Abstract
SARS-CoV-2, a member of the coronavirus family, is responsible for the current COVID-19 worldwide pandemic. We previously demonstrated that five nucleotide analogues inhibit the SARS-CoV-2 RNA-dependent RNA polymerase (RdRp), including the active triphosphate forms of Sofosbuvir, Alovudine, Zidovudine, Tenofovir alafenamide and Emtricitabine. We report here the evaluation of a library of nucleoside triphosphate analogues with a variety of structural and chemical features as inhibitors of the RdRps of SARS-CoV and SARS-CoV-2. These features include modifications on the sugar (2′ or 3′ modifications, carbocyclic, acyclic, or dideoxynucleotides) or on the base. The goal is to identify nucleotide analogues that not only terminate RNA synthesis catalyzed by these coronavirus RdRps, but also have the potential to resist the viruses' exonuclease activity. We examined these nucleotide analogues for their ability to be incorporated by the RdRps in the polymerase reaction and to prevent further incorporation. While all 11 molecules tested displayed incorporation, 6 exhibited immediate termination of the polymerase reaction (triphosphates of Carbovir, Ganciclovir, Stavudine and Entecavir; 3′-OMe-UTP and Biotin-16-dUTP), 2 showed delayed termination (Cidofovir diphosphate and 2′-OMe-UTP), and 3 did not terminate the polymerase reaction (2′-F-dUTP, 2′–NH2–dUTP and Desthiobiotin-16-UTP). The coronaviruses possess an exonuclease that apparently requires a 2′-OH at the 3′-terminus of the growing RNA strand for proofreading. In this study, all nucleoside triphosphate analogues evaluated form Watson-Crick-like base pairs. The nucleotide analogues demonstrating termination either lack a 2′-OH, have a blocked 2′-OH, or show delayed termination. Thus, these nucleotide analogues are of interest for further investigation to evaluate whether they can evade the viral exonuclease activity. Prodrugs of five of these nucleotide analogues (Cidofovir, Abacavir, Valganciclovir/Ganciclovir, Stavudine and Entecavir) are FDA-approved medications for treatment of other viral infections, and their safety profiles are well established. After demonstrating potency in inhibiting viral replication in cell culture, candidate molecules can be rapidly evaluated as potential therapies for COVID-19.
Keywords: COVID-19, SARS-CoV-2, RNA-Dependent RNA polymerase, Nucleotide analogues, Exonuclease
Highlights
-
•
Cidofovir triphosphate is a delayed terminator for SARS-CoV-2 RNA polymerase.
-
•
Abacavir, Ganciclovir, and Stavudine triphosphates inhibit SARS-CoV-2 polymerase.
-
•
2′-O-methylated UTP substantially terminates the SARS-CoV-2 polymerase reaction.
1. Introduction
The COVID-19 pandemic, caused by SARS-CoV-2, continues to have a devastating global impact. SARS-CoV-2 is a member of the Orthocoronavirinae subfamily (Zhu et al., 2020). Coronaviruses, HCV and the flaviviruses are all positive-sense single-strand RNA viruses that replicate their genomes using an RNA-dependent RNA polymerase (RdRp) (Zumla et al., 2016; Dustin et al., 2016).
Currently, there are no FDA-approved antiviral drugs for the treatment of human coronavirus infections, including COVID-19. The RdRp of coronaviruses is a well-established drug target; the active site of the RdRp is highly conserved among positive-sense RNA viruses (te Velthuis, 2014). These RdRps have low fidelity (Selisko et al., 2018), allowing them to recognize a variety of modified nucleotide analogues as substrates. Such nucleotide analogues may inhibit further RNA-polymerase catalyzed RNA replication making them important candidate anti-viral agents (McKenna et al., 1989; Öberg, 2006; Eltahla et al., 2015; De Clercq and Li, 2016). RdRps in SARS-CoV and SARS-CoV-2 have nearly identical sequences (Ju et al., 2020; Elfiky, 2020). Recently, the SARS-CoV-2 RdRp was cloned (Chien et al., 2020) [not peer-reviewed] and the RNA polymerase complex structure was determined (Gao et al., 2020), which will help guide the design and study of RdRp inhibitors.
Remdesivir, a phosphoramidate prodrug containing a 1′-cyano modification on the sugar, is converted in cells into an adenosine triphosphate analogue, which was shown to be an inhibitor of the RdRps of SARS-CoV and SARS-CoV-2 (Gordon et al., 2020a, Gordon et al., 2020b). It is currently in clinical trials in several countries as a therapeutic for COVID-19 infections and was recently approved for emergency use by the FDA. Remdesivir triphosphate was shown to be incorporated with higher efficiency than ATP by coronavirus RdRps, leading to delayed termination of RNA synthesis, thereby overcoming excision by the viral exonuclease (Gordon et al., 2020a, Gordon et al., 2020b). β-d-N4-hydroxycytidine is another prodrug targeting the coronavirus polymerase and was shown to have broad spectrum activity against coronaviruses, even in the presence of intact proofreading functions (Agostini et al., 2019; Sheahan et al., 2020).
1.1. Selection of candidate nucleoside triphosphates as inhibitors of the coronavirus RdRps
We previously demonstrated that five nucleotide analogues inhibit the SARS-CoV-2 RdRp, including the active triphosphates of Sofosbuvir, Alovudine, Zidovudine, Tenofovir alafenamide and Emtricitabine (Ju et al., 2020; Chien et al., 2020; Jockusch et al., 2020) [not peer-reviewed]. Emtricitabine and Tenofovir alafenamide are used in FDA-approved combination regimens for treatment of HIV/AIDS infections and as pre-exposure prophylaxis (PrEP) to prevent HIV infections (Anderson et al., 2011).
The fact that each of the previous five nucleotide analogues exhibited inhibition of the coronavirus polymerases indicates that the SARS-CoV-2 RdRp can accept a variety of nucleotide analogues as substrates. Here we evaluate additional nucleotide analogues with a larger variety of modifications to identify those with more efficient termination; we also consider the chemical or structural properties of these molecules that may help overcome the virus' proofreading function. These nucleotide analogues were selected based on one or more of the following criteria. First, they have structural and chemical properties such as (a) similarity in size and structure to natural nucleotides, including the ability to fit within the active site of the polymerase, (b) presence of a small 3′-OH capping group or absence of a 3′-OH group resulting in obligate termination of the polymerase reaction; or (c) modifications at the 2′ or other positions on the sugar or base that can potentially lead to delayed termination. We previously showed that nucleotides with substantial modifications on the base can be incorporated by DNA polymerases (Ju et al., 2006). The criteria above will provide structural and chemical features that we can explore to evade viral exonuclease activity (Minskaia et al., 2006). Second, if they have previously been shown to inhibit the polymerases of other viruses, even those with different polymerase types, they may have the potential to inhibit the SARS-CoV-2 RdRp, as we have previously shown for HIV reverse transcriptase (RT) inhibitors (Ju et al., 2020; Chien et al., 2020; Jockusch et al., 2020). Third, ideally, the inhibitors should display high selectivity for viral polymerases relative to cellular DNA or RNA polymerases. Fourth, there is an advantage in considering nucleotide analogues that are the active triphosphate forms of FDA-approved drugs, as these drugs are known to have acceptable levels of toxicity and are more likely to be tolerated by patients with coronavirus infections, including COVID-19.
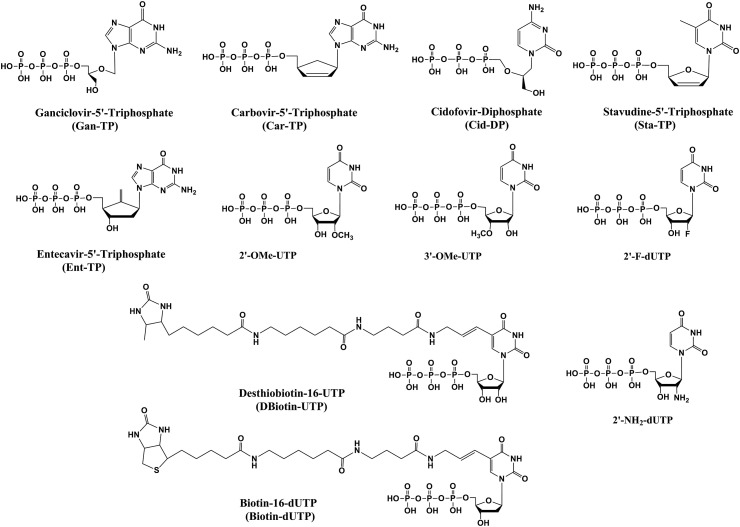
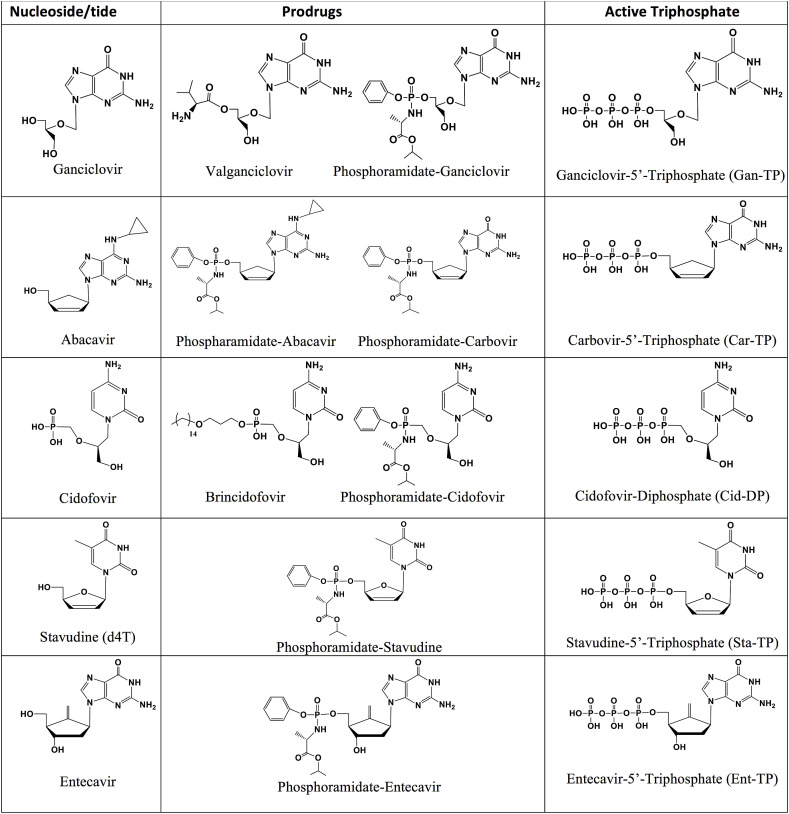
Using the criteria above, our study examines 11 nucleotide analogues with sugar or base modifications (structures shown in Fig. 1 ) for their ability to inhibit the SARS-CoV-2 or SARS-CoV RdRps: Ganciclovir 5′-triphosphate, Carbovir 5′-triphosphate, Cidofovir diphosphate, Stavudine 5′-triphosphate, Entecavir 5′-triphosphate, 2′-O-methyluridine-5′-triphosphate (2′-OMe-UTP), 3′-O-methyluridine-5′-triphosphate (3′-OMe-UTP), 2′-fluoro-2′-deoxyuridine-5′-triphosphate (2′-F-dUTP), desthiobiotin-16-aminoallyl-uridine-5′-triphosphate (Desthiobiotin-16-UTP), biotin-16-aminoallyl-2′-deoxyuridine-5′-triphosphate (Biotin-16-dUTP) and 2′-amino-2′-deoxyuridine-5′-triphosphate (2′–NH2–dUTP). The nucleoside and prodrug forms for the FDA-approved drugs are shown in Fig. 2 ; nucleoside and potential prodrug forms for three other nucleotide analogues are shown in Fig. S1.
Fig. 1.
Chemical structures of nucleoside triphosphate analogues used in this study.
Fig. 2.
Structures of viral nucleoside/nucleotide inhibitors, example prodrugs and active triphosphate forms. The compounds Ganciclovir, Abacavir, Cidofovir, Stavudine and Entecavir (left), example prodrug forms (middle) and their active triphosphate forms (right).
Some of the uridine analogues listed above have been previously shown to be substrates of viral polymerases (Arup et al., 1992; Lauridsen et al., 2012). The 2′-O-methyluridine triphosphate is of particular interest since 2′-O-methyl nucleotides can resist removal by the 3′-exonuclease found in coronaviruses (Minskaia et al., 2006). We describe the properties of 5 nucleotide analogues whose prodrug forms are FDA-approved for other virus infections as follows.
Ganciclovir triphosphate (Gan-TP) is an acyclic guanosine nucleotide (Fig. 1). The parent nucleoside Ganciclovir (Cytovene, Fig. 2) is used to treat AIDS-related cytomegalovirus (CMV) infections. The drug can inhibit herpesviruses and varicella zoster virus. The valyl ester prodrug Valganciclovir (Fig. 2) can be given orally. After cleavage of the valyl ester, Ganciclovir is converted to Ganciclovir triphosphate by viral and cellular enzymes to inhibit the viral polymerase (Matthews and Boehme, 1988; Akyürek et al., 2001).
Carbovir triphosphate (Car-TP) is a carbocyclic guanosine didehydro-dideoxynucleotide (Fig. 1). The parent prodrug, Abacavir (Ziagen, Fig. 2), is an FDA-approved nucleoside RT inhibitor for HIV/AIDS treatment (Faletto et al., 1997; Ray et al., 2002). It is taken orally and is well tolerated.
Cidofovir diphosphate (Cid-DP) is an acyclic cytidine nucleotide (Fig. 1). Its prodrug form Cidofovir (Vistide, Fig. 2) is an FDA-approved intravenous drug for the treatment of AIDS-related CMV retinitis and has been used off-label for a variety of DNA virus infections (De Clercq, 2002; Lanier et al., 2010). A second prodrug form of Cidofovir diphosphate, Brincidofovir (Fig. 2), is an oral antiviral drug with a lipid moiety masking the phosphate group and a candidate for treating smallpox infections. It is active against a wide range of DNA viruses in animals, including poxviruses, adenoviruses, herpesviruses and CMV (Trost et al., 2015; Cundy. 1999). Both Brincidofovir and a ProTide-based prodrug (Fig. 2) are expected to enter cells rapidly. Interestingly, although Cidofovir is incorporated into DNA in the polymerase reaction by vaccinia virus DNA polymerase, the termination of synthesis occurs after extension by an additional nucleotide, a delayed termination similar to that shown for Remdesivir for coronavirus RdRp; Cidofovir incorporated in the penultimate position of the DNA extension strand by the vaccinia virus polymerase is not removed by the viral 3′-exonuclease (Magee et al., 2005).
Stavudine triphosphate (Sta-TP, Fig. 1), a thymidine analogue, is the active triphosphate form of Stavudine (d4T, Zerit, Fig. 2), an antiviral used for the prevention and treatment of HIV/AIDS (Ho and Hitchcock, 1989) via inhibition of the HIV RT (Huang et al., 1992). The lack of a 3′-OH group makes it an obligate inhibitor.
Entecavir triphosphate (Ent-TP, Fig. 1), the active triphosphate form of the oral drug Entecavir (Baraclude, Fig. 2), is a guanosine nucleotide inhibitor of the hepatitis B virus polymerase (Matthews, 2006; Rivkina and Rybalow, 2002). It shows little if any inhibition of nuclear and mitochondrial DNA polymerases (Mazzucco et al., 2008) and has generally been shown to have low toxicity. Entecavir triphosphate is a delayed chain terminator of the HIV-1 reverse transcriptase, making it resistant to phosphorolytic excision (Tchesnokov et al., 2008).
We reasoned that once these nucleotide analogues are incorporated into a RNA primer in the polymerase reaction, the fact that they lack either a normal sugar ring configuration or the 2′- and/or 3′-OH groups would make them unlikely candidates for removal by the 3′-exonuclease involved in SARS-CoV-2 proofreading.
1.2. Coronaviruses have a proofreading exonuclease activity that must be overcome to develop effective SARS-CoV-2 RdRp nucleotide inhibitors
In contrast to many other RNA viruses, SARS-CoV and SARS-CoV-2 have very large genomes that encode a 3′-5′ exonuclease (nsp14) involved in proofreading (Ma et al., 2015; Shannon et al., 2020), the activity of which is enhanced by the cofactor nsp10 (Bouvet et al., 2012). This proofreading function increases replication fidelity by removing mismatched nucleotides (Ferron et al., 2018). Mutations in nsp14 led to reduced replication fidelity of the viral genome (Eckerle et al., 2010). Interestingly, while the nsp14/nsp10 complex efficiently excises single mismatched nucleotides at the 3′ end of the RNA chain, it is not able to remove longer stretches of unpaired nucleotides or 3′ modified RNA (Bouvet et al., 2012). For the nucleotide analogues to be successful inhibitors of the RdRps of these viruses, they need to overcome this proofreading function. The coronavirus exonuclease activity typically requires a 2′-OH group at the 3′ end of the growing RNA strand (Minskaia et al., 2006). However, in instances of delayed termination in which the offending nucleotide analogue is no longer at the 3′ end, they will also not be removed by the exonuclease (Bouvet et al., 2012; Gordon et al., 2020b, Gordon et al., 2020a). Nearly all the nucleotide analogues we selected lack the 2′-OH group, have modifications that block the 2′-OH group on the sugar, or are acyclic nucleotide derivatives. Such nucleotides will not likely be substrates for viral exonucleases.
2. Materials and Methods
2.1. Materials
Nucleoside triphosphates and nucleoside triphosphate analogues were purchased from TriLink BioTechnologies (Biotin-16-dUTP, Desthiobiotin-16-UTP, 2′-OMe-UTP, 3′-OMe-UTP, 2′-F-dUTP, 2′–NH2–dUTP, Cidofovir-DP, Ganciclovir-TP, dUTP, CTP, ATP and UTP), Santa Cruz Biotechnology (Stavudine-TP, Carbovir-TP), or Moravek, Inc. (Entecavir-TP). Oligonucleotides were purchased from Integrated DNA Technologies, Inc.
2.2. Extension reactions with SARS-CoV-2 RNA-dependent RNA polymerase
The primer and template (sequences shown in Fig. 3, Fig. 4, Fig. 5, S2-S9) were annealed by heating to 70 °C for 10 min and cooling to room temperature in 1x reaction buffer. The RNA polymerase mixture consisting of 6 μM nsp12 and 18 μM each of cofactors nsp7 and nsp8 (Chien et al., 2020) was incubated for 15 min at room temperature in a 1:3:3 ratio in 1x reaction buffer. Then 5 μl of the annealed template primer solution containing 2 μM template and 1.7 μM primer in 1x reaction buffer was added to 10 μl of the RNA polymerase mixture and incubated for an additional 10 min at room temperature. Finally 5 μl of a solution containing 2 mM 2′-OMe-UTP (Fig. 3 a), 2 mM Sta-TP (Fig. 3b), 2 mM Biotin-dUTP (Fig. 3c), 2 mM Cid-DP + 2 mM UTP +2 mM ATP (Fig. 4 ), 2 mM Car-TP + 2 mM UTP +2 mM ATP +2 mM CTP (Fig. 5 a), 2 mM Ent-TP + 2 mM UTP +2 mM ATP +2 mM CTP (Fig. 5b), 2 mM Gan-TP + 2 mM UTP +2 mM ATP +2 mM CTP (Fig. 5c), 0.2 mM Sta-TP (Fig. S2a), 0.2 mM Cid-DP + 0.4 mM UTP +0.4 mM ATP (Fig. S2b), 2 mM desthiobiotin-16-UTP + 2 mM ATP (Fig. S7), 2 mM 2′-OMe-UTP + 2 mM dUTP (Fig. S8), 1 mM UTP, 1 mM Biotin-dUTP and 1 mM dUTP (Fig. S9a), 1 mM 2′-F-dUTP, 1 mM 2′-OMe-UTP and 1 mM dUTP (Fig. S9b), or 1 mM 2′–NH2–dUTP, 1 mM 2′-OMe-UTP and 1 mM dUTP (Fig. S9c) in 1x reaction buffer was added and incubation was carried out for 2 h at 30 °C. The final concentrations of reagents in the 20 μl extension reactions were 3 μM nsp12, 9 μM nsp7, 9 μM nsp8, 425 nM RNA primer, 500 nM RNA template, 500 μM 2′-OMe-UTP (Fig. 3a), 500 μM Sta-TP (Fig. 3b), 500 μM Biotin-dUTP (Fig. 3c), 500 μM Cid-DP, 500 μM UTP and 500 μM ATP (Fig. 4), 500 μM Car-TP, 500 μM UTP, 500 μM ATP and 500 μM CTP (Fig. 5a), 500 μM Ent-TP + 500 μM UTP +500 μM ATP +500 μM CTP (Fig. 5b), 500 μM Gan-TP, 500 μM UTP, 500 μM ATP and 500 μM CTP (Fig. 5c), 50 μM Sta-TP (Fig. S2a), 50 μM Cid-DP + 100 μM UTP +100 μM ATP (Fig. S2b), 500 μM desthiobiotin-16-UTP + 500 μM ATP (Fig. S7), 500 μM 2′-OMe-UTP + 500 μM dUTP (Fig. S8), 250 μM UTP, 250 μM Biotin-dUTP and 250 μM dUTP (Fig. S9a), 250 μM 2′-F-dUTP, 250 μM 2′-OMe-UTP and 250 μM dUTP (Fig. S9b), and 250 μM 2′–NH2–dUTP, 250 μM 2′-OMe-UTP and 250 μM dUTP (Fig. S9c). The 1x reaction buffer contains the following reagents: 10 mM Tris-HCl pH 8, 10 mM KCl, 2 mM MgCl2 and 1 mM β-mercaptoethanol. Following desalting using an Oligo Clean & Concentrator (Zymo Research), the samples were subjected to MALDI-TOF-MS (Bruker ultrafleXtreme) analysis.
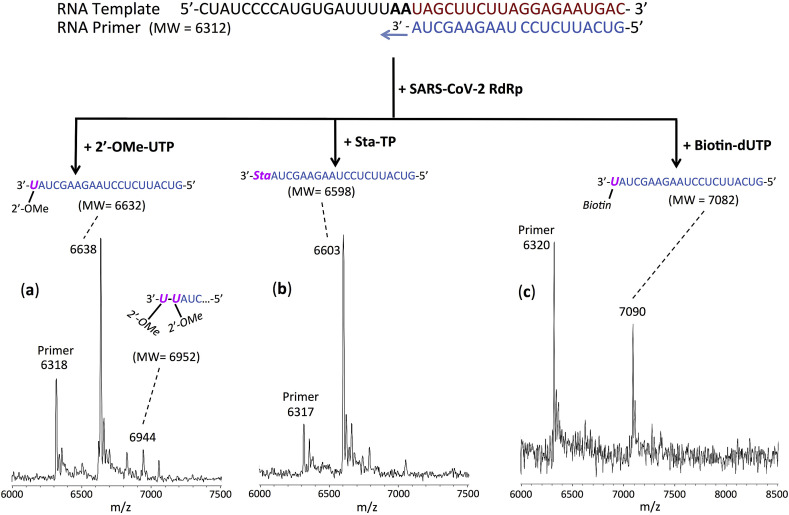
Fig. 3.
Incorporation of 2′-OMe-UTP, Sta-TP and Biotin-dUTP by SARS-CoV-2 RdRp to terminate the polymerase reaction. The sequences of the primer and template used for this extension reaction, which are at the 3′ end of the SARS-CoV-2 genome, are shown at the top of the figure. Polymerase extension reactions were performed by incubating 2′-OMe-UTP (a), Sta-TP (b) and Biotin-dUTP (c) with pre-assembled SARS-CoV-2 polymerase (nsp12, nsp7 and nsp8), the indicated RNA template and primer and the appropriate reaction buffer, followed by detection of reaction products by MALDI-TOF MS. The detailed procedure is shown in the Materials and Methods section. The accuracy for m/z determination is ±10 Da.
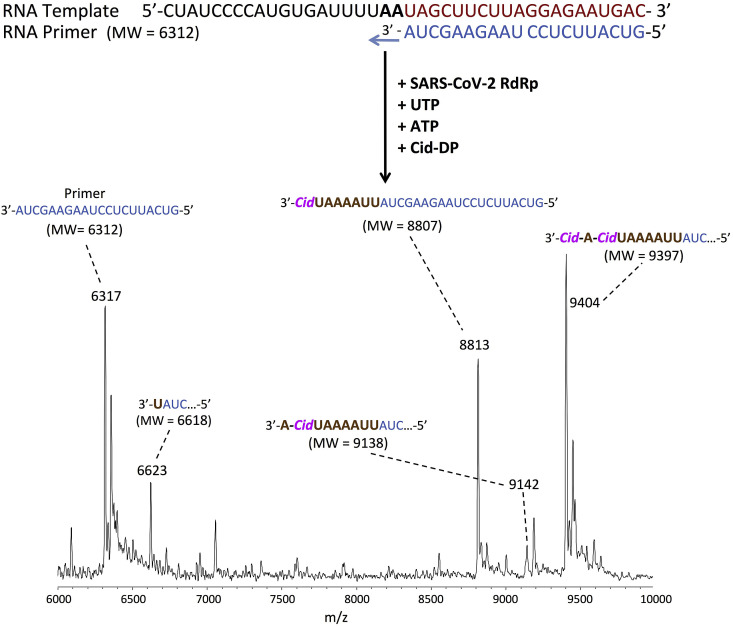
Fig. 4.
Incorporation of Cid-DP by SARS-CoV-2 RdRp to achieve delayed termination of the polymerase reaction. The sequences of the primer and template used for this extension reaction are shown at the top of the figure. The polymerase extension reaction was performed by incubating Cid-DP, UTP and ATP with pre-assembled SARS-CoV-2 polymerase (nsp12, nsp7 and nsp8), the indicated RNA template and primer and the appropriate reaction buffer, followed by detection of reaction products by MALDI-TOF MS. The accuracy for m/z determination is ±10 Da.
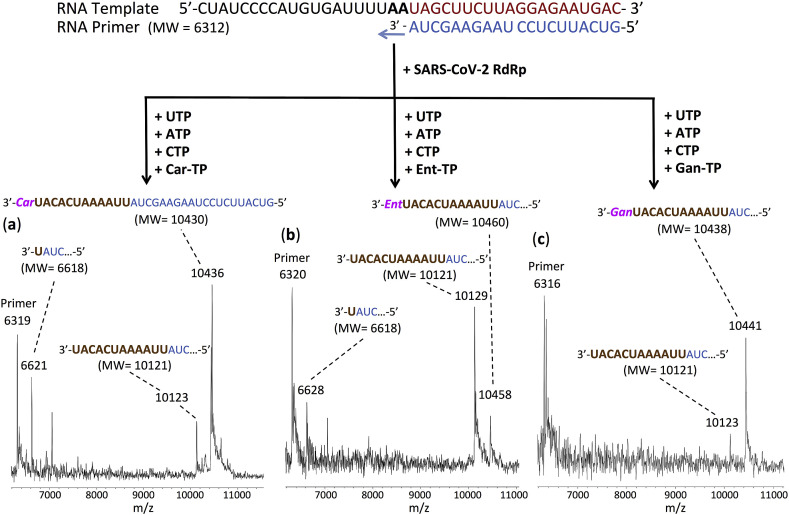
Fig. 5.
Incorporation of Car-TP, Ent-TP and Gan-TP by SARS-CoV-2 RdRp to terminate the polymerase reaction. The sequences of the primer and template used for this extension reaction are shown at the top of the figure. Polymerase extension reactions were performed by incubating Car-TP, UTP, ATP and CTP (a), Ent-TP, UTP, ATP and CTP (b), and Gan-TP, UTP, ATP and CTP (c) with pre-assembled SARS-CoV-2 polymerase (nsp12, nsp7 and nsp8), the indicated RNA template and primer and the appropriate reaction buffer, followed by detection of reaction products by MALDI-TOF MS. The accuracy for m/z determination is ±10 Da.
2.3. Extension reactions with SARS-CoV RNA-dependent RNA polymerase
The primer and template above were annealed by heating to 70 °C for 10 min and cooling to room temperature in 1x reaction buffer (described above). The RNA polymerase mixture consisting of 6 μM nsp12 and 18 μM each of cofactors nsp7 and nsp8 (Kirchdoerfer and Ward, 2019) was incubated for 15 min at room temperature in a 1:3:3 ratio in 1x reaction buffer. Then 5 μl of the annealed template primer solution containing 2 μM template and 1.7 μM primer in 1x reaction buffer was added to 10 μl of the RNA polymerase mixture and incubated for an additional 10 min at room temperature. Finally 5 μl of a solution containing 2 mM Cid-DP + 0.8 mM UTP +0.8 mM ATP (Fig. S3), 2 mM Car-TP + 0.8 mM UTP +0.8 mM ATP +0.8 mM CTP (Fig. S4a), or 2 mM Gan-TP + 0.8 mM UTP +0.8 mM ATP +0.8 mM CTP (Fig. S4b), 2 mM 2′-OMe-UTP (Fig. S5a), 2 mM 3′-OMe-UTP (Fig. S5b), or 2 mM 2′-F-dUTP (Fig. S6) in 1x reaction buffer was added and incubation was carried out for 2 h at 30 °C. The final concentrations of reagents in the 20 μl extension reactions were 3 μM nsp12, 9 μM nsp7, 9 μM nsp8, 425 nM RNA primer, 500 nM RNA template, 500 μM Cid-DP, 200 μM UTP and 200 μM ATP (Fig. S3), 500 μM Car-TP, 200 μM UTP, 200 μM ATP and 200 μM CTP (Fig. S4a), 500 μM Gan-TP, 200 μM UTP, 200 μM ATP and 200 μM CTP (Fig. S4b), 500 μM 2′-OMe-UTP (Fig. S5a), 500 μM 3′-OMe-UTP (Fig. S5b), and 500 μM 2′-F-dUTP (Fig. S6). Following desalting using an Oligo Clean & Concentrator, the samples were subjected to MALDI-TOF-MS analysis.
3. Results and discussion
We tested the ability of the active triphosphate forms of the nucleotide analogues (structures shown in Fig. 1) to be incorporated by the RdRp of SARS-CoV-2 or SARS-CoV. The RdRp of these coronaviruses, referred to as nsp12, and its two protein cofactors, nsp7 and nsp8, which were shown to be required for the processive polymerase activity of nsp12 in SARS-CoV (Subissi et al., 2014; Kirchdoerfer and Ward, 2019), were cloned and purified as described previously (Kirchdoerfer and Ward, 2019; Chien et al., 2020). We then performed polymerase extension assays with the library of nucleoside triphosphate analogues (Fig. 1) either alone or in combination with natural nucleotides: 2′-OMe-UTP, 3′-OMe-UTP, 2′-F-dUTP, 2′–NH2–dUTP, Biotin-UTP, desthiobiotin-16-UTP, Sta-TP, Cid-DP + UTP + ATP, Car-TP + UTP + ATP + CTP, Gan-TP + UTP + ATP + CTP, or Ent-TP + UTP + ATP + CTP, following the addition of a pre-annealed RNA template and primer to a pre-assembled mixture of the SARS-CoV and/or SARS-CoV-2 RdRp (nsp12) and the two cofactor proteins (nsp7 and nsp8). We also used combinations of nucleotide analogues in some cases to perform the polymerase reaction to compare their relative incorporation efficiencies. The polymerase reaction products were analyzed by MALDI-TOF mass spectrometry. The sequences of the RNA template and primer used for the polymerase extension assay, which correspond to the 3′ end of the SARS-CoV-2 genome, are indicated at the top of Fig. 3, Fig. 4, Fig. 5 and S2-S9.
In the case of the UTP and TTP analogues, because there are two A's in a row in the next available positions of the template for RNA polymerase extension downstream of the priming site, if they are indeed terminators of the polymerase reaction at the relatively high concentration used, the extension is expected to stop after incorporating one nucleotide analogue. If they do not serve as terminators, two base extension by the UTP or TTP analogue will be observed. In the case of Cid-DP which is a CTP analogue, UTP and ATP must be provided to allow extension to the point where there is a G in the template strand. If the Cid-DP is then incorporated and acts as a terminator, extension will stop; otherwise, additional incorporation events may be observed. Similarly, for Carbovir-TP, Ganciclovir-TP and Entecavir-TP, all of which are GTP analogues, UTP, ATP and CTP must be provided to allow extension to the point where there is a C in the template strand. If Car-TP, Gan-TP or Ent-TP is incorporated and acts as a terminator, extension will stop; otherwise additional incorporation events occur. Guided by polymerase extension results we obtained previously for the active triphosphate forms of Sofosbuvir, Alovudine, AZT, Tenofovir-DP and Emtricitabine-TP (Ju et al., 2020; Chien et al., 2020; Jockusch et al., 2020) [not peer-reviewed], various ratios of the nucleotides were chosen in the current study.
The results of the MALDI-TOF MS analysis of the primer extension reactions are shown in Fig. 3, Fig. 4, Fig. 5 and S2-S9. The observed peaks generally fit the nucleotide incorporation patterns described above, however, additional peaks assigned to intermediate stages of the extension reaction (and in some cases extension beyond the incorporation of the nucleotide analogue) were also observed. We describe the results for the SARS-CoV-2 polymerase catalyzed reaction in detail; we obtained similar results for the subset of nucleotide analogues tested with the SARS-CoV RdRp, as shown in the Supplementary Material.
The results for 2′-OMe-UTP, Sta-TP (a T analog) and Biotin-dUTP are presented in Fig. 3. In the case of extension with 2′-OMe-UTP (Fig. 3a), MS peaks representing incorporation by one 2′-OMe-UTP (6638 Da observed, 6632 Da expected) and to a lesser extent two 2′-OMe-UTPs (6944 Da observed, 6952 Da expected) were observed. Thus, 2′-OMe-UTP shows significant termination upon incorporation, indicating it can be a potential drug lead. 2′-O-methyl modification of RNA occurs naturally and therefore should have relatively low toxicity. In addition, ribose-2′-O-methylated RNA resists viral exonuclease activity (Minskaia et al., 2006). For Sta-TP (Fig. 3b), a single incorporation peak (6603 Da observed, 6598 Da expected) was seen, indicating that Sta-TP is very efficiently incorporated and achieves complete termination of the polymerase reaction. A 10-fold lower concentration of Sta-TP also resulted in termination of the polymerase reaction (Fig. S2a). Since Sta-TP is a dideoxynucleotide without any hydroxyl groups on the sugar moiety, it may resist exonuclease activity. In the case of Biotin-dUTP (Fig. 3c), a single incorporation peak was evident (7090 Da observed, 7082 Da expected), suggesting that Biotin-dUTP is also a terminator of the polymerase reaction under these conditions. This indicates that the presence of a modification on the base along with the absence of a 2′-OH group in this nucleotide analogue leads to termination of the polymerase reaction catalyzed by the SARS-CoV-2 RdRp.
The result for the CTP analogue Cid-DP, which has an OH group, is presented in Fig. 4. Major peaks were observed indicating incorporation of a Cid-DP at the 8th position from the initial priming site (8813 Da observed, 8807 Da expected) and a further 2 base extension by one ATP followed by one Cid-DP at the 10th position (9404 Da observed, 9397 Da expected). There is no further extension beyond this position, indicative of delayed termination by Cid-DP. A small intermediate peak was also observed indicating extension by an ATP at the 9th position from the initial priming site following the first Cid-DP incorporation (9142 Da observed, 9138 Da expected). A small partial UTP extension peak (6623 Da observed, 6618 Da expected) was also observed. A 10-fold lower concentration of Cid-DP also resulted in termination of the polymerase reaction (Fig. S2b). An essentially identical result was obtained with the SARS-CoV polymerase (Fig. S3). Delayed termination for Cid-DP has been described for a vaccinia virus DNA polymerase (Magee et al., 2005). The investigational drug Remdesivir, which is currently being used for the treatment of COVID-19 under emergency authorization, also displays delayed termination (Gordon et al., 2020a, Gordon et al., 2020b); this is a major factor in its ability to resist the nsp14 3′-5′ exonuclease activity. Thus, Cidofovir and its oral prodrugs are of interest for further investigation to evaluate whether they can evade the viral exonuclease activity. Based on these results, and if potency for viral inhibition in cell culture is demonstrated with limited toxicity, Cidofovir and its related prodrugs may be potential leads for COVID-19 treatment.
The results for the GTP analogues, Car-TP, Ent-TP and Gan-TP are presented in Fig. 5. In each case, extension to the first C position on the template occurs and further extension is blocked in the presence of ATP, UTP and CTP. In more detail, for Car-TP (Fig. 5a), the major peak observed indicates extension by UTP, ATP and CTP followed by complete termination with a Car-TP (10436 Da observed, 10430 Da expected). In addition, partial extension peaks were seen indicating a single UTP incorporation (6621 Da observed, 6618 Da expected) and extension up to but not including the Car-TP (10123 Da observed, 10121 Da expected). For Ent-TP (Fig. 5b), a peak was observed indicating extension by UTP, ATP and CTP followed by complete termination by a single Ent-TP (10458 Da observed, 10460 Da expected). Additional peaks represent a single UTP extension (6628 Da observed, 6618 Da expected) and a major peak indicating extension up to but not including the Ent-TP (10129 Da observed, 10121 Da expected), suggesting that Ent-TP is less efficiently incorporated than Car-TP. And for Gan-TP (Fig. 5c), a major peak observed indicated extension by UTP, ATP and CTP followed by complete termination with Gan-TP (10441 Da observed, 10438 Da expected). A small peak representing extension up to but not including Gan-TP (10123 Da observed, 10121 Da expected) was also seen. Similar results were obtained for Car-TP and Gan-TP using the SARS-CoV polymerase (Fig. S4). Both Car-TP and Ent-TP are carbocyclic nucleotides. Car-TP lacks the 2′- and 3′-OH groups, while Ent-TP lacks the 2′-OH group. Gan-TP is an acyclic nucleotide having an OH group but lacking a ribose ring. All three thus may resist the viral exonuclease activity. These results also indicate that Car-TP and Gan-TP are better terminators than Ent-TP.
Fig. S5 shows a side-by-side comparison of the results with 2′-OMe-UTP and 3′-OMe-UTP using the SARS-CoV polymerase. The results for 2′-OMe-UTP are practically identical to those with SARS-CoV-2 in Fig. 3a, indicating that 2′-OMe-UTP exhibits significant polymerase reaction termination. The 3′-OMe-UTP results are consistent with its being an obligate terminator, but with lower incorporation efficiency, represented by a small single-incorporation peak (6625 Da observed, 6632 Da expected).
In Fig. S6, the results are shown for incorporation of 2′-F-dUTP by SARS-CoV RdRp. 2′-F-dUTP was incorporated very efficiently, but also was incorporated opposite the Us in the template strand. This apparent mismatch incorporation may result from the high concentration of nucleotide analogues used and the relatively low fidelity of SARS-CoV RdRp.
The results for desthiobiotin-16-UTP are presented in Fig. S7. Desthiobiotin-16-UTP incorporation complementary to each A in the template was observed, just like a UTP. Thus, this nucleotide is incorporated and does not terminate the polymerase reaction. These results indicate that modification on the base of the UTP does not affect its incorporation by SARS-CoV-2 RdRp.
Fig. S8 presents the results of an experiment where both 2′-OMe-UTP and dUTP were added together at the same concentration. The major peak occurred at 6930 Da (6922 Da expected) representing incorporation by both dUTP and 2′-OMe-UTP in adjacent positions. Partial extension peaks of a single 2′-OMe-UTP (6626 Da observed, 6632 Da expected) and two dUTPs (6900 Da observed, 6892 Da expected) were found. The incorporation of a dUTP, a 2′-OMe-UTP, both of which lack a 2′-OH group, or their combination would enable them to potentially resist the nsp14 3′-5′ exonuclease activity.
Fig. S9 shows three mass spectra of the polymerase reaction products using equimolar combinations of nucleotide analogues, (a) biotin-dUTP, dUTP and UTP, (b) 2′-F-dUTP, 2′-OMe-UTP and dUTP and (c) 2′–NH2–dUTP, 2′-OMe-UTP and dUTP, to determine their relative incorporation efficiencies. Based on the results shown in Fig. S9a, biotin-dUTP and dUTP have lower incorporation efficiency than the natural UTP for SARS-CoV-2 RdRp, since peaks are only observed for UTP extension, either one UTP (6620 Da observed, 6618 Da expected) or two UTPs (6928 Da observed, 6924 Da expected). In Fig. S9b, it is seen that 2′-F-dUTP is incorporated far better than 2′-OMe-UTP and dUTP, with the only evident peaks in the spectrum at 6620 Da (6620 Da expected) for extension by one 2′-F-dUTP and at 6928 Da (6928 Da expected) for extension by two 2′-F-dUTPs. Finally, as shown in Fig. S9c, 2′–NH2–dUTP is more efficiently incorporated than 2′-OMe-UTP and dUTP as revealed by the presence of peaks only at 6623 Da (6617 Da expected) for extension by one 2′–NH2–dUTP and at 6929 Da (6922 Da expected) for extension by two 2′–NH2–dUTPs. Thus, 2′-F-dUTP and 2′–NH2–dUTP behave like UTP and do not terminate the polymerase reaction. Neither 2′-F-dUTP nor 2′–NH2–dUTP have a free 2′-OH group. It remains to be seen whether the RNAs produced by these two nucleotide analogues will resist exonuclease activity.
In summary, these results demonstrate that the library of nucleotide analogues we tested could be incorporated by the RdRps of SARS-CoV-2 and SARS-CoV. Of the 11 tested, 6 exhibited complete termination of the polymerase reaction (3′-OMe-UTP, Car-TP, Gan-TP, Sta-TP, Ent-TP, Biotin-dUTP), 2 showed incomplete or delayed termination (Cid-DP, 2′-OMe-UTP), and 3 did not terminate the polymerase reaction (2′-F-dUTP, 2′–NH2–dUTP and desthiobiotin-16-UTP) using the RdRp of SARS-CoV and/or SARS-CoV-2. Their prodrug versions (Figs. 2 and S1) are available or can be readily synthesized using the ProTide approach (Alanazi et al., 2019). The ProTide approach was used very successfully to develop Sofosbuvir and Remdesevir for treatment of HCV and COVID-19, respectively. It may be advantageous to use ProTide prodrug forms containing a phosphate masked by a hydrophobic phosphoramidate group for the five drugs whose structures are shown in Fig. 2, because such prodrugs can be delivered into cells and converted to the triphosphate more rapidly, and potentially improve the bioavailability and potency of these molecules. The five drugs (Ganciclovir/Valganciclovir, Cidofovir, Abacavir, Stavudine and Entecavir (Fig. 2)) are FDA-approved medications for treatment of other viral infections and their toxicity profile is well established, while Brincidofovir is an experimental oral antiviral drug. Thus, our results provide a molecular basis for further evaluation of these prodrugs in SARS-CoV-2 virus inhibition and animal models to test their efficacy for the development of potential COVID-19 therapeutics.
Funding
This research is supported by Columbia University, a grant from the Jack Ma Foundation, a generous gift from the Columbia Engineering Member of the Board of Visitors Dr. Bing Zhao, and Fast Grants to J.J. and a National Institute of Allergy and Infectious Disease grant AI123498 to R.N.K.
Author contributions
J.J. and R.N.K. conceived and directed the project; the approaches and assays were designed and conducted by J.J., X.L., S.K., S.J., J.J.R., M.C. and C.T. and SARS-CoV/SARS-CoV-2 polymerases (nsp12) and associated proteins (nsp 7 and 8) were cloned and purified by T.K.A. and R.N.K. Data were analyzed by all authors. All authors wrote and reviewed the manuscript.
Declaration of competing interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104857.
Contributor Information
Robert N. Kirchdoerfer, Email: rnkirchdoerf@wisc.edu.
Jingyue Ju, Email: dj222@columbia.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Agostini M.L. Small-molecule antiviral β-d-N4-hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance. J. Virol. 2019;93 doi: 10.1128/JVI.01348-19. e01348-e01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyürek L.M. Coexpression of guanylate kinase with thymidine kinase enhances prodrug cell killing in vitro and suppresses vascular smooth muscle cell proliferation in vivo. Mol. Ther. 2001;3:779–786. doi: 10.1006/mthe.2001.0315. [DOI] [PubMed] [Google Scholar]
- Alanazi A.S., James E., Mehellou Y. The ProTide prodrug technology: where next? ACS Med. Chem. Lett. 2019;10:2–5. doi: 10.1021/acsmedchemlett.8b00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P.L., Kiser J.J., Gardner E.M., Rower J.E., Meditz A., Grant R.M. Pharmacological considerations for tenofovir and emtricitabine to prevent HIV infection. J. Antimicrob. Chemother. 2011;66:240–250. doi: 10.1093/jac/dkq447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arup H., Williams D.M., Eckstein F. 2’-Fluoro and 2’-amino-2’-deoxynucleoside 5’-triphosphates as substrates for T7 RNA polymerase. Biochemistry. 1992;31:9636–9641. doi: 10.1021/bi00155a016. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase. BioRxiv. 2020 doi: 10.1101/2020.03.18.997585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cundy K.C. Clinical pharmacokinetics of the antiviral nucleotide analogues cidofovir and adefovir. Clin. Pharmacokinet. 1999;36:127–143. doi: 10.2165/00003088-199936020-00004. [DOI] [PubMed] [Google Scholar]
- Dustin L.B., Bartolini B., Capobianchi M.R., Pistello M. Hepatitis C virus: life cycle in cells, infection and host response, and analysis of molecular markers influencing the outcome of infection and response to therapy. Clin. Microbiol. Infect. 2016;22:826–832. doi: 10.1016/j.cmi.2016.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Cidofovir in the treatment of poxvirus infections. Antivir. Res. 2002;55:1–13. doi: 10.1016/S0166-3542(02)00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29:695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L.D. Infidelity of SARS-CoV nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A.A. Ribavirin, remdesivir, Sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltahla A.A., Luciani F., White P.A., Lloyd A.R., Bull R.A. Inhibitors of the hepatitis C virus polymerase; mode of action and resistance. Viruses. 2015;7:5206–5224. doi: 10.3390/v7102868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faletto M.B., Miller W.H., Garvey E.P., St Clair M.H., Daluge S.M., Good S.S. Unique intracellular activation of the potent anti-human immunodeficiency virus agent 1592U89. Antimicrob. Agents Chemother. 1997;41:1099–1107. doi: 10.1128/aac.41.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H.-T., Hitchcock M.J.M. Cellular pharmacology of 2’,3’-dideoxy-2’,3’-didehydrothymidine, a nucleoside analog active against human immunodeficiency virus. Antimicrob. Agents Chemother. 1989;33:844–849. doi: 10.1128/AAC.33.6.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Farquhar D., Plunkett W. Selective action of 2’,3’-didehydro-2’,3’-dideoxythymidine triphosphate on human immunodeficiency virus reverse transcriptase and human DNA polymerases. J. Biol. Chem. 1992;267:2817–2822. https://www.jbc.org/content/267/4/2817.full.pdf [PubMed] [Google Scholar]
- Jockusch S. Triphosphates of the two components in DESCOVY and TRUVADA are inhibitors of the SARS-CoV-2 polymerase. BioRxiv. 2020 doi: 10.1101/2020.04.03.022939. [DOI] [Google Scholar]
- Ju J. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19635–19640. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju J. Nucleotide analogues as inhibitors of SARS-CoV polymerase. BioRxiv. 2020 doi: 10.1101/2020.03.12.989186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier R. Development of CMX001 for the treatment of poxvirus infections. Viruses. 2010;2:2740–2762. doi: 10.3390/v2122740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauridsen L.H., Rothnagel J.A., Veedu R.N. Enzymatic recognition of 2’-modified ribonucleoside 5’-triphosphates: towards the evolution of versatile aptamers. Chembiochem. 2012;13:19–25. doi: 10.1002/cbic.201100648. [DOI] [PubMed] [Google Scholar]
- Ma Y. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee W.C., Hostetler K.Y., Evans D.H. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob. Agents Chemother. 2005;49:3153–3162. doi: 10.1128/AAC.49.8.3153-3162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews S.J. Entecavir for the treatment of chronic hepatitis B virus infection. Clin. Therapeut. 2006;28:184–203. doi: 10.1016/j.clinthera.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Matthews T., Boehme R. Antiviral activity and mechanism of action of ganciclovir. Rev. Infect. Dis. 1988;10:S490–S494. doi: 10.1093/clinids/10.supplement_3.s490. [DOI] [PubMed] [Google Scholar]
- Mazzucco C.E., Hamatake R.K., Colonno R.J., Tenney D.J. Entecavir for treatment of hepatitis B virus displays no in vitro mitochondrial toxicity or DNA polymerase gamma inhibition. Antimicrob. Agents Chemother. 2008;52:598–605. doi: 10.1128/AAC.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna C.E. Inhibitors of viral nucleic acid polymerases. Pyrophosphate analogues. ACS (Am. Chem. Soc.) Symp. Ser. 1989;401:1–16. doi: 10.1021/bk-1989-0401.ch001. (Chapter 1) [DOI] [Google Scholar]
- Minskaia E. Discovery of an RNA virus 3’→5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öberg B. Rational design of polymerase inhibitors as antiviral drugs. Antivir. Res. 2006;71:90–95. doi: 10.1016/j.antiviral.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Ray A.S., Basavapathruni A., Anderson K.S. Mechanistic studies to understand the progressive development of resistance in human immunodeficiency virus type 1 reverse transcriptase to abacavir. J. Biol. Chem. 2002;277:40479–40490. doi: 10.1074/jbc.M205303200. [DOI] [PubMed] [Google Scholar]
- Rivkina A., Rybalov S. Chronic hepatitis B: current and future treatment options. Pharma. 2002;22:721–737. doi: 10.1592/phco.22.9.721.34058. [DOI] [PubMed] [Google Scholar]
- Selisko B., Papageorgiou N., Ferron F., Canard B. Structural and functional basis of the fidelity of nucleotide selection by Flavivirus RNA-dependent RNA polymerases. Viruses. 2018;10:59. doi: 10.3390/v10020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.P. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 2008;283:34218–34228. doi: 10.1074/jbc.M806797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis A.J.W. Common and unique features of viral RNA-dependent polymerases. Cell. Mol. Life Sci. 2014;71:4403–4420. doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost L.C. The efficacy and pharmacokinetics of brincidofovir for the treatment of lethal rabbitpox virus infection: a model of smallpox disease. Antivir. Res. 2015;117:115–121. doi: 10.1016/j.antiviral.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Zhu N. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses – drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.