Abstract
Background
Streptococcus pneumoniae causes serious infections worldwide. The aim of this study was to determine the molecular characteristic, antibiotic resistance pattern and capsular types of invasive S. pneumoniae in Tehran, Iran.
Results
Of the 44 pneumococcal invasive isolates, 39 (89%) were isolated from children and 5 (11%) from adults. The results show that all pneumococcal isolates were susceptible to linezolid but had varying resistance to trimethoprim-sulfamethoxazole (86%), erythromycin (73%), tetracycline (66%), clindamycin (43%), penicillin (16%), chloramphenicol (14%) and levofloxacin (2%). The range of erythromycin, tetracycline and penicillin MICs were 2 - ≥ 256 μg/mL, 4 - ≥ 48 μg/mL, and 0.047 - ≥ 256 respectively. All of the penicillin resistant isolates were multidrug resistant (MDR) and in addition to penicillin were resistant to tetracycline, erythromycin and trimethoprim-sulfamethoxazole. The most common capsular types detected in 64% of the pneumococcal isolates was 6A/B, 19A, 15A, 23F. The multilocus sequence typing (MLST) of 10 pneumococcal isolates revealed 9 different sequence types (STs), including ST 15139 (capsular type 19A) and ST 15140 (capsular type 23F), which have not previously been reported.
Conclusions
The study revealed that the S. pneumoniae isolates belonged to diverse capsular types and clones with high rate of resistance to erythromycin, tetracycline, and penicillin.
Keywords: Invasive Streptococcus pneumoniae, Antibiotic resistance, MDR, MLST
Background
Streptococcus pneumoniae is the leading cause of invasive disease in young children, older adults and individuals with impaired immune systems [1, 2]. The invasive pneumococcal disease (IPD) is an important cause of morbidity and mortality worldwide [1]. IPD is described by isolation of S. pneumoniae from a normally sterile site, such as blood; cerebrospinal fluid (CSF), and pleural or ascitic fluid [1]. The polysaccharide capsule is the main virulence factor in IPD, providing protection the bacterium from the host’s immune system. To date 99 capsular types have been identified based on the antigenic capsular polysaccharide [3]. The introduction of pneumococcal conjugate vaccines (PCV7/10/13) has reduced the incidence of IPD. But, emerging nonvaccine serotypes, commonly 1, 7F, 12F, 15B/C, 22F, 24F, 23B, 33Fand 38 were related to an increase in IPD rates among children and adults [2, 4, 5].
The incredible capacity of S. pneumoniae to uptake genes has facilitated the spread of resistance in the pneumococcal population to penicillin and other antibiotics such as macrolides that used routinely to treat the disease [6–9]. The resistance mechanism to penicillin is structural modification in the penicillin binding proteins (PBPs) which have a major role in the synthesis of cell wall. Six PBPs have been identified in S. pneumoniae of which three PBPs (PBP2b, PBP2x and PBP1a) are the most often associated with penicillin resistance [9, 10]. While, macrolide resistance mechanisms in S. pneumoniae is conferred by two mechanisms. The major resistant determinant is acquisition of the ermB gene that encodes a methylase [9, 11, 12]. The second mechanism is acquisition of mefA/E genes that encoding an active efflux pump [9, 11, 12]. Noticeably, the majority of isolates that encode ermB exhibit the MLSB (Macrolide, lincosamide and streptogramin B) phenotype. While, the majority of isolates that carries mef reveal the M phenotype [9, 11–13]. Also the most common mechanism of resistance to tetracycline in S. pneumoniae is acquisition one of the two genes, tetM and less frequently the tetO genes [12, 14, 15] both of which located in mobile genetic elements such as transposons and encode ribosomal protection proteins [14, 15]. Interestingly, resistance to erythromycin and tetracycline is generally related to the insertion of the ermB gene into the transposons that contains tetM gene, raising worry about the role of tetracycline-resistant strains in the spread of macrolides -resistant strains. The high prevalence of tetracycline resistance among macrolide resistant S. pneumoniae has reported [9, 12]. As well, the transposons of the Tn916 or Tn917 family such as Tn6002, Tn3872, Tn6003 and Tn1545 have been described in pneumococci [9, 12]. Also, the main source of the tetM gene is Tn916 family [14].
Many molecular methods have been used to determine the genotypic background of S. pneumoniae. One of these methods is multilocus sequence typing (MLST), which relies on polymerase chain reaction (PCR) and sequencing of house-keeping genes [2, 6, 16, 17]. The most common sequence types (STs) in Canada were ST320 that is a frequently multidrug resistant (MDR) type and ST695, associated with susceptibility to all antibiotics except for clarithromycin [2]. The most prevalent STs reported in some Asian countries are ST81, ST283 and ST236 [18].
The purpose of the current study was to analyze the molecular characteristic, antibiotic resistance pattern and capsular types of invasive S. pneumoniae in Tehran, Iran.
Results
The pneumococcal isolates were obtained from blood cultures. Of the 44 pneumococcal invasive isolates, 39 (89%) were isolated from children and 5 (11%) from adults.
The antibiotic susceptibility pattern and molecular characteristics of pneumococcal isolates are summarized in Table 1. The results show that all isolates were susceptible to linezolid but had varying resistance to trimethoprim-sulfamethoxazole (86%), erythromycin (73%), tetracycline (66%), clindamycin (43%), penicillin (16%), chloramphenicol (14%) and levofloxacin (2%). The range of erythromycin, tetracycline and penicillin MICs were 2 - ≥ 256 μg/mL, 4 - ≥ 48 μg/mL, and 0.047 - ≥ 256, respectively.
Table 1.
Antimicrobial resistance pattern, antibiotic resistance genes, virulence genes, capsular type and sequence type of isolates
| Isolate | Resistance | MIC (μg/ml) | Tn | Virulence Factor Genes | Capsular type | ST | |||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | genes | P | E | T | |||||
| 1 | CD, E, Oxa, T, TS | ermB, mefA/E, tetM | ≥256 | ≥256 | 24 | – | cbpA, cpsA, lytA | 11A | 9533 |
| 2 | Oxa, TS | tetM | 0.75 | ND | ND | – | cbpA, cpsA, lytA, ply, pspA | – | – |
| 3 | CD, E, Oxa, T, TS | ermB, tetM | 1.5 | ≥256 | 16 | – | cbpA, cpsA, lytA, ply, pspA | 19A | 15,139 |
| 4 | Oxa, TS | mefA/E | 0.094 | ND | ND | – | cbpA, cpsA, lytA, ply, pspA | – | 12,224 |
| 5 | CD, E, Oxa, T, TS | ermB, mefA/E, tetM | 48 | ≥256 | 12 | – | cbpA, cpsA, lytA | 11A | 9533 |
| 6 | CD, E, Oxa, T, TS | ermB, mefA/E, tetM | 48 | ≥256 | 16 | – | cbpA, cpsA, lytA, ply | 15B/C | – |
| 7 | CD, E, Oxa, T, TS | ermB, mefA/E, tetM | 0.19 | ≥256 | 8 | – | cbpA, cpsA, lytA, ply, pspA | 19A | – |
| 8 | CD, CLR, E, Oxa, T, TS | ermB, mefA/E, tetM | 48 | ≥256 | 16 | – | cbpA, cpsA, lytA, ply | 23F | 15,140 |
| 9 | CD, CLR, E, Oxa, T, TS | ermB, tetM | 48 | ≥256 | 16 | – | cbpA, cpsA, lytA, ply, pspA | 23F | – |
| 10 | CD, CLR, E, Oxa, T, TS | ermB, tetM | 1.5 | ≥256 | 8 | – | cbpA, cpsA, lytA, ply, pspA | 23F | – |
| 11 | Oxa, T, TS | tetM | 1 | ND | 4 | – | cbpA, cpsA, lytA, ply, pspA | 15A | |
| 12 | E, Oxa, TS | mefA/E | 0.125 | 8 | ND | – | cbpA, cpsA, lytA, ply, pspA | 14 | – |
| 13 | CD, CLR, E, Oxa, T, TS | ermB, mefA/E, tetM | 32 | ≥256 | 24 | – | cbpA, cpsA, lytA, ply, pspA | 23F | – |
| 14 | E, Oxa, TS | mefA/E | 1 | 12 | ND | – | cbpA, cpsA, lytA, ply | 6A/B | – |
| 15 | Oxa, TS | – | 0.38 | ND | ND | – | cbpA, cpsA, lytA, ply | 6A/B | – |
| 16 | CD, E, Lvo, Oxa, T, TS | ermB, tetM | 0.5 | ≥256 | 8 | – | cbpA, cpsA, lytA, ply | 19A | – |
| 17 | TS | – | 1 | ND | ND | – | cbpA, cpsA, lytA, ply | 15A | – |
| 18 | CD, CLR, E, Oxa, T, TS | ermB, tetM | 2 | ≥256 | 12 | 6002 | cbpA, cpsA, lytA, ply | 23F | – |
| 19 | E, TS, Oxa | mefA/E | 0.047 | 2 | ND | – | cbpA, cpsA, lytA, ply, pspA | 6A/B | – |
| 20 | E, CD, T, Oxa, TS | ermB, tetM | 0.5 | ≥256 | 12 | 6002 | cbpA, cpsA, lytA | 15A | – |
| 21 | E, Oxa, TS | ermB, mefA/E | 0.19 | 3 | ND | – | cbpA, cpsA, lytA | 15B/C | 1888 |
| 22 | CD, E, Oxa, T, TS | ermB, tetM | 0.5 | ≥256 | 8 | 6002 | cbpA, cpsA, lytA | 15A | – |
| 23 | Oxa, TS | tetM | 1.5 | ND | ND | – | cbpA, cpsA, lytA, pspA | 15A | – |
| 24 | E, Oxa, T, TS | mefA/E, tetM | 0.5 | 12 | 12 | 2009 | cbpA, cpsA, lytA, ply | 9 V | – |
| 25 | Oxa, T | tetM | 1.5 | ND | 8 | 916 | cbpA, cpsA, lytA, pspA | 1 | – |
| 26 | E, Oxa, T, TS, | mefA/E, tetM | 16 | 2 | 24 | 2009 | cbpA, cpsA, lytA | 19A | 1339 |
| 27 | CD, E, Oxa, T, TS | ermB, tetM | 0.38 | ≥256 | 16 | – | cbpA, cpsA, lytA, ply, pspA | 14 | – |
| 28 | Oxa, TS | – | 1 | ND | ND | – | cbpA, cpsA, lytA, ply | 6A/B | – |
| 29 | CD, CLR, E, Oxa, T, TS | ermB, tetM | 2 | ≥256 | 24 | 6002 | cbpA, cpsA, lytA, ply | 23F | – |
| 30 | CD, E, Oxa, T, TS | ermB, tetM | 1 | 24 | 16 | – | cbpA, cpsA, lytA, ply | 19F | – |
| 31 | E, Oxa, TS | mefA/E | 0.75 | 3 | ND | – | cbpA, cpsA, lytA, ply, pspA | 6A/B | – |
| 32 | CD, E, T, TS | ermB, tetM | 0.094 | 4 | 8 | 3872 | cbpA, cpsA, lytA, ply | 19A | 12,888 |
| 33 | CD, E, T, TS | ermB, tetM | 0.38 | 12 | 24 | 6002 | cbpA, cpsA, lytA, ply | 6A/B | – |
| 34 | E, Oxa, TS | mefA/E | 0.75 | 4 | ND | – | cbpA, cpsA, lytA, ply | 9 V | – |
| 35 | E, Oxa, TS | mefA/E | 0.094 | 4 | ND | – | cbpA, cpsA, lytA, ply | 6A/B | – |
| 36 | Oxa | mefA/E | 0.75 | ND | ND | – | cbpA, cpsA, lytA | 19A | – |
| 37 | Oxa, T, TS | tetM | 0.75 | ND | 8 | 916 | cbpA, cpsA, lytA, ply | – | – |
| 38 | Oxa, T, TS | ermB, tetM | 0.5 | ND | 4 | 6002 | cbpA, cpsA, lytA, ply | 15A | – |
| 39 | Oxa, T, TS | tetM | 4 | ND | 8 | 916 | cbpA, cpsA, lytA, ply | 19A | – |
| 40 | E, Oxa, T | ermB, mefA/E, tetM | 1 | 3 | 16 | 1545/6003 + MEGA | cbpA, cpsA, lytA, pspA | 11A | – |
| 41 | E, Oxa, T | ermB, mefA/E, tetM | 0.75 | 2 | 16 | 1545/6003 + MEGA | cbpA, cpsA, lytA, pspA | 11A | – |
| 42 | E, Oxa, T | mefA/E, tetM | 0.094 | 3 | 48 | 2009 | cbpA, cpsA, lytA, pspA | 15A | – |
| 43 | CD, E, Oxa, T, TS | ermB, mefA/E, tetM | 0.75 | ≥256 | 6 | 2010 | cbpA, cpsA, lytA, ply | 19F | 2533 |
| 44 | E, Oxa | mefA/E | 0.19 | 4 | ND | – | cbpA, cpsA, lytA, ply, pspA | 6A/B | 1876 |
CD clindamycin, CLR chloramphenicol, E erythromycin, Lvo levofloxacin, Oxa oxacillin, P penicillin, T tetracycline, TS trimethoprim/sulfamethoxazole, ND non-determined, Tn transposon, ST sequence type
MIC Breakpoints: Penicillin: S ≤ 2; I = 4; R ≥ 8, Tetracycline: S ≤ 1; I = 2; R ≥ 4, Erythromycin: S ≤ 0.25; I = 0.5; R ≥ 1
The co resistance to erythromycin and clindamycin (the constitutive phenotype) was observed in 43% (19/44) of the isolates and resistance to erythromycin, but not to clindamycin (the M phenotype) was observed in 29.5% (13/44) of the isolates. As to macrolide resistant genes, the most prevalent gene was ermB found in 52% (23/44) of the isolates, followed by mefA/E found in 50% (22/44) of the isolates. The rate of coexistence of ermB and mefA/E was 23% (10/44). Analysis of resistance genes illustrated the significantly higher prevalence of ermB and tetM genes in MDR isolates (P = 0.0001).
The co resistance to erythromycin and tetracycline was found in 54.5% (24/44) of the isolates. The rate of tetM gene and coexistence of ermB and tetM were 68% (30/44) and 50% (22/44) respectively. Our result also showed that all of the penicillin resistant isolates were MDR and in addition to penicillin were resistant to tetracycline, erythromycin and trimethoprim-sulfamethoxazole.
In the present study, of 44 isolates 36% (16/44) were positive for transposon genes. The most prevalent transposon genes were Tn6002 14% (6/44) and Tn2009 7% (3/44) followed by Tn1545/6003 4.5% (2/44). No significant correlation was found between the type of transposon and the antibiotic resistance pattern (Table 1).
The pneumococcal isolates belonged to the following capsular types: 6A/B (18%), 19A (16%), 15A (16%), 23F (14%), 11A (9%), 14 (4.5%), 15B/C (4.5%), 19F (4.5%), 9 V (4.5%), 1 (2%) and noticeable (7%).
PCR analysis of virulence genes revealed that 100% of clinical isolates harbored the cbpA, cpsA, lytA genes, whereas 73% (32/44) the ply gene and 41% (18/44) the pspA gene.
The MLST typing of 10 pneumococcal isolates revealed 9 different ST types, including ST 15139 (capsular type 19A) and ST 15140 (capsular type 23F), which have not previously been reported (Fig. 1). The two ST 9533 isolates belonged to capsular type 11A. Both isolates were resistant to penicillin, erythromycin, clindamycin and tetracycline, but differed in the MIC levels. The MIC of isolate No.1 to penicillin, and tetracycline was > 256 μg/ml and 24 μg/ml, respectively, whilst isolate No.5 was 48 μg/ml and 12 μg/ml, respectively. No significant correlation was found between capsular types and STs (Table 1).
Fig. 1.
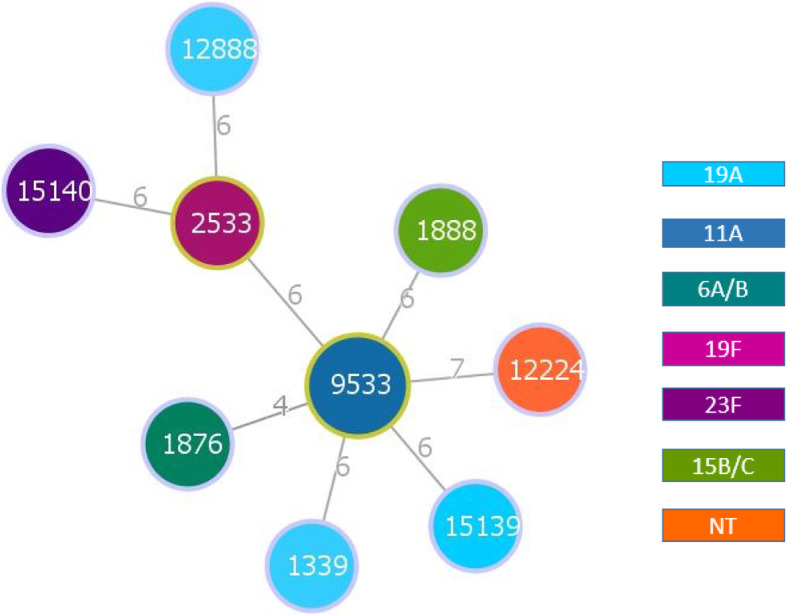
Minimum spanning tree of MLST sequence types of S. pneumoniae constructed by PHYLOViZ 2.0. Green outlines indicate a group founder; light blue outlines indicate relatedness to founder; the STs are displayed as circles; Numbers indicate the number of differences between the MLST profiles of the two connected circles; capsular types are characterized by circles different colors
Discussion
This study investigated molecular characteristics, antimicrobial resistance patterns and capsular types of S. pneumoniae isolated from invasive disease in Tehran, Iran. Of the 44 isolates, 73, 68 and 16% were resistant to erythromycin, tetracycline and penicillin, respectively. In a study conducted by Houri et al. in 2017, the percentages of resistance to erythromycin, tetracycline and penicillin in S. pneumoniae obtained from two children’s hospitals in Teheran were 71.4, 66.9 and 19.4%, respectively [19]. In another study, conducted by Talebi et al. in 2016, the percentages of resistance to tetracycline and penicillin in the erythromycin resistant S. pneumoniae isolates (ERSP) were 85 and 28% [20]. High resistance rates to erythromycin (80.2%), tetracycline (91.2%) and penicillin (47.3%) were reported from China [21]. In our study, the major mechanism conferring resistance to macrolide antibiotics was the constitutive phenotype (43%) mostly correlated with the ermB resistance gene (52%). The findings of the current study were in agreement with previous reports from our country, in which ermB (50%) was the most frequent genetic determinant among ERSP (20). In a study in Turkey, Kittana et al. reported that the majority of the ERSP isolates (88.2%) had a constitutive phenotype and 89.1 and 50% of the ERSP isolates harbored ermB and mefE genes, respectively [22]. In contrast to the findings in our study, the M phenotype encoded by mefA gene (53%) was more frequently found in Canada [23]. These differences in the prevalence of macrolide resistance and different phenotypes of resistance may be related to the dissemination of multiresistant clones and different patterns in use of macrolides, which led to the variation of resistant phenotypes [24]. In our study, 23% of strains harbored both ermB and mefA genes, which is in accordance with a report from Turkey (20%) [25]. The majority of macrolide-resistant strains 75% (24/32) were also resistant to tetracycline. This association is due to the insertion of ermB into composite transposons of the Tn916 family that contain tetM gene [9, 25, 26]. While, the existence of unexpressed tetM genes in tetracycline sensitive isolates showed that transposons of the Tn916 family may be more widespread in S. pneumoniae than expected to firmly associated with resistant tetracycline [26, 27].
Our study showed that 36% of isolates were positive for transposon genes and among them Tn6002 was more common, which accounts for 14%. Talebi et al. reported that 47% of clinical isolates of S. pneumonia were positive for transposon genes and the most predominant transposons were Tn2010 and Tn1545/6003 presented in 29 and 25% of the isolates, respectively [20]. Kittana et al. observed that 88.2% of clinical isolates of S. pneumonia harbored transposon genes and Tn2010 (37.2%) and Tn6002 (21.8%) were the most common transposon [22]. Generally, the distribution of pneumococcal transposons and the genes carried by them varies in different parts of the world [22]. One possible explanation for this discrepancy may be explained by the difference in the origin of the isolates as well as other factors.
This study showed that the most common capsular types in the order of frequency were 6A/B, 19A, 15A, 23F, which accounted for 64%. Houri et al. reported that the most common serotypes from blood/CSF were 23F, 19F, 19A and 9 V [19]. Talebi et al. found that serotypes 14 and 19F were the common serotypes isolated from patients with IPD [20]. Habibi Ghahfarokhi et al. observed that the common serotypes isolated from clinical samples were 23F, 14, 3, 19F and 19A [28]. In another study by Azarsa et al. in Iran, the common serotypes isolated from clinical samples were 23F, 19F, 14, 3 and 9 V [29]. In Japan, Sakata et al. evaluated 142 cases of IPD and observed that the most frequently serotypes were 6B, followed by 23F and 19F [30]. In study conducted by Percin et al. in Turkey, the most common invasive serotypes were 1, 19A, 19F, 3, 18C, 6A/B, 14, and 7F [31]. Compared to these reports, almost the same distribution of serotypes responsible for IPD was observed in the current study. The data revealed that serotypes 6A/B, 14, 18C, 19F, 19A, and 23F are the most common pneumococcal serotypes in Asia, particularly in non-vaccine areas [19].
Vaccination has been shown to be effective in reducing the rates of IPD associated with multiple drug resistance [4, 5]. But, vaccination against the most common serotypes of S. pneumoniae using PCVs is not still included to the routine immunization program in the Iran and only recommended for high-risk groups [28]. The present study, in addition to others from Iran, shows that the PCV13 could cover the majority of the invasive pneumococcal isolates [19, 20, 28, 29].
Capsular type 19A is frequent among MDR isolates which has already been described in many non-vaccinated regions such as Korea [32]. Researchers have formerly exhibited that the spread of MDR capsular types 19A isolates is due to antibiotic misuse in developing countries [33]. In our country, irrational use of antibiotics has contributed to the emergence of MDR isolates.
In current study, more than 70% (5/7) of capsular type 19A isolates were MDR and showed resistance to erythromycin (majority MIC≥256 μg/ mL), tetracycline (MIC ≥8 μg/ mL), clindamycin, and trimethoprim-sulfamethoxazole. One isolate of capsular type 19A in addition of mention antibiotics showed relatively high resistance to penicillin which carried Tn2009. As for, all (6/6) of capsular type 23F isolates and 50% (2/4) of capsular type 11A, 15B/C (1/2) isolates were MDR (high level MIC for erythromycin and tetracycline) as capsular type 19A isolates but 50% of 11A, 15B/C and 23F were resistance penicillin (MIC ≥32–48 μg/ mL).
Despite the importance of capsular type as an invasive determinant, other virulence determinants were also associated with invasive isolates [34]. As our results, the majority of the isolates contained the virulence determinants that probably indicate the essential of virulence determinants in the ability of an isolate to cause invasive disease [34]. Interestingly, pspA gene was encoded by the most pneumococcal isolates which only detected in 41% of our isolates. This is in accordance with the study suggesting that probable limitation of detection by conventional PCR and confirmed this hypothesis by a quantitative PCR assay at high level detection [35].
As for capsular type is assumed to be more important than genotype in the ability of an isolate led to invasive disease but also underline the role of genetic background in invasion [34, 36, 37]. Since pneumococcal isolates with diverse MLST profiles have showed various pathogenicity potential [36]. According to the other studies suggesting high capsular type and genetic diversities in IPD isolates [12, 37–39], there was important diversity among our isolates base on capsular types and different MLST profiles. In pneumococcal isolates, one of the important factor to selective pressure is use of antibiotics [39, 40], so the antibiotic selection pressure may be led to different genetic diversity of IPD isolates that observed in this study [39, 40]. However, causing agents associated with genetic diversity should be further studied.
Reliable and comprehensive data regarding antimicrobial resistance and genetic characteristics, S. pneumoniae are scarce, in Iran. This prompted our research. There were limitations to this study. One major bias is the low number of isolates analyzed, which led to no association was observed between the serotypes/transposons or serotypes/STs and STs/ antibiotic resistant phenotype
Conclusion
The study revealed that the S. pneumoniae isolates belonged to diverse capsular types and clones with high rate of resistance to erythromycin, tetracycline, and penicillin.
Methods
Bacterial isolates
A total of 44 invasive pneumococcal isolates were collected from hospitalized patients (inpatients) from 2 teaching hospitals (Imam Khomeini and Tehran Children’s Medical Center) affiliated with the Tehran University of Medical Sciences (TUMS), between October 2016 to September 2017. Only one isolate was investigated per patient. The organisms were identified to the species level using standard biochemical methods based on typical colony morphology, Gram staining, catalase, hemolysis, and optochin sensitivity testing (Difco, USA). To confirm the identification of the isolate as S. pneumoniae the lytA and ply genes were amplified by a PCR, using primers: LytA-F, 5′-CGGACTACCGCCTTTATATCG-3′; lytA-R, 5′-GTTTCAATCGTCAAGCCGTT-3′ [41] and ply-F, 5′-ATTTCTGTAACAGCTACCAACGA-3′; ply-R, 5′- GAATTCCCTGTCTTTTCAAAGTC-3′ [42].
Antibiotic susceptibility determination
Antimicrobial susceptibility testing was performed according to the Clinical Laboratory and Standards Institute (CLSI (guidelines. Disk agar diffusion (DAD) method was performed on Mueller-Hinton agar with 5% defibrinated sheep blood, incubated at 35 °C and 5% CO2 for 20–24 h, and zones of inhibition measured after incubation. All isolates were tested against Oxacillin (1 μg), tetracycline (30 μg), erythromycin (15 μg), levofloxacin (5 μg), chloramphenicol (30 μg), linezolid (30 μg), clindamycin (2 μg), trimethoprim/ sulfamethoxazole (1.25/23.75 μg). All of the antibiotic discs were purchased from Mast Diagnostics Ltd. (Merseyside, UK). Minimum inhibitory concentration (MIC) for erythromycin, tetracycline and penicillin were determined with E-test (0.016–256 μg/ml-Liofilchem, Via Scozia, Italy). The MIC was interpreted according to the CLSI breakpoints [43]. MDR was considered as resistance to three or more different classes of antimicrobial. S. pneumoniae ATCC 49619 was used for quality control strain to ensure the reliability of the results.
Capsular typing
The PCR were performed with capsular specific primers as described by Ahn et al. as Table 2 [44]. First of all the confirmed pneumococcal isolates were examined for amplification of cpsA gene (Table 3) [45]. Then, the capsular primers used to detect most common capsular types that were outlined in Table 2 [44].
Table 2.
Sequences of capsular primers
| Primer | Sequence | Amplicon size (bp) | Reference |
|---|---|---|---|
| 23 F |
5′-GTAACAGTTGCTGTAGAGGGAATTGGCTTTTC-3′ 5′-CACAACACCTAACACACGATGGCTATATGATTC-3’ |
384 | [44] |
| 19F |
5’-GTTAAGATTGCTGATCGATTAATTGATATCC-3′ 5′-GTAATATGTCTTTAGGGCGTTTATGGCGATAG-3’ |
304 | |
| 4 |
5’-CTGTTACTTGTTCTGGACTCTCGATAATTGG-3′ 5′-GCCCACTCCTGTTAAAATCCTACCCGCATTG-3’ |
430 | |
| 6A/B |
5’-AATTTGTATTTTATTCATGCCTATATCTGG-3′ 5′-TTAGCGGAGATAATTTAAAATGATGACTA-3’ |
250 | |
| 14 |
5’-CTTGGCGCAGGTGTCAGAATTCCCTCTAC-3′ 5′-GCCAAAATACTGACAAAGCTAGAATATAGCC-3’ |
208 | |
| 19A |
5’-GTTAGTCCTGTTTTAGATTTATTTGGTGATGT-3′ 5′-GAGCAGTCAATAAGATGAGACGATAGTTAG-3’ |
478 | |
| 3 |
5’-ATGGTGTGATTTCTCCTAGATTGGAAAGTAG-3′ 5′-CTTCTCCAATTGCTTACCAAGTGCAATAACG-3’ |
371 | |
| 15A |
5’-ATTAGTACAGCTGCTGGAATATCTCTTC-3′ 5′-GATCTAGTGAACGTACTATTCCAAAC-3’ |
434 | |
| 15B/C |
5’-TTGGAATTTTTTAATTAGTGGCTTACCTA-3′ 5′-CATCCGCTTATTAATTGAAGTAATCTGAACC-3’ |
496 | |
| 1 |
5’-CTCTATAGAATGGAGTATATAAACTATGGTTA-3′ 5′-CCAAAGAAAATACTAACATTATCACAATATTGGC-3’ |
280 | |
| 11A |
5’-GGACATGTTCAGGTGATTTCCCAATATAGTG-3′ 5′-GATTATGAGTGTAATTTATTCCAACTTCTCCC-3’ |
463 | |
| 9 V |
5’-CTTCGTTAGTTAAAATTCTAAATTTTTCTAAG-3′ 5′-GTCCCAATACCAGTCCTTGCAACACAAG-3’ |
753 | |
| 7F |
5’-CCTACGGGAGGATATAAAATTATTTTTGAG-3′ 5′-CAAATACACCACTATAGGCTGTTGAGACTAAC-3’ |
826 |
Table 3.
Sequences of oligonucleotide primers
| Primer | Sequence | Amplicon size (bp) | Reference |
|---|---|---|---|
| lytA |
5’-CGGACTACCGCCTTTATATCG-3′ 5′-GTTTCAATCGTCAAGCCGTT-3’ |
229 | [41] |
| ply |
5’-ATTTCTGTAACAGCTACCAACGA-3′ 5′-GAATTCCCTGTCTTTTCAAAGTC-3’ |
347 | [42] |
| cpsA |
5’- AGTGGTAACTGCGTTAGTCC − 3′ 5′- CTGCCAAGTAAGACGAACTC − 3’ |
362 | [45] |
| erm(B) |
5’-TGGTATTCCAAATGCGTAATG-3′ 5′-CTGTGGTATGGCGGGTAAGT-3’ |
745 | [46] |
| mef(A/E) |
5’-AGTATCATTAATCACTAGTGC-3′ 5′-TTCTTCTGGTACTAAAAGTGG-3’ |
346 | [47] |
| tetO |
5’-AACTTAGGCATTCTGGCTCAC-3′ 5′-TCCCACTGTTCCATATCGTCA-3’ |
515 | [13] |
| tetL |
5’-ATAAATTGTTTCGGGTCGGTAAT-3′ 5′-AACCAGCCAACTAATGACAATGAT-3’ |
1077 | [48] |
| tetK |
5’-GTAGCGACAATAGGTAATAGT-3′ 5′-GTAGTGACAATAAACCTCCTA-3’ |
361 | [49] |
| tetM |
5’-AGTGGAGCGATTACAGAA-3′ 5′-CATATGTCCTGGCGTGTCTA-3’ |
159 | [49] |
| aphA3 |
5’-GCCGATGTGGATTGCGAAAA-3′ 5′-GCTTGATCCCCAGTAAGTCA-3’ |
292 | [13] |
| int |
5’-GCGTGATTGTATCTCACT-3′ 5′-GACGCTCCTGTTGCTTCT-3’ |
1046 | [48] |
| xis |
5’-AAGCAGACTGAGATTCCTA-3′ 5′-GCGTCCAATGTATCTATAA-3’ |
194 | [48] |
| tnpR |
5’-CCAAGGAGCTAAAGAGGTCCC-3′ 5′-GTCCCGAGTCCCATGGAAGC-3’ |
1548 | [48] |
| tnpA |
5’-GCTTCCATGGGACTCGGGAC-3′ 5′-GCTCCCAATTAATAGGAGA-3’ |
2134 | [48] |
| tndX |
5’-ATGATGGGTTGGACAAAGA-3′ 5′-CTTTGCTCGATAGGCTCTA-3’ |
611 | [48] |
| pspA |
5’-CATAGACTAGAACAAGAGCTCAAA-3′ 5′-CTA CAT TAT TGT TTT CTT CAG CAG-3’ |
214 | [36] |
| cbpA |
5’-GCTAATGTAGCGACTTCAGATCAA-3′ 5′-AGCTTGGAAGAGTTTCTTCACCTA-3’ |
142 | [36] |
Detection of resistance genes, virulence genes and transposon profiles
The PCR assays were used to detect the macrolide resistance genes (ermB and mef A/E), tetracycline resistance genes (tetM, tetO, tetL and tetK), virulence genes including autolysin A (lytA), pneumolysin (ply), pneumococcal surface protein A (pspA), and Choline binding protein A (cbpA) with primers specific for each gene.
The transposons were detected using PCR assay for Tn916 and Tn917 transposon-related genes including xis, int, tndX, tnpR and tnpA. The resistance genes related to the different transposons were Tn2009 (tetM, int, xis, mef), Tn6002 (ermB, tetM, int, xis), Tn3872 (ermB, tetM, tnpA, tnpR), Tn2010 (ermB, tetM, int, xis, mef), Tn6003/Tn1545 (ermB, tetM, int, xis, aph3’-III), Tn6002+ MEGA (macrolide efflux genetic assembly) [12]. All primers are listed in Table 3.
MLST analysis
MLST was performed with selected isolates using the internal fragments of seven housekeeping gene including aroE, gdh, gki, recP, spi, xpt, and ddl as specified by Enright et al. [17].
The sequences types (STs) were determined by the comparison with those of corresponding allelic profiles at MLST database (http://pubmlst.org/spneumoniae/). Minimum spanning trees were produced using PHYLOViZ 2.0 software [50].
Statistical analysis
The statistical analysis of the difference in the frequency of the pneumococcal genes was evaluated by using the chi-square and Fisher’s as appropriate. The differences less than 0.05 were considered significant statistically.
Acknowledgements
We would like to thank Dr. Hamidreza Houri, who assisted with collection of the specimens used in this study.
Abbreviations
- IPD
Invasive pneumococcal disease
- CSF
Cerebrospinal fluid
- MLSB
Macrolide, lincosamide and streptogramin B
- MLST
Multilocus sequence typing
- MDR
Multidrug resistant
- MIC
Minimum Inhibitory Concentration
- PBPs
Penicillin binding proteins
- PCR
Polymerase chain reaction
- lytA
Autolysin A
- ply
Pneumolysin
- pspA
Pneumococcal surface protein A
- cbpA
Choline binding protein A
- MEGA
Macrolide efflux genetic assembly
- ST
Sequences type
- CLSI
Clinical Laboratory and Standards Institute
- DAD
Disk agar diffusion
- CD
Clindamycin
- CLR
Chloramphenicol
- E
Erythromycin (E)
- Lvo
Levofloxacin
- Oxa
Oxacillin
- P
Penicillin
- T
Tetracycline
- TS
Trimethoprim/sulfamethoxazole
- ND
Non-determined
- Tn
Transposon
- PCV
Pneumococcal conjugated vaccine
- ERSP
Eythromycin resistant S. pneumoniae
Authors’ contributions
MB conducted the experiments, collected data and drafted the manuscript. MMF, FBH and RB participated in the design of the study and advised in all parts of the study. ME and FJ supervised all parts of the study. ME and RB revised the manuscript. All authors read and approved the final manuscript.
Funding
This research has been supported by Tehran University of Medical Sciences and Health Services. Study grant no 96–01–30-34319. The funder played no role in the design of the study, nor in the collection, analysis, and interpretation of data, nor in writing the manuscript.
Availability of data and materials
All documents and additional data are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Tehran University of Medical Sciences (NO.34319). Consent to participate is not applicable for this study because the isolates included in the study were obtained from existing clinical collections routinely assembled as part of laboratory practices of university hospitals.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Song JY, Nahm MH, Moseley MA. Clinical implications of pneumococcal serotypes: invasive disease potential, clinical presentations, and antibiotic resistance. J Korean Med Sci. 2013;28(1):4–15. doi: 10.3346/jkms.2013.28.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golden AR, Adam HJ, Karlowsky JA, Baxter M, Nichol KA, Martin I, et al. Molecular characterization of predominant Streptococcus pneumoniae serotypes causing invasive infections in Canada: the SAVE study, 2011–15. J Antimicrob Chemother. 2018;73(suppl-7):vii20–vii31. doi: 10.1093/jac/dky157. [DOI] [PubMed] [Google Scholar]
- 3.Sempere J, de Miguel S, González-Camacho F, Yuste J, Domenech M. Clinical relevance and molecular pathogenesis of the emerging serotypes 22F and 33F of Streptococcus pneumoniae in Spain. Front Microbiol. 2020;11:309. doi: 10.3389/fmicb.2020.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yildirim I, Shea KM, Pelton SI. Pneumococcal disease in the era of pneumococcal conjugate vaccine. Infect Dis Clin N Am. 2015;29(4):679–697. doi: 10.1016/j.idc.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsells E, Guillot L, Nair H, Kyaw MH. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: A systematic review and meta-analysis. PloS one. 2017;12(5):e0177113. [DOI] [PMC free article] [PubMed]
- 6.McGee L, McDougal L, Zhou J, Spratt B, Tenover F, George R, et al. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J Clin Microbiol. 2001;39(7):2565–2571. doi: 10.1128/JCM.39.7.2565-2571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Cai L, Xiao M, Kong F, Oftadeh S, Zhou F, et al. Distribution of serotypes, genotypes, and resistance determinants among macrolide-resistant Streptococcus pneumoniae isolates. Antimicrob Agents Chemother. 2010;54(3):1152–1159. doi: 10.1128/AAC.01268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 2018;16(6):355. doi: 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornick J, Bentley S. Streptococcus pneumoniae: the evolution of antimicrobial resistance to beta-lactams, fluoroquinolones and macrolides. Microb infect. 2012;14(7–8):573–583. doi: 10.1016/j.micinf.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 10.El Moujaber G, Osman M, Rafei R, Dabboussi F, Hamze M. Molecular mechanisms and epidemiology of resistance in Streptococcus pneumoniae in the Middle East region. J Med Microbiol. 2017;66(7):847–858. doi: 10.1099/jmm.0.000503. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Fu J, Li S, Guo D, Chen Z, Chen S, et al. Phenotypic and molecular characterization of Streptococcus pneumoniae in pre-conjugate vaccine era: a Chinese hospital-based retrospective study. Vaccine. 2018;36(5):599–605. doi: 10.1016/j.vaccine.2017.12.063. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L, Ma X, Gao W, Yao K-H, Shen A-D, Yu S-J, et al. Molecular characteristics of erythromycin-resistant Streptococcus pneumoniae from pediatric patients younger than five years in Beijing, 2010. BMC Microbiol. 2012;12(1):228. doi: 10.1186/1471-2180-12-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochetti I, Tili E, Mingoia M, Varaldo PE, Montanari MP. Erm (B)-carrying elements in tetracycline-resistant pneumococci and correspondence between Tn1545 and Tn6003. Antimicrob Agents Chemother. 2008;52(4):1285–1290. doi: 10.1128/AAC.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ousmane S, Diallo B, Ouedraogo R. Genetic determinants of tetracycline resistance in clinical Streptococcus pneumoniae serotype 1 isolates from Niger. Antibiotics. 2018;7(1):19. doi: 10.3390/antibiotics7010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupien A, Gingras H, Bergeron MG, Leprohon P, Ouellette M. Multiple mutations and increased RNA expression in tetracycline-resistant Streptococcus pneumoniae as determined by genome-wide DNA and mRNA sequencing. Antimicrob Agents Chemother. 2015;70(7):1946–1959. doi: 10.1093/jac/dkv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klugman KP. The successful clone: the vector of dissemination of resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 2002;50(suppl_3):1–6. doi: 10.1093/jac/dkf500. [DOI] [PubMed] [Google Scholar]
- 17.Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144(11):3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 18.Jefferies JM, Yusof MYM, Sekaran SD, Clarke SC. Novel clones of Streptococcus pneumoniae causing invasive disease in Malaysia. PloS one. 2014;9(6):e97912. [DOI] [PMC free article] [PubMed]
- 19.Houri H, Tabatabaei SR, Saee Y, Fallah F, Rahbar M, Karimi A. Distribution of capsular types and drug resistance patterns of invasive pediatric Streptococcus pneumoniae isolates in Teheran. Iran Int J Infect Dis. 2017;57:21–26. doi: 10.1016/j.ijid.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Talebi M, Azadegan A, Sadeghi J, Ahmadi A, Ghanei M, Katouli M, et al. Determination of characteristics of erythromycin resistant Streptococcus pneumoniae with preferred PCV usage in Iran. PLoS One. 2016;11(12):e0167803. doi: 10.1371/journal.pone.0167803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang S, Liu X, Lao W, Zeng S, Liang H, Zhong R, et al. Serotype distribution and antibiotic resistance of Streptococcus pneumoniae isolates collected at a Chinese hospital from 2011 to 2013. BMC Infect Dis. 2015;15(1):312. doi: 10.1186/s12879-015-1042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kittana FNA, Mustak IB, Hascelik G, Saricam S, Gurler N, Diker KS. Erythromycin-resistant Streptococcus pneumoniae: phenotypes, genotypes, transposons and pneumococcal vaccine coverage rates. J Med Microbiol. 2019;68(6):874–881. doi: 10.1099/jmm.0.000995. [DOI] [PubMed] [Google Scholar]
- 23.McGeer D, Green A, Low D, Network TIBD. Macrolide resistance in bacteremic pneumococcal disease: implications for patient management. Clin Infect Dis. 2006;43(4):432–438. doi: 10.1086/505871. [DOI] [PubMed] [Google Scholar]
- 24.Calatayud L, Ardanuy C, Cercenado E, Fenoll A, Bouza E, Pallares R, et al. Serotypes, clones, and mechanisms of resistance of erythromycin-resistant Streptococcus pneumoniae isolates collected in Spain. Antimicrob Agents Chemother. 2007;51(9):3240–3246. doi: 10.1128/AAC.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telli M, Eyigör M, Gültekin B, Aydın N. Evaluation of resistance mechanisms and serotype and genotype distributions of macrolide-resistant strains in clinical isolates of Streptococcus pneumonia in Aydın. Turkey J Infect Chemother. 2011;17(5):658. doi: 10.1007/s10156-011-0238-x. [DOI] [PubMed] [Google Scholar]
- 26.Korona-Glowniak I, Maj M, Siwiec R, Niedzielski A, Malm A. Molecular epidemiology of Streptococcus pneumoniae isolates from children with recurrent upper respiratory tract infections. PLoS One. 2016;11(7):e0158909. doi: 10.1371/journal.pone.0158909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cochetti I, Tili E, Vecchi M, Manzin A, Mingoia M, Varaldo PE, et al. New Tn 916-related elements causing erm (B)-mediated erythromycin resistance in tetracycline-susceptible pneumococci. Antimicrob Agents Chemother. 2007;60(1):127–131. doi: 10.1093/jac/dkm120. [DOI] [PubMed] [Google Scholar]
- 28.Ghahfarokhi SH, Mosadegh M, Ahmadi A, Pourmand MR, Azarsa M, Rahbar M, et al. Serotype distribution and antibiotic susceptibility of Streptococcus pneumoniae isolates in Tehran, Iran: A Surveillance Study. Infect Drug Resist. 2020;13:333. doi: 10.2147/IDR.S234295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Azarsa M, Moghadam SO, Rahbar M, Baseri Z, Pourmand M. Molecular serotyping and genotyping of penicillin non-susceptible pneumococci: the introduction of new sequence types, Tehran, Iran. New Microbes New Infect. 2019;32:100597. [DOI] [PMC free article] [PubMed]
- 30.Sakata H. Invasive pneumococcal diseases in children in Hokkaido, Japan from April 2000, to March 2015. J Infect Chemother. 2016;22(1):24–26. [DOI] [PubMed]
- 31.Percin D, Altintop YA, Sumerkan B. Ten-year surveillance of invasive Streptococcus pneumoniae isolates in central Turkey prior to the introduction of a conjugate vaccine. J Infect Dev Ctries. 2010;4(09):560–5. [DOI] [PubMed]
- 32.Choi EH, Kim SH, Eun BW, Kim SJ, Kim NH, Lee J, et al. Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg Infect Dis. 2008;14(2):275. doi: 10.3201/eid1402.070807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SH, Song J-H, Chung DR, Thamlikitkul V, Yang Y, Wang H, et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for surveillance of resistant pathogens (ANSORP) study. Antimicrob Agents Chemother. 2012;56(3):1418–1426. doi: 10.1128/AAC.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donkor ES, Stabler RA, Hinds J, Adegbola RA, Antonio M, Wren BW. Comparative phylogenomics of Streptococcus pneumoniae isolated from invasive disease and nasopharyngeal carriage from west Africans. BMC Genomics. 2012;13(1):569. doi: 10.1186/1471-2164-13-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai F, Talekar SJ, Klugman KP, Vidal JE. Expression of Streptococcus pneumoniae virulence-related genes in the nasopharynx of healthy children. PLoS One. 2013;8(6):e67147. doi: 10.1371/journal.pone.0067147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melin M, Trzciński K, Meri S, Käyhty H, Väkeväinen M. The capsular serotype of Streptococcus pneumoniae is more important than the genetic background for resistance to complement. Infect Immun. 2010;78(12):5262–5270. doi: 10.1128/IAI.00740-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Cuyck H, Pichon B, Leroy P, Granger-Farbos A, Underwood A, Soullié B, et al. Multiple-locus variable-number tandem-repeat analysis of Streptococcus pneumoniae and comparison with multiple loci sequence typing. BMC Microbiol. 2012;12(1):241. doi: 10.1186/1471-2180-12-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YJ, Chen YS, Wang ZW, Li YQ, Wang DX, Shang Y, et al. Serological and molecular capsular typing, antibiotic susceptibility and multilocus sequence typing of Streptococcus pneumoniae isolates from invasive and non-invasive infections. Chin Med J. 2013;126(12):2296–303. [PubMed]
- 39.Zhou H, Guo J, Qin T, Ren H, Xu Y, Wang C, et al. Serotype and MLST-based inference of population structure of clinical Streptococcus pneumoniae from invasive and noninvasive pneumococcal disease. Infect Genet Evol. 2017;55:104–111. doi: 10.1016/j.meegid.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 40.Wyres KL, Lambertsen LM, Croucher NJ, McGee L, von Gottberg A, Liñares J, et al. Pneumococcal capsular switching: a historical perspective. J Infect Dis. 2012;207(3):439–449. doi: 10.1093/infdis/jis703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strålin K, Korsgaard J, Olcén P. Evaluation of a multiplex PCR for bacterial pathogens applied to bronchoalveolar lavage. Eur Respir J. 2006;28(3):568–575. doi: 10.1183/09031936.06.00006106. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki N, Yuyama M, Maeda S, Ogawa H, Mashiko K, Kiyoura Y. Genotypic identification of presumptive Streptococcus pneumoniae by PCR using four genes highly specific for S. pneumoniae. J Med Microbiol. 2006;55(6):709–714. doi: 10.1099/jmm.0.46296-0. [DOI] [PubMed] [Google Scholar]
- 43.Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018;56(4):e01934–e01917. doi: 10.1128/JCM.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn JG, Choi SY, Kim DS, Kim KH. Enhanced detection and serotyping of Streptococcus pneumoniae using multiplex polymerase chain reaction. Korean J Pediatr. 2012;55(11):424. doi: 10.3345/kjp.2012.55.11.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu DK, Wang DG, Liu Y, Liu CB, Yu LH, Qu Y, et al. Roles of virulence genes (PsaA and CpsA) on the invasion of Streptococcus pneumoniae into blood system. Eur J Med Res. 2013;18(1):14. [DOI] [PMC free article] [PubMed]
- 46.Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother. 2005;49(11):4798–4800. doi: 10.1128/AAC.49.11.4798-4800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takaya A, Kitagawa N, Kuroe Y, Endo K, Okazaki M, Yokoyama E, Wada A, Yamamoto T. Mutational analysis of reduced telithromycin susceptibility of Streptococcus pneumoniae isolated clinically in Japan. FEMS Microbiol Lett. 2010;307(1):87–93. doi: 10.1111/j.1574-6968.2010.01962.x. [DOI] [PubMed] [Google Scholar]
- 48.Brenciani A, Bacciaglia A, Vecchi M, Vitali LA, Varaldo PE, Giovanetti E. Genetic elements carrying erm (B) in Streptococcus pyogenes and association with tet (M) tetracycline resistance gene. Antimicrob Agents Chemother. 2007;51(4):1209–1216. doi: 10.1128/AAC.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in staphylococcus aureus. J Clin Microbiol. 2003;41(9):4089–4094. doi: 10.1128/JCM.41.9.4089-4094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, Vaz C. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. J Bioinform. 2016;33(1):128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All documents and additional data are available from the corresponding author upon reasonable request.


