Abstract
This study employed Bacillus spp. with α-amylase production isolated from Malaysian hot spring for domestic kitchen food waste treatment contained grains, vegetables, chicken and tuna that mimic the food waste discharge from domestic kitchens in Malaysian household. Results showed that Bacillus licheniformis HULUB1 and Bacillus subtilis SUNGB2 possess excellent amylolytic properties. Highest α-amylase activity was obtained when both isolates were cultivated at pH 6.0 and 65 °C with concentrations of 18.15 U/mL for HULUB1 and 22.14 U/mL for SUNGB2. Stability of α-amylase with significant levels of enzyme activity were recorded at 55–85 °C and pH 5.0–9.0. The extracted mixed α-amylase of HULUB1 and SUNGB2 showed greatest reduction were achieved at day 12 with 45% ± 0.03 solid content at 65 °C. While the mixed culture of HULUB1 and SUNGB2 displayed an enhanced effect on the food waste contents reduction with 43% ± 0.02 solid content at 45 °C after day 12. The findings showed that the combination of the two Bacillus spp. isolates possessed degradation of food wastes at faster rate than α-amylase. It was also pointed out that the standard food waste (SFW) and the treatment process assimilated for this study was suitable for the growth of Bacillus spp.
Keywords: Food science, Microbiology, α-amylase, Bacillus spp., Thermophilic, Biodegradation, Food waste
Food Science; Microbiology; α-amylase; Bacillus spp.; Thermophilic; Biodegradation; Food Waste
1. Introduction
Hot springs is an environment with different physical, chemical and nutritional properties that possesses biodiversity of microbes including thermostable bacteria [Chan et al., 2017,Msarah et al., 2018]. These sites include thermal fluids, microbial mats and sediments with potential microbes producing industrially important thermostable enzymes. The microbial populations existing in such environment produce valuable sources for numerous biotechnological products and applications (Sayeh et al., 2010). The importance of thermophilic Bacillus has increased because of their biotechnological significance as sources of thermostable enzymes. The enzymes from thermophilic microorganisms is extremely in high demand for industry such as biocatalysts, biotransformation and biodegradation. Thermostable enzymes are typically hard to denature at elevated temperature (>60 °C) while by increasing the temperature will disrupt the shape of the active site, which will reduce its activity of the enzyme (Brock, 2001). Enzymes from thermophilic bacteria offer several advantages compared to enzymes from mesophilic bacteria for examples longer useful shelf life, less contamination problems because the enzyme reaction takes place at high temperature and increased chemical resistance (Drejer et al., 2018).
Extracellular enzymes from thermophilic microorganisms are more resistant to extreme pH and chemical reagents in comparison to mesophilic microorganisms, which inspired an interest in the exploration of thermophiles. Amylases are enzymes that have attracted the interest of industries for their wide applications especially in food industries. This group of enzymes contribute to approximately 25% of the world enzyme market with great significance for biotechnology (Arikan, 2008). α-amylase has increased in demand for industrial applications due to its crucial role in starch hydrolysis processes utilised in food, paper, brewing and textile industries (Hmidet et al., 2009). According to Tanyildizi et al. (2005), microbial amylases retain a greater stability in comparison to animal and plant amylases. Bacteria that are renowned as an outstanding producer of thermostable α-amylase for examples are B. amyloliquefaciens, B. licheniformis, B. subtilis and B. stearothermophilus (Asghar et al., 2007; Prakash and Jaiswal, 2009).
Food waste and wastage is a global issue. According to a report published by the Food and Agricultural Organisation FAO (2015), about one-third of the global food production for human consumption (nearly 1.3 billion tons) is wasted annually at several stages, ranging from the initial production stage to the supply chain and final household consumption. Yang et al. (2016) highlighted that only 33% of food is consumed in the Southeast Asia region, while the remaining food is wasted. In Malaysia, the average daily food wastage from households is around 0.5–0.8 kg per day of leftover food and as the population of Malaysia is expected to reach 33.4 million by the year 2020, it is also expected that the problems associated with food wastes will equally increase in correspondence to the population growth and economic development (Bong et al. (2017). Food waste treatments using multiple microorganisms (bacterial consortium) by employing mesophilic aerobic and anaerobic fermentation performed by Kiran et al. (2014) and Kibler et al. (2018) showed that microbes can consume and decrease/biodegrade the total amount of food waste discharged from both commercial and domestic sources. An et al. (2018); Awasthi et al. (2017) and Awasthi et al. (2018) also acknowledged Bacillus spp. as a fast-growing bacterium that possessed strong extracellular enzymes which show different effects on hydrolysing organic compounds and decomposed of the food wastes facilitated by the degradative enzyme activity.
Food waste is organic waste discharged which can be found in domestic or/and commercial kitchens, restaurants and food processing factories. Microorganisms can consume and decrease food wastes because they contain various organic compounds. The most employed for treatment of food waste is bacteria owing to the natural presence of several bacteria in food wastes. Thermophilic enzymes are essential in bioremediation because treatment of wastes with higher temperatures have the advantage of increased solubility of the substrates and amylases are among those thermophilic enzymes with a great significance in modern industry. Therefore, this research is conducted to optimise the growth of the isolated thermophilic Bacillus spp. from selected Malaysian hot springs, to obtain α-amylase with a wide range of pH and temperature and to assess the ability of the isolated thermophilic Bacillus and the extracted α-amylase by these bacteria for biodegradation of food waste by using standardise food waste formula contained grains (cooked rice), vegetables (potatoes, onion), fruits (apples, oranges), chicken and tuna as food wastes with various organic compounds (carbohydrates, lipids, and proteins), that mimic the food waste discharge from domestic kitchens in Malaysian households.
2. Methodology
2.1. Identification of bacterial isolates from hot spring
Water samples were sourced from the Dusun Tua Hot Spring, Hulu Langat, Selangor (3°08′21.2″N101°50′10.9″E) and Sungai Klah Hot Spring, Perak (3°59′54.6″N 101°23′28.9″E). The physiochemical characterisation of water measured on site was carried out using a YSI 556 multi-probe system MPS. Isolation of bacteria was performed according to Adiguzel et al. (2009) through serial dilution method (10-fold), to estimate the number of bacterial colonies. One mL of hot spring water was added to 9 mL of sterile distilled water and then mixed by gentle vortexing. For each water sample, 10 mL were filtered through membrane filters (0.22 mm Millipore Corporation, Bedford). Subsequently, the filter paper was placed on Thermus agar enriched with NaCl (0.5%), beef extract (0.4%), peptone (0.5%) agar (2%), and yeast extract (0.2%) (Sikdar et al., 2015) to confirm the isolates are thermophiles. The agar was incubated at 45 °C for 24 to 48, and the purity of the colonies was checked microscopically.
Both biochemical and physiological tests were employed to identify and characterise the thermophiles following the techniques prescribed by the Bergey's Manual of Systematic Bacteriology (BMSB) (Kersters and Vancanneyt, 2005). The thermophilic strains were identified by relying on the specific phenotypic features of the Bacillus genus, as well as on the phylogenetic analysis of the 16S rDNA sequence. The process was performed using the TE boil extraction method (T method) and a slightly modified extraction method for bacterial DNA as described by Li et al. (2003).
A 1 mL bacterial cell (from the overnight culture at 45 °C on NB) was transferred to 1.5 mL micro-centrifuge tube and centrifuged at 10000 g for 1 min to obtain the pellet. The pellet was resuspended in TE buffer (200 μL) and the pH of the suspension was adjusted with conc. HCl to pH 8.0. The final volume was made up to 1 L with deionised water then 10 mmol/L Tris-HCl and 1 mmol/L added and mixed on a vortex mixer (Yang et al., 2008). The suspension was placed in a boiling water bath at 100 °C for 1 min, then freeze at -70 °C for 3 min, 100 °C for 2 min, -70 °C for 3 min, 100 °C for 2 min, and lastly -70 °C for 3 min before being centrifuged for 5 min at 9449 rpm. An aliquot (100 μL) of the clear supernatant for PCR testing was collected in a sterile tube and kept at -20 °C until further use.
The amplification of the 16S rRNA gene sequences was performed using the PROMEGA Go Taq® Green Mix, 2X. The process was performed using the universal bacterial primers: 8–27 (5′AGAGTTTGATCCTGGCTCAG3′) and 1492R (5′CTACGGCTACCTTGTTACGA3′) (Piterina et al., 2010). QIAEX® II Gel Extraction Kit (150) (QIAGEN) was used based on the recommendations by the manufacturer to purify the PCR products, while the NanoDrop Thermo Scientific™ NanoDrop-2000 full-spectrum U.V/ris spectrophotometer was used to determine the concentration of the purified DNA. The sequencing of the samples was done using the Biosystems Genetic Analyser BigDye® Terminator V3.1 cycle sequencing kit chemistry as earlier described by Sanger (1981). The extracted 16S rRNA gene sequence was analysed for bacterial identification using a Basic Local Alignment Search Tool (BLAST) which is available at http://blast.ncbi.nlm.nih.gov/Blast.cgi. The isolated sequences from the bacterial strains were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank) and were assigned unique accession numbers.
2.2. Carbohydrate utilisation assay
The experiments to determine the sugar utilisation capability of the isolates were performed using M9 Minimal medium enriched with vitamins and trace elements, in addition to diverse concentrations (0.5%, 1%, 1.5%) (w/v) of sugars (glucose, sucrose, starch) which served as the sole carbon source at pH 7.0. A 24 h overnight standardised culture of Bacillus isolate was added and then incubated at 45 °C for 48 h. Minimal medium was prepared by combining 250 mL of M9 salts, 20 mL of diverse concentrations (0.5%, 1%, 1.5%) (w/v) of sugars used (glucose, sucrose, starch) solution, 2.1 mL of trace minerals and 2.0 mL of vitamins containing only thiamine and biotin. The volume was brought to 1 L with distilled water, followed by pH adjustment to 7.0. The prepared medium was sterilised by filtration through a 0.45 µm pore-size filter.
2.3. Qualitative screening for α-amylase
In the qualitative test, the thermophilic isolates were inoculated in a medium containing 1 g/L yeast extract, 0.1 g/L MgSO4.7H2O, 7 g/L K2HPO4, 2 g/L KH2PO4, 1 g/L (NH4).2SO4, 5 g/L NaCl, 5 g/L starch and 15 g/L bacteriological agar. All ingredients were dissolved in deionised water and sterilised at 121 °C for 15 min. The plate was placed in the incubator and allowed for 24–48 h at 45 °C before pouring the iodine solution (1 g iodine, 2 g KI and 300 mL of distilled water) in the plates. The clear white zones against a blue background around the colonies indicated a positive result for amylase activity (Bragger et al., 1989).
2.4. Quantitative screening for α-amylase
The medium for the quantitative screening contains (g/L): soluble starch (10), peptone (20), MgSO4.7H2O (1.0), Na2HPO4 (3), FeSO4 (0.3) and NaCl (0.1), medium was sterilised then left to cool until 27 °C. Various culture condition parameters were assessed, a) the initial medium pH (3.0, 4.0,5.0, 6.0, 7.0 and 8.0) and b) the incubation temperatures (37, 40, 45 and 55 °C) in order to increase the amylase production by the isolated thermophilic Bacillus. One mL of 24 h culture (standardised 0.5 McFarland) was inoculated to 99 mL of cultivation medium in a 250 mL volumetric flask and allowed to incubate for 48 h at 45 °C with mild agitation (150 rpm). Every 6 h intervals incubation, the culture (100 mL) were harvested and filtered through Whatman No.1 filter paper. The obtained filtrate served as the source of enzyme during the amylase activity assay. The supernatant was collected in tubes and the amylase activity was estimated (Ajayi and Fagade, 2006).
The crude enzyme was further purified by ammonium sulphate precipitation (Divakaran et al., 2011). The modification of the DNS method according to Miller (1959) was applied to assess the amylase activity based on the release of reducing sugars from soluble starch with glucose as standard. The absorbance of the mixture was determined using Biochrom Libra S12 spectrophotometer Biochrom Ltd., Cambridge, England at 540 nm. One international unit (IU) of amylase activity was considered as the required amount of the enzyme to release 1 μmol of reducing sugar from the sugar source within one minute under standard experimental conditions. The α-amylase was partially purified by precipitation in ammonium sulphate. For the partial purification process, a 6 days broth culture of the organism was grown at the optimal growth conditions, then, centrifuged at 4 °C for 30 min at 7319 rpm. The obtained supernatant was then precipitated with saturated ammonium sulphate (80% saturation) before resuspending the precipitates in a pH 6.0 sodium phosphate buffer (20 mM), followed by the characterisation of the partially purified enzyme (Konsoula and Liakopoulou-Kyriakides, 2007). Enzymatic activity was carried out via conventional assay using Bradford method. Two parameters that affect the α-amylase activity and stability (the pH of the medium and the incubation temperature) were investigated and all assays were performed in triplicate.
2.4.1. Effect of temperature on the activity and stability of the enzyme
The partially purified enzymes were evaluated for enzymatic activity via incubation in 1% (w/v) starch solution at different temperatures (35, 45, 55, 65, 75 and 85 °C). The 1% starch solution was prepared in 20 mM sodium phosphate buffer (pH 6.0) (Behal et al., 2006). The incubation period of 10 min was allowed before assaying for the thermostability of the enzymes (Carvalho et al., 2008). Reaction without α-amylase was used as negative control. All experiments were carried out in triplicates in at least three different occasions.
2.4.2. Effect of pH on the activity of the enzyme
The effect of pH on the activity of the enzymes was assayed at various pH levels (3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0 and 11.0) using 1% starch as the carbon source. This assay was performed using different buffers, including acetate buffer (20 mM; pH 3.0–5.0), sodium phosphate buffer (20 mM; pH 5.0–8.0), and Tris/HCl (20 mM; pH 8.0–11.0) (Reyed, 2007). The incubation of the enzyme in each of these buffers lasted for 1 h at 55 °C before the assay for enzyme pH stability (Carvalho et al., 2008). Reaction without α-amylase was used as negative control. All experiments were carried out in triplicates in at least three different occasions.
2.4.3. Specific activity and protein determination
The Bradford method of protein assay was used to determine the concentration of protein at 595 nm (Bradford, 1976). One unit (U) of the enzyme under this method is defined as the amount of product liberated within one minute (expressed in μmol/min or nmol/min). It is assumed that the reaction rate is directly proportional to the amount of enzyme in the solution. The protein concentration was presented in mg/mL while the specific activity is determined by dividing the number of U/mL by the concentration of protein (mg/mL). The specific activity is expressed in μmol/min/mg or U/μg.
2.5. Treatment of standard food waste using Bacillus spp. and α-amylase
This study applied the established standard food waste (SFW) by The Korean Ministry of Environment Guideline (2013-179) with slight modification from An et al. (2018). Table 1 presents the detailed SFW formula. The preparation of the food waste was commenced by grinding all the food items listed in Table 1 using a blender. The blended food materials were stored at 4 °C before use. The subsequent SFW analysis was performed following standard methods recommended by The Association of the Official Analysis Chemist (AOAC). The pH of the SFW was determined by dissolving 5 g of the sample in distilled water (25 mL), followed by one hour mixing and 10 min centrifugation at 8000 rpm. The pH of the supernatant was determined using a portable pH meter (Accumet AP115, Fisher, UK). The following equation was used to determine the changes in the solid contents of the SFW:
where w0 indicates the weight of the dish (g); w1 indicates the weight of the sample and dish before drying (g); and w2 symbolises the weight of the sample and dish after drying (g). The SFW was dried in an oven to an average water content of about 80 ± 5% which was determined based on the weight difference between the wet and dry samples. After this process, the SFW was used within 24 h.
Table 1.
Formula and preparation of standard food waste (modified).
| Food Item | Total weight (g) | Food waste material (g) | Preparation size |
|---|---|---|---|
| Grains | 130 ± 15 | Cooked rice (120) | |
| Vegetables | 200 ± 30 | Potatoes (100) Onions (100) |
Cubed to 5 mm Cubed to 5 mm |
| Fruits | 70 ± 15 | Apples (35) Oranges (35) |
Split to 1/8 size with peel Split to 1/8 size with peel |
| Chicken/Fish | 100 ± 15 | Chicken (25) Tuna (75) |
Split to 30 mm (Fresh cut) Split to ¼ size (canned tuna) |
The treatment of the SFW was performed using the Bacillus spp. HULUB1 and SUNGB2. The ratios of HULUB1 and SUNGB2 mixtures were 0:0, 1:0, 0:1 and 0.5:0.5 (v/v). The bacterial culture/mixture (1 mL) with a colony count of 105 CFU/mL was inoculated into the SFW (100 g) and incubated for up to 12 days at 45 °C with agitation at 200 rpm. Initially, the bacterial concentration of the SFW ranged between 104 to 105 CFU/mL. Samples were withdrawn every 24 h during the cultivation period for pH, bacterial count and solid content analyses. The bacterial counts determination using a pour plating technique on nutrient agar of serial dilution of food waste samples were conducted and expressed as CFU/mL. The determination of the solid content of the SFW was done via a standard drying technique as follows: 5 g of the SFW was weighed and placed in a pre-weighed aluminium dish. The dish was placed in an oven and allowed to dry to a regular weight at 105 °C. The plate was weighed again after drying in order to determine the solid content (An et al., 2018). The pH of the SFW were recorded as described in the previous section.
For the treatment of the food waste, the partially purified α-amylase from HULUB1 and SUNGB2 and their mixtures was used. The ratios of partially purified α-amylase were 2:0, 0:2, 0:0, and 2:2 (U/U). An aliquot of 2 units of the individual partially purified α-amylase or partially purified α-amylase mixture was introduced into the SFW (100 g) and incubated in a shaking incubator at the optimum temperature at 45, 55 and 65 °C, pH 6.0 and 200 rpm for up to 12 days. After the incubation period, the SFW was analysed for solid content weight and pH.
2.6. Statistical analysis
The data obtained were subjected to ANOVA and all experiments were carried out in three replications based on a completely randomised design CRD (Gomez et al., 1984). Statistical analysis was performed using SPSS V25.0 software (SPSS Inc., USA), based on a single factor analysis of variance (ANOVA). Duncan multiple range tests used for meaningful comparison of the treatments at a P-value of 0.05.
3. Results and discussion
3.1. Identification of bacteria from hot spring
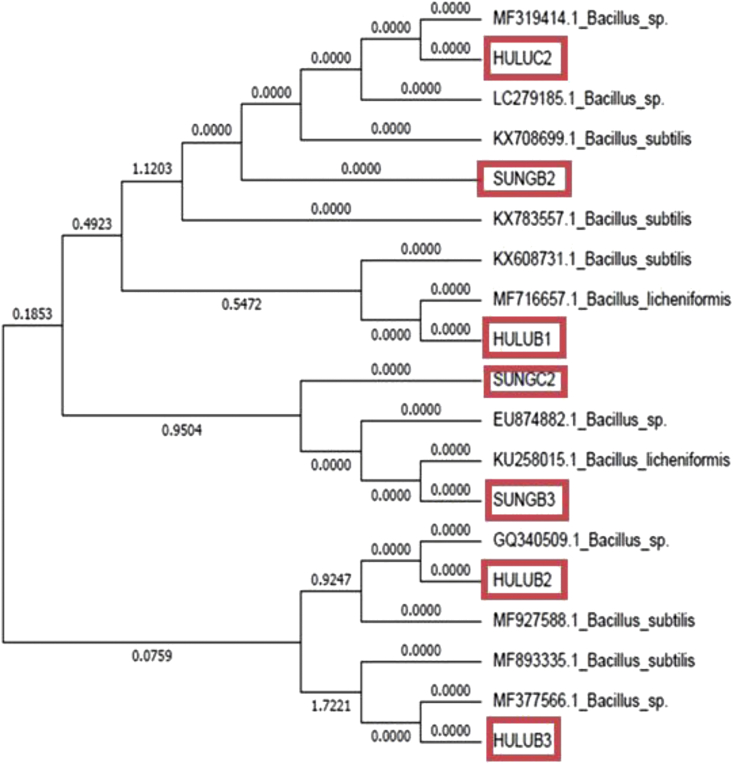
The isolates pre-identified on Thermus agar plates and subjected to various analyses such as cell morphology and Gram reaction. Preliminary tests showed that the isolates have similar properties of the genus Bacillus. The isolates were examined to determine the maximum and optimum temperature for growth and different NaCl concentrations. The results showed most of the isolates can grow at 30–55 °C. HULUB1 from Dusun Tua Hot Spring and SUNGB2 from Sungai Klah Hot Spring which showed the highest α-amylase activity were further identified by 16S rRNA sequencing. Both isolates were also used for further analyses on α-amylase activity and production. Sequence analysis indicated that all isolates belonged to the genus Bacillus with over 97% homology. HULUB1 strain identified as B. licheniformis with 99% homology. SUNGB2 strain identified as B. subtilis with a similarity of 100%. The two 16S rDNA sequences were deposited into the GenBank database under accession numbers MH062894 for B. licheniformis HULUB1 and MH062948 for B. subtilis SUNGB2 (Figure 1).
Figure 1.
Phylogenetic tree of B. licheniformis HULUB1 and B. subtilis SUNGB2 based on total 16S rDNA sequencing (software BioEdit 7.1.9, MEGA 7.0).
3.2. M9 Minimal Media sugar utilisation
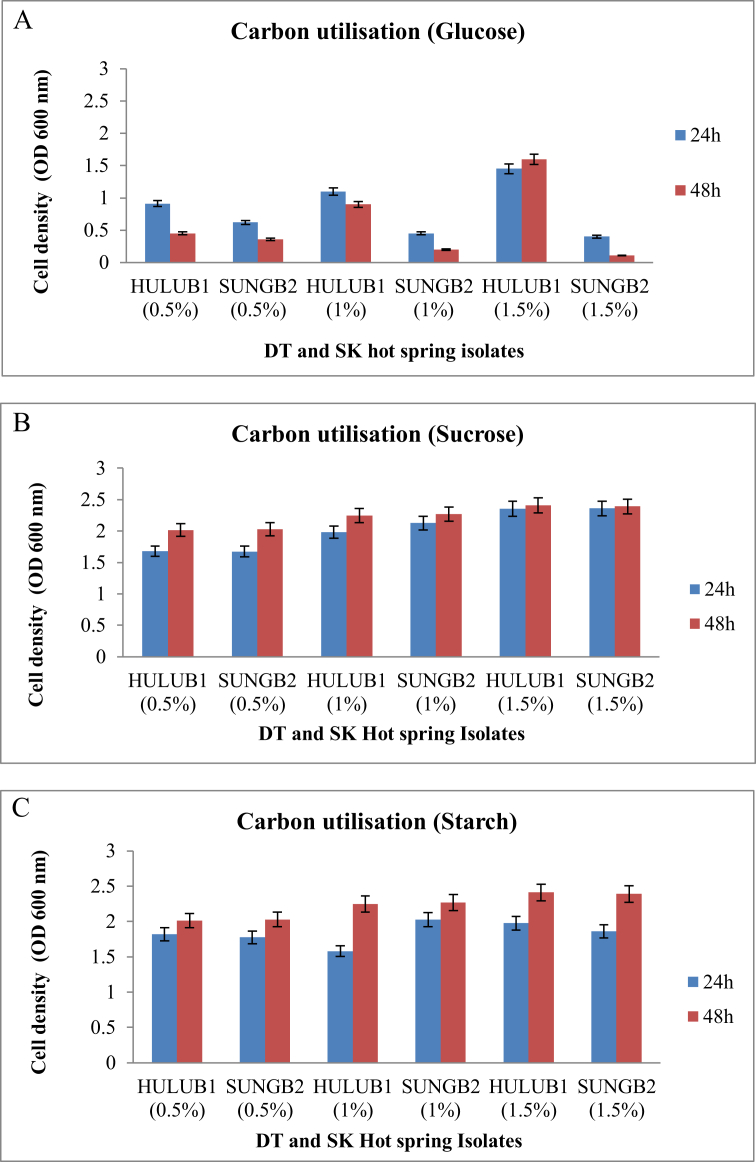
Results from sugar utilisation experiment carried out in M9 Minimal Media with the addition of 0.5%, 1% and 1.5% (w/v) of the sugars used (glucose, sucrose, starch) as the sole carbon source. B. licheniformis HULUB1 and B. subtilis SUNGB2 showed an excellent growth on sucrose and starch at a concentration range between 0.5-1.5% (w/v) when incubated at 45 °C for 24 h–48 h. The B. subtilis SUNGB2 showed a rapid decline in growth at 48 h with the increase of glucose concentration in comparison to B. licheniformis HULUB1 (Figure 2). Amylase production by Bacillus isolates (HULUB1 and SUNGB2) was found to be constant, as the enzyme biosynthesis proceeded in the presence of both starch and other carbon sources, which is similar to the previous findings, working with another strain of B. licheniformis (Swain et al., 2006).
Figure 2.
The growth of B. licheniformis HULUB1 and B. subtilis SUNGB2 on different concentrations of (A) glucose, (B) sucrose and (C) starch on M9 Medium.
The results revealed that B. licheniformis HULUB1 and B. subtilis SUNGB2 showed a preference for carbon sources in the order of highly preferred source of starch followed by sucrose and with glucose being the least preferable (Figure 2). The amylase production was influenced by the carbon source and concentrations (Divakaran et al., 2011). This trait may be exploited for growing B. subtilis and B. licheniformis in a medium containing starch for the production of industrially important products. The growth of both isolates were satisfactory in the presence of all sugars when incubated for 24–48 h at 45 °C.
3.3. Screening of α-amylase production
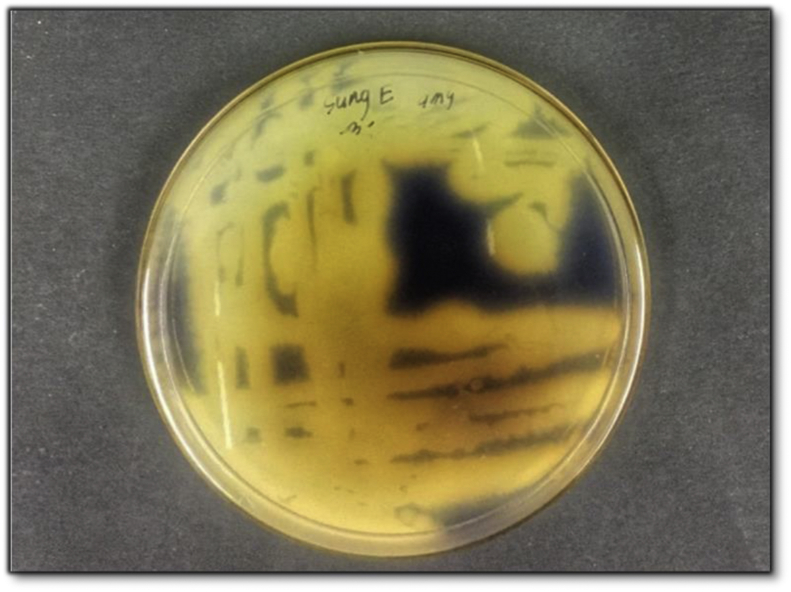
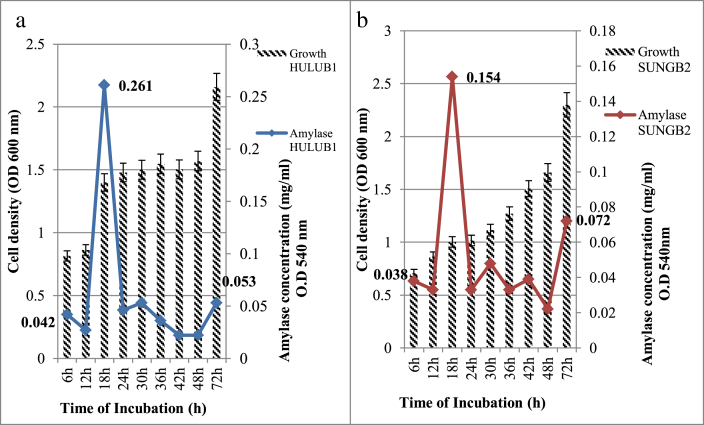
The thermophilic isolates were grown on selective media for the screening of amylase and incubated at 45 °C for 24 h. Positive activity was displayed by the size of the clearing zone (Figure 3). The bigger the size of the clearing zone, the higher the activity produced. According to Thebti et al. (2016) the activity of enzyme is revealed in the size of the clearing zone, measured by centimetres. The amylase activity was also assessed according to the glucose standard curve, which one IU of amylase activity represented by the total of enzyme required to release one U (μmol/min) of reducing sugar within one minute under ultimate experimental conditions. B. licheniformis HULUB1 and B. subtilis SUNGB2 were subjected to quantitative screening and growth profile Both strains B. subtilis SUNGB2 and B. licheniformis HULUB1 can produce α-amylase with different concentrations at different incubation times over a pH range of 3–11. The optimum bacterial growth and production of amylase were observed at pH 6.0 and at 18 h of growth, whereby B. licheniformis HULUB1 and B. subtilis SUNGB2 produced amylase 0.261 mg/mL and 0.154 mg/mL respectively.
Figure 3.
The amylase qualitative assay for the thermophilic isolates.
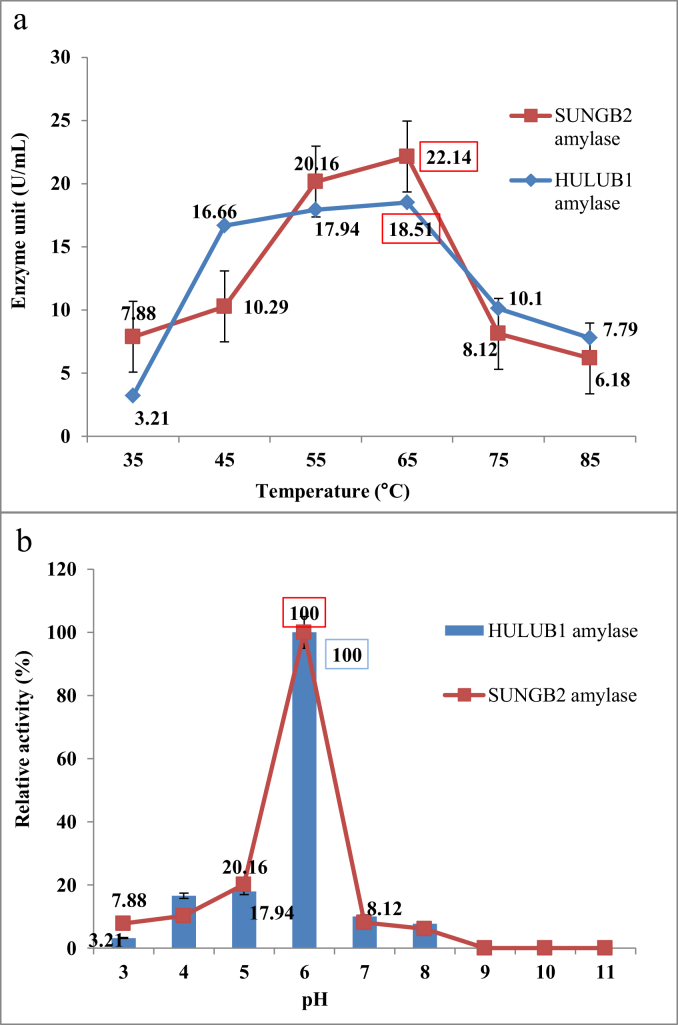
The α-amylase production was peaked at 45 °C. The amylase of the two isolates was partially purified by ammonium sulphate precipitation at 80% saturation, then further characterised by optimising the temperature and pH. The data revealed that the highest amount of amylase was produced at an initial pH of 6.0 at 65 °C with 18.15 U/mL for HULUB1, 22.14 U/mL for SUNGB2 and that other pH values yielded lower enzyme activity (Figure 4). While the highest amylase activity occurred at pH 6.0, the highest bacterial growth was observed at pH 8.0. The observation proposing that the enzymes synthesise occur in a wide range of 5.0–8.0 pH and was not growth dependent.
Figure 4.
Bacterial growth and amylase production by (a) HULUB1 and (b) SUNGB2 in amylase cultivation media.
Similar results were reported by Swain et al. (2006) in which the optimum activity of α-amylase extracted from B. subtilis CM3 was found at pH 5.0–9.0 and 50–70 °C. While Awasthi et al., 2018a, Awasthi et al., 2018b reported four isolates of Bacillus with maximum amylase activity at pH 5.3–8.2 and 60 °C. The level of amylase production was found to depend on the temperature effects on the growth of the organism. The temperature range of 35–80 °C has earlier been reported to favour the growth α-amylase -producing bacteria (Bajpai and Bajpai, 1989; Burhan et al., 2003; Ibrahim et al., 2013; Asad et al., 2014; Xu et al., 2019). However, temperatures above 65 °C were found to negatively affect the activity of enzymes while a further increase in temperature caused a decline in the activity of α-amylase (Figure 5) possibly due to enzyme denaturation which led to inhabitation (Anto et al., 2006).
Figure 5.
Activity and stability of B. licheniformis HULUB1 and B. subtilis SUNGB2 partially purified α-amylase at (a) different temperatures and (b) different pH.
The level of purity of an enzyme preparation determines its specific activity which is expressed as the ratio of the enzyme activity to the concentration of protein used during the enzymatic activity assay. The highest specific activity obtained by ammonium phosphate precipitation at 80% (w/v) was 10.66 U/μg for HULUB1 (10-fold increase) and U/μg for 5.40 SUNGB2 (21-fold increase), compared to crude enzymes, 1.04 U/μg for HULUB1 and 0.232 U/μg for SUNGB2.
3.4. Treatment and analysis of SFW
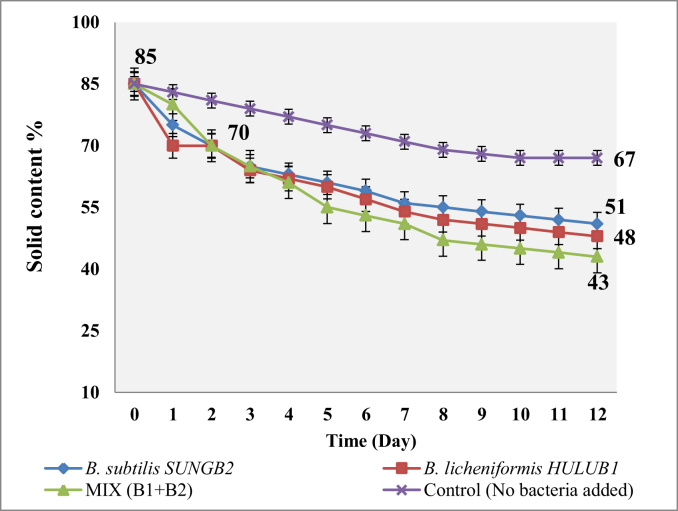
The two isolates and their extracted α-amylase were separately evaluated for biodegradation of food wastes. The tested Bacillus isolates were cultivated for 12 days in SFW where they were found to be increasing in number with increase in the incubation period until the 12th day when bacterial growth plateaued. B. licheniformis HULUB1 was observed to exhibit the maximum cell count after the 8 days of incubation. An initial decrease in the pH value of the SFW was observed until the 3rd day when it started to increase gradually and finally plateaued from the 4th day to the 8th day. Figure 6 presents the observed changes in the solid contents of the SFW after cultivation with the Bacillus isolates. These measurements were carried out in triplicates.
Figure 6.
Solid content of SFW treated with thermophilic B. licheniformis HULUB1 and B. subtilis SUNGB2 at 45 °C.
Naturally, there are several organic contents in food wastes which can serve as carbon sources to several microorganisms. Upon utilisation of these carbon sources by the microbes, the quantity of food wastes can be effectively reduced. Bacteria are the most considered microbes for the treatment of food waste owing to the natural presence of different indigenous bacteria in food wastes. Several studied have been dedicated to microorganisms-based food waste treatment via both aerobic and anaerobic processes of fermentation (Kwon et al., 2014; An et al., 2018). Among the thermophilic and mesophilic bacteria found in food waste include Pseudomonas spp., Stearothermophilus spp. and Bacillus spp. (An et al., 2018). The high growth rate of B. subtilis and B. licheniformis, as well as their ability to release several hydrolytic enzymes have made them as attractive industrial bacilli (Saxena et al., 2007; Parrado et al., 2014). Early reports by Seo et al. (2013), Parrado et al. (2014) and Sewalt et al. (2018) described B. licheniformis and B. subtilis strains produced several enzymes which can mediate several activities and can degrade several substrates and thrive under diverse conditions. B. subtilis and B. licheniformis exhibits strong cellulolytic and proteolytic activities which confer them the ability to strongly degrade food wastes (Awasthi et al., 2017; An et al., 2018). To our knowledge studies on using purified amylase from thermophilic Bacillus for the application of food waste treatment, is still novel.
The solid content of the SFW was found to be significantly reduced by the Bacillus species within the 8th days of cultivation. The mixed culture of HULUB1 and SUNGB2 exhibited the most significant reduction of 43% of the solid content. Several reports exist on the use of Bacillus species for effective food waste treatment at elevated temperatures (Yi et al., 2006; Kwon et al., 2014; Awasthi et al., 2017, 2018; Awasthi et al., 2018a, Awasthi et al., 2018b; An et al., 2018). When the SFW was treated with a mixed culture of HULUB1 and SUNGB2, there was a higher reduction of the solid content of the SFW compared to treatment with either of the Bacillus species. A 0.5:0.5 mixing ratio of the two species exhibited the maximum solid content reduction (dry weight) of around 43% possibly due to the additive activities of the host of multifunctional enzymes released by both species (HULUB1 and SUNGB2) on the SFW.
Most studies have used amylase from Bacillus isolated from kitchen waste as biodegradable materials (Mishra and Behera, 2008; Abdullah and Chin, 2010). Kitchen wastes mostly consist of starchy materials (similar to the food waste in this study); they are also a rich source of starch degrading microorganisms. Such microbes are also found in organic waste released from commercial kitchens and food processing factories (Chen et al., 2017). The volume of waste deposited in landfills can be effective reduced by converting such wastes into valuable products using microorganisms (Sakai et al., 2001). Both B. subtilis and B. licheniformis have also demonstrated robust capabilities to degrade food wastes aided by the cellulolytic and proteolytic activities (An et al., 2018). The Bacillus spp. can utilise keratinous wastes materials as a sole carbon and nitrogen source for their growth (Mazotto et al., 2011). Keratinase enzymes are a type of protease that possesses a biodegradative potential (Adetunji and Adejumo, 2018). Therefore, most proteases produced by Bacillus spp. remain active and will be responsible of the decrease solid contents of fish and chicken wastes.
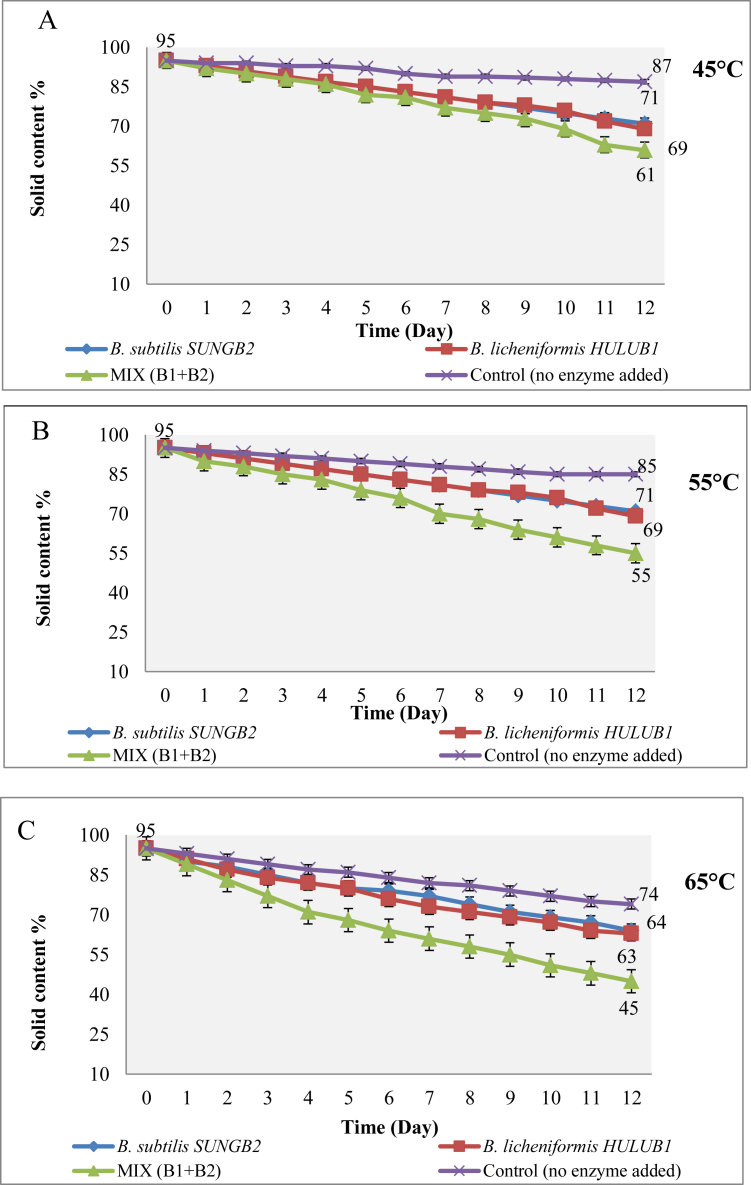
The food waste treated with α-amylase were incubated at different temperatures of 45 °C, 55 °C and 65 °C to obtain a maximum decrease in dry weight (Figure 7). The maximum weight reduction among the studied Bacillus spp. was exhibited by a mixed culture of B. licheniformis HULUB1 and B. subtilis SUNGB2 whereby 43% ± 0.02 of solid content remained after exposure at 45 °C for 12 days. The initial pH was 6.5 and at the end of the experiment it reduced to 4.9. While the greatest reduction in weight by using the amylase extracted was achieved by a mixed partially purified amylase B. licheniformis HULUB1 and B. subtilis SUNGB2, exposure to 65 °C with 45% ± 0.03 solid content remained after 12 days. The α-amylase at 45 °C resulted in a decrease in weight of about 1 g per day, whereby 61% solid content of total wastes remained after 12 days and 0.1 g for the control. Whereas, α-amylase at 55 °C resulted in a reduction in weight ranging from 4-6 g per day, which is 55% solid content and 1 g for the control. Moreover, α-amylase at 65 °C brought a reduction in weight (dry weight) ranging from 6-8 g per day which is 45% ± 0.03 solid content and 1.5 g for the control.
Figure 7.
The solid content of SFW treated with α-amylase extracted from B. licheniformis HULUB1 and B. subtilis SUNGB2 at different temperatures (A) 45 °C, (B) 55 °C and (C) 65 °C.
4. Conclusion
Thermophilic bacteria with amylolytic activity strains B. subtilis SUNGB2 and B. licheniformis HULUB1 were successfully isolated from different hot springs in Malaysia. Both strains were capable to have the maximum microbial growth and highest α-amylase enzyme production at pH 6.0 after 18 h of growth and the optimum temperature for optimum α-amylase production was observed at 45 °C. They were able to grow at high temperatures in an SFW mixture, causing in substantial reduction of the food materials. The utmost reduction in solid content of food waste was achieved by a mixed culture of B. licheniformis HULUB1 and B. subtilis SUNGB2 at temperature 45 °C, while the treatment using the mixed extracted α-amylase of HULUB1 and SUNGB2 showed the greatest reduction in solid content at 65 °C. The findings showed that the two strains displayed novel properties and a combination of the two Bacillus spp. were found to favour the degradation of food waste at a faster rate than α-amylase.
Declarations
Author contribution statement
Marwan J. Msarah: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Izyanti Ibrahim, Wan S. Aqma: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Aidil A. Hamid: Conceived and designed the experiments; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors gratefully acknowledge Universiti Kebangsaan Malaysia (UKM) for providing financial support (GUP-2018-112) and Department of Biological Sciences and Biotechnology laboratories for the support provided to this research project.
References
- Abdullah N., Chin N.L. Simplex-centroid mixture formulation for optimised composting of kitchen waste. Bioresour. Technol. 2010;101(21):8205–8210. doi: 10.1016/j.biortech.2010.05.068. [DOI] [PubMed] [Google Scholar]
- Adetunji C.O., Adejumo I.O. Efficacy of crude and immobilized enzymes from Bacillus licheniformis for production of biodegraded feather meal and their assessment. Environ. Technol. Innovat. 2018;11:116–124. [Google Scholar]
- Adiguzel A., Ozkan H., Baris O., Inan K., Gulluce M., Sahin F. Identification and characterisation of thermophilic bacteria isolated from hot springs in Turkey. J. Microbiol. Methods. 2009;79(3):321–328. doi: 10.1016/j.mimet.2009.09.026. [DOI] [PubMed] [Google Scholar]
- Ajayi A.O., Fagade O.E. Growth pattern and structural nature of amylases produced by some Bacillus species in starchy substrates. Afr. J. Biotechnol. 2006;5(5):440–444. https://www.ajol.info/index.php/ajb/article/view/137868 [Google Scholar]
- An B., Park M.K., Oh J.H. Food waste treatment using Bacillus species isolated from food wastes and production of air-dried Bacillus cell starters. Environ. Eng. Res. 2018;23(3):258–264. [Google Scholar]
- Anto H., Trivedi U., Patel K. Alpha amylase production by Bacillus cereus MTCC 1305 using solid-state fermentation. Food Technol. Biotechnol. 2006;44(2):241–245. https://hrcak.srce.hr/109861 [Google Scholar]
- Arikan B. Highly thermostable, thermophilic, alkaline, SDS and chelator resistant amylase from a thermophilic Bacillus sp. isolate A3-15. Bioresour. Technol. 2008;99(8):3071–3076. doi: 10.1016/j.biortech.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Asad W., Saleem F., Ajaz M., Rasool S.A. Optimisation of the growth conditions for amylase production by Bacillus licheniformis 208 isolated from local hot spring of Karachi. J. Chem. Soc. Pakistan. 2014;36(2):366–370. https://www.osti.gov/etdeweb/biblio/22299201 [Google Scholar]
- Asghar M., Asad M., Javaid, Rahman S.U., Legge R.L. A thermostable alpha amylase from a moderately thermophilic Bacillus subtilis strain for starch processing. J. Food Eng. 2007;79:950–955. [Google Scholar]
- Awasthi M.K., Selvam A., Chan M.T., Wong J.W. Bio-degradation of oily food waste employing thermophilic bacterial strains. Bioresour. Technol. 2018;248:141–147. doi: 10.1016/j.biortech.2017.06.115. [DOI] [PubMed] [Google Scholar]
- Awasthi M.K., Selvam A., Lai K.M., Wong J.W. Critical evaluation of post-consumption food waste composting employing thermophilic bacterial consortium. Bioresour. Technol. 2017;245:665–672. doi: 10.1016/j.biortech.2017.09.014. [DOI] [PubMed] [Google Scholar]
- Awasthi M.K., Wong J.W., Kumar S., Awasthi S.K., Wang Q., Wang M., Biodegradation of food waste using microbial cultures producing thermostable alpha amylase and cellulase under different pH and temperature. Bioresour. Technol. 2018;248:160–170. doi: 10.1016/j.biortech.2017.06.160. [DOI] [PubMed] [Google Scholar]
- Bajpai P., Bajpai P.K. High temperature alkaline alpha amylase from Bacillus licheniformis TCRDC-B13. Biotechnol. Bioeng. 1989;33(1):72–78. doi: 10.1002/bit.260330110. [DOI] [PubMed] [Google Scholar]
- Behal A., Singh J., Sharma M.K., Puri P., Batra N. Characterisation of alkaline alpha amylase from Bacillus sp. AB 04. Int. J. Agric. Biol. 2006;8:80–83. https://www.researchgate.net/profile/Arvind_Behal/publication/200447139 [Google Scholar]
- Bong C.P.C., Ho W.S., Hashim H., Lim J.S., Ho C.S., Tan W.S.P., Lee C.T. Review on the renewable energy and solid waste management policies towards biogas development in Malaysia. Renew. Sustain. Energy Rev. 2017;70:988–998. [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bragger J.M., Daniel R.M., Coolbear T., Morgan H.W. Very stable enzymes from extremely thermophilic archaebacteria and eubacteria. Appl. Microbiol. Biotechnol. 1989;31(5):556–561. [Google Scholar]
- Brock T.D. In: Thermophiles; Biodiversity, Ecology and Evolution. Reysenbach A.L., editor. Kluwer Academic/Plenum Publishers; New York: 2001. pp. 1–22. [Google Scholar]
- Burhan A., Nisa U., Gökhan C., Ömer C., Ashabil A., Osman G. Enzymatic properties of a novel thermostable, thermophilic, alkaline and chelator resistant amylase from an alkaliphilic Bacillus sp. isolate ANT-6. Process Biochem. 2003;38(10):1397–1403. [Google Scholar]
- Carvalho R.V.D., Côrrea T.L.R., Silva J.C.M.D., Mansur L.R.C.D.O., Martins M.L.L. Properties of an amylase from thermophilic Bacillus sp. Braz. J. Microbiol. 2008;39(1):102–107. doi: 10.1590/S1517-838220080001000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Chan K.G., Ee R., Hong K.W., Urbieta M.S., Donati E.R., Effects of physiochemical factors on prokaryotic biodiversity in Malaysian circumneutral hot springs. Front. Microbiol. 2017;8:1252. doi: 10.3389/fmicb.2017.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Jiang W., Yang Y., Yang Y., Man X. State of the art on food waste research: a bibliometrics study from 1997 to 2014. J. Clean. Prod. 2017;140:840–846. [Google Scholar]
- Divakaran D., Chandran A., Pratap Chandran R. Comparative study on production of alpha amylase from Bacillus licheniformis strains. Braz. J. Microbiol. 2011;42(4):1397–1404. doi: 10.1590/S1517-838220110004000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drejer E.B., Hakvåg S., Irla M., Brautaset T. Genetic tools and techniques for recombinant expression in thermophilic bacillaceae. Microorganisms. 2018;6(2):42. doi: 10.3390/microorganisms6020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO . 2015. Food Loss and Food Waste | FAO | Food and Agriculture Organisation of the United Nations.http://www.fao.org/food-loss-and-food-waste/en/ from. [Google Scholar]
- Gomez K.A., Gomez K.A., Gomez A.A. second ed. John Wiley and Sons; New York: 1984. Statistical Procedures for Agricultural Research; p. 680. [Google Scholar]
- Hmidet N., Ali N.E.H., Haddar A., Kanoun S., Alya S.K., Nasri M. Alkaline proteases and thermostable α-amylase co-produced by Bacillus licheniformis NH1: characterization and potential application as detergent additive. Biochem. Eng. J. 2009;47(1–3):71–79. [Google Scholar]
- Ibrahim D., Zhu H.L., Yusof N. Bacillus licheniformis BT5. 9 isolated from Changar Hot spring, Malang, Indonesia, as a potential producer of thermostable alpha-amylase. Trop. Life Sci. Res. 2013;24(1):71–84. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3799411/ [PMC free article] [PubMed] [Google Scholar]
- Kersters K., Vancanneyt M. Springer Verlag; 2005. Bergey's Manual of Systematic Bacteriology. [Google Scholar]
- Kibler K.M., Reinhart D., Hawkins C., Motlagh A.M., Wright J. Food waste and the food-energy-water nexus: a review of food waste management alternatives. Waste Manag. 2018;74:52–62. doi: 10.1016/j.wasman.2018.01.014. [DOI] [PubMed] [Google Scholar]
- Kiran E.U., Trzcinski A.P., Ng W.J., Liu Y. Bioconversion of food waste to energy: a review. Fuel. 2014;134:389–399. [Google Scholar]
- Konsoula Z., Liakopoulou-Kyriakides M. Co-production of alpha amylase and β-galactosidase by Bacillus subtilis in complex organic substrates. Bioresour. Technol. 2007;98(1):150–157. doi: 10.1016/j.biortech.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Kwon B.G., Na S.H., Lim H.J., Lim C.S., Chung S.Y. Slurry phase decomposition of food waste by using various microorganisms. J. Kor. Soc. Environ. Eng. 2014;36(5):303–310. [Google Scholar]
- Li M., Gong J., Cottrill M., Yu H., de Lange C., Burton J., Topp E. Evaluation of QIAamp® DNA Stool Mini Kit for ecological studies of gut microbiota. J. Microbiol. Methods. 2003;54(1):13–20. doi: 10.1016/s0167-7012(02)00260-9. [DOI] [PubMed] [Google Scholar]
- Mazotto A.M., Coelho R.R., Cedrola S.M., de Lima M.F., Couri S., Paraguai de Souza E., Vermelho A.B. Keratinase production by three Bacillus spp. using feather meal and whole feather as substrate in a submerged fermentation. Enzyme Res. 2011;523780 doi: 10.4061/2011/523780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31(3):426–428. [Google Scholar]
- Mishra S., Behera N. Amylase activity of a starch degrading bacteria isolated from soil receiving kitchen wastes. Afr. J. Biotechnol. 2008;7(18):3326–3331. https://www.ajol.info/index.php/ajb/article/view/59286 [Google Scholar]
- Msarah M.J., Yusoff M.F.M., Samion S.N.S., Prabhakaran P., Ibrahim I., Aqma W.S. Extreme environment: biofilms and microbial diversity. Malays. J. Microbiol. 2018;14(5):435–443. [Google Scholar]
- Parrado J., Rodriguez-Morgado B., Tejada M., Hernandez T., Garcia C. Proteomic analysis of enzyme production by Bacillus licheniformis using different feather wastes as the sole fermentation media. Enzym. Microb. Technol. 2014;57:1–7. doi: 10.1016/j.enzmictec.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Piterina A.V., Bartlett J., Pembroke J.T. Molecular analysis of bacterial community DNA in sludge undergoing auto-thermal thermophilic aerobic digestion (ATAD): pitfalls and improved methodology to enhance diversity recovery. Diversity. 2010;2(4):505–526. [Google Scholar]
- Prakash O., Jaiswal N. Alpha-amylase: an ideal representative of thermostable enzymes. Appl. Biochem. Biotechnol. 2009;160(8):2401–2414. doi: 10.1007/s12010-009-8735-4. [DOI] [PubMed] [Google Scholar]
- Reyed M.R. Biosynthesis and Properties of extracellular amylase by encapsulation Bifidobatrium bifidum in batch culture. Aust. J. Basic Appl. Sci. 2007;1(1):7–14. http://www.academia.edu/download/61672501/7-1420200103-114934-1b1e732.pdf [Google Scholar]
- Sakai K., Kawano H., Iwami A., Nakamura M., Moriguchi M. Isolation of a thermophilic poly-L-lactide degrading bacterium from compost and its enzymatic characterisation. J. Biosci. Bioeng. 2001;92(3):298–300. doi: 10.1263/jbb.92.298. [DOI] [PubMed] [Google Scholar]
- Sanger F. Determination of nucleotide sequences in DNA. Biosci. Rep. 1981;1(1):3–18. doi: 10.1007/BF01115145. [DOI] [PubMed] [Google Scholar]
- Saxena R.K., Dutt K., Agarwal L., Nayyar P. A highly thermostable and alkaline amylase from a Bacillus sp. PN5. Biores. Technol. 2007;98(2):260–265. doi: 10.1016/j.biortech.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Sayeh R., Birrien J.L., Alain K., Barbier G., Hamdi M. Microbial diversity in Tunisian geothermal springs as detected by molecular and culture-based approaches. Extremophiles. 2010;14:501–514. doi: 10.1007/s00792-010-0327-2. [DOI] [PubMed] [Google Scholar]
- Seo J.K., Park T.S., Kwon I.H., Piao M.Y., Lee C.H., Ha J.K. Characterisation of cellulolytic and xylanolytic enzymes of Bacillus licheniformis JK7 isolated from the rumen of a native Korean goat. Asian-Australas. J. Anim. Sci. 2013;26(1):50–58. doi: 10.5713/ajas.2012.12506. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4093055/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt V.J., Reyes T.F., Bui Q. Safety evaluation of two alpha amylase enzyme preparations derived from Bacillus licheniformis expressing an alpha amylase gene from Cytophaga species. Regul. Toxicol. Pharmacol. 2018;98:140–150. doi: 10.1016/j.yrtph.2018.07.015. [DOI] [PubMed] [Google Scholar]
- Sikdar A., Raziuddin A., Gupta K.K. Isolation and characterisation of thermophilic bacteria of a hot water spring source, Balbal. Int. J. Adv. Res. Biol. Sci. 2015;2(5):106–111. https://ijarbs.com/pdfcopy/may2015/ijarbs16.pdf [Google Scholar]
- Swain M.R., Kar S., Padmaja G., Ray R.C. Partial characterisation and optimisation of production of extracellular-amylase from Bacillus subtilis isolated from culturable cow dung microflora. Pol. J. Microbiol. 2006;55(4):289–296. https://www.ncbi.nlm.nih.gov/pubmed/17416065 [PubMed] [Google Scholar]
- Tanyildizi M.S., Özer D., Elibol M. Optimisation of alpha amylase production by Bacillus sp. using response surface methodology. Process Biochem. 2005;40(7):2291–2296. [Google Scholar]
- Thebti W., Riahi Y., Gharsalli R., Belhadj O. Screening and characterisation of thermo-active enzymes of biotechnological interest produced by thermophilic Bacillus isolated from hot springs in Tunisia. Acta Biochim. Pol. 2016;63(3) doi: 10.18388/abp.2016_1271. [DOI] [PubMed] [Google Scholar]
- Xu T.L., Peng J., Zhu Y.L., Li S., Zhou K., Cheng H., Zhou S. Yield enhancement of recombinant α-amylase s in Bacillus amyloliquefaciens by ARTP mutagenesis-screening and medium optimization. Sains Malays. 2019;48(5):965–974. http://journalarticle.ukm.my/13617/ [Google Scholar]
- Yang J.L., Wang M.S., Cheng A.C., Pan K.C., Li C.F., Deng S.X. A simple and rapid method for extracting bacterial DNA from intestinal microflora for ERIC-PCR detection. World J. Gastroenterol. 2008;14(18):2872. doi: 10.3748/wjg.14.2872. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2710730/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Koh S.K., Ng W.C., Lim R.C., Tan H.T., Tong Y.W., Wang C.H. Potential application of gasification to recycle food waste and rehabilitate acidic soil from secondary forests on degraded land in Southeast Asia. J. Environ. Manag. 2016;172:40–48. doi: 10.1016/j.jenvman.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Yi H.S., Jeong J.H., Park Y.M., Seul K.J., Ghim S.Y. Effect of thermophilic bacteria on degradation of food wastes. Microbiol. Biotechnol. Lett. 2006;34(4):363–367. http://www.koreascience.or.kr/article/JAKO200606141820477.do [Google Scholar]