Abstract
Progress in the identification of causal genes and understanding of the mechanism underlying kidney disease is hindered by the almost exclusive use of a few animal models with restrictive monogenic backgrounds that may be more resistant to kidney disease than humans and therefore poor models. Exploring the large genetic diversity in classical animal models such as mice and rats and leveraging species diversity will allow us to use the genetic advantages of zebrafish, fruit flies, and other species, to develop new animal models that are more relevant to the study of human kidney disease and develop potential therapies.
Keywords: Kidney, Animal model, Genetic Diversity
The Need for Animal Models to Study Kidney Disease
The past two decades have seen a tremendous gain in tools and approaches to study the genetic basis of kidney disease in humans. Genetics studies have moved from investigating genetic variation in single genes (i.e. candidate gene approach) using small populations (50–500 subjects) to large scale genetic studies [genome wide association study (GWAS)] involving the assay of millions of SNPs by microarray, exome sequencing, and even whole genome sequencing in populations including several hundred thousand or more subjects [1] [2]. Advances in sequencing technology have enabled more efficient and less expensive genotyping for human GWAS. These studies have now uncovered over 100 SNPs associated with kidney disease-defining traits, such as albuminuria (see Glossary), eGFR, and blood urea nitrogen (BUN) [3] [4] [5]. However, there are still a number of significant challenges in human studies, such as the inability to test causality, repeated sampling of the kidney, investigation of environmental influences, or developmental origins. In this context, animal models have and will continue to be useful in understanding the mechanisms underlying different forms of kidney disease. However, animal models do have their limitations in recapitulating human disease, for example, some rodents or lower vertebrates such as zebrafish may lack specific genes observed in humans, (e.g. lack of Apol) or a complex phenotype is not easily modeled using a single inbred strain. All individuals within an inbred strain are (nearly) genetically identical, which reduces the variation contributed to genetic factors and leads to more robust phenotypes. While there is much to gain in using inbred mice and rat models to study kidney disease, there is also vast genetic and species diversity that can be applied to study human kidney disease (Figure 1). In this review we discuss the richness of genetic variation in rodents that is available and how exploring this can lead to more insight and establishing better models. We also discuss some of the less traditional species that are used in studying the kidney and how they can make important contributions to our understanding of kidney disease.
Figure 1.
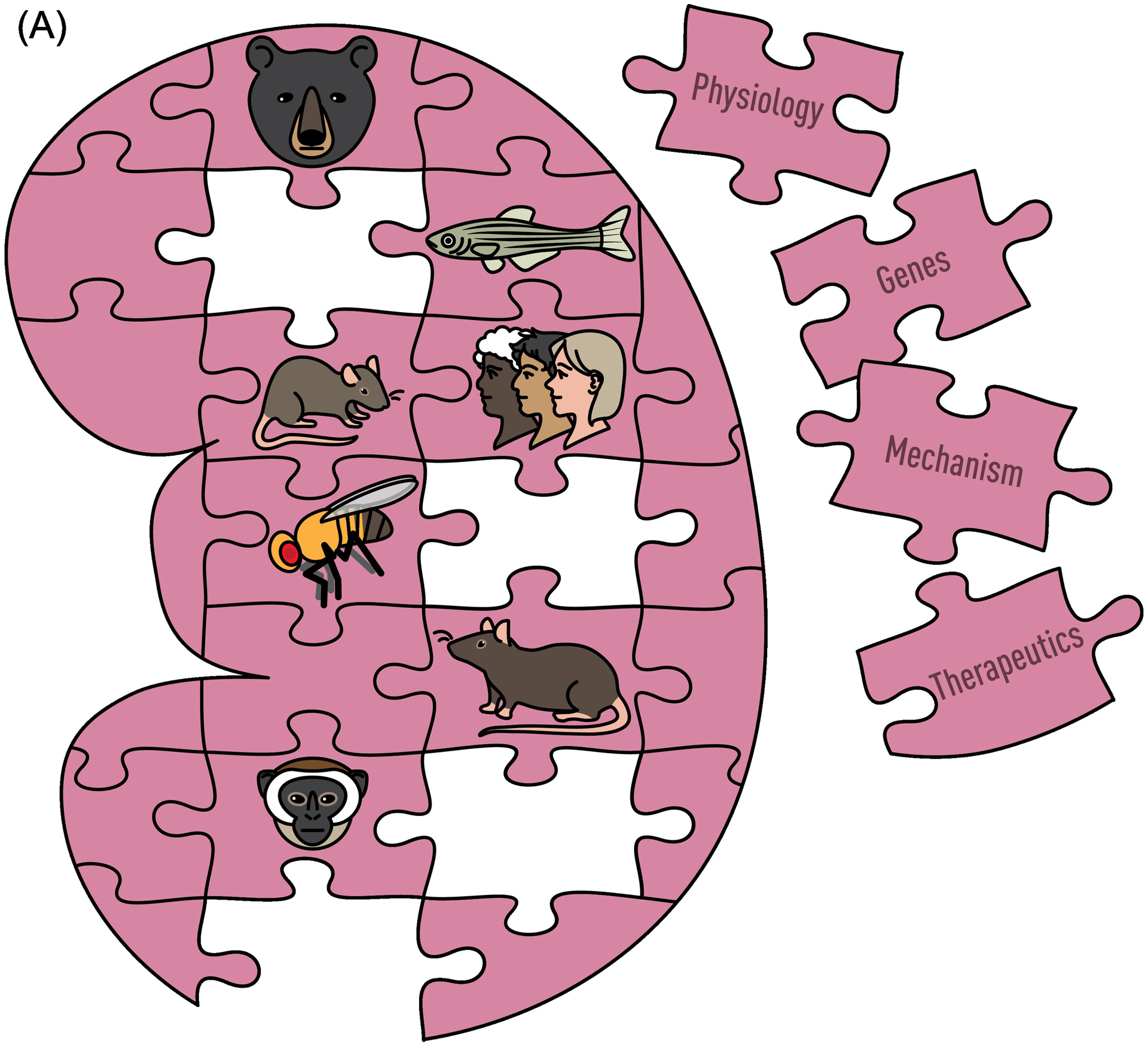
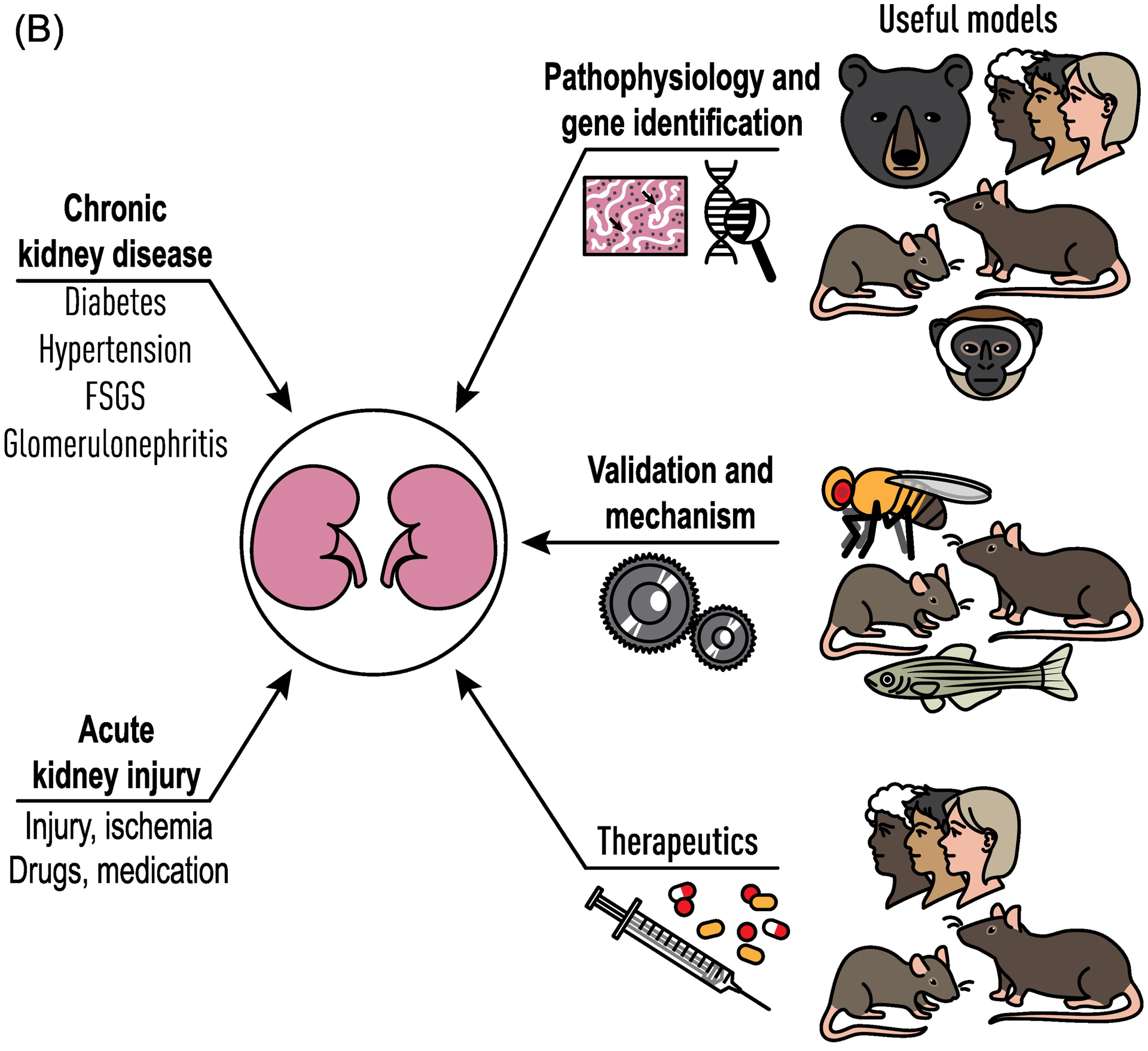
Genetic and Species Diversity to Study Human Kidney Disease. Animal models provide a rich source of both genetic and phenotypic variability, similar to the human population. (A) By taking advantage of the unique qualities of each animal model and genetic diversity within those models, advances in understanding physiology, mechanisms, and genes underlying kidney disease can lead to new therapeutics. (B) The integration of physiological and genetic information across all species will provide a more complete picture of kidney structure and function than the study of any one model, even humans.
The Mouse as a Model for Kidney Disease
The mouse has been an important model in biomedical research for more than a century. The most widely studied inbred strain is C57BL/6 (B6), developed in 1918 by Clarence Cook Little, which has almost exclusively been used in kidney research. This is mainly because the strain breeds well and is easy to maintain, there is a vast knowledge on this strain as it has been used by many investigators in the past 100 years, it is one of the few strains that allowed for genetic manipulation, and there is a vast amount of resources based on the B6 (e.g. complete genome sequence). However, a limitation of B6 is that, due to its genetic background, it only demonstrates a modest susceptibility to kidney disease on its own, requiring experimental intervention (e.g. nephrectomy, AngII, DOCA-salt, etc.). As a single human being with a unique genetic make-up is not expected to represent the complex presentation of kidney disease, nor can a single inbred strain be expected to perfectly model human disease. Similar to the human population, there is a great amount of genetic variation in mice (and rats) and exploiting this variation can allow researchers to develop better models to gain a deeper understanding of the molecular basis of kidney disease. To this end, there are many different mouse strains with different genetic backgrounds that can be investigated for susceptibility/resistance to kidney disease. There are currently 63 datasets comparing kidney phenotypes between mouse strains in the mouse phenome database (phenome.jax.org) which also illustrate the large diversity among them.
One example of the difference in effect of genetic background is the impact of lithium on the kidney. Lithium is used to treat bipolar disorder, but 20% of treated patients develop lithiopathies with nephrogenic diabetes insipidus, metabolic acidosis and hypercalcemia. B6 has been the mouse strain of choice and most that is known about the effect of lithium on the kidney is based on this inbred strain. However, a recent study comparing the effect of lithium exposure among 29 different inbred strains of mice showed that B6 may not be the best strain to study this disease. C3H/HeJ mice develop more severe nephrogenic diabetes insipidus than B6 mice and show hypercalcemia in response to lithium exposure, which is not observed in B6 [6]. A second example is diabetic nephropathy. Across the different experimental mouse models of diabetes, the B6 strain tends to be relatively resistant to the development of albuminuria and histopathological abnormalities, whereas the 129, FVB and DBA/2 strains are more permissive and therefore provide more robust platforms for the development of models for diabetic nephropathy [7]. This difference in susceptibility to diabetes due to differences in genetic background is also nicely illustrated in the study by Wu et al [8]. In their study, they crossed the DBA/2 strain that carried the diabetes causing Akita mutation in the Ins2 gene with 28 different inbred strains and compared the increase in albuminuria. Among these diabetic mice, albumin-to-creatinine-ratios varied widely with mean values ranging from 42 (SWR strain) to 750 ug/mg (CBA strain), and the effect of B6 being very moderate, showing the effect of the different alleles present in different inbred strains [8]. A third example is the severity of disease in models of Alport syndrome. This disease is caused by mutations in either COL4A3, COL4A4, or COL4A5 in humans and mouse models with mutations in either of these three genes have been developed [9] [10] [11] and are available at The Jackson Laboratory (www.jax.org). The mutations in these genes have been studied in different genetic backgrounds and show large differences in disease severity. 129X1/SvJ mice with a Col4a3 mutation develop end-stage renal failure much sooner (approx. 66 days) than C57BL/6J mice with the same mutation (approx. 194 days) [12], albuminuria and renal pathology is much more severe in DBA/2J mice with a Col4a4 mutation compared to 129S1/SvImJ [10], and moving a Col4a5 mutation from 129/SvJ mice to C57BL/6J mice and a different environment leads to a delay in renal failure (R. Korstanje, unpublished). A final example illustrates the important differences between substrains. Although many investigators talk about “B6” mice, there are many substrains of B6 (C57BL/6J and C57BL/6NJ being the best known and most used). Each substrain has different genetic variants leading to different phenotypes. This was illustrated by a study in which different substrains of B6 were compared for age-dependent nephropathy and C57BL/6J were shown to develop a more severe phenotype compared to C57BL/6JRj, C57BL/6JNia, or a substrain maintained at the University of Heidelberg [13]. Genomic comparisons and subsequent experimental follow-up strongly suggest that a variant in the Dhtkd1 gene unique to C57BL/6J is the cause of the more severe phenotype (R. Schmitt, in preparation).
Exploring genetic backgrounds other than B6 can be a tremendous improvement for modeling kidney disease in mice and with the introduction of the CRISPR/Cas9 system to manipulate the genome researchers are no longer restricted as previously had been with making knockouts and knockins, which was only possible in B6, 129, or FVB. For example, it is now possible to make precise edits in the genome and perform allele replacements in D2-InsAkita, a strain that is better suited to study diabetic nephropathy and MRL-Faslpr, a strain that develops lupus nephritis (Korstanje, unpublished).
A major strength of using mice (and rats) to study kidney disease, apart from the ability to better control experimental conditions and to manipulate the genome, is the ability to sample whole kidneys (and other relevant tissues) to measure gene expression, assess protein levels, and perform detailed histological analysis to identify changes in kidney structure under controlled conditions and at specific time points with age or disease progression. These strengths are tempered slightly by some differences in development, structure, and gene expression of the kidney between mice and humans [14] that may have an impact on disease. However, these findings are mostly based on B6 mice and a better exploration of other strains with different genetic backgrounds as illustrated above, may identify strains that better model human disease.
The Rat as a Model for Kidney Disease
In contrast with mice, a number of inbred rat genetic models exhibit a robust kidney disease phenotype, including the development of overt proteinuria, decline in renal function, and significant histological damage (glomerulosclerosis, tubulointerstial injury/fibrosis, and vascular hypertrophy), are available [15]. For example, there are several hypertensive models, such as the stroke-prone spontaneously hypertensive rat (SHRSP) [16], Dahl salt-sensitive (DSS) [17] [18], Fawn-hooded hypertensive rat (FHH) [19] [20], and Sabra hypertensive rat (SBH) [21] that develop significant renal injury associated with hypertension. These models originate from different outbred stocks/selection processes so likely represent a diverse set of alleles that contribute to the susceptibility to kidney disease. An exception are the SHR-A3 (SHR) and SHR-B2 (SHRSP) strains which are descended from a single founder pair of Wistar rats whose progeny were selectively bred to fix the trait of hypertension [22]. These two inbred lines both exhibit hypertension, but differ markedly in their susceptibility to end organ damage (SHRSP=stroke and kidney disease, whereas SHR do not). Whole genome sequencing of these lines have identified distinct haplotype blocks (e.g. 87% identical by descent, IBD) containing genes involved in immune response [23]. Haplotype blocks containing immunoglobulin heavy chain IgH has been linked with renal injury [24] as well as genetic variation in Stim1 which plays a role in lymphocyte calcium signaling that directs immune effector responses[23].
There is at least one rat model, the Milan normotensive strain (MNS) that is susceptible to glomerulosclerosis and proteinuria independent of hypertension [25]. Aside from hypertensive models, there are also several rat models that develops diabetic related nephropathy. For example, the Goto-Kakizaki (GK) rat develops early onset glucose intolerance and mild hyperglycemia, with development of renal injury later in life (>35 weeks of age) [26]. In contrast, the Otsuka Long-Evans Tokushima fatty (OLETF) [27] and type 2 diabetic nephropathy (T2DN) rat models develop earlier onset of diabetic phenotypes and more robust kidney injury [28]. The T2DN rat also develops a key histological feature, resembling Kimmelstiel-Wilson nodules, that are characteristic of human diabetic nephropathy [29].
Many of the genetic models noted above have undergone extensive linkage analysis, identifying a large number of loci associated with proteinuria [30]. For the most part, specific genes causing these phenotypes have not been identified in these strains with a few exceptions. For example, the DSS model has been used for forward genetic studies [e.g., Arhgef11, [31], [32]] and reverse genetic studies to validate genes identified in human GWAS studies, including Plekha7 [33], Sh2b3 [34], and Shroom3 [35]. In addition, there are more than two hundred mutant/knockout models of genes involved in hypertension/CKD available to the research community (rgd.mcw.edu/wg/gerrc/), mainly developed on the DSS, SHRSP, FHH, and T2DN genetic backgrounds.
Thus, physiologically, rat genetic models provide an advantage over current mice models as they better recapitulate what is observed in human kidney disease. A major limitation has been the inability to perform genetic modifications (e.g. knockout, allele specific knockin, tissue specific KO, etc) to the extent achieved in the mouse. Recent advances in gene-editing using CRISPR/Cas9 has provided some new tools for genetic modification in the rat [36]. However, gene editing in the mouse is still more advanced in terms of developing inducible and/or tissue/cell type specific modifications (i.e., availability of a large number of floxed genes and Cre recombinase strains), even though there are a few examples of investigators generating loxp/Cre rat models [37] [38]. Despite these limitations, rat inbred models provide a strong genetic diversity linked with phenotypic variability in kidney traits.
Using Genetic Diversity in Rodents to Identify Candidate Genes
Both mice and rats are excellent models to study a complex genetic trait such as kidney disease and identify the underlying genetic factors. As environmental factors such as diet and the microbiome can be controlled, the number of individuals needed is much smaller to have the same power of detection compared to humans. In the past few decades several genetic studies using crosses between inbred strains have been performed in mice and rats and we have reviewed these previously [30]. However, genetic crosses between two inbred strains provided limited genetic variation (at most two alleles) and low mapping resolution (25–50 Mb). For these reasons, other approaches have been developed that circumvent these challenges and have been shown to be successful for different traits, including kidney-related phenotypes.
One of these approaches is in silico QTL mapping or haplotype association mapping [39]. This method uses phenotyping data from a large number of inbred strains and when combined with the large amount of SNP genotype data available allows the association between the phenotype and a haplotype shared by multiple strains. The advantage of this method is that phenotype data is collected from multiple individuals of a strain and thereby a more robust phenotype average is obtained. The disadvantage of this approach is that the resolution depends on the size of the conserved haplotype block, which is highly variable, and which inbred strains are being used in the analysis. Nevertheless, this approach has been successfully used to identify candidate genes for albuminuria, mesangial matrix expansion and lithiopathies, which were confirmed to be causal by follow-up studies [40] [41] [6].
The Diversity Outbred (DO) panel [42] [43] was established at The Jackson Laboratory by outcrossing incipient inbred strains of the Collaborative Cross, a newly created panel of inbred mice derived from eight founder strains, including five classical lab strains (129S1/SvImJ, A/J, C57BL/6J, NOD/ShiLtJ, NZO/HILtJ) and three wild-derived strains (CAST/EiJ, PWK/PhJ and WSB/EiJ) (Figure 2) and was recently extensively discussed in TiG [44]. The wild-derived strains contribute ~75% of the genetic diversity. The eight founder genomes are fully sequenced [45], providing a catalog of variants that includes 38,261,117 SNPs, 5,376,127 small indels, and 228,286 structural variants. Among annotated exons and UTRs of protein coding genes, there are SNPs in 94% of genes, indels in 66% and structural variants in 12%. Accumulation of recombination breakpoints with each generation is the key to high-resolution genetic mapping in the DO. Using high-density array genotyping and haplotype reconstruction algorithms [46], the full genomic sequence of any DO animal can be accurately imputed.
Figure 2.
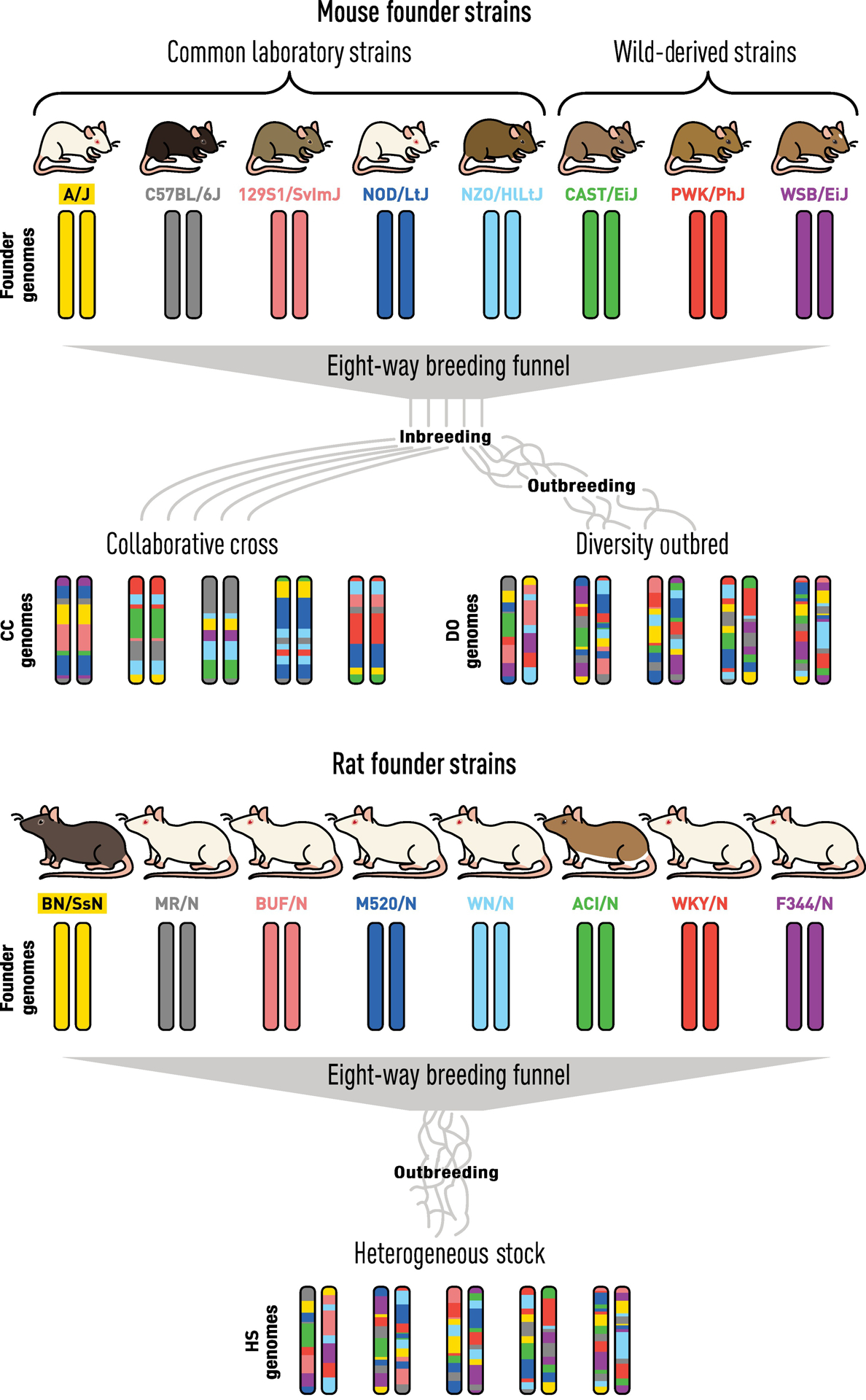
The Power of Genetic Mapping using Diversity Outbred (DO) Mice and Heterogeneous Stock (HS) Rats. Both DO and HS models offer unique resources to map traits involved in complex disease, including kidney disease. Each population was derived by selecting eight genetically diverse inbred strains and subsequent outbreeding for many generations. Each animal from this unique resource has a unique combination of the eight founder genome and both populations show a much greater phenotypic diversity than what we observe in the founder strains. As such, these outbred rodent populations can be viewed as similar to human populations as they exhibit diverse phenotypes, including susceptibility or resistance to kidney disease, with ability to define their underlying genetic basis with high resolution using GWAS techniques.
Similarly, the Heterogeneous Stock rat model (NIH-HS, denoted as HS) was created in 1984 by the National Institutes of Health as a source for genetically segregating animals [47]. The HS model was developed via an eight-way cross of different inbred strains including the BN/SsN, MR/N, BUF/N, M520/N, WN/N, ACI/N, WKY/N, and F344/N (Figure 2). As an outbred stock, the HS population demonstrates significant variation in many traits that allows for efficient fine-mapping [48], including; diabetes [49], adiposity [50], despair-like behavior [51], and HS have demonstrated phenotypic variation for multiple kidney traits, including proteinuria [52] and histological markers of kidney injury (manuscript in preparation). Like the mouse DO panel, the eight founder genomes that compose the HS are fully sequenced [53] and similarly provides a detailed catalog of sequence variants including 20,810,664 SNPs, 9,442,630 small indels, and 137,039 structural variants [54]. More recently, there has been an effort to develop a Hybrid Rat Diversity Panel (HRDP) composed of 96 inbred rat strains selected to maximize power to detect specific genetic loci associated with a complex trait [55] (rgd.mcw.edu/wg/hrdp_panel/). Once complete, the HRDP will provide another tool to identify genes involved in CKD as well as many other traits.
In summary, regardless of the rodent model, outbred populations provide great strength in mapping kidney disease phenotypes as they are genetically diverse and the accumulation of recombination events and small haplotype blocks result in high resolution mapping. Furthermore, the presence of eight or more alleles in the cohorts provides additional information that can lead to the identification of the causal gene or causal SNP.
The Advantages of the Zebrafish
In addition to the common rodent models, there are several other animal models that are very useful to study kidney disease. The zebrafish embryonic kidney (pronephros) shows histological and functional similarities to the mammalian adult nephron. This, combined with rapid ex utero development, transparent embryos, high fecundity, a well-characterized genome and tools for gene editing, has led to an exponential growth in the number of zebrafish models to study kidney disease. Several excellent reviews have been published recently describing the advantages and disadvantages of using the zebrafish in renal research [56] [57].
A particular powerful approach is the combination of rodent and zebrafish models to understand the role of genes in the kidney. One example is Kmo, encoding kynurenine 3-monooxygenase, this gene was identified as a candidate for a locus associated with albuminuria in a mouse genetic study [58]. The candidacy was tested by knockdown of kmo in zebrafish, which lead to edema, loss of circulating vitamin D binding protein, and podocyte effacement. The results from the zebrafish model were then confirmed with a knockout of Kmo in the mouse [58]. Another very nice example is Shroom3, a candidate gene for CKD in both human and rat with a possible dysfunctional allele in the FHH rat inbred strain compared to the BN strain. Knockdown of shroom3 in zebrafish indeed led to a severe renal phenotype that could be rescued by injection with BN Shroom3 mRNA, but not with FHH mRNA [35].
Drosophila as an Emerging Model
The Drosophila has a highly specialized filtration structure called the nephrocyte diaphragm, which shares similarities to the slit diaphragm structure of the mammalian glomerular podocyte [59]. Zhe Han has developed the fruit fly as a model to study podocyte biology and for large-scale genetic screens to identify and study novel genes that are involved in podocyte function and renal diseases. Similar to the experiment with Shroom3 in zebrafish, rescue of renal phenotypes in fruit fly with alleles from other species is possible as demonstrated in a knockdown of Cindr (the ortholog of CD2AP), which leads to a deficiency of ANF-RFP uptake and can be rescued by normal human CD2AP gene expression but not by a patient derived mutant allele [60].
In addition to these screens with RNAi libraries and gene knockdown, exploring the effect of natural genetic variation in Drosophila on renal phenotypes would be interesting. This could be accomplished by combining the tools developed by the Han lab with the resources established by Trudy Mackay: Similar to the mouse CC lines described above, the Drosophila melanogaster Genetic Reference Panel (DGRP) consists of fully sequenced inbred lines derived from a natural population [61]. Screening these lines, with or without a sensitizing mutation, for renal phenotypes will likely identify novel genes associated with these phenotypes and likely play a role in human kidney disease.
Unconventional Animal Models
There have been some studies that suggest felids, including tigers, leopards, lions, cheetahs, and cougars could provide some interesting insight into the pathophysiology of CKD. For example, a study of 38 deceased captive wild felids originating from eight German zoological gardens found that 33 animals or 87% demonstrated significant renal lesions [62]. In >50% of these animals, significant tubular changes, including (e.g. tubular epithelial degeneration, necrosis, and proteinaceous casts) were observed. In addition, glomerular lesions included glomerulonephritis (GN; 16/38 animals; 42%), glomerulosclerosis (32%) and amyloidosis (3%). While it is not practical to study kidney disease in these large animals, it may be feasible to use the domestic cat, which also demonstrates a high incidence of kidney disease (~3 in 10 older cats) [63]. For example, a recent study observed that cats (in comparison to canine) demonstrate a unique distribution pattern of kidney-infiltrating macrophages associated with cat-specific fibrotic processes [64].
While there are a number of experimentally induced models of hypertension and renal injury in baboons and rhesus monkeys [65] [66], the African Green monkey (AGM) that resides in the West Indies/Caribbean, is the only non-human primate that develops spontaneous hypertension and altered renal vascular and glomerular morphology. In a study of 424 AGM animals, the animals segregated into three groups: 37% (157/424) developed significant hypertension (SBP=172.0± 2.2 mmHg), 44% (187/424) normotensive (average SBP=99.6± 1.0 mmHg), and 18% with a modest increase in blood pressure (130.6 ± 0.6 mmHg) [67]. Hypertensive animals demonstrated greater vessel wall thickness in renal vasculature as well as structural changes in the glomeruli. The genetic basis of hypertension and kidney disease in this model has yet to be investigated, but it is an interesting model that could provide useful insight in genes associated with these diseases, as well as allow for testing/validation in an animal model closely related to humans.
A few years ago, Peter Stenvinkel highlighted the unique biology of the bear. Hibernating bears decrease their GFR by 70%, become anuric and glomeruli become partly sclerosed in early spring [68], but somehow are able to recover soon after hibernation. When comparing summer active and hibernating brown bears, he found that despite anuria and a 2.5-fold increase in creatinine levels, azotemia does not develop and inflammation does not ensue [69]. A recent study comparing gene expression profiles of black bears before and after hibernation identified 160 differentially expressed genes in the kidney with the noted absence of upregulation of inflammatory genes in the spring and instead upregulation of cytokines that decrease the inflammatory response [70]. Follow-up of these genes to see which of them are involved in the recovery of the kidney after hibernation will depend on rodent, zebrafish, and perhaps fly models.
Concluding Remarks and Future Perspectives
Despite the tremendous advances in human genetic studies, there is still a great need for animal models to study and better understand kidney disease and the genetic factors that are involved. The almost exclusive use of a few species (mouse and rat) with very restricted genetic backgrounds (e.g. mostly C57BL/6 mice) has limited these studies and contributed to the reputation that animal models are limited and do not reflect the human condition. Exploring the large genetic variation in rodent models and the unique features of other species can lead to the development of new models to study kidney disease and test renal therapies (See Outstanding Questions).
Outstanding Questions.
What makes some inbred strains of mice or rats more susceptible to kidney disease than others?
Why do species such as cats seem to be very susceptible to renal damage and have a high incidence of kidney disease while other species, such as black bears, are very resistant despite severely challenging their kidneys during hibernation?
The major developments in kidney organoid cultures are very exciting and in the short-term will complement animal experimentation, and eventually may even replace animal experiments to address some of the questions in the future. However, these organoids are currently still far away from truly recapitulating a functioning kidney as gene expression profiles suggest the organoids are similar to kidneys in the embryonic stage [71]. As most of the work in kidney organoids is limited to using a few human cell lines, similar to the use of inbred strains, there is a danger that findings based on cells with limited genetic variation may not be universally true as genetic background may play a significant role in genes or drugs being evaluated (i.e., similar to the single use of one inbred strain of mice). Thus, exploring differences in kidney organoids derived from different genetic backgrounds may be a powerful approach to gain deeper insight into kidney health and disease. Here also, resources like the DO or HS could help. Generating kidney organoids from a panel of stem cells derived from DO mice or HS rats would allow genetic analysis and the identification of genes associated with organoid development.
The tools and resources needed to make use of the available genetic and species diversity are already in place. What is needed is more awareness, better education, and detailed reporting of the exact animal model/strain utilized in scientific publications. For example, a better understanding and appreciation of differences between inbred strains and substrains (e.g., C57BL/6J and C57BL/6NJ demonstrate different susceptibility to kidney injury) will lead to better and more detailed reporting of the animals that were used in the study. Subsequently, this information can be captured in already existing repositories such as the Mouse Phenome Database and Rat Genome Database which will allow researchers to mine this valuable data and result in more informed choices of models to best address significant questions being posed.
Highlights.
Exploring the large diversity of genetic backgrounds in mice and rats can improve modeling of kidney disease.
The Diversity Outbred (DO) mouse population and Heterogeneous Stock (HS) rat population display high genetic and phenotypic variability and enables precise genetic mapping of complex genetic traits such as kidney disease.
Taking advantage of the genetic tools and natural occurring systems in species such as zebrafish, fruit flies, cats, and bears can provide more insight into the role of genes in the kidney.
Acknowledgements
This work was supported by grants from the National Institutes of Health (R01 HL137673 to MRG and P30 AG038070, U54 OD020351, and RO1 ES029916 to RK).
Glossary
- Albuminuria/proteinuria
the presence of albumin or protein in the urine which is an indicator of renal damage.
- Blood urea nitrogen (BUN)
an elevated BUN is indicative of reduced renal function.
- CRE recombinase
an enzyme that catalyzes the site-specific recombination of DNA between loxP sites (see floxed genes)
- CRISPR/Cas9
a genome editing system based on the ability of the Cas9 enzyme to cut DNA at a specific sequence as directed by a guide RNA sequence.
- eGFR
the estimated glomerular filtration rate which is used in humans as a measure of kidney function and is based on the amount of creatinine in the blood.
- Floxed gene
a gene in which a deletion is induced by first engineering loxP sites (usually flanking an exon) and subsequently exposing it to CRE recombinase.
- GWAS
genome-wide association study is an observational study of an association of a trait with a set of genetic variants covering the genome.
- Haplotype
a segment of DNA that is inherited as a block from a single parent
- Heterogeneous stock
a population derived from randomized matings of more than two founder strains, resulting in mosaic genomes with high recombination.
- Inbred strain
a strain derived from at least 20 generations of sibling-sibling matings, which effectively eliminates heterozygosity and results in isogenic offspring
- Linkage analysis
a genetic technique that searches for chromosomal regions or genes that cosegregate with a trait in a family
- Outbred stock
a stock derived from a genetically diverse source that is maintained through matings between unrelated individuals
- SNP
a single nucleotide polymorphism is a substitution of a nucleotide at a specific position in the genome.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wuttke M et al. (2019) A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 51, 957–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X et al. (2018) Molecular insights into genome-wide association studies of chronic kidney disease-defining traits. Nature Communications 9, 4800–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wuttke M and Köttgen A (2016) Insights into kidney diseases from genome-wide association studies. Nat Rev Nephrol 12, 549–562 [DOI] [PubMed] [Google Scholar]
- 4.Pattaro C et al. (2016) Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nature Communications 7, 10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorski M et al. (2017) 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep 7, 45040–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Groot T et al. (2019) Identification of Acer2 as a First Susceptibility Gene for Lithium-Induced Nephrogenic Diabetes Insipidus in Mice. J. Am. Soc. Nephrol DOI: 10.1681/ASN.2018050549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azushima K et al. (2018) Modelling diabetic nephropathy in mice. Nat Rev Nephrol 14, 48–56 [DOI] [PubMed] [Google Scholar]
- 8.Wu X et al. (2014) Genetic modulation of diabetic nephropathy among mouse strains with Ins2 Akita mutation. Physiol Rep 2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosgrove D et al. (1996) Collagen COL4A3 knockout: a mouse model for autosomal Alport syndrome. Genes Dev. 10, 2981–2992 [DOI] [PubMed] [Google Scholar]
- 10.Korstanje R et al. (2014) A mouse Col4a4 mutation causing Alport glomerulosclerosis with abnormal collagen α3α4α5(IV) trimers. Kidney International 85, 1461–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rheault MN et al. (2004) Mouse model of X-linked Alport syndrome. JASN 15, 1466–1474 [DOI] [PubMed] [Google Scholar]
- 12.Andrews KL et al. (2002) Quantitative trait loci influence renal disease progression in a mouse model of Alport syndrome. The American Journal of Pathology 160, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt R et al. (2009) Ageing mouse kidney--not always the SAME old story. Nephrol. Dial. Transplant 24, 3002–3005 [DOI] [PubMed] [Google Scholar]
- 14.Lindström NO et al. (2018) Conserved and Divergent Features of Human and Mouse Kidney Organogenesis. J. Am. Soc. Nephrol 29, 785–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz A and Kreutz R (2012) Mapping genetic determinants of kidney damage in rat models. Hypertens. Res 35, 675–694 [DOI] [PubMed] [Google Scholar]
- 16.Kato T et al. (2015) Blood pressure, renal biochemical parameters and histopathology in an original rat model of essential hypertension (SHRSP/Kpo strain). Biomed. Res 36, 169–177 [DOI] [PubMed] [Google Scholar]
- 17.Rapp JP and Dene H (1985) Development and characteristics of inbred strains of Dahl salt-sensitive and salt-resistant rats. Hypertension 7, 340–349 [PubMed] [Google Scholar]
- 18.Garrett MR et al. (2003) Time-course genetic analysis of albuminuria in Dahl salt-sensitive rats on low-salt diet. JASN 14, 1175–1187 [DOI] [PubMed] [Google Scholar]
- 19.Kuijpers MH and de Jong W (1986) Spontaneous hypertension in the fawn-hooded rat: a cardiovascular disease model. J Hypertens Suppl 4, S41–4 [PubMed] [Google Scholar]
- 20.de Keijzer MH et al. (1989) Proteinuria is an early marker in the development of progressive renal failure in hypertensive fawn-hooded rats. J. Hypertens 7, 525–528 [DOI] [PubMed] [Google Scholar]
- 21.Yagil C et al. (2002) Proteinuria and glomerulosclerosis in the Sabra genetic rat model of salt susceptibility. Physiological Genomics 9, 167–178 [DOI] [PubMed] [Google Scholar]
- 22.Doris PA (2017) Genetics of hypertension: an assessment of progress in the spontaneously hypertensive rat. Physiological Genomics 49, 601–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dhande IS et al. (2020) Stim1 Polymorphism Disrupts Immune Signaling and Creates Renal Injury in Hypertension. J Am Heart Assoc 9, e014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Garay ML et al. (2014) Diversity in the preimmune immunoglobulin repertoire of SHR lines susceptible and resistant to end-organ injury. Genes Immun. 15, 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandis A et al. (1986) Age-dependent glomerulosclerosis and proteinuria occurring in rats of the Milan normotensive strain and not in rats of the Milan hypertensive strain. Lab. Invest 55, 234–243 [PubMed] [Google Scholar]
- 26.Phillips AO et al. (2001) Association of prolonged hyperglycemia with glomerular hypertrophy and renal basement membrane thickening in the Goto Kakizaki model of non-insulin-dependent diabetes mellitus. Am. J. Kidney Dis 37, 400–410 [DOI] [PubMed] [Google Scholar]
- 27.Kawano K et al. (1992) Spontaneous long-term hyperglycemic rat with diabetic complications. Otsuka Long-Evans Tokushima Fatty (OLETF) strain. Diabetes 41, 1422–1428 [DOI] [PubMed] [Google Scholar]
- 28.Palygin O et al. (2019) Progression of diabetic kidney disease in T2DN rats. American Journal of Physiology-Renal Physiology 317, F1450–F1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nobrega MA et al. (2004) Initial characterization of a rat model of diabetic nephropathy. Diabetes 53, 735–742 [DOI] [PubMed] [Google Scholar]
- 30.Garrett MR et al. (2010) Integrating human and rodent data to identify the genetic factors involved in chronic kidney disease. J. Am. Soc. Nephrol 21, 398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams JM et al. (2012) Genetic variants in Arhgef11 are associated with kidney injury in the Dahl salt-sensitive rat. Hypertension 60, 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson AC et al. (2020) Loss of Arhgef11 in the Dahl Salt-Sensitive Rat Protects Against Hypertension-Induced Renal Injury. Hypertension 75, 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endres BT et al. (2014) Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc. Natl. Acad. Sci. U.S.A 111, 12817–12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudemiller NP et al. (2015) Mutation of SH2B3 (LNK), a genome-wide association study candidate for hypertension, attenuates Dahl salt-sensitive hypertension via inflammatory modulation. Hypertension 65, 1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo NC et al. (2015) Shroom3 contributes to the maintenance of the glomerular filtration barrier integrity. Genome Res. 25, 57–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko T (2017) Genome Editing of Rat. Methods Mol. Biol 1630, 101–108 [DOI] [PubMed] [Google Scholar]
- 37.Ma Y et al. (2017) CRISPR/Cas9-mediated targeting of the Rosa26 locus produces Cre reporter rat strains for monitoring Cre-loxP-mediated lineage tracing. FEBS J. 284, 3262–3277 [DOI] [PubMed] [Google Scholar]
- 38.Bryda EC et al. (2019) A novel conditional ZsGreen-expressing transgenic reporter rat strain for validating Cre recombinase expression. Sci Rep 9, 13330–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsaih S-W and Korstanje R (2009) Haplotype association mapping in mice. Methods Mol. Biol 573, 213–222 [DOI] [PubMed] [Google Scholar]
- 40.Noordmans GA et al. (2013) Genetic analysis of mesangial matrix expansion in aging mice and identification of Far2 as a candidate gene. J. Am. Soc. Nephrol 24, 1995–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backer G et al. (2018) FAR2 is associated with kidney disease in mice and humans. Physiological Genomics 50, 543–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svenson KL et al. (2012) High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Churchill GA et al. (2012) The Diversity Outbred mouse population. Mamm Genome 23, 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saul MC et al. (2019) High-Diversity Mouse Populations for Complex Traits. Trends Genet. 35, 501–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keane TM et al. (2011) Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gatti DM et al. (2014) Quantitative trait locus mapping methods for diversity outbred mice. G3 4, 1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen C and Spuhler K (1984) Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol. Clin. Exp. Res 8, 477–479 [DOI] [PubMed] [Google Scholar]
- 48.Rat Genome Sequencing and Mapping Consortium et al. (2013) Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat Genet 45, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solberg Woods LC et al. (2012) Fine-mapping diabetes-related traits, including insulin resistance, in heterogeneous stock rats. Physiological Genomics 44, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keele GR et al. (2018) Genetic Fine-Mapping and Identification of Candidate Genes and Variants for Adiposity Traits in Outbred Rats. Obesity (Silver Spring) 26, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holl K et al. (2018) Heterogeneous stock rats: a model to study the genetics of despair-like behavior in adolescence. Genes Brain Behav. 17, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solberg Woods LC et al. (2010) Heterogeneous stock rats: a new model to study the genetics of renal phenotypes. American Journal of Physiology-Renal Physiology 298, F1484–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramdas S et al. (2019) Extended regions of suspected mis-assembly in the rat reference genome. Sci Data 6, 39–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hermsen R et al. (2015) Genomic landscape of rat strain and substrain variation. BMC Genomics 16, 357–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dwinell M et al. (2019) Hybrid Rat Diversity Program (HRDP): A Rat Resource for Systems Genetics. The FASEB Journal [Google Scholar]
- 56.Elmonem MA et al. (2018) Genetic Renal Diseases: The Emerging Role of Zebrafish Models. Cells 7, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Outtandy P et al. (2019) Zebrafish as a model for kidney function and disease. Pediatr Nephrol 34, 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang W et al. (2012) Genome-wide association mapping of quantitative traits in outbred mice. G3 2, 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang F et al. (2013) An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J. Am. Soc. Nephrol 24, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Y et al. (2017) A Drosophila model system to assess the function of human monogenic podocyte mutations that cause nephrotic syndrome. Hum. Mol. Genet 26, 768–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang W et al. (2014) Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24, 1193–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Junginger J et al. (2015) Pathology in Captive Wild Felids at German Zoological Gardens. PLoS ONE 10, e0130573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown CA et al. (2016) Chronic Kidney Disease in Aged Cats: Clinical Features, Morphology, and Proposed Pathogeneses. Vet. Pathol 53, 309–326 [DOI] [PubMed] [Google Scholar]
- 64.Ohara Y et al. (2019) Renal Infiltration of Macrophages in Canine and Feline Chronic Kidney Disease. J. Comp. Pathol 170, 53–59 [DOI] [PubMed] [Google Scholar]
- 65.Makris A et al. (2016) Placental Growth Factor Reduces Blood Pressure in a Uteroplacental Ischemia Model of Preeclampsia in Nonhuman Primates. Hypertension 67, 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cox LA et al. (2013) Baboons as a model to study genetics and epigenetics of human disease. ILAR J 54, 106–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhoads MK et al. (2017) Renal vascular and glomerular pathologies associated with spontaneous hypertension in the nonhuman primate Chlorocebus aethiops sabaeus. Am. J. Physiol. Regul. Integr. Comp. Physiol 313, R211–R218 [DOI] [PubMed] [Google Scholar]
- 68.Stenvinkel P et al. (2013) Hibernating bears (Ursidae): metabolic magicians of definite interest for the nephrologist. Kidney International 83, 207–212 [DOI] [PubMed] [Google Scholar]
- 69.Stenvinkel P et al. (2013) Metabolic changes in summer active and anuric hibernating free-ranging brown bears (Ursus arctos). PLoS ONE 8, e72934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Srivastava A et al. (2019) Genome assembly and gene expression in the American black bear provides new insights into the renal response to hibernation. DNA Res. 26, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu H et al. (2018) Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell 23, 869–881.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]


