Abstract
Before it was molecularly cloned in 1994, acute-phase response factor or signal transducer and activator of transcription (STAT)3 was the focus of intense research into understanding the mammalian response to injury, particularly the acute-phase response. Although known to be essential for liver production of acute-phase reactant proteins, many of which augment innate immune responses, molecular cloning of acute-phase response factor or STAT3 and the research this enabled helped establish the central function of Janus kinase (JAK) family members in cytokine signaling and identified a multitude of cytokines and peptide hormones, beyond interleukin-6 and its family members, that activate JAKs and STAT3, as well as numerous new programs that their activation drives. Many, like the acute-phase response, are adaptive, whereas several are maladaptive and lead to chronic inflammation and adverse consequences, such as cachexia, fibrosis, organ dysfunction, and cancer. Molecular cloning of STAT3 also enabled the identification of other noncanonical roles for STAT3 in normal physiology, including its contribution to the function of the electron transport chain and oxidative phosphorylation, its basal and stress-related adaptive functions in mitochondria, its function as a scaffold in inflammation-enhanced platelet activation, and its contributions to endothelial permeability and calcium efflux from endoplasmic reticulum. In this review, we will summarize the molecular and cellular biology of JAK/STAT3 signaling and its functions under basal and stress conditions, which are adaptive, and then review maladaptive JAK/STAT3 signaling in animals and humans that lead to disease, as well as recent attempts to modulate them to treat these diseases. In addition, we will discuss how consideration of the noncanonical and stress-related functions of STAT3 cannot be ignored in efforts to target the canonical functions of STAT3, if the goal is to develop drugs that are not only effective but safe.
Significance Statement
Key biological functions of Janus kinase (JAK)/signal transducer and activator of transcription (STAT)3 signaling can be delineated into two broad categories: those essential for normal cell and organ development and those activated in response to stress that are adaptive. Persistent or dysregulated JAK/STAT3 signaling, however, is maladaptive and contributes to many diseases, including diseases characterized by chronic inflammation and fibrosis, and cancer. A comprehensive understanding of JAK/STAT3 signaling in normal development, and in adaptive and maladaptive responses to stress, is essential for the continued development of safe and effective therapies that target this signaling pathway.
I. Molecular and Cellular Biology of Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling
A. Canonical Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling
The Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signal transduction pathway is an evolutionarily conserved pathway present in Drosophila melanogaster through Homo sapiens (Hou et al., 2002). This pathway is activated in response to many protein ligands, including cytokines, growth factors, interferons (IFNs), and peptide hormones, where it regulates a wide range of cellular processes, including cell growth, proliferation, differentiation, and apoptosis (Rawlings et al., 2004; O’Shea et al., 2013). Protein ligands bind to the extracellular domains of their receptors, which transmit signals into the cytoplasm through a series of conformational changes and post-translational modifications, notably tyrosine phosphorylation, leading to reprogramming of the targeted cells. Most cytokine receptors lack intrinsic kinase activity; consequently, central to their signaling is a family of protein tyrosine kinases known as JAK that are constitutively associated with the cytoplasmic region of the receptors and provide tyrosine kinase activity. The binding of cytokines to cognate receptors leads to a conformational change within the receptor complex that repositions membrane-proximal, receptor-bound JAKs into an active orientation, resulting in mutual transphosphorylation that increases their activity toward tyrosine sites within the receptor. Specific phosphotyrosine (pY)–peptide motifs then act as recruitment sites for specific STAT proteins, via their Src homology 2 (SH2) domains, leading to their being phosphorylated at key tyrosine residue within a loop domain located immediately C-terminal to the SH2 domain, followed by their SH2-to-SH2 homodimerization. These activated homodimers accumulate in the nucleus, where they bind to promotor regions of many genes and activate their transcription.
1. Janus Kinases
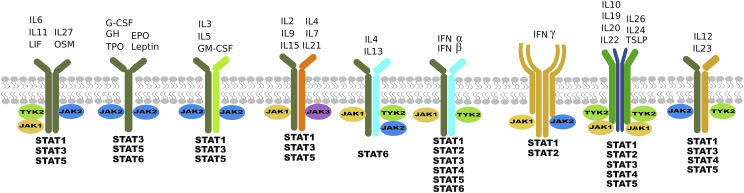
The human genome encodes four JAKs—JAK1, JAK2, JAK3, and tyrosine kinase 2 (TYK2)—that associate selectively (Fig. 1) with different receptors (Wilks, 1989; Firmbach-Kraft et al., 1990; Wilks et al., 1991; Harpur et al., 1992). Their essential role in developmental biology is underscored by the fact that deficiency in JAK1 and JAK2 is embryonically lethal due to neurologic defects and deficiencies in erythropoiesis, respectively, whereas deficiencies in JAK3 and TYK2 are associated with a variety of severe immunodeficiency syndromes in animal models and humans (Ghoreschi et al., 2009).
Fig. 1.
Schematic illustrating the complexity of cytokine signaling. Individual cytokines bind to more than one receptor complex, which associates with more than one JAK and activates one or more STAT proteins.
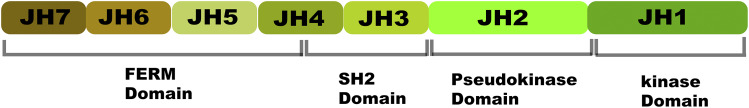
JAKs have a unique architecture (Fig. 2) that is distinguishable from other protein tyrosine kinases. Traditionally, JAK structure has been described based on its distinct regions of high homology consisting of seven JAK homology (JH) domains. Recent X-ray crystal structural studies have provided a clearer delineation of JAK structural architecture, with four distinct domains: 1) N-terminal band 4.1, ezrin, radixin, moesin (FERM) domain; 2) SH2 domain; 3) the catalytically active JH1 kinase domain located at the C-terminal end; and 4) the pseudo-kinase domain (JH2), located adjacent to it, which is enzymatically inactive but has a crucial regulatory role as a repressor of kinase activity (Lupardus et al., 2014). Not surprisingly, the majority of disease-causing mutations in JAKs are located in the JH2 domain. The N-terminal FERM and SH2 domains are tightly associated to form a single module that is required for the binding of JAKs to the cytoplasmic portion of cytokine receptors. The FERM–SH2 module has been shown to mediate specific association with two receptor motifs known as Box1 and Box2. Despite having structural data, it is still not clear how specificity for different groups of receptors is achieved (Yamaoka et al., 2004; Ferrao and Lupardus, 2017).
Fig. 2.
Schematic showing the seven sequence homology-based domains of JAK (JH1 through JH7) organized into four structural domains: FERM, SH2, pseudokinase, and kinase.
Many cytokines bind to receptors composed of heterologous polypeptide chains, and several of the polypeptide chains are common or shared. In addition, a single or combination of two to three JAKs selectively associates with each receptor (Lin et al., 1995; Baker et al., 2007; Kiu and Nicholson, 2012) (Fig. 2). The consequence is that a combination of different STATs can be activated by a single cytokine, resulting in a broad range of signaling outcomes.
2. Signal Transducer and Activator of Transcription Proteins
STAT3 is a member of a family of seven proteins known to play critical roles in cytokine and growth factor signaling (Darnell, 2002). More than 30 different cytokines in various tissue and cell systems have been implicated in the canonical signaling pathways of STAT3, involving nuclear translocation and activation of transcription of its target genes (Bharadwaj et al., 2016b). Similarly, multiple growth factors also signal via activation of STAT3 (Bharadwaj et al., 2016b). Latent STAT3 is predominantly localized in the cytoplasm as homotypic N-terminal domain (NTD)-to-NTD dimers. STAT3 activation is achieved when cytokines or peptide hormones bind to cell surface receptors, orchestrating a series of events, including phosphorylation at key sites, primarily tyrosine (Y) 705,which leads to activation or rearrangement of the dimer from NTD-to-NTD to SH2-to-SH2, nuclear localization, and transcriptional activation of specific genes by binding to respective DNA elements at their promoter sites.
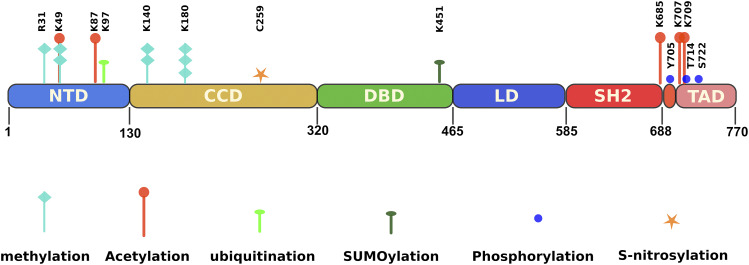
STAT3 is structured into seven functional domains (Fig. 3): 1) the globular N-terminal domain, 2) coil-coiled domain, 3) DNA-binding domain, 4) linker domain, 5) SH2 domain, 6) loop domain, and 7) transactivation domain, all of which play a role in STAT3 function (Shao et al., 2004). The N-terminal and SH2 domains are essential for the dimerization that, respectively, stabilizes the inactive and active conformations of STAT3. The SH2 domain is also required for docking to tyrosine-phosphorylated receptors (Vogt et al., 2011). In addition to recognizing specific DNA elements, the DNA-binding domain, in conjunction with the coil-coiled domain, participates in binding to the nuclear trafficking apparatus with both domains containing nuclear localization signals. The loop domain contains Y705, which loops into the SH2 domain of its dimerizing partner. The transactivation domain (TAD) plays a role in the recruitment of coactivators with histone acetyltransferase activity to facilitate transcriptional activation. This domain is missing in the major alternative isoform of STAT3, STAT3β, due to alternative mRNA splicing that shifts the reading frame and replaces the 55-amino-acid residue TAD with seven distinct residues that confer prolonged nuclear retention to STAT3β compared with STAT3βΔ (Shao et al., 2001).
Fig. 3.
Schematic showing amino acid residue sites within STAT3 that undergo post-translational modification.
3. Negative Regulation of Canonical Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling
Canonical STAT3 signaling is regulated by several feedback-inhibitory loops (Schmitz et al., 2000). The phosphatase tyrosine–protein phosphatase nonreceptor type 11, also known as protein–tyrosine phosphatase 1D or Src homology protein tyrosine phosphatase (SHP)-2, is recruited to the pYSTV motif of gp130 and phosphorylated on tyrosine residues by JAKs, which increases its tyrosine phosphatase activity for STAT3, which reduces pY-STAT3 levels (Schmitz et al., 2000). Other phosphatases have been implicated in the modulation of STAT3 signaling, for example, phosphatase and tensin homologue deleted on chromosome 10 (PTEN), which has both lipid and protein phosphatase activity, has been shown to act as a negative regulator of STAT3 (Sun and Steinberg, 2002; Mukhopadhyay et al., 2010), where its protein phosphatase activity negatively affects phosphorylation of STAT3. However, this modulation of STAT3 activity is context dependent, as it has been shown to also positively regulate STAT3 in certain tumors such as gliomas (de la Iglesia et al., 2008). Another inhibitory loop is mediated by suppressor of cytokine signaling (SOCS)3 (Yoshimura et al., 2007). SOCS3 mRNA is rapidly induced by activated STAT3 and STAT1, and SOCS3 protein interacts with pYSTV motif of gp130 through its SH2 domain and inhibits JAK activity through its kinase-inhibitory region, thereby restricting further STAT3 activation (Babon et al., 2012; Kershaw et al., 2013). The importance of SHP-2 goes beyond its tyrosine phosphatase activity, as it has been known to affect other signaling pathways, including extracellular signal-regulated kinase (ERK)1/2, ERK5, and Ak strain transforming (Akt)-mammalian target of rapamycin (mTOR) pathways (Kan et al., 2018) and is also known to be a proto-oncogene involved in K-RAS–mediated transformation (Kan et al., 2018).
4. Post-Translational Modifications of Signal Transducer and Activator of Transcription 3
In addition to tyrosine phosphorylation, STAT3 also undergoes a variety of other post-translational modifications, including methylation, acetylation, ubiquitination, IFN-stimulated gene (ISG)-ylation, and small ubiquitin-related modifiers (SUMOs)-conjugation (SUMOylation) (Kojima et al., 2013; You et al., 2015; Zouein et al., 2015; Avalle and Poli, 2018; Hu et al., 2018) with various effects on both transcriptional and nontranscriptional functions of STAT3, as stated below.
a. Serine 727 phosphorylation
In addition to phosphorylation on Y705, STAT3 can be phosphorylated on Ser727 within its TAD (Wen et al., 1995) by various serine/threonine kinases, e.g., mitogen-activated protein kinase (MAPK) like MAPK kinase/ERK (Chung et al., 1997; Stephens et al., 1998; Ng et al., 2011), c-Jun N-terminal kinase (JNK) (Zhang et al., 2001; Liu et al., 2012), p38 (Gollob et al., 1999; Lim and Cao, 1999; Turkson et al., 1999), casein kinase 2 (CK2) (Rozovski et al., 2015), protein kinase C (PKC)ε (Aziz et al., 2007a,b), mTOR (Kim et al., 2009), or cdk5 (Lam et al., 2015), depending on the following: 1) the stimulus, e.g., STAT3-activating cytokines/growth factors, e.g., interleukin (IL)-6 (Jain et al., 1999), other cellular stress signals, e.g., UV (Zhang et al., 2001; Liu et al., 2012), and nerve injury (Dubový et al., 2018); 2) the upstream signaling pathway, e.g., phosphatidylinositol 3-kinase (PI3K) (Kim et al., 2009) and Ras proteins (Saitoh et al., 2016); and 3) cell type (Boulton et al., 1995; Chung et al., 1997; Jain et al., 1999; Yokogami et al., 2000; Abe et al., 2001; Wierenga et al., 2003; Fu et al., 2004; Ohkawara et al., 2004; Kojima et al., 2005; Sato et al., 2005).
The effect of S727 phosphorylation (pS) on STAT3 gene transcription is varied and depends on the phosphorylating kinase, as well as the cell type, which likely dictates the availability of other transcription factors and proteins that help STAT3 bind to kinase (Avalle and Poli, 2018). Generally, pS is suggested to work with pY to activate STAT3-mediated transcription (Wen et al., 1995; Abe et al., 2001; Shen et al., 2004) most likely through recruiting coactivator proteins (Schuringa et al., 2001; Lufei et al., 2007; Lee et al., 2009a), e.g., JNK1/2-mediated pS-STAT3 following UVA irradiation, enhanced STAT3–DNA binding in epidermal JB6 cells (Zhang et al., 2001; Liu et al., 2012). In the human bronchial epithelial line, BEAS-2B, arsenic exposure induced vascular endothelial growth factor to promote cell migration through JNK-mediated Ser727 phosphorylation of STAT3 (Sun et al., 2014). However, there are instances when pS727 appears to counteract pY705 function. e.g., cdk5-mediated pS727 repressed FoxP3 upregulation, thereby limiting T regulatory cell (Treg) development (Lam et al., 2015), and reduction in pS727 by CK2–protein phosphatase 2A enhanced pY-STAT3–induced tumorigenic potential of glioma cells (Mandal et al., 2014). Similarly, pS727 mediated by UV-induced JNK activation (Lim and Cao, 1999), IL-6–mediated PKCδ activation (Jain et al., 1999), and JNK activation (Gkouveris et al., 2016) opposes Y705-mediated transcription. Mechanistically, pS727 in the nucleus is required for the recruitment of histone lysine methyltransferase SET9, which dimethylates STAT3 at Lys140, leading to pY705 inhibition (Yang et al., 2010). Furthermore, pS727 increases STAT3 binding to nuclear tyrosine phosphatase TC45 (Wakahara et al., 2012), leading to rapid dephosphorylation of pY705, thereby shortening the duration of STAT3 activity. Others have implicated pS-STAT3 in a distinct negative regulatory role (Chung et al., 1997; Yang et al., 2010) and in the mitochondrial function of Ras-transformed cells (Gough et al., 2009). STAT3 can also be activated independent of pY705 phosphorylation by simultaneous phosphorylation of Thr-714 and Ser727 (Waitkus et al., 2014) and upon engagement of CD44 through Lys-685 acetylation (Lee et al., 2009b).
The relative functional consequences of the two (pY and pS) phosphorylation events are discussed in following sections on STAT3 functions and its relevance in various diseases. Following is an example of how the information on these contrasting roles could be important. Constitutively activated pY-STAT3 seems to control oncogenic functions of most tumor cells (Bharadwaj et al., 2016b). Exceptionally, pS-STAT3, and not pY-STAT3, seems to be oncogenic in metastatic clear cell renal cell carcinoma (Lorente et al., 2019) and chronic lymphocytic leukemia (CLL) (Frank et al., 1997; Hazan-Halevy et al., 2010; Lupardus et al., 2014; Antosz et al., 2015; Rozovski et al., 2016, 2017a,b; Kondo et al., 2018). CK2, in the presence of increased levels of CD5 and B cell linker protein, both of which are low to absent in normal B cells, phosphorylates STAT3 at S727 (Rozovski et al., 2015) in CLL cells. In the hypothalamus, bradykinin, through epidermal growth factor receptor and ERK1/2, increased pS-STAT3 and resultantly STAT3-reporter activity, similar to IFN-γ–mediated pY-STAT3 (Breit et al., 2015), but the two differ in their specific functional consequences. Epidermal growth factor/ERK/pS-STAT3, but not IFN-γ/pY-STAT3, enhanced STAT3-regulated thyroglobulin-releasing hormone expression, while only transiently expressing SOCS3, which, in contrast, had a prolonged expression through IFN-γ/pY-STAT3, which failed to upregulate thyroglobulin-releasing hormone. pS-STAT3 also interacts with other transcription factors, e.g., nuclear factor κB (NF-κB), to transcribe differing but overlapping sets of genes (Kim et al., 2018; Coorens et al., 2019; Li et al., 2019), with varying transcriptional responses in endothelial cells thought to correlate with increasing stress and increased levels of pS727 phosphorylation (Kurdi and Booz, 2010). For example, in conditions of minimal stress, there are minimal levels of pS727, and pY705-mediated transcription dominates. In the case of mild inflammation as observed during oxidative stress or in the presence of tumor necrosis factor (TNF)-α, which activates NF-κB, the transcriptional output reflects that more pS727-STAT3 is associated with p65, and, with markedly increased pS727, it leads to the additional recruitment of transcription factor Sp1.
b. Methylation
Post-translational methylation of various lysine residues (K49, K140, and K180) in the NH2 terminus of STAT3 (Fig. 3) also is a mechanism by which STAT3 transcriptional activity is regulated in some cancer cells (Yang et al., 2010; Dasgupta et al., 2014). K49 and K140 methylation occurs after Y705 phosphorylation and binding to promoters (Yang et al., 2010; Stark et al., 2011; Dasgupta et al., 2015). Dimethylation of K49 and K140 by the histone-modifying enzymes EZH2 and SET9, respectively, regulate increased expression (K49) or decreased expression (K140) of many IL-6–dependent genes in colon cancer cells (Yang et al., 2010; Dasgupta et al., 2015). In this study, Y705 phosphorylation of STAT3 occurs first, and then S727 is phosphorylated, followed by binding of SET9 and dimethylation of K140. STAT3 is demethylated by lysine-specific demethylase 1 when it is bound to the SOCS3 promoter (Yang et al., 2010). Trimethylation of K180 by EZH2 enhanced pY-STAT3 in glioblastoma cells, presumably, by blocking access of a tyrosine phosphatase (Kim et al., 2013a). Methylation of the R31 arginine by protein arginine 2 contrastingly attenuated leptin-induced STAT3 activation in the hypothalamus (Iwasaki et al., 2010). Thus, the role of arginine methylation in regulating STAT3 activity is complex and cell-type specific (Komyod et al., 2005).
c. Acetylation
Reversible acetylation is also a contributor to STAT3 activity (Fig. 3). In addition to Y705 and S727 phosphorylation, cytokine treatment also acetylates STAT3 on the lysine residue, K685, within the SH2 domain (Yu et al., 2014). Acetylation at this site is mediated by the transcriptional coactivator and acetyltransferase, cAMP response element-binding protein-binding protein/p300, and is reversed by type I histone deacetylase (HDAC). Acetylation of two N-terminal lysines, K49 and K87, seems to be required for p300 binding and transcriptional activation by STAT3 (Ray et al., 2005; Hou et al., 2008), as well as HDAC1 binding and termination of transcription (Ray et al., 2008). Acetylated K685 also has been reported to be the initial contact of p300 with STAT3 (Ray et al., 2002, 2005, 2008; Hou et al., 2008). Furthermore, K685 acetylation has been suggested to be critical for forming stable STAT3 dimers that can efficiently bind to DNA and effectively transcribe cell growth–related genes in response to cytokines, e.g., oncostatin M (Yuan et al., 2005), as well as activation of the noncanonical NF-κB signaling pathway, by mediating the proteolytic processing of NF-κB p100 to p52 (Nadiminty et al., 2006). K685 acetylation also promotes STAT3 interaction with the nuclear exporter, exportin 7 (Herrmann Hua Yu, 2014), facilitating its nuclear egress; blocking acetylation results in nuclear retention of STAT3. K685 acetylation also has been implicated in gene silencing by STAT3 via targeting of DNA methyl transferase 1 to certain promoters (Lee et al., 2012; Thomas, 2012) because STAT3–DNA methyl transferase 1 binding is regulated by K685 acetylation (Lee et al., 2012). Although STAT3 deacetylation seems to be involved in termination of STAT3-mediated transcription, the overall impact of HDAC–STAT3 interactions remains controversial. HDAC5 directly regulates STAT3 localization and transcriptional activity via reciprocal STAT3 deacetylation at K685 and phosphorylation at Y705 in the hypothalamus (Kabra et al., 2016). Repression of STAT3 transcriptional activity by the HDAC, Sin3a, depends on deacetylation of K87, which is the main regulator of STAT3–Sin3a interaction (Icardi et al., 2012). Additionally, suppression by STAT3 of gluconeogenesis gene expression in the liver in the fed state is dependent on pY-STAT3 and acetylation of the C-terminal lysine residues K679, K685, K707, and K709, whereas it is opposed during the fasting state by the NAD-dependent deacetylase, sirtuin-1 (Nie et al., 2009). Therefore, the exact nature, extent, and effect of STAT3 acetylation may depend on tissue-specific interaction partners and differential transcriptional requirements for specific genes, as well as differential deacetylation kinetics (Hu et al., 2018).
d. Ubiquitination, ISGylation, and SUMOylation
Like most proteins, STAT3 also becomes conjugated to ubiquitin and undergoes proteasomal degradation through the ubiquitin–proteasome pathway (Aggarwal et al., 2009). The cysteinyl–aspartate proteases, i.e., caspases, also proteolytically process full-length STAT3α, thus reducing STAT3 expression and modulating its transcriptional activity via the formation of cleavage fragments (Darnowski et al., 2006). ISG15, a ubiquitin-like eukaryotic protein, can also modify the function and localization of its target proteins after covalent attachment (Seif et al., 2017). The ISGylation pathway is similar to ubiquitin conjugation, with differences being in the enzymes involved. However, in contrast to ubiquitination, ISGylation serves as a positive-feedback regulator of the JAK/STAT3 signaling pathway (Shuai and Liu, 2003). SUMO, another ubiquitin-related molecule that conjugates to STAT3 at lysine residue K451 in a process called SUMOylation, appears to negatively regulate the JAK/STAT3 pathway (Benekli et al., 2009) by promoting its binding to the phosphatase TC45, through the SUMO-interacting motif of TC45, leading to pY-STAT3 dephosphorylation and loss of activity (Hu et al., 2018). A major component of cigarette smoke and a potent carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, induced rapid generation of reactive oxygen species (ROS), which increased expression of the sentrin-specific protease 3 in head and neck squamous cell carcinoma (HNSCC) cell lines (Hecht, 2003). Sentrin-specific protease 3 deconjugated SUMO2/3 modification at the K451, thus weakening STAT3 binding to the nuclear phosphatase, TC45, which led to enhanced pY-STAT3.
B. Noncanonical Signal Transducer and Activator of Transcription 3 Activities
In addition to its canonical transcriptional activity, STAT3 has activities that are independent of its transcriptional role.
1. Phosphotyrosine 705–Unphosphorylated Signal Transducer and Activator of Transcription 3
A mutant of STAT3, in which the Y705 is replaced with phenylalanine (Y705F), was shown to shuttle between the nucleus and the cytoplasm (Liu et al., 2005) and transcribe multiple STAT3 target genes (Yang et al., 2005). S727 phosphorylation is not required for unphosphorylated STAT3 (U-STAT3)–p65 interaction (Yang et al., 2007a) and seems to be unimportant for U-STAT3–mediated gene transcription (Yang et al., 2007a; Yue et al., 2010). Not much is known about how U-STAT3 activates other target genes that do not require NF-κB (Cheon et al., 2011); presumably, binding of U-STAT3 to an IFN-γ activation site (GAS) or GAS-like element activates these genes. K685 acetylation is reported to enhance DNA binding, transactivation activity, and nuclear localization of STAT3 (Ray et al., 2002; Wang et al., 2005), although the conclusion that K685 is important for stable STAT3 dimer formation has been questioned based on structural considerations (O’Shea et al., 2005). Recently, evidence was provided that K685 acetylation is more important for gene expression by U-STAT3, rather than in response to tyrosine-phosphorylated STAT3 (Dasgupta et al., 2014). U-STAT3, which tends to form antiparallel dimers, is also speculated to be acting as a dominant negative of pY-STAT3 (Yue et al., 2010).
U-STAT3 can bind GAS elements as a monomer or dimer, but binding is much weaker than seen with pY-STAT3 (Timofeeva et al., 2012). However, U-STAT3 binds more strongly to AT-rich DNA sequence sites and sequences common in DNA elements referred to as scaffold/matrix attachment regions that are implicated in chromatin organization (Timofeeva et al., 2012). U-STAT3 also recognizes specific DNA structures, such as DNA nodes and four-way junctions that are involved in nucleosomal structure and assembly, reinforcing the idea that U-STAT3 influences chromatin organization. In fact, deletion of the Drosophila STAT homolog, Stat92E, disrupts heterochromatin integrity and allows transcriptional activation of genes that are not normally Stat92E targets (Yan et al., 2011), and Stat92E interacts with heterochromatin protein 1 to regulate histone 1 and histone 3 function (Shi et al., 2008; Xu et al., 2014b). This novel epigenetic role of Stat92E seems to be disrupted by its tyrosine phosphorylation by promoting its translocation to target genes (Shi et al., 2008). The observation that nuclei of various cell lines and primary cells contain substantial levels of U-STAT3 under nonstimulated conditions, i.e., ∼40% of total cellular STAT3 (Meyer et al., 2002; Liu et al., 2005), lends further credence to the noncanonical transcriptional role for U-STAT3 in the nucleus.
2. Mitochondrial Signal Transducer and Activator of Transcription 3
A mitochondrial pool of STAT3 has been reported to promote optimal functioning of the electron transport chain (Wegrzyn et al., 2009), support RAS-dependent oncogenic transformation (Gough et al., 2009), and inhibit the proapoptotic opening of the permeability transition pore (Boengler et al., 2010). Phosphorylation of STAT3 monomers on Ser727 leads to translocation into the mitochondria without dimerization (Garama et al., 2016; Yang and Rincon, 2016), where it associates with the inner mitochondrial membrane and the mitochondrial matrix. Nuclear translocation of pS-STAT3 may be mediated by gene associated with retinoid IFN-induced cell mortality 19 (Tammineni et al., 2013), heat shock protein 22 (Qiu et al., 2011), and/or the chaperone, TOM20 (Avalle and Poli, 2018).
Mitochondrial (mt) STAT3 promotes cell survival under stress, e.g., heart ischemia and Ras-mediated transformation (Gough et al., 2009; Wegrzyn et al., 2009; Szczepanek et al., 2011). pS-STAT3 interacts with electron transport chain (ETC) complex I (I) and ETCII to preserve optimal ETC activity, increase membrane polarization and ATP production, and enhance the activity of lactate dehydrogenase, thereby increasing aerobic glycolysis and decreasing ROS production (Avalle and Poli, 2018). mtSTAT3 also protects from apoptosis by inhibiting mitochondrial permeability transition pore opening, presumably through its interaction with cyclophilin D (Boengler et al., 2010), which heightens the requirement for calcium influx from the endoplasmic reticulum (ER) (Boengler et al., 2010). While increasing ETC activity, mtSTAT3 seems to reduce ROS production (Sarafian et al., 2010; Mantel et al., 2012; Yang et al., 2015), probably through formation of ETC supercomplexes, which, by optimizing coupling, are known to reduce electron leakage (Yang et al., 2015), and/or through increased synthesis of ROS scavengers, e.g., glutathione (Garama et al., 2015).
In addition to modulating its canonical activity, acetylation of STAT3 also seems to contribute to mitochondrial localization (Xu et al., 2016). STAT3 is acetylated in starved cells after serum reintroduction or insulin stimulation, and this acetylated STAT3 goes into the mitochondria, where it associates with the pyruvate dehydrogenase complex E1 and subsequently accelerates the conversion of pyruvate to acetyl-CoA, elevates the mitochondrial membrane potential, and promotes ATP synthesis (Xu et al., 2016). These observations suggest that STAT3 might control both energy metabolism and cell death through nontranscriptional effects; however, it should be noted that transcriptional regulation of both processes by STAT3 also has been suggested (Xu et al., 2016).
3. Scaffold Function in Platelets
A nontranscriptional role of pY-STAT3 in platelets during inflammation also has been suggested (Zhou et al., 2013). pY-STAT3 dimers acting as a protein scaffold have been shown to enhance collagen-induced intracellular signaling, resulting in platelet activation, calcium mobilization, and aggregation of IL-6/IL-6 receptor (R) (and probably IL-15/IL-15R). Activated pY-STAT3 dimers interact directly with spleen tyrosine kinase (Syk) and phospholipase C (PLC)γ2 to form a trimolecular complex, which enhances the catalytic interaction between Syk kinase and its substrate PLCγ2. Activated PLCγ2 hydrolyzes phosphatidylinositol 4,5-bisphosphate to produce inositol 1,4,5-triphosphate, leading to calcium mobilization. This novel nontranscriptional activity of STAT3 enhances collagen-induced signaling in platelets, making platelets hyperactive under inflammatory conditions and linking proinflammatory signals to increased hemostasis and thrombosis (Yuan et al., 2015a).
4. Signal Transducer and Activator of Transcription 3 in the Endoplasmic Reticulum
STAT3 has recently been found to translocate to the ER (Avalle et al., 2019), where it interacts with the calcium channel inositol 1,4,5-trisphosphate receptor, type 3 (IP3R3), facilitating its degradation, possibly through the ubiquitin E3 ligase FBXL2 (Kuchay et al., 2017). The decrease in IP3R3 reduces calcium efflux from ER to the cytoplasm and mitochondria. Because excessive cytoplasmic calcium causes mitochondrial calcium overload, the opening of the mitochondrial transition pore, and the initiation of the intrinsic apoptosis program, STAT3-mediated IP3R3 downregulation contributes to resistance to apoptosis induced by oxidative stress in STAT3-dependent mammary cells. Tyrosine phosphorylation of STAT3 does not seem to be required for STAT3 ER localization or IP3R3 interaction. However, pS727 appears to be involved, as mouse embryonic fibroblast cells expressing a STAT3 mutated at S727 displayed excessive calcium release and increased apoptotic cell death to H2O2 (Avalle and Poli, 2018).
II. Biologic Functions of Janus Kinase/Signal Transducer and Activator of Transcription 3 in Basal Conditions
The effect of global deletion of JAKs in mice and humans is outlined above. Similar to global deletion of JAK1 and JAK2, global deletion of STAT3 proved to be lethal at the embryonic stage day 6.5 (Takeda et al., 1997), indicating its prominent role in various key developmental processes. Generation of cell-specific STAT3 knockouts (KO) in mice (Table 1) has yielded information on contributions of STAT3 to cell- and organ-specific development, including contributions to the development of innate immunity (Welte et al., 2003); its role in the commitment of common lymphoid and myeloid (CMP) progenitors to the dendritic cell (DC) lineage during hematopoiesis (Laouar et al., 2003), Th17 cell development (Takeda et al., 1998; Yang et al., 2007b), B cell development (Chou et al., 2006; Fornek et al., 2006), and macrophage and neutrophil function (Takeda et al., 1999); and its role in liver regeneration (Inoue et al., 2004; Moh et al., 2007), heart muscle development and function (Jacoby et al., 2003; Hilfiker-Kleiner et al., 2004), development of neuronal cells (Gao et al., 2004), musculoskeletal system (Guadagnin et al., 2018), and mammary glands (Haricharan and Li, 2014). These KO studies also revealed the role of STAT3 in wound healing of all epithelial cells (Pickert et al., 2009), including cutaneous (Sano et al., 1999) and respiratory epithelium (Hokuto et al., 2004), and its role in lipid metabolism (Cernkovich et al., 2008) and reproduction (Robker et al., 2014). At the completion of embryogenesis and organ system development, the actions of STAT3 largely focus on stress, e.g., its role in cardiomyocyte resistance to inflammation and acute injury (Zouein et al., 2015) and in the pathogenesis of age-related diseases, such as those involving the heart (Jacoby et al., 2003; Zouein et al., 2015), as well as injury responses in other organs, as outlined below. Some of the functions of STAT3 are mediated by canonical pY-STAT3–dependent transcriptional activation of STAT-regulated genes, whereas others are modulated by other post-translational modifications, including Ser727 phosphorylation, acetylation, and sumoylation, which appear to work in tandem.
TABLE 1.
Summary of findings in cell-specific STAT3–KO mice
| STAT3 KO in Cell | Phenotype | Reference |
|---|---|---|
| All cells | Embryonic lethal at 7 days | Takeda et al., 1997 |
| Keratinocytes | Impaired hair cycle and skin wound healing | Sano et al., 1999 |
| Respiratory epithelium | Impaired injury tolerance | Hokuto et al., 2004 |
| Intestinal epithelium | Impaired epithelial wound healing | Pickert et al., 2009 |
| Cardiomyocyte | Increased presbycardia and MI susceptibility | Jacoby et al., 2003; Hilfiker-Kleiner et al., 2004 |
| Hepatocytes | Insulin resistance; impaired liver regeneration | Inoue et al., 2004; Moh et al., 2007 |
| Adipocyte | Increased body weight and adiposity | Cernkovich et al., 2008 |
| Neural cells | Obesity, diabetes, infertility, and thermal dysregulation | Gao et al., 2004 |
| Hematopoietic progenitors | Unaltered basal hematopoiesis; impaired stem cell regeneration after 5-FU | Lee et al., 2002; Chung et al., 2006 |
| Hematopoietic-derived cells | Impaired DC function: defective transition of CMP/CLP to common DC precursors | Laouar et al., 2003 |
| Bone marrow cells | Impaired myeloid cells resulting in Crohn’s disease-like pathology | Welte et al., 2003 |
| T cells | Increased apoptosis and impaired Th17 development | Takeda et al., 1998b; Yang et al., 2007b |
| B cells | Reduced B cell compartments and plasma cells | Chou et al., 2006; Fornek et al., 2006 |
| Macrophages and neutrophils | High susceptibility to endotoxin shock, ↑ TNF-α, IL-1, and IFN-γ | Takeda et al., 1999 |
CLP, common lymphoid progenitor; 5-FU, fluorouracil.
Clear insight into the contributions of STAT3 in humans is provided by reviewing the features of patients with autosomal-dominant hyper-IgE syndrome (AD-HIES) or Job’s syndrome (Yong et al., 2012; Zhang et al., 2018). AD-HIES is a rare multisystem disorder classified as a primary immunodeficiency disease, usually of children, in which patient’s cells have diminished STAT3 activity due to an autosomal-dominant, loss-of-function STAT3 mutation that leads to STAT3 protein instability and reduced half-life (Bocchini et al., 2016). Their primary immune defects are as follows: 1) impaired barrier functions, especially of the skin and respiratory tract, due to delayed wound healing (Table 1); 2) decreased generation of Th17 cells and production of IL-17 and IL-22, which reduces epithelial production of chemokines and defensins (Yong et al., 2012; Zhang et al., 2018); and 3) intrinsic impairments in neutrophil chemotaxis (Hill and Quie, 1974; Hill et al., 1974; Panopoulos et al., 2006; Mintz et al., 2010; Nguyen-Jackson et al., 2010; Mogensen, 2013). AD-HIES patients do not demonstrate quantitative defects in basal or stress hematopoiesis, including the production of neutrophils, total lymphocyte numbers, red blood cells, or platelets (Zhang et al., 2010). Patients most commonly present with moderate-to-severe eczema, bacterial pneumonia, staphylococcal skin abscesses, mucocutaneous candidiasis, elevated serum IgE levels, and eosinophilia, all consistent with the consequences of impaired mucosal barrier defenses. Infectious diseases form the main basis for morbidity and mortality, with death in most patients, resulting from chronic fungal infections of the lung. Management of infections includes careful maintenance of hygiene and use of preventive and treatment courses of antimicrobials. In addition to the immune system, AD-HIES patients have defects in the development and maintenance of bone, connective tissue, teeth, and vasculature. These defects manifest as asymmetric facies with prominent forehead, chin, and wide-set eyes, along with high arched palate, retention of primary teeth, and abnormalities of middle-sized arteries. The molecule basis for these abnormalities is not well understood and is managed symptomatically or, where necessary, surgically.
Following is a focused overview of the biologic functions of STAT3 under normal and stressed conditions. We have attempted to summarize the role of STAT3 in each of these functions and also to indicate, where information is available, whether it is mediated through canonical and/or noncanonical activities of STAT3. Table 2 indicates how an increase or decrease in these activities leads to disease, whether STAT3 augmentation or inhibition would be desirable to manage the disease, and at what stages in clinical development these interventions are. Additional information also is provided below.
TABLE 2.
STAT3 functions and perturbations causing disease
| Function | Localization: Modification | Activation/Repression Overall Role of STAT3 | Perturbation Leading to Disease | Suggested Intervention | Stage of Drug Development |
|---|---|---|---|---|---|
| Emergency granulopoiesis | N: pY, U | STAT3 positively regulates granulocytic progenitor proliferation, neutrophil release, and chemotaxis during emergency granulopoeisis following microbial infection (Hillmer et al., 2016) | STAT3 AD-HIES patients, having diminished STAT3 activity from AD-LOF STAT3 mutations, show neutrophil-migration impairments and increased risks of skin and lung infections (Zhang et al., 2010) | Augmentation | Preclinical |
| Platelet function | C: pY | Constitutive JAK3/STAT3, essential for platelet function. Pretreating platelets with JAK3i WHI-P131 ↓pY-STAT1/pY-STAT3 and platelet activation | Treatment of mice with Jak3i, WHI-P131 prolonged bleeding time and improved event-free survival in a mouse model of thromboplastin-induced generalized and fatal thromboembolism (Tibbles et al., 2001) | Inhibition | Preclinical |
| N: pY | IL-21/Jak3/STAT3 helps generation of megakaryocytes from CD34+ cells | IL-9/JAK2/STAT3 helps platelet function, promoting development of DVT (Feng et al., 2018) | Inhibition | Preclinical | |
| N: pY | STAT3-regulated TPO, through Jak2-pYSTAT3–mediated transcription, promotes platelet formation from megakaryocytes | Targeting JAK/STAT3 signaling could be an emerging strategy in the management of platelet-associated diseases | Inhibition | Preclinical | |
| C: pY | Collagen induces pYSTAT3 dimer-Syk-PLCγ2 complex, which accelerates catalytic interaction between the collagen-activated (phosphorylated) Syk and its substrate PLCγ2, thereby facilitating platelet activation, calcium mobilization, and aggregation, through hydrolysis of PIP2 to IP3 to mobilize calcium (Zhou et al., 2013) | This nontranscriptional STAT3 function enhances collagen-induced signaling in platelets, potentially making platelets hyperactive in conditions of inflammation, e.g., coronary artery diseases by linking proinflammatory cytokine signals to hemostasis/thrombosis. Repurposed STAT3i, piperlongumine-inhibited collagen-induced platelet activation, aggregation, and thrombus formation by blocking JAK2-STAT3 phosphorylation, reducing the pYSTAT3 dimer-Syk-PLCγ2 complex formation (Yuan et al., 2015a) | Inhibition | Preclinical | |
| DC function | N: pY | Flt3L-STAT3-Tcf4 positively regulates immature pDC and conventional DC development and functions, but activated STAT3 blocks DC maturation in cancer | Sustained IL-6/IL-10 activity in cancer could lead to more tolerogenic iDCs, contributing to loss of immune surveillance phenotype | Inhibition | Preclinical |
| Augmented LIF-STAT3 activity in DC prevents allogeneic transplant rejection (Barton, 2006) | Augmentation | Preclinical | |||
| Macrophage functions | N: pY | IL-10/STAT3 anti-inflammatory role in macrophages through suppression of TLR-NF-κB/MAPK axis (El Kasmi et al., 2007) | STAT3 AD-HIES patients with STAT LOF mutations, display ↑basal/TLR4-mediated proinflammatory cytokines in neutrophils and mononuclear cells (Holland et al., 2007) and severely impaired IL-10 responses | Augmentation | Preclinical |
| B cell functions | N: pY | STAT3- Flt3L promotes transition of Flt3+ pre-pro-B cell HPCs to subsequent precursors, e.g., CLPs (Chou et al., 2006) | Leptin/JAK2/STAT3 stimulates B cells to induce proinflammatory IL-6, TNF-α in obese individuals exacerbating adipose tissue inflammation and insulin resistance | Inhibition | Preclinical |
| N: pY | IL-21/STAT3-Blimp1 helps differentiation of later stage B-lineage–committed CD19+ precursors to IgG-secreting plasma cells | Naive B cells from AD-HIES patients fail to differentiate into antibody-secreting cells when activated with CD40L and IL-21 (Rincon and Pereira, 2018) | Augmentation | Preclinical | |
| CD4+ T cell function | N: pY, pS | IL-6/STAT3-mediated T cell survival independent of Bcl-2 | IL-6/STAT3-mediated resistance to apoptosis of lamina propria CD4+ T cells perpetuates IBD (Atreya et al., 2000) | Inhibition | Clinical |
| IL-6/TGF-β/IL-21 and IL-23/STAT3 mediate Th17 differentiation | Higher morbidity/mortality in ∼10% of asthma patients refractory to steroid treatment show Th17-driven phenotype (2000; McKinley et al., 2008; Al-Ramli et al., 2009) | Inhibition | Preclinical | ||
| N, pS | Cdk5-pS-STAT3 inhibits generation of CD4+ Treg from naive CD4+ precursors by suppressing Treg-specifying transcription factor Foxp3 in mature Tregs (Lam et al., 2015) | In the setting of GVHD, STAT3 deficiency promotes inducible Treg generation, restrains GVHD, and improves survival, suggesting that STAT3 blockade in CD4+ T cells may be useful in treating GVHD | Inhibition | Preclinical | |
| N: pY | STAT3, Foxp3 coregulate IL-10 in Tregs, maintaining ability of Foxp3+ Tregs to inhibit inflammatory Th17 cells | STAT3-HDAC6 transcriptionally ↑IL-10, preventing effective Ag-specific CD4+ T activation in tumor-bearing mice (Cheng et al., 2014), while using an HDACi, ↑effective T cell priming and antitumor response (Wang et al., 2011a; Cheng et al., 2014) | Inhibition | Preclinical | |
| CD8+ T cell function | N: pY | IL-10/IL-21/STAT3-Eomes/BCL6/Blimp1 helps generate stable, long-lived memory CD8+ T cells | STAT3 AD-HIES have ↓memory CD8+ T cells and memory CD4+ T cells, relative to healthy controls, impairing their ability to manage chronic infections | Augmentation | Preclinical |
| N, mt: pY, pS | IL-6/pY-STAT3 helps early transcription of IL-4/IL-21 in CD4 cells (Yang and Rincon, 2016); mtSTAT3 contributes to maintain their late expression through increases in mitochondrial Ca2+ levels and ATP (Yang et al., 2015b) | ||||
| Anaphylaxis | N, mt: pS | Clinical food allergy and anaphylaxis are decreased in AD-HIES patients due to defective mast cell degranulation and reduced endothelial cell permeability (Siegel et al., 2013; Hox et al., 2016), indicating a role of STAT3 in the positive regulation of these processes. STAT3 regulates endothelial permeability downstream of the proinflammatory cytokines IL-6 and TNF-α (Alsaffar et al., 2016) | Vascular inflammation–mediated endothelial STAT3 activation increases vascular leakage through downregulating tight junction proteins (Yun et al., 2017). mt-STAT3 is essential for immune-mediated degranulation of mast cells and basophils, thus making it a candidate for blocking in any allergy scenario (Erlich et al., 2014) | Inhibition | Preclinical |
| Lipid metabolism | N, Mt: pY, pS | Leptin-LepRb-Jak2-STAT3 regulates energy homeostasis, glucose and lipid metabolism, and immune function | Leptin-STAT3 causes immune aging, tumor progression, and PD1/PDL-mediated T cell dysfunction in obese cancer patients, making them better suited to PD1/PDL1-targeted immunotherapy (Wang et al., 2019) | Inhibition | Preclinical |
| N: pY, pS | STAT3-C/EBP/PPARγ regulates adipogenesis from preadipocytes. PPARγ agonist reverses STAT3 inhibition–induced adipogenesis | ||||
| N, Mt: pY, pS | Adipocyte-specific STAT3-KO mice have ↑body weight, ↑adipose tissue mass, but not adipocyte hyperplasia, hyperphagia, or reduced energy expenditure, implicating STAT3 in lipolysis, through ↑ATGL expression, ↓fatty acid synthase, ↓acetyl-CoA carboxylase, ↓AOX | CLL cells adopt to oxidize FFA with constitutively pS-STAT3, which transcriptionally upregulates LPL, which catalyzes the hydrolysis of triglycerides into FFA (Rozovski et al., 2015), making targeting STAT3 a viable strategy to kill CLL cells | Inhibition | Preclinical | |
| Glycometabolism | N, mt: pY, pS | Insulin/IL-6/pYSTAT3 ↓gluconeogenic genes, e.g., G6Pase and PEPCK (Inoue et al., 2004), thus maintaining glucose homeostasis by adjusting glucose production as per energy balance (Inoue, 2016) | Prolonged IL-6 exposure under chronic inflammation leads to insulin resistance and glucose intolerance in human adipocytes, hepatocytes (Gurzov et al., 2016), and skeletal muscles. Blocking IL-6 in diet-induced obese mice represses hepatic inflammation via inhibition of the IL-6/JAK2/STAT3 pathway (Park et al., 2010), thereby increasing insulin sensitivity and resultantly decreasing diet-induced obesity (Priceman et al., 2013) | Inhibition | Preclinical |
| The leptin-JAK2/STAT3 pathway inhibits insulin synthesis, whereas SOCS3 blocks the STAT3-dependent regulation of pre-proinsulin 1 gene | |||||
| RS | N, mt: pY, pS | Activated by RS. Protects from IR and non-IR, e.g., UV-induced cell death | Chronic UV- mediated STAT3 activation key step in UV-induced skin cancer | Inhibition | Clinical |
| OS | N, mt: pY, pS, Ac | Both activated and repressed by OS. Activation seems to be protected from ROS-mediated damage | Asbestos, cigarette smoke–activated pY-STAT3 protects cells from ROS-mediated apoptosis, leading to carcinogenesis | Inhibition | Clinical |
| pY-STAT3 protects from hyperoxia-induced acute lung injury resulting from increasing O2, used to treat lung failure, e.g., ARDS | Augment | Preclinical | |||
| GS | N, mt: pY, pS | Activated by GS. Augments DNA repair genes, e.g., MDC1-ATm-chk2 pathway helping in DNA repair and cell protection | Cancer cells escape IR through upregulation of pY-STAT3 that protects from IR leading to radio- or chemoresistance, e.g., HNSCC | Inhibition | Preclinical |
| Senescence | N, mt: pY, pS | JAK-STAT3 promotes replicative (Kojima et al., 2005) and oncogene-induced senescence (Kuilman et al., 2008) | Senescence is implicated in age-related diseases, including renal dysfunction, T2D, IPF, cardiovascular disease, and age-related cachexia (McHugh and Gil, 2018), suggesting use of STAT inhibition as therapy | Inhibition | Preclinical |
| IL-6/STAT3 activated by chemotherapy promoted drug-induced premature senescence in autocrine manner to help maintain a minimal residual tumor burden that could lead to relapse (Gilbert and Hemann, 2010) | Inhibition | Preclinical | |||
| Autophagy | N, mt: pY, pS | Nuclear STAT3 can be both proautophagic, e.g., LIF/STAT3 in mammary glandular cells through ↑PIK3R1/p55α and PIK3R1/p50α (Pensa et al., 2014) or antiautophagic through ↑antiautophagy-related genes, e.g., BCL2, BCL2L1, and MCL1 (Fukada et al., 1996; Bromberg et al., 1999; Bowman et al., 2000) | STAT3 increased autophagy in mouse models of cancer cachexia (Penna et al., 2013), suggesting inhibition as viable therapy | Inhibition | Preclinical |
| Loss of mtSTAT3, which interacts with complex I/II of the ETC to modulate their activities, results in ↑ROS (Szczepanek et al., 2012), inducing mitophagy (Scherz-Shouval and Elazar, 2007). | mtSTAT3 suppresses autophagy induced by oxidative stress and effectively preserves mitochondria from mitophagy, protecting from ischemia (Szczepanek et al., 2011) | Augment | Preclinical | ||
| Wound healing | N: pY, pS | STAT3 is involved both in the initial inflammatory stages following tissue injury (Dauer et al., 2005) as well as later re-epithelialization stage of wound healing (Sano et al., 1999). Both pY and pS (Shen et al., 2004; Lim et al., 2006; Dunkel et al., 2012) STAT3 involved | Persistently activated STAT3, independently as well as in conjunction with TGF-β, drives unregulated wound healing, leading to fibrosis through overexpression of ECM components, e.g., COL1A2 (Papaioannou et al., 2018), MMPs (Matsui et al., 2017), and promoting apoptosis resistance of fibroblasts (Moodley et al., 2003; Habiel and Hogaboam, 2014; Xu et al., 2014a; Milara et al., 2018), aberrant EMT (Kasembeli et al., 2018), etc. | Inhibition | Clinical |
Ac, acetylation; AOX, acyl-CoA oxidase; ARDS, adult respiratory distress syndrome; ATGL, adipose triglyceride lipase; C, cytoplasmic; COL1A2, collagen type I alpha 2 chain; CLP, common lymphoid progenitor; DVT, deep venous thrombosis; EMT, epithelial to mesenchymal transition; FFA, free fatty acids; FLT3L, FMS-like tyrosine kinase 3 ligand; G6Pase, glucose-6-phosphatase; GS, genotoxic stress; HPCs, hematopoietic progenitor cells; iDC, immature DC; IP3, inositol 1,4,5-triphosphate; IPF, idiopathic pulmonary fibrosis; LIF, leukemia inhibitory factor; LPL, lipoprotein lipase; N, nuclear; LOF, loss of function; OS, oxidative stress; pDC, plasmacytoid DC; PDC, pyruvate dehydrogenase complex; PEPCK, phosphoenolpyruvate carboxykinase; PIP2, phosphatidylinositol 4,5-bisphosphate; pS, pS727; pY, pY705; RS, radiation stress; T2D, type 2 diabetes; TPO, thrombopoietin.
A. Janus Kinase/Signal Transducer and Activator of Transcription 3 in Hematopoiesis and Immune Cell Function
The JAK/STAT3 signaling pathway is well characterized in hematopoiesis and immune cells, where it contributes to proliferation, differentiation, activation, and regulation of multiple types of hematopoietic and immune cells in both the normal and stress conditions.
1. Granulopoiesis
Although STAT3 was found not to be required for basal granulopoiesis (Lee et al., 2002), STAT3 was shown to regulate critical steps in emergency granulopoiesis (Hillmer et al., 2016). Specifically, granulocyte cell-stimulating factor–mediated activation of STAT3 induces granulocytic progenitor proliferation through transcriptional upregulation of CCAAT/enhancer-binding protein (C/EBP)β and c-Myc (Johansen et al., 2001; Hirai et al., 2006; Zhang et al., 2010). In addition, STAT3 activation mediates neutrophil mobilization in the bone marrow (Cheers et al., 1988; Lieschke et al., 1994; Boettcher et al., 2014), neutrophil release into the circulation (Panopoulos et al., 2006; Nguyen-Jackson et al., 2010), and neutrophil chemotaxis (Panopoulos et al., 2006; Nguyen-Jackson et al., 2010) by upregulating CXC chemokine receptor 2/Il8rb and macrophage-inflammatory protein-2/Cxcl2 (Nguyen-Jackson et al., 2010, 2012). These effects of STAT3 are mediated by the transcriptional activity of pY-STAT3. However, U-STAT3 and not pY-STAT3 (or pS-STAT3) recently was found to transcriptionally activate Fanconi C, a DNA repair protein that is expressed during early phases of granulopoiesis (Shah et al., 2018).
2. Platelets
Placentas from pregnancies complicated by hemolysis, elevated liver enzymes, and low platelet count were found to have very low expression of IL-6, STAT3α, STAT3β, and Bcl-2, a STAT3-regulated gene, which implicates the JAK/STAT3 pathway in aberrant platelet biology (Cecati et al., 2013). Several protein tyrosine kinases play major roles in platelet physiology (Melford et al., 1997; Ezumi et al., 1998; Laffargue et al., 1999; Pasquet et al., 2000), including JAK3, which is constitutively active in human platelets (Rodriguez-Linares and Watson, 1994; Witthuhn et al., 1999; Imada and Leonard, 2000), Thrombin treatment activates STAT1 and STAT3 (Tibbles et al., 2001); platelets from JAK3-deficient mice displayed reduced thrombin-induced increases in pY-STAT1 and pY-STAT3. Pretreatment of human platelets with the JAK3 inhibitor, WHI-P131, markedly diminished pY-STAT1 and pY-STAT3 levels and diminished all markers of platelet activation, including shape change, pseudopod formation, degranulation, serotonin release, as well as platelet aggregation. Importantly, in a mouse model of thromboembolism induced by administration of thromboplastin, treatment of mice with WHI-P131 prolonged bleeding time and improved event-free survival (Tibbles et al., 2001).
Thrombopoietin activates the JAK/STAT3 pathway in megakaryocytes (Broudy and Kaushansky, 1995; Kaushansky, 1995; Drachman and Kaushansky, 1997; Drachman et al., 1997), leading to upregulated expression of multiple genes required for platelet production (Xu et al., 2017). Thrombopoietin synthesis in the liver is regulated by Jak2/STAT3 (Grozovsky et al., 2015; Hoffmeister and Falet, 2016). In addition, the IL-21/JAK3/STAT3 signaling pathway promotes generation of megakaryocytes from CD34+ cells (Benbarche et al., 2017). As outlined above, STAT3 forms a complex with Syk and PLCγ2 to mediate platelet activation and aggregation (Zhou et al., 2013) that may also include JAK2 (Lu et al., 2014). IL-9 through a JAK2/STAT3 signaling pathway facilitated platelet function and promoted development of deep venous thrombosis (Feng et al., 2018). Several JAK inhibitors (Jakinibs), e.g., AG490 (Lu et al., 2014) and TG101348 (Lu et al., 2014), and several STAT3 inhibitors, e.g., piperlongumine (Yuan et al., 2016) and SCC99 (Xu et al., 2017), have been found to modulate platelet activation.
3. Dendritic Cells
STAT3 also plays major roles in immature DC development and function, affecting (Laouar et al., 2003) both professional IFN-I–producing plasmacytoid DCs and conventional DCs (Merad and Manz, 2009; Merad et al., 2013). Fms-related tyrosine kinase 3 ligand, the major DC growth factor, activates STAT3 to stimulate DC progenitor proliferation (Esashi et al., 2008; Li et al., 2012). Tcf4/E2-2, another transcriptional regulator of plasmacytoid DC development, is also transcriptionally upregulated by STAT3 (Panopoulos et al., 2002; D’Amico and Wu, 2003; Onai et al., 2006; Hegde et al., 2009). Contrastingly, IL-6– and IL-10–stimulated STAT3 is also known to suppress DC maturation and activation and promote immune tolerance (Cheng et al., 2003; Nefedova et al., 2004; Wang et al., 2004b; Bharadwaj et al., 2007; Lunz et al., 2007; Lin et al., 2010) through the following: 1) inhibition of major histocompatibility complex class II and costimulatory molecule expression; 2) upregulation of myeloid-related protein SA100A9, which suppresses DC function; 3) induction of inhibitory programmed death ligand-1 on DCs; 4) negatively regulating Toll-like receptor (TLR)–induced proinflammatory mediators (Park et al., 2004; Kitamura et al., 2005; Cheng et al., 2008; Melillo et al., 2010; Wölfle et al., 2011); and 5) increased tryptophan-catabolizing enzyme indoleamine 2,3-dioxygenase and resultant T cell apoptosis mediated by kynurenine, a tryptophan metabolite (Barton, 2006). These STAT3 effects are mediated by pY-STAT3; inhibition of JAK/STAT3 signaling improves mature DC function and augments antitumor immunity (Nefedova et al., 2004, 2005a,b), although attenuating STAT3 and NF-κB signaling in immature DCs leads to systemic DC dysfunction in non-small cell lung cancer (NSCLC) (Li et al., 2017).
4. Phagocyte Function
STAT3 suppresses TLR signaling in mature phagocytes (Takeda et al., 1999; Melillo et al., 2010) by suppressing NF-κB and MAPK, which majorly regulate inflammatory gene expression downstream of TLR activation (El Kasmi et al., 2007), through direct transcriptional repression of E2 ubiquitin–conjugating enzyme required for TLR signaling (Zhang et al., 2014). STAT3-deficient macrophages, neutrophils, and DCs (Takeda et al., 1999; Melillo et al., 2010) and hematopoietic-specific STAT3-KO mice have increased production of inflammatory cytokines (Takeda et al., 1999; Welte et al., 2003; Alonzi et al., 2004; Watowich and Liu, 2010), due to loss of regulation by IL-10 (Kühn et al., 1993; Takeda et al., 1999; Melillo et al., 2010), other transcriptional repressors (El Kasmi et al., 2007), or other anti-inflammatory effectors (Schaljo et al., 2009; Smith et al., 2011; Chan et al., 2012; Gaba et al., 2012; Hutchins et al., 2012, 2013; Curtale et al., 2013), and/or increased IL-12–activated T cell–induced IFN-γ (Takeda et al., 1999; Melillo et al., 2010). Macrophages produce both proinflammatory, e.g., IL-6, and anti-inflammatory cytokines, e.g., IL-10. Although both IL-6 and IL-10 activate STAT3, they confer opposite effects because of differential effects of SOCS3, induced by each (Yasukawa et al., 2003). SOCS3 blocks IL-6 signaling by binding to gp130, thus limiting the IL-6/pY-STAT3 activation, but it does not inhibit IL-10/pY-STAT3 axis, inducing prolonged STAT3 activation, thereby promoting its anti-inflammatory functions (Nakata et al., 2016).
5. Natural Killer Cell Function
Activation of STAT3 also promotes transcription of IL-2 and perforin genes, thereby promoting natural killer cell activation, which plays a major role in innate immunity (Zhao et al., 2003).
6. B Cells
STAT3 is required for developmental transition of pre-pro-B cell progenitors to subsequent precursor populations and their survival (Chou et al., 2006) through Fms-related tyrosine kinase 3 ligand in Flt3-positive hematopoietic precursor cells, including common lymphocyte progenitor cells (McKenna et al., 2000; Esashi et al., 2008). T cell–secreted IL-21 activates STAT3, which facilitates differentiation of later-stage B-lineage–committed CD19+ precursors to IgG-secreting plasma cells (Fornek et al., 2006). Leptin activates the JAK2/STAT3 signaling pathway in human B cells to induce IL-6 and TNF-α (Esashi et al., 2008), making it a target for reducing adipose tissue inflammation and insulin resistance in obese patients (Frasca et al., 2016).
7. CD4+ T Cells
Naive CD4+ T cells, responding to T cell receptor–mediated antigen presentation or other cues, develop into distinct effector subsets with unique functions, including CD8+ T cell activation, stimulation of innate immune cells, or induction of B cell responses (Kanno et al., 2012). T lymphocyte lineage-specific STAT3-KO mice studies revealed the crucial role of IL-6/STAT3-mediated T cell survival, independent of Bcl-2 (Schindler et al., 1992; Takeda et al., 1998). STAT3 is also essential for Th17 differentiation mediated by IL-6/transforming growth factor (TGF)-β/IL-21 (Laurence et al., 2007; Nurieva et al., 2007; Yang et al., 2007b; Zhou et al., 2007) and by IL-23/STAT3 (Ivanov et al., 2006; Yang et al., 2008b; Gaffen, 2009, 2011). Th17 express the transcription factors retinoic acid-related orphan receptor gamma t (RORγt) and RORα (Ivanov et al., 2006). STAT3 activated by several proinflammatory cytokines, e.g., IL-6, IL-21, and IL-23, upregulates RORγt, RORα, IL-21, IL-23R, and IL-17, thereby helping in the development and stabilization of Th17 cells (Laurence et al., 2007; Nishihara et al., 2007; Yang et al., 2007b, 2008b; Zhou et al., 2007). IL-6/TGF-β combination, in contrast, helps Th17 differentiation through upregulating the ectonucleotidases, CD39 and CD73 (IL-6/STAT3 action), which cleave extracellular ATP to produce immunosuppressive adenosine production and TGF-β–mediated downregulation of the transcriptional repressor growth factor independent-1 (Rébé et al., 2013). CD39-pos Th17 cells promotes tumor growth, suggesting their immunosuppressive role in cancer (Chalmin et al., 2012).
Th17 cells play crucial roles in host defense to bacteria and fungi and inflammatory and autoimmune disorders. Th17 produces IL-17, which stimulates production of antimicrobial peptides, chemokines, and granulopoietic cytokines (Gaffen, 2009, 2011). STAT3 also controls development of CXC chemokine receptor 5–expressing T follicular helper cells, their localization to the B cell follicle within germinal centers of secondary lymphoid organs, and their IL-21 secretion, which mediates B cell help in germinal centers by stimulating proliferation and antibody affinity maturation (Leonard et al., 2008; Ding et al., 2013). STAT3 also transcriptionally upregulates anti-inflammatory transforming growth factor (TGF)-β1 and IL-10 in CD4+ T cells (Kinjyo et al., 2006) and hence helps in IL-10–dependent induction of Th3 cells. Interestingly, STAT3 inhibits generation of CD4+ Tregs from naive CD4+ precursors by suppressing the Treg-specifying transcription factor Foxp3 in mature Tregs (Yang et al., 2008a; Laurence et al., 2012). In the setting of graft-versus-host disease (GVHD), STAT3 deficiency promotes inducible Treg generation, restrains GVHD, and improves mouse survival (Laurence et al., 2012). These data suggest STAT3 blockade in CD4+ T cells may be useful in treating GVHD. Moreover, STAT3 and Foxp3 appear to coregulate IL10 in T-regs (Chaudhry et al., 2011) through histone acetyl transferase-1/FoxP3-mediated epigenetic modification of the IL-10 promoter (Chaudhry et al., 2009, 2011; Chaudhry and Rudensky, 2013), thus maintaining the ability of Foxp3+ Tregs to inhibit inflammatory Th17 cells (Chaudhry et al., 2009, 2011; Chaudhry and Rudensky, 2013). STAT3–HDAC6 transcriptionally upregulates IL-10, preventing effective Ag-specific CD4+ T cell activation in tumor-bearing mice (Cheng et al., 2014); however, use of a HDAC inhibitor enabled effective T cell priming and antitumor response (Wang et al., 2011a; Cheng et al., 2014).
8. CD8+ T Cells
Cytotoxic CD8+ T cells are critical for clearing cells infected with intracellular pathogens, typically viruses, as well as cells expressing aberrant host proteins, e.g., oncoproteins. Functional cytotoxic CD8+ T cells in tumors are frequently associated with better prognosis and improved tumor clearance. Naive CD8+ T cells differentiate into armed effector cells, which in turn generate long-lived memory cells. IL-10– and IL-21–mediated STAT3 activation (Cui et al., 2011) helps generate stable, long-lived memory cells (Cui et al., 2011; Kaech and Cui, 2012) through regulating CD8+ T cell transcriptional regulators (Eomes, BCL6, and Blimp1). IL-6–activated pY-STAT3 transcriptionally induces early production of IL-4 and IL-21 in CD4+ T cells (Yang and Rincon, 2016), but mtSTAT3 contributes to maintaining their late expression through increases in mitochondrial calcium levels and ATP (Yang et al., 2015). AD-HIES patients demonstrate reduced numbers of memory CD8+ T cells, as well as fewer memory CD4+ T cells, relative to healthy controls (Siegel et al., 2013), contributing to their impaired ability to manage chronic infections (Siegel et al., 2013).
9. Anaphylaxis
The observations that clinical food allergy and anaphylaxis are decreased in patients with Job’s or AD-HIES, despite a significant burden of eczematous skin disease and increased levels of IgE, prompted Siegel et al. (2013) to explore these patients for aberrant mast cell degranulation and/or reduced endothelial cell permeability. Studies in the mut-Stat3 bearing two copies of the V463del STAT3 mutation frequently found in Job’s syndrome patients demonstrated both defective mast cell degranulation and reduced endothelial cell permeability (Siegel et al., 2013; Hox et al., 2016). In contrast, whereas human mast cell function in Job’s syndrome patients was affected somewhat by the reduced STAT3 signaling (Siegel et al., 2013), the STAT3-dependent transcriptional activity within endothelial cells that regulates critical components involved in the architecture and functional dynamics of endothelial junctions, i.e., microRNA17-92, PTEN, Src, E2F, and β-catenin, is a more important contributor to reduced endothelial cell permeability and anaphylaxis protection in these patients (Hox et al., 2016).
B. Janus Kinase/Signal Transducer and Activator of Transcription 3 and Metabolism
1. Lipid Metabolism
The anorexigenic hormone, leptin, secreted by adipose tissue, regulates energy homeostasis, glucose and lipid metabolism, immune function, and other systems (Hu et al., 2018). Leptin binds to its specific receptor, LepRb, on cells in the central nervous system (CNS) and peripheral tissues, including skeletal muscle, placenta, ovary, pituitary gland, and lymphoid tissue (Park and Ahima, 2014). Binding activates several intracellular signaling pathways, including JAK2, STAT3, insulin receptor substrate (IRS), PI3K, SHP-2, MAPK, 5′ adenosine monophosphate-activated protein kinase, and acetyl-CoA carboxylase. Leptin/LepRb/JAK2/STAT3 signaling plays major roles in energy homeostasis and neuroendocrine function, through upregulation of proopiomelanocortin (POMC), the precursor protein for melanocyte-stimulating hormones; corticotrophin; and β-endorphin, each of which increases appetite (Millington, 2007), and the down-regulation of agouti-related protein (AgRP), which also increases appetite and decreases metabolism and energy expenditure (Millington, 2007). As highlighted above, leptin/STAT3 activation in the CNS ultimately leads to decreases in food intake and body weight and an increase in energy expenditure. Thus, STAT3 deletion in neurons decreases POMC and increases AgRP and neuropeptide Y levels, leading to hyperphagia, obesity, infertility, and thermal dysregulation (Gao et al., 2004). Mice with homozygous knock-in substitution of LepRb tyrosine residue Tyr1138, which is phosphorylated by Jak2 upon Lep/LepRb engagement, had decreased POMC and increased AgRP mRNA levels in the hypothalamus and displayed hyperphagia and decreased energy expenditure, leading to massive early-onset obesity associated with increased serum leptin levels (Bates et al., 2003; Bates and Myers, 2004), which correlated with increased adipose mass in these mice and also indicated resistance to the normal energy homeostatic effects of leptin. The orexigenic hormone ghrelin acts in opposition to leptin to downregulate feeding behavior via the vagal afferent pathways by inducing exchange protein activated by cAMP–mediated increase in SOCS3 expression, which negatively affects leptin-induced pY-STAT3 and blocks neuronal firing in nodose ganglia neurons (Heldsinger et al., 2014). SOCS3 also regulates leptin/pY-Jak2/pY-STAT3 signaling in the hypothalamus, causing leptin resistance (Bjorbaek et al., 1999; Howard et al., 2004; Mori et al., 2004; de Lartigue et al., 2011; Metlakunta et al., 2011). Overall, STAT3 is indispensable for physiologic leptin actions, and STAT3 dysfunction in any direction could cause pathophysiological alterations in mice and humans (Akira et al., 1994; Wang et al., 2012; You et al., 2015; Dong et al., 2016; Liang et al., 2016; Villarino et al., 2017). The importance of the leptin/STAT3 pathway in immune aging, tumor progression, and T cell dysfunction in cancer is suggested by recent findings that obese cancer patients respond better than nonobese patients to checkpoint therapy, especially therapy directed against the programmed cell death protein 1 (PD1)/programmed death-ligand 1 (PD-L1) axis perhaps due to increased leptin signaling (Wang et al., 2019). Th17 cells were shown to be perturbed by the orexigenic adipocyte-derived hormone, Ghrelin (Xu et al., 2015), through its antagonization of Leptin/STAT3 signaling. Ghrelin activates exchange protein activated by cAMP, which, in turn, induces increased SOCS3 expression that negatively affects leptin/STAT3 signaling and neuronal firing in nodose ganglia neurons (Heldsinger et al., 2014).
STAT3 facilitates adipogenesis from preadipocytes (Wang et al., 2009; Kang et al., 2013), through epigenetic activation of peroxisome proliferator-activated receptor (PPAR)γ (Cristancho and Lazar, 2011), and transcriptional upregulation of C/EBPβ (Zhang et al., 2011), which binds to the PPARγ promoter to maintain adipocyte differentiation (Cristancho and Lazar, 2011). Interestingly, following induction of differentiation, relative levels of pS-mtSTAT3 are markedly reduced in contrast to elevated levels of total cellular pS-STAT3 (Kramer et al., 2015), leading to increased complex I activity and resultantly increased ROS, which in turn accelerates mitotic clonal expansion and increases the differentiation efficiency of preadipocytes during adipogenesis.
Adipocyte-specific Stat3-KO mice have increased body weight and more adipose tissue mass due to adipocyte hypertrophy, but do not show evidence of adipocyte hyperplasia, hyperphagia, or reduced energy expenditure (Cernkovich et al., 2008), suggesting a role of STAT3 in lipolysis. Moreover, adipocyte-specific JAK2-KO mice have impaired lipolysis and increased body weight through decreased STAT3 activation, which results in insulin resistance with ageing (Shi et al., 2014). Jak2/STAT3-mediated lipolysis is mediated through its ability to upregulate adipose triglyceride lipase expression (Li et al., 2010b) and reduced levels of fatty acid synthase, acetyl-CoA carboxylase, and acyl-CoA ocxidase (Xu et al., 2013). It was recently found that CLL cells modify their metabolism to oxidize free fatty acids with the help of constitutively activated STAT3 (mostly pS-STAT3) through transcriptional upregulation of lipoprotein lipase, which catalyzes the hydrolysis of triglycerides into free fatty acids (Rozovski et al., 2015). IL-10 protects against high-fat diet–induced inflammation and improves glucose tolerance through the activation of STAT3 (Grant et al., 2014).
2. Glucose Metabolism
STAT3 is involved in liver maintenance of glucose homeostasis by adjusting glucose production to achieve energy balance. Insulin increases IL-6 expression in Kupffer cells and activates STAT3 in hepatocytes (Inoue, 2016), which in turn regulates glucose homeostasis by transcriptionally suppressing (Ramadoss et al., 2009) expression of gluconeogenic genes, e.g., glucose-6-phosphatase and phosphoenolpyruvate carboxykinase (Inoue et al., 2004). The leptin/JAK2/STAT3 pathway inhibits insulin synthesis, whereas SOCS3 blocks the STAT3-dependent regulation of the preproinsulin 1 gene promoter (Laubner et al., 2005). STAT3 also plays a key role in amino acid–mediated dampening of insulin signaling in hepatic cells, primarily through a mTOR-mediated pS-STAT3 pathway that is independent of pY-STAT3 (Kim et al., 2009). The STAT3 pathway, thus, is essential for glucose homeostasis and may provide a novel therapeutic target for obesity.
III. Functions of Janus Kinase/Signal Transducer and Activator of Transcription 3 in Stress Conditions
In multicellular organisms, every cell constantly strives to maintain homeostasis in the face of destabilizing factors either external, e.g., toxic chemicals and changes in oxygen tension, or internal, e.g., alterations in pH or osmolarity due to normal cellular metabolism. The STAT group of proteins, especially STAT3, has evolved to be one of the major intracellular messengers involved in the maintenance of homeostasis in response to multiple types of stresses through both its canonical and noncanonical activities.
A. Radiation
Early experiments showed that UV light treatment of mouse embryonic fibroblasts resulted in phosphorylation of S727 in STAT1 via p38 MAPK (Kovarik et al., 1999; Ramsauer et al., 2002). STAT3 also is activated by UV through DNA damage and reactive oxygen species (Shen et al., 2001; Aziz et al., 2007a; Barry et al., 2010; Bito et al., 2010b). Although activated STAT1 seems to move cells toward apoptosis (Shen et al., 2001), STAT3 activation seems to be protective (Shen et al., 2001; Chan et al., 2004b; Sano et al., 2005; Aziz et al., 2007a; Barry et al., 2010; Bito et al., 2010a,b). Acutely, PKCε-mediated pS-STAT3 (Aziz et al., 2007a,b) and pY-STAT3 seem to work together to transcribe genes that are UV-protective, whereas persistently elevated pY-STAT3 levels mediated by chronic UV exposure are a key step in UV-induced skin cancer (Chan et al., 2004a,b; Aziz et al., 2007a,b; Barry et al., 2010). Levels of pY-STAT3 also are increased in in vitro models, as well as in patients with ionizing radiation (IR)–resistant tumors, by IR at doses used for radiotherapy (≥2 Gy), contributing to selection of IR-resistant cells in many different cancers (Ho et al., 2010; Liu et al., 2013; Pan et al., 2013; Yan et al., 2013; Gao et al., 2014; Klein et al., 2014; Lau et al., 2015; Zang et al., 2017; Lu et al., 2018). Interestingly, very low doses of radiation, e.g., 0.1 Gy, seem to reduce triple-negative breast cancer progression by deactivating pY-STAT3 through pJAK1 inhibition (Kaushik et al., 2017), leading to reduced numbers of CD44+/CD24− breast cancer cells, reduced ability of these cells to form mammospheres, and their reduced ability to self-renew and form metastasis (Kaushik et al., 2017). Recently, the importance of pS-STAT3 in radioresistance in glioblastoma multiforme also has been suggested (Ouédraogo et al., 2016).
B. Oxidative Stress
Reactive oxygen intermediates are produced when cells are subjected to oxidative stress. Although reactive oxygen intermediates are known to contribute to DNA damage and tumorigenesis, they also function as signal mediators in many normal cellular processes. Oxidative stress is implicated in the positive modulation of the activity of a number of protein tyrosine kinases, including those that activate STAT3. Peroxide treatment leads to phosphorylation of STAT3 (pY-STAT3) and its nuclear translocation in human lymphocytes (Carballo et al., 1999), and JAK2, STAT1, and STAT3 are activated by oxidized low-density lipoprotein (Maziere et al., 2001), suggesting a role of these proteins in cellular response to oxidative stress.
STAT3 itself can be subject to direct modification by oxidative stress, as 9 of the 14 cysteine residues within STAT3 have been reported to be redox sensitive. These modifications affect the transcriptional activity of STAT3 through inhibiting Y705 phosphorylation and/or DNA binding, as well as inducing formation of higher order complexes and interfering with the function of the TAD (Li and Shaw, 2004; Xie et al., 2009; Li et al., 2010a; Kurdi et al., 2012; Butturini et al., 2014; Sobotta et al., 2015). Similarly, thiol-targeting agents also inhibited IL-6–induced STAT3 activation by S-glutathionylation in HepG2 cells (Xie et al., 2009), human endothelial cells, neonatal rat cardiomyocytes, and adult mouse cardiac myocytes (Zgheib et al., 2012) presumably through a STAT3–peroxiredoxin-2 association (Sobotta et al., 2015) and/or steric hindrance. JAK2-mediated Y705 phosphorylation also was blocked (Butturini et al., 2014) by S-glutathionylation of C328 and C542 within the DNA binding domain and Linker domain of STAT3, respectively.
Peroxide treatment induced homodimerization of STAT3 formation through a disulfide bonding of cysteines within the amino terminus of STAT3 (Li and Shaw, 2004). Cysteine residues within the DNA binding domain and the transactivation domain appear to be involved in peroxide-induced STAT3 higher-order complex formation (Sobotta et al., 2015). ROS also may differentially affect STAT3 binding to various STAT3-binding elements, suggesting oxidative stress may alter the profile of genes activated by STAT3 (Li et al., 2010a). STAT3 activation in cardiac myocytes was impaired by glutathione depletion (Kurdi et al., 2012), which could be reversed by glutathione monoethyl ester, which is cleaved intracellularly to glutathione, as well as by the reducing agent, N-acetyl-cysteine.
S-nitrosylation is another redox-related post-translational modification that inhibits STAT3 activation. In microglial cells, endogenous nitric oxide (NO) produced by inducible NO synthase (iNOS) or by treatment with s-nitrosoglutathione led to s-nitrosylation of STAT3 on C259, which inhibited JAK2-STAT3–induced gene expression and cell proliferation (Kim et al., 2014).
Increasing oxygen concentration is a mainstay for therapy of lung failure, e.g., adult respiratory distress syndrome, but hyperoxia also may contribute to the pathogenesis of acute lung injury (Altemeier and Sinclair, 2007), which is characterized by noncardiogenic pulmonary edema, inflammation, and respiratory failure. Respiratory epithelial cell–specific STAT3-KO mice progressed more rapidly than STAT3 wild-type mice to lung injury after exposure to 95% oxygen (Hokuto et al., 2004), indicating a hyperoxia-protective role for STAT3. Levels of surfactant protein B, a lipid-associated protein essential for surfactant function that is STAT3 regulated (Yan et al., 2002; Yang et al., 2004), were reduced in the bronchial lavage fluid of the respiratory epithelial cell–specific STAT3-KO mice. Correspondingly, overexpressing of STAT3C (a constitutively active form of STAT3) in respiratory epithelial cells (Lian et al., 2005) improved animal survival following hyperoxia and protected the lung from inflammation and injury through reducing mRNA levels of matrix metalloproteases, e.g., matrix metalloproteinases (MMP)9 and MMP12, and levels of adhesion molecules, which facilitate migration of neutrophils into the lung. STAT3 also is essential for the protective effects of heme oxygenase-1 in oxidant-induced lung injury (Zhang et al., 2006). IL-11 and IL-6 pretreatment protected endothelial cells from H2O2-induced cell death reportedly through the activation of both MAPK and STAT3 signaling pathways (Waxman et al., 2003).
Many other toxins, e.g., asbestos, silica, cigarette smoke, airborne particulate matter, diesel exhaust, and ozone, cause lung injury through ROS generation (Mossman et al., 2006), which in turn induces production of chemokines or cytokines mediating inflammation. Cigarette smoke extract (CSE) induced IL-6 expression and increased levels of pY-STAT3 and STAT3-DNA binding in human bronchial epithelial cells (HBECs) (Liu, 2007), which protected these cells from apoptosis. Anti–IL-6 neutralizing antibody, as well as STAT3 small interfering RNA (siRNA), rendered HBECs sensitive to CSE-induced DNA damage, suggesting that STAT3 mediates HBEC survival in response to CSE-induced DNA damage (Liu, 2007).
C. Genotoxic Stress
The fact that STAT3 protects from apoptosis by DNA-damage agents, e.g., radiation (Shen et al., 2001; Chan et al., 2004b; Sano et al., 2005; Aziz et al., 2007a; Barry et al., 2010; Bito et al., 2010a,b) and ROS (Hokuto et al., 2004), suggests a direct protective role STAT3 from genotoxic stress. In fact, cells lacking STAT3 are less efficient in repairing damaged DNA due to reduced activity of ataxia-telangiectasia mutated (ATM)-Chk2 and ATM and RAD3‐related (ATR)-Chk1, two major pathways involved in sensing DNA damage (Barry et al., 2010). STAT3 transcriptionally regulates mediator of DNA damage checkpoint 1, the master regulator of the ATM-chk2 pathway (Barry et al., 2010). The epidermal growth factor receptor/STAT3 pathway upregulates the endonuclease, Eme1, to reduce DNA damage after topoisomerase I inhibition (Vigneron et al., 2008). JMJD2B, a hypoxia-inducible factor 1α–regulated master regulator of DNA repair genes, which maintains histone methylation balance important for the transcriptional activation of many oncogenes, acts through activating STAT3 (Chen et al., 2014).
Some of the DNA-protective functions of STAT3 now clearly are established to be dependent on pS-STAT3. Following DNA damage induced by topoisomerase I inhibition, STAT3 is phosphorylated by cdk5 at S727, but not Y705; pS-STAT3 transcriptionally upregulates Eme1, the endonuclease involved in DNA repair, which reduces DNA damage (Courapied et al., 2010).
D. Cell Senescence
Both normal and tumor cells undergo senescence in response to various insults that cause persistent DNA damage (Kuilman et al., 2010; Rodier and Campisi, 2011; Sikora et al., 2011; Ohtani and Hara, 2013); senescence is characterized by the inability to proliferate despite sufficient nutrients and mitogens while maintaining cell viability and metabolism (Kuilman et al., 2010; Rodier and Campisi, 2011; Sikora et al., 2011; Ohtani and Hara, 2013). There are three different types of senescence: replicative senescence, oncogene-induced senescence, and stress-induced senescence. Senescence is implicated in many age-related diseases, including renal failure, type 2 diabetes mellitus, idiopathic pulmonary fibrosis, cardiovascular disease, and age-related cachexia. There is a close relationship between elevated proinflammatory factors e.g., IL-1α, IL-6, TNF, and NF-κB, which tend to increase in tissues with senescence and age (McHugh and Gil, 2018).
JAK/STAT signaling has long been linked to senescence (Kojima et al., 2013). Induction of C/EBPβ and IL-6 was shown to be required for B-raf proto-oncogene serine/threonine-protein kinase-induced senescence (Kuilman et al., 2008). Also, embryonic lung fibroblast TIG3 cells, after 55 or more passages, expressed greater levels of IL-6, IL-6Rα, and pY-STAT3 compared with TIG3 cells at earlier passages (≤33). IL-6 plus soluble IL-6Rα treatment of young TIG3 cells (passage ≤33) resulted in growth arrest and senescence-associated-β-galactosidase activity, mimicking a p53/ROS driven DNA damage response. STAT3 was similarly described to be essential for both the early ROS increase, as well as the later senescence-associated-β-galactosidase activity, through STAT3-regulated soluble factor insulin-like growth factor binding protein 5 in both human fibroblasts (Udono et al., 2012) and vascular endothelial cells (Kim et al., 2007).
IL-6 and pY-STAT3 induced by chemotherapy promoted premature senescence in lymphoma cells in a paracrine manner, which maintained a minimal residual number of chemoresistant cells that could lead to relapse. Antibody against IL-6, gp130 siRNA, or dominant-negative STAT3 enhanced chemo-induced death in lymphoma (Gilbert and Hemann, 2010), as well as colon cancer (Yun et al., 2012), indicating that targeting IL-6/STAT3 signaling may be a viable strategy for bypassing senescence-induced chemoresistance. Interestingly, senescent breast cancer cells were better able to elicit antitumor immunity through cytotoxic natural killer cells and CD4+ T cells following treatment with STAT3 inhibitor (Tkach et al., 2012).
E. Autophagy
Autophagy is a process used by cells to disassemble unnecessary or dysfunctional organelles that are critical for cell survival and regulated by a network of signaling pathways, including STAT3 (Kroemer et al., 2010; Pietrocola et al., 2013). STAT3 transcriptionally activates genes of several anti-autophagy–related proteins e.g., BCL2, BCL2L1, and MCL1 (Fukada et al., 1996; Bromberg et al., 1999; Bowman et al., 2000). These proteins bind competitively to BECN1 and disrupt formation of BECN1/PIK3C3 complexes, which are essential for autophagy. Induction of autophagy occurs when these protein are competitively displaced by proapoptotic BCL-2 homology 3-containing proteins (Kroemer et al., 2010). Sorafenib, a small-molecule kinase insert domain receptor inhibitor, activates autophagy in multiple liver cancer cell lines through targeting STAT3 and reducing MCL1 expression (Tai et al., 2013). STAT3 also negatively regulates both PIK3C3 (Yamada et al., 2012) and BECN1 (Miao et al., 2014) through recruitment of HDAC3 on its promoter (Miao et al., 2014).
STAT3 also promotes autophagy, e.g., leukemia inhibitory factor/STAT3 and oncostatin M/STAT3 signaling promote autophagy in mammary gland cells by upregulating both the p55 and p50 isoforms of PI3K regulatory subunit 1, PIK3R1 (Pensa et al., 2014). Furthermore, STAT3 upregulates microRNAs that target autophagy-related genes, thus promoting autophagy, e.g., STAT3 upregulates MIR17HG (miR-17-92 cluster), members of which target autophagy-related genes ULK1, BECN1, and BCL2L11 (Wu et al., 2012; Chatterjee et al., 2014; Spaccarotella et al., 2014), through a highly conserved STAT3 binding site in the promoter (Brock et al., 2009).
In contrast to nuclear STAT3, cytoplasmic STAT3 inhibits autophagy by inhibiting eukaryotic translation initiation factor 2-α kinase 2 activity, by binding to its catalytic domain, thereby inhibiting its enzymatic activity, thereby preventing formation of phosphorylated eukaryotic translation initiation factor 2-α, a known autophagy activator (Shen et al., 2012). The loss of mtSTAT3 results in a significant increase in ROS (Szczepanek et al., 2012), which is implicated in the induction of autophagy, especially the selective autophagic degradation of mitochondria, or mitophagy (Scherz-Shouval and Elazar, 2007). Mitochondrial translocation of STAT3 also suppresses autophagy during ischemia by inhibiting cytochrome c, somatic release, and preventing opening of the mitochondrial permeability transition pore (Szczepanek et al., 2011).
F. Ischemia and Reperfusion Stress
Increased production of IL-6 and granulocyte cell–stimulating factor and activation of STAT3 was found to occur early in rat lungs following resuscitation from hemorrhagic shock, a whole-body ischemia–reperfusion injury (Hierholzer et al., 1998b,c; Meng et al., 2000, 2001). Examination of hemorrhagic shock in induced iNOS-KO mice or following administration of an iNOS inhibitor demonstrated that both NF-κB and STAT3 were activated in the lungs and livers in an iNOS-dependent manner (Hierholzer et al., 1998a), and that iNOS inhibition resulted in a marked reduction of lung and liver injury. In addition, administration of a NO scavenger also resulted in reduced lung injury and reduced activation of NF-κB and STAT3 after resuscitated hemorrhagic shock (Hierholzer et al., 2002), presumably through downregulation of acute inflammation (Hierholzer et al., 2002). Importantly, resuscitation from hemorrhagic shock also resulted in apoptosis of lung type I alveolar endothelial cells (Moran et al., 2009), liver hepatocytes (Moran et al., 2008), and cardiomyocytes (Thacker et al., 2013), which resulted in increased susceptibility to pneumonia (Thacker et al., 2014) and sepsis following intraperitoneal bacterial challenge (Arikan et al., 2006). Importantly, apoptosis of these critical parenchymal cells in the lung, liver, and heart and the increased susceptibility to pneumonia and bacterial peritonitis could be reversed by administration of IL-6 at the start of resuscitation; this action of IL-6 was mediated by activation of STAT3. Thus, STAT3 augmentation may be a viable strategy to prevent hemorrhagic shock–mediated pathologies, resulting in decrease in mortality, and its timing early in resuscitation may mitigate against it, which contributes to the deleterious acute inflammatory response that ensues following parenchymal cell apoptosis.
Ischemia–reperfusion (I/R) lung injury is a significant source of morbidity following cardio-pulmonary bypass and lung transplantation (Ng et al., 2006). Under ischemic conditions, activated neutrophils infiltrate the inflamed lung and produce ROS, leading to release of lysosomal enzymes, which contribute significantly to I/R lung injury (Ng et al., 2006). Ischemia followed by 15 minutes of reperfusion in the lung induces STAT3 and NF-κB nuclear translocation (Farivar et al., 2006). In studies of lung models of in vitro anoxia–reoxygenation and in vivo I/R, carbon monoxide attenuated endothelial cell apoptosis presumably through enhanced STAT3 activation via PI3K/Akt and p38 MAPK pathways (Zhang et al., 2003, 2005). IL-6 treatment also reduced endothelial disruption and neutrophil sequestration in lung and alveolar spaces, resulting in improved oxygenation after I/R (Zhang et al., 2016).
Leukemia inhibitory factor binding to its receptor and gp130 activates Akt, STAT3, and ERK1/2 and protects cardiomyocytes from oxidative and ischemic stress by acting on mitochondria and inducing gene transcription (Zouein et al., 2013b). JAK-induced pY-STAT3 and ERK1/2-activated pS-STAT3 seem to work together to increase transcription of antiapoptotic, antioxidative stress, and proangiogenic genes (Kurdi and Booz, 2007; Zouein et al., 2013a). Studies with cardiomyocyte-targeted STAT3-KO mice reveal that although cardiac STAT3 depletion did not affect infarct size, STAT3 is crucial for cardiac preconditioning (Smith et al., 2004). Following I/R injury (Hilfiker-Kleiner et al., 2004), cardiomyocyte-specific STAT3-KO mice had greater infarct size and cardiomyocyte apoptosis presumably due to increased mRNA levels of the proapoptotic and proautophagy gene, BNIP3, and decreased mRNA levels of the prosurvival gene, heat shock protein 70, as well as decreased mRNA encoding antiapoptotic proteins (Mcl-1, Bcl-xL, c-FLIPL, and c-FLIPS) and cardioprotective proteins (cyclo-oxygenase-2 and heme oxygenase-1) linked to cardiac preconditioning (Bolli et al., 2011). STAT3 in cardiomyocytes also helps heart remodeling during the subacute phase of myocardial infarction (MI); STAT3 deletion during days 11 to 24 after MI resulted in the following: 1) worsened cardiac function and increased mortality, 2) exacerbated cardiac fibrosis through upregulation of fibrosis-related genes, 3) increased death of cardiac myocytes, and 4) cardiac hypertrophy, which reduced capillary density in the border zone (Enomoto et al., 2015). However, persistently activated STAT3 within cardiomyocytes resulted in a worse outcome following MI due, in part, to increased inflammation (Hilfiker-Kleiner et al., 2010). Of note, pS-STAT3 optimizes mitochondrial respiration and limits ROS formation from complex I following cardiac ischemia (Szczepanek et al., 2012).
G. Hyperosmotic Stress
Hyperosmotic stress can also activate STATs. In the slime mold Dictyostelium, hyperosmotic stress led to increased levels of pY-STAT1 without any known involvement of JAK or MAPK (Araki et al., 2003). In mammalian cells, sorbitol-induced hyperosmolarity resulted in phosphorylation of JAK1, JAK2, and TYK2 and led to activation of STAT1 and STAT3, independent of gp130, leading to formation of STAT1/STAT3 heterodimer complexes and increased DNA binding (Gatsios et al., 1998). Hyperosmotic shock in COS-7 cells led to MKK6/p38 activation and pY-STAT1 (Bode et al., 1999). Prolonged elevated glucose could also signal as a cell stressor through multiple pathways, including hyperosmolarity (McGinn et al., 2003), protein kinase C (PKC) activation (Mandarino, 1992), and oxidative damage (Nishikawa et al., 2000), likely involving pS-STAT3, as well as pY-STAT3.
IV. Targeting Aberrant Janus Kinase/Signal Transducer and Activator of Transcription 3 Activity in Inflammatory, Fibrotic, Metabolic, and Oncological Diseases
As is evident from the above discussions and Table 2, diseases may result from both diminished and aberrantly augmented STAT3 activity in various cells, tissues, and organs. In the following sections, we discuss the following: 1) how JAKs and STAT3 are targeted to decrease their activity; 2) how augmented JAK/STAT3 activity leads to disease, focusing on asthma, inflammatory bowel disease, cachexia, fibrosis, metabolic disorders, and cancer; and 3) progress to this point in using strategies that target JAKs and STAT3 in treating these diseases.
A. Strategies to Target Janus Kinases
All Jakinibs in use or undergoing clinical trials to this point in time are ATP analogs that target the kinase (JH1) domain (Fig. 1). Each of these small-molecule inhibitors has been shown to bind in the ATP-binding cleft located between the β-sheet–rich N-lobe and α-helix–rich C-lobe of the JH1 domain (Alicea-Velázquez and Boggon, 2011). Although the pseudokinase (JH2) domain and FERM domain play an important role in regulating JAK activity and provide a potentially rich source of sites for targeting JAKs, there are no reports of attempts to reduce the activity of JAKs through targeting of these domains.
Excessive JAK activity has been linked to numerous disorders, including inflammatory diseases such as psoriasis and inflammatory bowel disease; myeloproliferative diseases, e.g., polycythemia vera (PV); and hematologic malignancies, including myeloid leukemia and multiple myeloma. The development of Jakinibs has focused mainly on treatment of hematologic disorders and inflammatory diseases, as outlined below.
B. Strategies to Target Signal Transducer and Activator of Transcription 3
Several approaches to reduce STAT3 activity have been proposed and include targeting of specific receptors or receptor-associated kinases, especially JAKs, upstream of STAT3, in addition to strategies to directly interfere with STAT3 activity by preventing its recruitment to receptors, dimerization, nuclear localization, and DNA binding [see Bharadwaj et al. (2016b)]. Other approaches currently underway involve modulation of autoregulators of STAT3 activity, such as strategies to activate protein phosphatases, members of the protein inhibitors of activated STAT family, or suppressor of cytokine signaling (SOCS) protein members (Heppler and Frank, 2017; Durham et al., 2019).
Substantial effort has focused on identifying a small molecule capable of blocking key residues within the STAT3 SH2 domain (Fig. 3) responsible for the binding of STAT3 to pY-peptide ligands located within ligand-activated receptors and within the loop domain of STAT3 itself. Such a molecule would inhibit the ability of STAT3 to modulate gene transcription by preventing several key steps in STAT3 activation, including its recruitment to activated receptor complexes, its phosphorylation by receptor-intrinsic or receptor-associated tyrosine kinases, its dimerization, its nuclear accumulation, and its DNA binding. Several groups have successfully identified small-molecule inhibitors using structure-based virtual ligand screens, biophysical techniques, and cell-based phenotypic screening that have focused on the STAT3 SH2 domain, either its pY-peptide–binding domain or dimerization interface. STAT3 inhibitors identified by structure-based high-throughput virtual screening include STA-21, Stattic (Stat three-inhibitory compound), S3I-201/NSC74859, BP-1-102, OPB-31121, TTI-101 (C188-9), and their analogs. Each demonstrates the ability to inhibit the growth of one or more cancer cell lines owing in some instances to their inhibition of STAT3 binding to its pY-peptide ligands and blocking IL-6–induced STAT3 activation, nuclear translocation, and transcriptional gene activation (Zhang et al., 2013c). It should, however, be noted that not all the molecules identified as STAT3 inhibitors are competitive inhibitors. Some, such as Stattic and some members of the S3I-201 family of compounds, inhibit STAT3 by covalently reacting with residues (largely cysteines) at key locations within STAT3, as well as other off-target proteins (Heidelberger et al., 2013; Ball et al., 2016).
Studies investigating the impact of disease-causing mutations on STAT3 suggest that there are long-range communications between domains of STAT3, implying that STAT3 activity may be subject to allosteric modulation (Zhang et al., 2000; Mertens et al., 2015; Namanja et al., 2016). Thus, in addition to orthosteric inhibitors, emerging evidence points to allostery as a mechanism of STAT3 inhibition observed in a number of small-molecule inhibitors. For example, hydrogen–deuterium exchange experiments and other studies show that STAT3 inhibitors—SF-1-066, BP-1-102, and MM206—may, in addition to binding to the SH2 domain, affect the function of the SH2 domain by binding to and perturbing the structure of domains of STAT3 at a distance from the SH2 domain, suggesting an allosteric destabilizing effect on the SH2 domain (Resetca et al., 2014; Minus et al., 2015). These findings suggest the possible utility of developing allosteric inhibitors of STAT3. Of note, compounds acting allosterically may be more specific as they tend to bind to less conserved regions of target proteins; this could be of potential benefit when attempting to specifically target a protein such as STAT3 that has a SH2 domain that is structurally very similar to other members of the STAT protein family. However, it is important to note that the toxicity of some STAT3 inhibitory compounds, as well as their antitumor activity, may arise from their ability to cause marked changes in STAT3 structure, leading to its aggregation within critical organelles. For instance, it has been reported that STAT3 forms proteotoxic aggregates in mitochondria when tumor cells are treated with OPB-51602, STA21, or WP1066 (Genini et al., 2017).
A number of inhibitors have been developed that appear to directly target the DNA-binding domain of STAT3. Nucleic acid–based drugs, including decoy oligonucleotides and, perhaps, G-quartet oligonucleotides, have been identified that inhibit STAT3-mediated gene expression and inhibit growth of different cancer cell lines (Leong et al., 2003; Lewis et al., 2008; Sun et al., 2008; Shen et al., 2009; Zhang et al., 2013b; Liu et al., 2014; Sen et al., 2014). Platinum IV compounds—CPA-1, CPA-7, IS3 295, and platinum (IV) tetrachloride—also can inhibit STAT3 DNA-binding activity and inhibit cancer cell line growth (Turkson et al., 2004). Other small-molecule inhibitors besides platinum-based compounds also have been identified, including InS3-54 and HO-3867, which specifically target the DNA-binding domain of STAT3, inhibit its binding to DNA, and block STAT3-mediated gene activation (Rath et al., 2014).
Strategies targeting STAT3 mRNA have been employed to inhibit STAT3 protein expression and thereby all of its activities. Approaches based on siRNA and short hairpin RNA targeting of STAT3 activity have resulted in the development of a naked antisense oligonucleotide–based drug, AZD9150 (IONIS-STAT3-2.5Rx or danvatirsen), which has entered Phase II clinical trials in cancer patients (see below).
C. Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling and Its Targeting in Asthma
Asthma has traditionally been associated with overactivation of T helper (Th)2 cells, infiltration of the lung with eosinophils (Wenzel, 2006; Fahy, 2009; Lin et al., 2013), and good responses to treatment with corticosteroids. However, higher morbidity and mortality are seen in approximately 10% of asthma patients who are refractory to corticosteroid treatment. These patients show a Th17 cell–driven phenotype and neutrophilic infiltration of the lung (American Thoracic Society, 2000; McKinley et al., 2008; Al-Ramli et al., 2009). STAT3 is known to regulate both Th2 cell recruitment during allergic inflammation and the production of Th17 cells and the expansion and recruitment of neutrophils (Chen et al., 2007; Ma et al., 2008; Milner et al., 2008). Targeting JAK/STAT3 signaling may be of benefit in the treatment of neutrophilic, steroid-resistant asthma. An early example of this approach involved the use of STAT1/3 decoy oligonucleotides in a mouse model of neutrophil-predominant asthma due to intranasal instillation of house dust mite (HDM) extract. Administration of STAT1/3 decoy oligonucleotides or the Jakinib, Tyr1A, was shown to mitigate airway inflammation and airway hyper-responsiveness in lungs of mice challenged with HDM (Simeone-Penney, 2008). Pharmacological blockade of STAT3 in HDM-challenged mice with TTI-101 (C188-9) inhibited allergen-induced STAT3 phosphorylation, resulting in concomitant reduction in airway inflammation and remodeling. These changes were accompanied by normalization of IL-4, IL-5, IL-13, and IL-17A cytokine levels and reduced expansion of Th2 cells and Th17 cells (Gavino et al., 2016). The data summarized above suggest that targeting JAK and STAT3 with small-molecule inhibitors may provide a superior anti-asthma treatment approach compared with standard therapies as, they would target both Th2- and Th17-associated asthmas (Choy et al., 2015).
D. Janus Kinase/Signal Transducer and Activator of Transcription Signaling and Its Targeting in Anaphylaxis
The Jakinib, ruxolitinib, was shown to reduce food allergy manifestations in mice through prevention of mast cell hyperplasia and inhibition of mast cell activation (Yamaki and Yoshino, 2014). Similar to findings in mut-Stat3 mice, Hox et al. (2016) found that hematocrit levels and local plasma transudate in response to the IgE/antigen anaphylaxis model and to histamine alone were significantly lower in TTI-101–treated mice compared with untreated mice. These data were supported by a decreased permeability induced by histamine and other vascular permeability–inducing agents in normal human umbilical vein endothelial cells treated in vitro with TTI-101. Together with the findings in the mut-Stat3 mice described above, these results suggest that STAT3 inhibition interferes with a common pathway regulating vascular permeability and raises the possibility of using JAK/STAT3 inhibition to prevent anaphylaxis in those suffering from severe allergies to peanuts or other allergens that are low-level contaminates of many foods.
E. Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling and Its Targeting in Inflammatory Bowel Disease
Ulcerative colitis (UC) and Crohn’s disease (CD) are the major forms of inflammatory bowel disease (IBD). Both are idiopathic chronic inflammatory diseases of the gastrointestinal (GI) tract that cause substantial morbidity, occasional mortality, and a 20- to 30-fold increased risk of developing colorectal cancer (CRC) (Kasembeli et al., 2018). UC and CD are distinct in the location of inflammation within the GI tract—the rectum and distal colon are typically involved in UC, whereas any part of the gut can be involved in CD—and the depth of tissue involvement (Kasembeli et al., 2018). Population-based, genome-wide–association studies followed by subsequent meta-analysis of population-based, genome-wide–association studies and immunochip data have identified more than 160 loci linked to IBD susceptibility (Jostins et al., 2012). Among these genes are included some related to intestinal mucosal immune responses, including STAT3 (Lees et al., 2011; Kasembeli et al., 2018). In addition, a number of cytokines found to be elevated in the plasma and tissue of patients with IBD are known to activate STAT3 and/or its downstream targets, including IL-1β, IL-2, IL-6, IL-12, IL-15, IL-21, IL-23, IL-17, IL-18, IL-10, IL-11, IL-22, IFN-α/β/γ and MMP9, TNF-α, and IFN-γ (Kasembeli et al., 2018). Increased levels of STAT3 protein have been detected in affected tissues in both UC and CD, which correlated with increased levels of pY-STAT3 and the histologic degree of inflammation (Mudter et al., 2005; Musso et al., 2005; Kasembeli et al., 2018); levels of pY-STAT3 were observed in both T cells and colonic epithelial cells. Studies undertaken to tease out the role played by CD4+ cells in IBD determined that activation of STAT3 in pathogenic T cells contributes to colitis by increasing their survival (Kasembeli et al., 2018). In contrast, genetic deletion of STAT3 in innate immune cells, such as myeloid cells (neutrophils and macrophages), and enterocytes resulted in chronic colitis (Takeda et al., 1999; Atreya et al., 2000; Yamamoto et al., 2000; Suzuki et al., 2001; Pickert et al., 2009).
Two isoforms of STAT3 (α, β), derived from one gene by alternative mRNA splicing, are expressed in most cells in a 4:1 ratio (α:β). STAT3α is proinflammatory and antiapoptotic, whereas STAT3β has opposing effects on STAT3α. Manifestations of dextran sulfate sodium–induced colitis were more severe in STAT3Δβ/Δβ transgenic mice, which express only STAT3α (Maritano et al., 2004; Marino et al., 2014), indicating that a STAT3 activity globally contributes to dextran sulfate sodium–induced colitis.
Humanized anti–IL-6R monoclonal antibody (tocilizumab), anti–IL-6R monoclonal antibody, anti–IL-6 avimer antibody (C326), and small-molecule IL-6R antagonist (TJ301) have shown beneficial effects in humans with CD (Ito et al., 2004; Kasembeli et al., 2018). Anti-TNF antibodies that disrupt the interaction of TNF-α with its receptor, including infliximab (a mouse–human chimeric anti–TNF-α antibody), adalimumab (a human monoclonal anti–TNF-α antibody), and certolizumab pegol (recombinant humanized anti–TNF-α antibody), appear to be beneficial in CD patients (Targan et al., 1997; Hanauer et al., 2002, 2006; Colombel et al., 2007; Schreiber et al., 2007; Sandborn et al., 2011). Vidofludimus, an immunosuppressive drug that inhibits dihydroorotate dehydrogenase involved in pyrimidine nucleoside biosynthesis, inhibits the proliferation of T cells and B cells, and the generation of IL-17A, IL-17F, and IFN-γ has shown to have clinical benefit in an open-label uncontrolled entrance study of patients with IBD (Herrlinger et al., 2013). Ustekinumab, a human monoclonal antibody that binds to the p40 subunit of both IL-12 and IL-23, causing disruption of their binding to receptor, induced a clinical response within 4 to 6 weeks of start of treatment in two clinical trials involving CD patients (Sandborn et al., 2008; Bruce, 2016). Also, antibodies against IL-12 and IL-23 have been evaluated in early Phase II studies, and these agents have demonstrated efficacy in moderate-to-severe CD patients who had previously failed infliximab (Sandborn et al., 2008; Bruce, 2016).
A selective, small-molecule inhibitor of JAK3, Janex-1, has shown promise as a therapeutic option for UC treatment (Durant et al., 2010; Anderson et al., 2011; De Vries et al., 2017). Furthermore, Phase II and III clinical studies of tofacitinib, an oral JAK3 inhibitor, have been Food and Drug Administration approved as the only nonsteroidal oral treatment that induces remission in moderate-to-severe UC (Sandborn et al., 2012; De Vries et al., 2017). In addition, filgotinib and upadacitinib (ABT-494, AbbVie), which selectively target JAK1, have demonstrated benefit in Phase II clinical studies in CD patients (https://clinicaltrials.gov/ct2/show/NCT02365649; Ananthakrishnan, 2017; De Vries et al., 2017).
F. Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling and Its Targeting in Cachexia
Proinflammatory cytokines, such as TNF-α, IL-6, and IFN-γ, drive catabolic signaling associated with cachexia (Mantovani et al., 2000; Miyamoto et al., 2016; Porporato, 2016). The link between IL-6/STAT3 signaling and muscle and adipose tissue wasting is well established in mouse models of cachexia (White et al., 2011; Silva et al., 2015) and has been reinforced preliminarily in clinical studies. A Phase II trial of the JAK1/2 inhibitor, ruxolitinib, has been shown to increase body weight, in patients with primary myelofibrosis (PMF) and metastatic pancreatic adenocarcinoma (Jatoi, 2013; Cerchione et al., 2017). In patients with myelofibrosis, changes in body weight were shown to closely correlate to changes in plasma levels of leptin and erythropoietin, both of which have been linked to metabolic regulation through STAT3 signaling (Mesa et al., 2015; Dey et al., 2016).
Recent studies have highlighted the direct role of JAK/STAT3 signaling in protein catabolism through muscle-specific proteolytic pathways. STAT3 directly binds to the promoter of caspase-3 in muscle cells, increasing levels of caspase-3 protein; caspase-3 cleaves actinomyosin and myofibrillar proteins, which triggers protein degradation via the ubiquitin/proteasome system. Another proteolytic pathway activated by JAK/STAT3 signaling involves pY-STAT3–mediated transcriptional activation of C/EBPδ, which, in turn, upregulates myostatin, leading to increased expression of the muscle-specific E3 ligases, MAFbx and Atrogin-1, and protein degradation via the ubiquitin/proteasome system. Emerging evidence suggests that STAT3 may also exert its procachexic effects in collaboration with other signaling networks; for instance, its activation has been shown to modulate IFN-γ/TNF-α–induced muscle wasting via the iNOS/NO pathway (Ma et al., 2017).
Accelerated loss of white adipose tissue is a salient characteristic of cachexia. IL-6 signaling through STAT3 has been shown to control this process, as evidenced by inhibition of white adipose tissue lipolysis in mice treated with anti–IL-6R antibody (Babaei et al., 2018; Wang et al., 2018a). Additionally, STAT3 is thought to be involved in cachexia-related adipose tissue breakdown by modulating enzymes involved in lipolysis. Results from experiments with bovine adipocytes treated with leptin suggest that there may be a link among lipolysis, adipose triglyceride lipase induction, and STAT3 signaling (Koltes et al., 2017). These observations are further strengthened by the results showing that STAT3 KO in adipose tissues of mice leads to obesity due to hypertrophy of adipose tissue (Cernkovich et al., 2008; Xu et al., 2013).
Levels of pY-STAT3 are increased in skeletal muscle in mouse models of cachexia, including models of chronic kidney disease (Zhang et al., 2013a) and cancer (Silva et al., 2015). Inhibition of STAT3 with TTI-101 reversed muscle loss in both cachexia models by suppressing the expression of MAFbx/Atrogin-1 and MuRF1 and reducing caspase-3 activity (Silva et al., 2015). Recent work by Guo et al. (2017) corroborates the findings of Zhang et al. (2013a) by demonstrating that pantoprazole inhibits JAK2/STAT3 signaling, which leads to reduced expression of MuRF1 and Fbx32 proteins, thereby mitigating the progression of tumor-associated muscle atrophy and protein degradation.
G. Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling and Its Targeting in Fibrosis
STAT3 plays a major role in many of the cellular processes that drive the development and progression of fibrosis, including fibroblast and myofibroblast cell proliferation, cell survival, induction of angiogenesis, and cell plasticity (Kasembeli et al., 2018). Fibrotic tissue samples from patients often show elevated levels of pY-STAT3 (Ogata et al., 2006). STAT3 is known to act both independently and in conjunction with other signaling networks such as TGF-β1, which is a well-known master regulator of fibrosis (Chakraborty et al., 2017). In fact, there is ample direct and indirect evidence indicating significant crosstalk between TGF-β1 signaling and STAT3. Studies have shown that TGF-β1 signaling can induce STAT3-dependent fibrotic responses by phosphorylation and activation of JAK2 (Zhang et al., 2015).
Genetic and pharmacological inhibition of STAT3 signaling has been shown to suppress fibrosis in many animal models, including models of lung fibrosis, in which haploinsufficiency of STAT3 eliminated the increase in lung collagen (hydroxproline) content in bleomycin-treated mice (O’Donoghue et al., 2012). pY-STAT3 activates transcription programs of various cellular processes to drive the development and progression of fibrosis. These include the following: 1) production of extracellular matrix (ECM) by upregulating collagen type I expression; 2) transcriptional control of MMPs, as well as their cognate inhibitor proteins—tissue inhibitors of MMPs—that also are necessary for the maintenance of ECM (Sun et al., 2008; Wang et al., 2011b; Giannandrea and Parks, 2014; Matsui et al., 2017); 3) resistance to apoptosis of ECM-producing cells, which slows resolution of the wound-healing process due to myofibroblast persistence (Kuhn and McDonald, 1991; Katzenstein and Myers, 1998; Habiel and Hogaboam, 2014; Xu et al., 2014a; Milara et al., 2018); and 4) contribution to fibroblast plasticity—a defining hallmark of fibrosis (Lamouille et al., 2014; Chakraborty et al., 2017; Liu et al., 2017; Pedroza et al., 2017).
Results from preclinical and clinical studies have confirmed the essential role of JAK/STAT3 signaling in the pathogenesis of fibrosis and generated considerable interest directed at targeting this pathway to develop novel treatments. Indeed, several Jakinibs are undergoing clinical testing in patients with fibrosis (Kasembeli et al., 2018), whereas preclinical studies examining small-molecule STAT3 inhibitors have shown promising results. Both TTI-101 and S3I-201 have been tested in several mouse models of fibrosis, including bleomycin-induced lung and skin fibrosis (scleroderma) and genetically-engineered models of scleroderma, as well as a model mouse of interstitial kidney fibrosis induced by unilateral ureteral obstruction, in which inhibition of STAT3 phosphorylation by these compounds blocks the development of organ fibrosis by reducing expression levels of proteins that mediate fibrosis and impairing myofibroblast differentiation (Pang et al., 2010; Pedroza et al., 2016, 2017; Chakraborty et al., 2017). In addition, cucurbitacin-B attenuates carbon tetrachloride–induced liver fibrosis, which was accompanied by reduced levels of pY-STAT3, along with reduced levels of hydroxyproline, collagen-1α, α-smooth muscle actin, and TGF-β in liver tissue (Sallam et al., 2018). Others have shown that sorafenib, and its derivative SC-1, can ameliorate liver fibrosis through STAT3 inhibition in hepatic stellate cells. In the thioacetamide liver fibrosis mouse model, administration of either sorafenib or SC-1 resulted in activation of SHP-1, which reduced levels of pY-STAT3 (Su et al., 2015). We demonstrated that TTI-101 administration to hepatocyte-specific Pten KO (HepPten-) mice, a model of nonalcoholic steatohepatitis (NASH) progressing to cirrhosis, reduced liver injury and fibrosis (Jung et al., 2017). S3I-201 also has shown activity in mouse preclinical models of carbon tetrachloride–induced fibrosis (Wang et al., 2018b). Zerumbone, a cyclic sesquiterpene derived from rhizomes of wild ginger, inhibits STAT3 phosphorylation and decreases mRNA expression of fibrotic genes such as α-SMA and TGF-β1, but not TGF-β2, in alkali-injured corneas of mice. Treatment with zerumbone reduced corneal neovascularization and fibrosis, thus promoting wound healing (Kim et al., 2017a).
Myeloproliferative neoplasms (MPNs) are a group of Philadelphia chromosome-negative chronic hematologic disorders that include PV, essential thrombocythemia (ET), and PMF. Dysregulation of bone marrow proinflammatory cytokine production leads to chronically elevated levels of circulating inflammatory cytokines, which drives clonal evolution in MPNs and leads to increased deposition of reticulin and collagen-containing bone marrow stromal fibers (Tefferi et al., 2011). Bone marrow fibrosis is a shared feature of all MPNs, but is the defining histopathological feature of PMF, a major diagnostic criterion (Zahr et al., 2016), and has been associated with poor outcomes (Kuter et al., 2007; Byrne et al., 2018).
Soluble factors associated with megakaryocytes and platelets known to signal through STAT3 have been implicated in the modulation of stromal cells and bone marrow fibrosis (Agarwal et al., 2016; Zahr et al., 2016). In addition, the JAK2 gain-of-function mutation (V617F) has been suggested to play a prominent role in MPNs (Baxter et al., 2005; Levine et al., 2005), being present in in ∼95% of PV, 50% to 60% of ET, and ∼50% of all PMF patients and associated with increased risk of mortality and splenomegaly (Verstovsek, 2010). JAK2 signaling through STAT5, which is constitutively active in JAK2 (V617F) mutant-positive cells, seems to mediate PV, as deletion of either STAT5A or STAT5B genes in JAK2V617F mice prevented the development of PV (Walz et al., 2012).
Currently, the only curative treatment option for PMF is allogenic hematopoietic cell transplantation, but long-term survival is usually hampered by disease relapse and/or graft-versus-host disease, leaving the unmet need to develop targeted therapies for PMF, e.g., JAK/STAT pathway inhibition. Ruxolitinib, a JAK1/JAK2 inhibitor, has demonstrated promising activity in PMF patients, showing increased benefit across several end points, e.g., reduction in splenomegaly, improvement in disease-related symptoms, and increased overall survival, when compared with placebo or best available therapy (Harrison et al., 2012; Verstovsek et al., 2012; Vannucchi et al., 2015). Pacritinib (a JAK2/FLT3 inhibitor) was even more effective than ruxolitinib in reducing splenomegaly and constitutional symptoms (Mascarenhas et al., 2018). Mometinib (a Jak1/2 inhibitor) and Itanicibib (a JAK1 inhibitor) also showed varying degrees of efficacy (Byrne et al., 2018). There is no clear correlation between the presence of a JAK2 V617F mutation and bone marrow fibrosis in MPN. However, when compared with best available therapy, Ruxolitinib and other Jakinibs appeared to reduce the odds of worsening bone marrow fibrosis and greater reduction in spleen size (Kvasnicka et al., 2018), suggesting that long-term JAK inhibition may be disease modifying.
It is important to note, however, that whereas JAK-STAT inhibitor treatment leads to improvement in constitutional symptoms and reduced splenomegaly, they show only modest benefit in reduction of clonal burden (Deininger et al., 2015), suggesting that the bulk of the observed clinical benefit of these inhibitors may arise from modulation of JAK-STAT signaling not associated with the expanded mutant clone(s). In fact, inhibition of JAK signaling in both wild-type and mutant cells was shown to be necessary to achieve improved clinical benefit in MPNs (Kleppe et al., 2015), and abnormal cytokine production continues to persist in plasma of many PMF patients after JAK2 inhibition with ruxolitinib (Fisher et al., 2019). Also, there is a word of caution regarding the use of Jakinibs in PMF as they may increase the risk of secondary malignancy, e.g., lymphoma (Porpaczy et al., 2018).
The BRD4/BET/NF-κB signaling pathway also has been suggested to contribute to MPN, as the addition of BET inhibitors to Jakinibs has been shown to markedly reduce serum levels of inflammatory cytokines, accompanied by reduction in disease burden, and reversal of bone marrow fibrosis in preclinical models (Kleppe et al., 2018). In addition, simultaneous inhibition of STAT5 phosphorylation on tyrosine and serine residues had synergistic effects against JAK2V617F mutant-positive cell lines, suggesting that approaches that target both post-translational modifications of STAT5 may be of benefit to patients with PMF (Bartalucci et al., 2017).
H. Janus Kinase/Signal Transducer and Activator of Transcription 3 Signaling and Its Targeting in Metabolic Disorders
Hepatic steatosis is an accumulation of fat in hepatocytes that can arise as a complication of obesity, type 2 diabetes mellitus, and alcohol intoxication. Treatment of mice in a murine model of chronic-binge ethanol feeding with STAT3-inducing cytokines, e.g., IL-6/IL-22, ameliorates fat accumulation in hepatocytes (Wang et al., 2004a; Ki et al., 2010). In morbidly obese individuals with congenital leptin deficiency, leptin replacement dramatically decreases insulin resistance, steatosis, dyslipidemia, and hyperglycemia (Farooqi et al., 2002; Licinio et al., 2004). Moreover, hepatocyte-specific gp130- or STAT3-KO mice exhibit exacerbated steatosis induced by a choline-deficient, methionine-supplemented diet; alcohol-containing diet; or a high-fat diet (Kroy et al., 2010). The antisteatogenic effect of hepatic STAT3 is suggested to be through direct inhibition of lipogenic sterol-regulatory element-binding protein-1 (a master transcription factor that controls lipid synthesis), acetyl-coenzyme A carboxylase, and fatty acid synthase, and stimulation of fatty acid oxidation genes, adenosine monophosphate-activated protein kinase, and carnitine palmitoyltransferase 1 (Inoue et al., 2004; Ueki et al., 2004; Horiguchi et al., 2008; Kroy et al., 2010). Inhibiting SOCS3 also increases pY-STAT3 levels, which ameliorates hepatic steatosis by downregulating sterol-regulatory element-binding protein-1 (Ueki et al., 2004). Interestingly, most of these mice models recapitulated alcoholic liver disease (ALD), which makes one speculate (Miller et al., 2011) that, probably in ALD, activation of STAT3 reduces hepatic steatosis. In fact, a current paper, which analyzed liver inflammatory and immune responses at early stages of alcoholic liver disease in a cohort of alcohol-dependent patients undergoing a highly standardized alcohol withdrawal program, concluded that in early stages of ALD, deficient IL-6/STAT3 signaling, high TLR7, and type I IFN production were risk factors for developing severe liver damage and fibrosis in these patients (Stärkel et al., 2019).
Recently, it was shown that an oxidative hepatic environment in obesity inactivates the STAT1 and STAT3 phosphatase T cell protein tyrosine phosphatase (TCPTP) and increases STAT1 and STAT3 signaling (Grohmann et al., 2018). Genetic deletion of TCPTP in hepatocytes also promoted T cell recruitment, NASH, and fibrosis, as well as hepatocellular carcinoma (HCC) in obese C57BL/6 mice that normally do not develop NASH, fibrosis, or HCC (Grohmann et al., 2018). Attenuating STAT1 signaling prevented T cell recruitment, NASH, and fibrosis, but did not prevent HCC. In contrast, correcting STAT3 signaling prevented HCC without affecting NASH and fibrosis. However, other studies have clearly established a role of STAT3 in non-alcoholic fatty liver disease. Pathogenesis of non-alcoholic fatty liver disease proceeds through lipid accumulation in hepatocytes that triggers a series of cytotoxic events that subsequently induces numerous pathologic processes, including insulin resistance, leptin deficiency, oxidative stress, fat accumulation, and liver tissue inflammation (Lonardo et al., 2016). STAT3, Krueppel-like factor 6, PPARs and methylenetetrahydrofolate reductase, and various proinflammatory cytokines are involved in the above processes leading to NASH (Sookoian et al., 2008). Activated IL-6/STAT3 in non-alcoholic fatty liver disease is associated with increased insulin resistance (Min et al., 2015). In a recent study (Belloni et al., 2018) targeting a phospho-STAT3/miR21 axis using either the STAT3 inhibitor S3I-201 or metformin, reduced STAT3 activation and improved vesicular hepatic steatosis in an in vitro fatty dHepaRG model (hepatic differentiated HepaRG cells treated with sodium oleate, S3I-201) as well as an in vivo high-fat diet induced age-dependent liver steatosis model (metformin).
In a hepatocyte-specific Pten KO mouse model of NASH-mediated HCC, TTI-101 treatment blocked hepatocellular carcinoma tumor development, through reducing liver steatosis, inflammation, and bile ductular reactions, thereby improving all pathologic lesions of NASH. It also greatly reduced liver injury, as measured by serum aspartate aminotransferase and alanine transaminase levels, as well as liver fibrosis, as indicated above. The reason for TTI-101 being able to reduce symptoms of both NASH/fibrosis and HCC might stem from the fact that TTI-101 treatment of HepPten− mice resulted in inhibition of both STAT1 and STAT3 pathways (Jung et al., 2017). Similarly, STAT3 inhibition in adipose tissue and, to a lesser extent, in hepatic tissues (Hua et al., 2018) was how known antilipemic agents (PPARγ agonists, gemfibrozil, and fenofibrate) attenuated obesity, high blood pressure, and insulin resistance in a PPARα-null mouse model of metabolic syndrome.
STAT3 also has been reported to play a definite detrimental role in development of insulin resistance. Prolonged IL-6 exposure leads to insulin resistance and glucose intolerance in human adipocytes, hepatocytes (Gurzov et al., 2016), and skeletal muscles. Blocking IL-6 in diet-induced obese mice represses hepatic inflammation via inhibition of the IL-6/JAK2/STAT3 pathway (Park et al., 2010), thereby increasing insulin sensitivity and resultantly decreasing diet-induced obesity (Priceman et al., 2013). TCPTP in the liver dephosphorylates the insulin receptor, JAK1, and STAT3 and negatively regulates both IFN regulatory factor/PI3K and STAT3 signaling pathways, thereby modulating expression of gluconeogenic genes and hepatic glucose output (Fukushima et al., 2010). Moreover, STAT3 silencing improves insulin sensitivity in IL-6–treated myotubes and prevents development of lipid-induced insulin resistance (Mashili et al., 2013; Gurzov et al., 2016). IL-6/STAT3 signaling in muscle during intense exercise, in contrast, plays a beneficial role in glucose homeostasis (Wunderlich et al., 2013). Thus, it seems, whereas transient IL-6/STAT3 seems to be helping maintain glucose homeostasis, chronic IL-6/STAT3 signaling impairs insulin action, leading to insulin resistance and inflammation-associated glucose intolerance, probably through an increase in basal SOCS3 levels in generally insulin-sensitive adipose tissue, liver, and skeletal muscles, by mechanism (Yang et al., 2012) similar to leptin/STAT3/SOCS3 axis in the CNS (Wunderlich et al., 2013).
IL-6/JAK/STAT3-induced SOCS3 impairs insulin action by binding to IRS-1 and IRS-2, resulting in their ubiquitination and degradation (Shi et al., 2004). Recent studies have reported that STAT3 is required for pancreatic β cell reprogramming and regeneration, although the downstream targets are unknown (Gurzov et al., 2016). Muscle-specific deletion of SOCS3 (Jorgensen et al., 2013) or STAT3-siRNA (Kim et al., 2017b) protects against the development of hyperinsulinemia and insulin resistance due to elevated IRS-1 and Akt phosphorylation that contribute to increased glucose uptake. Interestingly, hepatocyte-specific SOCS3–KO mice have improved insulin sensitivity resulting from increased pY-IRS-1, but develop systemic insulin resistance and obesity over a long period due to the hyperactivity of STAT3 and resultant chronic inflammation (Torisu et al., 2007), indicating a non-SOCS3–mediated mechanism of chronic STAT3-mediated insulin resistance.
Chronic inflammation has been known to accelerate insulin resistance and other metabolic abnormalities associated with obesity (Kalupahana et al., 2012; Wensveen et al., 2015). Obesity-induced inflammation also drives an influx of immune cells, including B cells and CD4+ and CD8+ T cells, leading to further release of proinflammatory cytokines, such as IL-6, from recruited inflammatory cells, such as macrophages (Pradhan et al., 2001; Hu et al., 2004; Wunderlich et al., 2013), which exacerbates unintended immune responses contributing to the pathology (Wensveen et al., 2015). Thus, the IL-6/JAK2/STAT3 signaling pathway is a valid target to treat NASH, non-alcoholic fatty liver disease, and insulin resistance, especially in the setting of obesity and type 2 diabetes.
I. Cancer
1. Janus Kinases in Cancer
There are several ways in which JAK activity contributes to cellular transformation and tumorigenesis. These include the following: 1) increased constitutive activation of upstream receptors due to overproduction of stimulating ligands; 2) increased receptor expression, or genetic alterations in cytokine receptors that constitutively induce JAK activation; 3) disruption of negative regulators of JAK/STAT3 signaling (SOCs, protein inhibitors of activated STAT, or protein phosphatases); and most prominently 4) mutations of JAKs that lead to their constitutive activation.
Chromosomal translocations resulting in JAK fusion proteins with hyperactive tyrosine kinase activities have long been linked to a variety of hematologic malignancies. For example, the fusion oncogene consisting of translocation ETS leukemia fused to JAK2 has been associated with several leukemias, including B cell and T cell childhood acute lymphocytic leukemias. The translocation ETS leukemia–JAK2 fusion oncoprotein has been shown to drive constitutive activation of JAK/STAT3, NF-κB, PI3K, and RAS/ERK signaling pathways. Other rearrangements, including BCR-JAK2, PMC1-JAK2, SSPBP-JAK2, PAX5-JAK2, and SEC3A-JAK2, also have been implicated in several myeloid and lymphoid tumors (Chen et al., 2012).
Gain-of-function mutations in JAKs also have been shown to lead to constitutive JAK-STAT3 pathway activation in hematologic malignancies. Mutations in JAK2 have been found in a large proportion of patients with chronic MPNs, with estimates ranging from 50% to 95% of patients with PV, ET, and PMF having activating V617F mutation in JAK2. Genetic alterations in JAK1, JAK2, and JAK3 also have been reported in both acute myeloid leukemia and acute lymphocytic leukemias (Chen et al., 2012; Vainchenker and Constantinescu, 2013).
The IL-6/JAK/STAT3 signaling pathway has also been implicated in many solid tumors, where it has been shown to promote tumor growth and progression while hindering antitumor immunity. Analysis of patient samples demonstrated that IL-6 promotes the development of many solid tumors, including breast, cervical, colorectal, esophageal, head-and-neck, ovarian, pancreatic, prostate, renal, and lung (NSCLC) cancers (Johnson et al., 2018). Taken all together, it is clear that JAK/STAT signaling pathway contributes to the development and progression of a wide range of both hematologic and solid cancers.
2. Targeting Janus Kinases in Cancer
Inhibitors of JAK/STAT3 signaling upstream of JAKs include monoclonal antibodies that neutralize IL-6 or that antagonize the IL-6R. Siltuximab (CNTO 328), the avimeric anti–IL-6 monoclonal antibody, has been investigated for treatment of various malignancies. For example, in a Phase I/II trial for metastatic renal cell cancer, siltuximab stabilized disease in more than 50% of patients and had a favorable toxicity. However, in patients with myelodysplastic syndrome and solid tumors like prostate cancer, ovarian cancer, and lung cancer, siltuximab as a monotherapy did not show significant clinical activity. For the treatment of patients with HIV-negative and human herpes virus-8–negative multicentric Castleman's disease, siltuximab received approval in the United States and European Union in 2014 (Chen and Chen, 2015), as treated patients showed a 5-year survival rate of 96.4% compared with 55% to 77% in those receiving standard of care treatment (Sitenga et al., 2018).
At present, four direct Jakinibs—tofacitinib, ruxolitinib, oclacitinib, and baricitinib—now are Food and Drug Administration approved for treatment of cancer. Several other drugs that target JAKs are at various stages of development and are being tested in various conditions, including precancer syndromes, e.g., MPNs, as well as in other noncancer diseases, psoriatic arthritis, and ulcerative colitis (O’Shea et al., 2015; Buchert et al., 2016; Bose et al., 2017). Although Jakinibs have shown encouraging therapeutic efficacy, due to the pleiotropic nature of JAK signaling, it is not surprising that there have been notable toxicities (Bose et al., 2017). For instance, Jakinibs are associated with immunosuppression, and common early adverse effects include diarrhea, nasopharyngitis, headache, and urinary tract infection, whereas prolonged treatment has been associated with upper respiratory tract infections, nasopharyngitis, and bronchitis. In addition, hematologic toxicities, such as thrombocytopenia and neutropenia, have been observed with the inhibition of JAKs (Kotyla, 2018). The limited effectiveness and toxicities of Jakinibs have provided an impetus to develop inhibitors of molecular targets downstream of JAK, such as STAT3.
3. Signal Transducer and Activator of Transcription 3 in Cancer
Hyperactivation of STAT3 is a salient attribute of many cancers (Bromberg et al., 1999; Sriuranpong et al., 2003; Bharadwaj et al., 2016b). Elevated levels of STAT3 have been observed in a large number of both solid and hematologic tumors, where its signaling underpins a majority of all known cardinal features of cancer (Yuan et al., 2015b), including growth and proliferation (Banerjee and Resat, 2016), antiapoptosis (Gritsko et al., 2006), cell transformation (Bromberg et al., 1999), angiogenesis (Doucette et al., 2012), metastasis (Xiong et al., 2012), and cancer stem cell maintenance (Sherry et al., 2009). Inhibition of STAT3 activation in many preclinical cancer models results in increased apoptosis and decreased proliferation. In addition, STAT3 plays an important role in the tumor microenvironment, as it modulates antitumor immune response, where it plays dual and complementary roles as an upregulator of tumor immune inhibitors and a suppressor of tumor immune activators (Yu et al., 2007; Marzec et al., 2008; Lee et al., 2014). Lastly, STAT3 has been implicated in chemotherapy resistance; there is a strong correlation between STAT3 activation and chemoresistance, and STAT3 inhibition has been shown to restore sensitivity of tumors to chemotherapeutic drugs (Real et al., 2002; Huang et al., 2012; Sims et al., 2013; Lee et al., 2014; Zhao et al., 2016).
4. Targeting Signal Transducer and Activator of Transcription 3 in Cancer
Many groups are developing STAT3 inhibitors with the goal of targeting STAT3 in cancer. For a recent in-depth review of STAT3 inhibitors for cancer treatment, the reader is referred to a book chapter by our group (Bharadwaj et al., 2016b). Summarized below are those STAT3 inhibitors currently in clinical trials.
a. Antisense oligonucleotide
The naked, antisense, modified oligonucleotide, AZD9150 (danvatirsen), targets STAT3 and demonstrated antitumor activity in a Phase 1 dose-escalation study in patients with highly treatment-refractory lymphoma and NSCLC (Hong et al., 2015). The maximum tolerated dose of danvatirsen in this study was determined to be 3 mg/kg. The major treatment-emergent adverse event (TEAE) was thrombocytopenia. Other adverse events (AEs) included elevation of serum aspartate and alanine aminotransferases. In a subsequent Phase IB trial, 30 patients were enrolled, of which 10 were given 2 mg/kg and 20 patients received 3 mg/kg. Twenty-seven patients had diffuse large B cell lymphoma; 2 of 27 patients with diffuse large B cell lymphoma demonstrated complete responses; and 2 demonstrated partial responses. Elevated serum aminotransferases were observed in approximately 40%, and thrombocytopenia was observed in approximately 30%. It was concluded that danvatirsen was efficacious, safe, and well tolerated at the doses administered (Reilley et al., 2018). Currently, there is an ongoing Phase 2 study of danvatirsen (3 mg/kg) plus durvalumab (anti–programmed death ligand-1 monoclonal antibody) in patients with HNSCC (NCT02499328) that is showing promising results.
b. Signal transducer and activator of transcription 3 decoy oligonucleotides
Expression levels of STAT3 target genes were decreased in head and neck cancer patients following intratumoral injection with the STAT3 decoy compared with tumors receiving saline control in a phase 0 trial (Sen et al., 2012). The future development of this agent may be as a radiation sensitizer for use in combination with radiation therapy in patients with HNSCC not associated with human papilloma virus.
c. Small-molecule inhibitors
Several promising small-molecule STAT3 inhibitors are in Phase I/II development, as outlined below:
i. OPB-31121
This molecule developed by Otsuka Pharmaceutical (Tokyo, Japan) showed a good pharmacokinetic profile (Hayakawa et al., 2013; Kim et al., 2013b; Bendell et al., 2014; Brambilla et al., 2015), but demonstrated limited antitumor activity in patients with HCC (Bendell et al., 2014). Furthermore, long-term dosing resulted in a high incidence of peripheral neuropathy (Bendell et al., 2014). A new generation of compounds developed by Otsuka that demonstrated efficacy in preclinical models has been evaluated in several clinical trials. Published findings of a Phase I trial for OPB-51602 in patients with refractory hematologic malignancies and solid tumors identified a maximum tolerated dose of 6 mg/day. The most common adverse side effects were nausea, vomiting, diarrhea, anorexia, peripheral neuropathy, and fatigue. Dose-limiting toxicities of severe lactic acidosis and peripheral neuropathy were observed in a number of the subjects. In this study, the authors reported tolerability issues with long-term dosing that led to termination of the trial for hematologic malignancies. Further exploration of less frequent dosing in a subsequent trial for patients with refractory solid malignancies was suggested (Ogura et al., 2015; Wong et al., 2015).
ii. OPB-111077
Most recently, a Phase I study of OPB-111077 that enrolled 145 patients was reported. OPB-111077 is another inhibitor of STAT3 and mitochondrial oxidative phosphorylation developed by Otsuka. Dose‐limiting toxicities of this compound were observed at 300 and 400 mg daily, and the maximum tolerated dose was defined as 250 mg daily. Drug-related AEs were reported as mild and manageable and included nausea, fatigue, and vomiting (Tolcher et al., 2018). Similar to OPB-31121, OPB-OPB‐111077 demonstrated only modest responses against unselected tumors with objective responses observed in only two patients with diffuse large B‐cell lymphoma. Both Otsuka compounds share similar toxicity profiles, suggesting that the adverse effects likely arise through a common mechanism. It was recently reported that cytotoxic effects of OPB-51602 and related STAT3 inhibitors were mainly due to drug-induced mitochondrial dysfunction (Genini et al., 2017). Of note, lactic acidosis is a clinical laboratory marker of mitochondrial dysfunction. In addition, there is a strong link between peripheral neuropathy and diminished mitochondrial function (Stacpoole, 1997). Thus, detailed understanding of these toxicities and how to mitigate them will be necessary to optimize the therapeutic potential of this line of STAT3-targeting molecules.
iii. Bardoxolone
This molecule is one of several derivatives of the synthetic triterpenoid C-28 methyl ester of the oleane triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO-Me) that was shown to have promising anticancer effects. This group of compounds is believed to have a covalent mode of action mediated by Michael addition to nucleophilic residues within proteins, mainly cysteine residues (Liby et al., 2007b). CDDO-Me inhibits constitutive and IL-6–induced JAK1 activity by directly binding to Cys1077 in the JAK1 kinase domain, thereby blocking its ability to phosphorylate STAT3 at Y705. Biochemical evidence shows that CDDO-Me also forms adducts with STAT3, disrupting its dimer structure (Ahmad et al., 2008) and decreasing levels of pY-STAT3. Preclinical findings in various cancer models, such as lung cancer, ER-negative breast cancer, pancreatic cancer, and hepatocellular carcinoma, have accumulated that establish CDDO-Me as a potent anticancer agent that acts, in part, by inhibiting STAT3 activity (Samudio et al., 2006; Liby et al., 2007a, 2008, 2010). However, it is noteworthy that bardoxolone has been found to be toxic to mitochondria, as it inhibits mitochondrial electron transport via perturbations in inner mitochondrial membrane integrity (Samudio et al., 2006), which, as with the Otsuka compounds described above, has implications for toxicity of this agent in patients.
In clinical trials, CDDO-Me showed therapeutic activities against several types of cancers. Of note, no toxicity attributed to CDDO-Me administration was observed in a Phase I clinical trial RTA 402-C-0702 for pancreatic adenocarcinomas in which gemcitabine was combined with oral administration of CDDO-Me at different doses ranging from 150 to 300 mg/day (Wang et al., 2014). In another Phase I trial of CDDO-Me in patients with advanced solid tumors and lymphomas, a complete tumor response occurred in one mantle cell lymphoma patient and a partial response was observed in an anaplastic thyroid carcinoma patient at a dose of 900 mg/day. Pharmacokinetic parameters suggest that CDDO-Me has a slow and saturable oral absorption, long terminal-phase half-life (39 hours at 900 mg/day), nonlinear dose-dependent exposures at high doses (600–1300 mg/day), and high interpatient variability. Dose-limiting toxicities were grade 3 liver transaminase elevations that were reversible. Overall, the data appeared encouraging enough to support continued development of other synthetic triterpenoids as anticancer agents (Hong et al., 2012).
Screening experiments show that CDDO-Me inhibits a wide range of targets in addition to STAT3, including PPARγ, kelch-like ECH-associated protein 1 (Keap1), and inhibitory NF-κB kinase. Inhibition of Keap1 leads to activation of the nuclear factor erythroid 2–related factor 2, an oxidative stress response transcription factor known to activate a variety of cytoprotective genes (Wang et al., 2014). Because targeting of Keap1 with CDDO-Me can induce nuclear factor erythroid 2–related factor 2, which plays a prominent role in kidney function (Tanaka et al., 2008), it has been evaluated in Phase I/II trials for clinical activity in patients with moderate to severe chronic kidney disease. Data from these studies demonstrated a significant increase from baseline in estimated glomerular filtration rate, a measure of improved kidney function, which suggested that CDDO-Me may be beneficial in these patients. However, a subsequent Phase III trial was terminated in chronic kidney disease patients due to a high rate of cardiovascular events when compared with placebo (Nezu et al., 2017).
iv. Napabucasin (BBI608)
Napabucasin is an inhibitor of cancer stemness currently undergoing Phase III clinical trials to treat several malignancies. It was first identified by its ability to inhibit properties of cancer stemness and STAT3 activity in gel-shift assays (US patent 8,877,803). It has been shown to downregulate expression of genes associated with cancer cell stemness and to kill cells exhibiting properties of cancer cell stemness in various types of cancers. Napabucasin treatment also appeared to inhibit multiple self-renewal pathways, suggesting that it may have activity toward targets other than STAT3 (Li et al., 2015).
Phase I and II trials showed that TEAE related to the administration of the napabucasin were generally mild and included predominantly grades 1 and 2 GI TEAE. Grade 3 TEAE were GI-related (7.3%) and fatigue (2.4%). Dose escalation was stopped because of pill burden, and no maximum tolerated dose was established. At daily dose of 400 mg, plasma levels of napabucasin were over its half-maximal inhibitory concentration (1.5 µM) for 8 hours (Jonker et al., 2014; Laurie et al., 2014). Preclinical studies in mouse models of several cancers show that napabucasin was effective both as monotherapy and in combination with other agents, particularly paclitaxel where synergy was observed. Phase I and II clinical studies demonstrated promising antitumor efficacy when napabucasin was combined with standard chemotherapy agents (Garcia et al., 2016). A Phase III clinical trial of napabucasin versus placebo in patients with advanced CRC showed no significant difference in unselected patients, with regard to overall survival, progression-free survival, or disease control rate. However, patients with pY-STAT3–positive disease demonstrated significantly improved overall survival when treated with napabucasin (median oxidative stress = 5.1 months) compared with placebo-treated patients (median oxidative stress = 3.0 months), even though patients with pY-STAT3–positive tumors had a poorer prognosis. There currently are three ongoing Phase III clinical trials involving napabucasin in combination with other standard chemotherapy agents in metastatic CRC, metastatic pancreatic ductal adenocarcinoma, and other advanced malignancies (Hubbard and Grothey, 2017). Notably, no new AE or pharmacokinetic interactions were observed when napabucasin was combined with paclitaxel.
v. Pyrimethamine (GLG-801)
Pyrimethamine currently is marketed for the treatment of parasitic diseases and is under evaluation in Phase I/II clinical trials in Europe for a number of cancer indications, including CLL and triple-negative breast cancer. It was identified as a STAT3 inhibitor using a cell-based functional screen of a Preswick library of compounds. Early studies demonstrated that GLG-801 inhibited STAT3 activation and renal cyst formation in a mouse model of polycystic kidney (Takakura et al., 2011). Subsequent studies published recently by Khan et al. (2018) showed that GLG-801 has direct antitumor activity in two breast cancer mouse models. A Phase I/II clinical study is underway in patients with metastatic triple-negative breast cancer (EU Clinical Trials Register, 2017). While demonstrating efficacy and the ability to block STAT3 activation in preclinical models, the mechanism by which GLG-801 inhibits STAT3 activation is not clear.
vi. WP-1066
WP-1066 is a potent inhibitor of JAK2 and downstream STAT3 signaling. A Phase I trial of WP-1066 in patients with glioblastoma multiforme and melanoma that has metastasized to the brain is currently underway at MD Anderson Cancer Center (Bharadwaj et al., 2016b). The primary objective of this study (NCT01904123) is to determine the maximum tolerated dose and dose-limiting side effects. There currently are no published results regarding the outcome of this study.
vii. TTI-101 (C188-9)
TTI-101 (C188-9) emerged from a structure-based, drug-design program that began with computer-based docking of 920,000 compounds into the SH2 domain of STAT3 and identified three hits (C3, C30, and C188) (Xu et al., 2009); each hit inhibited STAT3 binding to its pY-peptide ligand, thereby blocking two steps in STAT3 activation—recruitment to activated receptor complexes and homodimerization. Hit-to-lead strategies performed in collaboration with Tvardi Therapeutics (formerly StemMed), a clinical-stage drug-development company, focused on the most potent initial hit, C188 (Xu et al., 2009), and led to TTI-101 (Bharadwaj et al., 2016a). TTI-101 potently inhibits STAT3 binding to its pY-peptide ligand [Ki = 12.4 nM (Bharadwaj et al., 2016a)]. Pharmacokinetic studies in mice demonstrated tumor levels 2.6 times simultaneously measured plasma levels (Bharadwaj et al., 2016a). TTI-101 demonstrated potent antitumor activity in several preclinical cancer models, including NSCLC (Lewis et al., 2015), head and neck squamous cell carcinoma (Bharadwaj et al., 2016a), hepatocellular carcinoma (Jung et al., 2017), and ER-positive breast cancer resistant to inhibitors of cyclin-dependent kinases (Kettner et al., 2019).
Exposure of mice to TTI-101 has modest and expected effects on T cell subsets and myeloid cells. Specifically, daily administration of TTI-101 for 14 days skewed T cell development away from Th17 cells toward Th1 cells, thereby phenocopying mice that transgenically express a dominant-negative STAT3 mutation (Steward-Tharp et al., 2014). In addition, modestly increased numbers of peripheral blood mononuclear cells with myeloid markers were seen in treated animals, which phenocopied mice with myeloid-lineage–specific deletion of STAT3 (Lee et al., 2002).
The Food and Drug Administration approved an investigational new drug application for a Tvardi-sponsored Phase I study of TTI-101 in patients with advanced solid tumors, including NSCLC, at the University of Texas MD Anderson Cancer Center, which has completed enrollment at dose level 3. No serious active pharmaceutical ingredient–attributable AEs have been observed to date. Importantly, 5 of 10 patients demonstrated clinical benefit, with one patient experiencing a partial clinical response (40% reduction in tumor sizes according to response evaluation criteria in solid tumors) for 12 months.
V. Considerations for Future Development of Janus Kinase/Signal Transducer and Activator of Transcription 3 Inhibitors
Jakinibs have been successfully introduced into the clinic, as described above, and, for the foreseeable future, they will continue to be useful in the treatment of several inflammatory and neoplastic conditions. Their toxicities, although for the most part manageable, are the major impediment to their broader use, however.
The future of STAT3 inhibitors as therapies for inflammatory and fibrotic disorders and for cancer is promising, but caution is urged especially with regard to achieving sufficient target engagement, especially in the setting of cancer, and the avoidance of toxicity, especially in noncancer indications. The toxicities observed in Phase I/II studies of OPB-31121 that halted its further development resulted from its targeting of mtSTAT3 (Genini et al., 2017). It is of interest to note that targeting of mtSTAT3 to modify aberrant oxidative phosphorylation in cancer cells may have contributed to the anticancer effects of OPB-31121, similar to other agents undergoing clinical study that target this cancer cell vulnerability (Zhang et al., 2019). However, given the difficulty in identifying patients who may benefit from inhibition of oxidative phosphorylation at this point in time, drug-development programs focused on targeting canonical STAT3 activity would be wise to avoid targeting mtSTAT3 activity to minimize toxicity. Toward this end, we examined the effect of TTI-101 on mitochondrial function in DU145 and HS358 cells. Our studies showed that TTI-101 did not affect mitochondrial ATP generation or oxygen consumption, whereas studies performed in parallel with other STAT3 inhibitors previously shown to target mtSTAT3 (Genini et al., 2017) were confirmatory (data not shown).
The serious adverse side effects that resulted from targeting the mitochondrial activities of STAT3 underscore the importance of considering other potential unexpected toxicities that might be seen in current or new efforts focused on targeting canonical STAT3 signaling. These include consideration of effects on post-translational modifications of STAT3 other than Y705 phosphorylation and effects on noncanonical activities of STAT3 besides its mitochondrial activity already discussed. Also, the impact of targeting canonical STAT3 signaling on basal and stress-induced canonical functions needs to be considered in assessing STAT3 inhibitors for safety early in clinical development, as discussed below.
Most forms of post-translational modified STAT3 proteins, including those that that are phosphorylated on serine, methylated, acetylated, ubiquitinated, ISGylated, and SUMOylated, are modified after their phosphorylation at key tyrosine sites. Consequently, STAT3 inhibitors that reduce levels of pY-STAT3 would be expected to have similar effects on the levels and activities of these other post-translationally modified forms of STAT3. Similarly, because the scaffold function of STAT3 in platelet activation is mediated by homodimers of pY-STAT3, STAT3 inhibitors that reduce pY-STAT3 could potentially reduce platelet function in inflamed tissue, such as a ruptured atherosclerotic plaque. Importantly, however, these inhibitors would not be expected to reduce platelet activity early in hemostasis involving traumatically damaged vessels. In contrast, STAT3 inhibitors that reduced levels of pY-STAT3 would be expected to increase U-STAT3 levels, if only slightly, and would be expected to have minimal impact on the activity of U-STAT3. Similarly, because the activity of STAT3 in the ER does not require STAT3 to be phosphorylated at Y705, the impact of reduced pY-STAT3 on the ER function of STAT3 is expected to be minimal.
The above review of the important contributions of canonical STAT3 signaling in basal and stress conditions suggests that targeting STAT3 in adults might result in unacceptable toxicity, including hematopoietic and immune deficiencies, bleeding disorders, metabolic disorders, and a variety of impairments in stress responses. However, this may not be the case based on the more limited clinical manifestations observed in patients with Job’s syndrome or AD-HIES, as discussed above. Nevertheless, it would appear prudent to carefully monitor patients administered STAT3 inhibitors for development of symptoms and signs commonly reported in AD-HIES, e.g., eczema, bacterial pneumonia, skin abscesses, and mucocutaneous candidiasis. In addition, based on the critical role of STAT3 in wound healing, it would be advisable to avoid the use of STAT3 inhibitors in the period immediately prior to surgery or following traumatic injury until wound healing is complete.
Abbreviations
- AD-HIES
autosomal-dominant hyper-IgE syndrome
- AE
adverse event
- AgRP
agouti-related protein
- Akt
Ak strain transforming
- ALD
alcoholic liver disease
- C/EBP
CCAAT/enhancer-binding protein
- CD
Crohn’s disease
- CK2
casein kinase 2
- CLL
chronic lymphocytic leukemia
- CNS
central nervous system
- CRC
colorectal cancer
- CSE
cigarette smoke extract
- DC
dendritic cell
- ECM
extracellular matrix
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated kinase
- ET
essential thrombocythemia
- ETC
electron transport chain
- FERM
band four-point-one, ezrin, radixin, moesin
- GAS
IFN-γ activation site
- GI
gastrointestinal
- GVHD
graft-versus-host disease
- HBEC
human bronchial epithelial cell
- HCC
hepatocellular carcinoma
- HDAC
methyltransferase histone deacetylase
- HDM
house dust mite
- HNSCC
head and neck squamous cell carcinoma
- IBD
inflammatory bowel disease
- IFN
interferon
- IL
interleukin
- iNOS
inducible NO synthase
- IP3R3
inositol 1,4,5-trisphosphate receptor, type 3
- I/R
ischemia–reperfusion
- IR
ionizing radiation
- IRS
insulin receptor substrate
- ISG
IFN-stimulated gene
- JAK
Janus kinase
- Jakinib
JAK inhibitor
- JH
JAK homology
- JNK
c-Jun N-terminal kinase
- Keap1
kelch-like ECH-associated protein 1
- KO
knockout
- MAPK
mitogen-activated protein kinase
- MI
myocardial infarction
- MMP
matrix metalloproteinase
- MPN
myeloproliferative neoplasm
- mTOR
mammalian target of rapamycin
- mtSTAT
mitochondrial STAT
- NASH
nonalcoholic steatohepatitis
- NF-κB
nuclear factor κB
- NO
nitric oxide
- NSCLC
non-small cell lung cancer
- NTD
N-terminal domain
- PI3K
phosphatidylinositol 3-kinase
- PKC
protein kinase C
- PLC
phospholipase C
- PMF
primary myelofibrosis
- POMC
proopiomelanocortin
- PPAR
peroxisome proliferator-activated receptor
- pS
phosphorylation
- PTEN
phosphatase and tensin homologue deleted on chromosome 10
- PV
polycythemia vera
- pY
phosphotyrosine
- R
receptor
- ROS
reactive oxygen species
- SH2
Src homology 2
- SHP
Src homology protein tyrosine phosphatase
- siRNA
small interfering RNA
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- Syk
spleen tyrosine kinase
- TAD
transactivation domain
- TCPTP
T cell protein tyrosine phosphatase
- TEAE
treatment-emergent adverse event
- TGF
transforming growth factor
- Th
T helper
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- Treg
T regulatory cell
- U-STAT
unphosphorylated STAT
- UC
ulcerative colitis
- Y705
tyrosine 705
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Bharadwaj, Kasembeli, Robinson, Tweardy.
Footnotes
This work was supported by University of Texas for MD Anderson. This work was supported by the National Institutes of Health [Grant 1P50CA221707-01A1].
Baylor College and Medicine, with D.J.T. as inventor, has filed 19 patents covering the use of TTI-101, a small-molecule inhibitor of STAT3 cited in this review. These patents are exclusively licensed to Tvardi Therapeutics, which was founded and is co-owned by D.J.T.
References
- Abe K, Hirai M, Mizuno K, Higashi N, Sekimoto T, Miki T, Hirano T, Nakajima K. (2001) The YXXQ motif in gp 130 is crucial for STAT3 phosphorylation at Ser727 through an H7-sensitive kinase pathway. Oncogene 20:3464–3474. [DOI] [PubMed] [Google Scholar]
- Agarwal A, Morrone K, Bartenstein M, Zhao ZJ, Verma A, Goel S. (2016) Bone marrow fibrosis in primary myelofibrosis: pathogenic mechanisms and the role of TGF-β. Stem Cell Investig 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal BB, Kunnumakkara AB, Harikumar KB, Gupta SR, Tharakan ST, Koca C, Dey S, Sung B. (2009) Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann N Y Acad Sci 1171:59–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad R, Raina D, Meyer C, Kufe D. (2008) Triterpenoid CDDO-methyl ester inhibits the Janus-activated kinase-1 (JAK1)-->signal transducer and activator of transcription-3 (STAT3) pathway by direct inhibition of JAK1 and STAT3. Cancer Res 68:2920–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Nishio Y, Inoue M, Wang XJ, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. (1994) Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77:63–71. [DOI] [PubMed] [Google Scholar]
- Alicea-Velázquez NL, Boggon TJ. (2011) The use of structural biology in Janus kinase targeted drug discovery. Curr Drug Targets 12:546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonzi T, Newton IP, Bryce PJ, Di Carlo E, Lattanzio G, Tripodi M, Musiani P, Poli V. (2004) Induced somatic inactivation of STAT3 in mice triggers the development of a fulminant form of enterocolitis. Cytokine 26:45–56. [DOI] [PubMed] [Google Scholar]
- Al-Ramli W, Préfontaine D, Chouiali F, Martin JG, Olivenstein R, Lemière C, Hamid Q. (2009) T(H)17-associated cytokines (IL-17A and IL-17F) in severe asthma. J Allergy Clin Immunol 123:1185–1187. [DOI] [PubMed] [Google Scholar]
- Alsaffar H, Martino N, Vincent P, Adam A, Lowery A. (2016) STAT3 regulates endothelial permeability downstream of the pro-inflammatory cytokines IL6 and TNF-alpha. FASEB J 30. [Google Scholar]
- Altemeier WA, Sinclair SE. (2007) Hyperoxia in the intensive care unit: why more is not always better. Curr Opin Crit Care 13:73–78. [DOI] [PubMed] [Google Scholar]
- American Thoracic Society (2000) Proceedings of the ATS workshop on refractory asthma: current understanding, recommendations, and unanswered questions. American Thoracic Society. Am J Respir Crit Care Med 162:2341–2351. [DOI] [PubMed] [Google Scholar]
- Ananthakrishnan AN. (2017) Filgotinib for Crohn’s disease - expanding treatment options. Lancet 389:228–229. [DOI] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, et al. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 43:246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosz H, Wojciechowska K, Sajewicz J, Choroszyńska D, Marzec-Kotarska B, Osiak M, Pająk N, Tomczak W, Jargiełło-Baszak M, Baszak J. (2015) IL-6, IL-10, c-Jun and STAT3 expression in B-CLL. Blood Cells Mol Dis 54:258–265. [DOI] [PubMed] [Google Scholar]
- Araki T, Tsujioka M, Abe T, Fukuzawa M, Meima M, Schaap P, Morio T, Urushihara H, Katoh M, Maeda M, et al. (2003) A STAT-regulated, stress-induced signalling pathway in Dictyostelium. J Cell Sci 116:2907–2915. [DOI] [PubMed] [Google Scholar]
- Arikan AA, Yu B, Mastrangelo MA, Tweardy DJ. (2006) Interleukin-6 treatment reverses apoptosis and blunts susceptibility to intraperitoneal bacterial challenge following hemorrhagic shock. Crit Care Med 34:771–777. [DOI] [PubMed] [Google Scholar]
- Atreya R, Mudter J, Finotto S, Müllberg J, Jostock T, Wirtz S, Schütz M, Bartsch B, Holtmann M, Becker C, et al. (2000) Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in Crohn disease and experimental colitis in vivo. Nat Med 6:583–588. [DOI] [PubMed] [Google Scholar]
- Avalle L, Camporeale A, Morciano G, Caroccia N, Ghetti E, Orecchia V, Viavattene D, Giorgi C, Pinton P, Poli V. (2019) STAT3 localizes to the ER, acting as a gatekeeper for ER-mitochondrion Ca2+ fluxes and apoptotic responses. Cell Death Differ 26:932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalle L, Poli V. (2018) Nucleus, mitochondrion, or reticulum? STAT3 à La Carte. Int J Mol Sci 19:2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz MH, Manoharan HT, Sand JM, Verma AK. (2007a) Protein kinase Cepsilon interacts with Stat3 and regulates its activation that is essential for the development of skin cancer. Mol Carcinog 46:646–653. [DOI] [PubMed] [Google Scholar]
- Aziz MH, Manoharan HT, Verma AK. (2007b) Protein kinase C epsilon, which sensitizes skin to sun’s UV radiation-induced cutaneous damage and development of squamous cell carcinomas, associates with Stat3. Cancer Res 67:1385–1394. [DOI] [PubMed] [Google Scholar]
- Babaei R, Schuster M, Meln I, Lerch S, Ghandour RA, Pisani DF, Bayindir-Buchhalter I, Marx J, Wu S, Schoiswohl G, et al. (2018) Jak-TGFβ cross-talk links transient adipose tissue inflammation to beige adipogenesis. Sci Signal 11:eaa17838. [DOI] [PubMed] [Google Scholar]
- Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet IS, Norton RS, Nicola NA. (2012) Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity 36:239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SJ, Rane SG, Reddy EP. (2007) Hematopoietic cytokine receptor signaling. Oncogene 26:6724–6737. [DOI] [PubMed] [Google Scholar]
- Ball DP, Lewis AM, Williams D, Resetca D, Wilson DJ, Gunning PT. (2016) Signal transducer and activator of transcription 3 (STAT3) inhibitor, S3I-201, acts as a potent and non-selective alkylating agent. Oncotarget 7:20669–20679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Resat H. (2016) Constitutive activation of STAT3 in breast cancer cells: a review. Int J Cancer 138:2570–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry SP, Townsend PA, Knight RA, Scarabelli TM, Latchman DS, Stephanou A. (2010) STAT3 modulates the DNA damage response pathway. Int J Exp Pathol 91:506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartalucci N, Calabresi L, Balliu M, Martinelli S, Rossi MC, Villeval JL, Annunziato F, Guglielmelli P, Vannucchi AM. (2017) Inhibitors of the PI3K/mTOR pathway prevent STAT5 phosphorylation in JAK2V617F mutated cells through PP2A/CIP2A axis. Oncotarget 8:96710–96724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton BE. (2006) STAT3: a potential therapeutic target in dendritic cells for the induction of transplant tolerance. Expert Opin Ther Targets 10:459–470. [DOI] [PubMed] [Google Scholar]
- Bates SH, Myers MG. (2004) The role of leptin-->STAT3 signaling in neuroendocrine function: an integrative perspective. J Mol Med (Berl) 82:12–20. [DOI] [PubMed] [Google Scholar]
- Bates SH, Stearns WH, Dundon TA, Schubert M, Tso AW, Wang Y, Banks AS, Lavery HJ, Haq AK, Maratos-Flier E, et al. (2003) STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature 421:856–859. [DOI] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Cancer Genome Project (2005) Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365:1054–1061. [DOI] [PubMed] [Google Scholar]
- Belloni L, Di Cocco S, Guerrieri F, Nunn ADG, Piconese S, Salerno D, Testoni B, Pulito C, Mori F, Pallocca M, et al. (2018) Targeting a phospho-STAT3-miRNAs pathway improves vesicular hepatic steatosis in an in vitro and in vivo model. Sci Rep 8:13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbarche S, Strassel C, Angénieux C, Mallo L, Freund M, Gachet C, Lanza F, de la Salle H. (2017) Dual role of IL-21 in megakaryopoiesis and platelet homeostasis. Haematologica 102:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell JC, Hong DS, Burris HA, III, Naing A, Jones SF, Falchook G, Bricmont P, Elekes A, Rock EP, Kurzrock R. (2014) Phase 1, open-label, dose-escalation, and pharmacokinetic study of STAT3 inhibitor OPB-31121 in subjects with advanced solid tumors. Cancer Chemother Pharmacol 74:125–130. [DOI] [PubMed] [Google Scholar]
- Benekli M, Baumann H, Wetzler M. (2009) Targeting signal transducer and activator of transcription signaling pathway in leukemias. J Clin Oncol 27:4422–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj U, Eckols TK, Xu X, Kasembeli MM, Chen Y, Adachi M, Song Y, Mo Q, Lai SY, Tweardy DJ. (2016a) Small-molecule inhibition of STAT3 in radioresistant head and neck squamous cell carcinoma. Oncotarget 7:26307–26330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj U, Kasembeli MM, Tweardy DJ. (2016b) STAT3 inhibitors in cancer: a comprehensive update, in STAT Inhibitors in Cancer (Ward AC. 95–161, Springer International Publishing, Cham, Switzerland. [Google Scholar]
- Bharadwaj U, Li M, Zhang R, Chen C, Yao Q. (2007) Elevated interleukin-6 and G-CSF in human pancreatic cancer cell conditioned medium suppress dendritic cell differentiation and activation. Cancer Res 67:5479–5488. [DOI] [PubMed] [Google Scholar]
- Bito T, Izu K, Tokura Y. (2010a) Evaluation of toxicity and Stat3 activation induced by hydrogen peroxide exposure to the skin in healthy individuals. J Dermatol Sci 58:157–159. [DOI] [PubMed] [Google Scholar]
- Bito T, Sumita N, Masaki T, Shirakawa T, Ueda M, Yoshiki R, Tokura Y, Nishigori C. (2010b) Ultraviolet light induces Stat3 activation in human keratinocytes and fibroblasts through reactive oxygen species and DNA damage. Exp Dermatol 19:654–660. [DOI] [PubMed] [Google Scholar]
- Bjørbaek C, El-Haschimi K, Frantz JD, Flier JS. (1999) The role of SOCS-3 in leptin signaling and leptin resistance. J Biol Chem 274:30059–30065. [DOI] [PubMed] [Google Scholar]
- Bocchini CE, Nahmod K, Katsonis P, Kim S, Kasembeli MM, Freeman A, Lichtarge O, Makedonas G, Tweardy DJ. (2016) Protein stabilization improves STAT3 function in autosomal dominant hyper-IgE syndrome. Blood 128:3061–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode JG, Gatsios P, Ludwig S, Rapp UR, Häussinger D, Heinrich PC, Graeve L. (1999) The mitogen-activated protein (MAP) kinase p38 and its upstream activator MAP kinase kinase 6 are involved in the activation of signal transducer and activator of transcription by hyperosmolarity. J Biol Chem 274:30222–30227. [DOI] [PubMed] [Google Scholar]
- Boengler K, Hilfiker-Kleiner D, Heusch G, Schulz R. (2010) Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res Cardiol 105:771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher S, Gerosa RC, Radpour R, Bauer J, Ampenberger F, Heikenwalder M, Kopf M, Manz MG. (2014) Endothelial cells translate pathogen signals into G-CSF-driven emergency granulopoiesis. Blood 124:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolli R, Stein AB, Guo Y, Wang OL, Rokosh G, Dawn B, Molkentin JD, Sanganalmath SK, Zhu Y, Xuan YT. (2011) A murine model of inducible, cardiac-specific deletion of STAT3: its use to determine the role of STAT3 in the upregulation of cardioprotective proteins by ischemic preconditioning. J Mol Cell Cardiol 50:589–597. [DOI] [PubMed] [Google Scholar]
- Bose P, Abou Zahr A, Verstovsek S. (2017) Investigational Janus kinase inhibitors in development for myelofibrosis. Expert Opin Investig Drugs 26:723–734. [DOI] [PubMed] [Google Scholar]
- Boulton TG, Zhong Z, Wen Z, Darnell JE, Jr., Stahl N, Yancopoulos GD. (1995) STAT3 activation by cytokines utilizing gp130 and related transducers involves a secondary modification requiring an H7-sensitive kinase. Proc Natl Acad Sci USA 92:6915–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman T, Garcia R, Turkson J, Jove R. (2000) STATs in oncogenesis. Oncogene 19:2474–2488. [DOI] [PubMed] [Google Scholar]
- Brambilla L, Genini D, Laurini E, Merulla J, Perez L, Fermeglia M, Carbone GM, Pricl S, Catapano CV. (2015) Hitting the right spot: mechanism of action of OPB-31121, a novel and potent inhibitor of the signal transducer and activator of transcription 3 (STAT3). Mol Oncol 9:1194–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breit A, Besik V, Solinski HJ, Muehlich S, Glas E, Yarwood SJ, Gudermann T. (2015) Serine-727 phosphorylation activates hypothalamic STAT-3 independently from tyrosine-705 phosphorylation. Mol Endocrinol 29:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock M, Trenkmann M, Gay RE, Michel BA, Gay S, Fischler M, Ulrich S, Speich R, Huber LC. (2009) Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res 104:1184–1191. [DOI] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. (1999) Stat3 as an oncogene. Cell 98:295–303. [DOI] [PubMed] [Google Scholar]
- Broudy VC, Kaushansky K. (1995) Thrombopoietin, the c-mpl ligand, is a major regulator of platelet production. J Leukoc Biol 57:719–725. [DOI] [PubMed] [Google Scholar]
- Bruce ES. (2016) Inhibition of interleukin-12 and/or -23 for the treatment of inflammatory bowel disease. Gastroenterol Hepatol (N Y) 12:784–786. [PMC free article] [PubMed] [Google Scholar]
- Buchert M, Burns CJ, Ernst M. (2016) Targeting JAK kinase in solid tumors: emerging opportunities and challenges. Oncogene 35:939–951. [DOI] [PubMed] [Google Scholar]
- Butturini E, Darra E, Chiavegato G, Cellini B, Cozzolino F, Monti M, Pucci P, Dell’Orco D, Mariotto S. (2014) S-Glutathionylation at Cys328 and Cys542 impairs STAT3 phosphorylation. ACS Chem Biol 9:1885–1893. [DOI] [PubMed] [Google Scholar]
- Byrne M, Savani B, Savona MR. (2018) Leveraging JAK-STAT regulation in myelofibrosis to improve outcomes with allogeneic hematopoietic stem-cell transplant. Ther Adv Hematol 9:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo M, Conde M, El Bekay R, Martín-Nieto J, Camacho MJ, Monteseirín J, Conde J, Bedoya FJ, Sobrino F. (1999) Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J Biol Chem 274:17580–17586. [DOI] [PubMed] [Google Scholar]
- Cecati M, Sartini D, Pozzi V, Giannubilo SR, Ferretti F, Stortoni P, Saccucci F, Tranquilli AL, Emanuelli M. (2013) Clues to apoptosis pathway involvement in hemolysis, elevated liver enzyme, and low platelet (HELLP) syndrome and intrauterine growth restriction (IUGR). J Matern Fetal Neonatal Med 26:26–31. [DOI] [PubMed] [Google Scholar]
- Cerchione C, Peluso I, Nappi D, Pareto AE, Picardi M, Martinelli V, Pane F. (2017) Ruxolitinib rechallenge in combination with hydroxyurea is effective in reverting cachexia and reducing blood transfusion demand and splenomegaly symptoms in a patient with primary myelofibrosis. Ann Hematol 96:697–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernkovich ER, Deng J, Bond MC, Combs TP, Harp JB. (2008) Adipose-specific disruption of signal transducer and activator of transcription 3 increases body weight and adiposity. Endocrinology 149:1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty D, Šumová B, Mallano T, Chen C-W, Distler A, Bergmann C, Ludolph I, Horch RE, Gelse K, Ramming A, et al. (2017) Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat Commun 8:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmin F, Mignot G, Bruchard M, Chevriaux A, Végran F, Hichami A, Ladoire S, Derangère V, Vincent J, Masson D, et al. (2012) Stat3 and Gfi-1 transcription factors control Th17 cell immunosuppressive activity via the regulation of ectonucleotidase expression. Immunity 36:362–373. [DOI] [PubMed] [Google Scholar]
- Chan CS, Ming-Lum A, Golds GB, Lee SJ, Anderson RJ, Mui ALF. (2012) Interleukin-10 inhibits lipopolysaccharide-induced tumor necrosis factor-α translation through a SHIP1-dependent pathway. J Biol Chem 287:38020–38027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KS, Carbajal S, Kiguchi K, Clifford J, Sano S, DiGiovanni J. (2004a) Epidermal growth factor receptor-mediated activation of Stat3 during multistage skin carcinogenesis. Cancer Res 64:2382–2389. [DOI] [PubMed] [Google Scholar]
- Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, DiGiovanni J. (2004b) Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest 114:720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Chattopadhyay D, Chakrabarti G. (2014) miR-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. PLoS One 9:e95716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Rudensky AY. (2013) Control of inflammation by integration of environmental cues by regulatory T cells. J Clin Invest 123:939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. (2009) CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, Jack RS, Wunderlich FT, Brüning JC, Müller W, et al. (2011) Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity 34:566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheers C, Haigh AM, Kelso A, Metcalf D, Stanley ER, Young AM. (1988) Production of colony-stimulating factors (CSFs) during infection: separate determinations of macrophage-, granulocyte-, granulocyte-macrophage-, and multi-CSFs. Infect Immun 56:247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Staudt LM, Green AR. (2012) Janus kinase deregulation in leukemia and lymphoma. Immunity 36:529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Fu L, Kong X, Xu J, Wang Z, Ma X, Akiyama Y, Chen Y, Fang J. (2014) Jumonji domain-containing protein 2B silencing induces DNA damage response via STAT3 pathway in colorectal cancer. Br J Cancer 110:1014–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Chen B. (2015) Siltuximab (CNTO 328): a promising option for human malignancies. Drug Des Devel Ther 9:3455–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Laurence A, O’Shea JJ. (2007) Signal transduction pathways and transcriptional regulation in the control of Th17 differentiation. Semin Immunol 19:400–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Lienlaf M, Wang HW, Perez-Villarroel P, Lee C, Woan K, Rock-Klotz J, Sahakian E, Woods D, Pinilla-Ibarz J, et al. (2014) A novel role for histone deacetylase 6 in the regulation of the tolerogenic STAT3/IL-10 pathway in APCs. J Immunol 193:2850–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wang HW, Cuenca A, Huang M, Ghansah T, Brayer J, Kerr WG, Takeda K, Akira S, Schoenberger SP, et al. (2003) A critical role for Stat3 signaling in immune tolerance. Immunity 19:425–436. [DOI] [PubMed] [Google Scholar]
- Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, Ortiz M, Nacken W, Sorg C, Vogl T, et al. (2008) Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med 205:2235–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon H, Yang J, Stark GR. (2011) The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J Interferon Cytokine Res 31:33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou WC, Levy DE, Lee CK. (2006) STAT3 positively regulates an early step in B-cell development. Blood 108:3005–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy DF, Hart KM, Borthwick LA, Shikotra A, Nagarkar DR, Siddiqui S, Jia G, Ohri CM, Doran E, Vannella KM, et al. (2015) TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med 7:301ra129. [DOI] [PubMed] [Google Scholar]
- Chung J, Uchida E, Grammer TC, Blenis J. (1997) STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol 17:6508–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YJ, Park BB, Kang YJ, Kim TM, Eaves CJ, Oh IH. (2006) Unique effects of Stat3 on the early phase of hematopoietic stem cell regeneration. Blood 108:1208–1215. [DOI] [PubMed] [Google Scholar]
- Colombel JF, Sandborn WJ, Rutgeerts P, Enns R, Hanauer SB, Panaccione R, Schreiber S, Byczkowski D, Li J, Kent JD, et al. (2007) Adalimumab for maintenance of clinical response and remission in patients with Crohn’s disease: the CHARM trial. Gastroenterology 132:52–65. [DOI] [PubMed] [Google Scholar]
- Coorens M, Rao A, Gräfe SK, Unelius D, Lindforss U, Agerberth B, Mjösberg J, Bergman P. (2019) Innate lymphoid cell type 3-derived interleukin-22 boosts lipocalin-2 production in intestinal epithelial cells via synergy between STAT3 and NF-κB. J Biol Chem 294:6027–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courapied S, Sellier H, de Carné Trécesson S, Vigneron A, Bernard AC, Gamelin E, Barré B, Coqueret O. (2010) The cdk5 kinase regulates the STAT3 transcription factor to prevent DNA damage upon topoisomerase I inhibition. J Biol Chem 285:26765–26778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. (2011) Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol 12:722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. (2011) An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity 35:792–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. (2013) Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci USA 110:11499–11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico A, Wu L. (2003) The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J Exp Med 198:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JEJ., Jr. (2002) Transcription factors as targets for cancer therapy. Nat Rev Cancer 2:740–749. [DOI] [PubMed] [Google Scholar]
- Darnowski JW, Goulette FA, Guan YJ, Chatterjee D, Yang ZF, Cousens LP, Chin YE. (2006) Stat3 cleavage by caspases: impact on full-length Stat3 expression, fragment formation, and transcriptional activity. J Biol Chem 281:17707–17717. [DOI] [PubMed] [Google Scholar]
- Dasgupta M, Dermawan JK, Willard B, Stark GR. (2015) STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA 112:3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta M, Unal H, Willard B, Yang J, Karnik SS, Stark GR. (2014) Critical role for lysine 685 in gene expression mediated by transcription factor unphosphorylated STAT3. J Biol Chem 289:30763–30771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R, Enkemann S, Jove R, Haura EB. (2005) Stat3 regulates genes common to both wound healing and cancer. Oncogene 24:3397–3408. [DOI] [PubMed] [Google Scholar]
- Deininger M, Radich J, Burn TC, Huber R, Paranagama D, Verstovsek S. (2015) The effect of long-term ruxolitinib treatment on JAK2p.V617F allele burden in patients with myelofibrosis. Blood 126:1551–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. (2008) Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev 22:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lartigue G, Barbier de la Serre C, Espero E, Lee J, Raybould HE. (2011) Diet-induced obesity leads to the development of leptin resistance in vagal afferent neurons. Am J Physiol Endocrinol Metab 301:E187–E195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries LCS, Wildenberg ME, De Jonge WJ, D’Haens GR. (2017) The future of Janus kinase inhibitors in inflammatory bowel disease [published correction appears in J Crohns Colitis (2017) 11:1028]. J Crohn’s Colitis 11:885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Li X, Teng R, Alnaeeli M, Chen Z, Rogers H, Noguchi CT. (2016) Erythropoietin regulates POMC expression via STAT3 and potentiates leptin response. J Mol Endocrinol 56:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding BB, Bi E, Chen H, Yu JJ, Ye BH. (2013) IL-21 and CD40L synergistically promote plasma cell differentiation through upregulation of Blimp-1 in human B cells. J Immunol 190:1827–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong SS, Guo Y, Zhu DL, Chen XF, Wu XM, Shen H, Chen XD, Tan LJ, Tian Q, Deng HW, et al. (2016) Epigenomic elements analyses for promoters identify ESRRG as a new susceptibility gene for obesity-related traits. Int J Obes 40:1170–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucette TA, Kong L-Y, Yang Y, Ferguson SD, Yang J, Wei J, Qiao W, Fuller GN, Bhat KP, Aldape K, et al. (2012) Signal transducer and activator of transcription 3 promotes angiogenesis and drives malignant progression in glioma. Neuro-oncol 14:1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drachman JG, Kaushansky K. (1997) Dissecting the thrombopoietin receptor: functional elements of the Mpl cytoplasmic domain. Proc Natl Acad Sci USA 94:2350–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drachman JG, Sabath DF, Fox NE, Kaushansky K. (1997) Thrombopoietin signal transduction in purified murine megakaryocytes. Blood 89:483–492. [PubMed] [Google Scholar]
- Dubový P, Hradilová-Svíženská I, Klusáková I, Kokošová V, Brázda V, Joukal M. (2018) Bilateral activation of STAT3 by phosphorylation at the tyrosine-705 (Y705) and serine-727 (S727) positions and its nuclear translocation in primary sensory neurons following unilateral sciatic nerve injury. Histochem Cell Biol 150:37–47. [DOI] [PubMed] [Google Scholar]
- Dunkel Y, Ong A, Notani D, Mittal Y, Lam M, Mi X, Ghosh P. (2012) STAT3 protein up-regulates Gα-interacting vesicle-associated protein (GIV)/Girdin expression, and GIV enhances STAT3 activation in a positive feedback loop during wound healing and tumor invasion/metastasis. J Biol Chem 287:41667–41683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, et al. (2010) Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham GA, Williams JJL, Nasim MT, Palmer TM. (2019) Targeting SOCS proteins to control JAK-STAT signalling in disease. Trends Pharmacol Sci 40:298–308. [DOI] [PubMed] [Google Scholar]
- El Kasmi KC, Smith AM, Williams L, Neale G, Panopoulos AD, Watowich SS, Häcker H, Foxwell BMJ, Murray PJ. (2007) Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway [published correction appears in J Immunol (2008) 180:3612]. J Immunol 179:7215–7219. [DOI] [PubMed] [Google Scholar]
- Enomoto D, Obana M, Miyawaki A, Maeda M, Nakayama H, Fujio Y. (2015) Cardiac-specific ablation of the STAT3 gene in the subacute phase of myocardial infarction exacerbated cardiac remodeling. Am J Physiol Heart Circ Physiol 309:H471–H480. [DOI] [PubMed] [Google Scholar]
- Erlich TH, Yagil Z, Kay G, Peretz A, Migalovich-Sheikhet H, Tshori S, Nechushtan H, Levi-Schaffer F, Saada A, Razin E. (2014) Mitochondrial STAT3 plays a major role in IgE-antigen-mediated mast cell exocytosis. J Allergy Clin Immunol 134:460–469. [DOI] [PubMed] [Google Scholar]
- Esashi E, Wang YH, Perng O, Qin XF, Liu YJ, Watowich SS. (2008) The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity 28:509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EU Clinical Trials Register (2017) Phase I/II Open-Label Study to Evaluate the Safety, Tolerability and Recommended Phase II Dose (RP2D) of GLG-801 in patients with Advanced Solid Tumors (Phase I); and safety, tolerability and anticancer activity of GLG-801 in patients with Metastatic TNBC (Phase II)., 2017-005057-36 A
- Ezumi Y, Shindoh K, Tsuji M, Takayama H. (1998) Physical and functional association of the Src family kinases Fyn and Lyn with the collagen receptor glycoprotein VI-Fc receptor gamma chain complex on human platelets. J Exp Med 188:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy JV. (2009) Eosinophilic and neutrophilic inflammation in asthma: insights from clinical studies. Proc Am Thorac Soc 6:256–259. [DOI] [PubMed] [Google Scholar]
- Farivar AS, Merry HE, Fica-Delgado MJ, McCourtie AS, Mackinnon-Patterson BC, Mulligan MS. (2006) Interleukin-6 regulation of direct lung ischemia reperfusion injury. Ann Thorac Surg 82:472–478. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, Matarese G, Lord GM, Keogh JM, Lawrence E, Agwu C, Sanna V, Jebb SA, Perna F, Fontana S, et al. (2002) Beneficial effects of leptin on obesity, T cell hyporesponsiveness, and neuroendocrine/metabolic dysfunction of human congenital leptin deficiency. J Clin Invest 110:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yu M, Zhu F, Zhang S, Ding P, Wang M. (2018) IL-9 promotes the development of deep venous thrombosis by facilitating platelet function. Thromb Haemost 118:1885–1894. [DOI] [PubMed] [Google Scholar]
- Ferrao R, Lupardus PJ. (2017) The Janus kinase (JAK) FERM and SH2 domains: bringing specificity to JAK-receptor interactions. Front Endocrinol (Lausanne) 8:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmbach-Kraft I, Byers M, Shows T, Dalla-Favera R, Krolewski JJ. (1990) tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene 5:1329–1336. [PubMed] [Google Scholar]
- Fisher DAC, Miner CA, Engle EK, Hu H, Collins TB, Zhou A, Allen MJ, Malkova ON, Oh ST. (2019) Cytokine production in myelofibrosis exhibits differential responsiveness to JAK-STAT, MAP kinase, and NFκB signaling. Leukemia 33:1978–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornek JL, Tygrett LT, Waldschmidt TJ, Poli V, Rickert RC, Kansas GS. (2006) Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood 107:1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DA, Mahajan S, Ritz J. (1997) B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest 100:3140–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca D, Ferracci F, Diaz A, Romero M, Lechner S, Blomberg BB. (2016) Obesity decreases B cell responses in young and elderly individuals. Obesity (Silver Spring) 24:615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu AK, Fu WY, Ng AK, Chien WW, Ng YP, Wang JH, Ip NY. (2004) Cyclin-dependent kinase 5 phosphorylates signal transducer and activator of transcription 3 and regulates its transcriptional activity. Proc Natl Acad Sci USA 101:6728–6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. (1996) Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 5:449–460. [DOI] [PubMed] [Google Scholar]
- Fukushima A, Loh K, Galic S, Fam B, Shields B, Wiede F, Tremblay ML, Watt MJ, Andrikopoulos S, Tiganis T. (2010) T-cell protein tyrosine phosphatase attenuates STAT3 and insulin signaling in the liver to regulate gluconeogenesis. Diabetes 59:1906–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A, Grivennikov SI, Do MV, Stumpo DJ, Blackshear PJ, Karin M. (2012) Cutting edge: IL-10-mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production. J Immunol 189:2089–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen SL. (2009) Structure and signalling in the IL-17 receptor family. Nat Rev Immunol 9:556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen SL. (2011) Recent advances in the IL-17 cytokine family. Curr Opin Immunol 23:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Li FS, Chen XH, Liu QW, Feng JB, Liu QJ, Su X. (2014) Radiation induces phosphorylation of STAT3 in a dose- and time-dependent manner. Asian Pac J Cancer Prev 15:6161–6164. [DOI] [PubMed] [Google Scholar]
- Gao Q, Wolfgang MJ, Neschen S, Morino K, Horvath TL, Shulman GI, Fu XY. (2004) Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc Natl Acad Sci USA 101:4661–4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garama DJ, Harris TJ, White CL, Rossello FJ, Abdul-Hay M, Gough DJ, Levy DE. (2015) A synthetic lethal interaction between glutathione synthesis and mitochondrial reactive oxygen species provides a tumor-specific vulnerability dependent on STAT3. Mol Cell Biol 35:3646–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garama DJ, White CL, Balic JJ, Gough DJ. (2016) Mitochondrial STAT3: powering up a potent factor. Cytokine 87:20–25. [DOI] [PubMed] [Google Scholar]
- Garcia AA, Hays JL, Cote GM, Becerra C, Langleben A, Lau SK, Roman LD, McCormick CC, Richards DA, Braiteh FS, et al. (2016) A phase Ib/II study of cancer stemness inhibitor napabucasin (BB608) combined with weekly paclitaxel in platinum-resistant ovarian cancer. J Clin Oncol 34:5578. [Google Scholar]
- Gatsios P, Terstegen L, Schliess F, Häussinger D, Kerr IM, Heinrich PC, Graeve L. (1998) Activation of the Janus kinase/signal transducer and activator of transcription pathway by osmotic shock. J Biol Chem 273:22962–22968. [DOI] [PubMed] [Google Scholar]
- Gavino AC, Nahmod K, Bharadwaj U, Makedonas G, Tweardy DJ. (2016) STAT3 inhibition prevents lung inflammation, remodeling, and accumulation of Th2 and Th17 cells in a murine asthma model. Allergy 71:1684–1692. [DOI] [PubMed] [Google Scholar]
- Genini D, Brambilla L, Laurini E, Merulla J, Civenni G, Pandit S, D’Antuono R, Perez L, Levy DE, Pricl S, et al. (2017) Mitochondrial dysfunction induced by a SH2 domain-targeting STAT3 inhibitor leads to metabolic synthetic lethality in cancer cells. Proc Natl Acad Sci USA 114:E4924–E4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, O’Shea JJ. (2009) Janus kinases in immune cell signaling. Immunol Rev 228:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannandrea M, Parks WC. (2014) Diverse functions of matrix metalloproteinases during fibrosis. Dis Model Mech 7:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Hemann MT. (2010) DNA damage-mediated induction of a chemoresistant niche. Cell 143:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkouveris I, Nikitakis N, Karanikou M, Rassidakis G, Sklavounou A. (2016) JNK1/2 expression and modulation of STAT3 signaling in oral cancer. Oncol Lett 12:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollob JA, Schnipper CP, Murphy EA, Ritz J, Frank DA. (1999) The functional synergy between IL-12 and IL-2 involves p38 mitogen-activated protein kinase and is associated with the augmentation of STAT serine phosphorylation. J Immunol 162:4472–4481. [PubMed] [Google Scholar]
- Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. (2009) Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science 324:1713–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant L, Shearer KD, Czopek A, Lees EK, Owen C, Agouni A, Workman J, Martin-Granados C, Forrester JV, Wilson HM, et al. (2014) Myeloid-cell protein tyrosine phosphatase-1B deficiency in mice protects against high-fat diet and lipopolysaccharide-induced inflammation, hyperinsulinemia, and endotoxemia through an IL-10 STAT3-dependent mechanism. Diabetes 63:456–470. [DOI] [PubMed] [Google Scholar]
- Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, et al. (2006) Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res 12:11–19. [DOI] [PubMed] [Google Scholar]
- Grohmann M, Wiede F, Dodd GT, Gurzov EN, Ooi GJ, Butt T, Rasmiena AA, Kaur S, Gulati T, Goh PK, et al. (2018) Obesity drives STAT-1-dependent NASH and STAT-3-dependent HCC. Cell 175:1289–1306.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozovsky R, Giannini S, Falet H, Hoffmeister KM. (2015) Novel mechanisms of platelet clearance and thrombopoietin regulation. Curr Opin Hematol 22:445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadagnin E, Mázala D, Chen YW. (2018) STAT3 in skeletal muscle function and disorders. Int J Mol Sci 19:2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Wang C, Wang Q, Qiao Z, Tang H. (2017) Pantoprazole blocks the JAK2/STAT3 pathway to alleviate skeletal muscle wasting in cancer cachexia by inhibiting inflammatory response. Oncotarget 8:39640–39648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurzov EN, Stanley WJ, Pappas EG, Thomas HE, Gough DJ. (2016) The JAK/STAT pathway in obesity and diabetes. FEBS J 283:3002–3015. [DOI] [PubMed] [Google Scholar]
- Habiel DM, Hogaboam C. (2014) Heterogeneity in fibroblast proliferation and survival in idiopathic pulmonary fibrosis. Front Pharmacol 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanauer SB, Feagan BG, Lichtenstein GR, Mayer LF, Schreiber S, Colombel JF, Rachmilewitz D, Wolf DC, Olson A, Bao W, et al. ACCENT I Study Group (2002) Maintenance infliximab for Crohn’s disease: the ACCENT I randomised trial. Lancet 359:1541–1549. [DOI] [PubMed] [Google Scholar]
- Hanauer SB, Sandborn WJ, Rutgeerts P, Fedorak RN, Lukas M, MacIntosh D, Panaccione R, Wolf D, Pollack P. (2006) Human anti-tumor necrosis factor monoclonal antibody (adalimumab) in Crohn’s disease: the CLASSIC-I trial. Gastroenterology 130:323–333, quiz 591. [DOI] [PubMed] [Google Scholar]
- Haricharan S, Li Y. (2014) STAT signaling in mammary gland differentiation, cell survival and tumorigenesis. Mol Cell Endocrinol 382:560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpur AG, Andres AC, Ziemiecki A, Aston RR, Wilks AF. (1992) JAK2, a third member of the JAK family of protein tyrosine kinases. Oncogene 7:1347–1353. [PubMed] [Google Scholar]
- Harrison C, Kiladjian JJ, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V, McQuitty M, Hunter DS, Levy R, Knoops L, et al. (2012) JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med 366:787–798. [DOI] [PubMed] [Google Scholar]
- Hayakawa F, Sugimoto K, Harada Y, Hashimoto N, Ohi N, Kurahashi S, Naoe T. (2013) A novel STAT inhibitor, OPB-31121, has a significant antitumor effect on leukemia with STAT-addictive oncokinases. Blood Cancer J 3:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, Shanker S, Ferrajoli A, Keating MJ, Estrov Z. (2010) STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood 115:2852–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht SS. (2003) Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer 3:733–744. [DOI] [PubMed] [Google Scholar]
- Hegde S, Ni S, He S, Yoon D, Feng GS, Watowich SS, Paulson RF, Hankey PA. (2009) Stat3 promotes the development of erythroleukemia by inducing Pu.1 expression and inhibiting erythroid differentiation. Oncogene 28:3349–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberger S, Zinzalla G, Antonow D, Essex S, Basu BP, Palmer J, Husby J, Jackson PJ, Rahman KM, Wilderspin AF, et al. (2013) Investigation of the protein alkylation sites of the STAT3:STAT3 inhibitor Stattic by mass spectrometry. Bioorg Med Chem Lett 23:4719–4722. [DOI] [PubMed] [Google Scholar]
- Heldsinger A, Grabauskas G, Wu X, Zhou S, Lu Y, Song I, Owyang C. (2014) Ghrelin induces leptin resistance by activation of suppressor of cytokine signaling 3 expression in male rats: implications in satiety regulation. Endocrinology 155:3956–3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppler LN, Frank DA. (2017) Targeting oncogenic transcription factors: therapeutic implications of endogenous STAT inhibitors. Trends Cancer 3:816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrlinger KR, Diculescu M, Fellermann K, Hartmann H, Howaldt S, Nikolov R, Petrov A, Reindl W, Otte JM, Stoynov S, et al. (2013) Efficacy, safety and tolerability of vidofludimus in patients with inflammatory bowel disease: the ENTRANCE study. J Crohn’s Colitis 7:636–643. [DOI] [PubMed] [Google Scholar]
- Herrmann Hua Yu A. (2014) STAT3 nuclear egress requires exportin 7 via engaging lysine acetylation. MOJ Cell Sci Rep 1:9‒15. [Google Scholar]
- Hierholzer C, Harbrecht B, Menezes JM, Kane J, MacMicking J, Nathan CF, Peitzman AB, Billiar TR, Tweardy DJ. (1998a) Essential role of induced nitric oxide in the initiation of the inflammatory response after hemorrhagic shock. J Exp Med 187:917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierholzer C, Kalff JC, Billiar TR, Tweardy DJ. (1998b) Activation of STAT proteins in the lung of rats following resuscitation from hemorrhagic shock. Arch Orthop Trauma Surg 117:372–375. [DOI] [PubMed] [Google Scholar]
- Hierholzer C, Kalff JC, Omert L, Tsukada K, Loeffert JE, Watkins SC, Billiar TR, Tweardy DJ. (1998c) Interleukin-6 production in hemorrhagic shock is accompanied by neutrophil recruitment and lung injury. Am J Physiol 275:L611–L621. [DOI] [PubMed] [Google Scholar]
- Hierholzer C, Menezes JM, Ungeheuer A, Billiar TR, Tweardy DJ, Harbrecht BG. (2002) A nitric oxide scavenger protects against pulmonary inflammation following hemorrhagic shock. Shock 17:98–103. [DOI] [PubMed] [Google Scholar]
- Hilfiker-Kleiner D, Hilfiker A, Fuchs M, Kaminski K, Schaefer A, Schieffer B, Hillmer A, Schmiedl A, Ding Z, Podewski E, et al. (2004) Signal transducer and activator of transcription 3 is required for myocardial capillary growth, control of interstitial matrix deposition, and heart protection from ischemic injury. Circ Res 95:187–195. [DOI] [PubMed] [Google Scholar]
- Hilfiker-Kleiner D, Shukla P, Klein G, Schaefer A, Stapel B, Hoch M, Müller W, Scherr M, Theilmeier G, Ernst M, et al. (2010) Continuous glycoprotein-130-mediated signal transducer and activator of transcription-3 activation promotes inflammation, left ventricular rupture, and adverse outcome in subacute myocardial infarction. Circulation 122:145–155. [DOI] [PubMed] [Google Scholar]
- Hill HR, Ochs HD, Quie PG, Clark RA, Pabst HF, Klebanoff SJ, Wedgwood RJ. (1974) Defect in neutrophil granulocyte chemotaxis in Job’s syndrome of recurrent “cold” staphylococcal abscesses. Lancet 2:617–619. [DOI] [PubMed] [Google Scholar]
- Hill HR, Quie PG. (1974) Raised serum-IgE levels and defective neutrophil chemotaxis in three children with eczema and recurrent bacterial infections. Lancet 1:183–187. [DOI] [PubMed] [Google Scholar]
- Hillmer EJ, Zhang H, Li HS, Watowich SS. (2016) STAT3 signaling in immunity. Cytokine Growth Factor Rev 31:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG. (2006) C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol 7:732–739. [DOI] [PubMed] [Google Scholar]
- Ho JN, Kang GY, Lee SS, Kim J, Bae IH, Hwang SG, Um HD. (2010) Bcl-XL and STAT3 mediate malignant actions of gamma-irradiation in lung cancer cells. Cancer Sci 101:1417–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister KM, Falet H. (2016) Platelet clearance by the hepatic Ashwell-Morrell receptor: mechanisms and biological significance. Thromb Res 141 (Suppl 2):S68–S72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokuto I, Ikegami M, Yoshida M, Takeda K, Akira S, Perl AKT, Hull WM, Wert SE, Whitsett JA. (2004) Stat-3 is required for pulmonary homeostasis during hyperoxia. J Clin Invest 113:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, et al. (2007) STAT3 mutations in the hyper-IgE syndrome. N Engl J Med 357:1608–1619. [DOI] [PubMed] [Google Scholar]
- Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, Fowler N, Zhou T, Schmidt J, Jo M, et al. (2015) AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med 7:314ra185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DS, Kurzrock R, Supko JG, He X, Naing A, Wheler J, Lawrence D, Eder JP, Meyer CJ, Ferguson DA, et al. (2012) A phase I first-in-human trial of bardoxolone methyl in patients with advanced solid tumors and lymphomas. Clin Cancer Res 18:3396–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi N, Wang L, Mukhopadhyay P, Park O, Jeong WI, Lafdil F, Osei-Hyiaman D, Moh A, Fu XY, Pacher P, et al. (2008) Cell type-dependent pro- and anti-inflammatory role of signal transducer and activator of transcription 3 in alcoholic liver injury. Gastroenterology 134:1148–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou SX, Zheng Z, Chen X, Perrimon N. (2002) The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev Cell 3:765–778. [DOI] [PubMed] [Google Scholar]
- Hou T, Ray S, Lee C, Brasier AR. (2008) The STAT3 NH2-terminal domain stabilizes enhanceosome assembly by interacting with the p300 bromodomain. J Biol Chem 283:30725–30734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard JK, Cave BJ, Oksanen LJ, Tzameli I, Bjørbaek C, Flier JS. (2004) Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nat Med 10:734–738. [DOI] [PubMed] [Google Scholar]
- Hox V, O’Connell MP, Lyons JJ, Sackstein P, Dimaggio T, Jones N, Nelson C, Boehm M, Holland SM, Freeman AF, et al. (2016) Diminution of signal transducer and activator of transcription 3 signaling inhibits vascular permeability and anaphylaxis. J Allergy Clin Immunol 138:187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu FB, Meigs JB, Li TY, Rifai N, Manson JE. (2004) Inflammatory markers and risk of developing type 2 diabetes in women. Diabetes 53:693–700. [DOI] [PubMed] [Google Scholar]
- Hu W, Lv J, Han M, Yang Z, Li T, Jiang S, Yang Y. (2018) STAT3: the art of multi-tasking of metabolic and immune functions in obesity. Prog Lipid Res 70:17–28. [DOI] [PubMed] [Google Scholar]
- Hua H, Yang J, Lin H, Xi Y, Dai M, Xu G, Wang F, Liu L, Zhao T, Huang J, et al. (2018) PPARα-independent action against metabolic syndrome development by fibrates is mediated by inhibition of STAT3 signalling. J Pharm Pharmacol 70:1630–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Chen M, Shen Y, Shen W, Guo H, Gao Q, Zou X. (2012) Inhibition of activated Stat3 reverses drug resistance to chemotherapeutic agents in gastric cancer cells. Cancer Lett 315:198–205. [DOI] [PubMed] [Google Scholar]
- Hubbard JM, Grothey A. (2017) Napabucasin: an update on the first-in-class cancer stemness inhibitor. Drugs 77:1091–1103. [DOI] [PubMed] [Google Scholar]
- Hutchins AP, Diez D, Miranda-Saavedra D. (2013) The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges. Brief Funct Genomics 12:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins AP, Poulain S, Miranda-Saavedra D. (2012) Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages. Blood 119:e110–e119. [DOI] [PubMed] [Google Scholar]
- Icardi L, Mori R, Gesellchen V, Eyckerman S, De Cauwer L, Verhelst J, Vercauteren K, Saelens X, Meuleman P, Leroux-Roels G, et al. (2012) The Sin3a repressor complex is a master regulator of STAT transcriptional activity. Proc Natl Acad Sci USA 109:12058–12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K, Leonard WJ. (2000) The Jak-STAT pathway. Mol Immunol 37:1–11. [DOI] [PubMed] [Google Scholar]
- Inoue H. (2016) Central insulin-mediated regulation of hepatic glucose production [Review]. Endocr J 63:1–7. [DOI] [PubMed] [Google Scholar]
- Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, Hashimoto N, Kido Y, Mori T, Sakaue H, et al. (2004) Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med 10:168–174. [DOI] [PubMed] [Google Scholar]
- Ito H, Takazoe M, Fukuda Y, Hibi T, Kusugami K, Andoh A, Matsumoto T, Yamamura T, Azuma J, Nishimoto N, et al. (2004) A pilot randomized trial of a human anti-interleukin-6 receptor monoclonal antibody in active Crohn’s disease. Gastroenterology 126:989–996; discussion 947. [DOI] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. (2006) The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126:1121–1133. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Kovacic JC, Olive M, Beers JK, Yoshimoto T, Crook MF, Tonelli LH, Nabel EG. (2010) Disruption of protein arginine N-methyltransferase 2 regulates leptin signaling and produces leanness in vivo through loss of STAT3 methylation. Circ Res 107:992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby JJ, Kalinowski A, Liu MG, Zhang SSM, Gao Q, Chai GX, Ji L, Iwamoto Y, Li E, Schneider M, et al. (2003) Cardiomyocyte-restricted knockout of STAT3 results in higher sensitivity to inflammation, cardiac fibrosis, and heart failure with advanced age. Proc Natl Acad Sci USA 100:12929–12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Zhang T, Kee WH, Li W, Cao X. (1999) Protein kinase C delta associates with and phosphorylates Stat3 in an interleukin-6-dependent manner. J Biol Chem 274:24392–24400. [DOI] [PubMed] [Google Scholar]
- Jatoi A. (2013) Anti-inflammatory therapy: exploring exercise, serum-derived bovine immunoglobulin/protein isolates, and ruxolitinib for the cancer-associated weight loss syndrome. Curr Opin Support Palliat Care 7:339–341. [DOI] [PubMed] [Google Scholar]
- Johansen LM, Iwama A, Lodie TA, Sasaki K, Felsher DW, Golub TR, Tenen DG. (2001) c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol 21:3789–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DE, O’Keefe RA, Grandis JR. (2018) Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 15:234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker DJ, Stephenson J, Edenfield WJ, Supko JG, Li Y, Li W, Hitron M, Leggett D, Kerstein D, Li C. (2014) A phase I extension study of BBI608, a first-in-class cancer stem cell (CSC) inhibitor, in patients with advanced solid tumors. J Clin Oncol 32:2546. [Google Scholar]
- Jorgensen SB, O’Neill HM, Sylow L, Honeyman J, Hewitt KA, Palanivel R, Fullerton MD, Öberg L, Balendran A, Galic S, et al. (2013) Deletion of skeletal muscle SOCS3 prevents insulin resistance in obesity. Diabetes 62:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. International IBD Genetics Consortium (IIBDGC) (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Yoo W, Stevenson HL, Deshpande D, Shen H, Gagea M, Yoo SY, Wang J, Eckols TK, Bharadwaj U, et al. (2017) Multifunctional effects of a small-molecule STAT3 inhibitor on NASH and hepatocellular carcinoma in mice. Clin Cancer Res 23:5537–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabra DG, Pfuhlmann K, García-Cáceres C, Schriever SC, Casquero García V, Kebede AF, Fuente-Martin E, Trivedi C, Heppner K, Uhlenhaut NH, et al. (2016) Hypothalamic leptin action is mediated by histone deacetylase 5. Nat Commun 7:10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech SM, Cui W. (2012) Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalupahana NS, Moustaid-Moussa N, Claycombe KJ. (2012) Immunity as a link between obesity and insulin resistance. Mol Aspects Med 33:26–34. [DOI] [PubMed] [Google Scholar]
- Kan C, Yang F, Wang S. (2018) SHP2-mediated signal networks in stem cell homeostasis and dysfunction. Stem Cells Int 2018:8351374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Seo HA, Go Y, Oh CJ, Jeoung NH, Park KG, Lee IK. (2013) Dimethylfumarate suppresses adipogenic differentiation in 3T3-L1 preadipocytes through inhibition of STAT3 activity. PLoS One 8:e61411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. (2012) Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity, Annu Rev Immunol 30, pp 707–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasembeli MM, Bharadwaj U, Robinson P, Tweardy DJ. (2018) Contribution of STAT3 to inflammatory and fibrotic diseases and prospects for its targeting for treatment. Int J Mol Sci 19:2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenstein ALA, Myers JL. (1998) Idiopathic pulmonary fibrosis: clinical relevance of pathologic classification. Am J Respir Crit Care Med 157:1301–1315. [DOI] [PubMed] [Google Scholar]
- Kaushansky K. (1995) Thrombopoietin: the primary regulator of platelet production. Blood 86:419–431. [PubMed] [Google Scholar]
- Kaushik N, Kim MJ, Kim RK, Kumar Kaushik N, Seong KM, Nam SY, Lee SJ. (2017) Low-dose radiation decreases tumor progression via the inhibition of the JAK1/STAT3 signaling axis in breast cancer cell lines. Sci Rep 7:43361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw NJ, Murphy JM, Liau NP, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ. (2013) SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol 20:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner NM, Vijayaraghavan S, Durak MG, Bui T, Kohansal M, Ha MJ, Liu B, Rao X, Wang J, Yi M, et al. (2019) Combined inhibition of STAT3 and DNA repair in palbociclib-resistant ER-positive breast cancer. Clin Cancer Res 25:3996–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MW, Saadalla A, Ewida AH, Al-Katranji K, Al-Saoudi G, Giaccone ZT, Gounari F, Zhang M, Frank DA, Khazaie K. (2018) The STAT3 inhibitor pyrimethamine displays anti-cancer and immune stimulatory effects in murine models of breast cancer. Cancer Immunol Immunother 67:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ki SH, Park O, Zheng M, Morales-Ibanez O, Kolls JK, Bataller R, Gao B. (2010) Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology 52:1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. (2013a) Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23:839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Won JS, Singh AK, Sharma AK, Singh I. (2014) STAT3 regulation by S-nitrosylation: implication for inflammatory disease. Antioxid Redox Signal 20:2514–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Yoon MS, Chen J. (2009) Signal transducer and activator of transcription 3 (STAT3) mediates amino acid inhibition of insulin signaling through serine 727 phosphorylation. J Biol Chem 284:35425–35432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Jeong H, Yang MS, Lim CW, Kim B. (2017a) Therapeutic effects of zerumbone in an alkali-burned corneal wound healing model. Int Immunopharmacol 48:126–134. [DOI] [PubMed] [Google Scholar]
- Kim KS, Seu YB, Baek SH, Kim MJ, Kim KJ, Kim JH, Kim JR. (2007) Induction of cellular senescence by insulin-like growth factor binding protein-5 through a p53-dependent mechanism. Mol Biol Cell 18:4543–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Song K, Kim YS. (2017b) Alantolactone improves prolonged exposure of interleukin-6-induced skeletal muscle inflammation associated glucose intolerance and insulin resistance. Front Pharmacol 8:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Nam HJ, Kim HP, Han SW, Im SA, Kim TY, Oh DY, Bang YJ. (2013b) OPB-31121, a novel small molecular inhibitor, disrupts the JAK2/STAT3 pathway and exhibits an antitumor activity in gastric cancer cells. Cancer Lett 335:145–152. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Pham TH, Bak Y, Ryu HW, Oh SR, Yoon DY. (2018) Orientin inhibits invasion by suppressing MMP-9 and IL-8 expression via the PKCα/ERK/AP-1/STAT3-mediated signaling pathways in TPA-treated MCF-7 breast cancer cells. Phytomedicine 50:35–42. [DOI] [PubMed] [Google Scholar]
- Kinjyo I, Inoue H, Hamano S, Fukuyama S, Yoshimura T, Koga K, Takaki H, Himeno K, Takaesu G, Kobayashi T, et al. (2006) Loss of SOCS3 in T helper cells resulted in reduced immune responses and hyperproduction of interleukin 10 and transforming growth factor-β 1. J Exp Med 203:1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura H, Kamon H, Sawa S, Park SJ, Katunuma N, Ishihara K, Murakami M, Hirano T. (2005) IL-6-STAT3 controls intracellular MHC class II alphabeta dimer level through cathepsin S activity in dendritic cells. Immunity 23:491–502. [DOI] [PubMed] [Google Scholar]
- Kiu H, Nicholson SE. (2012) Biology and significance of the JAK/STAT signalling pathways. Growth Factors 30:88–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JD, Sano D, Sen M, Myers JN, Grandis JR, Kim S. (2014) STAT3 oligonucleotide inhibits tumor angiogenesis in preclinical models of squamous cell carcinoma. PLoS One 9:e81819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe M, Koche R, Zou L, van Galen P, Hill CE, Dong L, De Groote S, Papalexi E, Hanasoge Somasundara AV, Cordner K, et al. (2018) Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell 33:29–43.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe M, Kwak M, Koppikar P, Riester M, Keller M, Bastian L, Hricik T, Bhagwat N, McKenney AS, Papalexi E, et al. (2015) JAK-STAT pathway activation in malignant and nonmalignant cells contributes to MPN pathogenesis and therapeutic response. Cancer Discov 5:316–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Inoue T, Kunimoto H, Nakajima K. (2013) IL-6-STAT3 signaling and premature senescence. JAK-STAT 2:e25763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H, Sasaki T, Ishitani T, Iemura S, Zhao H, Kaneko S, Kunimoto H, Natsume T, Matsumoto K, Nakajima K. (2005) STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc Natl Acad Sci USA 102:4524–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltes DA, Spurlock ME, Spurlock DM. (2017) Adipose triglyceride lipase protein abundance and translocation to the lipid droplet increase during leptin-induced lipolysis in bovine adipocytes. Domest Anim Endocrinol 61:62–76. [DOI] [PubMed] [Google Scholar]
- Komyod W, Bauer UM, Heinrich PC, Haan S, Behrmann I. (2005) Are STATS arginine-methylated? J Biol Chem 280:21700–21705. [DOI] [PubMed] [Google Scholar]
- Kondo K, Shaim H, Thompson PA, Burger JA, Keating M, Estrov Z, Harris D, Kim E, Ferrajoli A, Daher M, et al. (2018) Ibrutinib modulates the immunosuppressive CLL microenvironment through STAT3-mediated suppression of regulatory B-cell function and inhibition of the PD-1/PD-L1 pathway. Leukemia 32:960–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotyla PJ. (2018) Are Janus kinase inhibitors superior over classic biologic agents in RA patients? BioMed Res Int 2018:7492904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarik P, Stoiber D, Eyers PA, Menghini R, Neininger A, Gaestel M, Cohen P, Decker T. (1999) Stress-induced phosphorylation of STAT1 at Ser727 requires p38 mitogen-activated protein kinase whereas IFN-gamma uses a different signaling pathway. Proc Natl Acad Sci USA 96:13956–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer AH, Edkins AL, Hoppe HC, Prinsloo E. (2015) Dynamic mitochondrial localisation of STAT3 in the cellular adipogenesis model 3T3-L1. J Cell Biochem 116:1232–1240. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Mariño G, Levine B. (2010) Autophagy and the integrated stress response. Mol Cell 40:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroy DC, Beraza N, Tschaharganeh DF, Sander LE, Erschfeld S, Giebeler A, Liedtke C, Wasmuth HE, Trautwein C, Streetz KL. (2010) Lack of interleukin-6/glycoprotein 130/signal transducers and activators of transcription-3 signaling in hepatocytes predisposes to liver steatosis and injury in mice. Hepatology 51:463–473. [DOI] [PubMed] [Google Scholar]
- Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, Florens L, Washburn MP, Collazo-Lorduy A, Castillo-Martin M, et al. (2017) PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature 546:554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn C, McDonald JA. (1991) The roles of the myofibroblast in idiopathic pulmonary fibrosis: ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am J Pathol 138:1257–1265. [PMC free article] [PubMed] [Google Scholar]
- Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. (1993) Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75:263–274. [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. (2010) The essence of senescence. Genes Dev 24:2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. (2008) Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133:1019–1031. [DOI] [PubMed] [Google Scholar]
- Kurdi M, Booz GW. (2007) Can the protective actions of JAK-STAT in the heart be exploited therapeutically? Parsing the regulation of interleukin-6-type cytokine signaling. J Cardiovasc Pharmacol 50:126–141. [DOI] [PubMed] [Google Scholar]
- Kurdi M, Booz GW. (2010) Deciphering STAT3 signaling in the heart: plasticity and vascular inflammation. Congest Heart Fail 16:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdi M, Sivakumaran V, Duhé RJ, Aon MA, Paolocci N, Booz GW. (2012) Depletion of cellular glutathione modulates LIF-induced JAK1-STAT3 signaling in cardiac myocytes. Int J Biochem Cell Biol 44:2106–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuter DJ, Bain B, Mufti G, Bagg A, Hasserjian RP. (2007) Bone marrow fibrosis: pathophysiology and clinical significance of increased bone marrow stromal fibres. Br J Haematol 139:351–362. [DOI] [PubMed] [Google Scholar]
- Kvasnicka HM, Thiele J, Bueso-Ramos CE, Sun W, Cortes J, Kantarjian HM, Verstovsek S. (2018) Long-term effects of ruxolitinib versus best available therapy on bone marrow fibrosis in patients with myelofibrosis. J Hematol Oncol 11:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffargue M, Ragab-Thomas JM, Ragab A, Tuech J, Missy K, Monnereau L, Blank U, Plantavid M, Payrastre B, Raynal P, et al. (1999) Phosphoinositide 3-kinase and integrin signalling are involved in activation of Bruton tyrosine kinase in thrombin-stimulated platelets. FEBS Lett 443:66–70. [DOI] [PubMed] [Google Scholar]
- Lam E, Choi SH, Pareek TK, Kim BG, Letterio JJ. (2015) Cyclin-dependent kinase 5 represses Foxp3 gene expression and Treg development through specific phosphorylation of Stat3 at Serine 727. Mol Immunol 67:317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. (2014) Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laouar Y, Welte T, Fu XY, Flavell RA. (2003) STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity 19:903–912. [DOI] [PubMed] [Google Scholar]
- Lau J, Ilkhanizadeh S, Wang S, Miroshnikova YA, Salvatierra NA, Wong RA, Schmidt C, Weaver VM, Weiss WA, Persson AI. (2015) STAT3 blockade inhibits radiation-induced malignant progression in glioma. Cancer Res 75:4302–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubner K, Kieffer TJ, Lam NT, Niu X, Jakob F, Seufert J. (2005) Inhibition of preproinsulin gene expression by leptin induction of suppressor of cytokine signaling 3 in pancreatic β-cells. Diabetes 54:3410–3417. [DOI] [PubMed] [Google Scholar]
- Laurence A, Amarnath S, Mariotti J, Kim YC, Foley J, Eckhaus M, O’Shea JJ, Fowler DH. (2012) STAT3 transcription factor promotes instability of nTreg cells and limits generation of iTreg cells during acute murine graft-versus-host disease. Immunity 37:209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, et al. (2007) Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26:371–381. [DOI] [PubMed] [Google Scholar]
- Laurie SA, Jonker DJ, Edenfield WJ, Stephenson J, Keller D, Hitron M, Li W, Li Y, Gada K, Gao Y, et al. (2014) A phase 1 dose-escalation study of BBI503, a first-in-class cancer stemness kinase inhibitor in adult patients with advanced solid tumors. J Clin Oncol 32:2527. [Google Scholar]
- Lee CK, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, DePinho RA, Levy DE. (2002) STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity 17:63–72. [DOI] [PubMed] [Google Scholar]
- Lee H, Herrmann A, Deng JH, Kujawski M, Niu G, Li Z, Forman S, Jove R, Pardoll DM, Yu H. (2009a) Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 15:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Zhang P, Herrmann A, Yang C, Xin H, Wang Z, Hoon DS, Forman SJ, Jove R, Riggs AD, et al. (2012) Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc Natl Acad Sci USA 109:7765–7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Zhuang G, Cao Y, Du P, Kim H-J, Settleman J. (2014) Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26:207–221. [DOI] [PubMed] [Google Scholar]
- Lee JL, Wang MJ, Chen JY. (2009b) Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J Cell Biol 185:949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees CW, Barrett JC, Parkes M, Satsangi J. (2011) New IBD genetics: common pathways with other diseases. Gut 60:1739–1753. [DOI] [PubMed] [Google Scholar]
- Leonard WJ, Zeng R, Spolski R. (2008) Interleukin 21: a cytokine/cytokine receptor system that has come of age. J Leukoc Biol 84:348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, et al. (2003) Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci USA 100:4138–4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. (2005) Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7:387–397. [DOI] [PubMed] [Google Scholar]
- Lewis HD, Winter A, Murphy TF, Tripathi S, Pandey VN, Barton BE. (2008) STAT3 inhibition in prostate and pancreatic cancer lines by STAT3 binding sequence oligonucleotides: differential activity between 5′ and 3′ ends. Mol Cancer Ther 7:1543–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KM, Bharadwaj U, Eckols TK, Kolosov M, Kasembeli MM, Fridley C, Siller R, Tweardy DJ. (2015) Small-molecule targeting of signal transducer and activator of transcription (STAT) 3 to treat non-small cell lung cancer. Lung Cancer 90:182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HS, Yang CY, Nallaparaju KC, Zhang H, Liu YJ, Goldrath AW, Watowich SS. (2012) The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood 120:4363–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cheung SH, Evans EL, Shaw PE. (2010a) Modulation of gene expression and tumor cell growth by redox modification of STAT3. Cancer Res 70:8222–8232. [DOI] [PubMed] [Google Scholar]
- Li L, Shaw PE. (2004) A STAT3 dimer formed by inter-chain disulphide bridging during oxidative stress. Biochem Biophys Res Commun 322:1005–1011. [DOI] [PubMed] [Google Scholar]
- Li R, Fang F, Jiang M, Wang C, Ma J, Kang W, Zhang Q, Miao Y, Wang D, Guo Y, et al. (2017) STAT3 and NF-κB are simultaneously suppressed in dendritic cells in lung cancer. Sci Rep 7:45395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Hong P, Zheng X. (2019) β-carotene attenuates lipopolysaccharide-induced inflammation via inhibition of the NF-κB, JAK2/STAT3 and JNK/p38 MAPK signaling pathways in macrophages. Anim Sci J 90:140–148. [DOI] [PubMed] [Google Scholar]
- Li W, Saud SM, Young MR, Colburn NH, Hua B. (2015) Cryptotanshinone, a Stat3 inhibitor, suppresses colorectal cancer proliferation and growth in vitro. Mol Cell Biochem 406:63–73. [DOI] [PubMed] [Google Scholar]
- Li YC, Zheng XL, Liu BT, Yang GS. (2010b) Regulation of ATGL expression mediated by leptin in vitro in porcine adipocyte lipolysis. Mol Cell Biochem 333:121–128. [DOI] [PubMed] [Google Scholar]
- Lian X, Qin Y, Hossain SA, Yang L, White A, Xu H, Shipley JM, Li T, Senior RM, Du H, et al. (2005) Overexpression of Stat3C in pulmonary epithelium protects against hyperoxic lung injury. J Immunol 174:7250–7256. [DOI] [PubMed] [Google Scholar]
- Liang Z, Wu G, Fan C, Xu J, Jiang S, Yan X, Di S, Ma Z, Hu W, Yang Y. (2016) The emerging role of signal transducer and activator of transcription 3 in cerebral ischemic and hemorrhagic stroke. Prog Neurobiol 137:1–16. [DOI] [PubMed] [Google Scholar]
- Liby K, Risingsong R, Royce DB, Williams CR, Yore MM, Honda T, Gribble GW, Lamph WW, Vannini N, Sogno I, et al. (2008) Prevention and treatment of experimental estrogen receptor-negative mammary carcinogenesis by the synthetic triterpenoid CDDO-methyl ester and the rexinoid LG100268. Clin Cancer Res 14:4556–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liby K, Royce DB, Williams CR, Risingsong R, Yore MM, Honda T, Gribble GW, Dmitrovsky E, Sporn TA, Sporn MB. (2007a) The synthetic triterpenoids CDDO-methyl ester and CDDO-ethyl amide prevent lung cancer induced by vinyl carbamate in A/J mice. Cancer Res 67:2414–2419. [DOI] [PubMed] [Google Scholar]
- Liby KT, Royce DB, Risingsong R, Williams CR, Maitra A, Hruban RH, Sporn MB. (2010) Synthetic triterpenoids prolong survival in a transgenic mouse model of pancreatic cancer. Cancer Prev Res (Phila) 3:1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liby KT, Yore MM, Sporn MB. (2007b) Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer 7:357–369. [DOI] [PubMed] [Google Scholar]
- Licinio J, Caglayan S, Ozata M, Yildiz BO, de Miranda PB, O’Kirwan F, Whitby R, Liang L, Cohen P, Bhasin S, et al. (2004) Phenotypic effects of leptin replacement on morbid obesity, diabetes mellitus, hypogonadism, and behavior in leptin-deficient adults. Proc Natl Acad Sci USA 101:4531–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler KJ, Basu S, Zhan YF, Dunn AR. (1994) Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood 84:1737–1746. [PubMed] [Google Scholar]
- Lim CP, Cao X. (1999) Serine phosphorylation and negative regulation of Stat3 by JNK. J Biol Chem 274:31055–31061. [DOI] [PubMed] [Google Scholar]
- Lim CP, Phan TT, Lim IJ, Cao X. (2006) Stat3 contributes to keloid pathogenesis via promoting collagen production, cell proliferation and migration. Oncogene 25:5416–5425. [DOI] [PubMed] [Google Scholar]
- Lin A, Schildknecht A, Nguyen LT, Ohashi PS. (2010) Dendritic cells integrate signals from the tumor microenvironment to modulate immunity and tumor growth. Immunol Lett 127:77–84. [DOI] [PubMed] [Google Scholar]
- Lin JX, Migone TS, Tsang M, Friedmann M, Weatherbee JA, Zhou L, Yamauchi A, Bloom ET, Mietz J, John S, et al. (1995) The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity 2:331–339. [DOI] [PubMed] [Google Scholar]
- Lin T-Y, Poon AH, Hamid Q. (2013) Asthma phenotypes and endotypes. Curr Opin Pulm Med 19:18–23. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen B, Lu Y, Guan Y, Chen F. (2012) JNK-dependent Stat3 phosphorylation contributes to Akt activation in response to arsenic exposure. Toxicol Sci 129:363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhong Y, Liu G, Zhang X, Xiao B, Huang S, Liu H, He L. (2017) Role of Stat3 signaling in control of EMT of tubular epithelial cells during renal fibrosis. Cell Physiol Biochem 42:2552–2558. [DOI] [PubMed] [Google Scholar]
- Liu L, McBride KM, Reich NC. (2005) STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci USA 102:8150–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang F, Wen Z, Shi M, Zhang H. (2014) Blockage of STAT3 signaling pathway with a decoy oligodeoxynucleotide inhibits growth of human ovarian cancer cells. Cancer Invest 32:8–12. [DOI] [PubMed] [Google Scholar]
- Liu SC, Tsang NM, Chiang WC, Chang KP, Hsueh C, Liang Y, Juang JL, Chow KP, Chang YS. (2013) Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J Clin Invest 123:5269–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. (2007) STAT3 activation inhibits human bronchial epithelial cell apoptosis in response to cigarette smoke exposure. Biochem Biophys Res Commun 353:121–126. [DOI] [PubMed] [Google Scholar]
- Lonardo A, Byrne CD, Caldwell SH, Cortez-Pinto H, Targher G. (2016) Global epidemiology of nonalcoholic fatty liver disease: meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64:1388–1389. [DOI] [PubMed] [Google Scholar]
- Lorente D, Trilla E, Meseguer A, Arevalo J, Nemours S, Planas J, Placer J, Celma A, Salvador C, Regis L, et al. (2019) The role of STAT3 protein as a prognostic factor in the clear cell renal carcinoma: systematic review. Actas Urol Esp 43:118–123. [DOI] [PubMed] [Google Scholar]
- Lu L, Dong J, Wang L, Xia Q, Zhang D, Kim H, Yin T, Fan S, Shen Q. (2018) Activation of STAT3 and Bcl-2 and reduction of reactive oxygen species (ROS) promote radioresistance in breast cancer and overcome of radioresistance with niclosamide. Oncogene 37:5292–5304. [DOI] [PubMed] [Google Scholar]
- Lu WJ, Lin KC, Huang SY, Thomas PA, Wu YH, Wu HC, Lin KH, Sheu JR. (2014) Role of a Janus kinase 2-dependent signaling pathway in platelet activation. Thromb Res 133:1088–1096. [DOI] [PubMed] [Google Scholar]
- Lufei C, Koh TH, Uchida T, Cao X. (2007) Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene 26:7656–7664. [DOI] [PubMed] [Google Scholar]
- Lunz JG, III, Specht SM, Murase N, Isse K, Demetris AJ. (2007) Gut-derived commensal bacterial products inhibit liver dendritic cell maturation by stimulating hepatic interleukin-6/signal transducer and activator of transcription 3 activity. Hepatology 46:1946–1959. [DOI] [PubMed] [Google Scholar]
- Lupardus PJ, Ultsch M, Wallweber H, Bir Kohli P, Johnson AR, Eigenbrot C. (2014) Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci USA 111:8025–8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, Fulcher DA, Tangye SG, Cook MC. (2008) Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med 205:1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JF, Sanchez BJ, Hall DT, Tremblay AK, Di Marco S, Gallouzi IE. (2017) STAT3 promotes IFNγ/TNFα-induced muscle wasting in an NF-κB-dependent and IL-6-independent manner. EMBO Mol Med 9:622–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal T, Bhowmik A, Chatterjee A, Chatterjee U, Chatterjee S, Ghosh MK. (2014) Reduced phosphorylation of Stat3 at Ser-727 mediated by casein kinase 2 - protein phosphatase 2A enhances Stat3 Tyr-705 induced tumorigenic potential of glioma cells. Cell Signal 26:1725–1734. [DOI] [PubMed] [Google Scholar]
- Mandarino LJ. (1992) Current hypotheses for the biochemical basis of diabetic retinopathy. Diabetes Care 15:1892–1901. [DOI] [PubMed] [Google Scholar]
- Mantel C, Messina-Graham S, Moh A, Cooper S, Hangoc G, Fu XY, Broxmeyer HE. (2012) Mouse hematopoietic cell-targeted STAT3 deletion: stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction, and a rapid aging-like phenotype. Blood 120:2589–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani G, Macciò A, Mura L, Massa E, Mudu MC, Mulas C, Lusso MR, Madeddu C, Dessì A. (2000) Serum levels of leptin and proinflammatory cytokines in patients with advanced-stage cancer at different sites. J Mol Med (Berl) 78:554–561. [DOI] [PubMed] [Google Scholar]
- Marino F, Orecchia V, Regis G, Musteanu M, Tassone B, Jon C, Forni M, Calautti E, Chiarle R, Eferl R, et al. (2014) STAT3β controls inflammatory responses and early tumor onset in skin and colon experimental cancer models. Am J Cancer Res 4:484–494. [PMC free article] [PubMed] [Google Scholar]
- Maritano D, Sugrue ML, Tininini S, Dewilde S, Strobl B, Fu X, Murray-Tait V, Chiarle R, Poli V. (2004) The STAT3 isoforms alpha and beta have unique and specific functions. Nat Immunol 5:401–409. [DOI] [PubMed] [Google Scholar]
- Marzec M, Zhang Q, Goradia A, Raghunath PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, et al. (2008) Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad Sci USA 105:20852–20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J, Hoffman R, Talpaz M, Gerds AT, Stein B, Gupta V, Szoke A, Drummond M, Pristupa A, Granston T, et al. (2018) Pacritinib vs best available therapy, including ruxolitinib, in patients with myelofibrosis: a randomized clinical trial. JAMA Oncol 4:652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashili F, Chibalin AV, Krook A, Zierath JR. (2013) Constitutive STAT3 phosphorylation contributes to skeletal muscle insulin resistance in type 2 diabetes. Diabetes 62:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui F, Babitz SK, Rhee A, Hile KL, Zhang H, Meldrum KK. (2017) Mesenchymal stem cells protect against obstruction-induced renal fibrosis by decreasing STAT3 activation and STAT3-dependent MMP-9 production. Am J Physiol Renal Physiol 312:F25–F32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazière C, Conte MA, Mazière JC. (2001) Activation of JAK2 by the oxidative stress generated with oxidized low-density lipoprotein. Free Radic Biol Med 31:1334–1340. [DOI] [PubMed] [Google Scholar]
- McGinn S, Poronnik P, King M, Gallery ED, Pollock CA. (2003) High glucose and endothelial cell growth: novel effects independent of autocrine TGF-beta 1 and hyperosmolarity. Am J Physiol Cell Physiol 284:C1374–C1386. [DOI] [PubMed] [Google Scholar]
- McHugh D, Gil J. (2018) Senescence and aging: causes, consequences, and therapeutic avenues. J Cell Biol 217:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, et al. (2000) Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood 95:3489–3497. [PubMed] [Google Scholar]
- McKinley L, Alcorn JF, Peterson A, Dupont RB, Kapadia S, Logar A, Henry A, Irvin CG, Piganelli JD, Ray A, et al. (2008) TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol 181:4089–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melford SK, Turner M, Briddon SJ, Tybulewicz VL, Watson SP. (1997) Syk and Fyn are required by mouse megakaryocytes for the rise in intracellular calcium induced by a collagen-related peptide. J Biol Chem 272:27539–27542. [DOI] [PubMed] [Google Scholar]
- Melillo JA, Song L, Bhagat G, Blazquez AB, Plumlee CR, Lee C, Berin C, Reizis B, Schindler C. (2010) Dendritic cell (DC)-specific targeting reveals Stat3 as a negative regulator of DC function. J Immunol 184:2638–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng ZH, Dyer K, Billiar TR, Tweardy DJ. (2000) Distinct effects of systemic infusion of G-CSF vs. IL-6 on lung and liver inflammation and injury in hemorrhagic shock. Shock 14:41–48. [DOI] [PubMed] [Google Scholar]
- Meng ZH, Dyer K, Billiar TR, Tweardy DJ. (2001) Essential role for IL-6 in postresuscitation inflammation in hemorrhagic shock. Am J Physiol Cell Physiol 280:C343–C351. [DOI] [PubMed] [Google Scholar]
- Merad M, Manz MG. (2009) Dendritic cell homeostasis. Blood 113:3418–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, Mortha A. (2013) The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting, Annu Rev Immunol 31, pp 563–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens C, Haripal B, Klinge S, Darnell JE. (2015) Mutations in the linker domain affect phospho-STAT3 function and suggest targets for interrupting STAT3 activity. Proc Natl Acad Sci USA 112:14811–14816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa RA, Verstovsek S, Gupta V, Mascarenhas JO, Atallah E, Burn T, Sun W, Sandor V, Gotlib J. (2015) Effects of ruxolitinib treatment on metabolic and nutritional parameters in patients with myelofibrosis from COMFORT-I. Clin Lymphoma Myeloma Leuk 15:214–221.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metlakunta AS, Sahu M, Yasukawa H, Dhillon SS, Belsham DD, Yoshimura A, Sahu A. (2011) Neuronal suppressor of cytokine signaling-3 deficiency enhances hypothalamic leptin-dependent phosphatidylinositol 3-kinase signaling. Am J Physiol Regul Integr Comp Physiol 300:R1185–R1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T, Gavenis K, Vinkemeier U. (2002) Cell type-specific and tyrosine phosphorylation-independent nuclear presence of STAT1 and STAT3. Exp Cell Res 272:45–55. [DOI] [PubMed] [Google Scholar]
- Miao LJ, Huang FX, Sun ZT, Zhang RX, Huang SF, Wang J. (2014) Stat3 inhibits Beclin 1 expression through recruitment of HDAC3 in nonsmall cell lung cancer cells. Tumour Biol 35:7097–7103. [DOI] [PubMed] [Google Scholar]
- Milara J, Hernandez G, Ballester B, Morell A, Roger I, Montero P, Escrivá J, Lloris JM, Molina-Molina M, Morcillo E, et al. (2018) The JAK2 pathway is activated in idiopathic pulmonary fibrosis. Respir Res 19:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AM, Horiguchi N, Jeong WI, Radaeva S, Gao B. (2011) Molecular mechanisms of alcoholic liver disease: innate immunity and cytokines. Alcohol Clin Exp Res 35:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millington GW. (2007) The role of proopiomelanocortin (POMC) neurones in feeding behaviour. Nutr Metab (Lond) 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, et al. (2008) Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452:773–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min HK, Mirshahi F, Verdianelli A, Pacana T, Patel V, Park CG, Choi A, Lee JH, Park CB, Ren S, et al. (2015) Activation of the GP130-STAT3 axis and its potential implications in nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol 308:G794–G803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz R, Garty BZ, Meshel T, Marcus N, Katanov C, Cohen-Hillel E, Ben-Baruch A. (2010) Reduced expression of chemoattractant receptors by polymorphonuclear leukocytes in hyper IgE syndrome patients. Immunol Lett 130:97–106. [DOI] [PubMed] [Google Scholar]
- Minus MB, Liu W, Vohidov F, Kasembeli MM, Long X, Krueger MJ, Stevens A, Kolosov MI, Tweardy DJ, Sison EA, et al. (2015) Rhodium(II) proximity-labeling identifies a novel target site on STAT3 for inhibitors with potent anti-leukemia activity. Angew Chem Int Ed Engl 54:13085–13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Hanna DL, Zhang W, Baba H, Lenz HJ. (2016) Molecular pathways: cachexia signaling-A targeted approach to cancer treatment. Clin Cancer Res 22:3999–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH. (2013) STAT3 and the hyper-IgE syndrome: clinical presentation, genetic origin, pathogenesis, novel findings and remaining uncertainties. JAKSTAT 2:e23435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moh A, Iwamoto Y, Chai GX, Zhang SS, Kano A, Yang DD, Zhang W, Wang J, Jacoby JJ, Gao B, et al. (2007) Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest 87:1018–1028. [DOI] [PubMed] [Google Scholar]
- Moodley YP, Misso NLA, Scaffidi AK, Fogel-Petrovic M, McAnulty RJ, Laurent GJ, Thompson PJ, Knight DA. (2003) Inverse effects of interleukin-6 on apoptosis of fibroblasts from pulmonary fibrosis and normal lungs. Am J Respir Cell Mol Biol 29:490–498. [DOI] [PubMed] [Google Scholar]
- Moran A, Akcan Arikan A, Mastrangelo MA, Wu Y, Yu B, Poli V, Tweardy DJ. (2008) Prevention of trauma and hemorrhagic shock-mediated liver apoptosis by activation of stat3alpha. Int J Clin Exp Med 1:213–247. [PMC free article] [PubMed] [Google Scholar]
- Moran A, Tsimelzon AI, Mastrangelo MA, Wu Y, Yu B, Hilsenbeck SG, Poli V, Tweardy DJ. (2009) Prevention of trauma/hemorrhagic shock-induced lung apoptosis by IL-6-mediated activation of Stat3. Clin Transl Sci 2:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, Torisu T, Chien KR, Yasukawa H, Yoshimura A. (2004) Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nat Med 10:739–743. [DOI] [PubMed] [Google Scholar]
- Mossman BT, Lounsbury KM, Reddy SP. (2006) Oxidants and signaling by mitogen-activated protein kinases in lung epithelium. Am J Respir Cell Mol Biol 34:666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudter J, Weigmann B, Bartsch B, Kiesslich R, Strand D, Galle PR, Lehr HA, Schmidt J, Neurath MF. (2005) Activation pattern of signal transducers and activators of transcription (STAT) factors in inflammatory bowel diseases. Am J Gastroenterol 100:64–72. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay UK, Mooney P, Jia L, Eves R, Raptis L, Mak AS. (2010) Doubles game: Src-Stat3 versus p53-PTEN in cellular migration and invasion. Mol Cell Biol 30:4980–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso A, Dentelli P, Carlino A, Chiusa L, Repici A, Sturm A, Fiocchi C, Rizzetto M, Pegoraro L, Sategna-Guidetti C, et al. (2005) Signal transducers and activators of transcription 3 signaling pathway: an essential mediator of inflammatory bowel disease and other forms of intestinal inflammation. Inflamm Bowel Dis 11:91–98. [DOI] [PubMed] [Google Scholar]
- Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. (2006) Stat3 activation of NF-kappaB p100 processing involves CBP/p300-mediated acetylation. Proc Natl Acad Sci USA 103:7264–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Yamamoto S, Okada T, Gantulga D, Okano H, Ozawa K, Yada T. (2016) IL-10 gene transfer upregulates arcuate POMC and ameliorates hyperphagia, obesity and diabetes by substituting for leptin. Int J Obes 40:425–433. [DOI] [PubMed] [Google Scholar]
- Namanja AT, Wang J, Buettner R, Colson L, Chen Y. (2016) Allosteric communication across STAT3 domains associated with STAT3 function and disease-causing mutation. J Mol Biol 428:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedova Y, Cheng P, Gilkes D, Blaskovich M, Beg AA, Sebti SM, Gabrilovich DI. (2005a) Activation of dendritic cells via inhibition of Jak2/STAT3 signaling. J Immunol 175:4338–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefedova Y, Huang M, Kusmartsev S, Bhattacharya R, Cheng P, Salup R, Jove R, Gabrilovich D. (2004) Hyperactivation of STAT3 is involved in abnormal differentiation of dendritic cells in cancer. J Immunol 172:464–474. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Nagaraj S, Rosenbauer A, Muro-Cacho C, Sebti SM, Gabrilovich DI. (2005b) Regulation of dendritic cell differentiation and antitumor immune response in cancer by pharmacologic-selective inhibition of the janus-activated kinase 2/signal transducers and activators of transcription 3 pathway. Cancer Res 65:9525–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezu M, Suzuki N, Yamamoto M. (2017) Targeting the KEAP1-NRF2 system to prevent kidney disease progression. Am J Nephrol 45:473–483. [DOI] [PubMed] [Google Scholar]
- Ng CSH, Wan S, Arifi AA, Yim APC. (2006) Inflammatory response to pulmonary ischemia-reperfusion injury. Surg Today 36:205–214. [DOI] [PubMed] [Google Scholar]
- Ng DC, Ng IH, Yeap YY, Badrian B, Tsoutsman T, McMullen JR, Semsarian C, Bogoyevitch MA. (2011) Opposing actions of extracellular signal-regulated kinase (ERK) and signal transducer and activator of transcription 3 (STAT3) in regulating microtubule stabilization during cardiac hypertrophy. J Biol Chem 286:1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Jackson H, Panopoulos AD, Zhang H, Li HS, Watowich SS. (2010) STAT3 controls the neutrophil migratory response to CXCR2 ligands by direct activation of G-CSF-induced CXCR2 expression and via modulation of CXCR2 signal transduction. Blood 115:3354–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Jackson HT, Li HS, Zhang H, Ohashi E, Watowich SS. (2012) G-CSF-activated STAT3 enhances production of the chemokine MIP-2 in bone marrow neutrophils. J Leukoc Biol 92:1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. (2009) STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol 11:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara M, Ogura H, Ueda N, Tsuruoka M, Kitabayashi C, Tsuji F, Aono H, Ishihara K, Huseby E, Betz UA, et al. (2007) IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int Immunol 19:695–702. [DOI] [PubMed] [Google Scholar]
- Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, et al. (2000) Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature 404:787–790. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, et al. (2007) Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448:480–483. [DOI] [PubMed] [Google Scholar]
- O’Donoghue RJ, Knight DA, Richards CD, Prêle CM, Lau HL, Jarnicki AG, Jones J, Bozinovski S, Vlahos R, Thiem S, et al. (2012) Genetic partitioning of interleukin-6 signalling in mice dissociates Stat3 from Smad3-mediated lung fibrosis. EMBO Mol Med 4:939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Holland SM, Staudt LM. (2013) JAKs and STATs in immunity, immunodeficiency, and cancer. N Engl J Med 368:161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Kanno Y, Chen X, Levy DE. (2005) Cell signaling: Stat acetylation--a key facet of cytokine signaling? Science 307:217–218. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. (2015) The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med 66:311–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Chinen T, Yoshida T, Kinjyo I, Takaesu G, Shiraishi H, Iida M, Kobayashi T, Yoshimura A. (2006) Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene 25:2520–2530. [DOI] [PubMed] [Google Scholar]
- Ogura M, Uchida T, Terui Y, Hayakawa F, Kobayashi Y, Taniwaki M, Takamatsu Y, Naoe T, Tobinai K, Munakata W, et al. (2015) Phase I study of OPB-51602, an oral inhibitor of signal transducer and activator of transcription 3, in patients with relapsed/refractory hematological malignancies. Cancer Sci 106:896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawara B, Shirakabe K, Hyodo-Miura J, Matsuo R, Ueno N, Matsumoto K, Shibuya H. (2004) Role of the TAK1-NLK-STAT3 pathway in TGF-beta-mediated mesoderm induction. Genes Dev 18:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Hara E. (2013) Roles and mechanisms of cellular senescence in regulation of tissue homeostasis. Cancer Sci 104:525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N, Obata-Onai A, Tussiwand R, Lanzavecchia A, Manz MG. (2006) Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon-producing and dendritic cell development. J Exp Med 203:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouédraogo ZG, Müller-Barthélémy M, Kemeny JL, Dedieu V, Biau J, Khalil T, Raoelfils LI, Granzotto A, Pereira B, Beaudoin C, et al. (2016) STAT3 serine 727 phosphorylation: a relevant target to radiosensitize human glioblastoma. Brain Pathol 26:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Zhou F, Zhang R, Claret FX. (2013) Stat3 inhibitor Stattic exhibits potent antitumor activity and induces chemo- and radio-sensitivity in nasopharyngeal carcinoma. PLoS One 8:e54565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang M, Ma L, Gong R, Tolbert E, Mao H, Ponnusamy M, Chin YE, Yan H, Dworkin LD, Zhuang S. (2010) A novel STAT3 inhibitor, S3I-201, attenuates renal interstitial fibroblast activation and interstitial fibrosis in obstructive nephropathy. Kidney Int 78:257–268. [DOI] [PubMed] [Google Scholar]
- Panopoulos AD, Bartos D, Zhang L, Watowich SS. (2002) Control of myeloid-specific integrin alpha Mbeta 2 (CD11b/CD18) expression by cytokines is regulated by Stat3-dependent activation of PU.1. J Biol Chem 277:19001–19007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos AD, Zhang L, Snow JW, Jones DM, Smith AM, El Kasmi KC, Liu F, Goldsmith MA, Link DC, Murray PJ, et al. (2006) STAT3 governs distinct pathways in emergency granulopoiesis and mature neutrophils. Blood 108:3682–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou I, Xu S, Denton CP, Abraham DJ, Ponticos M. (2018) STAT3 controls COL1A2 enhancer activation cooperatively with JunB, regulates type I collagen synthesis posttranscriptionally, and is essential for lung myofibroblast differentiation. Mol Biol Cell 29:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Österreicher CH, Takahashi H, Karin M. (2010) Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140:197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HK, Ahima RS. (2014) Leptin signaling. F1000Prime Rep 6:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Nakagawa T, Kitamura H, Atsumi T, Kamon H, Sawa S, Kamimura D, Ueda N, Iwakura Y, Ishihara K, et al. (2004) IL-6 regulates in vivo dendritic cell differentiation through STAT3 activation. J Immunol 173:3844–3854. [DOI] [PubMed] [Google Scholar]
- Pasquet JM, Quek L, Pasquet S, Poole A, Matthews JR, Lowell C, Watson SP. (2000) Evidence of a role for SHP-1 in platelet activation by the collagen receptor glycoprotein VI. J Biol Chem 275:28526–28531. [DOI] [PubMed] [Google Scholar]
- Pedroza M, Le TT, Lewis K, Karmouty-Quintana H, To S, George AT, Blackburn MR, Tweardy DJ, Agarwal SK. (2016) STAT-3 contributes to pulmonary fibrosis through epithelial injury and fibroblast-myofibroblast differentiation. FASEB J 30:129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroza M, To S, Assassi S, Wu M, Tweardy D, Agarwal SK. (2017) Role of STAT3 in skin fibrosis and transforming growth factor beta signalling. Rheumatology (Oxford) 57:1838–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penna F, Costamagna D, Pin F, Camperi A, Fanzani A, Chiarpotto EM, Cavallini G, Bonelli G, Baccino FM, Costelli P. (2013) Autophagic degradation contributes to muscle wasting in cancer cachexia. Am J Pathol 182:1367–1378. [DOI] [PubMed] [Google Scholar]
- Pensa S, Lloyd-Lewis B, Sargeant TJ, Resemann HK, Kahn CR, Watson CJ. (2014) Signal transducer and activator of transcription 3 and the phosphatidylinositol 3-kinase regulatory subunits p55α and p50α regulate autophagy in vivo. FEBS J 281:4557–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, et al. (2009) STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med 206:1465–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F, Izzo V, Niso-Santano M, Vacchelli E, Galluzzi L, Maiuri MC, Kroemer G. (2013) Regulation of autophagy by stress-responsive transcription factors. Semin Cancer Biol 23:310–322. [DOI] [PubMed] [Google Scholar]
- Porpaczy E, Tripolt S, Hoelbl-Kovacic A, Gisslinger B, Bago-Horvath Z, Casanova-Hevia E, Clappier E, Decker T, Fajmann S, Fux DA, et al. (2018) Aggressive B-cell lymphomas in patients with myelofibrosis receiving JAK1/2 inhibitor therapy. Blood 132:694–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porporato PE. (2016) Understanding cachexia as a cancer metabolism syndrome. Oncogenesis 5:e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan AD, Manson JE, Rifai N, Buring JE, Ridker PM. (2001) C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA 286:327–334. [DOI] [PubMed] [Google Scholar]
- Priceman SJ, Kujawski M, Shen S, Cherryholmes GA, Lee H, Zhang C, Kruper L, Mortimer J, Jove R, Riggs AD, et al. (2013) Regulation of adipose tissue T cell subsets by Stat3 is crucial for diet-induced obesity and insulin resistance. Proc Natl Acad Sci USA 110:13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Lizano P, Laure L, Sui X, Rashed E, Park JY, Hong C, Gao S, Holle E, Morin D, et al. (2011) H11 kinase/heat shock protein 22 deletion impairs both nuclear and mitochondrial functions of STAT3 and accelerates the transition into heart failure on cardiac overload. Circulation 124:406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadoss P, Unger-Smith NE, Lam FS, Hollenberg AN. (2009) STAT3 targets the regulatory regions of gluconeogenic genes in vivo. Mol Endocrinol 23:827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsauer K, Sadzak I, Porras A, Pilz A, Nebreda AR, Decker T, Kovarik P. (2002) p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc Natl Acad Sci USA 99:12859–12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath KS, Naidu SK, Lata P, Bid HK, Rivera BK, McCann GA, Tierney BJ, Elnaggar AC, Bravo V, Leone G, et al. (2014) HO-3867, a safe STAT3 inhibitor, is selectively cytotoxic to ovarian cancer. Cancer Res 74:2316–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings JS, Rosler KM, Harrison DA. (2004) The JAK/STAT signaling pathway. J Cell Sci 117:1281–1283. [DOI] [PubMed] [Google Scholar]
- Ray S, Boldogh I, Brasier AR. (2005) STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology 129:1616–1632. [DOI] [PubMed] [Google Scholar]
- Ray S, Lee C, Hou T, Boldogh I, Brasier AR. (2008) Requirement of histone deacetylase1 (HDAC1) in signal transducer and activator of transcription 3 (STAT3) nucleocytoplasmic distribution. Nucleic Acids Res 36:4510–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Sherman CT, Lu M, Brasier AR. (2002) Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity. Mol Endocrinol 16:824–836. [DOI] [PubMed] [Google Scholar]
- Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. (2002) Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene 21:7611–7618. [DOI] [PubMed] [Google Scholar]
- Rébé C, Végran F, Berger H, Ghiringhelli F. (2013) STAT3 activation: a key factor in tumor immunoescape. JAK-STAT 2:e23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilley MJ, McCoon P, Cook C, Lyne P, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, Fowler N, et al. (2018) STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: results of a phase 1b trial. J Immunother Cancer 6:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resetca D, Haftchenary S, Gunning PT, Wilson DJ. (2014) Changes in signal transducer and activator of transcription 3 (STAT3) dynamics induced by complexation with pharmacological inhibitors of Src homology 2 (SH2) domain dimerization. J Biol Chem 289:32538–32547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M, Pereira FV. (2018) A new perspective: mitochondrial Stat3 as a regulator for lymphocyte function. Int J Mol Sci 19:1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robker RL, Watson LN, Robertson SA, Dunning KR, McLaughlin EA, Russell DL. (2014) Identification of sites of STAT3 action in the female reproductive tract through conditional gene deletion. PLoS One 9:e101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Campisi J. (2011) Four faces of cellular senescence. J Cell Biol 192:547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Liñares B, Watson SP. (1994) Phosphorylation of JAK2 in thrombin-stimulated human platelets. FEBS Lett 352:335–338. [DOI] [PubMed] [Google Scholar]
- Rozovski U, Grgurevic S, Bueso-Ramos C, Harris DM, Li P, Liu Z, Wu JY, Jain P, Wierda W, Burger J, et al. (2015) Aberrant LPL expression, driven by STAT3, mediates free fatty acid metabolism in CLL cells. Mol Cancer Res 13:944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovski U, Harris DM, Li P, Liu Z, Jain P, Veletic I, Ferrajoli A, Burger J, O’Brien S, Bose P, et al. (2017a) Constitutive phosphorylation of STAT3 by the CK2-BLNK-CD5 complex. Mol Cancer Res 15:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovski U, Harris DM, Li P, Liu Z, Jain P, Veletic I, Ferrajoli A, Burger J, Thompson P, Jain N, et al. (2017b) Activation of the B-cell receptor successively activates NF-κB and STAT3 in chronic lymphocytic leukemia cells. Int J Cancer 141:2076–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovski U, Harris DM, Li P, Liu Z, Wu JY, Grgurevic S, Faderl S, Ferrajoli A, Wierda WG, Martinez M, et al. (2016) At high levels, constitutively activated STAT3 induces apoptosis of chronic lymphocytic leukemia cells. J Immunol 196:4400–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M, Endo K, Furuya S, Minami M, Fukasawa A, Imamura T, Miyazawa K. (2016) STAT3 integrates cooperative Ras and TGF-β signals that induce Snail expression. Oncogene 35:1049–1057. [DOI] [PubMed] [Google Scholar]
- Sallam AM, Esmat A, Abdel-Naim AB. (2018) Cucurbitacin-B attenuates CCl4 -induced hepatic fibrosis in mice through inhibition of STAT-3. Chem Biol Drug Des 91:933–941. [DOI] [PubMed] [Google Scholar]
- Samudio I, Konopleva M, Pelicano H, Huang P, Frolova O, Bornmann W, Ying Y, Evans R, Contractor R, Andreeff M. (2006) A novel mechanism of action of methyl-2-cyano-3,12 dioxoolean-1,9 diene-28-oate: direct permeabilization of the inner mitochondrial membrane to inhibit electron transport and induce apoptosis. Mol Pharmacol 69:1182–1193. [DOI] [PubMed] [Google Scholar]
- Sandborn WJ, Feagan BG, Fedorak RN, Scherl E, Fleisher MR, Katz S, Johanns J, Blank M, Rutgeerts P, Ustekinumab Crohn’s Disease Study Group (2008) A randomized trial of Ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with moderate-to-severe Crohn’s disease. Gastroenterology 135:1130–1141. [DOI] [PubMed] [Google Scholar]
- Sandborn WJ, Ghosh S, Panes J, Vranic I, Su C, Rousell S, Niezychowski W, Study A3921063 Investigators (2012) Tofacitinib, an oral Janus kinase inhibitor, in active ulcerative colitis. N Engl J Med 367:616–624. [DOI] [PubMed] [Google Scholar]
- Sandborn WJ, Schreiber S, Feagan BG, Rutgeerts P, Younes ZH, Bloomfield R, Coteur G, Guzman JP, D’Haens GR. (2011) Certolizumab pegol for active Crohn’s disease: a placebo-controlled, randomized trial. Clin Gastroenterol Hepatol 9:670–678.e3. [DOI] [PubMed] [Google Scholar]
- Sano S, Chan KS, Carbajal S, Clifford J, Peavey M, Kiguchi K, Itami S, Nickoloff BJ, DiGiovanni J. (2005) Stat3 links activated keratinocytes and immunocytes required for development of psoriasis in a novel transgenic mouse model. Nat Med 11:43–49. [DOI] [PubMed] [Google Scholar]
- Sano S, Itami S, Takeda K, Tarutani M, Yamaguchi Y, Miura H, Yoshikawa K, Akira S, Takeda J. (1999) Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J 18:4657–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafian TA, Montes C, Imura T, Qi J, Coppola G, Geschwind DH, Sofroniew MV. (2010) Disruption of astrocyte STAT3 signaling decreases mitochondrial function and increases oxidative stress in vitro. PLoS One 5:e9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Kawai T, Sugiyama K, Muromoto R, Imoto S, Sekine Y, Ishida M, Akira S, Matsuda T. (2005) Physical and functional interactions between STAT3 and ZIP kinase. Int Immunol 17:1543–1552. [DOI] [PubMed] [Google Scholar]
- Schaljo B, Kratochvill F, Gratz N, Sadzak I, Sauer I, Hammer M, Vogl C, Strobl B, Müller M, Blackshear PJ, et al. (2009) Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J Immunol 183:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Elazar Z. (2007) ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 17:422–427. [DOI] [PubMed] [Google Scholar]
- Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr. (1992) Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257:809–813. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Dahmen H, Grimm C, Gendo C, Müller-Newen G, Heinrich PC, Schaper F. (2000) The cytoplasmic tyrosine motifs in full-length glycoprotein 130 have different roles in IL-6 signal transduction. J Immunol 164:848–854. [DOI] [PubMed] [Google Scholar]
- Schreiber S, Khaliq-Kareemi M, Lawrance IC, Thomsen O, Hanauer SB, McColm J, Bloomfield R, Sandborn WJ, PRECISE 2 Study Investigators (2007) Maintenance therapy with certolizumab pegol for Crohn’s disease [published correction appears in N Engl J Med (2007) 357:1357]. N Engl J Med 357:239–250. [DOI] [PubMed] [Google Scholar]
- Schuringa JJ, Schepers H, Vellenga E, Kruijer W. (2001) Ser727-dependent transcriptional activation by association of p300 with STAT3 upon IL-6 stimulation. FEBS Lett 495:71–76. [DOI] [PubMed] [Google Scholar]
- Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. (2017) The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal 15:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen M, Paul K, Freilino ML, Li H, Li C, Johnson DE, Wang L, Eiseman J, Grandis JR. (2014) Systemic administration of a cyclic signal transducer and activator of transcription 3 (STAT3) decoy oligonucleotide inhibits tumor growth without inducing toxicological effects. Mol Med 20:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, Johnson JT, Duvvuri U, Lee J, Sahu N, Joyce S, et al. (2012) First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov 2:694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah CA, Broglie L, Hu L, Bei L, Huang W, Dressler DB, Eklund EA. (2018) Stat3 and CCAAT enhancer-binding protein β (C/ebpβ) activate Fanconi C gene transcription during emergency granulopoiesis. J Biol Chem 293:3937–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Quintero AJ, Tweardy DJ. (2001) Identification and characterization of cis elements in the STAT3 gene regulating STAT3 alpha and STAT3 beta messenger RNA splicing. Blood 98:3853–3856. [DOI] [PubMed] [Google Scholar]
- Shao H, Xu X, Mastrangelo MA, Jing N, Cook RG, Legge GB, Tweardy DJ. (2004) Structural requirements for signal transducer and activator of transcription 3 binding to phosphotyrosine ligands containing the YXXQ motif. J Biol Chem 279:18967–18973. [DOI] [PubMed] [Google Scholar]
- Shen J, Li R, Li G. (2009) Inhibitory effects of decoy-ODN targeting activated STAT3 on human glioma growth in vivo. In Vivo 23:237–243. [PubMed] [Google Scholar]
- Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, Souquere S, Mariño G, Lachkar S, Senovilla L, et al. (2012) Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell 48:667–680. [DOI] [PubMed] [Google Scholar]
- Shen Y, Devgan G, Darnell JE, Jr., Bromberg JF. (2001) Constitutively activated Stat3 protects fibroblasts from serum withdrawal and UV-induced apoptosis and antagonizes the proapoptotic effects of activated Stat1. Proc Natl Acad Sci USA 98:1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, Darnell JE., Jr. (2004) Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol 24:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry MM, Reeves A, Wu JK, Cochran BH. (2009) STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells 27:2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Tzameli I, Bjørbaek C, Flier JS. (2004) Suppressor of cytokine signaling 3 is a physiological regulator of adipocyte insulin signaling. J Biol Chem 279:34733–34740. [DOI] [PubMed] [Google Scholar]
- Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX. (2008) Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 10:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SY, Luk CT, Brunt JJ, Sivasubramaniyam T, Lu SY, Schroer SA, Woo M. (2014) Adipocyte-specific deficiency of Janus kinase (JAK) 2 in mice impairs lipolysis and increases body weight, and leads to insulin resistance with ageing. Diabetologia 57:1016–1026. [DOI] [PubMed] [Google Scholar]
- Shuai K, Liu B. (2003) Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 3:900–911. [DOI] [PubMed] [Google Scholar]
- Siegel AM, Stone KD, Cruse G, Lawrence MG, Olivera A, Jung MY, Barber JS, Freeman AF, Holland SM, O’Brien M, et al. (2013) Diminished allergic disease in patients with STAT3 mutations reveals a role for STAT3 signaling in mast cell degranulation [published correction appears in J Allergy Clin Immunol (2014) 133:1232]. J Allergy Clin Immunol 132:1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora E, Arendt T, Bennett M, Narita M. (2011) Impact of cellular senescence signature on ageing research. Ageing Res Rev 10:146–152. [DOI] [PubMed] [Google Scholar]
- Silva KAS, Dong J, Dong Y, Dong Y, Schor N, Tweardy DJ, Zhang L, Mitch WE. (2015) Inhibition of Stat3 activation suppresses caspase-3 and the ubiquitin-proteasome system, leading to preservation of muscle mass in cancer cachexia. J Biol Chem 290:11177–11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone-Penney MC. (2008) STAT3: A Novel Regulator of Airway Inflammation and Remodeling in Asthma in Sackler School of Graduate Biomedical Sciences, Tuft University (MA). ProQuest Dissertations Publishing 2008:3324688. [Google Scholar]
- Sims JT, Ganguly SS, Bennett H, Friend JW, Tepe J, Plattner R. (2013) Imatinib reverses doxorubicin resistance by affecting activation of STAT3-dependent NF-κB and HSP27/p38/AKT pathways and by inhibiting ABCB1. PLoS One 8:e55509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitenga J, Aird G, Ahmed A, Silberstein PT. (2018) Impact of siltuximab on patient-related outcomes in multicentric Castleman’s disease. Patient Relat Outcome Meas 9:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Qualls JE, O’Brien K, Balouzian L, Johnson PF, Schultz-Cherry S, Smale ST, Murray PJ. (2011) A distal enhancer in Il12b is the target of transcriptional repression by the STAT3 pathway and requires the basic leucine zipper (B-ZIP) protein NFIL3. J Biol Chem 286:23582–23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Suleman N, Lacerda L, Opie LH, Akira S, Chien KR, Sack MN. (2004) Genetic depletion of cardiac myocyte STAT-3 abolishes classical preconditioning. Cardiovasc Res 63:611–616. [DOI] [PubMed] [Google Scholar]
- Sobotta MC, Liou W, Stöcker S, Talwar D, Oehler M, Ruppert T, Scharf AN, Dick TP. (2015) Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol 11:64–70. [DOI] [PubMed] [Google Scholar]
- Sookoian S, Castaño G, Gianotti TF, Gemma C, Rosselli MS, Pirola CJ. (2008) Genetic variants in STAT3 are associated with nonalcoholic fatty liver disease. Cytokine 44:201–206. [DOI] [PubMed] [Google Scholar]
- Spaccarotella E, Pellegrino E, Ferracin M, Ferreri C, Cuccuru G, Liu C, Iqbal J, Cantarella D, Taulli R, Provero P, et al. (2014) STAT3-mediated activation of microRNA cluster 17∼92 promotes proliferation and survival of ALK-positive anaplastic large cell lymphoma. Haematologica 99:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. (2003) Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res 63:2948–2956. [PubMed] [Google Scholar]
- Stacpoole PW. (1997) Lactic acidosis and other mitochondrial disorders. Metabolism 46:306–321. [DOI] [PubMed] [Google Scholar]
- Stark GR, Wang Y, Lu T. (2011) Lysine methylation of promoter-bound transcription factors and relevance to cancer. Cell Res 21:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stärkel P, Schnabl B, Leclercq S, Komuta M, Bataller R, Argemi J, Palma E, Chokshi S, Hellerbrand C, Maccioni L, et al. (2019) Deficient IL-6/Stat3 signaling, high TLR7, and type I interferons in early human alcoholic liver disease: a triad for liver damage and fibrosis. Hepatol Commun 3:867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens JM, Lumpkin SJ, Fishman JB. (1998) Activation of signal transducers and activators of transcription 1 and 3 by leukemia inhibitory factor, oncostatin-M, and interferon-gamma in adipocytes. J Biol Chem 273:31408–31416. [DOI] [PubMed] [Google Scholar]
- Steward-Tharp SM, Laurence A, Kanno Y, Kotlyar A, Villarino AV, Sciume G, Kuchen S, Resch W, Wohlfert EA, Jiang K, et al. (2014) A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood 123:2978–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TH, Shiau CW, Jao P, Liu CH, Liu CJ, Tai WT, Jeng YM, Yang HC, Tseng TC, Huang HP, et al. (2015) Sorafenib and its derivative SC-1 exhibit antifibrotic effects through signal transducer and activator of transcription 3 inhibition. Proc Natl Acad Sci USA 112:7243–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yu M, Lu Y, Thakur C, Chen B, Qiu P, Zhao H, Chen F. (2014) Carcinogenic metalloid arsenic induces expression of mdig oncogene through JNK and STAT3 activation. Cancer Lett 346:257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Steinberg BM. (2002) PTEN is a negative regulator of STAT3 activation in human papillomavirus-infected cells. J Gen Virol 83:1651–1658. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang J, Wang L, Tian Z. (2008) Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett 262:201–213. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, Hoshino T, Kubo M, Yamashita A, Okabe M, Takeda K, et al. (2001) CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med 193:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanek K, Chen Q, Derecka M, Salloum FN, Zhang Q, Szelag M, Cichy J, Kukreja RC, Dulak J, Lesnefsky EJ, et al. (2011) Mitochondrial-targeted signal transducer and activator of transcription 3 (STAT3) protects against ischemia-induced changes in the electron transport chain and the generation of reactive oxygen species. J Biol Chem 286:29610–29620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanek K, Chen Q, Larner AC, Lesnefsky EJ. (2012) Cytoprotection by the modulation of mitochondrial electron transport chain: the emerging role of mitochondrial STAT3. Mitochondrion 12:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, Chen PJ, Chen KF. (2013) Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis 4:e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura A, Nelson EA, Haque N, Humphreys BD, Zandi-Nejad K, Frank DA, Zhou J. (2011) Pyrimethamine inhibits adult polycystic kidney disease by modulating STAT signaling pathways. Hum Mol Genet 20:4143–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Förster I, Akira S. (1999) Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10:39–49. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. (1998) Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice [published correction appears in J Immunol (2015) 194:3526]. J Immunol 161:4652–4660. [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. (1997) Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA 94:3801–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammineni P, Anugula C, Mohammed F, Anjaneyulu M, Larner AC, Sepuri NBV. (2013) The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J Biol Chem 288:4723–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Goedken MJ, Chen C, Reisman SA, Manautou JE, Klaassen CD. (2008) Coordinated induction of Nrf2 target genes protects against iron nitrilotriacetate (FeNTA)-induced nephrotoxicity. Toxicol Appl Pharmacol 231:364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targan SR, Hanauer SB, van Deventer SJ, Mayer L, Present DH, Braakman T, DeWoody KL, Schaible TF, Rutgeerts PJ. (1997) A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn’s disease: Crohn’s Disease cA2 Study Group. N Engl J Med 337:1029–1035. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A. (2011) Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol 29:1356–1363. [DOI] [PubMed] [Google Scholar]
- Thacker S, Moran A, Lionakis M, Mastrangelo MA, Halder T, del Pilar Huby M, Wu Y, Tweardy DJ. (2014) Restoration of lung surfactant protein D by IL-6 protects against secondary pneumonia following hemorrhagic shock. J Infect 68:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker SA, Robinson P, Abel A, Tweardy DJ. (2013) Modulation of the unfolded protein response during hepatocyte and cardiomyocyte apoptosis in trauma/hemorrhagic shock. Sci Rep 3:1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NS. (2012) The STAT3-DNMT1 connection. JAK-STAT 1:257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbles HE, Vassilev A, Wendorf H, Schonhoff D, Zhu D, Lorenz D, Waurzyniak B, Liu XP, Uckun FM. (2001) Role of a JAK3-dependent biochemical signaling pathway in platelet activation and aggregation. J Biol Chem 276:17815–17822. [DOI] [PubMed] [Google Scholar]
- Timofeeva OA, Chasovskikh S, Lonskaya I, Tarasova NI, Khavrutskii L, Tarasov SG, Zhang X, Korostyshevskiy VR, Cheema A, Zhang L, et al. (2012) Mechanisms of unphosphorylated STAT3 transcription factor binding to DNA. J Biol Chem 287:14192–14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M, Coria L, Rosemblit C, Rivas MA, Proietti CJ, Díaz Flaqué MC, Beguelin W, Frahm I, Charreau EH, Cassataro J, et al. (2012) Targeting Stat3 induces senescence in tumor cells and elicits prophylactic and therapeutic immune responses against breast cancer growth mediated by NK cells and CD4+ T cells. J Immunol 189:1162–1172. [DOI] [PubMed] [Google Scholar]
- Tolcher A, Flaherty K, Shapiro GI, Berlin J, Witzig T, Habermann T, Bullock A, Rock E, Elekes A, Lin C, et al. (2018) A first-in-human phase I study of OPB-111077, a small-molecule STAT3 and oxidative phosphorylation inhibitor, in patients with advanced cancers. Oncologist 23:658-e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisu T, Sato N, Yoshiga D, Kobayashi T, Yoshioka T, Mori H, Iida M, Yoshimura A. (2007) The dual function of hepatic SOCS3 in insulin resistance in vivo. Genes Cells 12:143–154. [DOI] [PubMed] [Google Scholar]
- Turkson J, Bowman T, Adnane J, Zhang Y, Djeu JY, Sekharam M, Frank DA, Holzman LB, Wu J, Sebti S, et al. (1999) Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol Cell Biol 19:7519–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkson J, Zhang S, Palmer J, Kay H, Stanko J, Mora LB, Sebti S, Yu H, Jove R. (2004) Inhibition of constitutive signal transducer and activator of transcription 3 activation by novel platinum complexes with potent antitumor activity. Mol Cancer Ther 3:1533–1542. [PubMed] [Google Scholar]
- Udono M, Kadooka K, Yamashita S, Katakura Y. (2012) Quantitative analysis of cellular senescence phenotypes using an imaging cytometer. Methods 56:383–388. [DOI] [PubMed] [Google Scholar]
- Ueki K, Kondo T, Tseng YH, Kahn CR. (2004) Central role of suppressors of cytokine signaling proteins in hepatic steatosis, insulin resistance, and the metabolic syndrome in the mouse. Proc Natl Acad Sci USA 101:10422–10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainchenker W, Constantinescu SN. (2013) JAK/STAT signaling in hematological malignancies. Oncogene 32:2601–2613. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Kantarjian HM, Kiladjian JJ, Gotlib J, Cervantes F, Mesa RA, Sarlis NJ, Peng W, Sandor V, Gopalakrishna P, et al. COMFORT Investigators (2015) A pooled analysis of overall survival in COMFORT-I and COMFORT-II, 2 randomized phase III trials of ruxolitinib for the treatment of myelofibrosis. Haematologica 100:1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstovsek S. (2010) Therapeutic potential of Janus-activated kinase-2 inhibitors for the management of myelofibrosis. Clin Cancer Res 16:1988–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, Catalano JV, Deininger M, Miller C, Silver RT, et al. (2012) A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med 366:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron A, Gamelin E, Coqueret O. (2008) The EGFR-STAT3 oncogenic pathway up-regulates the Eme1 endonuclease to reduce DNA damage after topoisomerase I inhibition. Cancer Res 68:815–825. [DOI] [PubMed] [Google Scholar]
- Villarino AV, Kanno Y, O’Shea JJ. (2017) Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol 18:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M, Domoszlai T, Kleshchanok D, Lehmann S, Schmitt A, Poli V, Richtering W, Müller-Newen G. (2011) The role of the N-terminal domain in dimerization and nucleocytoplasmic shuttling of latent STAT3. J Cell Sci 124:900–909. [DOI] [PubMed] [Google Scholar]
- Waitkus MS, Chandrasekharan UM, Willard B, Tee TL, Hsieh JK, Przybycin CG, Rini BI, Dicorleto PE. (2014) Signal integration and gene induction by a functionally distinct STAT3 phosphoform. Mol Cell Biol 34:1800–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakahara R, Kunimoto H, Tanino K, Kojima H, Inoue A, Shintaku H, Nakajima K. (2012) Phospho-Ser727 of STAT3 regulates STAT3 activity by enhancing dephosphorylation of phospho-Tyr705 largely through TC45. Genes Cells 17:132–145. [DOI] [PubMed] [Google Scholar]
- Walz C, Ahmed W, Lazarides K, Betancur M, Patel N, Hennighausen L, Zaleskas VM, Van Etten RA. (2012) Essential role for Stat5a/b in myeloproliferative neoplasms induced by BCR-ABL1 and JAK2(V617F) in mice. Blood 119:3550–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhou Y, Lei W, Zhang K, Shi J, Hu Y, Shu G, Song J. (2009) Signal transducer and activator of transcription 3 (STAT3) regulates adipocyte differentiation via peroxisome-proliferator-activated receptor gamma (PPARgamma). Biol Cell 102:1–12. [DOI] [PubMed] [Google Scholar]
- Wang H, Cheng F, Woan K, Sahakian E, Merino O, Rock-Klotz J, Vicente-Suarez I, Pinilla-Ibarz J, Wright KL, Seto E, et al. (2011a) Histone deacetylase inhibitor LAQ824 augments inflammatory responses in macrophages through transcriptional regulation of IL-10. J Immunol 186:3986–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lafdil F, Wang L, Yin S, Feng D, Gao B. (2011b) Tissue inhibitor of metalloproteinase 1 (TIMP-1) deficiency exacerbates carbon tetrachloride-induced liver injury and fibrosis in mice: involvement of hepatocyte STAT3 in TIMP-1 production. Cell Biosci 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Huang J, Jiang M, Lin H. (2012) Signal transducer and activator of transcription 2 (STAT2) metabolism coupling postmitotic outgrowth to visual and sound perception network in human left cerebrum by biocomputation. J Mol Neurosci 47:649–658. [DOI] [PubMed] [Google Scholar]
- Wang LH, Yang XY, Zhang X, Huang J, Hou J, Li J, Xiong H, Mihalic K, Zhu H, Xiao W, et al. (2004a) Transcriptional inactivation of STAT3 by PPARgamma suppresses IL-6-responsive multiple myeloma cells. Immunity 20:205–218. [DOI] [PubMed] [Google Scholar]
- Wang R, Cherukuri P, Luo J. (2005) Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J Biol Chem 280:11528–11534. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhang Q, Wang Q, Shen Q, Chen X, Li Z, Zhou Y, Hou J, Xu B, Li N, et al. (2018a) NEAT1 paraspeckle promotes human hepatocellular carcinoma progression by strengthening IL-6/STAT3 signaling. OncoImmunology 7:e1503913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. (2004b) Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med 10:48–54. [DOI] [PubMed] [Google Scholar]
- Wang YY, Yang YX, Zhe H, He ZX, Zhou SF. (2014) Bardoxolone methyl (CDDO-Me) as a therapeutic agent: an update on its pharmacokinetic and pharmacodynamic properties. Drug Des Devel Ther 8:2075–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Aguilar EG, Luna JI, Dunai C, Khuat LT, Le CT, Mirsoian A, Minnar CM, Stoffel KM, Sturgill IR, et al. (2019) Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat Med 25:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li J, Xiao W, Long J, Zhang H. (2018b) The STAT3 inhibitor S3I-201 suppresses fibrogenesis and angiogenesis in liver fibrosis. Lab Invest 98:1600–1613. [DOI] [PubMed] [Google Scholar]
- Watowich SS, Liu YJ. (2010) Mechanisms regulating dendritic cell specification and development. Immunol Rev 238:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman AB, Mahboubi K, Knickelbein RG, Mantell LL, Manzo N, Pober JS, Elias JA. (2003) Interleukin-11 and interleukin-6 protect cultured human endothelial cells from H2O2-induced cell death. Am J Respir Cell Mol Biol 29:513–522. [DOI] [PubMed] [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, et al. (2009) Function of mitochondrial Stat3 in cellular respiration. Science 323:793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte T, Zhang SSM, Wang T, Zhang Z, Hesslein DGT, Yin Z, Kano A, Iwamoto Y, Li E, Craft JE, et al. (2003) STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci USA 100:1879–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE., Jr. (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241–250. [DOI] [PubMed] [Google Scholar]
- Wensveen FM, Jelenčić V, Valentić S, Šestan M, Wensveen TT, Theurich S, Glasner A, Mendrila D, Štimac D, Wunderlich FT, et al. (2015) NK cells link obesity-induced adipose stress to inflammation and insulin resistance. Nat Immunol 16:376–385. [DOI] [PubMed] [Google Scholar]
- Wenzel SE. (2006) Asthma: defining of the persistent adult phenotypes. Lancet 368:804–813. [DOI] [PubMed] [Google Scholar]
- White JP, Baynes JW, Welle SL, Kostek MC, Matesic LE, Sato S, Carson JA. (2011) The regulation of skeletal muscle protein turnover during the progression of cancer cachexia in the Apc(Min/+) mouse. PLoS One 6:e24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga AT, Vogelzang I, Eggen BJ, Vellenga E. (2003) Erythropoietin-induced serine 727 phosphorylation of STAT3 in erythroid cells is mediated by a MEK-, ERK-, and MSK1-dependent pathway. Exp Hematol 31:398–405. [DOI] [PubMed] [Google Scholar]
- Wilks AF. (1989) Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc Natl Acad Sci USA 86:1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks AF, Harpur AG, Kurban RR, Ralph SJ, Zürcher G, Ziemiecki A. (1991) Two novel protein-tyrosine kinases, each with a second phosphotransferase-related catalytic domain, define a new class of protein kinase. Mol Cell Biol 11:2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthuhn BA, Williams MD, Kerawalla H, Uckun FM. (1999) Differential substrate recognition capabilities of Janus family protein tyrosine kinases within the interleukin 2 receptor (IL2R) system: Jak3 as a potential molecular target for treatment of leukemias with a hyperactive Jak-Stat signaling machinery. Leuk Lymphoma 32:289–297. [DOI] [PubMed] [Google Scholar]
- Wölfle SJ, Strebovsky J, Bartz H, Sähr A, Arnold C, Kaiser C, Dalpke AH, Heeg K. (2011) PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur J Immunol 41:413–424. [DOI] [PubMed] [Google Scholar]
- Wong AL, Soo RA, Tan DS, Lee SC, Lim JS, Marban PC, Kong LR, Lee YJ, Wang LZ, Thuya WL, et al. (2015) Phase I and biomarker study of OPB-51602, a novel signal transducer and activator of transcription (STAT) 3 inhibitor, in patients with refractory solid malignancies. Ann Oncol 26:998–1005. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang F, Hu S, Yin C, Li X, Zhao S, Wang J, Yan X. (2012) MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal 24:2179–2186. [DOI] [PubMed] [Google Scholar]
- Wunderlich CM, Hövelmeyer N, Wunderlich FT. (2013) Mechanisms of chronic JAK-STAT3-SOCS3 signaling in obesity. JAKSTAT 2:e23878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kole S, Precht P, Pazin MJ, Bernier M. (2009) S-glutathionylation impairs signal transducer and activator of transcription 3 activation and signaling. Endocrinology 150:1122–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Hong J, Du W, Lin YW, Ren LL, Wang YC, Su WY, Wang JL, Cui Y, Wang ZH, et al. (2012) Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem 287:5819–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Yin C, Wang S, Xiao Y. (2013) JAK-STAT in lipid metabolism of adipocytes. JAKSTAT 2:e27203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M-Y, Hu J-J, Shen J, Wang M-L, Zhang Q-Q, Qu Y, Lu L-G. (2014a) Stat3 signaling activation crosslinking of TGF-β1 in hepatic stellate cell exacerbates liver injury and fibrosis. Biochim Biophys Acta 1842:2237–2245. [DOI] [PubMed] [Google Scholar]
- Xu N, Emelyanov AV, Fyodorov DV, Skoultchi AI. (2014b) Drosophila linker histone H1 coordinates STAT-dependent organization of heterochromatin and suppresses tumorigenesis caused by hyperactive JAK-STAT signaling. Epigenetics Chromatin 7:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Kasembeli MM, Jiang X, Tweardy BJ, Tweardy DJ. (2009) Chemical probes that competitively and selectively inhibit Stat3 activation. PLoS One 4:e4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li Z, Yin Y, Lan H, Wang J, Zhao J, Feng J, Li Y, Zhang W. (2015) Ghrelin inhibits the differentiation of T helper 17 cells through mTOR/STAT3 signaling pathway. PLoS One 10:e0117081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YS, Liang JJ, Wang Y, Zhao XJ, Xu L, Xu YY, Zou QC, Zhang JM, Tu CE, Cui YG, et al. (2016) STAT3 undergoes acetylation-dependent mitochondrial translocation to regulate pyruvate metabolism. Sci Rep 6:39517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Xu YJ, Hao YN, Ren LJ, Zhang ZB, Xu X, Cao BY, Dai KS, Zhu L, Fang Q, et al. (2017) A novel STAT3 inhibitor negatively modulates platelet activation and aggregation. Acta Pharmacol Sin 38:651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada E, Bastie CC, Koga H, Wang Y, Cuervo AM, Pessin JE. (2012) Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Rep 1:557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki K, Yoshino S. (2014) Remission of food allergy by the Janus kinase inhibitor ruxolitinib in mice. Int Immunopharmacol 18:217–224. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yoshizaki K, Kishimoto T, Ito H. (2000) IL-6 is required for the development of Th1 cell-mediated murine colitis. J Immunol 164:4878–4882. [DOI] [PubMed] [Google Scholar]
- Yamaoka K, Saharinen P, Pesu M, Holt VE, III, Silvennoinen O, O’Shea JJ. (2004) The Janus kinases (Jaks). Genome Biol 5:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Naltner A, Martin M, Naltner M, Fangman JM, Gurel O. (2002) Transcriptional stimulation of the surfactant protein B gene by STAT3 in respiratory epithelial cells. J Biol Chem 277:10967–10972. [DOI] [PubMed] [Google Scholar]
- Yan S, Li Z, Thiele CJ. (2013) Inhibition of STAT3 with orally active JAK inhibitor, AZD1480, decreases tumor growth in neuroblastoma and pediatric sarcomas in vitro and in vivo. Oncotarget 4:433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan SJ, Lim SJ, Shi S, Dutta P, Li WX. (2011) Unphosphorylated STAT and heterochromatin protect genome stability. FASEB J 25:232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. (2005) Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 65:939–947. [PubMed] [Google Scholar]
- Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, et al. (2010) Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA 107:21499–21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. (2007a) Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev 21:1396–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lian X, Cowen A, Xu H, Du H, Yan C. (2004) Synergy between signal transducer and activator of transcription 3 and retinoic acid receptor-α in regulation of the surfactant protein B gene in the lung. Mol Endocrinol 18:1520–1532. [DOI] [PubMed] [Google Scholar]
- Yang R, Lirussi D, Thornton TM, Jelley-Gibbs DM, Diehl SA, Case LK, Madesh M, Taatjes DJ, Teuscher C, Haynes L, et al. (2015) Mitochondrial Ca2+ and membrane potential, an alternative pathway for interleukin 6 to regulate CD4 cell effector function. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Rincon M. (2016) Mitochondrial Stat3, the need for design thinking. Int J Biol Sci 12:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, et al. (2008a) Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. (2007b) STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem 282:9358–9363. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. (2008b) T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR α and ROR γ. Immunity 28:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hulver M, McMillan RP, Cai L, Kershaw EE, Yu L, Xue B, Shi H. (2012) Regulation of insulin and leptin signaling by muscle suppressor of cytokine signaling 3 (SOCS3). PLoS One 7:e47493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, et al. (2003) IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol 4:551–556. [DOI] [PubMed] [Google Scholar]
- Yokogami K, Wakisaka S, Avruch J, Reeves SA. (2000) Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol 10:47–50. [DOI] [PubMed] [Google Scholar]
- Yong PF, Freeman AF, Engelhardt KR, Holland S, Puck JM, Grimbacher B. (2012) An update on the hyper-IgE syndromes. Arthritis Res Ther 14:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Naka T, Kubo M. (2007) SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 7:454–465. [DOI] [PubMed] [Google Scholar]
- You L, Wang Z, Li H, Shou J, Jing Z, Xie J, Sui X, Pan H, Han W. (2015) The role of STAT3 in autophagy. Autophagy 11:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. (2007) Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 7:41–51. [DOI] [PubMed] [Google Scholar]
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. (2014) Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 14:736–746. [DOI] [PubMed] [Google Scholar]
- Yuan H, Houck KL, Tian Y, Bharadwaj U, Hull K, Zhou Z, Zhu M, Wu X, Tweardy DJ, Romo D, et al. (2015a) Piperlongumine blocks JAK2-STAT3 to inhibit collagen-induced platelet reactivity independent of reactive oxygen species [published correction appears in PLoS One (2016) 11:e0146626]. PLoS One 10:e0143964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Houck KL, Tian Y, Bharadwaj U, Hull K, Zhou Z, Zhu M, Wu X, Tweardy DJ, Romo D, et al. (2016) Correction: piperlongumine blocks JAK2-STAT3 to inhibit collagen-induced platelet reactivity independent of reactive oxygen species. PLoS One 11:e0146626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhang F, Niu R. (2015b) Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci Rep 5:17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZL, Guan YJ, Chatterjee D, Chin YE. (2005) Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307:269–273. [DOI] [PubMed] [Google Scholar]
- Yue H, Li W, Desnoyer R, Karnik SS. (2010) Role of nuclear unphosphorylated STAT3 in angiotensin II type 1 receptor-induced cardiac hypertrophy. Cardiovasc Res 85:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun JH, Park SW, Kim KJ, Bae JS, Lee EH, Paek SH, Kim SU, Ye S, Kim JH, Cho CH. (2017) Endothelial STAT3 activation increases vascular leakage through downregulating tight junction proteins: implications for diabetic retinopathy. J Cell Physiol 232:1123–1134. [DOI] [PubMed] [Google Scholar]
- Yun UJ, Park SE, Jo YS, Kim J, Shin DY. (2012) DNA damage induces the IL-6/STAT3 signaling pathway, which has anti-senescence and growth-promoting functions in human tumors. Cancer Lett 323:155–160. [DOI] [PubMed] [Google Scholar]
- Zahr AA, Salama ME, Carreau N, Tremblay D, Verstovsek S, Mesa R, Hoffman R, Mascarenhas J. (2016) Bone marrow fibrosis in myelofibrosis: pathogenesis, prognosis and targeted strategies. Haematologica 101:660–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Liu X, Li B, He Y, Jing S, He Y, Wu W, Zhang B, Ma S, Dai W, et al. (2017) IL-6/STAT3/TWIST inhibition reverses ionizing radiation-induced EMT and radioresistance in esophageal squamous carcinoma. Oncotarget 8:11228–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgheib C, Kurdi M, Zouein FA, Gunter BW, Stanley BA, Zgheib J, Romero DG, King SB, Paolocci N, Booz GW. (2012) Acyloxy nitroso compounds inhibit LIF signaling in endothelial cells and cardiac myocytes: evidence that STAT3 signaling is redox-sensitive. PLoS One 7:e43313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hu H, Greeley N, Jin J, Matthews AJ, Ohashi E, Caetano MS, Li HS, Wu X, Mandal PK, et al. (2014) STAT3 restrains RANK- and TLR4-mediated signalling by suppressing expression of the E2 ubiquitin-conjugating enzyme Ubc13. Nat Commun 5:5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Nguyen-Jackson H, Panopoulos AD, Li HS, Murray PJ, Watowich SS. (2010) STAT3 controls myeloid progenitor growth during emergency granulopoiesis. Blood 116:2462–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Guo W, Yang Y, Wu J. (2011) JAK2/STAT3 pathway is involved in the early stage of adipogenesis through regulating C/EBPβ transcription. J Cell Biochem 112:488–497. [DOI] [PubMed] [Google Scholar]
- Zhang L, Pan J, Dong Y, Tweardy DJ, Dong Y, Garibotto G, Mitch WE. (2013a) Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab 18:368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yao Y, Zhang S, Liu Y, Guo H, Ahmed M, Bell T, Zhang H, Han G, Lorence E, et al. (2019) Metabolic reprogramming toward oxidative phosphorylation identifies a therapeutic target for mantle cell lymphoma. Sci Transl Med 11:eaau1167. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Boisson B, Beziat V, Puel A, Casanova JL. (2018) Human hyper-IgE syndrome: singular or plural? Mamm Genome 29:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kee WH, Seow KT, Fung W, Cao X. (2000) The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol Cell Biol 20:7132–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qu X, Chen B, Snyder M, Wang M, Li B, Tang Y, Chen H, Zhu W, Zhan L, et al. (2016) Critical roles of STAT3 in β-adrenergic functions in the heart. Circulation 133:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu P, Zhang B, Mao H, Shen L, Ma Y. (2013b) Inhibitory effects of STAT3 decoy oligodeoxynucleotides on human epithelial ovarian cancer cell growth in vivo. Int J Mol Med 32:623–628. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shan P, Alam J, Fu XY, Lee PJ. (2005) Carbon monoxide differentially modulates STAT1 and STAT3 and inhibits apoptosis via a phosphatidylinositol 3-kinase/Akt and p38 kinase-dependent STAT3 pathway during anoxia-reoxygenation injury. J Biol Chem 280:8714–8721. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shan P, Jiang G, Zhang SSM, Otterbein LE, Fu XY, Lee PJ. (2006) Endothelial STAT3 is essential for the protective effects of HO-1 in oxidant-induced lung injury. FASEB J 20:2156–2158. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shan P, Otterbein LE, Alam J, Flavell RA, Davis RJ, Choi AMK, Lee PJ. (2003) Carbon monoxide inhibition of apoptosis during ischemia-reperfusion lung injury is dependent on the p38 mitogen-activated protein kinase pathway and involves caspase 3. J Biol Chem 278:1248–1258. [DOI] [PubMed] [Google Scholar]
- Zhang X, Sun Y, Pireddu R, Yang H, Urlam MK, Lawrence HR, Guida WC, Lawrence NJ, Sebti SM. (2013c) A novel inhibitor of STAT3 homodimerization selectively suppresses STAT3 activity and malignant transformation. Cancer Res 73:1922–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dees C, Beyer C, Lin NY, Distler A, Zerr P, Palumbo K, Susok L, Kreuter A, Distler O, et al. (2015) Inhibition of casein kinase II reduces TGFβ induced fibroblast activation and ameliorates experimental fibrosis. Ann Rheum Dis 74:936–943. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu G, Dong Z. (2001) MSK1 and JNKs mediate phosphorylation of STAT3 in UVA-irradiated mouse epidermal JB6 cells. J Biol Chem 276:42534–42542. [DOI] [PubMed] [Google Scholar]
- Zhao C, Li H, Lin H-J, Yang S, Lin J, Liang G. (2016) Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol Sci 37:47–61. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sun R, You L, Gao C, Tian Z. (2003) Expression of leptin receptors and response to leptin stimulation of human natural killer cell lines. Biochem Biophys Res Commun 300:247–252. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. (2007) IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol 8:967–974. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Gushiken FC, Bolgiano D, Salsbery BJ, Aghakasiri N, Jing N, Wu X, Vijayan KV, Rumbaut RE, Adachi R, et al. (2013) Signal transducer and activator of transcription 3 (STAT3) regulates collagen-induced platelet aggregation independently of its transcription factor activity. Circulation 127:476–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouein FA, Altara R, Chen Q, Lesnefsky EJ, Kurdi M, Booz GW. (2015) Pivotal importance of STAT3 in protecting the heart from acute and chronic stress: new advancement and unresolved issues. Front Cardiovasc Med 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouein FA, Kurdi M, Booz GW. (2013a) Dancing rhinos in stilettos: the amazing saga of the genomic and nongenomic actions of STAT3 in the heart. JAK-STAT 2:e24352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouein FA, Kurdi M, Booz GW. (2013b) LIF and the heart: just another brick in the wall? Eur Cytokine Netw 24:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]