Antibiotics constitute one of the cornerstones of modern medicine. However, individuals may succumb to a bacterial infection if a pathogen survives exposure to antibiotics. The ability of bacteria to survive bactericidal antibiotics results from genetic changes in the preexisting bacterial genome, from the acquisition of genes from other organisms, and from nonheritable phenomena that give rise to antibiotic tolerance. Nonheritable antibiotic tolerance can be exhibited by a large fraction of the bacterial population or by a small subpopulation referred to as persisters.
KEYWORDS: antibiotic tolerance, growth feedback regulation, persister
ABSTRACT
Antibiotics constitute one of the cornerstones of modern medicine. However, individuals may succumb to a bacterial infection if a pathogen survives exposure to antibiotics. The ability of bacteria to survive bactericidal antibiotics results from genetic changes in the preexisting bacterial genome, from the acquisition of genes from other organisms, and from nonheritable phenomena that give rise to antibiotic tolerance. Nonheritable antibiotic tolerance can be exhibited by a large fraction of the bacterial population or by a small subpopulation referred to as persisters. Nonheritable resistance to antibiotics has been ascribed to the activity of toxins that are part of toxin-antitoxin modules, to the universal energy currency ATP, and to the signaling molecule guanosine (penta) tetraphosphate. However, these molecules are dispensable for nonheritable resistance to antibiotics in many organisms. By contrast, nutrient limitation, treatment with bacteriostatic antibiotics, or expression of genes that slow bacterial growth invariably promote nonheritable resistance. We posit that antibiotic persistence results from conditions promoting feedback inhibition among core cellular processes, resulting phenotypically in a slowdown or halt in bacterial growth.
INTRODUCTION
The use of antibiotics as chemotherapeutic agents is an historic milestone in human achievement. Antibiotics have been used to control bacterial infections, decreasing morbidity and mortality, extending the length and quality of human life, and promoting economic growth (1–3). Antibiotics target essential bacterial processes, such as DNA supercoiling, transcription, translation, and cell wall biosynthesis (3, 4). Antibiotics are bactericidal when they promote bacterial killing and bacteriostatic when they inhibit bacterial growth.
Bacteria can survive and grow in the presence of bactericidal antibiotics through a variety of mechanisms. This antibiotic resistance can result from changes in genes encoding proteins that are targeted by antibiotics. Genetic changes may also result in an increased abundance of efflux pumps that decrease the cytoplasmic concentration of antibiotics below the threshold necessary for antibacterial action. In addition, bacteria can harbor products that destroy an antibiotic or that alter an antibiotic target so that it is no longer effectively inhibited by an antibiotic (Fig. 1). The latter products are often specified by genes in transmissible plasmids or transposable elements (4). Therefore, the acquisition of such genes can result in distantly related species harboring closely related antibiotic resistance determinants. The genes responsible for antibiotic resistance usually exhibit a limited phylogenetic distribution, most likely reflecting their association with species inhabiting niches with particular features that demand such a function and include recurrent antibiotic exposure (4, 5).
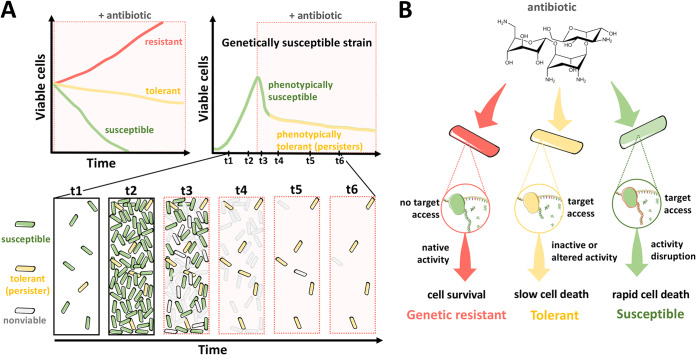
FIG 1.
Effect of a bactericidal antibiotic on killing kinetics among susceptible, tolerant, and resistant bacteria. (A) Schematic representation of antibiotic killing during the population growth of susceptible, tolerant, and resistant bacteria (top left). Resistant cells (red line) are unaffected and are able to grow in the presence of the antibiotic. Tolerant cells (yellow line) are susceptible to the antibiotic but display slower killing kinetics than susceptible cells (green line). Schematic representation of antibiotic killing during the population growth of genetically susceptible bacteria (top right). Cells within this population can be partitioned into phenotypically susceptible cells, which comprise the majority of the population (green line), and a small fraction of phenotypically tolerant, persister cells (yellow line). Cartoon representation of antibiotic killing during the population growth of a genetically susceptible bacterium (bottom). Phenotypically susceptible (green) and phenotypically tolerant, persister cells (yellow) are depicted. (B) Cartoon schematics depicting the inferred action of an antibiotic (kanamycin) on susceptible, tolerant, and resistant cells. Ribosome cartoons were modified from Servier Medical Art, licensed under a Creative Common Attribution 3.0 Generic License.
Genetically susceptible bacteria can also survive transient exposure to bactericidal antibiotics through a phenomenon known as tolerance. Tolerance is a behavior typically displayed by a large fraction of a bacterial population and results from genetic or nongenetic events. By contrast, persistence is a nongenetic form of tolerance exhibited by a small fraction of the bacterial population (Fig. 1A). Although the molecular basis for persistence is not well understood, such phenotypically resistant bacteria are thought to exist in a nonreplicative state in which the activity of their core biosynthetic machinery is transiently halted or diminished and, thus, protected from the lethal effects of antibiotics (Fig. 1B). In agreement with this notion, persister bacteria can withstand exposure to multiple antibiotics that target different core biochemical processes. In addition, when persisters are regrown in fresh media, they exhibit the same antibiotic susceptibility as the original culture (6–8). Unlike genetic resistance to antibiotics (4), nongenetic resistance has a broad phylogenetic distribution (8), suggesting that it is caused by a common attribute(s) central to diverse bacterial species and, perhaps, to all living organisms (9–11).
In this manuscript, we examine the physiological basis for nongenetic resistance to antibiotics. We posit that nongenetic antibiotic resistance results from a variety of mechanisms, all of which share the ability to dramatically decrease bacterial growth. A fundamental difference between genetic and nongenetic resistance to antibiotics is that the former relies on specific genes and pathways, whereas the latter does not. That eukaryotic organisms can display phenotypic resistance to a variety of drugs suggests that the underlying basis for nongenetic resistance to antimicrobial agents is conserved in evolution beyond bacteria (8–11).
The search for a genetic program responsible for nonheritable antibiotic resistance.
The clinical relevance of nonheritable resistance to antibiotics has led many laboratories to look for genetic determinants responsible for this phenomenon. Pioneering studies conducted with a laboratory strain of Escherichia coli established that mutations in the hipA gene (for high persistence) increased the frequency of persisters in the bacterial population >1,000-fold (12). Some of these mutations were later recovered in clinical isolates of E. coli and shown to confer increased antibiotic tolerance (13).
HipA is the toxin component of a toxin-antitoxin (TA) module that inhibits bacterial growth when not bound to the transcriptional regulator HipB (i.e., its cognate antitoxin) and the hipAB promoter (13). The toxin activity of HipA resides in its protein kinase domain, which phosphorylates the glutamyl aminoacyl tRNA synthase, thereby inhibiting protein synthesis (14, 15). Mutations in the hipA gene that result in high persistence map away from the regions specifying the kinase domain and the HipB binding region. These mutations decrease HipA’s ability to form homodimers, enabling monomeric HipA to promote persistence (13). The HipA/HipB pair is but one example of the many TA modules identified in bacteria (14–19). Despite the elegance of these and other studies that uncovered the biochemical activities of toxins and antitoxins (20, 21), as discussed below, TA modules are not necessary for tolerance/persistence to multiple antibiotics.
TA modules were first identified in plasmids as addiction modules that prevent plasmid loss by a mechanism referred to as postsegregation killing (see reference 22 for a recent review on TA modules). This mechanism involves an unstable antitoxin and a stable toxin that provokes the death of plasmid-free bacteria that fail to continuously produce the antitoxin. The vast majority of TA modules are encoded in bacterial chromosomes, and some species harbor dozens of TA modules (23), each consisting of a toxin that impairs cell growth and an antitoxin that inhibits the toxin (20, 21). However, the physiological role(s) that TA modules play in bacteria remains largely unclear (22).
Overexpression of toxins from TA modules can promote tolerance toward multiple antibiotics (8, 22, 24–27). For example, E. coli displayed tolerance to ciprofloxacin, ampicillin, and streptomycin when the tisB toxin gene was ectopically overexpressed from a plasmid (24). The tisB and istR genes form a TA module that is induced upon DNA damage by the bactericidal antibiotic ciprofloxacin. The TisB protein dissipates membrane potential, which decreases the concentration of ATP. This decrease is thought to increase tolerance to ciprofloxacin by preventing DNA synthesis. However, a different picture emerges when the tisB gene is expressed from its normal promoter and chromosomal location; partial tolerance is provided solely to its activating agent ciprofloxacin (24), thereby behaving similarly to an antibiotic resistance mechanism. Notably, tisB homologs are found only within selected members of the family Enterobacteriaceae, indicating that this TA module operates within restricted ecological contexts.
Tolerance toward multiple antibiotics can also be achieved upon overexpression of a variety of nontoxin genes (27–29). This is in spite of the fact that such genes encode proteins with different biochemical activities and subcellular locations and participate in a wide variety of cellular functions unrelated to those in which the toxins from TA modules act. These proteins include the chaperone DnaJ, the lipopolysaccharide-modifying PmrC, the transcriptional regulator Zur, the formate transporter FocA, the transhydrogenase PntA, and the sulfoquinovose isomerase YihS (27–29). The genes encoding these proteins share, along with toxin genes from TA modules, the ability to inhibit bacterial growth upon overexpression. Given that each of the proteins listed above inhibits growth by a different mechanism, we surmise that the multidrug tolerance phenotype associated with their overexpression results from the effects they have on cells aiming to maintain balanced growth.
TA modules were once believed to be required for antibiotic tolerance in several bacterial species. For example, the successive removal of TA module-encoding genes from the E. coli genome was reported to progressively decrease the level of persister organisms (30). However, that manuscript was later retracted (31), and the TA-mediated persistence originally reported was ascribed to contamination of the E. coli cultures with a bacteriophage (32). A different group ruled out a role for particular TA modules in E. coli tolerance by using bacterial cultures presumably free of the contaminating bacteriophage (33).
Surprisingly, 15-minute exposure of the facultative intracellular pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium) to bone marrow-derived macrophages was reported to increase the frequency of nonreplicating persister bacteria >100-fold relative to S. Typhimurium grown in Luria Bertani (LB) broth (34). Such an increase was also described for LB-grown S. Typhimurium pre-exposed to pH 4.5, a pH slightly lower than that S. Typhimurium experiences inside a macrophage phagosome (34). In addition, S. Typhimurium mutants deleted for particular TA module genes, including shpA/shpB and phD/doc, were defective in persister formation (34). However, the acidic pH induction of persisters could not be recapitulated by another group that used the same S. Typhimurium strain and growth conditions (28). Moreover, a collection of engineered S. Typhimurium strains lacking individual TA modules, sets of TA modules, or all known TA modules retained wild-type antibiotic tolerance (28). Furthermore, deletion of the shpA/shpB and phD/doc genes in a different S. Typhimurium genetic background did not affect S. Typhimurium replication rate during systemic infection in mice (35), when bacteria reside and form persisters in macrophages (36–38).
In the Gram-positive bacterium Staphylococcus aureus, removal of the three known TA modules had no effect on persister levels of either exponentially growing or stationary-phase bacteria (39). Taken together with the findings obtained with the Gram-negative species E. coli and S. Typhimurium discussed above, these findings provide direct genetic evidence against TA modules being required for antibiotic tolerance.
Reduced ATP abundance is associated with, but not required for, nonheritable antibiotic resistance.
The broad phylogenetic distribution of the persister phenotype (8) suggests that this phenomenon is implemented by a core, conserved feature of living cells and/or results from the action of a myriad of distinct cellular components that act on core cellular machines, impacting feedback control on such machines. The latter hypothesis finds experimental support from studies that link the defining property of persisters—high tolerance to multiple antibiotics—to a distinct cause or cellular perturbation. These perturbations range from mutations of particular genes to overexpression of various genes and also include exposure to chemicals or physical conditions that slow growth (12, 25–29, 39–45). For example, treatment of bacterial cultures with the ionophore m-chlorophenylhydrazone uncouples the electron transport chain from ATP synthesis, which results in ATP depletion, growth arrest, and multidrug tolerance (41).
Likewise, wild-type S. Typhimurium was 10,000 times more antibiotic tolerant when grown in defined media of low Mg2+ for 5 h than when grown in the same media for 2 h, possibly reflecting that ATP abundance was 15-fold lower at 5 h than at 2 h (28). Furthermore, expression of the soluble subunit of the ATP synthase resulted in a dramatic decrease in ATP abundance and a concomitant increase in S. Typhimurium survival in cefotaxime or ciprofloxacin (28). Cumulatively, these findings suggest that persister bacteria emerge as a consequence of a decrease in the cellular concentration of ATP (39, 42).
ATP is a universal chemical energy storage molecule utilized by all cells to fuel biosynthetic reactions, to transport solutes across biological membranes, and to synthesize proteins following its conversion into GTP. Therefore, changes in cellular ATP provide a unifying explanation for a decrease in the ATP concentration, driving tolerance across Gram-negative E. coli, Gram-positive S. aureus, and perhaps even eukaryotes. However, as detailed below, this notion fails to explain the root of the antibiotic persistence phenomenon.
First, at any given time, the cellular ATP concentration is determined by hundreds of anabolic and catabolic processes (46, 47). Therefore, fluctuations in ATP concentration must ultimately be explained in terms of changes in these processes. In agreement with this reasoning, correlations between low ATP concentrations and antibiotic tolerance have been attributed, in part, to the stochastic expression of the genes encoding tricarboxylic acid (TCA) cycle enzymes by a mechanism that remains unknown (48). Because the TCA cycle generates ATP (as well as other products), the abundance of TCA cycle enzymes would determine the ATP concentration in individual bacterial cells and, thus, antibiotic persistence.
Second, chemical treatments that result in ATP depletion promote multidrug tolerance to populations of E. coli and S. aureus (39, 41, 42). This observation is consistent with the hypothesis that low ATP concentration is the cause, or at least part, of a causal chain of events, leading to high, multiantibiotic tolerance. However, low ATP concentration is not necessary for tolerance to multiple bactericidal antibiotics. This is because treatment of wild-type S. Typhimurium with the bacteriostatic antibiotic chloramphenicol promoted tolerance despite dramatically increasing the ATP concentration (28). Chloramphenicol treatment increases the cellular ATP concentration because it is an inhibitor of translation, and translation is the most energy expensive activity cells undertake (49). Likewise, treatment of a laboratory strain of E. coli with tetracycline increased survival in ampicillin or ciprofloxacin >1,000-fold relative to bacteria that were not treated with tetracycline (41). Tetracycline is also a bacteriostatic protein synthesis inhibitor, but its mechanism of action is different from that of chloramphenicol (4). Because tetracycline and chloramphenicol inhibit bacterial growth, the antibiotic tolerance they promote can be ascribed to bacteria not growing.
Third, naturally emerging, phenotypically variant subpopulations of E. coli with rRNA promoter activity displayed tolerance to multiple antibiotics (42). Given that rRNA promoter activity increases with ATP concentration, bacteria exiting energy starvation conditions must have low ATP amounts (42). However, rRNA transcription increases not only when ATP concentration increases but also when growth rate increases. While the molecular basis responsible for the growth rate-dependent regulation of rRNA transcription is poorly understood, it is distinct from the control exerted by ATP abundance (50–55). Increases in growth rate lead to heightened rRNA transcription even though ATP amounts remain constant (53–55). In other words, the correlation between low rRNA transcription and subpopulations exhibiting multiantibiotic tolerance may reflect low ATP amounts, low growth rate, or both.
Fourth, subpopulations of S. aureus displaying multiantibiotic tolerance were presumed to have low ATP amounts by virtue of expressing genes normally induced at the stationary phase of growth and upon arsenate treatment (39). Because direct measurements of ATP amounts were not provided in this study and because the regulatory cascades and signals governing transcription of the tested genes are largely unknown, expression of the chosen reporter genes may reflect low ATP amounts, slow growth, or any other condition encountered during stationary phase or arsenate treatment.
Finally, when bacteria run out of ATP, they stop growing, which favors the formation of persisters. This in and of itself does not make low ATP concentration essential for persister formation because it is possible to achieve tolerance to multiple antibiotics by stopping growth while cells accumulate ATP to high concentrations.
Depending on growth conditions, the signaling molecules ppGpp and pppGpp are required, dispensable, or detrimental for nonheritable antibiotic resistance.
The stringent response is a collection of molecular responses mediated by the production of GDP 3′-diphosphate (ppGpp) and GTP 3′-diphosphate (pppGpp) by bacterial cells experiencing nutritional starvation (56). ppGpp and pppGpp [from here on abbreviated as (p)ppGpp] control a wide variety of cellular functions and are implicated in antibiotic tolerance.
(p)ppGpp accumulation was first identified in E. coli experiencing amino acid starvation (57), and it is now known that (p)ppGpp can accumulate in bacteria starving for other nutrients, such as carbon and phosphate (58–62). When E. coli starves for an amino acid, the corresponding uncharged tRNAs can bind to the A site of ribosomes, and the stalled ribosomes are detected by the protein RelA, which is responsible for the synthesis of (p)ppGpp (63). E. coli also harbors SpoT, a protein with lower (p)ppGpp synthase capacity than RelA but which, unlike RelA, can also hydrolyze (p)ppGpp. Bacterial species differ in the number and classes of proteins that can make and break down (p)ppGpp (i.e., monofunctional versus bifunctional) (56).
(p)ppGpp exerts its regulatory function by binding a wide variety of proteins (64, 65). In E. coli, (p)ppGpp binding to RNA polymerase directly changes transcription of ∼15% of the genome (66). Two of the most salient changes are a decrease in the transcription of rRNA genes and an increase in transcription of amino acid biosynthetic genes. It makes intuitive sense for a bacterium experiencing amino acid limitation to decrease the synthesis of components of its protein synthesis machine and to increase the production of the enzymes responsible for making amino acids. Even in organisms experiencing plenty of amino acids (i.e., the substrates of protein synthesis) and ATP (i.e., the energy source to run protein synthesis), low cytoplasmic Mg2+ decreases rRNA synthesis in a (p)ppGpp-dependent manner (67). This is because Mg2+ is necessary for the assembly of functional ribosomes, and bacteria decrease the synthesis of rRNA when the cytoplasmic Mg2+ concentration drops below a certain threshold (67). As proteins are essential for cell growth, a decrease in the number of ribosomes decreases the rate of protein synthesis, which in turn reduces growth rate. We posit that such a reduction in growth rate can give rise to antibiotic tolerance.
A role of (p)ppGpp in antibiotic tolerance has emerged from both the recovery of clinical isolates bearing large amounts of (p)ppGpp and laboratory studies conducted with different bacterial species. For example, isolates of the Gram-positive bacteria S. aureus and Enterobacter faecium bearing large amounts of (p)ppGpp due to mutations in the genes responsible for (p)ppGpp synthesis (68, 69) caused persistent infections and required prolonged antibiotic treatment for clearing (56). In the laboratory, starving E. coli for certain amino acids increased resistance to the bactericidal antibiotic penicillin (70). Resistance appears to be mediated, in part, by the stringent response because the protective effect of amino acid starvation was exhibited at later times in a relA mutant (70). Moreover, deletion of the relA and spoT genes eliminated the persistence phenotype of an E. coli strain bearing the hipA7 allele, which specifies a HipA variant that confers high persistence but is not toxic, unlike the wild-type HipA protein (71).
(p)ppGpp was proposed to promote antibiotic persistence in E. coli by increasing the fraction of free toxins from TA modules (72). According to this proposal, (p)ppGpp would inhibit the enzyme that degrades polyphosphate—termed exopolyphosphatase—resulting in the accumulation of polyphosphate, which in turn would activate the protease Lon to degrade the antitoxin from TA modules. The unbound toxins would then be free to inhibit their targets. In agreement with this notion, an E. coli mutant lacking the capacity to make (p)ppGpp due to a deletion of the relA and spoT genes was ∼100-fold less persistent to ciprofloxacin and ampicillin than the isogenic wild-type strain, and a lon null mutant was even less persistent than the relA spoT double mutant (72). However, the article describing these findings was later retracted (73).
In S. Typhimurium, a relA spoT double mutant and a lon mutant were reported to be ∼10 times and ∼100 times less tolerant to antibiotics than the wild-type strain, respectively, following bacterial internalization by macrophages (34). These phenotypes are reminiscent of those for E. coli in the retracted paper discussed above. In addition, the genes specifying 14 TA operons were reportedly upregulated 4- to 30-fold within 30 min of S. Typhimurium phagocytosis by macrophages (34). This upregulation was dependent on the relA and spoT genes and also observed upon acidification of the laboratory media, a condition that, as discussed above, was also reported to promote antibiotic persistence (34). By contrast, a group investigating the same S. Typhimurium strain and using the same experimental protocol found that the relA spoT double mutant retained wild-type tolerance to ciprofloxacin and cefotaxime in bacteria experiencing acidified LB medium (28). Moreover, (p)ppGpp was dispensable for the tolerance to cefotaxime displayed by an S. Typhimurium tryptophan auxotroph in medium lacking tryptophan (28).
Serine hydroxamate (Shx) is an amino acid analog often used to trigger the stringent response because it hinders the charging of seryl-tRNA with serine (74). It has been reported that Shx treatment of stationary-phase cultures of S. Typhimurium increased persistence ∼5-fold relative to bacteria grown in LB broth (34). This observation is puzzling because bacteria in stationary phase are already nutrient starved, which results in large amounts of (p)ppGpp (55, 61, 62, 75). This is why a different group testing the same S. Typhimurium strain established that Shx had no effect on tolerance when added to bacteria in the stationary phase but did render bacteria immune to cefotaxime when added to logarithmically growing bacteria (28). These sets of data argue that (p)ppGpp is dispensable for antibiotic tolerance elicited by a variety of conditions.
Actually, depending on the experimental conditions, the ability to make (p)ppGpp can hinder antibiotic tolerance. That is, a relA spoT double mutant survived cefotaxime treatment better than wild-type S. Typhimurium during growth in defined medium lacking amino acids (28). These results reflect that a relA spoT double mutant displays multiple auxotrophies and, thus, cannot grow in the absence of amino acids, whereas the wild-type strain can, rendering the latter, but not the former, susceptible to killing by bactericidal antibiotics (28, 62). In other words, the (p)ppGpp-promoted growth occurring in the absence of certain nutrients is what hinders tolerance in a wild-type strain but not in the (p)ppGpp-lacking relA spoT double mutant.
Cells coordinate the activities of their core machineries.
Living cells are composed of defined sets of components. These components are dynamic in nature. Therefore, the synthesis, degradation, and recycling of these components allow cells to maintain homeostasis despite changes in their surroundings. While the overall chemical composition of cells varies across cell types and growth conditions (50, 76, 77), all cells share a set of components that participate in processes responsible for growth and viability, namely, the machineries carrying out the three core cellular processes—DNA replication, transcription, and translation.
In bacteria, the three core cellular processes are coregulated (Fig. 2). Early studies established that global transcriptional and translational activities control chromosome replication (78–81). Similarly, the translational status of a cell conveys information to the transcriptional and DNA-replicating machineries, modulating their activities (51, 61–65, 82, 83). That ribosome activity is required for the initiation of chromosome replication is conserved in distantly related bacterial species, such as E. coli and Bacillus subtilis (78–83). Other vital cellular processes are also coordinated, namely cell division that is regulated by the DNA replication status (see reference 84 for a recent review on this topic). Given that imbalances in cellular components can hinder growth or lead to a loss of cellular viability (62, 67, 82, 83, 85–101), feedback regulatory mechanisms ensure that cellular constituents are synthesized within a stoichiometric range compatible with life. That is, the activities of cellular components are coordinately regulated to avoid deleterious effects resulting from inefficient resource allocation, which can lead to the accumulation of toxic metabolites, the depletion of vital biosynthetic precursors, stoichiometric imbalances of cellular components, and the establishment of artificial, inhibitory molecular interactions that disrupt cellular processes. This fundamental property can explain noninheritable antibiotic resistance, a phenomenon that, as discussed above, is conserved across distantly related bacterial species.
FIG 2.
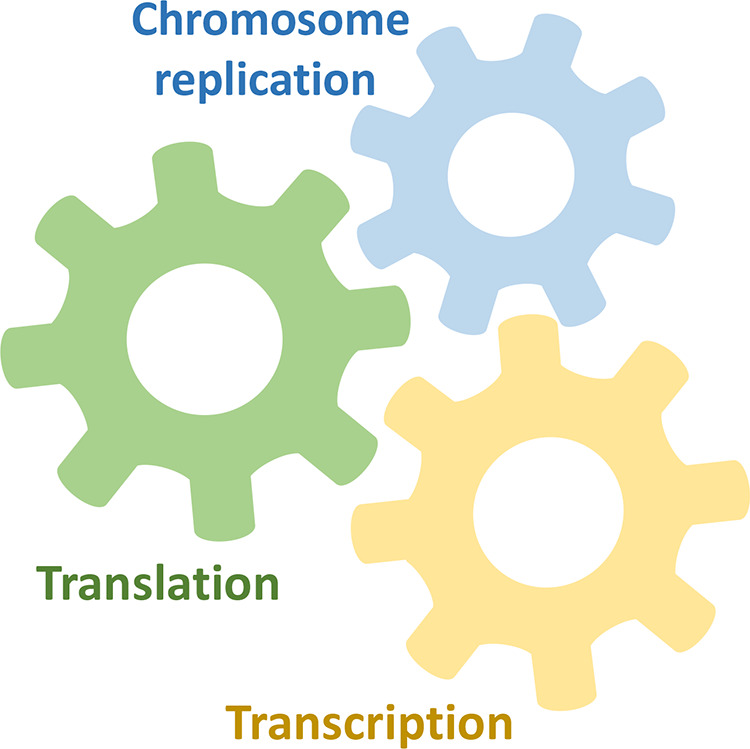
Cartoon depicting the three core cellular processes (chromosome replication, transcription, and translation) and coregulatory relationships among them.
In the laboratory, bacteria are typically grown under conditions favoring high catabolism, whereby ATP production from available substrates exceeds the energetic requirements of the cell. Under such conditions, bacteria dissipate excess ATP, and their growth rate is dependent on the availability of critical precursors rather than the energy to power nutrient transport and biosynthetic reactions (46, 47). Hence, under laboratory conditions, ATP amounts remain constant even when the growth rate is increased by nutritional up-shifts, which makes limiting precursors readily available, or by temperature increases, which speeds up biosynthetic reactions (46, 47, 53–55, 76, 102, 103). In other words, under typical laboratory conditions, ATP amounts are usually uncoupled from biosynthetic reactions, both of which are required for growth and targeted by antibiotics.
In addition to the intrinsic coregulation of DNA replication, transcription, and translation, ancillary proteins can also exert feedback control of the core machinery in response to environmental cues. For instance, the mgtC gene, which was acquired by horizontal gene transfer in S. Typhimurium (104), specifies a protein that reduces ATP concentration, thereby decreasing the synthesis of rRNA (67). MgtC is required when the S. Typhimurium cytoplasmic Mg2+ concentration drops below the threshold that compromises the assembly of functional ribosomes (67, 105–107). Therefore, MgtC decreases ATP amounts when the cytoplasmic Mg2+ concentration is low, which in turn decreases the transcription of rRNA genes, resulting in smaller amounts of ribosomes and a reduced rate of protein synthesis. In other words, S. Typhimurium adjusts the number of ribosomes to the Mg2+ amounts available for the assembly of functional ribosomes. Notably, while E. coli lacks an mgtC homolog, it also decreases rRNA synthesis in response to low cytoplasmic Mg2+ (67). Hence, E. coli and S. Typhimurium utilize different strategies to achieve the same goal, namely a reduction in ribosome numbers during Mg2+ limitation. In summary, bacterial species can control core biosynthetic activity in a like manner but often vary in how they implement feedback control in response to particular conditions.
Bacterial growth, rather than metabolic activity, governs nonheritable resistance to antibiotics.
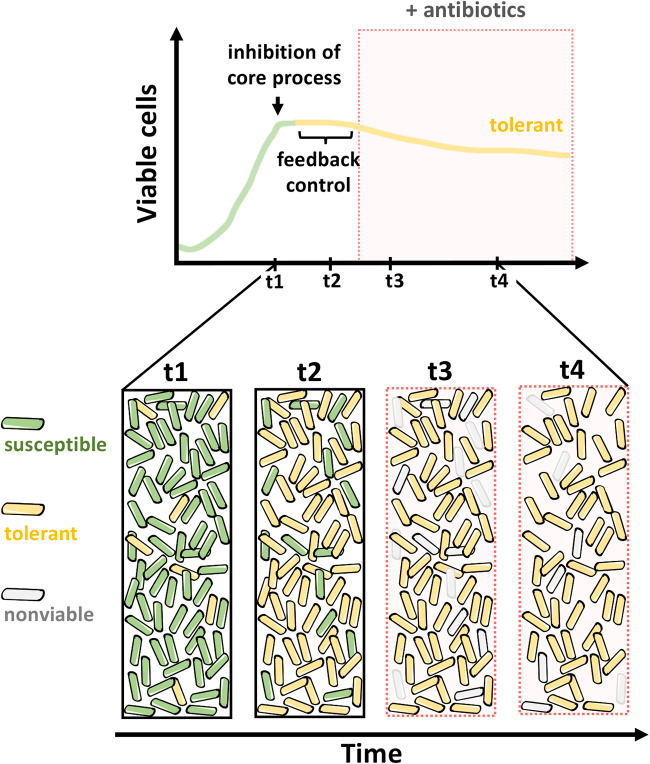
Experiments conducted over the last 70 years have established a strong correlation between growth rate and antibiotic susceptibility, in that rapidly dividing cells are more prone to antibiotic-mediated killing (28, 35, 37, 38, 108–117). This correlation holds for different bacterial species and for antibiotics that target different core processes. Feedback control on core cellular processes, which determines the rate of biosynthetic activities targeted by antibiotics and dictates growth rate, is most likely responsible for this phenomenon (Fig. 3).
FIG 3.
Schematic representation of antibiotic killing in a growing population of susceptible bacteria following inhibition of a core biosynthetic process (top). Feedback regulation promotes the inhibition of other major cellular processes, leading to multidrug tolerance. Cartoons depicting processes outlined in the top schematic (bottom).
Surprisingly, it has been argued that susceptibility to antibiotics is directly related to ATP amounts, even in conditions of energy abundance—when a large fraction of cellular ATP does not participate in biosynthetic reactions fostering growth—and apparently, growth rate no longer correlates with the efficacy of antibiotic killing (103). These results are hard to reconcile with the large body of work demonstrating a strong correlation between growth rate and antibiotic susceptibility (28, 35, 37, 38, 108–117), especially because these studies were largely conducted under high-energy conditions (28, 108–114, 116).
It has been known for over 60 years that the chemical composition of cells changes when growth rate is modulated by nutrient availability (50, 76, 77). Therefore, it is puzzling to see a recent publication examine the effects of growth rate and ATP amounts on antibiotic susceptibility by altering the nutrient content of the growth medium (103). The problem with the chosen experimental strategy is that many physiological parameters are not controlled under disparate growth conditions. For instance, changes in nutrient availability may affect membrane permeability, ultimately altering antibiotic susceptibility (118, 119). In this context, the association between ATP levels and antibiotic susceptibility would simply be an epiphenomenon. Interestingly, in this work, ATP amounts are similar in bacteria experiencing energy abundance, whether grown at 25°C or 37°C, despite the growth rate increasing with temperature (103). However, modulation of growth rate by temperature does not alter the chemical composition of bacteria (50, 76, 77). Actually, in direct contradiction to the proposed model, it was reported that bacteria rapidly growing at 37°C are killed faster by antibiotics than their slow-growing counterparts at 25°C (103).
Finally, if ATP abundance determines the efficacy of killing independently of growth rate and type of antibiotic, then two non-mutually exclusive conclusions would follow: (i) high ATP abundance should increase the rate of all biosynthetic processes targeted by antibiotics—translation, cell wall biosynthesis, and DNA replication—but the increase in the activity of these processes would not result in a concomitant increase in biomass (i.e., growth). This means that an increase in ATP abundance would lead to an increase in rRNA, resulting in a higher number of ribosomes and, thus, higher translation rates, but that no extra proteins would be synthesized. Also, (ii) antibiotics should kill bacteria independently of the activity of their respective targets but dependently on the amount of ATP in the cells. That is, the rate of killing by bactericidal antibiotics would depend on something other than their effect on the activity of their cellular targets. The latter conclusion is reminiscent of the proposal that a wide variety of classes of antibiotics mediate killing by the generation of reactive oxygen species (120), a notion disputed by several independent groups (121–123).
Conclusions.
In summary, we posit that there is no genetic program specifically devoted to nonheritable resistance to antibiotics. That is, a wide variety of growth and genetic conditions alter antibiotic resistance (124, 125). Therefore, a given gene or biochemical pathway may promote or inhibit resistance depending on the growth conditions. In this context, this hypothesis can explain both the inability of genetic screens to isolate mutations rendering cells unable to form persisters (126, 127) and the myriad of disruptive genetic and environmental contexts that lead to multidrug tolerance (8, 12, 13, 22, 24–29, 39–45).
We propose that the phenomenon of persister cell formation is the outcome of an evolved, essential property, namely feedback regulation of core cellular processes. Feedback regulation enables cells to buffer disturbances arising from external and internal perturbations that cause unbalanced growth and loss of viability. Typically, impairment of any major biosynthetic activity will lead to the inhibition of other vital cellular activities. Although a measurable outcome of this process is temporary immunity to multiple antibiotics, this is unlikely to be its evolved purpose.
Finally, the hypothesis presented above provides a conceptual framework that explains (128–130) and predicts that the disruption of feedback regulation should eradicate persisters. That is, persisters should be eliminated by imposing conditions that irreversibly abolish the core biosynthetic machinery when inactive core cellular components are degraded or the cytoplasmic environment is disrupted during a lethal antibiotic exposure (128–130). The identification of compounds that accomplish these tasks will result in the elimination of antibiotic persisters.
ACKNOWLEDGMENTS
We thank Jennifer Aronson for comments on the manuscript.
This work was supported by grants AI49561 and AI120558 from the National Institutes of Health to E.A.G. and funds from the Penn State College of Medicine to M.H.P.
Footnotes
Citation Pontes MH, Groisman EA. 2020. A physiological basis for nonheritable antibiotic resistance. mBio 11:e00817-20. https://doi.org/10.1128/mBio.00817-20.
REFERENCES
- 1.Aminov RI. 2010. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol 1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naylor NR, Atun R, Zhu N, Kulasabanathan K, Silva S, Chatterjee A, Knight GM, Robotham JV. 2018. Estimating the burden of antimicrobial resistance: a systematic literature review. Antimicrob Resist Infect Control 7:58. doi: 10.1186/s13756-018-0336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma C, Yang X, Lewis PJ. 2016. Bacterial transcription as a target for antibacterial drug development. Microbiol Mol Biol Rev 80:139–160. doi: 10.1128/MMBR.00055-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Hoek AH, Mevius D, Guerra B, Mullany P, Roberts AP, Aarts HJ. 2011. Acquired antibiotic resistance genes: an overview. Front Microbiol 2:203. doi: 10.3389/fmicb.2011.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet 17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 6.Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI, Brynildsen MP, Bumann D, Camilli A, Collins JJ, Dehio C, Fortune S, Ghigo JM, Hardt WD, Harms A, Heinemann M, Hung DT, Jenal U, Levin BR, Michiels J, Storz G, Tan MW, Tenson T, Van Melderen L, Zinkernagel A. 2019. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol 17:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilmaerts D, Windels EM, Verstraeten N, Michiels J. 2019. General mechanisms leading to persister formation and awakening. Trends Genet 35:401–411. doi: 10.1016/j.tig.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Ayrapetyan M, Williams T, Oliver JD. 2018. Relationship between the viable but nonculturable state and antibiotic persister cells. J Bacteriol 200:e00249-18. doi: 10.1128/JB.00249-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delarze E, Sanglard D. 2015. Defining the frontiers between antifungal resistance, tolerance and the concept of persistence. Drug Resist Updat 23:12–19. doi: 10.1016/j.drup.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Barrett MP, Kyle DE, Sibley LD, Radke JB, Tarleton RL. 2019. Protozoan persister-like cells and drug treatment failure. Nat Rev Microbiol 17:607–620. doi: 10.1038/s41579-019-0238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vera-Ramirez L, Hunter KW. 2017. Tumor cell dormancy as an adaptive cell stress response mechanism. F1000Res 6:2134. doi: 10.12688/f1000research.12174.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768–775. doi: 10.1128/JB.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher MA, Balani P, Min J, Chinnam NB, Hansen S, Vulić M, Lewis K, Brennan RG. 2015. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 524:59–64. doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germain E, Castro-Roa D, Zenkin N, Gerdes K. 2013. Molecular mechanism of bacterial persistence by HipA. Mol Cell 52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 15.Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. 2013. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun 4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 18.Winther KS, Gerdes K. 2011. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A 108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y, Pogliano J, Helinski DR, Konieczny I. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol 44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet 45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 21.Hayes F, Van Melderen L. 2011. Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol 46:386–408. doi: 10.3109/10409238.2011.600437. [DOI] [PubMed] [Google Scholar]
- 22.Fraikin N, Goormaghtigh F, Van Melderen L. 2020. Type II toxin-antitoxin systems: evolution and revolutions. J Bacterio 202:e00763-19. doi: 10.1128/JB.00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao Y, Harrison EM, Bi D, Tai C, He X, Ou HY, Rajakumar K, Deng Z. 2011. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res 39:D606–D611. doi: 10.1093/nar/gkq908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versées W, Hofkens J, Jansen M, Fauvart M, Michiels J. 2015. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell 59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y, Wood TK. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun 391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vázquez-Laslop N, Lee H, Neyfakh AA. 2006. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J Bacteriol 188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pontes MH, Groisman EA. 2019. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci Signal 12:eaax3938. doi: 10.1126/scisignal.aax3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chowdhury N, Kwan BW, Wood TK. 2016. Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci Rep 6:20519. doi: 10.1038/srep20519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.PNAS. 2018. Retraction for Maisonneuve et al., bacterial persistence by RNA endonucleases. Proc Natl Acad Sci U S A 115:E2901. doi: 10.1073/pnas.1803278115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K. 2017. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8:e01964-17. doi: 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goormaghtigh F, Fraikin N, Putrinš M, Hallaert T, Hauryliuk V, Garcia-Pino A, Sjödin A, Kasvandik S, Udekwu K, Tenson T, Kaldalu N, Van Melderen L. 2018. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio 9:e00640-18. doi: 10.1128/mBio.00640-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claudi B, Spröte P, Chirkova A, Personnic N, Zankl J, Schürmann N, Schmidt A, Bumann D. 2014. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 36.Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella Typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A 83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abshire KZ, Neidhardt FC. 1993. Growth rate paradox of Salmonella typhimurium within host macrophages. J Bacteriol 175:3744–3748. doi: 10.1128/jb.175.12.3744-3748.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser P, Regoes RR, Dolowschiak T, Wotzka SY, Lengefeld J, Slack E, Grant AJ, Ackermann M, Hardt WD. 2014. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol 12:e1001793. doi: 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K. 2016. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol 1:16051. doi: 10.1038/nmicrobiol.2016.51. [DOI] [PubMed] [Google Scholar]
- 40.Ocampo PS, Lázár V, Papp B, Arnoldini M, Abel Zur Wiesch P, Busa-Fekete R, Fekete G, Pál C, Ackermann M, Bonhoeffer S. 2014. Antagonism between bacteriostatic and bactericidal antibiotics is prevalent. Antimicrob Agents Chemother 58:4573–4582. doi: 10.1128/AAC.02463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwan BW, Valenta JA, Benedik MJ, Wood TK. 2013. Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother 57:1468–1473. doi: 10.1128/AAC.02135-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K. 2017. ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mBio.02267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 44.Bukhari AI, Taylor AL. 1971. Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J Bacteriol 105:844–854. doi: 10.1128/JB.105.3.844-854.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eidlic L, Neidhardt FC. 1965. Protein and nucleic acid synthesis in two mutants of Escherichia coli with temperature-sensitive aminoacyl ribonucleic acid synthetases. J Bacteriol 89:706–711. doi: 10.1128/JB.89.3.706-711.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell JB, Cook GM. 1995. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev 59:48–62. doi: 10.1128/MMBR.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marr AG. 1991. Growth rate of Escherichia coli. Microbiol Rev 55:316–333. doi: 10.1128/MMBR.55.2.316-333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zalis EA, Nuxoll AS, Manuse S, Clair G, Radlinski LC, Conlon BP, Adkins J, Lewis K. 2019. Stochastic variation in expression of the tricarboxylic acid cycle produces persister cells. mBio 10:e01930-19. doi: 10.1128/mBio.01930-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stouthamer AH. 1973. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek 39:545–565. doi: 10.1007/BF02578899. [DOI] [PubMed] [Google Scholar]
- 50.Schaechter M, Maaloe O, Kjeldgaard NO. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella Typhimurium. J Gen Microbiol 19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 51.Sarmientos P, Sylvester JE, Contente S, Cashel M. 1983. Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32:1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- 52.Gaal T, Gourse RL. 1990. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci U S A 87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petersen C, Møller LB. 2000. Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J Biol Chem 275:3931–3935. doi: 10.1074/jbc.275.6.3931. [DOI] [PubMed] [Google Scholar]
- 54.Schneider DA, Gourse RL. 2004. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J Biol Chem 279:8262–8268. doi: 10.1074/jbc.M311996200. [DOI] [PubMed] [Google Scholar]
- 55.Murray HD, Schneider DA, Gourse RL. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 56.Hobbs JK, Boraston AB. 2019. (p)ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS Infect Dis 5:1505–1517. doi: 10.1021/acsinfecdis.9b00204. [DOI] [PubMed] [Google Scholar]
- 57.Cashel M, Gallant J. 1969. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 58.Spira B, Silberstein N, Yagil E. 1995. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J Bacteriol 177:4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinella D, Albrecht C, Cashel M, D'Ari R. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol 56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 60.Seyfzadeh M, Keener J, Nomura M. 1993. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci U S A 90:11004–11008. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazzarini RA, Cashel M, Gallant J. 1971. On the regulation of guanosine tetraphosphate levels in stringent and relaxed strains of Escherichia coli. J Biol Chem 246:4381–4385. [PubMed] [Google Scholar]
- 62.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 63.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. 2002. Dissection of the mechanism for the stringent factor RelA. Mol Cell 10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 64.Kanjee U, Ogata K, Houry WA. 2012. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol 85:1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x. [DOI] [PubMed] [Google Scholar]
- 65.Gourse RL, Chen AY, Gopalkrishnan S, Sanchez-Vazquez P, Myers A, Ross W. 2018. Transcriptional responses to ppGpp and DksA. Annu Rev Microbiol 72:163–184. doi: 10.1146/annurev-micro-090817-062444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanchez-Vazquez P, Dewey CN, Kitten N, Ross W, Gourse RL. 2019. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc Natl Acad Sci U S A 116:8310–8319. doi: 10.1073/pnas.1819682116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pontes MH, Yeom J, Groisman EA. 2016. Reducing ribosome biosynthesis promotes translation during low Mg2+ stress. Mol Cell 64:480–492. doi: 10.1016/j.molcel.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Honsa ES, Cooper VS, Mhaissen MN, Frank M, Shaker J, Iverson A, Rubnitz J, Hayden RT, Lee RE, Rock CO, Tuomanen EI, Wolf J, Rosch JW. 2017. RelA mutant Enterococcus faecium with multiantibiotic tolerance arising in an immunocompromised host. mBio 8:e02124-16. doi: 10.1128/mBio.02124-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JI, Hartland EL, Stinear TP, Howden BP. 2010. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog 6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodell W, Tomasz A. 1980. Alteration of Escherichia coli murein during amino acid starvation. J Bacteriol 144:1009–1016. doi: 10.1128/JB.144.3.1009-1016.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korch SB, Henderson TA, Hill TM. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol 50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 72.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 73.Maisonneuve E, Castro-Camargo M, Gerdes K. 2018. Retraction notice to: (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 172:1135. doi: 10.1016/j.cell.2018.02.023. [DOI] [PubMed] [Google Scholar]
- 74.Tosa T, Pizer LI. 1971. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J Bacteriol 106:972–982. doi: 10.1128/JB.106.3.972-982.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, Rhee JH, Yoon H, Ryu S, Lim S, Choy HE. 2004. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem 279:34183–34190. doi: 10.1074/jbc.M313491200. [DOI] [PubMed] [Google Scholar]
- 76.Bremer H, Dennis PP. 1996. Modulation of chemical composition and other parameters of the cell by growth rate, p 1553–1569. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, vol 2 ASM Press, Washington, DC. [Google Scholar]
- 77.Neidhardt FC, Umbarger HE. 1996. Chemical composition of Escherichia coli, p 13–16. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 78.Lark KG. 1972. Evidence for the direct involvement of RNA in the initiation of DNA replication in Escherichia coli 15T−. J Mol Biol 64:47–60. doi: 10.1016/0022-2836(72)90320-8. [DOI] [PubMed] [Google Scholar]
- 79.Messer W. 1972. Initiation of deoxyribonucleic acid replication in Escherichia coli B/r: chronology of events and transcriptional control of initiation. J Bacteriol 112:7–12. doi: 10.1128/JB.112.1.7-12.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clewell DB. 1972. Nature of ColE1 plasmid replication in Escherichia coli in the presence of chloramphenicol. J Bacteriol 110:667–676. doi: 10.1128/JB.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murakami S, Inuzuka N, Yamaguchi M, Yamaguchi K, Yoshikawa H. 1976. Initiation of DNA replication in Bacillus subtilis. III. Analysis of molecular events involved in the initiation using a temperature-sensitive dna mutant. J Mol Biol 108:683–704. doi: 10.1016/s0022-2836(76)80112-x. [DOI] [PubMed] [Google Scholar]
- 82.Levine A, Vannier F, Dehbi M, Henckes G, Séror SJ. 1991. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J Mol Biol 219:605–613. doi: 10.1016/0022-2836(91)90657-r. [DOI] [PubMed] [Google Scholar]
- 83.Wang JD, Sanders GM, Grossman AD. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burby PE, Simmons LA. 2019. Regulation of cell division in bacteria by monitoring genome integrity and DNA replication status. J Bacteriol 202:e00408-19. doi: 10.1128/JB.00408-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pontes MH, Groisman EA. 2018. Protein synthesis controls phosphate homeostasis. Genes Dev 32:79–92. doi: 10.1101/gad.309245.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gray AN, Egan AJ, van't Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar AFM, Damen MJ, Huang KC, Simorre J-P, Breukink E, den Blaauwen T, Typas A, Gross CA, Vollmer W. 2015. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife 4:e07118. doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leavitt RI, Umbarger HE. 1962. Isoleucine and valine metabolism in Escherichia coli. XI. Valine inhibition of the growth of Escherichia coli strain K-12. J Bacteriol 83:624–630. doi: 10.1128/JB.83.3.624-630.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Menninger JR. 1979. Accumulation of peptidyl tRNA is lethal to Escherichia coli. J Bacteriol 137:694–696. doi: 10.1128/JB.137.1.694-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cohen SS, Barner HD. 1954. Studies on unbalanced growth in Escherichia coli. Proc Natl Acad Sci U S A 40:885–893. doi: 10.1073/pnas.40.10.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hosono R, Kuno S. 1974. Mechanism of inhibition of bacterial growth by adenine. J Biochem 75:215–220. doi: 10.1093/oxfordjournals.jbchem.a130388. [DOI] [PubMed] [Google Scholar]
- 91.Keating DH, Carey MR, Cronan JE Jr.. 1995. The unmodified (Apo) form of Escherichia coli acyl carrier protein is a potent inhibitor of cell growth. J Biol Chem 270:22229–22235. doi: 10.1074/jbc.270.38.22229. [DOI] [PubMed] [Google Scholar]
- 92.Lerner CG, Inouye M. 1991. Pleiotropic changes resulting from depletion of Era, an essential GTP-binding protein in Escherichia coli. Mol Microbiol 5:951–957. doi: 10.1111/j.1365-2958.1991.tb00770.x. [DOI] [PubMed] [Google Scholar]
- 93.Gong M, Gong F, Yanofsky C. 2006. Overexpression of tnaC of Escherichia coli inhibits growth by depleting tRNA2Pro availability. J Bacteriol 188:1892–1898. doi: 10.1128/JB.188.5.1892-1898.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Papp B, Pál C, Hurst LD. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 96.Bhattacharyya S, Bershtein S, Yan J, Argun T, Gilson AI, Trauger SA, Shakhnovich EI. 2016. Transient protein-protein interactions perturb E. coli metabolome and cause gene dosage toxicity. Elife 5:e20309. doi: 10.7554/eLife.20309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu YF, Amador-Noguez D, Reaves ML, Feng XJ, Rabinowitz JD. 2012. Ultrasensitive regulation of anapleurosis via allosteric activation of PEP carboxylase. Nat Chem Biol 8:562–568. doi: 10.1038/nchembio.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Puckett S, Trujillo C, Wang Z, Eoh H, Ioerger TR, Krieger I, Sacchettini J, Schnappinger D, Rhee KY, Ehrt S. 2017. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 114:E2225–E2232. doi: 10.1073/pnas.1617655114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu M, Dai X. 2019. Growth suppression by altered (p)ppGpp levels results from non-optimal resource allocation in Escherichia coli. Nucleic Acids Res 47:4684–4693. doi: 10.1093/nar/gkz211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Slagter-Jäger JG, Puzis L, Gutgsell NS, Belfort M, Jain C. 2007. Functional defects in transfer RNAs lead to the accumulation of ribosomal RNA precursors. RNA 13:597–605. doi: 10.1261/rna.319407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trinquier A, Ulmer JE, Gilet L, Figaro S, Hammann P, Kuhn L, Braun F, Condon C. 2019. tRNA maturation defects lead to inhibition of rRNA processing via synthesis of pppGpp. Mol Cell 74:1227–1238.e3. doi: 10.1016/j.molcel.2019.03.030. [DOI] [PubMed] [Google Scholar]
- 102.Farewell A, Neidhardt FC. 1998. Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J Bacteriol 180:4704–4710. doi: 10.1128/JB.180.17.4704-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lopatkin AJ, Stokes JM, Zheng EJ, Yang JH, Takahashi MK, You L, Collins JJ. 2019. Bacterial metabolic state more accurately predicts antibiotic lethality than growth rate. Nat Microbiol 4:2109–2117. doi: 10.1038/s41564-019-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blanc-Potard AB, Lafay B. 2003. MgtC as a horizontally-acquired virulence factor of intracellular bacterial pathogens: evidence from molecular phylogeny and comparative genomics. J Mol Evol 57:479–486. doi: 10.1007/s00239-003-2496-4. [DOI] [PubMed] [Google Scholar]
- 105.Soncini FC, García Véscovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol 178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spinelli SV, Pontel LB, García Véscovi E, Soncini FC. 2008. Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbiol Lett 280:226–234. doi: 10.1111/j.1574-6968.2008.01065.x. [DOI] [PubMed] [Google Scholar]
- 107.Cromie MJ, Shi Y, Latifi T, Groisman EA. 2006. An RNA sensor for intracellular Mg2+. Cell 125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 108.Hobby GL, Meyer K, Chaffee E. 1942. Observations on the mechanism of action of penicillin. Soc Exp Biol Med 50:281–285. doi: 10.3181/00379727-50-13773. [DOI] [Google Scholar]
- 109.Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A. 1986. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132:1297–1304. doi: 10.1099/00221287-132-5-1297. [DOI] [PubMed] [Google Scholar]
- 110.Lee AJ, Wang S, Meredith HR, Zhuang B, Dai Z, You L. 2018. Robust, linear correlations between growth rates and β-lactam-mediated lysis rates. Proc Natl Acad Sci U S A 115:4069–4074. doi: 10.1073/pnas.1719504115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haugan MS, Løbner-Olesen A, Frimodt-Møller N. 2018. Comparative activity of ceftriaxone, ciprofloxacin, and gentamicin as a function of bacterial growth rate probed by Escherichia coli chromosome replication in the mouse peritonitis model. Antimicrob Agents Chemother 63:e02133-18. doi: 10.1128/AAC.02133-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smirnova GV, Oktyabrsky ON. 2018. Relationship between Escherichia coli growth rate and bacterial susceptibility to ciprofloxacin. FEMS Microbiol Lett 365:10.1093/femsle/fnx254. doi: 10.1093/femsle/fnx254. [DOI] [PubMed] [Google Scholar]
- 113.Clerch B, Rivera E, Llagostera M. 1996. Identification of a pKM101 region which confers a slow growth rate and interferes with susceptibility to quinolone in Escherichia coli AB1157. J Bacteriol 178:5568–5572. doi: 10.1128/jb.178.19.5568-5572.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Greulich P, Scott M, Evans MR, Allen RJ. 2015. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol Syst Biol 11:796. doi: 10.15252/msb.20145949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Foerster S, Unemo M, Hathaway LJ, Low N, Althaus CL. 2016. Time-kill curve analysis and pharmacodynamic modelling for in vitro evaluation of antimicrobials against Neisseria gonorrhoeae. BMC Microbiol 16:216. doi: 10.1186/s12866-016-0838-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother 35:1824–1828. doi: 10.1128/aac.35.9.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 118.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Simpson BW, Trent MS. 2019. Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 17:403–416. doi: 10.1038/s41579-019-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 121.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 122.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mahoney TF, Silhavy TJ. 2013. The Cpx stress response confers resistance to some, but not all, bactericidal antibiotics. J Bacteriol 195:1869–1874. doi: 10.1128/JB.02197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnson PJ, Levin BR. 2013. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet 9:e1003123. doi: 10.1371/journal.pgen.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Levin BR, Concepción-Acevedo J, Udekwu KI. 2014. Persistence: a copacetic and parsimonious hypothesis for the existence of non-inherited resistance to antibiotics. Curr Opin Microbiol 21:18–21. doi: 10.1016/j.mib.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shan Y, Lazinski D, Rowe S, Camilli A, Lewis K. 2015. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6:e00078-15. doi: 10.1128/mBio.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cameron DR, Shan Y, Zalis EA, Isabella V, Lewis K. 2018. A genetic determinant of persister cell formation in bacterial pathogens. J Bacteriol 200:e00303-18. doi: 10.1128/JB.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gallo SW, Ferreira CAS, de Oliveira SD. 2017. Combination of polymyxin B and meropenem eradicates persister cells from Acinetobacter baumannii strains in exponential growth. J Med Microbiol 66:1257–1260. doi: 10.1099/jmm.0.000542. [DOI] [PubMed] [Google Scholar]
- 130.Chung ES, Ko KS. 2019. Eradication of persister cells of Acinetobacter baumannii through combination of colistin and amikacin antibiotics. J Antimicrob Chemother 74:1277–1283. doi: 10.1093/jac/dkz034. [DOI] [PubMed] [Google Scholar]