Control of gene expression by small noncoding RNA (sRNA) is well documented but underappreciated. Deep sequencing of mRNA preparations from Pseudomonas aeruginosa suggests that >500 sRNAs are generated. Few of those sRNAs have defined roles in gene expression. To address that knowledge gap, we constructed an sRNA expression library and identified sRNA 179 as a regulator of the type III secretion system (T3SS) and the cAMP-Vfr regulons. The T3SS- and cAMP-Vfr-controlled genes are critical virulence factors. Increased understanding of the signals and regulatory mechanisms that control these important factors will enhance our understanding of disease progression and reveal potential approaches for therapeutic intervention.
KEYWORDS: Pseudomonas aeruginosa, sRNA, Hfq, type III secretion, Vfr, ExsA, RsmA
ABSTRACT
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen causing skin and soft tissue, respiratory, and bloodstream infections. The type III secretion system (T3SS) is one important virulence factor. Production of the T3SS is controlled by ExsA, a transcription factor that activates expression of the entire T3SS regulon. Global regulators including Vfr, RsmA, and Hfq also contribute to regulation of the T3SS. Vfr is a cAMP-responsive transcription factor that activates exsA transcription. RsmA, an RNA-binding protein, inversely controls expression of the T3SS and the type VI secretion system (T6SS). Hfq is an RNA chaperone that functions by stabilizing small noncoding RNAs (sRNAs) and/or facilitating base pairing between sRNAs and mRNA targets. A previous study identified sRNA 1061, which directly targets the exsA mRNA and likely inhibits ExsA synthesis. In this study, we screened an sRNA expression library and identified sRNA 179 as an Hfq-dependent inhibitor of T3SS gene expression. Further characterization revealed that sRNA 179 inhibits the synthesis of both ExsA and Vfr. The previous finding that RsmA stimulates ExsA and Vfr synthesis suggested that sRNA 179 impacts the Gac/Rsm system. Consistent with that idea, the inhibitory activity of sRNA 179 is suppressed in a mutant lacking rsmY and rsmZ, and sRNA 179 expression stimulates rsmY transcription. RsmY and RsmZ are small noncoding RNAs that sequester RsmA from target mRNAs. Our combined findings show that Hfq and sRNA 179 indirectly regulate ExsA and Vfr synthesis by reducing the available pool of RsmA, leading to reduced expression of the T3SS and cAMP-Vfr regulons.
INTRODUCTION
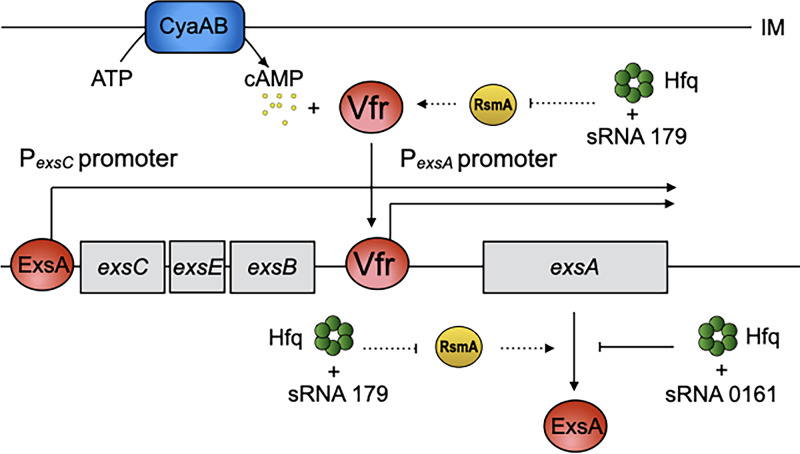
Pseudomonas aeruginosa is an important Gram-negative nosocomial pathogen that can cause skin and soft tissue, urinary tract, lung, and bloodstream infections (1). Many P. aeruginosa virulence factors are directly controlled by the cAMP-Vfr signaling (CVS) system (2, 3). The CVS consists of the CyaA and CyaB adenylate cyclases, a cAMP degrading phosphodiesterase, and the transcription factor Vfr (virulence factor regulator) (3). Vfr directly responds to increased intracellular cAMP pools to activate expression of the CVS regulon, which includes type IV pili, a type II secretion system, secreted factors (e.g., exotoxin A and protease IV), and the type III secretion system (T3SS) (4–11). The T3SS is an important virulence factor that contributes to the pathogenesis of human and animal infections (12–14). The T3SS is a needle-like apparatus used to translocate at least four effectors with antihost properties into eukaryotic target cells. The combined activities of the effector proteins promote phagocytic evasion and systemic spread of the microorganism (14). The T3SS consists of ∼40 genes that include structural components of the secretion and translocation machinery, the effectors and their cognate chaperones, and regulatory functions (15). ExsA is an AraC/XylS family protein that activates expression of the entire T3SS regulon (16). ExsA expression is highly regulated at both the transcriptional and posttranscriptional levels (17). ExsA autoregulates its own transcription through the PexsC promoter to generate a polycistronic mRNA encoding exsC, exsE, exsB, and exsA (Fig. 1) (16). A 297-bp intergenic region separates exsB and exsA and contains a Vfr-dependent promoter (PexsA) dedicated to exsA transcription (Fig. 1) (18). Several other factors contribute to exsA transcription, including PsrA, which stimulates transcription from the PexsC promoter, and MvaT/MvaU, VqsM, and Fis, which modulate PexsA promoter activity (19–21).
FIG 1.
Model for control of T3SS gene expression by ExsA, Vfr, RsmA, and Hfq.
Once transcribed, translation of ExsA appears to be inefficient, possibly owing to an inhibitory structure in the mRNA that reduces the efficiency of ribosomal recruitment (22). At least two RNA-binding proteins promote T3SS gene expression by stimulating ExsA synthesis. The RNA helicase DeaD is required for ExsA synthesis in vivo and stimulates ExsA translation in vitro (22). DeaD likely functions by denaturing an inhibitory structure in the exsA mRNA leader region to enhance ribosomal recruitment (22). RsmA, a member of the Escherichia coli CsrA family of RNA-binding proteins, can have both positive and negative effects on gene expression through direct interactions with target mRNAs (23). RsmA has positive effects on the synthesis of Vfr and ExsA through mechanisms that remain to be defined (Fig. 1) (24). The activity of RsmA is controlled by RsmY and RsmZ, small noncoding RNAs (sRNAs) with multiple RsmA binding sites. RsmY and RsmZ function by sequestering RsmA from target mRNAs (25). Transcription of rsmY and rsmZ is controlled by the GacAS two-component system (23). Increased levels of RsmY and RsmZ lead to RsmA sequestration and reduced T3SS gene expression.
Host factor for bacteriophage Qβ RNA replication (Hfq) was identified in E. coli for its essential role in bacteriophage replication (26). Hfq is an RNA-binding protein that functions as an RNA chaperone to stabilize sRNAs and/or facilitate imperfect base pairing between sRNAs and mRNA targets (27). The actions of Hfq and sRNA can affect translation and/or mRNA stability and result in negative or positive effects on gene expression. Hfq regulates ∼5% of the P. aeruginosa genome, including genes involved in stress responses, metabolism, and virulence (28–30). A P. aeruginosa hfq mutant has reduced alginate production, lipopolysaccharide (LPS), and quorum sensing and increased type III secreted products (29, 31). The mechanism of T3SS control by Hfq is poorly understood (31). sRNA PA0161 was recently identified using the GRIL-seq method and was proposed to interact with the exsA mRNA (32). In this study, we report that Hfq works with sRNA 0161 to directly inhibit ExsA synthesis and with sRNA 179 to indirectly regulate both exsA transcription and ExsA synthesis.
RESULTS
T3SS gene expression is derepressed in the absence of Hfq.
Disruption of hfq in E. coli results in pleiotropic phenotypes resulting in delayed growth, decreased negative supercoiling, increased cell size, and sensitivity to UV light (33, 34). Previous studies found that P. aeruginosa hfq insertion and deletion mutants in strain PAO1 have a growth defect when cultured in LB medium (28, 35). In the current study, we constructed an Δhfq deletion mutant (residues Δ2 to Δ82) in P. aeruginosa strain PA103 and observed a growth defect. In our subsequent studies with the PA103 Δhfq strain, emergence of growth suppressor mutants was common. A recent study by Hill et al. also noted the emergence of growth suppressor mutants with the PAO1 hfq deletion strain (35), and sequencing of several of the suppressor mutants demonstrated involvement of multiple genetic loci. Although the nature of the PA103 Δhfq suppressor mutants was not determined in the current study, each of the Δhfq mutant phenotypes reported below could be complemented by expressing hfq in trans from a plasmid. For this reason, we believe that the growth phenotype of the suppressor mutants is unrelated to Hfq effects on expression of the Vfr and T3SS regulons as described below.
Previous studies found that Hfq contributes to P. aeruginosa T3SS gene expression (29, 31), but a regulatory mechanism was not described. To examine the contribution of Hfq to T3SS gene expression, a PexsD-lacZ transcriptional reporter was integrated into the ΦCTX phage attachment site of wild-type (WT) strain PA103 and the Δhfq mutant. The PexsD-lacZ reporter is a measure of ExsA-dependent transcription and demonstrates strong induction when WT cells are cultured under inducing conditions for T3SS gene expression (low Ca2+, presence of EGTA) (Fig. 2) (36). PexsD-lacZ reporter activity increased significantly in the Δhfq mutant under both noninducing (high Ca2+, no EGTA) and inducing conditions (Fig. 2). Corresponding immunoblots showed an increase in ExsA protein levels under both noninducing and inducing conditions in the Δhfq mutant compared to the wild type. Whereas T3SS gene expression was derepressed in the hfq mutant, expression of Hfq from a plasmid inhibited PexsD-lacZ reporter activity as well as ExoU and ExsA production in both the WT and hfq mutant backgrounds (Fig. 2). Hfq protein was detectable in the WT strain, absent in the hfq mutant, and significantly elevated in strains carrying the Hfq expression plasmid (Fig. 2). These findings demonstrate that Hfq has a negative effect on T3SS gene expression.
FIG 2.
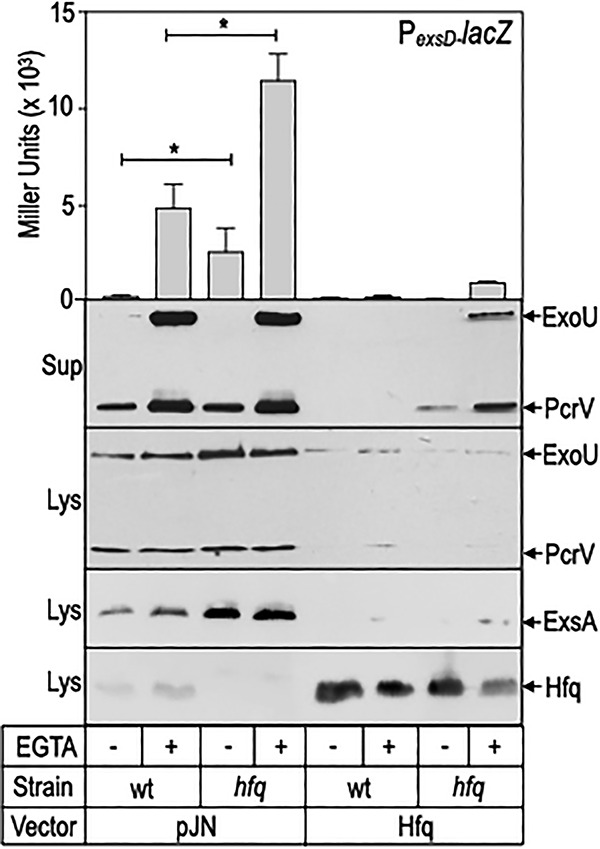
Hfq inhibits T3SS gene expression. Wild-type strain PA103 and an hfq mutant carrying an ExsA-dependent PexsD-lacZ transcriptional reporter were transformed with either a vector control (pJN105) or an Hfq expression vector (pHfq). Strains were cultured under noninducing (- EGTA) or inducing (+ EGTA) conditions for T3SS gene expression and assayed for PexsD-lacZ reporter activity. Beta-galactosidase activity is reported in Miller units with the standard error and represents the average of at least three independent experiments. P < 0.01. Cell lysate (Lys) or culture supernatant (Sup) fractions from the same cultures were immunoblotted for ExoU, PcrV, ExsA, and Hfq.
Hfq regulates T3SS-mediated cytotoxicity.
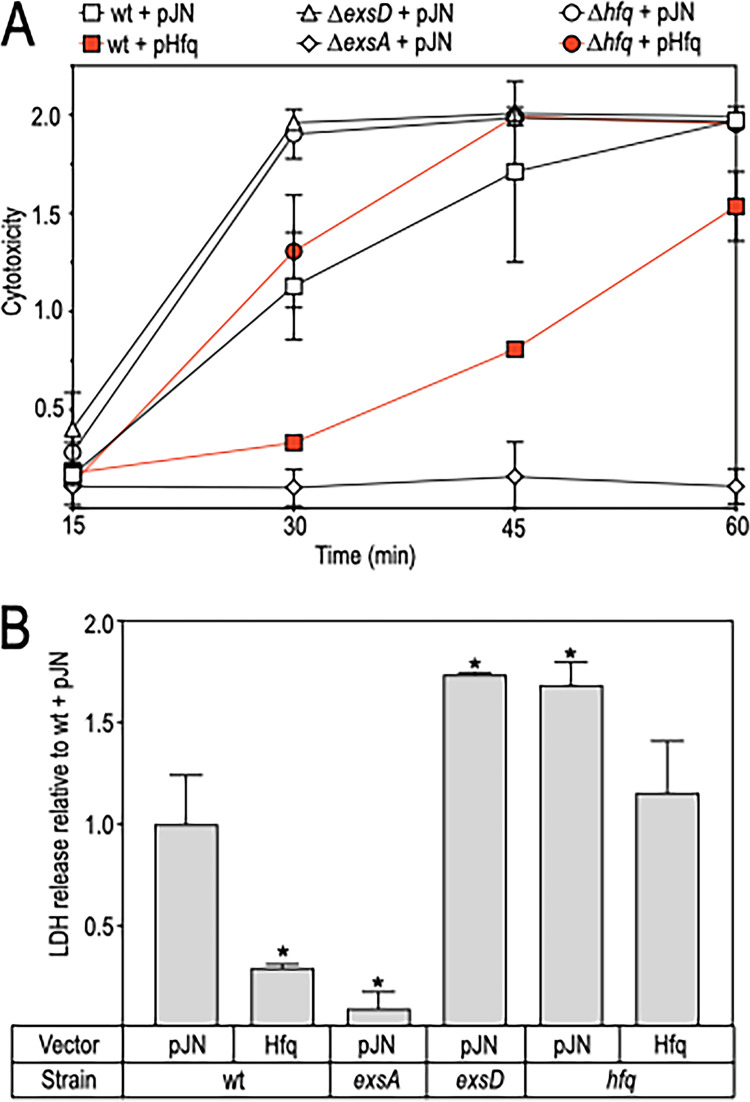
To verify that Hfq negatively regulates T3SS gene expression, we infected Chinese hamster ovary (CHO) cells with strains (wild type and Δhfq) carrying either a vector control or the Hfq expression vector. Previous studies have shown that coculturing CHO cells with P. aeruginosa strain PA103 results in acute cytotoxicity that is T3SS-dependent (37). In addition, the kinetics are advanced in strains derepressed for T3SS gene expression (e.g., an exsD mutant) (36). CHO cell lysis was measured by lactate dehydrogenase release following 15, 30, 45, and 60 min of coculture with P. aeruginosa (Fig. 3A). Relative to the parental strain, cytotoxicity was reduced with the exsA mutant, encoding a positive regulator of T3SS gene expression (Fig. 3A and B). In the absence of either exsD or hfq, cytotoxicity displayed enhanced kinetics, consistent with depressed T3SS gene expression. Expression of Hfq in the wild-type background resulted in an ∼4-fold reduction in cytotoxicity following 30 min of coculture relative to the vector control (Fig. 3A and B). Similarly, Hfq expression in the Δhfq deletion strain also reduced cytotoxicity compared to the vector control, but not to the same levels observed in the wild-type background (Fig. 3B). These results suggest that Hfq negatively regulates T3SS-mediated cytotoxicity.
FIG 3.
Hfq inhibits the T3SS-dependent toxicity of P. aeruginosa toward cultured cells. Chinese hamster ovary (CHO) cells were cocultured at an MOI of 10 with wild-type strain PA103, ΔexsA, ΔexsD, and an hfq mutant transformed with either a vector control (pJN) or an Hfq expression plasmid (pHfq). (A) Lactate dehydrogenase release as a marker of cytotoxicity was measured over a time course (reported in arbitrary units). The reported values represent the average of three wells with the standard error. (B) Lactate dehydrogenase release data from the 30-min time point in panel A relative to the wild-type PA103 vector control. *, P < 0.0001.
Identification of sRNAs that regulate type III secretion.
Hfq can inhibit protein synthesis by directly interacting with mRNA targets alone or by facilitating the base pairing of sRNAs with mRNA targets (27). To identify sRNAs that regulate the T3SS, we cloned and expressed a library of sRNAs in P. aeruginosa strain PAK and screened for effects on T3SS gene expression using the PexsD-lacZ transcriptional reporter. Gomez-Lozano et al. previously identified over 500 novel sRNAs in P. aeruginosa using three different methods to prepare transcript libraries (38). From this data set, we selected 240 sRNAs (referred to as Pants [P. aeruginosa novel transcripts]) in the original paper) that met two criteria, <200 nucleotides in length and known to originate from either the Watson or Crick strand of the genome (it has not been determined for all of the sRNAs). Each of the selected sRNAs was placed under the transcriptional control of an arabinose-inducible promoter. Six of the sRNAs (sRNA 18, 179, 182, 183, 214, and 351) inhibited PexsD-lacZ reporter activity greater than 2-fold in strain PAK (Fig. 4A and B).
FIG 4.
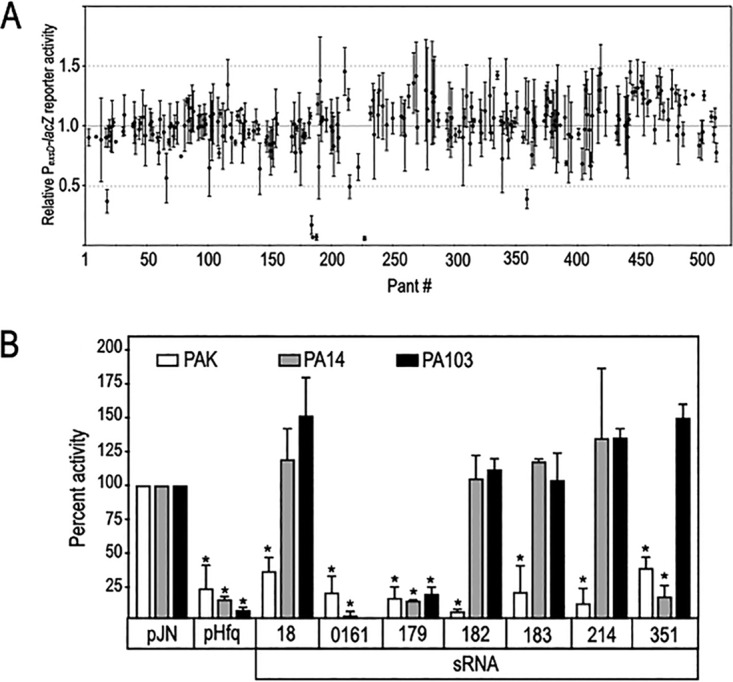
Identification of small noncoding RNAs that alter T3SS gene expression. (A) P. aeruginosa strain PAK carrying a PexsD-lacZ transcriptional reporter was transformed with a vector control and the sRNA expression vectors indicated in Table S1 (note that only 237 of the 512 putative sRNAs were tested). Strains were cultured under inducing (+ EGTA) conditions for T3SS gene expression in medium supplemented with 0.2% arabinose to induce sRNA expression and assayed for PexsD-lacZ reporter activity. The reporter values represent the activity of each sRNA relative to the vector control. (B) sRNAs that inhibited or stimulated PexsD-lacZ reporter activity greater than 2-fold (indicated by red lines in panel A) were selected for further analyses. Expression vectors for sRNA 18, 179, 182, 183, 214, 351, and 483 were introduced into strains PA14 and PA103 carrying the PexsD-lacZ reporter and assayed as described in panel A for reporter activity. sRNA 0161, previously shown to inhibit T3SS gene expression (32), and the Hfq expression vector were included as controls. The reported values represent the activity of each sRNA or pHfq relative to each parental strain carrying the vector control. The reported values with the standard error represent the average of at least three experiments. *, P < 0.01.
Strains and plasmids used in this study. Download Table S1, DOCX file, 0.03 MB (32.6KB, docx) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
As a secondary screen, the six sRNAs identified using strain PAK were tested for activity in strains PA14 and PA103 bearing the PexsD-lacZ reporter (Fig. 4B). We also included sRNA PA0161 as a positive control. sRNA PA0161 (not included in our original library) was recently identified using GRIL-Seq as an inhibitor of T3SS gene expression that directly targets the exsA mRNA (32). Although sRNAs 18, 182, 183, and 214 possess regulatory activity in strain PAK, they had no effect on PexsD-lacZ reporter activity when expressed in strains PA14 and PA103 (Fig. 4B). sRNA 351 inhibited PexsD-lacZ reporter activity in both the PAK and PA14 backgrounds but was inactive in strain PA103. The Hfq expression vector inhibited PexsD-lacZ reporter activity in all three genetic backgrounds. Since only Hfq, sRNA 0161, and sRNA 179 inhibited T3SS gene expression in all three genetic backgrounds, the remainder of this study focused on those factors.
sRNA 179 and Hfq function together to inhibit ExsA translation.
To determine whether the inhibitory activities of sRNAs 0161 and 179 are Hfq-dependent, both sRNAs were expressed in P. aeruginosa strain PA103 and the Δhfq mutant. Whereas expression of either RNA in WT cells resulted in strong inhibition of PexsD-lacZ reporter activity, ExsA production, and ExoU secretion, the inhibitory activities of sRNAs 0161 and 179 were largely suppressed in the Δhfq mutant (Fig. 5A). ExsA protein levels were reduced by ∼25% upon sRNA 0161 and 179 expression in the Δhfq mutant relative to the vector control. In contrast, ExsA protein was reduced by >90% upon sRNA 0161 and 179 expression in the WT background. ExoU protein levels followed a similar trend. These data are consistent with sRNA 0161 and 179, both functioning as typical sRNAs that depend upon Hfq for facilitated base pairing with target mRNA(s).
FIG 5.
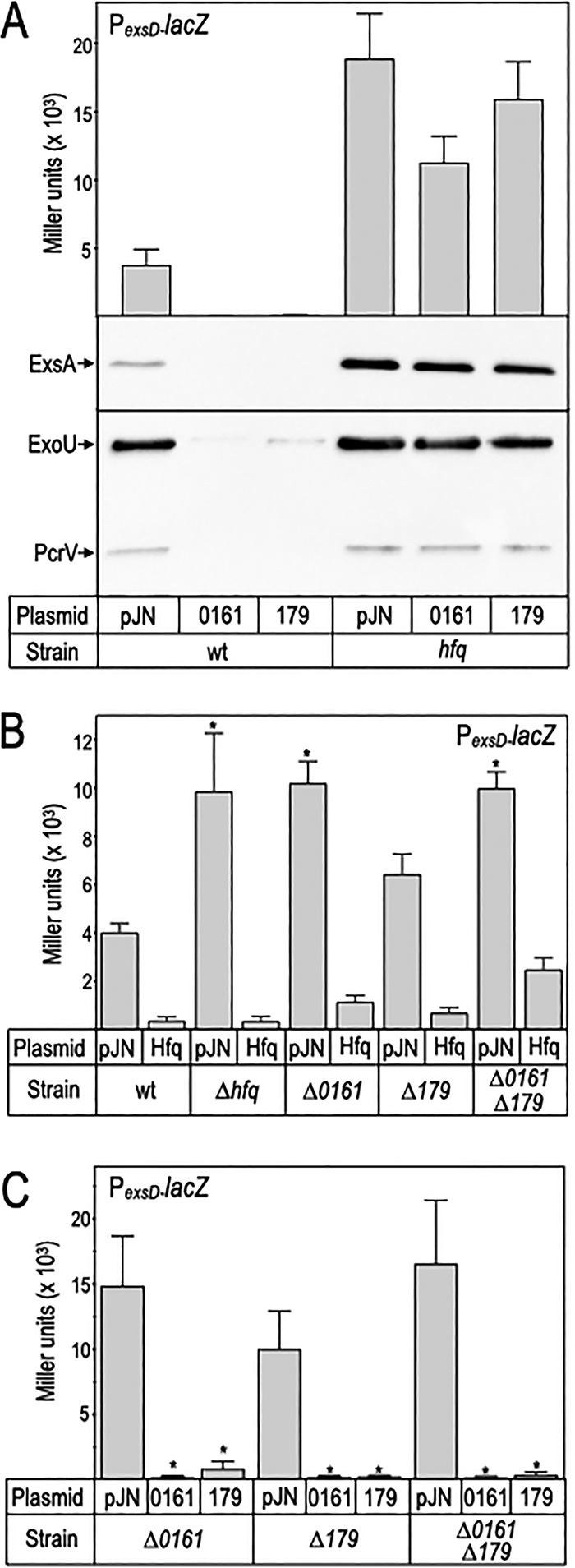
Inhibition of T3SS gene expression by sRNAs 0161 and 179 is Hfq-dependent. (A to C) The indicated strains carrying a PexsD-lacZ reporter were transformed with either the vector control (panels A to C), sRNA 0161, and 179 expression vectors (panel A and C) or an Hfq expression vector (panel B). Strains were cultured under inducing conditions for T3SS gene expression supplemented with 0.2% or 0.05% arabinose to induce sRNA or Hfq expression, respectively, and assayed for PexsD-lacZ reporter activity as reported in Miller units. Cell lysate or supernatant fractions from the samples in panel A were immunoblotted for ExsA or ExoU and PcrV, respectively. The reported values with the standard error represent the average of at least three experiments. *, P < 0.001 relative to the vector control in each case. (B) *, P < 0.005; (C) *, P < 0.05.
The data presented thus far show that sRNAs 0161 and 179 expressed from a plasmid possess inhibitory activity. To examine the effects of sRNA 0161 and 179 expressed from their native chromosomal promoters, we constructed deletion mutants for each and a double mutant lacking both sRNA 0161 and 179. Deletion mutants lacking hfq, sRNA 0161, and both sRNAs (0161 and 179) carrying a vector control (pJN105) demonstrated significant depression of PexsD-lacZ reporter activity (Fig. 5B). Although reporter activity was consistently elevated in the single Δ179 mutant, that difference did not meet a statistical test of significance. To determine whether sRNA 0161 and 179 account for all of the Hfq-dependent activity, the Hfq expression plasmid was introduced into each of the sRNA mutant backgrounds. Hfq expression resulted in significant inhibition of PexsD-lacZ reporter activity in the single and double sRNA 0161 and 179 mutants (Fig. 5B). Finally, complementation analyses demonstrate that plasmid-expressed 0161 and 179 are sufficient to repress PexsD-lacZ reporter activity in their cognate mutant backgrounds and in the double 0161 and 179 mutant (Fig. 5C). These combined findings demonstrate that (i) Hfq functions together with sRNAs 0161 and 179 to inhibit T3SS gene expression, (ii) sRNA 0161 and 179 activities are independent of one another, and (iii) Hfq also inhibits T3SS gene expression in a manner that does not rely upon sRNA 0161 or 179. Such activity could reflect sRNA-independent activity or involvement of an additional sRNA(s).
Hfq impacts ExsA synthesis at the posttranscriptional level.
Based on the previous observation that T3SS genes demonstrate increased mRNA levels in an hfq mutant (31), we hypothesized that Hfq, sRNA 0161, and/or sRNA 179, either directly or indirectly, regulate ExsA synthesis. Because ExsA autoregulates its own transcription, we established an experimental system to uncouple exsA transcription from ExsA synthesis. The native 101-nt upstream untranslated region (corresponding to the exsA transcription start site from the PexsA promoter) and coding sequence for exsA were placed under the transcriptional control of a rhamnose-inducible promoter (Prha) and integrated into the chromosome at the Tn7 transposon insertion site of an ΔexsA strain. Titration of rhamnose identified conditions (0.005%) where PexsD-lacZ reporter activity and ExsA expression levels were similar to the WT PA103 strain (Fig. 6A; see Fig. S2 in the supplemental material). Expression of the ExsA-dependent PexsD-lacZ reporter was entirely dependent upon addition of rhamnose to the culture medium and remained responsive to calcium chelation by EGTA (Fig. 6A), thus validating the system.
FIG 6.
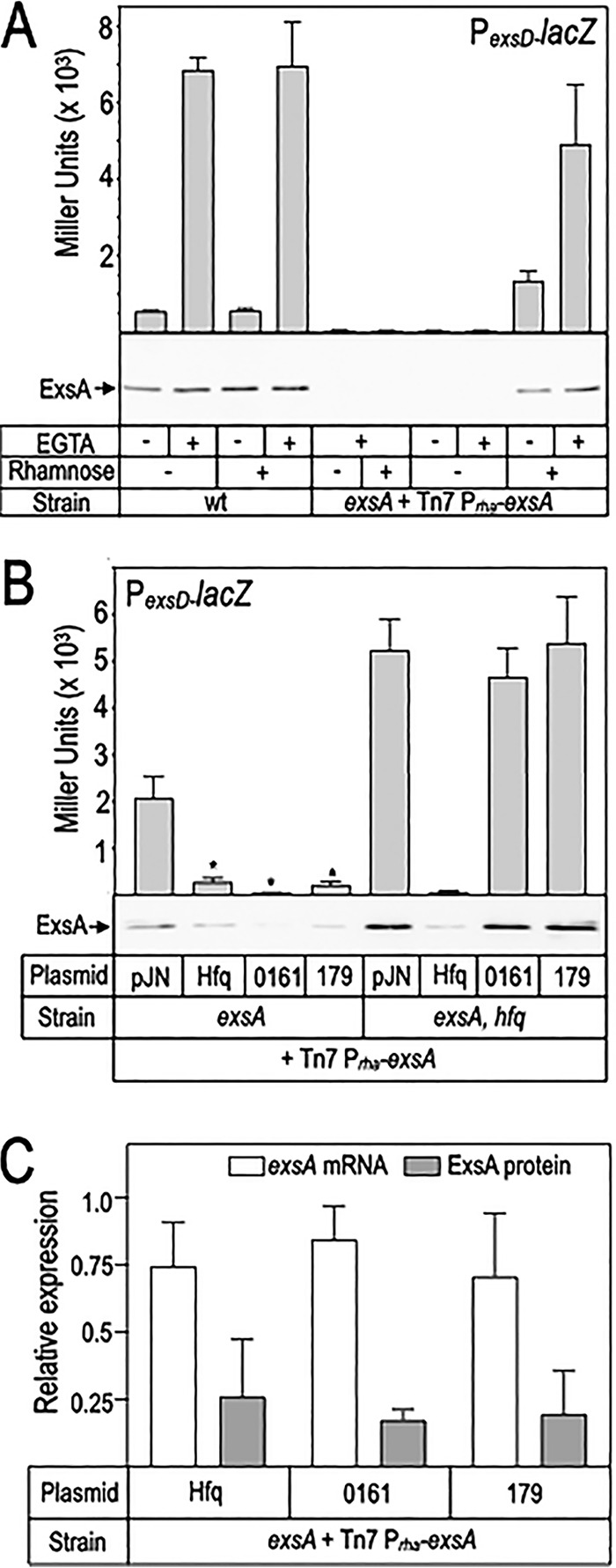
Hfq, sRNA 0161, and sRNA 179 inhibit ExsA synthesis. (A) WT PA103 and an exsA mutant carrying a rhamnose-inducible copy of exsA integrated at the Tn7 site were cultured under noninducing (- EGTA) or inducing (+ EGTA) conditions for T3SS gene expression. Rhamnose (0.005%) was included in the growth medium to induce exsA expression from the Tn7 integrant, and ExsA activity and protein levels were measured using the PexsD-lacZ reporter and ExsA immunoblots, respectively. (B) The indicated strains carrying the rhamnose-inducible copy of exsA were transformed with either the vector control, Hfq expression vector, sRNA 1061, or sRNA179 expression plasmids, cultured under T3SS inducing conditions, and assayed for PexsD-lacZ reporter activity and ExsA protein levels. (C) Comparison of exsA mRNA and ExsA protein levels in samples harvested from an exsA mutant carrying the rhamnose-inducible copy of exsA and either a vector control, pHfq, p0161, or p179. RNA samples were quantified for exsA using qRT-PCR and normalized to the rimM housekeeping gene. The reported values are relative to the exsA strain carrying the vector control.
(A) WT PA103 and an exsA mutant carrying a rhamnose-inducible copy of exsA integrated at the Tn7 site were cultured under inducing conditions for T3SS gene expression. Rhamnose was included as indicated to induce exsA expression from the Tn7 integrant (the concentrations used for the titration: 0.0025, 0.005, 0.01, 0.025, and 0.05%) ExsA activity was measured using the PexsD-lacZ reporter. Download FIG S1, TIF file, 0.3 MB (352KB, tif) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hfq interacts with sRNA 0161 and the exsA mRNA leader region. (A and B) Electrophoretic mobility shift assays were performed by incubating radiolabeled sRNA 0161 (A) or exsA mRNA leader region (B) with the indicated concentrations of Hfq. Radiolabeled sRNA 0161 was also incubated with unlabeled exsA mRNA leader region to detect formation of an RNA:RNA hybrid. The positions of the respective complexes are indicated with arrowheads. Download FIG S2, TIF file, 0.3 MB (317.7KB, tif) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The Tn7-rhamnose system was then used to determine whether Hfq, sRNA 0161, and/or sRNA 179 effect ExsA synthesis at the posttranscriptional level. Expression of Hfq, sRNA 0161, and sRNA 179 each resulted in strong inhibition of PexsD-lacZ reporter activity and ExsA protein levels (Fig. 6B, lanes 1 to 4), and the activity of the sRNAs was entirely dependent upon Hfq (lanes 5 to 8). To verify that the reduction in ExsA synthesis was unrelated to changes in exsA mRNA levels, qRT-PCR for exsA was performed on RNA harvested from the samples shown in lanes 1 to 4. Whereas ExsA protein levels were reduced ∼4-fold in the Hfq, 0161, and 179 expression strains relative to the vector control, exsA mRNA levels were only slightly reduced (∼25%) (Fig. 6C). Some degradation of the exsA mRNA was expected owing to the lack of ribosomal protection that occurs during active translation. These findings are consistent with Hfq and sRNAs 0161 and 179 controlling ExsA synthesis at a posttranscriptional level.
sRNA 0161 and the exsA mRNA interact with each other and Hfq.
Our finding that sRNA 0161 inhibits ExsA synthesis and the previous demonstration of a specific interaction between sRNA 0161 and the exsA mRNA by GRIL-seq (32) (Fig. 6B) was suggestive of a direct effect on ExsA translation. To measure the interaction in vitro, sRNA 0161 was in vitro synthesized, radiolabeled at the 5′ end, and incubated with an unlabeled portion of the exsA mRNA leader region prior to electrophoresis on a nondenaturing gel. The exsA probe (exsA 101) consists of the native 101 untranslated region and 20 bp downstream of the start codon. An sRNA 0161-exsA complex was observed when 20 nM unlabeled exsA mRNA leader region was added to the reaction (Fig. S2). We were unable to detect an interaction between sRNA 179 and the exsA mRNA using the same approach. We also assayed for Hfq binding and found that Hfq interacts with sRNA 0161 and/or the exsA mRNA (Fig. S3). These data are consistent with the previous GRIL-Seq findings of Zhang et al. (32) and suggest that Hfq directly interacts with sRNA 0161 and the 5′ untranscribed region (UTR) of exsA to reduce translation. Furthermore, the inability to detect an interaction between sRNA 179 and the exsA leader region suggested that the mechanism of inhibition by sRNA 179 may be indirect and distinct from the action of sRNA 0161.
Hfq inhibits the Vfr-cAMP signaling system.
We noted that Hfq appeared more effective at inhibiting PexsD-lacZ reporter activity in the WT background (Fig. 2) compared to the exsA mutant carrying the rhamnose-inducible exsA allele (Fig. 6B). One difference between these strains is that exsA transcription is no longer controlled by the Vfr-dependent PexsA promoter. This raised the possibility that Hfq also impacts the cAMP-Vfr system. Indeed, PexsA-lacZ reporter activity was significantly elevated in the Δhfq background and repressed upon Hfq or sRNA 179 overexpression (Fig. 7A). To confirm that the cAMP-Vfr system is altered by Hfq, we measured the activity of a vfr-lacZ translational reporter and Vfr protein levels by immunoblot analysis. Both reporter activity and Vfr protein levels were significantly elevated in the Δhfq mutant and were strongly reduced upon Hfq overexpression (Fig. 7B). In addition to controlling T3SS gene expression, Vfr also regulates type IV pili biogenesis and twitching motility (2). Relative to strains carrying a vector control, overexpression of Hfq resulted in significant inhibition of twitching motility in strains PA103, PAK, and PA14 (Fig. 7D). These combined data suggest that Hfq/sRNAs have two effects on T3SS gene expression, inhibition of Vfr synthesis resulting in reduced PexsA promoter activity and inhibition of ExsA synthesis.
FIG 7.
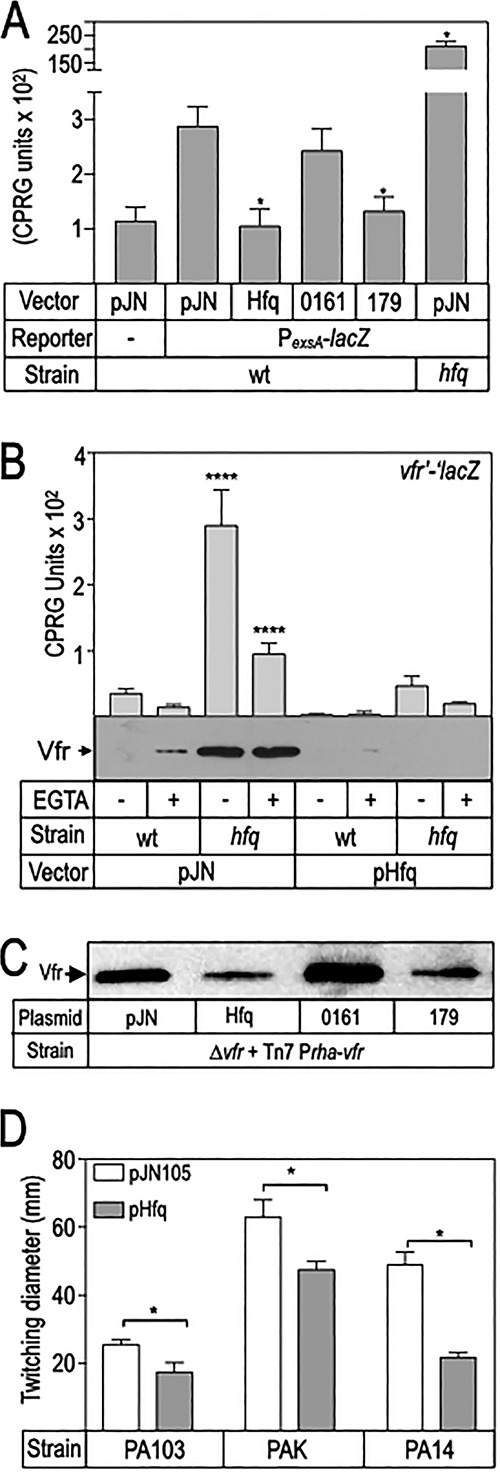
Hfq and sRNA 179 inhibit the cAMP-Vfr signaling system. (A) WT PA103 or an hfq mutant transformed with the indicated expression vectors were assayed for Vfr-dependent PexsA-lacZ transcriptional reporter activity. WT PA103 carrying a promoterless lacZ transcriptional reporter was included as a control for background activity. *, P < 0.05. (B) The indicated strains were transformed with a vector control or an Hfq expression vector and assayed for vfr’-’lacZ translational reporter activity and Vfr protein levels by immunoblot. *, P < 0.001. (C) Twitching motility zones for P. aeruginosa strains PA103, PAK, and PA14 carrying a vector control (pJN105) or an Hfq expression vector. *, P < 0.05.
sRNA 179 requires RsmY and RsmZ to control T3SS gene expression.
Previous studies have shown that the small RNA-binding protein RsmA is required for the synthesis of both Vfr and ExsA (39). Our finding that Hfq and sRNA 179 overexpression inhibits Vfr and ExsA synthesis suggested involvement of the Rsm system. RsmA availability is controlled by the small noncoding RNAs RsmY and RsmZ (40). Both RNAs possess multiple RsmA binding sites that serve to sequester RsmA from target mRNAs (40). To examine whether Hfq and sRNA 179 function through the Rsm system to control T3SS gene expression, we expressed Hfq, sRNA 0161, and sRNA 179 in ΔrsmY, ΔrsmZ, and ΔrsmYZ mutants and measured PexsD-lacZ reporter activity. Although no significant differences were observed between the WT strain and the ΔrsmY and ΔrsmZ single mutants, PexsD-lacZ reporter activity was significantly derepressed in the ΔrsmYZ mutant owing to increased RsmA availability (Fig. 8A). Overexpression of Hfq, sRNA 1061, and sRNA 179 resulted in strong inhibition of PexsD-lacZ reporter activity when expressed in each of the strains tested, with one exception; sRNA 179 lacked activity in the ΔrsmYZ mutant.
FIG 8.
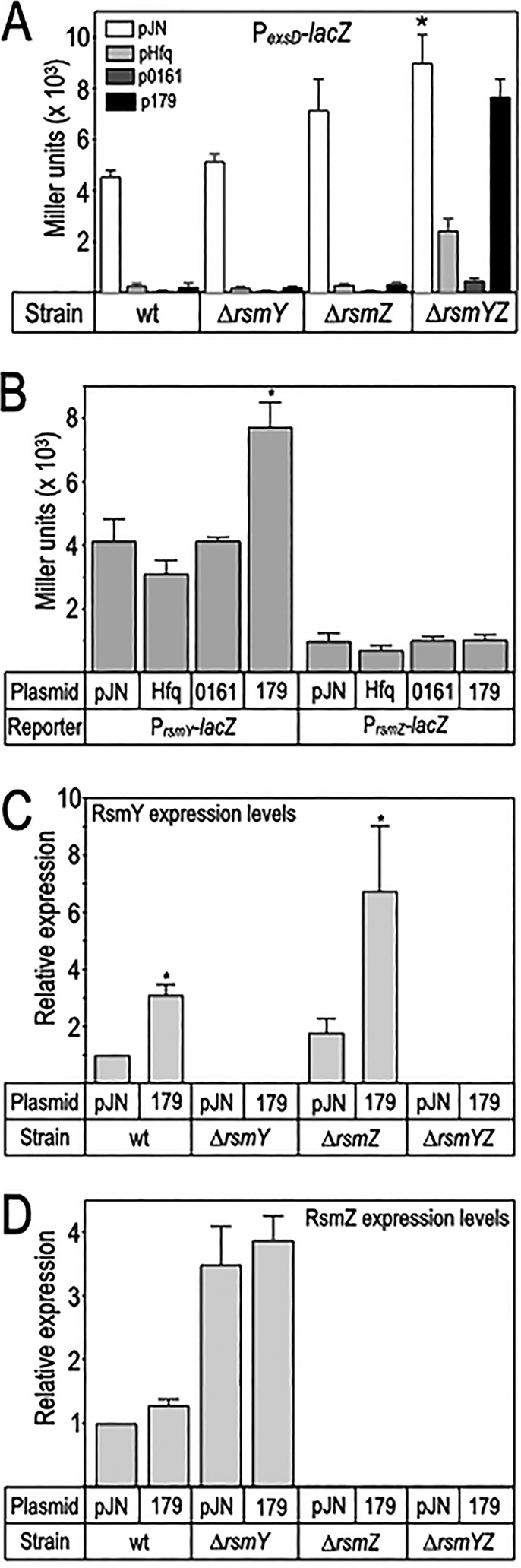
sRNA 179 modulates the Rsm system by increasing RsmY transcription. (A) The indicated strains carrying a PexsD-lacZ reporter were transformed with either the vector control (pJN), the Hfq expression vector, or sRNA 0161 and 179 expression vectors. Strains were cultured under inducing conditions for T3SS gene expression supplemented with 0.05% or 0.2% arabinose to induce Hfq or sRNA expression, respectively, and assayed for PexsD-lacZ reporter activity as reported in Miller units. The reported values with the standard error represent the average of at least three experiments. *, P < 0.05. (B) WT carrying either a PrsmY-lacZ or PrsmY-lacZ transcriptional reporter and the indicted expression vectors were assayed for reporter activity as in panel A. *, P < 0.005. (C and D) Comparison of RsmY (C) and RsmZ (D) levels in RNA samples harvested from the indicated strains. RNA samples were quantified by qRT-PCR and normalized to the rimM housekeeping gene. The reported values are relative to each strain carrying the vector control.
We next tested the hypothesis that sRNA 179 stimulates rsmY and/or rsmZ transcription. Both transcriptional reporter and reverse transcription-quantitative PCR (qRT-PCR) data demonstrate that sRNA 179 expression results in a significant increase in PrsmY-lacZ reporter and RsmY RNA levels (Fig. 8B to D). The effect of sRNA 179 is specific, as reporter activity and RsmY levels were unaffected by Hfq or sRNA 0161 expression, and Hfq, sRNA 0161, and sRNA 179 had no effect on PrsmZ-lacZ reporter and RsmZ RNA levels. These combined findings demonstrate that sRNA 179 has indirect effects on both exsA transcription and ExsA translation through modulation of RsmY levels, thus impacting RsmA availability, which itself has positive effects on Vfr and ExsA synthesis.
DISCUSSION
The previous finding that Hfq influences P. aeruginosa T3SS gene expression led us to investigate the mechanism (29, 30). Potential roles for Hfq include sRNA and mRNA stability control and mRNA translational control with sRNA-assisted targeting (41). We hypothesized the involvement of an sRNA(s) in the regulation of the T3SS. Screening an sRNA expression library resulted in the identification of sRNA 179 as an Hfq-dependent inhibitor of both the T3SS and CVS regulons. sRNA 179 is 134 nt long and located downstream of acp1, a putative acyl carrier protein. The reported 5′ end of sRNA 179 immediately follows the acp1 stop codon (38) and may be generated by a processing event. sRNA 179 lacks an intact open reading frame containing a stop codon. The finding that sRNA 179 activity is Hfq-dependent supports our model that the RNA is the relevant gene product and is consistent with a recent paper showing that sRNA 179 coprecipitates with Hfq (42). sRNA 179 indirectly inhibits ExsA and Vfr synthesis at the posttranscriptional level and likely works through the Rsm system. Data supporting this include (i) the lack of inhibitory activity when sRNA 179 is overexpressed in a ΔrsmYZ mutant (Fig. 8A), (ii) increased PrsmY-lacZ transcriptional reporter activity upon sRNA 179 overexpression (Fig. 8B), and (iii) increased RsmY RNA levels upon sRNA 179 overexpression (Fig. 8C). Our working model is that the sRNA 179-mediated increase in RsmY expression titrates RsmA from some target mRNAs, leading to reduced T3SS and CVS expression, as both ExsA and Vfr synthesis are dependent upon RsmA (Fig. 1) (39). The RsmA regulon consists of ∼500 target genes and includes the T6SS (43). To examine whether sRNA 179 has a general effect on the RsmA regulon, we immunoblotted culture supernatants for the T6SS secretion substrate Hcp1 upon sRNA 179 overexpression but observed no change relative to the parental strain (data not shown). This was not entirely unexpected, as we previously demonstrated that T3SS gene expression is more sensitive to changes in RsmA availability relative to T6SS gene expression (39).
sRNA 179 inhibits the T3SS when overexpressed in the WT, ΔrsmY, and ΔrsmZ single mutants but lacks activity when expressed in the ΔrsmYZ mutant (Fig. 8A). These data suggest involvement of both rsmY and rsmZ in T3SS gene expression by sRNA 179 and may reflect the redundant function that RsmY and RsmZ play in RsmA sequestration. The homeostatic relationship between RsmA availability and RsmY/RsmZ-mediated sequestration of RsmA is a complicating factor. Because RsmA has a positive effect on rsmY and rsmZ transcription (39), deletion of either may result in a compensatory effect wherein reduced sequestration of RsmA stimulates transcription of the remaining gene. Transcription of both rsmY and rsmZ is directly controlled by the GacAS two-component regulatory system (44). It is unclear how sRNA 179 stimulates rsmY transcription, but modulation of GacAS signaling seems unlikely since rsmZ transcription is unaffected by sRNA 179. In addition to GacAS, other factors, including HptB, MvaT, and MgtE, also influence rsmYZ transcription (45–47). HptB is a promising candidate, as both HptB and sRNA 179 effect rsmY transcription only. While it is clear that rsmYZ are required for sRNA 179 activity, they may not be sufficient. Our finding that PrsmZ-lacZ reporter activity and RsmZ levels are unchanged when sRNA 179 is overexpressed in the rsmY background (Fig. 8B and D) suggest additional mechanisms of inhibition by sRNA 179.
In addition to sRNA 179, sRNA 0161 was recently identified with GRIL-seq as another inhibitor of the T3SS (32), and we confirmed that sRNA 0161 is also Hfq-dependent (Fig. 4A). The GRIL-seq data suggest that sRNA 0161 directly targets the exsA leader region through imperfect base pairing and is consistent with our data showing that sRNA 0161 inhibits ExsA synthesis at the posttranscriptional level (Fig. 5B and C). In addition, Hfq coprecipitates with the exsA leader region (42). The simplest model to account for the inhibitory activity of sRNA 0161 is occlusion of the ribosome binding site. Alternative mechanisms may involve RsmA and DeaD, both positive regulators of ExsA synthesis (22, 39). Rather than blocking ribosome access, sRNA 0161 may instead prevent RsmA and/or DeaD from binding to the exsA leader region. Another possibility is that RsmA and/or DeaD activate by displacing/preventing sRNA 0161 from base pairing with the exsA leader region. Testing these models will be the subject of future studies.
sRNA 0161 and 179 function independently from one another, and either is sufficient to inhibit T3SS gene expression (Fig. 4C). The primary implication is that together they provide both direct (through the activity of sRNA 1061) and indirect (through sRNA 179) Hfq-dependent mechanisms to inhibit ExsA synthesis. Transcription and synthesis of exsA and ExsA are major inputs for upstream regulatory factors (17). Although seemingly redundant, sRNA 0161 and sRNA 179 may differentially coordinate and/or respond to distinct signals that influence T3SS expression. Future studies to understand transcriptional control of sRNA 0161 and sRNA 179 will be important.
Regulatory control of the T3SS by Hfq likely extends beyond the mechanisms described here. Expression of Hfq in an sRNA 0161 179 double mutant still resulted in significant inhibition of T3SS gene expression (Fig. 4B). This may reflect sRNA-independent activity or involvement of an additional sRNA(s) in control of the T3SS. There are over 500 predicted sRNAs in P. aeruginosa (38), only a small fraction of which have been characterized. Most sRNAs require a chaperone to facilitate interactions with their target mRNA. sRNAs that sequester Hfq (CrcZ) and RsmA (RsmY and RsmZ) result in indirect regulation of the T3SS and other virulence factors controlled by Hfq, RsmA, and Vfr (30, 48). Finally, Hfq is known to stabilize RsmY (49). Hfq stabilization protects RsmY from RNase E-mediated cleavage and thus enhances RsmA sequestration by RsmY. Regulation of the T3SS is complex, with numerous regulators that directly or indirectly alter exsA transcription and ExsA translation (18–20, 36). Hfq can now be added to that list of regulators, highlighting the complexity of regulatory mechanisms that tightly control T3SS gene expression.
MATERIALS AND METHODS
Strain and plasmid construction.
The P. aeruginosa strains and plasmids used in this study are listed in Table S1, and the cloning details and primers are provided in Tables S2 and S3. Routine cloning was performed with E. coli Top10 or DH5alpha cultured in LB-Lennox medium with tetracycline (12 μg/ml) or gentamicin (15 μg/ml) as required. Primers were used to amplify the sRNAs represented in the library (listed in Table S3), followed by cloning into the XbaI/SacI restriction sites of pJN105. The Δhfq mutant was generated by PCR amplification of 5′ and 3′ flanking regions using primers N1/N2 and C1/C2 using PA103 genomic DNA as a template. The C1/C2 and N1/N2 PCR products were sequentially cloned into the HindIII/EcoRI and EcoRI/BamHI sites, respectively, of pBluescript SK. The combined flanking regions were then excised from pBluescript SK by digestion with XmaI and cloned into pEX100T. The sRNA 0161 and 179 allelic exchange vectors were generated using the primer pairs listed in Tables S2 and S3 and cloned by isothermal assembly into pEXG2Tc (50). The Δhfq, Δ0161, and Δ179 mutants were generated by allelic exchange and sacB-mediated resolution as previously described (51). The hfq gene from P. aeruginosa from strain PA103 was cloned into the XbaI and SpeI sites of pJN105 (52).
Plasmid construction details. Download Table S2, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
sRNA sequences and primer pairs used for PCR cloning (the underlined sequence is the sRNA sequence followed by 100 nt of downstream sequence). Download Table S3, DOCX file, 0.1 MB (77.1KB, docx) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth curves.
PA103 strains were grown overnight at 37°C in LB containing gentamicin (80 μg/ml) as required. The strains were diluted to an absorbance (A600) of 0.01 in a 96-well plate. A600 was measured every hour using a Tecan plate reader (Tecan Trading AG, Switzerland) until the stationary phase. Background absorbance measured from wells with only LB was subtracted, and absorbance was plotted against time.
β-galactosidase assays and immunoblots.
PA103 strains were cultured at 37°C overnight in LB containing 80 μg/ml gentamicin as required. The next day, strains were diluted to an absorbance (A600) of 0.1 in tryptic soy broth (TSB) supplemented with 100 mM monosodium glutamate and 1% glycerol. Arabinose was added to induce Hfq (0.05%) or sRNA (0.2%) expression from the PBAD promoter, and rhamnose (0.005%) was added to induce exsA expression from the Prha promoter as appropriate. Cultures were incubated at 37°C and harvested when the A600 reached 1.0. β-galactosidase activity was determined as previously described with the substrates ortho-nitrophenyl-galactopyranoside (ONPG) (36) or chlorophenol red-β-d-galactopyranoside (CPRG) (18) as previously described. CPRG activity was determined by measuring product formation at 578 nM and defined as CPRG units (i.e., A578/culture A600/time [min]/culture vol [ml]) × 1,000. Statistical analyses were determined with one-way analysis of variance (ANOVA) using GraphPad Prism version 5.0c for Mac OS X (GraphPad, La Jolla, CA). Immunoblots using rabbit immune serum to ExsA, Hfq, ExoU, PcrV, and Hcp1 were performed as previously described (36).
Cytotoxicity assays.
Chinese hamster ovary (CHO) cells (ATCC CCL-61) were cultured in Ham’s F-12 medium (Invitrogen Corp., Carlsbad, California) supplemented with 10% fetal calf serum, 50 units/ml of penicillin and streptomycin, 2 mM l-glutamine, 0.12% sodium bicarbonate, and 2.5 mM HEPES at 37°C in 5% CO2. For cytotoxicity assays, CHO cells were seeded at 8 × 104 cells/well into 24-well tissue culture plates (80 to 85% confluence) and incubated for 18 to 24 h at 37°C in 5% CO2. P. aeruginosa strains were grown on Vogel Bonner Minimal medium plates overnight at 37°C, washed with PBS, diluted in prewarmed Ham’s F-12 medium, and added to the CHO cells to a multiplicity of infection (MOI) of 10. Plates were centrifuged (500 × g, 5 min, 25°C) and incubated at 37°C in 5% CO2 for the indicated times. At each time point the plates were centrifuged (500 × g, 5 min, 25°C) and 50 μl of the supernatant was transferred to a 96-well plate and assayed for lactate dehydrogenase (LDH) release using the CytoTox 96 nonradioactive cytotoxicity assay (Promega, Madison, WI). The percent cytotoxicity was calculated by subtracting the optical density at 490 nm (OD490) of an uninfected control from each sample and using WT PA103 as the positive control normalized to 100%.
Hfq purification.
E. coli Tuner(DE3) expressing histidine-tagged Hfq(pET23bHfq) was cultured overnight on LB with ampicillin (200 μg/ml). Cell suspensions were prepared and used to start a 2-liter culture at an A600 of approximately 0.1 in LB with ampicillin (200 μg/ml). At an A600 of 0.5, isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added at 1 mM to induce protein production. Once the culture reached an A600 of 3.0, cells were harvested by centrifugation and suspended in nickel nitrilotriacetic acid (Ni-NTA) binding buffer (20 mM Tris-HCl [pH 7.9], 300 mM NaCl, 5 mM imidazole) supplemented with 1 complete protease inhibitor cocktail (PIC) tablet (Roche). Cells were lysed by passage through a microfluidizer at 17,000 lb/in2 (Microfluidics, Westwood, MA). Cell lysates were cleared by centrifugation and immediately loaded onto a 1-ml HiTrap His column (GE Healthcare Life Sciences) and eluted with binding buffer containing 300 mM imidazole. The peak elution fractions were pooled and dialyzed against Ni-NTA binding buffer containing 5 mM dithiothreitol (DTT) for 4 h at 4°C. The resulting protein was flash-frozen in 1-ml aliquots. Two 1-ml frozen aliquots of Hfq were buffer-exchanged by diluting to 10 ml with Ni-NTA binding buffer and then concentrated to 1 ml using an Amicon Ultra centrifugal filter (nominal molecular weight limit, 10,000 Da) and repeated five times. The concentrated Hfq samples were then combined and exposed to 0.15 ml of preequilibrated Talon beads (Clontech) for 10 min, rocking at 4 degrees. Beads were then washed with 5 ml of Ni-NTA binding buffer three times and eluted with 10 ml of binding buffer containing 0.5 M imidazole. The eluted Hfq was concentrated again and buffer-exchanged three times to storage buffer (20 mM Tris-HCl [pH 7.9], 300 mM NaCl, 1 mM DTT). Protein concentration was determined using the Bradford assay.
Electrophoretic mobility shift assays.
RNA was generated from DNA templates encoding the exsA UTR region and sRNA 179 by in vitro transcription and end-labeled with [γ-32] ATP as previously described (24). Purified Hfqhis at the indicated concentrations was incubated with the RNA probes in 1× binding buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM KCl, 3.25 ng/μl total yeast tRNA [Life Technologies], 10 mM DTT, 5% [vol/vol] glycerol, and 0.1 unit RNaseOUT [Life Technologies]). Reaction mixtures were incubated at 37°C for 30 min and then mixed with 2 μl of gel loading buffer II (Life Technologies) and immediately subjected to electrophoresis on 10% (wt/vol) native polyacrylamide glycine gels (10 mM Tris-HCl [pH 7.5], 380 mM glycine, and 1 mM EDTA) at 4°C. Imaging was performed using an FLA-7000 phosphorimager (Fujifilm), and the images were analyzed using MultiGuage v3.0 software.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant number R01 AI055042-13 to T.L.Y. E.W.M. and L.D. were supported by National Institutes of Health training grants T32 AI007511-23 and T32 AI007343-30, respectively.
Special thanks go to University of Iowa undergraduate and summer students Matthew Ferry, Jessica Griffith, Trey Van Hemert, Emily Hoeper, Isabella Morbidelli, and Yuxun Zhang for assistance with the sRNA screen.
Footnotes
This article is a direct contribution from Timothy L. Yahr, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Daniel Wozniak, The Ohio State University, and James Slauch, University of Illinois at Urbana Champaign.
Citation Janssen KH, Corley JM, Djapgne L, Cribbs JT, Voelker D, Slusher Z, Nordell R, Regulski EE, Kazmierczak BI, McMackin EW, Yahr TL. 2020. Hfq and sRNA 179 inhibit expression of the Pseudomonas aeruginosa cAMP-Vfr and type III secretion regulons. mBio 11:e00363-20. https://doi.org/10.1128/mBio.00363-20.
REFERENCES
- 1.Stryjewski ME, Sexton DJ. 2003. Pseudomonas aeruginosa infections in specific types of patients and clinical settings, p 1–15. In Hauser AR, Rello J (ed), Severe infections caused by Pseudomonas aeruginosa. Perspectives on critical care infectious diseases vol 7 Springer, Boston, MA. [Google Scholar]
- 2.Wolfgang MC, Lee VT, Gilmore ME, Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell 4:253–263. doi: 10.1016/s1534-5807(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 3.Coggan KA, Wolfgang MC. 2012. Global regulatory pathways and cross-talk control pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol 14:47–70. [PubMed] [Google Scholar]
- 4.Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet 10:e1004518. doi: 10.1371/journal.pgen.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasil ML. 2007. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20:587–601. doi: 10.1007/s10534-006-9067-2. [DOI] [PubMed] [Google Scholar]
- 6.Koh AY, Mikkelsen PJ, Smith RS, Coggshall KT, Kamei A, Givskov M, Lory S, Pier GB. 2010. Utility of in vivo transcription profiling for identifying Pseudomonas aeruginosa genes needed for gastrointestinal colonization and dissemination. PLoS One 5:e15131. doi: 10.1371/journal.pone.0015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chugani S, Greenberg EP. 2007. The influence of human respiratory epithelia on Pseudomonas aeruginosa gene expression. Microb Pathog 42:29–35. doi: 10.1016/j.micpath.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisk A, Schurr JR, Wang G, Bertucci DC, Marrero L, Hwang SH, Hassett DJ, Schurr MJ. 2004. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect Immun 72:5433–5438. doi: 10.1128/IAI.72.9.5433-5438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs EL, Brutinel ED, Jones AK, Fulcher NB, Urbanowski ML, Yahr TL, Wolfgang MC. 2010. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J Bacteriol 192:3553–3564. doi: 10.1128/JB.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs EL, Brutinel ED, Klem ER, Fehr AR, Yahr TL, Wolfgang MC. 2010. In vitro and in vivo characterization of the Pseudomonas aeruginosa cyclic AMP (cAMP) phosphodiesterase CpdA, required for cAMP homeostasis and virulence factor regulation. J Bacteriol 192:2779–2790. doi: 10.1128/JB.00168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones AK, Fulcher NB, Balzer GJ, Urbanowski ML, Pritchett CL, Schurr MJ, Yahr TL, Wolfgang MC. 2010. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J Bacteriol 192:5709–5717. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Solh AA, Hattemer A, Hauser AR, Alhajhusain A, Vora H. 2012. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Med 40:1157–1163. doi: 10.1097/CCM.0b013e3182377906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peña C, Suarez C, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez F, Tubau F, Oliver A, Martínez-Martínez L, Spanish Network for Research in Infectious Diseases. 2013. Effect of adequate single-drug vs combination antimicrobial therapy on mortality in Pseudomonas aeruginosa bloodstream infections: a post hoc analysis of a prospective cohort. Clin Infect Dis 57:208–216. doi: 10.1093/cid/cit223. [DOI] [PubMed] [Google Scholar]
- 14.Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol 7:654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank DW. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol 26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 16.Yahr TL, Frank DW. 1994. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol 176:3832–3838. doi: 10.1128/jb.176.13.3832-3838.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams McMackin EA, Djapgne L, Corley JM, Yahr TL. 2019. Fitting pieces into the puzzle of Pseudomonas aeruginosa type III secretion system gene expression. J Bacteriol 201:e00209-19. doi: 10.1128/JB.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsden AE, Intile PJ, Schulmeyer KH, Simmons-Patterson ER, Urbanowski ML, Wolfgang MC, Yahr TL. 2016. Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 198:1442–1450. doi: 10.1128/JB.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X, Li M, Pan X, Zheng R, Liu C, Chen F, Liu X, Cheng Z, Jin S, Wu W. 2017. Fis regulates type III secretion system by influencing the transcription of exsA in Pseudomonas aeruginosa strain PA14. Front Microbiol 8:669. doi: 10.3389/fmicb.2017.00669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang H, Deng X, Li X, Ye Y, Wu M. 2014. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res 42:10307–10320. doi: 10.1093/nar/gku586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams McMackin EA, Marsden AE, Yahr TL. 2019. H-NS family members MvaT and MvaU regulate the Pseudomonas aeruginosa type III secretion system. J Bacteriol 201:e00054-19. doi: 10.1128/JB.00054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Intile PJ, Balzer GJ, Wolfgang MC, Yahr TL. 2015. The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 197:2664–2674. doi: 10.1128/JB.00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapouge K, Schubert M, Allain FH, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 24.Marden JN, Diaz MR, Walton WG, Gode CJ, Betts L, Urbanowski ML, Redinbo MR, Yahr TL, Wolfgang MC. 2013. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 110:15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen KH, Diaz MR, Golden M, Graham JW, Sanders W, Wolfgang MC, Yahr TL. 2018. Functional analyses of the RsmY and RsmZ small noncoding regulatory RNAs in Pseudomonas aeruginosa. J Bacteriol 200:e00736-17. doi: 10.1128/JB.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franze de Fernandez MT, Eoyang L, August JT. 1968. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 27.Sobrero P, Valverde C. 2012. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol 38:276–299. doi: 10.3109/1040841X.2012.664540. [DOI] [PubMed] [Google Scholar]
- 28.Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE, Blasi U. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog 35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- 29.Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Bläsi U. 2006. Hfq-dependent alterations of the transcriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Mol Microbiol 59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- 30.Sonnleitner E, Prindl K, Blasi U. 2017. The Pseudomonas aeruginosa CrcZ RNA interferes with Hfq-mediated riboregulation. PLoS One 12:e0180887. doi: 10.1371/journal.pone.0180887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shakhnovich EA, Davis BM, Waldor MK. 2009. Hfq negatively regulates type III secretion in EHEC and several other pathogens. Mol Microbiol 74:347–363. doi: 10.1111/j.1365-2958.2009.06856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YF, Han K, Chandler CE, Tjaden B, Ernst RK, Lory S. 2017. Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol Microbiol 106:919–937. doi: 10.1111/mmi.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsui HC, Leung HC, Winkler ME. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 34.Muffler A, Fischer D, Hengge-Aronis R. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev 10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 35.Hill IT, Tallo T, Dorman MJ, Dove SL. 2019. Loss of RNA chaperone Hfq unveils a toxic pathway in Pseudomonas aeruginosa. J Bacteriol 201:e00232-19. doi: 10.1128/JB.00232-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCaw ML, Lykken GL, Singh PK, Yahr TL. 2002. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol 46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 37.Finck-Barbancon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish JP, Fleiszig SM, Wu C, Mende-Mueller L, Frank DW. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol 25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 38.Gomez-Lozano M, Marvig RL, Molin S, Long KS. 2012. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol 14:2006–2016. doi: 10.1111/j.1462-2920.2012.02759.x. [DOI] [PubMed] [Google Scholar]
- 39.Intile PJ, Diaz MR, Urbanowski ML, Wolfgang MC, Yahr TL. 2014. The AlgZR two-component system recalibrates the RsmAYZ posttranscriptional regulatory system to inhibit expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 196:357–366. doi: 10.1128/JB.01199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papenfort K, Vogel J. 2010. Regulatory RNA in bacterial pathogens. Cell Host Microbe 8:116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Updegrove TB, Zhang A, Storz G. 2016. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol 30:133–138. doi: 10.1016/j.mib.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chihara K, Bischler T, Barquist L, Monzon VA, Noda N, Vogel J, Tsuneda S. 2019. Conditional Hfq association with small noncoding RNAs in Pseudomonas aeruginosa revealed through comparative UV cross-linking immunoprecipitation followed by high-throughput sequencing. mSystems 4:e00590-19. doi: 10.1128/mSystems.00590-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burrowes E, Baysse C, Adams C, O’Gara F. 2006. Influence of the regulatory protein RsmA on cellular functions in Pseudomonas aeruginosa PAO1, as revealed by transcriptome analysis. Microbiology 152:405–418. doi: 10.1099/mic.0.28324-0. [DOI] [PubMed] [Google Scholar]
- 44.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 45.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bordi C, Lamy MC, Ventre I, Termine E, Hachani A, Fillet S, Roche B, Bleves S, Mejean V, Lazdunski A, Filloux A. 2010. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol 76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chakravarty S, Melton CN, Bailin A, Yahr TL, Anderson GG. 2017. Pseudomonas aeruginosa magnesium transporter MgtE inhibits type III secretion system gene expression by stimulating rsmYZ transcription. J Bacteriol 199:e00268-17. doi: 10.1128/JB.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valverde C, Heeb S, Keel C, Haas D. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol Microbiol 50:1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x. [DOI] [PubMed] [Google Scholar]
- 49.Sorger-Domenigg T, Sonnleitner E, Kaberdin VR, Bläsi U. 2007. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem Biophys Res Commun 352:769–773. doi: 10.1016/j.bbrc.2006.11.084. [DOI] [PubMed] [Google Scholar]
- 50.Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, Berrington WR, Goddard AF, Zheng C, Angermeyer A, Brittnacher MJ, Kitzman J, Shendure J, Fligner CL, Mittler J, Aitken ML, Manoil C, Bruce JE, Yahr TL, Singh PK. 2015. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 52.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/s0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains and plasmids used in this study. Download Table S1, DOCX file, 0.03 MB (32.6KB, docx) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) WT PA103 and an exsA mutant carrying a rhamnose-inducible copy of exsA integrated at the Tn7 site were cultured under inducing conditions for T3SS gene expression. Rhamnose was included as indicated to induce exsA expression from the Tn7 integrant (the concentrations used for the titration: 0.0025, 0.005, 0.01, 0.025, and 0.05%) ExsA activity was measured using the PexsD-lacZ reporter. Download FIG S1, TIF file, 0.3 MB (352KB, tif) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Hfq interacts with sRNA 0161 and the exsA mRNA leader region. (A and B) Electrophoretic mobility shift assays were performed by incubating radiolabeled sRNA 0161 (A) or exsA mRNA leader region (B) with the indicated concentrations of Hfq. Radiolabeled sRNA 0161 was also incubated with unlabeled exsA mRNA leader region to detect formation of an RNA:RNA hybrid. The positions of the respective complexes are indicated with arrowheads. Download FIG S2, TIF file, 0.3 MB (317.7KB, tif) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid construction details. Download Table S2, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
sRNA sequences and primer pairs used for PCR cloning (the underlined sequence is the sRNA sequence followed by 100 nt of downstream sequence). Download Table S3, DOCX file, 0.1 MB (77.1KB, docx) .
Copyright © 2020 Janssen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.