Abstract
Cumulative evidence shows a linkage between gut microbiota pattern and depression through the brain-gut microbiome axis. The aim of this systematic review was to identify the alterations of the gut microbiota patterns in people with depression compared to healthy controls. A comprehensive literature search of human studies, published between January 2000 and June 2019, was reviewed. The key words included gastrointestinal microbiome, gut microbiome, microbiota, depression, depressive symptoms, and depressive disorder. The systematic review adhered to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. Nine articles met the eligibility criteria. Disparities in α-diversity and β-diversity of the microbiota existed in people with depression compared to healthy controls. At the phylum level, there were inconsistencies in the abundance of Firmicutes, Bacteroidetes, and Proteobacteria. However, high abundance in Actinobacteria and Fusobacteria phyla were observed in people with depression. On the family level, high abundance of Actinomycineae, Coriobacterineae, Bifidobacteriaceae, Clostridiales incertae sedis XI, Porphyromonadaceae, Clostridiaceae, Lactobacillaceae, Streptococcaceae, Eubacteriaceae, Thermoanaerobacteriaceae, Fusobacteriaceae, Nocardiaceae, Streptomycetaceae, and low abundance of Veillonellaceae, Prevotellaceae, Bacteroidaceae, Sutterellaceae, Oscillospiraceae, Marniabilaceae, and Chitinophagaceae were observed in people with depression. On the genus level, high abundance of Oscillibacter, Blautia, Holdemania, Clostridium XIX, Anaerostipes, Anaerofilum, Streptococcus, Gelria, Turicibacter, Parabacteroides, Eggerthella, Klebsiella, Paraprevotella, Veillonella, Clostridium IV, Erysipelotrichaceae incertae sedis, Eubacterium, Parvimonas, Desulfovibrio, Parasutterella, Actinomyces, Asaccharobacter, Atopobium, Olsenella and low abundance of Coprococcus, Lactobacillus, Escherichia/Shigella, Clostridium XlVa, Dialister, Howardella, Pyramidobacter, and Sutterella were found in people with depression. Alteration of gut microbiome patterns was evident in people with depression. Further evidence is warranted to allow for the translation of microbiome findings toward innovative clinical strategies that may improve treatment outcomes in people with depression.
Keywords: gastrointestinal microbiome, gut microbiome, microbiota, depression, major depressive disorder
Introduction
Depression affects more than 300 million people of all ages globally (1), and is one of the leading causes of psychiatric disability (2). Outcomes of current depression psychotherapies are suboptimal with treatment failures common (3). Research shows that one in two patients does not sufficiently respond to depression treatment, 40% of responders relapse, and many non-responders deteriorate despite treatment (2, 4). Failure to enhance the treatment outcomes reflects the need for further research related to the pathophysiology of depression.
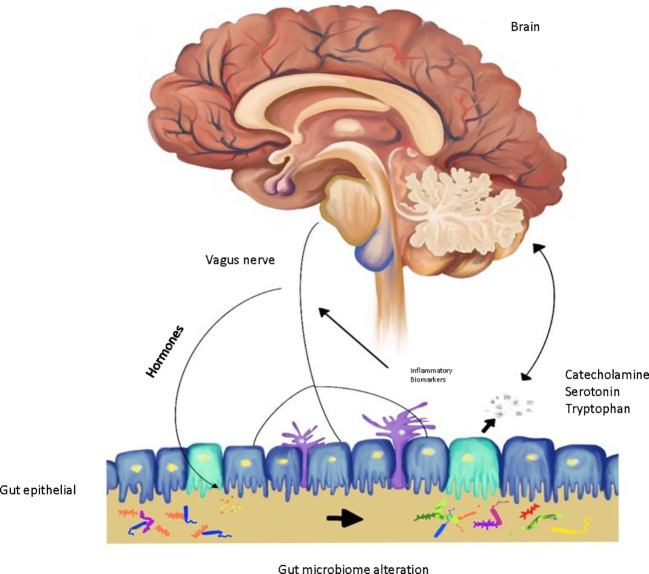
Traditionally, the theories behind the pathophysiology of depression are attributed to neurotransmitters, stress hormones, neurotrophic factors, and circadian rhythms (5). The monoaminergic system plays an important role in depression with multiple interactions in the central nervous system (CNS) function (6). In recent studies, the gut microbiota has been introduced as a novel area of investigation of depression pathophysiology (2). The gut microbiota is composed of over 100 trillion microorganisms that affect host homeostasis (7–10). Gut microbes in the brain-gut axis communicate to the CNS via endocrine, nervous, and immune signaling mechanisms. The brain may affect the community pattern and function of the gut microbiota throughout the autonomic nervous system (ANS) through modulation of the intestinal transit via secretion, regional gut motility, and gut permeability (11) (Figure 1).
Figure 1.
Gut-brain microbiome axis in depression.
Numerous studies have investigated the underlying mechanisms of depression through the brain-gut microbiota axis. Changes in gut microbiota patterns result in immune activation through the bidirectional interactions between the gut and the brain, potentially yielding the generation of various types of psychiatric symptoms (12). Some animal studies implicate fecal “depression microbiota” transplantation modulates depression-like behavior in germ-free (GF) mice (13). Moreover, human studies demonstrated the lower abundance of Lactobacillus and Bifidobacterium in persons with depression (14). Anti-inflammatory effects of Lactobacillus and Bifidobacterium may explain the role of gut microbiota in this trajectory (14). Despite current theories that propose the role of gut microbiota in the pathophysiology of depression, the mechanism by which gut microbiota modulate depression-like behaviors remains controversial.
Differences in the microbiota community at various taxonomic levels have been reported in study participants living with depression compared to those without depression (15–18). The studies observed various gut microbiota compositions in persons with depression. However, the reported results are inconclusive and require definitive distinctions (18). The purpose of this systematic review is to summarize the primary results from recent human studies and provide evidence for future research in finding the patterns of the gut microbiota in people with depression.
Materials and Methods
Study Selection Criteria
The targeted study population for this review was people with depression, depressive symptoms, or major depression. The primary outcome measures were gut microbiome composition, including the α-diversity and β-diversity, and the abundance of bacteria at phylum, family, and genus taxonomic levels. The articles eligible for this review paper were original research articles, human research studies conducted in adults, written or available in English, and published between January 2000 and June 2019. All study designs from any country were included. Manuscripts were excluded if they were letters, book chapters, reviews, theses/dissertations, secondary data analyses, or case studies. Studies of people with depression and other chronic or comorbid disorders such as irritable bowel syndrome, inflammatory bowel disease (IBD), anorexia nervosa, and/or cancer were excluded.
Information Sources and Search Strategies
The online databases of PubMed, CINAHL, Psych Info, and Scopus were searched for relations between depression and gut microbiome patterns with the terms gastrointestinal microbiome, gut microbiome, microbiota, gut flora, depression, depressive symptoms, depressive disorder, and major depressive disorder.
Study Selection
The irrelevant articles, based on the titles, were excluded and the remainder were screened for relevance after review of the abstract and full-text independently by two researchers.
Quality Control of Selected Studies
The methodological quality of the included studies was evaluated with a modified Newcastle Ottawa scale for cross-sectional studies (19). This scale was used to assess the quality of cross-sectional studies. The assessed criteria include selection, comparability, and outcomes. The maximum score that represents the highest possible quality is 10.
Data Analysis
The findings of gut microbiome patterns in persons with depression were categorized based on the bacteria diversity and taxonomic levels. Thus, the gut microbiome was grouped based on α-diversity (the richness and evenness of the microbial community), β-diversity (the compositional dissimilarity among the microbiome community), the abundance and proportion of bacteria at phylum, family, and genus levels.
Results
Selection of Literature
The initial online database search produced 616 articles, of which 98 were duplicates, and 440 excluded with no related outcome. The full-text articles assessed for eligibility were 78, of which 69 were ineligible because of comorbidity and nine full-text were eligible for the systematic review (Figure S1).
Characteristics of the Samples and Methodologies Used
The subjects of the studies were adults with major depressive disorder and healthy controls aged ≥18 years old. Depression was diagnosed clinically using Mini-International Neuropsychiatric Interview (MINI), Diagnostic and Statistical Manual of Mental Disorders (DSM-IV), Diagnostic and Statistical Manual of Mental Disorders Text Revision (DSM-IV-TR), and International Classification of Diseases (ICD-10). Depression severity was assessed with a variety of standardly accepted instruments including Hamilton Depression Rating Scale (HDRS), Montgomery–Åsberg Depression Rating Scale (MADRS), or Beck Depression Inventory. The selected studies compared the fecal gut microbiota composition between people with depression and healthy controls. Six studies were conducted in China, one in Norway, one in Ireland, and one in Japan. The study design for eight studies was cross-sectional, and one was a partially blinded observational study.
The microbiome analysis involved sequencing of hypervariable regions V1–V5 of the 16S rRNA genes. Different primers were used in the studies including bar-coded universal primers containing linker sequences for 454-pyrosequencing, 27F and 533R primers. Techniques including polymerase chain reaction (PCR), reverse transcription-quantitative PCR (RT-qPCR), pyrosequencing, Illumina sequencing, and Illumina sequencing MiSeq platform were utilized for DNA sequencing. Specifically, comparative meta-proteomics analysis by phylogenetic analysis of the bacterial peptides was utilized in one study (20). Sequential clustering of operational taxonomic units (OTUs) was identified at 95–100% nucleotide similarity to calculate the relative abundances of microbiota at different Levels (15–17, 21–25). More than 80% of the articles were published within the years 2016 to 2018.
Alteration of the Gut Microbiome Community in Depression
Altered Gut Microbiota Diversity and Richness in Depression
Microbial α-diversity was reported in six studies by using different indices and methods, such as Shannon index, Simpson index, phylogenetic diversity, total observed species and Chao 1 (26). Three studies reported contradictory results regarding α-diversity within bacterial community based on the Shannon index. Jiang et al. observed higher α-diversity, whereas Liu et al. reported lower α-diversity and Kelly et al. indicated no significant difference in this index (15, 16, 23). In terms of measuring α-diversity using other methods, Naseribafrouei et al. found no significant differences between people with depression and healthy controls with respect to Simpson’s index (25). Kelly et al. reported a decrease in total observed species and Chao 1 richness in people with depression (16). Zheng et al. and Chen et al. indicated no significant difference between people with depression and healthy people with respect to phylogenetic diversity (22, 24). However, Kelly et al. reported a decrease in phylogenetic diversity in people with depression (16).
Gut microbial β-diversity, measured using the Bray-Curtis Dissimilarity Index, was reported in three studies. Kelly et al. and Zheng et al. reported significant differences in β-diversity between people with depression and healthy controls. However, Jiang et al. did not obtain an estimate of this index due to a significant inter-individual variability (15, 16, 24). Therefore, no consistent directional alteration of the microbial diversity was found in people with depression compared to healthy controls.
Altered Gut Microbiota Composition in Depression
On the phylum level, five phyla including Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and Fusobacteria were reported in the seven studies with contradictory results in their abundance between people with depression and healthy control groups. Chen et al. and Lin et al. reported higher abundance of Firmicutes, whereas, Jiang et al. and Liu et al. found lower abundance of this phylum (15, 17, 22, 23). Interestingly, Zheng et al. reported no difference of Firmicutes phylum between people with depression and healthy controls (24). Bacteroidetes as another phylum was mostly reported in the studies. Jiang et al. and Liu et al. reported high abundance of this phylum, whereas Naseribafrouei et al., Zheng et al., Lin et al., Chen et al., and Chen et al. found lower abundance of this phylum in people with depression (15, 20, 22, 24, 25). Higher abundance of Actinobacteria were reported by Jiang et al., Zheng et al., Chen et al., and Chen et al. (15, 20, 22, 24) in people with depression. While, Jiang et al. reported high abundance of Proteobacteria, Chen et al. found lower abundance of this phylum in people with depression. Also, Jiang et al. reported high abundance of Fusobacteria in people with depression.
On the family level, studies reported higher abundance of 13 families including Actinomycineae, Coriobacterineae, Bifidobacteriaceae, Clostridiales incertae sedis XI, Porphyromonadaceae, Clostridiaceae, Lactobacillaceae, Streptococcaceae, Eubacteriaceae, Thermoanaerobacteriaceae, Fusobacteriaceae, Nocardiaceae, and Streptomycetaceae and lower abundance of seven families including Veillonellaceae, Prevotellaceae, Bacteroidaceae, Sutterellaceae, Oscillospiraceae, Marniabilaceae, and Chitinophagaceae in people with depression (15, 16, 20, 22, 24). Contradictory results were found on the abundance of six families, namely, Lachnospiraceae, Ruminococcaceae, Acidaminococcaceae, Enterobacteriaceae, Erysipelotrichaceae, and Rikenellacea in people with depression (15, 16, 20, 22, 24, 25). On the genus level, studies reported high abundance of 26 genera including Oscillibacter, Blautia, Holdemania, Clostridium XIX, Anaerostipes, Lachnospiracea incertae sedis, Anaerofilum, Streptococcus, Gelria, Turicibacter, Parabacteroides, Eggerthella, Klebsiella, Streptococcus, Paraprevotella, Veillonella, Clostridium IV, Erysipelotrichaceae incertae sedis, Eubacterium, Parvimonas, Desulfovibrio, Parasutterella, Actinomyces, Asaccharobacter, Atopobium, and Olsenella and a low abundance of eight genera including Coprococcus, Lactobacillus, Escherichia/Shigella, Clostridium XlVa, Dialister, Howardella, Pyramidobacter, and Sutterella in people with depression (15, 16, 22–24). However, a lack of congruence existed across investigations for the abundance of Bifidobacterium, Roseburia, Lachnospiracea incertaesedis, Megamonas, Clostridium XI, Bacteroides, Prevotella, Alistipes, Phascolarctobacterium, Faecalibacterium, and Ruminococcus (15, 17, 23, 24) (Tables 1 and 2).
Table 1.
Studies on gut microbiome composition in depression.
| Authors/Year Country |
Study Design | Study Subjects | Biospecimen | Diagnosis/Severity of Depression Scales | Mean severity of depression at baseline | Microbiological Analysis | Total score of quality assessment (0–10) |
|---|---|---|---|---|---|---|---|
| Jiang et al. (15) China |
Cross-sectional | n = 29 patients with depression [Active-Major Depressive Disorder, (A-MDD)] n = 17 patients during response to antidepressant treatment [Responded-Major Depressive Disorder (R-MDD)] n = 30 healthy controls Age: 18–40 years |
1) Fecal samples 2) Blood samples (Serum tumor necrosis factor-a (TNF-a), interleukin-1b (IL)-1b, IL-6, and brain-derived neurotrophic factor (BDNF) levels |
Diagnostic and Statistical Manual of Mental Disorders Fourth Edition (DSM IV), Hamilton’s Depression Scale (HAMDS), Montgomery–Asberg Depression Rating Scale (MADRS) |
HAMDS: 29.8 MADRS: 27.4 |
16S rRNA genes (V1–V3 regions), PCR, Pyrosequencing. α-diversity, ß-diversity, bacterial taxonomic composition | 7 |
| Kelly et al. (16) Ireland |
Cross-sectional | n = 34 patients with depression n = 33 healthy controls Mean age: 45.8 |
|
Diagnostic and Statistical Manual of Mental Disorders (DSM IV)/Hamilton rating scale for Depression (HAMD 17)/Beck Depression Inventory | HAMD: 19.5 Beck Depression: 32.4 |
16S rRNA genes, illumina MiSeq platform. α-diversity, ß-diversity, bacterial taxonomic composition | 6 |
| Aizawa et al. (21) Japan |
Cross-sectional | n = 43 patients with depression n = 57 healthy controls Mean age: 40 years |
Fecal samples | Diagnostic and Statistical Manual of Mental Disorders, 4th ed. (DSM-IV), Hamilton depression rating scale | HAM-D: 16.9 | 16S rRNA genes, Reverse Transcription quantitative PCR, bacterial taxonomic composition | 5 |
| Lin et al. (17) China |
Cross-sectional | n = 60 patients with depression n = 60 healthy controls Age: 20–85 years |
Fecal samples | Diagnostic and Statistical Manual of Mental Disorders (DSM-IVTR), Hamilton depression rating scale (HAM-D) scale | HAM-D ≥23 | 16S rRNA genes (V3-V4), Illumina MiSeq platform. bacterial taxonomic composition | 6 |
| Naseribafrouei et al. (25) Norway |
Partially blinded observational study |
n = 37, patients with depression n = 18 healthy controls Mean age: 48 years |
Fecal samples | ICD-10, Montgomery-Asberg Depression Rating Scale (MADRS) | MADRS: 26.3 |
16S rRNA genes, PCR, Illumina sequencing. α-diversity, ß-diversity, bacterial taxonomic composition | 5 |
| Chen et al. (20) China |
Cross-sectional | n = 10 people with depression n = 10 healthy controls Age:18–65 years |
Fecal samples | Diagnostic and Statistical Manual of Mental Disorders, 4th ed. (DSM-IV), Hamilton’s Depression Scale | HAMDS: 25.6 | Protein, Liquid chromatography-tandem mass spectrometry | 6 |
| Liu et al. (23) China |
Cross-sectional | n = 40 IBS-D patients n = 15 people with depression n = 25 comorbidity patients with both IBS and depression n = 20 healthy controls Age >18 years |
Fecal samples | Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV)/Self-Rating Depression Scale (SDS) | SDS:60 | 16S rRNA genes (V1 to V3 region), PCR, Pyro sequencing. α-diversity, ß-diversity, Bacterial taxonomic composition | 7 |
| Zheng et al. (24) China |
Cross-sectional | n = 58 patients with depression, n = 63 healthy controls; age >18 years | Fecal samples | DSM-IV-TR, Hamilton Depression Rating Scale | HDRS: 22.8 | 16S rRNA genes (V3-V5 regions), PCR, illumina sequencing. α-diversity, ß-diversity, bacterial taxonomic composition | 7 |
| Chen et al. (22) China |
Cross-sectional | n = 24 people with depression, n = 24 healthy controls; age >18 years old (18–60) | Fecal samples | Diagnostic and Statistical Manual of Mental Disorders IV, Hamilton Depression Rating Scale (HDRS-17) | HDRS: 23.04 | 16S rRNA genes (V3–V5 regions), PCR, Pyro sequencing. α-diversity, ß-diversity, Bacterial taxonomic composition | 6 |
Table 2.
Fecal bacteria diversity and abundance in people with depression.
| Alpha diversity | Beta diversity | Phylum | Family | Genus |
|---|---|---|---|---|
| Shannon index: Jiang ↑ Liu ↓ Kelly ↔ |
Jiang ↔ Kelly * Zheng * |
Firmicutes Jiang ↓ Zheng ↔ Lin ↑ Liu ↓ Zhi Chen ↑ |
Acidaminococcaceae Jiang ↑, Zheng ↓ |
Phascolarctobacterium Jiang ↑, Zheng ↓ |
| Erysipelotrichaceae Jiang ↓, Zhi Chen ↑ |
Holdemania Kelly ↑ |
|||
| Simpson index: Naseribafrouei ↔ Total observed species: Kelly ↓ |
Lachnospiraceae Jiang ↓, Naserabadi ↓, Zheng ↓, Zhi Chen ↑, Chen ↑ |
Clostridium XIX Jiang ↑ (A-MDD) Anaerostipes Zheng ↑, Chen ↑ Lachnospiracea incertae sedis Jiang ↑ (A-MDD) Roseburia Jiang ↑, Zheng ↓, Chen ↑ |
||
| Chao 1: Kelly ↓ Phylogenetic diversity: Zheng ↔, Chen ↔, Kelly ↓ |
Ruminococcaceae Jiang ↓, Zheng ↑, Zhi Chen ↑, Chen ↑ |
Faecalibacterium Jiang ↓, Zheng ↓, Zhi Chen ↓, Chen ↑ Ruminococcus Jiang ↓, Zheng ↑ Anaerofilum Kelly↑ |
||
|
Veillonellaceae Jiang ↓, Zheng ↓ |
||||
|
Lactobacillaceae Zheng ↑ |
Lactobacillus Aizawa ↓ Blautia Jiang ↑, Zheng ↑, Chen ↑ Clostridium XIX Jiang ↑ Lachnospiracea incertae sedis Jiang ↑, Liu ↓ Coprococcus Zheng ↓, Liu ↓ Clostridium XlVa Zheng ↓, Liu ↓ |
|||
|
Streptococcaceae Zheng ↑ |
Streptococcus Lin ↑ |
|||
|
Eubacteriaceae Zheng ↑ |
||||
|
Erysipelotrichaceae incertae sedis Zheng ↑ |
||||
|
Thermoanaerobacteriaceae Kelly ↑ |
Gelria Kelly ↑ |
|||
| Selenomonadaceae |
Megamonas Jiang ↑, Zheng ↓ |
|||
| Oscillospiraceae Zhi Chen ↓ |
Oscillibacter Jiang ↑ (A-MDD), Naserabadi ↑ |
|||
| Dialisteraceae |
Dialister Jiang ↓, Kelly ↓ |
|||
|
Clostridiaceae Zhi Chen ↑ |
Clostridium XI Lin ↑, Liu ↓ |
|||
| Turicibacteraceae |
Turicibacter Kelly ↑ |
|||
|
Bacteroidetes Jiang ↑, Liu↑, Naserabadi ↓, Zheng ↓, Lin ↓, Chen ↓, Zhi Chen ↓ |
Porphyromonadaceae Jiang ↑, Zhi Chen ↑ |
|||
|
Rikenellaceae Jiang ↑, Zheng ↓, Zhi Chen ↓ |
Alistipes Jiang ↑, Naserabadi ↑, Zheng ↓ |
|||
|
Bacteroidaceae Jiang ↓ (A-MDD), Zheng ↓ |
Bacteroides Jiang ↓ (A-MDD), Chen ↑, Liu↑ Prevotella Jiang ↓, Kelly ↓, Liu↑, Lin↑ Paraprevotella Liu↑, Kelly ↑ |
|||
|
Chitinophagacea Marniabilaceae Zhi Chen ↓ Prevotellaceae Jiang ↓, Kelly ↓, Zhi Chen ↓ |
||||
|
Clostridiales incertae sedis XI Zheng ↑ |
||||
|
Lactobacillus Aizawa ↓ |
||||
| Tannerellaceae |
Parabacteroides Jiang ↑ |
|||
|
Actinobacteria Jiang ↑, Chen ↑, Zheng ↑, Zhi Chen ↑ |
Actinomycineae Zheng ↑, Zhi Chen ↑ |
|||
|
Coriobacterineae Zheng ↑, Chen ↑ |
||||
|
Bifidobacteriaceae Zhi Chen ↑ |
Bifidobacterium Aizawa ↓, Chen ↑ |
|||
| Eggerthellaceae Nocardiaceae Zhi Chen ↑ Streptomycetaceae Zhi Chen ↑ |
Eggerthella Kelly ↑, Chen ↑ |
|||
|
Proteobacteria Jiang ↑, Zhi Chen ↓ |
Enterobacteriaceae Jiang ↑, Zhi Chen ↓ |
Escherichia/Shigella Jiang ↓ (R-MDD) Klebsiella Lin↑ |
||
|
Sutterellaceae Zheng ↓, Zhi Chen ↓ |
||||
| Burkholderiaceae |
Parasutterella Jiang ↑ (A-MDD) |
|||
|
Fusobacteria Jiang ↓ (R-MDD), ↑ (A-MDD) |
Fusobacteriaceae Jiang ↑ (A-MDD) |
↑, high abundance in people with depression; ↓, low abundance in people with depression; ↔, direction not deciphered; *, significant difference; A-MDD, active-MDD; R-MDD, recovering-MDD.
Altered Gut Microbiota Composition Association With Antidepressant Medications
Some studies addressed the effects of antidepressant medications on microbiome composition. Jiang et al. observed a high abundance of Bacteroidetes and Proteobacteria and a low abundance of Firmicutes, Actinobacteria, and Fusobacteria phyla in response to antidepressant treatment with selective serotonin reuptake inhibitors (SSRIs) or serotonin–norepinephrine reuptake inhibitors (SNRIs) (15). On the family level, higher abundance of Bacteroidaceae, Acidaminococcaceae, Porphyromonadaceae, Enterobacteriaceae, and Rikenellaceae were reported in response to anti-depressant medications in people with depression compared to healthy control group. Additionally, five genera, including Alistipes, Bacteroides, Parabacteroides, Phascolarctobacterium, and Roseburia, increased in in people with depression after antidepressant treatment (15). Lin et al. (17) evaluation of the bacterial composition at the phylum level in people with depression under treatment of escitalopram (SSRIs) across three visits for one month showed no differences in the microbial community during the three different visits. Also, Zheng et al. reported no significant correlation between antidepressant treatment and bacterial composition, although the majority of people with depression were drug-naïve and the remaining participants used SSRIs, SNRIs, or tricyclic antidepressants (TCAs) (24). Similarly, Aizawa et al. observed no significant correlation between anti-depressant medication (Imipramine) dosage and bacterial counts (21).
Discussion
The systematic review revealed some differences in the gut microbiota diversity, the richness and evenness of microbes were different in people living with depression as compared to healthy adults. No consensus in the α-diversity and β-diversity was evident. Additionally, different quantities of bacterial abundance were present at the family and genus levels. There was no consensus in abundance of Firmicutes even though most studies reported bacteria at family and genus levels belonged to the Firmicutes phylum among people with depression. Moreover, decrease in Bacteriodetes and increase in Actinobacteria abundance were observed in people with depression in the reviewed studies. The most common bacteria of interest were Firmicutes, followed closely at both family and genus level by Bacteriodetes and Actinobacteria phyla.
The findings from the systematic review illustrated inconsistencies in in the abundance of Firmicutes, a lower abundance of Bacteroidetes and a higher abundance of Actinobacteria phyla among people with depression. Firmicutes and Bacteriodetes, as the two major phyla in fecal microbial flora showed altered abundance and correlated to inflammatory conditions such as IBDs (17). A low abundance of Bacteroidetes has also been associated with obesity (25). Recently, research shows that colonization of GF mice from human depressed microbiota characterized by alterations in Firmicutes, Actinobacteria, and Bacteroidetes induced depression-like behavior in mice (24).
There was inconsistency among the abundance of bacteria at different taxonomic levels. High abundance of Oscillibacter, Parabacteroide, Klebsiella, Paraprevotella, Veillonella, Desulfovibrio, Parasutterella, and Paraprevotella as a gram-negative bacteria in people with depression may explain the contribution of microbiota in development/maintenance of depression. Gram-negative bacteria contain lipopolysaccharides (LPS) in the leaflet of the outer cell membrane (27). Studies show that LPS interacts with macrophages and stimulates the immune response through the release of pro-inflammatory cytokines (27). Some studies support this mechanism by reporting increased levels of proinflammatory cytokines interleukin, including IL-1β and IL-6, and decreased levels of anti-inflammatory cytokines, including IL-4 and IL-10 in people living with depression (28, 29). Thus, the gut microbiota alterations may modulate the inflammatory response in people with depression by attenuation of pro-inflammatory cytokines (30–33). Research indicate that the administration of probiotics such as Lactobacillus rhamnosus modulates the immune system through the prevention of the induction of IL-8 by TNF-α in human colon epithelial cell lines and modulates inflammation through the generation of regulatory T cells (34). Moreover, other studies suggest an anti-inflammatory effect of Bifidobacterium, Faecalibacterium, and Lactobacillus on stress responses and depressive disorders (15, 21, 35). Thus, underrepresentation of these genera in people with depression may verify elevated levels of inflammatory biomarkers in depressed patients. Current literature link microbiome dysbiosis with depression and related biomarkers, although the patterns of gut microbiome still requires definitive distinctions.
Traditionally, alteration of neurotransmitters may lead to depression (36). The gut microbiota are able to influence the body neurotransmitters levels by stimulate the CNS and the gut via the production of metabolites (32, 37). The produced metabolites may affect the body’s neurotransmitters and eventually lead to depression. For instance, short-chain fatty acids (SCFAs) can stimulate the release of serotonin in the gut (38). Also, Microbial metabolites may affect the central neurotransmitters by activating afferent nerve fibers (34). Thus, produced metabolites by gut microbiota may affect emotional behavior by influencing the body’s neurotransmitters either directly or indirectly.
The vast evidence from animal studies supports the hypothesis that gut microbiota plays a key role in CNS function, mainly through inflammation and the hypothalamic-pituitary-adrenal (HPA) axis (15), although the pathways linkage between the gut bacteria and the brain are not entirely understood, stress-induced altered intestinal permeability may play a role. Specifically, gut microbiota alterations and increased translocation of bacterial endotoxins due to a compromised gut barrier is linked to activation of the immune system and HPA axis (15). While a causal relationship is yet to be fully established, bacterial translocation products may lead to an increased production of inflammatory biomarkers (15, 35, 39).
Multiple potential drivers contributed to microbiome alteration in people with depression. These drivers may impact the bidirectional communication between the CNS and gut microbiota which eventually lead to depressive-like behavior (40). A few of drivers include environment, genetics, mode of birth and early exposure to stress, may play roles as potential drivers for alteration of gut microbiota pattern and differential signaling in the gut-brain axis (41, 42).
The other important factor is pattern of bacteria. It is important that the combination of the different taxa might enhance the pathogenic effect (27). The combination effects can be proved in animal studies. For instance, combination of E.coli and B. fragilis was needed for development of abscess in rats. Neither E.coli and B. fragilis alone cannot provoke abscess formation (43, 44). Thus, presence of various gram-negative bacteria at the same time in fecal microbiota of people with depression may be an indicator of depression.
The studies included herein were conducted in various geographic locations. Cultural and lifestyle differences may be factors that influence the pattern of the gut microbiota. Research shows that dietary differences between Eastern and Western countries may impact the bacterial composition and diversity (15). For instance, a high-fat diet may increase the abundance of the gram-negative bacteria in the gut which may lead to an increase in LPS concentration and stimulation of the immune system (27). Therefore, attention to the dietary pattern may also help to clarify the role of the gut microbiome in development/maintenance of depression.
Most of the included studies did not analyze the relationship between diet and gut microbial composition. Further studies are required to assess the role of dietary intake on gut microbiota pattern.
The use of diverse anti-depressant medications was observed to impose a variety of alterations in the bacterial community, which made it difficult to predict the bacterial community pattern in medicated depressed patients. Some animal studies have demonstrated both increases and decreases in the relative abundance of major phyla, notably, Firmicutes and Bacteroidetes during chronic antipsychotic medication administration (45). Hence, it remains unclear whether the gut pattern alternations are caused by depressive symptoms or anti-depressant medications.
Limitations of the Review
Results from the studies synthesized in this review indicated inconsistent results regarding gut microbiome patterns and diversity in people with depression compared to healthy controls. The variations in findings may be attributed to the difference in methodology and population of the included studies. Most of the studies used 16S rRNA sequencing; however, one study utilized phylogenetic analysis of bacterial peptides which influence comparability of the studies. The analytical methods were different across studies involving various regions (V1–V5), various cut off points for clustering OTUs and using different scales for diagnosis of depression which may affect the results. Difference in studies’ population, including age, diet, weight, the geographic location, host genetics, and behavioral factors may influence the pattern of the gut microbiome. Ultimately, the findings consistently demonstrated that depression is associated with marked alterations in gut microbiota composition, although the specific alterations still require further study.
Conclusion
The area of research on the role of microbiota in the brain-gut axis in development or maintenance of depression is still limited. The results of the included studies showed that the characteristics of gut microbiota in people with depression compared to healthy adults are inconsistent. There are conflicting reports on microbial diversity as well as the abundance of bacteria at phyla, family, and genus taxonomic levels in people with depression. The inconsistency suggests that there may be confounding factors within these complicated relationships. Furthermore, differences among taxonomic levels suggest that increased bacterial translocation and intestinal permeability may play a role in the pathophysiology of depression. Nonetheless, further studies are strongly suggested.
A higher bacterial diversity is potentially beneficial to human health, although the role in the gut-brain axis remains subjective to debate. The precise consequences of difference in bacterial diversity for people with depression thus remain unclear. The reported differences in diversity and composition of gut bacteria in this review paper are influenced by population characteristics including age, diet, health status, antibiotic treatment, as well as geographic location (15). The potential contributions of the demographic factors may highlight the role of psychological and social factors in the development of depression. The changes in the microbial patterns of fecal samples deserve more attention using a comprehensive approach in the future. Understanding the pattern of gut microbiota pattern may lead to novel strategies such as appropriate pre/probiotics in treatment of people with depression. Depression as a comorbid condition is among top 10 symptoms that is reported in chronic diseases such as cancer (46). Shedding light on the pattern of gut microbiota in depression may provide better insight in comorbidity between depression and other chronic conditions.
Author Contributions
ZB, XC, WH, AG, and AS contributed to writing and editing the manuscript. The authors agree to be accountable for the content of the work.
Funding
Funding was provided to the University of Connecticut, School of Nursing. 1P20NR016605-01 (PI: AS; pilot PI: XC), and RO1 NR016928 (PI: XC) and American Nurses Foundation (PI: ZB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge the support from the University of Connecticut, School of Nursing, Center for Advancement in Managing Pain. Support was also provided by the Center for Nursing Scholarship and Innovation, University of Connecticut, School of Nursing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2020.00541/full#supplementary-material
References
- 1. WHO. World Health Organization Depression 2018. Available from: http://www.who.int/en/news-room/fact-sheets/detail/depression.
- 2. Koopman M, El Aidy S. Depressed gut? The microbiota-diet-inflammation trialogue in depression. Curr Opin Psychiatry (2017) 30(5):369–77. 10.1097/YCO.0000000000000350 [DOI] [PubMed] [Google Scholar]
- 3. Hyman S. Mental health: depression needs large human-genetics studies. Nat News (2014) 515(7526):189. 10.1038/515189a [DOI] [PubMed] [Google Scholar]
- 4. Smith K. Mental health: a world of depression. Nature (2014) 515(7526):181. 10.1038/515180a [DOI] [PubMed] [Google Scholar]
- 5. Hasler G. Pathophysiology of depression: do we have any solid evidence of interest to clinicians? World Psychiatry (2010) 9(3):155–61. 10.1002/j.2051-5545.2010.tb00298.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brigitta B. Pathophysiology of depression and mechanisms of treatment. Dialog Clin Neurosci (2002) 4(1):7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. science (2006) 312(5778):1355–9. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wallace CJ, Milev R. The effects of probiotics on depressive symptoms in humans: a systematic review. Ann Gen Psychiatry (2017) 16(1):14. 10.1186/s12991-017-0141-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet (2003) 361(9356):512–9. 10.1016/S0140-6736(03)12489-0 [DOI] [PubMed] [Google Scholar]
- 10. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms (2019) 7(1):14. 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin CR, Osadchiy V, Kalani A, Mayer EA. The Brain-Gut-Microbiome Axis. Cell Mol Gastroenterol Hepatol (2018) 6(2):133–48. 10.1016/j.jcmgh.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohlsson L, Gustafsson A, Lavant E, Suneson K, Brundin L, Westrin Å, et al. Leaky gut biomarkers in depression and suicidal behavior. Acta Psychiatr Scandinavica (2019) 139(2):185–93. 10.1111/acps.12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo Y, Zeng B, Zeng L, Du X, Li B, Huo R, et al. Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Trans Psychiatry (2018) 8(1):187–. 10.1038/s41398-018-0240-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heym N, Heasman BC, Hunter K, Blanco SR, Wang GY, Siegert R, et al. The role of microbiota and inflammation in self-judgement and empathy: implications for understanding the brain-gut-microbiome axis in depression. Psychopharmacol (Berl) (2019) 236(5):1459–70. 10.1007/s00213-019-05230-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun (2015) 48:186–94. 10.1016/j.bbi.2015.03.016 [DOI] [PubMed] [Google Scholar]
- 16. Kelly JR, Borre Y, O’Brien C, Patterson E, El Aidy S, Deane J, et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res (2016) 82:109–18. 10.1016/j.jpsychires.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 17. Lin P, Ding B, Feng C, Yin S, Zhang T, Qi X, et al. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J Affect Disord (2017) 207:300–4. 10.1016/j.jad.2016.09.051 [DOI] [PubMed] [Google Scholar]
- 18. Liang S, Wu X, Hu X, Wang T, Jin F. Recognizing Depression from the Microbiota–Gut–Brain Axis. Int J Mol Sci (2018) 19(6):1592. 10.3390/ijms19061592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. OHRS OHRI Newcastle-Ottawa Scale adapted for cross-sectional studies 2018 [Available from: http://www.ohri.ca/programs/.
- 20. Chen Z, Li J, Gui S, Zhou C, Chen J, Yang C, et al. Comparative metaproteomics analysis shows altered fecal microbiota signatures in patients with major depressive disorder. Neuroreport (2018) 29(5):417–25. 10.1097/WNR.0000000000000985 [DOI] [PubMed] [Google Scholar]
- 21. Aizawa E, Tsuji H, Asahara T, Takahashi T, Teraishi T, Yoshida S, et al. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J Affect Disord (2016) 202:254–7. 10.1016/j.jad.2016.05.038 [DOI] [PubMed] [Google Scholar]
- 22. Chen J-J, Zheng P, Liu Y-Y, Zhong X-G, Wang H-Y, Guo Y-J, et al. Sex differences in gut microbiota in patients with major depressive disorder. Neuropsychiatr Dis Treat (2018) 14:647. 10.2147/NDT.S159322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y, Zhang L, Wang X, Wang Z, Zhang J, Jiang R, et al. Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clin Gastroenterol Hepatol (2016) 14(11):1602–11. e5. 10.1016/j.cgh.2016.05.033 [DOI] [PubMed] [Google Scholar]
- 24. Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry (2016) 21(6):786. 10.1038/mp.2016.44 [DOI] [PubMed] [Google Scholar]
- 25. Naseribafrouei A, Hestad K, Avershina E, Sekelja M, Linløkken A, Wilson R, et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil (2014) 26(8):1155–62. 10.1111/nmo.12378 [DOI] [PubMed] [Google Scholar]
- 26. Xu W, Judge MP, Maas K, Hussain N, McGrath JM, Henderson WA, et al. Systematic review of the effect of enteral feeding on gut microbiota in preterm infants. J Obstetric Gynecol Neonatal Nurs (2018) 47(3):451–63. 10.1016/j.jogn.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hakansson A, Molin G. Gut microbiota and inflammation. Nutrients (2011) 3(6):637–82. 10.3390/nu3060637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wong M, Inserra A, Lewis M, Mastronardi C, Leong L, Choo J, et al. Inflammasome signaling affects anxiety-and depressive-like behavior and gut microbiome composition. Mol Psychiatry (2016) 21(6):797. 10.1038/mp.2016.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berk M, Williams LJ, Jacka FN, O’Neil A, Pasco JA, Moylan S, et al. So depression is an inflammatory disease, but where does the inflammation come from? BMC Med (2013) 11(1):200. 10.1186/1741-7015-11-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience (2010) 170(4):1179–88. 10.1016/j.neuroscience.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 31. Zhou L, Foster JA. Psychobiotics and the gut–brain axis: in the pursuit of happiness. Neuropsychiatr Dis Treat (2015) 11:715. 10.2147/NDT.S61997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evrensel A, Ceylan ME. The gut-brain axis: the missing link in depression. Clin Psychopharmacol Neurosci (2015) 13(3):239. 10.9758/cpn.2015.13.3.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology (2009) 136(1):65–80. 10.1053/j.gastro.2008.10.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci (2011) 108(38):16050–5. 10.1073/pnas.1102999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci (2008) 105(43):16731–6. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nutt DJ. Relationship of neurotransmitters to the symptoms of major depressive disorder. J Clin Psychiatry (2008) 69:4–7. [PubMed] [Google Scholar]
- 37. Galland L. The gut microbiome and the brain. J Med Food (2014) 17(12):1261–72. 10.1089/jmf.2014.7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell (2015) 161(2):264–76. 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Foster JA, Neufeld K-AM. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci (2013) 36(5):305–12. 10.1016/j.tins.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 40. Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol: Q Publ Hellenic Soc Gastroenterol (2015) 28(2):203. [PMC free article] [PubMed] [Google Scholar]
- 41. Kelly JR, Minuto C, Cryan JF, Clarke G, Dinan TG. Cross talk: the microbiota and neurodevelopmental disorders. Front Neurosci (2017) 11:490. 10.3389/fnins.2017.00490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cong X, Henderson WA, Graf J, McGrath JM. Early life experience and gut microbiome: the brain-gut-microbiota signaling system. Adv Neonatal Care: Off J Natl Assoc Neonatal Nurses (2015) 15(5):314. 10.1097/ANC.0000000000000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Onderdonk AB. Animal models simulating anaerobic infections. Anaerobe (2005) 11(4):189–95. 10.1016/j.anaerobe.2004.12.001 [DOI] [PubMed] [Google Scholar]
- 44. Onderdonk AB, Bartlett JG, Louie T, Sullivan-Seigler N, Gorbach S. Microbial synergy in experimental intra-abdominal abscess. Infect Immun (1976) 13(1):22–6. 10.1128/IAI.13.1.22-26.1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davey K, Cotter P, O’sullivan O, Crispie F, Dinan T, Cryan J, et al. Antipsychotics and the gut microbiome: olanzapine-induced metabolic dysfunction is attenuated by antibiotic administration in the rat. Trans Psychiatry (2013) 3(10):e309. 10.1038/tp.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reilly CM, Bruner DW, Mitchell SA, Minasian LM, Basch E, Dueck AC, et al. A literature synthesis of symptom prevalence and severity in persons receiving active cancer treatment. Support Care Cancer (2013) 21(6):1525–50. 10.1007/s00520-012-1688-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.