Abstract
Background:
Extracellular matrix (ECM) affects cell behavior, and vice versa. How ECM changes after small bowel resection (SBR) to support adaptive cellular processes has not been described. Here we characterize changes in ECM following SBR and integrate this with concomitant transcriptional perturbations.
Methods:
A 50% proximal SBR or sham surgery was performed on mice. On post-operative day 7, ileal tissue was sequentially depleted of protein components to generate an ECM-enriched fraction. ECM was analyzed for protein composition using mass spectrometry with subsequent Ingenuity Pathway Analysis (IPA) to identify predicted pathways and upstream regulators. qPCR and RNA-sequencing (RNA-Seq) were performed to corroborate these predicted pathways.
Results:
3034 proteins were differentially regulated between sham and SBR, of which 95 were significant (P<0.05). IPA analysis predicted PPARα agonism to be an upstream regulator of the observed proteomic changes (P<0.001). qPCR and RNA-Seq with KEGG analysis confirmed significant engagement of the PPAR pathway (P<0.05).
Conclusion:
Transcriptional signatures of adapting bowel predict subsequent ECM changes after SBR. How ECM communicates with surrounding cells to drive adaptation and vice versa merits further investigation. Our findings thus far suggest ECM supports tissue hyperplasia and altered metabolic demand following SBR.
Keywords: Extracellular matrix, Small bowel resection, PPAR
Introduction:
There is mounting evidence that ECM is a critical component of the microenvironmental niche that influences cell behavior[1]. For example, processes of honing and differentiation/lineage restriction among cells seeded onto decellularized organ scaffolds (which largely retain ECM architecture and composition) are directly affected by location within the scaffold[2]. In other words, cells can “read” their ECM environment to determine their behavior, and even their identity. This makes ECM— defined not only by its structural proteins but perhaps, more importantly, by the signaling peptides they sequester [3–5]—a defining feature of organ biology. As such, better understanding of SI ECM during homeostasis and adaptive challenge will inform future SI regenerative medicine approaches including, but not limited to, tissue engineered small intestine (TESI).
One example of “adaptive challenge” in the SI is massive SBR. How the ECM responds to massive SBR is unknown. Given our recent study demonstrating shifted regional identity, or “proximalization” of ileal epithelium following proximal SBR, we were curious to know whether ECM may reflect or support these adaptive cellular changes. As such, the primary aim of this study was to characterize the “adaptation” of SI ECM to SBR. We hypothesized that—just as the epithelium undergoes a molecular adaptation to SBR[6]—so too would ECM. To test our hypothesis, we performed mass spectrometry (MS) with downstream IPA analysis on ileal ECM from mice that underwent either 50% proximal SBR or sham surgery (distal transection and anastomosis only), and corroborated our findings with transcriptional changes that accompany SBR.
Materials and Methods:
Small Bowel Resection:
50% proximal SBR was performed on male C57/B6 mice at 9 weeks of age, as previously described[7]. Sham surgery was performed as a control for exposure to anesthesia, laparotomy, and bowel transection. On post-operative day 7, ileal tissue distal to the anastomosis was submitted for histological analysis of structural adaptation. Day 7 was selected because this is a well-established time point in our lab by which structural and molecular adaptation are predicted to have occurred. All surgical and animal care procedures were approved by the Washington University Institutional Animal Care and Use Committee.
Extracellular Matrix Enrichment with Western Blot Validation:
ECM enrichment was performed according to previously published protocols[8, 9] using the commercially available CNMCS Compartmental Protein Extraction Kit (pke13011, CytoMol) on flash frozen ileal tissue from mice that demonstrated structural adaptation. Fractions generated at each stage of the protocol were as follows: cytoplasmic (C), nuclear (N), membrane (M), cytoskeletal (CS), and extracellular matrix (ECM), and these were flash frozen as they were derived. Appropriate fractionation was validated via Western Blotting (similar to previously described)[6] using antibodies against proteins expected within each fraction, as follows: anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (5174, Cell Signaling Technologies, used at 1:10,000), anti-epidermal growth factor receptor (EGFR) (06–847, Millipore, used at 1:2000), and anti-collagen1 (ab34710, Abcam, used at 1:1000).
Peptide Preparation, Isobaric Labeling, and nano-LC-MS/MS Analysis:
The samples were labeled with tandem mass tag reagents (TMT10) (Thermo Scientific) according to manufacturer’s protocol. The eluates were transferred to autosampler vials (200046, Sun-Sri), dried and stored at −80°C until LC-MS analysis. The samples in formic acid (1%) were loaded (2.5 μL) onto a 75 μm i.d. × 50 cm Acclaim® PepMap 100 C18 RSLC column (Thermo Fisher Scientific) on an EASY nanoLC (Thermo Fisher Scientific) at a constant pressure of 700 bar with 100% A (0.1%FA). Data was acquired using a Q-Exactive™ PLUS hybrid quadrupole Orbitrap™ mass spectrometer (Thermo Scientific™) in data-dependent acquisition mode.
Protein Identification and Relative Quantification:
The unprocessed data from the mass spectrometer were converted to peak lists using Proteome Discoverer (version 2.1.0.81, Thermo-Fischer Scientific) with the integration of reporter-ion intensities of TMT 10-plex at a mass tolerance of ±3.15 mDa[10]. The spectra were analyzed using Mascot software (Matrix Science, London, UK; version 2.5.1) [11] against a database of mouse and common contaminant proteins. Data processing and analysis were performed with the free software environment for statistical computing and graphics, R (R Core Team (2018). https://www.R-project.org/). Heat-map visualization of protein log2-ratios was performed using the R package: pheatmap. Volcano plot visualization was generated using VolcanoR (http://volcanor.bioinf.su).
Ingenuity Pathway Analysis:
The networks and functional analyses were generated using Ingenuity Pathway Analysis, or IPA (Build version 478438M, Content version 44691306) (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis)[12]. The top 100 enriched and depleted proteins were used as input.
Quantitative PCR (qPCR):
qPCR was performed as previously described[13] on RNA from ileal villus epithelium from sham and SBR mice at day 7 after surgery. A NanoDrop Spectrophotometer (ND-1000, NanoDrop Technologies, Wilmington, DE) was used to measure RNA concentration, and one-step TaqMan was used on the Applied Biosystems 7500 Fast Real-Time PCR System. Primer probes used were Cyp4a10 (Mm02601690_gH) with Actb as endogenous control (Mm04394036_g1), both from Applied Biosystems.
RNA-Seq data acquisition, quality control, and processing:
RNA isolation was performed per manufacturer instructions using the Total RNA Purification Kit with on-column DNA removal (37500 and 25710, Norgen Biotek Corp). RNA-sequencing was performed and analyzed at the Washington University Genome Technology Access Center (GTAC). Global transcriptomic changes in known KEGG terms were elucidated using with the R/Bioconductor packages GAGE and Pathview.
Results:
An ECM-Enriched Tissue Fraction Can Be Extracted from SI
50% proximal SBR (n=4) and sham surgery (n=4) were performed on mice that were cage, gender, age, and weight-matched (Figure 1A). On post-operative day 7, ileum was isolated and histologically analyzed. Structural adaptation in SBR mice—an expected compensatory mucosal hypertrophic response to resection— was confirmed, with villus length increasing by 42.3 ± 6% in SBR relative to sham (Figure 1B). ECM was extracted from flash frozen ileum of mice that demonstrated structural adaptation, and ECM enrichment was verified via Western blotting against Collagen I, a ubiquitous ECM protein (Figure 1C). Further, we verified depletion of membrane associated protein (EGFR) and total cellular protein (GAPDH) in our ECM fraction (Fig. 1C) prior to performing MS analysis.
Figure 1.
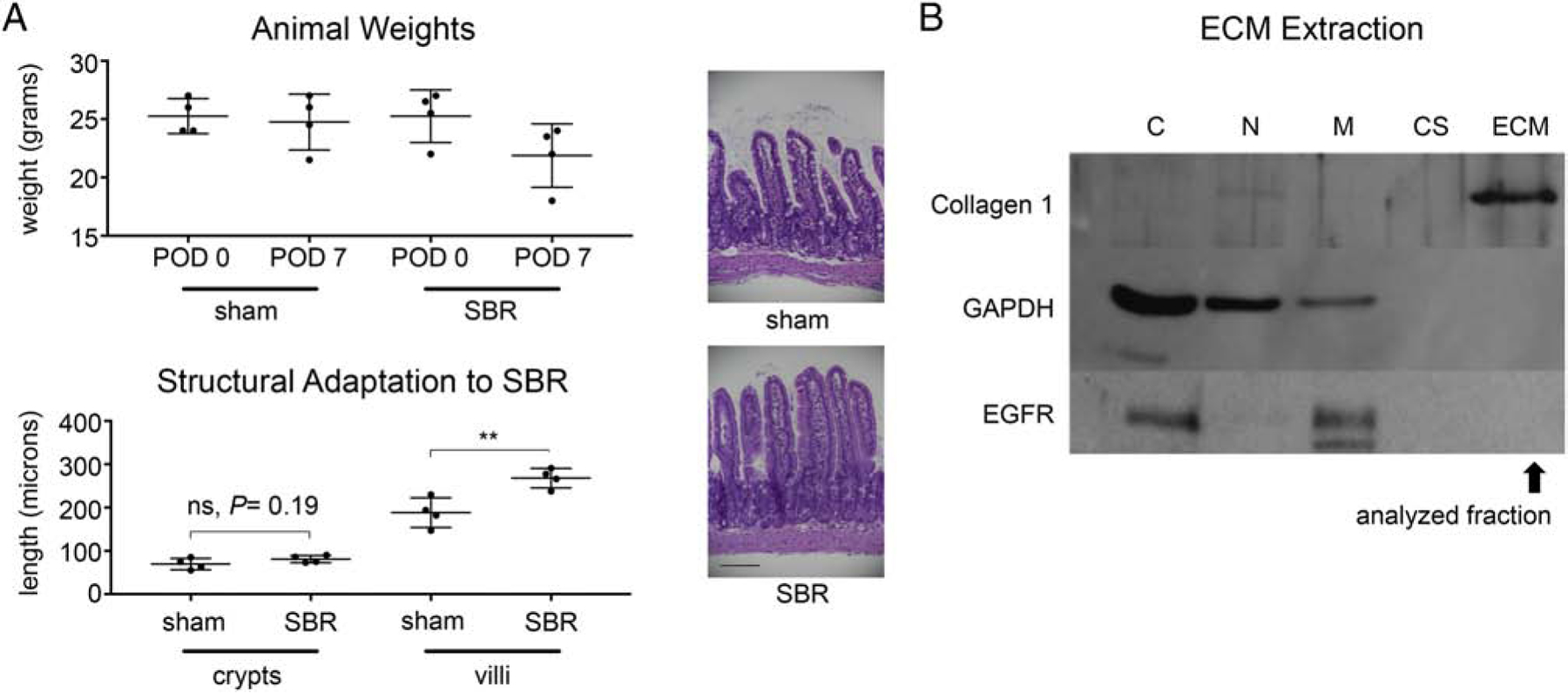
Structural adaptation and ECM enrichment of ileal tissue from sham surgery vs 50% proximal SBR mice. A. We controlled for metabolic factors such as age, gender, caging, and weight of mice. Graph depicts weights of sham vs SBR animals on post-operative day (POD) 0 (day of surgery) and 7 (time of tissue harvest). B. At POD 7 after either sham surgery or 50% proximal SBR, ileal tissue 2cm distal to the anastomosis was harvested and submitted for histological analysis to confirm structural adaptation (ie villus lengthening) in SBR mice (P< 0.05). Representative images of ileum from sham and SBR mice at post-operative day 7 are shown (40x, scale bar: 100 μm). B. Enrichment for ECM (which was extracted from flash-frozen ileal tissue using the CNMCS kit) was verified using Western Blotting for proteins expected within each fractionated component. C= cytoplasmic, N= nuclear, M= membrane, CS= cytoskeletal.
MS Analysis Reveals “Adaptation” of SI ECM to SBR
MS proteomic analysis of ileal ECM revealed 3034 differentially expressed proteins (heat map shown in Figure 2A), 95 of which reached statistical significance (P <0.05, 60 higher in sham and 35 higher in SBR), as shown in Figure 2B. MS protein enrichment results were analyzed using IPA, a widely utilized platform for interpreting proteomic data. IPA generated a list of pathways associated with Physiological System Development and Function, as well Canonical Pathways, and Upstream Regulators which were enriched in SBR, as shown in Table 1.
Figure 2.
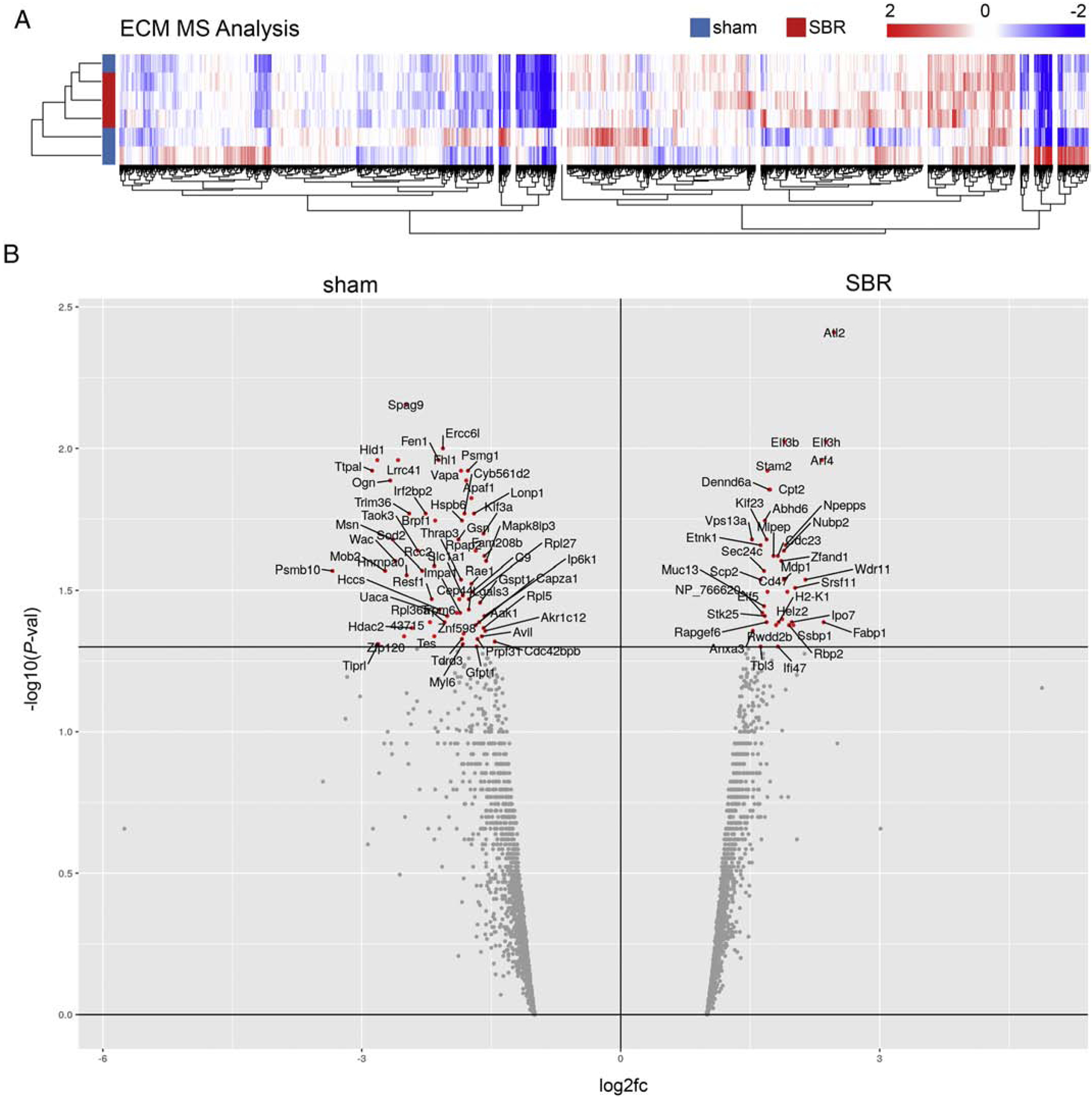
MS analysis of ECM from sham and SBR ileum reveals differences in composition. A. Heatmap shows protein expression in sham and SBR ECM, with 3034 genes differentially expressed. B. Volcano plotting of differential protein expression with proteins more highly expressed in sham (left) vs SBR (right) ECM. X axis indicates log2 fold-change (fc) in protein expression between the conditions, and Y axis indicates P-value of the expression difference (wherein -log10 P-value of >1.3 corresponds to a P-value of <0.05, as demarcated by bar, above which red data points indicate proteins with a P-value<0.05).
Table 1.
Ingenuity Pathway Analysis of Mass Spectrometry Results
| Top Physiological System Development and Function | |
|---|---|
| Name | p-value |
| Organismal Survival | <0.001 |
| Organismal Development | <0.001 |
| Digestive System Development and Function | <0.001 |
| Tissue Morphology | <0.001 |
| Top Canonical Pathways | |
| Name | p-value |
| GP6 Signaling Pathway | <0.001 |
| RhoA Signaling | <0.001 |
| Clathrin-mediated Endocytosis Signaling | <0.001 |
| mTOR Signaling | <0.001 |
| Fatty Acid-oxidation I | <0.001 |
| Top Upstream Regulators | |
| Name | p-value |
| TP53 | <0.001 |
| MYC | <0.001 |
| TGFB1 | <0.001 |
| pirinixic acid | <0.001 |
| HOXD3 | <0.001 |
PPAR Signaling is Globally Involved in Adaptation to SBR, including ECM Adaptation
We were interested to see pirinixic acid was a predicted upstream regulator of the observed ECM changes (Table 1) because pirinixic acid is a peroxisome proliferator-activated receptor alpha (PPAR∝) agonist[14]. In our prior single-cell analysis of intestinal epithelium following SBR, interactome analysis predicted the PPAR pathway influenced the observed transcriptional changes (which were primarily metabolic in nature) [15]. In that study, however, we were unable to capture an upregulation of epithelial Ppar∝ per say. That said, the present ECM analysis renewed our interest in exploring PPAR signaling during adaptation to SBR, and so we performed qPRC for Cyp4a10, a major downstream target of PPARα activation[16–18]. As shown in Figure 3A, there was a substantial and significant 6-fold increase (P <0.05) of Cyp4a10 expression in SBR as compared to sham.
Figure 3.
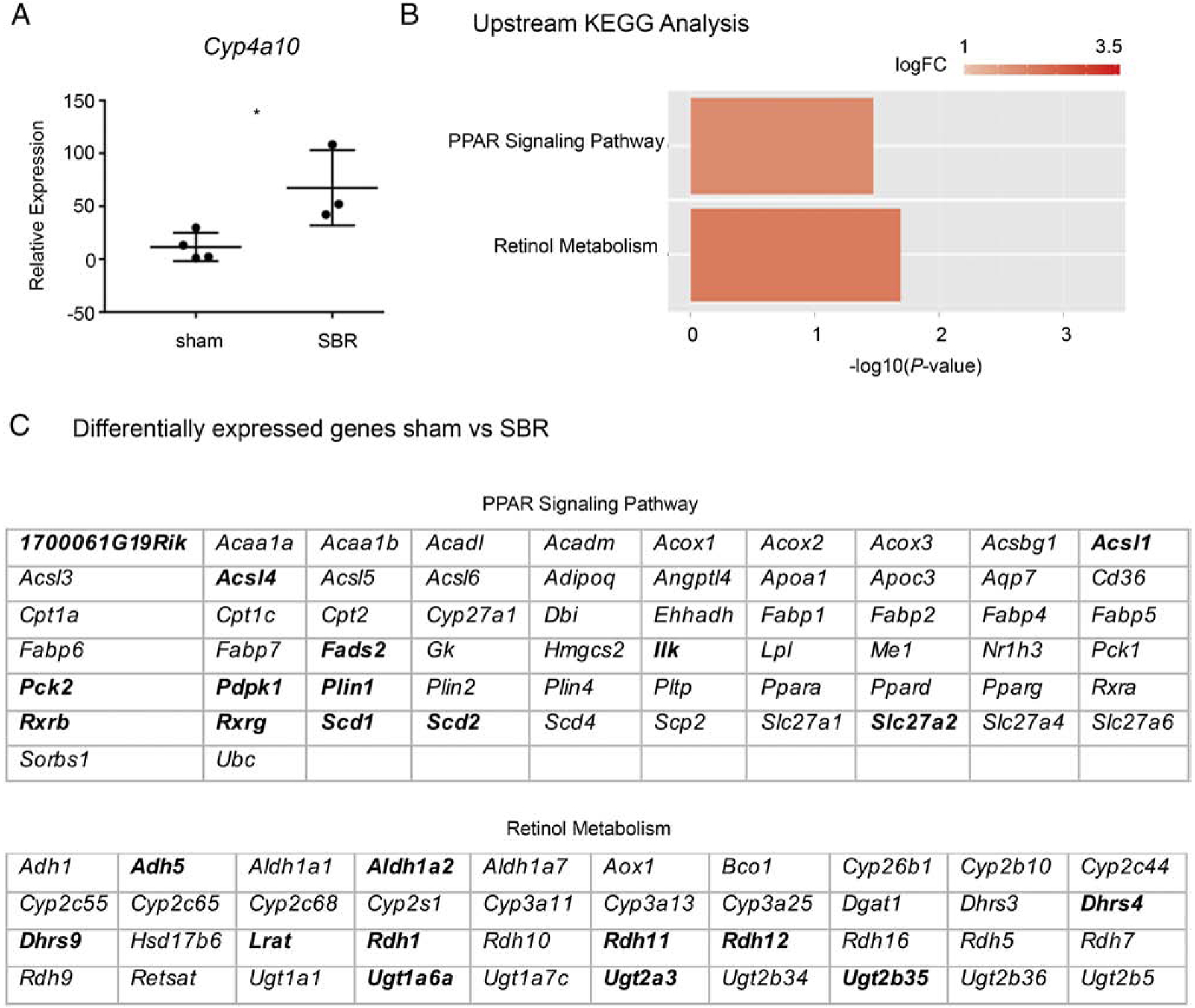
PPAR signaling is involved in multiple stages of adaptation to SBR. A. qPCR analysis of Cyp4a10 (a downstream indicator of PPAR activation) expression in villus epithelium of sham vs SBR animals (P<0.05). B. KEGG analysis of sub-epithelial tissue at an early time point after surgery reveals perturbed PPAR and retinol metabolism in SBR mice (P<0.05). Y axis indicates predicted perturbed pathways, and X axis indicates P-value of the prediction (wherein - log10 P-value of >1.3 corresponds to a P-value of <0.05). C. Genes that are differentially expressed in the SET of sham vs SBR mice. Bold indicates relatively higher expression in sham, non-bold indicates relatively higher expression in SBR.
After verifying PPAR signaling is active in adapted bowel from SBR mice, we next sought to determine whether PPAR signaling is also active at earlier stages of adaptation (and is thus an “Upstream Regulator”, as suggested in Table 1). We therefore performed RNA-Seq on sub-epithelial tissue (SET) at day 3 following sham or SBR surgery, with subsequent KEGG analysis. We utilized RNA-Seq because we wanted to capture global transcriptional changes which may be intrinsically linked with compositional changes in ECM after SBR. Further, we performed our analysis on SET in order to enrich for the primary ECM-producing cellular component of SI, and to avoid interference from the intestinal epithelium (which is highly metabolically active).
Ultimately, our analysis confirmed activation of the PPAR pathway within SET at this upstream timepoint (P <0.05, Figure 3B). Interestingly, we also noted increased Retinol Metabolism at this early time point (P <0.05, Fig. 3B). We highlight this because we have previously described a significant role for retinol metabolism during epithelial adaptation to SBR[6]. Presented in Figure 3C are genes differentially expressed between sham and SBR SET that were implicated in these KEGG pathways.
Discussion:
Here, we present data demonstrating altered composition and functional analysis of ECM (as assessed by MS and IPA)—with congruent upstream gene expression changes in ECM-producing tissue (as assessed by RNAseq and KEGG analysis of SET)— in mice that have undergone SBR vs sham surgery.
This study originated from the hypothesis that ECM must “adapt” in order to support altered structure and metabolic function of the ileum after SBR. We have previously demonstrated that SBR induces a shift in the metabolic signature of ileal epithelium, including significant upregulation of transcripts associated with lipid metabolism[6]. A stimulus for this shift is exposure to fatty acids and vitamin A (nutrients typically processed by proximal SI). These nutrients engage the PPAR and retinol metabolism pathways, respectively. We therefore expected to see involvement of PPAR and retinol signaling in our ECM, too.
As shown in Table 1, many of the pathways and regulators intrinsic to ECM’s adaptation to SBR are familiar within the SBR literature. For instance, our lab and others have previously described that mTOR[19] and TGF-beta[20, 21] signaling, perturbed TP53[22] and MYC[23] expression, and fatty acid oxidation[6] are pertinent to adaptation following SBR. And, while it has not been described to our knowledge, HOXD3, as well as other HoxD genes, are involved in gut development and maturation[24], which is challenged during adaptation to SBR.
Given our prior findings[6], we were excited to see that PPAR signaling is active not only in the epithelium, but also in the ECM and SET of adapted bowel. And, while retinoic acid was not a predicted upstream regulator of changes in ECM in Table 1, RhoA signaling was implicated in our analysis (Table 1), and it is known that retinoic acid can drive its effects via RhoA signaling[25]. Furthermore, nuclear PPARs form heterodimers with retinoid X receptors (RXRs)[26, 27] and the balance of PPAR to retinol signaling determines cellular behavior[28]. As such, their biology is intrinsically linked. Further study of the interaction between these two pathways—and how they interact to drive adaptation at both the epithelial and subepithelial level, including ECM, is warranted.
Our use of MS and RNA-Seq presents both advantages and disadvantages. One advantage is that all proteins and transcripts within samples were analyzed and accounted for. However, our findings largely lack tissue-architecture level resolution. Future studies—such as single cell level analysis of SET and complex imaging and immunolocalization analysis of ECM—will provide additional mechanistic insight. We hope this work establishes a foundation for such future study, as characterization of a “regenerative” or “adaptive” SI ECM could inform regenerative medicine and TESI approaches, wherein decellularized, ECM-enriched SI tissue may be used as a scaffolding substrate. The capacity for this has already been demonstrated in animal models[29]. Our prediction, moving forward, is that cells reading a “regenerative” matrix/scaffold will behave more favorably than cells reading a homeostatic matrix.
Conclusions:
Here we have described for the first time that ECM “adapts” following SBR. The major pathways and upstream regulators identified by our ECM analysis are largely consistent with known changes in ileal epithelial physiology following SBR. This includes major alterations in metabolic machinery, including significant upregulation of protein networks associated with PPAR signaling[6]. These findings indicate a previously undescribed role for ECM as an important component of adaptation. Future studies will provide additional structural and mechanistic insight, including whether the observed ECM changes are a necessary byproduct of adaptation or, perhaps more importantly, a driving force behind adaptation.
Acknowledgements:
This work was supported by the Children’s Discovery Institute of Washington University in St. Louis and St. Louis Children’s Hospital MI-F-2017–629 (K.M.S.); the Children’s Surgical Sciences Research Institute of the St. Louis Children’s Hospital (B.W.W.); the Association for Academic Surgery Foundation (K.M.S.), National Institutes of Health 4T32HD043010-14 (K.M.S). Select histological analyses were performed with support from the Washington University Digestive Diseases Research Core Center. The mass spectrometric experiments were performed at the Washington University Proteomics Shared Resource (WU-PSR), Director, R Reid Townsend MD.PhD. The expert technical assistance of Petra Erdmann-Gilmore, Yiling Mi and Rose Connors is gratefully acknowledged. The WU-PSR is supported in part by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR000448), the Mass Spectrometry Research Resource (NIGMS P41 GM103422) and the Siteman Comprehensive Cancer Center Support Grant (NCI P30 CA091842). We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1TR002345 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Daley WP, Peters SB, Larsen M. Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci 2008;121(Pt 3):255–64. [DOI] [PubMed] [Google Scholar]
- [2].Wang Y, Cui CB, Yamauchi M, Miguez P, Roach M, Malavarca R, et al. Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue-specific biomatrix scaffolds. Hepatology 2011;53(1):293–305. [DOI] [PubMed] [Google Scholar]
- [3].Brizzi MF, Tarone G, Defilippi P. Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr Opin Cell Biol 2012;24(5):645–51. [DOI] [PubMed] [Google Scholar]
- [4].Panayotou G, End P, Aumailley M, Timpl R, Engel J. Domains of laminin with growth-factor activity. Cell 1989;56(1):93–101. [DOI] [PubMed] [Google Scholar]
- [5].Wang X, Harris RE, Bayston LJ, Ashe HL. Type IV collagens regulate BMP signalling in Drosophila. Nature 2008;455(7209):72–7. [DOI] [PubMed] [Google Scholar]
- [6].Seiler KM, Waye SE, Kong W, Kamimoto K, Bajinting A, Goo WH, et al. Single-Cell Analysis Reveals Regional Reprogramming During Adaptation to Massive Small Bowel Resection in Mice. Cell Mol Gastroenterol Hepatol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Helmrath MA, VanderKolk WE, Can G, Erwin CR, Warner BW. Intestinal adaptation following massive small bowel resection in the mouse. J Am Coll Surg 1996;183(5):441–9. [PubMed] [Google Scholar]
- [8].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics 2012;11(4):M111014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Naba A, Clauser KR, Lamar JM, Carr SA, Hynes RO. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. Elife 2014;3:e01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Werner T, Sweetman G, Savitski MF, Mathieson T, Bantscheff M, Savitski MM. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal Chem 2014;86(7):3594–601. [DOI] [PubMed] [Google Scholar]
- [11].Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999;20(18):3551–67. [DOI] [PubMed] [Google Scholar]
- [12].Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30(4):523–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aladegbami B, Barron L, Bao J, Colasanti J, Erwin CR, Warner BW, et al. Epithelial cell specific Raptor is required for initiation of type 2 mucosal immunity in small intestine. Sci Rep 2017;7(1):5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Keller H, Devchand PR, Perroud M, Wahli W. PPAR alpha structure-function relationships derived from species-specific differences in responsiveness to hypolipidemic agents. Biol Chem 1997;378(7):651–5. [DOI] [PubMed] [Google Scholar]
- [15].Gajda AM, Storch J. Enterocyte fatty acid-binding proteins (FABPs): different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot Essent Fatty Acids 2015;93:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bunger M, van den Bosch HM, van der Meijde J, Kersten S, Hooiveld GJ, Muller M. Genome-wide analysis of PPARalpha activation in murine small intestine. Physiol Genomics 2007;30(2):192–204. [DOI] [PubMed] [Google Scholar]
- [17].de Vogel-van den Bosch HM, Bunger M, de Groot PJ, Bosch-Vermeulen H, Hooiveld GJ, Muller M. PPARalpha-mediated effects of dietary lipids on intestinal barrier gene expression. BMC Genomics 2008;9:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rakhshandehroo M, Knoch B, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res 2010;2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Barron L, Sun RC, Aladegbami B, Erwin CR, Warner BW, Guo J. Intestinal Epithelial-Specific mTORC1 Activation Enhances Intestinal Adaptation After Small Bowel Resection. Cell Mol Gastroenterol Hepatol 2017;3(2):231–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sukhotnik I, Lulu SB, Pollak Y, Bejar J, Coran AG, Mogilner JG. TGF-beta affects enterocyte turnover in correlation with TGF-beta receptor expression after massive small bowel resection. J Pediatr Gastroenterol Nutr 2012;55(6):721–7. [DOI] [PubMed] [Google Scholar]
- [21].Sukhotnik I, Mogilner JG, Shaoul R, Karry R, Lieber M, Suss-Toby E, et al. Responsiveness of intestinal epithelial cell turnover to TGF-alpha after bowel resection in a rat is correlated with EGF receptor expression along the villus-crypt axis. Pediatr Surg Int 2008;24(1):21–8. [DOI] [PubMed] [Google Scholar]
- [22].Cohran V, Managlia E, Bradford EM, Goretsky T, Li T, Katzman RB, et al. Epithelial PIK3R1 (p85) and TP53 Regulate Survivin Expression during Adaptation to Ileocecal Resection. Am J Pathol 2016;186(7):1837–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sukhotnik I, Roitburt A, Pollak Y, Dorfman T, Matter I, Mogilner JG, et al. Wnt/beta-catenin signaling cascade down-regulation following massive small bowel resection in a rat. Pediatr Surg Int 2014;30(2):173–80. [DOI] [PubMed] [Google Scholar]
- [24].Zakany J, Darbellay F, Mascrez B, Necsulea A, Duboule D. Control of growth and gut maturation by HoxD genes and the associated lncRNA Haglr. Proc Natl Acad Sci U S A 2017;114(44):E9290–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Namachivayam K, MohanKumar K, Arbach D, Jagadeeswaran R, Jain SK, Natarajan V, et al. All-Trans Retinoic Acid Induces TGF-beta2 in Intestinal Epithelial Cells via RhoA- and p38alpha MAPK-Mediated Activation of the Transcription Factor ATF2. PLoS One 2015;10(7):e0134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, et al. Structure of the intact PPAR-gamma-RXR-nuclear receptor complex on DNA. Nature 2008;456(7220):350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dawson MI, Xia Z. The retinoid X receptors and their ligands. Biochim Biophys Acta 2012;1821(1):21–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol 2008;8(9):685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kitano K, Schwartz DM, Zhou H, Gilpin SE, Wojtkiewicz GR, Ren X, et al. Bioengineering of functional human induced pluripotent stem cell-derived intestinal grafts. Nat Commun 2017;8(1):765. [DOI] [PMC free article] [PubMed] [Google Scholar]


