Abstract
Metabolomics has a critical need for better tools for mass spectral identification. Common metabolites may be identified by searching libraries of tandem mass spectra, which offers important advantages over other approaches to identification. But tandem libraries are not nearly complete enough to represent the full molecular diversity present in complex biological samples. We present a novel hybrid search method that can help identify metabolites not in the library by similarity to compounds that are. We call it “hybrid” searching because it combines conventional, direct peak matching with the logical equivalent of neutral-loss matching. A successful hybrid search requires the library to contain “cognates” of the unknown: similar compounds with a structural difference confined to a single region of the molecule, that does not substantially alter its fragmentation behavior. We demonstrate that the hybrid search is highly likely to find similar compounds under such circumstances.
Graphical Abstract

Mass spectral reference libraries are an indispensable tool for identifying molecules.1 When combined with gas2 or liquid chromatography (GC or LC), mass spectrometry (MS) can distinguish hundreds of components in complex mixtures. Less-polar molecules can be analyzed by GC-MS, using electron ionization (EI) to produce radical cations that often fragment extensively in the source, giving structural information useful for identification. EI fragmentation is highly reproducible, so most compounds can be represented by a single library spectrum.
Many metabolites are not volatile or stable enough for GC. The electrospray ionization (ESI) source used in LC-MS produces intact even-electron ions, often by [de]protonation. To obtain structural information, LC is coupled with “tandem” mass spectrometry (MS/MS or MS2). Tandem MS selects a precursor ion in MS1, fragments it by collision-induced dissociation (CID), then analyzes the product ions in MS2. Tandem libraries are more complicated than EI libraries.3 Electrospray often produces different ions from the same compound, so the library must include spectra from multiple precursors. And MS/MS fragmentation patterns vary strongly with collision energy, so the library must include spectra across a range of energies. The NIST library includes spectra obtained by ion-trap (IT) CID, which favors the lowest-energy dissociation channels and thus produces relatively simple spectra. It also includes beam-type collision-cell spectra (“HCD”) at various absolute collision energies (in eV, calculated from Thermo’s “normalized collision energy” and the precursor m/z).
Metabolomics has a critical need for better tools for mass spectral identification and expanded MS/MS libraries.4 Library searching offers key advantages. Libraries are empirical, so there is no need to predict spectra (in silico prediction works best for highly modular structures like lipids,5–7 but is less reliable for compounds with uncertain or complex fragmentation pathways). Libraries are also easily extended to higher MSn stages.8 But libraries may never be complete, especially considering the molecular diversity of complex biological samples.
Similarity searching—finding compounds sharing common structural features with the unknown—can expand the scope of library searching. The NIST software has long included a “simple” similarity search for EI spectra, plus support for substructure identification.9 We recently developed a novel, more powerful similarity search that can also be used for tandem spectra. We call it “hybrid” searching because it combines direct peak matching with the logical equivalent of neutral-loss matching. The algorithm elevates the scores of similar compounds by matching shifted peaks. With current libraries, hybrid searching has been shown10 to greatly increase the number of metabolites found in biological samples.
METHODS
Search Classes
The NIST software11 supports EI, small-molecule tandem, and peptide tandem spectra, each with their own libraries and search methods. EI assumes unit-mass resolution, while tandem searches accept high-resolution spectra using either relative (ppm) or absolute (m/z) tolerances. The hybrid search has been implemented for each. This paper focuses on high-resolution small-molecule tandem searches. Peptide tandem12 and EI13 hybrid searches are described elsewhere.
All NIST searches use the same basic steps: presearch, peak matching, “dot product” calculation, match factor calculation, and hit-list ranking/display. Presearching selects a subset of the library likely to score highly. The tandem hybrid search is an extension of tandem “in-source” and identity searches. Block diagrams for each of these searches are given in Figure S1 of the Supporting Information (SI).
Identity Searching
An “identity” search can succeed only if the library includes a spectrum of the unknown (from the same precursor) that is returned by the presearch. The tandem identity presearch is trivial: the precursor m/z must lie within tolerance of the unknown. We typically specify a 20 ppm precursor tolerance, which rarely returns more than 300 spectra.
For peak matching in small-molecule tandem searches, we typically use a 40 ppm product-ion tolerance and exclude peaks within 1.6 of the precursor m/z. Spectra are lists of (m/z, I) data points. The algorithm determines which unknown and library peaks lie within tolerance of each other. If multiple peaks match—common even with narrow tolerances—it attempts to select the best matching pair.
Scores for all NIST MS search types are based on a weighted “dot product” (cosine similarity) calculation.14 First, each abundance is transformed into a “weight” w = (m/z)mIn. All searches use square-root abundance weighting (n = 1/2), but m/z weightings vary. For small-molecule tandem searches, m = 0 and w = I 1/2. Next, let aL and aU denote “vectors” of all library and unknown weights. Their magnitudes are used to normalize the result between 0 (orthogonal) and 1 (colinear). Then, let mL and mU denote equal-length lists of weights for the subset of peaks returned by the matching algorithm:
| (1) |
Equation 1 is used for “forward” searches. In a “reverse” search, nonmatching unknown peaks are assumed to be contaminants, so mU replaces aU, and the result will be closer to unity. Only forward searches are considered here.
For all NIST searches, the “match factor” (MF) is calculated from cos θ by applying various empirical adjustments and rounding to an integer on a 999-point scale. For small-molecule tandem searches, the most important adjustments reduce the score when only a few peaks match (an unfortunately common situation, especially for ion-trap CID) to make it consistent with the decreased confidence an experienced analyst would have in such a hit. The detailed implementation is irrelevant—the hybrid search can be used with any reasonable metric.
“In-Source” Searching
It may not be possible to match the precursor m/z if, due to in-source dissociation (or adduction), the unknown spectrum comes from a different precursor than any library spectrum. So the in-source “identity” search (SI Figure S1b) replaces precursor matching with a presearch against indexes of the top 16 product-ion m/z values ranked by abundance and by mass-weighted abundance. This typically returns 200–4500 candidates. (NIST presearches use procedures first documented by Finnigan for searching EI libraries.15) It then proceeds like an identity search. The in-source search can be viewed as the hybrid search without peak shifting.
Hybrid Searching
Like in-source searching, the hybrid search (SI Figure S1c) does not require precursor matching. But the hybrid search can elevate scores for similar compounds by matching shifted product ion peaks. The core concept behind the hybrid similarity search—combining direct and neutral-loss peak matching—was originally used for substructure identification with EI spectra.9 Neutral losses were calculated from the molecular weight of the unknown, which is uncertain if the molecular ion is absent. Fortunately, in tandem MS (except for “data-independent” methods with broad isolation windows) the precursor m/z is always known.
Peaks may be shifted by “delta mass” (Δm), the mass of the unknown precursor minus that of the library spectrum. A related strategy (allowing ultrawide precursor tolerances) has been used to identify unanticipated modifications in peptides.16–19 Unlike these “open modification” searches, hybrid searching does not restrict the Δm range, nor does it attempt to predict spectra of modified compounds.
Effective hybrid searching requires the presearch to return spectra that are likely to benefit from neutral-loss matching. So we added an index of neutral losses corresponding to the most-abundant product ions. The presearch combines multiple queries against all three indexes, and typically returns 300–3000 candidates.
Next, a shifted library spectrum is created from each candidate by adding Δm to the mass of each fragment. (Singly charged ions are assumed unless otherwise annotated. Multiple charging is more common for peptides.12) An unknown fragment will match the shifted spectrum if it has the same neutral loss as the library peak. The original and shifted library spectra are separately compared to the unknown (again excluding precursor peaks), and the resulting match lists are merged.
The last step before scoring is constructing a “hybrid library spectrum.” Any fragment that matches after shifting but not before is replaced with its shifted version. If a peak matches before and after shifting, the shifted peak is added to the hybrid spectrum and the abundance is apportioned between the two. The dot product and MF are then calculated from the hybrid spectrum and the unknown. Since the number of matching peaks cannot decrease, the hybrid MF should be greater than or equal to the original MF.
A related peak-shifting strategy has been reported for low-resolution “molecular networking” of living microbial colonies20 and for high-resolution tandem MS of structurally related micropollutants.21 These papers compared pairs of experimental spectra from a relatively small pool—not against a much larger library—so no presearch was required. For molecular networking, peaks that matched before and after shifting were not split but assigned exclusively to whichever m/z gave the higher cosine similarity score. As shown below in Figure 1, there are many cases where all-or-nothing assignment is inappropriate, despite the difficulty of optimizing the apportionment.
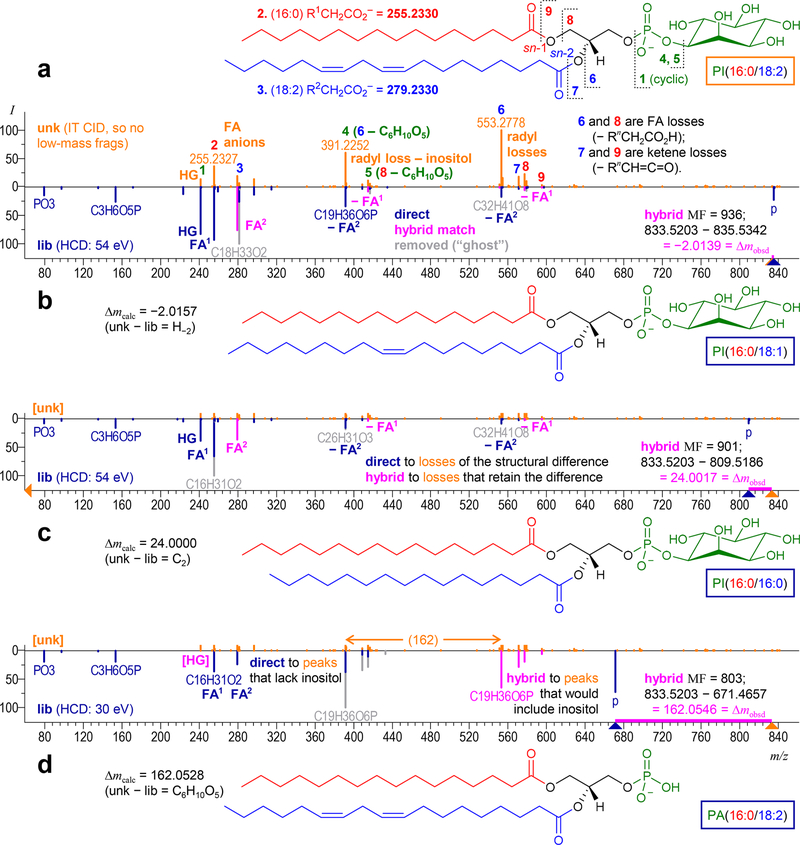
Figure 1.
Hybrid search results for “cluster_007290” from the “urine_HR_it_neg_rec” ARUS library. (a) Likely ID from inspecting the hybrid matches: PI(16:0/18:2), not in NIST17. Peak assignments for the unknown spectrum (orange) are numbered; FA = fatty acid, HG = headgroup. Hybrid library spectra for the (b) first, (c) second, and (d) third hits point down in the head-to-tail displays. Peaks that contain the differing group (gray) match after shifting by Δm (pink). The unknown spectrum is (mostly) cropped out of (c) and (d) to save space.
RESULTS AND DISCUSSION
Annotated Recurrent Unidentified Spectra (ARUS)
Our libraries include nearly every metabolite that can be purchased in pure form, but many more are unavailable or unknown. So we started using the hybrid search to annotate “recurrent unidentified spectra”1,22 observed in biological fluids. The hybrid search can be used to generate chemical class annotations for the large numbers of otherwise unidentified spectra typically observed in LC-MS/MS metabolomics experiments.10 Our ultimate goal is to build libraries of spectra identified from authentic samples, an approach that guarantees biological relevance.
Hybrid Search Example
Figure 1 shows hybrid matches to a consensus spectrum (“cluster_007290”) from the ARUS library “urine_HR_it_neg_rec” available on our website.23 The name given in the synonyms [“PI(16:0/16:0)” in LipidMaps24 notation] is not the unknown itself but a similar compound: the top hit from an earlier hybrid search using a prerelease library and software. The top three unique hits are shown in Figure 1b–d. Upon examination, we think that cluster_007290 is most likely the glycerophospholipid “PI-(16:0/18:2)” (Figure 1a). The fatty acid (FA) formulas are known here because the spectrum includes direct or loss peaks involving the acyl chains. Otherwise, we would notate total carbons and unsaturations as “PI(34:2).” More specific structural features (FA positions on the backbone, double-bond locations and stereochemistry, the absence of branching, and chirality) were assumed.
Interpreting Hybrid Spectra
A hybrid library spectrum is shown for each hit in Figure 1b–d. Unshifted peaks are blue, shifted pink, and removed “ghost” peaks gray. Nearly all shifted peaks also matched unshifted, so only portions of the original peaks are grayed out. Precursor m/z values for the unknown (orange) and library spectra are marked with triangles. A heavy pink line marks the difference (Δm).
Cognates
Successful hybrid searching requires the library to include similar compounds. To give a usable hybrid match, the structural difference must be confined to a single region of the molecule and must not substantially alter its fragmentation behavior. We call compounds that meet these criteria “cognates” of the unknown. In the first two hits in Figure 1, the difference is a single FA and Δm is due to differences in the number of carbons or unsaturations. Fragmentation is unchanged because the acyl group is lost as a unit. Unknown peaks having lost the differing group match directly, and fragments retaining it match shifted library peaks. In the third hit, the FAs are identical and the difference is in the headgroup: the cognate lacks the inositol. Unknown peaks without inositol match directly, and fragments with it match shifted.
Interpreting Delta Mass
Delta mass is the difference between distinct molecules: unknown – library. So unlike a neutral loss, Δm can be negative (Figure 1b) and does not have to represent a stable species (Figure 1c). The corresponding chemical formula may include negative subscripts. For example, deamidation (Δm = 0.9840) is H−1N−1O. (Note that the “odd-nitrogen rule” applies to nominal Δm values.) NIST MS Interpreter25 now allows negative subscripts and thus can be used to find formulas within tolerance of an experimental Δm. Another means of interpretation is to generate lookup tables of calculated Δm values for all structural differences that can be imagined for each chemical class. This gives helpful chemical descriptions of the structural difference (“deamidation” instead of just H−1N−1O). Once the difference is known, assigning shifted peaks can determine its location on the molecule.
Excluded-Query-Compound Analysis
To test the performance of the hybrid search, we executed a global “excluded-query-compound” (EQC) analysis on all high-resolution MS2 spectra in the NIST17 tandem library from compounds with known InChIKeys.26 NIST MSPepSearch27 (which also works for EI and small-molecule tandem MS) was used to search each spectrum against the rest of the EQC library, excluding all spectra from compounds with identical connectivity. Up to 100 hits were retained, but most hit lists were shorter (~30 on average) because lower-scoring spectra of the same compound from other precursors or collision energies were discarded. The output is a large, tab-separated, flat text file, which was processed in R28 using the data.table package.29 See the SI for technical details and a discussion of how EQC analysis differs from leave-one-out cross-validation.
Match Types
Most entries in a typical hybrid search hit list have scores (and thus ranks) that were elevated by peak shifting. But the software does not require this, or even that Δm be nonzero. For each hit, it returns the number of matching peaks and the “dot product” and MF from the hybrid library spectrum, plus “original” versions of these quantities without peak shifting. To assess the impact of peak shifting, we can classify hits using these criteria. We define three types: “Hyb,” “Ins,” and “ID” hits. Peak shifting can be detected by any change from the original MF, dot product, or number of matching peaks. ID hits have Δm within tolerance of zero and no shifted peaks, and should also appear in identity and in-source searches. Ins hits have nonzero Δm but no shifted peaks, and should also appear with the same score in an in-source search. And Hyb hits have both nonzero Δm and shifted peaks, and thus appear with elevated score only in a hybrid search (although they could also appear with their original score in an in-source search).
Global EQC Results
The success of any library search depends on the composition of the library. Match types from hybrid EQC searches are shown in Figure 2. If the library includes the actual unknown (excluded here), it should appear near the top of the hit list. But if the library includes isomers, these too can generate high-ranking ID hits, and there is no easy way to tell which is correct.
Figure 2.
Hybrid search “match types” by rank, excluding low-scoring hits (MF < 600). Percentages are for all ranks. (a) All spectra in the EQC library. Spectra from CID in ion-traps (b) and in beam-type collision cells at moderate (c) or higher (d) energies.
We have previously noted that, because compounds with similar structures often have similar mass spectra, most high-scoring incorrect hits in EI identity searches are actually correct “class identifications.”1 Tandem identity searches are constrained by precursor matching, but removing this constraint generates new hits to similar compounds. An in-source search will hit compounds having fragments in common with the unknown. (It only acts as an “identity” search when the library contains the unknown as a different precursor with similar fragmentation behavior.) And by including neutral-loss matching, the hybrid search elevates scores for cognates of the unknown (if present), promoting them toward the top of the hit list.30
Tandem searches also depend on the extent of fragmentation. Higher collision energies (Figure 2d) produce smaller fragments, which may directly match peaks from many different, often unrelated, library compounds. The resulting hit lists are longer, with more lower-scoring Ins hits. Conversely, ion-trap CID (Figure 2b) produces larger fragments that are more likely to contain the structural difference and thus match after peak shifting, generating higher-scoring Hyb hits. And since ion traps cannot retain smaller fragments, collision cells usually give more Ins hits even at lower energies (Figure 2c).
Accuracy
The output of any library search is a ranked hit list that must be interpreted by the user. For an identity search, interpretation could be as straightforward as assuming that the unknown is the top hit. Higher scores and a larger gap before lower-scoring entries31 increase confidence. Accuracy can be tested by checking if it correctly identifies known compounds.
For a similarity search, the most stringent definition of accuracy would be to enable an experienced analyst to identify the unknown within the limits of MS information. Identifying RUS requires accuracy in this sense. But interpretation requires class-specific expert knowledge and is difficult to automate. So we deem the hybrid search “accurate” when it returns a list of structurally similar compounds. This requires a workable definition of similarity. The micropollutants investigators21 computed substructure fingerprint similarities for 462 pairs of compounds. But our EQC searches generated over 2.4 million unique pairs, so we start with a simpler metric: we define chemical classes of interest and test each hit for membership.
Chemical Classes
The DataWarrior32 cheminformatics program was used to find all compounds in the EQC library containing class-defining substructures. Tested classes are listed in Table 1 and described in the SI. For some, we extended the class definition to additional compounds that were counted as “similar” but not used as query spectra. Hit rates depend on the breadth of these definitions, the depth of library coverage, and the distinctiveness of any class-specific fragmentation. Since the hybrid search works best at lower energies, we restricted queries for each precursor to its ion-trap spectrum and to the collision-cell spectrum obtained nearest 20 eV (within 10–30 eV; ignoring the effect of precursor m/z on energy deposition). We also restricted queries to [M + H]+ ions, the most common precursor type.
Table 1.
Excluded-Query-Compound Hit Rates (Score ≥600) for [M + H]+ by Chemical Class
| query data |
class-hit rate |
distribution of class-hits by match type |
||||||
|---|---|---|---|---|---|---|---|---|
| query class | query spectra | avg hits per query | % with >0 hits | % of hits to the combined class | % Hyb | % Ins combined | % ID combined | |
| query | extended | |||||||
| Ion-Trap CID Spectra | ||||||||
| amino acids | 974 | 11.8 | 84 | 90 | 96.7 | 0.5 | 2.8 | |
| nucleosides | 121 | 2.6 | 65 | 89 | 91.5 | 4.6 | 3.9 | |
| fentanyls | 37 | 12.0 | 89 | 81 | 90.0 | 3.3 | 6.7 | |
| flavonoidsa | 353 | 18.6 | 90 | 77 | 78.4 | 9.1 | 0.1 | 12.5 |
| carnitines | 30 | 8.0 | 93 | 92 | 96.8 | 0.5 | 2.7 | |
| sphingolipids | 48 | 4.9 | 81 | 61 | 100 | 0 | 0 | |
| glycerolipidsb | 31 | 12.3 | 87 | 55 | 88.6 | 6.6 | 0 | 4.7 |
| glycerophospholipidsc | 111 | 2.8 | 70 | 96 | 95.7 | 2.0 | 0.3 | 2.0 |
| hexuronidesd | 56 | 2.2 | 54 | 61 | 97.3 | 0 | 2.7 | |
| steroids | 329 | 22.9 | 89 | 77 | 95.1 | 1.2 | 3.7 | |
| glucuronide steroidse | 8 | 7.2 | 75 | 95 | 36.4 | 58.2 | 5.5 | 0 |
| Collision-Cell Spectra (∼20 eV) | ||||||||
| amino acids | 970 | 10.7 | 87 | 92 | 92.9 | 4.6 | 2.5 | |
| nucleosides | 124 | 4.9 | 76 | 94 | 93.9 | 4.4 | 1.8 | |
| fentanyls | 37 | 11.4 | 89 | 76 | 88.2 | 4.7 | 7.1 | |
| flavonoidsa | 346 | 11.9 | 87 | 82 | 81.9 | 5.3 | 0.3 | 12.5 |
| carnitines | 30 | 14.8 | 97 | 99+ | 97.5 | 0.9 | 1.6 | |
| sphingolipids | 42 | 6.0 | 88 | 74 | 93.1 | 6.4 | 0.5 | |
| glycerolipidsb | 32 | 13.2 | 84 | 53 | 84.7 | 7.7 | 3.2 | 4.5 |
| glycerophospholipidsc | 122 | 8.4 | 74 | 91 | 87.1 | 0.4 | 9.7 | 2.8 |
| hexuronidesd | 54 | 2.6 | 74 | 74 | 93.2 | 4.9 | 1.9 | |
| steroids | 291 | 15.2 | 89 | 87 | 95.2 | 1.7 | 3.2 | |
| glucuronide steroidse | 7 | 7.4 | 86 | 98 | 25.5 | 62.7 | 9.8 | 2.0 |
| Overall | 4153 | 12.0 | 84 | 85 | 91.5 | 1.7 | 2.0 | 4.8 |
The query class includes isoflavonoids; plus hits to an extended class of anthocyan[id]ins, [iso]flavan[one]s, and phenylcoumarins.
Plus hits to glycerophospholipids.
Plus hits to glycerolipids.
Mostly glucuronides with a few galacturonides.
Plus hits to steroids and hexuronides.
See the SI for structure templates and lists of compounds and spectra for each query and extended class.
Identity searching cannot be accurate if the presearch fails to return the correct compound. But similarity searching is more robust toward incomplete presearches: it does not require an exhaustive list of similar library compounds, just a representative one. The hybrid search can be run against the entire library, but this is impracticably slow for general use. We tested this for sphingolipids and glycerophospholipids and found negligible improvement.
Table 1 shows hit rates for the hybrid search by match type and chemical class. The score threshold (600, as in Figure 2) reasonably balances breadth (84% of queries yielded at least one hit) versus accuracy (85% class-hit rate). On average, the hybrid search returned 12.0 hits per query. Most of these (93.4%) were Hyb hits, while only 2.4% were Ins and 4.2% were ID hits (data not shown). The hybrid search converts many potential Ins hits to higher-scoring Hyb hits by matching shifted peaks, and unaltered Ins (and sometimes ID) hits may get displaced from the top 100 by greatly elevated Hyb hits.
The table also shows class-hit rates for each class. Overall, 85% of all hits were to the query class. The percent contribution of each match type to the class-hit rate is also shown. Contributions from the query and extended classes are listed separately for Hyb hits, but are combined for Ins and ID hits. Overall, 93.2% (91.5% + 1.7%) of class-hits resulted from peak shifting (Hyb hits), while only 2.0% were from Ins hits and 4.8% were from ID hits—mostly to isomers. Furthermore, the highest-ranked Hyb hit from each hit list was in the query class 93% of the time (data not shown). The hybrid search does an excellent job of finding similar compounds.
Class-hit rates were quite high for most classes tested, except sphingolipids, glycerolipids, and “hexuronides” (glucuronides plus a few galacturonides). Since glucuronidation is a broadly applicable metabolic process, glucuronides have only glucuronic acid in common. This is a facile neutral loss, leaving product ions from a variety of unrelated compounds. The highest scores will be for compounds similar to these fragments: for glucuronide steroids, adding steroids to the extended class roughly triples the class-hit rate.
Flavonoid Example
Even with an extended class, testing for membership in an arbitrarily defined class often underestimates the fraction of structurally similar hits. An example is shown in Figure 3 and Chart 1. The query class is [iso]flavonoids, and hits to anthocyan[id]ins, [iso]flavan[one]s, and phenylcoumarins are also counted as similar. The “unknown” query spectrum is from the [M + H]+ ion of isovitexin, fragmented in a beam-type collision cell at 19 eV. The spectrum is dominated by fragmentation of the 6-C-glycoside. While O-glycosides tend to lose the entire sugar moiety upon collisional activation,33 the C-glycosidic linkage is stronger, so the tandem spectrum of isovitexin shows a series of losses due to fragmentation of the sugar itself.34
Figure 3.
Hybrid EQC search for a 19 eV collision-cell spectrum of the [M + H]+ precursor of isovitexin, a 6-C-glycosidic flavonoid. The hit list shows the hybrid score (match factor) and number of matching peaks for each hit, paired with “original” values calculated without peak shifting. It also lists Δm and a description of each hit as it would appear in the NIST MS Search program. Score elevations for most hits in this example are very large, because most peaks match only after shifting (pink), as shown in the head-to-tail spectrum for the top hit.
Chart 1.
Isovitexin Peak Assignments and Hybrid Search Hit List
The EQC search hits 21 compounds: nine in the query class, two only in the extended class, and 10 in neither class. Structures (neglecting stereochemistry) are shown in Chart 1. Note that four of the “misses” (5, 6, 10, 17) have a C-glycoside linked to a flavonoid-like fused-ring core, and were not counted as flavonoids because they lack the phenyl substituent. Our class definitions did not capture this important structural similarity that produces similar fragmentation behavior.
The top four entries are Hyb hits to other 6-C-glycosidic flavonoids. The top hit (isoorientin) has one more O atom than isovitexin, on the phenyl ring. Its original spectrum matched only two minor peaks, giving a 0 score. But after shifting by −15.9949, a total of 15 library peaks matched, elevating the score to a near-perfect 990. The modification does not affect the pattern of losses produced by fragmentation of the 6-C-linked sugar.
Saponarin (4) is isovitexin plus an O-linked sugar. Its best match occurred at 29 eV, which cleanly removed the labile O-linked sugar and fragmented the 6-C-linked sugar. Its original score was high enough for an Ins hit, but a modest score elevation due to at least one partial peak shift (without changing the total number of matching peaks) made it a Hyb hit.
Vitexin (7) is an 8-C-isomer of isovitexin, so it generated an ID hit. Scores for it and other 8-C-glycosides (6, 8, 9, 13) are lower than the best Hyb hits to 6-C-glycosides because the position of the C-linkage alters the relative abundances of the loss peaks.34 Vitexin 4-O-glucoside (9) also gave an ID hit (despite its greater mass) because the library includes a spectrum with an in-source loss of the labile O-glucoside.
A 46 eV spectrum of vitexin-2″-O-rhamnoside (13) generated an Ins hit by directly matching smaller fragments produced after loss of the O-linked sugar.
Neohesperidin (18) generated the first Hyb hit to the extended class. Both it and 19 have a single bond at the 2-position, contributing −2H to the total Δm. But they also have differences in other locations, so neither is a true cognate. They matched anyway because the modifications are in a region that does not fragment significantly at these energies.
CONCLUSIONS
Our tests show that, for unknowns from typical chemical classes and with current libraries, the hybrid search is highly likely to return a list of similar compounds: 85% of all hits with a score of at least 600 were to compounds in the same class as the query. But as our flavonoid example shows, arbitrary class definitions often do not fully encompass the structural similarity evident in high-scoring hybrid hit lists. Also, valuable structural information can often be found deeper in the hit list (it is wasteful to consider only the top hit), and interpretation currently requires expert analysis, so a successful hybrid search does not generate a simple answer that is easy to check for accuracy. Our next step will be to use computational substructure analysis of hybrid search hit lists to partially automate unknown structure determination, toward our goal of identifying recurrent unidentified spectra from authentic biological samples.
Supplementary Material
ACKNOWLEDGMENTS
We thank Arun Moorthy and Tytus Mak for helpful discussions and Arun Moorthy for suggesting the word “cognate”. B.T.C. acknowledges UNC Charlotte and the Livingstone Foundation for supporting his leave during summer and fall 2016, and NIST Awards Nos. 70NANB17H165 and 70NANB18H167 for support during summer 2017 and 2018.
Footnotes
The authors declare no competing financial interest.
Certain commercial equipment, instruments, or materials are identified in this paper in order to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b03415.
Search algorithm block diagrams, EQC library construction and use, example MSPepSearch command lines for global EQC searches, and chemical class definitions (PDF)
Compound names and NIST spectrum identifiers for each defined class (XLSX)
REFERENCES
- (1).Stein S Anal. Chem. 2012, 84, 7274–7282. [DOI] [PubMed] [Google Scholar]
- (2).Stein SE J. Am. Soc. Mass Spectrom. 1999, 10, 770–781. [DOI] [PubMed] [Google Scholar]
- (3).Yang X; Neta P; Stein SE Anal. Chem. 2014, 86, 6393–6400. [DOI] [PubMed] [Google Scholar]
- (4).Doerr A Nat. Methods 2017, 14, 32–32. [Google Scholar]
- (5).Kind T; Liu KH; Lee DY; DeFelice B; Meissen JK; Fiehn O Nat. Methods 2013, 10, 755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kochen MA; Chambers MC; Holman JD; Nesvizhskii AI; Weintraub ST; Belisle JT; Islam MN; Griss J; Tabb DL Anal. Chem. 2016, 88, 5733–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Guijas C; Montenegro-Burke JR; Domingo-Almenara X; Palermo A; Warth B; Hermann G; Koellensperger G; Huan T; Uritboonthai W; Aisporna AE; Wolan DW; Spilker ME; Benton HP; Siuzdak G Anal. Chem. 2018, 90, 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Yang X; Neta P; Stein SE J. Am. Soc. Mass Spectrom. 2017, 28, 2280–2287. [DOI] [PubMed] [Google Scholar]
- (9).Stein SE J. Am. Soc. Mass Spectrom. 1995, 6, 644–655. [DOI] [PubMed] [Google Scholar]
- (10).Blazenovic I; Kind T; Sa MR; Ji J; Vaniya A; Wancewicz B; Roberts BS; Torbasinovic H; Lee T; Mehta SS; Showalter MR; Song H; Kwok J; Jahn D; Kim J; Fiehn O Anal. Chem. 2019, 91, 2155–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).The NIST Mass Spectral Search Program for the NIST/EPA/NIH Mass Spectral Library, ver. 2.3, 2017.
- (12).Burke MC; Mirokhin YA; Tchekhovskoi DV; Markey SP; Heidbrink Thompson J; Larkin C; Stein SE J. Proteome Res. 2017, 16, 1924–1935. [DOI] [PubMed] [Google Scholar]
- (13).Moorthy AS; Wallace WE; Kearsley AJ; Tchekhovskoi DV; Stein SE Anal. Chem. 2017, 89, 13261–13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Stein SE; Scott DR J. Am. Soc. Mass Spectrom. 1994, 5, 859–866. [DOI] [PubMed] [Google Scholar]
- (15).Sokolow S; Karnofsky J; Gustafson P The Finnigan Library Search Program; Finnigan Application Report 2, 1978. [Google Scholar]
- (16).Ye D; Fu Y; Sun RX; Wang HP; Yuan ZF; Chi H; He SM Bioinformatics 2010, 26, i399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ahrne E; Nikitin F; Lisacek F; Muller MJ Proteome Res. 2011, 10, 2913–2921. [DOI] [PubMed] [Google Scholar]
- (18).Ma CW; Lam HJ Proteome Res. 2014, 13, 2262–2271. [DOI] [PubMed] [Google Scholar]
- (19).Li Q; Shortreed MR; Wenger CD; Frey BL; Schaffer LV; Scalf M; Smith LM J. Proteome Res. 2017, 16, 1383–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Watrous J; Roach P; Alexandrov T; Heath BS; Yang JY; Kersten RD; van der Voort M; Pogliano K; Gross H; Raaijmakers JM; Moore BS; Laskin J; Bandeira N; Dorrestein PC Proc. Natl. Acad. Sci. U. S. A. 2012, 109, E1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Schollee JE; Schymanski EL; Stravs MA; Gulde R; Thomaidis NS; Hollender JJ Am. Soc. Mass Spectrom. 2017, 28, 2692–2704. [DOI] [PubMed] [Google Scholar]
- (22).Simon-Manso Y; Marupaka R; Yan X; Liang Y; Telu KH; Mirokhin Y; Stein SE Anal. Chem. 2019, 91, 12021–12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23). chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:nist17.
- (24).Sud M; Fahy E; Cotter D; Brown A; Dennis EA; Glass CK; Merrill AH Jr.; Murphy RC; Raetz CR; Russell DW; Subramaniam S Nucleic Acids Res. 2007, 35, D527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).The NIST MS Interpreter Program, ver. 3.4, 2019.
- (26).Heller S; McNaught A; Stein S; Tchekhovskoi D; Pletnev IJ Cheminf. 2013, 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).The NIST MSPepSearch Mass Spectral Library Search Program, ver. 0.96, 2019.
- (28).R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- (29).Dowle M; Srinivasan A data.table: Extension of ‘data.frame ’, 2019.
- (30).Jang I; Lee J.-u.; Lee J.-m.; Kim BH; Moon B; Hong J; Oh HB Anal. Chem. 2019, 91, 9119. [DOI] [PubMed] [Google Scholar]
- (31).Stein SE J. Am. Soc. Mass Spectrom. 1994, 5, 316–323. [DOI] [PubMed] [Google Scholar]
- (32).Sander T; Freyss J; von Korff M; Rufener CJ Chem. Inf. Model. 2015, 55, 460–473. [DOI] [PubMed] [Google Scholar]
- (33).Wojakowska A; Perkowski J; Goral T; Stobiecki MJ Mass Spectrom. 2013, 48, 329–339. [DOI] [PubMed] [Google Scholar]
- (34).Waridel P; Wolfender JL; Ndjoko K; Hobby KR; Major HJ; Hostettmann KJ Chromatogr. A 2001, 926, 29–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.