Abstract
Purpose:
Treatment with PD-(L)1 blockade can produce remarkably durable responses in non-small cell lung cancer (NSCLC) patients. However, a significant fraction of long-term responders ultimately progress and predictors of late progression are unknown. We hypothesized that circulating tumor DNA (ctDNA) analysis of long-term responders to PD-(L)1 blockade may differentiate those who will achieve ongoing benefit from those at risk of eventual progression.
Experimental Design:
In patients with advanced NSCLC achieving long-term benefit from PD-(L)1 blockade (PFS≥12 months), plasma was collected at a surveillance timepoint late during/after treatment to interrogate ctDNA by Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq). Tumor tissue was available for 24 patients and was profiled by whole-exome sequencing (n=18) or by targeted sequencing (n=6).
Results:
31 NSCLC patients with long-term benefit to PD-(L)1 blockade were identified and ctDNA was analyzed in surveillance blood samples collected at a median of 26.7 months after initiation of therapy. Nine patients also had baseline plasma samples available, and all had detectable ctDNA prior to therapy initiation. At the surveillance timepoint, 27 patients had undetectable ctDNA and 25 (93%) have remained progression-free; by contrast, all four patients with detectable ctDNA eventually progressed (Fisher’s p<0.0001; PPV 1 [95% CI 0.51-1]; NPV 0.93 [95% CI 0.80-0.99]).
Conclusions:
ctDNA analysis can noninvasively identify minimal residual disease in patients with long-term responses to PD-(L)1 and predict the risk of eventual progression. If validated, ctDNA surveillance may facilitate personalization of the duration of immune checkpoint blockade and enable early intervention in patients at high risk for progression.
Keywords: Immunotherapy, cfDNA, lung cancer, durable response, ctDNA
Introduction:
Recent trials have established PD-(L)1 blockade therapy as a routine component of first-line therapy for nearly all patients with advanced non-small cell lung cancer (NSCLC) (1–4). PD-(L)1 blockade is characterized by the potential for long-term benefit and relative safety of prolonged immunotherapy (5). However, the optimal duration of treatment remains unknown (6) and even among patients with long-term benefit, eventual progression can unfortunately occur (7,8). Current radiologic tools are inadequate (9,10) for differentiating patients with durable responses to PD-(L)1 blockade – who may have already achieved cure and may not need continued therapy – from those with residual disease – who are at risk for progression and may benefit from continued therapy and/or early intervention with additional treatment.
We hypothesized that circulating tumor DNA (ctDNA) analysis may permit more sensitive and specific detection of minimal residual disease among patients with metastatic NSCLC and long-term benefit from PD-(L)1 blockade. In early stage NSCLC, ctDNA has been shown to identify those at high risk of recurrence following definitive therapy (11,12). In patients with metastatic NSCLC, baseline ctDNA levels are higher and may be used to estimate tumor mutation burden (13) and on-treatment ctDNA dynamics within 4-8 weeks of treatment initiation can predict initial response to PD-L1 blockade (13–15).
Our group previously developed a next-generation sequencing-based method for ctDNA analysis called Cancer Personalized Profiling by Deep Sequencing (CAPP-Seq) that can sensitively track ctDNA burden while maintaining high specificity (16,17). It has been speculated that this approach could be useful to examine patients treated with immunotherapy to identify risk of ultimate progression (18), but no data have been reported to date. We examined this question using CAPP-Seq ctDNA analysis of patients with long-term benefit to PD-1 blockade and found that eventual progression is strongly associated with detectable residual ctDNA. Conversely, most patients with undetectable ctDNA late, during, or after treatment remained disease-free and may be approaching cure.
Materials and Methods:
Subject Details
Immunotherapy treated patients
All patients had stage IV non-small cell lung cancer (NSCLC) and were treated with PD-(L)1 blockade alone or in combination at Memorial Sloan Kettering Cancer Center (Figure 1A and Table S1). All patients initiated therapy between 05/16/2011 and 10/28/2016 dates. The study was conducted in accordance with the ethical principles set forward in the Declaration of Helsinki. All patients provided their written consent to participate in specimen collection and molecular analysis study approved by the Memorial Sloan Kettering Institutional Review Board. PD-L1 expression was assessed by immunohistochemistry. The median follow-up from the start of PD-(L)1 blockade was 38.7 months (range 14.3 – 81.7). Long-term benefit in this analysis was pre-defined as progression-free survival ongoing > 1 year.
Figure 1: Pre-treatment molecular profiles of tumor biopsies and cfDNA from long-term responders to PD-(L)1 blockade.
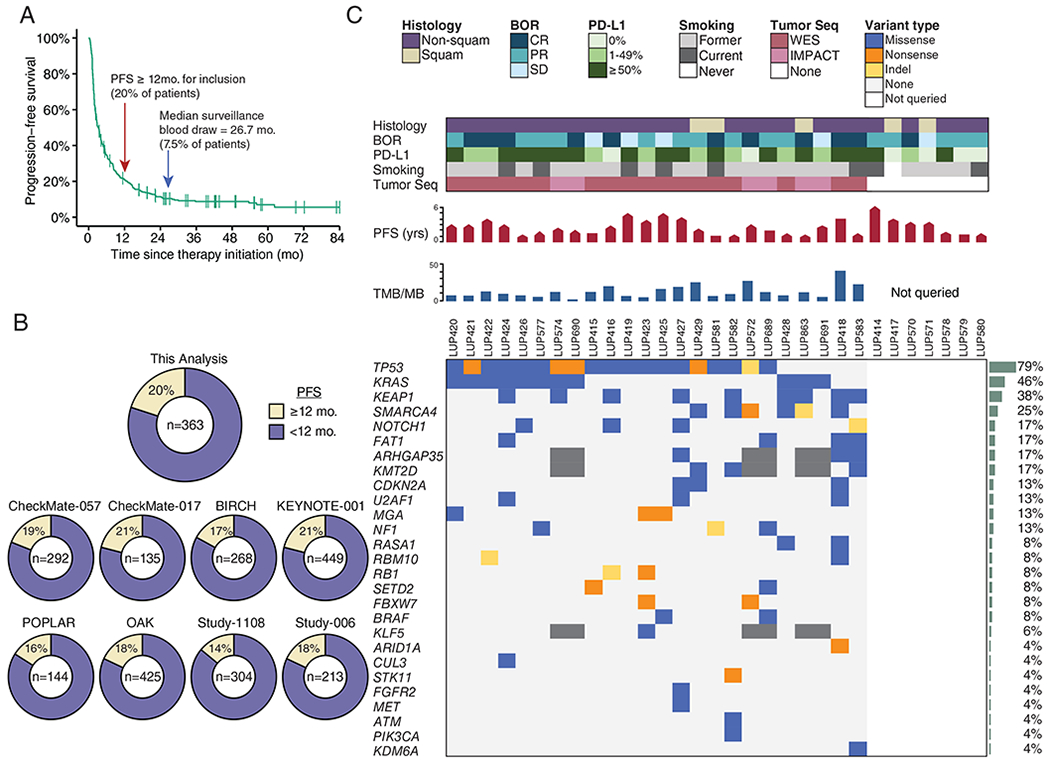
A) Progression-free survival of patients with NSCLC treated with PD-(L)1 blockade as part of initial clinical trials at MSKCC (n = 363). Arrows indicate cut-off for definition of long-term benefit (PFS ≥ 12 months), and the median surveillance plasma collection time (26.7 months).
B) Percent of patients who would be classified as achieving long-term benefit from (A) as well as, for context, other clinical trials with unselected NSCLC.
C) Clinical and molecular features of patients with advanced NSCLC experiencing long-term responses to PD-(L)1 blockade. Each column represents an individual patient. Boxes are color coded for tumor histology (squamous or non-squamous), smoking status (former, current, or never), and best overall response (BOR) by RECIST criteria (complete response [CR], partial response [PR], or stable disease [SD]) as indicated. Tumor PD-L1 expression is stratified as 0%, 1%-49%, or ≥50%. When available, pre-treatment tumor tissue was sequenced (Tumor Seq) by whole exome sequencing (WES) or a targeted panel (MSK-IMPACT). Patients with no Tumor Seq were unevaluable for TMB and individual tumor mutations as depicted on the right. PFS is depicted in months, where the pointed bars represent ongoing responses and the flat bars represent patients who have progressed. TMB is presented as the number of nonsynonymous mutations and indels per megabase of the coding exome. Nonsynonymous mutations and indels in genes recurrently mutated in NSCLC are shown in descending order of prevalence (35). Mutation recurrence rate in the cohort is depicted by bar graphs to the right.
Clinical efficacy analyses
Objective response was assessed by investigator-assessed RECIST v1.1. Partial and complete responses were confirmed by repeat imaging occurring at least 4 weeks after the initial identification of response; unconfirmed partial responses were considered stable disease. Progression-free survival was determined from the start of PD-(L)1 blockade, with outcomes determined or censored as of the 06/05/2018 database lock. Event-free survival was determined from the date of first surveillance plasma collection, with outcomes determined or censored as of the 06/05/2018 database lock.
Tumor and germline samples
24 of 31 patients had tumor tissue used for next-generation sequencing, either whole exome sequencing (n=18) or MSK-IMPACT (n=6). All tissue was obtained prior to treatment with immunotherapy. Germline DNA was obtained from peripheral blood mononuclear cells from all patients.
Plasma samples
Plasma was processed either from whole blood samples collected in sodium heparin CPT Cell Preparation Tubes (BD Biosciences) or Cell-Free DNA BCT tubes (STRECK). CPT tubes were centrifuged at 1500 x g at room temperature with brakes off for 20 minutes. After density gradient centrifugation, the plasma supernatant above the peripheral blood mononuclear cell monolayer was pipetted from the CPT tubes and distributed into cryovials as 1.5 ml aliquots and stored at −20 deg C. Cell-free DNA BCT tubes were centrifuged at 800 x g for 10 min at room temperature, separated plasma and buffy coat were saved in separate vials, and stored at −80°C.
Method Details:
cfDNA extraction
Cell-free DNA was extracted from 2 to 6 mL of plasma using the QiaAmp Circulating Nucleic Acid Kit according to manufacturer’s instructions. After isolation, DNA was quantified using the Qubit dsDNA High Sensitivity Kit. For samples collected in CPT tubes, cfDNA was subsequently treated with Heparinase II (Sigma) for 2 hours at 37°C and subsequently purified by 1.8X bead selection.
CAPP-Seq
Cancer Personalized Profiling by deep Sequencing (CAPP-Seq) was performed as previously described (16,17). In brief, a maximum of 32 ng of cfDNA or 32 ng of sonicated DNA from plasma-depleted whole blood (as a source of matched germline DNA) was utilized for library preparation with the KAPA HyperPrep Kit with some modifications to the manufacturer’s instructions as previously described (17,19). After library preparation, hybridization-based enrichment of specific sequences was performed using a custom-designed panel of biotinylated DNA oligonucleotides (17,19). Following enrichments, samples were sequenced on an Illumina HiSeq4000 and sequencing data were processed using a custom bioinformatics pipeline (16,17).
For tumor-informed CAPP-Seq analysis we: 1) limited variants to coding positions, 2) removed any variants with greater than 1 supporting read in the matched germline sample, 3) removed variants with more than 2 supporting reads in 5% of healthy control plasma samples (n = 54), 4) removed variants present in ≥0.05% of samples in the Genome Aggregation Database (20). Presence of ctDNA was determined by monitoring the tumor mutations overlapping with the CAPP-Seq selector and a previously described Monte Carlo–based ctDNA detection index with a significance cut-point of P≤0.05 (16). For patients with serial samples available, detection was defined by the last sample available.
Criteria for tumor-naïve ctDNA detection were defined using the 24 patients for whom tumor tissue was available and were then applied to patients for whom we did not have tumor tissue. Specifically, for tumor naïve calling we: 1) limited variants to coding positions, 2) removed any variants with greater than 0 reads in the matched germline sample, 3) removed variants with more than 2 reads in any healthy control plasma samples (n = 54), 4) removed variants present in ≥0.05% of samples in the Genome Aggregation Database (20). A sample with at least one variant identified was considered to be positive for ctDNA. For exploratory analyses of tumor-naïve ctDNA analysis, LUP425 was excluded due to interval development of a colorectal cancer that could have confounded tumor-naïve, but not tumor-informed ctDNA detection.
Tumor whole exome sequencing, alignment, assembly, and variant calling
Whole-exome capture libraries were constructed using the Agilent Sure-Select Human All Exon v2.0 (44Mb, n = 5), v4.0 (51Mb, n = 6), or Illumina’s Rapid Capture Exome (38Mb, n = 7) baited target kit. Enriched exome libraries were sequenced on a HiSeq 2000, 2500, or 4000 platform to generate paired-end reads (2x76bp). A BAM file was produced by aligning tumor and normal sequences to the hg19 human genome build using the Burrows-Wheeler Aligner (BWA) (21). Further indel realignment, base-quality score recalibration, and duplicate-read removal were performed using the Genome Analysis Toolkit (GATK) (22). Quality control metrics were computed using the Broad Institute Picard software. Fingerprint genotypes were used to verify match of tumor and normal samples. Artifacts produced by oxidation during DNA sequencing were removed using the OxoG3 filter (23). Samples with mean target coverage <60X in tumor or <30X in normal were excluded. Single nucleotide variants were identified using Mutect2 (24) with default parameters, filtered using FilterMutectCalls from the GATK, and annotated using Oncotator (25). Indelocator (http://archive.broadinstitute.org/cancer/cga/indelocator) was used to generate indel calls. Tumor mutation burden per megabase (TMB/MB) was calculated by dividing the total number of non-synonymous mutations by the coding region of each exome capture kit.
Tumor targeted next generation sequencing and variant calling
In cases where tumor whole exome sequencing was not available, targeted next-generation sequencing was performed using MSK-IMPACT, as previously described (26). Briefly, DNA was extracted from tumors and patient-matched blood samples. Barcoded libraries were generated and sequenced using a custom gene panel of 341 (n = 1), 410 (n = 2), or 468 (n = 3) genes. The TMB/MB was calculated by dividing the total number of nonsynonymous mutations by the coding region captured in each panel (8).
Quantification and Statistical Analyses
To determine a target sample size for our study, we extrapolated from a previous study in which we observed that detection of ctDNA molecular residual disease in localized NSCLC had a hazard ratio of approximately 40 for predicting event-free survival (12). Assuming 10% of patients with long-term responses to PD-(L)1 blockade will recur, that all of these patients will have detectable ctDNA molecular residual disease, and a similar hazard ratio as in our prior study, a cohort of ≥25 patients achieves ≥95% power to detect a difference in event-free survival between ctDNA positive and negative patients (one-sided two arm binomial with alpha = 0.05) (27). Fisher’s exact test was used to compare frequencies between two groups in 2×2 contingency tables. The Wilson method was used to compute 95% confidence intervals of proportions. For progression-free survival analysis, the log-rank test was used to compare Kaplan-Meier survival curves. To avoid guarantee-time bias (28), we only considered progression events following the plasma collection date as the landmark. Statistical analyses were performed using GraphPad Prism v.6 and R 3.3.2.
Results:
Clinical and molecular features of NSCLC patients with long-term benefit to PD-(L)1 blockade
To explore if ctDNA analysis can identify patients with long-term benefit to PD-(L)1 blockade who are at risk of eventual progression, we identified a cohort of 31 patients with advanced NSCLC who had sustained clinical benefit from PD-(L)1 blockade (progression free survival ≥12 months) and had a plasma sample that was collected ≥ 6 months after initiating treatment (“surveillance blood draw”). To assemble this cohort with necessary long-term follow-up (median 38.7 months, range 14.3 – 81.7), we focused on patients at MSKCC who were treated on initial clinical trials of PD-(L)1 blockade. In this cohort of clinical trial patients (n=363 patients), 20% (n=72) were progression-free at 12 months (Figure 1A), of whom 29 patients had plasma available for analysis. This rate of long-term benefit is similar to that observed in other clinical trials of PD-(L)1 therapy in patients with unselected NSCLC (Figure 1B) and highlights the relatively rarity of longer-term responders to PD-1 blockade in NSCLC. We also included an additional two patients who met the inclusion criteria and were treated with commercial PD-(L)1 inhibitors. Patients in our cohort received immune checkpoint inhibitors for a median of 20.4 months (range: 1.7-48.1) and the surveillance blood draw occurred at a median of 26.7 months (range: 8.3-61.8 months) after initiation of therapy. The time of surveillance blood draw relative to the start of treatment (Figure S1A) or the end of treatment (Figure S1B) was similar in those patients who did or did not eventually progress. As expected for a cohort consisting only of patients who achieved durable clinical benefit from PD-(L)1 blockade, the majority of tumors expressed PD-L1 by immunohistochemistry (22 of 31; 71%) and nearly all patients had been smokers (30 of 31, 97%; Figure 1C, Table S1). The frequency of driver mutations was as expected for a cohort of patients with advanced NSCLC (Figure 1C).
Tumor mutation profiling and pre-treatment ctDNA analysis
Tumor tissue was available for 24 patients and was profiled by whole exome sequencing (WES, n=18) or by targeted gene sequencing utilizing the MSK Integrated Mutation Profiling of Actionable Cancer Targets assay (MSK-IMPACT, n=6). Tumor mutation profiles included expected NSCLC driver mutations in genes such as TP53 and KRAS (Figure 1C, Table S6).
We performed CAPP-Seq ctDNA profiling as previously described (12,17). We applied either a “tumor-informed” or “tumor-naïve” approach to ctDNA detection. Tumor-informed CAPP-Seq ctDNA detection leverages prior knowledge of tumor mutations from sequencing of tumor tissues and matched leukocytes and queries these in plasma cell-free DNA (cfDNA) using a Monte Carlo-based algorithm (16). When tumor tissue was not available, we employed tumor-naïve CAPP-Seq ctDNA detection, which identifies ctDNA mutations in plasma cell-free DNA without consideration of prior knowledge of mutations present in a patient’s tumor. Samples were considered positive for ctDNA by the tumor-naïve approach if any mutations were detected. To minimize the risk of erroneously considering mutations due to clonal hematopoiesis, both ctDNA detection approaches included sequencing of matched cellular DNA derived from leukocytes and filtering out of mutations detected in both samples. Moreover, variants for both approaches were filtered for presence in the Genome Aggregation Database to further exclude any potential single nucleotide polymorphisms (20). Tumor-informed CAPP-Seq allows for greater sensitivity by decreasing multiple hypothesis testing and has ctDNA detection limit of ~0.002% while tumor-naïve analysis has a detection limit of ~0.1% (17).
To begin, we analyzed pre-treatment ctDNA levels in nine patients for whom baseline plasma samples were available. We detected ctDNA before anti-PD-(L)1 therapy in all nine patients, eight via tumor-informed CAPP-Seq and one (for whom no tumor tissue was available) via tumor-naïve CAPP-Seq. Applying tumor-naïve detection to all nine pre-treatment samples resulted in a modest decrease in sensitivity to 89%, since the ctDNA concentration in one sample detected using the tumor-informed approach was below the tumor-naïve detection limit (Figure 2A). The patients for whom we had baseline plasma available did not have different tumor burden than remaining patients, suggesting they are representative of the cohort as a whole and routine ctDNA detectability at baseline was not a result of disproportionately high tumor burden (Figure S2A). The median allele fraction of mutations in baseline plasma samples was 0.29% (range: 0.07% - 6.62%), and the median allele fractions were 40-fold higher in corresponding tumor biopsies (range: 3.68% - 59.09%; Figure 2B, Tables S3 and S5). These results are similar to recently published data demonstrating that the majority of patients with untreated, advanced NSCLC have detectable ctDNA (13,29).
Figure 2: ctDNA analysis identifies patients at risk of eventual progression after long-term response to PD-(L)1 blockade.

A) ROC analysis of pre-treatment ctDNA detection using CAPP-Seq in either the tumor-naïve (red, n = 9) or tumor-informed (green, n = 8) context.
B) Comparisons of pre-treatment variant allele (%) for plasma ctDNA by CAPP-Seq (right) versus corresponding tumor biopsies by WES (left) are shown for nine patients with baseline plasma available. Tumor-informed CAPP-Seq was performed when tumor tissue was available (n=8) and tumor-naïve CAPP-Seq was performed when it was not (n=1; LUP417). N-values depict the number of mutant genes detected by WES in tumor biopsies and monitored by CAPP-Seq in plasma. ND = not detected.
C) Percent of patients eventually experiencing progression based on presence or absence of detectable ctDNA in the surveillance plasma sample. P < 0.0001 (one-sided Fisher’s Exact Test).
D) Comparison of event-free survival after surveillance cfDNA collection, stratified by ctDNA status using tumor-informed detection (detection limit ~0.002%) for patients with pre-treatment tumor tissue (n = 24) and tumor-naïve detection (detection limit ~0.1%) for patients without (n = 7). P < 0.0001 (log-rank test).
ctDNA analysis can identify patients at risk of eventual progression
We next asked whether detection of ctDNA in surveillance blood draws in patients with long-term responses to PD-1 blockade correlates with the development of eventual progression. To maximize sensitivity of ctDNA detection, we applied tumor-informed CAPP-Seq analysis when tumor tissue was available and tumor-naïve analysis if it was not. ctDNA was not detected in 27 patients, 25 of whom have not progressed (median event-free survival since plasma collection 16.96 months [range 4.76-24.21 months]). Conversely, ctDNA was detected at the surveillance timepoint in four patients and each of these patients ultimately progressed (Fisher’s Exact Test, p < 0.0001; PPV 1 [95% CI 0.51-1]; NPV 0.93 [95% CI 0.80-0.99]) (Figure 2C, Figure 3B, Tables S2 and S4). Patients with undetectable ctDNA had significantly longer freedom from progression than patients in whom ctDNA was detectable (Figure 2D). Tumor sequencing was available for all patients with detectable ctDNA during surveillance. There was no difference in the time of surveillance blood draw collection relative to the start (Figure S2B) or the end (Figure S2C) of treatment in patients with ctDNA detected or not detected. Moreover, residual tumor burden and number of metastatic sites were similar in patients with ctDNA detected and not detected, indicating that ctDNA detection was not driven by visible tumor burden differences (Figure S3A–B). Thus, detection of ctDNA during surveillance of patients with extended responses to PD-(L)1 blockade portends a high risk of recurrence while undetectable ctDNA is encouraging of continued durable response.
Figure 3: Tumor-naïve and tumor-informed ctDNA analyses are largely concordant if leukocyte variants are considered.
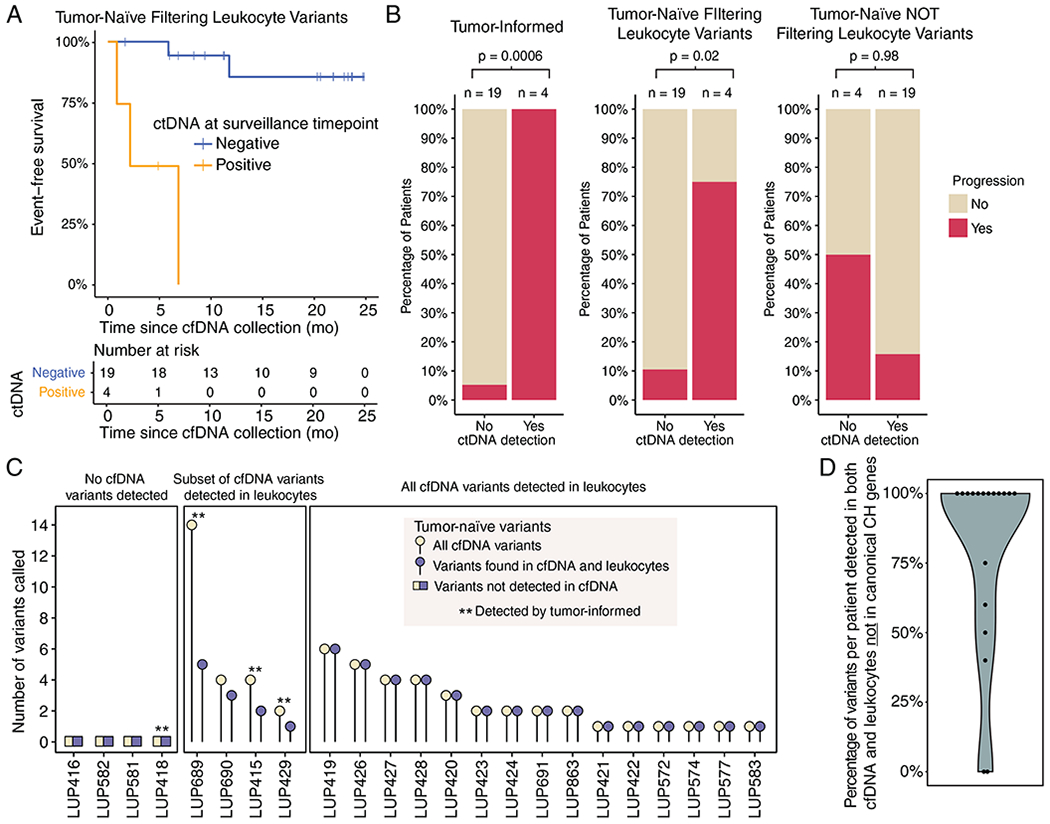
A) Event-free survival after surveillance cfDNA collection in the subset of patients with tumor tissue available using tumor-naïve detection (n = 23). P < 0.0001 (log-rank test).
B) Concordance levels between surveillance sample ctDNA detection status (x-axis) and ultimate progression status (y-axis, colored bars) are depicted as a function of 3 ctDNA genotyping strategies. The tumor-informed strategy (left) demonstrates the best predictive performance (one-sided Fisher’s Exact Test P = 0.0006), followed by the tumor naïve strategy after excluding leukocyte variants (one-sided Fisher’s Exact Test P = 0.02, middle). In the absence of genotyping of leukocyte-derived variants (right), the tumor-naïve strategy fails to significantly predict progression risk (one-sided Fisher’s Exact Test P = 0.98, right). Data are for the same 23 patients with available tumor tissues in other panels of this figure.
C) Relationship between the total number of mutations called by a tumor-naïve strategy (y-axis) in each patient (x-axis), as a function of evidence for mutations in leukocytes (3 large boxes). Patients detected by tumor-informed ctDNA analysis are indicated by the asterisks (n = 4). Data are for the same 23 patients with available tumor tissues in other panels of this figure.
D) Percentage of variants present in both cfDNA and leukocytes that were not in genes previously implicated in clonal hematopoiesis (n = 19).
We next asked if baseline ctDNA levels might similarly inform risk of eventual progression in patients with PFS≥12 months on PD-(L)1 blockade. Since we had baseline plasma samples for only a subset of our cohort, we analyzed published results from NSCLC patients receiving the anti-PD-L1 monoclonal antibody atezolizumab (13). Focusing on the 75 patients in this study with baseline ctDNA measurement and PFS ≥ 12 mo, we found that there was no difference in baseline variant allele fraction between patients who ultimately progressed versus those who remained progression-free (Figure S2D).
Next, we performed an exploratory analysis to investigate if we could have achieved similar outcome stratification using only our tumor-naïve detection approach. We therefore applied tumor-naïve ctDNA detection to all patients who had available tumor tissue and compared results to those of tumor-informed analysis in the same patients. It is important to note that since these same cases were used to establish the tumor-naïve calling thresholds, the performance of tumor-naïve calling may be overfit in these cases. Tumor-naïve calling was discordant with tumor-informed calling in two out of 23 (9%) patients, but the difference in event-free survival between ctDNA positive and negative patients was nearly as striking (Figure 3A–B). Importantly, achieving high concordance between the tumor-informed and -naïve ctDNA approaches required sequencing of matched leukocyte DNA to remove variants that were found in both the leukocyte and cell-free compartments (Figure 3B–C). Mutations present in matched leukocytes were found in the cfDNA of 19 (83%) patients and would have led to misclassification of response in 18 (78%) patients if not eliminated. However, exclusion of leukocyte-derived variants reduced misclassification when using the tumor-naïve approach to 3 (13%) patients (Figure 3B–C). Notably, the majority of these mutations were not in genes previously implicated in clonal hematopoiesis (30,31) (mean per patient = 81%) and thus could not have been eliminated using gene level filtering (Figure 3D, Table S7).
Among 27 patients with undetectable ctDNA in the surveillance sample, two eventually progressed (Figure 4A). Progression occurred 5.7 and 9.1 months after the blood draw and plasma was not available more proximally to the time of clinical progression. In four patients with detectable ctDNA in the surveillance blood draw, ctDNA was detected a median of 4.4 months (range: 0.9 – 11.5 months) prior to identification of progression by imaging (Figure 4B).
Figure 4: Presence of ctDNA during surveillance precedes radiologic progression and informs disease status of patients undergoing PD-(L)1 blockade.
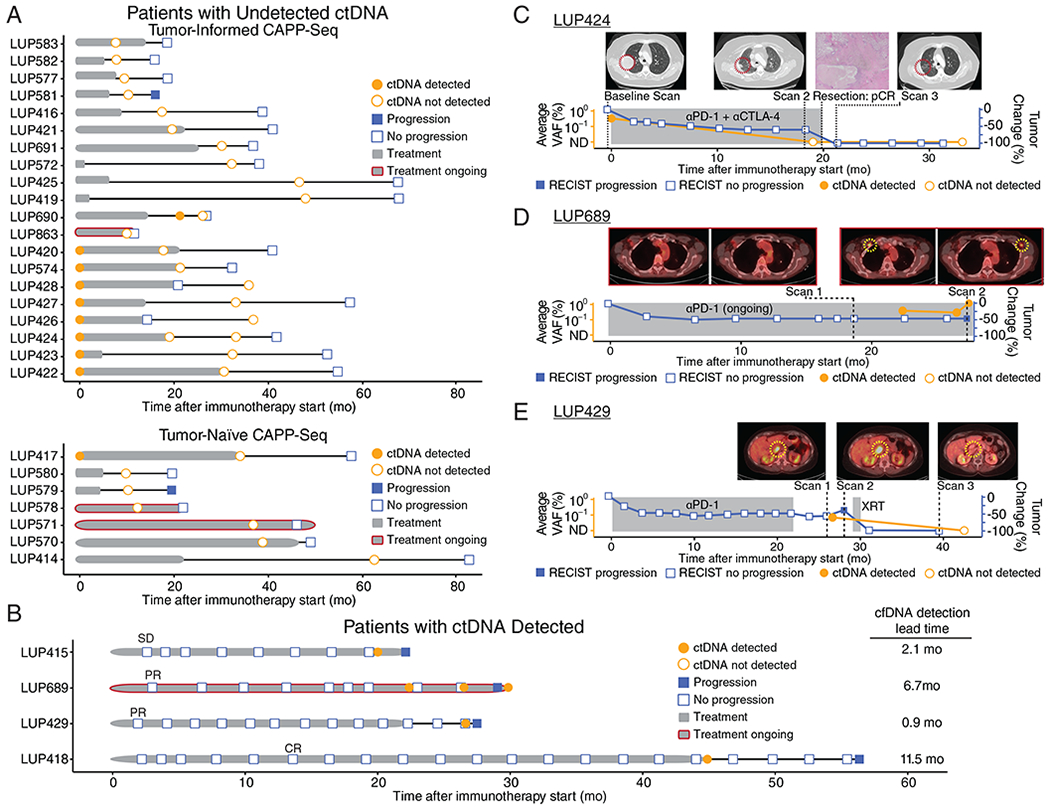
A) Event chart for patients without ctDNA detected (n = 27) and B) patients with detectable ctDNA at the surveillance timepoint (n=4). Chart depicts RECIST v1.1 status at last follow-up (filled blue squares = progression; open blue squares = no progression), ctDNA detection status by tumor-informed CAPP-Seq (filled orange circles = detected; open orange circles = not detected), duration of PD-(L)1 blockade treatment (grey bar, red outline = ongoing treatment), and PFS after treatment discontinuation (black bar). In patients with ctDNA detected, the earliest scan with the best overall response is indicated.
C) An exemplar patient treated with PD-1 plus CTLA-4 blockade who achieved a −60% reduction in tumor volume by RECIST v1.1. cfDNA collected 19 months after initiating treatment showed undetectable ctDNA. These findings were confirmed by resections of both the adrenal and lung lesions that both showed complete pathological response. This patient remains progression-free with undetectable ctDNA at 34 months from treatment initiation. ctDNA levels are shown as average variant allele fraction of all variants monitored.
D) A second exemplar patient who achieved a −45% response to PD-1 blockade with progression after ~29 months by RECIST v1.1. ctDNA was initially detected 22 months into treatment, prior to progression by PET-CT. ctDNA levels are shown as average variant allele fraction of all variants monitored.
E) A third exemplar patient treated with PD-1 blockade with detectable ctDNA 25 months after starting treatment, confirmed by imaging which revealed an isolated recurrence that was treated with radiotherapy. ctDNA was not detectable ~1 year following localized radiation to the progressing aortocaval lymph node. ctDNA levels are shown as average variant allele fraction of all variants monitored.
ctDNA informs disease status among patients with long-term response to PD-(L)1 blockade
Our findings suggest that ctDNA detection might be useful for clarifying radiographic findings at during surveillance and to inform the duration of therapy. For example, patient LUP424 received PD-1 plus CTLA-4 blockade for 20 months, achieving a −60% radiologic response by RECIST (Figure 4C). At baseline, ctDNA was detectable (average AF 0.33%). At the surveillance timepoint (19.0 months), ctDNA was undetectable by tumor-informed CAPP-Seq analysis although the patient continued to have residual lesions visible on radiologic imaging. The patient experienced Grade 3 pneumonitis, so treatment was discontinued shortly thereafter and surgical resection was pursued to remove the residual lesion in the right lung. Pathology demonstrated a complete pathologic response with no viable tumor. Continued follow-up scans after discontinuation of therapy have continued to show no evidence of recurrent disease, and a repeat follow-up plasma draw at 33.1 months confirmed the absence of ctDNA. This case illustrates that ctDNA may allow distinguishing between patients in whom residual radiologic lesions contain viable cells versus those in whom all cancer cells have been eliminated.
Similarly, ctDNA analysis may contribute to early detection of progression and aid in interpretation of equivocal radiologic findings. For example, patient LUP689 had been receiving PD-1 blockade for two years and scans showed partial response (−45% by RECIST; Figure 4D). A CT scan 7 months into treatment revealed a new, small right pectoral lymph node that, at the time, was felt to be non-specific and not a new site of disease. A PET scan at 19 months showed ongoing response without progression. At 22 months into treatment, ctDNA was detected in a plasma sample (average AF 0.39%) although a CT scan (Figure S4A–B) at that time showed a decrease in size of the right pectoral lymph node. A repeat plasma sample taken at 26.5 months again confirmed the presence of ctDNA (average AF 0.30%). At a third timepoint 2.5 months later, ctDNA was again detected and had further increased (average AF 1.16%) and a concurrent PET scan now demonstrated progressive disease. Thus, ctDNA analysis can aid in the interpretation of equivocal radiologic findings and may provide a window of opportunity for therapeutic interventions to improve outcomes.
Lastly, ctDNA analysis may also be useful in monitoring the efficacy of interventions after recurrence. For example, patient LUP429 received PD-1 blockade for 22.4 months and achieved a partial response but stopped treatment due to late-onset colitis (Figure 4E). However, a plasma sample collected 2.6 months after stopping treatment was positive for ctDNA (average AF 0.07%) and repeat imaging ~1 month later revealed an isolated recurrence in an abdominal lymph node. The patient received localized radiation (3300 cGy in 5 fractions) to the site of recurrence and has been disease-free since. A plasma sample collected over one year later remained negative for ctDNA and the patient remains disease-free to date.
Discussion:
In this study we examined a cohort of patients with long-term benefit to PD-(L)1 blockade to evaluate the role of ctDNA to predict the risk of recurrence. We found ctDNA analysis during surveillance can be highly sensitive for detecting minimal residual disease and predicting risk of eventual progression. Additionally, ctDNA was undetectable in all patients with ongoing long-term benefit to PD-(L)1 blockade, which may be a molecular reflection of eradication of all tumor cells by PD-(L)1 blockade that exceeds the insight gained from routine scans.
To our knowledge, our report is the first to demonstrate the potential utility of ctDNA analysis during surveillance of patients with ≥12 months of PFS after initiating PD-(L)1 blockade. Recent studies assessing on-treatment ctDNA changes 4-8 weeks after initiation of PD-(L)1 blockade have demonstrated concordance with initial radiologic response, but the majority of these patients later progress (14,15) suggesting early on-treatment ctDNA changes do not optimally predict longer term outcomes. Similarly, we found that baseline ctDNA levels did not discriminate long-term response. Based on our results, we envision that surveillance ctDNA analysis could be used in conjunction with standard imaging in patients with long-term benefit to immune checkpoint blockade in order to facilitate early identification of progression.
In patients with long-term benefit to PD-(L)1 blockade, it is challenging to determine whether there is active residual disease and/or when it might be appropriate to discontinue treatment, especially in the context of emerging evidence for late immunological sequalae that can have ambiguous radiographic correlates (2,9,11). In clinical trials of PD-(L)1 blockade in NSCLC, radiologic complete response is rare, with rates ranging from 0.5-3.7% (1,2,4). However, the radiologic assessment of true complete response may be imprecise and ctDNA can provide additional insight. In the current study, we found that 19 patients have undetectable ctDNA and ongoing long-term benefit from PD-(L)1 blockade despite persistently measurable disease by CT imaging. One patient with −60% response by RECIST and undetectable ctDNA was found to have complete pathologic response at the time of resection.
Similarly, in a report of neoadjuvant PD-1 blockade, three patients achieved complete pathologic response but none were evident by CT imaging; one patient even had apparent tumor growth radiologically (9). ctDNA analysis may therefore help to identify those patients who have achieved durable elimination of the malignant clone through PD-(L)1 blockade and avoid the need for invasive biopsy or resections to further investigate residual lesions. Additionally, we hypothesize that ctDNA may inform a personalized approach to duration of PD-(L)1 treatment in patients with long-term response wherein those with undetectable ctDNA may be able to safely discontinue treatment. Additional work will be needed to determine the optimal timepoint (e.g. 12-24 months after initiating treatment) at which to query ctDNA to guide such a decision.
Although we analyzed a single surveillance plasma timepoint for most patients, it is possible that serial ctDNA analysis may maximize sensitivity and specificity of residual disease detection in patients undergoing PD-(L)1 blockade. Three patients in our cohort had serial surveillance samples available without an intervening change in therapy. In two of these (LUP424 and LUP689), ctDNA detection results were the same in both samples. In the third patient (LUP690), ctDNA was detected in the first sample taken 6.7 months after stopping PD-(L)1 blockade based on a single cfDNA molecule containing a KRAS G12A mutation at an allele frequency of 0.03% that was present in the patient’s tumor and not present in the matched leukocyte sample (tumor-informed Monte Carlo p = 0.017). In contrast, in the 4 cases scored as detected, each were detected with at least 2 mutant reads (median = 10.5, range = 2-26), and a detected allele frequency of at least 0.07% (median = 0.16%, range = 0.07-0.39%). A follow-up sample collected 4.8 months later was sequenced 1.24X deeper, and contained no mutant reads. Since the confirmatory draw was negative we scored the patient as being ctDNA negative and the patient has not recurred at last follow-up. There are several potential explanations for the single mutation-containing molecule detected in the first but not second sample. First, this could represent a technical false positive, a low rate of which we expect to find given our method is tuned to 95% specificity (16,17). Second, this could be evidence of late tumor clearance as there could have been tumor deposits remaining that were eradicated by the immune system between the two blood draws. Third, the second blood draw could be a false negative and the patient may have residual disease that has not yet manifested on imaging. Given this result, we envision that serial ctDNA analysis may be useful for confirming positive results, particularly in cases where ctDNA detection is based on a single mutant read.
As we and others have previously demonstrated, maximal sensitivity of ctDNA detection can be achieved by having prior knowledge of tumor mutations and then applying tumor-informed ctDNA analysis (11,16,17). However, in an exploratory analysis we observed that in our cohort, CAPP-Seq-based tumor-naïve ctDNA analysis yielded similar clinical results as the tumor-informed approach. It is critical to note that this result required sequencing of matched leukocytes in order to eliminate mutations due to clonal hematopoiesis (30–34). Our data suggests that similar performance is unlikely to be achieved with commercially-available ctDNA tests that do not genotype matched leukocytes.
Limitations of our study include a relatively modest cohort size, varying PD-(L)1 treatment regimens, and non-uniform timing of blood collection. However, our cohort represents a substantial effort to interrogate an uncommon but important clinical phenotype. Moreover, we believe our results are robust to the non-uniform timing of blood collection as there was no difference in time of blood collection relative to the start or end of therapy in patients who did or did not ultimately progress, nor in those in whom ctDNA was or was not detectable. Additionally, baseline plasma was not available for all patients in order to confirm the presence of detectable ctDNA prior to beginning treatment. However, we demonstrated that ctDNA was detectable in all nine patients in whom baseline plasma was available and prior reports by our group and others have demonstrated that ctDNA is detectable in the vast majority of patients with metastatic NSCLC (13,17,19,29). All of the nine patients who had detectable ctDNA at baseline had no detectable ctDNA in the surveillance sample and have ongoing benefit from PD-(L)1 blockade, confirming the ability of ctDNA to determine the in vivo disease state. We believe the findings of our study warrant validation in a prospective clinical trial where patients receive uniform PD-(L)1 blockade regimens and samples are collected uniformly both prior to therapy and at surveillance timepoints.
If validated, we envision at least two potential applications of ctDNA surveillance to personalize treatment in patients with long-term response to PD-(L)1 blockade. For patients in whom ctDNA is undetectable at a surveillance landmark (the optimal timing remains to be determined as our results here did not prespecify the timepoint, but 12 or 24 months since initiating treatment may be reasonable), we hypothesize that it may be possible to discontinue therapy and to monitor closely thereafter, with the expectation of continued durable response. By contrast, among patients who are ctDNA detectable, additional imaging (including PET scan) could be performed to identify residual disease and guide early therapeutic interventions (e.g. radiation or surgery for oligoresidual lesions or systemic therapy if more diffuse disease) to proactively intervene upon impending resistance and maximize continued long-term response. Importantly, prospective trials will need to be performed to test these potential applications.
In summary, we demonstrate that analysis of ctDNA in advanced NSCLC patients undergoing PD-(L)1 blockade can distinguish patients at risk for eventual progression from those who may have achieved elimination of disease. We therefore envision that ctDNA analysis will be useful for surveilling of patients receiving immune checkpoint blockade and might allow personalization of the duration and early interventions during therapy.
Supplementary Material
Statement of Translational Relevance:
Long-term response to PD-(L)1 blockade is uncommon in non-small cell lung cancer (NSCLC); even among patients with initial response, a substantial fraction ultimately progress. We hypothesized that circulating tumor DNA (ctDNA) could be used to distinguish long-term responders to PD-(L)1 blockade who were likely to remain progression-free from those who were at highest risk for eventual progression. In this study, we collected plasma at late surveillance timepoints from NSCLC patients achieving long-term benefit from PD-(L)1 blockade and ctDNA was interrogated using CAPP-Seq. Nearly all patients with undetectable ctDNA at the surveillance blood draw remained disease free, while all of the patients with detectable ctDNA eventually progressed. Therefore, ctDNA analysis may inform personalization of the duration of immune checkpoint blockade and permit early intervention in those likely to develop acquired resistance.
Acknowledgements:
This work was supported by grants from the National Cancer Institute (M. Diehn and A.A. Alizadeh; R01CA188298), the US National Institutes of Health Director’s New Innovator Award Program (M. Diehn; 1-DP2-CA186569), the Virginia and D.K. Ludwig Fund for Cancer Research (M. Diehn and A.A. Alizadeh), the CRK Faculty Scholar Fund (M. Diehn), V-Foundation (A.A. Alizadeh). the Damon Runyon Cancer Research Foundation (M.D. Hellmann), Ludwig Trust, Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748), Parker Institute for Cancer Immunotherapy, Druckenmiller Center for Lung Cancer Research at MSKCC, Swim Across America, and Stand Up to Cancer-American Cancer Society Lung Cancer Dream Team Translational Research Grant (SU2C-AACR-DT17-15). Stand Up to Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. Four investigators (M.D. Hellmann, D.K. Wells, T. Merghoub, A.A. Alizadeh) are members of the Parker Institute for Cancer Immunotherapy. M.D. Hellmann is a Damon Runyon Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI-98-18). B.Y. Nabet is a Stanford Cancer Systems Biology Scholar and supported by the NIH (5R25CA180993). B.Y. Nabet is supported by the Postdoctoral Research Fellowship (134031-PF-19-164-01-TBG) from the American Cancer Society. A.A. Chaudhuri was funded by a Radiological Society of North America Resident/Fellow Grant, and by a Conquer Cancer Foundation ASCO Young Investigator Award supported by Takeda Pharmaceuticals. Any opinions, findings and conclusions expressed in this material are those of the authors and do not necessarily reflect those of the American Society of Clinical Oncology® or Conquer Cancer®, or Takeda®.
Conflicts of Interest Statement:
M.D.H. reports paid consultancy from Bristol-Myers Squibb, Merck, Genentech, AstraZeneca/MedImmune, Nektar, Syndax, Mirati Therapeutics, Blueprint Medicines, Immunai, Shattuck Labs; travel/honoraria from Bristol-Myers Squibb and AstraZeneca; research funding from Bristol-Myers Squibb; and a patent has been filed by MSK related to the use of tumor mutation burden to predict response to immunotherapy (PCT/US2015/062208), which has received licensing fees from Personal Genome Diagnostics. A.A.C. reports speaker honoraria and travel support from Roche Sequencing Solutions, Varian Medical Systems, and Foundation Medicine; a research grant from Roche Sequencing Solutions, and has served as a paid consultant for Oscar Health. J.J.C. reports paid consultancy from Lexent Bio Inc. J.L.S. reports stock or other ownership of Merck, Chemed Corporation, and Thermo Fisher Scientific. H.A.W. has received honoraria from Novartis and AstraZeneca and has participated on the advisory boards of Xcovery, Janssen, and Mirati. S.K.P. reports grant support from EpicentRx, Forty Seven Inc, Bayer, and Boehringer Ingelheim and serves in a consulting or advisory role for AstraZeneca, AbbVie, G1 Therapeutics, and Pfizer. J.W.N. reports research support from Genentech/Roche, Merck, Novartis, Boehringer Ingelheim, Exelixis, Takeda Pharmaceuticals, Nektar Therapeutics, Adaptimmune, and GSK, and has served in a consulting or advisory role for AstraZeneca, Genentech/Roche, Exelixis Inc, Jounce Therapeutics, Takeda Pharmaceuticals, and Eli Lilly and Company. J.E.C. reports paid consultancy from Genentech/Roche, AstraZeneca/MedImmune, Merck, Bristol-Myers Squibb; research funding from Genentech/Roche, Bristol-Myers Squibb, AstraZeneca/MedImmune. M.G.K. reports paid consultancy from AstraZeneca, Pfizer, and Regeneron. C.M.R. reports paid consultancy from AbbVie, Amgen, Ascentage, Astra Zeneca, BMS, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Loxo, and Pharmar, and is on the scientific advisory board for Elucida and Harpoon. T.M. is a co-founder of Imvaq. B.T.L. reports paid consultancy for Biosceptre International, Roche/Genentech, Thermo-Fisher Scientific, Mersana Therapeutics, Guardant Health, and Hengrui Therapeutics. A.A.A. reports ownership interest in CiberMed and FortySeven, patent filings related to cancer biomarkers, and paid consultancy from Genentech, Roche, Chugai, Gilead, and Celgene. M.D. reports research funding from Varian Medical Systems, ownership interest in CiberMed, patent filings related to cancer biomarkers, and paid consultancy from Roche, AstraZeneca, and BioNTech. The remaining authors declare no potential conflicts of interest.
References:
- 1.Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus Chemotherapy in Metastatic Non–Small-Cell Lung Cancer. N Engl J Med. 2018;378:2078–92. [DOI] [PubMed] [Google Scholar]
- 2.Hellmann MD, Ciuleanu T-E, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med. 2018;378:2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N Engl J Med. 2016;375:1823–33. [DOI] [PubMed] [Google Scholar]
- 4.Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N Engl J Med. 2018;378:2288–301. [DOI] [PubMed] [Google Scholar]
- 5.Gettinger S, Horn L, Jackman D, Spigel D, Antonia S, Hellmann M, et al. Five-Year Follow-Up of Nivolumab in Previously Treated Advanced Non–Small-Cell Lung Cancer: Results From the CA209–003 Study. J Clin Oncol. 2018;36:1675–84. [DOI] [PubMed] [Google Scholar]
- 6.Spigel DR, McLeod M, Hussein MA, Waterhouse DM, Einhorn L, Horn L, et al. Randomized results of fixed-duration (1-yr) vs continuous nivolumab in patients with advanced non-small cell lung cancer (NSCLC). Ann Oncol. 2017;28. [Google Scholar]
- 7.Gettinger SN, Wurtz A, Goldberg SB, Rimm D, Schalper K, Kaech S, et al. Clinical Features and Management of Acquired Resistance to PD-1 Axis Inhibitors in 26 Patients With Advanced Non–Small Cell Lung Cancer. J Thorac Oncol. 2018;13:831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, et al. Molecular Determinants of Response to Anti–Programmed Cell Death (PD)-1 and Anti–Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non–Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol. 2018;36:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forde PM, Chaft JE, Smith KN, Anagnostou V, Cottrell TR, Hellmann MD, et al. Neoadjuvant PD-1 Blockade in Resectable Lung Cancer. N Engl J Med. 2018;378:1976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell. 2018;33:843–852.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545:446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24:1441–8. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg SB, Narayan A, Kole AJ, Decker RH, Teysir J, Carriero NJ, et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin Cancer Res. 2018;24:1872–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, et al. Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non–Small Cell Lung Cancer. Cancer Res. 2019;79:1214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Bratman SV, To J, Wynne JF, Eclov NCW, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cabel L, Proudhon C, Romano E, Girard N, Lantz O, Stern M-H, et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat Rev Clin Oncol. 2018;15:639–50. [DOI] [PubMed] [Google Scholar]
- 19.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costello M, Pugh TJ, Fennell TJ, Stewart C, Lichtenstein L, Meldrim JC, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67–e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: Cancer Variant Annotation Tool. Hum Mutat. 2015;36:E2423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT). J Mol Diagnostics. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleiss JL, Tytun A, Ury HK. A Simple Approximation for Calculating Sample Sizes for Comparing Independent Proportions. Biometrics. [Wiley, International Biometric Society]; 1980;36:343–6. [PubMed] [Google Scholar]
- 28.Anderson JR, Cain KC, Gelber RD. Analysis of survival by tumor response. J Clin Oncol. 1983;1:710–9. [DOI] [PubMed] [Google Scholar]
- 29.Zill OA, Banks KC, Fairclough SR, Mortimer SA, Vowles J V, Mokhtari R, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res. 2018;24:3528–38. [DOI] [PubMed] [Google Scholar]
- 30.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N Engl J Med. 2014;371:2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N Engl J Med. 2017;377:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin Cancer Res. 2018;24:4437–43. [DOI] [PubMed] [Google Scholar]
- 33.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman P V, Mar BG, et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N Engl J Med. 2014;371:2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ptashkin RN, Mandelker DL, Coombs CC, Bolton K, Yelskaya Z, Hyman DM, et al. Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors. JAMA Oncol. 2018;4:1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173:371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


