The work described here further elucidates how microorganisms sense and adapt to changes in their environment to establish infections in the human host. Specifically, we uncover a novel mechanism by which an opportunistic human fungal pathogen, Cryptococcus neoformans, responds to increases in extracellular pH in order to survive and thrive within the relatively alkaline environment of the human lung. This mechanism, which is intimately linked with fungal membrane sterol homeostasis, is independent of the previously well-studied alkaline response Rim pathway. Furthermore, this ergosterol-dependent alkaline pH response is present in Candida albicans, indicating that this mechanism spans diverse fungal species. These results are also relevant for novel antimicrobial drug development as we show that currently used ergosterol-targeting antifungals are more active in alkaline environments.
KEYWORDS: Cryptococcus neoformans, ergosterol, fungal genetics, membrane, pH homeostasis
ABSTRACT
The ability for cells to maintain homeostasis in the presence of extracellular stress is essential for their survival. Stress adaptations are especially important for microbial pathogens to respond to rapidly changing conditions, such as those encountered during the transition from the environment to the infected host. Many fungal pathogens have acquired the ability to quickly adapt to changes in extracellular pH to promote their survival in the various microenvironments encountered during a host infection. For example, the fungus-specific Rim/Pal alkaline response pathway has been well characterized in many fungal pathogens, including Cryptococcus neoformans. However, alternative mechanisms for sensing and responding to host pH have yet to be extensively studied. Recent observations from a genetic screen suggest that the C. neoformans sterol homeostasis pathway is required for growth at elevated pH. This work explores interactions among mechanisms of membrane homeostasis, alkaline pH tolerance, and Rim pathway activation. We find that the sterol homeostasis pathway is necessary for growth in an alkaline environment and that an elevated pH is sufficient to induce Sre1 activation. This pH-mediated activation of the Sre1 transcription factor is linked to the biosynthesis of ergosterol but is not dependent on Rim pathway signaling, suggesting that these two pathways are responding to alkaline pH independently. Furthermore, we discover that C. neoformans is more susceptible to membrane-targeting antifungals under alkaline conditions, highlighting the impact of microenvironmental pH on the treatment of invasive fungal infections. Together, these findings further connect membrane integrity and composition with the fungal pH response and pathogenesis.
INTRODUCTION
Diverse cell types, from simple unicellular microorganisms to complex multicellular eukaryotes, interpret alterations in extracellular pH as a common signal for changes in the external environment. Pathogenic microorganisms are often uniquely exposed to wide fluctuations in pH as they move between various microenvironments in the human host. Among these, fungi that cause invasive fungal infections (IFIs) have acquired the ability to rapidly adapt to changes in extracellular pH to promote their survival during an infection. The shift of a fungal pathogen from an acidic external environment to the neutral/alkaline pH of the mammalian host is associated with the activation of the fungus-specific Rim/Pal signaling pathway, triggering cellular changes important for survival under these new conditions. These changes include alterations in the cell wall, often accompanied by larger morphological transitions that promote host colonization. In the common fungal pathogen Candida albicans, pH-directed cellular responses include the ability to transition between yeast-like growth and invasive hyphal forms (1–3). The opportunistic human fungal pathogen and basidiomycete yeast Cryptococcus neoformans similarly activates Rim signaling to respond to changes in pH. Because C. neoformans initially colonizes the human lung, which is often relatively more alkaline than its natural environmental reservoirs, this signaling pathway is activated in the setting of infection. In fact, the C. neoformans Rim101 transcription factor, the terminal component of the Rim pathway, is among the most highly induced transcripts in vivo (4).
Given its pH-dependent activation, as well as its important role in the adaptation of fungal cells to elevated pH, the Rim signaling cascade is often considered to be the major alkaline pH response pathway in fungi. However, other cellular processes and pathways are required for fungal growth under conditions of extreme pH (both acidic and alkaline). These processes include the production of glycosphingolipids (GSLs) that associate with proteins in the outer leaflet of fungal plasma membranes to form lipid rafts and maintain membrane fluidity and organization (5–7). Recent studies have demonstrated that mutations resulting in reduced or absent GSLs render fungi such as Kluyveromyces lactis, Neurospora crassa, and C. neoformans unable to grow in alkaline environments (8–11). The connection between membrane composition and the ability for fungal cells to grow in alkaline environments has been associated with defects in cytokinesis and altered activity of plasma membrane proton pumps, as well as an altered lipid profile (10). Furthermore, reduced ergosterol content in membranes has been linked to salt stress sensitivity in Saccharomyces cerevisiae (12, 13) and to aberrant V-ATPase regulation of pH gradients in Candida albicans (13, 14).
Recent observations from our genetic screen suggest that C. neoformans sterol homeostasis might also be required for growth at elevated pH (15). The sterol homeostasis pathway (SREBP pathway) has been extensively studied in both mammalian and fungal cells (16–20). Proteins in this pathway regulate the production and delivery of sterols to the plasma membrane to maintain appropriate cell homeostasis (17, 21, 22). In several fungal species, including C. neoformans, the Sre1 transcription factor (the terminal transcription factor in this sterol homeostasis pathway) is activated in response to low-oxygen conditions (21, 23–26). In addition to hypoxia, the C. neoformans Sre1 transcriptional response is necessary for tolerance to low iron and to antifungals that target sterols in the membrane (21). Upon activation of the C. neoformans sterol homeostasis pathway, the basidiomycete-specific Stp1 protease cleaves Sre1, freeing its N terminus to release from the membrane of the endoplasmic reticulum and translocate to the nucleus (22). This cleavage is induced in an O2-dependent manner and is important for the transcription of many ergosterol biosynthesis genes (23, 25). However, the association between the sterol homeostasis pathway and pH adaptation has not yet been explored.
Here, we define potential interactions among fungal sterol homeostasis, alkaline pH tolerance, and Rim pathway activation. We find that the sterol homeostasis pathway is indeed necessary for growth in an alkaline environment and that an elevated pH is sufficient to induce Sre1 cleavage and activation. This pH-mediated activation of the Sre1 transcription factor is not dependent on Rim pathway signaling, suggesting that these two pathways are responding to alkaline pH independently. Furthermore, we demonstrate that Sre1-mediated ergosterol biosynthesis is linked to the response to alkaline pH and relevant in biologically diverse fungi. Finally, we discover that C. neoformans is more susceptible to membrane-targeting antifungals under alkaline conditions, highlighting the impact of microenvironmental pH on the treatment of this invasive fungal infection. Together, these findings connect a highly conserved pathway involved in membrane homeostasis and sterol maintenance to the adaptive response to changes in extracellular pH.
RESULTS
Convergent and divergent phenotypes of the sre1Δ and rim101Δ mutants.
A recent forward genetic screen identified two elements of the C. neoformans sterol homeostasis pathway, the Sre1 transcription factor and its activating protease Stp1, as proteins required for growth of this pathogenic fungus at an alkaline pH (15). To confirm this observation, we generated and acquired multiple, independent C. neoformans sre1Δ mutants and verified that all demonstrated a severe growth defect at high pH (Fig. 1A). We performed detailed phenotypic comparisons between mutants in the alkaline-responsive Rim pathway and the sterol homeostasis pathway, as exemplified by the rim101Δ and sre1Δ transcription factor mutant strains, respectively. Both mutant strains grew similarly to wild type (WT) on a rich growth medium at pH 5.5 (yeast extract-peptone-dextrose [YPD] medium). These two mutants also displayed similar growth defects as the wild type on growth medium buffered to a pH greater than 7 (Fig. 1A). Importantly, the sre1Δ alkaline pH-sensitive mutant phenotype was rescued by the reintroduction of the wild-type SRE1 allele (see Fig. S1A in the supplemental material).
FIG 1.
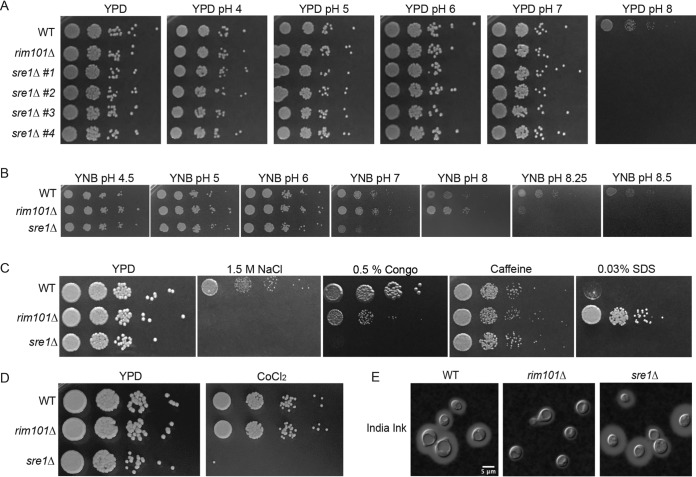
Stress response phenotypes of the sre1Δ and rim101Δ mutant strains. (A) Four independent sre1Δ mutant strains were serially diluted onto YPD medium and YPD pH 4 to 8. Growth was compared to wild type (WT) and a rim101Δ mutant known to have alkaline pH sensitivity. Growth was assessed after 3 days. sre1Δ #1 (HEB5) is shown for all subsequent phenotyping and analysis. (B) The sre1Δ and rim101Δ mutant strains are unable to grow at increasing pH levels on minimal medium (YNB). Strains were spotted in serial dilutions onto YNB medium buffered to pH 4.5 to 8.5, and growth was compared to WT after 3 days. (C) The sre1Δ and rim101Δ mutant strains display distinct and overlapping phenotypes to cell stressors. Strains were serially diluted and spotted to either YPD, YPD plus 1.5 M NaCl, YPD plus 0.5% Congo red, YPD plus 1 mg/ml caffeine, or YPD plus 0.03% SDS. Growth was compared to WT and assessed after 3 days. (D) The sre1Δ mutant strain displays a growth defect in response to hypoxia-mimicking growth conditions (7 mM CoCl2). Strains were spotted in serial dilutions onto YPD at 30°C and YES plus 7 mM CoCl2 at 30°C. Growth was assessed after 3 days and compared to WT and the rim101Δ mutant. (E) The sre1Δ mutant strain does not have the same capsule deficiency as the rim101Δ mutant strain. Strains were incubated in CO2-independent medium for 3 days before imaging using India ink exclusion counterstaining. Capsule is noted as a halo of clearing around the yeast cells.
The sre1Δ mutant phenotypes are rescued by the reintroduction of the wild-type SRE1 allele, demonstrating a distinct growth defect from the rim101Δ mutant under microaerophilic conditions. (A) Strains were spotted in serial dilutions onto YPD pH 5.5, YPD pH 8, and YES plus 7 mM CoCl2. Growth was assessed after 3 days of growth and compared to both the sre1Δ mutant and WT strains. (B) The sre1Δ and stp1Δ mutant strains displayed an expected growth rate and colony size reduction in response to microaerophilic growth conditions. The rim101Δ mutant was able to grow in this environment similarly to wild-type levels. Strains were spotted in serial dilutions onto YPD at 30°C and YPD at 30°C and incubated under microaerophilic conditions. Download FIG S1, TIF file, 1.0 MB (1,021.3KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To account for a potential confounding effect on growth by exogenous lipids in the yeast extract-rich medium, we also assessed the ability for these mutant strains to grow on a minimal medium (yeast nitrogen base [YNB]) buffered to a range of pH values. The sre1Δ and rim101Δ mutants were able to grow on YNB medium buffered to pH 4 through pH 7. At more alkaline pH, the growth defect of the sre1Δ mutant strain was more severe than that of the rim101Δ mutant, with the sre1Δ mutant unable to grow at pH 8 and the rim101Δ strain displaying complete growth inhibition only at pH >8 (Fig. 1B).
Given the established role of Sre1 in mediating growth in hypoxia, we compared growth rates of these mutant strains in the presence of cobalt chloride (CoCl2), an agent that disrupts many biochemical pathways, including the ergosterol biosynthesis pathway and cellular respiration (27–30). Consistent with previous reports (21, 24, 25, 31), the sre1Δ mutant is unable to grow under these conditions (Fig. 1D and Fig. S1A). The rim101Δ mutant did not share this growth defect and was not sensitive to CoCl2 (Fig. 1D). We also compared growth rates of all mutant strains in a microaerophilic chamber to more directly test phenotypes in response to reduced oxygen. The sterol homeostasis pathway mutants displayed a lower growth rate under conditions of reduced oxygen concentration (Fig. S1B). The rim101Δ mutant grew to similar levels as the wild type (Fig. S1B). Therefore, although sharing a similar alkaline growth defect, the rim101Δ and sre1Δ mutants display distinct growth patterns under hypoxia-like conditions.
We also tested the sensitivity of the sre1Δ and rim101Δ mutant strains to cell wall stressors such as Congo red (interferes with beta glucan-chitin linkages), caffeine (affects cell wall integrity), high salt (osmotically stresses the cell wall), and SDS (stresses the cell membrane) (32, 33). Similarly to alkaline pH, high salt resulted in complete growth inhibition for both mutant strains (Fig. 1C). In contrast, caffeine did not affect the growth of either mutant (Fig. 1C). The sre1Δ mutant strain was unable to grow in the presence of Congo red, whereas the rim101Δ mutant strain showed only a subtle growth defect due to this chitin polymer inhibitor (Fig. 1C). Also, SDS completely inhibited growth of the sre1Δ strain, whereas the rim101Δ strain appeared to be hyperresistant to the membrane-targeting effects of SDS, as evident in the more robust growth of this strain than of the wild type (32, 34) (Fig. 1C).
Sensitivities of mutant strains to cell surface stressors can indicate alterations in the cell wall structure and/or integrity. In addition to providing a protective barrier for the cell, the cell wall serves as an anchor for the attachment of the polysaccharide capsule that can further protect the fungal cells during a human infection (35). The rim101Δ mutant strain is known to have a disorganized cell wall and thus a decrease in attached capsule (36, 37). In contrast, the sre1Δ mutant strain revealed intact capsule formation (38) (Fig. 1E). Overall, these phenotypic comparisons distinguish the rim101Δ mutant from the sre1Δ mutant in the distinct responses of these strains to cell wall and membrane stress.
Independent signaling of the Rim and sterol homeostasis pathways.
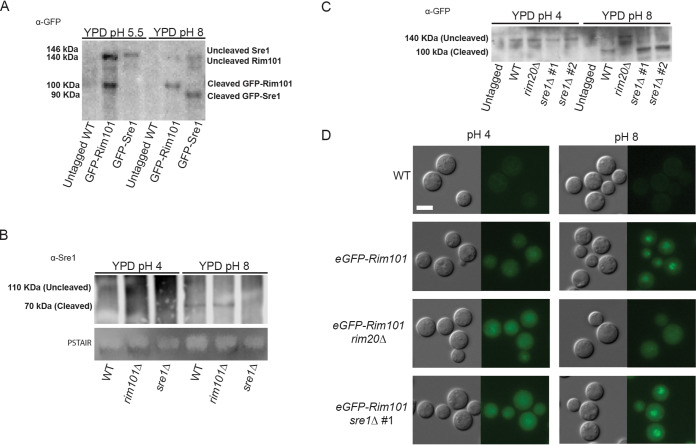
To determine whether the C. neoformans sterol homeostasis pathway is specifically activated in response to alkaline pH, we assessed the pH dependence of the proteolytic cleavage of the Sre1 transcription factor, a marker of pathway activation (21, 23–25, 38). At pH 5.5, the green fluorescent protein (GFP)-Sre1 fusion protein remains uncleaved in a 140-kDa form (Fig. 2A). In contrast, incubating this strain in the same growth medium buffered to pH 8 results in GFP-Sre1 protein cleavage to a 90-kDa form, similar to its proteolytic activation in response to hypoxia (Fig. 2A) (21). There was no defect in Sre1 cleavage in the rim101Δ mutant strain background (Fig. 2B). Therefore, the C. neoformans sterol homeostasis pathway is specifically activated by an alkaline pH signal and in a manner that is independent of the Rim alkaline response pathway.
FIG 2.
Sre1 activation is dependent on alkaline pH but not Rim signaling. (A) Western blot of both Sre1 and GFP-Rim101 protein processing under low-pH and high-pH growth conditions. The GFP-Rim101 fusion protein is cleaved from its 140-kDa form to its active 100-kDa form at pH 8. Similarly, the GFP-Sre1 fusion protein is proteolytically processed from 146 kDa to approximately 90 kDa in response to alkaline pH. Indicated strains were incubated for 60 min in either pH 5.5 or pH 8 YPD medium prior to lysing. Rim101 and Sre1 protein processing was determined using a GFP-trap pulldown and Western blotting using an anti-GFP antibody. Protein levels were normalized prior to loading. (B) Western blot analysis of the Sre1 protein in different genetic backgrounds revealed the cleavage and processing (from 110 kDa to approximately 90 kDa) of the Sre1 transcription factor in the WT and rim101Δ mutant backgrounds. Indicated strains were incubated for 60 min in either pH 4 or pH 8 YPD medium prior to lysing. Protein processing was determined through protein A pulldown and Western blotting using a polyclonal anti-Sre1 antibody. Total protein levels are represented by a PSTAIR loading control. (C) The Sre1 protein is cleaved in response to alkaline pH. The eGFP-RIM101 allele was expressed in the WT, rim20Δ mutant, and two independent sre1Δ mutant strains (sre1Δ #1 and sre1Δ #2). The untagged WT strain and the eGFP-Rim101-expressing strains were incubated in YPD medium pH 4 or pH 8 for 60 min. Rim101 processing was assessed using a GFP-trap pulldown and Western blotting using an anti-GFP antibody. Protein levels were normalized prior to loading. (D) The indicated strains (the same as in panel C) were incubated in synthetic complete medium buffered to pH 4 or pH 8 for 60 min. Rim101 localization was assessed by epifluorescence microscopy, and alkaline-induced nuclear localization was compared to the eGFP-Rim101 positive control. White scale bars indicate 5 μm.
To further define the interaction between the Sre1 and Rim101 signaling pathways, we assessed whether the Sre1 transcription factor is necessary for activation of the Rim pathway as measured by the pH-dependent proteolytic processing and subcellular localization of the Rim101 transcription factor (39). In both the wild-type and sre1Δ mutant strains, we observed intact Rim101 processing and cleavage at elevated pH (Fig. 2C). Similarly, GFP-Rim101 nuclear localization was enhanced at activating pH in both strain backgrounds (Fig. 2D). In contrast, we confirmed both defective protein cleavage and impaired nuclear localization of the Rim101 transcription factor in the rim20Δ mutant, a strain lacking a known upstream Rim signaling component (Fig. 2C and D). These data indicate that the sterol homeostasis pathway is not required for Rim pathway activation.
The cell wall organization of the sre1Δ mutant and its in vitro immune phenotypes.
The sre1Δ mutant strain is avirulent in a mouse model of C. neoformans infection (25, 38), whereas the rim101Δ mutant strain and other Rim pathway mutants have paradoxical hypervirulent phenotypes in the same model (36). In previous work, we demonstrated by transmission electron microscopy that the rim101Δ mutant has an aberrant, thick, and disorganized cell wall in comparison to wild-type cells (36). We probed the rim101Δ and sre1Δ mutant strains with calcofluor white (CFW) and wheat germ agglutinin (WGA) to assess total and exposed levels of chitin, respectively. In both mutant strains, we noted similar increases in cell wall chitin levels as measured by CFW staining. However, the level of exposed chitin (WGA) was increased only in the rim101Δ strain. The intensity of the WGA fluorescence was quantified by measuring brightness intensity (Fiji) in photomicrographs (Fig. 3A) as well as by flow cytometry (Fig. S2). The observed increase in total chitin levels can be a nonspecific response to cell stress (40). However, increased chitin exposure, as assessed by intensity of WGA staining, has been previously demonstrated to correlate with the degree of macrophage activation in vitro (36, 41). Together, these cell wall analyses suggest that the Rim pathway and sterol homeostasis pathway induce distinct microbial physiological responses to host-like conditions. Specifically, the Sre1-mediated response to host stress does not include increased exposure of chitin.
FIG 3.
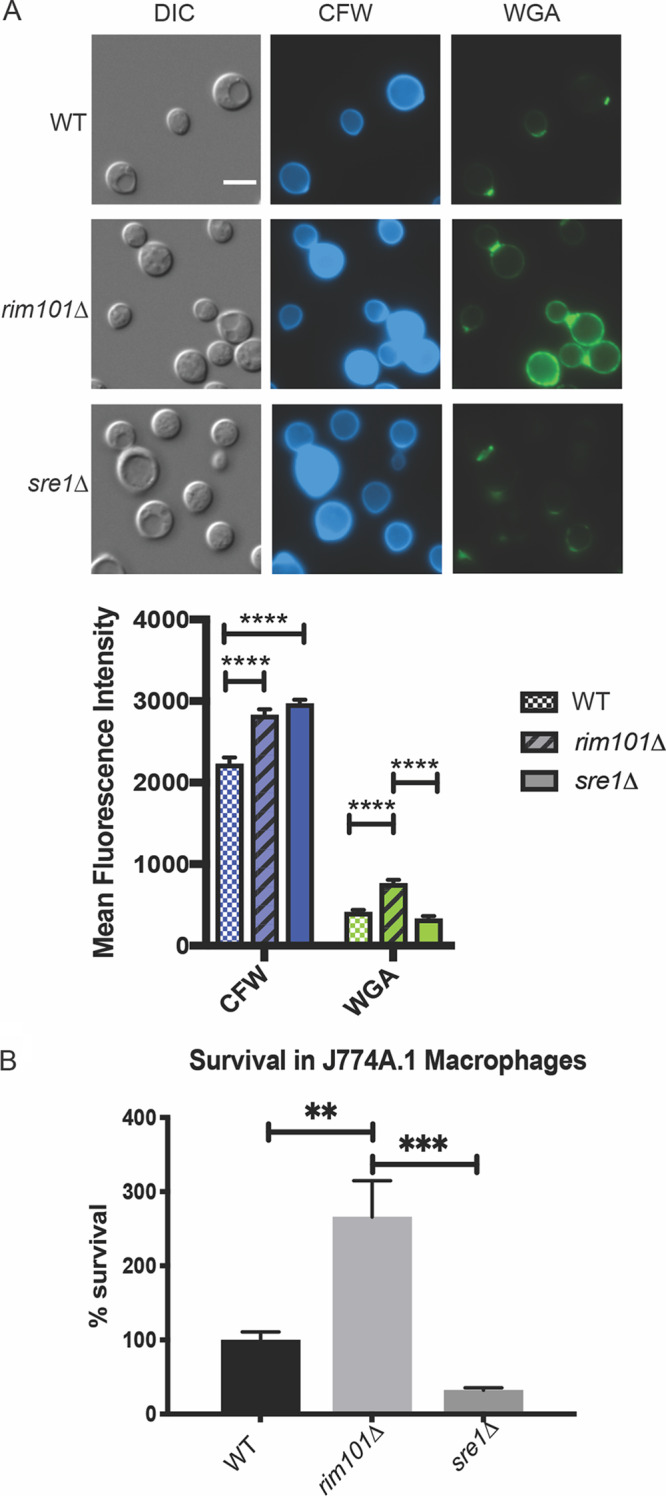
sre1Δ and rim101Δ mutant strains have varied changes in cell wall chitin exposure and interactions with host immune cells. (A) Staining of rim101Δ, sre1Δ, and wild-type cells with calcofluor white (CFW) and wheat germ agglutinin (WGA). Cells were incubated in CO2-independent medium for 18 h at 37°C. Cells were stained with FITC-conjugated WGA and CFW and incubated in the dark for 35 min and 10 min, respectively. Mean fluorescence intensity was quantified for each strain and each condition. Two-way ANOVA and Tukey’s multiple-comparison test were run to determine statistical significance. White scale bars indicate 5 μm. ****, P value < 0.0001. DIC, differential inference contrast. (B) When grown in the presence of J774A.1 macrophages, the rim101Δ mutant strain can survive significantly better than both the wild-type and the sre1Δ mutant strain. Indicated strains were coincubated with macrophages for 24 h, and survival was determined by quantitative cultures. One-way ANOVA and Tukey’s multiple-comparison tests were run to assess statistical significance between fungal cell survival percentages. Six biological replicates of each strain were analyzed. **, P value < 0.003; ***, P value < 0.0002.
Quantitative analysis of exposed chitin levels in rim101Δ and sre1Δ mutant cell walls. The rim101Δ cell wall has increased exposed chitin staining by flow cytometry compared to WT and the sre1Δ mutant cell walls. (A and C) All strains were incubated for 16 to 18 h at 30°C in YPD medium (A) or 37°C in TC medium (C), fixed, labeled, and analyzed by flow cytometry. Wheat germ agglutinin (WGA) was used to stain the cell walls for exposed chitin. (A and C) Positive events were gated in the forward scatter (FSC)/side scatter (SSC) plots and are represented as histograms with cell counts on the y axis and mean fluorescence on the x axis. Unstained cells were sorted as controls to determine positive events. (B and D) The geometric means from positive events for each strain. Download FIG S2, TIF file, 1.0 MB (1,016.2KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To further define the extent to which these cell wall epitopes may affect virulence, we assessed macrophage interactions with the sre1Δ mutant compared to the rim101Δ mutant strain. Macrophages are among the first immune cells encountered by this pathogen when infecting its host in the human lung. We therefore quantified fungal survival after coculturing stimulated J774A.1 murine macrophage-like cells with the wild-type, rim101Δ, and sre1Δ strains. Following coincubation with macrophages, the rim101Δ mutant strain displayed increased survival compared to wild type, as has been shown previously (37) (Fig. 3B). The sre1Δ mutant strain displayed a moderate, reproducible reduction in viability in the presence of macrophages compared to the wild-type strain. This result was consistent with the previously reported attenuated virulence of the sre1Δ mutant strain in animal models of infection (25, 38). The significantly different patterns of macrophage interaction of the sre1Δ and rim101Δ mutant strains further suggest that distinct downstream cellular processes are controlled by these alkaline-responsive pathways (Fig. 3B).
Ergosterol biosynthesis is required for growth at alkaline pH in C. neoformans and other fungal pathogens.
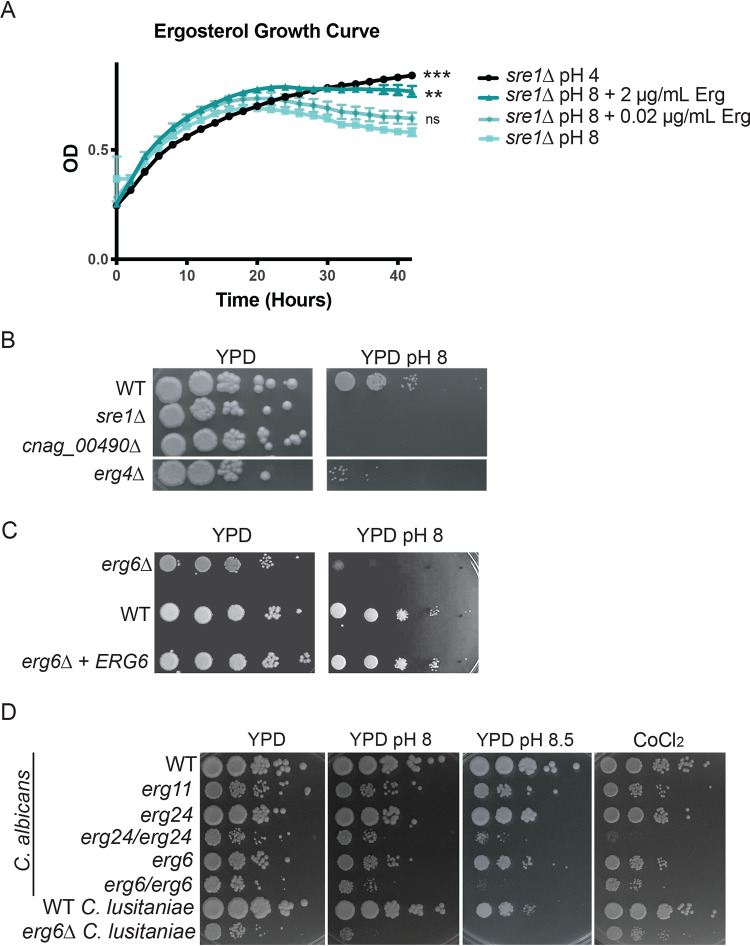
Our data support that the Rim and sterol homeostasis pathways are independent cell signaling pathways that each mediate adaptive responses to alkaline stress. Given the established role of fungal Sre1 orthologs in the regulation of membrane sterol content, we hypothesized that alterations in minor membrane lipids, especially ergosterol, might be involved in the adaptive response to alkaline pH. Previous work in C. neoformans sterol homeostasis documented decreased ergosterol levels in the sre1Δ mutant strain (22, 25). The sre1Δ alkaline pH sensitivity was rescued by the addition of exogenous ergosterol to the growth medium in a dose-dependent manner (Fig. 4A). Importantly, addition of exogenous sterols did not affect the pH of the growth medium. This observation is similar to prior investigations showing growth rescue of various S. cerevisiae ergosterol biosynthesis mutants by supplementation with exogenous ergosterol (42). These data suggest that intact ergosterol induction and homeostasis are specifically required for fungal adaptation to alkaline pH.
FIG 4.
Altered ergosterol content renders strains sensitive to alkaline pH. (A) The reduced growth rate of the sre1Δ mutant strain in liquid growth medium at pH 8 can be rescued through the addition of exogenous ergosterol in a dose-dependent manner. Growth rate of indicated strains was assessed by changes in OD595 in biological triplicate every 10 min for 42 h at 30°C. Ergosterol was added as indicated. One-way ANOVA and Dunnett’s multiple-comparison test were run on the last time point under each condition compared to the pH 8-alone condition to determine statistical significance. **, P value < 0.003; ***, P value < 0.0005; ns, not significant. (B) Other sterol-related mutants exhibit alkaline pH sensitivity. Two deletion mutants related to ergosterol biosynthesis in C. neoformans (erg4Δ and cnag_00490Δ) display a pH sensitivity when grown on pH 8 growth medium. Indicated strains were serially diluted onto YPD medium and YPD-150 mM HEPES pH 8. Growth was compared to WT and assessed after 3 days. (C) erg6Δ C. neoformans mutant also exhibits alkaline pH sensitivity when grown on pH 8 medium. Indicated strains were serially diluted onto YPD medium and YPD-150 mM HEPES pH 8. Growth was compared to WT and reconstituted strains and assessed after 3 days. (D) Candida species ergosterol mutants reveal similar pH-sensitive phenotypes. C. albicans and C. lusitaniae wild-type strains and strains with mutations in various components of ergosterol biosynthesis were serially diluted onto YPD medium and YPD-150 mM HEPES pH 8 and 8.5 as well as YES medium with 7 mM CoCl2. Growth was compared to WT and assessed after 2 days.
To further explore the role of ergosterol biosynthesis in the alkaline pH response, we tested three C. neoformans ergosterol-related mutants for growth at pH 8, and all shared an alkaline pH growth defect (Fig. 4B). Many steps in ergosterol biosynthesis are essential for growth under routine conditions, limiting the availability of ERG gene mutants. The nonessential ERG4 and ERG6 genes encode terminal enzymes in the ergosterol biosynthesis pathway (22, 43). Compared to wild type, the erg4Δ and erg6Δ mutants displayed a specific growth defect at alkaline pH (Fig. 4B and C) (15). Similarly, the CNAG_00490 locus encodes a putative acetyl coenzyme A (acetyl-CoA) acetyltransferase, as does the ERG10 (CNAG_02918) gene. The loss-of-function cnag_00490Δ mutant also displays alkaline pH sensitivity (Fig. 4B). The pH sensitivity of the CNAG_00490 mutant as well as the predictive function of its gene product suggests that it might participate in the conversion of acetyl-CoA to squalene, an early step in sterol synthesis.
Ergosterol is a major component of most fungal membranes, including those of distantly related fungal pathogens in the ascomycete phylum. To further explore the association between sterol homeostasis and alkaline pH response, we tested alkaline pH survival for ergosterol biosynthesis mutants in two Candida species, C. albicans and C. lusitaniae (Fig. 4D). The homozygous diploid C. albicans erg6/erg6 and erg24/erg24 mutants displayed severe growth defects at high pH that were not evident under more acidic conditions (Fig. 4D). Similarly, the haploid C. lusitaniae erg6 mutant had impaired growth compared to wild type under alkaline conditions (Fig. 4D). These results suggest a conserved requirement for efficient sterol maintenance in the adaptation to alkaline pH among highly divergent fungal species.
Sre1 regulates membrane-associated transcripts under alkaline growth conditions.
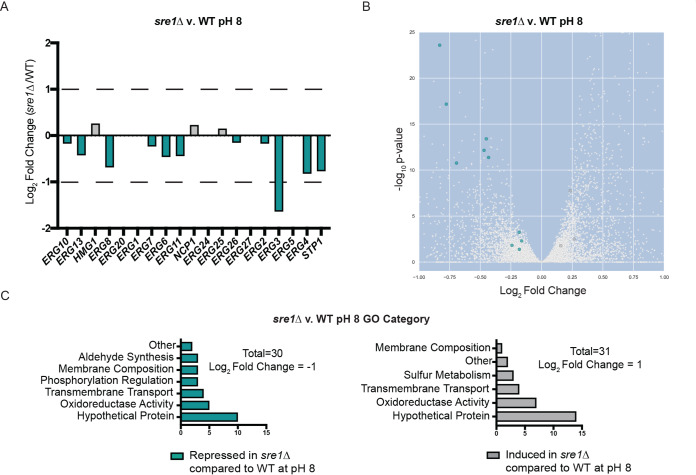
The C. neoformans SRE1-dependent transcriptome has been defined in the context of the cellular response to low oxygen (21, 22, 38). These prior studies revealed that Sre1 is required for the induction of genes involved in ergosterol homeostasis in an oxygen-dependent manner. However, given the novel role for Sre1 pathway activation at alkaline pH, we defined the pH-responsive Sre1-regulated transcriptional response. Comparison of the transcriptomes of the sre1Δ mutant and wild type after 1.5 h of growth in alkaline pH revealed 2,655 transcripts that were differentially regulated in a statistically significant manner (adjusted P value of <0.05) (Fig. S3A and Table S1). This represents approximately one-quarter of the C. neoformans genome, indicating that Sre1 has a major impact on the cell in response to pH stress. Similar to the transcriptome studies in hypoxia, transcript abundance of the majority of the ERG genes (13/18) and the STP1 activating protease was differentially regulated at alkaline pH (Fig. 5A and B). The stp1Δ mutant strain displays a pH-sensitive mutant phenotype similar to the sre1Δ mutant strain (15). Importantly, ERG3 transcript levels had the highest relative fold change in the sre1Δ mutant at high pH compared to wild type (Fig. 5A and B). ERG3 encodes a component of the ergosterol biosynthesis pathway and displays similar Sre1-dependent expression under low-oxygen conditions (22).
FIG 5.
Transcriptomic analysis of the sre1Δ and wild-type strains in response to alkaline pH. WT and sre1Δ cells were incubated in YPD medium pH 4 or pH 8 for 90 min. This experiment was conducted with six biological replicates for each strain and condition. Total RNA was extracted, mRNA was isolated, and libraries were prepared and finally sequenced using an Illumina NextSeq 500 sequencer. GO-term analysis was performed using FungiDB. (A) The majority of the known genes in C. neoformans ergosterol biosynthesis were significantly differentially expressed in the sre1Δ versus wild-type transcriptome at pH 8. ERG genes that were significantly differentially expressed have an adjusted P value of <0.016 (teal, repressed in the sre1Δ mutant compared to wild type; gray, induced in the sre1Δ mutant compared to wild type). (B) Volcano plot displaying the significantly regulated transcripts in the sre1Δ versus wild-type transcriptome at pH 8 (adjusted P value of <0.05) (teal, repressed in the sre1Δ mutant compared to wild type; gray, induced in the sre1Δ mutant compared to wild type). The full volcano plot (zoomed out) is shown in Fig. S2. (C) GO-term analysis of the sre1Δ versus wild-type differentially expressed genes following a 90-min shift from YPD pH 4 to YPD pH 8. These transcripts were selected based on a strict cutoff of log2 fold change of ±1. Teal, biological processes repressed in sre1Δ mutant compared to wild type at high pH; gray, biological processes induced in sre1Δ mutant compared to wild type.
Full transcriptome analyses and WT GO terms. (A) Volcano plot of the full transcriptome as analyzed from the data set for sre1Δ mutant versus WT at pH 8 (cropped version in Fig. 5B). (B) Volcano plot of the wild-type transcriptome. (C and D) GO-term analysis of the wild-type differentially expressed genes that are induced (C) and repressed (D) following a 90-min shift from YPD pH 4 to YPD pH 8. These transcripts were selected based on a strict cutoff of log2 fold change of +1 for induced transcripts and log2 fold change of −3 for repressed transcripts based on the uneven distribution of total repressed transcripts shown on the volcano plot (B). Download FIG S3, TIF file, 0.3 MB (361.2KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-seq analysis. Page 1. sre1Δ versus wild-type differentially expressed genes at pH 8 (repressed). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < −1. Page 2. sre1Δ versus wild-type differentially expressed genes at pH 8 (induced). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < 1. Page 3. sre1Δ versus wild-type entire transcriptome at pH 8. Raw data. Page 4. Wild-type pH 4 versus wild-type pH 8 entire transcriptome. Raw data. Page 5. Wild-type pH 4 versus wild-type pH 8 differentially expressed genes (induced). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < 1. Page 6. Wild-type pH 4 versus wild-type pH 8 differentially expressed genes (repressed). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < −3. Download Table S1, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Due to the large number of differentially expressed transcripts identified in this analysis, we performed a modified Gene Ontology (GO)-term analysis using FungiDB on genes with a 2-fold or greater change in transcript abundance in the sre1Δ mutant compared to wild type (44). Genes repressed in the sre1Δ mutant at high pH are enriched for biological processes such as aldehyde synthesis, cellular respiration/oxidoreduction, membrane composition, phosphorylation regulation, and transmembrane transport. Genes that are induced in this mutant background under alkaline conditions are involved in cellular respiration/oxidoreduction, membrane composition, sulfur metabolism, and transmembrane transport (Fig. 5C and Table S1). Interestingly, although some of these GO terms are shared with the previously published SRE1 transcriptome under 3% oxygen conditions, the majority of the Sre1-dependent transcripts differ between the two experimental inducing conditions: hypoxia versus alkaline pH (22) (Fig. S4). Using the same fold change values to compare these transcript data sets, only nine genes are induced under both conditions, the majority of which are related to ergosterol biosynthesis: SRE1, ERG3, ERG11, ERG6, ERG4, and ERG13 (Fig. S4). This transcriptome analysis supports the central role for ergosterol biosynthesis genes as potential Sre1-dependent effectors of both hypoxia and the response to alkaline pH. We also documented that different inducing conditions mediate distinct Sre1-dependent transcriptional responses.
Compared transcriptomics. Venn diagram showing the overlap between three different transcriptome data sets in C. neoformans serotype A (H99). Bien et al. (22) performed a comparative transcriptomics microarray study using the sre1Δ mutant strain and the sre1Δ + SRE1 complemented strain in low-oxygen growth environments (3% O2) (dark blue). A 1.4-fold-change cutoff was applied to this data set, and the transcripts included in this comparison were increased in the reconstituted strain compared to the mutant. Our data set looking at the sre1Δ mutant compared to wild type at low pH (4) and high pH (8) is shown in teal. We originally applied a fold change cutoff of 2 to our data set to analyze our GO-term categories (Fig. 5C and D) but lowered the cutoff to 1.4 to match the work of Bien (2009) and included only those transcripts that were repressed in the sre1Δ mutant compared to wild type. The overlap of our data with the previously published data set revealed many ergosterol-related transcripts: SRE1, ERG3, ERG4, ERG11, ERG6, and ERG13. Download FIG S4, TIF file, 0.7 MB (768.2KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We were also able to define groups of genes in the wild-type strain that are either induced or repressed following the shift from low to high pH. These groups include a significant portion of membrane-associated transcripts, including integral membrane components, composition regulators, and membrane transporters (Fig. S3C and D and Table S1). Transcripts with increased abundance in response to alkaline pH include many of the known Rim pathway regulators (RIM101 and RIM23) and pathway outputs (ENA1, CIG1, and SKN1) (Fig. S3C and Table S1). Consistent with prior reports of the involvement of Sre1 in iron homeostasis (21, 45), we also identified an iron transporter (CNAG_00815), suggesting a conserved role for iron regulation to adapt to changes in extracellular pH (Table S1). Furthermore, many genes involved in membrane composition, glucose/complex carbohydrate metabolism, and regulation of protein phosphorylation were induced under alkaline conditions (Fig. S3C). Complex carbohydrates are major components of the fungal cell wall, supporting previous findings that the Rim-mediated pH response is linked to the reorganization of the cell wall (36). GO-term analysis of transcripts with reduced abundance at high pH revealed genes involved in membrane transport, potentially in an effort to regulate import of extracellular ions into the cell (Fig. S3D). This analysis revealed no clear repression of membrane composition transcripts at high pH (Fig. S3D and Table S1).
pH affects efficacy of membrane-targeting antifungals.
Given our observation of a correlation between fungal sterols and growth at alkaline pH, we tested the pH-dependent efficacy of antifungal agents targeting different aspects of membrane ergosterol homeostasis. Amphotericin B (AMB) is a polyene antifungal that removes ergosterol from fungal membranes (46). We observed a dramatic reduction in the AMB MIC for wild-type C. neoformans cells grown on YPD pH 8 (0.25 μg/ml) compared to YPD pH 5.5 (2 μg/ml) (Fig. 6A). Furthermore, the time-dependent killing of fungal cells by AMB increased in a pH-dependent manner, further supporting that this drug has a higher efficacy under alkaline growth conditions (Fig. 6B). We also found that AMB was significantly more efficacious against the sre1Δ strain (MIC = 0.00125 μM) than the wild type when the cells were grown at low pH (pH 4 to 6) (Fig. 6B). The significant increase in AMB activity against this mutant strain with reduced ergosterol content is consistent with our model that disruption in fungal sterols leads to pH sensitivity. Furthermore, in a drug disc diffusion assay using pyrifenox, a drug used to treat phytopathogens through inhibition of ergosterol biosynthesis (47), there was a significantly greater zone of clearance and inhibition of growth of wild-type C. neoformans cells when grown on medium buffered to pH 8 compared to pH 5.5 (Fig. 6A).
FIG 6.
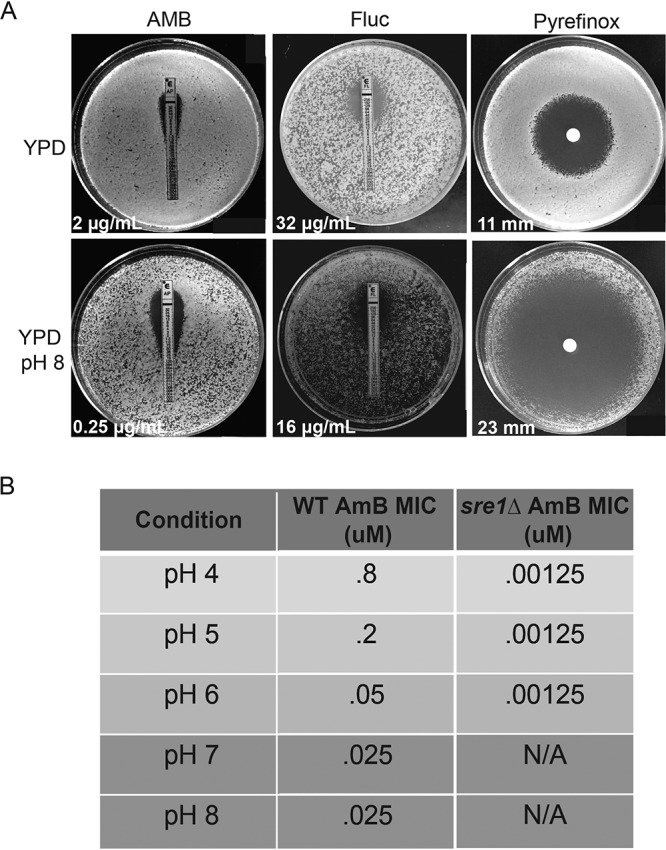
Membrane-targeting antifungals are more active at alkaline pH. (A) Assessing MICs and the zones of inhibition (white values) of membrane-targeting drugs (amphotericin B [AMB], fluconazole [Fluc], and pyrifenox) on wild-type cells grown on YPD or alkaline (YPD pH 8) medium. Measurements were taken after 5 days of growth for AMB and pyrifenox and 3 days of growth for Fluc. All plates were incubated at 30°C. (B) MIC of AMB for wild-type and the sre1Δ mutant C. neoformans strains grown under increasingly alkaline conditions. MIC was determined after 48 h of growth at 30°C by broth microdilution. MIC values could not be determined (N/A) for sre1Δ mutant at pH >6 due to the inability of this strain to grow under these more alkaline conditions.
Fluconazole is an antifungal that inhibits the activity of Erg11, an important component of the ergosterol biosynthesis pathway. We hypothesized that removing ergosterol from the cell membrane in this way would cause a similar sensitivity to alkaline pH as we observed with the ergosterol mutant strains in various fungal pathogens (Fig. 4). In contrast to the major pH-dependent activity of AMB and pyrifenox, we observed a reproducible but more subtle effect of pH on fluconazole efficacy. The fluconazole MIC was 2-fold lower for wild type at pH 8 (16 μM) compared to YPD pH 5.5 (32 μM) (Fig. 6A). The azoles and polyenes have been shown in other organisms, such as Aspergillus fumigatus, to have variable activity against invasive fungal infections depending on the pH of the growth environment (48). Similar to the findings in A. fumigatus, these data support that increases in alkalinity allow for higher efficacy of specific polyenes and azoles against C. neoformans. Our data reveal that reduction of ergosterol, either genetically or biochemically using known antifungals, leads to reduced growth in alkaline environments. Altogether, these results further inform the connection between fungal plasma membrane homeostasis, the molecular interactions that drive environment sensing, and the ability for a biologically diverse group of fungi to grow in increasingly alkaline environments, including their human host.
DISCUSSION
Novel, Rim-independent pH-sensing pathway in C. neoformans.
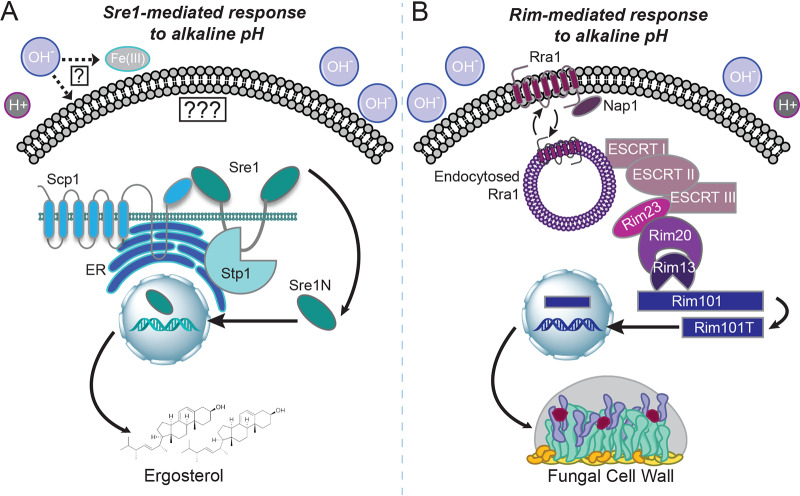
These experiments support a model in which several cell processes and signaling pathways work together to allow microbial growth under stress conditions such as elevated pH. The Rim signaling pathway has been identified in multiple fungal species including C. neoformans, C. albicans, and S. cerevisiae as a major signaling response to increases in extracellular pH (1–3, 15, 36, 37, 39, 49–53) (Fig. 7). Its primary function appears to be translating extracellular alkaline pH signals to control adaptive changes in the fungal cell wall (36) (Fig. 7B). Data presented in this study identified the sterol homeostasis pathway as a unique mechanism that responds to alkaline pH in a Rim-independent way (Fig. 7).
FIG 7.
Model of the Sre1-mediated and Rim-mediated distinct responses to physiological pH. (A) The activating sensor for the sterol homeostasis pathway is unknown and could be linked to alkaline pH-induced reductions in ergosterol/membrane alterations or bioavailable iron. In response to alkaline pH, the Sre1 transcription factor is cleaved, activated, and localized to the nucleus to aid in the transcription of many genes involved in ergosterol biosynthesis and membrane homeostasis. This cleavage and activation are dependent on both the conserved transmembrane protein Scp1 and the basidiomycete-specific protease Stp1. (B) The Rim alkaline response pathway is signaled through the transmembrane pH sensor Rra1 and its interaction with the plasma membrane. At elevated pH levels, Rra1 is endocytosed, allowing it to interact with the downstream components of the pathway and propagate the signal to the endosomal membrane complex (ESCRT components, Rim23, and Rim20) and activate the Rim13 protease. This protease cleaves the Rim101 transcription factor, allowing it to translocate to the nucleus and induce the expression of genes involved in cell wall and surface remodeling.
The sterol homeostasis pathway has been implicated in the response to alterations in oxygen availability, membrane ergosterol levels, and various stressors in diverse fungal species. In the fission yeast, Schizosaccharomyces pombe, the induction of ergosterol biosynthesis genes by the Sre1 transcription factor and its chaperone proteins (Scp1 and Ins1) has been well characterized in response to hypoxia (18, 20, 54). C. neoformans, similarly to S. pombe, has a well characterized Sre1-mediated response to hypoxia that results in the induction of ergosterol biosynthesis genes to maintain membrane homeostasis (21, 25, 27, 34, 38). However, in C. neoformans, a basidiomycete-specific protease has been identified that specifically activates Sre1 in response to hypoxia (Fig. 7A) (22, 38). Elements of this pathway have also been identified in the filamentous fungal pathogen A. fumigatus. The Sre1 homolog, SrbA, is essential for the ability of this pathogen to grow in environments with limited oxygen or low iron or in the presence of membrane-targeting antifungals (19, 23, 45, 55, 56). This hypoxic response is required for survival in the infected host in which hypoxic microenvironments exist, especially in poorly viable tissue such as necrotic tumors and wounds (57). The dimorphic fungal pathogen Histoplasma capsulatum also contains a homolog of Sre1 (Srb1) that is essential for the response to hypoxia as well as for virulence (58–60). Other yeasts such as C. albicans and S. cerevisiae do not contain genes in their sequenced genomes encoding obvious SREBP homologs. Instead, these species respond to hypoxic stress through the activation of a different transcription factor, Upc2, which directs the induction of ergosterol biosynthesis genes (61, 62). However, the C. albicans Cph2 protein binds SRE1-like elements in the genome, and it may therefore be a functional ortholog of Sre1 (63).
The identification of a new role for the sterol homeostasis pathway is informative to better conceptualize and target fungal pathogenesis in general and cryptococcal pathogenesis in particular for several reasons. First, the sterol pathway in C. neoformans has a basidiomycete-specific Stp1 protease that is required for cleavage and activation of Sre1 (22, 25, 38). Genes encoding a similar protease are found in the genomes of other basidiomycete fungi such as Cryptococcus gattii, Malassezia globosa, and Mucor circinelloides (44) and not in those of more distantly related fungi or higher eukaryotes. This fungal specificity and distinction from the mammalian sterol homeostasis pathway (16, 17, 64) may provide an interesting future target for novel antifungals. Second, understanding the extracellular cues that activate this pathway may elucidate more detailed signaling mechanisms controlling sterol homeostasis, potentially revealing some currently unknown upstream components. Presently, it is not known if a common signal in hypoxia or alkaline pH initiates Sre1 signaling, or if multiple upstream Sre1 activators are present (Fig. 7A). The C. neoformans sterol homeostasis pathway is lacking an obvious INSIG homolog as well as a site-1 protease (24). Elucidating the Sre1-mediated response to alkaline pH through further analysis of our forward genetic screen may uncover either functional orthologs of these proteins or novel pathway components that mediate specific stress responses in C. neoformans.
The transcriptional analysis of the sre1Δ mutant strain at high pH provided further support for the distinct activation of the Sre1 transcription factor in response to increases in extracellular pH. This type of analysis has been conducted for the C. neoformans sre1Δ mutant strain previously but with conditions of low and high oxygen availability (22). When comparing our transcriptomics data to this previously published microarray analysis, the majority of the transcripts were nonoverlapping, suggesting independent downstream effectors of Sre1 in response to specific stress (see Fig. S4 in the supplemental material). Furthermore, there was no overlap between the Sre1-associated transcriptome at high pH and the previously published Rim101-associated transcriptome at a similar pH, further supporting the distinct nature of these two pH response mechanisms and the specificity of the Sre1-mediated response to alkaline pH stress (data not shown and Fig. 7) (15).
Ergosterol biosynthesis is essential for the ability of fungal pathogens to grow in an alkaline environment.
The generation of ergosterol for overall fungal membrane integrity has been well studied in the response to extracellular stresses such as hypoxia and low iron (23, 24, 26, 38, 45, 55, 56). Ergosterol controls the fluidity and structure of fungal cells (65), and it is needed for the formation of microdomains within the membrane containing ion pumps and transmembrane proteins necessary for cellular growth and signaling (12, 13, 65–67). In this study, we have demonstrated that supplementing pH-sensitive mutant strains with ergosterol can rescue the pH-sensitive mutant phenotype, suggesting that the sre1Δ mutant pH sensitivity is specifically linked to its ergosterol deficiency.
Our studies further supported this link between ergosterol and the pH response through analysis of the effects of alkaline pH on the biosynthesis of ergosterol at the transcriptional level. In response to a shift in pH, the majority of the known C. neoformans ergosterol biosynthesis genes were differentially regulated in the sre1Δ strain compared to wild type. These results support our model and implicate Sre1-mediated membrane homeostasis as a direct response to alkaline stress (Fig. 7A). Furthermore, C. neoformans and C. albicans strains with mutations in known and predicted ergosterol synthetic processes were unable to grow at alkaline pH. These results indicate that ergosterol levels and membrane homeostasis are important in the pH response mechanisms of many fungal species. This broadens these findings from Sre1-specific regulation of ergosterol affecting pH growth of a basidiomycete fungal pathogen to general ergosterol maintenance affecting the pH response in many different fungal pathogens across phyla.
In addition to establishing a link between alkaline pH and membrane sterols, our data also support emerging data on the interplay between the pH of the external environment and iron homeostasis (Fig. 7A). In divergent cell types, bioavailable iron concentrations are often reduced at alkaline pH (68). Our data demonstrate the induction of an iron transporter transcript in response to alkaline pH (Table S1), further suggesting that the cell is responding to reduced iron availability under this condition. In A. fumigatus, supplementing ΔsrbA mutants with exogenous iron rescues growth defects in low oxygen and during azole treatment (45). Also, in the dimorphic fungal pathogen H. capsulatum, Sre1 signaling mediates the ability of this fungus to survive under hypoxic conditions as well as to control iron regulation. Each of these processes may mediate separate roles in fungal virulence (59, 60) Furthermore, prior investigations have also demonstrated that the C. neoformans Sre1-mediated stress response is linked to iron availability (21). Future studies will determine if exogenous iron will fully or partially suppress the C. neoformans sre1Δ mutant pH growth defects in a similar manner as exogenous sterols.
Ergosterol-depleting antifungals render cryptococcal cells sensitive to alkaline pH.
Our results have shown not only that genetic manipulation of fungal membrane homeostasis and ergosterol biosynthesis can increase the sensitivity of C. neoformans to alkaline pH but also that biochemical and pharmaceutical interventions have the same effect. We tested relevant antifungals that prevent sterol production or directly deplete sterols from fungal membranes and demonstrated that the activity of these drugs improves in neutral/alkaline environments. AMB, an antifungal that directly disrupts the plasma membrane through sequestration of ergosterol (46), was significantly more potent with increases in the pH of the growth environment. Similarly, fluconazole and pyrifenox, drugs that inhibit the ergosterol biosynthesis pathway (47, 69), were also more effective at alkaline pH. These results reflect similar findings in Aspergillus species treated with itraconazole and AMB (48). Similar studies using ketoconazole, AMB, and flucytosine (5-FC) against Candida species showed that the in vitro drug activity increases as a function of pH (70, 71). Interestingly, there has also been one study demonstrating increased efficacy of 5-FC against C. neoformans at higher pH (72). The fact that flucytosine does not directly target the cell membrane, together with the subtle alterations in fluconazole activity as a function of pH, suggests that multiple factors control this phenomenon. However, our findings that known ergosterol-targeting antifungals render diverse fungi more vulnerable to growth environments with increasing pH further support our leading hypothesis that ergosterol homeostasis is a central contributor to the alkaline pH response of many fungal pathogens.
Translating basic investigations in the role of pH modulation in human disease into potential clinical applications has precedent in cancer biology. In mammalian cells, studies of pH regulation in tumor metastasis demonstrated an association between the pH within a tumor and the degree of tumor cell apoptosis, survival, and proliferation (73). The preference among certain malignant cells for more acidic external environments has prompted the exploration of “buffer therapy,” in which site-directed pH modulation is used as an adjunctive therapy to limit tumor growth (74). This type of therapy is also effective against microbial infections that colonize the airways and intestines, such as Pseudomonas aeruginosa and Escherichia coli, respectively (75–77). If these interventions can be used against bacterial infections, one might imagine how similar pH modulation could be specifically applied to combat the acidic, necrotic core of many established invasive fungal infections, including cryptococcal lesions (57, 78, 79). Understanding pH-mediated microbial changes in various host microniches will allow for the development of optimized antifungal activity at the site of infection.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains generated and/or utilized in this study are shown in Table 1. Each mutant, reconstituted strain, and fluorescent strain were generated in the C. neoformans H99 MATα genetic background and incubated in either yeast-peptone-dextrose medium (YPD) (1% yeast extract, 2% peptone, and 2% dextrose) or yeast nitrogen base medium (YNB). The pH 4, 5, 5.5, 6, 7, and 8 media were made by adding 150 mM HEPES buffer to YPD or YNB medium, adjusting the pH with concentrated HCl (for pH <5.5) or NaOH (for pH >5.5), prior to autoclaving. Medium was supplemented with 20% glucose following autoclaving unless otherwise noted. Cell wall stress phenotypes were assessed by growth on various stress medium agar plates as previously described (32). Congo red (0.5%) and NaCl (1.5 M) were added to YPD medium prior to autoclaving. Caffeine (1 mg/ml) and SDS (0.03%) were filter sterilized and added to YPD medium following autoclaving. Cobalt chloride plates were made by adding 7 mM (90.89 mg/liter) CoCl2 solution to autoclaved YES medium (glucose, yeast extract, adenine, uracil, histidine, leucine, lysine, and agar) (80, 81). Capsule induction and analyses were completed as previously described (32). Briefly, strains were incubated overnight in YPD medium and then diluted in tissue culture medium (CO2-independent tissue culture medium [TC]; Gibco) for 72 h with shaking at 37°C and then counterstained with India ink. The microaerophilic conditions were generated using a sealed chamber (BD GasPak) and two activated packs of GasPak EZ Campy container system (containing campylobacter) to reduce oxygen levels. YPD plates with serial dilutions of normally grown strains were placed in the chamber for 24 h at 37°C (microaerophilic) or outside the chamber for 24 h at 37°C (ambient air).
TABLE 1.
Strain list
| Strain | Genotype | Reference or source |
|---|---|---|
| H99 | MATα | 95 |
| TOC35 | rim101Δ::NAT | 37 |
| HEB5 | sre1::NEO MATα (#1) | 15 |
| HEB6 | sre1::NEO MATα (#2) | This study |
| YSB2493 | sre1::NAT MATα (#3) | 34 |
| YSB2494 | sre1::NAT MATα (#4) | 34 |
| HEB94 | sre1::NEO + His-SRE1(NAT) MATα | This study |
| HEB71 | His-GFP-Sre1 MATα | This study |
| KS91 | His-GFP-Rim101 MATα | 96 |
| TOC106 | eGFP-Rim101 MATα | 96 |
| HEB13 | eGFP-Rim101 + sre1::NEO MATα 1 | This study |
| HEB14 | eGFP-Rim101 + sre1::NEO MATα 2 | This study |
| KS118-2 | rim20::NAT eGFP-Rim101 MATα | 39 |
| KS33 | rim13::NEO MATα | 39 |
| HM.5-F6a | erg4Δ::NAT MATα | 97 |
| HM.21-E12a | cnag_00490Δ::NAT MATα | 97 |
| erg6Δ | erg6::HPH (hygromycin resistance) | 43 |
| SC5314 | WT Candida albicans | 98 |
| 4A | erg11/ERG11 Candida albicans | 99 |
| NJ25-1 | erg24/ERG24 Candida albicans | 100 |
| NJ51-2 | erg24/erg24 Candida albicans | 100 |
| KPC1 | erg6/ERG6 Candida albicans | 99 |
| KPC8 | erg6/erg6 Candida albicans | 99 |
| ATCC 42720 | Candida lusitaniae | 101 |
| CL130 | erg6 Candida lusitaniae | 102 |
Strains obtained from the 2015 and 2016 Madhani plates. Designated HM.#-xx for plate number (#) and well (xx).
The ergosterol supplementation and growth curve analysis were conducted in a 96-well plate. Strains were incubated overnight (∼18 h) at 30°C with 150-rpm shaking. Cells were then pelleted and resuspended in either pH 4 or pH 8 synthetic complete medium buffered with McIlvaine’s buffer (39). Resuspended strains were added to wells containing the same-pH synthetic complete medium with either 2 μg/ml or 0.02 μg/ml of ergosterol (Sigma)-Tween 80-ethanol (2-mg/ml stock as previously described in reference 82). Growth was then measured at an absorbance of 595 nm every 10 min for 42 h with shaking between readings and incubation at 30°C. Control wells containing vehicle alone (ethanol and Tween) were also measured in order to ensure that any growth rate change detected was due to the addition of ergosterol. One-way analysis of variance (ANOVA) and Dunnett’s multiple-comparison test were run on the last time point under each condition compared to the pH 8-alone condition to determine statistical significance. The pH of the medium in the wells was tested following the experiment to ensure that the medium remained buffered.
To generate the sre1Δ deletion and eGFP-Rim101 + sre1Δ deletion and tagged deletion constructs, respectively, we performed the previously described double-joint PCR with split drug resistance marker method to make targeted gene deletions (15, 83). In brief, we generated the following two PCR products: 5′ flanking region of the target locus (1,000 bp) with a truncated drug resistance cassette and the remainder of the drug resistance cassette with the 3′ flanking region of the target locus (1,000 bp). We then used biolistics to transform these two amplicons into either the wild-type C. neoformans strain (H99) or the C. neoformans strain that contains endogenously expressed GFP-Rim101 (84). Transformants were selected for the presence of the construct on YPD medium plus neomycin (NEO). To generate the fluorescently tagged His-GFP-Sre1 strain, we used In-Fusion (Clontech) to clone the SRE1 gene and terminator into the HGNAT (pCN19) plasmid, containing the GFP sequence and the nourseothricin (NAT) resistance marker (85). This plasmid was then biolistically transformed into the H99, wild-type (WT) strain. To generate the SRE1 reconstituted strain, we cloned the SRE1 gene and terminator into the pCH233 plasmid, containing the nourseothricin (NAT) resistance marker (86). This plasmid was then biolistically transformed into the sre1Δ (HEB5) strain. The primers used to generate each strain are listed in Table 2. Primers used to validate all sre1Δ mutants through Southern analysis (data not shown) are also listed in Table 2. Transformants were selected on YPD medium containing NAT (fluorescent strain) or NAT/NEO (reconstituted strain). Plasmids used in this study to amplify markers and clone new plasmids are listed in Table 3.
TABLE 2.
Primers used in this study
| Primer type and name | Primer sequence | Primer descriptiona |
|---|---|---|
| Deletion constructs | ||
| AA4950 | AGGATTTGGGCAAATCGAGA | SRE1 ko primer 1 |
| AA4951 | GTCATAGCTGTTTCCTGGGGAAAGAATCGTCTCATCA | SRE1 ko primer 2 |
| AA4952 | ACTGGCCGTCGTTTTACAGGCGATGCTATCTATGGGT | SRE1 ko primer 3 |
| AA4953 | GGAACCAATAAAGCGACCCA | SRE1 ko primer 4 |
| M13F | GTAAAACGACGGCCAGT | NEO cassette flank (F) |
| M13R | CAGGAAACAGCTATGAC | NEO cassette flank (R) |
| AA3935 | CCTGAATGAACTGCAGGA | NEO internal cassette (R) |
| AA3934 | TCGATGCGATGTTTCGCT | NEO internal cassette (F) |
| Reconstitution constructs | ||
| AA5546 | CGTCGCACTAGTGAGAGGGAGAAAGCTGGC | SRE1 complement (F) |
| AA5547 | CGTCGCACTTTTGGTGGACGGGCATTAATA | SRE1 complement (R) |
| Southern probes | ||
| AA4975 | GGAACTGGCCAAATACGCAG | SRE1 Southern probe (F) |
| AA4976 | TTCCATGGTCCCTATCCATT | SRE1 Southern probe (R) |
| Fluorescent constructs | ||
| AA5514 | GTACGGATCCACTAGTATGGCCTCATTACAGGACAAGATGC | HIS-GFP-SRE1 (F) 1 |
| AA5517 | GGCGGCCGTTACTAGTACATCACGTACGTACATACAGC | HIS-GFP-SRE1 (R) 2 |
Abbreviations: ko, knockout; F, forward; R, reverse.
TABLE 3.
Plasmids used in this study
Microscopy.
To analyze GFP-Rim101 localization in the WT, rim20Δ, and sre1Δ backgrounds, strains were incubated overnight (∼18 h) at 30°C with 150-rpm shaking. Cells were then pelleted and resuspended in either pH 4 or pH 8 synthetic complete medium buffered with McIlvaine’s buffer. Strains were shaken at 150 rpm and 30°C for 60 min as this has been shown to be sufficient time to observe the nuclear localization of Rim101 in WT cells (15). Fluorescent images were captured using a Zeiss Axio Imager A1 fluorescence microscope equipped with an Axio-Cam MRM digital camera. Images were created using ImageJ software (Fiji) (87).
Protein extraction, immunoprecipitation, and Western blotting.
Protein extracts were prepared in a similar manner to what was previously described (15). Briefly, strains were incubated for ∼18 h at 30°C with 150-rpm shaking in YPD medium buffered to pH 4 or 5.5 with HEPES and HCl. Cells were then pelleted and resuspended in YPD medium buffered to pH 8 with HEPES and NaOH. These cells were incubated for 60 min and immediately pelleted and flash frozen on dry ice. Lysis was performed by bead beating (0.5 ml of 3-μm glass beads in a Mini-BeadBeater-16 [BioSpec] for 6 cycles of 30 s each with a 1-min ice incubation between bead-beating cycles for cell recovery). Supernatants were washed 3 times with 0.4 ml of lysis buffer (2× protease inhibitors [Complete, Mini, EDTA-free; Roche], 1× phosphatase inhibitors [PhosStop; Roche], and 1 mM phenylmethanesulfonyl fluoride [PMSF]). The crude pellet was pelleted through centrifugation at 15,000 rpm, 4°C, for 5 min, and the supernatant (cell lysate) was transferred (∼1 ml) to a new tube. For Western blotting assays assessing the presence of protein by probing for GFP, 50 μl of lysate was saved as whole lysate and 25 μl of GFP-trap (Chromotek) resin (equilibrated and resuspended in lysis buffer) was added to the remaining lysate. The lysates containing GFP-trap were incubated at 4°C for 2 h, rotating. Following incubation, lysates were spun down (2,500 × g for 2 min at 4°C) and washed 3 times with detergent-free buffer containing 2× protease inhibitors (Complete, Mini, EDTA-free; Roche), 1× phosphatase inhibitors (PhosStop; Roche), and 1 mM phenylmethanesulfonyl fluoride (PMSF). GFP-trap resin was resuspended in 4× NuPage lithium dodecyl sulfate (LDS) loading buffer and 10× NuPage reducing agent. Western blotting assays were performed using a 4% to 12% NuPage BisTris gel. To probe and detect GFP-Rim101 and GFP-Sre1, immunoblots were incubated with an anti-GFP primary antibody (using a 1/10,000 dilution; Roche), followed by a secondary anti-mouse peroxidase-conjugated secondary antibody (using a 1/25,000 dilution; Jackson Laboratory). Proteins were detected by enhanced chemiluminescence (ECL Prime Western blotting detection reagent; GE Healthcare).
For Western blotting assays assessing the presence of cleaved and uncleaved Sre1 using the polyclonal anti-Sre1, lysates were prepared in the same way as previously described. Following lysis and initial centrifugation of the crude pellet, 500 μl of lysates was precleared with 30 μl protein A-agarose (Sigma) and rotated for 1 h at 4°C. Lysates were incubated with 5 μl of anti-Sre1 polyclonal antibody (generously given to us by the Espenshade lab [21]) for 1 h. Protein A (60 μl/sample) was washed twice in lysis buffer and resuspended in equal volumes. Equilibrated protein A was then added to each lysate and incubated at 4°C for 1 h, rotating. Following incubation, lysates were spun down (2,500 × g for 2 min at 4°C) and washed twice with lysis buffer, once with lysis buffer plus 1 M NaCl, and twice with lysis buffer. Protein A resin was then resuspended in 4× NuPage lithium dodecyl sulfate (LDS) loading buffer and 10× NuPage reducing agent. Western blot assays were performed using a 3% to 8% NuPage Tris-acetate gel, with Tris-acetate running buffer. To probe for and detect Sre1, immunoblots were incubated in anti-Sre1 primary antibody (using a 1/200 dilution [21]) and then in anti-rabbit peroxidase-conjugated secondary antibody (using a 1/50,000 dilution; Jackson Laboratory). Proteins were detected in the same way as described above.
Cell wall staining and flow cytometry.
For chitin and exposed chitin detection, cell wall staining with wheat germ agglutinin (WGA) and calcofluor white (CFW) was assessed as previously described (32). Briefly, overnight YPD cultures were diluted 1:10 in CO2-independent liquid medium and incubated (∼18 h) at 37°C with 150-rpm shaking. Cells were stained with 100 μg/ml of fluorescein isothiocyanate (FITC)-conjugated WGA and 25 μg/ml CFW and incubated in the dark for 35 min and 10 min, respectively. Quantitative analysis using ImageJ software was performed as previously described (32, 41).
For flow cytometry analysis, cells were incubated similarly as described above and fixed with 3.7% formaldehyde for 5 min at room temperature. Cells were then slowly pelleted and washed twice with phosphate-buffered saline (PBS). Cells were stained with 100 μg/ml FITC-conjugated wheat germ agglutinin (WGA; Molecular Probes). Cells stained with WGA were incubated in the dark at room temperature for 35 min. Cells were then slowly pelleted and washed twice with PBS. Cells from each strain were stained and resuspended in PBS at a concentration of 107 cells/ml. Cells at 106/ml were submitted to the Duke Cancer Institute Flow Cytometry Shared Resource for analysis using a BD FACSCanto II flow cytometer. Data were analyzed by FlowJo v10.6.1 software (FlowJo, LLC). Unstained cells were used as negative controls, and positive events were gated in the forward scatter/side scatterplots and represented as histograms. Geometric means were calculated based on the mean fluorescence intensity (x axis of histogram) of all cells quantified for each strain (y axis of histogram).
Macrophage survival assay.
J774A.1 cells were incubated in a humidified 37°C incubator with 5% CO2, passaged twice weekly, and kept in tissue culture flasks in 20 to 25 ml of macrophage medium (Dulbecco’s modified Eagle’s medium [DMEM], heat-inactivated fetal bovine serum [FBS], penicillin-streptomycin [Gibco 15140-122], and minimal essential medium [MEM] nonessential amino acid solution [Gibco 11140-050]). Survival of C. neoformans strains within alveolar macrophage-like J744A.1 cells was assessed by aliquoting 100 μl of 105 viable cells into a 96-well plate, avoiding edges as previously described (88). The plates were incubated overnight in a 37°C incubator with 5% CO2. Macrophages were then activated with 10 nM phorbol myristate acetate (PMA) and incubated at 37°C, 5% CO2, for 1 h. Fungal cells were incubated overnight (∼18 h) at 30°C with 150-rpm shaking. Cells were then pelleted, washed twice in PBS, and resuspended in macrophage medium. Fungal cells (106 cells/ml) were opsonized with monoclonal antibody (Mab) 18B7 (1 μg/ml) for 1 h at 37°C. Cell concentrations were verified with quantitative culture. Macrophage medium was removed from the 96-well plate, and 100 μl of opsonized fungal cells was added to each well. The cocultures were incubated for 1 h at 37°C incubator with 5% CO2. Each well was then washed 3 times with PBS to remove extracellular yeast. One hundred microliters of macrophage medium was added to each well and incubated for 24 h at 37°C with 5% CO2. Following incubation, macrophage killing was determined by adding 200 μl sterile distilled water (dH2O) to each well, incubating at room temperature for 5 min, and assessing by quantitative cultures. One-way ANOVA and Tukey’s multiple-comparison tests were run to assess statistical significance between fungal cell survival percentages. Six biological replicates of each strain were analyzed.
RNA-sequencing preparation and analyses.
WT and sre1Δ cells were incubated at 30°C with 150-rpm shaking in YPD medium to mid-logarithmic phase. Approximately 1 × 109 cells from each strain were pelleted and resuspended in YPD medium buffered to pH 4 or pH 8 and incubated at 30°C for 90 min with 150-rpm shaking. All cells were pelleted, flash frozen on dry ice, and lyophilized overnight. This experiment was conducted with six biological replicates for the WT strain and the sre1Δ strain under both pH 4 and pH 8 conditions (24 samples total). RNA was isolated using the Qiagen RNeasy Plant minikit with optional on-column DNase digestion (Qiagen, Valencia, CA). RNA quantity and quality were measured using the Agilent 2100 Bioanalyzer. The NEBNext poly(A) mRNA magnetic isolation module was used to enrich for mRNA, and the NEBNext Ultra II directional RNA library prep kit for Illumina was used to prepare libraries (New England Biolabs, Ipswich, MA). Libraries were submitted to the Duke Sequencing and Genomic Technologies Shared Resource for sequencing on the Illumina NextSeq 500 with 75-bp, single-end reads.
Reads were mapped to the C. neoformans H99 reference genome (obtained from NCBI, accessed July 2019) using STAR alignment software (89). Differential expression analyses were performed in R using an RNA‐Seq Bioconductor workflow (90, 91) followed by the DESeq2 package with a false-discovery rate (FDR) of 5% (92). Genes were considered statistically differentially expressed if they had an adjusted P value of <0.05.
A modified Gene Ontology-term (GO-term) analysis using the FungiDB database was performed to identify genes that were significantly regulated in a given process as previously reported (15, 93). The differentially expressed genes in each category were determined based on two criteria: P value of <0.05 and base mean value of >20. Further differentiation was made based on the log2 fold change values. For the sre1Δ versus wild-type data set, we used a log2 fold change of ±1. For the positively regulated genes in the wild-type pH 4 versus pH 8 data set, we used a log2 fold change of 1, and for the negatively regulated genes in the wild-type data set, we used a log2 fold change of −3 due to the large amount of genes in this set. Fold change graphs were generated in GraphPad Prism (GraphPad Prism version 8.00 for Mac, GraphPad Software, San Diego, CA, USA), and Seaborn was used to visualize the DESeq2 results in a volcano plot (94). A complete list of the transcriptome sequencing (RNA-seq) data sets containing differentially expressed genes in each strain and associated with the appropriate GO-term category can be found in Table S1 in the supplemental material.
Antifungal susceptibility tests.
For fluconazole and amphotericin B (AMB) Etest assays and pyrifenox disc diffusion, fungal cells were incubated overnight (∼18 h) at 30°C with 150-rpm shaking in YPD. Cells were normalized to an optical density at 600 nm (OD600) of 0.6 and diluted 1:10 in PBS, and 100 μl was plated to either YPD pH 5.5 or YPD pH 8 agarose plates. For the fluconazole and AMB Etest assay, an Etest strip (bioMérieux) containing a gradient of drug concentrations was placed on top of the plated fungal lawn. Plates were then incubated at 30°C for 72 (AMB) and 120 (fluconazole) h. Pyrifenox susceptibility was assessed by standard disc diffusion assays using 5 μl pyrifenox (Sigma-Aldrich; CAS number 88283-41-4; final concentration of 1.2 g/ml). Plates were then incubated at 30°C for 72 h. Zones of inhibition were determined as a surrogate of antifungal activity.
MIC testing of AMB against a pH gradient was performed by broth microdilution. AMB resuspended in dimethyl sulfoxide (DMSO) was serially diluted in synthetic complete medium buffered to pH 4, 5, 6, 7, or 8 with McIlvaine’s buffer in a 96-well plate with the highest concentration being 3.2 μg/ml. Fungal cells were incubated overnight (∼18 h) at 30°C with 150-rpm shaking in YPD. Cells were then normalized and diluted in synthetic complete medium buffered to pH 4, 5, 6, 7, or 8 with McIlvaine’s buffer and added to the corresponding pH well containing AMB. Plates were incubated at 30°C for 48 h, and the MIC was determined to be the lowest concentration of drug that led to no fungal cell growth.
Data availability.
All raw and analyzed RNA-sequencing data have been submitted to the NCBI GEO database under accession no. GSE147109 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147109).
ACKNOWLEDGMENTS
We acknowledge the Duke Sequencing and Genomic Technologies Shared Resource for their assistance with the various projects in this study. We thank Josh Granek and the other staff of the High Throughput Sequencing Course held through the Department of Biostatistics and Bioinformatics at Duke University for their assistance with our RNA sequencing library prep and analysis. We thank the Espenshade Laboratory for their generous gifting of the anti-Sre1 polyclonal antibody. We also thank the Madhani Laboratory for the erg4Δ and cnag_00490Δ deletion strains available through the Fungal Genetics Stock Center. We thank Max Moskovitz for his assistance with phenotyping sterol-related deletion strains. We thank the Heitman Laboratory for sharing the various Candida deletion strains.
We acknowledge our own NIH funding (R01 AI074677, 1F31A140427-01A1, P01 AI104533) that made these studies possible. We also thank the NIH funding (R01AI100272) for the erg4Δ and cnag_00490Δ deletion strains available through the Fungal Genetics Stock Center.
The authors have no relevant conflicts of interest.
H.E.B., C.L.T., J.W.S., and J.A.A. were involved with the conception and design of experiments and the writing process. H.E.B., C.L.T., and L.F. were involved in the acquisition of the data. All authors participated in the analysis and interpretation of the data.
Footnotes
Citation Brown HE, Telzrow CL, Saelens JW, Fernandes L, Alspaugh JA. 2020. Sterol-response pathways mediate alkaline survival in diverse fungi. mBio 11:e00719-20. https://doi.org/10.1128/mBio.00719-20.
REFERENCES
- 1.Davis D, Edwards JE, Mitchell AP, Ibrahim AS. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun 68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selvig K, Alspaugh JA. 2011. pH response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology 39:249–256. doi: 10.5941/MYCO.2011.39.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garnaud C, García-Oliver E, Wang Y, Maubon D, Bailly S, Despinasse Q, Champleboux M, Govin J, Cornet M. 2018. The Rim pathway mediates antifungal tolerance in Candida albicans through newly identified Rim101 transcriptional targets including Hsp90 and Ipt1. Antimicrob Agents Chemother 62:e01785-17. doi: 10.1128/AAC.01785-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Toffaletti DL, Tenor JL, Litvintseva AP, Fang C, Mitchell TG, McDonald TR, Nielsen K, Boulware DR, Bicanic T, Perfect JR. 2014. The Cryptococcus neoformans transcriptome at the site of human meningitis. mBio 5:e01087-13. doi: 10.1128/mBio.01087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes CM, Goldman GH, Del Poeta M. 2018. Biological roles played by sphingolipids in dimorphic and filamentous fungi. mBio 9:e00642-18. doi: 10.1128/mBio.00642-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luberto C, Toffaletti DL, Wills EA, Tucker SC, Casadevall A, Perfect JR, Hannun YA, Del Poeta M. 2001. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev 15:201–212. doi: 10.1101/gad.856001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Hennig M, Luberto C, Del Poeta M. 2006. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest 116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber A, Oemer G, Malanovic N, Lohner K, Kovács L, Salvenmoser W, Zschocke J, Keller MA, Marx F. 2019. Membrane sphingolipids regulate the fitness and antifungal protein susceptibility of Neurospora crassa. Front Microbiol 10:605. doi: 10.3389/fmicb.2019.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito K, Takakuwa N, Ohnishi M, Oda Y. 2006. Presence of glucosylceramide in yeast and its relation to alkali tolerance of yeast. Appl Microbiol Biotechnol 71:515–521. doi: 10.1007/s00253-005-0187-3. [DOI] [PubMed] [Google Scholar]
- 10.Munshi MA, Gardin JM, Singh A, Luberto C, Rieger R, Bouklas T, Fries BC, Del Poeta M. 2018. The role of ceramide synthases in the pathogenicity of Cryptococcus neoformans. Cell Rep 22:1392–1400. doi: 10.1016/j.celrep.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raj S, Nazemidashtarjandi S, Kim J, Joffe L, Zhang X, Singh A, Mor V, Desmarini D, Djordjevic J, Raleigh DP, Rodrigues ML, London E, Del Poeta M, Farnoud AM. 2017. Changes in glucosylceramide structure affect virulence and membrane biophysical properties of Cryptococcus neoformans. Biochim Biophys Acta Biomembr 1859:2224–2233. doi: 10.1016/j.bbamem.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kodedová M, Sychrová H. 2015. Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10:e0139306. doi: 10.1371/journal.pone.0139306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang YQ, Gamarra S, Garcia-Effron G, Park S, Perlin DS, Rao R. 2010. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog 6:e1000939. doi: 10.1371/journal.ppat.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang YQ, Rao R. 2010. Beyond ergosterol: linking pH to antifungal mechanisms. Virulence 1:551–554. doi: 10.4161/viru.1.6.13802. [DOI] [PubMed] [Google Scholar]
- 15.Brown HE, Ost KS, Esher SK, Pianalto KM, Saelens JW, Guan Z, Alspaugh JA. 2018. Identifying a novel connection between the fungal plasma membrane and pH-sensing. Mol Microbiol 109:474–493. doi: 10.1111/mmi.13998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown MS, Goldstein JL. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 17.Espenshade PJ, Hughes AL. 2007. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet 41:401–427. doi: 10.1146/annurev.genet.41.110306.130315. [DOI] [PubMed] [Google Scholar]
- 18.Hughes AL, Todd BL, Espenshade PJ. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog 4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. 2006. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol 26:2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang YC, Bien CM, Lee H, Espenshade PJ, Kwon-Chung KJ. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol 64:614–629. doi: 10.1111/j.1365-2958.2007.05676.x. [DOI] [PubMed] [Google Scholar]
- 22.Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, Espenshade PJ. 2009. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol Microbiol 74:672–690. doi: 10.1111/j.1365-2958.2009.06895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung D, Barker BM, Carey CC, Merriman B, Werner ER, Lechner BE, Dhingra S, Cheng C, Xu W, Blosser SJ, Morohashi K, Mazurie A, Mitchell TK, Haas H, Mitchell AP, Cramer RA. 2014. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog 10:e1004487. doi: 10.1371/journal.ppat.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bien CM, Espenshade PJ. 2010. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell 9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang YC, Ingavale SS, Bien C, Espenshade P, Kwon-Chung KJ. 2009. Conservation of the sterol regulatory element-binding protein pathway and its pathobiological importance in Cryptococcus neoformans. Eukaryot Cell 8:1770–1779. doi: 10.1128/EC.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart EV, Lloyd S-A, Burg JS, Nwosu CC, Lintner RE, Daza R, Russ C, Ponchner K, Nusbaum C, Espenshade PJ. 2012. Yeast sterol regulatory element-binding protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. J Biol Chem 287:672–681. doi: 10.1074/jbc.M111.317370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H, Bien CM, Hughes AL, Espenshade PJ, Kwon-Chung KJ, Chang YC. 2007. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol Microbiol 65:1018–1033. doi: 10.1111/j.1365-2958.2007.05844.x. [DOI] [PubMed] [Google Scholar]
- 28.Gluhcheva Y, Pavlova E, Atanasov V, Vladov I. 2014. Cobalt chloride treatment and iron metabolism in immature mice. Ecol Eng Environ Prot 2014:18–23. [Google Scholar]
- 29.Zhao Y, Cao C, Liu Y, Wang J, Li S, Li J, Deng Y. 2020. Genetic analysis of oxidative and endoplasmic reticulum stress responses induced by cobalt toxicity in budding yeast. Biochim Biophys Acta 1864:129516. doi: 10.1016/j.bbagen.2020.129516. [DOI] [PubMed] [Google Scholar]
- 30.Huang BW, Miyazawa M, Tsuji Y. 2014. Distinct regulatory mechanisms of the human ferritin gene by hypoxia and hypoxia mimetic cobalt chloride at the transcriptional and post-transcriptional levels. Cell Signal 26:2702–2709. doi: 10.1016/j.cellsig.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Florio A, Ferrari S, De Carolis E, Torelli R, Fadda G, Sanguinetti M, Sanglard D, Posteraro B. 2011. Genome-wide expression profiling of the response to short-term exposure to fluconazole in Cryptococcus neoformans serotype A. BMC Microbiol 11:97. doi: 10.1186/1471-2180-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esher SK, Ost KS, Kohlbrenner MA, Pianalto KM, Telzrow CL, Campuzano A, Nichols CB, Munro C, Wormley FL, Alspaugh JA. 2018. Defects in intracellular trafficking of fungal cell wall synthases lead to aberrant host immune recognition. PLoS Pathog 14:e1007126. doi: 10.1371/journal.ppat.1007126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ram AFJ, Klis FM. 2006. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat Protoc 1:2253–2256. doi: 10.1038/nprot.2006.397. [DOI] [PubMed] [Google Scholar]
- 34.Jung K-W, Yang D-H, Maeng S, Lee K-T, So Y-S, Hong J, Choi J, Byun H-J, Kim H, Bang S, Song M-H, Lee J-W, Kim MS, Kim S-Y, Ji J-H, Park G, Kwon H, Cha S, Meyers GL, Wang LL, Jang J, Janbon G, Adedoyin G, Kim T, Averette AK, Heitman J, Cheong E, Lee Y-H, Lee Y-W, Bahn Y-S. 2015. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun 6:6757. doi: 10.1038/ncomms7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reese AJ, Doering TL. 2003. Cell wall α-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol 50:1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- 36.O’Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. 2013. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio 4:e00522-12. doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog 6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun CD, Liu OW, Madhani HD. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog 3:e22. doi: 10.1371/journal.ppat.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ost KS, O’Meara TR, Huda N, Esher SK, Alspaugh JA. 2015. The Cryptococcus neoformans alkaline response pathway: identification of a novel Rim pathway activator. PLoS Genet 11:e1005159. doi: 10.1371/journal.pgen.1005159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown HE, Esher SK, Alspaugh JA. 2019. Chitin: a “hidden figure” in the fungal cell wall. Curr Top Microbiol Immunol doi: 10.1007/82_2019_184. [DOI] [PubMed] [Google Scholar]
- 41.Ost KS, Esher SK, Leopold Wager CM, Walker L, Wagener J, Munro C, Wormley FL, Alspaugh JA. 2017. Rim pathway-mediated alterations in the fungal cell wall influence immune recognition and inflammation. mBio 8:e02290-16. doi: 10.1128/mBio.02290-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gachotte D, Pierson CA, Lees ND, Barbuch R, Koegel C, Bard M. 1997. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc Natl Acad Sci U S A 94:11173–11178. doi: 10.1073/pnas.94.21.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliveira FFM, Paes HC, Peconick LDF, Fonseca FL, Marina CLF, Bocca AL, Homem-de-Mello M, Rodrigues ML, Albuquerque P, Nicola AM, Alspaugh JA, Felipe MSS, Fernandes L. 2020. Erg6 affects membrane composition and virulence of the human fungal pathogen Cryptococcus neoformans. Fungal Genet Biol 140:103368. doi: 10.1016/j.fgb.2020.103368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ, Roos DS. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blatzer M, Barker BM, Willger SD, Beckmann N, Blosser SJ, Cornish EJ, Mazurie A, Grahl N, Haas H, Cramer RA. 2011. SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet 7:e1002374. doi: 10.1371/journal.pgen.1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray KC, Palacios DS, Dailey I, Endo MM, Uno BE, Wilcock BC, Burke MD. 2012. Amphotericin primarily kills yeast by simply binding ergosterol. Proc Natl Acad Sci U S A 109:2234–2239. doi: 10.1073/pnas.1117280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silva VKA, May RC, Rodrigues ML. 2020. Pyrifenox, an ergosterol inhibitor, differentially affects Cryptococcus neoformans and Cryptococcus gattii. Med Mycol doi: 10.1093/mmy/myz132. [DOI] [PubMed] [Google Scholar]
- 48.Te Dorsthorst DTA, Mouton JW, van den Beukel CJP, van der Lee HAL, Meis J, Verweij PE. 2004. Effect of pH on the in vitro activities of amphotericin B, itraconazole, and flucytosine against Aspergillus isolates. Antimicrob Agents Chemother 48:3147–3150. doi: 10.1128/AAC.48.8.3147-3150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pianalto KM, Ost KS, Brown HE, Alspaugh JA. 2018. Characterization of additional components of the environmental pH-sensing complex in the pathogenic fungus Cryptococcus neoformans. J Biol Chem 293:9995–10008. doi: 10.1074/jbc.RA118.002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucena-Agell D, Hervás-Aguilar A, Múnera-Huertas T, Pougovkina O, Rudnicka J, Galindo A, Tilburn J, Arst HN, Peñalva MA, Peñalva MA. 2016. Mutational analysis of the Aspergillus ambient pH receptor PalH underscores its potential as a target for antifungal compounds. Mol Microbiol 101:982–1002. doi: 10.1111/mmi.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mira NP, Lourenço AB, Fernandes AR, Becker JD, Sá-Correia I. 2009. The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res 9:202–216. doi: 10.1111/j.1567-1364.2008.00473.x. [DOI] [PubMed] [Google Scholar]
- 52.Nishino K, Obara K, Kihara A. 2015. The C-terminal cytosolic region of Rim21 senses alterations in plasma membrane lipid composition: insights into sensing mechanisms for plasma membrane lipid asymmetry. J Biol Chem 290:30797–30805. doi: 10.1074/jbc.M115.674382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamb TM, Xu W, Diamond A, Mitchell AP. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276:1850–1856. doi: 10.1074/jbc.M008381200. [DOI] [PubMed] [Google Scholar]
- 54.Hughes AL, Lee CYS, Bien CM, Espenshade PJ. 2007. 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J Biol Chem 282:24388–24396. doi: 10.1074/jbc.M701326200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung D, Haas H, Cramer R. 2012. Coordination of hypoxia adaptation and iron homeostasis in human pathogenic fungi. Front Microbiol 3:381. doi: 10.3389/fmicb.2012.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grahl N, Shepardson KM, Chung D, Cramer RA. 2012. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot Cell 11:560–570. doi: 10.1128/EC.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wezensky SJ, Cramer RA. 2011. Implications of hypoxic microenvironments during invasive aspergillosis. Med Mycol 49(Suppl 1):S120–S124. doi: 10.3109/13693786.2010.495139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DuBois JC, Pasula R, Dade JE, Smulian AG. 2016. Yeast transcriptome and in vivo hypoxia detection reveals Histoplasma capsulatum response to low oxygen tension. Med Mycol 54:40–58. doi: 10.1093/mmy/myv073. [DOI] [PubMed] [Google Scholar]
- 59.DuBois JC, Smulian AG. 2016. Sterol regulatory element binding protein (Srb1) is required for hypoxic adaptation and virulence in the dimorphic fungus Histoplasma capsulatum. PLoS One 11:e0163849. doi: 10.1371/journal.pone.0163849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hwang LH, Seth E, Gilmore SA, Sil A. 2012. SRE1 regulates iron-dependent and -independent pathways in the fungal pathogen Histoplasma capsulatum. Eukaryot Cell 11:16–25. doi: 10.1128/EC.05274-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoot SJ, Oliver BG, White TC. 2008. Candida albicans UPC2 is transcriptionally induced in response to antifungal drugs and anaerobicity through Upc2p-dependent and -independent mechanisms. Microbiology 154:2748–2756. doi: 10.1099/mic.0.2008/017475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vik A, Rine J. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol 21:6395–6405. doi: 10.1128/mcb.21.19.6395-6405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lane S, Zhou S, Pan T, Dai Q, Liu H. 2001. The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via Tec1. Mol Cell Biol 21:6418–6428. doi: 10.1128/mcb.21.19.6418-6428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Espenshade PJ. 2006. SREBPs: sterol-regulated transcription factors. J Cell Sci 119:973–976. doi: 10.1242/jcs02866. [DOI] [PubMed] [Google Scholar]
- 65.Rodrigues ML. 2018. The multifunctional fungal ergosterol. mBio 9:e01755-18. doi: 10.1128/mBio.01755-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mollinedo F. 2012. Lipid raft involvement in yeast cell growth and death. Front Oncol 2:140. doi: 10.3389/fonc.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baumann NA, Sullivan DP, Ohvo-Rekilä H, Simonot C, Pottekat A, Klaassen Z, Beh CT, Menon AK. 2005. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry 44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- 68.Hsieh E-J, Waters BM, Takahashi H. 2016. Alkaline stress and iron deficiency regulate iron uptake and riboflavin synthesis gene expression differently in root and leaf tissue: implications for iron deficiency chlorosis. J Exp Bot 67:5671–5685. doi: 10.1093/jxb/erw328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pianalto KM, Alspaugh JA. 2016. New horizons in antifungal therapy. J Fungi (Basel) 2:26. doi: 10.3390/jof2040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Odds FC, Webster CE, Abbott AB. 1984. Antifungal relative inhibition factors: BAY 1-9139, bifonazole, butoconazole, isoconazole, itraconazole (R 51211), oxiconazole, Ro 14-4767/002, sulconazole, terconazole and vibunazole (BAY n-7133) compared in vitro with nine established antifungal agents. J Antimicrob Chemother 14:105–114. doi: 10.1093/jac/14.2.105. [DOI] [PubMed] [Google Scholar]
- 71.Minagawa H, Kitaura K, Nakamizo N. 1983. Effects of pH on the activity of ketoconazole against Candida albicans. Antimicrob Agents Chemother 23:105–107. doi: 10.1128/AAC.23.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viviani MA, Esposto MC, Cogliati M, Tortorano AM. 2003. Flucytosine and cryptococcosis: which in vitro test is the best predictor of outcome? J Chemother 15:124–128. doi: 10.1179/joc.2003.15.2.124. [DOI] [PubMed] [Google Scholar]
- 73.Damaghi M, Wojtkowiak JW, Gillies RJ. 2013. pH sensing and regulation in cancer. Front Physiol 4:370. doi: 10.3389/fphys.2013.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raghunand N, Altbach MI, Van Sluis R, Baggett B, Taylor CW, Bhujwalla ZM, Gillies RJ. 1999. Plasmalemmal pH-gradients in drug-sensitive and drug-resistant MCF-7 human breast carcinoma xenografts measured by 31P magnetic resonance spectroscopy. Biochem Pharmacol 57:309–312. doi: 10.1016/s0006-2952(98)00306-2. [DOI] [PubMed] [Google Scholar]
- 75.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanhauteghem D, Janssens GPJ, Lauwaerts A, Sys S, Boyen F, Cox E, Meyer E. 2013. Exposure to the proton scavenger glycine under alkaline conditions induces Escherichia coli viability loss. PLoS One 8:e60328. doi: 10.1371/journal.pone.0060328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobay O, Laub K, Stercz B, Kéri A, Balázs B, Tóthpál A, Kardos S, Jaikumpun P, Ruksakiet K, Quinton PM, Zsembery Á. 2018. Bicarbonate inhibits bacterial growth and biofilm formation of prevalent cystic fibrosis pathogens. Front Microbiol 9:2245. doi: 10.3389/fmicb.2018.02245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Farnoud AM, Bryan AM, Kechichian T, Luberto C, Del Poeta M. 2015. The granuloma response controlling cryptococcosis in mice depends on the sphingosine kinase 1-sphingosine 1-phosphate pathway. Infect Immun 83:2705–2713. doi: 10.1128/IAI.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McQuiston T, Luberto C, Del Poeta M. 2010. Role of host sphingosine kinase 1 in the lung response against cryptococcosis. Infect Immun 78:2342–2352. doi: 10.1128/IAI.01140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cold Spring Harbor Laboratory Press. 2016. Yeast extract with supplements (YES). Cold Spring Harb Protoc doi: 10.1101/pdb.rec091355. [DOI] [Google Scholar]
- 81.Ingavale SS, Chang YC, Lee H, McClelland CM, Leong ML, Kwon-Chung KJ. 2008. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog 4:e1000155. doi: 10.1371/journal.ppat.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hickman MJ, Spatt D, Winston F. 2011. The Hog1 mitogen-activated protein kinase mediates a hypoxic response in Saccharomyces cerevisiae. Genetics 188:325–338. doi: 10.1534/genetics.111.128322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim MS, Kim S-Y, Yoon JK, Lee Y-W, Bahn Y-S. 2009. An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochem Biophys Res Commun 390:983–988. doi: 10.1016/j.bbrc.2009.10.089. [DOI] [PubMed] [Google Scholar]
- 84.Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol 175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nichols CB, Ferreyra J, Ballou ER, Alspaugh JA. 2009. Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot Cell 8:181–189. doi: 10.1128/EC.00351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McDade HC, Cox GM. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol 39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- 87.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Telzrow CL, Nichols CB, Castro-Lopez N, Wormley FL, Alspaugh JA. 2019. A fungal arrestin protein contributes to cell cycle progression and pathogenesis. mBio 10:e02682-19. doi: 10.1128/mBio.02682-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Love MI, Anders S, Kim V, Huber W. 2015. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Res 4:1070. doi: 10.12688/f1000research.7035.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 92.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brandão F, Esher SK, Ost KS, Pianalto K, Nichols CB, Fernandes L, Bocca AL, Poças-Fonseca MJ, Alspaugh JA. 2018. HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci Rep 8:5209. doi: 10.1038/s41598-018-21965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Waskom M, Botvinnik O, Ostblom J, Lukauskas S, Hobson P, Gelbart M, Gemperline DC, Augspurger T, Halchenko Y, Cole JB, Warmenhoven J, de Ruiter J, Pye C, Hoyer S, Vanderplas J, Villalba S, Kunter G, Quintero E, Bachant P, Martin M, Meyer K, Swain C, Miles A, Brunner T, O’Kane D, Yarkoni T, Williams ML, Evans C. 2020. mwaskom/seaborn: v0.10.0 (January 2020).
- 95.Perfect JR, Lang SD, Durack DT. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol 101:177–194. [PMC free article] [PubMed] [Google Scholar]
- 96.O’Meara TR, Xu W, Selvig KM, O’Meara MJ, Mitchell AP, Alspaugh JA. 2014. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol 34:673–684. doi: 10.1128/MCB.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chun CD, Madhani HD. 2010. Applying genetics and molecular biology to the study of the human pathogen Cryptococcus neoformans. Methods Enzymol 470:797–831. doi: 10.1016/S0076-6879(10)70033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 99.Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, Perfect JR, McCusker JH, Heitman J. 2002. Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J 21:546–559. doi: 10.1093/emboj/21.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jia N, Arthington-Skaggs B, Lee W, Pierson CA, Lees ND, Eckstein J, Barbuch R, Bard M. 2002. Candida albicans sterol C-14 reductase, encoded by the ERG24 gene, as a potential antifungal target site. Antimicrob Agents Chemother 46:947–957. doi: 10.1128/aac.46.4.947-957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Butler G, Rasmussen MD, Lin MF, Santos MAS, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJP, Brunke S, Costanzo MC, Fitzpatrick DA, De Groot PWJ, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KAT, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MPH, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NAR, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Young LY, Hull CM, Heitman J. 2003. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob Agents Chemother 47:2717–2724. doi: 10.1128/aac.47.9.2717-2724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fraser JA, Subaran RL, Nichols CB, Heitman J. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell 2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The sre1Δ mutant phenotypes are rescued by the reintroduction of the wild-type SRE1 allele, demonstrating a distinct growth defect from the rim101Δ mutant under microaerophilic conditions. (A) Strains were spotted in serial dilutions onto YPD pH 5.5, YPD pH 8, and YES plus 7 mM CoCl2. Growth was assessed after 3 days of growth and compared to both the sre1Δ mutant and WT strains. (B) The sre1Δ and stp1Δ mutant strains displayed an expected growth rate and colony size reduction in response to microaerophilic growth conditions. The rim101Δ mutant was able to grow in this environment similarly to wild-type levels. Strains were spotted in serial dilutions onto YPD at 30°C and YPD at 30°C and incubated under microaerophilic conditions. Download FIG S1, TIF file, 1.0 MB (1,021.3KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantitative analysis of exposed chitin levels in rim101Δ and sre1Δ mutant cell walls. The rim101Δ cell wall has increased exposed chitin staining by flow cytometry compared to WT and the sre1Δ mutant cell walls. (A and C) All strains were incubated for 16 to 18 h at 30°C in YPD medium (A) or 37°C in TC medium (C), fixed, labeled, and analyzed by flow cytometry. Wheat germ agglutinin (WGA) was used to stain the cell walls for exposed chitin. (A and C) Positive events were gated in the forward scatter (FSC)/side scatter (SSC) plots and are represented as histograms with cell counts on the y axis and mean fluorescence on the x axis. Unstained cells were sorted as controls to determine positive events. (B and D) The geometric means from positive events for each strain. Download FIG S2, TIF file, 1.0 MB (1,016.2KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Full transcriptome analyses and WT GO terms. (A) Volcano plot of the full transcriptome as analyzed from the data set for sre1Δ mutant versus WT at pH 8 (cropped version in Fig. 5B). (B) Volcano plot of the wild-type transcriptome. (C and D) GO-term analysis of the wild-type differentially expressed genes that are induced (C) and repressed (D) following a 90-min shift from YPD pH 4 to YPD pH 8. These transcripts were selected based on a strict cutoff of log2 fold change of +1 for induced transcripts and log2 fold change of −3 for repressed transcripts based on the uneven distribution of total repressed transcripts shown on the volcano plot (B). Download FIG S3, TIF file, 0.3 MB (361.2KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA-seq analysis. Page 1. sre1Δ versus wild-type differentially expressed genes at pH 8 (repressed). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < −1. Page 2. sre1Δ versus wild-type differentially expressed genes at pH 8 (induced). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < 1. Page 3. sre1Δ versus wild-type entire transcriptome at pH 8. Raw data. Page 4. Wild-type pH 4 versus wild-type pH 8 entire transcriptome. Raw data. Page 5. Wild-type pH 4 versus wild-type pH 8 differentially expressed genes (induced). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < 1. Page 6. Wild-type pH 4 versus wild-type pH 8 differentially expressed genes (repressed). Annotation of GO terms and putative processes/functions included. P < 0.05 and log2 fold change < −3. Download Table S1, XLSX file, 1.2 MB (1.2MB, xlsx) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Compared transcriptomics. Venn diagram showing the overlap between three different transcriptome data sets in C. neoformans serotype A (H99). Bien et al. (22) performed a comparative transcriptomics microarray study using the sre1Δ mutant strain and the sre1Δ + SRE1 complemented strain in low-oxygen growth environments (3% O2) (dark blue). A 1.4-fold-change cutoff was applied to this data set, and the transcripts included in this comparison were increased in the reconstituted strain compared to the mutant. Our data set looking at the sre1Δ mutant compared to wild type at low pH (4) and high pH (8) is shown in teal. We originally applied a fold change cutoff of 2 to our data set to analyze our GO-term categories (Fig. 5C and D) but lowered the cutoff to 1.4 to match the work of Bien (2009) and included only those transcripts that were repressed in the sre1Δ mutant compared to wild type. The overlap of our data with the previously published data set revealed many ergosterol-related transcripts: SRE1, ERG3, ERG4, ERG11, ERG6, and ERG13. Download FIG S4, TIF file, 0.7 MB (768.2KB, tif) .
Copyright © 2020 Brown et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All raw and analyzed RNA-sequencing data have been submitted to the NCBI GEO database under accession no. GSE147109 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147109).