Abstract
Proper cell-cycle progression is essential for the self-renewal and differentiation of human pluripotent stem cells (hPSCs). The fluorescent ubiquitination-based cell-cycle indicator (FUCCI) has allowed the dual-color visualization of the G1 and S/G2/M phases in various dynamic models, but its application in hPSCs is not widely reported. In addition, lineage-specific FUCCI reporters have not yet been developed to analyze complex tissue-specific cell-cycle progression during hPSC differentiation. Desiring a robust tool for spatiotemporal reporting of cell-cycle events in hPSCs, we employed the CRISPR/Cas9 genome editing tool and successfully knocked the FUCCI reporter into the AAVS1 safe harbor locus of hPSCs for stable and constitutive FUCCI expression, exhibiting reliable cell-cycle-dependent fluorescence in both hPSCs and their differentiated progeny. We also established a cardiac-specific TNNT2-FUCCI reporter for lineage-specific cell-cycle monitoring of cardiomyocyte differentiation from hPSCs. This powerful and modular FUCCI system should provide numerous opportunities for studying human cell-cycle activity, and enable the identification and investigation of novel regulators for adult tissue regeneration.
Keywords: cell cycle, CRISPR/Cas9, FUCCI, gene editing, heart regeneration, human pluripotent stem cells
1 |. INTRODUCTION
Human pluripotent stem cells (hPSCs), both induced pluripotent stem cells (iPSCs) and embryonic stem cells (hESCs), offer innovative biomedical applications and have significantly advanced human medicine (Avior, Sagi, & Benvenisty, 2016; Sakaue-Sawano et al., 2008; Shi, Inoue, Wu, & Yamanaka, 2017; Z. Zhu & Huangfu, 2013) due to their unique properties: unlimited self- renewal and the ability to differentiate into any somatic cell type (Takahashi et al., 2007; Trounson & DeWitt, 2016; Yu et al., 2007). A major goal of stem cell research is to identify factors regulating the cell cycle and use them to promote cell-cycle re-entry for regenerating damaged tissues, including (Hirai, Chen, & Evans, 2016). The cell cycle itself plays critical roles in pluripotency maintenance and differentiation of hPSCs (Hindley & Philpott, 2013; Kapinas et al., 2013). At the pluripotent stage, hPCSs are highly proliferative and have an altered cell cycle that enables them to quickly change from DNA synthesis to cell division by shortening the intervening gap phases (Boward, Wu, & Dalton, 2016), resulting in a distinct cell-cycle machinery without G1 checkpoint regulation (Kalkan et al., 2017). On the other hand, the differentiation of hPSCs is associated with an irreversible exit from the original cell cycle at the G phase to acquire definitive phenotypic features (Ng et al., 2004; Trounson & DeWitt, 2016; Vermeulen, Van Bockstaele, & Berneman, 2003). For instance, Pauklin and Vallier (2013) demonstrated that the cell fate decisions of stem cells are tightly related to cell-cycle machinery modulation. However, the discerning lineage-specific cell-cycle profile has been challenging because of the mixed highly proliferative cell types during hPSC differentiation. Thus, a versatile cell-cycle reporter for robust measurement of the cell-cycle progression in the pluripotent and differentiating hPSCs should shed light on stem cell biology and have implications for cell-based therapies.
Recently, novel molecular tools, most of which rest upon the expression of a chimeric transgene that undergoes cell-cycle-dependent stability or distribution (Bajar et al., 2016), have been developed for the convenient and accurate identification of cell-cycle stages of fixed and live cells (Calder et al., 2013). To date, the most commonly used tool is the fluorescence ubiquitination cell-cycle indicator (FUCCI) system, which combines two distinct fluorescently tagged human genes, CDT1 fused to monomeric Kusabira-orange 2 (mKO2), an orange-emitting fluorescent protein, and GMNN fused to mAG, a green-emitting fluorescent protein (Sakaue-Sawano et al., 2008). Both CDT1 and GMNN-encoded proteins are direct substrates of Anaphase Promoting Complex (APCCdh1) and Skp 1-Cullin 1-F-box protein (SCFskp2), two mutually antagonistic E3 ligases that are reciprocally active during the cell cycle (Wei et al., 2004). As a result, the CDT1 protein is highly expressed and enriched during G1 phase, but at the beginning of the S phase, it is degraded by the SCFskp2 and thus disappears during the course of S/G2/M phases. In contrast, geminin protein, encoded by GMNN gene, is degraded by the APCCdh during the G1 phase, but accumulates in S/G2/M phases. Thus, the FUCCI probes allow real-time reporting of not only the distinct G1 (orange) and S/G2/M (green) phases, but also the phase transitions, including early the S phase (yellow) and late M/early G1 phases (nonfluorescent), during cell-cycle progression. The FUCCI reporter has been employed in several dynamic systems, including zebrafish, mice, and hPSCs, and has provided new insights into stem cell biology (Abe et al., 2013; Bouldin & Kimelman, 2014; Bouldin, Snelson, Farr, & Kimelman, 2014; Nakajima, Kuranaga, Sugimura, Miyawaki, & Miura, 2011).
Despite its broad application, a lineage-specific hPSC-FUCCI reporter line has not yet been generated to discern lineage-specific cell-cycle profiles and identify novel factors to promote cell-cycle re-entry for regenerating damaged tissues. Here, we exploited the CRISPR/Cas9 system to insert an improved FUCCI system that employs Clover-Geminin and mKO2-Cdt1 (Bajar et al., 2016), into the AAVS1 safe harbor locus. When differentiated into the three germ layer lineages, that is mesoderm, ectoderm, and endoderm, the hPSC-FUCCI reporters displayed dynamic and distinct cell-cycle profiles. Importantly, we further equipped the FUCCI system with a cardiac-specific promoter, enabling lineage-specific cell-cycle visualization of cardiomyocyte (CM) differentiation from hPSCs. Overall, we established a powerful hPSC-FUCCI system, which can be re-engineered for other tissue-specific cell-cycle reporting. Using this system, we illustrated its applications in human stem cell biology, which will allow us to address previously intractable questions and identify novel genes or drugs for cardiac and other tissue regeneration.
2 |. MATERIALS AND METHODS
2.1 |. Donor plasmid construction
The donor plasmids targeting AAVS1 locus were constructed as previously described (Bao et al., 2019). Briefly, to generate the CAG-FUCCI plasmid, the Clover-Geminin (1–110)-IRES-mKO2-Cdt (30–120) fragment was amplified from Addgene plasmid #83841 and then cloned into the AAVS1-Pur-CAG-EGFP donor plasmid (Addgene; #80945), replacing the EGFP. For TNNT2-FUCCI plasmid, the cTnT promoter was polymerase chain reaction (PCR) amplified from TroponinT-GCaMP5-Zeo (Addgene; #46027), and then cloned into the CAG-FUCCI plasmid via Gibson Assembly (NEB; #2611S), replacing the CAG promoter. Both FUCCI plasmids were sequenced and submitted to Addgene (#136934 and #136935).
2.2 |. hPSC maintenance and differentiation
H9 hPSCs were purchased from WiCell and maintained on Matrigel- coated six-well plates in mTeSR plus or mTeSR1 medium at 37°C in a humidified incubator with 5% CO2. To differentiate hPSCs into mesoderm (Lian et al., 2013), hPSCs were singularized with Accutase, then seeded onto a Matrigel-coated 12-well plate in mTeSR plus or mTeSR1 with 5 μM Y27632 (day-3) overnight. hPSCs were then cultured and expanded in pluripotent medium for another 48 hr. To initiate cardiac differentiation (Day 0), pluripotent medium was replaced by the RPMI basal medium with 6 μM CHIR99021 (CHIR), followed by a medium change with RPMI/B27 minus insulin after 24 hr. Day 3 differentiated cultures were treated with 2 μM Wnt-C59 (Cayman Chemical), followed by a medium change on Day 5. Starting from Day 7, cells were cultured in RPMI/B27 with medium change every 3 days until analysis.
To induce ectoderm differentiation, cells were continuously cultured in LaSR basal medium (Lian et al., 2014) with daily medium change until analysis. Endoderm lineage differentiation was performed with a modified protocol (C. Du et al., 2018). To initiate endoderm differentiation, 0.5% dimethyl sulfoxide (DMSO) was added to the pluripotent medium (Day 0). On Day 1, the medium was switched to RPMI/B27 medium, containing 3 μM CHIR9902, followed by a medium change with RPMI/B27 after 24 hr. For the next 5 days, the differentiated cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium containing 0.5 mM A83–01 (Cayman Chemical), 2.5 mM sodium butyrate (Sigma) and 0.5% DMSO, with a daily medium change. From Day 8 and onwards, the medium was switched to DMEM/F12 medium with 5 small molecules: 15 μM FH1, 0.5 μM A83–01, 100 nM dexamethasone, 15 μM FPH1, and 10 nM hydrocortisone. Medium change was performed every other day.
2.3 |. Nucleofection and transfection
To increase cell viability, 10 μM Y27632 was used to treat hPSCs 3–4 hr before nucleofection or overnight. Cells were then singularized by Accutase for 8–10 min, and 1–2.5 × 106 hPSCs were nucleofected with 3 μg AAVS1 gRNA T2 (Addgene; #41818), 4.5 μg pCas9 GFP (Addgene; #44719), and 6 μg donor plasmids in 100 μl human stem cell nucleofection solution (Lonza; #VAPH-5012) or 200 μl room temperature PBS−/− using program B-016 in a Nucleofector 2b. The nucleofected cells were seeded into one well of a Matrigel-coated 6-well plate in 3ml pre-warmed mTeSR plus or mTeSR1 with 10 μM Y27632. 24 hr later, the medium was changed with fresh mTeSR plus or mTeSR1 containing 5 μM Y27632, followed by a daily medium change. When cells were more than 80% confluent, drug selection was performed with 1 μg/ml puromycin (Puro) for approximately 1 week, and individual clones were picked using a microscope inside a tissue culture hood and expanded for 2–5 days in each well of a 96-well plate pre-coated with Matrigel, followed by a PCR genotyping. The plasmid transfection of hPSC-CMs was performed according to a published method (Tan et al., 2019). Briefly, 1 μg cyclin D2 (CCND2) plasmid (Addgene; #8958) was added to 100 μl DMEM/F12 medium containing 1 μl Lipofectamine Stem Transfection Reagent, mixed well, and incubated for 10 min. A pellet of 0.5 million CMs were resuspended in the resulting 100 μl plasmid mixture and incubated for 10 min at room temperature before being seeded into Matrigel-coated 24-well plates in 10% fetal bovine serum medium.
2.4 |. Genomic DNA extraction and genotyping
The genomic DNA of single clone-derived hPSCs was extracted by scraping cells into 40 μl QuickExtract™ DNA Extraction Solution (Epicentre; #QE09050). 2×GoTaq Green Master Mix (Promega; #7123) was used to perform the genomic DNA PCR. For positive genotyping, the following primer pair with an annealing temperature of 65°C was used: CTGTTTCCCCTTCCCAGGCAGGTCC and TCG TCGCGGGTGGCGAGGCGCACCG. For homozygous screening, we used the following set of primer sequences: CGGTTAATGTGGCTC TGGTT and GAGAGAGATGGCTCCAGGAA with an annealing temperature of 60°C.
2.5 |. Immunostaining analysis
To fix the hPSCs and differentiated cells, room temperature PBS−/− was used to wash the cells, followed by 4% paraformaldehyde treatment for 15 min. Cells were then washed twice with PBS−/−, and stained with primary and secondary antibodies in PBS−/− containing 5% nonfat dry milk blocker and 0.4% Triton X-100. Primary antibodies anti-beta III tubulin (Abcam;,’ ab18207), HNF4A (DSHB; PCRP-HNF4A-2A8), Troponin I (Santa Cruz; sc-365446), MF20 (DSHB; MF20), and Lab Vision™ Troponin T (Thermo Fisher Scientific; MS-295-P1) were used in this study. 4’,6-Diamidino-2-phenylindole was used to stain the cell nuclei. Leica DMi-8 fluorescent microscope was used for imaging. Images were then processed in ImageJ, and the same threshold setting across different backgrounds for quantification may accidentally remove weak signals (Figures S1B, S1D, S2A–C, S3B, S4B–D, and S5B).
2.6 |. Flow-cytometry analysis
Cells were singularized with Accutase for 10 min and pelleted by centrifuging for 5 min. Then the cells were resuspended in PBS−/− and filtered through a 100μm strainer before flow analysis. Data was collected on a BD Accuri C6 plus flow cytometer or an Invitrogen Attune NxT flow cytometer and analyzed with FlowJo software.
2.7 |. Reverse transcription PCR (RT-PCR) analysis
Cells cultured on a 24-well plate were rinsed with cold PBS−/− once and lysed in 500 μl TRIzol™ reagent (Invitrogen). Total RNA was then prepared with the Direct-zol RNA miniprep kit (Zymo) with in-column DNase treatment following the manufacturer’s instruction. cDNA was reverse-transcribed from 1 μg RNA with ProtoScript First Strand cDNA Synthesis Kit (NEB) and used for RT-PCR with GoTaq Green Master Mix (Promega). GAPDH was used as an endogenous housekeeping control, and the primer pairs for targeted genes were listed in Table S1.
2.8 |. Statistical analysis
Data are presented as mean±standard error of the mean (SEM). Student’s t test (two-tail) was used to determine the statistical significance (P<0.05) between two groups, and one-way analysis of variance was used for three or more groups together with Student’s t test. For each group, 3–6 samples from at least three biological replicates are used.
3 |. RESULTS
3.1 |. Generation of constitutive FUCCI expressing hPSCs
Obtaining stable transgene expression in hPSCs can be challenging and the promoters used are of great importance due to the high likelihood of transgene silencing in hPSCs (Z. W. Du, Hu, Ayala, Sauer, & Zhang, 2009). To generate a stable hPSC-FUCCI reporter line, we employed the CRISPR/Cas9-mediated homology-directed repair (HDR) and knocked the all-in-one FUCCI construct CAG-Clover-Geminin-IRES-mKO2-Cdt1 (Figure 1a) into the AAVS1 safe harbor locus, a well-known site for stable transgene expression (Bao et al., 2019; Smith et al., 2008). After puromycin selection for about 2 weeks, we isolated two Clover fluorescent protein-expressing single cell-derived hPSC clones and confirmed transgene presence by PCR genotyping (Figure 1b). The dominant expression of Clover indicated that the hPSCs are primarily at the S/G2 phase (Figure 1c,d), which is consistent with a previous report (Singh, Trost, Boward, & Dalton, 2016). Importantly, the engineered hPSCs retained strong expression of pluripotency markers, stage-specific embryonic antigen-4 (SSEA-4; Figure 1e) and octamer-binding transcription factor 4 (OCT-4; Figure 1f). Taken together, these results demonstrated the successful generation of FUCCI knock-in hPSCs without losing their pluripotent status.
FIGURE 1.
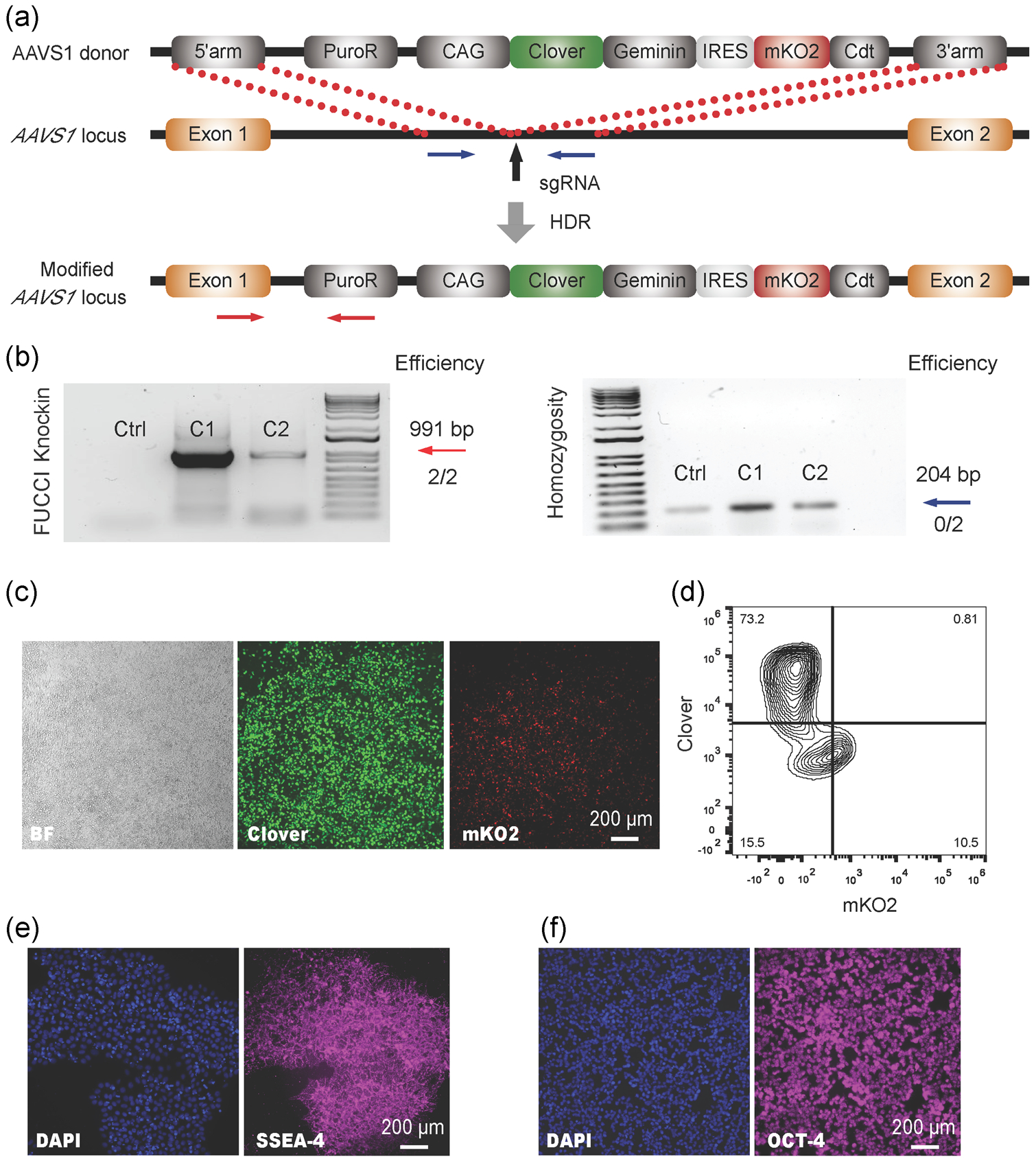
Construction of the FUCCI reporter knock-in H9 hPSC line using Cas9 nuclease. (a) Schematic illustration of the homology-directed repair (HDR)-based knock-in strategy at the AAVS1 safe harbor locus. Vertical black arrow indicates the sgRNA T2 targeting site. (b) The representative PCR genotyping of hPSC clones after puromycin selection is shown, and the expected PCR product for correctly targeted AAVS1 site is 991bp (red arrow) with an efficiency of two clones from a total of two. A homozygosity assay was performed on the knock-in clones, and those without ~204bp PCR products were homozygous (blue arrow). Representative live-cell images (c) and flow-cytometry analysis (d) of Clover and mKO2 of successfully targeted FUCCI reporter H9 hPSCs were shown. Representative fluorescent images of SSEA-4 (e) and OCT-4 (f) of H9 FUCCI cells were shown. Scale bar=200μm. FUCCI, fluorescent ubiquitination-based cell-cycle indicator; HDR, homology-directed repair; hPSC, human pluripotent stem cell; OCT-4, octamer-binding transcription factor 4; PCR, polymerase chain reaction; SSEA-4, stage-specific embryonic antigen-4
3.2 |. Continuous and quantitative visualization of cell-cycle progression during hPSC differentiation
Cell cycle progression is of great importance to stem cell biology. The FUCCI system has been broadly used in mammalian cells to understand the roles of the cell cycle during differentiation, morphogenesis and cell death (Abe et al., 2013; Bouldin & Kimelman, 2014; Calder et al., 2013; Choi et al., 2013; Coronado et al., 2013; Jovic et al., 2013; Singh et al., 2013); however its application in visualizing the spatiotemporal dynamics of cell-cycle progression has not been extensively reported in hPSCs. To survey the utility of our FUCCI reporter for visualizing multi-lineage cell-cycle progression, the FUCCI knock-in hPSCs were differentiated into mesodermal, ectodermal, and endodermal lineages (Figure 2a). Mesoendoderm marker Brachyury (Figure S1A,B) and endoderm marker FoxA2 (Figure S1C–D) were detected along mesodermal and endodermal differentiation. As expected, the expression of green Clover (S/G2 phase) decreased while red mKO2 increased (G1 phase) during hPSC differentiation (Figure S2A–C). The fate of terminally differentiated mesodermal, endodermal, and ectodermal cells was confirmed by the immunostaining analysis of Troponin T (cTnT) (Figure 2b), HNF4A (Figure 2c), and β-III tubulin (Figure 2d), as well as RT-PCR analysis of MYL7, NKX2-5, AFP, ALB, PAX6 and NEUROD1 (Figure S2D), respectively. Additional molecular analysis was also performed during cardiac differentiation. Importantly, the expression of mKO2 was upregulated along with the increasing expression of cTnT during cardiac differentiation (Figure S3A–C), and the cardiac cell fate was further confirmed by MF20 and cTnI expression (Figure S3D). Collectively, this data shows that the FUCCI knock-in hPSC system constructed by CRISPR/Cas9 technology exhibits reliable cell-cycle-dependent fluorescence function in both undifferentiated hPSCs and their differentiated progeny.
FIGURE 2.
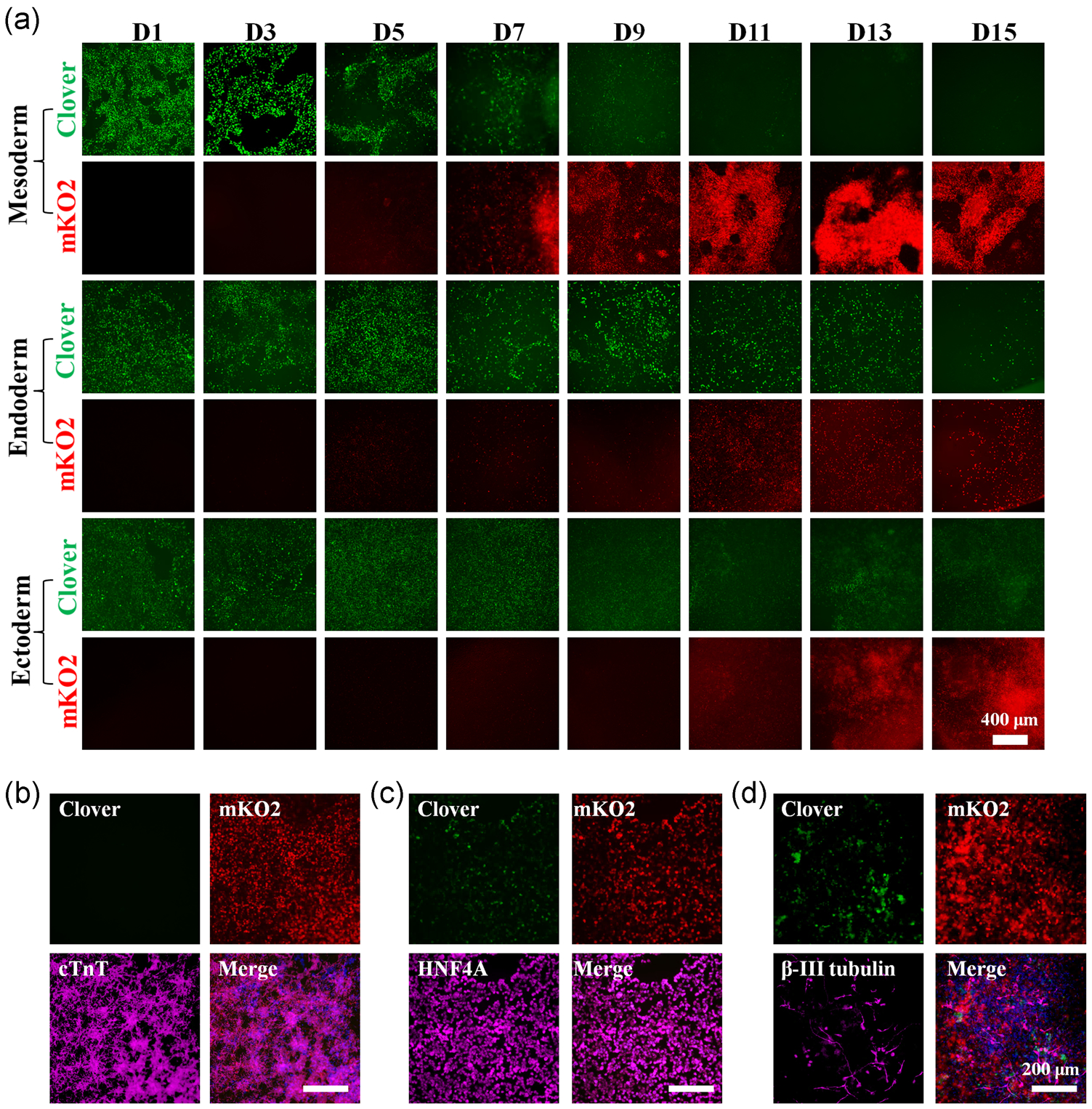
Continuous visualization of cell-cycle progression during hPSC differentiation. (a) Representative fluorescent images of Clover and mKO2 during mesoderm, endoderm, and ectoderm differentiation (Day 1 to Day 15). Scale bar=400μm. The fate of terminally differentiated cells was confirmed by immunostaining of mesodermal (b), endodermal (c), and ectodermal (d) lineage-specific markers, respectively. Scale bar=200μm. hPSC, human pluripotent stem cell; mKO2, monomeric Kusabira-orange 2
3.3 |. TNNT2-FUCCI system for cardiac-specific cell-cycle visualization
Cell cycle regulation is a key factor in the regeneration of damaged tissues, including infarcted myocardium. Thus, understanding CM cell-cycle regulation would greatly impact the development of regenerative therapies for heart disease, and has been an active area of study in the cardiac field. However, discerning CM cell cycle is challenging due to the mixed highly proliferative cell types within the heart including cardiac fibroblasts, epicardial cells, smooth muscle cells, and endothelial cells. To facilitate studies of lineage-specific cell proliferation, we replaced the CAG promoter with a cardiac-specific TNNT2 promoter (Figure 3a) to enable the lineage-dependent analysis of cell-cycle behavior. Once again, we employed the CRISPR/Cas9-mediated HDR to knock-in the new TNNT2-FUCCI construct to the AAVS1 safe harbor locus. After puromycin selection, we picked eight single cell-derived hPSC clones and confirmed that all of them were successfully targeted based on PCR genotyping analysis (Figure 3b). Of the eight clones, six were homozygous and we selected clone one, which retained the strong expression of SSEA4 (Figure 3c) and OCT-4 (Figure 3d), for future experiments. The cells also retained a normal karyotype (Figure S4A). Importantly, no Clover or mKO2 expression was observed at the pluripotent stage, suggesting the lineage-specific activity of TNNT2-FUCCI.
FIGURE 3.
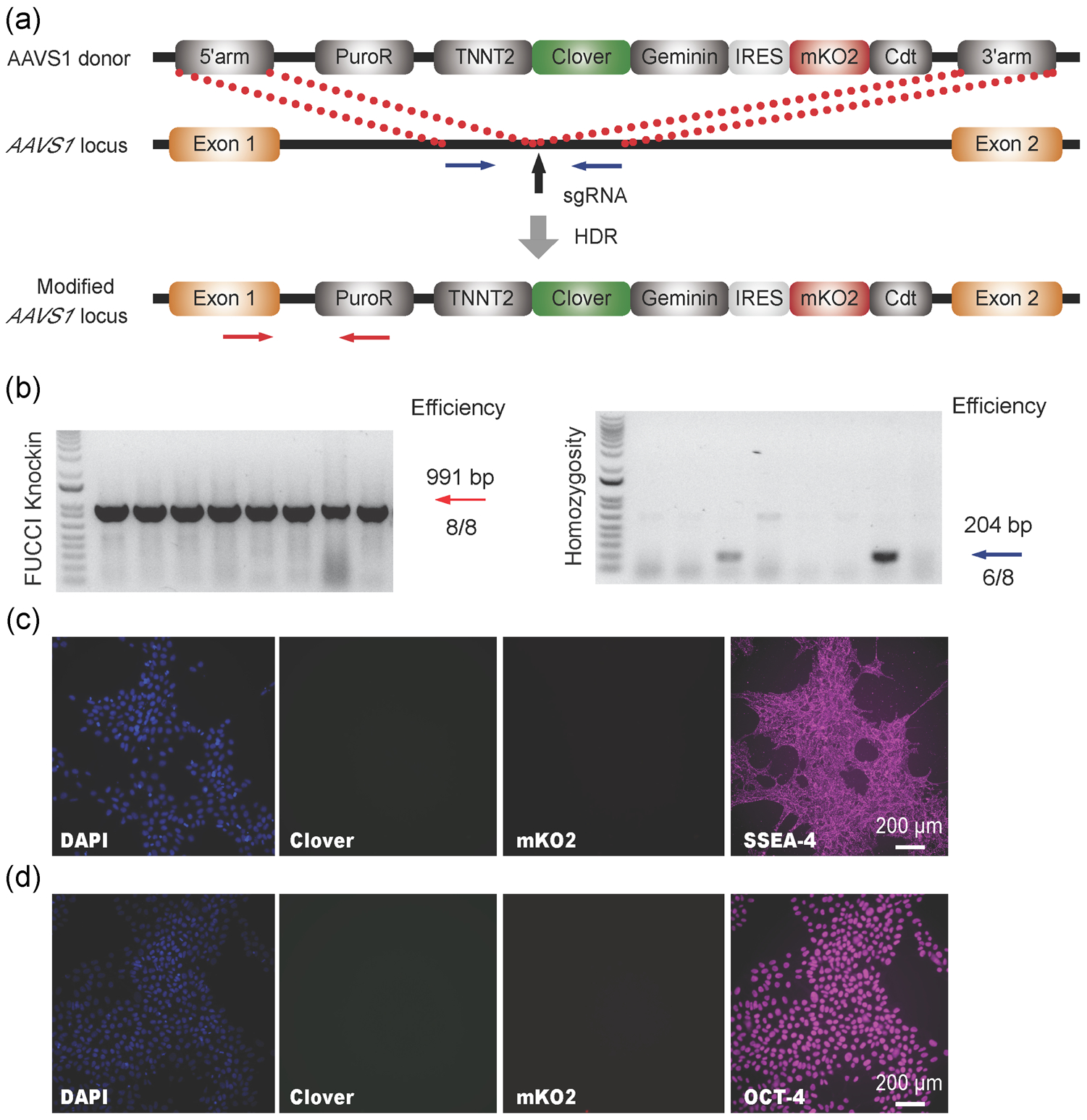
Construction of cardiac-specific FUCCI reporter knock-in H9 hPSC line using Cas9 nuclease. (a) Schematic illustration of the HDR-based knock-in strategy at the AAVS1 safe harbor locus. Vertical black arrow indicates the sgRNA T2 targeting site. (b) The representative PCR genotyping of hPSC clones after puromycin selection is shown, and the expected PCR product for correctly targeted AAVS1 site is ~991bp (red arrow) with an efficiency of eight clones from a total of eight. A homozygosity assay was performed on the knock-in clones, and those without ~204bp PCR products were homozygous with an efficiency of 6 clones from a total of 8 (blue arrow). Representative fluorescent images of Clover, mKO2, SSEA-4 (c) and OCT-4 (d) of successfully targeted cardiac FUCCI reporter H9 hPSCs were shown. Scale bar=200μm. FUCCI, fluorescent ubiquitination-based cell-cycle indicator; HDR, homology-directed repair; hPSC, human pluripotent stem cell; OCT-4, octamer-binding transcription factor 4; PCR, polymerase chain reaction; sgRNA, single guide RNA; SSEA-4, stage-specific embryonic antigen-4
To investigate the cardiac cell-dependent behavior of this new FUCCI system, we differentiated the engineered hPSCs into all three germ layers. As expected, TNNT2-FUCCI was not expressed in the endoderm and ectoderm differentiated cultures (Figure 4a), while the mKO2 expression was selectively and gradually increased along the cardiac differentiation of hPSCs (Figure S4B–D), demonstrating the lineage-specific activity of TNNT2-FUCCI. Furthermore, these differentiated cardiac cells exhibited significantly greater red fluorescent (G1 phase) intensity than green fluorescent (S/G2 phase), indicating most of the hPSCs- derived cardiac cells were nonproliferative. The fate of terminally differentiated mesodermal, endodermal, and ectodermal cells was confirmed by the immunostaining analysis of Troponin T (cTnT) (Figure 4b), HNF4A (Figure 4c), and β-III tubulin (Figure 4d). As expected, the expression of Brachyury was detected during cardiac differentiation of TNNT2-FUCCI hPSCs (Figure S5A,B) and the differentiation culture on Day 15 contained ~70% CMs as confirmed by the expression of cTnT, MF20, and cTnI (Figure S5C–E). Collectively, this data shows that the proliferation rate of CMs gradually decreases with progressive development, consistent with previous finding in both mice (de Boer, van den Berg, de Boer, Moorman, & Ruijter, 2012; Hirai et al., 2016; Soonpaa, Kim, Pajak, Franklin, & Field, 1996) and humans (Kaba et al., 2001; Villanueva et al., 2002).
FIGURE 4.
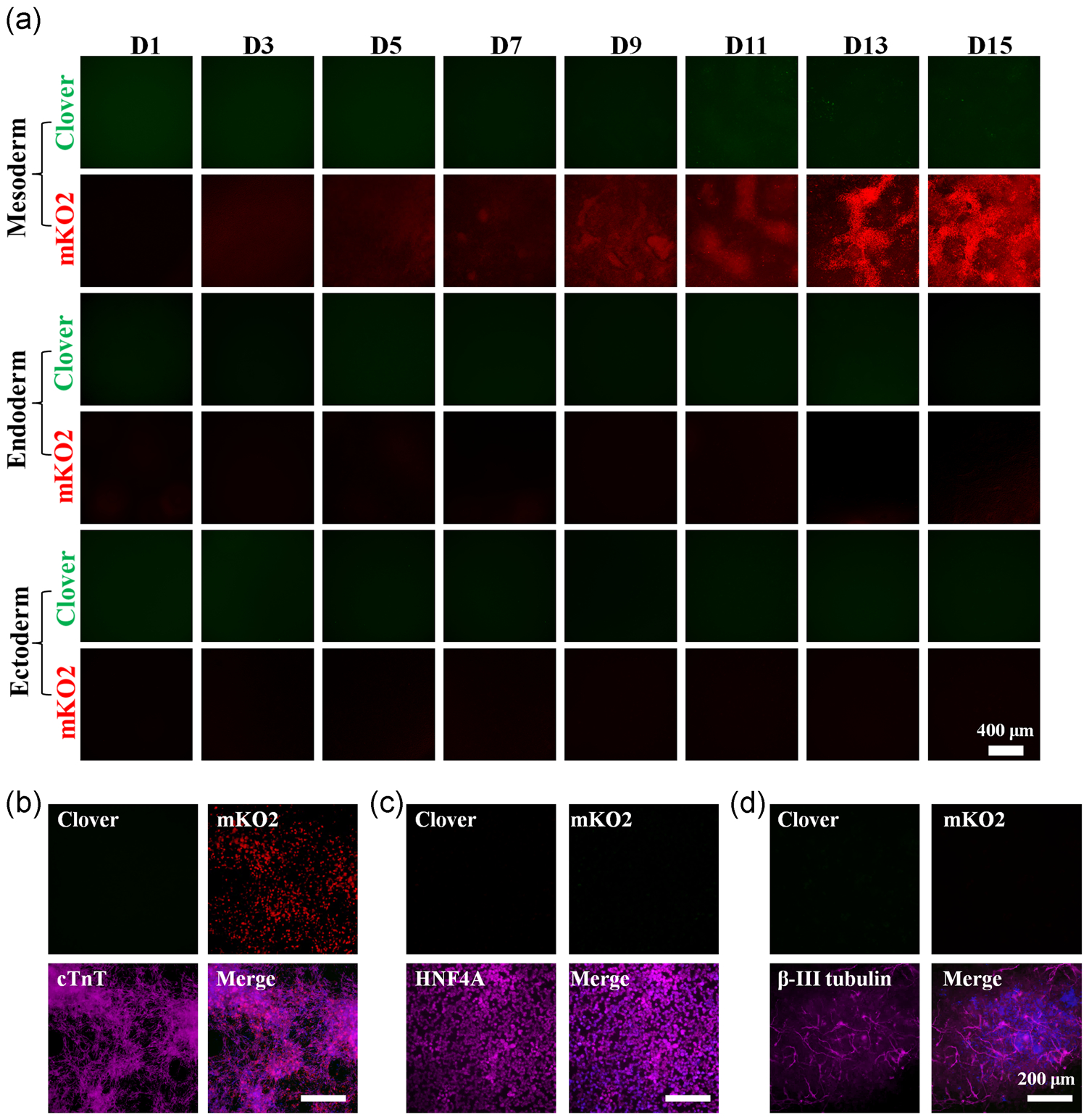
Lineage-dependent activity of cardiac FUCCI reporter during hPSC differentiation. (a) Representative fluorescent images of Clover and mKO2 expression during mesoderm, endoderm, and ectoderm differentiation. Scale bar=400μm. The fate of terminally differentiated cells was confirmed by immunostaining of mesodermal (b), endodermal (c), and ectodermal (d) lineage-specific markers, respectively. Scale bar=200μm. FUCCI, fluorescent ubiquitination-based cell-cycle indicator; hPSC, human pluripotent stem cell; mKO2, monomeric Kusabira-orange 2
Since our TNNT2-FUCCI reporter can accurately recapitulate the cell-cycle behavior of human CMs, the cells can be easily adopted to identify novel chemicals or genes for heart regeneration in a high-throughput manner. To test this function, we transfected Day 20 TNNT2-FUCCI hPSC-derived CMs with CCND2 expressing plasmid as CCND2 overexpression has been shown to increase CM cell-cycle activity (W. Zhu, Zhao, Mattapally, Chen, & Zhang, 2018). As expected, CCND2 overexpression significantly increased the Clover expression in the TNNT2-FUCCI hPSC-CMs (Figure 5a,b). Consistent with a previous report (Tan et al., 2019), transfection efficiency of ~70% was achieved in wild-type H9 CMs with an eGFP plasmid (Figure S5F), and the propidium iodide (PI) staining showed the CM cycle profile changed significantly with CCND2 transfection (Figure S5G). In summary, our TNNT2-FUCCI reporter works in a lineage-specific manner and allows for the sensitive spatiotemporal detection of cycling CMs during hPSC differentiation, enabling the identification of novel regulators for heart regeneration.
FIGURE 5.
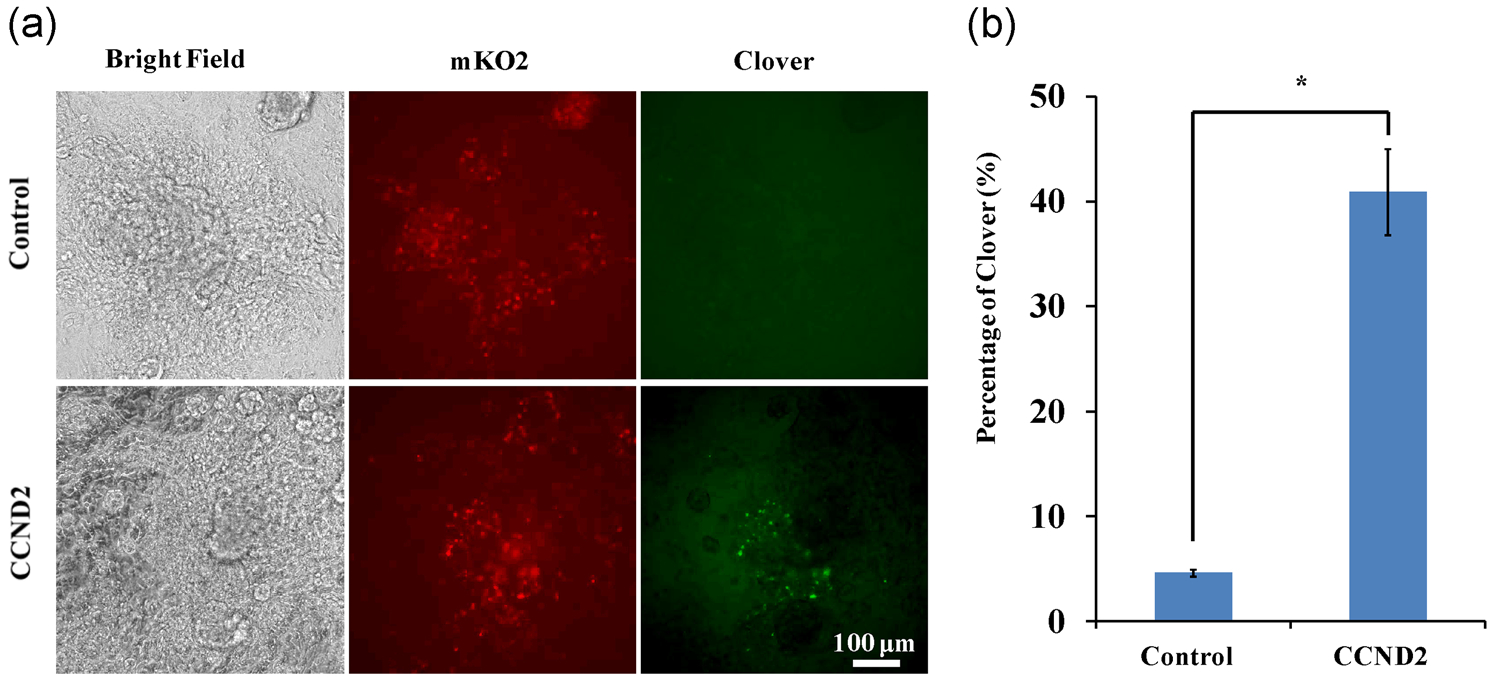
CCND2 overexpression increased the Clover expression in cardiac FUCCI reporter. (a) Representative fluorescent images of Clover and mKO2 expression in hPSC-FUCCI CMs with or without CCND2 overexpression. Scale bar=100μm. Quantitative analysis of Clover signal was shown in (b). CCND2, cyclin D2; FUCCI, fluorescent ubiquitination-based cell-cycle indicator; hPSC, human pluripotent stem cell; mKO2, monomeric Kusabira-orange 2
4 |. DISCUSSION
hPSCs have a unique cell-cycle comprised of short G1 and G2 gap phases, while their differentiation remodels the cell-cycle profile with increased G1- and G2-phases and a decreased rate of cell division. This atypical cell- cycle regulation in hPSCs is closely related to the maintenance of their pluripotent state, and has attracted intense interest (Soufi & Dalton, 2016; Yiangou et al., 2019). Recently, novel factors have been identified to regulate the cell cycle and used to promote cell-cycle re-entry for regenerative medicine (Cheah & Xu, 2016; Kipreos & van den Heuvel, 2019; Naumov et al., 2017). Due to the importance of cell-cycle regulation for both the self-renewal and differentiation of hPSCs, a stable and robust cell-cycle visualization system could effectively provide more valuable information to broaden the applications of hPSCs. Although the FUCCI system has been widely used in many dynamic models, including zebrafish (Choi et al., 2013), mouse (Abe et al., 2013), and stem cells (Calder et al., 2013; Coronado et al., 2013; Jovic et al., 2013; Roccio et al., 2013; Singh et al., 2013; Singh et al., 2016), a lineage-specific hPSC-FUCCI reporter has not yet been generated for versatile and high-throughput analysis of cell-cycle behavior during hPSC self-renewal and differentiation. Here, we have established and validated a robust and universally applicable FUCCI system in hPSCs for continuous and lineage-specific reporting of cell-cycle behavior. The constitutive mode of these FUCCI probes can accurately track cell-cycle patterns during mesoderm, endoderm, and ectoderm differentiation from hPSCs, while the cardiac TNNT2-FUCCI reporter allows lineage-specific visualization of the cell-cycle profile during CM differentiation. Importantly, our FUCCI platform is designed to be adaptable and can therefore readily be reengineered to support other tissue-specific reporting of the cell-cycle phase transitions for studying developmental biology and tissue regeneration, including neural, hematopoietic, pancreatic, hepatic, etc.
The implemented hPSC-FUCCI reporter also provides insights into the design of future cell-cycle-related research. First, our hPSC-FUCCI platform, particularly the lineage-specific FUCCI hPSCs, could be used as a high-throughput fluorescent reporter during unbiased, high-content chemical or genome-wide gene screening approaches, similar to the small molecule-screening for cardiac regeneration using the zebrafish FUCCI system (Choi et al., 2013). Secondly, hPSC-FUCCI allows the accurate isolation of actively proliferating (Clover expressing) cells via fluorescence-activated cell sorting (FACS) for future assays, such as genomic DNA isolation, gene profiling or cell transplantation. In line with this idea, Singh et al. (2016) has recently profiled the cell-cycle regulated gene expression in pluripotent cells purified by FACS with a similar FUCCI reporter. Collectively, the hPSC-FUCCI platform represents a versatile and robust toolkit for the real-time visualization of human cell-cycle dynamics, enabling the design of advanced studies and large-scale screening strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Bao and Lian groups for their critical discussion of this manuscript. We also thank the startup funding from the Davidson School of Chemical Engineering and the College of Engineering at Purdue University to X.B., NIH Trailblazer Award R21EB026035 to X.L.L. and NSF CAREER Award 1943696 to X.L.L..
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
DATA AVAILABILITY STATEMENT
The data sets, plasmids and cell lines generated and used in this study are available upon request submitted to the corresponding author.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- Abe T, Sakaue-Sawano A, Kiyonari H, Shioi G, Inoue KI, Horiuchi T, … Fujimori T (2013). Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Dev, 140, 237–246. [DOI] [PubMed] [Google Scholar]
- Avior Y, Sagi I, & Benvenisty N (2016). Pluripotent stem cells in disease modelling and drug discovery. Nature Reviews Molecular Cell Biology, 17, 170–182. [DOI] [PubMed] [Google Scholar]
- Bajar BT, Lam AJ, Badiee RK, Oh YH, Chu J, Zhou XX, … Lin MZ (2016). Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nature Methods, 13, 993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Adil M, Muckom R, Zimmermann J, Tran A, Suhy N, … Schaffer D (2019). Gene editing to generate versatile human pluripotent stem cell reporter lines for analysis of differentiation and lineage tracing: Tracing cell lineages in hPSCs with Cre reporter. Stem Cells, 37, 1556–1566. [DOI] [PubMed] [Google Scholar]
- deBoer BA, van denBerg G, deBoer PAJ, Moorman AFM, & Ruijter JM (2012). Growth of the developing mouse heart: An interactive qualitative and quantitative 3D atlas. Developmental Biology, 368, 203–213. [DOI] [PubMed] [Google Scholar]
- Bouldin CM, & Kimelman D (2014). Dual fucci: A new transgenic line for studying the cell cycle from embryos to adults. Zebrafish, 11, 182–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouldin CM, Snelson CD, Farr GH, & Kimelman D (2014). Restricted expression of cdc25a in the tailbud is essential for formation of the zebrafish posterior body. Genes and Development, 28, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boward B, Wu T, & Dalton S (2016). Concise review: Control of cell fate through cell cycle and pluripotency networks. Stem Cells, 34, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder A, Roth-Albin I, Bhatia S, Pilquil C, Lee JH, Bhatia M, … Draper JS (2013). Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells and Development, 22, 279–295. [DOI] [PubMed] [Google Scholar]
- Cheah KSE, & Xu PX (2016). SOX2 in neurosensory fate determination and differentiation in the inner ear Kondoh H & Lovell-Badge R. Sox2 Biol. Role Dev. Dis, 263–280. Elsevier Science; 10.1016/B978-0-12-800352-7.00015-3 [DOI] [Google Scholar]
- Choi W-Y, Gemberling M, Wang J, Holdway JE, Shen M-C, Karlstrom RO, & Poss KD (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development, 140, 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado D, Godet M, Bourillot P-Y, Tapponnier Y, Bernat A, Petit M, … Savatier P (2013). A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Research, 10, 118–131. [DOI] [PubMed] [Google Scholar]
- Du C, Feng Y, Qiu D, Xu Y, Pang M, Cai N, … Zhang Q (2018). Highly efficient and expedited hepatic differentiation from human pluripotent stem cells by pure small-molecule cocktails. Stem Cell Research & Therapy, 9, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZW, Hu BY, Ayala M, Sauer B, & Zhang SC (2009). Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines. Stem Cells, 27, 1032–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley C, & Philpott A (2013). The cell cycle and pluripotency. Biochemical Journal, 451, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M, Chen J, & Evans SM (2016). Tissue-specific cell cycle indicator reveals unexpected findings for cardiac myocyte proliferation. Circulation Research, 118, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic D, Sakaue-Sawano A, Abe T, Cho C-S, Nagaoka M, Miyawaki A, & Akaike T (2013). Direct observation of cell cycle progression in living mouse embryonic stem cells on an extracellular matrix of E-cadherin. SpringerPlus, 2, 585 http://www.ncbi.nlm.nih.gov/pubmed/25674414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaba RA, Coppen SR, Dupont E, Skepper JN, Elneil S, Haw MP, … Severs NJ (2001). Comparison of connexin 43, 40 and 45 expression patterns in the developing human and mouse hearts. Cell Adhesion and Communication, 8(4–6), 339–343. 10.3109/15419060109080750 [DOI] [PubMed] [Google Scholar]
- Kalkan T, Olova N, Roode M, Mulas C, Lee HJ, Nett I, … Smith A (2017). Tracking the embryonic stem cell transition from ground state pluripotency. Dev, 144, 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinas K, Grandy R, Ghule P, Medina R, Becker K, Pardee A, … Stein G (2013). The abbreviated pluripotent cell cycle. Journal of Cellular Physiology, 228, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos ET, & van denHeuvel S (2019). Developmental control of the cell cycle: Insights from Caenorhabditis elegans. Genetics, 211, 797–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Bao X, Al-Ahmad A, Liu J, Wu Y, Dong W, … Palecek SP (2014). Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem Cell Reports, 3, 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, … Palecek SP (2013). Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nature Protocols, 8, 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y. -i, Kuranaga E, Sugimura K, Miyawaki A, & Miura M (2011). Nonautonomous apoptosis is triggered by local cell cycle progression during epithelial replacement in drosophila. Molecular and Cellular Biology, 31, 2499–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov A, Kratzer S, Ting LM, Kim K, Suvorova ES, & White MW (2017). The Toxoplasma centrocone houses cell cycle regulatory factors. mBio, 8(4). 10.1128/mbio.00579-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SC, Chen N, Yip WY, Liow SL, Tong GQ, Martelli B, … Martelli P (2004). The first cell cycle after transfer of somatic cell nuclei in a non-human primate. Development, 131, 2475–2484. [DOI] [PubMed] [Google Scholar]
- Pauklin S, & Vallier L (2013). XThe cell-cycle state of stem cells determines cell fate propensity. Cell, 155, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccio M, Schmitter D, Knobloch M, Okawa Y, Sage D, & Lutolf MP (2013). Predicting stem cell fate changes by differential cell cycle progression patterns. Development, 140, 459–470. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, … Miyawaki A (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell, 132, 487–498. [DOI] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, & Yamanaka S (2017). Induced pluripotent stem cell technology: A decade of progress. Nature reviews. Drug discovery, 16, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Chappell J, Trost R, Lin L, Wang T, Tang J, … Dalton S (2013). Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Reports, 1, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Trost R, Boward B, & Dalton S (2016). Utilizing FUCCI reporters to understand pluripotent stem cell biology. Methods, 101, 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Maguire S, Davis LA, Alexander M, Yang F, Chandran S, … Pedersen RA (2008). Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells, 26, 496–504. http://www.ncbi.nlm.nih.gov/pubmed/18024421 [DOI] [PubMed] [Google Scholar]
- Soonpaa MH, Kim KK, Pajak L, Franklin M, & Field LJ (1996). Cardiomyocyte DNA synthesis and binucleation during murine development. American Journal of Physiology-Heart and Circulatory Physiology, 271, H2183–H2189. [DOI] [PubMed] [Google Scholar]
- Soufi A, & Dalton S (2016). Cycling through developmental decisions: How cell cycle dynamics control pluripotency, differentiation and reprogramming. Dev, 143, 4301–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, & Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Tan S, Tao Z, Loo S, Su L, Chen X, & Ye L (2019). Non-viral vector based gene transfection with human induced pluripotent stem cells derived cardiomyocytes. Scientific Reports, 9, 14404 http://www.nature.com/articles/s41598-019-50980-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A, & DeWitt ND (2016). Pluripotent stem cells progressing to the clinic. Nature Reviews Molecular Cell Biology, 17, 194–200. [DOI] [PubMed] [Google Scholar]
- Vermeulen K, Van Bockstaele DR, & Berneman ZN (2003). The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Proliferation, 36, 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva MP, Aiyer AR, Muller S, Pletcher MT, Liu X, Emanuel B, … Reeves RH (2002). Genetic and comparative mapping of genes dysregulated in mouse hearts lacking the Hand2 transcription factor gene. Genomics, 80, 593–600. [DOI] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang G-J, Kirschner MW, & Kaelin WG (2004). Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature, 428, 194–198. http://www.ncbi.nlm.nih.gov/pubmed/15014503 [DOI] [PubMed] [Google Scholar]
- Yiangou L, Grandy RA, Morell CM, Tomaz RA, Osnato A, Kadiwala J, … Vallier L (2019). Method to synchronize cell cycle of human pluripotent stem cells without affecting their fundamental characteristics. Stem Cell Reports, 12, 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, … Thomson JA (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhao M, Mattapally S, Chen S, & Zhang J (2018). CCND2 overexpression enhances the regenerative potency of human induced pluripotent stem cell-derived cardiomyocytes: Remuscularization of injured ventricle. Circulation Research, 122, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, & Huangfu D (2013). Human pluripotent stem cells: An emerging model in developmental biology. Dev, 140, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


