Abstract
Cancer and organ injury—such as that occurring in the perioperative period, including acute lung injury, myocardial infarction, and acute gut injury—are among the leading causes of death in the United States and impose a significant impact on quality of life. MicroRNAs (miRNAs) have been studied extensively during the last two decades for their role as regulators of gene expression, their translational application as diagnostic markers, and their potential as therapeutic targets for disease treatment. Despite promising preclinical outcomes implicating miRNA targets in disease treatment, only a few miRNAs have reached clinical trials. This likely relates to difficulties in the delivery of miRNA drugs to their targets to achieve efficient inhibition or overexpression. Therefore, understanding how to efficiently deliver miRNAs into diseased tissues and specific cell types in patients is critical. This review summarizes current knowledge on various approaches to deliver therapeutic miRNAs or miRNA inhibitors and highlights current progress in miRNA-based disease therapy that has reached clinical trials. Based on ongoing advances in miRNA delivery, we believe that additional therapeutic approaches to modulate miRNA function will soon enter routine medical treatment of human disease, particularly for cancer or perioperative organ injury.
Significance Statement
MicroRNAs have been studied extensively during the last two decades in cancer and organ injury, including acute lung injury, myocardial infarction, and acute gut injury, for their regulation of gene expression, application as diagnostic markers, and therapeutic potentials. In this review, we specifically emphasize the pros and cons of different delivery approaches to modulate microRNAs, as well as the most recent exciting progress in the field of therapeutic targeting of microRNAs for disease treatment in patients.
I. Introduction
Considered together, cancer and perioperative organ injury are among the leading causes of death in Western countries. According to the Center for Disease Control and Prevention’s National Vital Statistics, cancer alone accounts for 21% of deaths and is the second leading cause of mortality in the United States. Coupled with the economic burden of care, which is predicted to increase to 173 billion dollars by 2020, cancer poses a significant impact on society (Mariotto et al., 2011). The identification of therapeutic targets for cancer treatment has been an area of intense research for several decades. Advancements in cancer immunotherapy have improved outcomes for certain subsets, such as melanoma, bladder cancer, kidney cancer, and non-Hodgkin lymphoma. However, there is still an urgent need for the development of new therapeutic targets for cancer treatment. Perioperative organ injury, including acute respiratory distress syndrome (ARDS), myocardial infarction, and acute gut injury, account for the third leading cause of death in the United States if considered as a separate catalog (Bartels et al., 2013). For instance, ARDS alone accounts for more than an estimated 200,000 deaths per year in the United States when extrapolated from the 2017 American Heart Association Annual Survey (Bellani et al., 2016). Thus, the development of novel therapeutic measures to prevent or treat perioperative organ injury is imperative to reduce mortality rates during the perioperative period.
In both cases, cancer and perioperative organ injury are often the results of dysregulation of genetic information as a response to environmental changes. In the last 2 decades, accumulating evidence has indicated that genetic and epigenetic alterations are not limited to protein-coding genes. Rather, noncoding RNAs, including microRNAs (miRNAs), have risen as a major player in mediating the regulation of gene expression into biologic phenotypes together with proteins. Noncoding RNAs (ncRNAs) are endogenously transcribed into functional RNA species but are not translated further into proteins, as they lack an open reading frame with start and stop codons. Although they are not used as templates for protein synthesis, ncRNAs have been found to play diverse functional roles in many biologic processes and disease development (Cech and Steitz, 2014). In general, ncRNAs can be divided into two groups based on the length of their final product. RNAs longer than 200 nucleotides are long noncoding RNAs, whereas RNAs that are shorter than 200 nucleotides are referred to as short noncoding RNAs.
Approximately 22 nucleotides in length, miRNAs are the smallest members of ncRNAs, and they are highly conserved evolutionarily (Bartel, 2009). Since the discovery of abnormal cell LINeage-14 (lin-14) from Caenorhabditis elegans in 1993, miRNAs have been the most heavily studied among short noncoding RNAs (Lee et al., 1993; Wightman et al., 1993) and have been identified in nearly every eukaryote, including in humans. Both studies during this time found that a small piece of RNA transcribed from the lin-4 gene regulates the expression of genes through sequence-specific binding. However, its impact on medical science has only been appreciated in the last 20 years, since the discovery of post-transcriptional gene silencing activity by short interfering RNAs (siRNAs) in plants in 1999 (Hamilton and Baulcombe, 1999). In 2001, RNA interference (RNAi) was also demonstrated in mammalian cells with synthetic artificial siRNAs (Elbashir et al., 2001). According to miRBase (http://www.mirbase.org/), there are approximately 2588 miRNAs in humans, and the number is still growing as new miRNAs are discovered. It has been estimated that more than 60% of human genes, involved in various biologic processes, are regulated by miRNAs (Bartel, 2009), such as during the cell cycle (Hatfield et al., 2005), differentiation and development (Chen et al., 2004), and metabolism (Boehm and Slack, 2006). miRNAs are also involved in the oncogenesis, progression, and metastasis phases of multiple types of cancer (Calin and Croce, 2006). Besides, many miRNAs have been identified as major regulators during the pathogenesis of perioperative organ injury, cardiovascular disease, inflammation, sepsis, anesthetic neurotoxicity, and infectious diseases, in which miRNA expression levels may play a role as useful perioperative biomarkers and pharmacologic targets (Neudecker et al., 2016). Interestingly, the same miRNAs of pathologic importance in certain chronic and inflammatory diseases seem to play similar functions in both cancer and perioperative organ injury. For example, miR-155, one of the most extensively studied oncogenic miRNAs in various types of cancer, is found to be overexpressed in human carotid plaques and promotes the proinflammatory activity of macrophages to exaggerate arteriosclerosis in cardiovascular diseases (Nazari-Jahantigh et al., 2012). A substantial body of evidence shows that each disease displays a unique profile of miRNA expression, which is distinct from miRNA expression in disease-free, normal tissue. These differentially regulated miRNAs are called “signature miRNAs.” Signature miRNAs are believed to serve as diagnostic or prognostic markers, which may improve the conventional detection methods currently in use. In addition, some, if not all, of the signature miRNAs may serve as useful therapeutic targets in the development of antidisease strategies. This review intends to describe the current knowledge of miRNAs in various human diseases by highlighting the most recent studies on perioperative organ injury and cancer. In addition, it will focus on novel approaches to alter miRNA function in human disease—an approach that has unequivocally important translational implications.
II. Biology, Regulation, and Detection of MicroRNA
A. Biogenesis and Working Mechanism of MicroRNA
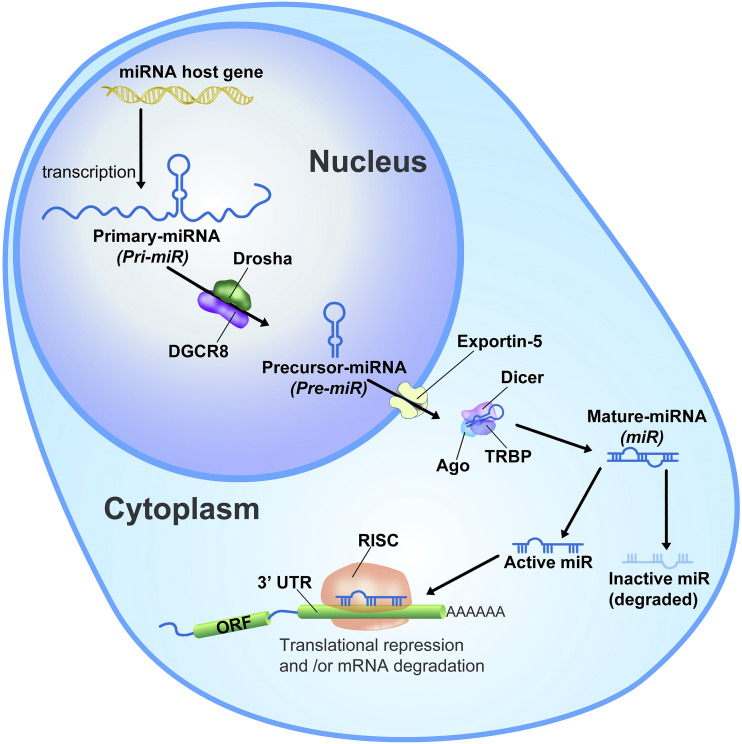
Approximately 70% of miRNA coding genes are associated with protein-coding host genes and are cotranscribed. Only 30% of miRNAs are transcribed from their own open reading frames located in intergenic areas (Bartel, 2009, 2018; Ha and Kim, 2014). In either case, during the expression process, miRNA coding sequences are first transcribed by RNA polymerase II into a long primary transcript of up to 10 kb, called primary-miRNA (pri-miRNA) (Lee et al., 2004) (Fig. 1). The pri-miRNA transcript is then processed by a nuclear RNase III called Drosha into a shorter length of transcript (about 70 bases), forming a hairpin-like structure, called precursor-miRNA (pre-miR) (Lee et al., 2003). The processed pre-miR is exported to the cytoplasm through nuclear membrane channel protein Exportin-5 (XPO5) (Yi et al., 2003). A recent study showed that XPO5 activity can be regulated by pre-miR phosphorylation. The phosphorylation by extracellular signal-regulated kinase suppresses the pre-miR export through XPO5 in cancers (Sun et al., 2016). Once translocated into the cytoplasm, the terminal hairpin loop of the pre-miR is further cleaved by Dicer, a cytoplasmic RNase III, to produce a 22-nucleotide–long mature miRNA (Hutvágner et al., 2001). Interestingly, it was suggested that intact RNAi machinery is critical to maintaining stem cell populations during early development. Studies in which Dicer was lost had a lethal effect, showing stem cell depletion in embryos during the early developmental stages of mice (Bernstein et al., 2003).
Fig. 1.
Biogenesis of miRNAs is a multistep process (Lee et al., 2003, 2004; Calin and Croce, 2006; Ha and Kim, 2014; Bartel, 2018). miRNA host genes are located in intragenic or intergenic regions and are primarily transcribed into a long, capped (Bartel, 2009), and polyadenylated transcript (pri-miRNAs) by RNA polymerase II, which is often longer than several kilobases (Lee et al., 2004). The pri-miRNAs are first processed in the nucleus into a shorter hairpin-structured transcript (pre-miRNAs) by the nuclear enzyme Drosha (Lee et al., 2003). The hairpin-loop pre-miRNAs are exported into the cytoplasm through a nuclear member channel protein, Exportin-5 (Yi et al., 2003). In the cytoplasm, pre-miRNAs are further processed by Dicer to a hairpin-free duplex form of miRNA called mature miRNAs (Hutvágner et al., 2001; Ketting et al., 2001). The duplex miRNAs bind with Argonaute 2 (Ago2) and transactivation-responsive RNA-binding protein (TRBP) to form RISC, and then a functional strand of the duplex miRNA remains in RISC until it binds to its target mRNA while the unfunctional strand is degraded. The eight-base-long seed sequences on the mature miRNA recognize and bind to their partial complementary sequences on the 3′ UTR of target gene mRNAs. The complex between miRNA and mRNA rapidly represses the translation of mRNA into proteins and eventually leads to mRNA degradation (Bartel, 2009, 2018; Eichhorn et al., 2014). DGCR8, Drosha and DiGeorge syndrome chromosome region; ORF, open reading frame.
The mature miRNA binds to Argonaute 2 and transactivation-responsive RNA-binding protein to form the RNA-induced silencing complex (RISC) (Redfern et al., 2013). Although associated with RISC, inactive strands, mostly 3′ end, are degraded, leaving the other active strand with the RISC until guided to the 3′ untranslated region (3′ UTR) of target mRNAs (Ha and Kim, 2014). siRNA is also processed, in a similar way to miRNAs, to a double-stranded RNA by Dicer and loaded onto RISC to bind to its target mRNA. The binding between the target mRNA and siRNA is fully complementary (Carthew and Sontheimer, 2009). Unlike the working mechanism of siRNA, miRNAs recognize their target mRNAs through partial complementary sequences between the second and the eighth nucleotide, called “seed sequences.” Once the seed sequence region of miRNA forms the partial base pairing with the 3′ UTR of target mRNA, the miRNA/mRNA complexes are unable to associate with active ribosomal complexes to complete protein synthesis (Bartel, 2009, 2018). This partial complementary base pairing between miRNAs and 3′ UTR of target mRNAs can vary depending on the target mRNA. Therefore, in theory, a single miRNA can bind to hundreds of different target mRNAs. There are several computer-based prediction algorithms to predict miRNA targeting. However, because of the complex nature of partial base pairing, it is still unsatisfactory to predict potent miRNA binding sites with reliable accuracy. In addition to the translational repression role of miRNAs (Olsen and Ambros, 1999), it has also been found that miRNAs can cause target mRNA degradation through the deadenylation of the target mRNA poly-A tail (Bagga et al., 2005). David Bartel’s laboratory proposed from their global steady-state measurements that the irreversible process of mRNA destabilization by miRNAs often overwhelms miRNA-based translation repression in a rapid, albeit relatively weak, manner (Eichhorn et al., 2014). Through these two major mechanisms of miRNAs, aberrantly regulated miRNAs can reprogram a significant amount of intracellular signal pathways over a threshold limit to cause disease development including human cancers (Bartel, 2018).
B. Regulation of MicroRNA Expression
Since mature miRNAs regulate the expression of multiple target genes, dysregulation of miRNA expression process can lead to abnormal gene expression profiles in cells, which in turn can provide a favorable environment for the development of organ injuries or even cancers (Ferrari et al., 2016; Neudecker et al., 2016, 2017c). Such deregulations of miRNA expression can take place at different stages of miRNA biogenesis. Transcription factors that regulate protein-encoding genes also regulate miRs, and some of these transcription factors are over- or underexpressed in diseases to modulate miR expression. For example, hypoxia-inducible factor (HIF) is stabilized during hypoxia and inflammation (Haeberle et al., 2008; Eltzschig and Carmeliet, 2011) and has been shown to be involved in the dysregulated expression of miRNAs in certain disease conditions (Eltzschig and Eckle, 2011; Choudhry et al., 2016). The center core region of tumor tissues becomes hypoxic because of the limited supply of oxygen via poor vasculatures and thus results in HIF-1α stabilization. In patients with breast cancer, miR-210 was induced by HIF-1α and showed an inverse correlation with disease-free and overall survival (Camps et al., 2008). Another hypoxia or ischemia-triggered transcription factor is specificity protein 1 (Sp1), which is stabilized during conditions of perioperative organ injury (Eltzschig et al., 2009; Hart et al., 2010). In cancers, Sp1 triggers a transcriptional activation of miRNAs, such as miR-205 in esophageal squamous cell carcinoma, to result in tumor progression and drug resistance (Pan et al., 2017). Besides, miRNAs also regulate Sp1 and other transcription factors and thereby serve as tumor suppressor–like molecules (Fulciniti et al., 2016; Hedrick et al., 2016; Li et al., 2018; Lv and Wang, 2018). Several studies demonstrated the cross talk between pro-oncogenic microRNAs and oncogene cellular myelocytomatosis (c-Myc) (Zheng et al., 2014; Chen et al., 2017; Yarushkin et al., 2017). The regulation of microRNAs by transcription factors will confer alternative strategies to target microRNAs as potential therapeutic interventions. Transcriptional inactivation of the miRNA-encoding gene through promoter hypermethylation has also been observed in many human diseases, including cancers. For example, the miR-127 expression can be silenced by promoter hypermethylation in bladder cancer cell lines and patients, and hypomethylating agents restored its expression level (Saito et al., 2006). miRNAs themselves can also modulate DNA methylation in cancers by interfering with the DNA methylation machinery (Holubekova et al., 2017). Single polymorphism (SNP) is another mechanism to produce variations on miRNA expressions and functions. Polymorphic genetic mutations of miRNA-binding sites in 3′ UTR of target mRNAs were proposed to be causative variants for human cancers (Chen and Rajewsky, 2006; Saunders et al., 2007). When the regions harboring two microRNA-binding sites in the proto-oncogene c-KIT gene from patients with papillary thyroid carcinoma were sequenced, a polymorphism of G>A SNP (rs17084733) was found in the proto-oncogene c-KIT 3′ UTR complementary sequences to the seed sequences of two miRNAs: miR-221/222 (He et al., 2005). In 5 out of 10 cases, this germline mutation increased the association between miR-221/222 and the target gene that was believed to contribute to cancer progression. Furthermore, when over 100 human cancer samples and cancer cell lines were examined by sequence variation comparison, it was found that the SNP (G>A mutation) at a 19th base pair of Lethal-7 (Let-7)e miRNA dramatically reduced the expression level of Let-7e (Wu et al., 2008b). On the other hand, SNP in miRNA sequences can also contribute to tumorigenesis. For instance, it was found that miR-16-1 carries germline mutations at a high frequency in chronic lymphocytic leukemia (CLL) (Calin et al., 2005). Overall, the frequency of SNPs in miRNA-binding sites or SNPs in miRNA sequences was predicted to be less than 1% because of the size of miRNA and its binding site (Saunders et al., 2007). However, successfully pinpointing such SNPs in miRNAs and its targets will provide a fine-tuned personalized miRNA signature for each cancer patient.
C. MicroRNA Detection
In the early days of miRNA research, a Northern blotting method using a specifically designed RNA probe, unlike Southern blotting, which uses DNA probes to hybridize to target DNA sequences, was widely used to detect miRNAs, similar to mRNA detection (Hunt et al., 2015). In Northern blotting, total RNAs were separated on urea-containing denatured PAGE and transferred to a positively charged nylon membrane. Then, a target RNA sequence is hybridized with a probe labeled with either radioactive isotopes (32P) or chemiluminescent enzymes, such as alkaline phosphatase or horseradish peroxidase. To better detect miRNAs by Northern blotting, the probes are usually generated with locked nucleic acids (LNAs), which give at least 10-fold higher sensitivity than DNA probes (Valoczi et al., 2004). Because of the high specificity and sensitivity of LNA-based probes, fluorescently labeled LNA probes are also often used to visualize specific miRNAs directly from tissue or cell samples by fluorescence in situ hybridization (Urbanek et al., 2015). Although Northern blotting is a reliable RNA detection method in terms of sensitivity and selectivity, it is a time-consuming procedure and involves the potential use of radioactive isotope–labeled probes. Therefore, it was critical to develop new methods that are optimized for high-throughput detection with comparable sensitivity and selectivity. The laboratory of Thomas Schmittgen applied quantitative real-time polymerase chain reaction (qRT-PCR) using fluorescent SyBr-Green dye to detect 23 pre-miRs from six human cancer cell lines (Schmittgen et al., 2004), which has launched the quantitative comparison of miRNA expressions at high-throughput levels. As more miRNAs and their significant roles in human diseases were identified, global screening at high throughput levels was needed. A custom-built microarray platform using oligonucleotide probes against miRNA precursors and mature miRNAs was introduced in 2004 for screening the genome-wide miRNA expression profiling in human cancer specimens (Calin et al., 2004a). This platform has been used to identify several sets of miRNA signatures from various types of human cancer specimens. qRT-PCR has also been widely used to detect individual miRNA signatures from human cancer samples. For example, Megaplex reverse transcription format has been employed to use stem-loop–specific primers for qRT-PCR–based miRNA arrays after preamplified cDNA (Mestdagh et al., 2008), which significantly reduced the amount of input RNA to 10 ng. Unlike a one-way hybridization-based microarray, the qRT-PCR–based array was able to track the changes of miRNA expression on different stages of miRNA maturation. qRT-PCR–based arrays on a set of primary effusion lymphomas found that 68 primary effusion lymphoma–specific signature miRNAs are amplified at three levels of expression: gene alteration, transcription (pre-miRNA), and processing (mature miRNA) (O’Hara et al., 2008). Since the qRT-PCR–based array uses a set of primers for preselected miRNAs, it is relatively more flexible to customize the array for a specific range of pathways or disease conditions (Chugh et al., 2010). These advancements in the detection of miRNAs enabled high-throughput profiling on numerous human specimens obtained from patients, leading to the discovery of signature miRNAs in the disease types as useful biomarkers.
III. MicroRNAs in Human Cancers
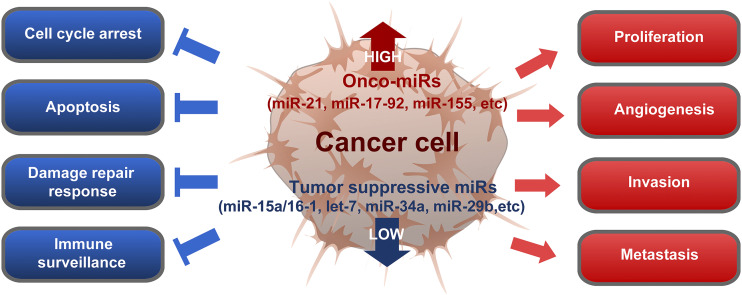
As a pivotal controller of cell cycle regulation, cell differentiation, immune responses, and inflammation, the functional role of miRNA in cancer biology has been intensively explored in the last decades. A series of seminal discoveries have identified the link between miRNAs and human cancers. These studies highlighted the differential expression of microRNA in CLL and its association with cancer prognosis and progression. Since then, miRNAs have been explored extensively as a signature and crucial regulator for cancer progression, which will be summarized and discussed in this review. In general, miRNAs that are downregulated in comparison with normal tissue are considered to be tumor suppressors, whereas upregulated miRNAs are oncogenic or provide a favorable environment to cancer cells for proliferation, progression, and metastasis (Fig. 2).
Fig. 2.
Functional orchestration to show how deregulated miRNAs can affect human cancers during malignant transformation, progression, invasion, and metastasis. Certain miRNAs are found to be downregulated in cancers, referred to as tumor-suppressive miRNAs (Calin et al., 2002, 2004a,b, 2005; Calin and Croce, 2006; Yanaihara et al., 2006). When those tumor-suppressive miRNAs, such as miR-15a/16-1 (Kasar et al., 2012), let-7 (Yu et al., 2007; Trang et al., 2010), miR-34a (Lodygin et al., 2008; Li et al., 2009, 2013a; Pang et al., 2010; Silber et al., 2012; Yamamura et al., 2012; Cosco et al., 2015; Gaur et al., 2015; Beg et al., 2017), and miR-29b (Huang et al., 2013; Wu et al., 2013; Xu et al., 2014), are reintroduced to the cancer cells, they induce cell cycle arrest, apoptosis, DNA damage response, and/or immune surveillance to inhibit cancer growth. On the other hand, some miRNAs, including miR-21 (Meng et al., 2007; Hatley et al., 2010; Ren et al., 2010; Gaur et al., 2011; Fabbri et al., 2012; Griveau et al., 2013; Pfeffer et al., 2015; Huo et al., 2017; Lee et al., 2017), miR-17-92 cluster (He et al., 2005; Sylvestre et al., 2007; Xiao et al., 2008; Olive et al., 2013), or miR-155 (Eis et al., 2005; Costinean et al., 2009; Wang et al., 2009; Fabani et al., 2010; Jurkovicova et al., 2014; Cheng et al., 2015), are often upregulated in cancer cells, referred to as onco-miRs, and they are responsible for cancer cell proliferation, angiogenesis, invasion, and metastasis. Therefore, the inhibition of active onco-miRs or suppression of onco-miR expression in cancer cells can also lead to cancer regression. The consequence of such miRNA deregulation is to reprogram the severe level of multiple cell signaling pathways enough to transform the fate of affected cells since single miRNA can repress the expression of multiple target genes. Development of a therapeutic approach based on targeting those deregulated miRNAs is expected to re-reprogram cellular function of cancer cells to execute suicide or sensitize to other conventional therapeutics by releasing target genes from the miRNA-based suppression.
A. MicroRNAs as Tumor Suppressors
The loss of function of certain miRNAs due to genomic deletions, mutations, epigenetic silencing, and/or miRNA processing alterations can cause malignant transformation of normal cells. In this case, those miRNAs are considered to normally play a tumor-suppressive role during cancer development and progression. Two signature miRNAs in CLL, miR-15a and miR-16-1, were discovered from a 30-kb deleted region between exons 2 and 5 of the deleted in lymphocytic leukemia 2 (DLEU2) gene from the 13q14.2 region. Frequent hemizygous or homozygous deletions of this region occur in more than 50% of CLL cases (Calin et al., 2002, 2004a, 2005), and these miRNAs were found to target antiapoptotic protein B-cell lymphoma 2 (BCL-2). As a result of the epigenetic deletion of the miR-15a and miR-16-1, BCL-2 is widely overexpressed in a subset of patients with CLL. In myelodysplasia and therapy-related acute myeloid leukemia (AML), the miR-29 family, miR-29b-1/miR-29a, was found in chromosome 7q32, which is a frequently deleted region in AML (Garzon et al., 2008, 2009). It was also inversely correlated with upregulated oncogenic proteins BCL-2 and myeloid cell leukemia 1 (MCL-1) (Xu et al., 2014). In lung cancers, the miR-29 family is also frequently downregulated and targets DNA methyltransferase (DNMT) 3A and DNMT3B (de novo methyltransferases), known to be highly upregulated in lung cancers with poor prognosis (Fabbri et al., 2007). In sarcoma, it was suggested that miR-29 play as a decoy for human antigen R by preventing the binding of human antigen R to the 3′ UTR of tumor suppressor A20 transcript (Balkhi et al., 2013). MiR-34a has also been recognized as a tumor-suppressive miRNA since it is highly expressed in most normal tissues but downregulated in most types of cancer through methylation on its 5′-C-phosphate-G-3′ island (Lodygin et al., 2008). It is also directly activated by p53 (He et al., 2007). Ectopic overexpression of miR-34a in cancer cells causes cell cycle arrest by targeting tyrosine-protein kinase Met, c-Myc, Cyclin D1, and cyclin-dependent kinase 6 (CDK6) (Sun et al., 2008; Li et al., 2009; Yamamura et al., 2012) or apoptosis by targeting notch receptor 1 and Bcl-2 (Pang et al., 2010; Li et al., 2013a). Interestingly, miR-34a was shown to directly target sirtuin 1, a known inhibitor of p53, leading to activation of p53 as a positive-feedback loop (Yamakuchi et al., 2008). The let-7 family consists of 12 miRNAs and is frequently found to be correlated to the development of lung, breast, urothelial, and cervical cancers. Because of its location at a fragile site and frequent downregulation in cancer, it is thought that the Let-7 family member miRNAs function as tumor suppressors (Calin et al., 2004b). For example, loss of Let-7 expression in lung cancer leads to the upregulation of one of its targets, Rat sarcoma (Ras) (Johnson et al., 2005). Interestingly, many of the tumor suppressor miRNAs (miR-15-a/16-1, miR-29s, and let-7) have multiple genomic location that expresses identical mature miRNAs, which can be the conserved mechanism through evolution to preserve the function of important miRNAs. Global miRNA profiling on mouse embryonic stem cells has revealed stem cell–specific signature miRNAs, which function in the regulation of cell development and the maintenance of pluripotency (Houbaviy et al., 2003). In cancer cell populations, some of the miRNAs seemed to involve cancer stem cell (CSC) formation and maintenance. Expression of tumor-suppressive let-7 was found to be downregulated in breast cancer stem cells, leading to the overexpression of H-RAS and high-mobility group AT-hook 2, which is responsible for stem cell–like self-renewal and reduced differentiation properties (Yu et al., 2007). In breast cancers, miR-200b was found as a cancer-specific signature miRNA and was epigenetically lost during CSC formation (Iliopoulos et al., 2010). The loss of miR-200b released the expression of its direct target gene, suppressor of zeste 12 protein homolog, which epigenetically modulates polycomb-mediated repression of the E-cadherin gene. Since CSCs have been proposed to initiate tumor formation and induce cancer recurrences, miRNA signatures enriched in CSC populations may become attractive therapeutic targets or serve as diagnostic/prognostic markers.
B. MicroRNAs as Oncogenes
In contrast to the tumor-suppressive miRNAs, a certain number of miRNAs are significantly upregulated in human cancers. They are considered to possess oncogenic roles and are often called onco-miRs. For example, miR-155 was identified from a transcript of a noncoding RNA gene, B-cell integration cluster, located in chromosome 21q23 in human B-cell lymphomas (Eis et al., 2005). MiR-155 is highly expressed in many other human cancers, including pediatric Burkitt lymphoma, Hodgkin disease, primary mediastinal non-Hodgkin lymphoma, CLL, AML, lung cancer, and breast cancer (Jurkovicova et al., 2014). Mice with B-cell targeted miR-155 overexpression showed a polyclonal preleukemia pre-B cell proliferation phenotype accompanied by a high level of B cell malignancy (Costinean et al., 2006). A later study in mice identified Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein β, both previously implicated in the interleukin-6 (IL-6) signaling pathway, as direct targets of MiR-155 (Costinean et al., 2009). Laboratory of Kalpana Ghoshal also found that nonalcoholic steatohepatitis is associated with a high level of miR-155 expression, leading to the development of hepatocellular carcinoma (HCC) (Wang et al., 2009). Single cistronic cluster miR-17-92 is located within 800 bp of the noncoding gene chromosome 13 in open reading frame 25 at 13q31.3. It is frequently amplified in follicular lymphoma and diffuse large B-cell lymphoma and produces six mature miRNAs, including miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1 (Olive et al., 2013). The expression levels of the miR-17-92 cluster are significantly upregulated in hematologic malignancies such as lymphomas, and in various solid tumors such as breast, colon, lung, pancreas, prostate, and stomach tumors (Olive et al., 2013). In a mouse model, miR-17-92 promoted tumor proliferation and angiogenesis while inhibiting tumor cells by modulating E2F transcription factor 1 (E2F1) expression (O’Donnell et al., 2005). Both E2F1 and E2F transcription factor 3 (E2F3) activate the miR-17-92 cluster as a regulatory loop (Sylvestre et al., 2007). Additionally, miR-21 is the most frequently upregulated miRNA in human cancer, including both hematopoietic and solid organs (Pfeffer et al., 2015). It is one of the most extensively studied miRNAs, with several known targets, including phosphatase and tensin homolog (Meng et al., 2007), programmed cell death 4 (Gaur et al., 2011), and BTG antiproliferation factor 2 (Liu et al., 2009). Although miR-21 expression has been found at a high level in most human cancers, transgenic mice overexpressing miR-21 throughout the whole body did not develop tumor formation. However, providing a second hit by crossing the miR-21 transgenic mice to activated k-Ras mutant mice increased tumor incidents in lung cancer by negatively regulated multiple tumor suppressors, including Sprouty homolog 1, Sprouty homolog 2, BTG antiproliferation factor 2, and programmed cell death 4, through activation of the Ras/mitogen-activated protein kinase kinase/extracellular signal-regulated kinase pathway (Hatley et al., 2010). Interestingly, overexpression of miR-21 in the spleen has been shown to lead to lethal B-cell lymphoma in mice, and more importantly, the malignancy was related to the expression level of miR-21 (Medina et al., 2010). This addiction of tumorigenesis to a specific miRNA was also observed in a lymphoma study with miR-155 overexpressing mice (Cheng et al., 2015). These oncogenic miRNAs have been suggested as promising therapeutic targets since their downregulation or knockdown induces apoptosis, cell cycle arrest, and inhibition of invasion and metastasis of cancer cells. Further investigation into onco-miRs may benefit the development of cancer therapeutic strategies, since targeting onco-miRs may be sufficient to inhibit tumor initiation and progression.
C. Circulating MicroRNAs as Biomarkers of Human Cancers
Early detection is one of the key factors in disease diagnosis that leads to a positive outcome. This is especially true in cancer because of the higher chance of metastasis associated with late diagnosis. Global profiling of miRNAs is time- and cost-consuming; thus, it is essential to narrow down to a limited number of “disease signature” microRNAs. In this regard, defining accurate biomarkers and affordable detection methods for different types of cancer is critical for patients’ long-term outcomes. The various types of human cancer have allowed researchers to pinpoint the most critical miRNAs. For early diagnosis, the discovery of miRNA signatures can be useful biomarkers in the determination of disease diagnosis and prognosis. Profiling on circulating RNAs extracted from plasma or serum has been proposed as a diagnostic and prognostic marker since this method is relatively simple, noninvasive, low-cost, and rapid (Tsang and Lo, 2007). The first attempt to use a profile of circulating miRNAs as a diagnostic biomarker was in serum RNA samples from patients with diffuse large B-cell lymphoma (DLBCL). Three signature miRNAs, miR-155, miR-210, and miR-21, were highly elevated in DCBCL compared with normal sera (Lawrie et al., 2008). At that time, the miRNA profiles seemed to match those in their primary tumor cells. In colorectal cancer (CRC), onco-miR miR-17-92 cluster members were found as an miRNA signature in serum, and these RNAs were similar to their primary CRC cells (Ng et al., 2009). In HCC, a profiling result performed in HCC tissue miRNAs identified miR-21 as the most enriched miRNA (Meng et al., 2007), and miR-21 was also highly enriched in circulating miRNAs (Tomimaru et al., 2012). However, some other studies also reported that miRNA signatures are not always correlated between primary cancer cells and serum. For example, the serum miRNA signature from patients with breast cancer overlapped with only seven miRNAs from primary breast cancer tissues. More importantly, 16 of the top 20 miRNAs in the serum miRNA signature were not found in the tissue miRNA profile (Chan et al., 2013). The discrepancy may be caused by the heterogeneous nature of cancer origins and genetic backgrounds in each patient or may simply be due to the methodology of sampling procedures, microarray methods, and validation strategies (Pritchard et al., 2012; Moldovan et al., 2014). The accuracy of using the circulating miRNA profiles as diagnostic and prognostic markers is still debatable.
IV. MicroRNAs in Perioperative Organ Injury
Perioperative organ injury is considered the third leading cause of death in the USA and has a significant impact on surgical patient’s long-term mortality and morbidity (Bartels et al., 2013). Despite substantial improvement in the management of surgical patients, single or multiorgan dysfunction still significantly contributes to inpatient mortality after noncardiac high-risk surgeries (Lobo et al., 2011). MicroRNAs have been studied extensively in the setting of perioperative organ injury for the identification of potential biomarkers and therapeutic targets. This is particularly important, especially for cancer patients, as elective surgery is quite common in this patient cohort as a treatment of cancer. The elective nature of surgeries for patients with cancer makes them the perfect candidate for prophylactic intervention to prevent perioperative organ injury. In this review, we will focus on three types of acute perioperative organ injuries: ARDS, myocardial infarction, and acute gut injury.
A. Acute Respiratory Distress Syndrome
ARDS is clinically defined by acute pulmonary onset after direct or indirect insult (such as pneumonia and sepsis), profound arterial hypoxemia, and bilateral radiographic opacities consistent with pulmonary edema (Ranieri et al., 2012). The pathogenesis of ARDS is characterized by exaggerated pulmonary inflammation, infiltration of immune cells, disrupted alveolar structure, and attenuated gas exchanges (Thompson et al., 2017). The incidence of ARDS is quite high in critically ill patients, as a recent study indicated that 23% of mechanically ventilated patients in the intensive care unit developed ARDS, with mortality up to 46% (Bellani et al., 2016). Patients who have recovered from ARDS also face long-term physiologic and psychologic challenges, with high risk for lifelong disability and decreased quality of life (Herridge et al., 2011). Previous studies demonstrated the importance of HIF-1A in the dampening of inflammation during acute lung injury (ALI) via transcriptional upregulation of the A2B adenosine receptor (Poth et al., 2013; Eckle et al., 2014). CD73 and adenosine receptors were also shown to be crucial in regulatory T cells during the resolution of ALI (Ehrentraut et al., 2012, 2013). Moreover, one of the mucin family members, Mucin 5AC, was identified as a proinflammatory mediator during ventilator-induced lung injury (VILI) (Koeppen et al., 2013). However, up to now, there is no pharmacologic therapy for the prevention and treatment of ARDS, whereas low tidal volume ventilation and conservative fluid management are the main lung-protective strategies in the management of patients with ARDS (Thompson et al., 2017). Thus, the discovery of a new molecular mechanism, such as the involvement of microRNAs, is essential for the development of novel therapeutic targets. Previous studies have identified several miRNAs that are involved in the resolution of inflammation through the regulation of resolvins, lipid mediators enzymatically generated to promote the crucial resolution process (Spite et al., 2009; Norling and Serhan, 2010). In particular, Dr. Charles Serhan’s laboratory found that resolvin D1 (RvD1) selectively upregulates miRNAs such as miR-21, miR-146b, and miR-219 and downregulates miR-208a, resulting in the reduction in the activity of nuclear factor kappa B and mothers against decapentaplegic family in macrophages (Recchiuti et al., 2011). They also found that the RvD1-triggered resolution of acute inflammation is mediated through two G protein–coupled receptors, formyl peptide receptor 2 and G protein–coupled receptor 32, which elevate the expression of miR-208a, resulting in interleukin-10 production (Krishnamoorthy et al., 2012). Interestingly, it was found that miR-4661 is overexpressed in neutrophils to initiate inflammation, whereas its overexpression in macrophages increases the production of specialized proresolving mediators, such as RvD1 and resolvin D5, to maintain homeostasis through accelerating the resolution process (Li et al., 2013b). Moreover, several studies have conducted screening experiments to identify differentially regulated pulmonary miRNAs during murine models of ARDS and in vitro mechanical stretch (Cai et al., 2012; Dong et al., 2012; Vaporidi et al., 2012; Yehya et al., 2012; Huang et al., 2014; Xiao et al., 2015; Mao et al., 2017). A recent study described the temporal changes in miRNA in peripheral blood mononuclear cells from patients with ARDS and identified 25 miRNAs differentially regulated in ARDS (Narute et al., 2017). However, because of the heterogeneity of human ARDS, a more comprehensive discovery study in patients with ARDS is still in dire need to identify potential biomarkers and therapeutic targets.
MiR-155 is one of the most profoundly upregulated microRNAs post–pulmonary infection and endotoxin exposure with a proinflammatory role to promote ALI (Rao et al., 2014; McAdams et al., 2015; Wang et al., 2016). Elevation of miR-155 is observed upon exposure to Staphylococcal enterotoxin B (SEB), a toxin known to cause food poisoning in humans (Rao et al., 2014). SEB exposure in mice significantly upregulates miR-155 levels in the lung-infiltrating mononuclear cells, and miR-155−/− mice display attenuated pulmonary inflammation and histopathological injury. Suppressor of cytokine signaling (SOCS-1), a negative regulator of immune responses, was identified as a potential target of miR-155, as a gain of function of miR-155 inhibits its expression, whereas the loss of function of miR-155 enhances its expression. Besides SEB, intratracheal instillation of lipopolysaccharide (LPS) also induced the expression of miR-155, and its increased expression is mainly contributed to by alveolar macrophages (Wang et al., 2016). Similar to previously described SEB-induced ALI, miR-155–deficient animals are protected from LPS-induced lung injury marked by reduced lung permeability and dampened inflammatory responses. The same study identifies and confirms that miR-155 directly targets the 3′ UTR of SOCS-1 and that the inhibition of SOCS-1 by miR-155 allows the initiation of inflammatory responses in macrophages upon LPS stimulation. Additional study of fetal lung injury in nonhuman primates further suggests the proinflammatory role of miR-155 during Streptococcal infection (McAdams et al., 2015). The expression of miR-155 increases after group B Streptococcal–exposed fetal lung and in immortalized human fetal airway epithelial cells after exposure to IL-6 and tumor necrosis factor-α. In fetal airway epithelial cells, overexpression of miR-155 raises the expression level of chemokine (C-C motif) ligand 5, IL-6, and C-X-C motif chemokine ligand 10. However, besides its proinflammatory function, miR-155 also directly targets the 3′ UTR of fibroblast growth factor 9, implicating a potential role for miR-155 in fetal lung development. Summarized from the studies above, pharmacological inhibition of miR-155 could potentially benefit patients with ARDS to dampen pulmonary inflammation.
Besides proinflammatory roles, microRNAs also contribute to the protection against ALI. For example, miR-146a is upregulated in LPS-stimulated THP-1 cells (human monocytic), LPS-induced rat ALI, and acid-induced murine ALI (Nahid et al., 2009; Zeng et al., 2013; Vergadi et al., 2014). Nahid et al. (2009) were the first to report miR-146a as a toll-like receptor 4 responsive microRNA in human monocytic cell lines, which gradually increases after 4 hours of LPS challenge and then peaks at 24 hours post–LPS challenge. Interestingly, the elevated expression of miR-146a correlated with dampening of tumor necrosis factor-α production, and the following analysis suggests that miR-146a is the only microRNA upregulated in the tolerated state of LPS challenge after prolonged exposure. Functionally, THP-1 cells with overexpression of miR-146a are less responsive to LPS stimulation, representing a tolerance state, whereas inhibition of miR-146a dampens the LPS tolerance effect. After extensive studies of miR-146a during LPS stimulation in vitro, the upregulation of miR-146a is also demonstrated in rat LPS-induced ALI (Zeng et al., 2013). However, the anti-inflammatory role of miR-146a still needs to be further studied in LPS-induced ALI. In vivo function of miR-146a is further implicated by Vergadi et al. (2014) in murine hydrochloric acid aspiration–induced ALI. MiR-146a levels have been observed to be induced upon hydrochloric acid aspiration and to be significantly higher in AKT serine/threonine kinase 2 deficient (Akt2−/−) mice, which are protected from acid-induced lung injury. MiR-146a mimic treatment significantly reduced acid-induced inducible NO synthase generation in alveolar macrophages, suggesting an anti-inflammatory function of miR-146a during aseptic ALI. In summary, the lung-protective role of miR-146a during ARDS/ALI has been suggested by several studies, and therapeutic enhancement of miR-146a could potentially improve the outcome of patients with ARDS.
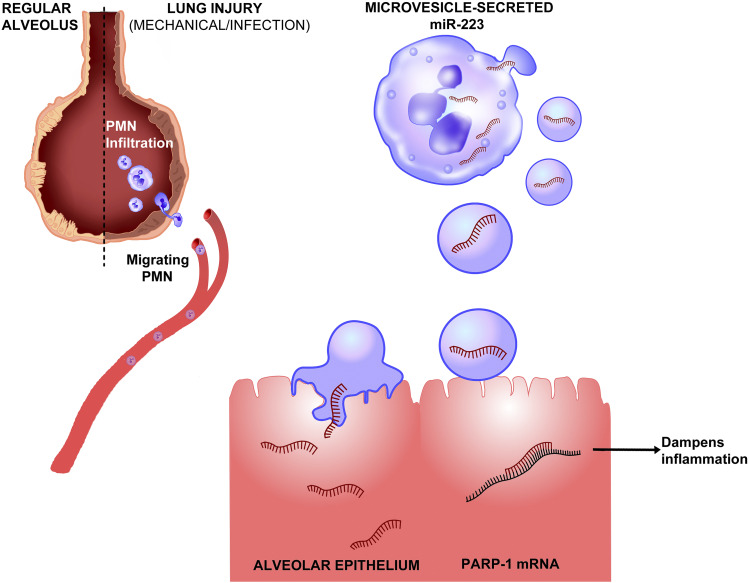
Another microRNA and a new mechanism that is capable of dampening inflammation in ARDS is the shuttling of miR-223 from infiltrating neutrophils to alveolar epithelial cells (Neudecker et al., 2017a). Neudecker et al. (2017a) defined a novel lung-protective role of miR-223 shuttling in ARDS in a recent study (Fig. 3). The upregulation of miR-223 has been observed in both patients with ARDS and mice exposed to VILI. The function of miR-223 in ARDS was addressed by exposing mice deficient in miR-223 (miR-223-/y mice) to VILI. Compared with wild-type counterparts, miR-223-/y mice demonstrated exaggerated inflammation and tissue injury, characterized by increased bronchoalveolar lavage albumin levels to and elevated IL-6 and chemokine (C-X-C motif) ligand 1 levels in the lung. Further mechanistic studies identified the interaction between the shuttling of miR-223 from polymorphonuclear neutrophils (PMNs) to alveolar epithelial cells in vitro and in vivo during murine ARDS. Specifically, miR-223 is released from PMNs via microvesicles, which are transferred into alveolar epithelial cells and dampen inflammatory gene expression and lung inflammation. A similar shuttling mechanism for miR-21 and miR-29a between primary cancer cells and normal recipient cells via exosomes has been previously reported during prometastatic inflammatory responses (Fabbri et al., 2012). Besides the VILI model of ARDS, miR-223-/y mice demonstrated exaggerated susceptibility to pulmonary bacterial infection, indicating a protective function of miR-223 in ALI. Moreover, poly (ADP-ribose) polymerase 1 (PARP-1) was identified as a target of miR-223 in this study, as the 3′ UTR of PARP-1 was directly targeted by miR-223. To confirm the functional relevance of the miR-223-PARP-1 interaction, pharmaceutical inhibition of PARP-1 in vivo attenuated the severity of VILI in miR-223-/y mice. Subsequently, nanoparticle delivery of miR-223 before the onset of murine ALI dampened pulmonary inflammation and tissue injury. To conclude, the shuttling of miR-223 between neutrophils and alveolar epithelial cells during ALI established a unique mechanism for microRNA-mediated lung protection during ARDS, and direct targeting of alveolar epithelial cells to deliver miR-223 mimetics could potentially dampen pulmonary inflammation in patients with ARDS. Inhalation of nanoparticle-based therapy to achieve overexpression or inhibition of microRNA in the lung would lead to substantial advances in the treatment and prevention of ARDS.
Fig. 3.
Shuttling of miR-223 protects against acute lung injury. Infection or mechanical ventilation can result in acute lung injury, during which neutrophils will transmigrate through the vasculature to inflamed alveolae (Dengler et al., 2013). Recruited neutrophils release microvesicles containing miR-223, a microRNA highly expressed in myeloid lineage cells. MiR-223–containing microvesicles are shuttled to pulmonary alveolar type II cells, resulting in the transcriptional repression of its inflammatory target gene PARP-1. Inhibition of PARP-1 leads to the attenuation of pulmonary inflammation and tissue injury. Therapeutically, nanoparticle delivery of miR-223 to pulmonary epithelial cells could potentially dampen pulmonary inflammation to prevent and treat acute lung injury (Neudecker et al., 2017a,c).
B. Acute Myocardial Infarction
Acute myocardial infarction (AMI) is clinically diagnosed by abnormally high levels of plasma cardiac troponin concentration, ischemia, EKG abnormality, imaging of intracoronary thrombus, and/or wall motion abnormality (Thygesen et al., 2012; Anderson and Morrow, 2017). The incidence of MI ranges from two to four cases per 1000 person-years globally and with 750,000 episodes of MI each year in the United States (Yusuf et al., 2014; Mozaffarian et al., 2016). Multicenter international cohort studies by Devereaux and colleagues demonstrated that in perioperative settings, AMI results in about five-times-higher mortality rates, posing a significant impact on surgical outcome (Devereaux et al., 2005, 2011). Clinical management of AMI includes percutaneous coronary interventions and supplemental oxygen. Pharmaceutical interventions for MI include a combination of antiplatelet and anticoagulant agents, β-blockers, nitrates, and statins. Although the mortality of AMI has been reduced significantly by traditional management and pharmaceutical interventions, it is still crucial for the development of novel therapeutic targets to prevent perioperative AMI and to improve the long-term prognosis of AMI. Previous studies have indicated the important role of extracellular nucleotide and nucleoside signaling in vascular inflammation and ischemic preconditioning (Eltzschig et al., 2006, 2013; Hart et al., 2008, 2010; Riegel et al., 2011; Idzko et al., 2014). Furthermore, extensive studies have been conducted to understand the regulation of microRNAs during AMI in patients and murine models (Hullinger et al., 2012; Martinez et al., 2017). Here, we will summarize microRNAs identified to have a detrimental or protective role in AMI.
Several microRNAs and microRNA families have been identified to be detrimental in AMI. For example, the miR-15 family, including miR-15a, miR-15b, miR-16-1, miR-16-2, miR-195, and miR-497, are upregulated in the infarction area 24 hours after the onset of AMI (Hullinger et al., 2012). The same study showed that inhibition of miR-15 by anti-miRs reduced infarct size and improved cardiac function in the murine AMI model, suggesting the therapeutic potential of miR-15 inhibition. Another important microRNA, miR-34a, was first identified in a discovery screening of microRNA changes during cardiomyocyte aging and is involved in the recovery of AMI (Boon et al., 2013). The level of miR-34a increases as the heart ages, and its inhibition prevents age-associated cardiomyocyte death while facilitating the recovery of AMI. Furthermore, protein phosphatase 1 regulatory subunit 10 was identified as a direct target of miR-34a, and its main function includes reduction of aging damage and apoptosis. Additionally, the miR-17-92 cluster (miR-92a especially) is highly expressed in endothelial cells to modulate the growth of new vessels. In vivo silencing of miR-92a improved the functional recovery of murine AML via direct targeting of integrin subunit α5 (Bonauer et al., 2009). The therapeutic potential of miR-92a has been further evaluated in the large-animal AMI model (Hinkel et al., 2013). Indeed, only regional administration of miR-92a LNA reduced infarct size, whereas systemic delivery of miR-92a failed to render protection, thus implicating the necessity of precise delivery of miR-92a–based therapy. Moreover, a recent study by Martinez et al. (2017) identified a progressive response of miR-31 after the onset of rat AMI. Pharmaceutic miR-31 inhibition enhances the expression of its target genes, cardiac troponin-T, E2F transcription factor 6 (E2f6), nuclear receptor subfamily 3 group C member 2, and metalloproteinase inhibitor 4 mRNAs, and leads to improved recovery of cardiac function. To summarize, therapeutic inhibition of those abovementioned microRNAs could potentially improve the recovery of AMI.
In contrast to the miRNAs described in the previous paragraph, several microRNAs have been found to provide cardiac protection during AMI. For example, higher levels of miR-210 were detected in live cardiomyocytes when compared with apoptotic cells, and overexpression of miR-210 in vitro prevents apoptosis (Hu et al., 2010). Intramyocardial injections of nanoparticles carrying miR-210 precursor significantly improve cardiac function after recovery from murine AMI. The mechanistic study identified Eph-related receptor tyrosine kinase ligand 3 and protein-tyrosine phosphatase 1B as potential targets for miR-210. Interestingly, miR-210 was identified as one of the hypoxia-miRs, a group of microRNAs induced by hypoxia and dependent on HIF (Camps et al., 2008; Gou et al., 2012). Previous studies have identified a tissue-protective role of HIF1a and HIF2a in the setting of murine myocardial infarction (Eckle et al., 2008, 2012; Koeppen et al., 2018). Thus miR-210 could serve as an HIF responsive element and a hypoxia adaptive mechanism that facilitates HIF-elicited tissue protection during myocardial infarction. The tight link between HIF and miR-210 could also potentially serve as a mechanism to tie hypoxia and inflammation during ischemia/reperfusion injury, which was discussed in detail in our previous reviews (Eltzschig and Carmeliet, 2011; Eltzschig et al., 2014). Another microRNA with cardiac protective potential is miR-132, which is highly expressed and secreted by saphenous vein–derived pericyte progenitor cells (SVPs) (Katare et al., 2011). The authors demonstrate that SVP-conditioned media facilitates endothelial tube formation while inhibiting fibrosis. SVP transplantation improves cardiac function via inhibition of cardiomyocyte apoptosis and dampening of interstitial fibrosis, whereas miR-132 inhibition attenuates this effect. Following along the line of secreted microRNAs by progenitor cells, another study by Barile et al. (2014) identified miR-210, miR-132, and miR-146 as highly enriched microRNAs in extracellular vesicles (EVs) secreted by human cardiac progenitor cells (CPCs). In this study, most EVs are around 30–90 nm in diameter, which is similar to the size of exosomes, as measured by transmission electron microscopy and nanoparticle tracking analysis. Consistent with the other two reports, in vivo, functional studies suggest that injection of EV secreted from CPCs inhibits cardiomyocyte apoptosis and improves cardiac function. In conclusion, the delivery of EVs from CPCs and pharmaceutic enhancement of miR-210 and miR-132 could potentially attenuate cardiomyocyte apoptosis and aid the recovery from AMI. Notably, it is essential for the delivery of microRNA mimics or inhibitors in the heart, which could pave the way for important improvement in therapy for AMI.
C. Acute Gut Injury
The prevention of acute gut injury (AGI) in perioperative patients is pivotal to impede the development of multiorgan failure and sepsis (Chen et al., 2013; Yuan et al., 2018b). Several factors contribute to the development of AGI, including translocation of bacteria and/or their endotoxin products from the intestinal lumen into the systemic circulation or other tissues/organs across the intestinal mucosal barrier and activation of intestinal immunity and mesenteric ischemia resulting from the low-flow state commonly encountered in high-risk surgeries (Ohri and Velissaris, 2006; Deitch, 2010). AGI occurs in 0.3%–6.1% of patients undergoing high-risk surgery, such as lung transplant and cardiopulmonary bypass surgeries, and the mortality of AGI ranges from 18% to 58% (Rodriguez et al., 2010; Lahon et al., 2011). AGI involves disrupted intestinal barrier function and intensified intestinal inflammation, which can manifest into systemic inflammation in patients at risk. For example, A2B adenosine receptor signaling is beneficial in the protection against acute colitis and intestinal ischemia/reperfusion injury (Hart et al., 2009; Grenz et al., 2011; Aherne et al., 2015). Additionally, neuronal guidance molecule netrin-1 has also been shown to be important in the attenuation of acute experimental colitis (Aherne et al., 2012). Moreover, Charles Serhan’s laboratory found that the resolution of inflammation also plays a crucial role to reduce long-term tissue injury, and several microRNAs have been reported to function in this process (Fredman et al., 2012; Krishnamoorthy et al., 2012; Li et al., 2013b). Furthermore, in the attempt to understand the mechanism of AGI and the development of intestinal inflammation, several screening studies demonstrate differentially regulated microRNAs in intestinal inflammation (Wu et al., 2008a, 2010; Takagi et al., 2010; Neudecker et al., 2017b). Here, we will discuss several microRNAs that have been studied extensively in intestinal inflammation.
Several microRNAs have been identified to be detrimental in AGI. MiR-146a has been observed to be upregulated in human patients with inflammatory bowel disease (IBD) by several studies. MiR-146a is commonly known by its function in the regulation of T cells, especially T helper (Th) 1, Th17, regulatory T cells (Treg), and follicular helper T cells (Lu et al., 2010; Runtsch et al., 2015). MiR-146a−/− mice have increased Th17 and Treg population in the lamina propria and increased follicular helper T cells in the germinal center (Runtsch et al., 2015). Increased T-cell and B-cell responses predict enhanced inflammation. Although surprisingly, miR-146a−/− mice are protected in dextran sulfate sodium (DSS)-induced intestinal inflammation, suggesting a detrimental role of miR-146a in the intestinal barrier function. Additionally, the increased level of miR-214 has been observed in colon biopsies of patients with active colitis (Polytarchou et al., 2015). The upregulation of miR-214 is mediated by interleukin-6–induced signal transducer and activator of transcription 3 activation in colon tissues, and chemical inhibition of miR-214 attenuated the severity of DSS-induced murine intestinal inflammation. The above studies suggest that pharmacological inhibition of miR-146 and miR-214 could potentially benefit the outcome of patients with AGI by limiting intestinal inflammation and improving barrier functions.
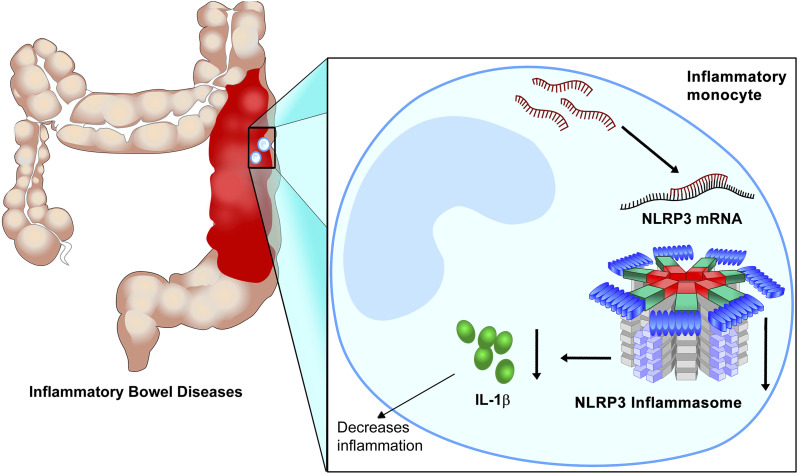
In contrast to the miRNAs described in the previous paragraph, miR-223 is anti-inflammatory during intestinal inflammation (Zhou et al., 2015; Neudecker et al., 2017b). Increased levels of miR-223 have been observed in intestinal biopsies from patients with IBD and colon tissues in a murine model of intestinal inflammation (Neudecker et al., 2017b). In mouse models of colitis, Zhou et al. (2015) demonstrated that miR-223-/y mice exhibit exaggerated intestinal inflammation. CCAAT enhancer-binding protein β was identified in this study as a proinflammatory target of miR-223. Further study also suggested the role of miR-223 in the protection of murine intestinal inflammation (Neudecker et al., 2017b). The function of miR-223 in intestinal inflammation was addressed by exposing miR-223-/y mice to DSS-induced colitis. Compared with wild-type counterparts, miR-223-/y mice demonstrated exacerbated intestinal inflammation marked by more weight loss and exaggerated tissue injury. Based on previously reported roles of miR-223 in myeloid cell compartments, bone marrow chimeric mice were generated to identify whether the myeloid or nonmyeloid compartment was crucial for the function of miR-223 during murine colitis. The importance of miR-223 in murine colitis lies within the myeloid compartment, as an miR-223 deficiency in the myeloid compartment showed a similar phenotype of miR-223-/y mice. To identify the importance of different subtypes of myeloid cell in miR-223–mediated intestinal protection, neutrophils or monocytes in miR-223-/y mice were deleted by antibody, and monocyte deletion disrupted the protective effect of miR-223. Furthermore, NLR family pyrin domain containing 3 (NLRP3) was identified as a functional target of miR-223 during murine DSS-induced colitis (Fig. 4). In support of the direct targeting in vivo, targeted deletion of the miR-223 binding site in the 3′ UTR region of NLRP3 in mice resulted in increased severity of DSS-induced colitis, which resembled that of miR-223-/y mice. To investigate the therapeutic potential of miR-223 during intestinal inflammation, miR-223 mimic was delivered systemically in mice, and the delivery of miR-223 resulted in attenuation of DSS-induced colitis. To conclude, the protective role of miR-223 was suggested in experimental colitis, and this protection stems from innate immune responses controlled by the myeloid cell. Therapeutic enhancement of miR-223 in patients with IBD could serve as a potential novel treatment in the control of access intestinal inflammation. It is critical to achieving the overexpression of microRNA such as miR-223 in the intestinal tract; thus, the oral delivery of packaged miRNA mimic or inhibitor would be a feasible way to treat or prevent acute gut injury.
Fig. 4.
MiR-223 in inflammatory monocytes dampens intestinal inflammation. DSS-induced intestinal injury leads to the upregulation of MiR-223 in inflammatory monocytes. Increased miR-223 levels result in the inhibition of its target gene NLRP3, and dampened NLRP3 sequesters inflammasome activity and downstream production of inflammatory cytokines, such as IL-1β. Turning down IL-1β attenuates intestinal inflammation and histologic signs of intestinal injury. Pharmacological enhancement of miR-223 could potentially alleviate acute gut injury and intestinal inflammation in the perioperative period (Neudecker et al., 2016, 2017b,c; Yuan et al., 2018a).
V. Pharmacologic Approaches for MicroRNA Modulation
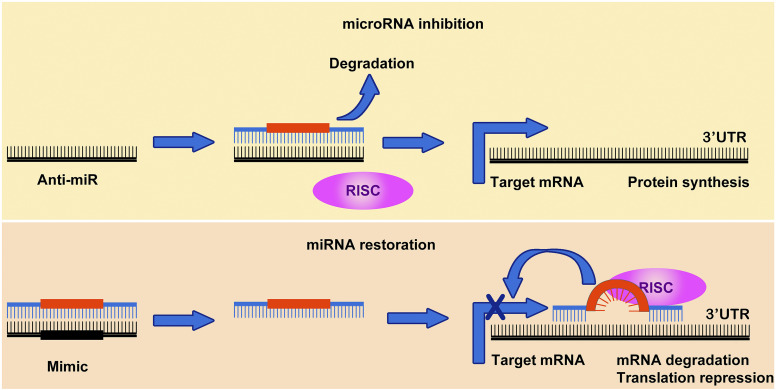
As noted earlier, introducing mimic or inhibiting sequences of a specific miRNA into diseased cells requires a specific strategy to protect the miRNA mimics or inhibitors from self-hydrolysis or RNase-mediated enzymatic degradation. When administered into a body without such protection, the half-life of naked RNA mimics or inhibitors is less than 30 minutes, which significantly dampens the potential effect of microRNAs. At the same time, it should also be noted that most difficulties in those clinical trials came from the toxicities or nonspecific cell-targeting nature of carriers. Restoration or inhibition of specific miRNAs in the target diseased cells can be achieved by several strategies (Table 1): 1) delivery of miRNA mimics or inhibitors by using modified nucleic acids, 2) modification of miRNAs using viral delivery vector systems, 3) delivery of miRNA mimics or inhibitors by using nonviral delivery vector systems, and 4) small molecule drugs targeting microRNAs.
TABLE 1.
Pharmacologic approaches for miRNA-based human disease therapy
| Type of Vector | Structure of Base Vector Material | Target miR | Therapeutic Purpose | Target Disease | References | ||
|---|---|---|---|---|---|---|---|
| Antisense modified oligonucleotides | 2′-O-Me | 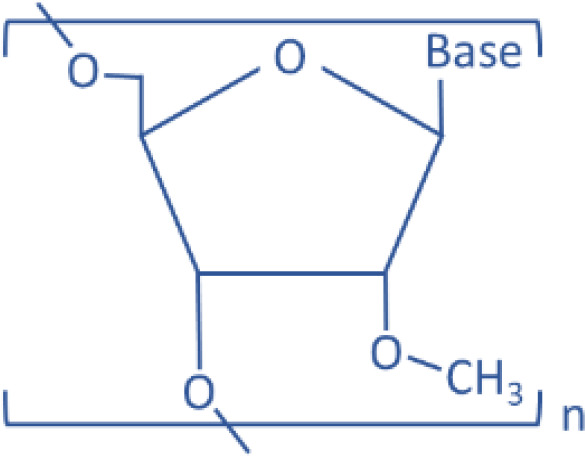 |
miR-122 | Antisense | Liver disease | Krützfeldt et al., 2005 | |
| 2′-F | 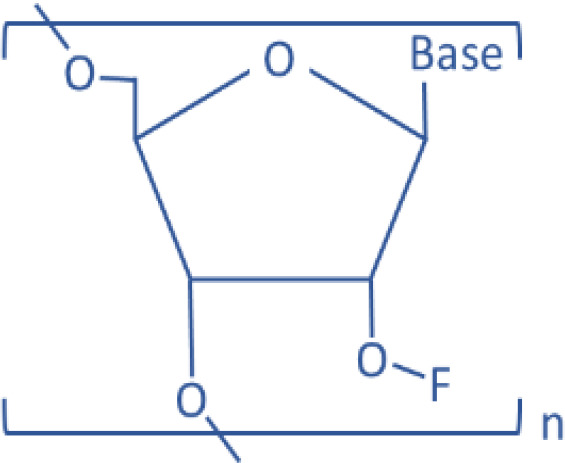 |
miR-122 | Antisense | Liver disease | Davis et al., 2009 | ||
| LNA | 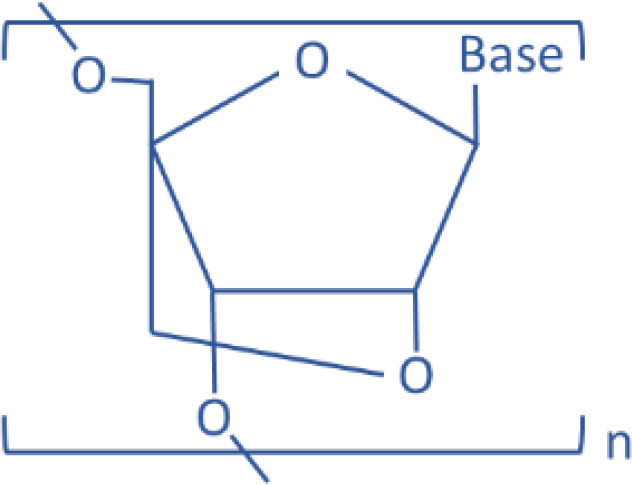 |
miR-122 | Antisense | Liver disease | Elmen et al., 2008 | ||
| miR-122 | Antisense | HCV | Lanford et al., 2010 | ||||
| miR-21 | Antisense | Glioma | Griveau et al., 2013 | ||||
| miR-221 | Antisense | MM | Di Martino et al., 2014 | ||||
| PNA | 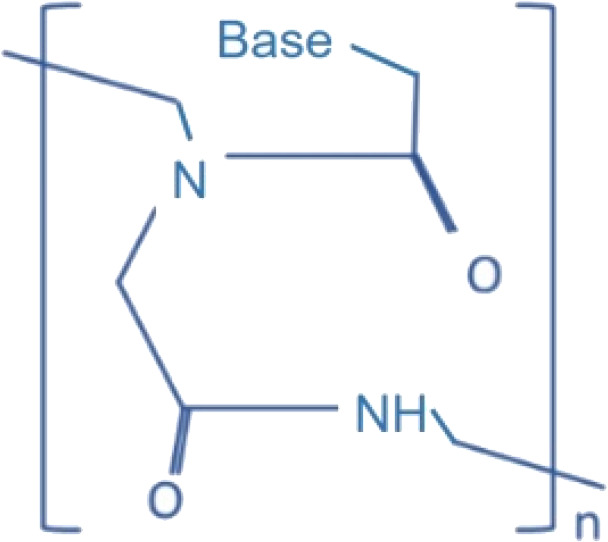 |
miR-155 | Antisense | Primary B cells | Fabani et al., 2010 | ||
| Viral vectors | Lentiviruses |  |
miR-146a | Overexpression | Diabetes | Zhuang et al., 2017 | |
| miR-199a | Overexpression | HCC | Jia et al., 2012 | ||||
| Let-7 | Overexpression | NSCLC | Trang et al., 2010 | ||||
| miR-15a/16 | Overexpression | CLL | Kasar et al., 2012 | ||||
| AAV |  |
miR-221 | Inhibition | HCC | Moshiri et al., 2014 | ||
| miR-26a | Overexpression | HCC | Kota et al., 2009 | ||||
| Nonviral vectors | Lipids | Cationic | DOTMA:cholesterol:TPGS | miR-29b | Mimic | NSCLC | Wu et al., 2013 |
| DDAB:cholesterol:TPGS | miR-107 | Mimic | HNSCC | Piao et al., 2012 | |||
| Anionic | DOPE:linoleic acid:DMG-PEG | miR-29b | Mimic | AML | Huang et al., 2013 | ||
| Neutral | MaxSuppressor© (1,2-dioleoyl-sn-glycero-3-phosphocholine, squalene oil, polysorbate 20) | miR-34a, let-7 | Mimic | NSCLC | Trang et al., 2011 | ||
| miR-34a | mimic | DLBCL | Craig et al., 2012 | ||||
| Polymer | PEI | 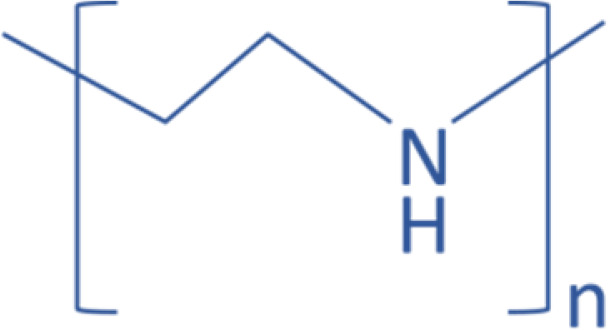 |
miR-145 | Mimic | CRC | Ibrahim et al., 2011 | |
| miR-33a | |||||||
| miR-145 | Mimic | GBM | Yang et al., 2012 | ||||
| miR-145 | Mimic | NSCLC | Chiou et al., 2012 | ||||
| miR-135 | Mimic | GBM | Liang et al., 2017 | ||||
| PLGA | 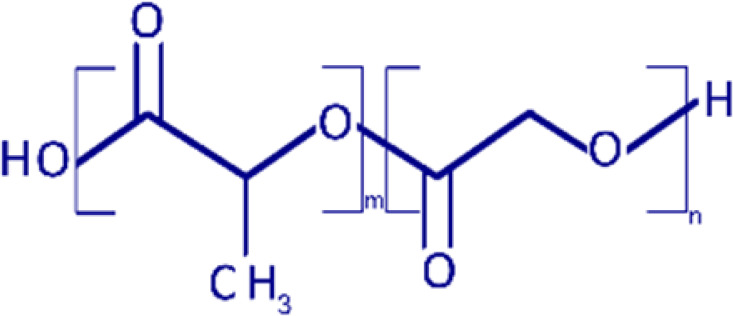 |
miR-21 | Antisense | TNBC | Devulapally et al., 2015 | ||
| miR-10b | |||||||
| miR-34a | Mimic | MM | Cosco et al., 2015 | ||||
| miR-99a | Mimic | HCC | Cai et al., 2017 | ||||
| miR-204 | Mimic | CRC | Zheng et al., 2018 | ||||
| Dendrimer/PAMAM | 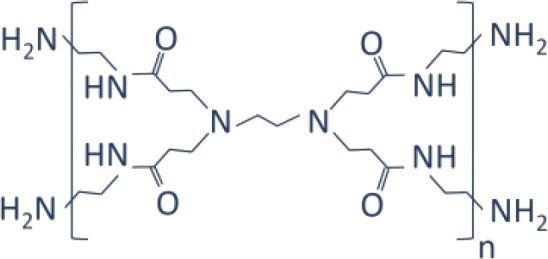 |
miR-21 | Antisense | BC | Ren et al., 2016 | ||
| Chitosan | 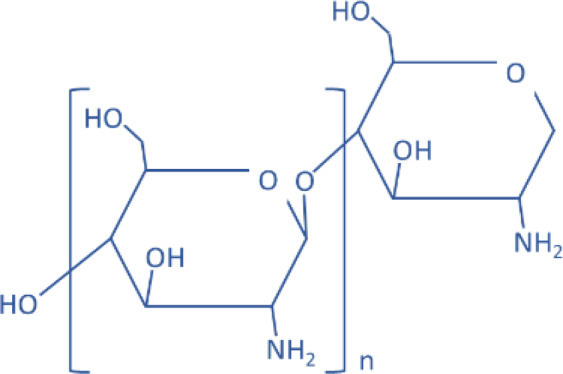 |
miR-34a | Mimic | Prostate cancer | Gaur et al., 2015 | ||
| Liposomes | 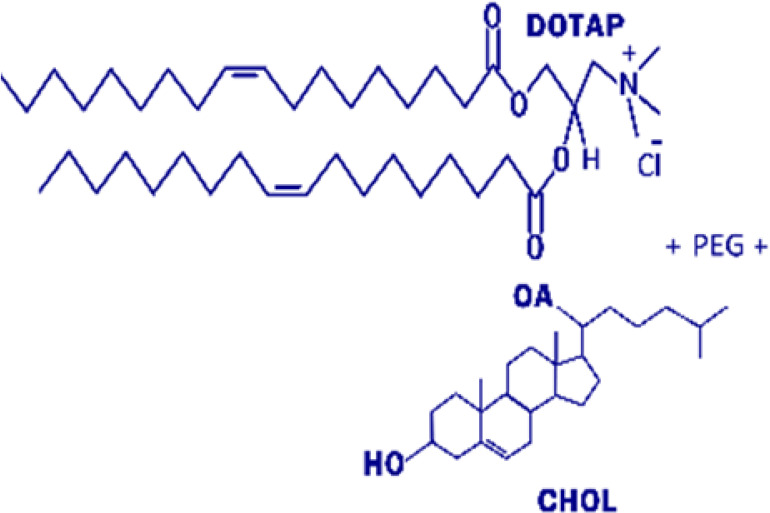 |
miR-101 | Mimic | HCC | Xu et al., 2017 | ||
| Exosomes | miR-124a | Mimic | Glioma | Lang et al., 2018 | |||
| Inorganic | Gold nanoparticle |  |
miR-29b | Antisense | CC | Kim et al., 2011 | |
| miR-375 | Mimic | HCC | Xue et al., 2016 | ||||
| miR-145 | Mimic | BC | Ekin et al., 2014 | ||||
| Magnetic nanoparticle |  |
Let-7 | Mimic | GBM | Yin et al., 2014a | ||
| miR-145 | Mimic | PC | Setua et al., 2017 | ||||
| miR-16 | Mimic | GC | Sun et al., 2014 | ||||
| Diamond nanoparticle |  |
miR-203 | Mimic | EC | Cao et al., 2013 | ||
| Nucleic acid nanoparticle | RNA nanoparticle | 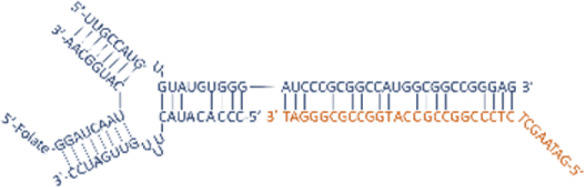 |
miR-21 | Antisense | TNBC | Shu et al., 2015 | |
| miR-21 | Antisense | GBM | Lee et al., 2017 | ||||
BC, breast cancer; CC, cervical cancer; DDAB, dimethyl dioctadecyl ammonium bromide; DMG, dimyristolglycerol; DOPE, dioleoylphosphatidylethanolamine; DOTMA, 1,2-di-O-octadecenyl-3-trimethylammonium propane; EC, esophageal cancer; GBM, glioblastoma; GC, gastric cancer; HNSCC, head and neck squamous cell carcinoma; MM, multiple myeloma; PAMAM, polyamidoamine; PC, pancreatic cancer; PEG, polyethylene glycol; PNA, peptide nucleic acids; TPGS, α-tocopheryl polyethylene glycol 1000 succinate; 2′-O-Me, 2′-O-methylation.
A. Modification of Nucleotide Analogs to Increase Stability
RNA contains a 5-carbon sugar, ribose, with a hydroxyl group on its 2′ position (2′-OH), whereas DNA has a deoxyribose sugar (2′-H) without the hydroxyl group on the 2′ position. The 2′-OH is crucial for the degradation of RNA by self-hydrolysis and by endogenous RNases. Therefore, the removal and modification of the 2′-OH group from mimic or inhibitor RNAs will make them resistant to both self-hydrolysis or RNase-mediated enzymatic degradation. This approach includes direct modification of the sugar-phosphate backbone of RNA or using nucleic acid analogs that are chemically mimicking RNA molecules (Prakash and Bhat, 2007; Lennox and Behlke, 2011; Prakash, 2011). These approaches generate more chemically stable RNA analogs; therefore, they are suitable for designing anti-miRNA oligonucleotides (AMOs) to reduce a specific miRNA level. For example, phosphorothioate (PS) RNA is an RNA analog generated by replacing nonbridging oxygen with sulfur in the phosphodiester bond of the RNA strand, because of which the PS RNA is more stable (Lennox et al., 2006). Since PS RNAs lack phosphodiester bonds, which are attacked by RNases, PS AMOs are more resistant to the RNase-mediated enzymatic degradation. Replacement of the 2′-OH group on RNA ribose ring with a chemically inert group has also been studied by many different approaches to increase the AMO stability. The main idea is to deprive the hydrogen on the 2′-OH group to avoid the pH or enzyme-dependent deprotonation. For example, 2′-O-Methylation AMO duplexes were designed to specifically target mouse liver abundant miR-122 and were injected through mouse tail vein. When the miR-122 expression level was measured by Northern blotting 24 hours after the last injection, the hepatic miR-122 was completely degraded in the mouse group injected with 240 mg/kg per day (Krützfeldt et al., 2005). Davis et al. (2009) used 2′-fluoro/2′-methoxyethyl AMO to inhibit the endogenous miR-122 in mice. The 2′-O-MOE modification makes AMO form more stable binding with target RNAs than 2′-O-methyl by increasing 2°C of melting temperature (Tereshko et al., 1998), therefore increasing their tissue half-life up to 10-fold higher than PS RNA (Geary et al., 2001). 2′-Fluoro (2′-F) modification also increases the stability of AMO. However, combining the 2′-F modification with PS RNA ensures RNase resistance.
Besides the modification of 2′-OH, several other strategies have been employed to facilitate the delivery of RNA oligonucleotide in vivo. Peptide bond–based nucleic acid (PNA) mimics is another promising candidate for the targeting of microRNAs in vivo. The sugar-phosphodiester backbone of PNA is replaced with N-(2-aminoethyl)-glycine units, which present no surface charge, so that naked PNA molecules can directly penetrate cell membranes without the aid of transfection agent. When a PNA technology–based anti-miR against miR-155 was intraperitoneally given to mice, the endogenous miR-155 level in primary B cells was successfully inhibited, and the gene expression profiles from the treated mice were similar to that from miR-155–deficient mice (Fabani et al., 2010). Besides PNAs, locked nucleic acid (LNA) is another artificial RNA analog to facilitate the targeting of microRNAs in vivo. The 2′-oxygen and 4′-carbon on the ribose of LNA are chemically locked by a bridge. This closed conformation physically inhibits the binding of RNase, resulting in high stability in serum, but is still capable of pairing with complementary RNA strand. For this reason, LNAs are frequently used to either detect specific miRNAs for fluorescence staining or inhibit target miRNAs by complementarily binding to functional miRNA strands (Wahlestedt et al., 2000). Whereas unmodified oligonucleotides are degraded within 1.5 hours in serum, the half-life of LNA is up to 15 hours in serum, enabling longer intervals between administrations (Kurreck et al., 2002). Compared with PNA, LNA is superior in affinity to a target sequence than PNA. For example, the antisense efficiency of PNA against human telomerase mRNA was almost 200-fold lower than that of LNA (Elayadi et al., 2002). LNA is more cost-effective and has higher cellular uptake than PNA; therefore, LNA has been more frequently used as miRNA inhibitors than PNA.
B. Viral Delivery of MicroRNAs
Besides the modification of nucleotide to prevent degradation, viruses are also powerful genetic tools that have been studied extensively to use as a gene-editing vector system in mammalian cells. In general, two modifications were applied to those viruses, such as retroviruses, lentiviruses, adenoviruses, or adeno-associated viruses (AAVs), to control their cell type–specific expression of inquired genes and harmful side effects: 1) insertion of cell type–specific promoter sequences and 2) removal of pathologically harmful genes, except for the genes that are essential for viral entry and replication. Over the years, it has been shown that siRNAs or miRNAs can be expressed by these genetically engineered viruses to target disease cells (Davidson and Harper, 2005). Virus-mediated vector systems are especially useful to overexpress miRNAs that are found to be downregulated in diseased cells since the restoration of such downregulated miRNAs in the targeted cells is expected to reverse already ongoing pathologic phenomena. Viruses are suitable for continuous expression of miRNA by using a constitutive promoter. Thus, it enables sufficient overexpression of miRNA in the virus-infected cells with a single dose of administration. Because of the high specificity and transduction efficiency of viral vectors, high levels of miRNA expression can be achieved with no further chemical modification of miRNA sequences to protect the miRNAs from serum degradation.
Lentiviruses are useful to deliver the various sizes of genes since they are flexible for the cloning of a long size of DNA. Upon host cell infection, lentiviruses integrate into host genomes. The host gene integration enables the maximal and stable expression of the gene of interest by a single administration. However, the chromosomal integration of exogenous DNA sequences by lentivirus-based delivery may cause harmful genomic mutations when applied to long-term clinical trials (Milone and O’Doherty, 2018). AAVs, on the other hand, do not integrate into the host genomes, which reduces the chance of genomic mutations. AAVs are commonly used to deliver small noncoding RNAs, such as short hairpin RNAs or miRNAs, because their small packaging capacity is still large enough for small noncoding RNAs and because they are relatively safer than other viruses because of their lack of pathogenicity (Grimm, 2009). AAVs are physically stable in a wide range of temperatures and pH’s (Penaud-Budloo et al., 2018). Although the viral delivery of miRNA seems to be quite efficient for sustained expression of miRNAs, using such viral vector systems is often challenging because of their potential activation of oncogenes (depending on their integration location), systemic toxicity, or immunogenic response due to the nature of the viral infection. For these reasons, the Food and Drug Administration (FDA) recommends a level of residual cellular foreign DNA less than 10 ng per parenteral dose to limit the genotoxic risk of recombinant AAVs after systemic administration, which corresponds to 1011 vector genomes (Penaud-Budloo et al., 2018). One of the most groundbreaking advancements in the AAV-mediated delivery for gene therapy is the FDA approval of Luxturna (voretigene neparvovec-rzyl) as a treatment of an inherited form of vision loss in children and adult patients. In clinical trials, Russell et al. (2017) have demonstrated that Luxturna (AAV2-hourRPE65v2) gene replacement significantly improved functional vision in retinal pigment epithelium-specific 65 KDa protein–mediated inherited retinal dystrophy. As the first directly administered gene therapy to be approved by the FDA, although Luxturna is approved for injection into the eye, a relatively immune-privileged organ, it opens many doors for the design of future gene therapy with AAV-mediated delivery of microRNA-based approaches. For example, Wang et al. (2019) have reported that AAV-mediated delivery of anti-miR-214 (AAV-anti-miR-214) successfully prevented the collapse of the femoral head in a rat model of osteonecrosis by rescuing the miR-214 targets activating transcription factor 4 and phosphatase and tensin homolog. Spark Therapeutics, the company that developed Luxturna, has recently announced that they evaluated the Luxturna-based miRNA delivery for the knockdown of Huntingtin gene to treat Huntington disease.
C. Nonviral Delivery of MicroRNAs
Using virus-based vector systems to target miRNAs is one of the most efficient methods because of their high level of expression controlled by strong promoters. However, some of the concerns and safety issues, as mentioned above, are still challenging to resolve. To overcome the limitation of viral delivery, delivering miRNA mimics or miRNA inhibitors by formulating with three major nonviral materials—polymers, lipids, and inorganic nanoparticles—seems to be a reasonable and promising approach. These nonviral delivery systems prevent serum degradation, endosomal capture, lysosomal degradation, and immunogenic response. Besides, many of the nonviral molecules are chemically flexible for further conjugation with various ligand molecules, which presents a great potential for cell-specific targeted delivery of miRNAs. All of these nonviral vector-based delivery approaches have shown adequate therapeutic capability and potential for clinical applications with a satisfactory safety profile compared to the viral vector systems. However, additional studies are needed to evaluate in vivo pharmacology of these nonviral particles to facilitate future clinical trials.
1. Polymers
Polymers have been used as drug carriers for peptides or proteins because of their ease of formulation with positively charged biomaterials and low cytotoxicity from easy biodegradation. For the same reasons, small noncoding RNAs, such as siRNAs or miRNAs, can also be delivered by these polymeric vectors. For example, many previous studies showed that positively charged low–molecular weight polyethyleneimine (PEI) is an efficient carrier for miRNA delivery in animal models because of its moderate level of toxicity compared with high–molecular weight PEI, which has poor biodegradation and prolonged cellular accumulation (Kunath et al., 2003). Poly lactic-co-glycolic acid (PLGA) has also been used for miRNA deliveries (Malik and Bahal, 2019); however, the negatively charged polymer needs further modification to increase the positive charges by including more amine-like cationic residues or another type of positively charged polymers, such as chitosan or PEI. Chitosan itself can also be formulated into nanoparticles for miRNA delivery. Since chitosan is biocompatible with natural polysaccharides, chitosan-based nanoparticles have reduced immunogenicity and toxicity (Denizli et al., 2017).
2. Lipids
Lipid-based miRNA delivery systems are biocompatible and biodegradable so that they allow the easy penetration of phospholipidic human cell membranes. In addition, advancement in organic chemistry enabled an extensive application of various lipids to construct nucleic acid carriers. Since nucleic acids including miRNAs are negatively charged, cationic lipids are frequently used to construct lipid-based nanoparticles (Campani et al., 2016). For examples, Piao et al. (2012) constructed lipid-based cationic nanoparticles by mixing dimethyl dioctadecyl ammonium bromide, cholesterol, and α-tocopheryl polyethylene glycol 1000 succinate at a molar ratio of 60:35:5 and encapsulated precursors of miR-107 with the cationic lipid nanoparticles. The miR-107 encapsulated cationic lipid nanoparticles were spherical, with a mean diameter of 150.1 ± 3.5 nm, a ζ potential of +10.3 ± 0.4 mV, and 98.9% ± 1.5% encapsulation efficiency of pre-miR-107. When 1 nmol of the miR-107 encapsulated cationic lipid nanoparticles was intravenously injected into athymic nude mice with head and neck squamous cell carcinoma xenograft via tail veins twice a week, expression levels of miR-107 direct target genes, such as ε isoform of protein kinase C, HIF1-β, CDK6, Nanog, SRY-box transcription factor 2, and octamer-binding transcription factor three-fourths, were reduced by 45%–75%, leading to tumor grown inhibition (Piao et al., 2012). Another lipid-based nanoparticle that has been implemented in the clinical setting is 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC). Dr. Anil Sood and colleagues demonstrated the great safety and efficacy of the delivery of RNAi in vivo via packaging in DOPC in preclinical studies (Landen et al., 2005; Wagner et al., 2017). Currently, DOPC packaged ephrin type-A receptor 2 siRNAs are being tested in phase I clinical trials to treat patients with advanced or recurrent solid tumors (NCT01591356). Besides, recent advancements in a lipid nanoparticle (LNP) have provided an exciting new opportunity for the clinical use of lipid delivery of microRNAs (Payne, 2019). In September 2017, APOLLO phase 3 study of ONPATTRO (Patisiran, or ALN-TTR02), an LNP-based RNAi therapeutic developed by Alnylam, has met its primary efficacy endpoint and all secondary endpoints for patients with hereditary amyloid transthyretin (ATTR) amyloidosis with polyneuropathy (NCT01960348) (Adams et al., 2018; Kristen et al., 2019). ONPATTRO/Patisiran was subsequently approved by the FDA for the treatment of patients with amyloid transthyretin amyloidosis with polyneuropathy in August 2018. The LNP technology used by ONPATTRO/Patisiran could potentially be employed in targeting microRNAs in cancers and perioperative organ injury.
3. Inorganic Nanoparticles
Inorganic nanoparticles, such as gold, silica, or magnetic materials, were used to formulate miRNA-carrying complexes. For example, anti-miRNA oligonucleotides against oncogenic miR-29b were delivered into HeLa cells by using functionalized gold nanoparticles and rescued the expression of MCL-1, which was a target of miR-29b (Kim et al., 2011). Yin et al. (2014a) formulated magnetic zinc-doped iron oxide nanoparticles (ZnFe2O4) to deliver tumor-suppressive let-7a into brain tumor cells. Interestingly, the magnetic nanoparticle itself can also induce cytotoxic magnetic hyperthermia via Neel and Brownian relaxation when the internalized magnetic nanoparticles were exposed to the alternating magnetic field. Although formulation with such inorganic compounds is relatively easy, their pharmacokinetics after systemic administration may elevate concerns regarding toxicity, clearance, and lack of specific targeting.
4. Exosomes
Exosomes are another emerging field as carriers of small RNAs including miRNAs. Exosomes are defined as small EVs with a diameter of 30–150 nm and are usually generated by the endocytic pathway (Sun et al., 2018). Although exosomes belong to EVs, which are defined as lipid bilayer–enclosed extracellular structures, they are a subcategory of EVs that are distinguished from larger microvesicles, for which sizes range to submicron (Mathieu et al., 2019). Endosomal sorting complex required for transport is an important player for exosome biogenesis by promoting the maturation of early endosomes into late endosomes or multivesicular bodies (MVBs) (Hessvik and Llorente, 2018). Moreover, the intracellular trafficking of MVBs for exosome release is highly dependent on Rab27a/b, as silencing of those two molecules inhibits docking of MVBs to the plasma membrane and thus dampens exosome release (Ostrowski et al., 2010). Exosomes contain a variety of intracellular components, including proteins, lipids, DNAs, mRNAs, miRNAs, and long noncoding RNAs. Compared with artificial liposomes, endogenous exosomes are stable in serum, less toxic, and less immunogenic, which makes them an attractive drug delivery system for both local and systemic application. For instance, Dr. Charles Serhan’s laboratory used human neutrophil-derived endogenous microparticles to engineer humanized nano-proresolving medicine (NPRM) to contain aspirin-triggered resolvin D1 (AT-RvD1) or a lipoxin A4 analog (Norling et al., 2011). The size of the NPRMs was around 198 nm, as determined by flow cytometry and confirmed by electronic microscopy. NPRMs containing AT-RvD1 accelerate the wound healing process in vitro, and intravenous injection of AT-RvD1 NPRMs significantly reduced PMN influx in murine peritonitis and complete Freund’s adjuvant-induced inflamed temporomandibular joint. Also, tumor cell–secreted exosomes containing a high level of mature miR-21 are transferred into macrophages through toll-like receptors to activate immune responses (Fabbri et al., 2012). Inspired by this finding, Yin et al. (2014b) treated CD4+ cells with HEK293 cell–derived exosomes overexpressing antisense RNAs against miR-214 and observed inactivation of miR-214, which induces regulatory T cells to form an immune evasion environment. Exosomes, similar to liposome-based delivery, are unable to achieve targeted therapy because of the lack of cell specificity, which may endanger adjacent cells from the cell toxic miRNA deliveries. Besides, a standard procedure has not been established to achieve exosome identification and purification to evaluate their pharmacological quality. Recently, surface modification technology evolved to modify the exosome surface with cell-targeting specific moieties to enable targeted therapy. For example, systemic administration of Survivin siRNA-carrying exosomes that were chemically conjugated with epidermal growth factor receptor (EGFR)-targeting RNA aptamers specifically delivered the Survivin-targeting siRNAs into the breast cancer cells and reduced the tumor growth (Pi et al., 2018). Using this method, the specific delivery of therapeutic miRNA-loaded exosomes can be achieved.
5. RNA Nanoparticles
Delivering miRNAs with a formulation of non-RNA molecules including lipids requires an accurate design of drug-to-lipid ratio, size, charge, and biocompatibility for the successful delivery of cargo RNAs. Using naked RNAs as a delivery backbone has been recently introduced after the remarkable advance in RNA nanotechnology over the last decade (Guo, 2010; Shu et al., 2014; Jasinski et al., 2017). The laboratory of Peixuan Guo developed three-way junction (3WJ) RNA nanoparticles in 2011 by artificially engineering packaging RNA of the DNA packaging motor system in bacteriophage Phi29 (Shu et al., 2011). When the 3WJ RNA nanoparticles were conjugated with a cancer cell–targeting ligand, folate, to target folate receptors that are highly overexpressed in most cancer cells, it showed the capability of siRNA delivery into cancer cells, including the brain tumor model (Lee et al., 2015a,b). Biodistribution analysis and in vivo fluorescence imaging revealed that the 3WJ RNA nanoparticles were found in mouse brain tumors 15 hours after a single systemic injection, while there was no significant accumulation in other major internal organs. Chemically synthesized 2′-F–modified RNA nanoparticles demonstrated a favorable pharmacokinetic profile in mice when systemically administered via tail vein: 5–10 hours of half-life, less than 0.13 l/kg per hour of clearance, and 1.2 l/kg of the volume of distribution (Abdelmawla et al., 2011). The RNA nanoparticles also showed a favorable safety profile, with low induction of interferon response and inflammatory cytokine production, as well as the lack of any detectable toxicity. This approach provides a great opportunity in the future of miRNA delivery, since the RNA nanoparticle can be easily conjugated with cargo molecules, including miRNAs, siRNAs, and aptamers.
D. Small Molecule Drugs Targeting MicroRNAs
So far, we have outlined several approaches using oligonucleotides and other agents to target miRNAs for disease therapy. There is an important and novel strategy using small molecule drugs to directly target microRNAs or transcription factors that regulate miRNA expression (Wen et al., 2015; Safe et al., 2018). Small molecule drugs are capable of inhibiting or inducing microRNAs in disease settings. Lee et al. (2016) reported that Kenpaullone derivatives, small molecule inducers of miR-182, significantly improved heart function after ischemia/reperfusion injury in the rat. Another small molecule, enoxacin (Penetrex), is reported to promote the biogenesis of endogenous miRNAs (Shan et al., 2008). Moreover, neomycin B, an FDA-approved small molecule drug, can selectively inhibit the production of miRNA-525 and, in turn, inhibit the invasion of the HCC cell line (Childs-Disney and Disney, 2016). Small molecule drugs can also modulate the transcription factors to regulate and inhibit the constitutive expression of miRNAs. As mentioned above, c-Myc has been shown to activate the expression of several pro-oncogenic microRNAs. Thus, the downregulation of c-Myc by reactive oxygen species–inducing agents such as hydrogen peroxide, isothiocyanates, histone deacetylase inhibitors, penfluridol, piperlongumine, and many other natural products can target the critical reactive oxygen species-c-Myc-OncomiR interaction, resulting in downregulation of Sp transcription factors and pro-oncogenic Sp-regulated genes (Jutooru et al., 2010, 2014; Chintharlapalli et al., 2011; Chadalapaka et al., 2012; Kasiappan et al., 2016; Karki et al., 2017). To summarize, besides delivering microRNA mimics or inhibitors, small molecule compounds could serve as an opportunity to safely and effectively target microRNAs in cancer and perioperative organ injury.
VI. MicroRNA Modulation for Disease Therapy
Since single miRNA, in theory (but not all miRNAs), can regulate the protein expression of multiple direct target genes, changing the level of the aberrantly expressed miRNA can affect many aspects of cell fate, such as proliferation, invasion, and apoptosis, during disease development and progression. Taking advantage of the fact that miRNAs affect many pathways can benefit their therapeutic application, since many diseases, including perioperative infection and cancer, are the results of abnormal changes in multiple pathways (Hanahan and Weinberg, 2011). Therefore, it has been suggested that targeting a dysregulated miRNA may rescue several affected pathways, giving maximum therapeutic efficacy. Targeting microRNAs is hugely beneficial compared with conventional chemo drugs or siRNAs, which can only target a single gene product. Therefore, many researchers have attempted to develop a new therapeutic strategy to modulate the aberrantly expressed miRNAs in the diseased cells by introducing miRNA mimics or inhibitors (Fig. 5). However, the targeting of miRNAs in the diseased organs or abnormal cells has been hampered due to the lack of safe and effective delivery approaches. To date, there are significantly increasing numbers of microRNA-based therapies that have entered into phase 2 clinical trials in 2018–2019, which is very encouraging for translating microRNA-based therapy into the clinic (Table 2). Moreover, recent advances and approval of LNPs and AAV-based gene therapy are quite encouraging for the safe and effective targeting of microRNAs in cancers and perioperative organ injury.
Fig. 5.
The mechanistic model of action in miRNA-based therapeutic modulation in the application for human diseases. Deregulated miRNA expression can be overcome either by the restoration of those downregulated tumor-suppressive miRNAs or inhibition of those upregulated onco-miRs in the diseased cells. Top: inhibition of upregulated miRNAs can be attained by the introduction of AMOs. Chemically modified nucleic acids LNA (Elayadi et al., 2002; Kurreck et al., 2002; Valoczi et al., 2004; Griveau et al., 2013; Di Martino et al., 2014) or PNA (Hanvey et al., 1992; Demidov et al., 1994; McMahon et al., 2002; Fabani et al., 2010) are used as single-stranded inhibitors. These are often further formulated into lipid or inorganic nanoparticles to protect them from serum degradation and/or enable a targeted delivery. Once internalized into the diseased cells, the complementary sequence of AMO recognizes and binds to its target onco-miR, of which double-stranded complex will be degraded and release the target gene mRNAs from the repression of protein synthesis by the onco-miR. Bottom: restoration of downregulated tumor-suppressive miRNAs in diseased cells can be achieved by the introduction of mimic sequences into the diseased cells. Viral vectors, such as (Jia et al., 2012; Kasar et al., 2012; Huo et al., 2017) or AAVs (Kota et al., 2009), are often used to induce overexpression of such downregulated miRNAs. Mimic miRNA sequences can also be delivered to the diseased cells in a formulation of liposomes (Rai et al., 2011; Campani et al., 2016; Beg et al., 2017; Xu et al., 2017) or inorganic nanoparticles (Kim et al., 2011; Sun et al., 2014; Ren et al., 2016; Xue et al., 2016; Setua et al., 2017) for protection from serum degradation.
TABLE 2.
Current ongoing or recently completed clinical trials for therapeutic targeting of miRNAs
| Drug (Company) | Delivery Strategy | Target miR | Therapeutic Purpose | Target Disease | Status | Clinical Trial Gov Identifier | References |
|---|---|---|---|---|---|---|---|
| Miravirsen (Santaris Pharma) | Anti-miR LNA | miR-122 | Inhibition | HCC | Phase 2a | NCT01200420 | Janssen et al., 2013 |
| Completed (2010–2012) | |||||||
| MRX34 (Mirna Therapeutics) | Mimic miR in liposome | miR-34a | Restoration | Primary liver cancer | Phase 1 | NCT01829971 | Beg et al., 2017 |
| SCLC | Terminated (2013–2016) | ||||||
| Lymphoma | |||||||
| Melanoma | |||||||
| MM | |||||||
| RCC | |||||||
| NSCLC | |||||||
| TargomiRs (EnGeneIC) | Mimic miR in EDV with EGFR antibody | miR-16 | Restoration | MPM | Phase 1 | NCT02369198 | van Zandwijk et al., 2017 |
| NSCLC | Completed (2015–2017) | ||||||
| Cobomarsen/MRG106 (MiRagen) | Anti-miR LNA | miR-155 | Inhibition | MF | Phase 1 | NCT02580552 | NA |
| CLL | |||||||
| DLBCL | Ongoing (2015–) | ||||||
| ATLL | |||||||
| Cobomarsen/MRG106 (MiRagen) | Anti-miR LNA | miR-155 | Inhibition | Cutaneous T-Cell lymphoma/MF | Phase 2 | NCT03713320 | NA |
| Ongoing (2018–) | |||||||
| Cobomarsen/MRG106 (MiRagen) | Anti-miR LNA | miR-155 | Inhibition | Cutaneous T-Cell lymphoma/MF | Phase 2 | NCT03837457 | NA |
| Ongoing (2019–) | |||||||
| Remlarsen/MRG201 (MiRagen) | Mimic miR | miR-29 | Restoration | Healthy volunteers | Phase 1 | NCT02603224 | NA |
| Completed (2015–2017) | |||||||
| Remlarsen/MRG201 (MiRagen) | Mimic miR | miR-29 | Restoration | Keloid | Phase 2 | NCT03601052 | NA |
| Ongoing (2018–) | |||||||
| MRG-110 (MiRagen) | Anti-miR LNA | miR-92a | Inhibition | Healthy volunteers | Phase 1 | NCT03603431 | NA |
| Completed (2018–2019) | |||||||
| ABX464 (Abivax S.A.) | Small molecule | miR-124 | Overexpression | Healthy volunteers | Phase 1 | NCT02731885 | Scherrer et al., 2016 |
| Completed (2014–2015) | |||||||
| ABX464 (Abivax S.A.) | Small molecule | miR-124 | Overexpression | HIV infection | Phase 2 | NCT02735863 | Rutsaert et al., 2019 |
| Completed (2016–2017) | |||||||
| ABX464 (Abivax S.A.) | Small molecule | miR-124 | Overexpression | HIV infection; healthy volunteers | Phase 1/2 | NCT02990325 | NA |
| Completed (2016–2017) | |||||||
| ABX464 (Abivax S.A.) | Small molecule | miR-124 | Overexpression | Ulcerative colitis | Phase 2 | NCT03093259 | NA |
| Completed (2017–2018) | |||||||
| ABX464 (Abivax S.A.) | Small molecule | miR-124 | Overexpression | Ulcerative colitis | Phase 2b | NCT03760003 | NA |
| Ongoing (2018–) | |||||||
| ABX464 (Abivax S.A.) | Small molecule | miR-124 | Overexpression | Rheumatoid arthritis | Phase 2 | NCT03813199 | NA |
| Ongoing (2019–) | |||||||
| ABX464 (Abivax S.A.) | Small molecule | miR-124 | Overexpression | Crohn disease | Phase 2 | NCT03905109 | NA |
| Ongoing (2019–) |
ATLL, adult T-cell leukemia/lymphoma; EDV, nonliving bacterial minicell nanoparticles; MF, mycosis fungoides; MM, multiple myeloma; MPM, malignant pleural mesothelioma; NA, not available; RCC, renal cell carcinoma; SCLC, small-cell lung cancer.
A. MicroRNA Restoration
As described above, many miRNAs, such as tumor-suppressive miRNAs, are downregulated in cancer. In these cases, introducing high levels of such miRNAs will restore the capability of the diseased cells by inhibiting the expression of pathogenic or oncogenic target genes. Virus-mediated vector systems are especially useful to overexpress miRNAs that are found to be downregulated in diseased cells since the restoration of such downregulated miRNAs in the targeted cells is expected to reverse already ongoing pathologic phenomena. In addition, viruses are suitable for continuous expression of miRNA by using a constitutive promoter. It enables a single dose of administration to be enough to maintain a high level of miRNA expression in the virus-infected cells. Kota et al. (2009) were able to overexpress miR-26a in an Myc-induced HCC-bearing mouse model by using an engineered AAV as a delivery vector system. Another tumor-suppressive miRNA, let-7, was also delivered into breast cancer cells through lentiviruses and reduced proliferation, mammosphere formation, tumor formation, and metastasis in nonobese diabetic/severe combined immunodeficiency mice (Yu et al., 2007). Let-7–carrying lentiviruses were also shown to slow non–small-cell lung cancer (NSCLC) tumor growth in mice (Trang et al., 2010). Although the viral delivery vectors were proven to be efficient in overexpressing miRNAs in the infected cells, none of these preclinical studies has made it to clinical trials because of challenges including virus clearance, aberrant immune responses, and unwanted genomic mutations originated by genome integration. Cationic lipid-based nanoparticles were also used to deliver miRNA mimics since their positive charges make it convenient to carry negatively charged miRNA mimics. Wu et al. delivered tumor-suppressive miR-29b mimics into lung cancer cells via a systemic injection after formulation into 1,2-di-O-octadecenyl-3-trimethylammonium propane:cholesterol:TSGA lipoflex nanoparticles (Wu et al., 2013). They observed a 5-fold increase of miR-29b in the tumor region after systemic delivery and downregulation of target gene expressions including CDK6, DNMT3B, and MCL-1 by ∼57.4%, ∼40.5%, and ∼52.4%, respectively. As a result, the tumor growth was significantly inhibited by ∼60% compared with negative control miRNA carrying nanoparticles (Wu et al., 2013). Liposomes have replaced viral vector systems to restore downregulated miRNAs because of fewer safety concerns and have made it to clinical trials. In lung cancer cells, restoration of let-7 through an overexpressing plasmid DNA incorporated into liposome-sensitized EGFR inhibitor–resistant lung cancer cells by suppressing EGFR expression (Rai et al., 2011). The laboratory of Anil Sood generated chitosan-based nanoparticle to deliver miR-34a mimics into prostate cancer cells and observed the downregulation of tyrosine-protein kinase Met and tyrosine-protein kinase receptor UFO and c-Myc, which are responsible for the proliferation of prostate cancer cells (Gaur et al., 2015). Remlarsen/MRG-201, a synthetic mimic of microRNA-29 developed by miRagen for the potential treatment of patients with fibrotic diseases, is currently in phase 2 clinical trials for the treatment of keloid (NCT03601052). This study was built on phase 1 data which indicated the favorable safety profile of MRG-201 in healthy subjects (NCT02603224). Besides, small molecule compounds can also mediate the overexpression/restoration of microRNAs, and this therapeutic approach has been translated into clinical studies. According to Abivax, one of the small molecule compounds, ABX464, delivers its anti-inflammatory effect via potentiation of the splicing of a long, noncoding RNA, resulting in overexpression of miR-124. ABX464 is currently undergoing multiple phase 2 clinical trials to treat multiple diseases (Table 2). For instance, phase 2 trials have been completed to test the safety and efficacy of ABX464 in human immunodeficiency virus (HIV) infection and ulcerative colitis (NCT02735863; NCT03093259) (Rutsaert et al., 2019). The technology used in the new generation of microRNA mimics, as well as small molecule–mediated overexpression of microRNAs, is crucial for the therapeutic targeting of microRNAs by overexpression in cancers and perioperative organ injuries.
B. MicroRNA Inhibition
Upregulated miRNAs have been implicated in the pathogenesis of many human diseases, which makes aberrantly overexpressed miRNAs attractive targets for therapeutic interventions. Recently, many approaches have been employed to inhibit upregulated miRNAs. Elmen et al. (2008) systemically injected LNA-anti-miRs against miR-122 to nonhuman primates (African green monkeys) and observed successful uptake of the LNA-anti-miR-122 in hepatocytes and depletion of endogenous miR-122 through stable heteroduplexes formed between the LNA-anti-miR-122 and miR-122. Because of the stability in serum and profound ability to bind to target miRNAs, LNA-based specific miRNA inhibition has been extensively investigated in preclinical studies and clinical trials. For example, miR-122 inhibition by complementary LNAs suppressed hepatitis C virus (HCV) proliferation in chimpanzee models without serious cytotoxicity in hepatocytes (Lanford et al., 2010). LNA-based inhibition of onco-miRs can also successfully induce cancer cell suppression. For example, LNA targeting miR-221, known to be an onco-miR in many cancer types, inhibited the proliferation of multiple myeloma cells (Di Martino et al., 2014). In glioma models, LNA-based miR-21 inhibition inhibited cell proliferation and sensitivity to radiation (Griveau et al., 2013). In clinical applications, Santaris Pharma A/S, based in Denmark, has generated a pharmacological miR-122–inhibiting LNA named Miravirsen, which has completed phase 2 clinical trials in patients with HCV (NCT01200420) (Janssen et al., 2013). Additionally, miRagen Therapeutics has another LNA-based anti-miR drug candidate inhibiting miR-155 (Cobomarsen/MRG106), which is currently undergoing phase 2 clinical trial for patients with cutaneous T-cell lymphoma of the mycosis fungoides subtype who are given multiple dosages of MRG106 through intratumoral injection (NCT03713320). For the treatment of chronic ischemic disorders, miRagen is currently developing and testing the safety and efficacy of inhibiting miR-92a (MRG-110), a microRNA linked to the regulation of blood vessel growth (NCT03603431). It is speculated that the anti-miR-92a could potentially accelerate the revascularization to facilitate the healing process. To summarize, the recent implementation of multiple phase 2 clinical trials based on microRNA inhibition is an important step toward the future of microRNA-based therapy for cancers and perioperative organ injuries.
C. Challenges and Limitations
Recently, the focus of drug delivery has moved toward targeted therapy using specific strategies to recognize target cells. Without such specific targeting methods, miRNA delivery methods can cause unwanted side effects because of the delivery of therapeutic miRNAs into nondiseased cells. For example, the phase I clinical trial of MRX34 harboring miR-34a was terminated early because of the lack of a tissue-specific delivery method (NCT01829971). The liposome-based formula was likely delivered into many cells throughout the body, and there is little preclinical evidence regarding the effect of excess miR-34a when introduced to normal cells in different organs. This concern calls for further modification and optimization of current delivery methods to enable tissue-specific targeting. To achieve cell/tissue-specific targeting, the carrier molecules need to be modifiable to include targeting moieties. In the clinical trial of TargomiRs for patients with recurrent thoracic cancer, EGFR-targeting antibodies were included in the liposome formulations, which enabled the miR-16–containing drug to be specifically delivered to cancer cells that overexpress EGFR on their cell surface (Reid et al., 2016). Folate (FA) is another targeting moiety for the specific delivery of many miRNAs. For example, polymer nanoparticles made from PLGA/poly(L-lactide)-block-poly(ethylene glycol) loaded with miR-204 were conjugated with FA to target folate receptors on colon cancer cells. When injected into mice bearing xenograft tumor, the FA-conjugated miR-204–carrying nanoparticles caused the regression of the tumors by inducing apoptosis (Zheng et al., 2018). 3WJ RNA nanoparticles derived from the packaging RNA of the DNA packaging motor system in bacteriophage phi29 have also been chemically conjugated with FA to specifically target the brain tumor tissues that highly overexpress folate receptors (Shu et al., 2011). This cancer cell–specific FA ligand conjugation was especially beneficial for targeted delivery of miRNAs in brain tumor since already differentiated normal brain neuronal cells do not express folate receptors, whereas glioblastoma cells overexpress folate receptors (Lee et al., 2015a,b, 2017). Moreover, recently emerging RNA aptamer technologies could potentially replace antibodies to reduce the antibody-related host immune response. Many RNA aptamers have been developed for recognizing specific cell surface markers (Nimjee et al., 2017). For example, targeted nanoparticle delivery of miR-29b to NSCLC was mediated by MUC-1 aptamer conjugation, which enabled selective delivery of an miR-29b mimic in vivo to cancer cells (Perepelyuk et al., 2018). When 3WJ RNA nanoparticles were loaded with EGFR-targeting aptamer together with antisense inhibitor sequences against miR-21, systemic delivery of the EGFR aptamer and anti-miR-21–carrying RNA nanoparticles successfully inhibited the tumor growth of triple-negative breast cancer (TNBC) (Shu et al., 2015). Recently, surface modification technology has evolved to modify the exosome surface with cell-targeting specific moieties to enable targeted therapy. For example, systemic administration of siRNA to breast cancer–bearing mice with siRNA against Survivin-carrying exosomes that were chemically conjugated with epidermal growth factor receptor (EGFR)-targeting RNA aptamers specifically delivered the Survivin-targeting siRNAs into the breast cancer cells and reduced tumor growth (Pi et al., 2018). Using this method, the delivery of therapeutic miRNA-loaded exosomes specifically into cancer cells can be achieved.
VII. Perspectives
Ever since the discovery of miRNA and its profound function in almost every pathway in cellular signaling, the small noncoding molecules opened a new field—not only in cancer research but also in various types of human diseases, including perioperative organ injuries. Global miRNA profiling from several patient cohorts has identified certain miRNA signatures that are aberrantly expressed in the diseased cells compared with those in normal cells. One way to reverse the impact of the dysregulated miRNAs was to modulate the expression level of such miRNAs to rescue the malfunctioning cell signaling pathways in a hope to cure the diseases. Not to mention the impact on cancers, miRNA-based targeted treatment of perioperative organ injury, especially in the form of prophylactic approaches, would greatly benefit the outcome of surgical patients, including patients with cancer with elective surgeries. It is also crucial to identify surgical patients at risk of perioperative organ injury, which could be achieved by more-extensive profiling of microRNA signatures in a different type of organ injury. Delivery of either miRNA mimics or inhibitors has been attempted extensively by simple chemical modification of miRNAs to the development of viral or nonviral delivery vector. Although many of those studies seemed therapeutically promising in preclinical stages, only a few miRNA-based therapies successfully translated into clinical trials, and none of them made it to final approval for commercial markets by the FDA. The main concerns include unsatisfactory delivery efficiency and potential toxicity. However, the emerging development of miRNA-based disease therapy with new strategies for cell- and tissue-specific delivery, such as RNA-based nanoparticle or exosomes, is a promising candidate to provide efficient and safe therapeutic targeting. Additional research studies are warranted to increase the stability of miRNA modulators and to reduce toxicity to achieve safety and therapeutic efficacy. miRNA-based therapy is a promising candidate to surpass or complement conventional treatment options to improve patient outcomes.
Acknowledgments
We appreciate Senior Manager Beth Ardoin Watson and Medical Illustrator Cynthia Reyna in Multimedia Scriptorium of Academic Technology at the University of Texas Health Science Center at Houston for their assistance in the preparation of illustration artworks in figures and tables.
Abbreviations
- AAV
adeno-associated virus
- AGI
acute gut injury
- ALI
acute lung injury
- AMI
acute myocardial infarction
- AML
acute myeloid leukemia
- AMO
antisense modified oligonucleotide
- ARDS
acute respiratory distress syndrome
- AT-RvD1
aspirin-triggered resolvin D1
- BCL-2
B-cell lymphoma 2
- CDK6
cyclin-dependent kinase 6
- CLL
chronic lymphocytic leukemia
- c-Met
cellular mesenchymal epithelial transition factor
- c-Myc
cellular myelocytomatosis
- CPC
cardiac progenitor cell
- CpG
5′-C-phosphate-G-3′
- CRC
colorectal cancer
- CSC
cancer stem cell
- DLBCL
diffuse large b-cell lymphoma
- DNMT
DNA methyltransferase
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DSS
dextran sodium sodium
- E2F
E2F transcription factor
- EGFR
epidermal growth factor receptor
- EV
extracellular vesicle
- 2′-F
2′-fluoro
- FA
folate
- FDA
Food and Drug Administration
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HIF
hypoxia-inducible factor
- HIV
human immunodeficiency virus
- IBD
inflammatory bowel disease
- IL
interleukin
- Let-7
Lethal-7
- lin-14
abnormal cell LINeage-14
- LNA
locked nucleic acid
- LNP
lipid nanoparticle
- LPS
lipopolysaccharide
- MCL-1
myeloid cell leukemia 1
- MI
myocardial infarction
- miR
microRNA
- miRNA
microRNA
- MVB
multivesicular body
- ncRNA
noncoding RNA
- NLRP3
NLR family pyrin domain containing 3
- NPRM
nano-proresolving medicine
- NSCLC
non–small-cell lung cancer
- onco-miR
oncogenic miRNA
- PARP-1
poly (ADP-ribose) polymerase 1
- PEI
polyethyleneimine
- PLGA
poly lactic-co-glycolic acid
- PMN
polymorphonuclear neutrophil
- PNA
peptide bond–based nucleic acid
- pre-miR
precursor-miRNA
- pri-miRNA
primary-miRNA
- PS
phosphorothioate
- qRT-PCR
quantitative real-time polymerase chain reaction
- Ras
rat sarcoma
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RvD1
resolvin D1
- SEB
Staphylococcal enterotoxin B
- siRNA
short interfering RNA
- SNP
single polymorphism
- SOCS-1
suppressor of cytokine signaling
- Sp1
specificity protein 1
- SVP
saphenous vein–derived pericyte progenitor cell
- Th
T helper
- TNBC
triple-negative breast cancer
- Treg
regulatory T cell
- 3′ UTR
3′ untranslated region
- VILI
ventilator-induced lung injury
- 3WJ
three-way junction
- XPO5
Exportin-5
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Lee, Yuan, Kerr, Yoo, Kim, Kaur, Eltzschig.
Footnotes
This work was supported in part by the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) [Grants R01-DK097075 and R01-DK109574] and the NIH National Heart, Lung, and Blood Institute (NHLBI) [Grants P0I-HL114457, R01-HL109233, R01-HL119837, and R01-HL133900] to H.K.E.; Pilot/Feasibility Program from Texas Medical Center Digestive Diseases Center through NIH NIDDK [Grant P30 DK056338] to T.J.L.; the American Thoracic Society Unrestricted Grant, American Heart Association Career Development Award [19CDA34660279], American Lung Association Catalyst Award [CA-622265], and The Center for Clinical and Translational Sciences, McGovern Medical School Pilot Award [1UL1TR003167–01], to X.Y.; American Cancer Society Research Scholar Grants [RSG-19-185-01-MPC] to J.Y.Y.; NIH National Cancer Institute (NCI) [Grant R01 CA150153 and P01 CA163205] and NIH National Institute of Neurologic Disorders and Stroke (NINDS) [Grant R01 NS064607] to B.K.; and NIH NINDS [Grant R01 NS104280] to D.H.K.
References
- Abdelmawla S, Guo S, Zhang L, Pulukuri SM, Patankar P, Conley P, Trebley J, Guo P, Li QX. (2011) Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery [published correction appears in Mol Ther (2011) 19:1748]. Mol Ther 19:1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, Tournev I, Schmidt HH, Coelho T, Berk JL, et al. (2018) Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med 379:11–21. [DOI] [PubMed] [Google Scholar]
- Aherne CM, Collins CB, Masterson JC, Tizzano M, Boyle TA, Westrich JA, Parnes JA, Furuta GT, Rivera-Nieves J, Eltzschig HK. (2012) Neuronal guidance molecule netrin-1 attenuates inflammatory cell trafficking during acute experimental colitis. Gut 61:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aherne CM, Saeedi B, Collins CB, Masterson JC, McNamee EN, Perrenoud L, Rapp CR, Curtis VF, Bayless A, Fletcher A, et al. (2015) Epithelial-specific A2B adenosine receptor signaling protects the colonic epithelial barrier during acute colitis. Mucosal Immunol 8:1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JL, Morrow DA. (2017) Acute myocardial infarction. N Engl J Med 376:2053–2064. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122:553–563. [DOI] [PubMed] [Google Scholar]
- Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, London CA, Kraybill W, Perrotti D, Croce CM, et al. (2013) miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR [published correction appears in Sci Signal (2013) 6:er6]. Sci Signal 6:ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM, Torre T, Siclari F, Moccetti T, Vassalli G. (2014) Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res 103:530–541. [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. (2018) Metazoan microRNAs. Cell 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels K, Karhausen J, Clambey ET, Grenz A, Eltzschig HK. (2013) Perioperative organ injury. Anesthesiology 119:1474–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 35:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, Gattinoni L, van Haren F, Larsson A, McAuley DF, et al. LUNG SAFE Investigators; ESICM Trials Group (2016) Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 315:788–800. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. (2003) Dicer is essential for mouse development. Nat Genet 35:215–217. [DOI] [PubMed] [Google Scholar]
- Boehm M, Slack FJ. (2006) MicroRNA control of lifespan and metabolism. Cell Cycle 5:837–840. [DOI] [PubMed] [Google Scholar]
- Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, et al. (2009) MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324:1710–1713. [DOI] [PubMed] [Google Scholar]
- Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Tréguer K, Carmona G, Bonauer A, et al. (2013) MicroRNA-34a regulates cardiac ageing and function. Nature 495:107–110. [DOI] [PubMed] [Google Scholar]
- Cai C, Xie Y, Wu L, Chen X, Liu H, Zhou Y, Zou H, Liu D, Zhao Y, Kong X, et al. (2017) PLGA-based dual targeted nanoparticles enhance miRNA transfection efficiency in hepatic carcinoma. Sci Rep 7:46250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai ZG, Zhang SM, Zhang Y, Zhou YY, Wu HB, Xu XP. (2012) MicroRNAs are dynamically regulated and play an important role in LPS-induced lung injury. Can J Physiol Pharmacol 90:37–43. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6:857–866. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99:15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. (2005) A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353:1793–1801. [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al. (2004a) MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA 101:11755–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. (2004b) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campani V, Salzano G, Lusa S, De Rosa G. (2016) Lipid nanovectors to deliver RNA oligonucleotides in cancer. Nanomaterials (Basel) 6:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. (2008) hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14:1340–1348. [DOI] [PubMed] [Google Scholar]
- Cao M, Deng X, Su S, Zhang F, Xiao X, Hu Q, Fu Y, Yang BB, Wu Y, Sheng W, et al. (2013) Protamine sulfate-nanodiamond hybrid nanoparticles as a vector for MiR-203 restoration in esophageal carcinoma cells. Nanoscale 5:12120–12125. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157:77–94. [DOI] [PubMed] [Google Scholar]
- Chadalapaka G, Jutooru I, Safe S. (2012) Celastrol decreases specificity proteins (Sp) and fibroblast growth factor receptor-3 (FGFR3) in bladder cancer cells. Carcinogenesis 33:886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, Tan PH, Ho GH, Lee AS. (2013) Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 19:4477–4487. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86. [DOI] [PubMed] [Google Scholar]
- Chen JF, Eltzschig HK, Fredholm BB. (2013) Adenosine receptors as drug targets--what are the challenges? Nat Rev Drug Discov 12:265–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. (2006) Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet 38:1452–1456. [DOI] [PubMed] [Google Scholar]
- Chen P, Guo X, Zhang L, Zhang W, Zhou Q, Tian Z, Zheng Y, Liao Q, Wang H, Li G, et al. (2017) MiR-200c is a cMyc-activated miRNA that promotes nasopharyngeal carcinoma by downregulating PTEN. Oncotarget 8:5206–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, et al. (2015) MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature 518:107–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs-Disney JL, Disney MD. (2016) Small molecule targeting of a microRNA associated with hepatocellular carcinoma. ACS Chem Biol 11:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintharlapalli S, Papineni S, Lei P, Pathi S, Safe S. (2011) Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer 11:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou GY, Cherng JY, Hsu HS, Wang ML, Tsai CM, Lu KH, Chien Y, Hung SC, Chen YW, Wong CI, et al. (2012) Cationic polyurethanes-short branch PEI-mediated delivery of Mir145 inhibited epithelial-mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. J Control Release 159:240–250. [DOI] [PubMed] [Google Scholar]
- Choudhry H, Harris AL, McIntyre A. (2016) The tumour hypoxia induced non-coding transcriptome. Mol Aspects Med 47–48:35–53. [DOI] [PubMed] [Google Scholar]
- Chugh P, Tamburro K, Dittmer DP. (2010) Profiling of pre-micro RNAs and microRNAs using quantitative real-time PCR (qPCR) arrays. J Vis Exp (46):2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosco D, Cilurzo F, Maiuolo J, Federico C, Di Martino MT, Cristiano MC, Tassone P, Fresta M, Paolino D. (2015) Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer treatment of multiple myeloma. Sci Rep 5:17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR, et al. (2009) Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood 114:1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA 103:7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig VJ, Tzankov A, Flori M, Schmid CA, Bader AG, Müller A. (2012) Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia 26:2421–2424. [DOI] [PubMed] [Google Scholar]
- Davidson BL, Harper SQ. (2005) Viral delivery of recombinant short hairpin RNAs. Methods Enzymol 392:145–173. [DOI] [PubMed] [Google Scholar]
- Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C. (2009) Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res 37:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitch EA. (2010) Gut lymph and lymphatics: a source of factors leading to organ injury and dysfunction. Ann N Y Acad Sci 1207 (Suppl 1):E103–E111. [DOI] [PubMed] [Google Scholar]
- Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sönnichsen SH, Nielsen PE. (1994) Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol 48:1310–1313. [DOI] [PubMed] [Google Scholar]
- Dengler V, Downey GP, Tuder RM, Eltzschig HK, Schmidt EP. (2013) Neutrophil intercellular communication in acute lung injury. Emerging roles of microparticles and gap junctions. Am J Respir Cell Mol Biol 49:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denizli M, Aslan B, Mangala LS, Jiang D, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK. (2017) Chitosan nanoparticles for miRNA delivery. Methods Mol Biol 1632:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereaux PJ, Goldman L, Yusuf S, Gilbert K, Leslie K, Guyatt GH. (2005) Surveillance and prevention of major perioperative ischemic cardiac events in patients undergoing noncardiac surgery: a review. CMAJ 173:779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereaux PJ, Xavier D, Pogue J, Guyatt G, Sigamani A, Garutti I, Leslie K, Rao-Melacini P, Chrolavicius S, Yang H, et al. POISE (PeriOperative ISchemic Evaluation) Investigators (2011) Characteristics and short-term prognosis of perioperative myocardial infarction in patients undergoing noncardiac surgery: a cohort study. Ann Intern Med 154:523–528. [DOI] [PubMed] [Google Scholar]
- Devulapally R, Sekar NM, Sekar TV, Foygel K, Massoud TF, Willmann JK, Paulmurugan R. (2015) Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano 9:2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino MT, Gullà A, Gallo Cantafio ME, Altomare E, Amodio N, Leone E, Morelli E, Lio SG, Caracciolo D, Rossi M, et al. (2014) In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One 9:e89659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Carey WA, Abel S, Collura C, Jiang G, Tomaszek S, Sutor S, Roden AC, Asmann YW, Prakash YS, et al. (2012) MicroRNA-mRNA interactions in a murine model of hyperoxia-induced bronchopulmonary dysplasia. BMC Genomics 13:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle T, Hartmann K, Bonney S, Reithel S, Mittelbronn M, Walker LA, Lowes BD, Han J, Borchers CH, Buttrick PM, et al. (2012) Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia. Nat Med 18:774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle T, Kewley EM, Brodsky KS, Tak E, Bonney S, Gobel M, Anderson D, Glover LE, Riegel AK, Colgan SP, et al. (2014) Identification of hypoxia-inducible factor HIF-1A as transcriptional regulator of the A2B adenosine receptor during acute lung injury. J Immunol 192:1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckle T, Köhler D, Lehmann R, El Kasmi K, Eltzschig HK. (2008) Hypoxia-inducible factor-1 is central to cardioprotection: a new paradigm for ischemic preconditioning. Circulation 118:166–175. [DOI] [PubMed] [Google Scholar]
- Ehrentraut H, Clambey ET, McNamee EN, Brodsky KS, Ehrentraut SF, Poth JM, Riegel AK, Westrich JA, Colgan SP, Eltzschig HK. (2013) CD73+ regulatory T cells contribute to adenosine-mediated resolution of acute lung injury. FASEB J 27:2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut H, Westrich JA, Eltzschig HK, Clambey ET. (2012) Adora2b adenosine receptor engagement enhances regulatory T cell abundance during endotoxin-induced pulmonary inflammation. PLoS One 7:e32416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villén J, Bartel DP. (2014) mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol Cell 56:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102:3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekin A, Karatas OF, Culha M, Ozen M. (2014) Designing a gold nanoparticle-based nanocarrier for microRNA transfection into the prostate and breast cancer cells. J Gene Med 16:331–335. [DOI] [PubMed] [Google Scholar]
- Elayadi AN, Braasch DA, Corey DR. (2002) Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry 41:9973–9981. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498. [DOI] [PubMed] [Google Scholar]
- Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. (2008) LNA-mediated microRNA silencing in non-human primates. Nature 452:896–899. [DOI] [PubMed] [Google Scholar]
- Eltzschig HK, Bonney SK, Eckle T. (2013) Attenuating myocardial ischemia by targeting A2B adenosine receptors. Trends Mol Med 19:345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Bratton DL, Colgan SP. (2014) Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat Rev Drug Discov 13:852–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Carmeliet P. (2011) Hypoxia and inflammation. N Engl J Med 364:656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Eckle T. (2011) Ischemia and reperfusion--from mechanism to translation. Nat Med 17:1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Köhler D, Eckle T, Kong T, Robson SC, Colgan SP. (2009) Central role of Sp1-regulated CD39 in hypoxia/ischemia protection. Blood 113:224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Weissmüller T, Mager A, Eckle T. (2006) Nucleotide metabolism and cell-cell interactions. Methods Mol Biol 341:73–87. [DOI] [PubMed] [Google Scholar]
- Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KG, Enright AJ, Gait MJ, Vigorito E. (2010) Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res 38:4466–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. (2007) MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA 104:15805–15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, et al. (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 109:E2110–E2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari D, Bianchi N, Eltzschig HK, Gambari R. (2016) MicroRNAs modulate the purinergic signaling network. Trends Mol Med 22:905–918. [DOI] [PubMed] [Google Scholar]
- Fredman G, Li Y, Dalli J, Chiang N, Serhan CN. (2012) Self-limited versus delayed resolution of acute inflammation: temporal regulation of pro-resolving mediators and microRNA. Sci Rep 2:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulciniti M, Amodio N, Bandi RL, Cagnetta A, Samur MK, Acharya C, Prabhala R, D’Aquila P, Bellizzi D, Passarino G, et al. (2016) miR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J 6:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, et al. (2008) Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA 105:3945–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. (2009) MicroRNA 29b functions in acute myeloid leukemia. Blood 114:5331–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur AB, Holbeck SL, Colburn NH, Israel MA. (2011) Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro-oncol 13:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur S, Wen Y, Song JH, Parikh NU, Mangala LS, Blessing AM, Ivan C, Wu SY, Varkaris A, Shi Y, et al. (2015) Chitosan nanoparticle-mediated delivery of miRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy. Oncotarget 6:29161–29177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, Sioufi NB, Sasmor H, Manoharan M, Levin AA. (2001) Pharmacokinetic properties of 2′-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J Pharmacol Exp Ther 296:890–897. [PubMed] [Google Scholar]
- Gou D, Ramchandran R, Peng X, Yao L, Kang K, Sarkar J, Wang Z, Zhou G, Raj JU. (2012) miR-210 has an antiapoptotic effect in pulmonary artery smooth muscle cells during hypoxia [published correction appears in Am J Physiol Lung Cell Mol Physiol (2012) 303:L1011]. Am J Physiol Lung Cell Mol Physiol 303:L682–L691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenz A, Homann D, Eltzschig HK. (2011) Extracellular adenosine: a safety signal that dampens hypoxia-induced inflammation during ischemia. Antioxid Redox Signal 15:2221–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. (2009) Small silencing RNAs: state-of-the-art. Adv Drug Deliv Rev 61:672–703. [DOI] [PubMed] [Google Scholar]
- Griveau A, Bejaud J, Anthiya S, Avril S, Autret D, Garcion E. (2013) Silencing of miR-21 by locked nucleic acid-lipid nanocapsule complexes sensitize human glioblastoma cells to radiation-induced cell death. Int J Pharm 454:765–774. [DOI] [PubMed] [Google Scholar]
- Guo P. (2010) The emerging field of RNA nanotechnology. Nat Nanotechnol 5:833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524. [DOI] [PubMed] [Google Scholar]
- Haeberle HA, Dürrstein C, Rosenberger P, Hosakote YM, Kuhlicke J, Kempf VA, Garofalo RP, Eltzschig HK. (2008) Oxygen-independent stabilization of hypoxia inducible factor (HIF)-1 during RSV infection. PLoS One 3:e3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950–952. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. [DOI] [PubMed] [Google Scholar]
- Hanvey JC, Peffer NJ, Bisi JE, Thomson SA, Cadilla R, Josey JA, Ricca DJ, Hassman CF, Bonham MA, Au KG, et al. (1992) Antisense and antigene properties of peptide nucleic acids. Science 258:1481–1485. [DOI] [PubMed] [Google Scholar]
- Hart ML, Gorzolla IC, Schittenhelm J, Robson SC, Eltzschig HK. (2010) SP1-dependent induction of CD39 facilitates hepatic ischemic preconditioning. J Immunol 184:4017–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart ML, Jacobi B, Schittenhelm J, Henn M, Eltzschig HK. (2009) Cutting edge: A2B adenosine receptor signaling provides potent protection during intestinal ischemia/reperfusion injury. J Immunol 182:3965–3968. [DOI] [PubMed] [Google Scholar]
- Hart ML, Much C, Gorzolla IC, Schittenhelm J, Kloor D, Stahl GL, Eltzschig HK. (2008) Extracellular adenosine production by ecto-5′-nucleotidase protects during murine hepatic ischemic preconditioning. Gastroenterology 135:1739–1750.e3. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. (2005) Stem cell division is regulated by the microRNA pathway. Nature 435:974–978. [DOI] [PubMed] [Google Scholar]
- Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, Olson EN. (2010) Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell 18:282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. (2007) A microRNA component of the p53 tumour suppressor network. Nature 447:1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. (2005) A microRNA polycistron as a potential human oncogene. Nature 435:828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick E, Cheng Y, Jin UH, Kim K, Safe S. (2016) Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget 7:22245–22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge MS, Tansey CM, Matté A, Tomlinson G, Diaz-Granados N, Cooper A, Guest CB, Mazer CD, Mehta S, Stewart TE, et al. Canadian Critical Care Trials Group (2011) Functional disability 5 years after acute respiratory distress syndrome. N Engl J Med 364:1293–1304. [DOI] [PubMed] [Google Scholar]
- Hessvik NP, Llorente A. (2018) Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 75:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkel R, Penzkofer D, Zühlke S, Fischer A, Husada W, Xu QF, Baloch E, van Rooij E, Zeiher AM, Kupatt C, et al. (2013) Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation 128:1066–1075. [DOI] [PubMed] [Google Scholar]
- Holubekova V, Mendelova A, Jasek K, Mersakova S, Zubor P, Lasabova Z. (2017) Epigenetic regulation by DNA methylation and miRNA molecules in cancer. Future Oncol 13:2217–2222. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. (2003) Embryonic stem cell-specific microRNAs. Dev Cell 5:351–358. [DOI] [PubMed] [Google Scholar]
- Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, et al. (2010) MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation 122 (11 Suppl):S124–S131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xiao X, Chintagari NR, Breshears M, Wang Y, Liu L. (2014) MicroRNA and mRNA expression profiling in rat acute respiratory distress syndrome. BMC Med Genomics 7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Schwind S, Yu B, Santhanam R, Wang H, Hoellerbauer P, Mims A, Klisovic R, Walker AR, Chan KK, et al. (2013) Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: a novel therapeutic strategy in acute myeloid leukemia. Clin Cancer Res 19:2355–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, Dalby CM, Robinson K, Stack C, Latimer PA, et al. (2012) Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 110:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt EA, Broyles D, Head T, Deo SK. (2015) MicroRNA detection: current technology and research strategies. Annu Rev Anal Chem (Palo Alto, Calif) 8:217–237. [DOI] [PubMed] [Google Scholar]
- Huo W, Zhao G, Yin J, Ouyang X, Wang Y, Yang C, Wang B, Dong P, Wang Z, Watari H, et al. (2017) Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer 8:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838. [DOI] [PubMed] [Google Scholar]
- Ibrahim AF, Weirauch U, Thomas M, Grünweller A, Hartmann RK, Aigner A. (2011) MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res 71:5214–5224. [DOI] [PubMed] [Google Scholar]
- Idzko M, Ferrari D, Riegel AK, Eltzschig HK. (2014) Extracellular nucleotide and nucleoside signaling in vascular and blood disease. Blood 124:1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. (2010) Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell 39:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. (2013) Treatment of HCV infection by targeting microRNA. N Engl J Med 368:1685–1694. [DOI] [PubMed] [Google Scholar]
- Jasinski D, Haque F, Binzel DW, Guo P. (2017) Advancement of the emerging field of RNA nanotechnology. ACS Nano 11:1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia XQ, Cheng HQ, Qian X, Bian CX, Shi ZM, Zhang JP, Jiang BH, Feng ZQ. (2012) Lentivirus-mediated overexpression of microRNA-199a inhibits cell proliferation of human hepatocellular carcinoma. Cell Biochem Biophys 62:237–244. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. (2005) RAS is regulated by the let-7 microRNA family. Cell 120:635–647. [DOI] [PubMed] [Google Scholar]
- Jurkovicova D, Magyerkova M, Kulcsar L, Krivjanska M, Krivjansky V, Gibadulinova A, Oveckova I, Chovanec M. (2014) miR-155 as a diagnostic and prognostic marker in hematological and solid malignancies. Neoplasma 61:241–251. [DOI] [PubMed] [Google Scholar]
- Jutooru I, Chadalapaka G, Abdelrahim M, Basha MR, Samudio I, Konopleva M, Andreeff M, Safe S. (2010) Methyl 2-cyano-3,12-dioxooleana-1,9-dien-28-oate decreases specificity protein transcription factors and inhibits pancreatic tumor growth: role of microRNA-27a. Mol Pharmacol 78:226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutooru I, Guthrie AS, Chadalapaka G, Pathi S, Kim K, Burghardt R, Jin UH, Safe S. (2014) Mechanism of action of phenethylisothiocyanate and other reactive oxygen species-inducing anticancer agents. Mol Cell Biol 34:2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki K, Hedrick E, Kasiappan R, Jin UH, Safe S. (2017) Piperlongumine induces reactive oxygen species (ROS)-dependent downregulation of specificity protein transcription factors. Cancer Prev Res (Phila) 10:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasar S, Salerno E, Yuan Y, Underbayev C, Vollenweider D, Laurindo MF, Fernandes H, Bonci D, Addario A, Mazzella F, et al. (2012) Systemic in vivo lentiviral delivery of miR-15a/16 reduces malignancy in the NZB de novo mouse model of chronic lymphocytic leukemia. Genes Immun 13:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasiappan R, Jutooru I, Karki K, Hedrick E, Safe S. (2016) Benzyl isothiocyanate (BITC) induces reactive oxygen species-dependent repression of STAT3 protein by down-regulation of specificity proteins in pancreatic cancer. J Biol Chem 291:27122–27133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katare R, Riu F, Mitchell K, Gubernator M, Campagnolo P, Cui Y, Fortunato O, Avolio E, Cesselli D, Beltrami AP, et al. (2011) Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132. Circ Res 109:894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Yeom JH, Ko JJ, Han MS, Lee K, Na SY, Bae J. (2011) Effective delivery of anti-miRNA DNA oligonucleotides by functionalized gold nanoparticles. J Biotechnol 155:287–292. [DOI] [PubMed] [Google Scholar]
- Koeppen M, Lee JW, Seo SW, Brodsky KS, Kreth S, Yang IV, Buttrick PM, Eckle T, Eltzschig HK. (2018) Hypoxia-inducible factor 2-alpha-dependent induction of amphiregulin dampens myocardial ischemia-reperfusion injury. Nat Commun 9:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen M, McNamee EN, Brodsky KS, Aherne CM, Faigle M, Downey GP, Colgan SP, Evans CM, Schwartz DA, Eltzschig HK. (2013) Detrimental role of the airway mucin Muc5ac during ventilator-induced lung injury. Mucosal Immunol 6:762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, et al. (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy S, Recchiuti A, Chiang N, Fredman G, Serhan CN. (2012) Resolvin D1 receptor stereoselectivity and regulation of inflammation and proresolving microRNAs. Am J Pathol 180:2018–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristen AV, Ajroud-Driss S, Conceição I, Gorevic P, Kyriakides T, Obici L. (2019) Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener Dis Manag 9:5–23. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. (2005) Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438:685–689. [DOI] [PubMed] [Google Scholar]
- Kunath K, von Harpe A, Fischer D, Petersen H, Bickel U, Voigt K, Kissel T. (2003) Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release 89:113–125. [DOI] [PubMed] [Google Scholar]
- Kurreck J, Wyszko E, Gillen C, Erdmann VA. (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res 30:1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahon B, Mordant P, Thabut G, Georger JF, Dauriat G, Mal H, Lesèche G, Castier Y. (2011) Early severe digestive complications after lung transplantation. Eur J Cardiothorac Surg 40:1419–1424. [DOI] [PubMed] [Google Scholar]
- Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK. (2005) Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res 65:6910–6918. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. (2010) Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter D, Shahar T, Yamashita S, Parker Kerrigan B, Fueyo J, et al. (2018) Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro-oncol 20:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, et al. (2008) Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 141:672–675. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854. [DOI] [PubMed] [Google Scholar]
- Lee SY, Lee S, Choi E, Ham O, Lee CY, Lee J, Seo HH, Cha MJ, Mun B, Lee Y, et al. (2016) Small molecule-mediated up-regulation of microRNA targeting a key cell death modulator BNIP3 improves cardiac function following ischemic injury. Sci Rep 6:23472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Haque F, Shu D, Yoo JY, Li H, Yokel RA, Horbinski C, Kim TH, Kim SH, Kwon CH, et al. (2015a) RNA nanoparticle as a vector for targeted siRNA delivery into glioblastoma mouse model. Oncotarget 6:14766–14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Haque F, Vieweger M, Yoo JY, Kaur B, Guo P, Croce CM. (2015b) Functional assays for specific targeting and delivery of RNA nanoparticles to brain tumor. Methods Mol Biol 1297:137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ, Yoo JY, Shu D, Li H, Zhang J, Yu JG, Jaime-Ramirez AC, Acunzo M, Romano G, Cui R, et al. (2017) RNA nanoparticle-based targeted therapy for glioblastoma through inhibition of oncogenic miR-21. Mol Ther 25:1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA. (2011) Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther 18:1111–1120. [DOI] [PubMed] [Google Scholar]
- Lennox KA, Sabel JL, Johnson MJ, Moreira BG, Fletcher CA, Rose SD, Behlke MA, Laikhter AL, Walder JA, Dagle JM. (2006) Characterization of modified antisense oligonucleotides in Xenopus laevis embryos. Oligonucleotides 16:26–42. [DOI] [PubMed] [Google Scholar]
- Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. (2013a) MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med 13:109–117. [DOI] [PubMed] [Google Scholar]
- Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. (2009) miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett 275:44–53. [DOI] [PubMed] [Google Scholar]
- Li Y, Dalli J, Chiang N, Baron RM, Quintana C, Serhan CN. (2013b) Plasticity of leukocytic exudates in resolving acute inflammation is regulated by microRNA and proresolving mediators. Immunity 39:885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu Y, Sun Z, Wang R, Ma D. (2018) MicroRNA-376a inhibits cell proliferation and invasion in glioblastoma multiforme by directly targeting specificity protein 1. Mol Med Rep 17:1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Sun W, He H, Zhang B, Ling C, Wang B, Huang T, Hou B, Guo Y. (2017) Antitumor effect of a new nano-vector with miRNA-135a on malignant glioma. Int J Nanomedicine 13:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wu H, Liu T, Li Y, Wang F, Wan H, Li X, Tang H. (2009) Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res 19:828–837. [DOI] [PubMed] [Google Scholar]
- Lobo SM, Rezende E, Knibel MF, Silva NB, Páramo JA, Nácul FE, Mendes CL, Assunção M, Costa RC, Grion CC, et al. (2011) Early determinants of death due to multiple organ failure after noncardiac surgery in high-risk patients. Anesth Analg 112:877–883. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J, Hermeking H. (2008) Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7:2591–2600. [DOI] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. (2010) Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142:914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Wang X. (2018) MicroRNA-296 targets specificity protein 1 to suppress cell proliferation and invasion in cervical cancer. Oncol Res 26:775–783. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Malik S, Bahal R. (2019) Investigation of PLGA nanoparticles in conjunction with nuclear localization sequence for enhanced delivery of antimiR phosphorothioates in cancer cells in vitro. J Nanobiotechnology 17:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Li J, Huang Y, Wu S, Pang X, He W, Liu X, Slutsky AS, Zhang H, Li Y. (2017) MicroRNA-19b mediates lung epithelial-mesenchymal transition via phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase in response to mechanical stretch. Am J Respir Cell Mol Biol 56:11–19. [DOI] [PubMed] [Google Scholar]
- Mariotto AB, Yabroff KR, Shao Y, Feuer EJ, Brown ML. (2011) Projections of the cost of cancer care in the United States: 2010-2020. J Natl Cancer Inst 103:117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez EC, Lilyanna S, Wang P, Vardy LA, Jiang X, Armugam A, Jeyaseelan K, Richards AM. (2017) MicroRNA-31 promotes adverse cardiac remodeling and dysfunction in ischemic heart disease. J Mol Cell Cardiol 112:27–39. [DOI] [PubMed] [Google Scholar]
- Mathieu M, Martin-Jaular L, Lavieu G, Théry C. (2019) Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21:9–17. [DOI] [PubMed] [Google Scholar]
- McAdams RM, Bierle CJ, Boldenow E, Weed S, Tsai J, Beyer RP, MacDonald JW, Bammler TK, Liggitt HD, Farin FM, et al. (2015) Choriodecidual group B streptococcal infection induces miR-155-5p in the fetal lung in Macaca nemestrina. Infect Immun 83:3909–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon BM, Mays D, Lipsky J, Stewart JA, Fauq A, Richelson E. (2002) Pharmacokinetics and tissue distribution of a peptide nucleic acid after intravenous administration. Antisense Nucleic Acid Drug Dev 12:65–70. [DOI] [PubMed] [Google Scholar]
- Medina PP, Nolde M, Slack FJ. (2010) OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 467:86–90. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J. (2008) High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res 36:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milone MC, O’Doherty U. (2018) Clinical use of lentiviral vectors. Leukemia 32:1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. (2014) Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med 18:371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri F, Callegari E, D’Abundo L, Corrà F, Lupini L, Sabbioni S, Negrini M. (2014) Inhibiting the oncogenic mir-221 by microRNA sponge: toward microRNA-based therapeutics for hepatocellular carcinoma. Gastroenterol Hepatol Bed Bench 7:43–54. [PMC free article] [PubMed] [Google Scholar]
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, Fullerton HJ, et al. Writing Group Members; American Heart Association Statistics Committee; Stroke Statistics Subcommittee (2016) Heart disease and stroke statistics-2016 update: a report from the American Heart Association [published correction appears in Circulation (2016) 133:e599]. Circulation 133:e38–e360. [DOI] [PubMed] [Google Scholar]
- Nahid MA, Pauley KM, Satoh M, Chan EK. (2009) miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J Biol Chem 284:34590–34599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narute P, Seam N, Tropea M, Logun C, Cai R, Sun J, Shelhamer JH, Meduri GU, Suffredini AF. (2017) Temporal changes in microrna expression in blood leukocytes from patients with the acute respiratory distress syndrome. Shock 47:688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazari-Jahantigh M, Wei Y, Noels H, Akhtar S, Zhou Z, Koenen RR, Heyll K, Gremse F, Kiessling F, Grommes J, et al. (2012) MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest 122:4190–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker V, Brodsky KS, Clambey ET, Schmidt EP, Packard TA, Davenport B, Standiford TJ, Weng T, Fletcher AA, Barthel L, et al. (2017a) Neutrophil transfer of miR-223 to lung epithelial cells dampens acute lung injury in mice. Sci Transl Med 9:eaah5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker V, Brodsky KS, Kreth S, Ginde AA, Eltzschig HK. (2016) Emerging roles for microRNAs in perioperative medicine. Anesthesiology 124:489–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker V, Haneklaus M, Jensen O, Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS, Gerich ME, et al. (2017b) Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J Exp Med 214:1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudecker V, Yuan X, Bowser JL, Eltzschig HK. (2017c) MicroRNAs in mucosal inflammation. J Mol Med (Berl) 95:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58:1375–1381. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, White RR, Becker RC, Sullenger BA. (2017) Aptamers as therapeutics. Annu Rev Pharmacol Toxicol 57:61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norling LV, Serhan CN. (2010) Profiling in resolving inflammatory exudates identifies novel anti-inflammatory and pro-resolving mediators and signals for termination. J Intern Med 268:15–24. [DOI] [PubMed] [Google Scholar]
- Norling LV, Spite M, Yang R, Flower RJ, Perretti M, Serhan CN. (2011) Cutting edge: humanized nano-proresolving medicines mimic inflammation-resolution and enhance wound healing. J Immunol 186:5543–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839–843. [DOI] [PubMed] [Google Scholar]
- O’Hara AJ, Vahrson W, Dittmer DP. (2008) Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood 111:2347–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri SK, Velissaris T. (2006) Gastrointestinal dysfunction following cardiac surgery. Perfusion 21:215–223. [DOI] [PubMed] [Google Scholar]
- Olive V, Li Q, He L. (2013) mir-17-92: a polycistronic oncomir with pleiotropic functions. Immunol Rev 253:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. (1999) The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol 216:671–680. [DOI] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12:19–30; sup pp 11–13. [DOI] [PubMed] [Google Scholar]
- Pan F, Mao H, Bu F, Tong X, Li J, Zhang S, Liu X, Wang L, Wu L, Chen R, et al. (2017) Sp1-mediated transcriptional activation of miR-205 promotes radioresistance in esophageal squamous cell carcinoma. Oncotarget 8:5735–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, Lee KF, Yeung WS. (2010) MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis 31:1037–1044. [DOI] [PubMed] [Google Scholar]
- Payne D. (2019) RNA therapies. Nature 574:S1. [DOI] [PubMed] [Google Scholar]
- Penaud-Budloo M, François A, Clément N, Ayuso E. (2018) Pharmacology of recombinant adeno-associated virus production. Mol Ther Methods Clin Dev 8:166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelyuk M, Sacko K, Thangavel K, Shoyele SA. (2018) Evaluation of MUC1-aptamer functionalized hybrid nanoparticles for targeted delivery of miRNA-29b to nonsmall cell lung cancer. Mol Pharm 15:985–993. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR, Yang CH, Pfeffer LM. (2015) The role of miR-21 in cancer. Drug Dev Res 76:270–277. [DOI] [PubMed] [Google Scholar]
- Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, Li H, Haque F, Wang S, Croce CM, et al. (2018) Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol 13:82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao L, Zhang M, Datta J, Xie X, Su T, Li H, Teknos TN, Pan Q. (2012) Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Mol Ther 20:1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polytarchou C, Hommes DW, Palumbo T, Hatziapostolou M, Koutsioumpa M, Koukos G, van der Meulen-de Jong AE, Oikonomopoulos A, van Deen WK, Vorvis C, et al. (2015) MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology 149:981–992.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poth JM, Brodsky K, Ehrentraut H, Grenz A, Eltzschig HK. (2013) Transcriptional control of adenosine signaling by hypoxia-inducible transcription factors during ischemic or inflammatory disease. J Mol Med (Berl) 91:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash TP. (2011) An overview of sugar-modified oligonucleotides for antisense therapeutics. Chem Biodivers 8:1616–1641. [DOI] [PubMed] [Google Scholar]
- Prakash TP, Bhat B. (2007) 2′-Modified oligonucleotides for antisense therapeutics. Curr Top Med Chem 7:641–649. [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Cheng HH, Tewari M. (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Takigawa N, Ito S, Kashihara H, Ichihara E, Yasuda T, Shimizu K, Tanimoto M, Kiura K. (2011) Liposomal delivery of microRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol Cancer Ther 10:1720–1727. [DOI] [PubMed] [Google Scholar]
- Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, Camporota L, Slutsky AS, ARDS Definition Task Force (2012) Acute respiratory distress syndrome: the Berlin definition. JAMA 307:2526–2533. [DOI] [PubMed] [Google Scholar]
- Rao R, Rieder SA, Nagarkatti P, Nagarkatti M. (2014) Staphylococcal enterotoxin B-induced microRNA-155 targets SOCS1 to promote acute inflammatory lung injury [published correction appears in Infect Immun (2014) 82:3986]. Infect Immun 82:2971–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, Serhan CN. (2011) MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J 25:544–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfern AD, Colley SM, Beveridge DJ, Ikeda N, Epis MR, Li X, Foulds CE, Stuart LM, Barker A, Russell VJ, et al. (2013) RNA-induced silencing complex (RISC) proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad Sci USA 110:6536–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Kao SC, Pavlakis N, Brahmbhatt H, MacDiarmid J, Clarke S, Boyer M, van Zandwijk N. (2016) Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 8:1079–1085. [DOI] [PubMed] [Google Scholar]
- Ren Y, Kang CS, Yuan XB, Zhou X, Xu P, Han L, Wang GX, Jia Z, Zhong Y, Yu S, et al. (2010) Co-delivery of as-miR-21 and 5-FU by poly(amidoamine) dendrimer attenuates human glioma cell growth in vitro. J Biomater Sci Polym Ed 21:303–314. [DOI] [PubMed] [Google Scholar]
- Ren Y, Wang R, Gao L, Li K, Zhou X, Guo H, Liu C, Han D, Tian J, Ye Q, et al. (2016) Sequential co-delivery of miR-21 inhibitor followed by burst release doxorubicin using NIR-responsive hollow gold nanoparticle to enhance anticancer efficacy. J Control Release 228:74–86. [DOI] [PubMed] [Google Scholar]
- Riegel AK, Faigle M, Zug S, Rosenberger P, Robaye B, Boeynaems JM, Idzko M, Eltzschig HK. (2011) Selective induction of endothelial P2Y6 nucleotide receptor promotes vascular inflammation. Blood 117:2548–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R, Robich MP, Plate JF, Trooskin SZ, Sellke FW. (2010) Gastrointestinal complications following cardiac surgery: a comprehensive review. J Card Surg 25:188–197. [DOI] [PubMed] [Google Scholar]
- Runtsch MC, Hu R, Alexander M, Wallace J, Kagele D, Petersen C, Valentine JF, Welker NC, Bronner MP, Chen X, et al. (2015) MicroRNA-146a constrains multiple parameters of intestinal immunity and increases susceptibility to DSS colitis. Oncotarget 6:28556–28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, Wittes J, Pappas J, Elci O, McCague S, et al. (2017) Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutsaert S, Steens JM, Gineste P, Cole B, Kint S, Barrett PN, Tazi J, Scherrer D, Ehrlich HJ, Vandekerckhove L. (2019) Safety, tolerability and impact on viral reservoirs of the addition to antiretroviral therapy of ABX464, an investigational antiviral drug, in individuals living with HIV-1: a phase IIa randomised controlled study. J Virus Erad 5:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Abbruzzese J, Abdelrahim M, Hedrick E. (2018) Specificity protein transcription factors and cancer: opportunities for drug development. Cancer Prev Res (Phila) 11:371–382. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. (2006) Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9:435–443. [DOI] [PubMed] [Google Scholar]
- Saunders MA, Liang H, Li WH. (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA 104:3300–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer D, Rouzier R, Cardona M, Barrett PN, Steens JM, Gineste P, Murphy RL, Tazi J, Ehrlich HJ. (2016) Randomized trial of food effect on pharmacokinetic parameters of ABX464 administered orally to healthy male subjects. Antimicrob Agents Chemother 61:e01288-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L. (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setua S, Khan S, Yallapu MM, Behrman SW, Sikander M, Khan SS, Jaggi M, Chauhan SC. (2017) Restitution of tumor suppressor microRNA-145 using magnetic nanoformulation for pancreatic cancer therapy. J Gastrointest Surg 21:94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, Faghihi MA, Khalil AM, Lu L, Paroo Z, et al. (2008) A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol 26:933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Li H, Shu Y, Xiong G, Carson WE, III, Haque F, Xu R, Guo P. (2015) Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano 9:9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. (2011) Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat Nanotechnol 6:658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Pi F, Sharma A, Rajabi M, Haque F, Shu D, Leggas M, Evers BM, Guo P. (2014) Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv Drug Deliv Rev 66:74–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Jacobsen A, Ozawa T, Harinath G, Pedraza A, Sander C, Holland EC, Huse JT. (2012) miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One 7:e33844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spite M, Norling LV, Summers L, Yang R, Cooper D, Petasis NA, Flower RJ, Perretti M, Serhan CN. (2009) Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature 461:1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. (2008) Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 582:1564–1568. [DOI] [PubMed] [Google Scholar]
- Sun HL, Cui R, Zhou J, Teng KY, Hsiao YH, Nakanishi K, Fassan M, Luo Z, Shi G, Tili E, et al. (2016) ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell 30:723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Luo JD, Jiang H, Duan DD. (2018) Tumor exosomes: a double-edged sword in cancer therapy. Acta Pharmacol Sin 39:534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Song X, Li X, Su T, Qi S, Qiao R, Wang F, Huan Y, Yang W, Wang J, et al. (2014) In vivo multimodality imaging of miRNA-16 iron nanoparticle reversing drug resistance to chemotherapy in a mouse gastric cancer model. Nanoscale 6:14343–14353. [DOI] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. (2007) An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282:2135–2143. [DOI] [PubMed] [Google Scholar]
- Takagi T, Naito Y, Mizushima K, Hirata I, Yagi N, Tomatsuri N, Ando T, Oyamada Y, Isozaki Y, Hongo H, et al. (2010) Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J Gastroenterol Hepatol 25 (Suppl 1):S129–S133. [DOI] [PubMed] [Google Scholar]
- Tereshko V, Portmann S, Tay EC, Martin P, Natt F, Altmann KH, Egli M. (1998) Correlating structure and stability of DNA duplexes with incorporated 2′-O-modified RNA analogues. Biochemistry 37:10626–10634. [DOI] [PubMed] [Google Scholar]
- Thompson BT, Chambers RC, Liu KD. (2017) Acute respiratory distress syndrome. N Engl J Med 377:1904–1905. [DOI] [PubMed] [Google Scholar]
- Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD, Thygesen K, Alpert JS, White HD, Jaffe AS, et al. Joint ESC/ACCF/AHA/WHF Task Force for Universal Definition of Myocardial Infarction; Authors/Task Force Members Chairpersons; Biomarker Subcommittee; ECG Subcommittee; Imaging Subcommittee; Classification Subcommittee; Intervention Subcommittee; Trials & Registries Subcommittee; Trials & Registries Subcommittee; Trials & Registries Subcommittee; Trials & Registries Subcommittee; ESC Committee for Practice Guidelines (CPG); Document Reviewers (2012) Third universal definition of myocardial infarction. J Am Coll Cardiol 60:1581–1598.22958960 [Google Scholar]
- Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K, et al. (2012) Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol 56:167–175. [DOI] [PubMed] [Google Scholar]
- Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB, et al. (2010) Regression of murine lung tumors by the let-7 microRNA. Oncogene 29:1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. (2011) Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther 19:1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang JC, Lo YM. (2007) Circulating nucleic acids in plasma/serum. Pathology 39:197–207. [DOI] [PubMed] [Google Scholar]
- Urbanek MO, Nawrocka AU, Krzyzosiak WJ. (2015) Small RNA detection by in situ hybridization methods. Int J Mol Sci 16:13259–13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válóczi A, Hornyik C, Varga N, Burgyán J, Kauppinen S, Havelda Z. (2004) Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey DL, et al. (2017) Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol 18:1386–1396. [DOI] [PubMed] [Google Scholar]
- Vaporidi K, Vergadi E, Kaniaris E, Hatziapostolou M, Lagoudaki E, Georgopoulos D, Zapol WM, Bloch KD, Iliopoulos D. (2012) Pulmonary microRNA profiling in a mouse model of ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol 303:L199–L207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergadi E, Vaporidi K, Theodorakis EE, Doxaki C, Lagoudaki E, Ieronymaki E, Alexaki VI, Helms M, Kondili E, Soennichsen B, et al. (2014) Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J Immunol 192:394–406. [DOI] [PubMed] [Google Scholar]
- Wagner MJ, Mitra R, McArthur MJ, Baze W, Barnhart K, Wu SY, Rodriguez-Aguayo C, Zhang X, Coleman RL, Lopez-Berestein G, et al. (2017) Preclinical mammalian safety studies of EPHARNA (DOPC nanoliposomal EphA2-targeted siRNA). Mol Cancer Ther 16:1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hökfelt T, Broberger C, Porreca F, Lai J, Ren K, et al. (2000) Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci USA 97:5633–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K, Jacob ST. (2009) Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology 50:1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sun W, Ling S, Wang Y, Wang X, Meng H, Li Y, Yuan X, Li J, Liu R, et al. (2019) AAV-anti-miR-214 prevents collapse of the femoral head in osteonecrosis by regulating osteoblast and osteoclast activities. Mol Ther Nucleic Acids 18:841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Liu Z, Su J, Chen WS, Wang XW, Bai SX, Zhang JZ, Yu SQ. (2016) Macrophage micro-RNA-155 promotes lipopolysaccharide-induced acute lung injury in mice and rats. Am J Physiol Lung Cell Mol Physiol 311:L494–L506. [DOI] [PubMed] [Google Scholar]
- Wen D, Danquah M, Chaudhary AK, Mahato RI. (2015) Small molecules targeting microRNA for cancer therapy: promises and obstacles. J Control Release 219:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75:855–862. [DOI] [PubMed] [Google Scholar]
- Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, Brant SR, Kwon JH. (2010) Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis 16:1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. (2008a) MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 135:1624–1635.e24. [DOI] [PubMed] [Google Scholar]
- Wu M, Jolicoeur N, Li Z, Zhang L, Fortin Y, L’Abbe D, Yu Z, Shen SH. (2008b) Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis 29:1710–1716. [DOI] [PubMed] [Google Scholar]
- Wu Y, Crawford M, Mao Y, Lee RJ, Davis IC, Elton TS, Lee LJ, Nana-Sinkam SP. (2013) Therapeutic delivery of microRNA-29b by cationic lipoplexes for lung cancer. Mol Ther Nucleic Acids 2:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. (2008) Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol 9:405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Tang J, Chen Q, Tang D, Liu M, Luo M, Wang Y, Wang J, Zhao Z, Tang C, et al. (2015) miR-429 regulates alveolar macrophage inflammatory cytokine production and is involved in LPS-induced acute lung injury. Biochem J 471:281–291. [DOI] [PubMed] [Google Scholar]
- Xu F, Liao JZ, Xiang GY, Zhao PX, Ye F, Zhao Q, He XX. (2017) MiR-101 and doxorubicin codelivered by liposomes suppressing malignant properties of hepatocellular carcinoma. Cancer Med 6:651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Jing Z, Wang X, Zha X, Zeng C, Chen S, Yang L, Luo G, Li B, Li Y. (2014) Altered expression pattern of miR-29a, miR-29b and the target genes in myeloid leukemia. Exp Hematol Oncol 3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue HY, Liu Y, Liao JZ, Lin JS, Li B, Yuan WG, Lee RJ, Li L, Xu CR, He XX. (2016) Gold nanoparticles delivered miR-375 for treatment of hepatocellular carcinoma. Oncotarget 7:86675–86686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA 105:13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Chang I, Tanaka Y, Gupta A, Dahiya R. (2012) MicroRNA-34a suppresses malignant transformation by targeting c-Myc transcriptional complexes in human renal cell carcinoma. Carcinogenesis 33:294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189–198. [DOI] [PubMed] [Google Scholar]
- Yang YP, Chien Y, Chiou GY, Cherng JY, Wang ML, Lo WL, Chang YL, Huang PI, Chen YW, Shih YH, et al. (2012) Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials 33:1462–1476. [DOI] [PubMed] [Google Scholar]
- Yarushkin AA, Kazantseva YA, Kobelev VS, Pustylnyak YA, Pustylnyak VO. (2017) Peroxisome proliferator-activated receptor γ activation inhibits liver growth through miR-122-mediated downregulation of cMyc. Eur J Pharmacol 797:39–44. [DOI] [PubMed] [Google Scholar]
- Yehya N, Yerrapureddy A, Tobias J, Margulies SS. (2012) MicroRNA modulate alveolar epithelial response to cyclic stretch. BMC Genomics 13:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin PT, Shah BP, Lee KB. (2014a) Combined magnetic nanoparticle-based microRNA and hyperthermia therapy to enhance apoptosis in brain cancer cells. Small 10:4106–4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Cai X, Chen X, Liang H, Zhang Y, Li J, Wang Z, Chen X, Zhang W, Yokoyama S, et al. (2014b) Tumor-secreted miR-214 induces regulatory T cells: a major link between immune evasion and tumor growth. Cell Res 24:1164–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. (2007) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131:1109–1123. [DOI] [PubMed] [Google Scholar]
- Yuan X, Berg N, Lee JW, Le TT, Neudecker V, Jing N, Eltzschig H. (2018a) MicroRNA miR-223 as regulator of innate immunity. J Leukoc Biol 104:515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Lee JW, Bowser JL, Neudecker V, Sridhar S, Eltzschig HK. (2018b) Targeting hypoxia signaling for perioperative organ injury. Anesth Analg 126:308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf S, Rangarajan S, Teo K, Islam S, Li W, Liu L, Bo J, Lou Q, Lu F, Liu T, et al. PURE Investigators (2014) Cardiovascular risk and events in 17 low-, middle-, and high-income countries. N Engl J Med 371:818–827. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Gong H, Li Y, Jie K, Ding C, Shao Q, Liu F, Zhan Y, Nie C, Zhu W, et al. (2013) Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp Lung Res 39:275–282. [DOI] [PubMed] [Google Scholar]
- Zheng B, Chen L, Pan CC, Wang JZ, Lu GR, Yang SX, Xue ZX, Wang FY, Xu CL. (2018) Targeted delivery of miRNA-204-5p by PEGylated polymer nanoparticles for colon cancer therapy. Nanomedicine (Lond) 13:769–785. [DOI] [PubMed] [Google Scholar]
- Zheng GX, Do BT, Webster DE, Khavari PA, Chang HY. (2014) Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol 21:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xiao J, Wu N, Liu C, Xu J, Liu F, Wu L. (2015) MicroRNA-223 regulates the differentiation and function of intestinal dendritic cells and macrophages by targeting C/EBPβ. Cell Rep 13:1149–1160. [DOI] [PubMed] [Google Scholar]
- Zhuang P, Muraleedharan CK, Xu S. (2017) Intraocular delivery of miR-146 inhibits diabetes-induced retinal functional defects in diabetic rat model. Invest Ophthalmol Vis Sci 58:1646–1655. [DOI] [PubMed] [Google Scholar]