Abstract
SARS coronavirus (COVID-19) is a real health challenge of the 21st century for scientists, health workers, politicians, and all humans that has severe cause epidemic worldwide. The virus exerts its pathogenic activity through by mechanism and gains the entry via spike proteins (S) and Angiotensin-Converting Enzyme 2 (ACE2) receptor proteins on host cells. The present work is an effort for a computational target to block the residual binding protein (RBP) on spike proteins (S), Angiotensin-Converting Enzyme 2 (ACE2) receptor proteins by probiotics namely Plantaricin BN, Plantaricin JLA-9, Plantaricin W, Plantaricin D along with RNA-dependent RNA polymerase (RdRp). Docking studies were designed in order to obtain the binding energies for Plantaricin metabolites. The binding energies for Plantaricin W were −14.64, −11.1 and −12.68 for polymerase, RBD and ACE2 respectively comparatively very high with other compounds. Plantaricin W, D, and JLA-9 were able to block the residues (THR556, ALA558) surrounding the deep grove catalytic site (VAL557) of RdRp making them more therapeutically active for COVID-19. Molecular dynamics studies further strengthen stability of the complexes of plantaricin w and SARS-CoV-2 RdRp enzyme, RBD of spike protein, and human ACE2 receptor. The present study present multi-way options either by blocking RBD on S proteins or interaction of S protein with ACE2 receptor proteins or inhibiting RdRp to counter any effect of COVID-19 by Plantaricin molecules paving a way that can be useful in the treatment of COVID-19 until some better option will be available.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, spike proteins, RNA dependent RNA polymerase, ACE2, probiotics, plantaricin
Introduction
It is high time since the emergence and spread of Coronavirus (COVID-19) from Wuhan, China has gained much attention without any prophylactic or therapeutic treatment as of now (Huang et al., 2020). The situation has already taken a pandemic form with new cases even after the closing of all the air routes for travelers returning to their native land from China and other countries (World Health Organization (WHO)), 2020). As on date, a total number of cases crossed is approximately. 4.03 million from 195 countries with 278 892 deaths. There are no specific therapeutic agents or vaccines which are available for COVID-19 (WHO, 2020). There are various drugs such as Favipiravir and Remdesivir along with Convalescent plasma are under investigation but antiviral activity on COVID-19 for these drugs is yet to be confirmed (Chen et al., 2020; Enayatkhani et al., 2020; Joshi et al., 2020; Shen et al., 2020; Wahedi et al., 2020). Therefore finding a potent and effective anti-COVID-19 agent to control and prevent further infection has become the need of an hour. Several computational methods (Elfiky, 2020; Elfiky, 2020; Elmezayen et al., 2020; Enmozhi et al., 2020; Islam et al., 2020; Muralidharan et al., 2020, Sinha et al., 2020) have been put forward that could decipher the possible mechanism for COVID-19 treatment (Boopathi et al., 2020; Khan et al., 2020) through drug repositioning targeting at the various active sites of SARS-CoV-2. These targets sites are including protease, (Joshi et al., 2020) different enzymes (Gupta et al., 2020; Khan et al., 2020), protein terminals, (Sarma et al., 2020) proteins including various polymerase (Elfiky & Azzam, 2020) and small peptides (Pant et al., 2020) can be a good option. Probiotics are living organism with health-promoting benefits when consumed adequate amount either in the form of diet or drugs (Novik & Savich, 2020), is beneficial for health. Lactobacillus bacterial species plays a vital role in the treatment of gastrointestinal disorder and are known to possess antibacterial (Di Cerbo et al., 2016) and anti-viral (Sunmola et al., 2019) properties. Inhibition of H1N1, HIV, Gastric Corona and Rotavirus in vitro along with a marked reduction in viral (Hasan et al., 2020) load in vivo is well established and documented (Al Kassaa, 2016; Ismail, 2016). Various metabolites of Lactobacillus plantarumsecretes such as Plantaricin, lactic acid, acetic acid, and gamma-aminobutyric acid can enhance the antiviral immunity (Albarracin et al., 2017). In order to infect any host, receptor recognition is the first step by a virus and is a critical aspect of the host cell and tissue tropism. Lie et al. in 2005 gave the concept of binding affinity between SARS-CoV and hACE2 that can correlate the viral transmissibility and disease intensity in humans (Li et al., 2005). COVID-19 entry in a host cell is mediated by transmembrane spike (S) glycoprotein forming homotrimers expressed from a viral surface (Tortorici & Veesler, 2019; Wan et al., 2020). S glycoprotein binds with high affinity to Angiotensin-Converting Enzyme 2 (ACE2) receptor in humans (Walls et al., 2020). We targeted both spike (S) glycoprotein through blocking of Residual binding domain (RBD) and ACE2 by Plantaricin group of probiotic metabolites to establish the molecular, computational role in inhibiting the entry of COVID-19 by these two mechanisms followed by blocking the RNA dependent RNA polymerase (RdDp) for the virus which might get the entry into the alveolar cell. We selected four metabolic products of Lactobacillus plantarum from Plantaricin category viz., Plantaricin BN, Plantaricin JLA-9, Plantaricin W, Plantaricin D to designed computer-based antiviral computational product for COVID-19 that can be consumed as probiotics to retard, inhibit and kill COVID-19 in humans either by blocking or inactivation of RdRp preventing the rise of newly budded progeny virus or by blocking RBD of S protein or interfering with ACE2 receptor proteins and could be useful in the treatment of COVID-19 until some better option is available.
Methods
Protein modelling and quality score
The three-dimensional structure of the SARS-CoV-2 RNA-dependent-RNA polymerase (RdRp) enzyme was built by the Swiss-Model server. Angiotensin-converting enzyme 2 (ACE2) receptor (6VW1) and SARS-CoV-2 spike protein model (6VSB) structures were obtained from the RCSB PDB database (Berman et al., 2002). Ramachandran plots (Furnham et al., 2006) used to assess generated model quality parameters.
Ligands preparation
Four Plantaricin compounds (Plantaricin BN CID:380907, Plantaricin JLA-9 CID_132535900, Plantaricin D CID_139586697, and Plantaricin W CID_139586573) were obtained from PubChem. Compounds energy minimizedand prepared for docking by using Molecular Operating Environment (MOE) software (MOE, 2018).
Docking
Prior docking proteins 3D structure subjected to energy minimization, atoms partially charged and protonated using MOE. Docking software default parameters were used to find the best fit poses in the protein cavities. Chimera software will be used for visualization and calculations of ligands and protein interaction and estimation of H-bonds distances.
Ligand protein interaction and generation of images were rendered using Discovery studio,(Visualizer, 2005) the UCSF Chimera package, (Pettersen et al., 2004) and PLIP webserver (Salentin et al., 2015). Compounds with docking energy ≤ −6.5 kcal/mol are considered auspicious and proceeded for further analysis (Neira et al., 2017).
Molecular dynamics simulations
By using of Groningen Machine for Chemicals Simulations (GROMACS) 5.1.5 package, time-dependent molecular (MD) simulation was carried out for protein-ligand complexes. Molecules scored the best docking results (plantaricin w) were selected for MD simulation. A water solvation step was carried out by AMBER 18 package, as described by Wang et al. (2019) (Wang et al., 2019). System charges was then neutralized by addition of ions, then the energy minimization step was carried out (nsteps = 50,000) using the steepest descent approach (1,000 ps) with maximum force < 1000.0 kJ/mol/nm. Particle Mesh Ewald (PME) method was employed for calculation of electrostatic interaction, then by using of leap-frog integrator system was equilibrated at time step of 2 fs (nsteps= 50,000) using a constant number, pressure and temperature (NPT), and a constant number, volume and temperature (NVT) ensembles. Molecular dynamic simulation finally runs into 10 ns, time step 0.002ps, and 5000,000 steps. Xmgarce (Turner, 2005) was used for visualization of the graphs of Root Mean Square Deviation (RMSD) relative to the structure present in the minimized, equilibrated system (Lu et al., 2010).
Results and discussion
Molecular docking results
Three compounds (Plantaricin W, Plantaricin JLA-9, and Plantaricin D) significantly interacted with RdRp, RBD, and ACE2, scoring the lowest binding energy (≤−6.5 kcal/mol) (Table 1). Antiviral activity of Lactobacillus plantarum probiotics bacteria has been documented previously on the influenza virus, (Rodrigo-Torres et al., 2019) supports our finding.
Table 1.
Docking scores and PubChem CIDs of the four Plantaricin compounds.
| Molecule | PubChem CID | RdRp |
RBD |
ACE2 |
|||
|---|---|---|---|---|---|---|---|
| S (kcal/mol) | RMSD(Å) | S (kcal/mol) | RMSD(Å) | S (kcal/mol) | RMSD(Å) | ||
| Plantaricin W | 139586573 | −14.7 | 3.87 | −11.1 | 2.5 | −12.7 | 4.3 |
| Plantaricin JLA-9 | 132535900 | −11.4 | 4.1 | −8.0 | 1.3 | −9.1 | 2.6 |
| Plantaricin D | 139586697 | −10.1 | 1.9 | −8.6 | 1.9 | −8.5 | 3.2 |
| Plantaricin BN | 380907 | −6.4 | 1.8 | −5.4 | 2.2 | −6.0 | 1.47 |
Molecular docking of RdRp
Plantaricin W, D, and JLA-9 are able to bind RdRp tightly, with binding energies of −14.7, −11.4, and −10.1ACE2, respectively. The following residues in RpRp active site showed different types of interactions as shown in Table 2: GLU167, PRO169, HIS439, ASP452, TYR455, TYR456, ARG457, ASN497, LYS500, ALA550, LYS551, ARG553, THR556, ALA558, ARG569, TYR619, LYS621, ASP623, ARG624, THR680, SER682, ASP684, ALA688, ASN760, ALA797, and LYS798. These three compounds were able to block the residues (THR556, ALA558) surrounding the deep grove catalytic site (VAL557) of RdRp (Lung et al., 2020).
Table 2.
Interaction of compounds with RdRp, RBD and ACE2.
| Molecule | PubChem ID | RdRp |
RBD |
ACE2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bond | Residue | AA | Distance (Å) | Residue | AA | Distance (Å) | Residue | AA | Distance (Å) | ||
| Plantaricin W | 139586573 | Hydrogen Bonds | 167 | GLU | 2.26 | 345 | THR | 2.74 | 26 | LYS | 2.79 |
| 439 | HIS | 2.72 | 345 | THR | 3.28 | 33 | ASN | 3.7 | |||
| 456 | TYR | 2.32 | 347 | PHE | 2.2 | 37 | GLU | 1.95 | |||
| 456 | TYR | 2.05 | 441 | LEU | 2.6 | 90 | ASN | 2.7 | |||
| 457 | ARG | 2.87 | 446 | GLY | 3.14 | 90 | ASN | 2.81 | |||
| 550 | ALA | 2.38 | 446 | GLY | 3.2 | 319 | GLY | 2.54 | |||
| 551 | LYS | 2.17 | 446 | GLY | 2.3 | 353 | LYS | 2.9 | |||
| 553 | ARG | 2.96 | 451 | TYR | 2.58 | 354 | GLY | 2.63 | |||
| 553 | ARG | 2.34 | 383 | MET | 2.28 | ||||||
| 621 | LYS | 1.9 | 393 | ARG | 2.47 | ||||||
| 623 | ASP | 3.45 | 548 | THR | 2.61 | ||||||
| 624 | ARG | 2.62 | 26 | LYS | 2.79 | ||||||
| 624 | ARG | 1.96 | 33 | ASN | 3.7 | ||||||
| 682 | SER | 2.27 | |||||||||
| 691 | ASN | 2.58 | |||||||||
| 760 | ASP | 2.9 | |||||||||
| 797 | ALA | 2.74 | |||||||||
| 798 | LYS | 2.67 | |||||||||
| Hydrophobic Interactions | 169 | PRO | 3.77 | 446 | GLY | 3.99 | 29 | LEU | 3.41 | ||
| 555 | ARG | 3.94 | 447 | GLY | 3.95 | 321 | PRO | 3.81 | |||
| 558 | ALA | 3.9 | 386 | ALA | 3.91 | ||||||
| 389 | PRO | 3.89 | |||||||||
| 555 | PHE | 3.74 | |||||||||
| Salt Bridges | 623 | ASP | 4.07 | 37 | GLU | 3.86 | |||||
| 760 | ASP | 3.87 | |||||||||
| Plantaricin JLA-9 | 132535900 | Hydrogen Bonds | 452 | ASP | 2.05 | 446 | GLY | 3.5 | 26 | LYS | 3.07 |
| 452 | ASP | 2.3 | 494 | SER | 2.1 | 30 | ASP | 1.89 | |||
| 497 | ASN | 2.29 | 351 | TYR | 2.59 | 92 | THR | 2.75 | |||
| 500 | LYS | 2.24 | 491 | GLN | 3.50 | 96 | GLN | 2.27 | |||
| 553 | ARG | 2.14 | 352 | GLY | 2.9 | ||||||
| 556 | THR | 2.51 | 352 | GLY | 3.03 | ||||||
| 619 | TYR | 2.6 | 353 | LYS | 3.07 | ||||||
| 621 | LYS | 3.08 | 354 | GLY | 2.32 | ||||||
| 623 | ASP | 2.3 | 355 | ASP | 2.91 | ||||||
| 624 | ARG | 2.56 | 387 | ALA | 2.19 | ||||||
| 680 | THR | 2.83 | 388 | GLN | 2.37 | ||||||
| 682 | SER | 2.05 | |||||||||
| 682 | SER | 2.83 | |||||||||
| 688 | ALA | 2.6 | |||||||||
| Hydrophobic Interactions | 455 | TYR | 3.78 | 451 | TYR | 3.60 | 26 | LYS | 3.73 | ||
| 455 | TYR | 3.47 | 351 | TYR | 2.95 | ||||||
| 553 | ARG | 3.59 | |||||||||
| 623 | ASP | 3.5 | |||||||||
| Salt Bridges | 500 | LYS | 3.36 | ||||||||
| 569 | ARG | 5.01 | |||||||||
| π-Cation Interactions | 798 | LYS | 5.19 | 26 | LYS | 3.32 | |||||
| Plantaricin D | 139586697 | Hydrogen Bonds | 500 | LYS | 3.74 | 415 | THR | 3.23 | 96 | GLN | 2.45 |
| 553 | ARG | 2.44 | 449 | TYR | 2.1 | 350 | ASP | 2.66 | |||
| 621 | LYS | 2.83 | 453 | TYR | 2.71 | 353 | LYS | 2.47 | |||
| 682 | SER | 3.11 | 496 | GLY | 3.13 | 354 | GLY | 3.07 | |||
| 684 | ASP | 2.27 | 383 | MET | 2.34 | ||||||
| 688 | ALA | 2.86 | 393 | ARG | 3.49 | ||||||
| 760 | ASP | 2.27 | |||||||||
| Hydrophobic Interactions | 455 | TYR | 3.75 | 403 | ARG | 3.52 | 386 | ALA | 3.73 | ||
| 455 | TYR | 3.5 | 421 | TYR | 3.72 | ||||||
| 624 | ARG | 3.77 | 505 | TYR | 3.62 | ||||||
Plantaricin Winteracted with RdRp active site residues by 18 hydrogen bonds with RdRp enzyme with mean distance 2.7 Å and three hydrophobic bonds, which contribute significantly in binding (Table 2) (Figures 1, 2, and 3). The activity of Plantaricins W, d, and JLA-9 on catalytic site of RdRp is better than the activity of other molecules on RdRp: Lung and his colleagues found thataflavin interacts with RdRp with energy −9.11 (Lung et al., 2020), and (Elfiky, 2020) found IDX-184 interact with binding energy −9.3 to RdRp enzyme.
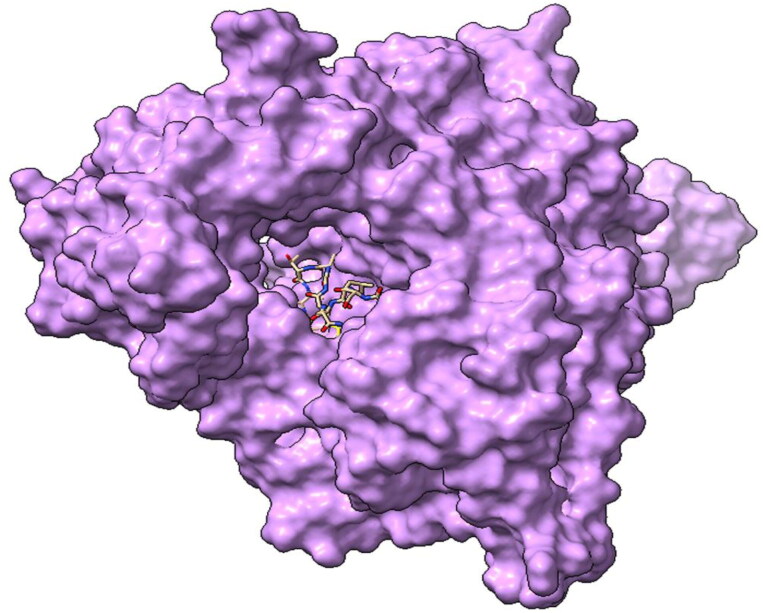
Figure 1.
Three-dimensional structure of RNA-dependent-RNA polymerase (RdRp) enzymes with Plantaricin W blocking the active site cavity.
Figure 2.
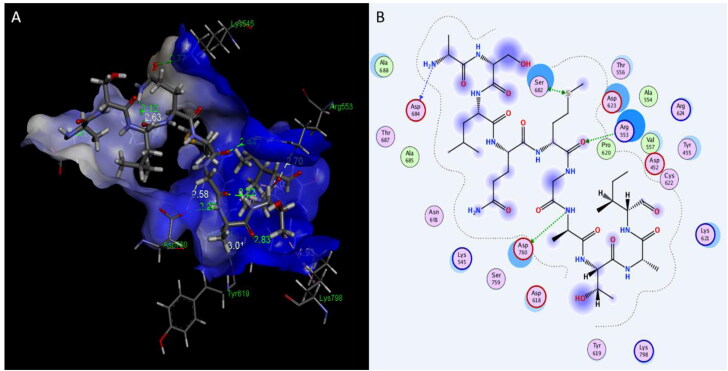
Interaction of Plantaricin D and RNA-dependent-RNA polymerase(RdRp) enzymes. (A) The three-dimensional structure of ligand is surrounded d by the hydrophobic surface of RdRp. (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines.
Figure 3.
Interaction of Plantaricin JLA and RNA-dependent-RNA polymerase (RdRp) enzymes. (A) The three-dimensional structure of ligand is surrounded d by the hydrophobic surface of RdRp. (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines.
Molecular docking of RBD
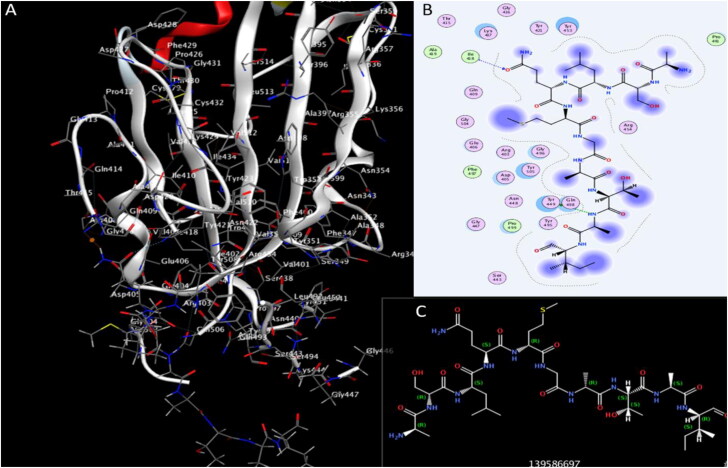
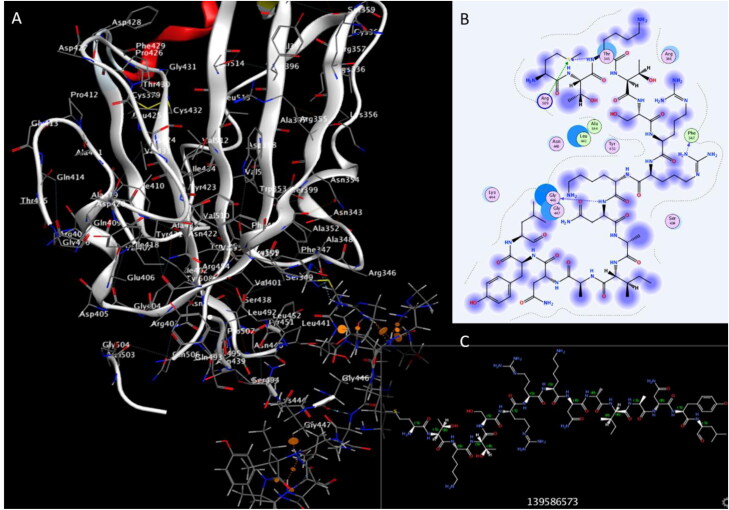
We investigated the potential activity of the four Plantaricinpeptides against RBD of SARS-CoV-2. Plantaricin W, Plantaricin D, and Plantaricin JLA-9 exhibited a strong docking affinity to the RBD, with binding energies −11.1, −8.6, and −8.0 Å respectively (Table 1). They formed different hydrogen, hydrophobic, and salt bridges bonds with the following residues of RBD THR345, PHE347, TYR351, ARG403, THR415, TYR421, LEU441, GLY446, GLY447, TYR449, TYR451, TYR453, GLN491, SER494, GLY496and TYR505 as presented in Table 2. These compounds showed strong hydrogen bonds with key residues (GLY446, TYR449, and TYR453) of RBD, that have a role in ACE2 binding (Yan, Zhang, Guo, et al., 2020; Yan, Zhang, Li, et al., 2020) (Figure 4, 5, and 6).
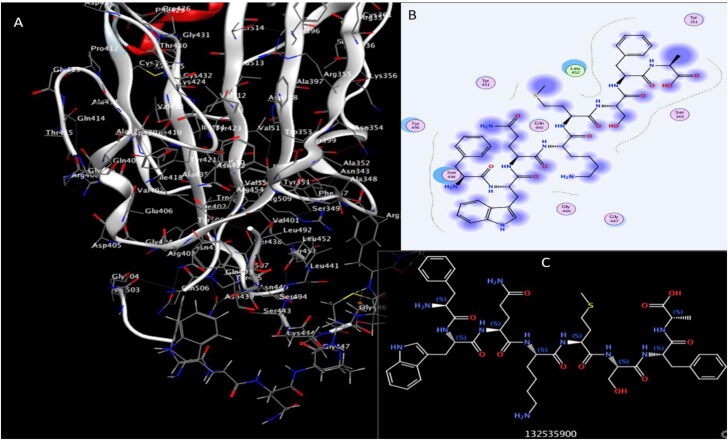
Figure 4.
Interaction of Plantaricin JLA and SARS-CoV-2 receptor-binding domain (RBD). (A) The three-dimensional structure of ligand and amino acids, the RBD is coloured white. (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines. (C) Two-dimensional structure and PubChem ID of Plantaricin JLA ligand.
Figure 5.
Interaction of Plantaricin D and SARS-CoV-2 receptor-binding domain (RBD). (A) The three-dimensional structure of ligand and amino acids, the RBD is coloured white. (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines. (C) Two-dimensional structure and PubChem ID of Plantaricin D ligand.
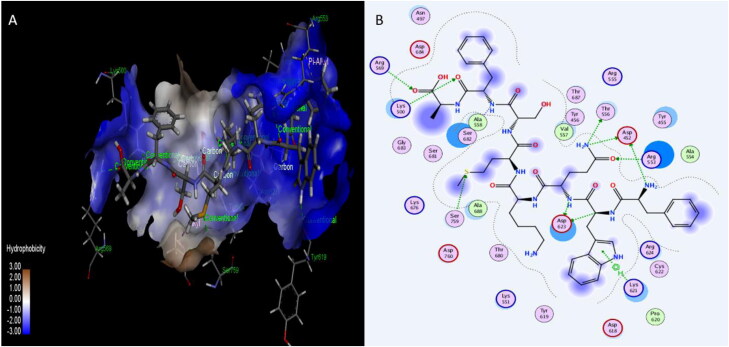
Figure 6.
Interaction of Plantaricin W and SARS-CoV-2 receptor-binding domain (RBD). (A) The three-dimensional structure of ligand and amino acids, the RBD is coloured white. (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines. (C) Two-dimensional structure and PubChem ID of Plantaricin W ligand.
Molecular docking of ACE2
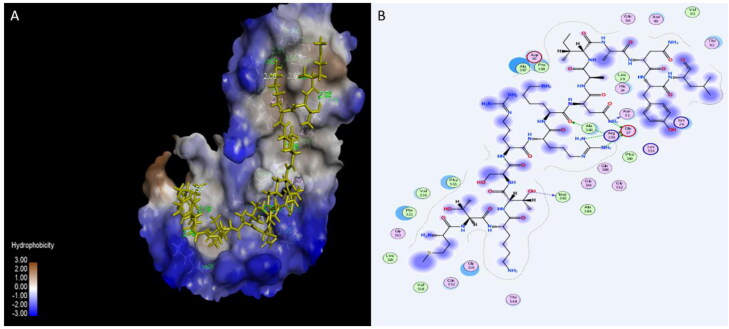
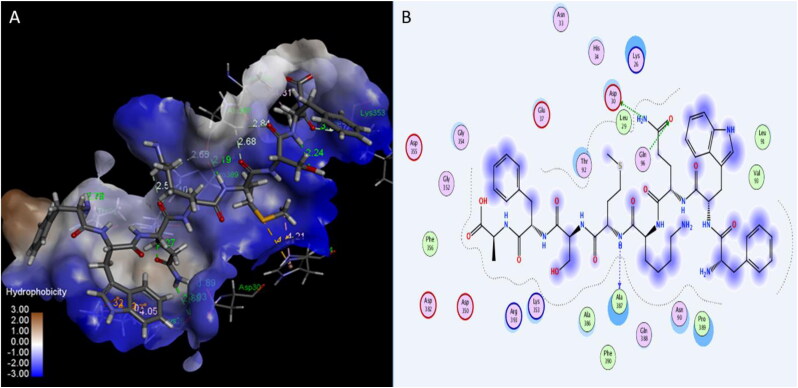
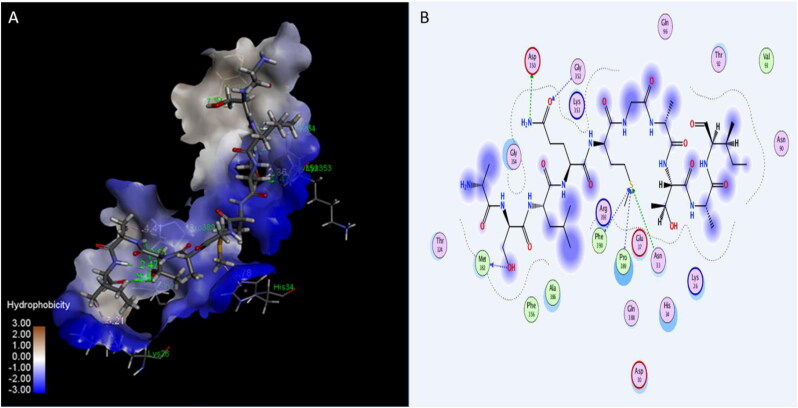
Many published studies suggested that the blocking of viral attachment site in the ACE2 receptor could be useful for patients with COVID-19 (Gurwitz, 2020). In this study, three peptides: Plantaricin W, Plantaricin JLA-9, Plantaricin D showed significant activity on human ACE2 receptor, with binding energies −12.7, −9.1, and -8.5 Å respectively (Table 1). The following residues involving in the interaction with ACE2 receptor:LYS26, LEU29, ASP30, ASN33, GLU37, ASN90, THR92, GLN96, GLY319, PRO321, ASP350, GLY352, LYS353, GLY354, MET383, ALA386, ALA387, GLN388, ARG393, THR548, and PHE555 (Table 2).Plantaricin JLA-9 form strong hydrogen bond (with distance 1.89 Å) with ASP30, and Plantaricin W forms another hydrogen bond (2.9 Å) with LYS353 key residues in interaction with RBD of SARS-CoV-2 (Yan, Zhang, Guo, et al., 2020; Yan, Zhang, Li, et al., 2020) (Figures 7, 8, and 9).
Figure 7.
Interaction of Plantaricin W and angiotensin-converting enzyme 2 (ACE2) receptor. (A) The three-dimensional structure of ligand is surrounded d by the hydrophobic surface of ACE2. (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines.
Figure 8.
Interaction of Plantaricin JLA-9 and angiotensin-converting enzyme 2 (ACE2) receptor. (A) The three-dimensional structure of ligand is surrounded d by the hydrophobic surface of ACE2 (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines.
Figure 9.
Interaction of Plantaricin D and angiotensin-converting enzyme 2 (ACE2) receptor. (A) The three-dimensional structure of ligand is surrounded d by the hydrophobic surface of ACE2. (B) Two-dimensional interaction shows hydrogen bonds in green dotted lines.
Lactobacilli species produce hydrogen peroxide, acides, biosurfactants and bacteriocins giving sufficient protection to host cell (La Fata et al., 2018; Mousavi Khaneghah et al., 2020). Several lactic acid bacteria can synthesize Exopolyscacharides that can modulate the health benefits such as antitumor, antibiofilm and antioxidant activity along with immunomodulatory effects (Silva et al., 2019) such as interference of HIV attachment to target host cells (Martenot et al., 2019).
Lactobacillus plantarum metabolic products established earlier as antiviral metabolites 8 is now computationally proved to inhibit the COVID-19 at various levels. Assessment of our results on COVID-19 through computational studies provides a better glimpse of inhibition, there may be other mechanisms by which these plant products might be working on COVID-19 that need to be deciphered for experiments. Further, these metabolites may also be involved in indirect stimulation of innate and other immunity. Our study can be used as one of the anti-COVID-19 mechanistic approaches through probiotic intake until some effective treatment or vaccine is developed.
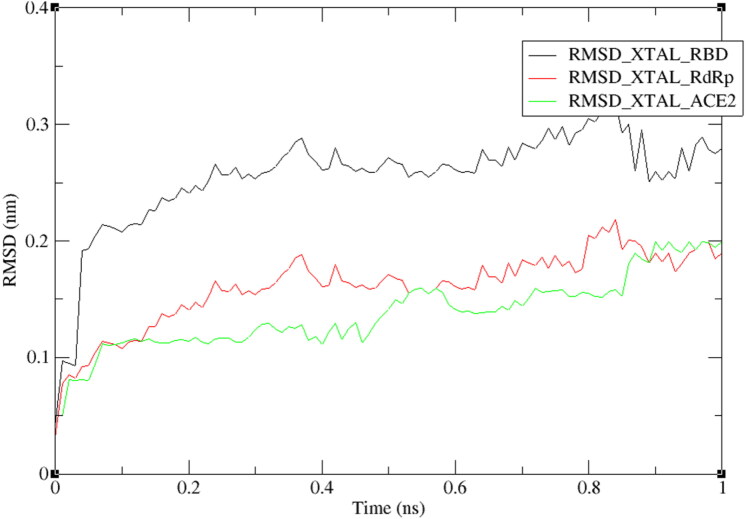
Molecular dynamic simulations
The molecular dynamic simulation was used to check the stability of the complexes of plantaricin w and SARS-CoV-2 RdRp enzyme, RBD of spike protein, and human ACE2 receptor. Root-mean-square deviation (RMSD) of the protein backbones was calculated to study the stability of complexes over a period of time. From RMSD trajectories, as shown in Figure 10, all complexes scored values ranged from 1 nm to 3 nm, indicating their high stability. RdRP and ACE2 complexes showed RMSD level near to 0.1 nm (1 Å) after a short period of time (0.1 ns) indicating their best stability (Vyas et al., 2017), this finding agrees with that they have scored the best docking energies (−14.7, and −12.7 respectively) stating their strong interaction.
Figure 10.
RMSD plot of RBD, RdRp, and ACE2 proteins backbones in complex with plantaricin w, over one nanosecond period.
Conclusion
The present computational representation and molecular dynamics study clearly demonstrates the antiviral activity (Aanouz et al., 2020) of Plantiricin compounds, through multiple mechanistic approach by metabolic product of Lactobacillus plantarum blocks the entry by binding with RdRp, RBD, and ACE2. The blocking of main structural protein S is one of the essential accessory protien, playing a vital role in the life cycle of SARS-CoV-2 can prove to be one of the best target for other molecules. The claim is substantiated by Molecular dynamics model that further strengthen stability of the complexes of plantaricin w and SARS-CoV-2 RdRp enzyme, RBD of spike protein, and human ACE2 receptor. Molecular dynamics and computational model are the tools that can precisely predict to reposition the drugs without going in the clinical phase for the effective treatment of COVID-19. Further, it is difficult to find the drug that has a better safety profile than these macrolides, with a cheaper cost, which can be linked with the clinical data for further studies. The present study can state that best possible way about these metabolites that can cease the infection if taken under proper medical supervision. The three antibiotics Azithromycin, Clarithromycin and Erythromycin can be used against new strain of coronavirus with promising result with three way specific target against COVID-19. As of now the plantaricin metabolites can be the alternate till new antiviral drug specific for COVID-19 is discovered.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aanouz I., Belhassan A., El Khatabi K., Lakhlifi T., El Idrissi M., & Bouachrine M. (2020). Moroccan medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure & Dynamics, 6, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Kassaa I. (2016). New insights on antiviral probiotics: From research to applications. New Insights on Antiviral Probiotics: From Research to Applications. 10.1007/978-3-319-49688-7 [DOI]
- Albarracin L., Kobayashi H., Iida H., Sato N., Nochi T., Aso H., Salva S., Alvarez S., Kitazawa H., & Villena J. (2017). Transcriptomic analysis of the innate antiviral immune response in porcine intestinal epithelial cells: Influence of immunobiotic lactobacilli. Frontiers in Immunology, 8, 57 10.3389/fimmu.2017.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Battistuz T., Bhat T. N., Bluhm W. F., Bourne P. E., Burkhardt K., … Zardecki C. (2002). The protein data bank. Acta Crystallographica Section D: Biological Crystallography. 10.1107/S0907444902003451 [DOI] [PubMed]
- Boopathi S., Poma A. B., & Kolandaivel P. (2020). Novel 2019 Coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure & Dynamics, 30, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiong J., Bao L., & Shi Y. (2020). Convalescent plasma as a potential therapy for COVID-19. The Lancet. Infectious Diseases, 20(4), 398–400. 10.1016/S1473-3099(20)30141-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cerbo A., Palmieri B., Aponte M., Morales-Medina J. C., & Iannitti T. (2016). Mechanisms and therapeutic effectiveness of lactobacilli. Journal of Clinical Pathology, 69(3), 187–203. 10.1136/jclinpath-2015-202976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. a). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure & Dynamics, 5, 1–10. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. b). Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences, 253, 117592 10.1016/j.lfs.2020.117592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A., & Azzam E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure & Dynamics, 27, 1-9. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A. A. (2020. c). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure & Dynamics, 6, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen A. D., Al-Obaidi A., Şahin A. T., & Yelekçi K. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure & Dynamics, 26, 1-13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani M., Hasaniazad M., Faezi S., Guklani H., Davoodian P., Ahmadi N., Einakian M. A., Karmostaji A., & Ahmadi K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 2, 1–16. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi S. K., Raja K., Sebastine I., & Joseph J. (2020). Andrographolide as a potential Inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 5, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnham N., Doré A. S., Chirgadze D. Y., de Bakker P. I. W., DePristo M. A., & Blundell T. L. (2006). Knowledge-based real-space explorations for low-resolution structure determination. Structure (London, England : 1993), 14(8), 1313–1320. 10.1016/j.str.2006.06.014 [DOI] [PubMed] [Google Scholar]
- Gupta M. K., Vemula S., Donde R., Gouda G., Behera L., & Vadde R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure & Dynamics, 15, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D. (2020). Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Development Research, 4 10.1002/ddr.21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A. A., & Falahati M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 22, 1–9. 10.1080/07391102.2020.1754293 https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., … Cao B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Parves R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. D., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure & Dynamics, 12, 1–12. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail B. (2016). The use of probiotics as vaccine vectors to prevent viral infections. In New Insights on Antiviral Probiotics: From Research to Applications. 10.1007/978-3-319-49688-7_2 [DOI]
- Joshi R. S., Jagdale S. S., Bansode S. B., Shankar S. S., Tellis M. B., Pandya V. K., Chugh A., Giri A. P., & Kulkarni M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease $. Journal of Biomolecular Structure and Dynamics, 5, 1-16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R. J., Jha R. K., Amera G. M., Jain M., Singh E., Pathak A., … Singh A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2’-O-ribosemethyltransferase. Journal of Biomolecular Structure & Dynamics, 20, 1-14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fata G., Weber P., & Mohajeri M. H. (2018). Probiotics and the gut immune system: Indirect regulation. Probiotics and Antimicrobial Proteins, 10(1), 11–21. 10.1007/s12602-017-9322-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J. H., Moore M. J., Luo S., Wong S.-K., Huang I.-C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W. A., Guan Y., Choe H., & Farzan M. (2005). Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. The EMBO Journal, 24(8), 1634–1643. 10.1038/sj.emboj.7600640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. Y., Jiang Y. J., Lv J., Wu T. X., Yu Q., Sen., & Zhu W. L. (2010). Molecular docking and molecular dynamics simulation studies of GPR40 receptor-agonist interactions. Journal of Molecular Graphics & Modelling, 28(8), 766–774. 10.1016/j.jmgm.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Lung J., Lin Y. ‐S., Yang Y. ‐H., Chou Y. ‐L., Shu L. ‐H., Cheng Y. ‐C., Liu H. T., & Wu C. ‐Y. (2020). The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. Journal of Medical Virology, 92(6), 693–697. 10.1002/jmv.25761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung J., Lin Y. ‐S., Yang Y. ‐H., Chou Y. ‐L., Shu L. ‐H., Cheng Y. ‐C., Liu H. T., & Wu C. ‐Y. (2020). The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. Journal of Medical Virology, 92(6), 693–697. 10.1002/jmv.25761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martenot C., Faury N., Morga B., Degremont L., Lamy J. B., Houssin M., & Renault T. (2019). Exploring first interactions between ostreid herpesvirus 1 (OsHV-1) and its host, Crassostrea gigas: Effects of specific antiviral antibodies and dextran sulfate. Frontiers in Microbiology, 10, 1128 10.3389/fmicb.2019.01128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi Khaneghah A., Abhari K., Eş I., Soares M. B., Oliveira R. B. A., Hosseini H., Rezaei M., Balthazar C. F., Cruz A. G., Ranadheera S., & Sant’Ana A. S. (2020). Interactions between probiotics and pathogenic microorganisms in hosts and foods: A review. Trends in Food Science and Technology, 95, 205–218. 10.1016/j.tifs.2019.11.022 [DOI] [Google Scholar]
- Muralidharan N., Sakthivel R., Velmurugan D., & Gromiha M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure & Dynamics, 16, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Neira J. L., Bintz J., Arruebo M., Rizzuti B., Bonacci T., Vega S., Lanas A., Velázquez-Campoy A., Iovanna J. L., & Abián O. (2017). Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Scientific Reports, 7, 39732 10.1038/srep39732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neira J. L., Bintz J., Arruebo M., Rizzuti B., Bonacci T., Vega S., Lanas A., Velázquez-Campoy A., Iovanna J. L., & Abián O. (2017). Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Scientific Reports, 7, 39732 10.1038/srep39732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novik G., & Savich V. (2020). Beneficial microbiota. Microbes and Infection, 22(1), 8–18. 10.1016/j.micinf.2019.06.004 [DOI] [PubMed] [Google Scholar]
- Pant S., Singh M., Ravichandiran V., Murty U. S. N., & Srivastava H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure & Dynamics, 6, 1–10. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., & Ferrin T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Rodrigo-Torres L., Yépez A., Aznar R., & Arahal D. R. (2019). Genomic insights into five strains of Lactobacillus plantarum with biotechnological potential isolated from chicha, a traditional maize-based fermented beverage from Northwestern Argentina. Frontiers in Microbiology, 10, 2232. 10.3389/fmicb.2019.02232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentin S., Schreiber S., Haupt V. J., Adasme M. F., & Schroeder M. (2015). PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Research, 43(W1), W443–W447. 10.1093/nar/gkv315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma P., Sekhar N., Prajapat M., Avti P., Kaur H., Kumar S., Singh S., Kumar H., Prakash A., Dhibar D. P., & Medhi B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure & Dynamics, 18, 1–9. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., Wei J., Xiao H., Yang Y., Qu J., Qing L., Chen L., Xu Z., Peng L., Li Y., … Liu L. (2020). Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA, 323(16), 1582 10.1001/jama.2020.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva L. A., Lopes Neto J. H. P., & Cardarelli H. R. (2019). Exopolysaccharides produced by Lactobacillus plantarum: Technological properties, biological activity, and potential application in the food industry. Annals of Microbiology, 69(4), 321–328. 10.1007/s13213-019-01456-9 [DOI] [Google Scholar]
- Sinha S. K., Shakya A., Prasad S. K., Singh S., Gurav N. S., Prasad R. S., & Gurav S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 13, 1-12. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunmola A. A., Ogbole O. O., Faleye T. O. C., Adetoye A., Adeniji J. A., & Ayeni F. A. (2019). Antiviral potentials of Lactobacillus plantarum, Lactobacillus amylovorus, and Enterococcus hirae against selected Enterovirus. Folia Microbiologica, 64(2), 257–264. 10.1007/s12223-018-0648-6 [DOI] [PubMed] [Google Scholar]
- Tortorici M. A., Veesler D. (2019). Structural insights into coronavirus entry. In advances in virus research. 10.1016/bs.aivir.2019.08.002 [DOI] [PMC free article] [PubMed]
- Turner P. (2005). XMGRACE, Version 5.1. 19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology; 10.1163/_q3_SIM_00374 [DOI] [Google Scholar]
- Visualizer D. S. (2005). v4. 0.100. 13345. In Accelrys Software Inc.
- Vyas V. K., Shah S., & Ghate M. (2017). Generation of new leads as HIV-1 integrase inhibitors: 3D QSAR, docking and molecular dynamics simulation. Medicinal Chemistry Research, 26(3), 532–550. 10.1007/s00044-016-1772-y [DOI] [Google Scholar]
- Wahedi H. M., Ahmad S., & Abbasi S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure & Dynamics, 12, 1–10. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Walls A. C., Park Y. J., Tortorici M. A., Wall A., McGuire A. T., & Veesler D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R. S., & Li F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology, 94(7), e00127-20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Alekseenko A., Kozakov D., & Miao Y. (2019). Improved modeling of peptide-protein binding through global docking and accelerated molecular dynamics simulations. Frontiers in Molecular Biosciences, 6, 112. 10.3389/fmolb.2019.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) (2020). Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases. Interim Guidance. [Google Scholar]
- Yan R., Zhang Y., Guo Y., Xia L., & Zhou Q. (2020). Structural basis for the recognition of the 2019-nCoV by human ACE2. BioRxiv. 10.1101/2020.02.19.956946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., & Zhou Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]