Abstract
A Staphylococcus aureus strain deleted for the c‐di‐AMP cyclase gene dacA is unable to survive in rich medium unless it acquires compensatory mutations. Previously identified mutations were in opuD, encoding the main glycine‐betaine transporter, and alsT, encoding a predicted amino acid transporter. Here, we show that inactivation of OpuD restores the cell size of a dacA mutant to near wild‐type (WT) size, while inactivation of AlsT does not. AlsT was identified as an efficient glutamine transporter, indicating that preventing glutamine uptake in rich medium rescues the growth of the S. aureus dacA mutant. In addition, GltS was identified as a glutamate transporter. By performing growth curves with WT, alsT and gltS mutant strains in defined medium supplemented with ammonium, glutamine or glutamate, we revealed that ammonium and glutamine, but not glutamate promote the growth of S. aureus. This suggests that besides ammonium also glutamine can serve as a nitrogen source under these conditions. Ammonium and uptake of glutamine via AlsT and hence likely a higher intracellular glutamine concentration inhibited c‐di‐AMP production, while glutamate uptake had no effect. These findings provide, besides the previously reported link between potassium and osmolyte uptake, a connection between nitrogen metabolism and c‐di‐AMP signalling in S. aureus.
Keywords: AlsT, amino acid transport, c‐di‐AMP, GltS, glutamate transporter, glutamine transporter, OpuD, osmolyte transport, Staphylococcus aureus
A large number of amino acid transporters and oligopeptide permeases are encoded in bacterial genomes. However, their actual substrate specificity and functions are hard to predict bioinformatically. In this study, we report that GltS and AlsT are main glutamate and glutamine transporters in Staphylococcus aureus, respectively, and show that glutamine and ammonium uptake inhibit the production of the nucleotide signalling molecule c‐di‐AMP.
1. INTRODUCTION
In the human host, Staphylococcus aureus can grow in various tissues such as kidneys, bones, heart, soft tissues and lungs (Fridkin et al., 2005; Kluytmans, Belkum, & Verbrugh, 1997). Sensitive regulatory mechanisms enable this organism to rapidly respond to external stimuli and environmental changes. Amongst others, this allows bacteria to adapt their metabolism and utilize different carbon and nitrogen sources available in each specific niche (Crooke et al., 2013; Fridkin et al., 2005; Fuller et al., 2011; Halsey et al., 2017; Lehman et al., 2019; Richardson, Libby, & Fang, 2008; Spahich, Vitko, Thurlow, Temple, & Richardson, 2016; Vitko, Spahich, & Richardson, 2015).
Glucose is the preferred carbon source for S. aureus, but it can be limiting during infection due to the host immune response (Halsey et al., 2017; Kelly & O'Neill, 2015; Lehman et al., 2019; Spahich et al., 2016). In glucose‐limiting conditions, S. aureus instead catabolizes secondary carbon sources and amino acids, particularly glutamate and proline, which serve as major carbon sources during growth in the absence of glucose (Halsey et al., 2017). However, not much is known about amino acid uptake and catabolism in S. aureus and how the availability of certain nutrients can affect virulence factor expression and invasion of the host. While a large number of amino acid transporters and oligopeptide permeases can be identified bioinformatically, their actual substrate specificities and functions in S. aureus have not yet been studied in detail. Predicting the substrates for transporters bioinformatically remains challenging and hence such questions need to be addressed experimentally.
Secondary messenger molecules are crucial in allowing bacteria to rapidly adapt to different environmental and host cell niches (Hengge, 2009; Römling, 2008). There is now considerable evidence that one of these messengers, cyclic di‐adenosine monophosphate (c‐di‐AMP) plays a key role in osmoregulation in bacteria (Bai et al., 2014; Corrigan, Abbott, Burhenne, Kaever, & Gründling, 2011; Devaux et al., 2018; Fahmi, Faozia, Port, & Cho, 2019; Gundlach, Commichau, & Stülke, 2017; Gundlach, Herzberg, Hertel, et al., 2017; Pham et al., 2018; Pham & Turner, 2019; Quintana et al., 2019; Rocha, Teixeira‐Duarte, Jorge, & Morais‐Cabral, 2019; Teh, Dramsi, Tolker‐Nielsen, Yang, & Givskov, 2019; Whiteley et al., 2017; Whiteley, Pollock, & Portnoy, 2015; Witte et al., 2013; Zarrella, Metzger, & Bai, 2018; Zeden et al., 2018). c‐di‐AMP binds to and negatively regulates a number of different potassium and osmolyte importers (Chin et al., 2015; Corrigan et al., 2013; Devaux et al., 2018; Gundlach, Commichau, et al., 2017; Gundlach, Herzberg, Hertel, et al., 2017; Gundlach, Herzberg, Kaever, et al., 2017c; Huynh et al., 2016; Kim et al., 2015; Moscoso et al., 2015; Pham et al., 2018; Pham & Turner, 2019; Quintana et al., 2019; Rocha et al., 2019; Schuster et al., 2016; Zarrella et al., 2018). In many Firmicutes, c‐di‐AMP is essential for bacterial growth under standard rich medium growth conditions, but it is also toxic at high levels. Hence, the cellular levels must be tightly regulated (Corrigan et al., 2011; Corrigan, Bowman, Willis, Kaever, & Gründling, 2015; Gundlach et al., 2015; Mehne et al., 2013; Witte et al., 2013; Woodward, Iavarone, & Portnoy, 2010). In S. aureus, Streptococcus agalactiae and Listeria monocytogenes, deletion of dacA (also referred to as cdaA in many bacteria), coding for the diadenylate cyclase enzyme and responsible for the synthesis of c‐di‐AMP, was only possible in chemically defined medium (Devaux et al., 2018; Whiteley et al., 2015; Zeden et al., 2018), whereas in Bacillus subtilis all three c‐di‐AMP cyclases could only be inactivated in minimal medium also containing low amounts of potassium (Gundlach, Herzberg, Hertel, et al., 2017).
Previously, we found that inactivation of the main glycine betaine transporter OpuD (SAUSA300_1245) as well as the predicted amino acid transporter AlsT (SAUSA300_1252) allows an S. aureus dacA mutant to grow in rich medium in the absence of c‐di‐AMP (Zeden et al., 2018). In several other Firmicutes, including B. subtilis, Lactococcus lactis, Streptococcus pneumoniae, S. agalactiae, Streptococcus pyogenes and L. monocytogenes, inactivating mutations have also been identified in osmolyte and potassium transport systems that allow these bacteria to grow in the absence of c‐di‐AMP (Bai et al., 2014; Corrigan et al., 2011; Devaux et al., 2018; Fahmi et al., 2019; Gundlach, Commichau, et al., 2017; Gundlach, Herzberg, Hertel, et al., 2017; Pham et al., 2018; Pham & Turner, 2019; Quintana et al., 2019; Rocha et al., 2019; Teh et al., 2019; Whiteley et al., 2017, 2015; Witte et al., 2013; Zarrella et al., 2018; Zeden et al., 2018). This suggests that potassium and osmolyte transporters are more active in the absence of c‐di‐AMP, resulting in the accumulation of toxic levels of potassium and osmolytes in the cell. Consistent with a key function of c‐di‐AMP in regulating the osmotic balance in the cell, we found that S. aureus cells show significant differences in cell size depending on their intracellular c‐di‐AMP levels (Corrigan et al., 2011; Zeden et al., 2018). Cells of strain LAC*gdpP, which have high c‐di‐AMP levels, show a decrease in cell size, while cells of strain LAC*dacAG206S, containing low levels of c‐di‐AMP, show an increase in cell size (Corrigan et al., 2011; Zeden et al., 2018). As c‐di‐AMP negatively regulates potassium and osmolyte uptake (Corrigan et al., 2013; Moscoso et al., 2015; Schuster et al., 2016), the increase in cell size is consistent with the hypothesis that an increase in potassium and osmolyte uptake and retention of water at reduced c‐di‐AMP levels leads to the observed increase in cell size. c‐di‐AMP levels affecting bacterial cell size has also been observed for other bacteria such as S. pneumoniae and L. monocytogenes (Bai et al., 2013; Commichau, Heidemann, Ficner, & Stülke, 2019).
AlsT is a predicted amino acid transporter and a correlation between cellular levels of c‐di‐AMP and the amino acids glutamate and glutamine has been reported for B. subtilis and L. monocytogenes (Gundlach, Commichau, et al., 2017; Gundlach et al., 2015; Sureka et al., 2014; Whiteley et al., 2017). A twofold increase in cellular c‐di‐AMP levels was observed in B. subtilis when bacteria where grown in Spizizen minimal medium with glutamate (Glu) as compared to growth in the same medium with glutamine (Gln) as a nitrogen source (Gundlach et al., 2015). In L. monocytogenes, c‐di‐AMP was identified as a negative regulator of the key TCA cycle enzyme pyruvate carboxylase (Sureka et al., 2014). Depletion of c‐di‐AMP resulted in an increased flux into the TCA cycle and as a consequence an increase in the cellular glutamine/glutamate pool (Sureka et al., 2014). In a citZ mutant, which lacks the TCA cycle enzyme citrate synthase and thus has an early block in the TCA cycle, the depletion of c‐di‐AMP no longer resulted in the accumulation of glutamate/glutamine in the cell (Sureka et al., 2014). Interestingly, an L. monocytogenes dacA/citZ double mutant was again viable in rich medium (Sureka et al., 2014; Whiteley et al., 2017).
As part of this study, we further investigated why inactivation of the main glycine‐betaine transporter OpuD and the predicted amino acid transporter AlsT allows S. aureus to grow in the absence of c‐di‐AMP in rich medium. We show that AlsT is a main glutamine transporter in S. aureus and that AlsT‐mediated glutamine uptake, and hence likely a high intracellular glutamine concentration, represses c‐di‐AMP production. Similarly, growth in ammonium‐containing defined medium but not in glutamate‐containing medium, repressed c‐di‐AMP production. The repression of c‐di‐AMP production was independent of the activity of the c‐di‐AMP phosphodiesterase GdpP and the predicted cyclase regulator YbbR. With this study, we not only provide a further link between the c‐di‐AMP signalling network and osmotic regulation in bacterial cells but also with the uptake of specific nitrogen sources and amino acids in S. aureus.
2. RESULTS
2.1. Inactivation of OpuD but not AlsT reduces the cell size of an S. aureus dacA mutant
In previous work, we reported a correlation between the cell size and c‐di‐AMP levels in S. aureus: bacteria with high c‐di‐AMP level are smaller, whereas bacteria with low c‐di‐AMP levels (strain LAC*dacAG206S) are larger as compared to wild‐type (WT) bacteria (Corrigan et al., 2011; Zeden et al., 2018). We also reported that inactivating mutations in opuD (SAUSA300_1245) coding for the main glycine betaine osmolyte transporter and alsT (SAUSA300_1252) coding for a predicted amino acid sodium symporter, rescue the growth defect observed for the c‐di‐AMP negative S. aureus strain LAC*dacA::kan in the rich medium TSB (Zeden et al., 2018). Here, we investigated further the mechanism by which the growth defect of the dacA mutant strain is rescued in the LAC*dacA/opuD and LAC*dacA/alsT suppressor strains. Initially, we compared the cell size of bacteria from the suppressor strains LAC*dacA/opuD and LAC*dacA/alsT to that of WT LAC* and the low c‐di‐AMP level strain LAC*dacAG206S after growth in the rich medium TSB. As expected, the bacteria with low levels of c‐di‐AMP showed an increase in cell size as compared to WT bacteria (Figure 1a,b). While a similar increase in cell size was still observed for bacteria of strain LAC*dacA/alsT, the cell size of LAC*dacA/opuD bacteria, while still increased as compared to the WT, was significantly smaller as compared to the low‐level LAC*dacAG206S strain (Figure 1a,b). Because regular TSB medium is not suitable for the growth of the c‐di‐AMP null strain LAC*dacA::kan, bacterial cell sizes were also determined following growth in TSB medium supplemented with 0.4 M NaCl, which is permissive for the growth of the dacA mutant (Figure 1c–f). Similar to what was observed for the low c‐di‐AMP level dacAG206S mutant strain, the size of bacteria from the c‐di‐AMP null strain LAC*dacA::kan was significantly increased compared to WT bacteria. As observed before, the cell size was not rescued for bacteria of the LAC*dacA/alsT suppressor strain (Figure 1c–f). On the other hand, the size of LAC*dacA/opuD bacteria was similar to that of WT bacteria (Figure 1c–f). Taken together, the observed differences in cell size indicate that the underlying molecular mechanisms enabling the opuD and alsT mutant strains to survive in the absence of c‐di‐AMP in rich medium might be different.
Figure 1.

Inactivation of the glycine betaine transporter OpuD rescues the cell size of S. aureus dacA mutant bacteria. (a, c, e) Microscopy images of S. aureus cells stained with BODIPY‐labelled vancomycin. Cultures of S. aureus LAC* (WT), LAC*dacA G206S (dacA G206S) (panels A and C only), LAC*dacA::kan (dacA) (panel E only) and the suppressor strains LAC*dacA/alsT (dacA/alsT) and LAC*dacA/opuD (dacA/opuD) were grown in (A) TSB or (C and E) TSB 0.4 M NaCl medium and subsequently stained with fluorescently labelled vancomycin. The bacteria were then viewed using a fluorescent microscope and representative images are shown. Scale bars are 1 µm. (b, d, f) Bacterial cell diameter measurements. The diameters of nondividing bacterial cells were measured as described in the Materials and Method section for S. aureus strains grown in (b) TSB or grown in (d and f) TSB 0.4 M NaCl medium. The diameters of 50 cells were determined and the average diameter calculated. The experiment was performed in triplicate (b and f) or quadruplicate (d) and the averages and SDs of the average cell diameters plotted. For statistical analysis, one‐way ANOVAs followed by Dunnett's multiple comparison tests were performed (ns = not significant, * = p < .01, ** = p < .001, *** = p < .0001)
2.2. AlsT is a glutamine transporter in S. aureus
AlsT (SAUSA300_1252) is a predicted amino acid transporter and annotated in the InterPro database (www.ebi.ac.uk/interpro) as alanine/sodium symporter. However, in a previous study no difference in the uptake of radiolabelled alanine was detected between a WT and the LAC*dacA/alsT mutant strain (Zeden et al., 2018), indicating that AlsT is not an alanine transporter. To identify potential substrates for the S. aureus AlsT transporter, we initially followed the depletion of different amino acids from the culture supernatant during the growth of the WT LAC* strain and the isogenic alsT transposon mutant strain LAC*alsT::tn in TSB medium, where both strains exhibit similar growth rates (Figure S1a). However, no drastic differences in the uptake of the different amino acids could be observed between the WT and alsT mutant strains (Figure S1). Of note, using this method, tryptophan uptake cannot be measured and it is also not possible to distinguish between glutamine/glutamate or asparagine/aspartate utilization. Next, a more detailed bioinformatics analysis was performed to identify potential AlsT substrates. A BlastP search against the B. subtilis 168 genome led to the identification of four proteins showing significant homology to the S. aureus AlsT (SAUSA300_1252) protein, namely AlsT (e‐value: e‐166), GlnT (e‐value: e‐149), YrbD (e‐value: e‐117) and YflA (e‐value: 2e‐72). Also in S. aureus, a second AlsT homologue, SAUSA300_0914 (e‐value 9e‐108), could be identified (Figure S2), which is encoded at a different chromosomal region. AlsT is annotated in B. subtilis as a potential glutamine sodium symporter (Zhu & Stülke, 2018), but to the best of our knowledge, this has not yet been experimentally verified. To test if S. aureus AlsT is a potential glutamine or glutamate transporter, uptake assays were performed with radiolabelled glutamine and glutamate using the WT S. aureus strain LAC*, the alsT mutant LAC*alsT::tn piTET and the complementation strain LAC*alsT::tn piTET‐alsT. Uptake of glutamine, but not of glutamate, was severely reduced in the alsT mutant when compared to the WT strain (Figure 2a,b). This defect was restored upon expression of alsT in the complementation strain (Figure 2b). To confirm that alsT functions as main glutamine transporter also in the LAC*dacA/alsT suppressor strain, uptake assays were also performed with strain LAC*dacA/alsT along with the WT LAC* and LAC*dacA::kan control strains (Figure 2c,d). Similar as observed for the alsT single mutant, glutamate uptake was only marginally affected in strain LAC*dacA/alsT (Figure 2c), whereas glutamine uptake was severely reduced in strain LAC*dacA/alsT when compared to the control strains (Figure 2d). These data suggest that under the uptake assay conditions tested, AlsT functions as the main glutamine transporter in S. aureus. Our data further suggest that S. aureus cells that are unable to produce c‐di‐AMP can survive in rich medium such as TSB, when glutamine uptake is reduced or blocked. However, we cannot formally exclude that AlsT is able to transport other amino acid or substrates present in rich medium.
Figure 2.

AlsT is a glutamine transporter in S. aureus. (a–d) Amino acid uptake assays. (a, b) S. aureus strain LAC* (WT), the alsT mutant LAC*alsT::tn piTET (alsT::tn piTET) and the complementation strain LAC*alsT::tn piTET‐alsT (alsT::tn piTET‐alsT) were grown to mid‐log phase in the glucose defined medium GDM + Glu+NH3, which was supplemented with 200 ng/ml Atet for the strains containing plasmids. Subsequently, radiolabelled (a) glutamate or (b) glutamine was added to culture aliquots, samples removed and filtered at the indicated time points and the radioactivity accumulated in the cells measured. The average values and SDs from three (b, d) or four (a, c) experiments were plotted. (c, d) the same uptake assay experiment was performed as described in (a, b) but using S. aureus strains LAC*dacA::kan (dacA) and LAC*dacA/alsT (dacA/alsT). The amino acid uptake curves for the LAC* (WT) strain are the same as shown in panels A and B, as all strains were grown and processed at the same time
2.3. Investigating the contribution of SAUSA300_0914 and GlnQ to glutamine and glutamate transport in S. aureus
S. aureus SAUSA300_0914 codes for a predicted amino acid symporter, which shows 41% identity to the S. aureus AlsT protein. After assigning AlsT a function as glutamine transporter, we wanted to test if SAUSA300_0914 might also play a role in glutamine or glutamate transport. To this end, strain LAC*0914::tn was constructed by transducing the genomic region from the NMTL strain NE1463 (Fey et al., 2013) containing a transposon insertion in SAUSA300_0914 into the S. aureus LAC* background. Subsequently, the uptake of radiolabelled glutamine and glutamate was assessed (Figure 3a,b). No significant differences in the uptake of these amino acids was observed between WT LAC* and strain LAC*0914::tn, showing that SAUSA300_0914 does not function as a major glutamine or glutamate transporter under our assay conditions (Figure 3a,b). AlsT and SAUSA300_0914 are members of the amino acid/sodium symporter family of transporters, which are single, multimembrane spanning proteins. Besides this type of transporter, GlnPQ‐type ABC transporters play a major role in glutamine and glutamate transport in other bacteria (Schuurman‐Wolters & Poolman, 2005). S. aureus contains a glnPQ (SAUSA300_1808–SAUSA300_1807) operon with glnP coding for a substrate‐binding domain‐permease fusion protein and glnQ coding for the cytoplasmic nucleotide‐binding ATPase domain. The results from a previous study suggested that this transporter functions as glutamine transporter in S. aureus, as a glnP mutant was more resistant to the toxic glutamine analogue γ‐L‐glutamyl hydrazide (Zhu et al., 2009). To assess the contribution of the GlnPQ transporter to glutamine and glutamate transport in S. aureus LAC* under our assay and growth conditions, the strain LAC*glnQ::tn was generated by transducing the glnQ::tn region from the NMTL strain NE153 (Fey et al., 2013) into the LAC* background. The resulting glnQ mutant strain LAC*glnQ::tn displayed no difference in glutamine or glutamate uptake compared to WT LAC* (Figure 3c,d). This indicates that the ABC transporter GlnPQ does not function under our assay conditions and in the S. aureus LAC* strain background as a main glutamate or glutamine transporter. However, we cannot exclude that SAUSA300_0914 and GlnPQ could still function as glutamine or glutamate transporters under different growth conditions.
Figure 3.

LAC*0914::tn and LAC*glnQ::tn strains do not show a defect in glutamine or glutamate uptake. Amino acid uptake assays. (a, b) S. aureus strains LAC* (WT), LAC*0914::tn and LAC*alsT::tn were grown to mid‐log phase in GDM + Glu+NH3. Subsequently, radiolabelled (a) glutamine or (b) glutamate was added to culture aliquots, samples removed and filtered at the indicated time points and the radioactivity accumulated in the cells measured. The average values and SDs from three experiments were plotted. (c, d) Amino acid uptake assays were performed and the data plotted as described in panels (a) and (b), but using S. aureus strains LAC* (WT) and LAC*glnQ::tn (glnQ::tn)
2.4. Inactivation of AlsT but not SAUSA300_0914 or GlnQ reduces the susceptibility of S. aureus to the toxic glutamine analogue γ‐L‐glutamyl hydrazide
To further validate the findings from the uptake assays and verify that AlsT is the main glutamine transporter, we performed growth curves in the presence of increasing concentrations of the toxic glutamine analogue γ‐L‐glutamyl hydrazide with the WT and LAC*alsT::tn mutant strains. Strains LAC*0914::tn and LAC*glnQ::tn were also included in these assays, to uncover a potential low‐level glutamine uptake activity for the SAUSA300_0914 and GlnPQ transporters. Strains defective in taking up this glutamine analogue are expected to show reduced susceptibility to this toxic compound. In the absence of the compound, all strains grew similarly in the chemically defined medium used for this assay (Figure 4a). As expected, addition of γ‐L‐glutamyl hydrazide reduced the growth of the WT LAC* strain, in a dose‐deponent manner (Figures 4b and S3). Similar growth inhibition curves to that of the WT strain were obtained for strains LAC*0914::tn and LAC*glnQ::tn, while strain LAC*alsT::tn showed increased resistance to the compound (Figures 4b and S3). These findings support our earlier conclusion that AlsT is the main glutamine transporter in S. aureus LAC* under our assay conditions, while GlnPQ and SAUSA300_0914 are either unable to take up glutamine or play only a minor role in its uptake under our growth conditions.
Figure 4.
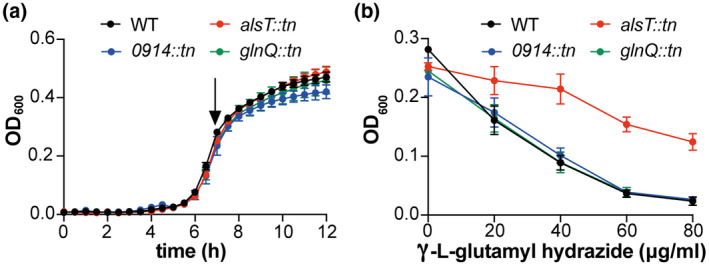
LAC*alsT::tn shows increased resistance to the toxic glutamine analogue γ‐L‐glutamyl hydrazide. (a) Bacterial growth curves. S. aureus strains LAC* (WT), LAC*alsT::tn, LAC*0914::tn and LAC*glnQ::tn were grown in GDM + NH3 medium in 96‐well plates and OD600 reading determined over 12 hr. The average OD600 values and SDs from three independent biological replicates were plotted. (b) γ‐L‐glutamyl hydrazide susceptibility assay. The same S. aureus strains as in (a) were grown in GDM + NH3 medium in the absence or presence of γ‐L‐glutamyl hydrazide at a final concentration of 20, 40, 60 or 80 μg/ml. OD600 reading were determined over 12 hr and the complete growth curves are shown in Figure S3. In this graph, the average OD600 values from the 7 hr time point (marked with an arrow in (a)) and SDs from three biological replicates were plotted against the γ‐L‐glutamyl hydrazide concentration in the growth medium
2.5. GltS (SAUSA300_2291) is a glutamate transporter in S. aureus
S. aureus does not only take up glutamine but also shows robust glutamate uptake (Figures 2 and 3). However, none of the transporters (AlsT, SAUSA300_0914 and GlnPQ) investigated so far plays a major role in glutamate uptake under our growth conditions. In B. subtilis GltT, belonging to the dicarboxylate/amino acid cation symporter (DAACS) family of proteins, is a major high‐affinity Na+‐coupled glutamate/aspartate symporter and can also mediate the uptake of glyphosate (Wicke et al., 2019; Zaprasis, Bleisteiner, Kerres, Hoffmann, & Bremer, 2015). Two paralogs, DctP and GltP, are found in B. subtilis of which GltP has also been shown to be a glutamate transporter (Tolner, Ubbink‐Kok, Poolman, & Konings, 1995). The S. aureus protein SAUSA300_2329 (from here on referred to as GltT) shows a high degree of similarity (52% identity) to the B. subtilis GltT protein. In addition, SAUSA300_2291 (from here on referred to as GltS) is annotated in UniProt (www.uniprot.org) as a potential glutamate transporter in S. aureus. To experimentally test if GltT or GltS impact glutamate transport in S. aureus, strains LAC*gltT::tn and LAC*gltS::tn were constructed by moving the respective gltT and gltS transposon insertion regions from the NMTL strains NE566 and NE560 (Fey et al., 2013) into the LAC* strain background. Next, the uptake of radiolabelled glutamine and glutamate was assessed for WT LAC* and strains LAC* gltT::tn and LAC* gltS::tn. No difference in the uptake of glutamine was observed between the strains (Figure 5a) and in the case of LAC*gltT::tn, also no difference in the uptake of glutamate was observed (Figure 5b). However, a significant reduction in glutamate uptake was observed for strain LAC*gltS::tn when compared to the WT strain (Figure 5b). The glutamate uptake defect could be restored in a complementation strain harbouring plasmid piTET‐gltS allowing for inducible gltS expression (Figure 5c). Indeed, increased glutamate uptake was observed in the complementation strain, indicating increased gltS expression in the complementation strain as compared to the WT strain. Taken together, these data indicate that under the growth conditions tested, GltS is the main glutamate transporter in S. aureus.
Figure 5.

GltS is a glutamate transporter in S. aureus. Amino acid uptake assays. (a,b) S. aureus strains LAC* (WT), LAC*gltT::tn and LAC*gltS::tn were grown to mid‐log phase in GDM + Glu+NH3. Subsequently, radiolabelled (a) glutamine or (b) glutamate was added to culture aliquots, samples removed and filtered at the indicated time points and the radioactivity accumulated in the cells measured. (c) Same as (b) but using S. aureus strains LAC* (WT), LAC*gltS::tn piTET and the complementation strain LAC*gltS::tn piTET‐gltS and supplementing the GDM + Glu+NH3 medium with 200 ng/µl Atet. The average values and SDs from three experiments were plotted
2.6. Ammonium and glutamine but not glutamate stimulate the growth of S. aureus in defined medium containing glucose as carbon source
Glutamine and glutamate are important amino acids that can serve, together with ammonium, as nitrogen sources for the synthesis of many other cellular metabolites. Since S. aureus is phenotypically auxotroph for many amino acids and at the same time can use several amino acids as carbon and nitrogen sources, it is not possible to grow this organism in any of the typical minimal media that are used to assess the ability of bacteria to specifically use ammonium, glutamine or glutamate as nitrogen sources. However, to begin to examine the effect of these compounds on the growth of S. aureus, growth curves were performed with the WT LAC* strain in glucose containing defined medium (GDM), containing essential vitamins, metals and 17 amino acids but lacking ammonium, glutamine and glutamate as potential nitrogen/amino acid sources (see Table S1 for medium composition). In addition, the WT LAC* strain was also grown in GDM containing glutamine (GDM + Gln), glutamate (GDM + Glu), ammonium (GDM + NH3), glutamine and ammonium (GDM + Gln + NH3) or glutamate and ammonium (GDM + Glu+NH3). The addition of glutamine or ammonium alone or in combinations stimulated the growth of the WT LAC* strain as compared to its growth in GDM (Figure 6a). On the other hand, no growth improvement was seen in the presence of glutamate (GDM + Glu) (Figure 6a). To examine the contribution of the glutamine and glutamate transporters AlsT and GltS, additional growth curves were performed in the different media with WT LAC* as well as strains LAC*alsT::tn and LAC*gltS::tn (Figure S4). Similar growth profiles were observed for all strains in the different media (Figure S4), except in GDM + Gln, in which the alsT mutant strain exhibited reduced growth compared to the WT and gltS mutant strains (Figure S4b). The growth defect could be restored in the alsT complementation strain harbouring plasmid piTET‐alsT (Figure 6b). Taken together, these data indicate that ammonium and glutamine are preferred over glutamate for the growth of S. aureus. The observation that the addition of ammonium improves the growth of S. aureus indicates that our base medium is likely nitrogen limiting and suggests that glutamine but not glutamate can likely also serve as nitrogen source under these growth conditions. Finally, these data further confirm the importance of AlsT for glutamine uptake in S. aureus.
Figure 6.
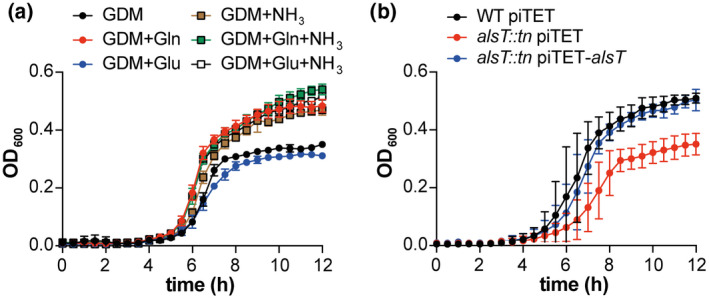
Addition of glutamine and ammonium but not glutamate stimulates the growth of S. aureus in glucose‐containing defined medium. (a) Bacterial growth curves. The WT S. aureus strain LAC* was grown in 96‐well plates in glucose‐containing defined medium (GDM) or in GDM supplemented with glutamine (Gln), glutamate (Glu), ammonium (NH3) or combinations thereof as specified in the legend. OD600 readings were determined every 30 min and the average and SDs of three biological replicates plotted. (b) Bacterial growth curves. S. aureus strains LAC* piTET (WT piTET), the alsT mutant strain LAC*alsT::tn piTET and the complementation strain LAC*alsT::tn piTET‐alsT were grown in GDM + Gln medium supplemented with 200 ng/µl Atet. OD600 readings were determined every 30 min and the average and SDs of three biological replicates plotted
2.7. Ammonium and glutamine uptake lead to a reduction in c‐di‐AMP levels in S. aureus
For B. subtilis, it has been reported that the addition of glutamine, glutamate or ammonium to a defined growth medium can affect cellular c‐di‐AMP levels (Gundlach et al., 2015). It was further proposed that glutamate uptake and to some extent also ammonium uptake leads to an activation of c‐di‐AMP synthesis in this organism (Gundlach et al., 2015). To assess if the presence of glutamine, glutamate or ammonium would also affect c‐di‐AMP levels in S. aureus, the intracellular c‐di‐AMP concentrations were determined for the WT S. aureus strain LAC* following growth in GDM, GDM + Gln, GDM + Glu, GDM + NH3, GDM + Gln+NH3 and GDM + Glu+NH3. Using a competitive ELISA assay, c‐di‐AMP could be readily detected in bacteria grown in GDM, our base medium (Figure 7a). Similar amounts of c‐di‐AMP were detected in bacteria grown in the glutamate‐containing medium (GDM + Glu); however, the c‐di‐AMP levels were significantly lower in bacteria grown in medium containing either ammonium or glutamine (GDM + Gln, GDM + NH3, GDM + Gln+NH3, GDM + Glu+NH3) (Figure 7a). To verify that the addition of glutamine reduces c‐di‐AMP production and to investigate the contribution of the glutamine transporter AlsT to this inhibition, c‐di‐AMP levels were determined for WT piTET, the alsT mutant LAC*alsT::tn piTET and the complementation strain LAC*alsT::tn piTET‐alsT following growth in the glutamine containing medium GDM + Gln. While again low c‐di‐AMP levels were detected for the WT strain, the c‐di‐AMP levels increased significantly in the alsT mutant and were restored back to WT levels in the complementation strain (Figure 7b). A similar experiment was performed with the gltS mutant and complementation strain in the glutamate containing medium GDM + Glu. High and similar c‐di‐AMP levels were detected for all strains (Figure 7c), indicating that neither the addition of glutamate to the medium nor its uptake impacts c‐di‐AMP production in S. aureus under our test conditions. Taken together, these data highlight that ammonium as well as AlsT‐mediated glutamine uptake, which likely results in a higher intracellular glutamine concentration, represses c‐di‐AMP production in S. aureus.
Figure 7.

Ammonium and glutamine uptake inhibit c‐di‐AMP production in S. aureus. (a–c) Cellular c‐di‐AMP levels. (a) The WT S. aureus strain LAC* was grown in GDM or GDM supplemented with glutamine (Gln), glutamate (Glu), ammonium (NH3), or combinations thereof as indicated on the x axis. Cell extracts were prepared, and c‐di‐AMP concentrations measured using a competitive ELISA assay. The average values and SDs from three biological replicates were plotted as ng c‐di‐AMP/mg protein. For statistical analysis, one‐way ANOVAs followed by Dunnett's multiple comparison tests were performed to identify statistically significant differences between the different media as compared to GDM (ns = not significant, ** = p < .001, *** = p < .0001). (b) S. aureus strains LAC* piTET (WT piTET), LAC*alsT::tn piTET and the complementation strain LAC*alsT::tn piTET‐alsT were grown in GDM + Gln supplemented with 200 ng/µl Atet and c‐di‐AMP levels determined and plotted as described in (a). (c) S. aureus strains LAC* piTET (WT piTET), LAC*gltS::tn piTET and the complementation strain LAC*gltS::tn piTET‐gltS were grown in GDM + Glu supplemented with 200 ng/µl Atet and c‐di‐AMP levels determined and plotted as described in (a)
2.8. The inhibition of the c‐di‐AMP production by glutamine and ammonium is not mediated by GdpP or YbbR
The observed reduction of c‐di‐AMP levels in the presence of glutamine or ammonium could potentially be achieved through an increase in the activity of the c‐di‐AMP‐specific phosphodiesterase GdpP. To investigate this, cellular c‐di‐AMP levels were compared between WT LAC* and the isogenic gdpP mutant strain LAC*gdpP::kan. As previously reported for strain LAC*gdpP::kan following growth in TSB medium (Corrigan et al., 2011), the c‐di‐AMP levels were also increased in the gdpP mutant compared to the WT strain following growth in GDM, the glucose containing defined medium used as part of this study (Figure 8a). However, a significant reduction in the cellular c‐di‐AMP levels was also seen for the gdpP mutant following the addition of glutamine or ammonium to the medium (Figure 8a). This indicates that the reduction in c‐di‐AMP levels upon addition of glutamine or ammonium is likely due to decreased synthesis by DacA and not increased degradation by GdpP. We next tested the involvement of YbbR, a proposed c‐di‐AMP cyclase regulator, by comparing the cellular c‐di‐AMP levels produced by WT LAC* and strain LAC*ΔybbR. Similar c‐di‐AMP levels were detected in the WT and ybbR mutant in GDM medium (Figure 8b). The addition of glutamine or ammonium to the medium led also to a large reduction in the cellular c‐di‐AMP in the ybbR mutant strain (Figure 8b). These data suggest that the observed reduction of c‐di‐AMP production in the presence of glutamine and ammonium is neither mediated by GdpP nor YbbR, and hence involves a different regulator protein, or that the cellular glutamine and nitrogen levels are directly sensed by the cyclase DacA.
Figure 8.
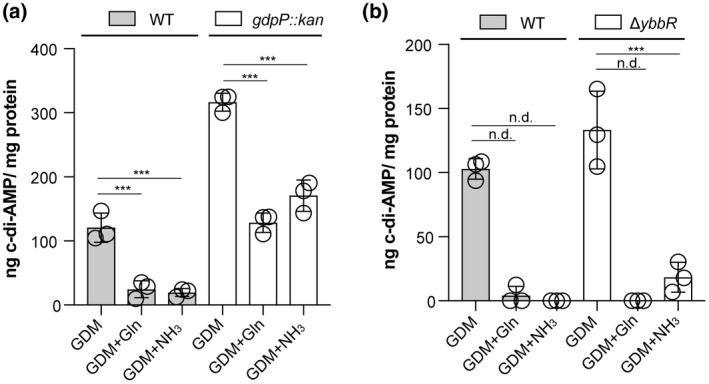
The inhibition of c‐di‐AMP production by glutamine and ammonium is independent of GdpP and YbbR. (a, b) Cellular c‐di‐AMP levels. (a) S. aureus strains LAC* (WT) and LAC*gdpP::kan (gdpP::kan) were grown in GDM or in GDM containing glutamine (GDM + Gln) or ammonium (GDM + NH3). Cell extracts were prepared, and c‐di‐AMP concentrations measured using a competitive ELISA assay. The average values and SDs from three biological replicates were plotted as ng c‐di‐AMP/ mg protein. For statistical analysis one‐way ANOVAs followed by Dunnett's multiple comparison tests were performed to identify statistically significant differences between the different media as compared to the GDM medium (n.d. = not determined. Statistical analysis was not performed due to values being below our detection limit; these values were set to 0, ** = p < .001, *** = p < .0001). (b) Same as in (a) but using S. aureus strains LAC* (WT) and LAC*ΔybbR (ΔybbR)
3. DISCUSSION
Over the last decade, considerable evidence has emerged that c‐di‐AMP plays a major role in osmotic regulation in bacteria, primarily by positively regulating potassium export or negatively regulating potassium and osmolyte uptake (Chin et al., 2015; Corrigan et al., 2013; Devaux et al., 2018; Gundlach, Commichau, et al., 2017; Gundlach, Herzberg, Hertel, et al., 2017; Gundlach, Herzberg, Kaever, et al., 2017c; Gundlach et al., 2019; Huynh et al., 2016; Kim et al., 2015; Moscoso et al., 2015; Pham et al., 2018; Pham & Turner, 2019; Quintana et al., 2019; Rocha et al., 2019; Schuster et al., 2016; Zarrella et al., 2018). However, individual c‐di‐AMP target proteins identified thus far are themselves not essential. Therefore, the essentiality of c‐di‐AMP is likely due to its ability to regulate multiple target proteins simultaneously. Furthermore, in the absence of this molecule, many transporters are activated rather than inactivated, likely leading to accumulation of toxic levels of metabolites, such as potassium and osmolytes. Consistent with this idea, inactivating mutations in potassium uptake systems, oligopeptide and osmolyte transporters have been reported to rescue the growth defect of bacteria unable to produce c‐di‐AMP (Devaux et al., 2018; Gundlach, Herzberg, Hertel, et al., 2017; Gundlach, Herzberg, Kaever, et al., 2017c; Pham et al., 2018; Whiteley et al., 2017, 2015; Zeden et al., 2018). We have previously shown that inactivation of the main glycine betaine transporter OpuD bypasses the requirement of c‐di‐AMP for the growth of S. aureus in rich medium (Zeden et al., 2018). We hypothesize that inactivation of OpuD might help a c‐di‐AMP null strain survive by allowing bacteria to re‐establish their osmotic balance. Bacteria of the dacA/opuD mutant strain, which cannot produce c‐di‐AMP but are also defective in glycine betaine transport, are similar in size to WT bacteria (Figure 1). At this point, it is not known if c‐di‐AMP can directly bind to and regulate the activity of the S. aureus OpuD protein. We attempted to address this question; however, despite using multiple different approaches, we were unable to produce sufficient amounts of the full‐length OpuD membrane protein to perform protein/nucleotide interaction studies. On the other hand, a direct role for c‐di‐AMP in the control of glycine betaine or betaine transporters has been reported in S. agalactiae and L. lactis. In these organisms, c‐di‐AMP binds to the transcriptional regulator BusR, which controls the expression of the predicted glycine betaine or betaine transporter BusAB (Devaux et al., 2018; Pham et al., 2018).
Bacteria of the dacA/alsT suppressor strain, which survive in the absence of c‐di‐AMP, remained enlarged, indicating that the essentiality of c‐di‐AMP is bypassed in this strain potentially through a different mechanism. Here, we show that AlsT is an efficient glutamine transporter in S. aureus (Figure 2). These findings indicate that eliminating or reducing the ability of S. aureus to take up glutamine from rich growth medium rescues the growth of an S. aureus unable to produce c‐di‐AMP. There are several (not mutually exclusive) possibilities how preventing glutamine uptake could rescue the growth of a c‐di‐AMP null strain in rich medium. Glutamine as well as proline have been shown to accumulate in S. aureus under NaCl stress conditions (Anderson & Witter, 1982). While it has been suggested that the glutamine accumulation is due to synthesis rather than uptake (Anderson & Witter, 1982), these data highlight that glutamine likely plays an important role in osmotic regulation in S. aureus. Despite the cell size not being restored in the dacA/alsT suppressor strain, blocking glutamine uptake could potentially still help bacteria to better balance their cellular osmolality during growth in rich medium in the absence of c‐di‐AMP. Another possible explanation how eliminating glutamine uptake could allow S. aureus to grow in the absence of c‐di‐AMP could be connected to changes in metabolism and TCA cycle activity. In L. monocytogenes, an increased flux of pyruvate into the TCA cycle has been described for a strain unable to produce c‐di‐AMP (Sureka et al., 2014). As a consequence, an accumulation of citrate and increased carbon flux into glutamine/glutamate was observed, which resulted in a metabolic imbalance and growth defect (Sureka et al., 2014). Perhaps similar to the observations in L. monocytogenes, the absence of c‐di‐AMP could also boost TCA cycle activity in S. aureus, thus leading to glutamine accumulation and a metabolic imbalance. Hence, the lack of c‐di‐AMP combined with active glutamine uptake could fuel the bacterial metabolism and the resulting metabolic imbalance might become toxic to the cell, similar as observed for L. monocytogenes (Sureka et al., 2014; Whiteley et al., 2017).
In a recent study investigating genetic determinants required for eDNA release during biofilm formation, it was found that inactivation of GdpP as well as of AlsT, resulted in a significant decrease in eDNA release and in an increase in resistance to Congo red (DeFrancesco et al., 2017). Therefore, inactivation of AlsT and preventing/reducing glutamine uptake might lead to alterations in the bacterial cell wall that make bacteria more resistant to cell lysis. Such changes could also be an advantage during osmotic stress or c‐di‐AMP deficiency. Indeed, we have recently shown a correlation between specific changes in the peptidoglycan structure and the NaCl stress resistance in S. aureus (Schuster et al., 2019). In addition, since the cellular c‐di‐AMP levels are significantly higher in the gdpP as well as the alsT mutant strains compared to WT (Figures 7b and 8a), the underlying mechanistic bases for the decrease in eDNA release observed for the gdpP and alsT mutant strains might be related.
The actual stimuli and underlying molecular mechanisms that regulate c‐di‐AMP production in bacterial cells are at the moment poorly understood. As part of this study, we show that ammonium and AlsT‐mediated glutamine uptake (and likely a reduction in the cellular glutamine levels) but not GltS‐mediated glutamate uptake negatively impacts c‐di‐AMP production (Figure 7). Changes in cellular c‐di‐AMP levels depending on the presence of ammonium, glutamine or glutamate have already been reported for B. subtilis (Gundlach et al., 2015). For B. subtilis it has been suggested that glutamine and to some extent ammonium uptake stimulates c‐di‐AMP production (Gundlach et al., 2015). Here, we show that in S. aureus ammonium and glutamine uptake leads to an inhibition of c‐di‐AMP production rather than glutamate promoting its synthesis (Figure 7). The decrease in c‐di‐AMP production in the presence of ammonium or glutamine is likely achieved by reducing the activity of the c‐di‐AMP cyclase DacA and not by activation of the c‐di‐AMP‐specific phosphodiesterase GdpP. This conclusion is based on our observation that the cellular c‐di‐AMP levels are also decreased in a gdpP mutant strain upon addition of glutamine or ammonium (Figure 8a). Current evidence suggests that the activity of DacA can be regulated through the interaction with two proteins: the membrane anchored and proposed DacA regulator protein YbbR (also named as CdaR in other bacteria) and the phosphoglucosamine mutase enzyme GlmM (Gundlach et al., 2015; Pham, Liang, Marcellin, & Turner, 2016; Tosi et al., 2019; Zhu et al., 2016). We could exclude that the observed reduction in cellular c‐di‐AMP levels in the presence of ammonium or glutamine is mediated by YbbR, as a ybbR mutant showed a similar decrease in the c‐di‐AMP levels as observed for the WT strain (Figure 8b). GlmM has been shown to be a negative regulator of DacA activity both in vivo and in vitro (Pham et al., 2018; Tosi et al., 2019). However, since GlmM is likely an essential enzyme in S. aureus, we were unable to construct a glmM mutant and test its involvement in the observed repression of c‐di‐AMP synthesis in the presence of ammonium or glutamine as we did for the gdpP and ybbR mutant strains. Nevertheless, with this work, we not only identified main glutamine and glutamate transporters in S. aureus, but we also linked the c‐di‐AMP signalling network to central nitrogen metabolism in S. aureus. It will be interesting to determine in future studies the mechanistic bases for the observed changes in cellular c‐di‐AMP levels depending on ammonium and glutamine uptake and the involvement of GlmM or other factors in this process.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains and culture conditions
Bacterial strains used in this study are listed in Table 1. S. aureus strains were grown in tryptic soy broth (TSB), where indicated supplemented with 0.4 M NaCl, tryptic soy agar (TSA) or glucose defined medium (GDM). GDM was prepared similar to the chemically defined medium (CDM) reported in an earlier study (Zeden et al., 2018), with some modifications. The detailed content of the GDM (which contains essential vitamins, trace metals, amino acids and glucose as a carbon source but lacks ammonium, glutamine or glutamate as a potential nitrogen or amino acid source) is shown in Table S1. In addition to the GDM, GDM containing glutamine (GDM + Gln), glutamate (GDM + Glu), ammonium (GDM + NH3), glutamine and ammonium (GDM + Gln+NH3) or glutamate and ammonium (GDM + Glu+NH3) were used as part of this study (for exact composition see Table S1). Escherichia coli strains were grown in lysogeny broth (LB). Where appropriate, antibiotics and/or inducers were added to the media at the following concentration: 200 ng/ml anhydrotetracycline (Atet), 90 µg/ml Kanamycin (Kan), 10 µg/ml Erythromycin (Erm), 7.5 or 10 µg/ml Chloramphenicol (Cam) and Ampicillin (Amp) 100 µg/ml.
Table 1.
Bacterial strains used in this study
| Unique ID | Strain name and resistance | Source |
|---|---|---|
| Escherichia coli strains | ||
| ANG284 | XL1‐Blue piTET; AmpR | Gründling and Schneewind (2007) |
| ANG2154 | DH10B pIMAY; CamR | (Monk, Shah, Xu, Tan, & Foster, 2012) |
| ANG3724 | IM08B | Monk, Tree, Howden, Stinear, and Foster (2015) |
| ANG3928 | IM08B piTET; AmpR | Zeden et al. (2018) |
| ANG3937 | XL1‐Blue piTET‐alsT; AmpR | This study |
| ANG3955 | IM08B piTET‐alsT; AmpR | This study |
| ANG5494 | XL1‐Blue piTET‐gltS; AmpR | This study |
| ANG5495 | IM08B piTET‐gltS; AmpR | This study |
| Staphylococcus aureus strains | ||
| AH1263 | LAC* Erm sensitive CA‐MRSA USA300 strain (ANG1575) | Boles, Thoendel, Roth, and Horswill (2010) |
| ANG1961 | LAC*gdpP::kan; KanR | Corrigan et al. (2011) |
| ANG3301 | LAC*ΔybbR | Bowman et al. (2016) |
| ANG3664 | LAC*dacAG206S; KanR | Bowman et al. (2016) |
| ANG3666 | LAC*dacA::kan (dacA) KanR | Zeden et al. (2018) |
| ANG3835 | LAC*dacA::kan‐S7 (LAC*dacA/opuD); KanR | Zeden et al. (2018) |
| ANG3838 | LAC*dacA::kan‐S10 (LAC*dacA/alsT)]; KanR | Zeden et al. (2018) |
| ANG3940 | NE142 (alsT::tn)—NMTN strain | Fey et al. (2013) |
| ANG4054 | LAC* piTET; CamR | Zeden et al. (2018) |
| ANG4803 | LAC*alsT::tn; ErmR | This study |
| ANG4854 | LAC*alsT::tn piTET‐alsT; ErmR CamR | This study |
| ANG4853 | LAC*alsT::tn piTE; ErmR CamR | This study |
| ANG4968 | NE1463 (JE2 SAUSA300_0914::tn)—NMTN strain | Fey et al. (2013) |
| ANG5070 | NE153 (JE2 glnQ::tn)—NMTN strain | Fey et al. (2013) |
| ANG5141 | LAC*0914::tn; ErmR | This study |
| ANG5242 | LAC*glnQ::tn; ErmR | This study |
| ANG5309 | NE566 (JE2 gltT::tn)—NMTN strain | Fey et al. (2013) |
| ANG5310 | NE560 (JE2 gltS::tn)—NMTN strain | Fey et al. (2013) |
| ANG5366 | LAC*gltT::tn; ErmR | This study |
| ANG5367 | LAC*gltS::tn; ErmR | This study |
| ANG5492 | LAC*gltS::tn piTET; ErmR CamR | This study |
| ANG5493 | LAC*gltS::tn piTET‐gltS; ErmR CamR | This study |
4.2. Bacterial strain construction
All strains used in this study are listed in Table 1 and primers used in this study are listed in Table 2. The transposon insertion sites in the Nebraska transposon mutant library (NTML) strains (Fey et al., 2013) used as part of this study were confirmed by PCR and sequencing. The transposon and surrounding regions were moved by phage transduction using phage 85 into the S. aureus LAC* strain background. This resulted in the generation of S. aureus strains LAC*alsT::tn (SAUSA300_1252::tn; ANG4803), LAC*0914::tn (SAUSA300_0914::tn; ANG5141), LAC*glnQ::tn (SAUSA300_1807::tn; ANG5070), LAC*gltT::tn (SAUSA300_2329::tn; ANG5366) and LAC*gltS::tn (SAUSA300_2291::tn; ANG5367). The transposon insertion in the respective gene was again confirmed by PCR and sequencing. For complementation analysis, the Atet inducible single‐copy integration plasmids piTET‐alsT and piTET‐gltS were constructed. To this end, alsT (SAUSA300_1252) and gltS (SAUSA300_2291) were amplified using LAC* chromosomal DNA and primers ANG2250/ANG2251 and ANG3209/ANG3210 respectively. The products as well as piTET were digested with AvrII and SacII and then ligated. Plasmid piTET‐alsT was recovered in E. coli strain XL1‐Blue (yielding strain ANG3937), shuttled through E. coli strain IM08B (strain ANG3955) and then introduced into LAC*alsT::tn (ANG4803), yielding strain LAC*alsT::tn piTET‐alsT (ANG4854). As a control, plasmid piTET was also introduced into LAC*alsT::tn (ANG4803) yielding strain LAC*alsT::tn piTET (ANG4853). Plasmid piTET‐gltS was transformed into E. coli XL1‐Blue (yielding strain ANG5494), shuttled through E. coli IM08B (yielding strain ANG5495) and transformed into LAC*gltS::tn, yielding the complement strain LAC*gltS::tn piTET‐gltS (ANG5493). As a control, the piTET plasmid was transformed into LAC*gltS::tn strain, yielding the strain LAC*gltS:tn piTET (ANG5492). Correct plasmid integration into the geh locus was confirmed by PCR and the sequences of all plasmid inserts were confirmed by fluorescent automated sequencing.
Table 2.
Cloning primers used in this study
| Primer ID | Name | Sequence |
|---|---|---|
| ANG2250 | 5‐AvrII‐alsT | AGTCCCTAGGCGGTCTAATTTTATAGAAGG |
| ANG2251 | 3‐SacII‐alsT | TCCCCGCGGGGTTTATTTGATTTTTATATAATGAATCG |
| ANG3209 | 5‐AvrII‐gltS | ATACCTAGGAGGGAGAGGGATATTCAACAAGGGGGATTTG |
| ANG3210 | 3‐SacII‐gltS | GCCCGCGGTTTAACTAAACCATTGTATGAATCCCATAATG |
4.3. Bacterial growth curves and amino acid analysis in culture supernatants
S. aureus strains LAC* and LAC*alsT::tn were grown overnight in TSB supplemented with 10 µg/ml erythromycin where appropriate. Overnight cultures were then diluted to an OD600 of 0.01 into 50 ml of fresh TSB. Cultures were incubated at 37°C with aeration, and OD600 values determined every hour. The experiment was performed with three biological replicates and the average OD600 values and standard deviations (SDs) were plotted. Using the same cultures, supernatant samples were prepared at the 0, 6, 10 and 12 hr time points and amino acid levels determined as previously described using an amino acid analyser (Halsey et al., 2017). For measuring the growth of S. aureus strains LAC*, LAC* piTET, LAC*alsT::tn, LAC*alsT::tn piTET, LAC*alsT::tn piTET‐alsT and LAC*gltS::tn in GDM, GDM + Gln, GDM + Glu, GDM + NH3, GDM + Gln+NH3 or GDM + Glu+NH3, the bacteria were grown overnight in TSB medium supplemented with chloramphenicol and erythromycin where appropriate. Next day, bacteria from a 1 ml aliquot were washed twice with PBS and diluted to an OD600 of 0.005 in the indicated GDM. LAC* WT, LAC*alsT::tn and LAC*gltS::tn were grown in GDM, GDM + Gln, GDM + Glu, GDM + NH3, GDM + Gln+NH3 and GDM + Glu+NH3, while LAC* piTET, LAC*alsT::tn piTET and LAC*alsT::tn piTET‐alsT were grown in GDM + Gln supplemented with 200 ng/ml Atet. One hundred microlitres of the diluted cultures (six technical replicates) were transferred into wells of a 96‐well plate and the plate was then incubated with shaking (500 rpm) in a plate reader and OD600 readings determined every 30 min. The average values of the technical replicates were determined for each strain. The experiment was performed three times and the average readings and standard deviations were plotted.
4.4. γ‐L‐glutamyl hydrazide susceptibility assay
The susceptibility of S. aureus LAC*, LAC*alsT::tn (ANG4803), LAC*0914::tn (ANG5141) and LAC*glnQ::tn (ANG5242) to the toxic glutamine analogue γ‐L‐glutamyl hydrazide (Alfa Aesar, MA, USA) was determined using a similar method as previously reported (Zhu et al., 2009). Briefly, the different strains were grown overnight at 37°C in 5 ml TSB medium, supplemented with 10 μg/ml erythromycin where appropriate. Next day, the bacteria were washed twice with PBS, diluted to an OD600 of 0.005 in GDM + NH3. Next, 10 μl of water (0 mM control) or 10 μl of a γ‐L‐glutamyl hydrazide solution dissolved in water was added to 0.99 ml aliquots of these bacterial suspension to give a final concentration of 20, 40, 60 or 80 μg/ml, respectively. One hundred microlitres were subsequently transferred in four replicates into wells of a 96‐well plates and the plate incubated at 37°C with shaking (500 rpm) in a plate reader and OD600 readings determined every 10 min for 12 hr. The experiment was performed three times and the average OD600 values of the three experiments presented as growth curves. The average values and SDs of the OD600 values from the 7 hr time point were also plotted against the different γ‐L‐glutamyl hydrazide concentrations.
4.5. Microscopic analysis and cell size measurements
The microscopic analysis to determine bacterial cell sizes was performed essentially as previously described (Zeden et al., 2018). Briefly, S. aureus strains LAC*, LAC*dacA::kan, LAC*dacA G206S, LAC*dacA/opuD (ANG3835) and LAC*dacA/alsT (ANG3838) were grown overnight at 37°C in TSB or TSB supplemented with 0.4 M NaCl where stated. Next day, the cultures were diluted to an OD600 of 0.01 and grown for 3 hr at 37°C to mid‐exponential phase (OD600 of 0.5–0.9). One hundred microlitres of these cultures were then stained for 20 min at 37°C with Vancomycin‐BODIPY FL used at a final concentration of 2 µg/ml. One and a half microlitres of each sample were spotted onto a thin 1.5% agarose gel patch prepared in H2O or in 0.4 M NaCl and the bacteria subsequently imaged at 1,000× magnification using an Axio Imager A2 Zeiss microscope equipped with a GFP filter set. Images were acquired using the ZEN 2012 (blue edition) software. The bacterial cell diameters were determined using the Fiji software. Only nondividing cells (cells without any obvious fluorescent dots or lines at the mid‐cell), were used for cell diameter measurements. The cell diameters of 50 cells were measured and the average cell diameter determined. The experiment was conducted three or four times (as indicated in the figure legend) and the averages and standard deviations of the average cell diameters plotted.
4.6. Uptake assays using 14C‐labelled amino acids
Uptake assays were conducted as previously described with some minor modifications (Zeden et al., 2018). Briefly, S. aureus strains were streaked on TSA or TSA 0. 4M NaCl plates with appropriate antibiotics and the plates incubated overnight at 37°C. Bacteria were subsequently scraped off from the plates and suspended in 1 ml PBS pH 7.4 buffer and the OD600 determined. Fifty ml of GDM + Glu+NH3 (where indicated with 200 ng/ml of the inducer Atet added) were inoculated with the appropriate bacterial suspensions to an OD600 of 0.05. The cultures were grown at 37°C to an OD600 between 0.4 and 0.9 and bacteria from an OD600 equivalent of 8 were harvested by centrifugation for 10 min at 19,000 x g at RT. Supernatants were discarded and the bacterial pellets were suspended in 2 ml of GDM + NH3. The OD600 of the cell suspensions were measured and the cells diluted to an OD600 of approximately 1. The OD600 was re‐measured and this measurement used for normalization purposes. Five hundred and fifty microlitres of these cell suspensions were aliquoted into 50 ml conical tubes and 100 µl used to measure the background radiation, by filtering the cells onto a nitrocellulose membrane filter, followed by a wash step with 16 ml PBS. Then, 6.2 µl of glutamine, L‐[14C(U)] (Hartmann Analytic, MC1124) or glutamic acid, L‐[14C(U)] (Hartmann Analytic, MC156) was added to the remaining 450 µl sample. One hundred microlitres aliquots were filtered 0, 3, 6 and 9 min after addition of the radiolabelled amino acid and the filters were then washed two times with 16 ml of PBS pH 7.4. The filters were subsequently dissolved in 9 ml of Filter Count scintillation cocktail (Perkin Elmer) and the radioactivity measured in counts per minute (CPM) using a Wallac 1,409 DSA liquid scintillation counter. The CPMs were then normalized to the OD600 reading of the final cell suspension and the means and standard deviations of the CPM/ml OD600 = 1 of three or four (as indicated in the figure legends) independent experiments were plotted.
4.7. Determination of cellular c‐di‐AMP levels by competitive ELISA
Intracellular c‐di‐AMP levels in WT LAC* and the indicated S. aureus mutant strains were determined using a previously described competitive ELISA method (Underwood, Zhang, Metzger, & Bai, 2014) and a slightly modified method for the preparation of S. aureus samples (Bowman, Zeden, Schuster, Kaever, & Gründling, 2016). Briefly, single colonies of the WT LAC* strain were picked from TSA plates and used to inoculate 5 ml of GDM, GDM + Gln, GDM + Glu, GDM + NH3, GDM + Gln+NH3 and GDM + Glu+NH3. Colonies of the strains LAC*gdpP::kan and LAC*ΔybbR were inoculated into 5 ml of GDM, GDM + Glu and GDM + Gln. Colonies of strains LAC* piTET, LAC*alsT::tn piTET and LAC*alsT::tn piTET‐alsT were inoculated into GDM + Gln containing 200 ng/ml Atet and colonies of strains LAC* piTET, LAC*gltS::tn piTET and LAC*gltS::tn piTET‐gltS were inoculated into GDM + Glu supplemented with 200 ng/ml Atet. All cultures were incubated for 18 hr at 37°C with shaking. Next, bacteria from 4.5 ml culture were collected by centrifugation, washed three times with PBS and subsequently suspended in 0.75 to 1 ml 50 mM Tris pH 8 buffer supplemented with 20 ng/ml lysostaphin and the cells were lysed by bead beating. The lysates were cleared by centrifugation for 5 min at 17,000× g and the supernatant transferred to a new tube. A small sample aliquot was removed, and the protein concentration determined for normalization purposes using a Pierce BCA protein assay kit (Thermo Scientific, Waltham, MA, USA). The remainder of the sample was heated to 95°C for 10 min. For the competitive ELISA assay, the samples were diluted to a protein concentration of 100, 200, 400 or 500 μg/ml as, appropriate. ELISA plates were prepared by adding 100 μl of coating buffer (50 mM Na2C03, 50 mM NaHCO3, pH 9.6) containing 10 μg/ml of the c‐di‐AMP binding protein CpaASP to each well of a 96 well NUNC MaxiSorp plate (Thermo Scientific, Waltham, MA, USA) and the plate was incubated for approximately 18 hr at 4°C. Next, the plate was washed three times with 200 μl PBST pH 7.4 (10 mM Na2HPO4, 1.8 mM KH2PO4 137 mM NaCl, 2.7 mM KCl, 0.05% (v/v) Tween 20), blocked for 1 hr at 18°C with 150 μl blocking solution (1% BSA in PBS pH 7.4) and washed three times with 200 μl PBST. Fifty microlitres of the samples (three biological replicates and three technical replicates) or standards (two technical replicates) were mixed with 50 μl of a 50 nM biotinylated c‐di‐AMP solution prepared in 50 mM Tris pH 8 buffer. For the standard curve, c‐di‐AMP standards were prepared in 50 mM Tris pH 8 buffer at concentrations of 0, 12.5, 25, 37.5, 50, 75, 100 and 200 nM. Following the addition of the samples and the standards, the plate was incubated for 2 hr at 18°C and then washed three times with PBST. Next, 100 μl of a high‐sensitivity streptavidin‐HRP solution (Thermo Scientific, Waltham, MA, USA) diluted 1:500 in PBS was added to each well and the plate was incubated for 1 hr at 18°C. The plate was washed again 3 x with 200 μl PBST and 100 μl of a developing solution (0.103 M NaHPO4, 0.0485 M citric acid, 500 mg/L o‐phenylenediamine dihydrochloride, 0.03% H2O2) was added to each well and the plate incubated for 15 min at 18°C. The reaction was then stopped by adding 100 μl of 2 M H2SO4 solution. The absorbance was measured in a plate reader at a wavelength of 490 nm and c‐di‐AMP concentrations were calculated as ng c‐di‐AMP/ mg protein.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
MSZ, IK and AG designed the study; MSZ, IK and CFS acquired the data; MSZ, IK, CFS, VCT, PDF and AG designed experiments, analysed and interpreted the data; MSZ, IK and AG prepared the figures and wrote the original draft of the manuscript. All authors approved the final version of the manuscript.
Supporting information
ACKNOWLEDGEMENTS
This work was funded by the Wellcome Trust grants 100289/Z/12/Z and 210671/Z/18/Z to AG and the National Institutes of Health (NIH)/ National Institute of Allergy and Infectious Diseases (NIAID) grants P01AI083211 and R01AI125588 to PDF and VCT, respectively. MSZ was supported by a Medical Research Council Centre for Molecular Bacteriology and Infection (MRC CMBI) studentship. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Zeden MS, Kviatkovski I, Schuster CF, Thomas VC, Fey PD, Gründling A. Identification of the main glutamine and glutamate transporters in Staphylococcus aureus and their impact on c‐di‐AMP production. Mol Microbiol. 2020;113:1085–1100. 10.1111/mmi.14479
Merve S. Zeden and Igor Kviatkovski contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study will be openly available.
REFERENCES
- Anderson, C. B. , & Witter, L. D. (1982). Glutamine and proline accumulation by Staphylococcus aureus with reduction in water activity. Applied and Environment Microbiology, 43, 1501–1503. 10.1128/AEM.43.6.1501-1503.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y. , Yang, J. , Eisele, L. E. , Underwood, A. J. , Koestler, B. J. , Waters, C. M. , … Bai, G. (2013). Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di‐AMP phosphodiesterase activity and affect bacterial growth and virulence. Journal of Bacteriology, 195, 5123–5132. 10.1128/JB.00769-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y. , Yang, J. , Zarrella, T. M. , Zhang, Y. , Metzger, D. W. , & Bai, G. (2014). Cyclic di‐AMP impairs potassium uptake mediated by a cyclic di‐AMP binding protein in Streptococcus pneumoniae . Journal of Bacteriology, 196, 614–623. 10.1128/JB.01041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles, B. R. , Thoendel, M. , Roth, A. J. , & Horswill, A. R. (2010). Identification of genes involved in polysaccharide‐independent Staphylococcus aureus biofilm formation. PLoS ONE, 5, e10146 10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, L. , Zeden, M. S. , Schuster, C. F. , Kaever, V. , & Gründling, A. (2016). New insights into the cyclic di‐adenosine monophosphate (c‐di‐AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus . Journal of Biological Chemistry, 291, 26970–26986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, K. H. , Liang, J. M. , Yang, J. G. , Shih, M. S. , Tu, Z. L. , Wang, Y. C. , … Chou, S. H. (2015). Structural insights into the distinct Binding mode of cyclic di‐AMP with SaCpaA_RCK. Biochemistry, 54, 4936–4951. [DOI] [PubMed] [Google Scholar]
- Commichau, F. M. , Heidemann, J. L. , Ficner, R. , & Stülke, J. (2019). Making and breaking of an essential poison: The cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di‐AMP in bacteria. Journal of Bacteriology, 201, e00462–e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan, R. M. , Abbott, J. C. , Burhenne, H. , Kaever, V. , & Gründling, A. (2011). c‐di‐AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Path, 7, e1002217 10.1371/journal.ppat.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan, R. M. , Bowman, L. , Willis, A. R. , Kaever, V. , & Gründling, A. (2015). Cross‐talk between two nucleotide‐signaling pathways in Staphylococcus aureus . Journal of Biological Chemistry, 290, 5826–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan, R. M. , Campeotto, I. , Jeganathan, T. , Roelofs, K. G. , Lee, V. T. , & Gründling, A. (2013). Systematic identification of conserved bacterial c‐di‐AMP receptor proteins. Proceedings of the National Academy of Sciences, 110, 9084–9089. 10.1073/pnas.1300595110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke, A. K. , Fuller, J. R. , Obrist, M. W. , Tomkovich, S. E. , Vitko, N. P. , & Richardson, A. R. (2013). CcpA‐independent glucose regulation of lactate dehydrogenase 1 in Staphylococcus aureus . PLoS ONE, 8, e54293 10.1371/journal.pone.0054293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFrancesco, A. S. , Masloboeva, N. , Syed, A. K. , DeLoughery, A. , Bradshaw, N. , Li, G. W. , … Losick, R. (2017). Genome‐wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus . Proceedings of the National Academy of Sciences, 114, E5969–E5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux, L. , Sleiman, D. , Mazzuoli, M. V. , Gominet, M. , Lanotte, P. , Trieu‐Cuot, P. , … Firon, A. (2018). Cyclic di‐AMP regulation of osmotic homeostasis is essential in Group B Streptococcus . PLoS Genetics, 14, e1007342 10.1371/journal.pgen.1007342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmi, T. , Faozia, S. , Port, G. , & Cho, K. H. (2019). The second messenger c‐di‐AMP regulates diverse cellular pathways involved in stress response, biofilm formation, cell wall homeostasis, SpeB expression and virulence in Streptococcus pyogenes . Infection and Immunity, 87(6), e00147‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey, P. D. , Endres, J. L. , Yajjala, V. K. , Widhelm, T. J. , Boissy, R. J. , Bose, J. L. , & Bayles, K. W. (2013). A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio, 4, e00537–00512 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridkin, S. K. , Hageman, J. C. , Morrison, M. , Sanza, L. T. , Como‐Sabetti, K. , Jernigan, J. A. , … Farley, M. M. (2005). Methicillin‐resistant Staphylococcus aureus disease in three communities. New England Journal of Medicine, 352, 1436–1444. [DOI] [PubMed] [Google Scholar]
- Fuller, J. R. , Vitko, N. P. , Perkowski, E. F. , Scott, E. , Khatri, D. , Spontak, J. S. , … Richardson, A. R. (2011). Identification of a lactate‐quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Frontiers in Cellular and Infection Microbiology, 1, 19 10.3389/fcimb.2011.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling, A. , & Schneewind, O. (2007). Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus . Journal of Bacteriology, 189, 2521–2530. 10.1128/JB.01683-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach, J. , Commichau, F. M. , & Stülke, J. (2017). Perspective of ions and messengers: An intricate link between potassium, glutamate, and cyclic di‐AMP. Current Genetics. 10.1007/s00294-017-0734-3 [DOI] [PubMed] [Google Scholar]
- Gundlach, J. , Herzberg, C. , Hertel, D. , Thurmer, A. , Daniel, R. , Link, H. , & Stülke, J. (2017). Adaptation of Bacillus subtilis to life at extreme potassium limitation. Mbio, 8, e00861‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach, J. , Herzberg, C. , Kaever, V. , Gunka, K. , Hoffmann, T. , Weiss, M. , … Stülke, J. (2017c). Control of potassium homeostasis is an essential function of the second messenger cyclic di‐AMP in Bacillus subtilis . Science Signalling, 10, eaal3011. [DOI] [PubMed] [Google Scholar]
- Gundlach, J. , Krüger, L. , Herzberg, C. , Turdiev, A. , Poehlein, A. , Tascon, I. , … Stülke, J. (2019). Sustained sensing in potassium homeostasis: Cyclic di‐AMP controls potassium uptake by KimA at the levels of expression and activity. Journal of Biological Chemistry, 294, 9605–9614. 10.1074/jbc.RA119.008774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach, J. , Mehne, F. M. , Herzberg, C. , Kampf, J. , Valerius, O. , Kaever, V. , & Stülke, J. (2015). An essential poison: Synthesis and degradation of cyclic di‐AMP in Bacillus subtilis . Journal of Bacteriology, 197, 3265–3274. 10.1128/JB.00564-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey, C. R. , Lei, S. , Wax, J. K. , Lehman, M. K. , Nuxoll, A. S. , Steinke, L. , … Fey, P. D. (2017). Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. Mbio, 8, e01434‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge, R. (2009). Principles of c‐di‐GMP signalling in bacteria. Nature Reviews Microbiology, 7, 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Huynh, T. N. , Choi, P. H. , Sureka, K. , Ledvina, H. E. , Campillo, J. , Tong, L. , & Woodward, J. J. (2016). Cyclic di‐AMP targets the cystathionine beta‐synthase domain of the osmolyte transporter OpuC. Molecular Microbiology. 10.1111/mmi.13456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, B. , & O'Neill, L. A. (2015). Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Research, 25, 771–784. 10.1038/cr.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. , Youn, S. J. , Kim, S. O. , Ko, J. , Lee, J. O. , & Choi, B. S. (2015). Structural studies of potassium transport protein KtrA regulator of conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c‐di‐AMP). Journal of Biological Chemistry, 290, 16393–16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluytmans, J. , van Belkum, A. , & Verbrugh, H. (1997). Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clinical Microbiology Reviews, 10, 505–520. 10.1128/CMR.10.3.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, M. K. , Nuxoll, A. S. , Yamada, K. J. , Kielian, T. , Carson, S. D. , & Fey, P. D. (2019). Protease‐mediated growth of Staphylococcus aureus on host proteins is opp3 dependent. Mbio, 10, e02553‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehne, F. M. , Gunka, K. , Eilers, H. , Herzberg, C. , Kaever, V. , & Stülke, J. (2013). Cyclic di‐AMP homeostasis in Bacillus subtilis: Both lack and high level accumulation of the nucleotide are detrimental for cell growth. Journal of Biological Chemistry, 288, 2004–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, I. R. , Shah, I. M. , Xu, M. , Tan, M.‐W. , & Foster, T. J. (2012). Transforming the untransformable: Application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis . Mbio, 3(2), e00277‐11 10.1128/mBio.00277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk, I. R. , Tree, J. J. , Howden, B. P. , Stinear, T. P. , & Foster, T. J. (2015). Complete bypass of restriction systems for major Staphylococcus aureus lineages. MBio, 6, e00308‐15 10.1128/mBio.00308-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso, J. A. , Schramke, H. , Zhang, Y. , Tosi, T. , Dehbi, A. , Jung, K. , & Gründling, A. (2015). Binding of cyclic di‐AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter. Journal of Bacteriology, 198, 98–110. 10.1128/JB.00480-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, H. T. , Nhiep, N. T. H. , Vu, T. N. M. , Huynh, T. N. , Zhu, Y. , Huynh, A. L. D. , … Turner, M. S. (2018). Enhanced uptake of potassium or glycine betaine or export of cyclic‐di‐AMP restores osmoresistance in a high cyclic‐di‐AMP Lactococcus lactis mutant. PLoS Genetics, 14, e1007574 10.1371/journal.pgen.1007574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, H. T. , & Turner, M. S. (2019). Onwards and [K(+)]upwards: A new potassium importer under the spell of cyclic‐di‐AMP. Journal of Bacteriology, 201(10), e00150‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, T. H. , Liang, Z. X. , Marcellin, E. , & Turner, M. S. (2016). Replenishing the cyclic‐di‐AMP pool: Regulation of diadenylate cyclase activity in bacteria. Current Genetics, 62, 731–738. 10.1007/s00294-016-0600-8 [DOI] [PubMed] [Google Scholar]
- Quintana, I. M. , Gibhardt, J. , Turdiev, A. , Hammer, E. , Commichau, F. M. , Lee, V. T. , … Stülke, J. (2019). The KupA and KupB proteins of Lactococcus lactis IL1403 are novel c‐di‐AMP receptor proteins responsible for potassium uptake. Journal of Bacteriology, 201(10), e00028‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, A. R. , Libby, S. J. , & Fang, F. C. (2008). A nitric oxide‐inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science, 319, 1672–1676. 10.1126/science.1155207 [DOI] [PubMed] [Google Scholar]
- Rocha, R. , Teixeira‐Duarte, C. M. , Jorge, J. M. P. , & Morais‐Cabral, J. H. (2019). Characterization of the molecular properties of KtrC, a second RCK domain that regulates a Ktr channel in Bacillus subtilis . Journal of Structural Biology, 205, 34–43. 10.1016/j.jsb.2019.02.002 [DOI] [PubMed] [Google Scholar]
- Römling, U. (2008). Great times for small molecules: C‐di‐AMP, a second messenger candidate in Bacteria and Archaea. Science Signalling, 1, pe39 10.1126/scisignal.133pe39 [DOI] [PubMed] [Google Scholar]
- Schuster, C. F. , Bellows, L. E. , Tosi, T. , Campeotto, I. , Corrigan, R. M. , Freemont, P. , & Gründling, A. (2016). The second messenger c‐di‐AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus . Science Signalling, 9, ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, C. F. , Wiedemann, D. M. , Kirsebom, F. C. M. , Santiago, M. , Walker, S. , & Gründling, A. (2019). High‐throughput transposon sequencing highlights the cell wall as an important barrier for osmotic stress in methicillin resistant Staphylococcus aureus and underlines a tailored response to different osmotic stressors. Molecular Microbiology, 1–19. 10.1111/mmi.14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurman‐Wolters, G. K. , & Poolman, B. (2005). Substrate specificity and ionic regulation of GlnPQ from Lactococcus lactis. An ATP‐binding cassette transporter with four extracytoplasmic substrate‐binding domains. Journal of Biological Chemistry, 280, 23785–23790. [DOI] [PubMed] [Google Scholar]
- Spahich, N. A. , Vitko, N. P. , Thurlow, L. R. , Temple, B. , & Richardson, A. R. (2016). Staphylococcus aureus lactate‐ and malate‐quinone oxidoreductases contribute to nitric oxide resistance and virulence. Molecular Microbiology, 100, 759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka, K. , Choi, P. H. , Precit, M. , Delince, M. , Pensinger, D. , Huynh, T. N. , … Woodward, J. J. (2014). The cyclic dinucleotide c‐di‐AMP is an allosteric regulator of metabolic enzyme function. Cell, 158, 1389–1401. 10.1016/j.cell.2014.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh, W. K. , Dramsi, S. , Tolker‐Nielsen, T. , Yang, L. , & Givskov, M. (2019). Increased intracellular cyclic di‐AMP levels sensitize Streptococcus gallolyticus subsp. gallolyticus to osmotic stress and reduce biofilm formation and adherence on intestinal cells. Journal of Bacteriology, 201, e00597‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolner, B. , Ubbink‐Kok, T. , Poolman, B. , & Konings, W. N. (1995). Characterization of the proton/glutamate symport protein of Bacillus subtilis and its functional expression in Escherichia coli . Journal of Bacteriology, 177, 2863–2869. 10.1128/JB.177.10.2863-2869.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi, T. , Hoshiga, F. , Millership, C. , Singh, R. , Eldrid, C. , Patin, D. , … Gründling, A. (2019). Inhibition of the Staphylococcus aureus c‐di‐AMP cyclase DacA by direct interaction with the phosphoglucosamine mutase GlmM. PLoS Path, 15, e1007537 10.1371/journal.ppat.1007537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood, A. J. , Zhang, Y. , Metzger, D. W. , & Bai, G. (2014). Detection of cyclic di‐AMP using a competitive ELISA with a unique pneumococcal cyclic di‐AMP binding protein. Journal of Microbiol Methods, 107, 58–62. 10.1016/j.mimet.2014.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitko, N. P. , Spahich, N. A. , & Richardson, A. R. (2015). Glycolytic dependency of high‐level nitric oxide resistance and virulence in Staphylococcus aureus . Mbio, 6(2), e00045‐15 10.1128/mBio.00045-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley, A. T. , Garelis, N. E. , Peterson, B. N. , Choi, P. H. , Tong, L. , Woodward, J. J. , & Portnoy, D. A. (2017). c‐di‐AMP modulates Listeria monocytogenes central metabolism to regulate growth, antibiotic resistance and osmoregulation. Molecular Microbiology, 104, 212–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley, A. T. , Pollock, A. J. , & Portnoy, D. A. (2015). The PAMP c‐di‐AMP is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host & Microbe, 17, 788–798. 10.1016/j.chom.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke, D. , Schulz, L. M. , Lentes, S. , Scholz, P. , Poehlein, A. , Gibhardt, J. , … Commichau, F. M. (2019). Identification of the first glyphosate transporter by genomic adaptation. Environmental Microbiology, 21, 1287–1305. 10.1111/1462-2920.14534 [DOI] [PubMed] [Google Scholar]
- Witte, C. E. , Whiteley, A. T. , Burke, T. P. , Sauer, J. D. , Portnoy, D. A. , & Woodward, J. J. (2013). Cyclic di‐AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. MBio, 4, e00282‐00213 10.1128/mBio.00282-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, J. J. , Iavarone, A. T. , & Portnoy, D. A. (2010). c‐di‐AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science, 328, 1703–1705. 10.1126/science.1189801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaprasis, A. , Bleisteiner, M. , Kerres, A. , Hoffmann, T. , & Bremer, E. (2015). Uptake of amino acids and their metabolic conversion into the compatible solute proline confers osmoprotection to Bacillus subtilis . Applied and Environment Microbiology, 81, 250–259. 10.1128/AEM.02797-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrella, T. M. , Metzger, D. W. , & Bai, G. (2018). Stress suppressor screening leads to detection of regulation of cyclic di‐AMP homeostasis by a Trk family effector protein in Streptococcus pneumoniae . Journal of Bacteriology, 200, e00045‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeden, M. S. , Schuster, C. F. , Bowman, L. , Zhong, Q. , Williams, H. D. , & Gründling, A. (2018). Cyclic di‐adenosine monophosphate (c‐di‐AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. Journal of Biological Chemistry, 293, 3180–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B. , & Stülke, J. (2018). SubtiWiki in 2018: From genes and proteins to functional network annotation of the model organism Bacillus subtilis . Nucleic Acids Research, 46, D743–D748. 10.1093/nar/gkx908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Pham, T. H. , Nhiep, T. H. , Vu, N. M. , Marcellin, E. , Chakrabortti, A. , … Turner, M. S. (2016). Cyclic‐di‐AMP synthesis by the diadenylate cyclase CdaA is modulated by the peptidoglycan biosynthesis enzyme GlmM in Lactococcus lactis . Molecular Microbiology, 99, 1015–1027. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Xiong, Y. Q. , Sadykov, M. R. , Fey, P. D. , Lei, M. G. , Lee, C. Y. , … Somerville, G. A. (2009). Tricarboxylic acid cycle‐dependent attenuation of Staphylococcus aureus in vivo virulence by selective inhibition of amino acid transport. Infection and Immunity, 77, 4256–4264. 10.1128/IAI.00195-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study will be openly available.


