Abstract
Background -
Atrial fibrillation is a common cardiovascular disorder, characterized by irregular electrical activity in the upper chambers of the heart. Both chronic cardiometabolic risk factors and genetics have been shown to contribute to the development of atrial fibrillation. Birthweight has also been associated with risk of atrial fibrillation.
Methods -
In the current study, we utilized a genetic approach to study the effect of birthweight on atrial fibrillation. We used two-sample Mendelian randomization to consider the impact of birthweight on incident atrial fibrillation using summary data from the Early Growth Genetics Consortium GWAS of birthweight and a large biobank-based GWAS of atrial fibrillation.
Results -
Using the framework of 2-sample Mendelian randomization, we found that a 1-SD genetic elevation of birthweight was associated with increased risk of atrial fibrillation (OR 1.27; 95% CI 1.14–1.41; p = 1×10−5) with sensitivity analyses demonstrating robustness of this result.
Conclusions -
Our findings clarify the directionality of the relationship between birthweight and atrial fibrillation, supporting the growing body of evidence that intrauterine growth has a lifelong impact on cardiovascular health.
Journal Subject Terms: Genetics, Atrial Fibrillation, Cardiovascular Disease, Risk Factors
Keywords: genetics, atrial fibrillation, Mendelian randomization, birthweight
Introduction
Atrial fibrillation is a common arrhythmia, characterized by irregular electrical activity in the upper chambers of the heart (atria). Large population studies estimate a worldwide prevalence of approximately 0.5% with more than 33 million affected individuals.1 The irregular cardiac conduction resulting from atrial fibrillation can lead to significant complications, including cardioembolic stroke, heart failure, and death. Many chronic cardiometabolic risk factors contribute to the development of atrial fibrillation, including hypertension, diabetes, heart failure, thyroid disease, chronic kidney disease, smoking, obesity, cardiac surgery, coronary heart disease, and valvular heart disease.2 Genetic risk factors also contribute to the development of atrial fibrillation. Heritability of atrial fibrillation is estimated at 20%, and family linkage studies and genome-wide association studies have identified both rare, coding, large-effect monogenic variants, and more common, small-effect variants that contribute to the development of atrial fibrillation.3–10
Birthweight has been identified as a risk factor for a number of cardiometabolic diseases, including atrial fibrillation, coronary heart disease, obesity, hypertension, and diabetes.11–14 The relationship between birthweight and risk of atrial fibrillation has previously been investigated in a number of population studies, including the Women’s Health Study15, Atherosclerosis Risk In Communities (ARIC)16, and the Helsinki Birth Cohort Study (HBCS).17 However, the cumulative results from this collection of studies have been conflicting. Both low and high birthweight have been associated with increased risk for atrial fibrillation (Supplemental Table 1).15–18 We hypothesized that a Mendelian randomization approach may help clarify the relationship between these two traits.
Birthweight, like many anthropometric traits, is highly heritable. Large-scale birthweight GWAS have identified over 60 genome-wide significant loci and have explained 15% of the variation in birthweight.11,12,19 Genetic studies of birthweight and other health-related traits have identified a negative genetic correlation between birthweight and type 2 diabetes, systolic blood pressure, and coronary artery disease, and a positive association with anthropometric and obesity-related traits such as body mass index (BMI).13
In the current study, we utilized birthweight genetics to study the impact of birthweight on atrial fibrillation. We used two-sample Mendelian randomization to consider the impact of birthweight on incident atrial fibrillation using summary data from the Early Growth Genetics Consortium GWAS of birthweight (N=153,781) and a large biobank-based GWAS of atrial fibrillation (n=60,620 cases and n=970,216 controls).4,12
Methods
The data, analytic methods, and study materials may be made available to other researchers for purposes of reproducing the results or replicating the procedure. The study was approved by the Institutional Review Board of the University of Pennsylvania. Full Methods are available in the in the Data Supplement of the article.
Results
We performed two-sample Mendelian randomization (Supplemental Tables 2 and 3), using summary statistics from a genome-wide association study of atrial fibrillation including more than >1 million individuals.4 Mendelian randomization leverages genetic instrumental variables to provide unconfounded causal estimates of the effects of one or exposure on another. We constructed a genetic instrument composed of 42 SNPs that had previously been strongly associated with birthweight at a genome-wide level of significance (p < 5×10−8) (Supplemental Table 4). After removal of heterogenous variants (n=3), inverse variance-weighted modeling revealed a significant (OR 1.27; 95% CI 1.14–1.41; p = 1×10−5) association between increasing birthweight and increased risk of developing atrial fibrillation (Figure 1; Supplemental Figures 1–3). This association remained significant in sensitivity analysis using weighted median and weighted mode. The intercept from Egger regression was −0.001 (p=0.8), suggesting lack of significant pleiotropic bias. In additional sensitivity analysis, an instrument composed of 39 SNPs associated with birthweight was investigated for association with atrial fibrillation using summary statistics from the AFGen consortium genome-wide association study of atrial fibrillation, with similar results (Supplemental Table 5).
Figure 1:
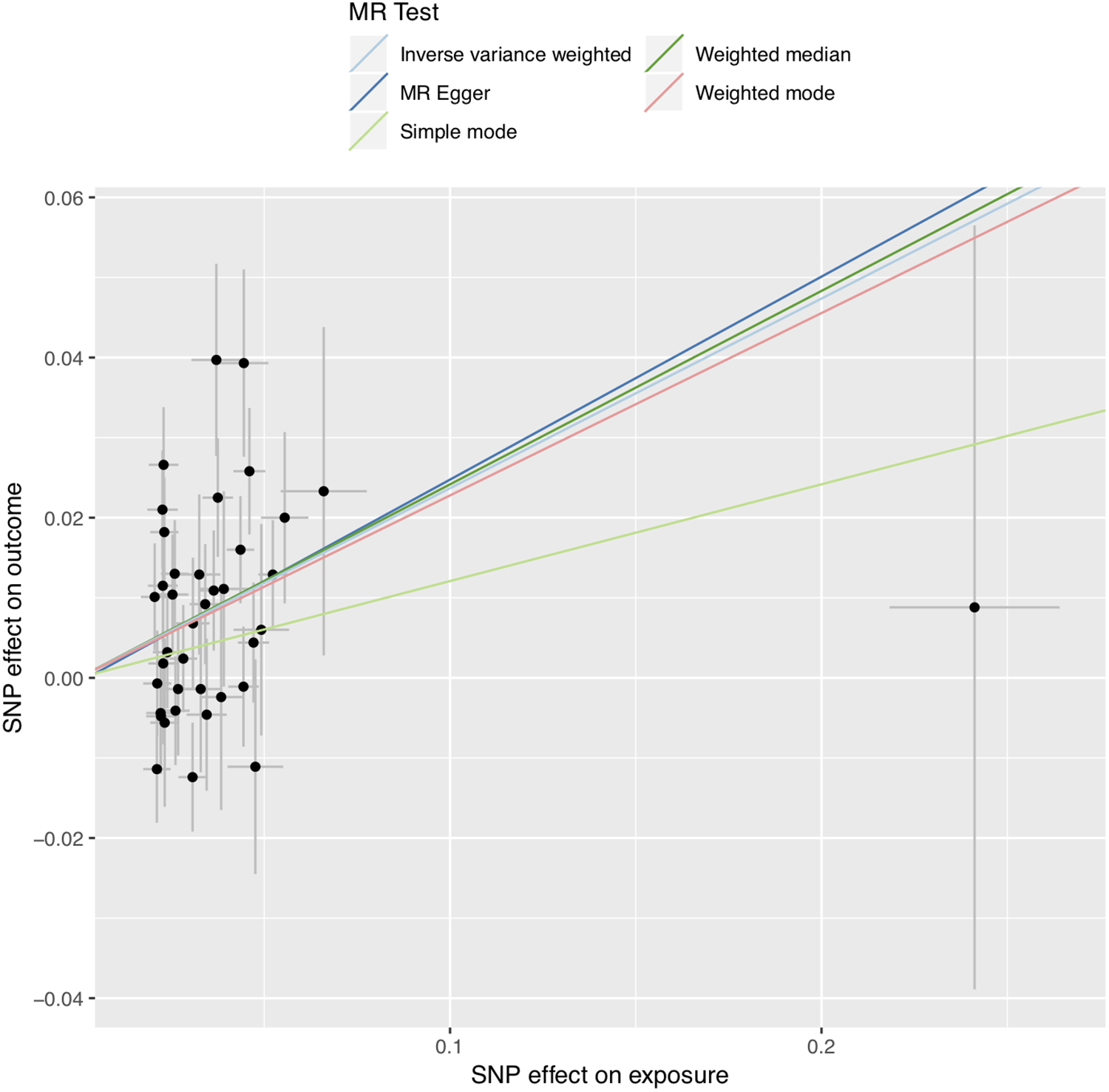
Mendelian Randomization tests. Mendelian Randomization tests for birthweight on atrial fibrillation. Inverse variance weighted, MR Egger, Simple, Weighted Median, and Weighted mode are shown.
Asymmetry analysis was performed to detect any bias from reverse causality. A genetic instrument comprised of 98 SNPs strongly associated with atrial fibrillation (p < 5×10−8) was tested for association with birthweight. No significant associations were identified by any Mendelian randomization method (inverse variance-weighted, weighted median, weighted mode, simple mode, and MR-Egger).
We queried published GWAS studies to determine whether variants included in the genetic instrument for birthweight were associated with risk factors for atrial fibrillation. Variants included in the instrument had previously been associated with anthropometric, metabolic, lipid, and pulmonary traits, among others (Supplemental Table 6).
A prior Mendelian randomization study13 previously identified associations between birthweight and known atrial fibrillation risk factors including coronary heart disease,20 diabetes,21 BMI,22 and LDL-cholesterol.23 We performed multivariable Mendelian randomization to determine whether shared genetic risk among these traits may mediate the relationship between birthweight and atrial fibrillation. After adjustment for shared genetic risk factors, birthweight remained significantly associated with risk of atrial fibrillation (OR 1.32; 95% CI 1.2–1.5; p = 5×10−6 (Figure 2).
Figure 2:
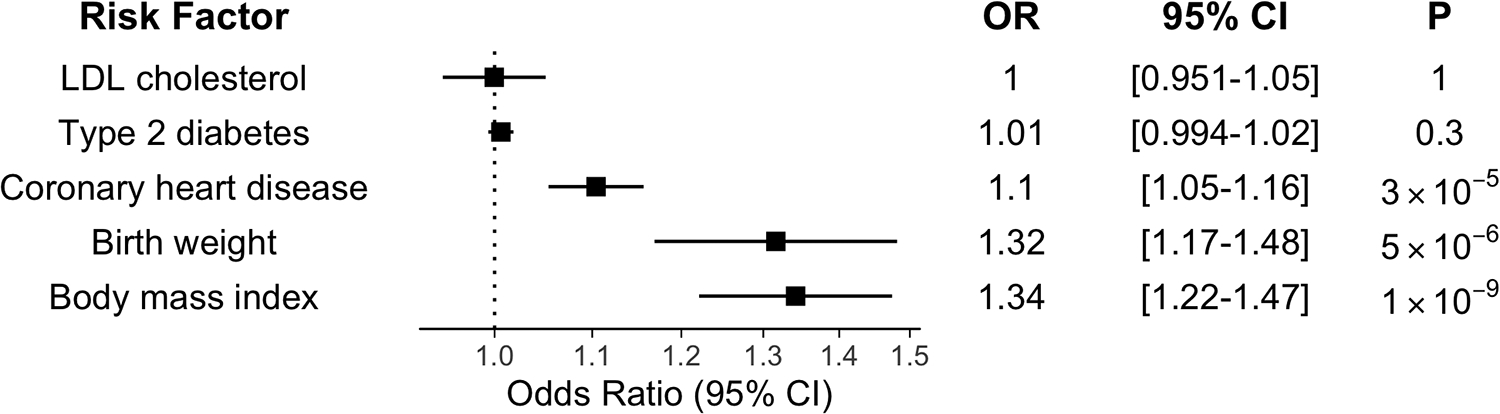
Multivariable Mendelian Randomization. Multivariable Mendelian randomization demonstrated birthweight is associated with atrial fibrillation, after adjustment for cardiometabolic risk factors previously associated with birthweight by MR (Type 2 diabetes, LDL cholesterol, coronary heart disease, and body mass index). Dots represent estimate of odds ratio (OR), and whiskers represent 95% confidence interval (CI).
Discussion
The current study highlights the use of human genetics to clarify prior epidemiologic observations that birthweight is significantly associated with the risk of developing of atrial fibrillation. A number of large epidemiologic studies including ARIC, WHS, and HBCS previously investigated the relationship between birthweight and atrial fibrillation. We used a genetic approach to further refine this association by employing 2 sample Mendelian randomization using summary statistics from large genome wide association studies of birthweight and atrial fibrillation including >1 million individuals. Our results show that the genetic risk of increased birthweight, as assessed using a genetic instrument composed of 42 independent genome-wide significant SNPs, was strongly associated with an increased risk of atrial fibrillation.
Our findings are consistent with those of the 2010 analysis of the WHS. Analysis of data from 27,982 women with median follow-up of 14.5 years identified a significant linear relationship between self-reported birthweight and atrial fibrillation, with women in the highest birthweight category (>4.5kg) at 71% increased risk of incident atrial fibrillation compared to women in the lowest birthweight category (<2.5kg), after adjustment for age, hypercholesterolemia, smoking, exercise, alcohol consumption, education, race/ethnicity, and hormone replacement therapy.15
Mendelian randomization analysis applied to birthweight and cardiovascular outcomes has previously been investigated in the UK BioBank, finding strong associations between low birthweight and type II diabetes mellitus and coronary artery disease.13 In that same study, Mendelian randomization was also used to examine the association between birthweight and atrial fibrillation, but the genetic instruments and outcomes were derived from older genome-wide associations of birthweight and atrial fibrillation.12,24 Using summary statistics from larger studies of birthweight and atrial fibrillation, we refine the magnitude and confidence around the effect estimate (IVW OR 1.27; 95% CI 1.14–1.41) vs. (IVW OR 1.15; 95% CI 0.95–1.39).
Our results must be interpreted with caution. In MR-Egger and MR-PRESSO analysis, used to assess for and account for violations of the horizontal pleiotropy assumption of Mendelian randomization, we found no significant evidence of pleiotropic bias.25–27 However, because of the complex relationship between parental genetics and diseases/risk factors associated with atrial fibrillation, birthweight may not necessarily be the causal exposure through which our genetic instrument increases the risk of atrial fibrillation.28 Although we did not find evidence of horizontal pleiotropy in our MR analysis, our query of published GWAS results revealed associations between birthweight variants and multiple anthropometric and cardiometabolic risk factors which may in part mediate the relationship between birthweight and atrial fibrillation. A combined observational and Mendelian randomization study of birthweight and cardiometabolic disease in UK Biobank found that increased birthweight protects from some traditional atrial fibrillation risk factors like coronary artery disease and diabetes, while also being positively associated with other atrial fibrillation risk factors like BMI.13 In multivariable Mendelian randomization analysis, birthweight remained significantly associated with atrial fibrillation despite adjusting for the shared genetic component among these risk factors. While horizontal pleiotropic effects may plausibly exist between birthweight and atrial fibrillation, we were unable to identify these effects despite extensive Mendelian randomization sensitivity analyses. In sum, these results highlight the complex relationship between the genetic and clinical risks birthweight, cardiometabolic, and anthropometric risk factors that converge to increase atrial fibrillation risk.
Birthweight is only one proxy for fetal growth and the intrauterine environment. Instrumental variables reflecting more specific intrauterine exposures would help refine our understanding of the developmental origins of atrial fibrillation.29 As maternal and fetal genetic variation are correlated, further study of maternal-fetal pairs with both genetic and longitudinal outcome data of the offspring may further clarify this association. Recent work by Warrington and colleagues to identify the contributions of fetal and maternal genetic influences on birthweight may also help parse the direct genetic effect of variants contributing to both birthweight and atrial fibrillation from the indirect maternal effects.30,31
Our Mendelian randomization findings remained robust to sensitivity analysis, and showed no evidence of horizontal pleiotropy.25,26 Our findings were limited to individuals of European ancestry due to lack of large-scale genome-wide association studies of birthweight in populations of diverse ancestry, which may also limit generalizability. Studies in these populations are clearly needed.
Overall, we used genetic analysis to provide clarity into the relationship between birthweight and risk of atrial fibrillation, finding a robust positive association between these traits. Our results provide further support to the growing body of evidence that intrauterine growth has a lifelong impact on health.
Supplementary Material
Sources of Funding:
Genotyping was performed in collaboration with Regeneron Genetics Center; individual scientific contributions by Regeneron Genetics Center personnel are listed in the Supplemental Material. SMD is supported by the U.S. Department of Veterans Affairs (IK2-CX001780). This publication does not represent the views of the Department of Veterans Affairs or the United States government. BFV is supported by a grant from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK101478), and a Linda Pechenik Montague Investigator Award. We acknowledge and thank the participants of the Penn Medicine BioBank.
Footnotes
Disclosures: PTE has consulted with Bayer AG, Novartis and Quest Diagnostics. SMD receives research grants to the institution from CytoVAS, LLC, and RenalytixAI. BFV currently serves as an associate editor at Circulation: Genomic and Precision Medicine.
References:
- 1.Chugh SS, Roth GA, Gillum RF, Mensah GA. Global burden of atrial fibrillation in developed and developing nations. Glob Heart. 2014;9:113–119. doi: 10.1016/j.gheart.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 2.January CT, Wann LS, Alpert JS, Calkins H, Cigarroa JE, Cleveland JC, Conti JB, Ellinor PT, Ezekowitz MD, Field ME, et al. 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation. J Am Coll Cardiol. 2014. doi: 10.1016/j.jacc.2014.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson CD, Aragam KG, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50:1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50:1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulit SL, Weng L-C, McArdle PF, Trinquart L, Choi SH, Mitchell BD, Rosand J, de Bakker PI, Benjamin EJ, Ellinor PT, et al. Atrial Fibrillation Genetic Risk Differentiates Cardioembolic Stroke from other Stroke Subtypes. Neurol Genet. 2018; 4:e293. doi: 10.1212/NXG.0000000000000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Helgadottir A, Gretarsdottir S, Benonisdottir S, Magnusdottir A, Davidsson OB, Rajamani S, Roden DM, et al. A Missense Variant in PLEC Increases Risk of Atrial Fibrillation. J Am Coll Cardiol. 2017;70:2157–2168. doi: 10.1016/j.jacc.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, Magnusson OT, Helgason A, Saemundsdottir J, Gylfason A, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–20. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV., Hjartarson E, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–44. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 9.Thorolfsdottir RB, Sveinbjornsson G, Sulem P, Nielsen JB, Jonsson S, Halldorsson GH, Melsted P, Ivarsdottir EV., Davidsson OB, Kristjansson RP, et al. Coding variants in RPL3L and MYZAP increase risk of atrial fibrillation. Commun Biol. 2018;1:68. doi: 10.1038/s42003-018-0068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi SH, Weng L-C, Roselli C, Lin H, Haggerty CM, Shoemaker MB, Barnard J, Arking DE, Chasman DI, Albert CM, et al. Association Between Titin Loss-of-Function Variants and Early-Onset Atrial Fibrillation. J Am Med Assoc. 2018;320:1–11. doi: 10.1001/jama.2018.18179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ, Bradfield JP, St Pourcain B, Evans DM, Charoen P, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horikoshi M, Beaumont RN, Day FR, Warrington NM, Kooijman MN, Fernandez-Tajes J, Feenstra B, Van Zuydam NR, Gaulton KJ, Grarup N, et al. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538:248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanetti D, Tikkanen E, Gustafsson S, Priest JR, Burgess S, Ingelsson E. Birthweight, Type 2 Diabetes and Cardiovascular Disease: Addressing the Barker Hypothesis with Mendelian randomization. Circ Genom Precis Med. 2017;11:e002054. doi: 10.1161/CIRCGEN.117.002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hales CN, Barker DJP. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Int J Epidemiol. 2013;42:1215–1222. doi: 10.1093/ije/dyt133. [DOI] [PubMed] [Google Scholar]
- 15.Conen D, Tedrow UB, Cook NR, Buring JE, Albert CM. Birth weight is a significant risk factor for incident atrial fibrillation. Circulation. 2010;122:764–770. doi: 10.1161/CIRCULATIONAHA.110.947978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larsson SC, Drca N, Jensen-Urstad M, Wolk A. Incidence of atrial fibrillation in relation to birth weight and preterm birth. Int J Cardiol. 2015;178:149–152. doi: 10.1016/j.ijcard.2014.10.138. [DOI] [PubMed] [Google Scholar]
- 17.Johnson LSB, Salonen M, Kajantie E, Conen D, Healey JS, Osmond C, Eriksson JG. Early Life Risk Factors for Incident Atrial Fibrillation in the Helsinki Birth Cohort Study. J Am Heart Assoc. 2017;6. doi: 10.1161/JAHA.117.006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawani SO, Demerath EW, Lopez FL, Soliman EZ, Huxley RR, Rose KM, Alonso A. Birth weight and the risk of atrial fibrillation in whites and African Americans: the Atherosclerosis Risk In Communities (ARIC) study. BMC Cardiovasc Disord. 2014;14:69. doi: 10.1186/1471-2261-14-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaumont RN, Warrington NM, Cavadino A, Tyrrell J, Nodzenski M, Horikoshi M, Geller F, Myhre R, Richmond RC, Paternoster L, et al. Genome-wide association study of offspring birth weight in 86 577 women identifies five novel loci and highlights maternal genetic effects that are independent of fetal genetics. Hum Mol Genet. 2018;27:742–756. doi: 10.1093/hmg/ddx429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MCY, Prokopenko I, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, Lin H, Arking DE, Smith AV., Albert CM, et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 2017;49:946–952. doi: 10.1038/ng.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1101/157552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden J, Smith GD, Burgess S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195–R208. 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies NM, Holmes MV., Davey Smith G. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. 2018;362. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freathy RM. Can genetic evidence help us to understand the fetal origins of type 2 diabetes? Diabetologia. 2016;59:1850–1854. doi: 10.1007/s00125-016-4057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warrington NM, Freathy RM, Neale MC, Evans DM. Using structural equation modelling to jointly estimate maternal and fetal effects on birthweight in the UK Biobank. Int J Epidemiol. 2018;47:1229–1241. doi: 10.1093/ije/dyy015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland Ø, Laurin C, Bacelis J, Peng S, Hao K, Feenstra B, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


