Abstract
Helicobacter pylori (H. pylori) infection is associated with some gastric diseases, such as gastritis, peptic ulcer, and gastric cancer. CagA and VacA are known virulence factors of H. pylori, which play a vital role in severe clinical outcomes. Additionally, the expression of outer membrane proteins (OMPs) helps H. pylori attach to gastric epithelial cells at the primary stage and increases the virulence of H. pylori. In this review, we have summarized the paralogs of H. pylori OMPs, their genomic loci, and the different receptors of OMPs identified so far. We focused on five OMPs, BabA (HopS), SabA (HopP), OipA (HopH), HopQ, and HopZ, and one family of OMPs: Hom. We highlight the coexpression of OMPs with other virulence factors and their relationship with clinical outcomes. In conclusion, OMPs are closely related to the pathogenic processes of adhesion, colonization, persistent infection, and severe clinical consequences. They are potential targets for the prevention and treatment of H. pylori–related diseases.
Keywords: Helicobacter pylori, Outer membrane proteins, Virulence factors
Introduction
Helicobacter pylori (H. pylori) colonizes mainly the surface of the gastric epithelium and is an essential human pathogen that was discovered in the twentieth century. The prevalence of H. pylori is probably 44.3% of the entire human population [1]. According to the statistics, its prevalence is 34.7% in developed countries and 50.8% in developing countries, and the worldwide annual recurrence rate is 4.3% [2]. Although the global infection rate of H. pylori is high, a large proportion of infected people have no apparent symptoms and only show gastritis under an endoscope [3, 4]. However, these asymptomatic or endoscopic gastritis patients may develop changes in their condition. More severe conditions include peptic ulcer (PU), gastric cancer (GC), and mucosa-associated lymphoid tissue (MALT) lymphoma [4]. The outcome of infection depends mainly on the interactions among H. pylori, the stomach, and the environment [5, 6]. For developing countries with high infection rates and low economic levels, eradication of H. pylori with antibiotics to prevent serious diseases is the most effective and relatively cheaper method being currently applied [7].
The outer membrane is the outer barrier of Gram-negative bacteria, which consists of two highly asymmetric layers—the inner monolayer contains only phospholipids and the outer monolayer consists mainly of outer membrane proteins (OMPs) that are resistant to the external environment [8]. OMPs have a variety of biological functions, not only in maintaining the outer membrane structure and guaranteeing the material transportation but also in playing an essential role in the process of contact with the host [9]. The study of H. pylori OMPs will contribute to the development of vaccine and drug targets [10, 11]. OMPs in H. pylori mainly include lipoproteins, porins, iron-regulated proteins, efflux pump proteins, and adhesins [12]. The expression of OMPs in different strains is related to the virulence of H. pylori. The pathogenicity of the OMPs may be achieved through the following mechanisms: (1) adhesion, (2) penetration of the bacteria through the defense barrier, and (3) evasion of the immune system.
Several strains of H. pylori have been subjected to whole-genome sequencing, and approximately 4% of their genes were found to encode OMPs. We further divided them into five paralogous gene families according to their functions [12]. Hop (outer membrane porins) and Hor (Hop-related proteins) proteins are the best-known family. The second and third are Hof (H. pylori OMP) and Hom (H. pylori outer membrane) proteins, respectively. Iron-regulated OMPs are the fourth family, and efflux pump OMPs are the fifth family. The other types of OMPs that do not belong to any of these families have different alleles, regional distribution characteristics, and distinct correlations with other virulence factors. Allelic variation and phase variation are the most common methods used to regulate the expression of OMPs [13–15]. Usually, there are multiple alleles for each OMP, and the functions of proteins encoded by each allele are somewhat different. Additionally, due to geographical differences among strains, the expression of OMPs is also variable. In this review, we summarize recent progress in our understanding of the five best characterized OMPs: BabA (HopS), SabA (HopP), OipA (HopH), HopQ, and HopZ, and one family of OMPs: Hom and their receptors, as well as their relationship with other virulence factors and clinical outcomes.
Main OMPs
BabA (HopS)
BabA belongs to the Hop protein family, also called blood-group-antigen-binding adhesin. So far, three genomic loci have been found in bab, namely, babA, babB, and babC [16]. BabA and its babB allelic gene are irrelevant to each other according to phylogenetic analysis, and the expression of the two genes differs geographically. The multiple synonymous and nonsynonymous substitutions of the babA gene region indicate that it predates babB in phylogeny [17]. However, the babA diversity region may have more functional constraints, and there is a clear distinction between the US and Asian strains [3, 18].
The primary role of BabA in the pathogenesis of H. pylori is adhesion. Ilver et al. were the first to isolate BabA and put forward the theory that BabA adheres to the fucosylated LewisB histo-blood group antigen for host selectively [19, 20]. Backstrom found the strain 17,875 had two babA alleles, which were babA1 (silence) and babA2 (expression) [21]. BabA2-cam was a derivative gene of babA2, which was obtained by recombination of the previously silent babA1 gene with the expressed and partially homologous babB locus. Then, they confirmed that BabA2-cam could bind to LewisB. Similarly, the binding affinity of chimeric BabB/A adhesin to LewisB was similar to that of BabA adhesin. However, the expression level of BabB/A was low, and phase variation could occur through a slipped-strand mispairing mechanism [21, 22]. These phenomena all indicate that their transferability and heterogeneity contribute to the adaptation of the bacteria, and some strains have the potential to periodically activate and inactivate their virulence according to the host’s response to infection. The expression of BabA is acid-sensitive and has nothing to do with the interpretation of LewisB or the binding affinity of LewisB [23]. Bugaytsova et al. believe that the loss expression of BabA is not related to adaptive immunity or the toll-like receptor signaling system, indicating that BabA has some other functions besides being an adhesin [24].
After the calculation and analysis of the X-ray diffraction pattern of BabA, it was found that BabA has three structural domains for combining with LewisB, including two diversity loops (DL1 and DL2) and one conserved loop (CL2) [16]. The specific binding region is the hydrogen bond network structure formed between four residues of LewisB and eight amino acids of BabA [25]. Previous research suggested that H. pylori had blood group binding preferences, and the specific strains only combined with O antigen residues, resulting in people with type O blood being more likely to suffer from PU than those with other blood types [16]. According to the recent study of BabA and LewisB structures, the prevalence of PU is higher in type O blood, probably because it overexpresses LewisB.
Multiple studies on the relationship between H. pylori and clinical diseases have shown that, compared with the control group, patients with PU and GC have higher BabA expression. This indicates that BabA is positively correlated with severe clinical diseases caused by H. pylori [26, 27]. Possible mechanism to explain this phenomenon is that the combination of BabA and LewisB triggers the type IV secretion system (T4SS), which helps CagA to enter the gastric mucosal epithelial cells [28]. Cag pathogenicity island (cag PAI) is a large (usually 20–100 kb) DNA fragment encoding many virulence-related genes on the H. pylori chromosome, which often contains repetitive or inserted sequences on both sides. Cag PAI mediates the expression of CagA and is closely associated with the occurrence of gastritis, PU, GC, and other diseases. Cag PAI also mediates the expression of T4SS [29], through which the vital virulence factor CagA is delivered into attached cells. CagA will induce the production of inflammatory factors, leading to intestinal metaplasia and precancerous lesions [28].
SabA (HopP)
The sab gene has two alleles, sabA and sabB. SabA is the second most commonly reported OMP in H. pylori. It is named sialic-acid-binding adhesin according to its binding receptors and it belongs to the Hop family, also known as HopP or OMP17. SabB is known as HopO or OMP16 [12]. However, genomic analysis showed that the strains often both have sabA and sabB genes, which may selectively express SabA during the colonization of H. pylori in the host. The specific mechanism may be a phase variation caused by slipped-strand mispairing in a 5′ dinucleotide repeat region, which affects the on-off states of sabA and sabB [30]. Like BabA, the expression of SabA is rapidly regulated in response to changes in the gastric environment, such as inflammation [31]. On the one hand, the expression of SabA can be regulated at the gene level through phase variation; on the other hand, H. pylori can also adjust the expression of genes at the cell level through its two-component signal transduction (TCST) system. The TCST can adapt to the changes in the environment at the transcriptional level to selectively express SabA. This is also known as acid-responsive signaling because the pH of the environment is one of the factors that activates the signal [32, 33].
Pang et al. analyzed X-ray diffraction patterns after the extraction of SabA and found that the adhesion domain of SabA was highly similar to the tetratricopeptide repeat fold family, which is often gathered into multiprotein complex-mediated protein interactions. The adhesion domain of SabA is mainly a barrel transmembrane domain. Its N-terminal and C-terminal form right angles from the head domain and create a shaft together. This group also conducted experiments in vitro and found that the SabA adhesion domain could bind to sialyl-LewisX, sialyl-LewisA, and LewisX, but not to other Lewis antigens, such as LewisA, LewisB, or LewisY [34]. The expression of sialyl-LewisX can be further regulated, after the combination of SabA and sialyl-LewisX. This process relies on specific glycosyltransferase β3GlcNAcT5, which can upregulate the expression of sialyl-LewisX, enhancing the combination of SabA and sialyl-LewisX, which increases the colonization ability of H. pylori [35]. SabA can also combine with ganglioside in addition to sialyl-LewisX and LewisX. Benktander et al. extracted two gangliosides sialyl-neolactohexaosylceramide and sialyl-neolactooctaosylceramide that combined with SabA [36].
The expression of SabA is called its “on” state, while the nonexpression is called its “off” state. The clinical strains of H. pylori SabA have a frequent “on/off” state, which means that SabA can be rapidly regulated in response to the changes in the gastric ecological environment [37]. When the pH in the stomach decreases, the expression of SabA is increased, which reflects the ability of H. pylori to adapt to the host. Meanwhile, the combination of SabA and ganglioside also mediates the chronic infection process of H. pylori [38]. A study in Japan showed a close relationship between SabA and GC [39]. However, another study reported that Asian strains with SabA-positive status had little influence on the clinical outcomes [40], so the relationship between SabA and clinical diseases is still controversial.
OipA (HopH)
Outer inflammatory protein A (OipA) is also a member of the Hop family, sometimes referred to as HopH. The location of oipA on the H. pylori chromosome is approximately 100 kb from the cag PAI [41]. There are no available studies on the structure of OipA and its receptors so far.
The oip gene also has an “on/off” state, and when OipA is expressed, CagA is usually positive, which means these two proteins have a close link [42, 43]. Except for CagA, OipA seems to have no significant interactions with the other virulence factors. Several studies have reported that OipA can increase the secretion of interleukin-8 (IL-8) and other inflammatory factors to cause neutrophil infiltration, aggravating the inflammation in the stomach and helping H. pylori to colonize, and this is why it is called an outer inflammatory protein [42, 44]. OipA also inhibits the apoptosis of gastric cells [45], reducing the incidence of β-catenin nuclear translocation and cancer when the oip gene is switched off [46]. However, an in vitro study showed that OipA did not induce an increase in IL-8 expression [42]. Therefore, a more extensive sample study is needed to confirm the relationship between OipA and IL-8. OipA also plays a vital role in the activation of focal adhesion kinase (FAK). FAK is a cytoplasmic nonreceptor tyrosine kinase that can regulate the shape of cells, mediate cell movement, and play an essential role in the occurrence and invasive growth of tumors [47–49].
Many studies have suggested that the expression of OipA was positively correlated with PU, GC, and MALT in both Asian and Western strains [50–52]. Mahboubi et al. inoculated mice with OipA vaccine and the amount of H. pylori colonized in the stomach was significantly reduced, as well as the inflammation after some time. Therefore, immunogenic OipA could be used as an H. pylori vaccine to make up for the increasing antibiotic resistance of H. pylori [53].
HopQ
HopQ is encoded by the hopQ (omp27) gene and also belongs to the pore protein Hop family. Studies have reported that hopQ has two alleles of I and II [12]. A study reported that, among H. pylori strains, hopQ I type was the most common (72.5%), followed by hop Q II type (15.4%), and chimeric I type and II type were the rarest [54]. Further studies on Asian and Western strains showed that the Asian strains were dominated by the hopQ I type, while the hopQ II strains were sporadic [55].
We mentioned above that BabA mediates the activation of T4SS and facilitates the translocation of virulence factor CagA. Studies have found that HopQ also plays an essential role in this process [13, 56]. HopQ can exploit the carcinoembryonic antigen–related cell adhesion molecule family (CEACAMs) as host cell receptors, mainly CEACAM1, CEACAM3, CEACAM5, and CEACAM6 [56, 57]. HopQ I type or II type targets the β-strands G, F, and C in the N-terminal domain (C1ND) of CEACAM. It has also been reported that HopQ binds to the IgV-like domain at the N-terminal of CEACAMs to facilitate the transfer of crucial pathogenic factor CagA to host cells [57]. In vitro animal experiments have also confirmed that HopQ-CEACAM interactions provide a pathway for H. pylori to adhere to the host and trigger signal transduction, and then, it produces long-term effects on humans [58, 59].
At present, although there are no additional reports on the relationship between HopQ and disease, in both Western and Asian strains, the hopQ I type genotype was significantly linked with vacA s1 and m1 genotypes and cagA-positive status, suggesting that HopQ may be an essential virulence factor related to gastroduodenal diseases [55].
HopZ
Among the Hop family, the hopZ gene has also been well characterized. HopZ has two allelic variants, HopZ-I and HopZ-II. The apparent difference between HopZ-I and HopZ-II is a conserved sequence of only 20 amino acids in HopZ-I. Furthermore, the conservatism of HopZ-II is higher than that of HopZ-I, and the change frequency of the amino acid sequence is lower [60].
The host receptors of HopZ have not been well characterized yet. Nonetheless, HopZ may play a vital role in adhesion [61]. By analyzing the hopZ gene sequences of 15 strains of H. pylori, Peck found that the slipped-strand mispairing mechanism of the CT dinucleotide repeats in the signal peptide coding region of the HopZ was correlated with the expression of hopZ [62]. After analyzing the bacteria extracted from the stomach of infected patients, Kennemann et al. found that the expression of the gene hopZ also depended on the on/off switch mediated by phase variation during the early colonization of H. pylori, which indicated that the gene hopZ had a strong selectivity in vivo [13]. The HopZ gene can be expressed stably in the stomach without mixed infection [63]. At present, there is not enough evidence to prove that HopZ has a relationship with other virulence factors [64]. Moreover, after analysis of 63 patients with chronic infection, it was found that there was no significant correlation between HopZ and chronic atrophic gastritis, and the relationship between HopZ and other clinical diseases is not clear [13].
Hom
The Hom family consists of four OMPs: HomA, HomB, HomC, and HomD. HomA and homB are highly homologous, with 90% similarity in their nucleotide sequences, with two identical conserved loci [12]. HomA, homB, and homC sequences have considerable geographic heterogeneity, but homD is highly conserved [65]. Oleastro et al. distinguished homA and homB at the nucleotide and amino acid levels, and found they had six allelic variants named AI–AVI [66]. Three allelic variants were observed for homA (AII, AIII, and AIV). In contrast, five distinct alleles were observed for homB (AI, AII, AIII, AV, and AVI), suggesting that homA has more significant heterogeneity. The expressions of the homA and homB genes are different in strains all over the world, and there are significant differences between Western and Asian strains [66]. Kim et al. found that homC variations in different geographical origins were related to bab [67]. They identified and isolated three polymorphic forms of homC: homCS, homCL, and homCM. HomCL was found to be most closely related to bab after the detection of different populations.
Similar to OipA, HomB can improve the adhesion of H. pylori to the host and promote the secretion of IL-8 and other inflammatory factors. A Portuguese study found that HomB had a significant positive correlation with the occurrence and development of PU in children and adolescents [68], and HomA was associated with nonulcerative gastritis [69]. At the same time, some studies also confirmed that there was no significant relationship between HomB and GC [70]. Therefore, whether HomB was positive or not could be used as an important virulence factor to distinguish GC from PU [71].
OMPs as treatment targets
The mechanism of drug resistance of bacteria mainly includes three aspects: producing corresponding hydrolytic and modifying enzymes to destroy drug activity; changing the structure of the drug target so that the drug cannot be recognized; blocking drug contact with target sites, including mechanisms of regulating membrane permeability and the antibiotic expulsion system [9]. With increasing numbers of antibiotic-resistant strains of H. pylori, people are devoting much effort to finding other treatments besides antibiotic treatment, and the development of an H. pylori vaccine is currently a hot research topic [72, 73].
The whole cell of the bacterium vaccine was first investigated in 1992. Chen et al. [74] used ultrasonic grinding of the bacteria and immunized mice with these antigens and the adjuvant cholera toxin, which provided nearly 100% of immune protection. Due to the complex antigen components of H. pylori and the long cycle required for whole cell of the bacterium vaccine production, low production of some strains, their ease of contamination, and poor storage stability, whole cell of the bacterium vaccines was abandoned. Then, came the development of recombinant vaccines that combined protective antigens with immune adjuvants. Immune adjuvants can enhance the body’s ability to respond to antigens, especially in the early stages of immunity. Vaccines lacking adjuvants have low or no protective effects [75]. The commonly used immune adjuvants are cholera toxin [76], E. coli heat-labile enterotoxin [76], synthetic oligodeoxynucleotides containing unmethylated CPG motifs [77], and aluminum hydroxides [78]. The most commonly used protective antigens include CagA, VacA, urease, catalase, neutrophil-activating protein, Hsp60, BabA, SabA, and OipA [79].
Proteomic studies on E. coli, Leptospira, and P. aeruginosa have proven that OMPs are related to bacterial resistance to antibiotics [80–82]. The adhesion of OMPs can stimulate the immune response of the host cell and promote the intracellular signal transduction of the proinflammatory cells, so OMPs can be used as an immunizing antigen [83]. It is also thought that mucosal immunity, rather than systemic immunity with the production of IgG, plays a greater role in adhesin-based vaccines [84]. There are also many studies using proteomics to evaluate suitable OMPs as possible candidates for vaccines [85]. BabA can be recognized as a candidate vaccine, and the recombinant BabA stimulated the humoral and cellular immunity against H. pylori infection [75]. OipA also plays a crucial role in the first step of H. pylori colonization-adhesion, so it may be considered as a suitable candidate for vaccine development [53, 86]. Other OMPs, HopB, HopC, and HopZ, are being studied further.
However, the ability of H. pylori to vary its OMPs limits the effectiveness of vaccines or therapeutics that target any single one of these components [87]. Peck et al. [88] found four members of OMPs, namely, HopV, HopW, HopX, and HopY. They are highly conserved in the strains analyzed, which suggests they have the potential for vaccine development. No indications of phase variation such as homopolymeric dinucleotide repeats or monomeric G or C tracts were found in the gene promoter regions corresponding to these four proteins, which means that H. pylori can continuously express these proteins during all stages of chronic infection, and they are stable as immune antigens. Therefore, the research direction of OMPs as immune antigens is to look for OMPs that are not prone to phase variation or to recombine OMPs with antigens such as CagA, VacA, urease, and catalase to achieve a higher immune protection rate. For asymptomatic infected patients, analyzing the strains isolated from them to determine their expression of OMPs is vital. This will achieve individualized treatment and reduce the drug resistance rate.
Conclusion
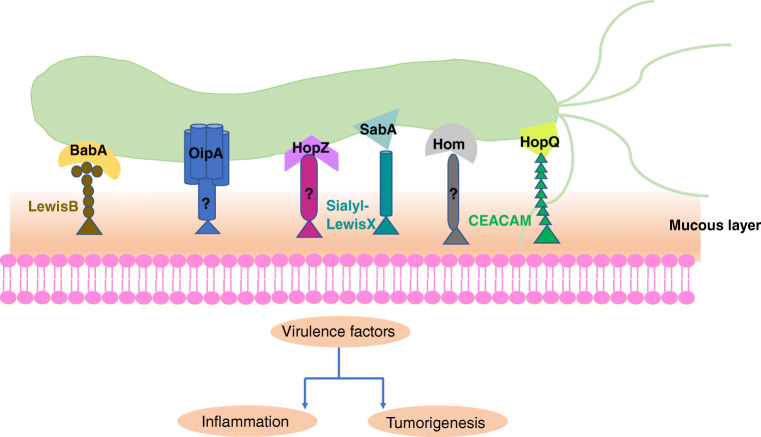
OMPs play a vital role in the colonization of H. pylori. The adhesion of H. pylori to gastric epithelial cells is a complex process, involving a variety of adhesins and target receptors (Table 1). Diverse OMPs can also rapidly regulate themselves in response to changes in gastric inflammation and pH, to adapt to changes in their environment. OMPs not only mediate the adhesion between the bacteria and the gastric epithelial cells but also cooperate with other virulence factors such as CagA and VacA to increase the release of inflammatory factors, leading to different clinical outcomes (Fig. 1). Therefore, OMPs at least partially determine the virulence of H. pylori infection. Understanding the receptor interaction and mechanisms of pathogenesis of OMPs is essential for the prevention and treatment of H. pylori infections. Moreover, the potential of OMPs as vaccines should be further explored as an alternative therapeutic option. We expect that further investigations in this direction would help against H. pylori–mediated resistance and improve the clinical outcomes of H. pylori infections.
Table 1.
Main OMPs and their receptors and functions
| OMPs | Gene no. | Protein length (amino acids) | Receptors identified | Localization | Suggested function |
|---|---|---|---|---|---|
| BabA (HopS) | 26695:1243 | 733 | Lewis B blood group antigens [19] | Gastric epithelia | Adhesion to host cell, enhancing translocation of CagA via the T4SS |
| Terminal fucose residues on blood group O (H antigen), A and B antigens salivary nonmucin glycoprotein [89, 90] | Gastric epithelia | ||||
| Salivary mucin MUC5B and proline-rich glycoprotein [90] | Saliva | ||||
| Proline-rich glycoprotein containing Fucα1-2Galβ motif [90] | Saliva | ||||
| Secretory immunoglobulin A containing fucose-oligosaccharide motifs [91, 92] | Saliva | ||||
| Salivary agglutinin DMBT1 [93] | Saliva | ||||
| Mucin MUC5AC with N-Acetylgalactosamine-β-1,4-nacetylglucosamine [94, 95] | Gastric mucus | ||||
| Mucin MUC1 [96] | Gastric mucus | ||||
| Mucin MUC2 [97] | Gastric mucus | ||||
| SabA (HopP) | 26695:0725 | 653 | Sialyl-Lewis X, sialyl-Lewis A, Lewis X [37] | Gastric epithelia | Establishment of the T4SS to enhance inflammatory response |
| Salivary mucin MUC7, MUC5B [90, 98] | Saliva | ||||
| Salivary glycoproteins (carbonic anhydrase VI, secretory component, heavy chain of secretory IgA1, parotid secretory protein, zinc α2 glycoprotein) [90] | Saliva | ||||
| Sialylated moieties on the extracellular matrix protein laminin [98] | Gastric epithelia | ||||
| Sialylated structures on the surface of erythrocytes [99] | Erythrocytes | ||||
| Sialylated carbohydrates on neutrophils [100] | Neutrophils | ||||
| HopQ | 26695:1177 | 641 | CEACAMs family [58, 101] | Leukocytes, granulocytes, endothelial and epithelial cells. | Adhesion to host cell, translocation of CagA via the T4SS |
| OipA (HopH) | 26695:0638 | 305 | Not clear | Not clear | Adhesion, induction of inflammatory cytokine production [44] |
| HopZ | 26695:0009 | 672 | Not clear | Not clear | Involved in adhesion |
| HomB | J99:0870 | 669 | Not clear | Not clear | HomB promotes the secretion of proinflammatory cytokine IL-8 and increases the adhesion to host cells |
OMPs, outer membrane proteins; T4SS, type IV secretion system; MUC, mucin; CEACAM, carcinoembryonic antigen–related cell adhesion molecule; IL-8, interleukin-8
Fig. 1.
BabA, SabA, and HopQ bind to respective receptors on mucous layer while the receptors of OipA, HopZ, and Hom remain to be determined. These OMPs promote the adhesion of H. pylori to host cells
Authors’ contributions
Shunfu Xu conceived the study; Chenjing Xu drafted the manuscript; Djaleel Muhammad Soyfoo and Yao Wu modified and polished the manuscript, and all authors commented on the drafts of the manuscript and approved the final draft of the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
There are no ethical issues.
Informed consent
There is no informed consent required for this review article.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chenjing Xu, Email: xuchenjing@njmu.edu.cn.
Djaleel Muhammad Soyfoo, Email: 1241034624@qq.com.
Yao Wu, Email: dr_water@163.com.
Shunfu Xu, Email: xushfu@njmu.edu.cn.
References
- 1.Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology. 2017;153(2):420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Zamani M, Ebrahimtabar F, Zamani V, Miller WH, Alizadeh-Navaei R, Shokri-Shirvani J. Systematic review with meta-analysis: the worldwide prevalence of Helicobacter pylori infection. Aliment Pharmacol Ther. 2018;47(7):868–876. doi: 10.1111/apt.14561. [DOI] [PubMed] [Google Scholar]
- 3.Kim A, Servetas SL, Kang J, Kim J, Jang S, Cha HJ, Lee WJ, Kim J, Romero-Gallo J, Peek RM, Merrell DS. Helicobacter pylori bab paralog distribution and association with cagA, vacA, and homA/B genotypes in American and South Korean clinical isolates. PLoS One. 2015;10(8):e0137078. doi: 10.1371/journal.pone.0137078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalach N, Bontems P (2017) Helicobacter pylori infection in children. Helicobacter. 10.1111/hel.12414 [DOI] [PubMed]
- 5.Boehnke KF, Brewster RK, Sánchez BN, Valdivieso M, Bussalleu A, Guevara M, Saenz CG, Alva SO, Gil E. An assessment of drinking water contamination with Helicobacter pylori in Lima, Peru. Helicobacter. 2018;23(2):e12462. doi: 10.1111/hel.12462. [DOI] [PubMed] [Google Scholar]
- 6.Siavoshi F, Sahraee M, Ebrahimi H, Sarrafnejad A. Natural fruits, flowers, honey, and honeybees harbor Helicobacter pylori-positive yeasts. Helicobacter. 2018;23(2):e12471. doi: 10.1111/hel.12471. [DOI] [PubMed] [Google Scholar]
- 7.Chen Q, Liang X, Long X, Yu L, Liu W. Cost-effectiveness analysis of screen-and-treat strategy in asymptomatic Chinese for preventing Helicobacter pylori-associated diseases. Helicobacter. 2019;24(2):e12563. doi: 10.1111/hel.12563. [DOI] [PubMed] [Google Scholar]
- 8.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511(7507):108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 9.Egan AJF. Bacterial outer membrane constriction. Mol Microbiol. 2018;107(6):676–687. doi: 10.1111/mmi.13908. [DOI] [PubMed] [Google Scholar]
- 10.Waskito LA, Salama NR (2018) Pathogenesis of Helicobacter pylori infection. Helicobacter:e12516. 10.1111/hel.12516 [DOI] [PubMed]
- 11.Koebnik R, Locher KP. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. 2000;37(2):239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 12.Alm RA, Bina J, Andrews BM, Doig P, Hancock RE. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect Immun. 2000;68(7):4155–4168. doi: 10.1128/iai.68.7.4155-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennemann L, Brenneke B, Andres S, Engstrand L, Meyer TF, Aebischer T, Josenhans C. In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori. Infect Immun. 2012;80(12):4364–4373. doi: 10.1128/iai.00977-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oleastro M, Cordeiro R, Ménard A, Yamaoka Y, Queiroz D, Mégraud F. Allelic diversity and phylogeny of homB, a novel co-virulence marker of Helicobacter pylori. BMC Microbiol. 2009;9:248. doi: 10.1186/1471-2180-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey VC, Acio CR, Bredehoft AK, Zhu L, Hallinger DR, Quinlivan-Repasi V, Harvey SE. Repetitive sequence variations in the promoter region of the adhesin-encoding gene sabA of Helicobacter pylori affect transcription. J Bacteriol. 2014;196(19):3421–3429. doi: 10.1128/jb.01956-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moonens K, Gideonsson P, Subedi S, Bugaytsova J, Romaõ E, Mendez M, Nordén J, Fallah M, Rakhimova L, Shevtsova A, Lahmann M, Castaldo G, Brännström K, Coppens F, Lo AW, Ny T, Solnick JV, Vandenbussche G, Oscarson S, Hammarström L, Arnqvist A, Berg DE, Muyldermans S, Borén T. Structural insights into polymorphic ABO glycan binding by Helicobacter pylori. Cell Host Microbe. 2016;19(1):55–66. doi: 10.1016/j.chom.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pride DT, Meinersmann RJ, Blaser MJ. Allelic variation within Helicobacter pylori babA and babB. Infect Immun. 2001;69(2):1160–1171. doi: 10.1128/iai.69.2.1160-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansari S, Kabamba ET, Shrestha PK, Aftab H, Myint T, Tshering L, Sharma RP, Ni N, Aye TT, Subsomwong P, Uchida T, Ratanachu-Ek T, Vilaichone RK, Mahachai V, Matsumoto T, Akada J. Helicobacter pylori bab characterization in clinical isolates from Bhutan, Myanmar, Nepal and Bangladesh. PLoS One. 2017;12(11):e0187225. doi: 10.1371/journal.pone.0187225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilver D, Arnqvis A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279(5349):373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 20.Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002;316(3):640–642. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- 21.Bäckström A, Lundberg C, Kersulyte D, Berg DE, Borén T. Metastability of Helicobacter pylori bab adhesin genes and dynamics in Lewis b antigen binding. Proc Natl Acad Sci U S A. 2004;101(48):16923–16928. doi: 10.1073/pnas.0404817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nell S, Kennemann L, Schwarz S, Josenhans C. Dynamics of Lewis b binding and sequence variation of the babA adhesin gene during chronic Helicobacter pylori infection in humans. mBio. 2014;5(6):e02281–e02214. doi: 10.1128/mBio.02281-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kable ME, Hansen LM, Styer CM, Deck SL, Rakhimova O, Shevtsova A, Eaton KA, Martin ME, Gideonsson P, Borén T. Host determinants of expression of the Helicobacter pylori BabA adhesin. Sci Rep. 2017;7:46499. doi: 10.1038/srep46499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bugaytsova JA, Björnham O, Chernov YA, Gideonsson P, Henriksson S, Mendez M, Sjöström R, Mahdavi J, Shevtsova A, Ilver D, Moonens K, Quintana-Hayashi MP, Moskalenko R, Aisenbrey C, Bylund G, Schmidt A, Åberg A, Brännström K, Königer V, Vikström S, Rakhimova L, Hofer A, Ögren J, Liu H, Goldman MD, Whitmire JM, Ådén J, Younson J, Kelly CG, Gilman RH, Chowdhury A, Mukhopadhyay AK, Nair GB, Papadakos KS, Martinez-Gonzalez B, Sgouras DN, Engstrand L, Unemo M, Danielsson D, Suerbaum S, Oscarson S, Morozova-Roche LA, Olofsson A, Gröbner G, Holgersson J, Esberg A, Strömberg N, Landström M, Eldridge AM, Chromy BA, Hansen LM, Solnick JV, Lindén SK, Haas R, Dubois A, Merrell DS, Schedin S, Remaut H, Arnqvist A, Berg DE. Helicobacter pylori adapts to chronic infection and gastric disease via pH-responsive BabA-mediated adherence. Cell Host Microbe. 2017;21(3):376–389. doi: 10.1016/j.chom.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hage N, Howard T, Phillips C, Brassington C, Overman R, Debreczeni J, Gellert P, Stolnik S, Winkler GS. Structural basis of Lewis(b) antigen binding by the Helicobacter pylori adhesin BabA. Sci Adv. 2015;1(7):e1500315. doi: 10.1126/sciadv.1500315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odenbreit S, Swoboda K, Iris B, Stefan R, Rainer H. Outer membrane protein expression profile in Helicobacter pylori clinical isolates. Infect Immun. 2009;77(9):3782–3790. doi: 10.1128/IAI.00364-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang WL, Yeh YC, Sheu BS. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci. 2018;25(1):68. doi: 10.1186/s12929-018-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Borén T, Haas R, Sasakawa C. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem. 2011;286(28):25256–25264. doi: 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillemin K, Salama NR, Tompkins LS. Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc Natl Acad Sci U S A. 2002;99(23):15136–15141. doi: 10.1073/pnas.182558799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talarico S, Whitefield SE, Fero J, Haas R. Regulation of Helicobacter pylori adherence by gene conversion. Mol Microbiol. 2012;84(6):1050–1061. doi: 10.1111/j.1365-2958.2012.08073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaoka Y. Increasing evidence of the role of Helicobacter pylori SabA in the pathogenesis of gastroduodenal disease. J Infect Dev Ctries. 2008;2(3):174. doi: 10.3855/jidc.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael P, Patricia D, Jennifer S, Beier D. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol Lett. 2004;234(1):51–61. doi: 10.1016/j.femsle.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 33.Loh JT, Gupta SS, Friedman DB, Krezel AM. Analysis of protein expression regulated by the Helicobacter pylori ArsRS two-component signal transduction system. J Bacteriol. 2010;192(8):2034–2043. doi: 10.1128/jb.01703-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang SS, Nguyen ST, Perry AJ, Day CJ, Panjikar S, Tiralongo J, Whisstock JC, T K. The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori. J Biol Chem. 2014;289(10):6332–6340. doi: 10.1074/jbc.M113.513135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcos NT, Magalhães A, Ferreira B, Oliveira MJ, Carvalho AS, Mendes N, Gilmartin T, Head SR, Figueiredo C, David L, Santos-Silva F. Helicobacter pylori induces beta3GnT5 in human gastric cell lines, modulating expression of the SabA ligand sialyl-Lewis x. J Clin Invest. 2008;118(6):2325–2336. doi: 10.1172/jci34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benktander J, Barone A, Johansson MM. Helicobacter pylori SabA binding gangliosides of human stomach. Virulence. 2018;9(1):738–751. doi: 10.1080/21505594.2018.1440171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, Altraja S, Wadström T, Kersulyte D, Berg DE, Dubois A, Petersson C, Magnusson KE, Norberg T, Lindh F, Lundskog BB, Arnqvist A, Hammarström L. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297(5581):573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehours P, Ménard A, Dupouy S, Bergey B, Richy F, Zerbib F, Ruskoné-Fourmestraux A, Delchier JC. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low-grade gastric mucosa-associated lymphoid tissue lymphoma. Infect Immun. 2004;72(2):880–888. doi: 10.1128/iai.72.2.880-888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaoka Y, Ojo O, Fujimoto S, Odenbreit S, Haas R, Gutierrez O, El-Zimaity HM, Reddy R, Arnqvist A. Helicobacter pylori outer membrane proteins and gastroduodenal disease. Gut. 2006;55(6):775–781. doi: 10.1136/gut.2005.083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanai A, Maeda S, Hikiba Y, Shibata W, Ohmae T, Hirata Y, Ogura K, Yoshida H, Omata M. Clinical relevance of Helicobacter pylori sabA genotype in Japanese clinical isolates. J Gastroenterol. 2007;22(12):2228–2232. doi: 10.1111/j.1440-1746.2007.04831.x. [DOI] [PubMed] [Google Scholar]
- 41.Yamaoka Y, Dong HK, Graham DYA. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci U S A. 2000;97(13):7533–7538. doi: 10.1073/pnas.130079797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horridge DN, Begley AA, Kim J, Aravindan N, Fan K, Forsyth MH (2017) Outer inflammatory protein a (OipA) of Helicobacter pylori is regulated by host cell contact and mediates CagA translocation and interleukin-8 response only in the presence of a functional cag pathogenicity island type IV secretion system. Pathog Dis 75(8). 10.1093/femspd/ftx113 [DOI] [PMC free article] [PubMed]
- 43.Farzi N, Yadegar A, Aghdaei HA, Yamaoka Y, Zali MR. Genetic diversity and functional analysis of oipA gene in association with other virulence factors among Helicobacter pylori isolates from Iranian patients with different gastric diseases. Infect Genet Evol. 2018;60:26–34. doi: 10.1016/j.meegid.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 44.Yamaoka Y, Kikuchi S, el-Zimaity HM, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123(2):414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 45.Al-Maleki AR, Loke MF, Lui SY, Ramli NSK, Khosravi Y, Ng CG, Venkatraman G, Goh KL, Ho B, Vadivelu J (2017) Helicobacter pylori outer inflammatory protein A (OipA) suppresses apoptosis of AGS gastric cells in vitro. Cell Microbiol 19(12). 10.1111/cmi.12771 [DOI] [PubMed]
- 46.Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, Wroblewski LE, Piazuelo MB, Correa P, Peek RM. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68(2):379–387. doi: 10.1158/0008-5472.Can-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su B, Ceponis PJM, Sherman PM. Cytoskeletal rearrangements in gastric epithelial cells in response to Helicobacter pylori infection. J Med Microbiol. 2003;52:861–867. doi: 10.1099/jmm.0.05229-0. [DOI] [PubMed] [Google Scholar]
- 48.Moese S, Selbach M, Kwok T, Brinkmann V, König W, Meyer TF. Helicobacter pylori induces AGS cell motility and elongation via independent signaling pathways. Infect Immun. 2004;72(6):3646–3649. doi: 10.1128/iai.72.6.3646-3649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabassam FH, Graham DY, Yamaoka Y. OipA plays a role in Helicobacter pylori-induced focal adhesion kinase activation and cytoskeletal re-organization. Cell Microbiol. 2008;10(4):1008–1020. doi: 10.1111/j.1462-5822.2007.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dabiri H, Maleknejad P, Yamaoka Y, Feizabadi MM, Jafari F, Rezadehbashi M, Nakhjavani FA, Mirsalehian A, Zali MR. Distribution of Helicobacter pylori cagA, cagE, oipA and vacA in different major ethnic groups in Tehran, Iran. J Gastroenterol Hepatol. 2009;24(8):1380–1386. doi: 10.1111/j.1440-1746.2009.05876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, He C, Chen M, Wang Z, Xing C, Yuan Y. Association of presence/absence and on/off patterns of Helicobacter pylori oipA gene with peptic ulcer disease and gastric cancer risks: a meta-analysis. BMC Infect Dis. 2013;13:555. doi: 10.1186/1471-2334-13-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Braga LLBC, Batista MHR, de Azevedo OGR, da Silva Costa KC, Gomes AD, Rocha GA, Queiroz DMM. oipA “on” status of Helicobacter pylori is associated with gastric cancer in North-Eastern Brazil. BMC Cancer. 2019;19(1):48. doi: 10.1186/s12885-018-5249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahboubi M, Falsafi T, Sadeghizadeh M, Mahjoub F. The role of outer inflammatory protein A (OipA) in vaccination of theC57BL/6 mouse model infected by Helicobacter pylori. Turk J Med Sci. 2017;47(1):326–333. doi: 10.3906/sag-1505-108. [DOI] [PubMed] [Google Scholar]
- 54.Yakoob J, Abbas Z, Jafri W, Khan R, Salim SA, Awan S, Abid S, Hamid S, Ahmad Z. Helicobacter pylori outer membrane protein and virulence marker differences in expatriate patients. Epidemiol Infect. 2016;144(10):2200–2208. doi: 10.1017/s095026881600025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohno T, Sugimoto M, Nagashima A, Ogiwara H, Vilaichone RK, Mahachai V, Graham DY, Yamaoka Y. Relationship between Helicobacter pylori hopQ genotype and clinical outcome in Asian and Western populations. J Gastroenterol Hepatol. 2009;24(3):462–468. doi: 10.1111/j.1440-1746.2008.05762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N, Salama N, Churin Y. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell Microbiol. 2013;15(11):1896–1912. doi: 10.1111/cmi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tegtmeyer N, Harrer A, Schmitt V, Singer BB. Expression of CEACAM1 or CEACAM5 in AZ-521 cells restores the type IV secretion deficiency for translocation of CagA by Helicobacter pylori. Cell Microbiol. 2019;21(1):e12965. doi: 10.1111/cmi.12965. [DOI] [PubMed] [Google Scholar]
- 58.Moonens K, Hamway Y, Neddermann M, Reschke M, Tegtmeyer N, Kruse T, Kammerer R, Mejías-Luque R, Singer BB, Backert S, Gerhard M (2018) Helicobacter pylori adhesin HopQ disrupts dimerization in human CEACAMs. EMBO J 37(13). 10.15252/embj.201798665 [DOI] [PMC free article] [PubMed]
- 59.Königer V, Holsten L, Harrison U, Busch B, Loell E, Zhao Q, Bonsor DA, Roth A, Kengmo-Tchoupa A, Smith SI, Mueller S, Sundberg EJ, Zimmermann W, Fischer W, Hauck CR. Erratum: Helicobacter pylori exploits human CEACAMs via HopQ for adherence and translocation of CagA. Nat Microbiol. 2016;2:16233. doi: 10.1038/nmicrobiol.2016.233. [DOI] [PubMed] [Google Scholar]
- 60.Teymournejad O, Mobarez AM, Hassan ZM. Binding of the Helicobacter pylori OipA causes apoptosis of host cells via modulation of Bax/Bcl-2 levels. Sci Rep. 2017;7(1):8036. doi: 10.1038/s41598-017-08176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheu BS, Yang HB, Yeh YC. Helicobacter pylori colonization of the human gastric epithelium: a bug’s first step is a novel target for us. J Gastroenterol Hepatol. 2010;25(1):26–32. doi: 10.1111/j.1440-1746.2009.06141.x. [DOI] [PubMed] [Google Scholar]
- 62.Peck B, Ortkamp M, Diehl KD, Hundt E. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 1999;27(16):3325–3333. doi: 10.1093/nar/27.16.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, Falush D. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci U S A. 2011;108(12):5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Odenbreit S, Kavermann H, Püls J, Haas R. CagA tyrosine phosphorylation and interleukin-8 induction by Helicobacter pylori are independent from alpAB, HopZ and bab group outer membrane proteins. Int J Med Microbiol. 2002;292:257–266. doi: 10.1078/1438-4221-00205. [DOI] [PubMed] [Google Scholar]
- 65.Servetas SL, Kim A, Su H, Cha JH. Comparative analysis of the Hom family of outer membrane proteins in isolates from two geographically distinct regions: the United States and South Korea. Helicobacter. 2018;23(2):e12461. doi: 10.1111/hel.12461. [DOI] [PubMed] [Google Scholar]
- 66.Oleastro M, Cordeiro R, Ménard A, Gomes JP. Allelic diversity among Helicobacter pylori outer membrane protein genes homB and homA generated by recombination. J Bacteriol. 2010;192(15):3961–3968. doi: 10.1128/jb.00395-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim A, Servetas SL, Kang J, Kim J, Jang S, Choi YH, Su H, Jeon YE, Hong YA, Yoo YJ, Merrell DS. Helicobacter pylori outer membrane protein, HomC, shows geographic dependent polymorphism that is influenced by the Bab family. J Microbiol. 2016;54(12):846–852. doi: 10.1007/s12275-016-6434-8. [DOI] [PubMed] [Google Scholar]
- 68.Oleastro M, Monteiro L, Lehours P, Mégraud F. Identification of markers for Helicobacter pylori strains isolated from children with peptic ulcer disease by suppressive subtractive hybridization. Infect Immun. 2006;74(7):4064–4074. doi: 10.1128/iai.00123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oleastro M, Cordeiro R, Yamaoka Y, Queiroz D, Mégraud F, Monteiro L. Disease association with two Helicobacter pylori duplicate outer membrane protein genes, homB and homA. J Gut pathogens. 2009;1(1):12. doi: 10.1186/1757-4749-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jung SW, Sugimoto M, Graham DY. homB status of Helicobacter pylori as a novel marker to distinguish gastric cancer from duodenal ulcer. J Biol Chem. 2009;47(10):3241–3245. doi: 10.1128/jcm.00293-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang J, Jones KR, Jang S, Olsen CH, Yoo YJ, Merrell DS. The geographic origin of Helicobacter pylori influences the association of the homB gene with gastric cancer. J Clin Microbiol. 2012;50(3):1082–1085. doi: 10.1128/jcm.06293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng M, Mao XH, Li JX, Tong WD, Wang B, Zhang YJ, Guo G, Zhao ZJ, Li L, Wu DL, Lu DS, Tan ZM, Liang HY, Wu C, Li DH, Luo P, Zeng H, Zhang WJ, Zhang JY, Guo BT, Zhu FC. Efficacy, safety, and immunogenicity of an oral recombinant Helicobacter pylori vaccine in children in China: a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2015;386(10002):1457–1464. doi: 10.1016/s0140-6736(15)60310-5. [DOI] [PubMed] [Google Scholar]
- 73.Fallone CA, Moss SF, Malfertheiner P. Reconciliation of recent Helicobacter pylori treatment guidelines in a time of increasing resistance to antibiotics. Gastroenterology. 2019;157(1):44–53. doi: 10.1053/j.gastro.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 74.Minhu C, Adrian L, Stuart H (1992) Immunisation against gastric helicobacter infection in a mouse/Helicobacter felis model. Lancet. 10.1016/0140-6736(92)90720-N
- 75.Mirzaei N, Poursina F, Moghim S, Rashidi N, Safaei HG. The study of H. pylori putative candidate factors for single- and multi-component vaccine development. Crit Rev Microbiol. 2017;43(5):631–650. doi: 10.1080/1040841X.2017.1291578. [DOI] [PubMed] [Google Scholar]
- 76.Larena M, Holmgren J, Lebens M, Terrinoni M. Cholera toxin, and the related nontoxic adjuvants mmCT and dmLT, promote human Th17 responses via cyclic AMP-protein kinase A and inflammasome-dependent IL-1 signaling. J Immunol. 2015;194(8):3829–3839. doi: 10.4049/jimmunol.1401633. [DOI] [PubMed] [Google Scholar]
- 77.Maisonneuve C, Bertholet S, Philpott DJ, De GE. Unleashing the potential of NOD- and Toll-like agonists as vaccine adjuvants. Proc Natl Acad Sci U S A. 2014;111(34):12294–12299. doi: 10.1073/pnas.1400478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He P, Zou Y, Hu Z. Advances in aluminum hydroxide-based adjuvant research and its mechanism. Hum Vaccin Immunother. 2015;11(2):477–488. doi: 10.1080/21645515.2014.1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keikha M, Eslami M, Yousefi B, Ghasemian A. Potential antigen candidates for subunit vaccine development against Helicobacter pylori infection. J Cell Physiol. 2019;234(12):21460–21470. doi: 10.1002/jcp.28870. [DOI] [PubMed] [Google Scholar]
- 80.Ebbensgaard A, Mordhorst H, Aarestrup FM, Hansen EB. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front Microbiol. 2018;9:2153. doi: 10.3389/fmicb.2018.02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barbosa AS, Monaris D, Silva LB, Morais ZM, Vasconcellos SA, Cianciarullo AM, Isaac L. Functional characterization of LcpA, a surface-exposed protein of Leptospira spp. that binds the human complement regulator C4BP. Infect Immun. 2010;78(7):3207–3216. doi: 10.1128/iai.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu C, Pan X, Xia B, Chen F, Jin Y, Bai F, Priebe G, Cheng Z, Jin S. Construction of a protective vaccine against lipopolysaccharide-heterologous Pseudomonas aeruginosa strains based on expression profiling of outer membrane proteins during infection. Front Immunol. 2018;9:1737. doi: 10.3389/fimmu.2018.01737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tohidi F, Teymournejad O, Taravati A, Ahmadi KJA. Analysis of important H. pylori outer membrane proteins by detection of common sequences in exposed areas; in silico study. Biosci Biotechnol Res Asia. 2015;12(Spl. Edn. 2):135–143. doi: 10.13005/bbra/2020. [DOI] [Google Scholar]
- 84.Ofek I, Hasty DL, Sharon N. Anti-adhesion therapy of bacterial diseases: prospects and problems. Med Microbiol Immunol. 2003;38(3):181–191. doi: 10.1016/s0928-8244(03)00228-1. [DOI] [PubMed] [Google Scholar]
- 85.Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794(5):808–816. doi: 10.1016/j.bbapap.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen J, Li N, She F. Helicobacter pylori outer inflammatory protein DNA vaccine-loaded bacterial ghost enhances immune protective efficacy in C57BL/6 mice. Vaccine. 2014;32(46):6054–6060. doi: 10.1016/j.vaccine.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 87.George L, Goldberg JB (2012) Outer membrane biogenesis in Escherichia coli, Neisseria meningitidis, and Helicobacter pylori: paradigm deviations in H. pylori. Front Cell Infect Microbiol 2:–29. 10.3389/fcimb.2012.00029 [DOI] [PMC free article] [PubMed]
- 88.Peck B, Ortkamp M, Nau U, Niederweis M, Hundt E. Characterization of four members of a multigene family encoding outer membrane proteins of Helicobacter pylori and their potential for vaccination. Microbes Infect. 2001;3(3):171–179. doi: 10.1016/s1286-4579(01)01377-6. [DOI] [PubMed] [Google Scholar]
- 89.Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikström S, Sjöström R, Lindén S, Bäckström A, Lundberg C, Arnqvist A, Mahdavi J, Nilsson UJ, Velapatiño B, Gilman RH, Gerhard M, Alarcon T, López-Brea M, Nakazawa T, Fox JG, Correa P, Dominguez-Bello MG, Perez-Perez GI, Blaser MJ, Normark S, Carlstedt I, Oscarson S, Teneberg S, Berg DE. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science. 2004;305(5683):519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- 90.Walz A, Odenbreit S, Stühler K, Wattenberg A, Meyer HE, Mahdavi J, Borén T. Identification of glycoprotein receptors within the human salivary proteome for the lectin-like BabA and SabA adhesins of Helicobacter pylori by fluorescence-based 2-D bacterial overlay. Proteomics. 2009;9(6):1582–1592. doi: 10.1002/pmic.200700808. [DOI] [PubMed] [Google Scholar]
- 91.Borén T, Falk P, Roth KA, Larson G. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262(5141):1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 92.Royle L, Roos A, Harvey DJ, Wormald MR, van Gijlswijk-Janssen D, Redwan e-RM, Wilson IA, Daha MR, Dwek RA. Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J Biol Chem. 2003;278(22):20140–20153. doi: 10.1074/jbc.M301436200. [DOI] [PubMed] [Google Scholar]
- 93.Issa S, Moran AP, Ustinov SN, Lin JH, Ligtenberg AJ. O-linked oligosaccharides from salivary agglutinin: Helicobacter pylori binding sialyl-Lewis x and Lewis b are terminating moieties on hyperfucosylated oligo-N-acetyllactosamine. Glycobiology. 2010;20(8):1046–1057. doi: 10.1093/glycob/cwq066. [DOI] [PubMed] [Google Scholar]
- 94.Lindén S, Mahdavi J, Hedenbro J, Borén T, Carlstedt I. Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins. Biochem J. 2004;384:263–270. doi: 10.1042/bj20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kenny DT, Skoog EC, Lindén SK, Struwe WB, Rudd PM. Presence of terminal N-acetylgalactosamineβ1-4N-acetylglucosamine residues on O-linked oligosaccharides from gastric MUC5AC: involvement in Helicobacter pylori colonization? Glycobiology. 2012;22(8):1077–1085. doi: 10.1093/glycob/cws076. [DOI] [PubMed] [Google Scholar]
- 96.Lindén SK, Sheng YH, Every AL, Miles KM, Skoog EC, Florin TH, Sutton P. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009;5(10):e1000617. doi: 10.1371/journal.ppat.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cohen M, Drut R, Cueto RE. SIALYL-Tn antigen distribution in Helicobacter pylori chronic gastritis in children: an immunohistochemical study. Pediatr Pathol Mol Med. 2003;22(2):117–129. doi: 10.1080/pdp.22.2.117.129. [DOI] [PubMed] [Google Scholar]
- 98.Walz A, Odenbreit S, Mahdavi J, Borén T. Identification and characterization of binding properties of Helicobacter pylori by glycoconjugate arrays. Glycobiology. 2005;15(7):700–708. doi: 10.1093/glycob/cwi049. [DOI] [PubMed] [Google Scholar]
- 99.Aspholm M, Olfat FO, Nordén J, Sondén B, Lundberg C, Sjöström R, Altraja S, Odenbreit S, Haas R, Wadström T, Engstrand L, Semino-Mora C, Liu H, Dubois A, Teneberg S, Arnqvist A. SabA is the H. pylori hemagglutinin and is polymorphic in binding to sialylated glycans. PLoS Pathog. 2006;2(10):e110. doi: 10.1371/journal.ppat.0020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Unemo M, Aspholm-Hurtig M, Ilver D, Bergström J, Borén T, Danielsson D. The sialic acid binding SabA adhesin of Helicobacter pylori is essential for nonopsonic activation of human neutrophils. J Biol Chem. 2005;280(15):15390–15397. doi: 10.1074/jbc.M412725200. [DOI] [PubMed] [Google Scholar]
- 101.Matsuo Y, Kido Y, Yamaoka Y (2017) Helicobacter pylori outer membrane protein-related pathogenesis. Toxins 9(3). 10.3390/toxins9030101 [DOI] [PMC free article] [PubMed]