Abstract
Aggression is a quantitative trait deeply entwined with individual fitness. Mapping the genomic architecture underlying such traits is complicated by complex inheritance patterns, social structure, pedigree information, and gene pleiotropy. Here, we leveraged the pedigree of a reintroduced population of gray wolves in Yellowstone National Park, Wyoming USA to examine the heritability of and the genetic variation associated with aggression. Since their reintroduction, many ecological and behavioral aspects have been documented, providing unmatched records of aggressive behavior across multiple generations of a wild population of wolves. Using a linear mixed model, a robust genetic relationship matrix, 12,288 SNPs, and 111 wolves, we estimated the SNP-based heritability of aggression to be 37% and an additional 14% of the phenotypic variation explained by shared environmental exposures. We identified 598 SNP genotypes from 425 gray wolves to resolve a consensus pedigree that was included in a heritability analysis of 141 individuals with SNP genotype, meta-data, and aggression data. The pedigree-based heritability estimate for aggression is 14%, and an additional 16% of the phenotypic variation explained by shared environmental exposures. We find strong effects of breeding status and relative pack size on aggression. Through an integrative approach, these results provide a framework for understanding the genetic architecture of a complex trait that influences individual fitness, with linkages to reproduction, in a social carnivore. Along with few other studies, we present here the incredible utility of a pedigreed natural population for dissecting a complex, fitness-related behavioral trait.
Keywords: Aggression, canid, behavior, heritability, RAD-seq
Introduction
Aggressive behavior across species is correlated with two central aspects of fitness, fecundity and reproductive success (Réale et al., 2007; Wolf et al., 2007), shaped by the interaction of hormones, neurotransmitters, genetic variation, and the environment (Nelson & Trainor, 2007). This quantitative and continuous trait is found to vary within natural populations (e.g. Brodkin et al., 2002), yet little is known about the genetic components of aggression in natural populations (de Boer et al., 2003). Quantifying the extent to which genetic variation contributes towards aggression can enhance our understanding of the evolutionary constraints on, or the plasticity of, this fitness-related behavior (reviewed by Anholt & Mackay, 2012). Research on the molecular mechanisms of aggressive behavior has historically focused on neurochemicals and their associated receptors, which are known to have a central role in regulating behaviors (e.g. Mandel et al., 1981; Haller et al., 1998; Takahashi & Miczek, 2014). Further, domesticated species have been successfully used for discovering some of the underlying molecular components of complex traits, particularly in dogs (e.g. aggression, Vage et al., 2010; Eo et al., 2013; Proskura et al., 2013; sociability, vonHoldt et al., 2017). Although these studies have provided insights into the genetic basis of complex behavioral traits, their interpretations are limited to systems that have been artificially modified and controlled. Here, we suggest an extension to study aggression in an extensively monitored gray wolf (Canis lupus) population in North America.
After six decades of extirpation, 41 gray wolves were reintroduced to Yellowstone National Park (YNP) in 1995 and 1996, and nearly every aspect of their recovery documented (e.g. life history traits, Stahler et al., 2013; genetics, vonHoldt et al., 2008, 2010; pigmentation, Anderson et al., 2009). Further, behavioral studies of wolf intra-specific aggression have been successful in northern YNP as wolves are highly visible in this region, which contains a high density of overlapping wolf territories and elk populations (Supplemental Note). Such spatial overlap can result in higher intra-specific mortality rates (Cubaynes et al., 2014; Cassidy et al., 2015, 2017) and disease (Almberg et al., 2009). Here, we harness the integrative power of these past studies, field observations, a quantified behavioral trait, pedigree information, and newly collected genetic data to investigate the following facets of aggression.
Among canines, aggression can significantly impact fitness in individual interactions relating to territory defense, social dominance, predation events, and mate acquisition and thus reproduction (Mech & Boitani, 2000). Wolves live in territorial, cooperative, kin-structured groups called packs that vary tremendously in structure, ranging from a single monogamous breeding pair to multiple mating pairs of subordinate ranks (Mech & Boitani, 2000; vonHoldt et al., 2008). Not surprisingly, aggressive territorial behavior is prominent in group-living social mammals and is used as a means of territorial defense and dominance establishment (Maher & Lott, 2000). Levels of aggression are expected to influence reproductive success and likely evolve under balancing selection (Anholt & Mackay, 2012). Given the social hierarchy in wolf packs, breeding is cooperative, with rank and reproductive access often assumed to be correlated with aggression and are population density-dependent (Sands & Creel, 2004; Cubaynes et al., 2014) where successful inter-pack aggression leads to better access to resources such as territory, prey, and pup-rearing space (Smith et al., 2015). These factors influence pack size, individual survival, and fitness, which have important impacts on female reproductive success (Stahler et al., 2013). Wolves in YNP exhibit natural variation in aggressive interactions both within and between packs that is density-dependent and correlated with survival (Cubaynes et al., 2014; Cassidy et al., 2015). Sex-based differences in aggression are also known to occur in dogs, with males exhibiting higher aggression levels than females, and positively correlated with age in males but not females (Proskura et al., 2013). Further, aggression is also more likely to be exhibited among same-sex interactions (Eo et al., 2013). The relative numbers of wolves in each pack also has a strong effect on pack success and an individual’s degree of aggression (Cassidy et al., 2015, 2017).
Previous research has found a clear association between aggression and neurotransmitter-related genes across taxa (Comai et al., 2012; Pavlov et al., 2012), including domestic dogs (Vage et al., 2010). Further, a 3-bp mutation in the canine beta-defensin 103 (CBD103) gene is responsible for melanism and segregates as a Mendelian trait in gray wolves (Anderson et al., 2009). Although melanism is typically associated with increased aggression in a variety of taxa (Ducrest et al., 2008; Roulin & Ducrest 2011; Beziers et al., 2017), Cassidy et al. (2015) found that gray-coated wolves were more aggressive than melanistic individuals during inter-pack conflict. The capacity for the CBD103 gene to competitively bind other melanocortin receptors (Candille et al., 2009) that modulate aggressive behavior (Ducrest et al., 2008) may decrease aggression in melanistic wolves. Support of this mechanism comes from evidence that black-coated dogs have lower aggression rates than non-melanistic dogs (Houpt & Willis, 2001; Amat et al., 2009).
Here, we explore the inheritance and stability of aggression in a pedigreed gray wolf population. We hypothesize that: 1) individuals who have reproduced will exhibit increased levels of aggression, and 2) due to familial aggregation, aggression will display a positive, narrow-sense heritability. We employed a restriction site associated DNA marker sequencing approach to generate genome-wide SNP genotypes to infer (or confirm) pedigree relationships (following vonHoldt et al., 2008). Using linear mixed models, we explored the relationship of inter-pack aggression (herein, aggression) with life history traits as fixed effects and shared environment (i.e. natal packs) as a random effect to determine the degree of heritability of aggression. Our investigation will provide both a perspective on the success of future gene mapping efforts to uncover possible universalities of genes and pathways, as well as further insights into the environmental or correlated factors that shape behaviors in a social canine.
Methods
Sample collection and DNA extraction
Since their reintroduction, gray wolves are annually monitored in YNP. During winter captures using helicopter darting techniques following protocols approved by NPS (IACUC #IMR_YELL_Smith_wolves_2012), blood is collected in EDTA vacutainers, along with radio-collaring and morphometric data collection on age, sex, and breeding status. Further, YNP collects tissue specimens from carcasses to ensure a high representation of individuals in the curated collections. We extracted high molecular weight genomic DNA from blood and tissue samples collected from YNP since 1995 until the present using the DNeasy Blood and Tissue Kit (Qiagen) or the BioSprint 96 DNA Blood Kit in conjunction with a KingFisher Flex Purification platform (Thermo Fisher Scientific) following manufacturers’ protocols. DNA was visualized on a 2% agarose gel with a 2-log DNA ladder (New England Biolabs) for degradation, quantified using either PicoGreen or Qubit 2.0 fluorometry, and standardized to a concentration of 5ng/μL.
Life history traits and behavioral data
We accessed the extensive collection of phenotype data for 205 individual wolves in YNP with at least one observation contributing to the individual aggression score (IAS), with higher IAS values indicative that an individual consistently displays higher levels of aggression (see Supplemental Note). We limited our inclusion for individuals with at least three IAS and full covariate information, which resulted in 141 individuals. These individuals do not necessarily also have paired RAD-seq genotype data. Cohorts based on date-of-birth and natal pack were utilized for family-based analyses in addition to a life history data table, documented for every individual in the study. Static life history data (annually documented) include sex, date of birth and death, lifespan, cause of death, breeding status, genetically confirmed parentage (vonHoldt et al., 2008), and coat color (melanistic/gray) phenotypes. We used molecular methods to assign sex when field observations were unavailable (DeCandia et al., 2016). Individuals were considered “breeding” if they have genetically confirmed offspring at any point in their life, otherwise they were considered “non-reproductive”.
In 1995, YNP embarked on a 16-year effort to document direct observations of inter-pack interactions in northern YNP among individually recognizable wolves (Cassidy et al., 2015, 2017) (Supplemental Note). For all observations, the pack or group affiliation was recorded for each wolf. Inter-pack aggressive conflicts occurred when one wolf chased and physically displaced another wolf with some cases in which not all interacting partners have been identified (Cassidy et al., 2015). In some cases, the aggressive conflict escalated to an attack that was defined by physical contact between individuals, some with a mortality outcome, where at least one wolf was killed or fatally wounded. Inter-pack aggression was summarized on an ordinal scale for each interaction as a per individual wolf, ranging from 1 (flee) to 10 (led a chase which resulted in a kill). Because a single individual score is not a good indication of underlying aggressive tendency, individual scores were then averaged by the total number of observations per individual (Supplemental Note). To reduce the effects of viewer subjectivity and effects due differences wolf pack compositions, we required a minimum of three documented inter-pack interactions per individual for all subsequent analyses, with each individual average IAS derived from data collected across all documented interactions. We used these average IAS data to estimate the heritability of aggressive behavior and explore genetic associations with aggressive inter-pack interactions. As differences in pack size is a strong predictor of aggression (Cassidy et al., 2017), we also recorded the relative pack size at each inter-pack interaction and assigned every individual an average relative pack size by averaging over the documented interactions in which this individual was present. Note that if the average relative pack size for a wolf is greater than zero, then that wolf was on average in the larger of the two packs interacting and if the average relative pack size is less than zero that wolf was on average in the smaller of the two packs interacting.
Restriction site associated DNA marker sequencing (RAD-seq) and data processing
We prepared 75ng of high molecular weight DNA from 589 samples representing 468 unique wolves for a modified RAD-seq protocol as described by Ali et al. (2016) (Supplemental Table S1). Briefly, genomic DNA was digested with sbfI followed by ligation of a unique 8bp-barcoded biotinylated adapter to ultimately allow for pooled sequencing of 96 individuals. Pooled DNA was randomly sheared on a Covaris LE220 to 400bp, with subsequent enrichment for adapter ligated DNA fragments through a Dynabeads M-280 streptavidin bead assay (Thermo Fisher Scientific). We prepared each enriched genomic library using the NEBnext Ultra II DNA Library Prep Kit following manufacturer’s instructions for paired-end sequencing (2×150nt) on a rapid flowcell of the Illumina HiSeq 2500 at Princeton University’s Lewis-Sigler Institute for Integrative Genomics core facility. We conducted a size selection to retain genomic fragments between 300–400bp using Agencourt AMPure XP magnetic beads.
After sequencing, both the forward and reverse raw sequencing reads were aligned using a custom perl script (flip_trim_sbf1_150821.pl, see supporting information) to identify and then retain reads that contained the sbf1 cut site along with a barcode. We demultiplexed pooled libraries using the process_radtags function and allowed two mismatches in STACKS v1.42 (Catchen et al., 2013). We discarded reads with >2bp barcode mismatches or quality scores below 90% within the sliding window (set to 15% of the read) and removed PCR duplicates using default parameters in the clone_filter program. We mapped all samples with >500,000 reads to the reference dog CanFam3.1 genome assembly (Lindblad-Toh et al., 2005) using the paired-end mapping feature in STAMPY v1.0.21 (Lunter & Goodson, 2011). We sorted and filtered mapped reads based on a minimum quality score (MAPQ>96) and converted files to bam format in SAMtools v0.1.18 (Li et al., 2009). We then implemented the updated gstacks pipeline in STACKS v2.2, due to its ability to confidently identify and genotype SNPs from low coverage, paired-end data using the Marukilow model (Rochette et al., 2019). This model implements a maximum-likelihood method that incorporates population-level genotype frequencies and error rates to assess the statistical likelihood of each polymorphic site and individual genotype call (Maruki & Lynch 2017). When paired with the previous clone-filtering step, this model removes the need for subsequent coverage filtering, as a posteriori removal of statistically significant alleles may introduce more bias and allelic dropout than it corrects.
We implemented the populations module twice to remove duplicate samples and minimize missing data in the final SNP dataset. We first included 485 high-quality samples that passed clone-filtering and set the --write_single_snp flag (which retains only the first SNP per locus) as the only filtering parameter. We used PLINK (Purcell et al., 2007) to assess missingness per sample, and removed duplicate samples with a higher percentage of missing loci and any sample with missingness >80%. We then ran the populations module a second time with a reduced sample set containing 423 unique wolves and an additional filtering parameter that removed loci genotyped in fewer than 90% of individuals (–r 0.90). We conducted principal component analysis (PCA) of the genotype data using flashPCA (Abraham & Inouye 2016) and the PCs used in subsequent analyses where appropriate.
Parentage and pedigree construction
We conducted parentage testing to assign relationships using a multi-pronged approach that integrated past observational and parentage information from vonHoldt et al. (2008), which was previously constructed using 26 tetra-nucleotide microsatellite loci, with genome-wide SNP data obtained from RAD-seq methods to both update and resolve challenging relationships. For SNP-based analyses, we applied strict data filtering parameters that are optimized for pedigree reconstruction (Huisman 2017). We removed problematic individuals (e.g. putative monozygotic twins) and filtered the remaining wolves (n=413) to retain SNPs that segregated two alleles (--biallelic-only --snps-only), had a minor allele frequency of 0.45 (--maf 0.45), were in Hardy-Weinberg equilibrium (--hwe 0.001), and were pruned to obtain loci in statistical linkage equilibrium using genotypic correlation (r<0.2) as a proxy metric (--indep-pairwise 50 5 0.2) in PLINK (Purcell et al., 2007). We assessed the degree of missingness per sample to remove those with missing values higher than two standard deviations above the mean (referred to as the pruned dataset).
To perform parentage analyses at a finer scale, we additionally created annual datasets for 1995 through 2018 and retained life history data regarding which individuals were reproductively mature (≥1 year old) as candidate parents with no a priori preferences for parentage testing based on pack affiliation, social rank, or copulatory/mating behaviors. We filtered the dataset to reflect annual mortality events and removed individuals with missingness higher than two standard deviations above the annual mean. For complicated families, we later included observations on reproductive access or the display of copulatory mating behaviors to manually resolve candidate parents.
We used the R package related to calculate relatedness coefficients between each pair of wolves using both the 413 and 384 wolf datasets (Pew et al., 2015). Using the coancestry function, we implemented the dyadic likelihood estimator (dyadml=1; Milligan 2003) with allowance for inbreeding (allow.inbreeding=TRUE) on our parentage datasets. We selected this relatedness measure for its inbreeding allowance, computational efficiency, and low error rate with SNP datasets, as prior simulations implemented in related returned similar results for both moment (e.g. Wang 2002) and likelihood (e.g. dyadml) estimators (Milligan 2003; Wang 2011).
We next used the R package sequoia to reconstruct pedigrees with our parentage-informative pruned dataset, and separately the annual datasets that contained individuals alive in each year (Huisman 2017). This program implemented a heuristic hill-climbing algorithm to optimize the likelihood of unrelated, first-, second-, and third-degree relationships in the dataset. It subsequently assigned parent-offspring pairs, half-siblings sharing a “dummy” parent, and grandparents that sired unsampled “dummies” to build multigenerational pedigrees. We used default parameter settings with the estimated genotyping error rate (Err) relaxed to 1e-03 for each analysis. We then analyzed the pruned dataset a second time with Err set to 1×10−2 to enable additional parent-offspring assignments.
We merged results from related, sequoia, and previous microsatellite analyses (vonHoldt et al., 2008) to create a consensus pedigree. We first assigned parent-offspring pairs when all three analyses were consistent in the identification of the same individual parent (our “gold standard of high support”). We next considered parent-offspring pairs that were supported by two of three analyses pairs: 1) related and sequoia relationship assignments inferred from RAD-seq SNP genotypes; or 2) related inferences based on SNP genotypes and previous microsatellite data. When sequoia and microsatellite assignments mismatched, we assigned the parent-offspring pair with the higher relatedness value based on SNP genotypes.
Estimating the heritability of inter-pack aggression
The kinship for two wolves i and j was defined as the probability that a gene selected randomly from an autosomal locus originating in the genome of wolf i and a gene selected randomly at the same locus from the genome of wolf j are identical by descent (IBD) (Malécot, 1948). To estimate the kinship matrix needed for SNP-based heritability estimates, we further filtered the LD-pruned full SNP set to exclude loci with genotyping success rates less than 95%, significant deviations from Hardy-Weinberg equilibrium (P<1×10−7), or minor allele frequency MAF<0.05, and any individuals with more than 12.5% missing SNP data. We estimated the kinship matrix for the resulting wolves with genotype, covariate, and phenotype data using a robust genetic relationship matrix (VanRaden, 2008; Wang et al., 2017) as implemented in the SNPArrays package of OpenMendel (Zhou et al., 2019) using only the autosomal SNPs remaining after the above filtering. To address any differences that may be obtained from global kinship estimates derived from the GRM versus those from the pedigree, we also estimated heritability using the theoretical kinship values using the pedigree structure and Jacquard’s recurrence formulas (Emik & Terrill, 1949; Lange, 2002; Zhou et al., 2019).
We used a REML-based linear mixed model as implemented by the VC test routine (Zhou et al., 2017) of the OpenMendel package (Lange et al., 1976, 1983; Bauman et al., 2005; Lange et al., 2013; Zhou et al., 2016; Zhou et al., 2019) to estimate both fixed and random effects. Our most general model is:
In all analyses, the X matrix includes sex (male as the reference group), breeding status (non-reproductive as the reference group), coat color (gray as the reference group), and average relative pack size as fixed effects; β denotes the corresponding vector of coefficients. The Vpheno matrix is composed of the additive genetic variance vA, natal pack variance vpack, independent environmental variance ve, dominance genetic variance vD and maternal effect variance vM. These effects are treated as random. The design matrices were: 1) I, a matrix with 1 on the diagonal and 0 elsewhere; 2) K, the kinship matrix; 3) D, a matrix of probabilities of sharing two genes IBD; and 4) H, a matrix of ones and zeros with 1 denoting wolves i and j from the same natal pack and 0 otherwise. Heritability is defined as the fraction of phenotypic variation that is due to genetic effects, 5) M, a matrix of ones and zeros with 1 denoting wolves i and j as having the same mother and 0 otherwise. Typically, narrow sense heritability (h2) is estimated as the fraction of phenotypic variance due to the alleles acting independently. As an example, when dominance and pack are also included as random effects:
We calculated the fraction of phenotypic variance that is due to the natal pack. For the same example:
We similarly used year of birth to define a common environment effect of birth year. As our goal was to assess the degree to which genetic effects may influence aggression, we used a stepwise approach to determine if any additional variance components significantly improved the model when additive genetic variance was also included. Similarly, in order to address the possibility of any residual population substructure, we tested whether the inclusion of the first three PCs as fixed effects improved the model fit when additive genetic variance was also included using the GRM as the estimate of kinship coefficient matrix.
Pedigree-based genetic associations with inter-pack aggression
With the acknowledgement that this analysis is likely under-powered, we assessed genome-wide association of SNP variants with IAS. We filtered SNP to retain sites with a maximum of 20% missing data per individual and a minor allele frequency of 1%. We employed a linear mixed model (LMM) in gemma (Zhou & Stephens, 2014) and included a kinship relatedness matrix estimated for 391 individuals in related (see above section for more details). We included IAS phenotypes for 121 individuals that had a minimum of three inter-pack aggression interactions observed, with individuals lacking such observational support excluded from the LMM analysis. We included sex, coat color, and breeding status as covariates in the LMM. We assessed the significance of the association using the likelihood ratio test (LRT) and inferred significance using an adjustment for multiple testing (B-Y modified; Benjamini & Yekutieli, 2001). We used an experiment-wide B-Y false discovery threshold of . Our rationale is to acknowledge that the dataset is expected to be underpowered and our goal was to minimize erroneous inference of genotype associations. All sites were catalogued with Ensembl’s Variant Effect Predictor for their predicted impact (McLaren et al., 2016). We further conducted functional profiling using g:GOSt in g:Profiler to determine if outlier SNPs that were catalogued as genic were enriched in specific gene ontological categories using the Benjamini-Hochberg FDR of 0.05 (Benjamini & Hochberg, 1995; Raudvere et al., 2019). We searched only annotated genes and included all default data sources from ontology, biological pathways, and regulatory motifs databases.
Data Accessibility
Mapped bam files for these 413 wolves are available on NCBI’s public Sequence Read Archive (PRJNA577957). Additional meta-data for each individual wolf can be found in Supplemental Table S1.
Results
RAD-seq data processing and parentage analyses
We discovered 212,667 SNP variants in the 485 samples that passed initial clone filtering. After excluding duplicates, putative monozygotic twins, and samples with low sequence coverage and low quality data (n=72), we retained 413 wolves with 120,327 SNPs for strict filtering and pedigree analysis. Our final parentage datasets consisted of 598 uncorrelated neutral SNPs with MAF>45% (i.e. HWE P>0.001). We assessed missingness per individual and removed an additional 29 wolves with missingness higher than two standard deviations above the mean, which produced two datasets of 598 SNPs for parentage analysis: 1) the full dataset containing 413 wolves; and 2) the pruned dataset containing 384 wolves. We additionally created annual datasets of wolves living in YNP between 1995 and 2018 to assign parentage within smaller subsets of wolves, where possible parents were restricted to individuals recorded to be alive in each year.
We conducted PCAs using 598 parentage-filtered SNPs across each of these datasets (full, pruned, and annual), which revealed two components of the demographic history of wolves in YNP. First, PC1 reveals an axis that is polarized by the Nez Perce pack (low PC1 values), a pack that received translocated pups from the Sawtooth pack of northwestern Montana that represents a distinct genotypes. These individuals eventually contributed to the genetic diversity of YNP wolves through gene flow. Second, PC2 differentiates the two source populations from which wolves were originally translocated (high PC2 values, 1995 relocation from Alberta; low PC2 values, 1996 relocation from British Columbia) (reviewed in vonHoldt et al., 2010) (Supplemental Figs. S1–S3).
Pedigree construction and confirmation
We assigned SNP-based parentage results from sequoia when also supported by concordant relatedness estimates across the full, pruned, and annual datasets. In total, 505 parent-offspring (PO) assignments are supported: 264 PO pairs are supported by all SNP and microsatellite analyses (“gold standard of high support”); 140 PO pairs are supported by only SNP-based analyses; and 101 PO pairs are supported by only SNP-based relatedness estimates (parentage inference was not conclusive) and microsatellite analyses.
Linear mixed model estimates for inter-pack aggression
After filtering, 111 wolves with full covariate information, at least three IAS values and 12,288 SNPs were used to calculate the robust GRM and to estimate the heritability of inter-pack aggression (Table 1). When additive genetic variance is included in the model, the first three principal components do not improve the model fit (Table 2). The best fitting model by Akaike Information Criterion (AIC) included variance components for additive genetic effects, natal pack as a common environmental effect, and residual independent environment. The SNP narrow-sense heritability of aggression is h2=0.369 and the proportion of the phenotypic variance explained by natal pack membership is fpack=0.134 (Table 3). The inclusion of dominance genetic effects, maternal effects, and year of birth cohort effects did not improve the fit of a model that included additive genetic effects. We note that in our best fitting model, sex, and coat color are not significantly associated (P=0.448 and 0.637), although their trends are in the previously observed direction where females tended towards lower IAS values than males, while melanistic wolves also tended towards lower IAS values than gray-coated wolves. Breeding status and average relative pack size are significantly associated (P=1.37×10−4 and 7.93×10−8) with breeding individuals having an IAS 0.713 higher than a non-breeding individual, and a unit increase in average relative pack size increasing IAS by 0.111. Interpretation of these results is challenging because on a per interaction level the IAS value is ordinal, but clearly an individual that has reproduced will, on average, display a higher level of aggression relative to an equivalent non-breeding individual. Similarly, average aggression levels are increased when individuals tend to travel in a larger pack, likely due to an advantage over any opponents in relatively smaller packs (e.g. Cassidy et al., 2015).
Table 1.
Summary statistics of individuals included in the linear mixed model analyses.
| Sample | Percent males | Percent melanism | Percent Reproductive | Mean IAS (SD)* | Mean relative pack size (SD; Range) | Mean number of interactions (SD)** | Number of natal packs represented |
|---|---|---|---|---|---|---|---|
| GRM (n=111) | 47.7 | 47.7 | 37.8 | 4.50 (1.2) | 5.87 (5.8; −8.4–22.2) | 15.72 (14.5) | 14 |
| Pedigree (n=141) | 51.1 | 51.8 | 37.8 | 4.55 (1.2) | 5.68 (6.1; −11.3–23.5) | 15.56 (14.5) | 17 |
(Abbreviations: GRM, genetic relatedness matrix; IAS, individual aggression score; n, sample size; SD, standard deviation)
The range of IAS was 1–7
The range for the number of interactions was 3–78
Table 2.
Log-likelihoods and Akaike Information Criterion (AICs) for linear mixed models examined. All models include fixed effects of sex, breeding status, coat color and average relative pack size. The fixed effects of the PCs were not included in the pedigree-based models because of extensive missing data.
| Model | df | GRM Log-likelihood | GRM AIC | Pedigree Log-likelihood | Pedigree AIC |
|---|---|---|---|---|---|
| Ve | 6 | −153.9397 | 319.8794 | −170.7956 | 353.5912 |
| Ve; PCs | 9 | −152.5409 | 323.0818 | ||
| Ve, Va | 7 | −151.6879 | 317.3758 | −165.7513 | 345.5026 |
| Ve, Va; PCs | 10 | −151.0484 | 322.0968 | ||
| Ve, Va, Vd | 8 | −150.7475 | 317.4950 | −165.6432 | 347.2864 |
| Ve, Va, Vmat | 8 | −151.0995 | 318.1990 | −165.9977 | 347.9954 |
| Ve, Va, VYOB | 8 | −151.6714 | 319.3428 | −165.3743 | 346.7486 |
| Ve,Va, VPack | 8 | −150.2500 | 316.5000 | −164.6324 | 345.2648 |
| Ve, VPack | 7 | −151.6069 | 317.2138 | −164.9119 | 343.8238 |
| Ve, Va, Vpack; PCs | 11 | −149.4536 | 320.9072 | ||
(Abbreviations: df, degrees of freedom; GRM, genetic relatedness matrix; PCs, the first three principal components; Ve, variance due to independent environmental effects; Va, additive genetic variance; Vd, dominance genetic variance; VPack, variance due to natal pack treated as a random effect; VYOB, variance due to year of birth cohort treated as a random effect; Vmat, variance due to shared maternal effect)
Table 3.
Coefficients for the best-fitting linear mixed models for the SNP-based analysis by Akaike Information Criterion and corresponding model for Pedigree-based analysis.
| Parameter | Mean | Sex | Coat color | Breeding status |
Relative pack size | vA | vpack | ve | h2 | fpack |
|---|---|---|---|---|---|---|---|---|---|---|
| Robust GRM-based estimates | 3.749 | −0.143 | −0.092 | 0.713 | 0.111 | 0.383 | 0.139 | 0.517 | 0.369 | 0.134 |
| Standard error | 0.220 | 0.188 | 0.195 | 0.187 | 0.021 | 0.232 | 0.082 | 0.119 | 0.224 | 0.079 |
| P-value | 0.448 | 0.637 | 1.37×10−4 | 7.93×10−8 | 4.97×10−2 | 4.49×10−2 | ||||
| Pedigree-based estimates | 3.807 | −0.175 | −0.036 | 0.824 | 0.112 | 0.142 | 0.165 | 0.725 | 0.138 | 0.160 |
| Standard error | 0.223 | 0.166 | 0.173 | 0.169 | 0.016 | 0.190 | 0.110 | 0.163 | 0.158 | 0.107 |
| P-value | 0.139 | 0.837 | 1.11×10−6 | 2.14×10−12 | 0.227 | 6.73×10−2 | ||||
(Abbreviations: fpack, the proportion of the phenotypic variance explained by natal pack membership; Va, additive genetic variance; Ve, variance due to independent environmental effects; Vpack, variance due to natal pack treated as a random effect)
Using the consensus pedigree, 141 wolves with full covariate and at least three behavioral observations are included in the analysis (Table 1). These data, along with the theoretical kinships, were used to estimate the heritability of inter-pack aggression in a second linear mixed model analysis. Again, we used a stepwise approach to determine the variance components that lead to the best fitting model by AIC, keeping in mind the hierarchical nature of some of the random effects (Table 2). The model including additive genetic variance provided a better fit than one with only the independent environmental effect (P=0.01236). As with the GRM estimate of kinship, a model that included a dominance genetic effect along with an additive genetic effect or maternal effects was not the best fitting model. Similarly, the year of birth cohort effect explained only 2% of the total variance and was not included in the best fitting model. One major difference from the first analysis is that neither the additive genetic variance nor the natal pack variance are significantly greater than zero when the other effect is included in the model (P=0.2273 and P=0.0673, respectively). Indeed, when assessing model fits with AIC, the best fitting model included only the natal pack effect with the next best model being the one with both additive genetic and natal pack effects (Table 2).
For comparison with the analysis using the robust GRM we consider this model further. The narrow sense heritability of aggression, h2, is 0.138 and the proportion of the phenotypic variance explained by natal pack membership, fpack is 0.160 (Table 3). As with the robust GRM, sex and coat color are not significantly associated (P=0.1386 and 0.8365), whereas breeding status and average relative pack size are significant (P=1.106×10−6 and 2.141×10−12). Breeding individuals are predicted to have a 0.8243 higher IAS value than non-breeding individuals have and a unit increase in relative pack size is expected to increase IAS by 0.1124.
Pedigree-based association
This dataset included 391 wolves for 56K SNPs after filtering for 10% missing data per locus, a minor allele frequency of 1%, and excluding individuals with more than 20% missing data across all loci. Of these loci, 31,491 SNPs were informative for the model association. We restricted our association analysis to a total of 121 wolves that have a minimum of three behavioral observations and included sex, coat color, and breeding status as covariates. We identified 45 SNPs with alleles significantly associated with IAS (LRT, adjusted P<9.145×10−4) (Supplemental Table S3). Using Ensembl’s Variant Effect Predictor, all 45 sites are catalogued as having a putatively “modifier” (non-coding variant) impact. Of these, only 17 are categorized as ‘genic’ with associated genes (Supplemental Table S4), two of which (NOCT and EDC3) belong to a single ontological category passed the FDR (cytoplasmic mRNA processing body assembly, GO: 0033962. Padj=4.112×10−2). Despite our underpowered study, we do observe associations of genetic variation in the genes MYO9A and TRAK1. Although these gene functional categories do not surpass the FDR, their respective functions remain relevant and include involvement in neuronal growth and the regulation of endocytic trafficking of GABA-A receptors, respectively (O’Connor et al., 2016; Barel et al., 2017).
Discussion
To explore the life history, ecological, and molecular factors associated with inter-pack aggression, we investigated behavioral and genetic data across a 16-year study of a pedigreed population of gray wolves. Overall, we found that aggression is heritable and subject to common environmental effects that are captured by natal pack. Aggression is predicted by breeding status, relative pack size and a small subset of functionally relevant genes. Our analyses suggest that aggression demonstrates moderate levels of narrow sense heritability with additive genetic effects explaining 14–37% of variation in aggressive behavior in gray wolves in YNP. The estimate of heritability based on theoretical kinship matrix derived from the pedigree is 14%, which is substantially lower than the heritability estimate of 37% based on the correlation among the SNPs using the robust GRM matrix. Both estimates are potentially biased, albeit in different directions. We further suggest that our variance models also are unlikely to fully capture the total evolutionary potential of aggression, and the role of indirect effects will likely add to our understanding of aggression in this wild pedigreed population of gray wolves (e.g. Camerlink et al., 2013; Alemu et al., 2014).
As is often the case with wild populations, the pedigree is not exactly known and relatedness inaccuracies among even a few founders can bias heritability estimates towards zero (Wilson et al., 2010). In contrast, using SNP correlations to estimate the kinship among closely related individuals can lead to inflated heritability (Zaitlen at al., 2013). We therefore suspect that the true estimate of IAS heritability is bounded by the pedigree and GRM-based estimates. Because we used the average effect of IAS estimated over a minimum of three interactions, we are not accounting for within-individual variability, which may inflate heritability estimates. As estimates of additive genetic variance can include other effects when the models are too simplistic, we explored models that might partially lead to apparent heritability. We found no evidence of maternal effects, dominance effects, residual population substructure, or year of birth cohort effects, although our sample size is likely underpowered unless these effects are relatively large. We do find evidence suggestive of a natal pack effect that explains 14–16% of the total variation in IAS. This effect likely reflects shared exposure to environmental conditions and behavioral experiences as a group-living, territorial species, as well as potentially capturing a dominance effect as siblings are a major component of the natal pack membership.
Our heritability estimates for aggression in gray wolves are comparable to those reported for domestic animal temperament (~30%) (e.g. Morris et al., 1994; Le Neindre et al., 1995, 1996; Dingemanse et al., 2002; Lovedahl et al., 2005; Pérez-Guisado et al., 2006; Chervet et al., 2011). Studies of parent-offspring trait correlations and applications of the animal model approach in wild populations have confirmed the low to moderate estimates of aggression (e.g. North American red squirrel h2=0.08–0.12, Taylor et al., 2012; western bluebirds h2=0.34, Duckworth & Kruuk 2009; laboratory zebrafish h2=0.36, Ariyomo et al., 2013). Experimental simulations of territorial intrusions and regression modeling also estimated comparable estimates of heritability for aggressive behavior in male great tits (h2=0.260–0.266 Araya-Ajoy & Dingemanse, 2017). Further, experimental breeding studies offer evidence that behavioral and complex traits have a heritable genetic component (Takahashi & Miczek, 2014). For example, the fear-selected line of silver foxes (Vulpes vulpes) exhibit a strong and heritable aggression response after generations of selection (Trut, 1980; Kukekova et al., 2011). A caveat of heritability estimates is that they are difficult to directly compare, as heritability models often vary in their components and depend on the amount of environmental variation. We included fixed effects in both analyses, which found that only breeding status and relative pack size were strongly significantly associated with aggression. Thus, our heritability analyses included sex and grey/melanism as covariates despite not being significant to allow better comparison to other studies of similar wolf populations.
Gray wolves rely upon aggression for acquisition and maintenance of both territories and mates. However, access to these resources central to individual fitness is often variable, may be density dependent, and experience annual fluctuations. For example, by 2002, wolves in northern YNP had one of the highest densities ever recorded in North America at 98 wolves/1000 km2 (Paquet & Carbyn, 2003). Since 2008, it has stabilized on average at about 39 wolves/1000 km2 (Smith et al., 2019). The Druid Peak (1996–2009), Slough Creek (2003–2008), and Agate Creek (2002–2012) packs maintained territories concurrently in the northern range for >6 years and each were observed intensively for thousands of hours by biologists. The genetic relationship composition within and between packs was annually augmented due to changing memberships at the pack level. Slough Creek and Agate Creek packs were formed by females dispersing from Druid Peak and joining males from other packs (Fig. 1). All three packs alternated in being the largest and most dominant pack in the area. Inter-pack aggressive conflicts were common, with at least 14 mortalities documented. All three packs eventually disintegrated after a significant loss of a key member during an aggressive conflict with neighboring packs.
Figure 1.
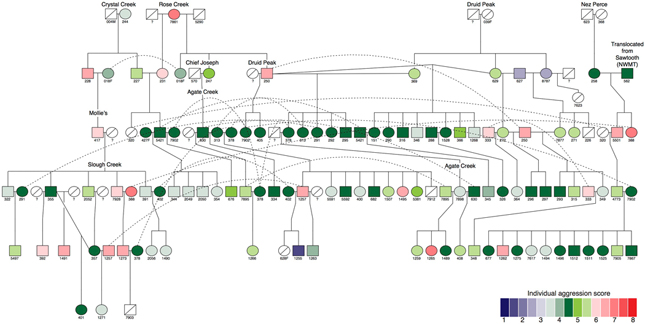
An example pedigree for a subset of YNP wolves with SNP genotypes and symbols (male, square; female, circle) shaded to represent their individual aggression score (IAS) level. This pedigree is of the Druid Peak, Slough Creek, and Agate Creek packs. Symbols with a diagonal line indicate the lack of data for the aggression behavioral phenotype. Dashed lines indicate where an individual was involved in parentage events across disparate sections of the pedigree. Pack names are indicated at the place in the pedigree when the pack was established. (Abbreviations: NWMT, northwest Montana)
Similarly, Cubaynes et al. (2014) showed that wolf survival was dependent upon wolf density. Intra-specific aggressive behavior is presumably expected to serve as the primary mechanism for resource acquisition and defense, although aggression may also be influenced by group size and composition (Cassidy et al., 2015, 2017), or modulated at the individual level relative to their environment or social composition. The maintenance of the aggression trait is likely under stabilizing selection, where strongly or weakly aggressive behaviors are likely to lower individual fitness through decreased access to critical resources or mortality, respectively. Consequently, plasticity in aggression may be constrained by underlying molecular mechanisms (e.g. epigenetic gene regulation), which may have resulted in the evolution of evident genetic polymorphisms for aggressive behavior in this species.
We explored the genetic association of aggression in gray wolves and it is unclear whether these genetic variants play a direct role in regulating gene expression. We found suggestive evidence that changes in neuronal growth and the GABA-A receptors may influence aggression levels, with the latter playing a well-established role in aggression (Miyakawa et al., 2003; Bannai et al., 2009; Takahashi & Miczek, 2014) with similar findings recently reported in heritable aggression in dogs (MacLean, Snyder-Mackler, et al., 2019).
Taken together, our results suggest that aggression is influenced by heritable genetic variation. The long-term pedigree, combined with robust behavioral observations, provides an unprecedented opportunity to integrate trait and genome-wide molecular data to discover associations with a complex, fitness-related trait in a natural population of a social canine. This study provides a new foundation that can support future studies that aim to expand upon explicit evaluations of individual-level fitness, ecological models, and explorations of natural selection in a pedigreed natural population.
Supplementary Material
Acknowledgements
The authors would like to thank the Princeton University Computational Science & Engineering Support (CSES) group for providing computational assistance for multiple components of our work. This study was supported in part by the National Science Foundation (DEB-1245373 and DMS 1264153), the NIH (GM053275), Yellowstone National Park, and many donors through Yellowstone Forever. This material was also partially supported by the National Science Foundation Graduate Research Fellowship (DGE1656466).
References
- Abraham G, & Inouye M. (2016). Fast principal component analysis of large-scale genome-wide data. PLoS ONE 9(4), e93766. DOI: 10.1371/journal.pone.0093766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemu SW, Bijma P, Moller SH, Janss L, & Berg P. (2014) Indirect genetic effects contribute substantially to heritable variation in aggression-related traits in group-housed mink (Neovison vison). Genetics Selection Evolution 46(1), 30 DOI: 10.1186/1297-9686-46-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali OA, O’Rourke SM, Amish SJ, Meek MH, Luikart G, Jeffres C, & Miller MR (2016). RAD capture (Rapture): Flexible and efficient sequence-based genotyping. Genetics 202, 389–400. DOI: 10.1534/genetics.115.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almberg ES, Mech LD, Smith DW, Sheldon JW, & Crabtree RL (2009). A serological survey of infectious disease in Yellowstone National Park’s canid community. PLoS One 4, e7042 DOI: 10.1371/journal.pone.0007042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amat M, Manteca X, Mariotti VM, de la Torre JLR, & Fatjo J. (2009). Aggressive behavior in the English cocker spaniel. Journal of Veterinary Behavior – Clinical Applications and Research 4, 111–117. DOI: 10.1016/j.veb.2008.08.010 [DOI] [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, Stahler DR, Smith DW, Padhukasahasram B, Randi E, Leonard JA, Bustamante CD, Ostrander EA, Tang H, Wayne RK, & Barsh GS (2009). Molecular and evolutionary history of melanism in North American gray wolves. Science 323, 1339–1343. DOI: 10.1126/science.1165448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt RRH, & Mackay TFC (2012) Genetics of aggression. Annual Review of Genetics 46, 145–164. DOI: 10.1146/annurev-genet-110711-155514 [DOI] [PubMed] [Google Scholar]
- Araya-Ajoy YG, & Dingemanse NJ (2017) Repeatability, heritablity, and age-dependence of seasonal plasticity in aggressiveness in a wild passerine bird. Journal of Animal Ecology 86, 227–238. DOI: 10.1111/1365-2676.12621 [DOI] [PubMed] [Google Scholar]
- Ariyomo TO, Carter M, & Watt PJ (2013). Heritability of boldness and aggressiveness in the zebrafish. Behavioral Genetics 43, 161–167. DOI: 10.1007/s10519-013-9585-y [DOI] [PubMed] [Google Scholar]
- Bannai H, Lévi S, Schweizer C, Inoue T, Launey T, Racine V, Sibarita J-B, & Mikoshiba, Triller A. (2009). Activity-dependent tuning of inhibitory neurotransmission based on GABAAR diffusion dynamics. Neuron 62, 670–682. DOI: 10.1016/j.neuron.2009.04.023 [DOI] [PubMed] [Google Scholar]
- Barel O, Malicdan MCV, Ben-Zeev B, Kandel J. Pri-Chen H, Stephen J, Castro IG, Metz J, Atawa O, Moshkovitz S, Ganelin E, Barshack I, Polak-Charcon S, Nass D, Marek-Yagel D, Amariglio N, Shalva N, Vilboux T, Ferreira C, Pode-Shakked B, Heimer G, Hoffman C, Yardeni T, Nissenkorn A, Avivi C, Eyal E, Kol N, Glick Saar E, Wallace DC, Gahl WA, Rachavi G, Shrader M, Eckmann DM, & Anikster Y. (2017). Deleterious variants in TRAK1 disrupt mitochondrial movement and cause fetal encephalopathy. Brain 140, 568–581. DOI: 10.1093/brain/awx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman L, Almasy L, Blangero J, Duggirala R, Sinsheimer JS, & Lange K. (2005). Fishing for pleiotropic QTLs in a polygenic sea. Annals of Human Genetics 69, 590–611. DOI: 10.1111/j.1529-8817.2005.00181.x [DOI] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 289–300. DOI: http://jstor.org/stable/2346101 [Google Scholar]
- Benjamini Y, & Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 29(4), 1165–1188. DOI: 10.1214/aos/1013699998 [DOI] [Google Scholar]
- Beziers P, Ducrest A-L, Simon C, & Roulin A. (2017). Circulating testosterone and feather-gene expression of receptors and metabolic enzymes in relation to melanin-based colouration in the barn owl. General and Comparative Endocrinology 250, 36–45. DOI: 10.1016/j.ygcen.2017.04.015 [DOI] [PubMed] [Google Scholar]
- Brodkin ES, Goforth SA, Keene AH, Fossella JA, & Silver LM (2002). Identification of quantitative trait loci that affect aggressive behavior in mice. The Journal of Neuroscience 22, 1165–1170. DOI: 10.1523/JNEUROSCI.22-03-01165.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerlink I, Turner SP, Bijma P, & Bolhuis JE (2013) Indirect genetic effects and housing conditions in relation to aggressive behaviours in pigs. PLoS ONE 8(6), e65136. DOI: 10.1371.journal.pone.0065136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, Kerns JA, Schmutz SM, Milhauser GL, & Barsh GS (2007). A β-defensin mutation causes black coat color in domestic dogs. Science 318, 1418–1423. DOI: 10.1126/science.1147880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy KA, MacNulty DR, Stahler DR, Smith DW, & Mech LD (2015). Group composition effects on aggressive interpack interactions of gray wolves in Yellowstone National Park. Behavioral Ecology 26, 1352–1360. DOI: 10.1093/beheco/arcv081 [DOI] [Google Scholar]
- Cassidy KA, Mech LD, MacNulty DR, Stahler DR, & Smith DW (2017). Sexually dimorphic aggression indicates male gray wolves specialize in pack defense against conspecific groups. Behavioral Processes 136, 64–72. DOI: 10.1016/j.beproc.2017.01.011 [DOI] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, & Cresko WA (2013). Stacks: an analysis tool set for population genomics. Molecular Ecology 22, 3124–3140. DOI: 10.1111/mec.12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervet N, Zöttle M, Schürch R, Taborsky M, & Heg D. (2011). Repeatability and heritability of behavioural types in social cichlid. International Journal of Evolutionary Biology 321729, 1–15. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai S, Tau M, & Gobbi G. (2012). The psychopharmacology of aggressive behavior: a translational approach: part 1: neurobiology. Journal of Clinical Psychopharmacology 32, 83–94. DOI: 10.1097/JCP.0b013e31823f8770 [DOI] [PubMed] [Google Scholar]
- Cubaynes S, MacNulty DR, Stahler DR, Quimby KA, Smith DS, & Coulson T. (2014). Density-dependent intraspecific aggression regulates survival in northern Yellowstone wolves (Canis lupus). Journal of Animal Ecology 83(6), 1344–1356. DOI: 10.1111/1365-2656.12238 [DOI] [PubMed] [Google Scholar]
- de Boer S, van der Vegt B, & Koolhaas J. (2003). Individual variation in aggression of feral rodent strains: A standard for the genetics of aggression and violence? Behavior Genetics 33, 485–501. DOI: 10.1023/A:1025766415 [DOI] [PubMed] [Google Scholar]
- DeCandia AL, Gaughran S, Caragiulo A, & Amato G. (2016). A novel molecular method for noninvasive sex identification of order Carnivora. Conservation Genetics Resources 8(2), 119–121. DOI: 10.1007/s12686-016-0525-z [DOI] [Google Scholar]
- Dingemanse NJ, Both C, Drent PJ, van Oers K, & van Noordwijk AJ (2002). Repeatability and heritablity of exploratory behaviour in great tits from the wild. Animal Behaviour 64(6), 929–938. DOI: 10.1098/rspb.2003.2518 [DOI] [Google Scholar]
- Duckworth RA, & Kruuk LEB (2009). Evolution of genetic integration between dispersal and colonization ability in a bird. Evolution 63-4, 968–977. DOI: 10.1111/j.1558-5646.2009.00625.x [DOI] [PubMed] [Google Scholar]
- Ducrest A-L, Keller K, & Roulin A. (2008). Pleiotropy in the melanocortin system, coloration and behavioral syndromes. Trends in Ecology and Evolution 23(9), 502–510. DOI: 10.1016/j.tree.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Eo J, Choi B-H, Jung Y-D, Kwon Y-J, Kim T-H, Seong H-H, & Kim H-S (2013). Polymorphism analysis of tyrosine hydroxylase (TH) in military working dogs. Genes & Genomics 35, 817–821. DOI: 10.1007/s13258-013-0156-7. [DOI] [Google Scholar]
- Emik L, & Terrill CE (1949). Systematic procedures for calculating inbreeding coefficients. Journal of Heredity 40, 51–55. DOI: 10.1093/oxfordjournals.jhered.a105986 [DOI] [PubMed] [Google Scholar]
- Haller J, Abrahám I, Zelena D, Juhász G, Makara GB, & Kruk MR (1998). Aggressive experience affects the sensitivity of neurons towards pharmacological treatment in the hypothalamic attack area. Behaviorial Pharmacology 9, 469–475. DOI: 10.1097/00008877-199809000-00010 [DOI] [PubMed] [Google Scholar]
- Houpt KA, & Willis M. (2001). Genetics of behaviour In: Ruvinsky A, Sampson J(Eds.), The genetics of the dog. CABI Publishing, Wallingford, pp. 371–700. [Google Scholar]
- Huisman J. (2017). Pedigree reconstruction from SNP data: parentage assignment, sibship clustering and beyond. Molecualr Ecology Resources 17(5), 1009–1024. DOI: 10.1111/1755-0998.12665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukekova AV, Trut LN, Chase K, Kharlamova AV, Johnson JL, Temnykh S, Oskina IN, Gulevich RG, Vladimirova AV, Klebanov S, Shepeleva DV, Shikhevich SG, Acland GM, & Lark KG (2011). Mapping loci for fox domestication: deconstruction/reconstruction of a behavioral phenotype. Behavioral Genetics 41(4), 593–606. DOI: 10.1007/s10519-010-9418-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Westlake J, & Spence MA (1976). Extensions to pedigree analysis. III. Variance components by the scoring method. Annals of Human Genetics 39,485–491. DOI: 10.1111/j.1469-1809.1976.tb00156.x [DOI] [PubMed] [Google Scholar]
- Lange K, & Boehnke M. (l983). Extensions to pedigree analysis. IV. Covariance components models for multivariate traits. American Journal of Medical Genetics 14, 513–524. DOI: 10.1002/ajmg.1320140315 [DOI] [PubMed] [Google Scholar]
- Lange K. (2002). Mathematical and statistical methdos for genetic analysis. 2nd Edition Springer, New York, New York. [Google Scholar]
- Lange K, Papp JC, Sinsheimer JS, Sripracha R, Zhou H, & Sobel EM (2013). Mendel: The Swiss army knife of genetic analysis programs. Bioinformatics 29, 1568–1570. DOI: 10.1093/bioinformatics/btt187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Neindre P, Trillat G, Sapa J, Menissier F, Bonnet JN, & Chupin JM (1995). Individual differences in docility in limousin cattle. Journal of Animal Science 73, 2249–2253. DOI: 10.2527/1995.7382249x [DOI] [PubMed] [Google Scholar]
- Le Neindre. P., Boivin X, & Boissy A. (1996). Handling of extensively kept animals. Applied Animal Behaviour Science 49, 73–81. DOI: 10.1016/0168-1591(95)00669-9 [DOI] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, & 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16), 2078–2079. DOI: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O’Neill B, O’Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, & Lander ES (2005). Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438(7069), 803–819. DOI: 10.1038/nature04338 [DOI] [PubMed] [Google Scholar]
- Lovedahl P, Damgaard LH, Nielsen BL, Thodberg K, Su G, & Rydhmer L. (2005). Aggressive behaviour of sows at mixing and maternal behaviour are heritable and genetically correlated traits. Livestock Science 93(1), 73–85. DOI: 10.1016/j.livprodsci.2004.11.008 [DOI] [Google Scholar]
- Lunter G, & Goodson M. (2011). Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Research 21(6), 936–939. DOI: 10.1101/gr.111120.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean EL, Snyder-Mackler N, vonHoldt BM, & Serpell JA (2019). Highly heritable and fucntionally relevant breed differences in dog behaviour. Proceedings of the Royal Society B 286, 20190716. DOI: 10.1098/rspb.2019.0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CR, & Lott DF (2000). A review of ecolgoical determinants of territoriality within vertebrate species. The American Midland Naturalist 143(1), 1–29. DOI: 10.1674/0003-0031(2000)143[0001:AROEDO]2.0.CO;2 [DOI] [Google Scholar]
- Malécot G. (1948). Les Mathématiques de l’Hérédité. Masson et Cie, Paris. [Google Scholar]
- Mandel P, Ciesielski L,, Maitre M, Simler S, Kempf E, & Mack G. (1981). Inhibitory amino acids, aggresiveness, and convulsions. Advances in Biochemical Psychopharmacology 29, 1–9. [PubMed] [Google Scholar]
- Maruki T, & Lynch M. (2017). Genotype calling from population-genomic sequencing data. G3 7, 1393–1404. DOI: 10.1534/g3.117.039008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HD, Ritchie GR, Thormann A, Flicek P, Cunningham F. (2016). The Ensembl variant effect predictor. Genome Biology 17(1), 122 DOI: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mech LD, & Boitani L. (2003). Wolves: Behavior, Ecology, and Conservation: Behavior, Ecology, and Conservation University of Chicago Press. [Google Scholar]
- Mech LD, & Barber-Meyer S (2015). Yellowstone wolf (Canis lupus) density predicted by elk (Cervus elaphus) biomass. Canadian Journal of Zoology 93, 499–502. DOI: 10.1139/cjz-2015-0002 [DOI] [Google Scholar]
- Milligan BG (2003). Maximum-likelihood estimation of relatedness. Genetics 163, 1153–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa T, Leiter LM, Gerber DJ, Gainetdinov RR, Sotnikova TD, Zeng H, Caron MG, & Tonegawa S. (2003). Conditional calcineurin knockout mice exhibit multiple abnormal behaviors related to schizophrenia. Proceedings of the National Academy of Sciences of the United States of America 100(15), 8987–8992. DOI: 10.1073/pnas.1432926100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CA, Cullen NG, Kilgour R, & Bremner KJ (1994). Some genetic factors affecting temperament in Bos taurus cattle. New Zealand Journal of Agricultural Research 37, 167–175. DOI: 10.1080/00288233.1994.9513054 [DOI] [Google Scholar]
- Nelson RJ, & Trainor BC (2007). Neural mechanisms of aggression. Nature Reviews Neuroscience 8, 536–546. DOI: 10.1038/nrn2174 [DOI] [PubMed] [Google Scholar]
- O’Connor E, Topf A, Muller JS, Cox D, Evangelista T, Colomer J. Abicht A, Senderek J, Hasselmann O, Yaramix A, Laval SH, & Lochmueller H. (2016). Identification of mutations in the MYO9A gene in patients with congentical myasthenic syndrome. Brain 139, 2143–2153. DOI: DOI: 10.1093/brain/aww130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet PC, & Carbyn LN (2003). Gray wolf (Canis lupus) and allies In Wild Mammals of North America: Biology, Management, and Conservation. Second Edition, Feldhamer GA, Thompson BC and Chapman JA (eds). pp. 482–510. Baltimore: Johns Hopkins University Press. [Google Scholar]
- Pavlov KA, Chistiakov DA, & Chekhonin VP (2012). Genetic determinants of aggression and impulsivity in humans. Journal of Applied Genetics 53, 61–82. DOI: 10.1007/s13353-011-0069-6 [DOI] [PubMed] [Google Scholar]
- Pew J, Muir PH, Wang J, & Frasier TR (2015). related: an R package for analysing pairwise relatedness from codominant molecular markers. Molecular Ecology Resources 15(3), 557–561. DOI: 10.1111/1755-0998.12323 [DOI] [PubMed] [Google Scholar]
- Pérez-Guisado J, Lopoz-Rodríguez R, & Muñoz-Serrano A. (2006). Heritablity of dominant-aggressive behaviour in English Cocker Spaniels. Applied Animal Behaviour Science 100, 219–227. DOI: 10.1016/j.applanim.2005.11.005 [DOI] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, & Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81, 559–575. DOI: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proskura WS, Frost A, Gugała L, Dybus A, Grzesiak W, Wawrzyniak J, & Uchman S. (2013). Genetic background of aggressive behaviour in dogs. Acta Veterinaria Brno 82, 441–445. DOI: 10.2754/avb201382040441 [DOI] [Google Scholar]
- Raudvere U, Kohberg L, Kuzmin I, Arak T, Adler P, Peterson H, & Vilo J. (2019). g:Profiler: a web server for functional enrichment anlaysis and conversions of gene lists. Nucleic Acids Research 47(W1), W191–W198. DOI: 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réale D, Reader SM, Sol D, McDougall PT, & Dingemanse NJ (2007). Integrating animal temperament within ecology and evolution. Biological Reviews of the Cambridge Philosophical Society 82, 291–318. DOI: 10.1111/j.1469-185X.2007.00010.x [DOI] [PubMed] [Google Scholar]
- Rochette NC, Rivera-Colón AG, & Catchen JM (2019). Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. bioRxiv 615385. DOI: 10.1101/615385 [DOI] [PubMed] [Google Scholar]
- Roulin A, & Ducrest A-L (2011). Association between melanism, physiology and beahvior: A role for the melanocortin system. European Journal of Pharmacology 660, 226–233. DOI: 10.1016/j.ejphar.2011.01.036 [DOI] [PubMed] [Google Scholar]
- Sands J, & Creel S. (2004) Social dominance, aggression and faecal glucocoticoid levels in a wild population of wolves, Canis lupus. Animal Behaviour 67, 387–396. DOI: 10.1016/j.anbehav.2003.03.019 [DOI] [Google Scholar]
- Smith DW, Metz M, Cassidy KA, Stahler EE, McIntyre RT, Almberg ES, Stahler DR (2015) Infanticide in wolves: seasonality of mortalities and attacks at dens support evolution of territoriality. Journal of Mammalogy 96, 1174–1183. DOI: 10.1093/jmammal/gyv125 [DOI] [Google Scholar]
- Smith D, Stahler D, Cassidy K, Stahler E, Metz M, Cassidy B, Koitzsch L, Cato L, Meyer C, Loggers E, Rabe J, Tatton N, Thomas-Kuzilik R, & Koitzsch K. (2019). Yellowstone National Park Wolf Project Annual Report 2018. National Park Service, Yellowstone Center for Resources, Yellowstone National Park, WY, USA, YCR-2019–05. [Google Scholar]
- Stahler DR, MacNulty DR, Wayne RK, vonHoldt B, & Smith DW (2013). The adaptive value of morphological, behavioural and life-history traits in reproductive female wolves. Journal of Animal Ecology 82, 222–234. DOI: 10.1111/j.1365-2656.2012.02039.x [DOI] [PubMed] [Google Scholar]
- Takahashi A, & Miczek KA (2014). Neurogenetics of aggressive behavior – Studies in rodents. Current Topics in Beahvioral Neuroscience 17, 3–44. DOI: 10.1007/7854_2013_263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Boon AK, Dantzer B, Réale D. Humphries MM, Boutin S, Gorrell JC, & Coltman DW, & McAdam AG. (2012). Low heritabilities, but genetic and maternal correlations between red squirrel behaviours. Journal of Evolutionary Biology 25, 614–624. DOI: 10.1111/j.1420-9101.2012.02456.x [DOI] [PubMed] [Google Scholar]
- Trut LN (1980). The genetics and phenogenetics of domestic behaviour. Problems in General Genetics (Proceeding of the XIV International Congress of Genetics); Moscow: Mir Publishers, p123–136. [Google Scholar]
- Vage J, Wade C, Biagi T, Fatjó J, Amat M, Lindblad-Toh K, & Lingaas F. (2010). Association of dopamine- and serotonin-related genes with canine aggression. Genes, Brain, and Behavior 9, 372–378. DOI: 10.1111/j.1601-183X.2010.00568.x [DOI] [PubMed] [Google Scholar]
- VanRaden PM (2008). Efficient methods to compute genomic predictions. J. Dairy Sci 91: 4414–4423. DOI: 10.3169/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Stahler DR, Smith DW, Earl DA, Pollinger JP, & Wayne RK (2008). The genealogy and genetic viability of reintroduced Yellowstone grey wolves. Molecular Ecology 17, 252–274. DOI: 10.1111/j.1365-294X.2007.03468.x [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Stahler DR, Bangs EE, Smith DW, Jimenez MD, Mack CM, Niemeyer CC, Pollinger JP, & Wayne RK (2010). A novel assessment of population structure and gene flow in grey wolf populations of the Northern Rocky Mountains of the United States. Molecular Ecology 19(20), 4412–4427. DOI: 10.1111/j.1365-294X.2010.04769.x [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Shuldiner E, Janowitz Koch I, Kartzinel RY, Hogan A, Brubaker L, Wanser S, Stahler D, Wynne CDL, Ostrander EA, Sinsheimer JS, & Udell MAR (2017). Structural variants in genes associated with human Williams-Beuren syndrome underlie stereotypical hypersociablity in domestic dogs. Science Advances 3(7), e1700398. DOI: 10.1126/sciadv.1700398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. (2002). An estimator for pairwise relatedness using molecular markers. Genetics 160(3), 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. (2011). Coancestry: A program for simulating, estimating and analyzing relatedness and inbreeding coefficients. Molecular Ecology Resources 11(1), 141–145. DOI: 10.1111/j.1755-0998.2010.02885.x [DOI] [PubMed] [Google Scholar]
- Wang B, Sverdlov S, & Thompson E. (2017). Efficient Estimation of Realized Kinship from Single Nucleotide Polymorphism Genotypes. Genetics 205, 1063–1078. doi: 10.1534/genetics.116.197004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AJ, Réale D, Clements MN, Morrissey MM, Postma E, Walling CA, Kruuk LE, & Nussey DH (2010). An ecologist’s guide to the animal model. Journal of Animal Ecology 79, 13–26. DOI: 10.1111/j.1365-2656.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- Wolf M, van Doorn GS, Leimar O, & Weissin FJ (2007). Life-history trade-offs favour the evolution of animal personalities. Nature 447, 581–584. DOI: 10.1038/nature05835 [DOI] [PubMed] [Google Scholar]
- Zaitlen N, Kraft P, Patterson N, Pasaniuc B, Bhatia G, Pollack S, & Price AL (2013). Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genetics 9, e1003520. DOI: 10.1371/journal.pgen.1003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Hu T, Qiao D, Cho MH, & Zhou H. (2016). Boosting gene mapping power and efficiency with efficient exact variance component tests of single nucleotide polymorphism sets. Genetics 204, 921–931. DOI: 10.1534/genetics.116.190454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Blangero J, Dyer TD, Chan KK, Lange K, & Sobel EM (2017). Fast genome-wide QTL association mapping on pedigree and population data. Genetic Epidemiology 41(3), 174–186. DOI: 10.1002/gepi.21988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sinsheimer JS, Bates DM, Chu BB, German CA, Ji SS, Keys KL, Kim J, Ko S, Mosher GD, Papp JC, Sobel EM, Zhai J, Zhou JJ, & Lange K. (2019). OPENMENDEL: a cooperative programming project for statistical genetics. Human Genetics DOI: 10.1007/s00439-019-02001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. & Stephens M. (2014). Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nature Methods 11, 407–709. DOI: 10.1038/nmeth.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mapped bam files for these 413 wolves are available on NCBI’s public Sequence Read Archive (PRJNA577957). Additional meta-data for each individual wolf can be found in Supplemental Table S1.


