Abstract
Nuclear factor erythroid 2‐related factor 2 (NRF2) is a master transcriptional regulator of genes whose products defend our cells for toxic and oxidative insults. Although NRF2 activation may reduce cancer risk by suppressing oxidative stress and tumor-promoting inflammation, many cancers exhibit elevated NRF2 activity either due to mutations that disrupt the negative control of NRF2 activity or other factors. Importantly, NRF2 activation is associated with poor prognosis and NRF2 has turned out to be a key activator of cancer-supportive anabolic metabolism. In this review, we summarize the diverse roles played by NRF2 in cancer focusing on metabolic reprogramming and tumor-promoting inflammation.
Introduction
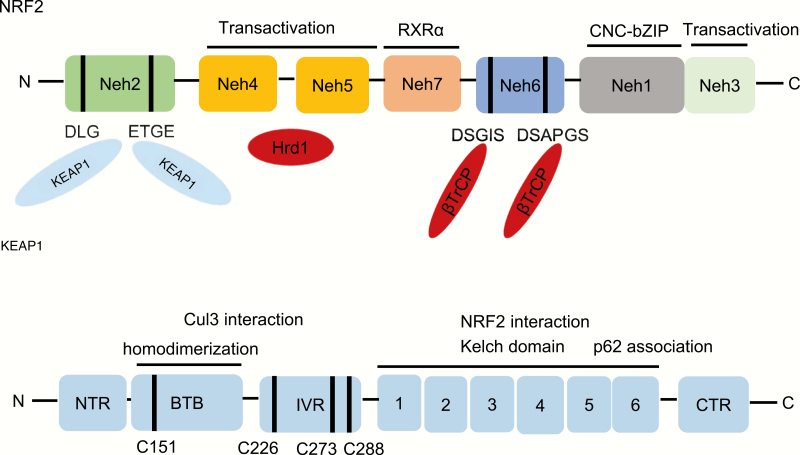
Nuclear factor erythroid 2‐related factor 2 (NRF2), encoded by NFE2L2 gene (Nuclear Factor, Erythroid 2 Like 2), belongs to the Cap’n’Collar (CNC) subfamily of basic leucine zipper (bZip) transcription factors, which comprises Nuclear Factor Erythroid-derived 2 (NFE2) and NRF1, NRF2 and NRF3 (1,2). NRF2 forms heterodimers with the small musculoaponeurotic fibrosarcoma proteins (MAFs) K, G and F, which allows it to bind antioxidant response elements and activate gene transcription (3). NRF2 possesses seven conserved NRF2‐ECH homology (Neh) domains with different functions (Figure 1). The bZip in the Neh1 domain heterodimerizes with the MAFs (3), whereas the Neh2 domain contains two ETGE and DLG motifs that specifically bind to Kelch domain of Kelch‐like‐ECH‐associated protein 1 (KEAP1), which mediates NRF2 ubiquitination and degradation (3). The C-terminal Neh3 domain acts as a transcriptional activation domain (1). The Neh4 and Neh5 also support transcriptional activation by interacting with CREB‐binding protein (CBP), which has histone acetyltransferase activity (1). The Neh6 domain is rich in serine residues and contains two β-transducin repeat-containing protein (β-TrCP) degrons DSGIS and DSAPGS, involved in NRF2 degradation (4–7). The Neh7 domain interacts with retinoic X receptor alpha (RXRα), which inhibits the cytoprotective activity of NRF2 (8). NRF2 is best known as the master regulator of a battery of antioxidant response element-containing cytoprotective genes whose expression is induced in response to cell stress. The products of these genes form a network of cooperating enzymes involved in phase I, II and III drug detoxification reactions and metabolic elimination of pro-oxidants (3). Phase I enzymes mediate the oxidation, reduction and hydrolysis of various xenobiotics and include aldo-keto reductases (AKRs), carbonyl reductases (CBRs), aldehyde dehydrogenase 1 (ALDH1), NAD(P)H quinone oxidoreductase 1 (NQO1) and various cytochrome P450s (CYPs), such as CYP2a5 and CYP2b6 (9,10). The metabolites generated by phase I enzymes can be further modified by phase II conjugation enzymes, which attach endogenous hydrophilic molecules to the phase I reaction products to increase their solubility and promote their excretion. Typical phase II enzymes include glutathione S-transferases (GSTs), UDP-glucuronosyltransferases (UGTs) and heme oxygenase-1 (HO-1) (9,11). Phase III enzymes include several drug efflux transporters, such as multidrug resistance-associated proteins (MDR), breast cancer resistant protein (BCRP), adenosine triphosphate (ATP)-binding cassette g5 (ABCG5) and g8 (ABCG8) (9,11). Key antioxidant pathways induced by NRF2 include enzymes that catalyze glutathione (GSH) synthesis and reduction, as well as reduced nicotinamide adenine dinucleotide phosphate (NADPH) generation. More recent studies have identified numerous new NRF2 target genes and had revealed several new functions of NRF2 that go beyond its redox-regulating capacity. NRF2 has become a prime target for research in the field of inflammation, cancer metabolism, cancer prevention and cancer treatment. This great expansion in the regulatory capacity of NRF2 presents new challenges for understanding its biological function, which seems to be context-dependent, especially in respect to tumor biology.
Figure 1.
The architecture of NRF2 and its negative regulator KEAP1. NRF2 contains seven conserved NRF2-ECH homology (Neh) domains, Neh1–Neh7. Neh1 has a bZip motif, where the basic region is responsible for DNA binding and the Zip dimerizes with small MAF proteins. Neh2 contains ETGE and DLG motifs, which are required for binding to KEAP1 and subsequent KEAP1-dependent polyubiquitination and degradation of NRF2. Neh3-5 are transactivation domains and Neh4 and Neh5 domains also interact with Hrd1. Neh6 contains DSGIS and DSAPGS motifs responsible for the β-TrCP-mediated ubiquitination and degradation. The Neh7 domain interacts with RXRα, which inhibits NRF2 activity. KEAP1 contains five domains, an amino terminal region (NTR), a broad complex, tramtrack, bric-a-brac (BTB) domain, an intervening region (IVR), six Kelch motifs and the C-terminal region (CTR). The Kelch motifs and the CTR mediate interactions with NRF2 and p62. The BTB domain mediates homodimerization and contributes to its interaction of the IVR with Cul3. Several functionally important cysteine residues (C151, C226, C273 and C278) sense ROS and electrophiles and are needed in their reduced state for KEAP1 function.
Regulation of NRF2 activity
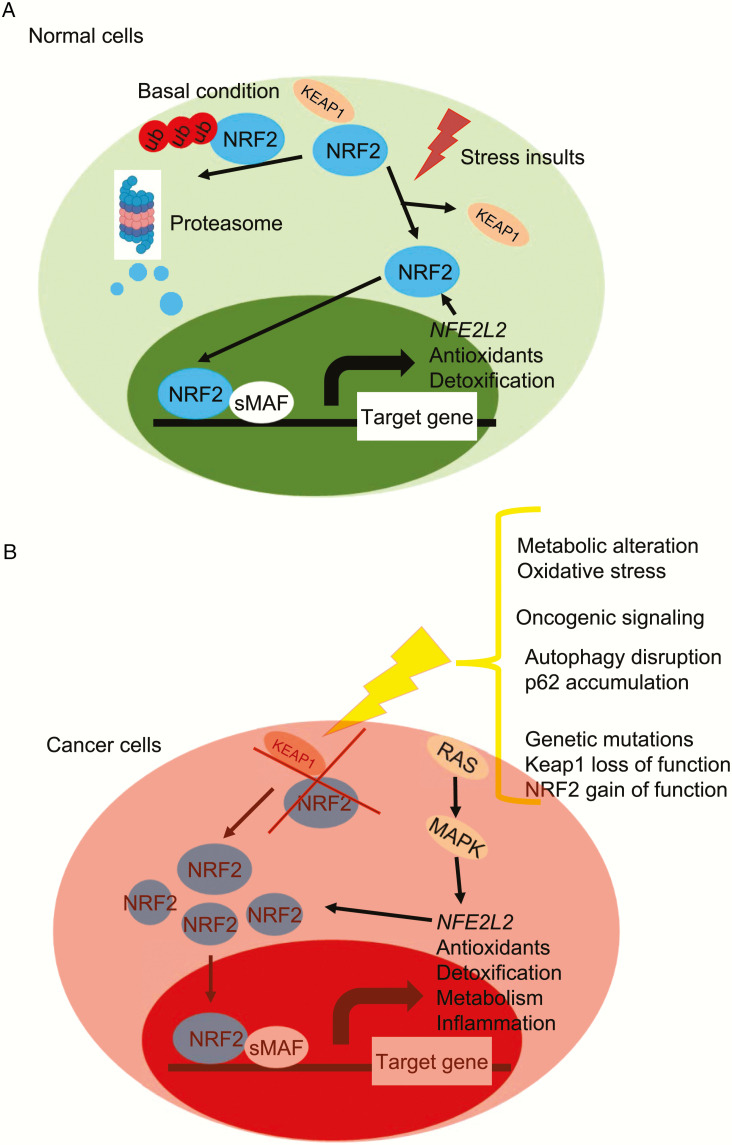
NRF2 is expressed in all cell types and its basal level is usually low in unstressed cells due to well-established KEAP1-mediated proteasomal degradation (Figure 2). KEAP1 is a redox-regulated adaptor for the Cullin (Cul)3-RING-box protein (Rbx)1 ubiquitin ligase complex that binds NRF2 as a dimer through its C-terminal Kelch domain, which interacts with the DLG and ETGE motifs in the Neh2 domain of NRF2 (1). During oxidative stress, electrophiles and reactive oxygen species (ROS) lead to oxidation and modification of KEAP1 sensor cysteines, especially cysteine 151 (C151), C226, C273 and C288, to induce a conformational change, which inhibits KEAP1-mediated NRF2 degradation (12–15). As a result, newly synthesized NRF2 can accumulate in the nucleus and lead to activation of cytoprotective and metabolic genes (1,12,16,17). Interestingly, NRF2 can also activate its own (i.e. NFE2L2) transcription, which also responds to other oncogenic inputs, such as RAS-RAF-MAPK signaling, leading to activation of AP-1 and other transcription factors (18,19). Another mechanism that leads to NRF2 stabilization is activated in response to disruption of autophagy. p62/sequestosome 1 (SQSTM1) is a cargo-adaptor that binds to ubiquitinated organelles and protein aggregates to mediate their autophagic degradation (20). Disruption of autophagy results in p62 accumulation and binding of p62 via its KIR motif to KEAP1, resulting in KEAP1 sequestration and NRF2 stabilization (20–22). Similar to p62, dipeptidyl peptidase 3 (DPP3) (23), Wilms tumor gene on X chromosome (WTX) (24) and Partner and Localizer of BRCA2 (PALB2) (25) all contain KIR-like ETGE motifs and can thereby disrupt the KEAP1–NRF2 complex by binding to KEAP1. Another NRF2 stabilizer p21Cip1/WAF1, which is upregulated to protect cells from oxidative damage and can directly interact with the DLG and ETGE motifs of NRF2 through its KRR motifs, thereby attenuating KEAP1-mediated ubiquitination of NRF2 (26). Breast Cancer Type 1 Susceptibility Protein (BRCA1) can also interact with the ETGE motif of NRF2 and interfere with the KEAP1–NRF2 interaction, making it another sensor of oxidative stress with important implications to BRCA1-related tumorigenesis (27).
Figure 2.
Regulation of NRF2 stability and activation. (A) In normal cells, under basal conditions, NRF2 amounts are low due to its continuous KEAP1-mediated proteasomal degradation. Under stressed condition, cellular NRF2 amounts are temporarily increased upon exposure to toxicants and ROS that inactivate KEAP1 and allow newly synthesized NRF2 to accumulate. Part of this response may be mediated by liberated NRF2 auto-activating NFE2L2 gene transcription. (B) In cancer cells, oncogenic signaling, genetic mutations, autophagy disruption or metabolic alterations results in loss of KEAP1 function and NRF2 stabilization. NRF2 expression can be further increased by RAS-MAPK signaling. NRF2 activation results in metabolic reprogramming and entrenchment of the transformed state.
KEAP1-independent proteasomal degradation of NRF2 has also been reported, involving the β-TrCP-SKP1-CUL1-RBX1, CR6-interacting factor 1 (CRIF1) (28), WDR-23 (29) and HRD1 (30) ubiquitin ligase complexes. The Neh6 domain of NRF2 contains two motifs, DSGIS and DSAPGS, which can be recognized by the F box of the WD40 substrate adaptor β-TrCP. Glycogen synthase kinase-3β (GSK3β) phosphorylates the DSGIS motif and increases the affinity of β-TrCP for NRF2, thereby stimulating NRF2 degradation (1,7). Both CRIF1 and WDR-23 contribute to NRF2 proteasome degradation both under normal and stressed conditions (28,29). Although the Neh2 domain of NRF2 mediates its interaction with CRIF1 and WDR-23, this interaction is KEAP1 independent (28,29). Interestingly, both CRIF1 and WDR-23 isoform 2 are mainly nuclear and their activity may contribute to termination of NRF2-mediated transcriptional response, whereas KEAP1 mainly acts in the cytoplasm. HRD1 is an endoplasmic reticulum (ER) membrane-associated E3 ubiquitin ligase involved in ER-associated degradation. In response to ER stress, HRD1 interacts with the Neh 4–5 domains of NRF2 to mediates NRF2 ubiquitylation and degradation in cirrhotic liver (31). Certain cellular metabolites can also influence NRF2 stability and transcriptional activity. In hepatocellular carcinoma triggered by MYC and KEAP1 inactivation, NRF2 activity and oncogenic function depend on its deglycation mediated by fructosamine-3-kinase (FN3K) (32).
NRF2 in cancer metabolism
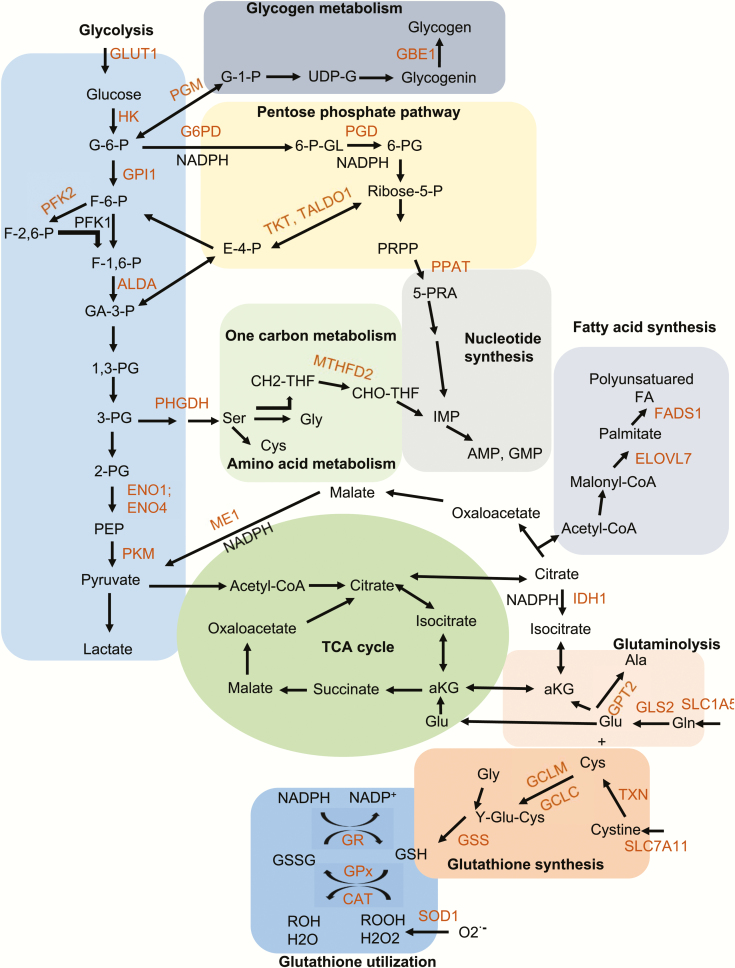
Cancer cells take up nutrients, including glucose and glutamine, and reprogram their energy metabolism to support cell growth and proliferation, which require upregulation of anabolic metabolism and macromolecular biosynthesis (33). Ample evidence support a key role for NRF2 in the reprogramming of cancer metabolism (33). Chromatin Immunoprecipitation Sequencing (ChIP-Seq) analyses of NRF2 target genes in mouse A549 cells, mouse embryonic fibroblasts, lymphoid cells and the mouse esophagus resulted in identification of numerous NRF2 target genes that participate in the regulation of glycolysis, pentose phosphate pathway (PPP), fatty acid metabolism, glutamine metabolism and glutathione metabolism (Figure 3) (34–37). Correspondently, NRF2 is recognized as a key transcriptional regulation of metabolism. Under aerobic conditions, normal cells process glucose, first to pyruvate via glycolysis in the cytosol and then to carbon dioxide via oxidative phosphorylation in mitochondria, resulting in generation of 36 ATP molecules per each molecule of glucose. However, due to Warburg effect cancer cells use aerobic glycolysis to mainly produce anabolic precursors to support rapid tumor growth, but resulting in much less efficient production of two ATP molecules per each molecule of glucose (33). Cancer cells compensate for the lower efficiency of ATP production by increasing glucose import into the cytoplasm and NRF2 induces expression of glucose transporter GLUT1 (34). Aerobic glycolysis allows cancer cells to divert glycolytic intermediates into various biosynthetic pathways, responsible for generation of nucleosides, amino acids and lipids. NRF2 induces the expression of several genes encoding key glycolytic enzymes, including hexokinase 1 (HK1), HK2, glucose phosphate isomerase 1 (GPI1), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFK2), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFK4), fructose-bisphosphate aldolase A (ALDA), enolase 1 (ENO1), enolase 4 (ENO4), pyruvate kinase, muscle (PKM) to increase glycolytic flow and maintain pool sizes of glycolytic intermediates for anabolic reactions (34). The first glycolytic intermediate is glucose-6-phosphate, which can be diverted to the PPP by glucose-6-phosphate dehydrogenase (G6PD). NRF2 upregulates several genes encoding the PPP enzymes, including G6PD, phosphogluconate dehydrogenase (PGD), transketolase (TKT) and transaldolase 1 (TALDO1), as well as enzymes for de novo nucleotide synthesis enzymes, such as phosphoribosyl pyrophosphate amidotransferase (PPAT) and methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) (34,35). Glucose-6-phosphate could also be converted to glucose-1-phosphate by phosphoglucomutase (PGM) for glycogen metabolism and NRF2 activates the expression of PGM5, 1,4-alpha-glucan branching enzyme 1 (GBE1) and glucosidase alpha, acid (GAA) (34,35). The liver and skeletal muscle are the two major organs responsible for glycogen synthesis and storage, but they respond differently to NRF2. Conditional NRF2 activation in liver increases glycogen storage (38), but skeletal muscle-specific Keap1-knockout mice show decreased glycogen content in skeletal muscle (39). Another glycolytic metabolite 3-phosphoglycerate can be diverted to de novo serine synthesis and serine can be used for cysteine and glycine synthesis via one carbon metabolism (folate cycle) and methionine cycle to support protein and nucleic acid synthesis. Genes encoding enzymes of the one carbon metabolism, such as MTHFD2, are also activated by NRF2. In addition to their enzymatic function, some NRF2 targets, such as HK2, PFK2 and PKM also have non-metabolic functions whereby they promote tumor progression upon nuclear translocation and interaction with transcription factors and coactivators (40).
Figure 3.
NRF2 reprograms cellular metabolism. NRF2 regulates expression of genes encoding the denoted enzymes involved in glycolysis, the pentose phosphate pathway, one carbon metabolism, nucleotide biosynthesis, glutaminolysis, fatty acid synthesis, and glutathione synthesis and utilization (colored in red). Enzyme reactions that produce NADPH are highlighted. Enzyme abbreviations: ELOVL7, elongation of very long chain fatty acids protein 7; FADS1, fatty acid desaturase 1; G6PD, glucose-6-phosphate dehydrogenase; GCLC, glutamate-cysteine ligase, catalytic subunit; GCLM, glutamate-cysteine ligase, modifier subunit; GLS2, glutaminase 2; GPT2, glutamic pyruvate transaminase 2; GSS, glutathione synthetase; IDH1, isocitrate dehydrogenase 1; ME1, malic enzyme 1; MTFHD2, methylenetetrahydrofolate dehydrogenase 2; PGD, 6-phosphogluconate dehydrogenase; PHGDH, phosphoglycerate dehydrogenase; PPAT, phosphoribosyl pyrophosphate amidotransferase; TALDO, transaldolase; TKT, transketolase; TXN, thioredoxin; SLC7A11, solute carrier family 7, member 11; SLC1A5, solute carrier family 1, member 5; HK1, hexokinase 1; GPI1, glucose phosphate isomerase 1; PFK2, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2; ALDA, fructose-bisphosphate aldolase A; ENO1, enolase 1; ENO4, enolase 4; PKM, pyruvate kinase, muscle; GR, glutathione reductase; GPx, glutathione peroxidase; SOD1, superoxide dismutase 1; CAT, catalase; PGM, phosphoglucomutase; GBE1, glycogen branching enzyme 1. Metabolite abbreviations: G-6-P, glucose 6-phosphate; G-1-P, glucose 1-phosphate; F-6-P, fructose 6-phosphate; F-1,6-BP, fructose 1,6-bisphosphate; F-2,6-BP, fructose 2,6-bisphosphate; GA-3-P, glyceraldehyde 3-phosphate; 1,3-PG, 1,3-phosphoglycerate; 3-PG, 3-phosphoglycerate; 2-PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; UDP-G, uracil-diphosphate glucose; 6-P-GL, 6-phosphogluconolactone; 6-PG, 6-phosphogluconate; R-5-P, ribulose 5-phosphate; 5-PRA, phosphoribosylamine; PRPP, 5-phosphoribosyl-α-1-pyrophosphate; THF, tetrahydrofolate; CH2-THF, 5,10-methylene-tetrahydrofolate; CHO-THF, 10-formyl-tetrahydrofolate; IMP, inosine monophosphate; AMP, adenosine monophosphate; GMP, guanosine monophosphate. Gly, glycine; Ala, alanine; Cys, cysteine; Glu, glutamate; Gln, glutamine; aKG, a-ketoglutarate; Y-Glu-Cys, gamma-glutamyl cysteine; Acetyl-CoA, acetyl-coenzyme A; NADPH, nicotinamide adenine dinucleotide phosphate, reduced.
NRF2 also activates genes encoding for several key enzymes in lipid metabolism, including elongation of very long chain fatty acids protein 7 (ELOVL7), fatty acid desaturase 1 (FADS1), acyl-CoA synthetase short-chain family member 2 (ACSS2), acyl-CoA thioesterase 7 (ACOT7), acyl-Coenzyme A dehydrogenase family member 10 (ACAD10) and acyl-Coenzyme A dehydrogenase family member 12 (ACAD12) (34,35). The response of lipid metabolism to NRF2 activation varies in different cell types and is context dependent (34,36,41). Of note, esophagus of Keap1-null mice shows elevated phospholipids and long chain free fatty acid (34), whereas conditional NRF2 stabilization in hepatocytes results in liver triglyceride accumulation (38). However, liver-specific Keap1-knockout mice, which are also hypomorphic for KEAP1 in other cell types, exhibit reduced liver lipids (41), while Keap1 knockdown enhances adipogenesis in 3T3-L1 cells (42). NRF2 also stimulates expression of enzymes that synthesize NADPH, which is needed for de novo lipogenesis, such as malic enzyme 1 (ME1), G6PD and isocitrate dehydrogenase 1 (IDH1) (34,35). In addition to lipogenesis, NADPH is required as a cofactor for amino acid and nucleic acid syntheses, as well as the reduction of glutathione and redox cycling enzymes, including glutathione reductase (GR), glutathione peroxidase 2 (GPx2), glutathione peroxidase 4 (GPx4), superoxide dismutase 1 (SOD1) and catalase (CAT). Nfe2l2 knockdown leads to a dramatic decrease in NADPH levels, whereas high NRF2 activity in Keap1-knockout cells, increases NADPH amounts and the NADPH/NADP+ ratio (35).
In non-proliferating cells, the glycolysis end product pyruvate enters the trichloroacetic acid (TCA) cycle to maximize ATP production via oxidation of substrates to CO2. In proliferating cells, however, the TCA cycle serves as an important source of biosynthetic precursors in addition to providing ATP. For example, the TCA cycle intermediate citrate is used for lipid biosynthesis, whereas oxaloacetate and α-ketoglutarate are used to synthesize four non-essential amino acids (aspartate, asparagine, glutamate and proline). Glutamate is required for synthesis of glutathione in response to NRF2 activation, which also being secreted via Xc- or xCT antiporter system (43). As a result, TCA cycle intermediates must be replenished via a process called anaplerosis. Glutamine, the most abundant amino acid in human plasma (44), is a major contributor together with glucose to anaplerotic flux and is also an important carbon source and nitrogen donor. The glutamine transporter solute carrier family 1, member 5 (SLC1A5) that mediates glutamine uptake is upregulated by NRF2 (45). NRF2 also activates the genes encoding for the glutaminolysis enzymes glutaminase (GLS2) and glutamic pyruvate transaminase 2 (GPT2) thereby controlling the production of glutamate, aspartate, alanine and α-ketoglutarate, which are needed for nucleotides and non-essential amino acids synthesis in cancer cells (34). NRF2 also regulates de novo serine biosynthesis in cooperation with ATF4, leading to synthesis of serine-derived glycine and cysteine via the methionine cycle and one carbon metabolism (30,46). Cysteine and glycine are used for 7-glutathione synthesis, catalyzed by glutamate-cysteine ligase (GCLC/GCLM) and glutathione synthetase (GSS), both of which are encoded by NRF2 target genes (34). Glutamate is also an obligate exchange molecule for the NRF2-regulated glutamate-cystine antiporter solute carrier family 7, member 11 (SLC7A11), which controls the intracellular availability of cysteine (34,43).
Some of the first NRF2 target genes identified code for antioxidant and detoxification drug-metabolism enzymes including glutathione S-transferases alpha 4 (GSTA4), glutathione S-transferase alpha 3 (GSTA3), glutathione transferase zeta 1 (GSTZ1), glutathione S-transferase pi 1 (GSTP1), glutathione S-transferase pi 2 (GSTP2), glutathione S-transferase omega 2 (GSTO2), microsomal glutathione S-transferase 2 (MGST2), glutathione S-transferase mu 7 (GSTM7), glutathione S-transferase mu 6 (GSTM6), glutathione S-transferase mu 3 (GSTM3), glutathione S-transferase mu 2 (GSTM2) and glutathione S-transferase mu 1 (GSTM1), all of which are involved in phase II drug detoxification (34). These enzymes act in concert with phase I enzymes, such as AKR1B10, AKR1C1, AKR1C2, AKR1C3, ALDHs, CBRs, CYPs as well as phase III enzymes to allow both normal and transformed cells to metabolize and inactivate various xenobiotics (9,47,48). NRF2-regulated glycolysis and glutaminolysis support cancer cells growth and proliferation. However, increased protein production under oxidative stress or nutrient deprivation can lead to protein misfolding and proteotoxic stress (30,33). To avoid such stress, NRF2 also control expression of various proteasome subunits, including PSMA1, PSMA4, PSMA5, PSMB3, PSMB6, PSMC1, PSMC3, PSMD4 and PSMD14 to promote the degradation of misfolded proteins (30,49). NRF2 also stimulates the expression of various autophagy-related genes encoding for p62/SQSTM1, CALCOCO2, ULK1, ATG5 and GABARAPL1 to further promote the degradation of denatured and aggregated proteins (50). NRF2 also induces the unfolded protein response, which alleviates the ER stress (38,51).
NRF2 and inflammation
Inflammation is triggered when innate immune cells detect infectious agents or tissue injury, resulting in recruitment and activation of neutrophils, monocytes, macrophages and other immune cells (52,53). Acute inflammation is self-limiting and beneficial to the host as it eliminates dead cells, denatured extracellular matrix component or infectious agents and initiates tissue repair. However, uncontrolled inflammation can injure the host, resulting in so-called collateral damage. The failure to properly terminate the inflammatory response can also result in chronic inflammation, which is well known to be tumor-promoting (54). Activation of inflammatory cells results in the induction of cyclooxygenase (COX)-2 and inducible nitric oxide synthase (iNOS), which produce a number of inflammatory electrophiles, such as 15-deoxy-delta-12,14-prostaglandin J2 (15d-PGJ2), prostaglandin A2 (PGA2), 8-nitroguanosine 3′,5′-cyclic monophosphate (8-nitro-cGMP), all of which readily react with cysteine residues (55–57). When such mediators react with KEAP1, they lead to NRF2 activation, which alleviates inflammation-associated oxidative stress (56). By decreasing oxidative stress NRF2 can prevent tissue and cell damage, decreasing the production of danger-associated molecular patterns that are released by necrotic cells and amplify the inflammatory response. Accordingly, Nfe2l2-null mice display more severe lung inflammation and damage upon exposure to cigarette smoke (58) and hyperoxia (59) than wild-type mice. Nfe2l2-deficient mice are also highly susceptible to drug-induced liver injury, alcoholic liver disease and non-alcoholic fatty liver disease (60). Nfe2l2-deficient mice of certain genetic background also exhibit age-dependent autoimmune phenotypes (61,62). Conversely, genetic or pharmacological amplification of NRF2 inhibits acute inflammatory liver injury in a model of T-cell-mediated acute inflammatory liver injury (63). Myeloid cell-specific Nfe2l2 ablation enhances the production of inflammatory mediators and increase susceptibility to sepsis in response to microbial infection, while Keap1 ablation has the opposite effect (64). Pathogen-associated molecular patterns and danger-associated molecular patterns activate pattern recognition receptors that are mainly expressed by immune cells to activate inflammatory nuclear factor kappa B (NF-κB) signaling (65). Curiously, NF-κB activation is enhanced in Nfe2l2-deficient mice, resulting in severe inflammation (66). Conversely, the NRF2 activator, sulforaphane, inhibits inflammation by decreasing the expression of NF-κB-activated proinflammatory cytokines and inflammatory mediators, such as tumor necrosis factor (TNF), interleukin (IL)-1β, COX-2 and iNOS (67). The endogenous metabolite itaconate directly alkylates the redox sensitive cysteines of KEAP1 leading to NRF2 activation and the inhibition of IL-1β and interferon (IFN)-β production by macrophages (68). Likewise, fumarate mediates the succination of KEAP1 and activates NRF2 to control IL-6 induction (69). Furthermore, NRF2 activation decreases STING expression by destabilizing its mRNA, resulting in decreased production of type I IFN and increased susceptibility to infection with DNA viruses, whereas the silencing of NRF2 decreases virus infectivity (70). Similarly, Nfe2l2 -null mice show better survival and reduced viral replication after marburg virus (MARV) infection (71). However, NRF2 activation was reported to induce IL-17D and thereby potentiate antiviral defense against vaccina virus (VV) and mouse cytomegalovirus (MCMV) (72). Likewise, mice treated with the NRF2 agonist tert-butylhydroquinone are protected from MCMV infection (73) and the NRF2 activator RA839 was found to reduce rotavirus (RV) production in MA104 cells (74). In primary human fibroblast NRF2 activation induces an antiviral program to restrict HSV1 infection (75) and NRF2 activation in human alveolar epithelial cells decreases influenza A virus (IAV) infection and replication (76). Hepatitis B virus (HBV)-induced NRF2 activation protects infected cells against oxidative damage and maintains the integrity of the human and the viral genome (77). The basis for these highly discordant effects on antiviral immunity is not known, underscoring the necessity for a more mechanistic approach to understand the effects of NRF2 on the antiviral immunity.
NRF2 indirectly inhibits the production of pro‐inflammatory mediators through the induction of HO-1, a rate-limiting enzyme that catalyzes the degradation of heme into carbon monoxide (CO), free iron and biliverdin. HO-1 induction inhibits TNF-dependent NF-κB activation and induces the expression of E-selectin, intercellular adhesion molecule-1 (ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) (78). NQO1 has both positive and negative effects on NF-κB. In human monocytes, NRF2-induced NQO1 expression downregulates lipopolysaccharide (LPS)-induced expression of TNF and IL-1, while silencing NRF2 or NQO-1 enhances LPS-induced inflammatory responses (79). NQO1-deficient mice show decreased circulating lymphocytes, myeloid hyperplasia and increased susceptibility to autoimmune disease (80). A cytosine to thymidine (C→T) polymorphism in exon 6 of the human NQO1 gene results in a proline to serine (P187S) mutation that destabilizes NQO1 and inactivates its enzymatic activity and is associated with increased risk of multiple sclerosis and acute leukemia (81,82). p62/SQSTM1 is directly induced by NRF2 and NF-κB to prevent excessive inflammation and restrain NLRP3-inflammasome activation by inducing mitophagy of damaged mitochondria, the source of fragmented oxidized mitochondrial DNA (mtDNA), the NLRP3 ligand and the activator (83,84). ROS-mediated NRF2-dependent superinduction of ATF3 protects mice against endotoxic shock by suppressing IL-6 expression, but causes high susceptibility to bacterial and fungal infections and leads sepsis-associated immunosuppression (85,86).
NRF2 was also described to directly block the transcription of the proinflammatory genes. Chromatin immunoprecipitation sequencing analysis revealed that NRF2 directly binds to the promoter proximal regions of IL-6 and IL-1β genes to disrupt RNA polymerase II recruitment and blocks gene induction upon exposure to LPS (87). This NRF2-mediated transcriptional interference is different from its activatory functions on metabolism and antioxidation. Furthermore, activated NRF2 was also reported to directly induce IL-6 transcription in hepatocytes (88) and in persistent polyclonal B-cell lymphocytosis B cells (69). NRF2 also inhibits production of IL-17 and suppresses autoimmune encephalomyelitis (89). In addition, NRF2 directly upregulates the MARCO gene, encoding a scavenger receptor required for phagocytosis of bacteria, thereby improving bacterial clearance by alveolar macrophages in patients with chronic obstructive pulmonary disease (90). NRF2 has a dual role in controlling inflammasome activation, which mediates caspase-1-dependent production of IL-1β and the induction of pyroptosis. NLRP3-inflammasome activation depends on the production of ROS-oxidized mtDNA (91). NRF2 can inhibit inflammation by reducing ROS production, but NRF2 activation was also reported to promote activation of the NLRP3 and AIM2 inflammasomes, but not the NLRC4 inflammasome in mouse macrophages (92). The mechanism underlying these divergent effects is far from clear.
NRF2 can either inhibit or promote anticancer immunity in a cell type and disease context-dependent manner. Immune surveillance, involving both innate and adaptive immunity, is responsible for recognizing and eliminating the vast majority of incipient cancer cells and plays a big role in immune system’s antitumor activities (93,94). The tumor microenvironment harbors both tumor-promoting and tumor-antagonizing immune cells with antitumor immunity being mediated by CD8+ cytotoxic T lymphocytes, CD4+ T helper (Th) 1 cells and natural killer cells (65). NRF2 activation in macrophages can enhance cytotoxic T lymphocyte function by providing them with GSH and cysteine to support their activation and proliferation (95). Conversely, Nfe2l2 gene ablation or pharmacological inhibition of γ-GCS-dependent GSH synthesis as well as knockdown of SLC7A11 in bone marrow–derived macrophages attenuates cytotoxic T lymphocyte activation (64). Activated NRF2 in cancer cells also induces IL-17D expression that that leads to recruitment of natural killer cells, which can promote tumor regression (72). However, in myeloid-derived suppressor cells NRF2 activation reduces ROS accumulation and increases cell survival and expansion leading to attenuation of T-cell function (96). KEAP1 deficiency in Scurfy mice attenuates effector T-cell activation and decreased IFN-γ production by effector Th1 and CD8+ T cells (97). NRF2 activation decreases IFN-γ production and increases IL-4, IL-5 and IL-13 production in CD4+ cells and skews them toward Th2 differentiation, thereby inducing antitumor immunity (98). In human ovarian cancer, the lack of NRF2-dependent antioxidant defense in Treg cells leads to apoptosis in high oxidative stress tumor microenvironment and the apoptotic Treg cells inhibit antitumor immunity via the adenosine receptor pathway (99). Two of the NRF2 transcriptional targets, p62 and NDP52, promote autophagy, which can potentiate antitumor immunity by enhancing antigen presentation by professional antigen presenting cells and increasing cytotoxic T lymphocyte infiltration into the tumor (100). T-cell functionality paradoxically depends not only on ROS, but also on the ability to limit ROS accumulation. T-cell activation is accompanied by rapid ROS generation and ROS can amplify signaling pathways involved in antigen-stimulated T-cell activation and expansion. However, excessive ROS production compromise cell survival. Accordingly, NRF2 expression is upregulated in tumor infiltrating T cells but NRF2 expression and its target genes are decreased by T-cell receptor (TCR) activation (101). Considering that many of the above results are derived from the use of whole body NRF2-knockout mice or pharmacological NRF2 activators of questioned specificity, the function of NRF2 in specific immune cell subsets needs to be more critically evaluated. Given that NRF2 activation in cancer cells is protumorigenic, it is safe to assume that it attenuates antitumor immunity. Although it may be premature to arrive at any definite conclusions regarding the role of NRF2 activation in T cells, it appears that the majority of reports listed above suggest that it attenuates T-cell-mediated antitumor immunity, in part through a decrease in IFN-γ production.
NRF2 in tumor progression
As already mentioned above, NRF2 has a Janus-like functions in cancer cells. On the one hand, NRF2 suppresses cell damage, oxidative stress and exerts an anti-inflammatory function resulting in suppression of tumor initiation. Accordingly, Nfe2l2−/− mice are more susceptible to chemical- and radiation-induced tumorigenesis and NRF2 activators were reported to reduce the burdens of gastric cancer (102), prostate cancer (103), breast cancer (104), colon cancer (105), bladder cancer (106) and liver cancer (107,108). However, strong NRF2 activation due to genetic alteration or other causes has been observed in numerous cancers and it was shown to enable cancer cells to adapt to a hostile microenvironment, modify their metabolism to a more anabolic metabolism that supports rapid cancer cell proliferation, tumor growth and invasion (30). Thus, once tumor has been established, NRF2 is a clear tumor promoter (109). Although, NRF2 activators have been suggested to provide chemoprotection, it seems that the adverse effects of NRF2 activation outweigh its benefits. It is therefore crucial to identify the turning point at which NRF2 changes from being a tumor suppressor to a tumor promoter.
NRF2-activating mutations, together with loss of function mutations in KEAP1 and Cul3, frequently occur in lung cancer (110), liver cancer (111), ovarian cancer (112), head and neck cancer, kidney cancer (113), breast cancer (114) and esophageal cancer (3). About 600 somatic mutations in the NFE2L2 gene have been reported in diverse cancers (115). Most of them affects in the DLG and ETGE motifs and abolish NRF2 interaction with KEAP1 (116), thus resulting in NRF2 stabilization and constitutive activation. In general, NRF2 activation is an adverse prognostic indicator.
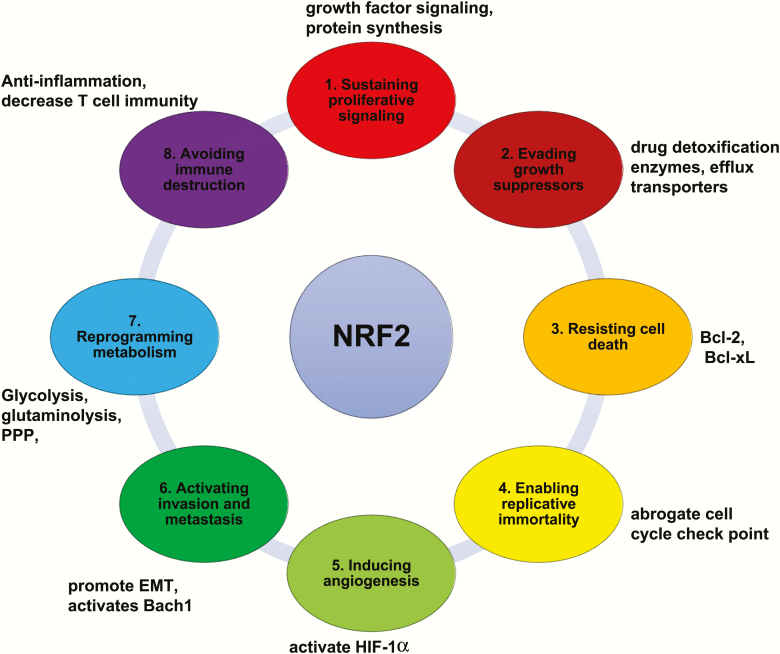
Obviously, NRF2 activation contributes to many of the hallmarks of cancer, a topic that was comprehensively and recently reviewed by Rojo de la Vega et al. (30). Here we just summarize some of the key cancer-supportive function of NRF2 (Figure 4). (i) NRF2 stimulates cancer cell proliferation through effects on epidermal growth factor receptor signaling and through upregulation of anabolic metabolism (35,117). Several oncogenic proteins, such as KRASG12D, BRAFV619E and MYC, increase NRF2 expression and antioxidant defenses (19). NRF2 also directly stimulates gene transcription of growth factors, resulting in activation of AKT signaling (38,118). In addition, NRF2 induces transcription of NOTCH1 to promote tissue regeneration and cancer cell proliferation (119). (ii) NRF2 helps cancer cells evade growth suppressors and senescence inducers by maintaining redox homeostasis and survival. Highly activated NRF2 confers resistance to chemotherapy by inducing expression of drug detoxification enzymes and efflux transporters (120). In lung cancer NRF2 also confers resistance to receptor tyrosine kinase (RTK) and mitogen-activated protein kinase (MAPK) inhibitors (121). (iii) NRF2 confers resistance to apoptosis by inducing expression of ROS‐scavenging enzymes and antiapoptotic B‐cell lymphoma 2 (Bcl‐2) family members, such as Bcl-2 and B-cell lymphoma-extra large (Bcl-xL) (122). By inducing key enzymes of glutathione synthesis and metabolism, NRF2 also confers resistance to ferroptosis (123). (iv) NRF2 induces MDM2 expression, which acts through p53-dependent and -independent mechanisms to abrogate checkpoints that prevent conversion of differentiated acinar cells to proliferative ductal progenitors in pancreatic cancers (124). In addition, NRF2 protects telomeres by reducing oxidative DNA damage and suppresses p53-induced senescence (30). (v) NRF2 induces angiogenesis and vasculogenesis the NRF2 targets NQO1 and TRX1 were reported to activate hypoxia-inducible factor 1-alpha (HIF-1α) in cancer cells (58,125). (vi) NRF2 activates invasion and metastasis by promoting epithelial-to-mesenchymal transition (EMT) via downregulation of E-cadherin expression (126). The NRF2 target HO-1 promotes lung cancer metastasis by stabilizing transcription factor BTB and CNC homology 1 (Bach1), an activator of prometastatic genes (127,128). (vii) NRF2 controls metabolic reprogramming and cellular energetics through induction of the anabolic and redox-maintaining genes discussed above. (viii) NRF2 helps cancer cells avoid immune destruction and its expression in immune cells inhibits the production of antitumor cytokines, such as IFN-γ (97,98). In addition, NRF2 activation drives macrophage polarization toward an M2-like tumor-promoting phenotype and can further enhance cancer cell EMT via vascular endothelial growth factor induction (129).
Figure 4.
NRF2 promotes and supports cancer hallmarks. NRF2 targets directly enhance cancer hallmarks or modulate oncogenic signaling to indirectly enhance cancer hallmarks.
NRF2 as a therapeutic target
Given its dual role in tumor initiation and progression, both NRF2 activator and inhibitory have been considered in cancer prevention and therapy. Unfortunately, it is difficult to specifically activate NRF2 in normal tissues, while using other drugs to inhibit its activity in established cancers.
In various animal models, NRF2 activators were found to enhance carcinogen detoxification and protect different tissues, especially the liver. Most NRF2 inducers are electrophiles or redox-active compounds that react with KEAP1 cysteine residues and stabilize NRF2. Several such NRF2 activators are at various stages of clinical development (109). Sulforaphane, present in cruciferous vegetables, such as broccoli, protects mice from tobacco-induced lung carcinogenesis, carcinogen-induced pancreatic cancer and skin cancer (2). Oltipraz is a synthetic NRF2 activator that was shown to inhibit the formation of various cancers in rodent models and can also attenuate the progression of non-alcoholic steatohepatitis (3). Dimethyl fumarate (DMF) is another NRF2 activator that has been approved by the Food and Drug Administration for the treatment of multiple sclerosis in 2013 and may also have tumor preventative activity (2). Of note, electrophilic NRF2 activators lack the specificity to target only KEAP1 and they can target most other nucleophilic cysteines residues in proteins, such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (130), protein kinase Cθ (PKCθ) (131) and IL-1 receptor-associated kinase 4 (IRAK4) (132). More selective drug candidates are non-electrophilic compounds that disrupt NRF2:KEAP1 protein–protein interactions (PPIs) or KEAP1:CUL3 PPIs, as well as molecules with KEAP1-independent modes of action (109). CPUY192018 is a high-affinity KEAP1 ligand that exerts anti-inflammatory activity in mouse model of ulcerative colitis (133,134). Another such compound is KI-696, with nanomolar affinity for KEAP1, which activates NRF2 and reduces ozone-induced lung inflammation in rats (135).
Pharmacological NRF2 inhibition has also emerged as a promising approach for cancer therapy, especially for NRF2-addicted cancers. However, only several NRF2 inhibitors have been developed so far and none of them has yielded strong and practicable results. Brusatol, isolated from the Brucea javanica plant, decreases NRF2 protein levels and attenuates target gene expression, thus enhancing the cytotoxic effect of chemotherapeutic agents such as cisplatin, paclitaxel, etoposide and 5-fluorouracil (2,136). However, brusatol is not a specifically NRF2 inhibitor and it seems to function as a global protein synthesis inhibitor (137). All-trans-retinoic acids (ATRAs) and other RA receptor α (RARα) and RXRα agonists were shown to inhibit basal and inducible NRF2 activity in both in vitro and in vivo models by forming an inactive NRF2:RARα complex (2). The flavonoid luteolin inhibits NRF2 activity by increasing NRF2 mRNA turnover and can sensitize cancer cells toward chemotherapeutic agents (138). Interestingly, luteolin also has antioxidant activity by activating NRF2-HO-1 signaling in RAW 264.7 cells and human colorectal cancer HCT116 cells (139,140). Halofuginone, a bioactive component of the traditional Chinese medicinal herb Dichroa febrifuga, decreases NRF2 protein synthesis by inhibiting prolyl-tRNA synthetase and enhances the effects of conventional anticancer drugs, such as cisplatin or doxorubicin, in xenograft tumor models (3,141). Obviously, halofuginone is not a specific NRF2 inhibitor and it also inhibits collagen type-I synthesis (142). While direct pharmacological NRF2 inhibition is challenging, alternative approaches for NRF2 suppression include targeting proteins that confer dependence on NRF2 and represent specific vulnerabilities that NRF2 activation impose on cancer cells. NRF2-addicted lung cancer growth and proliferation depend on increased glutaminolysis and decreased sulfitolysis (45,143).Correspondingly, glutaminase inhibitor CB-839 suppresses the growth of various cancers harboring KEAP1 or NRF2 mutations, including melanoma, lung cancer, colon cancer and urinary tract cancers (43,45). NR0B1 is an atypical orphan nuclear receptor that participates in a multimeric protein complex to regulate transcription NRF2-related genes. The NR0B1 ligands BPK-26 and BKP29 block the NRF2-dependent cell growth and proliferation in KEAP1-mutated non-small-cell lung cancers cells (144).
Given their low specific, most NRF2 activators and inhibitors produce high off-target toxic effects that limit their clinical applicability. Future studies should focus on better understanding of NRF2 activation and on the development of much more specific NRF2 targeting agents. The context in which NRF2 is activated is also important in terms of therapeutic development. The impact of NRF2-activating genetic mutations in cancer cells is different from that caused by pharmacological activation of NRF2 in normal cells. KEAP1 and NFE2L2 mutations, especially in the background of RAS and RAF mutations lead to unrestrained NRF2 activity, while pharmacological NRF2 activators cause much more restrained and transient NRF2 activation. Nonetheless, it is important to determine potential cancer risk posed by NRF2 activators, especially in high risk individuals, who already harbor premalignant lesions, whose progression is likely to accelerate on NRF2 activation. Although meta-analysis of a phase III trial of DMF has shown no difference in the cancer rate between the placebo and DMF-treated patients with multiple sclerosis (145), the result could be due to the inhibition of GAPDH by DMF (130) and may be entirely different in high risk individuals who are the target population for chemoprevention.
Concluding remarks and a future perspective
Overall, the role of NRF2 activation in cancer cells and the tumor microenvironment is complex, cell type- and context-dependent. Much more research is needed. It is quite clear that NRF2 activation has a clear strong protumorigenic effect manifested at many different levels, of which the reprogramming of cancer cell metabolism is likely to be of foremost importance. Undoubtedly, effective and specific NRF2 inhibition in established cancers is likely to slow down malignant progression. However, systemic NRF2 inhibition is bound to mouse susceptibility to toxic challenges. The major future challenge is to find better ways of inhibiting NRF2 activation only in cancer cells without affecting its essential activation in response to toxic challenges in tissues, such as the liver and kidney.
Acknowledgements
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- ATP
adenosine triphosphate
- ER
endoplasmic reticulum
- IL
interleukin
- IFN
interferon
- KEAP1
Kelch‐like‐ECH‐associated protein 1
- Neh
NRF2‐ECH homology
- NFE2
nuclear factor erythroid-derived 2
- NFE2L2
nuclear factor, enrythroid 2 like 2
- NF-κB
nuclear factor kappa B
- NRF2
nuclear factor erythroid 2-related factor 2
- ROS
reactive oxygen species
- TNF
tumor necrosis factor
Funding
F.H. was supported by Lilly innovation fellowship award. Research was supported by grants from the National Institutes of Health and C3 Pedal the Cause grant to M.K., who is an American Cancer Society Research Professor and holder of the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases. We apologize to the authors whose work could not be cited due to space constraints.
References
- 1. Canning P., et al. (2015) Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med., 88(Pt B), 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jaramillo M.C., et al. (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev., 27, 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taguchi K., et al. (2017) The KEAP1-NRF2 system in cancer. Front. Oncol., 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McMahon M., et al. (2003) Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem., 278, 21592–21600. [DOI] [PubMed] [Google Scholar]
- 5. McMahon M., et al. (2004) Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem., 279, 31556–31567. [DOI] [PubMed] [Google Scholar]
- 6. Rada P., et al. (2011) SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol., 31, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chowdhry S., et al. (2013) Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene, 32, 3765–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang H., et al. (2013) RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res., 73, 3097–3108. [DOI] [PubMed] [Google Scholar]
- 9. Wu K.C., et al. (2012) Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS One, 7, e39006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X.H., et al. (2017) High fat diet-induced hepatic 18-carbon fatty acids accumulation up-regulates CYP2A5/CYP2A6 via NF-E2-related factor 2. Front. Pharmacol., 8, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaw P., et al. (2020) Nrf2-ARE signaling in cellular protection: mechanism of action and the regulatory mechanisms. J. Cell. Physiol., 235, 3119–3130. [DOI] [PubMed] [Google Scholar]
- 12. Zhang D.D., et al. (2003) Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol., 23, 8137–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McMahon M., et al. (2010) Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA, 107, 18838–18843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y., et al. (2012) Mechanism of chemical activation of Nrf2. PLoS One, 7, e35122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baird L., et al. (2013) Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc. Natl. Acad. Sci. USA, 110, 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Itoh K., et al. (2003) Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells, 8, 379–391. [DOI] [PubMed] [Google Scholar]
- 17. Zhang D.D., et al. (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol., 24, 10941–10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tonelli C., et al. (2018) Transcriptional regulation by Nrf2. Antioxid. Redox Signal., 29, 1727–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DeNicola G.M., et al. (2011) Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature, 475, 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taniguchi K., et al. (2016) p62/SQSTM1-Dr. Jekyll and Mr. Hyde that prevents oxidative stress but promotes liver cancer. FEBS Lett., 590, 2375–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Komatsu M., et al. (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol., 12, 213–223. [DOI] [PubMed] [Google Scholar]
- 22. Umemura A., et al. (2016) p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell, 29, 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hast B.E., et al. (2013) Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res., 73, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Camp N.D., et al. (2012) Wilms tumor gene on X chromosome (WTX) inhibits degradation of NRF2 protein through competitive binding to KEAP1 protein. J. Biol. Chem., 287, 6539–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma J., et al. (2012) PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol. Cell. Biol., 32, 1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen W., et al. (2009) Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell, 34, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gorrini C., et al. (2013) BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J. Exp. Med., 210, 1529–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kang H.J., et al. (2010) CR6-interacting factor 1 (CRIF1) regulates NF-E2-related factor 2 (NRF2) protein stability by proteasome-mediated degradation. J. Biol. Chem., 285, 21258–21268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lo J.Y., et al. (2017) WDR23 regulates NRF2 independently of KEAP1. PLoS Genet., 13, e1006762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rojo de la Vega M. et al. (2018) NRF2 and the hallmarks of cancer. Cancer Cell, 34, 21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu T., et al. (2014) Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev., 28, 708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanghvi V.R., et al. (2019) The oncogenic action of NRF2 depends on de-glycation by fructosamine-3-kinase. Cell, 178, 807–819.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 34. Fu J., et al. (2019) Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J. Biol. Chem., 294, 327–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitsuishi Y., et al. (2012) Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell, 22, 66–79. [DOI] [PubMed] [Google Scholar]
- 36. Chorley B.N., et al. (2012) Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res., 40, 7416–7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malhotra D., et al. (2010) Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res., 38, 5718–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He F. et al. (2020) NRF2 activates growth factor genes and downstream AKT signaling to induce mouse and human hepatomegaly. J. Hepatol., in press. doi: 10.1016/j.jhep.2020.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uruno A., et al. (2016) Nrf2-mediated regulation of skeletal muscle glycogen metabolism. Mol. Cell. Biol., 36, 1655–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu X., et al. (2017) Non-metabolic functions of glycolytic enzymes in tumorigenesis. Oncogene, 36, 2629–2636. [DOI] [PubMed] [Google Scholar]
- 41. Yates M.S., et al. (2009) Genetic versus chemoprotective activation of Nrf2 signaling: overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis, 30, 1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pi J., et al. (2010) Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem., 285, 9292–9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sayin V.I. et al. (2017) Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer. Elife, 6, e28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lunt S.Y., et al. (2011) Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol., 27, 441–464. [DOI] [PubMed] [Google Scholar]
- 45. Romero R., et al. (2017) Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med., 23, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. DeNicola G.M., et al. (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet., 47, 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacLeod A.K., et al. (2009) Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis, 30, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Agyeman A.S., et al. (2012) Transcriptomic and proteomic profiling of KEAP1 disrupted and sulforaphane-treated human breast epithelial cells reveals common expression profiles. Breast Cancer Res. Treat., 132, 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kapeta S., et al. (2010) Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J. Biol. Chem., 285, 8171–8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pajares M., et al. (2016) Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy, 12, 1902–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mukaigasa K., et al. (2018) Nrf2 activation attenuates genetic endoplasmic reticulum stress induced by a mutation in the phosphomannomutase 2 gene in zebrafish. Proc. Natl. Acad. Sci. USA, 115, 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang D., et al. (2015) Immunosuppression associated with chronic inflammation in the tumor microenvironment. Carcinogenesis, 36, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Karin M., et al. (2016) Reparative inflammation takes charge of tissue regeneration. Nature, 529, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grivennikov S.I., et al. (2010) Immunity, inflammation, and cancer. Cell, 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Groeger A.L., et al. (2010) Cyclooxygenase-2 generates anti-inflammatory mediators from omega-3 fatty acids. Nat. Chem. Biol., 6, 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kobayashi E., et al. (2013) Roles Nrf2 plays in myeloid cells and related disorders. Oxid. Med. Cell. Longev., 2013, 529219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Forrester S.J., et al. (2018) Reactive oxygen species in metabolic and inflammatory signaling. Circ. Res., 122, 877–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iizuka T., et al. (2005) Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells, 10, 1113–1125. [DOI] [PubMed] [Google Scholar]
- 59. Reddy N.M., et al. (2009) Disruption of Nrf2 impairs the resolution of hyperoxia-induced acute lung injury and inflammation in mice. J. Immunol., 182, 7264–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tang W., et al. (2014) Role of Nrf2 in chronic liver disease. World J. Gastroenterol., 20, 13079–13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yoh K., et al. (2001) Nrf2-deficient female mice develop lupus-like autoimmune nephritis. Kidney Int., 60, 1343–1353. [DOI] [PubMed] [Google Scholar]
- 62. Ma Q., et al. (2006) Multiorgan autoimmune inflammation, enhanced lymphoproliferation, and impaired homeostasis of reactive oxygen species in mice lacking the antioxidant-activated transcription factor Nrf2. Am. J. Pathol., 168, 1960–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Osburn W.O., et al. (2008) Genetic or pharmacologic amplification of nrf2 signaling inhibits acute inflammatory liver injury in mice. Toxicol. Sci., 104, 218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kong X., et al. (2011) Enhancing Nrf2 pathway by disruption of Keap1 in myeloid leukocytes protects against sepsis. Am. J. Respir. Crit. Care Med., 184, 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Taniguchi K., et al. (2018) NF-kappaB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol., 18, 309–324. [DOI] [PubMed] [Google Scholar]
- 66. Thimmulappa R.K., et al. (2006) Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Invest., 116, 984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lin W., et al. (2008) Sulforaphane suppressed LPS-induced inflammation in mouse peritoneal macrophages through Nrf2 dependent pathway. Biochem. Pharmacol., 76, 967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mills E.L., et al. (2018) Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature, 556, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Burgener A.V., et al. (2019) SDHA gain-of-function engages inflammatory mitochondrial retrograde signaling via KEAP1-Nrf2. Nat. Immunol., 20, 1311–1321. [DOI] [PubMed] [Google Scholar]
- 70. Olagnier D., et al. (2018) Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun., 9, 3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Page A., et al. (2014) Marburgvirus hijacks nrf2-dependent pathway by targeting nrf2-negative regulator keap1. Cell Rep., 6, 1026–1036. [DOI] [PubMed] [Google Scholar]
- 72. Saddawi-Konefka R., et al. (2016) Nrf2 induces IL-17D to mediate tumor and virus surveillance. Cell Rep., 16, 2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Seelige R., et al. (2018) Interleukin-17D and Nrf2 mediate initial innate immune cell recruitment and restrict MCMV infection. Sci. Rep., 8, 13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Patra U., et al. (2019) RA-839, a selective agonist of Nrf2/ARE pathway, exerts potent anti-rotaviral efficacy in vitro. Antiviral Res., 161, 53–62. [DOI] [PubMed] [Google Scholar]
- 75. Wyler E., et al. (2019) Single-cell RNA-sequencing of herpes simplex virus 1-infected cells connects NRF2 activation to an antiviral program. Nat. Commun., 10, 4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kosmider B., et al. (2012) Nrf2 protects human alveolar epithelial cells against injury induced by influenza A virus. Respir. Res., 13, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hagen T.M., et al. (1994) Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA, 91, 12808–12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seldon M.P., et al. (2007) Heme oxygenase-1 inhibits the expression of adhesion molecules associated with endothelial cell activation via inhibition of NF-kappaB RelA phosphorylation at serine 276. J. Immunol., 179, 7840–7851. [DOI] [PubMed] [Google Scholar]
- 79. Rushworth S.A., et al. (2008) Lipopolysaccharide-induced expression of NAD(P)H:quinone oxidoreductase 1 and heme oxygenase-1 protects against excessive inflammatory responses in human monocytes. J. Immunol., 181, 6730–6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Iskander K., et al. (2006) NQO1 and NQO2 regulation of humoral immunity and autoimmunity. J. Biol. Chem., 281, 30917–30924. [DOI] [PubMed] [Google Scholar]
- 81. Smith M.T., et al. (2001) Low NAD(P)H:quinone oxidoreductase 1 activity is associated with increased risk of acute leukemia in adults. Blood, 97, 1422–1426. [DOI] [PubMed] [Google Scholar]
- 82. Stavropoulou C., et al. (2011) The C609T inborn polymorphism in NAD(P)H:quinone oxidoreductase 1 is associated with susceptibility to multiple sclerosis and affects the risk of development of the primary progressive form of the disease. Free Radic. Biol. Med., 51, 713–718. [DOI] [PubMed] [Google Scholar]
- 83. Jain A., et al. (2010) p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem., 285, 22576–22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhong Z., et al. (et al. ) NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell, 164, 896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hoetzenecker W., et al. (2011) ROS-induced ATF3 causes susceptibility to secondary infections during sepsis-associated immunosuppression. Nat. Med., 18, 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bambouskova M., et al. (2018) Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta-ATF3 inflammatory axis. Nature, 556, 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kobayashi E.H., et al. (2016) Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun., 7, 11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wruck C.J., et al. (2011) Nrf2 induces interleukin-6 (IL-6) expression via an antioxidant response element within the IL-6 promoter. J. Biol. Chem., 286, 4493–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pareek T.K., et al. (2011) Triterpenoid modulation of IL-17 and Nrf-2 expression ameliorates neuroinflammation and promotes remyelination in autoimmune encephalomyelitis. Sci. Rep., 1, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Harvey C.J., et al. (2011) Targeting Nrf2 signaling improves bacterial clearance by alveolar macrophages in patients with COPD and in a mouse model. Sci. Transl. Med., 3, 78ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhong Z., et al. (2018) New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature, 560, 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhao C., et al. (2014) Nuclear factor E2-related factor-2 (Nrf2) is required for NLRP3 and AIM2 inflammasome activation. J. Biol. Chem., 289, 17020–17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gajewski T.F., et al. (2013) Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol., 14, 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hou J., et al. (2020) The immunobiology of hepatocellular carcinoma in humans and mice: basic concepts and therapeutic implications. J. Hepatol., 72, 167–182. [DOI] [PubMed] [Google Scholar]
- 95. Sha L.K., et al. (2015) Loss of Nrf2 in bone marrow-derived macrophages impairs antigen-driven CD8(+) T cell function by limiting GSH and Cys availability. Free Radic. Biol. Med., 83, 77–88. [DOI] [PubMed] [Google Scholar]
- 96. Ohl K., et al. (2018) Nrf2 is a central regulator of metabolic reprogramming of myeloid-derived suppressor cells in steady state and sepsis. Front. Immunol., 9, 1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Suzuki T. et al. (2017) Systemic activation of NRF2 alleviates lethal autoimmune inflammation in Scurfy mice. Mol. Cell. Biol., 37, e00063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yarosz E.L., et al. (2018) The role of reactive oxygen species in regulating T cell-mediated immunity and disease. Immune Netw., 18, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Maj T., et al. (2017) Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol., 18, 1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhong Z., et al. (2016) Autophagy, inflammation, and immunity: a troika governing cancer and its treatment. Cell, 166, 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jo Y. et al. (2016) Nrf2 expression is upregulated in tumor infiltrating T cells and induces T cell anergy. J. Immunol., 96(suppl 1), 143.15. [Google Scholar]
- 102. Ramos-Gomez M., et al. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. USA, 98, 3410–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Frohlich D.A., et al. (2008) The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene, 27, 4353–4362. [DOI] [PubMed] [Google Scholar]
- 104. Kim E.H., et al. (2012) CDDO-methyl ester delays breast cancer development in BRCA1-mutated mice. Cancer Prev. Res. (Phila), 5, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Long M., et al. (2015) Nrf2-dependent suppression of azoxymethane/dextran sulfate sodium-induced colon carcinogenesis by the cinnamon-derived dietary factor cinnamaldehyde. Cancer Prev. Res. (Phila), 8, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Iida K., et al. (2007) Nrf2 and p53 cooperatively protect against BBN-induced urinary bladder carcinogenesis. Carcinogenesis, 28, 2398–2403. [DOI] [PubMed] [Google Scholar]
- 107. Johnson N.M., et al. (2014) Complete protection against aflatoxin B(1)-induced liver cancer with a triterpenoid: DNA adduct dosimetry, molecular signature, and genotoxicity threshold. Cancer Prev. Res. (Phila), 7, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kitamura Y., et al. (2007) Increased susceptibility to hepatocarcinogenicity of Nrf2-deficient mice exposed to 2-amino-3-methylimidazo[4,5-f]quinoline. Cancer Sci., 98, 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cuadrado A., et al. (2019) Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov., 18, 295–317. [DOI] [PubMed] [Google Scholar]
- 110. Padmanabhan B., et al. (2006) Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell, 21, 689–700. [DOI] [PubMed] [Google Scholar]
- 111. Eichenmüller M., et al. (2014) The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J. Hepatol., 61, 1312–1320. [DOI] [PubMed] [Google Scholar]
- 112. Konstantinopoulos P.A., et al. (2011) Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res., 71, 5081–5089. [DOI] [PubMed] [Google Scholar]
- 113. Ooi A., et al. (2013) CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res., 73, 2044–2051. [DOI] [PubMed] [Google Scholar]
- 114. Nioi P., et al. (2007) A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem. Biophys. Res. Commun., 362, 816–821. [DOI] [PubMed] [Google Scholar]
- 115. Gao J., et al. (2017) 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med., 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zehir A., et al. (2017) Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med., 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chio I.I.C., et al. (2016) NRF2 promotes tumor maintenance by modulating mRNA translation in pancreatic cancer. Cell, 166, 963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Liu D., et al. (2016) Activation of AKT pathway by Nrf2/PDGFA feedback loop contributes to HCC progression. Oncotarget, 7, 65389–65402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wakabayashi N., et al. (2010) Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci. Signal., 3, ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Maher J.M., et al. (2005) Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab. Dispos., 33, 956–962. [DOI] [PubMed] [Google Scholar]
- 121. Krall E.B. et al. (2017) KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. Elife, 6, e18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Niture S.K., et al. (2012) Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J. Biol. Chem., 287, 9873–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Fan Z., et al. (2017) Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis, 6, e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Todoric J., et al. (2017) Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell, 32, 824–839.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Malec V., et al. (2010) HIF-1 alpha signaling is augmented during intermittent hypoxia by induction of the Nrf2 pathway in NOX1-expressing adenocarcinoma A549 cells. Free Radic. Biol. Med., 48, 1626–1635. [DOI] [PubMed] [Google Scholar]
- 126. Arfmann-Knübel S., et al. (2015) The crosstalk between Nrf2 and TGF-β1 in the epithelial-mesenchymal transition of pancreatic duct epithelial cells. PLoS One, 10, e0132978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lignitto L., et al. (2019) Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell, 178, 316–329.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wiel C., et al. (2019) BACH1 stabilization by antioxidants stimulates lung cancer metastasis. Cell, 178, 330–345.e22. [DOI] [PubMed] [Google Scholar]
- 129. Feng R., et al. (2018) Nrf2 activation drive macrophages polarization and cancer cell epithelial-mesenchymal transition during interaction. Cell Commun. Signal., 16, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kornberg M.D., et al. (2018) Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science, 360, 449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Blewett M.M., et al. (2016) Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci. Signal., 9, rs10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zaro B.W., et al. (2019) Dimethyl fumarate disrupts human innate immune signaling by targeting the IRAK4-MyD88 complex. J. Immunol., 202, 2737–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lu M.C., et al. (2016) An inhibitor of the Keap1-Nrf2 protein-protein interaction protects NCM460 colonic cells and alleviates experimental colitis. Sci. Rep., 6, 26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lu M.C., et al. (2019) CPUY192018, a potent inhibitor of the Keap1-Nrf2 protein-protein interaction, alleviates renal inflammation in mice by restricting oxidative stress and NF-κB activation. Redox Biol., 26, 101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Davies T.G., et al. (2016) Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein-protein interaction with high cell potency identified by fragment-based discovery. J. Med. Chem., 59, 3991–4006. [DOI] [PubMed] [Google Scholar]
- 136. Ren D., et al. (2011) Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA, 108, 1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Harder B., et al. (2017) Brusatol overcomes chemoresistance through inhibition of protein translation. Mol. Carcinog., 56, 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Tang X., et al. (2011) Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic. Biol. Med., 50, 1599–1609. [DOI] [PubMed] [Google Scholar]
- 139. Song Y.S., et al. (2014) Luteolin and luteolin-7-O-glucoside strengthen antioxidative potential through the modulation of Nrf2/MAPK mediated HO-1 signaling cascade in RAW 264.7 cells. Food Chem. Toxicol., 65, 70–75. [DOI] [PubMed] [Google Scholar]
- 140. Zuo Q., et al. (2018) The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. J. Cell. Biochem., 119, 9573–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Tsuchida K., et al. (2017) Halofuginone enhances the chemo-sensitivity of cancer cells by suppressing NRF2 accumulation. Free Radic. Biol. Med., 103, 236–247. [DOI] [PubMed] [Google Scholar]
- 142. Pines M., et al. (1997) Halofuginone, a specific inhibitor of collagen type I synthesis, prevents dimethylnitrosamine-induced liver cirrhosis. J. Hepatol., 27, 391–398. [DOI] [PubMed] [Google Scholar]
- 143. Kang Y.P. et al. (2019) Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer. Elife, 8, e45572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bar-Peled L., et al. (2017) Chemical proteomics identifies druggable vulnerabilities in a genetically defined cancer. Cell, 171, 696–709.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Pakpoor J., et al. (2015) No evidence for higher risk of cancer in patients with multiple sclerosis taking cladribine. Neurol. Neuroimmunol. Neuroinflamm., 2, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]