 10.1016/j.rbmo.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monteleone P.A.A., Nakano M., Lazar V., Gomes A.P., de Martin H., Bonetti T.C. A review of initial data on pregnancy during the COVID-19 out-break: implications for assisted reproductive treatments. JBRA Assist Reprod. 2020;24:219–225. doi: 10.5935/1518-0557.20200030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laopaiboon M., Lumbiganon P., Intarut N., Mori R., Ganchimeg T., Vogel J.P., et al. Advanced maternal age and pregnancy outcomes: a multicountry assessment. BJOG. 2014;121:49–56. doi: 10.1111/1471-0528.12659. [DOI] [PubMed] [Google Scholar]
- 13.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy Eur J Allergy Clin Immunol. Published online February 27, 2020. Available at: 10.1111/all.14238. [DOI] [PubMed]
- 14.Huang Y, Tu M, Wang S, Chen S, Zhou W, Chen D, et al. Clinical characteristics of laboratory confirmed positive cases of SARS-CoV-2 infection in Wuhan, China: a retrospective single center analysis. Travel Med Infect Dis. Published online February 27, 2020. Available at: 10.1016/j.tmaid.2020.101606. [DOI] [PMC free article] [PubMed]
- 15.Nikolich-Zugich J., Knox K.S., Rios C.T., Natt B., Bhattacharya D., Fain M.J. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience. 2020;42:505–514. doi: 10.1007/s11357-020-00186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H., Guo J., Wang C., Luo F., Yu X., Zhang W., et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Mascio D., Khalil A., Saccone G., Rizzo G., Buca D., Liberati M., et al. Outcome of coronavirus spectrum infections (SARS, MERS, COVID-19) during pregnancy: a systematic review and meta-analysis. Am J Obstet Gynecol MFM. 2020;2 doi: 10.1016/j.ajogmf.2020.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gajbhiye R, Modi D, Mahale S. Pregnancy outcomes, newborn complications and maternal-fetal transmission of SARS-CoV-2 in women with COVID-19: a systematic review. medRxiv. Published online May 5, 2020. Available at: 10.1101/2020.04.11.20062356. [DOI]
- 19.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kai H., Kai M. Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors—lessons from available evidence and insights into COVID-19. Hypertens Res. 2020;43:648–654. doi: 10.1038/s41440-020-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benigni A., Cassis P., Remuzzi G. Angiotensin II revisited: new roles in inflammation, immunology and aging. EMBO Mol Med. 2010;2:247–257. doi: 10.1002/emmm.201000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwai M., Horiuchi M. Devil and angel in the renin-angiotensin system: ACE-angiotensin II-AT1 receptor axis vs. ACE2-angiotensin-(1–7)-Mas receptor axis. Hypertens Res. 2009;32:533–536. doi: 10.1038/hr.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Dhindsa RS, Povysil G, Zoghbi A, Joshua E, Hostyk JA, et al. Transcriptional inhibition of host viral entry proteins as a therapeutic strategy for SARS-CoV-2. Preprints. Published online April 28, 2020. Available at: 10.20944/preprints202003.0360.v1.
- 25.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv. Published online March 14, 2020. Available at: 10.1101/2020.03.14.988345. [DOI]
- 26.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain M, Jabeen N, Amanullah A, Baig AA, Aziz B, Shabbir S, et al. Structural basis of SARS-CoV-2 spike protein priming by TMPRSS2. bioRxiv. Published online April 22, 2020. Available at: 10.1101/2020.04.21.052639. [DOI]
- 28.Zang R, Castro MFG, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 mediate SARS-CoV-2 infection of human small intestinal enterocytes. bioRxiv. April 23, 2020. Available at: 10.1101/2020.04.21.054015. [DOI] [PMC free article] [PubMed]
- 29.Wang Q, Qiu Y, Li JY, Zhou ZJ, Liao CH, Ge XY. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol Sin. Published online March 20, 2020. Available at: 10.1007/s12250-020-00212-7. [DOI] [PMC free article] [PubMed]
- 30.Mei Z, Bingpeng L, Hongbin G, Xinhong W, Kaibin W, Mingxiao L, et al. Significant expression of FURIN and ACE2 on oral epithelial cells may facilitate the efficiency of 2019-nCov entry. bioRxiv. Published online April 18, 2020. Available at: 10.1101/2020.04.18.047951. [DOI]
- 31.Bhattacharyya C, Das C, Ghosh A, Singh AK, Mukherjee S, Majumder PP, et al. Global spread of SARS-CoV-2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. bioRxiv. Published online May 5, 2020. Available at: 10.1101/2020.05.04.075911. [DOI]
- 32.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan L., Mu M., Yang P., Sun Y., Wang R., Yan J., et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China. Am J Gastroenterol. 2020;115:766–773. doi: 10.14309/ajg.0000000000000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan C, Li K, Ding Y, Lu WL, Wang J. ACE2 expression in kidney and testis may cause kidney and testis damage after 2019-nCoV infection. medRxiv. Published online February 13, 2020. Available at: 10.1101/2020.02.12.20022418. [DOI]
- 35.Wang Z., Xu X. scRNA-seq Profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, Leydig and Sertoli cells. Cells. 2020;9:920. doi: 10.3390/cells9040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li M.-Y., Li L., Zhang Y., Wang X.-S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou L, Niu Z, Jiang X, Zhang Z, Zheng Y, Wang Z, et al. Systemic analysis of tissue cells potentially vulnerable to SARS-CoV-2 infection by the protein-proofed single-cell RNA profiling of ACE2, TMPRSS2 and Furin proteases. bioRxiv. Published online April 10, 2020. Available at: 10.1101/2020.04.06.028522. [DOI]
- 38.Stanley KE, Thomas E, Leaver M, Wells D. Coronavirus disease (COVID-19) and fertility: viral host entry protein expression in male and female reproductive tissues. Fertil Steril. Published online May 8, 2020. Available at: 10.1016/j.fertnstert.2020.05.001. [DOI] [PMC free article] [PubMed]
- 39.Noyes R.W., Hertig A.T., Rock J. Dating the endometrial biopsy. Am J Obstet Gynecol. 1975;122:262–263. doi: 10.1016/s0002-9378(16)33500-1. [DOI] [PubMed] [Google Scholar]
- 40.Sebastian-Leon P., Garrido N., Remohí J., Pellicer A., Diaz-Gimeno P. Asynchronous and pathological windows of implantation: two causes of recurrent implantation failure. Hum Reprod. 2018;33:626–635. doi: 10.1093/humrep/dey023. [DOI] [PubMed] [Google Scholar]
- 41.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navani S. The human protein atlas. J Obstet Gynecol India. 2011;61:27–31. [Google Scholar]
- 43.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 44.National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/ Available at:
- 45.Moher D., Liberati A., Tetzlaff J., Altman D.G., Altman D., Antes G., et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villanueva R.A.M., Chen Z.J. 2nd ed. Springer-Verlag; New York: 2019. ggplot2: Elegant graphics for data analysis. [Google Scholar]
- 49.Tajti F., Kuppe C., Antoranz A., Ibrahim M.M., Kim H., Ceccarelli F., et al. A functional landscape of CKD entities from public transcriptomic data. Kidney Int Reports. 2020;5:211–224. doi: 10.1016/j.ekir.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 51.Freedman D., Pisani R., Purves R. 4th ed. W.W. Norton; New York: 2007. Statistics. [Google Scholar]
- 52.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A language and environment for statistical computing. Version 3.0.1.http://www.R-project.org Available at: [Google Scholar]
- 53.Talbi S., Hamilton A.E., Vo K.C., Tulac S., Overgaard M.T., Dosiou C., et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology. 2006;147:1097–1121. doi: 10.1210/en.2005-1076. [DOI] [PubMed] [Google Scholar]
- 54.Altmäe S., Koel M., Võsa U., Adler P., Suhorutšenko M., Laisk-Podar T., et al. Meta-signature of human endometrial receptivity: a meta-analysis and validation study of transcriptomic biomarkers. Sci Rep. 2017;7:10077. doi: 10.1038/s41598-017-10098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sigurgeirsson B., Åmark H., Jemt A., Ujvari D., Westgren M., Lundeberg J., et al. Comprehensive RNA sequencing of healthy human endometrium at two time points of the menstrual cycle. Biol Reprod. 2016;96:24–33. doi: 10.1095/biolreprod.116.142547. [DOI] [PubMed] [Google Scholar]
- 56.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;2016:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 57.Carson D.D. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol Hum Reprod. 2002;8:871–879. doi: 10.1093/molehr/8.9.871. [DOI] [PubMed] [Google Scholar]
- 58.Meng T, Cao H, Zhang H, Kang Z, Xu D, Gong H, et al. The insert sequence in SARS-CoV-2 enhances spike protein cleavage by TMPRSS. bioRxiv. Published online February 16, 2020. Available at: 10.1101/2020.02.08.926006. [DOI]
- 59.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shan T., Shang W., Zhang L., Zhao C., Chen W., Zhang Y., et al. Effect of angiotensin-(1–7) and angiotensin II on the proliferation and activation of human endometrial stromal cells in vitro. Int J Clin Exp Pathol. 2015;8:8948–8957. [PMC free article] [PubMed] [Google Scholar]
- 61.Li X.F., Ahmed A. Dual role of angiotensin II in the human endometrium. Hum Reprod. 1996;11:95–108. doi: 10.1093/humrep/11.suppl_2.95. [DOI] [PubMed] [Google Scholar]
- 62.Vaz-Silva J., Carneiro M.M., Ferreira M.C., Pinheiro S.V.B., Silva D.A., Silva Filho A.L., et al. The vasoactive peptide angiotensin-(1–7), its receptor Mas and the angiotensin-converting enzyme type 2 are expressed in the human endometrium. Reprod Sci. 2009;16:247–256. doi: 10.1177/1933719108327593. [DOI] [PubMed] [Google Scholar]
- 63.Jing Y, Run-Qian L, Hao-Ran W, Hao-Ran C, Ya-Bin L, Yang G, et al. Potential influence of COVID-19/ACE2 on the female reproductive system. Mol Hum Reprod. Published online May 4, 2020. Available at: 10.1093/molehr/gaaa030. [DOI] [PMC free article] [PubMed]
- 64.Richards B, ed. COVID-19 host genetics initiative. Quebec: McGill University. Available at: https://www.covid19hg.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relation of activation and repression among viral genes for each cycle phase. Pearson correlation R2 values are shown for each pair of genes through endometrial progression. Grayscale represents the magnitude of activation or repression between viral genes. High values are colored darker, and lower are colored lighter.