Graphical abstract
Keywords: Brassica napus, Drought tolerance, Genome-wide association studies (GWAS), Specific length amplified fragments (SLAFs), Candidate genes
Abstract
Drought seriously curtails growth, physiology and productivity in rapeseed (Brassica napus). Although drought tolerance is a complex trait, efficient phenotyping and genotyping has led to the identification of novel marker-trait associations underlying drought tolerance. A diverse panel of 228 Brassica accessions was phenotyped under normal (without stress) and water-stress conditions, simulated by polyethylene glycol (PEG-6000) (15% PEG stress) at the seedling stage; stress tolerance index (STI) and stress susceptibility index (SSI) values were acquired. Genome-wide association studies (GWAS) using 201 817 high quality SNPs identified 314 marker-trait associations strongly linked with drought indices and distributed across all nineteen chromosomes in both the A and C genomes. None of these quantitative trait loci (QTL) had been previously identified by other studies. We identified 85 genes underlying these QTL (most within 100 kb of associated SNPs) which were orthologous to Arabidopsis genes known to be associated with drought tolerance. Our study provides a novel resource for breeding drought-tolerant Brassica crops.
Introduction
Abiotic stress is now been considered to be a leading threat to agricultural productivity worldwide, with up to 70% of the current yield of staple crops potentially impacted by different abiotic stresses [1]. Of these abiotic stresses, drought is a major threat to crop productivity. Water deficit decreases the percentage and rate of seed germination, and can hinder seedling growth and establishment [2]. Selection for drought tolerance is a desirable breeding goal. However, drought tolerance is a complex trait which is difficult to select for directly, necessitating adoption of indirect selection criteria. Recently, there has been a move to dissect the complex set of interrelated traits which make up drought tolerance through the use of drought tolerance indices. Of these indices, stress tolerance index (STI) and stress susceptibility index (SSI) have proven to be reliable measures of plant survival under drought stress conditions. More importantly, each of these indexes provides a different perspective and a distinct focus on different aspects of drought tolerance. A better understanding of and selection for drought tolerance is becoming ever more realistic with the emergence of new phenotyping technologies and robust methodologies that allow for non-destructive, simultaneous and high-throughput measurements of numerous traits.
The genus Brassica comprises approximately 100 species, many of which are used agriculturally as condiments and as oilseed, leaf and root crops for human and animal feed. The most economically important Brassica crop is B. napus, which produces canola or rapeseed oil, the third most consumed edible oil in the world (after palm and soybean). Brassica napus is an allotetraploid species (AACC genome, 2n = 4x = 38 chromosomes), derived from interspecific hybridization between ancestors of extant diploid species B. rapa (AA genome, 2n = 2x = 20 chromosomes) and B. oleracea (CC genome, 2n = 2x = 18 chromosomes). Rapeseed (B. napus) is cultivated across the world in diverse environments, and contains spring-type, semi-winter and winter crop ecotypes. Regardless of growing environment, drought stress is a major constraint on rapeseed production globally, as rapeseed is very sensitive to water deficit conditions at all growth stages, from germination to seed setting and ripening [3]. Improvement of drought tolerance in rapeseed would allow for both increased cultivation area and improved crop yields.
Genetic improvement of crops with respect to drought tolerance requires the examination of possible mechanisms of tolerance at the seedling stage, as well as the identification of genetic variation for drought tolerance within a species [4]. Genome wide association studies (GWAS) are a powerful tool for characterizing the genetic constitution of the traits and identifying potential candidate genes associated with desired traits [5], [6], [7]. To date, although major advances have been made and some drought resistant varieties developed, the molecular bases for drought tolerance are still poorly understood [8]. GWAS has been effectively used for the genetic dissection of drought resistance in many crop species, including rice [9], Arabidopsis [10], barley [11] and wheat [12]. In B. napus, GWAS has already identified important genetic loci underlying agronomic traits such as oil content [13], seedling metabolism [14], yield [15], and flowering time [16]. However, only a few studies report the use of GWAS to genetically dissect mechanisms of drought tolerance in canola, many of which were based on low marker density mapping approaches which may not be able to capture the global genetic diversity of drought tolerance mechanisms [17]. Previously, Zhang et al. [18] reported 16 loci, 79 candidate genes and 8 putative candidate genes significantly associated with water stress response in canola. GWAS has also been applied to assess germination percentage and germination index under drought stress in 520B. napus lines [19].
In the present study, we phenotyped a panel of 228 lines under normal (without drought stress) and drought stress (imposed by PEG 6000) conditions; drought stress was measured by STI and SSI. GWAS was performed using combination of drought tolerance indices with high-quality SNPs developed using SLAF-seq [20]. Single nucleotide polymorphisms (SNPs) are the marker of choice in most species for GWAS, and specific locus amplified fragment sequencing (SLAF-seq) is an excellent method for developing high-throughput SNP markers [21]. We aimed to identify SNP markers and candidate genes that were significantly associated with either drought susceptibility or drought tolerance during the seed germination and early seedling establishment stage in canola, and subsequently to better our understanding of the mechanisms of drought tolerance in B. napus seedlings.
Materials and methods
Plant materials and growth conditions
In the present study, an association panel consisting of 228 accessions of B. napus was selected on the basis of pedigree, region of cultivar release, agronomic performance, economic importance and cultivated area [20]. Of these accessions, 197 were semi-winter types, 9 were spring types and 22 were winter types. In total, 206 lines were collected from China and the remaining 22 accessions were from different countries, including the United States, Canada, Japan and various European countries. These accessions were used to investigate and understand the mechanism of drought tolerance at germination and seedling growth stage. Before all accessions were screened for drought tolerance during seed germination and seedling growth, a pilot study was carried out to identify optimal PEG concentrations for inducing suitable levels of drought stress.
Pilot experiment to obtain optimum PEG concentrations with which to assess drought tolerance
Initially, a pilot study was designed to determine the optimum concentration of drought stress imposed by polyethylene glycol (PEG-6000). For this, 6 lines were randomly selected and exposed to various PEG treatments. These were 0% PEG (control with distilled water), 10%, 15%, 20%, 25% and 30% PEG on the first day, followed 2 ml on alternate days for 7 days. Thirty seeds from each line were placed in Petri dishes containing two layers of filter paper. Each treatment was carried out in three replications using a completely randomized design. The effects of various PEG treatments were assessed on germination and seedling traits. Major variation for germination and seedling traits was observed under various PEG concentrations. The seed germination and seedling growth parameters under 10% PEG concentration were similar to those of the control (without PEG), which suggests that this concentration was too low to induce a visible drought response. Concentrations of 20% and 25% PEG allowed seed germination but severely stunted seedling growth, hindering measurements of seedling length and weight. No seeds germinated under 30% PEG concentration. The maximum variation in seed germination and seedling growth parameters was observed under 15% PEG concentration. This concentration was therefore used as the drought stress treatment for phenotyping the whole population, along with the 0% PEG control treatment.
Experimental set-up and drought stress treatment
Seeds from the 228 accessions were initially surface-sterilized by treatment with 4% NaClO followed by rinsing with distilled water by several times. Thirty of these surface-sterilized seeds per accession were selected based on uniformity in color, weight and size, and were placed on Petri dishes (9 cm diameter) on double-layered filter paper (Whatman™, Malaga, WA, Australia) for germination under the two treatment conditions (0% PEG and 15% PEG solution) with no seeds touching each other. The experiment was laid out in a completely randomized design with three replicates in each treatment. The experiment was conducted between 4 and 6 pm so scoring of radicle emergence could start in the morning of the next day. The Petri dishes were placed in a growth chamber with a constant temperature of 21 °C under 16 h/8 h light/dark conditions. The seeds were considered germinated when radicle protrusion had occurred. All experiments were conducted in the Key Laboratory of Crop Physiology, Ecology and Genetics Breeding, Jiangxi Agricultural University, Nanchang, P.R China.
Phenotypic trait measurements
Germinated seeds were counted daily based on the emergence of the radicles of each accession in each Petri dish. After the 7th day of treatment, the total numbers of germinated and non-germinated seeds were counted. Final seed germination rate was calculated as GR = total number of germinated seeds on the last day/total number of seeds grown × 100. On the 8th day of treatment, all cultivars were surveyed, seedlings were harvested and 10 seedlings were randomly selected: roots and shoots of these seedlings were separated and root length (RL) was measured by a ruler. Thereafter, the root fresh weight (RFW) was measured from the same seedlings after removing surface water by blotting. Seedling vigor index (SVI) was calculated as: SVI = germination percentage (GP) × sum of seedling length.
Additionally, the response of genotypes to drought was evaluated by calculating the stress tolerance index (STI) and the stress susceptibility index (SSI) for all evaluated traits. The results obtained from the SSI and STI for all traits were used for GWAS. STI was calculated according to [22] and SSI according to [23] using the following formulas:
where Ysi = performance of a genotype under stress; Ypi = performance of a genotype under no stress; SI/D (stress intensity) = 1- (mean of all genotypes under stress/mean of all genotypes under no stress); Yp2 = square root of all genotypes for a trait under non-stress conditions.
Statistical analysis of phenotypic data
The phenotypic data collected from the experiment was analysed statistically. The two-way analysis of variance (ANOVA) for germination and seedling traits for all accessions was calculated using the Minitab v.18 software (Pennsylvania State University, PA, USA) using general linear modelling with p < 0.01 significance level; differences between groups were further validated using Tukey’s honest significant differences (HSD) test. The frequency distribution of traits and descriptive statistics such as coefficient of variation, mean, standard deviation, minimum, maximum, skewness and kurtosis of traits were calculated with SPSS v.20 (SPSS, Chicago, IL, USA). In addition, Pearson’s correlation coefficient between seedling traits was estimated with the Statistix 8.1 software package using a significance level of p < 0.01. Boxplots were drawn using the “ggplot2” package in R (The R Project for Statistical Computing, Vienna, Austria). The broad sense heritability was estimated as follows: h2 = Vg (Vg + Ve/r), where Vg is the genetic variance, Ve is the error variance and r is the replication.
DNA extraction, enzyme selection and SLAF library construction
Young fresh leaf samples were collected from one plant from each of the 228 selected accessions [20] for DNA extraction using the cetyltrimethylammonium bromide (CTAB) method [24] with minor modifications. DNA concentration and quality were assessed using a Nanodrop 2000 UVV spectrophotometer (NanoDrop, Wilmington, DE, USA). Then, quantified DNA was diluted to 100 ngµl − 1 for SLAF sequencing. SLAF sequencing success was predicted using the reference rapeseed genome “Darmor-bzh” [25]; http://www.genoscope.cns.fr/brassicanapus/data/). In order to maximise the number of SLAF tags obtained, we decided on four criteria for SLAF-seq: (1) restriction segments must have a low percentage of repetitive sequences; (2) restriction fragments should be uniformly distributed across all chromosomes; (3) simulated segments should align uniquely to the reference genome; and (4) a high number of SLAF tags should obtained which meet previously defined criteria. On the basis of these criteria, the restriction enzyme combinations Rsal and HaeIII (NEB, Ipswich, MA, USA) with sizes of 341–414 bp were selected.
SLAF sequencing, genotyping and SNP calling
Clean genomic DNA was digested using the enzyme combinations to attain SLAF tags, and fragment end compensation, PCR amplification, dual index paired-end adapter ligation, and targeted fragment selection were performed step-by-step. Subsequently, high-throughput SLAF sequencing was performed using an Illumina HiseqTM 2400 (Illumina, Inc; San Diego, CA, USA) at the Biomarker Technologies Corporation in Beijing. Raw SLAF sequencing data was then processed using the software Dual index [26]. After removal of adapter reads, the quality of sequences generated was evaluated by estimating guanine-cytosine (GC) content and with the Q30 ratio (Q = −10*loge10; (indicating a 0.1% chance of an error and thus 99.9% confidence) in the raw reads. Thereafter, paired-end sample reads were grouped according to sequence similarity using the BLAT software [27]. SLAF tags which showed high sequence polymorphism across the accessions as well as high quality scores were then mapped to the B. napus reference genome [25] using the Burrow’s Wheeler Alignment Tool (BWA) software [28].
Genome-wide association study (GWAS) haplotype block analyses
To dissect the genetic basis underlying the natural variation of drought tolerance, 201 817 SNPs generated from the 228 rapeseed accessions and results obtained from the SSI and STI calculations for each trait were used to perform GWAS using TASSEL version 5.0 [29] software with general linear models (GLM) and mixed linear models (MLM). The fixed effects were calculated with a Q matrix (population structure) and random effects were estimated with a K matrix (Kinship). Only the Q matrix was considered in the GLM while both Q and K matrices were used in the MLM. The software Admixture [30] was employed to calculate the Q matrix and the K matrix (genetic relationship) between the 228 lines was predicted using the software SPAGeDi [31]. The p-values of SNPs associated with traits were calculated according to following formula:
where Y is the phenotype, X represents genotype, Qβ is the fixed effects and Kµ represents random effects. The QQ plot (quantile-quantile) was generated using the software package “ggplot2” in R [32] and the Manhattan plots for each trait were drawn using the software QQman [33]. The -log10 (P) threshold value was set as -log10 0.1 / 201 817 SNPs [P < 4.96E10-7, -log10 (P) value is equal to 6.3] to identify true SNP-trait associations using the false discovery rate (FDR) test value; a true marker–trait association should show an FDR < 0.05, and only FDR scores < 0.01 meet the criteria for extremely significant marker-trait associations [P < 4.96E10−8, −log10 (P) value is equal to 7.3]. Haplotype block analysis was carried out with the software Haploview v. 4.2 [34]. The data of SNPs was used to calculate the pairwise LD between the SNPs using a cutoff values of 1%; which means if any addition of SNP in to the block that results in a recombination allele a frequency exceeds 1%, the SNP will not be included in the block. These haplotype blocks were constructed according to four gamete method [35].
Identification of superior allelic variation for seedling traits
The phenotypic effect of each allelic variant for each marker significantly associated with drought tolerance was calculated using the EAM method [16]. Combined allelic variant scores for traits were calculated by summing up positive (trait score increase) and negative (trait score decrease) effects of all variants. Thus, the average allelic effect (AAE) was estimated as follows:
where ac represents the magnitude of the positive or negative effect of an allele for a particular SNP and nc is the number of alleles with a positive or negative effect. The number of positive (superior) and negative (inferior) effect alleles in each rapeseed line were then counted.
For SSI-based analysis, SNPs with negative allelic effect values for any trait were considered as the superior alleles, while those which had positive effect values were considered as inferior alleles. By contrast, in STI-based analysis SNPs with positive allelic effects for the traits were considered as superior and SNPs with negative allelic effects for a trait were considered as inferior alleles.
SNP validation through kompetitive allele specific PCR (KASP) markers
For the validation of SNPs identified through SLAF-seq, we randomly selected 25 SNPs included in our GWAS analysis and 50 lines from the panel of 228 accessions. DNA was extracted from these lines on the eighth day using the same procedure as previously described above. After checking the quality and concentration of DNA, samples were sent to the LGC Genomics laboratory in Beijing for KASP marker development. The DNA sequences of identified SNP markers were used to produce KASP markers by LGC Genomics (LGC, Beijing, China) and these KASP markers were subsequently screened on the 50 lines of known phenotype. The list of these SNPs, the sequences of their allele-specific primers (allele FAM and allele HEX) and common primers along with their CG ratio, are presented in Supplementary Table S1. Polymerase chain reaction (PCR) was performed according to the manufacturer’s recommendations. KASP reactions were run on a 96-well DNA plate using 5 μL template DNA, 5 μL 2 × KASP Master mix standard ROX and 5 μL KASP Assay mix (LGC Genomic, Beijing, China). The PCR thermocycling protocol for the KASP marker assay was 94 °C for 15 min, followed by 10 cycles of 94 °C for 20 s and 61 °C for 1 min (dropping −0.6 °C per cycle to achieve an the annealing temperature of 55 °C) followed by 26 cycles of 94 °C for 20 s and 55 °C for 60 s. After amplification, data was imported and analysed using the genotype cluster analysis software package KlusterCaller (LGC) (www.lgcgroup.com/software) to identify SNP alleles.
Candidate gene prediction for drought tolerance
Using linkage disequilibrium (LD) analysis methods as described in our previous study [20], LD blocks where the significant marker trait associations were located in which flanking markers had strong LD (r2 > 0.6) [36] were described as candidate gene regions (extending from the leftmost unrelated SNP to the rightmost unrelated SNP). All genes within the same LD block (r2 > 0.6) containing a significant marker-trait association were assessed as possible candidate genes. In addition, candidate genes outside the LD blocks but within 100 kb of a significantly associated marker were also interrogated as possible candidates [16], [20], [25]. Candidate genes were selected based on matches to gene ontology (GO) terms such as dehydration, proline, osmotic pressure, cell desiccation, cell dryness and stomata closure. BLASTX alignment was then performed against the Arabidopsis genome to identify orthologous genes associated with drought tolerance in Arabidopsis.
Validation of candidate genes through quantitative real-time PCR
In order to further validate candidate genes obtained via GWAS, we determined the expression levels of ten candidate genes in two lines (one drought-sensitive and one drought tolerant) under drought conditions by qRT-PCR analysis. For this, seedlings of the two lines were grown for seven days with 15% PEG-6000. The samples were collected on the 7th day and stored at −80 °C after freezing in liquid nitrogen. The total RNA was isolated from plants using TRIzol reagent RNA extraction kits (TaKaRa, Japan) according to the manufacturer’s protocols. The concentration and quality of the extracted RNA was assessed using the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, USA) and the integrity of RNA samples was assessed via 0.8% agarose gel electrophoresis. Consequently, cDNA synthesis was done in 20 µL reaction mixture using the Prime Script 1st strand cDNA synthesis plant kit according to manufacturer’s instructions. The SYBR-based real time quantitative PCR reactions (SYBR Green I, Osaka, Japan) were then performed using ABI VII@7 using the following cycling conditions: 50 °C for 2 min, 95 °C for five minutes, 40 cycles at 95 °C for 15 s and 60 °C for 34 s. All qRT-PCR reactions were performed in three independent biological replicates for each line, and the relative gene expression results were analyzed using the system’s relative quantification software (ver.1.5) based on the 2-ΔΔCT method. The details of the primer sequences used are listed in Supplementary Table S2.
Results
Phenotypic variation for germination and seedling traits under drought stress
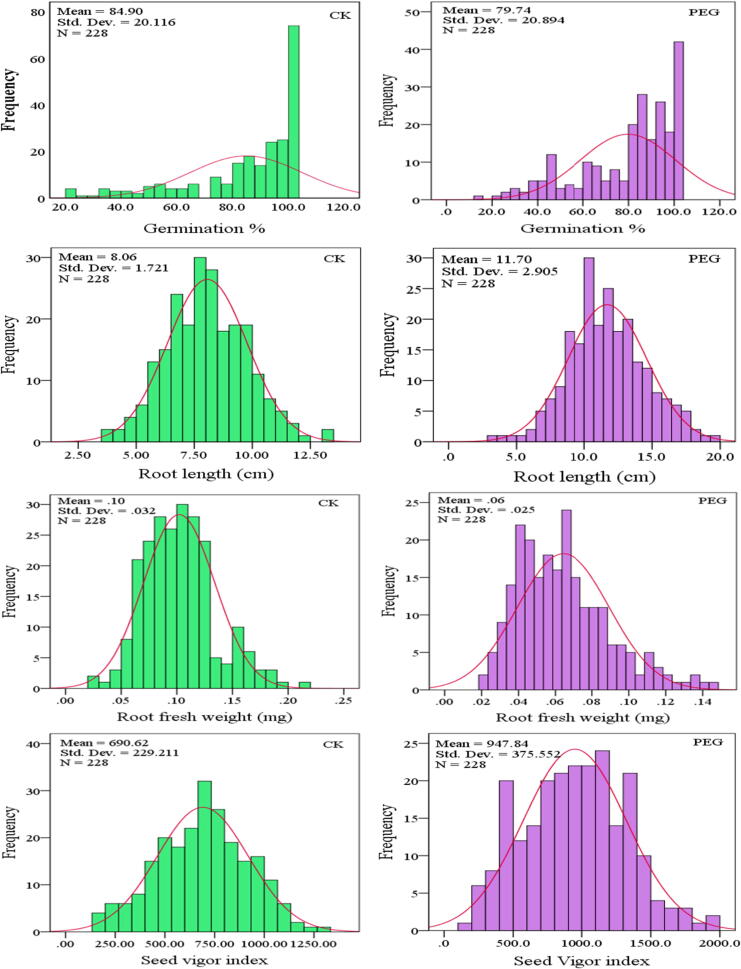
To investigate the phenotypic variation in seedling growth response to drought, the performance of four seedling traits was recorded under normal and drought stress conditions. Significant differences were found for all phenotypic traits across the accession panel (p < 0.01, (Table 1). The mean values of all measured traits were significantly reduced under the drought treatment compared to the control treatment (Table 1), with similar variation (coefficient of variance) observed between the control and drought stress condition for all four traits (21–40%, average 30%), and good fit to the normal distribution (Fig. 1, Table 1).Two-way analysis of variance revealed that phenotypic variation between accessions, treatments, and line × treatment interactions were all highly significant (Table 2). Hence, all the evaluated traits were significantly affected by drought. The seed vigor index had the maximum mean square values (459874, 23,815,912 and 148,115) in lines, treatments and L × T interaction.
Table 1.
Descriptive statistics values for germination and seedling traits of rapeseed natural population.
| Trait | Trt. | Mean | Min. | Max. | SD | CV (%) | Skewness | Kurtosis |
|---|---|---|---|---|---|---|---|---|
| Germination | CK | 84.8964 | 20.00 | 100.00 | 20.11599 | 23.619 | −1.524 | 1.433 |
| PEG | 79.7436 | 14.330 | 100.00 | 20.89432 | 26.221 | −1.048 | 0.081 | |
| Root Length | CK | 8.0602 | 3.607 | 13.410 | 1.72064 | 21.347 | 0.181 | 0.100 |
| PEG | 11.695 | 3.403 | 19.820 | 2.90469 | 25.242 | 0.112 | 0.078 | |
| Root fresh weight | CK | 0.1021 | 0.0270 | 0.213 | 0.03209 | 31.430 | 0.597 | 0.531 |
| PEG | 0.0643 | 0.0210 | 0.1480 | 0.02503 | 39.014 | 0.848 | 0.558 | |
| Seed vigor index | CK | 690.6227 | 154.10 | 1270.12 | 229.21122 | 33.439 | −0.077 | −0.388 |
| PEG | 947.8439 | 136.50 | 1984.80 | 375.55230 | 39.997 | 0.150 | −0.462 | |
Trt. = Treatment, Min. = Minimum, Max. = Maximum, SD = Standard deviation CV = Coefficient of variance, CK = non-stress condition, PEG = Stress condition.
Fig. 1.
Distribution of seedling growth-related phenotypes in 228B. napus accessions. Histograms of the distribution of the different phenotypes measured in control (CK) and drought-stressed plants (PEG 15%) in 228 accessions with 3 replicates.
Table 2.
Mean squares of various seedling growth related traits revealed by Analysis of Variance (ANOVA).
| Source of Variation | Replication (R) | Lines (L) | Treatment (T) | L × T | Error | Heritability (bs) |
|---|---|---|---|---|---|---|
| D.F. = 2 | D.F. = 227 | D.F. = 1 | D.F. = 1367 | D.F. = 227 | ||
| GR | 432.15 | 2233.03** | 9372.45** | 271.55** | 43.55 | 98.050 |
| RL | 0.82 | 23.01** | 4746.24** | 13.63** | 0.32 | 98.609 |
| RFW | 0.000021 | 0.003342** | 0.485520** | 0.001748** | 0.000078 | 97.666 |
| SVI | 97,895 | 459,874** | 23,815,912** | 148,115** | 7873 | 98.288 |
GR = germination, RL- root length, RFW root fresh weight, SVI = seed vigor index.
= Significant at P < 0.01.
Broad-sense heritability (h2 b.s) estimates of the four traits showed highly stable inheritance, with average heritability of 98.1%, ranging from 97.7 to 98.6% (Table 2). In addition, high degree of phenotypic changing trends for RL and SVI traits accounted for a high percentage, which represent the drought response independent of seedling growth in control conditions (Supplementary Fig. S1). Percentage seed germination was not significantly associated with root fresh weight or with root length under drought stress, but was significantly associated with root length in the control condition (Supplementary Table S3). Root length and root fresh weight showed highly significant correlation under both drought and control conditions, and seedling vigor index had positive and highly significant correlations with all other traits in both control and stress conditions.
GWAS revealed marker-trait associations for drought tolerance
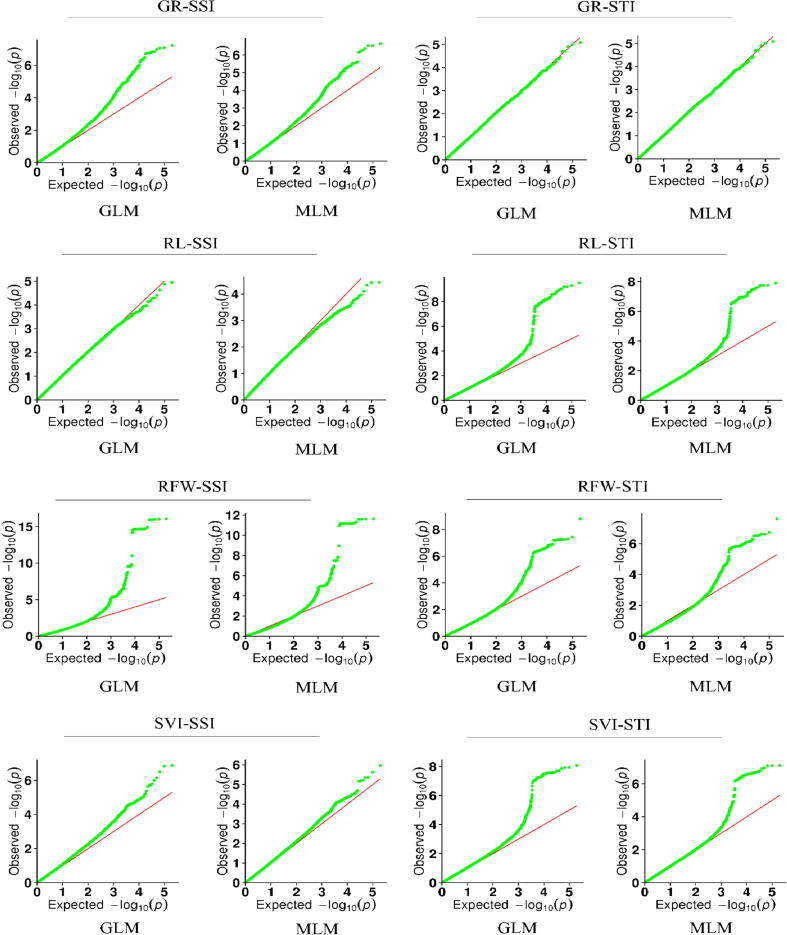
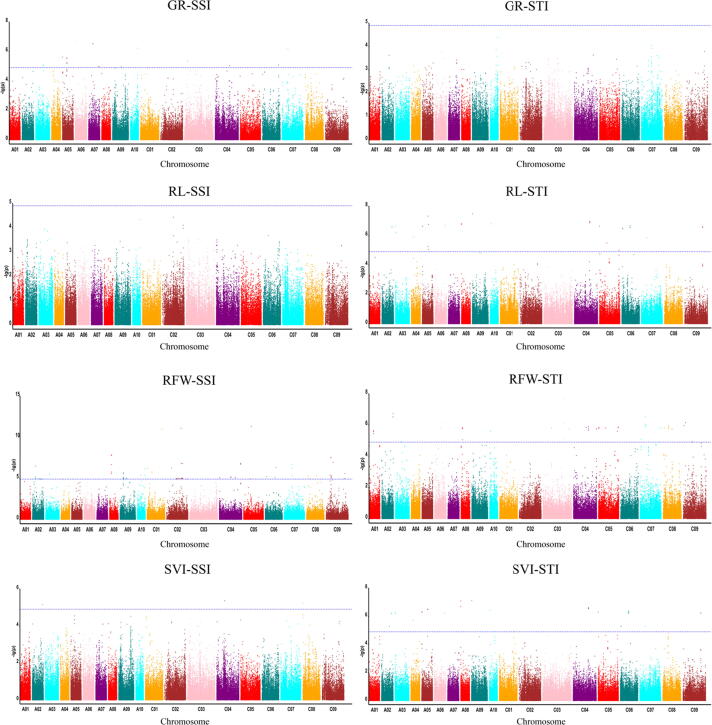
We calculated STI and SSI on germination and seedling traits and performed GWAS separately on each of these indices to identify genomic regions significantly associated with drought tolerance, after assessing the fit of the data to the general linear model (GLM) and standard mixed linear model (MLM) the ideal models present uniformity among the observed –log10 (P) values vs expected–log10 (P) values in the quantile–quantile QQ plot (Fig. 2). We identified significant SNP-trait associations using both MLM (Fig. 3) and GLM (Supplementary Fig S2), with 314 SNPs mapped to the 19B. napus chromosomes (108 in the A and 206 in the C genome) using both models, the four seedling traits and the SSI and STI values (Supplementary Table S4). GLM analysis identified 314 SNPs, while MLM analysis identified 183 SNPs (Supplementary Table S4). Chromosome C02 harbored the largest number of SNPs (52), while A04 harbored only one SNP associated with root length and seedling vigor at position Bn-A04-p4263955. More SNPs were significantly associated with the SSI phenotype values (192/314) than the STI (122/314) (Fig. 4).
Fig. 2.
Quantile-quantile plots from GWAS of stress susceptible index (SSI) and stress tolerance index (STI) under a general linear model (GLM) and mixed linear model (MLM). The green curve indicates the observed negative log p-values (Y-axes) of marker-trait association and the red line represents expected p-values (X-axes). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Manhattan plots representing the SNP markers-traits associations (MLM) under stress susceptible index (SSI) and stress tolerance index (STI) indices. The X-axes indicates the nineteen chromosomes from (left to right) and the Y-axes represents -log10 (p) values of the SNP marker. The blue dashed horizontal line depicts the genome-wide significance threshold. Different colors displayed each chromosome. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
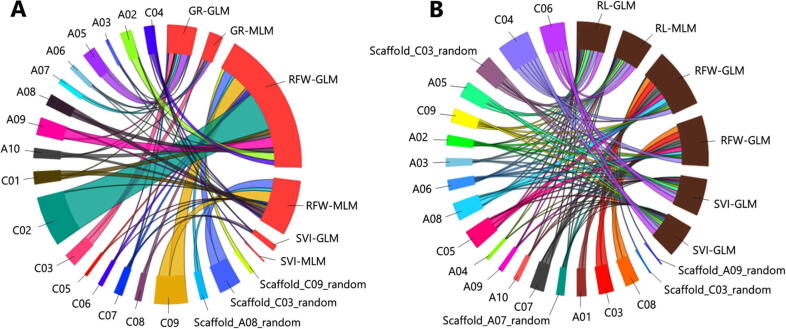
Comparison of the proportion of SNPs among SSI and STI based on GLM and MLM. (A) Association of homologous loci on drought-related traits based on SSI index among GLM and MLM. (B) Association of homologous loci based on the STI index among GLM and MLM. Salmon color bars represent SSI index while saddle brown color bars represent STI index among GLM and MLM models. SNP markers distributed in the genome are depicted in different colors bars. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
With respect to SNPs for individual traits, 32 SNP markers on 11 chromosomes were associated with germination rate; 16 of these were detected by GLM and 16 were detected by both the GLM and MLM analyses (Supplementary Table S5). All these SNPs were identified in the SSI GWAS analysis. For the root fresh weight, 230 SNPs were identified on fifteen chromosomes (Supplementary Table S6). Of these, 106 SNPs were detected only by GLM and 124 SNPs were detected by both GLM and MLM models. Of the 230 SNPs associated with root fresh weight, 154 were identified by SSI GWAS and 76 were identified by STI GWAS analysis. For root length 44 SNPs on the thirteen chromosomes were identified; two SNPs were detected only by GLM and 42 were detected by both GLM and MLM models (Supplementary Table S7). All root length-associated SNPs were identified on the basis of STI GWAS analysis. Forty-nine SNPs on fifteen chromosomes were detected for seedling vigor; six SNPs were detected only by GLM and 43 SNPs were detected by both models (Supplementary Table S8). Forty-three of these SNPS were identified in the STI GWAS analysis and 6 were identified in the SSI GWAS analysis.
Haplotype block analysis
Haplotype block analysis has revealed that several haplotype blocks were found across genome. Because of the large number of SNPs in our study, we drawn haplotype blocks based on single SNPs (Supplementary Fig S3). Most of the blocs has covered very less genomic regions in size (<100 kb).
Identification of alleles positively associated with drought tolerance based on STI and SSI
SSI-GLM analysis was more effective at identifying superior alleles (59.4%) than inferior alleles (39.6%) for three seedling traits (Supplementary Table S9a). Of these, the highest percentage of superior alleles was associated with root fresh weight. SSI-MLM analysis also identified more superior alleles (66.0%) than inferior alleles (34.0%) (Supplementary Table S9b), where highest number of superior alleles was linked to the root fresh weight and the highest number of inferior alleles was associated with germination rate. Although SSI-based GWAS found more significant marker-trait associations (192), more superior alleles were detected via STI-GWAS analysis than via SSI-GWAS analysis. In total, 164 marker-trait associations were identified using STI-GLM for three traits (no marker-trait association was detected for germination rate), with 234 superior alleles (71.3%) (Supplementary Table S9c). Of these, the highest number of superior alleles (32.6%) were for root fresh weight, followed by root length and seedling vigor (20.7% and 18.90% respectively). STI-MLM analysis also revealed a high percentage (80.8%) of superior alleles for the three traits (again excluding germination rate) compared to inferior alleles (19.2%), where the highest numbers of superior alleles were linked with the root fresh weight trait (Supplementary Table S9d). Overall, the highest number of superior alleles was detected for the root fresh weight, and the lowest number of superior alleles were detected for germination rate (Fig. 5).
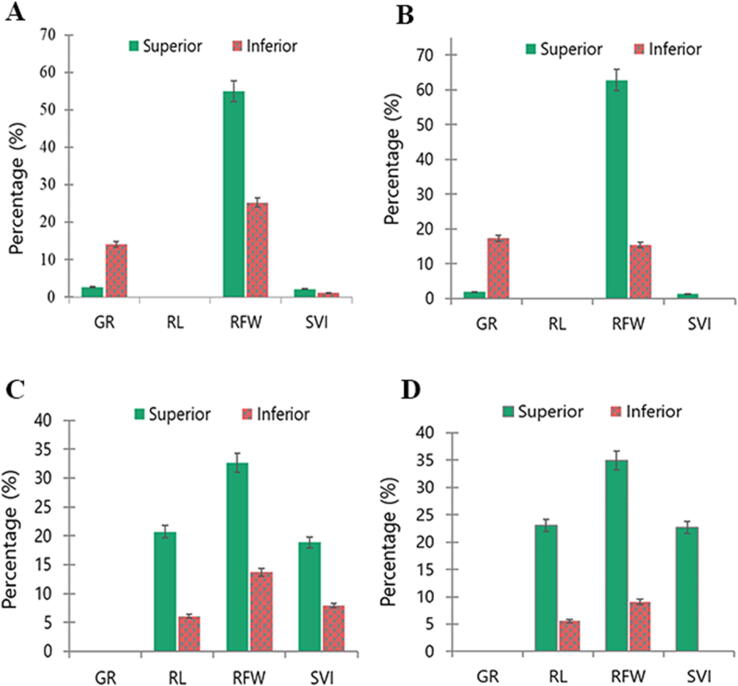
Fig. 5.
Percentage of superior and inferior alleles for four seedling traits related to drought stress under SSI and STI indices. The green bars represent the superior alleles, while red bars represent inferior alleles in each aspect of GWAS analysis; (A) SSI_GLM, (B), SSI_MLM, (C) STI_GLM, (D) STI_MLM. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Several regions of the genome showed pleiotropic associations with drought tolerance. In total, 37 SNPs distributed across 13 chromosomes were found to be associated with more than one trait (root length and seedling vigor) (Supplementary Table S10). For 14 SNPs identified via GLM and 6 SNPs identified through MLM analysis, one of the associated alleles had a positive effect and the other allele had a negative effect. Some SNPs showed three alleles with positive effects and one allele with a negative effect. For example, a T allele at the Bn-A05-p750174 position had a positive effect on both root length and seedling vigor, but a G allele at the same position increased root length but decreased seedling vigor. Some alleles also had contrasting effects on traits depending on the model used (Supplementary Table S10). For example, a T allele at the position Bn-A05-p750094 decreased root length and seedling vigor in the GLM-based analysis, but increased root length and seedling vigor under the MLM model. More SNPs were observed for which all alleles had positive effects (43 in the GLM analysis and 47 SNPs in the MLM analysis). The number of lines carrying elite alleles for each SNP varied from 1 to 195 lines per SNP locus.
Discovering candidate genes for drought tolerance
We identified possible candidate genes underlying significant marker-trait associations by investigating all genes within shared LD blocks (r2 > 0.6) or with physical proximity within 100 kb either side of the SNP. A total of 3569 genes meeting these criteria were annotated using GO analysis. Of these, 102 genes closely associated with 55 SNPs were putatively related to drought tolerance on the basis of GO annotation. These genes were distributed across all chromosomes, with 67 genes in the A genome and 35 genes in the C genome. Chromosome A05 had the highest number of genes and C08 harboured only one gene (Supplementary Table S11). These 102 gene sequences were also aligned using BLAST to the Arabidopsis reference genome: 85 genes were orthologous with Arabidopsis genes putatively associated with drought response, including NAC genes, PIP2, HB7, CPK1, MYC2, and CDPK (Fig. 6; Supplementary Table S12). Of these genes, 13 genes on 7 chromosomes were associated with GR-SSI, 18 genes on 9 chromosomes were associated with RL-STI, 13 genes on 7 chromosomes were associated with RFW-SSI, 21 genes on 9 chromosomes were associated with RFW-STI, 2 genes on A02 were associated with SVI-SSI and 18 genes on 8 chromosomes were associated with SVI-STI. Of these Arabidopsis gene orthologs, more genes were identified by STI-based analysis (57) than SSI (28). Furthermore, the genes identified could be divided into several groups: transcription factors, NAC domain, protein kinases, and DNA-binding proteins, among others.
Fig. 6.
Distribution pattern of candidate genes and their corresponding SNPs on the chromosomes associated with drought tolerance. The abbreviations of orthologous genes in Arabidopsis thaliana are shown in brackets after the candidate genes. SNPs are marked in red. Numbers represent the relative distances in the genome, 1 = 1 kb.
Two genes identified in this study (BnaC06g14590D and BnaA03g27130D) were orthologous to the PIP1 and PIP2 genes in Arabidopsis, which belong to the aquaporin proteins group. Aquaporins (AQPs) are important membrane proteins that play critical roles in the transport of water and several other solutes across cell membranes, hydraulic conductivity, regulation of plant water uptake, water loss and regulation of tissue and whole plant water relations [37]. Four genes (BnaA03g33890D, BnaA07g30680D, BnaA03g01250D, and BnaA03g01250D) identified in our study belong to the NAC domain group. These genes were orthologous to the NAC3, NASCENT, ANAC077 and NAC77 genes in Arabidopsis. These genes have been reported to play vital roles in drought tolerance in Arabidopsis: NAC domain proteins have been implicated functionally in a variety of stress-related responses, such as to drought, salinity and biotic stresses, and also are thought to be involved in plant development [38].
SNP validation through kompetitive allele specific PCR (KASP) markers
For the validation of SNPs detected by our GWAS study, we selected 25 SNP markers from and 50 accessions. When comparing SNPs obtained from GWAS with KASP markers, KASP analysis has also revealed the presence of these SNPs on the same accessions as their original SNPs among the accessions. KASP analysis has retained 21 SNPs which showed homozygosity, and four SNPs (Bn-A05-p13327224, Bn-A06-p3580313, Bn-C06-p17371730, Bn-C09-p38094742) were not confirmed by KASP analysis (Supplementary Table S13). However, few of these SNPs has shown heterozygosity in different accessions. Overall, KASP analysis revealed that majority of our SNPs are true and original, hence these markers are very important to utilize them in drought breeding programs in rapeseed.
Expression profile of putative candidate genes
To validate the GWAS data, the expression of ten genes was determined by quantitative real-time polymerase chain reaction (qRT-PCR) in a drought tolerant and a drought susceptible line under drought-stress conditions (Supplementary Fig S4). The expression level of eight genes (BnA10g03880D, BnC06g14590D, BnaA03g33890D, BnaA05g01330D, BnaA05g17490D, BnaC04g50970D, BnaA02g31890D, and BnaA04g05410D) was significantly higher in the drought tolerant line than in the drought sensitive line (Supplementary Fig S4). The other two genes (BnaC01g03570D and BnaA03g01490D) showed significantly higher expression in the drought sensitive line relative to the drought tolerant line.
Discussion
Germination and seedling growth affected by drought
Drought tolerance is a very complex quantitative trait which is affected by the timing and severity of the stress relative to plant development and growth, but which nevertheless causes negative effects on every stage of plant development. B. napus is very sensitive to the drought stress during all growth stages from seed germination to seed setting. Drought stress can not only inhibit germination and seedling establishment, but it also has adverse effects on yield in rapeseed. Hence, improvement of drought tolerance in rapeseed is a major breeding goal. In the present study, we genotyped 228 B. napus accessions and phenotyped seeds and seedlings under normal and PEG-mediated drought stress conditions using different drought phenotyping indices. Significant diversity for all four traits was observed under both control and drought stress across the association panel, and drought significantly affected the measured traits (Supplementary Fig S1). All four traits assessed (root length, root fresh weight, germination rate and seedling vigor) were significantly correlated, with the exception of germination rate which was only associated with root fresh weight under drought stress. This may suggest that different mechanisms affect germination rate and root development under drought stress compared to under non-drought stress conditions. Since, seed vigor index combines germination and seedling growth, and STI is an efficient indicator of investigating drought tolerance status of lines, we used them to differentiate the tolerance levels of accessions. All the accessions used in this study has shown wide range of variability towards drought tolerance (Supplementary Fig S5). All the measured traits showed high heritability (97.7–98.1%), suggesting both that drought tolerance at the seedling stage in rapeseed is primarily determined by genetics, and also that we captured most of this genetic variation using SNPs in our association panel. Variation (CV) in seedling vigor index was however high relative to that of the other traits: this may be due to the fact that this index combines seedling and root length and seed germination, where seed germination is very strongly influenced by drought stress. Both STI and SSI phenotyping indices provided suitable, high quality data for phenotyping drought tolerance.
Identification of SNPs significantly associated with drought tolerance
We employed GWAS based on the SSI and STI values to uncover the genetic basis of drought tolerance at the seedling stage. We identified 314 SNPs across all chromosomes (108 in the A genome and 206 in the C genome) which were significantly associated with the four seedling traits. Of these SNPs, more than half (192/314) were detected through SSI-GWAS analysis, and 122/314 were detected based on STI-GWAS analysis. Hence, both drought indices (STI and SSI) proved to be good measures for the GWAS. However, In addition, we compared our SNP positions with previously reported QTLs, and all SNPs identified in the present study were novel relative to published results. This could be due to our experimental design: most previously published studies relied on yield under field conditions (e.g. in rainout shelters) as a proxy for drought tolerance, which is likely to pick up entirely different mechanisms than those operating at the seedling stage in our study.
Identifying superior allelic variants is an important requirement for breeding for drought tolerance. Overall, our association analysis identified a higher frequency of superior alleles than inferior alleles, with the highest proportion of superior alleles associated with germination rate (Fig. 5). Interestingly, although more SNP markers related to drought tolerance were identified via SSI-based analysis, a higher number of superior alleles were identified through STI-based analysis. These alleles are of particular importance for marker-assisted selection to improve drought tolerance in rapeseed breeding programs.
Genes associated with drought tolerance
Although a number of studies have been conducted to uncover the genetic factors controlling drought tolerance in B. napus [18], [19], our understanding of the mechanisms underlying drought tolerance is still limited. A key step in genomic assisted breeding for drought tolerance in canola involves characterization of functional drought stress tolerance genes or identification of markers closely associated with these genes [39]. Our GWAS study identified 85 candidate genes linked with 52 significant SNPs associated with four drought-tolerance traits. In many cases, genes identified were very close to the SNPs: for instance, BnaA10g03880D and BnaC06g14590D genes encoding putative orthologs of POM1 and PIP2 in Arabidopsis were found to be linked with SNPs only 8.65 and 5.72 kb away respectively. Homeobox domain genes BnaA07g30820D, BnaA05g01190D and BnaC04g50970D were identified, and are putatively orthologous to the HDG11 and HB-7 genes of Arabidopsis which are known to be involved in drought stress responses. The gene HB-7 belongs to the homeodomain-leucine zipper subfamily I (HD-Zip I) which is responsible for the control of plant development and abiotic responses [40]. This gene has also been reported to be induced by water stress conditions and ABA response in Arabidopsis [41], and to influence plant phenotype when over-expressed, affecting plant growth especially in the leaves and inflorescence stems [42]. HDG11 is the member of a homeodomain-START transcription factor family known to play a vital role in drought stress response and plant development: HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis [43]. When over-expressed, it was found to enhance drought tolerance in Arabidopsis, and also to confer drought tolerance and subsequently increase yield in transgenic rice [44]. Our study also identified two MYB-like binding genes (BnaA01g17750D and BnaC07g27330D) that are homologous to the ARR2 and MYB78 genes in Arabidopsis: MYB genes play an important role in many aspects of plant development and growth, and in response to abiotic and biotic stresses [45]. Several transcription factor-related genes were identified: BnaC03g23420D, BnaA05g01330D, BnaA05g18020D, BnaA03g18200D, BnaA10g17670D, BnaC04g01070D, BnaC05g42130D, BnaC07g27220D and BnaC07g44670D, encoding homologs of the TFIIB, AIB, MYC2, NF-YB1, HSF3, GBF3, DREB2B, BZIP1, ABF3 genes in Arabidopsis. TFIIB genes were found to be involved in pollen and endosperm development in Arabidopsis [46], while gene AIB is upregulated under drought and oxidative stress in Arabidopsis [47]. ABF3 is a member of the basic leucine zipper (bZIP) sub-family of transcription factors in Arabidopsis, and it is induced by both drought and abscisic acid [48]. It is well documented that MYC2 is the major regulator of the both the jasmonic acid and abscisic acid signalling pathways, which indicates that MYC2 is involved in the regulation of the crosstalk between these two pathways in drought stress response [49].
Two genes (BnaA03g01490D and BnaA10g14790D) were identified as belonging to the calcium dependent kinase group and to be homologous to the CDPK1 and CDKP-9 genes in Arabidopsis. Several lines of evidence suggest that CDPK genes are involved in drought stress response [50]. CDPKI genes have been found to play an important role in stomatal closure, and to show induced expression in response to the plant hormones salicylic acid, abscisic acid and methyl jasmonate [51]. Beside this, Mittal et al. [50] reported the involvement of ZmCDPK1 in regulating leaf, root and shoot development under drought stress, and ZmCDPK9 showed higher drought-related expression in leaves compared to shoots. These results clearly indicate that this CDPKI is involved in the plant defense response. Beside this, we identified several other important genes which play vital roles in drought tolerance and seedling development. These genes include BnaC03g23350D, BnaA04g05410D, BnaA06g25550D, BnaA10g25000D, BnaC01g15850D and BnaC05g01410D, with homologs of CID1, PUB22, ERD7, DREB2A, NLP7 and DR in Arabidopsis. We suggest that these genes identified as previously associated with drought response in other species as well as novel genes identified in this study as putatively associated with drought tolerance responses should be considered for further studies on drought tolerance in rapeseed.
Identification of hotspot/gene cluster regions on the chromosomes
Clustering of QTLs for different traits is a common phenomenon in crops (references). We identified ten clusters on six chromosomes carrying multiple SNPs associated with drought tolerance: four clusters on chromosome C02, two clusters on C09 and additional clusters on chromosomes A05, A08, C06 and C03 (Table 3). These clusters contained from 6 to 18 significant SNP-trait associations. Eight of these clusters were associated only with root fresh weight, while the remaining two were associated with germination rate, root length, root fresh weight, and seedling vigor index. The largest cluster was on C02, and harbored 18 SNPs within a 2.12 kb region associated with root fresh weight. Several candidate genes within these cluster regions were identified. Four genes (BnaA05g14890D, BnaC09g14910D, BnaA01g17750D and BnaA01g17870D) which play important roles in drought tolerance were detected in these hotspot regions, and these genes were homologous to BGL1, MATRIXIN, ARR2 and MAP70-5 in Arabidopsis. Hotspot/cluster regions may be of major interest and utility for Marker Assisted Breeding (MAS) programs, and provide a way forward to improve and understand drought tolerance in B. napus.
Table 3.
Hot spot regions for drought tolerance on various chromosomes.
| Cluster name | Position of the cluster | Size of the cluster (Kb) | No. of associations within cluster | Name of the SNPs found within cluster | Traits associated with cluster |
|---|---|---|---|---|---|
| A08-cluster | 4107072–4107139 | 0.67 | 6 | Bn-A08-p4107072 | RFW |
| Bn-A08-p4107084 | |||||
| Bn-A08-p4107097 | |||||
| Bn-A08-p4107113 | |||||
| Bn-A08-p4107120 | |||||
| Bn-A08-p4107139 | |||||
| C03-cluster SCAFFOLDC03_RANDOM | 4725238–4725547 | 0.399 | 12 | Bn-SC03-p4725238 | RFW |
| Bn-SC03-p4725248 | |||||
| Bn-SC03-p4725249 | |||||
| Bn-SC03-p4725251 | |||||
| Bn-SC03-p4725260 | |||||
| Bn-SC03-p4725287 | |||||
| Bn-SC03-p4725292 | |||||
| Bn-SC03-p4725295 | |||||
| Bn-SC03-p4725301 | |||||
| Bn-SC03-p4725317 | |||||
| Bn-SC03-p4725518 | |||||
| Bn-SC03-p4725547 | |||||
| A05-cluster | 9415440–9438724 | 23.284 | 15 | Bn-A05-p9415440 | GR, RFW |
| Bn-A05-p9415445 | |||||
| Bn-A05-p9415483 | |||||
| Bn-A05-p9415605 | |||||
| Bn-A05-p9415635 | |||||
| Bn-A05-p9415638 | |||||
| Bn-A05-p9415646 | |||||
| Bn-A05-p9415660 | |||||
| Bn-A05-p9415692 | |||||
| Bn-A01-p9438473 | |||||
| Bn-A01-p9438519 | |||||
| Bn-A01-p9438520 | |||||
| Bn-A01-p9438709 | |||||
| Bn-A01-p9438712 | |||||
| Bn-A01-p9438724 | |||||
| C09-cluster-1 | 10939116–10939426 | 0.31 | 11 | Bn-C09-p10939116 | RFW |
| Bn-C09-p10939123 | |||||
| Bn-C09-p10939129 | |||||
| Bn-C09-p10939131 | |||||
| Bn-C09-p10939134 | |||||
| Bn-C09-p10939150 | |||||
| Bn-C09-p10939160 | |||||
| Bn-C09-p10939163 | |||||
| Bn-C09-p10939193 | |||||
| Bn-C09-p10939198 | |||||
| Bn-C09-p10939426 | |||||
| C09-cluster-2 | 11492981–11493070 | 0.89 | 6 | Bn-C09-p11492981 | RFW |
| Bn-C09-p11492983 | |||||
| Bn-C09-p11492997 | |||||
| Bn-C09-p11493057 | |||||
| Bn-C09-p11493062 | |||||
| Bn-C09-p11493070 | |||||
| C06-cluster | 17371522–17371765 | 0.243 | 6 | Bn-C06-p17371522 | RL, SVI |
| Bn-C06-p17371576 | |||||
| Bn-C06-p17371585 | |||||
| Bn-C06-p17371730 | |||||
| Bn-C06-p17371761 | |||||
| Bn-C06-p17371765 | |||||
| C02-cluster-1 | 21679637–21681743 | 2.106 | 18 | Bn-C02-p21679637 | RFW |
| Bn-C02-p21679661 | |||||
| Bn-C02-p21679692 | |||||
| Bn-C02-p21679715 | |||||
| Bn-C02-p21679741 | |||||
| Bn-C02-p21679896 | |||||
| Bn-C02-p21679898 | |||||
| Bn-C02-p21679901 | |||||
| Bn-C02-p21679915 | |||||
| Bn-C02-p21679927 | |||||
| Bn-C02-p21679973 | |||||
| Bn-C02-p21679979 | |||||
| Bn-C02-p21681428 | |||||
| Bn-C02-p21681467 | |||||
| Bn-C02-p21681665 | |||||
| Bn-C02-p21681701 | |||||
| Bn-C02-p21681736 | |||||
| Bn-C02-p21681743 | |||||
| C02-cluster-2 | 32264521–32264802 | 0.281 | 9 | Bn-C02-p32264521 | RFW |
| Bn-C02-p32264526 | |||||
| Bn-C02-p32264534 | |||||
| Bn-C02-p32264549 | |||||
| Bn-C02-p32264556 | |||||
| Bn-C02-p32264559 | |||||
| Bn-C02-p32264594 | |||||
| Bn-C02-p32264783 | |||||
| Bn-C02-p32264802 | |||||
| C02-cluster-3 | 32580540–32580866 | 0.326 | 9 | Bn-C02-p32580540 | RFW |
| Bn-C02-p32580541 | |||||
| Bn-C02-p32580572 | |||||
| Bn-C02-p32580585 | |||||
| Bn-C02-p32580601 | |||||
| Bn-C02-p32580624 | |||||
| Bn-C02-p32580789 | |||||
| Bn-C02-p32580799 | |||||
| Bn-C02-p32580866 | |||||
| C02-cluster-4 | 34530033–34643074 | 113.041 | 8 | Bn-C02-p34530033 | RFW |
| Bn-C02-p34642787 | |||||
| Bn-C02-p34642871 | |||||
| Bn-C02-p34642967 | |||||
| Bn-C02-p34642972 | |||||
| Bn-C02-p34642992 | |||||
| Bn-C02-p34643071 | |||||
| Bn-C02-p34643074 | |||||
Conclusions
The panel of B. napus accessions used for GWAS in our study showed large natural variation for drought tolerance as assessed by four seedling traits. Benefitting from the relatively high resolution of GWAS, our study identified 314 SNPs/biomarkers significantly influencing drought stress responses based on SSI and STI phenotyping indices. Comprehensive drought profiling followed by GWAS mapping using this panel of accessions and facilitated large-scale candidate gene identification. Via BLAST alignment and GO analysis, we identified 85 candidate genes homologous to Arabidopsis genes known to be associated with drought stress. We also identified superior alleles conferring increased drought tolerance in the accessions used. Although more significant marker-trait associations were detected using STI, SSI proved more useful in identifying alleles conferring improved drought tolerance, suggesting that these indices may elucidate slightly different mechanisms. Overall, our results provide a valuable reference for the study of drought tolerance in B. napus. These SNP loci, superior alleles, accessions carrying these desired alleles and candidate genes will be useful for future drought tolerance breeding programs in rapeseed. Besides this, the integrative approach of using SSI and STI described here seems to comprise a viable potential strategy for interactive functional genomics studies to elucidate overall genetic regulation of drought response in crop improvement.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The research is financially supported by the National Natural Science Foundation of China (31860417). ASM is supported by DFG Emmy Noether grant MA6473/1-1.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.05.019.
Contributor Information
Qinghong Zhou, Email: qinghongzhou@126.com.
Donghui Fu, Email: fudhui@163.com.
Yingjin Huang, Email: yjhuang_cn@126.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Mantri N., Patade V., Penna S., Ford R., Pang E. Springer; 2012. Abiotic stress responses in plants: present and future. Abiotic stress responses in plants; pp. 1–19. [Google Scholar]
- 2.Macar T.K., Turan Ö., Ekmekçi Y. Effects of water deficit induced by PEG and NaCl on chickpea (Cicer arietinum L.) cultivars and lines at early seedling stages. Gazi Univ J Sci. 2009 [Google Scholar]
- 3.Zhu M., Monroe J.G., Suhail Y., Villiers F., Mullen J., Pater D. Molecular and systems approaches towards drought-tolerant canola crops. New Phytol. 2016;210:1169–1189. doi: 10.1111/nph.13866. [DOI] [PubMed] [Google Scholar]
- 4.Dhanda S., Sethi G., Behl R. Indices of drought tolerance in wheat genotypes at early stages of plant growth. J Agron Crop Sci. 2004;190:6–12. [Google Scholar]
- 5.Huang X., Zhao Y., Li C., Wang A., Zhao Q., Li W. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2012;44:32. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 6.Li H., Peng Z., Yang X., Wang W., Fu J., Wang J. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 2013;45:43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 7.Xu L., Hu K., Zhang Z., Guan C., Chen S., Hua W. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.) DNA Res. 2016;23:43–52. doi: 10.1093/dnares/dsv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P., Yang C., Chen H., Song C., Zhang X., Wang D. Transcriptomic basis for drought-resistance in Brassica napus L. Sci Rep. 2017;7:40532. doi: 10.1038/srep40532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Guo Z., Lv Y., Cen X., Ding X., Wu H. Genetic control of the root system in rice under normal and drought stress conditions by genome-wide association study. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bac-Molenaar J.A., Granier C., Keurentjes J.J., Vreugdenhil D. Genome-wide association mapping of time-dependent growth responses to moderate drought stress in Arabidopsis. Plant Cell Environ. 2016;39:88–102. doi: 10.1111/pce.12595. [DOI] [PubMed] [Google Scholar]
- 11.Wehner G.G., Balko C.C., Enders M.M., Humbeck K.K., Ordon F.F. Identification of genomic regions involved in tolerance to drought stress and drought stress induced leaf senescence in juvenile barley. BMC Plant Biol. 2015;15:125. doi: 10.1186/s12870-015-0524-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin P., Lin Y., Hu Y., Liu K., Mao S., Li Z. Genome-wide association study of drought-related resistance traits in Aegilops tauschii. Genet Mol Biol. 2016;39:398–407. doi: 10.1590/1678-4685-GMB-2015-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Fan C., Li J., Cai G., Yang Q., Wu J. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor Appl Genet. 2016;129:1203–1215. doi: 10.1007/s00122-016-2697-z. [DOI] [PubMed] [Google Scholar]
- 14.Bus A., Korber N., Parkin I.A., Samans B., Snowdon R.J., Li J. Species- and genome-wide dissection of the shoot ionome in Brassica napus and its relationship to seedling development. Front Plant Sci. 2014;5:485. doi: 10.3389/fpls.2014.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai D., Xiao Y., Yang W., Ye W., Wang B., Younas M. Association mapping of six yield-related traits in rapeseed (Brassica napus L.) Theor Appl Genet. 2014;127:85–96. doi: 10.1007/s00122-013-2203-9. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q., Han D., Mason A.S., Zhou C., Zheng W., Li Y. Earliness traits in rapeseed (Brassica napus): SNP loci and candidate genes identified by genome-wide association analysis. DNA Res. 2017;25:229–244. doi: 10.1093/dnares/dsx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher R.S., Mullen J.L., Heiliger A., McKay J.K. QTL analysis of root morphology, flowering time, and yield reveals trade-offs in response to drought in Brassica napus. J Exp Bot. 2015;66:245–256. doi: 10.1093/jxb/eru423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Mason A.S., Wu J., Liu S., Zhang X., Luo T. Identification of Putative Candidate Genes for Water Stress Tolerance in Canola (Brassica napus) Front Plant Sci. 2015;6:1058. doi: 10.3389/fpls.2015.01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan M., Liao F., Hou L., Wang J., Wei L., Jian H. Genome-wide association analysis of seed germination percentage and germination index in Brassica napus L. under salt and drought stresses. Euphytica. 2017;213:40. [Google Scholar]
- 20.Zhou Q., Zhou C., Zheng W., Mason A.S., Fan S., Wu C. Genome-wide SNP markers based on SLAF-seq uncover breeding traces in rapeseed (Brassica napus L.). Front Plant Sci. 2017;8:648. doi: 10.3389/fpls.2017.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun X., Liu D., Zhang X., Li W., Liu H., Hong W. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0058700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez GC, editor Effective selection criteria for assessing plant stress tolerance. In: Proceeding of the International Symposium on Adaptation of Vegetables and other Food Crops in Temperature and Water Stress, Aug 13–16, Shanhua, Taiwan, 1992; 1992.
- 23.Fischer R., Maurer R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust J Agric Res. 1978;29:897–912. [Google Scholar]
- 24.Murray M.G., Thompson W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalhoub B., Denoeud F., Liu S., Parkin I.A., Tang H., Wang X. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 26.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 30.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy O.J., Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- 32.Ginestet C. ggplot2: elegant graphics for data analysis. J Roy Stat Soc Ser A (Stat Soc) 2011;174:245–246. [Google Scholar]
- 33.Turner S.D. qqman: an R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv. 2014 [Google Scholar]
- 34.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Wang N., Akey J.M., Zhang K., Chakraborty R., Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet. 2002;71:1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raman H., Raman R., Coombes N., Song J., Prangnell R., Bandaranayake C. Genome-wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 2016;39:1228–1239. doi: 10.1111/pce.12644. [DOI] [PubMed] [Google Scholar]
- 37.Chaumont F., Tyerman S.D. Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 2014;164:1600–1618. doi: 10.1104/pp.113.233791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hickman R., Hill C., Penfold C.A., Breeze E., Bowden L., Moore J.D. A local regulatory network around three NAC transcription factors in stress responses and senescence in A rabidopsis leaves. The Plant J. 2013;75:26–39. doi: 10.1111/tpj.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Y., Warburton M.L., Sawkins M., Zhang X., Setter T., Xu Y. Genome-wide association analysis for nine agronomic traits in maize under well-watered and water-stressed conditions. Theor Appl Genet. 2013;126:2587–2596. doi: 10.1007/s00122-013-2158-x. [DOI] [PubMed] [Google Scholar]
- 40.Re D.A., Capella M., Bonaventure G., Chan R.L. Arabidopsis AtHB7 and AtHB12 evolved divergently to fine tune processes associated with growth and responses to water stress. BMC Plant Biol. 2014;14:150. doi: 10.1186/1471-2229-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Söderman E., Mattsson J., Engström P. The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 1996;10:375–381. doi: 10.1046/j.1365-313x.1996.10020375.x. [DOI] [PubMed] [Google Scholar]
- 42.Hjellström M., Olsson A.S., Engström P., Söderman E. Constitutive expression of the water deficit-inducible homeobox gene ATHB7 in transgenic Arabidopsis causes a suppression of stem elongation growth. Plant, Cell Environ. 2003;26:1127–1136. [Google Scholar]
- 43.Xu P., Cai X.T., Wang Y., Xing L., Chen Q., Xiang C.B. HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. J Exp Bot. 2014;65:4285–4295. doi: 10.1093/jxb/eru202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162:1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Zhou J.-J., Liang Y., Niu Q.-K., Chen L.-Q., Zhang X.-Q., Ye D. The Arabidopsis general transcription factor TFIIB1 (AtTFIIB1) is required for pollen tube growth and endosperm development. J Exp Bot. 2013;64:2205–2218. doi: 10.1093/jxb/ert078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Babitha K.C., Ramu S.V., Pruthvi V., Mahesh P., Nataraja K.N., Udayakumar M. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2013;22:327–341. doi: 10.1007/s11248-012-9645-8. [DOI] [PubMed] [Google Scholar]
- 48.Fujita Y., Yoshida T., Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol Plant. 2013;147:15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- 49.Liu S., Lv Z., Liu Y., Li L., Zhang L. Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet Mol Biol. 2018;41:624–637. doi: 10.1590/1678-4685-GMB-2017-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mittal S., Mallikarjuna M.G., Rao A.R., Jain P.A., Dash P.K., Thirunavukkarasu N. Comparative analysis of CDPK family in maize, arabidopsis, rice, and sorghum revealed potential targets for drought tolerance improvement. Front Chem. 2017;5:115. doi: 10.3389/fchem.2017.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nie L., Wang R., Xia Y., Li G. CDPK1, an Arabidopsis thaliana calcium-dependent protein kinase, is involved in plant defense response. Russ J Plant Physiol. 2015;62:866–874. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.